Summary
Plants have served as a preeminent study system for photoperiodism due to their propensity to flower in concordance with the seasons. A nearly singular focus on understanding photoperiodic flowering has prevented discovery of other photoperiod measuring systems necessary for vegetative health. Here we use bioinformatics to identify photoperiod-induced genes in Arabidopsis. We show that one, PP2-A13, is expressed exclusively in, and required for plant fitness in, short winter-like photoperiods. We create a real-time photoperiod reporter, using the PP2-A13 promoter driving luciferase, and show that photoperiodic regulation is independent of the canonical CO/FT mechanism for photoperiodic flowering. We then reveal that photosynthesis combines with circadian clock-controlled starch production to regulate cellular sucrose levels to control photoperiodic expression of PP2-A13. This work demonstrates the existence of a photoperiod measuring system housed in the metabolic network of plants that functions to control seasonal cellular health.
eTOC blurb
Liu et al. identify PP2-A13 as a representative winter photoperiod-induced gene necessary for winter plant fitness. The authors demonstrate the existence of a photoperiod measuring system controlled by the plant metabolic network, independent of the canonical CO/FT mechanism that regulates flowering.
Graphical Abstract
Introduction
Earth’s obliquity results in day and night durations (photoperiods) that change throughout the year. Photoperiod is a highly predictable signal that can help organisms anticipate seasonal changes (Nelson, et al., 2010). Photoperiod measuring mechanisms are found in fungi (Tan, et al., 2004; Roenneberg and Merrow, 2001), plants (Shim and Imaizumi, 2015; Song, et al., 2015), and animals and coordinate seasonal developmental programs to mitigate damage from less predictable stresses (Walker, et al., 2019). They also act to align growth and reproduction with seasons optimal for organismal fitness. Furthermore, human syndromes, such as seasonal affective disorder and its comorbidities, are under photoperiodic control (Garbazza and Benedetti, 2018).
Plants are a preeminent study system for understanding photoperiod measurement mechanisms because flowering time is easily observable and is often regulated by photoperiod. In the early part of the 20th century, Erwin Bünning used flowering time studies to postulate a two-state model for photoperiod measuring systems (Saunders, 2005; Bunning, 1969). In the first part of the 24-hour day, organisms are in a photophilic (light-loving) state and then later in the day they switch to a skotophilic (dark-loving) state. This underlying two-state mechanism allows the organism to enact different developmental programs depending on whether dusk coincides with either state. For instance, winter dusk occurs in the photophilic state and the organism has one developmental outcome (i.e. vegetative growth in a “long day” flowering plant). Conversely, summer dusk occurs in the skotophilic state and a different outcome occurs (i.e. flowering in a “long day” flowering plant). These criteria allow for a so-called “true photoperiod measuring mechanism” that counts the number of hours of light or dark each day, irrespective of light intensity, within a range of intensities (Thomas and Vince-Prue, 1997). Subsequent to the discovery of the circadian clock, Bünning’s general model was refined to what is called the “external coincidence” model. This model, posits that the circadian clock sets a light-inducible phase each day that coincides with the period of the day when lights are present or absent depending on the season.
With seasonal flowering, the external coincidence model was upheld by molecular and genetic studies. Photoperiodic flowering relies on circadian clock-controlled transcription of a gene called CONSTANS (CO) (Putterill, et al., 1995). In Arabidopsis, CO mRNA expression is phased to the latter portion of the 24-hour day, thus low and high CO mRNA levels define the photophilic and skotophilic states, respectively (Yanovsky and Kay, 2002). Dusk timing is then sensed through light-mediated stabilization of CO protein. When day length is extended into the inducible skotophilic phase, CO protein is stable and activates transcription of FLOWERING LOCUS T (FT), the tissue-mobile florigen (Jang, et al., 2008; An, et al., 2004; Valverde, et al., 2004; Kardailsky, et al., 1999).
Outside of photoperiod-controlled flowering, less is known about other photoperiod-controlled processes in plants. Along with lower average temperatures and changes in water availability, winter poses a unique challenge for plants due to the lower average amount of photosynthetic light (Vitasse, et al., 2014; Oquist and Huner, 2003). Despite potential danger, winter is necessary for survival in many plants and provides them with a yearly “memory” to distinguish between identical photoperiods in different seasons (Bouche, et al., 2017; Henderson, et al., 2003). Perennial trees serve as models for winter photoperiod-induced dormancy and growth cessation, and recent work predicts that a variation of the CO/FT module used for flowering is likely repressing winter photoperiod transcripts in long summer-like days (Cubas, 2020; Azeez and Sane, 2015; Bohlenius, et al., 2006). However, the gene regulatory networks that control induction of winter photoperiod transcripts have not been studied in detail, and it has been postulated that winter photoperiod induced biological processes could simply be activated by the absence of summer photoperiod repressive mechanisms. Alternatively, it is possible that there is a wholly separate winter photoperiod transcript induction system. It is likely that we have yet to make this distinction due to a lack of genetic and molecular tools and sparse knowledge of the genes and cellular processes induced in plants in winter photoperiods.
To address this gap, we analyzed genome-wide expression data using daily expression integral calculations to identify transcripts whose expression are induced in short winter-like photoperiods in Arabidopsis. We found one prevailing dark biphasic expression pattern associated with transcripts that are induced by winter photoperiods. We characterized the function of one gene, PHLOEM PROTEIN2-A13 (PP2-A13), showing that it is necessary for cellular health and reproduction in short winter-like photoperiods and functions in parallel to autophagy to prevent premature senescence of vegetative tissue in plants. We created a PP2-A13promoter::luciferase transgenic plant, that acts as a real-time photoperiod reporter, and define the properties of the transcript induction system demonstrating that its regulation is independent of the CO/FT photoperiod measuring system. We then show that PP2-A13 expression is controlled by a photoperiod measuring system that relies on daily photosynthetic sucrose levels being governed by circadian clock controlled starch production and breakdown. This allows the plant to respond to darkness early in the 24-hour day with rapid and sustained induction of PP2-A13, and late in the 24-hour day with slow induction. Together, we show that metabolic networks allow plants to measure day length to drive winter photoperiod transcripts critical for plant fitness.
Results
Calculating relative daily expression integrals to identify photoperiod-induced transcripts and biological processes
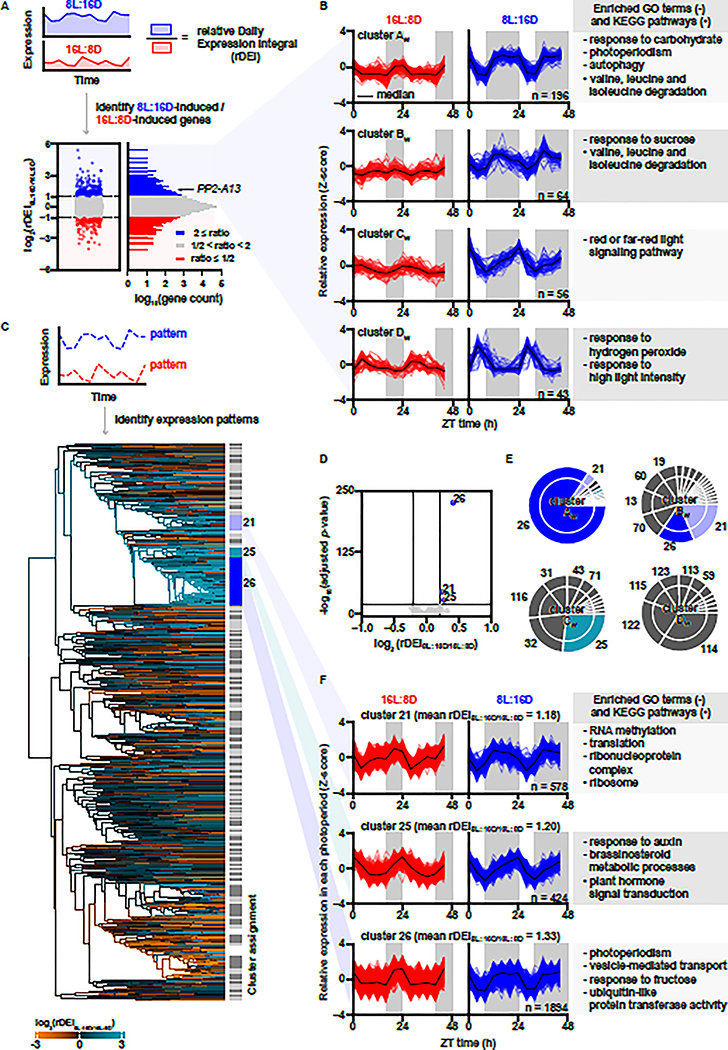
The well-studied photoperiod-induced flowering time gene, FT, has a daily expression rhythm in Arabidopsis with high amplitude in 16 hours light:8 hours dark (16L:8D) growth conditions, and low or no amplitude in 8 hours light:16 hours dark (8L:16D) (Yanovsky and Kay, 2002; Suarez-Lopez, et al., 2001). We surmised that other photoperiod-induced transcripts may also be identified through a photoperiod-specific daily rhythm. We estimated daily expression induction by calculating a relative daily expression integral (rDEI = sum of 24 hours of expression in condition one / sum of 24 hours of expression in condition two) (Figure 1A). To find photoperiod-induced transcripts we calculated rDEI using gene expression data from plants grown in 8L:16D or 16L:8D growth conditions (rDEI8L:16D/16L:8D) (Figure 1A and Table S1) (Michael, et al., 2008b; Mockler, et al., 2007). 359 transcripts are induced greater than two-fold in an 8L:16D photoperiod, and another 194 transcripts in a 16L:8D photoperiod. Clustering analyses revealed 4 co-expression clusters in the 8L:16D-induced transcripts and 4 in the 16L:8D-induced transcripts (Figure 1B, S1 and Table S2, S3). Approximately 88% of these transcripts are phased to the dark part of the photoperiod, suggesting that nighttime expression is important for an 8L:16D-induced gene expression signature (316/359; 8L:16D clusters AW-CW; Figure 1B). Conversely, 73% of the 16L:8D-induced transcripts are phased to the light part of the photoperiod (141/194; 16L:8D Clusters AS and BS; Figure S1).
Figure 1. Induced gene expression in an 8L:16D photoperiod is correlated to rhythmic expression patterns with nighttime phasing.
(A) Distribution of rDEI8L:16D/16L:8D (n = 22810). Blue: 8L:16D-induced transcripts with rDEI8L:16D/16L:8D > 2.0; red: 16L:8D-induced transcripts with rDEI8L:16D/16L:8D < 0.5. (B) Normalized expression of 8L:16D-induced transcripts grouped by k-means clustering. Black lines indicate median expression level. Grey areas indicate darkness. Top enriched GO terms (−) and KEGG pathways (•) are listed. (C) Hierarchical clustering of all transcripts by 16L:8D and 8L:16D patterns (Table S4). Dendrogram edges are colored by the average rDEI8L:16D/16L:8D of transcripts in the node. Light grey and dark grey indicate clusters, and white indicates transcripts unassigned to any cluster. (D) Identification of photoperiod-induced clusters (average rDEI8L:16D/16L:8D > 1.15 or average rDEI8L:16D/16L:8D < 0.87; adjusted p-value < 1.0 × 10−20 one-sample Wilcoxon test with Bonferroni correction). Blue: 8L:16D-induced clusters. (E) Overlap between clusters of 8L:16D-induced transcripts and clusters of transcripts showing an 8L:16D-induction correlated pattern. (F) Expression pattern of transcripts in clusters 21, 25, and 26 normalized in each photoperiod (Table S4).
We next performed enrichment tests of Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways from the 8L:16D-induced transcripts (Figure 1B and Table S2, S3) (Hvidsten, et al., 2001; Kanehisa and Goto, 2000; Ogata, et al., 1998). Supporting the validity of our approach, “photoperiod” and “red/far red light signaling” are enriched GO terms from clusters AW-CW. Furthermore, the “response to carbohydrate,” “response to sucrose,” and “autophagy” GO terms and the “valine, leucine and isoleucine degradation” KEGG pathway are also enriched, highlighting that 8L:16D photoperiods signal the induction of energy response and nutrient conservation and scavenging pathways (Figure 1B and Table S2, S3). We also searched for examples of photoperiod-specific function for the genes. HOMOGENTISATE 1,2-DIOXYGENASE (HGO-AT5G54080) from cluster AW is an enzyme involved in tyrosine catabolism, specifically in 8L:16D (Zhi, et al., 2016; Han, et al., 2013), and MALATE SYNTHASE (MLS-AT5G03860) from cluster CW is a gene that is necessary for establishing true leaves in 8L:16D (Cornah, et al., 2004). Perhaps the clearest example of a gene that is important for 8L:16D development is TEMPRANILLO1 (TEM1- AT1G25560) in cluster AW, a transcriptional regulator that blocks flowering in short winter-like photoperiods by repressing FT expression directly, in competition with CO (Figure 1B) (Johansson and Staiger, 2014; Castillejo and Pelaz, 2008).
Defining an expression pattern for transcripts induced in 8L:16D
To determine whether the dark-phased expression pattern of 8L:16D-induced genes is linked to a high rDEI8L:16D/16L:8D, we normalized the expression patterns to each photoperiod and performed hierarchical clustering for all transcripts from the 8L:16D and 16L:8D microarray experiments (Figure 1C and Table S4). This identified 131 expression pattern clusters. Three large clusters, numbered 21, 25, and 26 had expression patterns similar to clusters AW, BW, and CW (Figure 1B) and also have higher rDEI8L:16D/16L:8D when compared to other transcripts in the microarray (Figure 1D). In particular, >85% of transcripts from cluster AW fall within cluster 26, a large cluster of >1800 transcripts (Figure 1E–F). This congruence suggests that the temporal expression pattern represented by cluster 26 is correlated to higher rDEI8L:16D/16L:8D. We performed GO and KEGG analyses on clusters 21, 25, and 26 (Figure 1F). Cluster 26 contains terms similar to those found in clusters AW and BW, including “photoperiodism”, “response to fructose”, and “vesicle-mediated transport” (a broader term containing “autophagy”). Cluster 26 also included the GO term “ubiquitin-like protein transferase activity” suggesting that the ubiquitin proteasome system is being induced in 8L:16D.
PP2-A13 is essential for Arabidopsis fitness in 8L:16D
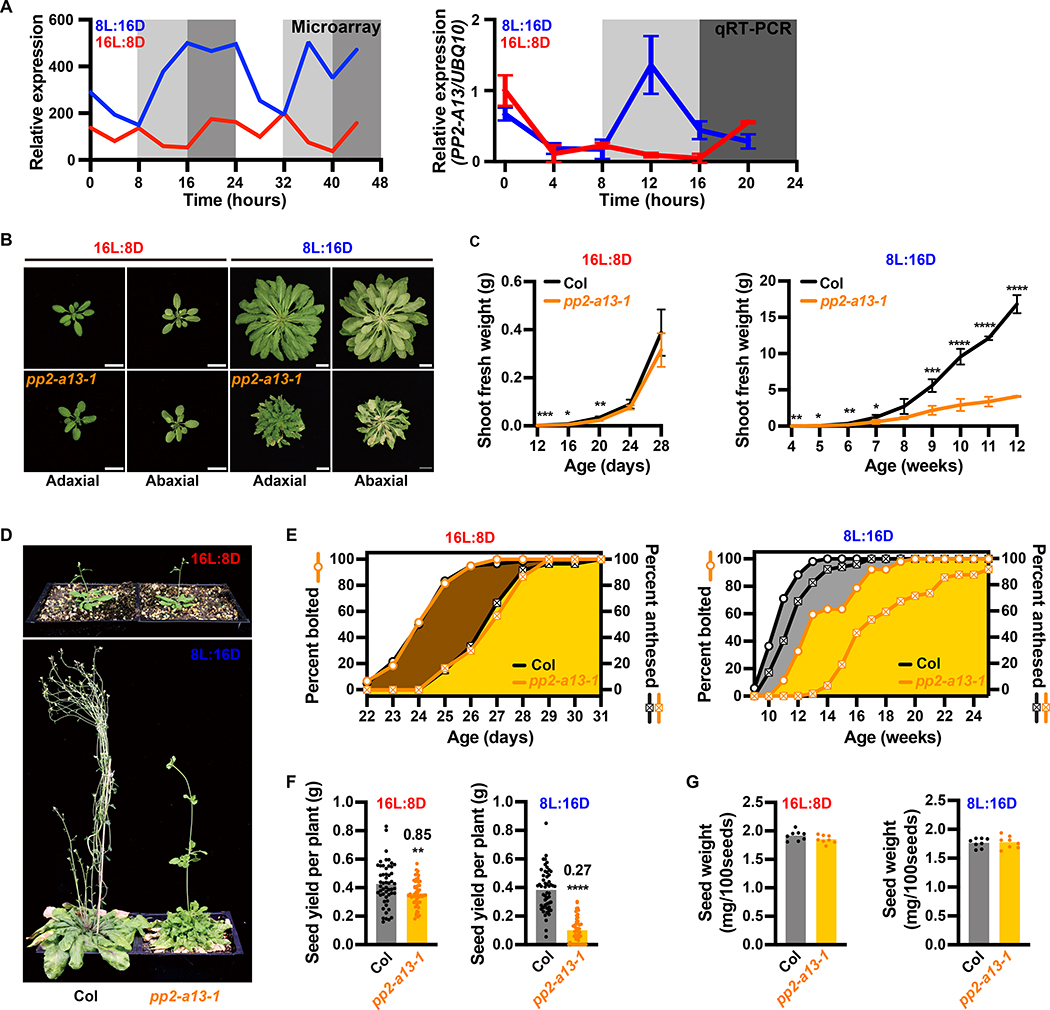
We previously curated a large group of genetic resources for F-box-type E3 ubiquitin ligases (Feke, et al., 2020; Feke, et al., 2019; Lee, et al., 2019; Lee, et al., 2018), which are part of the “ubiquitin-like protein transferase activity” GO term (Figure 1F). Here, we chose to further study one F-box gene from our library, PP2-A13, because it is strongly transcriptionally induced in 8L:16D and had not been studied in detail previously. PP2-A13 shares sequence similarity with the human lectin-containing F-box gene F-BOX ONLY 2 (FBXO2, also known as Fbs1/Nfb42/Fbx2/Fbg1) which is critical for cytoplasmic glycoprotein quality (Yoshida, et al., 2005; Dinant, et al., 2003; Yoshida, et al., 2003). The microarray data indicate that PP2-A13 follows a dark-phased expression pattern similar to cluster AW which was confirmed when we tested this by qRT-PCR (Figure 2A). PP2-A13 expression is qualitatively different between the 8L:16D and 16L:8D growth regimes. A dominant 8L:16D-specific expression peak appears at about 4 hours after dusk but is absent from 16L:8D, reminiscent of the FT expression peak that is present in 16L:8D but absent in 8L:16D.
Figure 2. Disruption of the PP2-A13 gene causes 8L:16D-specific fitness defects.
(A) Microarray expression and qRT-PCR of PP2-A13 from 12-day-old plants grown in 8L:16D (blue) and 16L:8D (red). UBQ10 was used as an internal control. Error bars indicate SD, n = 3. (B) Representative wild type (Col) and pp2-a13–1 mutant plants grown for 24 days in 16L:8D or 11 weeks in 8L:16D. Adaxial and abaxial views of the rosettes are presented. Scale bar = 3 cm. (C) Aerial fresh weight of wild-type (Col, black) and pp2-a13–1 mutant (orange) plants grown in 16L:8D and 8L:16D. Error bar indicates SD. *, p⩽0.05; **, p⩽0.01; ***, p⩽0.001; ****, p⩽0.0001 (Welch’s t-test). (D) Representative wild-type (Col) and pp2-a13–1 mutant plants grown for 28 days in 16L:8D or 14 weeks in 8L:16D. (E) Percentage of wild-type (Col) and pp2-a13–1 mutant plants that are bolting or anthesed. Plants were grown in 16L:8D (left) and 8L:16D (right). n = 52–60. (F) Total seed yield from wild-type (Col) and pp2-a13–1 mutant plants grown in 8L:16D and 16L:8D. Numbers on the column indicate the fold change. n = 52–60. **, p⩽0.01; ****, p⩽0.0001 (Welch’s t-test). (G) Seed weight in mg/100 seeds from wild-type (Col) and pp2-a13–1 mutant plants grown in 8L:16D and 16L:8D. n = 8.
We next identified a transgenic line (pp2-a13–1) containing a T-DNA insertion in PP2-A13 that compromises expression (Figure S2A–B). Assessing development in 8L:16D and 16L:8D growth conditions (Figure 2B–G and S2C–G), we found the leaves of the pp2-a13–1 mutant senesce prior to flowering, exclusively in 8L:16D, a qualitative reversal of these important developmental processes (Figure 2B and S2D). In 8L:16D, the pp2-a13–1 mutant is unable to maintain generation of biomass prior to flowering, while in 16L:8D the mutant is only partially compromised early in vegetative development and recovers later (Figure 2C, S2C, and S2E). Expression of the full length PP2-A13 driven by the native promoter complements the mutant phenotype suggesting defects are caused by the T-DNA (Figure S2H).
We then noted altered inflorescence morphology, bolting time, and anthesis in the pp2-a13–1 mutant exclusively in 8L:16D (Figure 2D–E and S2F–G). Furthermore, in 8L:16D, 4 out of 52 (7.7%) mutant plants never underwent anthesis and did not produce seeds, while an additional 9 mutant plants produced no viable seeds (17.3%). We also found that the mutant plants in 16L:8D had a slight defect in seed yield while the 8L:16D grown mutant seeds were severely compromised, but neither growth condition caused a differential effect on weight per 100 seeds (Figure 2F–G) indicating that seed filling is not compromised. In sum, these results show that PP2-A13 is necessary for Arabidopsis cellular health and reproduction in 8L:16D growth conditions.
PP2-A13 and autophagy prevent plant senescence in winter photoperiods
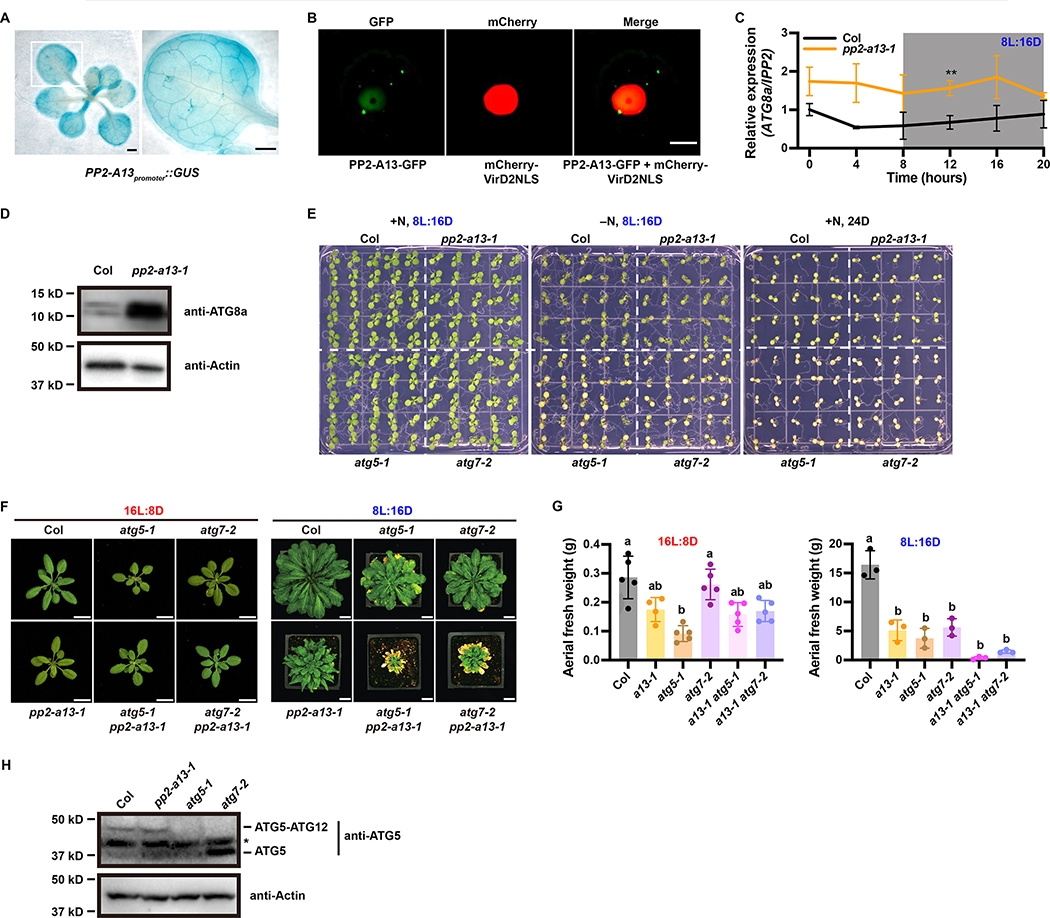
We determined the spatial expression pattern of PP2-A13 using a transgenic line expressing β-glucuronidase under the PP2-A13 promoter (PP2-A13promoter::GUS) (Figure 3A). PP2-A13 is expressed widely and does not seem to be tissue-specific. Subcellular localization of the PP2-A13 protein, using transient expression of PP2-A13 fused to GFP in Arabidopsis protoplasts, (Figure 3B) showed diffuse localization in the nucleus but also foci outside of the nucleus.
Figure 3. PP2-A13 works in parallel to autophagy.
(A) GUS staining of the PP2-A13promoter::GUS transgenic line. Right: zoom-in view of the white box area in the left image. Scale bar = 1mm. (B) PP2-A13-GFP was co-expressed with a nuclear marker mCherry-VirD2NLS in Arabidopsis protoplasts. Scale bar = 10 μm. (C) qRT-PCR of ATG8a from 6-week-old wild-type (Col, black) and pp2-a13–1 mutant (orange) grown in 8L:16D. IPP2 was used as an internal control. Error bars indicate SD (n = 3). **, p⩽0.01 (Welch’s t-test) (D) Immunoblot analysis of the pp2-a13–1 mutant. Crude protein extracts of 11-week-old wide-type (Col) and pp2-a13–1 mutant were subjected to SDS-PAGE and immunoblot analysis with anti-ATG8a antibody. Equal protein loads were confirmed by immunoblot analysis with anti-Actin antibody. (E) Phenotypes of Col (WT), pp2-a13–1, atg5–1, and atg7–2 mutants in response to nitrogen starvation and continuous darkness. One-week-old Col (WT), pp2-a13–1, atg5–1, and atg7–2 seedlings germinated on 1/2 MS media plates were transferred to nitrogen rich (+N) or nitrogen deficient (−N) media and grown in 8L:16D or 24D growth conditions. The seedlings were photographed at 7 days after treatment. (F) Representative images of wild-type (Col), pp2-a13–1, atg5–1, atg7–2, atg5–1 pp2-a13–1, and atg7–2 pp2-a13–1 mutant plants grown in 16L:8D for 28 days or 8L:16D for 87 days. Scale bar = 2 cm in 16L:8D and 3 cm in 8L:16D. (G) Aerial fresh weight of wild-type (Col), pp2-a13–1, atg5–1, atg7–2, atg5–1 pp2-a13–1, and atg7–2 pp2-a13–1 mutant plants grown in 16L:8D and 8L:16D. Different letters indicate significant differences as determined by one-way ANOVA followed by Dunnett’s T3 multiple comparison test; p⩽0.05. Error bars indicate SD (n = 3–5). (H) Immunoblot analysis of ATG5 in wild-type (Col), pp2-a13–1, atg5–1, and atg7–2 mutant plants. Asterisk indicates protein cross-reacting with ATG5 antibody.
The observable phenotypic effects of the pp2-a13–1 mutant are reminiscent of the effects of autophagy mutants grown in short winter-like photoperiods with early senescence and decreased biomass (Figure 2B–C S2C–D). In concordance, pp2-a13–1 mutant plants have higher expression of ATG8a mRNA and increased accumulation of the ATG8a protein, similar to the effects seen in autophagy mutants (Figures 3C–D) (Phillips, et al., 2008). Despite these similarities, the pp2-a13–1 mutant shows wild-type levels of survival in darkness or on nitrogen deficient media, contrary to the increased mortality of the autophagy mutants (Figure 3E). To test if the pp2-a13–1 phenotypes are due to defects in autophagy using a genetic approach, we crossed the pp2-a13–1 mutant with the atg5–1 and atg7–2 mutants and observed the phenotypes of the double mutants (Figures 3F–G). The double mutants showed defects in growth that were more severe than either single mutant alone, exclusively in 8L:16D, indicating that PP2-A13 has some functions that are parallel to autophagy in plant cells. Furthermore, the pp2-a13–1 mutant showed no defect in dimerization of ATG5 and ATG12, an event that is disrupted by autophagy mutants (Figure 3H) (Marshall and Vierstra, 2018). Together these results suggest that PP2-A13 and autophagy have important but independent functions in preventing senescence in 8L:16D but also show some level of crosstalk.
PP2-A13 expression is photoperiodically induced
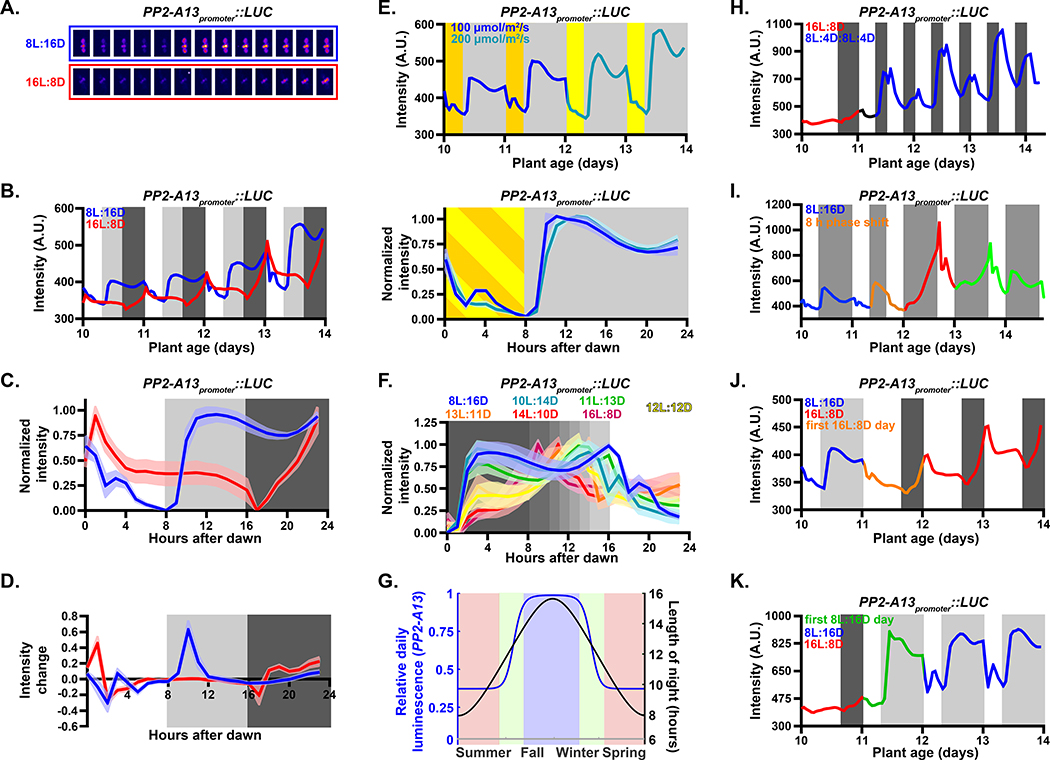
We next generated transgenic plants expressing the Luciferase gene under the control of the PP2-A13 promoter (PP2-A13promoter::Luciferase) (Figure 4A). We measured luminescence from the PP2-A13promoter::Luciferase plants under 8L:16D and 16L:8D conditions (Figure 4B). The patterns generated from this experiment were similar to those seen in the qRT-PCR and microarray experiments (Figure 2A) with the 8L:16D-specific expression peak occurring after dusk, the dawn expression peak in 8L:16D and 16L:8D, and repression by light exposure. To examine the daily expression shape and compare across experiments, we normalized the data to the trough and peak levels. While this removes amplitude information, it gives a clearer view of the comparative expression pattern shapes (Figure 4C). We also calculated the rate of change in intensity (“intensity change”) (Figure 4D). These analyses show that PP2-A13 expression rises rapidly after dusk in 8L:16D and slowly in 16L:8D.
Figure 4. A photoperiod measuring mechanism controls winter photoperiod gene expression.
(A-C) PP2-A13promoter∷Luciferase expression from plants grown under 8L:16D and 16L:8D photoperiods. Grey shading represents the dark period. Lines represent the intensity traces and shading indicates SD. (A) False color images of representative plants taken every two hours from ZT0 to ZT24. (B) Average from traces of raw luciferase intensity. (C) Normalized traces of the daily pattern. (D) Average rate of change in expression. (E) Traces of plants grown under 8L:16D with 100 μM m−2 s−1 light (dark yellow) were transferred into 200 μM m−2 s−1 light (light yellow). (F) Determination of the critical photoperiod. Traces are from plants grown in indicated conditions. (G) Night lengths in Landsberg, Germany (black) and estimated yearly expression pattern (red) of PP2-A13promoter:Luciferase as calculated from the normalized expression in figure 3F. (H) Traces of plants grown under 16L:8D were transferred to double dusk (8L:4D:8L:4D) on day 11. (I) Traces of plants grown under 8L:16D until day 10. On day 11, plants underwent a dawn phase advance of 8 hours. (J) Traces of plants grown under 8L:16D were transferred into 16L:8D on day 11. (K) Traces of plants grown under 16L:8D were transferred into 8L:16D on day 11.
We next tested whether PP2-A13 expression is under the control of a “true” photoperiodic measuring system and functions independent of light intensity within a range. We grew the plants in 8L:16D at 100 μM m−2 s−1 (8L100:16D) and then on day 12 we doubled the light intensity to 200 μM m−2 s−1 (8L200:16D) (Figure 4E and S3A), matching the daily light integral of the 16L:8D experiment in figure 4B. The pattern of PP2-A13 expression was nearly unchanged after doubling the light intensity. We also performed the entire experiment with plants grown in 8L100:16D and 8L200:16D and did not detect a difference in the pattern of PP2-A13 expression (Figure S3B).
To determine the critical photoperiod we imaged the reporter plants in photoperiods ranging from 4L:20D to 20L:4D (Figure 4F, S3C, and S4A–C). Plants grown in photoperiods with longer nights, akin to fall and winter (8L:16D, 10L:14D, and 11L:13D), exhibit the hallmark PP2-A13 8L:16D expression signature. Plants grown in photoperiods with days at least one hour longer than night, akin to late spring and early summer (14L:10D, 16L:8D), exhibit summer photoperiod-like expression patterns. These trends continue in more extreme photoperiods (4L:20D and 20L:4D) (Figure S4B–C). In plants grown in photoperiods with days that are equal to or slightly longer than nights, akin to spring or fall equinox and early spring or late summer (12L:12D, 13L:11D), the expression pattern appears to be in a transitional state with a small expression “shoulder” early in the night, suggesting that these are near the critical photoperiod. Using area under the curve (Figure S4A) and comparing to night lengths over one year in central Germany, where the Columbia ecotype was first isolated (Latitude 48° N) (Figure S4D), we calculated a predicted expression level for PP2-A13 over one full year (Figure 4G). The data shows that the expression pattern of PP2-A13 is not linear with the night length, supporting the idea that a photoperiodic switch controls the observed changes in expression levels.
Using the real-time reporter, we can observe post-dusk induction rates before and after the critical photoperiod in the same 24-hour period (a “double dusk” experiment), which tests for the inducible and non-inducible phases in the same 24-hour day (Vince-Prue, 1975). We performed this experiment by growing the reporter plants in 16L:8D and then exchanging the light cycle with 8L:4D:8L:4D, maintaining the same daily light integral as 16L:8D but providing one dusk prior to the critical photoperiod and one after the critical photoperiod (Figure 4H and S3D). Supporting the idea that the inducible and non-inducible phases occur in the same 24-hour period, the rate of induction and expression peak are higher in the first dark period than the second dark period.
Circadian clock or hourglass-like timers function in photoperiodic measurement systems (Bradshaw and Holzapfel, 2010; Saunders, 2005; Saunders, 1997). A circadian clock-like mechanism takes time to re-entrain to a new dawn after a phase shift while an hourglass, by nature, resets immediately to a new dawn. We grew the plants in 8L:16D and then advanced the phase of dawn by eight hours (Figure 4I and S3E). On day one after the phase advance (Figure 4I, red trace), we observe a PP2-A13 expression pattern that is different than any daily expression pattern observed in previous experiments. On day two and three after the phase shift (Figure 4I, green trace) the expression pattern is returning to the standard 8L:16D pattern seen previously, suggesting PP2-A13 expression is under the control of a circadian clock-like timer.
To determine if PP2-A13 photoperiodic expression is measuring the length of day or length of night, we performed photoperiod shift experiments. We grew plants in 8L:16D and then changed the light cycle to 16L:8D and vice versa (Figure 4J–K and S3F–G). In both experiments, on the first day after the shift the expression patterns reset to the new photoperiod. The plants were able to readjust the post-dusk expression pattern after only experiencing one light period, suggesting that this process counts the number of hours of light.
PP2-A13 photoperiodic expression is independent of the canonical CO/FT photoperiod measuring system
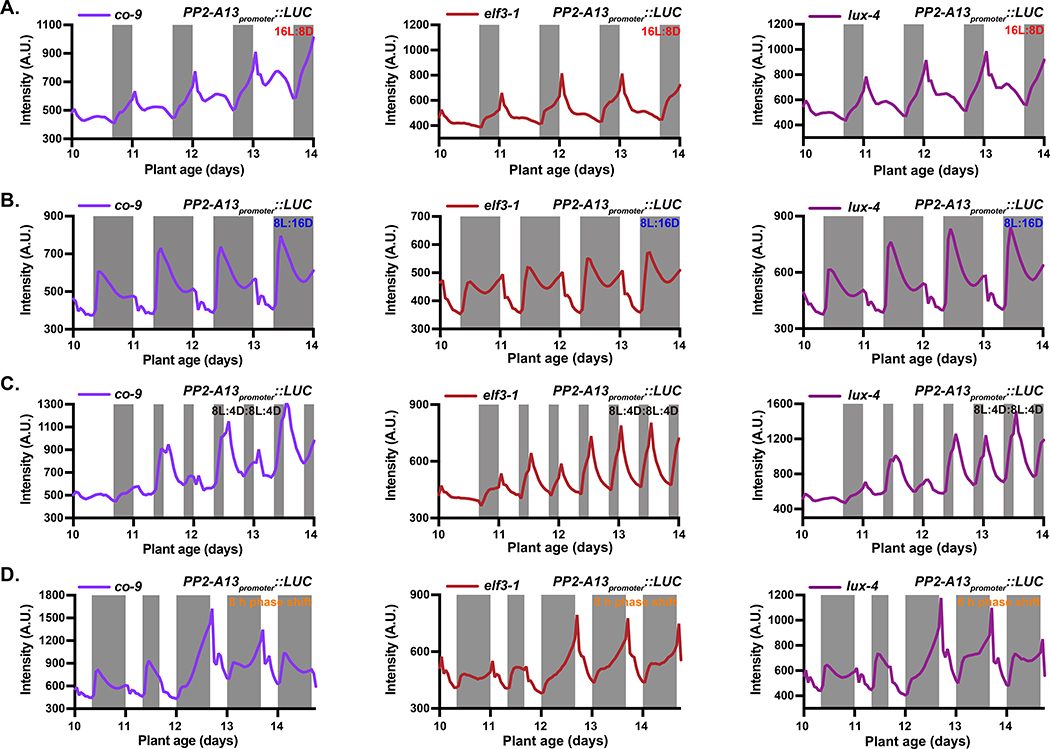
We tested whether the CO photoperiod measuring system controls PP2-A13 expression. We crossed the co-9 mutant into our reporter and grew the plants in 16L:8D and 8L:16D for imaging (Figure 5A–B). We also performed the double dusk (8L:4D8L:4D; Figure 5C), phase shift (Figure 5D), and photoperiod flip experiments (Figure S5A–B). On the whole, the expression pattern of the PP2-A13 reporter was similar between the wild-type and co-9 mutant plants (Figure 4B, 4H, 4I, 4J and 4K), despite the co-9 mutant plants flowering late. Under these conditions, CO seemingly plays little role in the regulation of PP2-A13 expression, but this does not exclude the possibility that communication between winter photoperiod-induced genes and the floral induction pathway exist, as was seen with TEM1 and CO co-regulating FT (Castillejo and Pelaz, 2008).
Figure 5. The circadian clock, independent of CONSTANS, is required for photoperiodic induction of PP2-A13.
(A-B) PP2-A13promoter∷Luciferase traces from co-9, elf3–1, and lux-4 mutant plants grown under 16L:8D (A) and 8L:16D (B). (C) Traces of plant mutants grown under 16L:8D and transferred to double dusk (8L:4D:8L:4D) on day 11. (D) Traces of plant mutants grown under 8L:16D until day 10. On day 11, plants underwent a dawn phase advance of 8 hours but kept in 8L:16D for the remainder of the experiment.
The evening complex, necessary for circadian clock function, is required for the winter-photoperiod measuring mechanism
The transcription factor LUX ARRHYTHMO (LUX) and the associated protein EARLY FLOWERING 3 (ELF3) are part of the evening complex that regulates clock function at the end of the day into the early night (Helfer, et al., 2011; Nusinow, et al., 2011). We crossed our PP2-A13promoter::Luciferase reporter into the lux-4 and elf3–1 mutants that cause arrhythmicity of the circadian clock (Figures 5A–D, Figure S5A–B) (Hazen, et al., 2005; Hicks, et al., 1996). Under 8L:16D and 16L:8D the lux-4 mutant showed wild-type patterns of PP2-A13 expression (Figure 5A–B). The elf3–1 mutant showed a slightly altered pattern in some experiments with a seemingly higher expression peak at dawn (Figure 5A–B). On the third day after a phase shift, wild-type plants showed a pattern of PP2-A13 expression that was identical to the pattern prior to the shift, while at the same time the mutants had not achieved the pre-shift expression patterns (Figure 5D). Interestingly, the elf3–1 mutant also had trouble quickly establishing a new seasonal expression pattern in photoperiod flip experiments, while lux-4 was able to adjust similar to the wild-type plants (Figure S5A–B). Both clock mutants caused disruption in the pattern of PP2-A13 expression in the double dusk experiment (8L:4D:8L:4D) (Figure 5C). lux-4 and elf3–1 were not able to maintain the phased response of PP2-A13 expression to darkness. Combined with the phase shift experiment (Figure 5D) these results suggest that the circadian clock is central to maintenance of PP2-A13 photoperiodic expression.
Light/dark transitions are communicated by the photosynthetic apparatus to control PP2-A13 expression
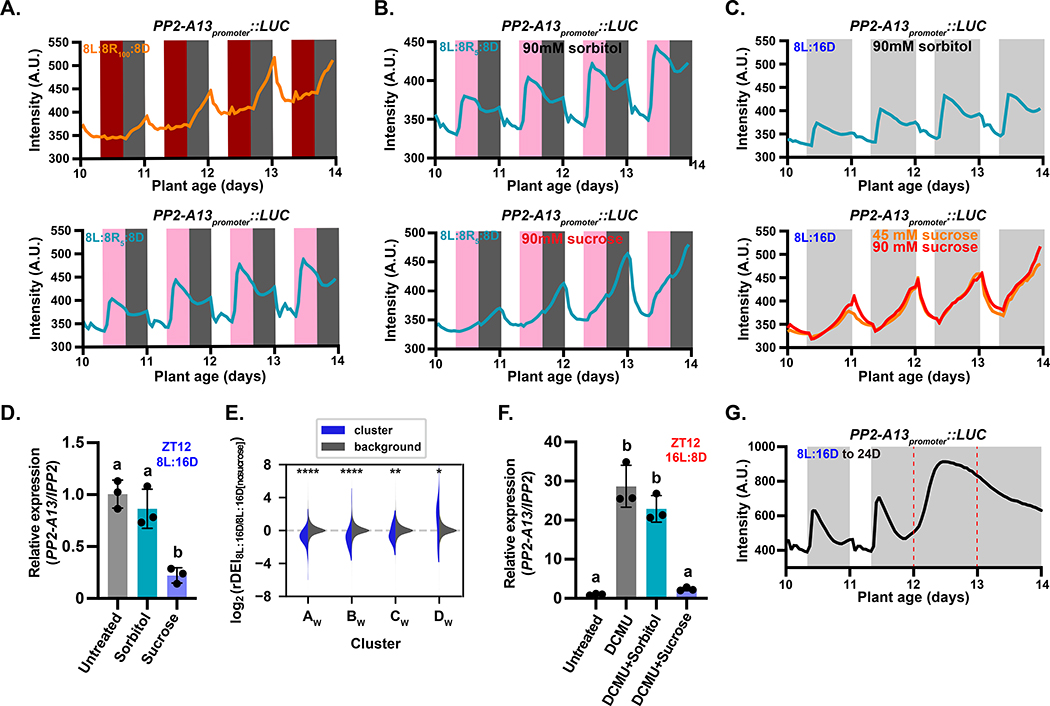
To determine how light/dark transitions are sensed to control PP2-A13 expression, we replaced the first eight hours of darkness in an 8L:16D growth condition with red light (635 nm), a single photosynthetically active wavelength that is sensed by phytochromes, red-light photoreceptors, in plants. This regime was performed at two red light intensities, one at 100 μM m−2 s−1 in which phytochrome signaling is presumably saturated and the intensity is well above the light compensation point (8L:8R100:8D), and the second at 5 μM m−2 s−1 in which phytochrome signaling should be active but is well below the light compensation point for Arabidopsis (the 8L:8R5:8D) (Figure 6A and S6A) (Moraes, et al., 2019). In the 8L:8R100:8D condition, PP2-A13 expression remains low when the lights change to red, similar to the pattern seen in 16L:8D and showing that high red light is sufficient to mimic white light in control of PP2-A13 expression. However, in the 8L:8R5:8D condition, the expression pattern is similar to the 8L:16D expression pattern. This indicates that light intensities above the compensation point are needed to repress PP2-A13 making it likely that the photosynthetic apparatus senses light/dark transitions to control photoperiodic expression of PP2-A13.
Figure 6. The photosynthetic apparatus is necessary for determining light/dark transitions for PP2-A13 photoperiodic expression.
(A-C) PP2-A13promoter ∷Luciferase trace data from plants grown in (A) 8L:8R100:8D (top panel) and 8L:8R5:8D (bottom panel), (B) 8L:8R5:8D treated with 90mM sorbitol (top panel) and 90mM sucrose (bottom panel), (C) Traces of plants grown in 8L:16D treated with 90mM sorbitol (top panel) and 90mM sucrose (bottom panel). (D) qRT-PCR of PP2-A13 from 12-day-old plants grown in 8L:16D. The indicated treatment started at ZT0 and samples were collected at ZT12. IPP2 was used as an internal control. (E) The rDEI8L:16D/8L:16D no sucrose of 8L:16D-induced transcripts (blue) compared to the rDEI8L:16D/8L:16D no sucrose of all other transcripts (grey). rDEI8L:16D/8L:16D no sucrose is calculated as the rDEI of the DIURNAL “shortday” time course divided by that of the “LER_SD” time course. Asterisks: *, p⩽0.05; **; p⩽0.01; ***, p⩽0.0005; ****, p⩽0.0001 (Welch’s t-test). (F) qRT-PCR of PP2-A13 from 12-day-old plants grown in 16L:8D. The indicated treatment started at ZT0 and samples were collected at ZT12. IPP2 was used as an internal control. In both (D) and (F), different letters indicate statistically significant differences as determined by one-way ANOVA followed by Dunnett’s T3 multiple comparison test; p⩽0.05. Error bars indicate SD (n = 3). (G) PP2-A13promoter∷Luciferase traces from plants grown under 8L:16D and transferred to continuous darkness (24D) on day 12. Red dashed lines indicate prospective dawn of day 12 and day 13.
To test if sucrose, a main product of photosynthesis, can alter the PP2-A13 photoperiodic response, we performed imaging experiments in 8L:8R5:8D in the presence of exogenously supplied sucrose (Figure 6B and S6B). The winter photoperiod expression peak of PP2-A13 is nearly ablated when sucrose is supplied to the plants and begins to resemble the expression pattern seen in summer photoperiods. We also tested this in white light with two concentrations of sucrose, both of which suppressed the winter photoperiod expression peak (Figure 6C and S6C). We next grew plants in 8L:16D and treated them with sucrose or sorbitol starting at ZT0 (Zeitgeber time 0). We collected tissue at ZT12 (4 hours post-dusk in 8L:16D), and measured PP2-A13 expression using qRT-PCR (Figure 6D). We found that the sorbitol treatment had little effect on PP2-A13 expression while the sucrose repressed expression, similar to what we found with the reporter. Furthermore, the three night-phased clusters of 8L:16D-induced genes, AW, BW, and CW (Figure 1B), are all repressed by the presence of sucrose in the growth media (Figure 6E).
Next, we grew plants in 16L:8D but blocked photosynthesis using a specific chemical inhibitor of photosystem II called 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU). Technical challenges with the reporter necessitated the use of qRT-PCR. In 16L:8D we treated the plants with DCMU at ZT0 of day 12 (Figure 6F) and collected tissue at ZT12 corresponding to low expression of PP2-A13. In the presence of DCMU, PP2-A13 expression is induced, despite being in the light. This effect was reversed upon the addition of sucrose. To determine if PP2-A13 8L:16D expression pattern is the same as a simple starvation response, we grew the plants in 8L:16D and then transferred them to constant dark (Figure 6G). After the first prospective dawn (day 12) in constant dark, the levels of PP2-A13 rise to a high level, and then diminish as the plants progress to the second day of dark (day 13) different than the pattern of expression seen in 8L:16D.
Starch synthesis and breakdown is required for the winter-photoperiod measuring mechanism
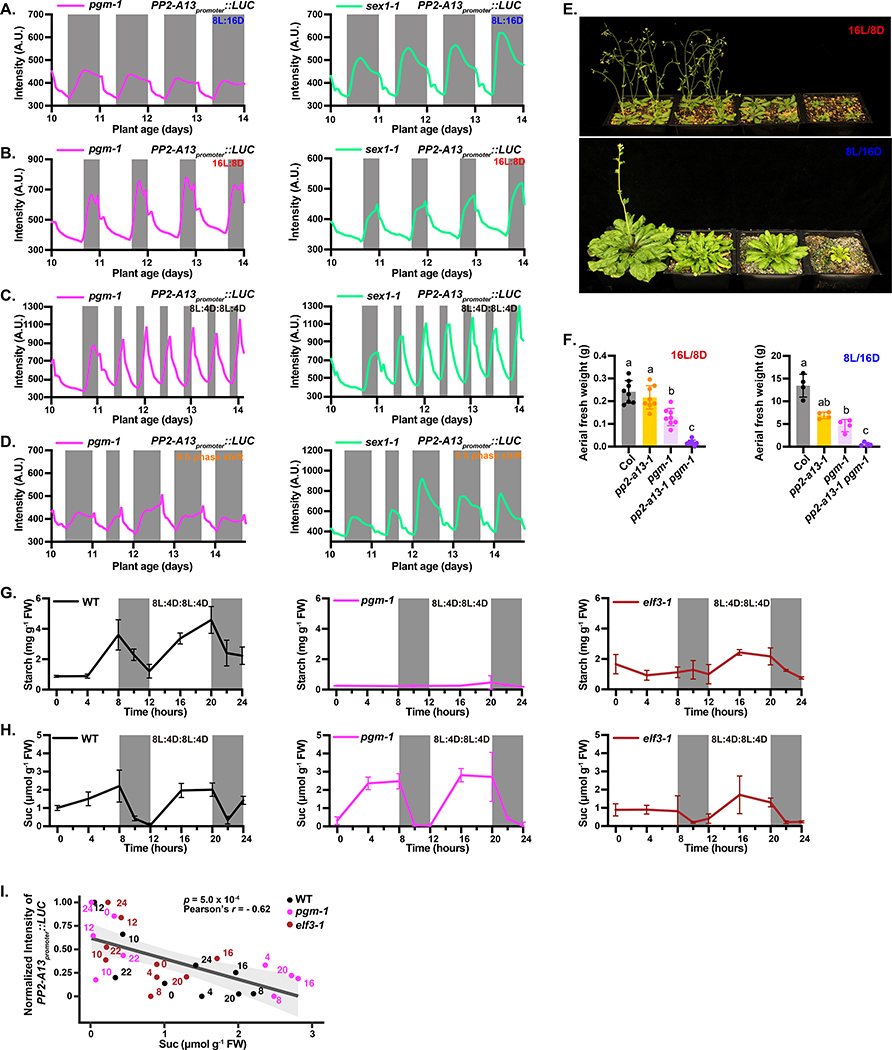
Extensive studies have shown that starch production and breakdown is circadian clock and photoperiod regulated and is critical for maintaining daily and seasonal cellular levels of sucrose produced through photosynthesis (Moraes, et al., 2019; Fernandez, et al., 2017; Kim, et al., 2017; Mengin, et al., 2017; Flis, et al., 2016; Sulpice, et al., 2014; Pal, et al., 2013; Stitt and Zeeman, 2012; Graf, et al., 2010; Gibon, et al., 2009). To test whether starch production is required for photoperiodic induction of PP2-A13 expression, we crossed the PP2-A13promoter::Luciferase reporter into a starchless mutant, phosphoglucomutase (pgm-1) (Caspar, et al., 1985), and starch breakdown mutant, starch excess1 (sex1–1) (Caspar, et al., 1991), and monitored expression in 8L:16D, 16L:8D, double dusk, phase shift, and photoperiod flip experiments (Figure 7A–D and S7A–F).
Figure 7. Starch synthesis and degradation are required for PP2-A13 photoperiodic gene expression.
(A-D) PP2-A13promoter∷Luciferase traces from pgm-1 and sex1–1 mutant plants grown in (A) 8L:16D, (B) 16L:8D, (C) 8L:4D:8L:4D, and (D) phase shift. (E) Representative wild type (Col), pp2-a13–1, pgm-1, and pgm-1 pp2-a13–1 double mutants grown for 31 days in 16L:8D or 11 weeks in 8L:16D. (F) Aerial fresh weight of wild type (Col), pp2-a13–1, pgm-1, and pgm-1 pp2-a13–1 double mutants grown for 25 days in 16L:8D or 11 weeks in 8L:16D. Different letters indicate significant differences as determined by one-way ANOVA followed by Dunnett’s T3 multiple comparison test; p⩽0.05. Error bars indicate SD (n = 4–8). (G-H) Starch (G) and sucrose (H) levels from wild type (Col), pgm-1, and elf3–1 mutants. Note: n = 2 for the starch level at ZT10 in wild type and for the sucrose level at ZT8, 10, and 12 in wild-type and ZT0 in the elf3–1 mutant. In those cases, the error bar indicates the two sample values. n = 3 for all the other time points, error bar indicates SD. (I) Correlation between sucrose levels from wild type (Col), pgm-1, and elf3–1 mutants (Fig. 7H) and averaged normalized levels of PP2-A13promoter∷Luciferase from day 12 to day.14 (WT: Fig. S7E; pgm-1: Fig. 7C; elf3–1: Fig. 5C). Significance was determined by Pearson’s correlation test. Numbers at each data point indicate the number of hours from dawn. Grey line and shading indicate line of best fit and 95% confidence interval, respectively.
In 8L:16D the winter photoperiod expression peak is delayed in both mutants (Figure 7A and S7A–B), while in 16L:8D PP2-A13 expression is more rapidly induced and has two peaks of expression, similar to wild type in short winter-like days (Figure 7B, S7C–D). Similarly, to sucrose suppression in 8L:16D, this first peak in expression seen in 16L:8D is also suppressed by sucrose (Figure S7G–H). In the double dusk condition the mutants are unable to maintain the dark inducible and non-inducible phases of PP2-A13 expression, similar to what was seen in the lux-4 and elf3–1 mutants. One notable difference is the loss of the phases occurs on the first day of 8L:4D:8L:4D (Figure 7C and S7E–F), whereas the clock mutants lose the phased response later in the 8L:4D:8L:4D regime.
The loss of the circadian clock makes it difficult for PP2-A13 to quickly re-entrain to a new dawn (Figure 5D). Conversely, in the phase shift experiment with the starch mutants, the expression pattern of PP2-A13 quickly re-entrains to the mutant 8L:16D photoperiodic expression pattern (Figure 7D). The shortened re-entrainment time indicates that the impact of the circadian clock on PP2-A13 has been diminished. Additionally, the photoperiod flip experiments also show that the mutants quickly adapt to a new dusk (Figure S7I–J).
The starch mutants are inappropriately activating PP2-A13 in 16L:8D (Figure 7B). To test this genetically, we crossed the pgm-1 mutant with the pp2-a13–1 mutant and found growth defects in both 8L:16D and 16L:8D in the double mutant plants (Figure 7E–F). This indicates that proper starch production is necessary for Arabidopsis to restrict winter photoperiod gene function to short days.
Starch regulated sucrose levels correlate with PP2-A13 expression
Previous studies show that photoperiod strongly influences the levels of daily sucrose by controlling the synthesis and breakdown of starch (Mengin, et al., 2017; Martins, et al., 2013; Matsoukas, et al., 2013; Pal, et al., 2013; Blasing, et al., 2005). To determine if sucrose levels correlate with PP2-A13 expression, we measured starch and sucrose in the double dusk growth condition in the wild type, pgm-1, and elf3–1 mutants (Figure 5C and 7C). From these experiments it appeared that low levels of sucrose correlate with high levels of PP2-A13 expression and vice versa (Figure 7G–H). To confirm this, we performed correlation analysis (Figure 7I) of the sucrose levels (Figure 7H) and PP2-A13 expression (Figure 5C, 7C, S7E). The levels of sucrose negatively correlate with the expression of PP2-A13 showing that sucrose can repress expression of PP2-A13. The expression of PP2-A13 also corresponds well with published data from other groups showing the levels of sucrose across a wide range of photoperiods in wild-type plants (Mengin, et al., 2017; Sulpice, et al., 2014; Lu, et al., 2005). Briefly, the levels of sucrose drop to low levels after dusk when photoperiods are shorter than 12L:12D. Conversely, the levels of sucrose remain at higher levels, post-dusk, when photoperiods are 12L:12D or longer.
Discussion
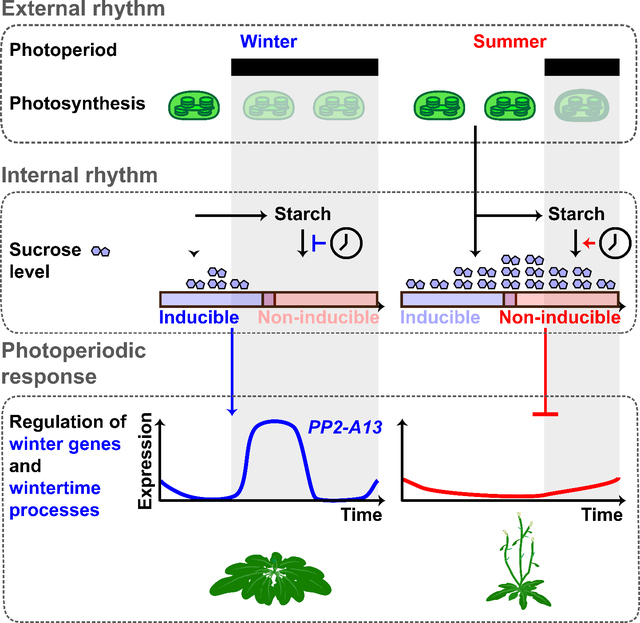
The visually stunning transition from vegetative growth to flowering, which is often under tight control of day length, has made plants a premier system for understanding photoperiod measuring systems (Lumsden and Millar, 1998; Vince-Prue, 1975; Bunning, 1969). Winter often appears to be a time of biological inactivity, but here we show that plants are actively promoting the expression of genes to maintain fitness in winter photoperiods. We adopted the nomenclature of winter photoperiod-induced genes rather than short day-induced genes to distinguish these genes from those controlled by short day floral inductive programs. This nomenclature may in fact be more accurate because the maximal expression of these genes is centered on the winter solstice and the critical photoperiod is not centered directly on the equinoxes.
The expression patterns of the PP2-A13promoter:Luciferase reporter follow the rules of the external coincidence model of photoperiodic measuring mechanisms. Despite similarities with the photoperiodic control of FT expression, we find that the canonical CO/FT photoperiodic measuring system has little effect on PP2-A13 expression. Instead, photoperiodic control of winter photoperiod gene expression relies on a system where the timing of light dark transition is interpreted by the metabolic systems of the plant. Specifically, light and dark are sensed by the photosynthetic apparatus and converted to a chemical signal in the form of sucrose. Circadian clock-controlled starch synthesis and breakdown act to control cellular sucrose levels during the 24-hour day resulting in two different responses to darkness across the day. These two responses reflect the probability of reaching adequately low levels of sucrose after dark to induce PP2-A13 induction. Rapid induction of PP2-A13 and other winter photoperiod-induced genes occurs when dusk coincides with the early part of a 24-hour day, starch degradation is slowed to maintain energy levels across the long night, and sucrose quickly drops to low levels. Slow PP2-A13 induction occurs when dusk coincides with the later part of a 24-hour day, starch degradation rate is high, and sucrose levels remain high through the short night (Figure S7K) (Seaton, et al., 2018; Fernandez, et al., 2017; Mengin, et al., 2017; Flis, et al., 2016; Lu, et al., 2005).
While we have established some of the cellular systems that drive this system, it will be critical to continue exploring additional component parts. We have explored the role of sucrose in this process, but it remains to be seen whether other metabolites can impact winter photoperiod gene expression. Additionally, sucrose seems to play a dominant role in controlling winter photoperiod gene expression, but we have not eliminated the possibility that redox signaling emanating from the photosynthetic apparatus could also participate in this process. We believe that our data indicate that sucrose is regulated in a photoperiodic manner to control downstream genes, such as PP2-A13. Although, it is possible that the clock integrates into this system at additional points such as controlling sensitivity to, transport of, or metabolism of sucrose differentially across the day. It will be important to determine how sucrose is converted into a transcriptional signal in this system, and if other metabolites can participate in winter photoperiod gene control. Our luciferase reporter, akin to the real-time circadian clock reporters (Millar, et al., 1995a; Millar, et al., 1995b; Millar, et al., 1992), will pave the way for identifying these components using a host of genetic and reverse genetic approaches.
We chose to focus our attention on PP2-A13 because the insertion mutant line has striking and easily observable developmental defects (Figure 2 and S2). Here we show that PP2-A13 functions in a plant cellular pathway that is parallel to autophagy but seemingly equally important to plant survival in winter photoperiods. PP2-A13 is a lectin-containing F-box protein, and it will now be important to further define the sugar-binding specificity and scope of potential targets of PP2-A13 to refine our understanding of its function expand our knowledge of the cellular pathways that it controls. It will also be important to further explore other winter photoperiod-induced genes and their role in vegetative health and reproduction.
Plants predict both adverse and beneficial seasonal changes by measuring photoperiod, but climate change is rapidly decoupling photoperiod from important seasonal cues such as temperature and water availability (Inoue, et al., 2020; Walker, et al., 2019; Stromme, et al., 2017; Fournier-Level, et al., 2016; Diez, et al., 2014). Importantly to our work, climate change has a disproportionately large effect on winter (Kreyling, 2010) and many plants need winter signals for proper reproductive and vegetative development, making it important that we expand our knowledge of photoperiodism in plants.
Limitations of the study
There are a few limitations of the luciferase reporter system used here. It does not report on post-transcriptional changes and does not contain introns, exons or the 3’UTR of PP2-A13 that may play a role in regulation of transcription. Additionally, the microarray experiment used for the rDEI analysis is not optimal. An RNA-seq experiment covering multiple photoperiods is necessary to determine a fuller complement of photoperiod-controlled genes.
In this study we did not define how sucrose is converted into transcription of downstream genes. It is possible that this is mediated by direct sucrose signaling, signaling by sugars that are by-products of sucrose, or through sensing pf physiological changes in cells caused by variations in sucrose concentration.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Actin antibody (rabbit) | Millipore-Sigma | Cat#SAB4301137 |
| anti-ATG5 antibody (rabbit) | Agrisera | Cat#AS15 3060 |
| anti-ATG8a antibody (rabbit) | Abcam | Cat#ab77003 |
| Bacterial and virus strains | ||
| Agrobacterium tumefaciens | Widely distributed | Strain GV3101 |
| Chemicals, peptides, and recombinant proteins | ||
| 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) | Sigma-Aldrich | Cat#D2425 |
| 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid, cyclohexylammonium salt (X-gluc) | Thermo Fisher Scientific | Cat#R0851 |
| Agar | AmericanBio | Cat#AB01185 |
| cOmplete EDTA-free Protease Inhibitor Cocktail | Roche | Cat#11873580001 |
| D-luciferin | Cayman Chemical Company | Cat#115144-35-9 |
| Murashige & Skoog Basal Medium (MS) | Cassion Laboratories | Cat#MSP01 |
| Murashige & Skoog Basal Medium without nitrogen | PhytoTech Labs | Cat#M531 |
| Phenylmethylsulfonyl fluoride (PMSF) | AmericanBio | Cat#AB01620-00005 |
| TRIzol™ | Thermo Fisher Scientific | Cat#15596026 |
| Critical commercial assays | ||
| iScript™ Reverse Transcription Supermix | Bio-Rad | Cat#1708841 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat#1725121 |
| LR Clonase II | Invitrogen | Cat#11791020 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 |
| RNeasy Plant Mini kit | Qiagen | Cat#74904 |
| RNase-Free DNase Set | Qiagen | Cat#79254 |
| Sucrose/D-Fructose/D-Glucose Assay Kit | Megazyme | Cat#K-SUFRG |
| TOPO cloning kit | Invitrogen | Cat#K240020 |
| Total Starch Assay Kit | Megazyme | Cat#K-TSHK |
| Deposited data | ||
| Arabidopsis genome annotation | TAIR10 | www.arabidopsis.org |
| Microarray “longday” and “shortday” datasets | (Michael, et al., 2008b) | E-MEXP-1304 |
| Microarray “LER_SD” dataset | (Michael, et al., 2008a) | E-MEXP-1299 |
| Time course microarray data | DIURNAL Project | diurnal.mocklerlab.org |
| Experimental models: Organisms/strains | ||
| Arabidopsis thaliana: Col-0 wild type | Widely distributed | N/A |
| Arabidopsis: pp2-a13-1 | ABRC | SALK_101611 |
| Arabidopsis: pgm-1 | ABRC | CS210 |
| Arabidopsis: sex1-1 | ABRC | CS3093 |
| Arabidopsis: co-9 | ABRC | CS870084 |
| Arabidopsis: atg5-1 | ABRC | CS39993 |
| Arabidopsis: atg7-2 | ABRC | CS69859 |
| Arabidopsis: elf3-1 | (Nusinow, et al., 2011) | N/A |
| Arabidopsis: lux-4 | (Nusinow, et al., 2011) | N/A |
| Arabidopsis: PP2-A13promoter::Luciferase | This paper | N/A |
| Arabidopsis: PP2-A13promoter::gPP2-A13 in pp2-a13-1 | This paper | N/A |
| Arabidopsis: atg5-1 pp2-a13-1 | This paper | N/A |
| Arabidopsis: atg7-2 pp2-a13-1 | This paper | N/A |
| Arabidopsis: pgm-1 pp2-a13-1 | This paper | N/A |
| Arabidopsis: PP2-A13promoter::Luciferase in co-9 | This paper | N/A |
| Arabidopsis: PP2-A13promoter::Luciferase in pgm-1 | This paper | N/A |
| Arabidopsis: PP2-A13promoter::Luciferase in sex1-1 | This paper | N/A |
| Arabidopsis: PP2-A13promoter::Luciferase in elf3-1 | This paper | N/A |
| Arabidopsis: PP2-A13promoter::Luciferase in lux-4 | This paper | N/A |
| Oligonucleotides | ||
| See Table S5 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pENTR-D-TOPO | Thermo Fisher Scientific | Cat#K240020 |
| Plasmid: pFlash | (Gendron, et al., 2012) | N/A |
| Plasmid: pGWB16 | (Nakagawa, et al., 2007) | N/A |
| Plasmid: pMDC164 | (Curtis and Grossniklaus, 2003) | N/A |
| Plasmid: pSAT6-mCherry-VirD2NLS | (Citovsky, et al., 2006) | CD3-1106 |
| Plasmid: pENTR- PP2-A13promoter (genomic) | This paper | N/A |
| Plasmid: pFlash-PP2-A13promoter::Luciferase (genomic) | This paper | N/A |
| Plasmid: pGWB16-PP2-A13promoter::gPP2-A13-4XMYC (genomic) | This paper | N/A |
| Plasmid: 35S::PP2-A13-GFP (cDNA) | This paper | N/A |
| Plasmid: pMDC164- A13promoter::GUS (genomic) | This paper | N/A |
| Software and algorithms | ||
| Code used is attached to this paper | This paper | N/A |
| Fiji | (Schindelin, et al., 2012) | imagej.net/software/fiji |
| ImageJ | (Schneider, et al., 2012) | imagej.nih.gov/ij |
| ggplot2 R package | (Wickham, 2009) | N/A |
| ggpubr R package | (Kassambara, 2020) | N/A |
| PRISM 8 | GraphPad | www.graphpad.com |
| Python (3.8.3) | Python Software Foundation | N/A |
| R(4.0.1) | The R Project for Statistical Computing | N/A |
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Joshua M. Gendron (joshua.gendron@yale.edu).
Materials Availability
Plasmids generated in this study will be made available on request with completion of an MTA for thirdparty components. Seed lines generated in this study will be made available on request with completion of appropriate governmental regulatory paperwork.
Data and Code Availability
The accession numbers for the datasets reported in this paper are ArrayExpress E-MEXP-1304 (Michael, et al., 2008b) and E-MEXP-1299 (Michael, et al., 2008a). The data are also available on http://diurnal.mocklerlab.org/. All code used to generate and analyze all datasets in this study are provided in supplementary materials.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Arabidopsis materials
The Arabidopsis seeds of Col-0, pp2-a13–1 (SALK_101611), pgm-1 (CS210), sex1–1 (CS3093), co-9 (CS870084), atg5–1 (CS39993), and atg7–2 (CS69859) were obtained from ABRC. The elf3–1 and lux-4 mutant seeds were obtained from Dr. Dmitri Nusinow (Nusinow, et al., 2011). The PP2-A13promoter::Luciferase transgenic line was generated by transformation of agrobacteria GV3101 harboring PP2-A13promoter::Luciferase construct into Arabidopsis Col-0 background and the transgenic plants were selected by gentamicin antibiotic and genotyping. The PP2-A13 complementation line was generated by transformation of agrobacteria GV3101 harboring PP2-A13promoter::gPP2-A13 construct into the pp2-a13–1 mutant background and the transgenic plants were selected by hygromycin and genotyping. The pp2-a13–1 mutant was also crossed to atg5–1, atg7–2, and pgm-1 mutant plants and the atg5–1 pp2-a13–1, atg7–2 pp2-a13–1, and pgm-1 pp2-a13–1 double mutants were identified by genotyping. The PP2-A13promoter::Luciferase transgenic line was crossed to co-9, pgm-1, sex1–1, elf3–1, and lux-4 mutant plants and the homozygous lines were identified by genotyping and bioluminescence imaging. The pgm-1 allele was genotyped as described by (Veley, et al., 2012). The sex1–1 and elf3–1 alleles were genotyped by PCR followed by StyI and HincII digestion, respectively. The lux-4 allele was genotyped by PCR followed by sequencing. The primers used for genotyping are listed in table S5.
Arabidopsis growth conditions
Arabidopsis thaliana seeds from Col-0, mutants, and transgenic lines were surface sterilized for 20 minutes in 70% ethanol with 0.1% Triton X-100 then sown on freshly poured 1/2 MS plates, pH 5.7, (Cassion Laboratories, cat. # MSP01) and 0.8% bacteriological agar (AmericanBio cat. # AB01185) without sucrose. The seeds were stratified in the dark for two days at 4 °C then transferred into 22°C, 12L:12D illuminated by white fluorescent lamps at 150 μmol m2 s−1 for seven days. The seven-day-old seedlings were then transferred to different photoperiod for given experiments as indicated. For soil grown plants, after two days stratification, seeds were germinated and grown in Fafard-2 mix at 22°C in 16L:8D or 8L:16D with light intensity of 100 μmol m2 s−1 for the indicated duration.
METHOD DETAILS
Plasmid construction
To generate the PP2-A13promoter::LUC construct, a 2233 bp promoter sequence upstream of the PP2-A13 coding sequence, including 5’ UTR, was obtained by PCR and inserted into pENTR/D-TOPO vector (Invitrogen, cat. # K240020) and then transferred into the pFLASH destination vectors to drive the luciferase (Gendron, et al., 2012).
For the PP2-A13 complementation plasmids, the PP2-A13promoter::gPP2-A13 fragment was generated from PCR using Col-0 genomic DNA as the template, inserted into pENTR/D-TOPO and then transferred into pGWB16 destination vectors using LR recombination (Nakagawa, et al., 2007).
To generate the PP2-A13promoter::GUS construct, the 2233 bp promoter sequence was subcloned from entry vector pENTR-PP2-A13promoter to the destination vector pMDC164 by LR recombination (Curtis and Grossniklaus, 2003).
The primers used for cloning are listed in table S5.
Luciferase imaging and analysis
Seven-day old seedlings grown in 12L:12D at 22°C were transferred onto a 10 × 10 grid freshly poured 100 mm square MS plates with or without added sugars as indicated for a given experiment. Seedlings were then treated with 5 mM D-luciferin (Cayman Chemical Company, cat. # 115144–35-9) dissolved in 0.01% TritonX-100 and imaged at 22°C under the indicated conditions. Under light conditions, lights were on for 52 minutes of every hour: the lights are off for two minutes prior to a five-minute exposure collected on an Andor iKon-M CCD camera, and then remain off for one minute following the exposure. During the dark period, images were taken during the same five-minute time period. Light was provided by two LED light panels (Heliospectra L1) with light fluence rate of 100–150 μmol m−2 s−1, unless otherwise indicated. The CCD camera was controlled using Micromanager, using the following settings: binning of 2, pre-amp gain of 2, and a 0.05 MHz readout mode (Edelstein, et al., 2014). Using this setup, up to 400 seedlings are simultaneously imaged across four plates. Images are acquired each hour for approximately six and a half days. Data was collected for the entire imaging period (the end of day 7 through the dawn of day 14 or 15). The mean intensity of each seedling at each time point was calculated using ImageJ (Schneider et al., 2012). These raw values are presented as raw trace plots.
Normalization of luciferase imaging data
For normalization, the maximum and minimum expression values in a 25 hour period (defined as either one hour before dawn to the subsequent dawn or one hour before dusk to the subsequent dusk, as indicated for each experiment) were calculated. The minimum expression value was subtracted from each expression value, then this value was divided by the difference in expression between the maximum and minimum expression within that 24 hour period.
The mean and standar within an experiment of the same light conditions, unless otherwise indicated. Only the normalized expression values from dawn to dawn or dusk to dusk are plotted. The rate of c also calculated from the normalized expression values by calculating the difference between the expression at time t and the expression at time t–1. Because of the nature of this calculation, only 24 rate values are calculated. The mean and standard deviation of these rate values were calculated for all days within an experiment of the same light conditions, unless otherwise indicated.
Estimation of yearly expression of PP2-A13promoter::Luciferase
The total PP2-A13promoter::Luciferase intensity is first determined by taking the area under the curve, using the trapezoidal rule for numerical integration, for the six different light/dark conditions in figure S4. Since the plots in figure S4 are averaged over multiple days, a correction in the total PP2-A13promoter::Luciferase intensity for the growth of the plant should be included. This is done by taking the intensity value at dusk and at 23 hours after dusk, connecting these points with a straight line, evaluating the resulting area under the curve (area of a triangle), then subtracting the total area under the curve by that triangular area. The area correction helps diminish the effects of plant growth. These corrected areas are then divided by the largest value (the 8L:16D condition) to obtain the normalized PP2-A13promoter::Luciferase intensity. The normalized intensities are then fit with an approximately sigmoid function
The built-in non-linear data fitting tool in Xmgrace was used to determine the best fit parameters to the data are c1 = 0.62, c2 = 26.27, c3 = 12.67, and c4 = 0.37.
Using the sigmoidal fit from figure S5, the expression of PP2-A13promoter::Luciferase over the course of a year is estimated. Since Arabidopsis Columbia ecotype was first isolated in Landsberg, Germany (https://peerj.com/preprints/26931v5/) (latitude, ~ 48°N), the length of the night for each day in 2019 in Landsberg, Germany (https://www.timeanddate.com/sun/germany/landsberg-am-lech) was used to estimate the daily normalized expression of PP2-A13promoter::Luciferase.
qRT-PCR
For qRT-PCR experiments, RNA extraction was performed with two different methods. For figures 2A, total RNA was extracted from Arabidopsis seedlings grown in indicated conditions using TRIzol™ reagent (ThermoFisher, cat. # 15596026); for the remaining figures 3C, 6D, 6F, and S2B, extraction was performed with RNeasy Plant Mini Kit (QIAGEN cat. # 74904). In both methods, the resulted RNA was subsequently treated with DNase (QIAGEN, cat. # 79254). The subsequent reverse-transcription and conditions for qRT-PCR reactions were described previously with minor modifications (Lee, et al., 2018). Briefly, four hundred nanograms of total RNA were used for reverse-transcription using iScript™ Reverse Transcription Supermix for RT-qPCR (Bio-Rad, cat. # 1708841). iTaq Universal SYBR Green Supermix was used for qRT-PCR reaction (Bio-Rad, cat. # 1725121). IPP2 (AT3G02780) or UBQ10 (AT4G05320) was used as an internal control as indicated. The relative expression represents means of 2(−ΔCT) from three biological replicates, in which ΔCT = (CT of Gene of Interest – CT of internal control). The primers used are listed in table S5.
Clustering analysis
The time-course microarray dataset was downloaded from the DIURNAL database (ftp://www.mocklerlab.org/diurnal) (Michael, et al., 2008b; Mockler, et al., 2007). Relative daily expression integral for a transcript was calculated as: (sum of expression values in the DIURNAL “shortday” 8L:16D condition) / (sum of expression values in the DIURNAL “longday” 16L:8D condition). For the k-means clustering by both 16L:8D and 8L:16D expression values (Figure 1B), we performed log2-transformation followed by Z-score transformation in a gene-wise manner across both 16L:8D and 8L:16D expression values. We performed k-means clustering with the ‘kmeans’ function from scikit-learn python package (Pedregosa, et al., 2011) and determined the number of clusters using the elbow method with inertia.
For the hierarchical clustering analysis (Figure 1C), we performed log2-transformation of the data followed by Z-score transformation in a gene-wise manner separately for each time course to obtain the pattern. Principal components amounting to just above 90% of the total variance were used for clustering using the ‘factoextra’ R package (Kassambara and Mundt, 2020). factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package version 1.0.7. https://CRAN.R-project.org/package=factoextra). Gene-wise Pearson correlation was used as similarity measure for hierarchical clustering using the R ‘hclust’ function with average linkage. The ‘cutreeDynamic’ function from the ‘dynamicTreeCut’ R package (Langfelder, et al., 2008) was used to identify clusters from the dendrogram, with the parameters: method=“hybrid”, minClusterSize=50, deepSplit=1, pamStage=FALSE. For figure 1D, clusters of strongly photoperiodic expression were identified by testing the mean log2 (rDEI8L:16D/16L:8D) of the cluster against zero using the one-sample Wilcoxon signed rank test. All three identified clusters with −log10 (adjusted p-value) > 20 (Bonferroni correction) were 8L:16D-induced. All code used for clustering analysis are provided in the supplementary materials.
Functional enrichment analysis
Only clusters that have at least 40 transcripts were tested for enrichment of functional annotations. Enrichment analysis of Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was performed with the R package ‘clusterProfiler’, using the enrichGO function and the enrichKEGG function with the parameters: pAdjustMethod = “BH”, pvalueCutoff = 0.05, qvalueCutoff = 0.05, respectively (Yu, et al., 2012; Hvidsten, et al., 2001; Kanehisa and Goto, 2000; Ogata, et al., 1998). Highly similar GO terms were merged with the ‘simplify’ function with the parameters: cutoff = 0.5, measure = ‘Wong’, by=‘p.adjust’. Since redundant annotations were still present after merging, notable annotations were manually selected for figure 1B. The full list of annotations is available in supplemental tables 2 and 3.
GUS histochemical analysis
For GUS assay, the PP2-A13promoter::GUS transgenic plant was grown in 12L:12D for 12 days and then transferred to 8L:16D for 3 more days. The plant was freshly harvested and stained at 37 °C over night with 2 mM 5-bromo-4-chloro-3-indolyl-beta-D-glucuronic acid (X-gluc) in 100 mM potassium phosphate buffer, pH 7.0, containing 0.1% (v/v) Triton X-100, 1 mM K3Fe(CN)6 and 10 mM EDTA. Tissues were cleared before observation by washing with 75% (v/v) ethanol.
Subcellular localization
For subcellular localization studies, the coding sequences of the PP2-A13 gene were recombined into pGW-GFP vector which harbors an in-frame C-terminal GFP and is driven by the Cauliflower mosaic virus (CaMV) 35S promoter. The 35S::PP2-A13-GFP construct was co-transformed with 35S::mCherry-VirD2NLS as a nuclear marker (Citovsky, et al., 2006). Arabidopsis protoplast transfection was performed as previously described (Yoo, et al., 2007) and the subcellular localization of the fluorescent-tagged protein was detected with a Nikon ECLIPSE Ti confocal microscope system.
Immunoblotting
To detect ATG8a by immunoblotting analysis, 11-week-old soil grown wild-type (Col-0) and pp2-a13–1 mutant plants were harvested and ground in liquid nitrogen. Crude proteins were extracted with SII buffer (100 mM sodium phosphate, pH 8.0, 150 mM NaCl, 5 mM EDTA, and 0.1% (v/v) Triton X-100) with cOmplete EDTA-free Protease Inhibitor Cocktail (Roche, cat. # 11873580001) and 1 mM phenylmethylsulfonyl fluoride (PMSF). Protein concentration was quantified with a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, cat. # 23225). Approximately 50 μg of total protein was loaded and separated on 12% (w/v) SDS-PAGE for immunoblot analyses. ATG8a levels were detected with anti-ATG8a antibody (1:1000; abcam, ab77003). To detect ATG5 and ATG5-ATG12 complex, wild-type (Col-0), pp2-a13–1, atg5–1, and atg7–2 mutant plants were grown in 8L:16D for 14 days. Crude proteins from the plants were extracted as above. Approximately 80 μg of total protein was loaded for immunoblot analyses. ATG5 and ATG5-ATG12 levels were detected with anti-ATG5 antibody (1:3000; Agrisera, AS15 3060). Equal protein loads were confirmed by immunoblot analysis with anti-Actin antibody (1:3000; Millipore-Sigma, SAB4301137).
Sucrose and starch measurement
Sucrose and starch measurement were followed as described previously with minor modifications (Karlsson, et al., 2015). Briefly, wild-type (Col-0), pgm-1, and elf3–1 mutant plants were grown under 16L:8D conditions and transferred to double dusk (8L:4D:8L:4D) on day 11. Around 100 mg of Arabidopsis seedlings were harvested at day 12 from the indicated time points in the 8L:4D:8L:4D cycles and frozen immediately in liquid nitrogen. Soluble sugars were extracted three times at 80°C for 30 min each, first time with 800 μL of 95% (v/v) ethanol and 2 mM HEPES (pH 7.5) at a ratio of 4:1 and second time with 400 μL of 95% (v/v) ethanol and 2 mM HEPES (pH 7.5) at a ratio of 1:1. Third time with 200 μL of 95% (v/v) ethanol and 2 mM HEPES (pH 7.5) at a ratio of 4:1. Supernatants after three extractions were pooled, dried with Vacufuge Concentrator (Eppendorf 5301) and then dissolved again with water. Soluble sucrose was measured by spectrophotometry with a Sucrose/Fructose/Glucose kit (Megazyme, K-SUFRG) according to the manufacturer’s manual. The pellets were also dried and analyzed by enzymatic hydrolysis to glucose and assay with a Total Starch kit (Megazyme, K-TSHK) according to the manufacturer’s manual.
QUANTIFICATION AND STATISTICAL ANALYSIS
All details of the statistical analysis of the data from this paper are provided in the respective part of the STAR Methods section and all statistical parameters are indicated in the figure legends. The correlation analysis between sucrose content (Figure 7H) and luciferase intensity of the PP2-A13promoter::Luciferase reporter (Figure 5C, 7C, S7E) was performed using R. Line of best fit and the confidence interval were calculated with the “geom_smooth(method = ‘lm’)” function from the R package “ggplot2” (Wickham, 2009). Correlation test was performed with the “stat_cor()” function from the R package “ggpubr” (Kassambara, 2020). GraphPad PRISM 8 was also used for plotting graphs and performing statistical analyses.
Supplementary Material
Table S1: Relative daily expression integral (rDEI) of the probes of ATH1 microarray (Related to Figure 1).
Table S2: Description and enriched annotations of 8L:16D-induced clusters (Related to Figure 1).
Table S3: Description and enriched annotations of 16L:8D-induced clusters (Related to Figure 1 and Figure S1).
Table S4: Description and enriched annotations of clusters identified by hierarchical clustering (Related to Figure 1).
Highlights.
Winter photoperiod-induced genes were identified in Arabidopsis
PP2-A13 is expressed and required for plant fitness in winter
Photoperiodic control of PP2-A13 is independent of the CO/FT mechanism
Photoperiodic expression of PP2-A13 is mediated by plant metabolic systems
ACKNOWLEDGEMENTS
We would like to thank Christopher Adamchek and Suyuna Eng Ren for their technical support. We would also like to thank Sandra Pariseau for administrative support. Additionally, we would like to thank Chris Bolick, Eileen Williams, and the staff at Marsh Botanical Gardens for their support in maintaining plant growth spaces. We would like to thank Dr. Shirin Bahmanyar for help with confocal microscopy. We would also like to thank Dr. Shirin Bahmanyar, Dr. Qingqing Wang, Christopher Adamchek, Harper Lowrey, and Lilijana Oliver for critical reading of the manuscript. We would like to thank Dr. Joel Greenwood for invaluable technical support for our automated luciferase imaging system. This work was supported by the National Science Foundation (EAGER #1548538) and the National Institutes of Health (R35 GM128670) to J.M.G; and by the National Institutes of Health (T32 GM007499), the Gruber Foundation, and the National Science Foundation (GRFP DGE-1122492) to A.F. W.L. was supported by the Forest BH and Elizabeth DW Brown Fund Fellowship. DT was supported by the National Institutes of Health (T32GM007223–44).
Footnotes
DECLARATION OF INTERESTS
All authors claim no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pineiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development (Cambridge, England) 131, 3615–26. [DOI] [PubMed] [Google Scholar]
- Azeez A, and Sane AP (2015). Photoperiodic growth control in perennial trees. Plant signaling & behavior 10, e1087631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, and Stitt M (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17, 3257–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, and Nilsson O (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science (New York, N.Y.) 312, 1040–3. [DOI] [PubMed] [Google Scholar]
- Bouche F, Woods DP, and Amasino RM (2017). Winter Memory throughout the Plant Kingdom: Different Paths to Flowering. Plant physiology 173, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw WE, and Holzapfel CM (2010). What season is it anyway? Circadian tracking vs. photoperiodic anticipation in insects. J Biol Rhythms 25, 155–65. [DOI] [PubMed] [Google Scholar]
- Bunning E (1969). Common features of photoperiodism in plants and animals. Photochem Photobiol 9, 219–28. [DOI] [PubMed] [Google Scholar]
- Caspar T, Huber SC, and Somerville C (1985). Alterations in Growth, Photosynthesis, and Respiration in a Starchless Mutant of Arabidopsis thaliana (L.) Deficient in Chloroplast Phosphoglucomutase Activity. Plant physiology 79, 11–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T, Lin TP, Kakefuda G, Benbow L, Preiss J, and Somerville C (1991). Mutants of Arabidopsis with altered regulation of starch degradation. Plant physiology 95, 1181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C, and Pelaz S (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Current biology : CB 18, 1338–43. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, and Tzfira T (2006). Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362, 1120–31. [DOI] [PubMed] [Google Scholar]
- Cornah JE, Germain V, Ward JL, Beale MH, and Smith SM (2004). Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. The Journal of biological chemistry 279, 42916–23. [DOI] [PubMed] [Google Scholar]
- Cubas P (2020). Plant Seasonal Growth: How Perennial Plants Sense That Winter Is Coming. Current biology : CB 30, R21–R23. [DOI] [PubMed] [Google Scholar]
- Curtis MD, and Grossniklaus U (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant physiology 133, 462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez JM, Ibanez I, Silander JA Jr., Primack R, Higuchi H, Kobori H, Sen A, and James TY (2014). Beyond seasonal climate: statistical estimation of phenological responses to weather. Ecol Appl 24, 1793–802. [DOI] [PubMed] [Google Scholar]
- Dinant S, Clark AM, Zhu Y, Vilaine F, Palauqui JC, Kusiak C, and Thompson GA (2003). Diversity of the superfamily of phloem lectins (phloem protein 2) in angiosperms. Plant physiology 131, 114–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, and Stuurman N (2014). Advanced methods of microscope control using μManager software. Journal of Biological Methods 1, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feke A, Liu W, Hong J, Li MW, Lee CM, Zhou EK, and Gendron JM (2019). Decoys provide a scalable platform for the identification of plant E3 ubiquitin ligases that regulate circadian function. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feke AM, Hong J, Liu W, and Gendron JM (2020). A Decoy Library Uncovers U-Box E3 Ubiquitin Ligases That Regulate Flowering Time in Arabidopsis. Genetics 215, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez O, Ishihara H, George GM, Mengin V, Flis A, Sumner D, Arrivault S, Feil R, Lunn JE, Zeeman SC, et al. (2017). Leaf Starch Turnover Occurs in Long Days and in Falling Light at the End of the Day. Plant Physiol 174, 2199–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flis A, Sulpice R, Seaton DD, Ivakov AA, Liput M, Abel C, Millar AJ, and Stitt M (2016). Photoperiod-dependent changes in the phase of core clock transcripts and global transcriptional outputs at dawn and dusk in Arabidopsis. Plant, cell & environment 39, 1955–81. [DOI] [PubMed] [Google Scholar]
- Fournier-Level A, Perry EO, Wang JA, Braun PT, Migneault A, Cooper MD, Metcalf CJ, and Schmitt J (2016). Predicting the evolutionary dynamics of seasonal adaptation to novel climates in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 113, E2812–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbazza C, and Benedetti F (2018). Genetic Factors Affecting Seasonality, Mood, and the Circadian Clock. Front Endocrinol (Lausanne) 9, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, and Kay SA (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci U S A 109, 3167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Hohne M, Gunther M, and Stitt M (2009). Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant, cell & environment 32, 859–74. [DOI] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, and Smith AM (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc Natl Acad Sci U S A 107, 9458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Ren C, Zhi T, Zhou Z, Liu Y, Chen F, Peng W, and Xie D (2013). Disruption of fumarylacetoacetate hydrolase causes spontaneous cell death under short-day conditions in Arabidopsis. Plant physiology 162, 1956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, and Kay SA (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proceedings of the National Academy of Sciences of the United States of America 102, 10387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, and Kay SA (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Current biology : CB 21, 126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Shindo C, and Dean C (2003). The need for winter in the switch to flowering. Annu Rev Genet 37, 371–92. [DOI] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carre IA, Somers DE, Straume M, Meeks-Wagner DR, and Kay SA (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science (New York, N.Y.) 274, 790–2. [DOI] [PubMed] [Google Scholar]
- Hvidsten TR, Komorowski J, Sandvik AK, and Laegreid A (2001). Predicting gene function from gene expressions and ontologies. Pac Symp Biocomput, 299–310. [DOI] [PubMed] [Google Scholar]
- Inoue S, Dang QL, Man R, and Tedla B (2020). Photoperiod and CO2 elevation influence morphological and physiological responses to drought in trembling aspen: implications for climate change-induced migration. Tree Physiol 40, 917–927. [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, and Coupland G (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27, 1277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, and Staiger D (2014). SRR1 is essential to repress flowering in non-inductive conditions in Arabidopsis thaliana. Journal of experimental botany 65, 5811–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, and Weigel D (1999). Activation tagging of the floral inducer FT. Science (New York, N.Y.) 286, 1962–5. [DOI] [PubMed] [Google Scholar]
- Karlsson PM, Herdean A, Adolfsson L, Beebo A, Nziengui H, Irigoyen S, Unnep R, Zsiros O, Nagy G, Garab G, et al. (2015). The Arabidopsis thylakoid transporter PHT4;1 influences phosphate availability for ATP synthesis and plant growth. The Plant journal : for cell and molecular biology 84, 99–110. [DOI] [PubMed] [Google Scholar]
- Kassambara A (2020). ggpubr: ‘ggplot2’ Based Publication Ready Plots. R package version 0.4.0. https://CRAN.R-project.org/package=ggpubr. [Google Scholar]
- Kassambara A, and Mundt F (2020). factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package version 1.0.7. https://CRAN.R-project.org/package=factoextra. [Google Scholar]
- Kim JA, Kim HS, Choi SH, Jang JY, Jeong MJ, and Lee SI (2017). The Importance of the Circadian Clock in Regulating Plant Metabolism. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreyling J (2010). Winter climate change: a critical factor for temperate vegetation performance. Ecology 91, 1939–48. [DOI] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, and Horvath S (2008). Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–20. [DOI] [PubMed] [Google Scholar]
- Lee CM, Feke A, Li MW, Adamchek C, Webb K, Pruneda-Paz J, Bennett EJ, Kay SA, and Gendron JM (2018). Decoys Untangle Complicated Redundancy and Reveal Targets of Circadian Clock F-Box Proteins. Plant physiology 177, 1170–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Li MW, Feke A, Liu W, Saffer AM, and Gendron JM (2019). GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nature communications 10, 3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, and Sharkey TD (2005). Daylength and circadian effects on starch degradation and maltose metabolism. Plant physiology 138, 2280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden PJ, and Millar AJ (1998). Biological rhythms and photoperiodism in plants (Oxford Washington, D.C. Herndon, VA: Bios Scientific Publishers; Bios Scientific Publishers distributor; ) [Google Scholar]
- Marshall RS, and Vierstra RD (2018). Autophagy: The Master of Bulk and Selective Recycling. Annu Rev Plant Biol 69, 173–208. [DOI] [PubMed] [Google Scholar]
- Martins MC, Hejazi M, Fettke J, Steup M, Feil R, Krause U, Arrivault S, Vosloh D, Figueroa CM, Ivakov A, et al. (2013). Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant physiology 163, 1142–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsoukas IG, Massiah AJ, and Thomas B (2013). Starch metabolism and antiflorigenic signals modulate the juvenile-to-adult phase transition in Arabidopsis. Plant, cell & environment 36, 1802–11. [DOI] [PubMed] [Google Scholar]
- Mengin V, Pyl ET, Alexandre Moraes T, Sulpice R, Krohn N, Encke B, and Stitt M (2017). Photosynthate partitioning to starch in Arabidopsis thaliana is insensitive to light intensity but sensitive to photoperiod due to a restriction on growth in the light in short photoperiods. Plant, cell & environment 40, 2608–2627. [DOI] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, and Chory J (2008a). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6, e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008b). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4, e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, and Kay SA (1995a). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science (New York, N.Y.) 267, 1161–3. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, and Kay SA (1992). A novel circadian phenotype based on firefly luciferase expression in transgenic plants. The Plant cell 4, 1075–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua NH, and Kay SA (1995b). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science (New York, N.Y.) 267, 1163–6. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, and Chory J (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72, 353–63. [DOI] [PubMed] [Google Scholar]
- Moraes TA, Mengin V, Annunziata MG, Encke B, Krohn N, Hohne M, and Stitt M (2019). Response of the Circadian Clock and Diel Starch Turnover to One Day of Low Light or Low CO2. Plant physiology 179, 1457–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, and Kimura T (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Denlinger DL, and Somers DE (2010). Photoperiodism : the biological calendar (Oxford; New York: Oxford University Press; ) [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, and Kay SA (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Goto S, Fujibuchi W, and Kanehisa M (1998). Computation with the KEGG pathway database. Biosystems 47, 119–28. [DOI] [PubMed] [Google Scholar]
- Oquist G, and Huner NP (2003). Photosynthesis of overwintering evergreen plants. Annu Rev Plant Biol 54, 329–55. [DOI] [PubMed] [Google Scholar]
- Pal SK, Liput M, Piques M, Ishihara H, Obata T, Martins MC, Sulpice R, van Dongen JT, Fernie AR, Yadav UP, et al. (2013). Diurnal changes of polysome loading track sucrose content in the rosette of wild-type arabidopsis and the starchless pgm mutant. Plant physiology 162, 1246–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. (2011). Scikit-learn: Machine Learning in Python. J Mach Learn Res 12, 2825–2830. [Google Scholar]
- Phillips AR, Suttangkakul A, and Vierstra RD (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, and Coupland G (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–57. [DOI] [PubMed] [Google Scholar]
- Roenneberg T, and Merrow M (2001). Seasonality and photoperiodism in fungi. J Biol Rhythms 16, 403–14. [DOI] [PubMed] [Google Scholar]
- Saunders DS (1997). Insect circadian rhythms and photoperiodism. Invert Neurosci 3, 155–64. [DOI] [PubMed] [Google Scholar]
- Saunders DS (2005). Erwin Bunning and Tony Lees, two giants of chronobiology, and the problem of time measurement in insect photoperiodism. J Insect Physiol 51, 599–608. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton DD, Graf A, Baerenfaller K, Stitt M, Millar AJ, and Gruissem W (2018). Photoperiodic control of the Arabidopsis proteome reveals a translational coincidence mechanism. Molecular systems biology 14, e7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JS, and Imaizumi T (2015). Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis. Biochemistry 54, 157–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Shim JS, Kinmonth-Schultz HA, and Imaizumi T (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol 66, 441–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, and Zeeman SC (2012). Starch turnover: pathways, regulation and role in growth. Current opinion in plant biology 15, 282–92. [DOI] [PubMed] [Google Scholar]
- Stromme CB, Julkunen-Tiitto R, Olsen JE, Nybakken L, and Tognetti R (2017). High daytime temperature delays autumnal bud formation in Populus tremula under field conditions. Tree Physiol 37, 71–81. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, and Coupland G (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–20. [DOI] [PubMed] [Google Scholar]
- Sulpice R, Flis A, Ivakov AA, Apelt F, Krohn N, Encke B, Abel C, Feil R, Lunn JE, and Stitt M (2014). Arabidopsis coordinates the diurnal regulation of carbon allocation and growth across a wide range of photoperiods. Molecular plant 7, 137–55. [DOI] [PubMed] [Google Scholar]
- Tan Y, Merrow M, and Roenneberg T (2004). Photoperiodism in Neurospora crassa. J Biol Rhythms 19, 135–43. [DOI] [PubMed] [Google Scholar]
- Thomas B, and Vince-Prue D (1997). Photoperiodism in plants (San Diego, Calif.: Academic Press; ) [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, and Coupland G (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science (New York, N.Y.) 303, 1003–6. [DOI] [PubMed] [Google Scholar]
- Veley KM, Marshburn S, Clure CE, and Haswell ES (2012). Mechanosensitive channels protect plastids from hypoosmotic stress during normal plant growth. Current biology : CB 22, 408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vince-Prue D (1975). Photoperiodism in plants (London; New York: McGraw-Hill; ) [Google Scholar]
- Vitasse Y, Lenz A, and Korner C (2014). The interaction between freezing tolerance and phenology in temperate deciduous trees. Front Plant Sci 5, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker WH 2nd, Melendez-Fernandez OH, Nelson RJ, and Reiter RJ (2019). Global climate change and invariable photoperiods: A mismatch that jeopardizes animal fitness. Ecol Evol 9, 10044–10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2009). ggplot2: Elegant Graphics for Data Analysis. Use R, 1–212. [Google Scholar]
- Yanovsky MJ, and Kay SA (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–12. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, and Sheen J (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2, 1565–72. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Adachi E, Fukiya K, Iwai K, and Tanaka K (2005). Glycoprotein-specific ubiquitin ligases recognize N-glycans in unfolded substrates. EMBO Rep 6, 239–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Tokunaga F, Chiba T, Iwai K, Tanaka K, and Tai T (2003). Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. The Journal of biological chemistry 278, 43877–84. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi T, Zhou Z, Huang Y, Han C, Liu Y, Zhu Q, and Ren C (2016). Sugar suppresses cell death caused by disruption of fumarylacetoacetate hydrolase in Arabidopsis. Planta 244, 557–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Relative daily expression integral (rDEI) of the probes of ATH1 microarray (Related to Figure 1).
Table S2: Description and enriched annotations of 8L:16D-induced clusters (Related to Figure 1).
Table S3: Description and enriched annotations of 16L:8D-induced clusters (Related to Figure 1 and Figure S1).
Table S4: Description and enriched annotations of clusters identified by hierarchical clustering (Related to Figure 1).
Data Availability Statement
The accession numbers for the datasets reported in this paper are ArrayExpress E-MEXP-1304 (Michael, et al., 2008b) and E-MEXP-1299 (Michael, et al., 2008a). The data are also available on http://diurnal.mocklerlab.org/. All code used to generate and analyze all datasets in this study are provided in supplementary materials.