Abstract
Trichosporon asahii is a pathogenic fungus that causes severe, deep-seated fungal infections in neutropenic patients. Elucidating the infection mechanisms of T. asahii based on genetic studies requires a specific gene-targeting system. Here, we established an efficient gene-targeting system in a highly pathogenic T. asahii strain identified using the silkworm infection model. By comparing the pathogenicity of T. asahii clinical isolates in a silkworm infection model, T. asahii MPU129 was identified as a highly pathogenic strain. Using an Agrobacterium tumefaciens-mediated gene transfer system, we obtained a T. asahii MPU129 mutant lacking the ku70 gene, which encodes the Ku70 protein involved in the non-homologous end-joining repair of DNA double-strand breaks. The ku70 gene-deficient mutant showed higher gene-targeting efficiency than the wild-type strain for constructing a mutant lacking the cnb1 gene, which encodes the beta-subunit of calcineurin. The cnb1 gene-deficient mutant showed reduced pathogenicity against silkworms compared with the parental strain. These results suggest that an efficient gene-targeting system in a highly pathogenic T. asahii strain is a useful tool for elucidating the molecular mechanisms of T. asahii infection.
Subject terms: Gene targeting, Microbiology techniques, Fungal genetics, Fungal pathogenesis
Introduction
Trichosporon asahii is a basidiomycete yeast that is widely distributed in the environment and is often isolated from human blood, sputum, skin, feces, and urine1–6. T. asahii causes severe, deep-seated fungal infections in neutropenic patients7–9. Deep mycoses caused by T. asahii has a twofold higher mortality rate than those caused by Candida albicans (80% vs 40%)10. Since T. asahii is resistant to echinocandin antifungals, patients treated with micafungin are susceptible to the development of severe infections11. T. asahii strains resistant to amphotericin B and azole antifungals such as fluconazole have also been isolated from patients12,13. Moreover, T. asahii forms a biofilm, a three-dimensional structure comprising microbe aggregates and extracellular matrix, on catheter surfaces in patients14. The T. asahii cells within biofilms are resistant to antifungal drugs13. T. asahii has morphological forms: yeast form, hyphae (filament form) and arthroconidia (chains of cells and asexual spores)4. Furthermore, arthroconidia of T. asahii may play a key role in biofilm formation by promoting cellular adhesion15. T. asahii is therefore a highly problematic clinical pathogen9. Since the technology to construct gene-deficient mutants of T. asahii has not been established, it has not been possible to study pathogenicity and drug resistance in T. asahii using a gene-deficient mutant.
In general, mammals such as mice are used as experimental models in studies of infectious diseases16. The use of mammalian animals in infection experiments requires specialized experimental facilities, and the large number of animals required for these studies is a severe limitation due to ethical issues regarding animal welfare17. T. asahii infection experiments are not easy to perform in mice because immunosuppressive drugs must be administered18,19. To address these issues, we established a silkworm infection model for elucidating the mechanisms of T. asahii infection20. Compared with mammals such as mice, the use of invertebrate silkworms is advantageous because they are less costly to house and easier to rear in large numbers in simple facilities, and fewer ethical problems are associated with their use. Therefore, the use of silkworms as an experimental animal enhances the feasibility of performing large-scale, in vivo screening using a large number of individuals16.
Novel virulence genes in the pathogenic bacterium Staphylococcus aureus were identified using a silkworm infection model and a library of gene-deficient strains21,22. Silkworm infection models have also been used to identify virulence genes of the pathogenic fungi C. albicans and Candida glabrata23,24. In Cryptococcus neoformans, a basidiomycete yeast like T. asahii, a strain that is highly pathogenic to mice is also highly pathogenic to silkworms25. Moreover, a C. neoformans strain lacking the gene encoding the calcineurin subunit, which contributes to the pathogenicity against mice, was less virulent against silkworms25. Since the silkworm can be used to evaluate differences in the pathogenicity between strains of pathogenic fungi, the silkworm infection model with T. asahii may be useful for elucidating the infection mechanisms of T. asahii with the gene-deficient strains. We successfully established a T. asahii strain that expresses green fluorescent protein using an Agrobacterium tumefaciens-mediated gene transfer (ATMT) system20. A method for generating a gene-deficient strain of T. asahii, however, has not yet been established.
Homologous recombination (HR), a repair mechanism for DNA double-strand breaks (DSBs), is required to introduce mutations into a gene-targeting system using homologous DNA fragments26,27. Another repair mechanism is the non-homologous end joining (NHEJ) of DSBs26. These 2 main repair mechanisms affect gene-targeting efficiency by introducing homologous DNA fragments. NHEJ repair mediates the insertion of introduced homologous DNA fragments into genome sites that are different from the target region, thereby reducing the gene-targeting efficiency for generating a gene-deficient strain26. Therefore, gene-targeting efficiency can be increased by inhibiting NHEJ repair26,28. Ku70 and Ku80 proteins form heterodimers and are involved in the NHEJ repair for DSBs29. In several fungi, deletion of the genes encoding these proteins led to the increase of gene-targeting efficiency for generating gene-deficient strains28,30,31. In C. neoformans, gene-deficient strains could be generated in the ku80 gene-deficient strain, but not in the wild-type strain, by electroporation, a gene transfer method32. Therefore, strains with inhibited NHEJ repair due to disruption of the gene encoding Ku proteins are useful parental strains for promoting genetic studies.
In this study, we identified the T. asahii MPU129 strain, a clinical isolate that is highly pathogenic to silkworms, and generated a T. asahii MPU129 mutant deficient in the ku70 gene. Gene-targeting efficiency to obtain the ku70 gene-deficient strain was higher than that in the wild-type strain. Our findings suggest that a T. asahii strain showing high gene-targeting efficiency and the silkworm infection model are useful tools for studying infectious diseases as a preliminary step to conducting experiments in mice.
Results
Comparison of the pathogenicity of T. asahii strains using a silkworm infection model
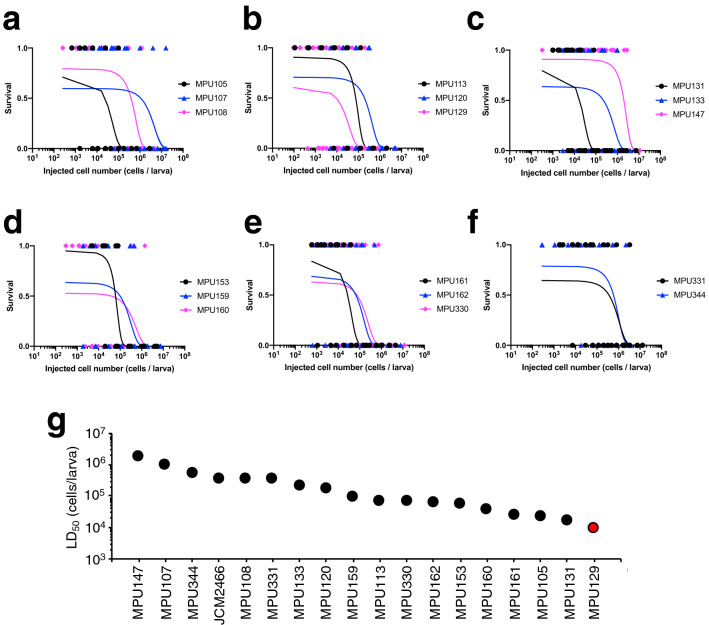
Highly pathogenic strains are useful for understanding the molecular mechanisms of pathogens because several pathogenic strains obtain virulence genes by horizontal gene transfer and gene mutation33,34. First, we identified T. asahii strains that are highly pathogenic to silkworms. Using a silkworm infection model, we determined the median lethal dose (LD50 values) on the basis of curves drawn by a simple logistic regression model (Fig. 1). The LD50 values of the 17 clinical isolates were 9.3 × 103–2.0 × 106 cells/larva and the LD50 value of the MPU129 strain was the lowest, more than tenfold lower than that of the JCM2466 strain (Fig. 1g). The result suggests that the pathogenicity of the MPU129 strain against silkworms is highest among these T. asahii strains.
Figure 1.
Comparison of pathogenicity of T. asahii strains against silkworms. (a–f) The number of surviving silkworms under a rearing condition at 37 °C was determined 48 h after administration of the fungal cells (1 × 102 to 2 × 107 cells/larva) into the hemolymph of silkworms. Survived and dead silkworms are indicated as 1 and 0, respectively. n = 4/group. The curves were drawn from the combined data of 2–6 independent experiments by a simple logistic regression model. (g) LD50 values of T. asahii strains. Data for JCMM2466 are cited from Matsumoto et al.20.
Generation of the ku70 gene-deficient mutant in the T. asahii MPU129 strain
We next obtained a ku70 gene-deficient mutant of the MPU129 strain using the ATMT system. The targeting plasmid, pAg1-5′UTR (ku70)-nptII-3′UTR (ku70), contained the nptII gene that leads to resistance against G418, an aminoglycoside used as a selective agent for eukaryotic cells (Fig. 2a). Colonies on Sabouraud dextrose agar containing G418 were obtained using the ATMT system (Fig. 2b). In the genome of the 414th candidate colony, polymerase chain reaction (PCR) amplification revealed DNA fragments of the predicted size (Fig. 2c,d). The results suggest that the ku70 gene-deficient mutant in the T. asahii MPU129 strain was obtained using the ATMT system.
Figure 2.
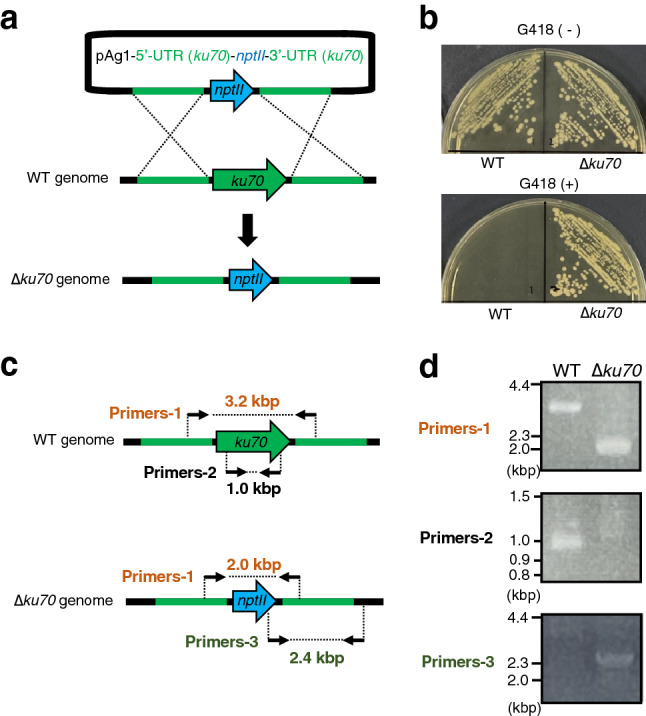
Construction of the ku70 gene-deficient mutant in the T. asahii MPU129 strain by the ATMT system. (a) Structure of the plasmid used to construct the ku70 gene-deficient mutant and the predicted genome of the ku70 gene-deficient mutant. (b) The wild-type (MPU129) and ku70 gene-deficient candidate were spread on Sabouraud agar medium with or without G418 (50 µg/ml) and incubated at 27 °C for 2 days. (c) Location of the primers for confirming the genome structure of the ku70 gene-deficient candidate by PCR. (d) Confirmation of the ku70 gene-deficiency of the ku70 gene-deficient candidate by PCR using extracted genome DNA. Cropped blots were used. Full-length blots are presented in Supplementary Fig. 1.
Effect of ku70 gene deficiency on growth and pathogenicity of the T. asahii MPU129 strain
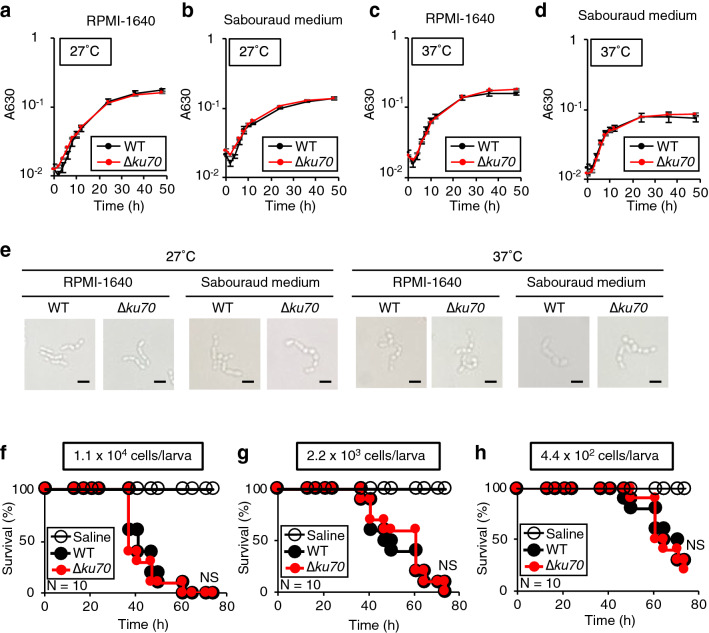
We investigated whether the deficiency of ku70 gene in the T. asahii MPU129 strain affected its growth on nutrient media and its pathogenicity in silkworms. The growth of the ku70 gene-deficient mutant in RPMI-1640 or Sabouraud liquid medium was similar to that of wild-type at either 27 °C or 37 °C (Fig. 3a–d). Moreover, microscopic analysis did not reveal significant differences in the morphology (Fig. 3e). Furthermore, the time required for the ku70 gene-deficient mutant to kill all the silkworms was similar to that of the wild-type strain (Fig. 3f–h). These results demonstrated that ku70 gene deficiency in the T. asahii MPU129 strain did not significantly affect its growth on nutrient media or its pathogenicity to silkworms.
Figure 3.
Effects of ku70 gene-deficiency in the T. asahii MPU129 mutant on growth, morphology, and pathogenicity against silkworms. (a–d) The wild-type (MPU129) and ku70 gene-deficient mutant were inoculated on RPMI-1640 medium and Sabouraud medium and incubated at 27 °C or 37 °C. Absorbance 630 nm of the culture was monitored. Data are shown as means ± standard error of the mean (SEM). (e) The wild-type (MPU129) and ku70 gene-deficient mutant were incubated at 2 days after inoculation and observed with a microscope. (f–h) Saline, T. asahii MPU129 (WT), or the ku70 gene-deficient mutant (∆ku70) [1.1 × 104 cells/larva (f), 2.2 × 103 cells/larva (g), 4.4 × 102 cells/larva (h)] were injected into the silkworm hemolymph and incubated at 37 °C. The survival of the silkworms was monitored for 74 h. The significance of differences between groups in silkworm infection experiments was calculated by the log-rank test based on the curves by the Kaplan–Meier method. NS Not significant (P > 0.05). n = 10/group.
Increased gene-targeting efficiency in the ku70 gene-deficient T. asahii mutant
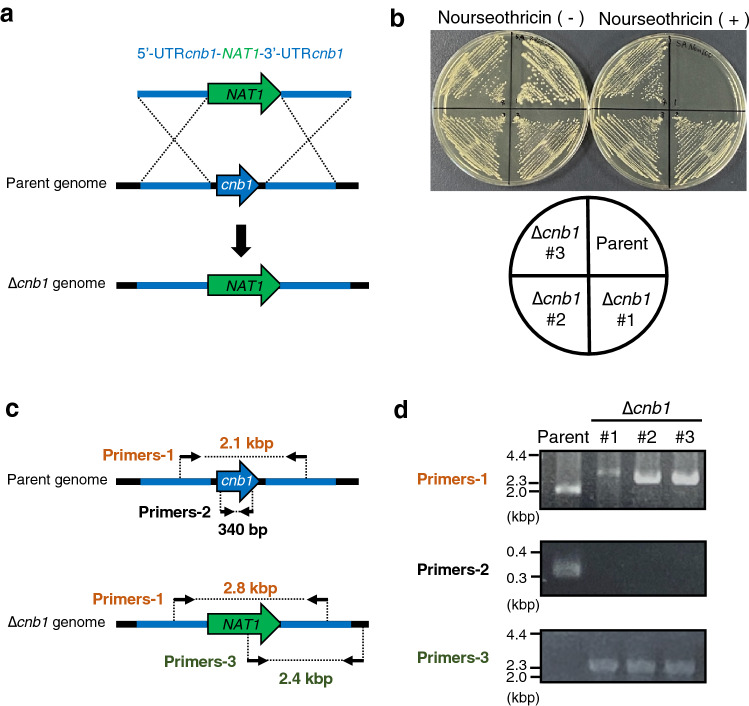
We examined whether gene-targeting efficiency was increased in the ku70 gene-deficient mutant by determining the ratio of the strain lacking the cnb1 gene, which encodes the β-subunit of calcineurin. Since gene-targeting efficiency in ku80 gene-deficient mutant of C. neoformans was tested by electroporation, a faster and simpler gene transfer method32, we also used electroporation to investigate T. asahii. A DNA fragment, 5'UTR (cnb1)-NAT1-3'UTR (cnb1), was introduced to delete the cnb1 gene by electroporation. Nourseothricin-resistant strains were obtained, and each colony was confirmed by PCR to be deficient in the cnb1 gene (Fig. 4a–d). Of the 21 nourseothricin-resistant colonies obtained by introducing the 5'UTR (cnb1)-NAT1-3'UTR (cnb1) into the ku70 gene-deficient mutant, 4 were deficient for the cnb1 gene (Table 1). On the other hand, none of the 120 nourseothricin-resistant colonies obtained by introducing the 5'UTR (cnb1)-NAT1-3′UTR (cnb1) into the wild-type was deficient for the cnb1 gene (Table 1). These results suggest that the deficiency of the ku70 gene in the MPU129 strain increases the gene-targeting efficiency for generating a gene-deficient mutant by electroporation.
Figure 4.
Construction of cnb1 gene-deficient mutant in the ku70 gene-deficient mutant of T. asahii MPU129 strain by electroporation. (a) Structure of the DNA fragment for construction of the cnb1 gene-deficient mutant and the predicted genome of the cnb1 gene-deficient mutant. (b) The parent strain (MPU129 ∆ku70) and cnb1 gene-deficient candidates (∆cnb1 #1, #2, and #3) were spread on Sabouraud agar medium with or without nourseothricin (100 µg/ml) and incubated at 27 °C for 2 days. (c) Location of the primers for confirming the genome structure of the cnb1 gene-deficient candidate by PCR. (d) Confirmation of the cnb1 gene-deficiency of the cnb1 gene-deficient candidate by PCR using extracted genome DNA. Cropped blots were used. Full-length blots are presented in Supplementary Fig. 1.
Table 1.
Efficiency of homologous replacement on cnb1 gene region.
| Strain | Total transformants | Homologous replacement (∆cnb1) | Efficiency (%) (∆cnb1/total transformants) |
|---|---|---|---|
| Wild type | 120 | 0 | 0% |
| ∆ku70 | 21 | 4 | 19% |
Attenuated pathogenicity of the cnb1 gene-deficient mutant against silkworms
In C. neoformans, the pathogenicity of the cnb1 gene-deficient mutant against silkworms was reduced25. We examined whether the cnb1 gene-deficient mutants of T. asahii had reduced pathogenicity against silkworms. The survival time of silkworms injected with the cnb1 gene-deficient mutants was longer than that of the parental strain (Fig. 5a). The LD50 values of the cnb1 gene-deficient mutants were 89-fold higher than that of the parent strain (Fig. 5b). The result suggests that pathogenicity against silkworms was reduced by cnb1 gene deficiency in T. asahii.
Figure 5.
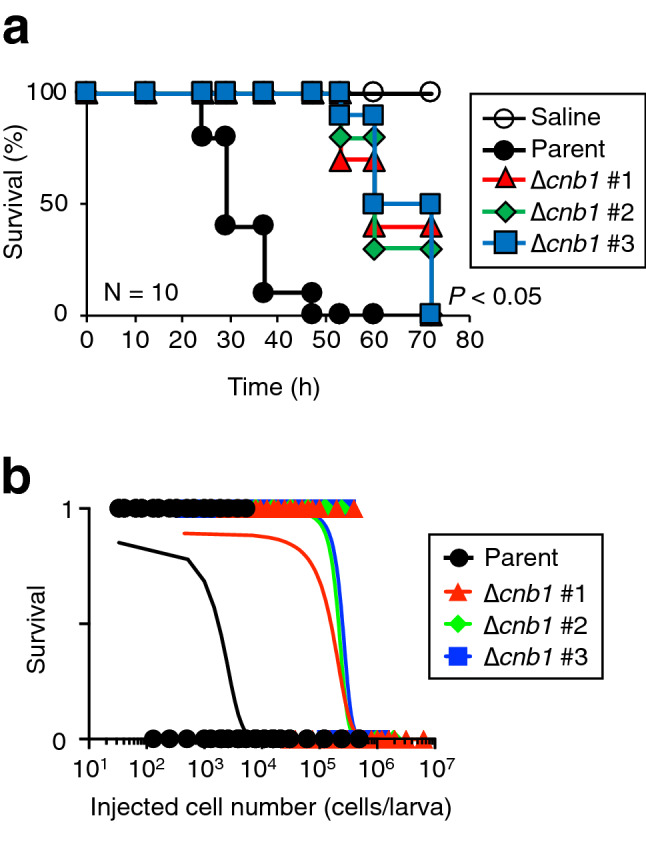
Attenuated pathogenicity of T. asahii against silkworms by cnb1 gene-deficiency. (a) Saline, T. asahii MPU129 ∆ku70 (parent strain) (5.4 × 104 cells/larva), or the cnb1 gene-deficient mutants [∆cnb1 #1 (7.1 × 104 cells/larva), #2 (5.2 × 104 cells/larva), and #3 (2.5 × 104 cells/larva)] were injected into the silkworm hemolymph and the silkworms were incubated at 37 °C. The survival of the silkworms was monitored for 72 h. The significance of differences between parent strain group and the cnb1 gene-deficient mutant groups was calculated by the log-rank test based on the curves by the Kaplan–Meier method. n = 10/group. (b) The number of surviving silkworms under a rearing condition at 37˚C was determined at 48 h after administration of the fungal cells (3.3 × 102 to 6.2 × 106 cells/larva) into the hemolymph of silkworms. Survived and dead silkworms were indicated as 1 and 0, respectively. n = 4/group. The curves were drawn from combined data of 2–3 independent experiments by simple logistic regression model.
Discussion
In this study, we identified a T. asahii strain that is highly pathogenic against silkworms and established a platform for generating a gene-deficient mutant. The cnb1 gene-deficient mutant obtained using the technique showed decreased pathogenicity against silkworms. To our knowledge, this is the first report of a method for obtaining a gene-deficient mutant of T. asahii. Our results suggest that the calcineurin pathway is involved in the pathogenicity of T. asahii.
In the silkworm infection model with T. asahii, the MPU129 strain showed high pathogenicity among clinical isolates used in this study. We assumed that the MPU129 strain can adapt to the host environments and appropriately regulate the pathogenicity compared with other isolates. To reveal the relationship between clinical information and the pathogenicity in the silkworm infection model among the clinical isolates will be an important study.
Silkworms are suitable experimental animals for performing large-scale in vivo evaluations because they are relatively inexpensive and few ethical issues are associated with their use. Therefore, silkworm infection models are suitable for quantitative evaluation of the pathogenicity of microorganisms based on the calculation of LD50 values. Using the silkworm infection model and a gene-deficient mutant library of S. aureus, we previously identified the virulence genes that contribute to pathogenicity against mice21. The gene-targeting system in T. asahii developed in the present study will facilitate the construction of a gene-deficient mutant library of T. asahii. It is expected that a gene-deficient mutant library of T. asahii for application to silkworm infection models will help elucidate the molecular mechanisms of T. asahii infection. The contribution of candidate virulence genes to pathogenicity that is identified using the silkworm infection model should be confirmed by infection experiments in mice.
Gene-targeting efficiency by electroporation was higher in the ku70 gene-deficient mutant, while growth on nutrient media and pathogenicity to silkworms remained unaltered. Therefore, the ku70 gene-deficient mutant is useful as a parental strain for elucidating the infection mechanism of T. asahii based on genetic studies. In the pathogenic fungus Aspergillus fumigatus, the pathogenicity of the ku80 gene-deficient mutant did not differ from that of wild-type against mice35. We considered that NHEJ repair of DSBs might not be greatly involved in the pathogenicity of T. asahii or A. fumigatus. Although electroporation is a faster and simpler gene transfer method than the ATMT system for obtaining gene-deficient mutants, homologous recombination by electroporation occurs at low frequency32,36. When using electroporation, no cnb1 gene-deficient mutants were obtained in the wild-type, but a 19% ratio of mutants was obtained in the ku70 gene-deficient mutant. The result suggests that NHEJ of DSBs occurs at a high frequency in T. asahii. Therefore, we reasoned that the generation of target gene-deficient mutants by electroporation requires the ku70 gene-deficient mutant that lacks NHEJ repair activity. When we obtain fungal colonies grown on a drug-containing agar medium, both strains with mutations in the targeted gene region and strains with non-specific gene insertions caused by NHEJ were obtained. Therefore, NHEJ may contribute to obtaining the strains with a non-specific gene inserted mutants by selecting drug-resistant strains. It can be constructed the gene-deficient mutants in T. asahii within two weeks by using the TR129 ku70 gene-deficient mutant with electroporation method. Construction of a gene-deficient mutant library of T. asahii using the ku70 gene-deficient mutant as a parent strain is thus planned for future studies.
Although we tried to obtain a cnb1 gene-deficient mutant using the ku70 gene-deficient mutant as the parent strain with the ATMT system, we did not obtain a drug-resistant candidate. Optimization of the ATMT system using the ku70 gene-deficient mutant is needed to obtain target gene-deficient mutants. Moreover, CRISPR-CAS9 technology was applied for gene editing in fungi including C. neoformans37,38. The establishment of the CRISPR-CAS9 mediated gene-editing method for T. asahii will be a future subject.
In C. neoformans, the calcineurin pathway is involved in capsule production and melanin synthesis, which are responsible for evading host immunity39. A calcineurin-deficient strain of C. neoformans showed decreased pathogenicity against mice and silkworms25,40,41. The present study also showed that the calcineurin pathway is involved in the pathogenicity of T. asahii against silkworms. The calcineurin in C. neoformans regulates gene expression via the dephosphorylation of the transcription factors39. Therefore, we assumed that the calcineurin in T. asahii also regulates the virulence-related gene expression. To reveal the role of calcineurin in the T. asahii pathogenicity will be an important subject. Further studies are needed to investigate the generation of a revertant strain obtained by reintroducing the cnb1 gene into the cnb1 gene-deficient mutant and to perform a detailed functional analysis using the cnb1 gene-deficient mutant and its revertant strain.
In conclusion, we established a simple method for generating a gene-deficient T. asahii strain that is highly pathogenic against silkworms. The ku70 gene-deficient mutant in the T. asahii MPU129 strain is useful as a parental strain for genetic studies and an important tool for studying infectious diseases of T. asahii.
Methods
Reagents
Kanamycin, cefotaxime, and chloramphenicol were purchased from Wako Pure Chemical Industries (Osaka, Japan). Nourseothricin and G418 were purchased from Jena Bioscience (Dortmund, Germany) and Enzo Life Science, Inc. (Farmingdale, NY, USA), respectively.
Culture of T. asahii
The T. asahii strains (MPU105, MPU107, MPU108, MPU113, MPU120, MPU129, MPU131, MPU133, MPU147, MPU153, MPU159, MPU160, MPU161, MPU162, MPU330, MPU331, and MPU344) used in this study were stocked in the previously reported MPU library15. T. asahii strains were grown on Sabouraud dextrose agar (1% hipolypepton [Nihon Pharmaceutical Co., Ltd., Tokyo, Japan], 4% dextrose and 1.5% agar [both from FUJIFILM Wako Pure Chemical Industries, Osaka, Japan]) and incubated at 27 °C for 2 days.
For growth on liquid medium, RPMI-1640 (RPMI medium 1640 [Life Technologies Ltd., Paisley, UK] containing 165 mM 3-(N-morpholino) propanesulfonic acid [Dojindo Laboratories, Kumamoto, Japan], pH 7.0) and Sabouraud liquid medium (1% hipolypeptone, 4% dextrose) were used in this study. Suspensions of wild-type T. asahii (MPU129 strain) and ku70 gene-deficient T. asahii mutants were prepared with RPMI-1640 or Sabouraud medium and adjusted to 0.01–0.02 on absorbance at 630 nm. The T. asahii suspensions were incubated at 27 °C or 37 °C for 48 h and absorbance at 630 nm was measured using a microplate reader (iMark™ microplate reader; Bio-Rad Laboratories Inc., Hercules, CA, USA). After incubation for 2 days, the T. asahii cells were observed with a light microscope (CH30; Olympus, Tokyo, Japan).
Silkworm infection experiments
Silkworm infection experiments were performed according to a previous report20. Eggs of silkworms (Hu・Yo × Tukuba・Ne) were purchased from Ehime-Sanshu Co., Ltd. (Ehime, Japan), disinfected, and hatched at 25–27 °C. The silkworms were fed an artificial diet, Silkmate 2S, containing antibiotics purchased from Ehime-Sanshu Co., Ltd. Fifth instar larvae were used in the infection experiments. Silkworm fifth instar larvae were fed the artificial diet (Silkmate 2S; Ehime-Sanshu Co., Ltd.) overnight. T. asahii grown on Sabouraud agar plates was suspended in physiologic saline solution (0.9% w/v NaCl) and filtered through a 40-μm cell strainer (Corning Inc., Corning, NY, USA). A 50-µl suspension of T. asahii cells was administered into the silkworm hemolymph by injecting the silkworm dorsally using a 1-ml tuberculin syringe (Terumo Medical Corporation, Tokyo, Japan). Silkworms injected with T. asahii cells were placed in an incubator and their survival was monitored.
LD50 measurement
The dose of T. asahii required to kill half of the silkworms (LD50) was determined according to the previous report20. T. asahii strains (1 × 102 to 2 × 107 cells/50 µl) were injected into the silkworm hemolymph and the silkworms were incubated at 37 °C. Survival of the silkworms (n = 4/group) at 48 h was monitored. The LD50 was determined from the combined data of 2–3 independent experiments by simple logistic regression model using Prism 9.1.2 (GraphPad Software, LLC, San Diego, CA, USA, https://www.graphpad.com/scientific-software/prism/).
Construction of gene-deficient mutants in T. asahii
The plasmid for gene-deficient mutants in T. asahii was constructed according to a previous report30. To generate the ku70 gene-deficient mutant, the 5′-UTR of the ku70 gene, neomycin phosphotransferase gene (nptII) cassette, and 3′-UTR of the ku70 gene were introduced into a pAg1 vector42. To generate the cnb1 gene-deficient mutant, the 5′-UTR and 3′-UTR of the cnb1 gene were introduced into a pAg1-NAT1 vector42. Cloning was performed by the infusion method according to the general method (In-Fusion HD Cloning Kit, Takara, Shiga, Japan). The primers used for PCR amplification of each DNA region are shown in Table 2. The pAg1-5′UTR(ku70)-nptII-3′UTR(ku70) was introduced into the T. asahii MPU129 strain using the A. tumefaciens-mediated transformation method described previously20. The pAg1-5′UTR(ku70)-nptII-3′UTR(ku70) was introduced into the A. tumefaciens EHA105 strain by electroporation and transformants were grown on 2 × YT agar (1.6% tryptone and 1% yeast extract [both from Becton, Dickinson, and Company, NJ, USA], 0.5% NaCl, and 1.5% agar) containing chloramphenicol (25 μg/ml) and kanamycin (50 μg/ml). The A. tumefaciens EHA105 strain harboring the plasmid introduces the 5′UTR(ku70)-nptII-3′UTR(ku70) fragment into the nucleus of T. asahii cells via the DNA transport proteins. The transformant was co-cultured with the T. asahii MPU129 strain at 27 °C for 2 days. The candidates were isolated as colonies grown on Sabouraud dextrose agar containing G418 (50 μg/ml) and cefotaxime (100 μg/ml). Introduction of the mutation into the genome of the candidate strains was confirmed by PCR using the primers shown in Table 2.
Table 2.
Primers used in this study.
| Primers | Nucleic acid sequence |
|---|---|
| pAg1-ku70(3′UTR)-nptII-ku70(5′UTR) for cloning | |
| F ku70(5′UTR) | GCGGTACTAGTCGCCACCACGGTAGCGGTA |
| R ku70(5′UTR) | ACAAGATCTTGACGTCCTTTGGATGTTGCT |
| F nptII | ATGATTGAACAAGATGGATTGC |
| R nptII | TCAGAAGAACTCGTCAAGAAG |
| F ku70(3′UTR) | GCGGATCCAGTGTACTAGCGTGACGCTAGA |
| R ku70(3′UTR) | CTGGCGGTACCGCTGGCCGACCCACTCGTA |
| pAg1-cnb1(3′UTR)-NAT1-cnb1(5′UTR) for cloning | |
| F cnb1(5′UTR) | TGAACTAGTCCGTGATCTGCTGCACGTTCGGGTCC |
| R cnb1(5′UTR) | AAAGGGCCCAAGATCTAGTGATAGATGTGTGGAGA |
| F cnb1(3′UTR) | CTGGGATCCGCGCGCACACACGGATGTGAGCGTAA |
| R cnb1(3′UTR) | CGCGGTACCACTGTTCACCTCTGGCATTGTTACGA |
| Genotyping | |
| Primers-1 for ku70 genotyping | |
| F ku70 gene locus | TCGAGGTCGCGACTTTGTTATTGCCAGGTCCTGA |
| R ku70 gene locus | AGAGCTGCGATCGTGGGCTGATCCGTCC |
| Primers-2 for ku70 genotyping | |
| F ku70 gene ORF | TTTCAGCAACTCCGTCAGATCAGCGCCGAAGACA |
| R ku70 gene ORF | ATCTGCGAAAGAGCGGCCGGGCC |
| Primers-3 for ku70 genotyping | |
| F ku70 gene outside | TTCATCGACTGTGGCCGGCTGGGTGTGG |
| R ku70 gene outside | GGACGAGATGGCCGGGGACCGGCTC |
| Primers-1 for cnb1 genotyping | |
| F cnb1 gene locus | GGAGTGAAGAAGGGCAGAGAGCAACAACAGCGGT |
| R cnb1 gene locus | CCGTGATCGCATGGGGCGTGCACAAAGTG |
| Primers-2 for cnb1 genotyping | |
| F cnb1 gene ORF | CGGCTCGGGTACGGTAGACTTCCAGGAGTTTGTCG |
| R cnb1 gene ORF | AACAGGTCCTCGAGCGTCATCTGCTTGACGATGT |
| Primers-3forcnb1genotyping | |
| F cnb1 gene outside | GGACGGCGAGCAGGCGCTCTACATGAGC |
| R cnb1 gene outside | CTGAGTCCCATCGGCCCTTGCCTTCAAGCTACC |
| Amplification of cnb1 cassette for electroporation | |
| F cnb1-cassette | CCGTGATCTGCTGCACGTTCGGGTCCG |
| R cnb1-cassette | CTGTTCACCTCTGGCTACGACCCCCTCCTC |
To prepare competent cells for electroporation, T. asahii MPU129 strain was spread on a Sabouraud dextrose agar plate and cultured at 27 °C for 3 days. T. asahii cells on the agar were suspended by physiologic saline solution (2 ml), and the suspension was transferred to a 1.5-ml tube. The fungal cells were collected by centrifugation at 8000 rpm for 3 min (TOMY-MX100, TOMY Digital Biology Co. Ltd, Tokyo, Japan) and suspended by adding 1 ml of ice-cold water and centrifuged at 8000 rpm for 3 min. This washing process was repeated 4 times. The washed cells were suspended by adding 1 ml of 1.2 M sorbitol solution and centrifuged at 8000 rpm for 3 min. The obtained fungal cells were suspended with 0.2 ml of 1.2 M sorbitol solution as competent cells. The PCR-amplified 5′-UTR (cnb1) -NAT1-3′-UTR (cnb1) fragment (180 ng/2 µl) was added to the T. asahii competent cells (40 µl) and placed on ice for 15 min. The suspension was added to a 0.2-cm gap cuvette (Bio-Rad Laboratories, Inc.) and electroporated (Time constant protocol: 1800 V, 5 ms) using a Gene Pulser Xcell (Bio-Rad Laboratories, Inc.). The cells were suspended by adding 500 µl YPD containing 0.6 M sorbitol and incubated at 27 °C for 3 h. After incubation, the cells were collected by centrifugation at 10,000 rpm for 5 min and suspended in 100 µl of physiologic saline solution and applied to Sabouraud dextrose agar containing nourseothricin (300 µg/ml). The cells were incubated at 27 °C for 3 days and the growing colonies were isolated as cnb1 gene-deficient strain candidates. Introduction of the mutation into the genome of the candidate strains was confirmed by PCR using the primers shown in Table 2 and the extracted genome as a template DNA.
Statistical analysis
All experiments were performed at least twice and the representative results were shown. The significance of differences between groups in silkworm infection experiments was calculated by the log-rank test based on the curves by the Kaplan–Meier method using Prism 9.1.2.
Supplementary Information
Acknowledgements
We thank Yu Sugiyama, Eri Sato, and Asuka Toshima (Meiji Pharmaceutical University) for their technical assistance rearing the silkworms. This study was supported by JSPS KAKENHI Grant number JP20K07022 (Scientific Research (C) to Y.M.) and in part by the Research Program on Emerging and Re-emerging Infectious Diseases of the Japan Agency for Medical Research and Development, AMED (Grant to T.S.).
Author contributions
Study conception and design: Y.M. Acquisition of data: Y.M., T.N., A.Y., H.Y., Y.Y., T.Y.; Analysis and interpretation of data: Y.M. Drafting of manuscript: Y.M. Critical revision: Y.M., T.N., A.Y., H.Y., Y.Y., T.Y., T.S. All authors have read and approved the final version of the manuscript.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-97287-3.
References
- 1.Sugita T, Nishikawa A, Ichikawa T, Ikeda R, Shinoda T. Isolation of Trichosporon asahii from environmental materials. Med. Mycol. 2000;38:27–30. doi: 10.1080/mmy.38.1.27.30. [DOI] [PubMed] [Google Scholar]
- 2.Sugita T, et al. Genetic diversity and biochemical characteristics of Trichosporon asahii isolated from clinical specimens, houses of patients with summer-type-hypersensitivity pneumonitis, and environmental materials. J. Clin. Microbiol. 2001;39:2405–2411. doi: 10.1128/JCM.39.7.2405-2411.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang E, Sugita T, Tsuboi R, Yamazaki T, Makimura K. The opportunistic yeast pathogen Trichosporon asahii colonizes the skin of healthy individuals: Analysis of 380 healthy individuals by age and gender using a nested polymerase chain reaction assay. Microbiol. Immunol. 2011;55:483–488. doi: 10.1111/j.1348-0421.2011.00341.x. [DOI] [PubMed] [Google Scholar]
- 4.Colombo AL, Padovan ACB, Chaves GM. Current knowledge of Trichosporon spp. and Trichosporonosis. Clin. Microbiol. Rev. 2011;24:682–700. doi: 10.1128/CMR.00003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gouba N, Raoult D, Drancourt M. Eukaryote culturomics of the gut reveals new species. PLoS One. 2014;9:e106994. doi: 10.1371/journal.pone.0106994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho O, Matsukura M, Sugita T. Molecular evidence that the opportunistic fungal pathogen Trichosporon asahii is part of the normal fungal microbiota of the human gut based on rRNA genotyping. Int. J. Infect. Dis. 2015;39:87–88. doi: 10.1016/j.ijid.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Walsh TJ, et al. Experimental Trichosporon infection in persistently granulocytopenic rabbits: Implications for pathogenesis, diagnosis, and treatment of an emerging opportunistic mycosis. J. Infect. Dis. 1992;166:121–133. doi: 10.1093/infdis/166.1.121. [DOI] [PubMed] [Google Scholar]
- 8.Walsh TJ, Melcher GP, Lee JW, Pizzo PA. Infections due to Trichosporon species: New concepts in mycology, pathogenesis, diagnosis and treatment. Curr. Top. Med. Mycol. 1993;5:79–113. [PubMed] [Google Scholar]
- 9.Duarte-Oliveira C, et al. The cell biology of the Trichosporon-host interaction. Front. Cell Infect. Microbiol. 2017;7:118. doi: 10.3389/fcimb.2017.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krcmery V, et al. Hematogenous trichosporonosis in cancer patients: Report of 12 cases including 5 during prophylaxis with itraconazol. Support Care Cancer. 1999;7:39–43. doi: 10.1007/s005200050221. [DOI] [PubMed] [Google Scholar]
- 11.Kimura M, et al. Micafungin breakthrough fungemia in patients with hematological disorders. Antimicrob. Agents Chemother. 2018;62:324. doi: 10.1128/AAC.02183-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toriumi Y, Sugita T, Nakajima M, Matsushima T, Shinoda T. Antifungal pharmacodynamic characteristics of amphotericin B against Trichosporon asahii, using time-kill methodology. Microbiol. Immunol. 2002;46:89–93. doi: 10.1111/j.1348-0421.2002.tb02663.x. [DOI] [PubMed] [Google Scholar]
- 13.Iturrieta-González IA, Padovan ACB, Bizerra FC, Hahn RC, Colombo AL. Multiple species of Trichosporon produce biofilms highly resistant to triazoles and amphotericin B. PLoS One. 2014;9:e109553. doi: 10.1371/journal.pone.0109553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Bonaventura G, et al. Biofilm formation by the emerging fungal pathogen Trichosporon asahii: Development, architecture, and antifungal resistance. Antimicrob. Agents Chemother. 2006;50:3269–3276. doi: 10.1128/AAC.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurakado S, et al. Role of arthroconidia in biofilm formation by Trichosporon asahii. Mycoses. 2021;64:42–47. doi: 10.1111/myc.13181. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto Y. Facilitating drug discovery in human disease models using insects. Biol. Pharm. Bull. 2020;43:216–220. doi: 10.1248/bpb.b19-00834. [DOI] [PubMed] [Google Scholar]
- 17.Flecknell P. Replacement, reduction and refinement. Altex. 2002;19:73–78. [PubMed] [Google Scholar]
- 18.Gokaslan A, Anaissie E. A novel murine model of disseminated trichosporonosis. Infect. Immun. 1992;60:3339–3344. doi: 10.1128/iai.60.8.3339-3344.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montoya AM, et al. In vivo pathogenicity of Trichosporon asahii isolates with different in vitro enzymatic profiles in an immunocompetent murine model of systemic trichosporonosis. Med. Mycol. 2018;56:434–441. doi: 10.1093/mmy/myx057. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto Y, et al. A novel silkworm infection model with fluorescence imaging using transgenic Trichosporon asahii expressing eGFP. Sci. Rep. 2020;10:10991–11011. doi: 10.1038/s41598-020-67841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaito C, et al. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol. Microbiol. 2005;56:934–944. doi: 10.1111/j.1365-2958.2005.04596.x. [DOI] [PubMed] [Google Scholar]
- 22.Kaito C, Murakami K, Imai L, Furuta K. Animal infection models using non-mammals. Microbiol. Immunol. 2020;64:585–592. doi: 10.1111/1348-0421.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanaoka N, et al. Identification of the putative protein phosphatase gene PTC1 as a virulence-related gene using a silkworm model of Candida albicans infection. Eukaryot. Cell. 2008;7:1640–1648. doi: 10.1128/EC.00129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto Y, Sekimizu K. Silkworm as an experimental animal to research for fungal infections. Microbiol. Immunol. 2019 doi: 10.1111/1348-0421.12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumoto Y, et al. Quantitative evaluation of cryptococcal pathogenesis and antifungal drugs using a silkworm infection model with Cryptococcus neoformans. J. Appl. Microbiol. 2012;112:138–146. doi: 10.1111/j.1365-2672.2011.05186.x. [DOI] [PubMed] [Google Scholar]
- 26.Ding Y, et al. Increasing the homologous recombination efficiency of eukaryotic microorganisms for enhanced genome engineering. Appl. Microbiol. Biotechnol. 2019;103:4313–4324. doi: 10.1007/s00253-019-09802-2. [DOI] [PubMed] [Google Scholar]
- 27.Gusa A, Jinks-Robertson S. Mitotic recombination and adaptive genomic changes in human pathogenic fungi. Genes (Basel) 2019;10:901. doi: 10.3390/genes10110901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U.S.A. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fell VL, Schild-Poulter C. The Ku heterodimer: Function in DNA repair and beyond. Mutat. Res. Rev. Mutat. Res. 2015;763:15–29. doi: 10.1016/j.mrrev.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Yamada T, et al. Enhanced gene replacements in Ku80 disruption mutants of the dermatophyte, Trichophyton mentagrophytes. FEMS Microbiol. Lett. 2009;298:208–217. doi: 10.1111/j.1574-6968.2009.01714.x. [DOI] [PubMed] [Google Scholar]
- 31.Carvalho NDSP, Arentshorst M, Jin Kwon M, Meyer V, Ram AFJ. Expanding the ku70 toolbox for filamentous fungi: Establishment of complementation vectors and recipient strains for advanced gene analyses. Appl. Microbiol. Biotechnol. 2010;87:1463–1473. doi: 10.1007/s00253-010-2588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin X, Chacko N, Wang L, Pavuluri Y. Generation of stable mutants and targeted gene deletion strains in Cryptococcus neoformans through electroporation. Med. Mycol. 2015;53:225–234. doi: 10.1093/mmy/myu083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Das B, Kumar N. Vibrio pathogenicity island-1: The master determinant of Cholera pathogenesis. Front. Cell Infect. Microbiol. 2020;10:561296. doi: 10.3389/fcimb.2020.561296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaito C, et al. Non-pathogenic Escherichia coli acquires virulence by mutating a growth-essential LPS transporter. PLoS Pathog. 2020;16:e1008469. doi: 10.1371/journal.ppat.1008469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Silva Ferreira ME, et al. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell. 2006;5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang YC, Kwon-Chung KJ. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morio F, Lombardi L, Butler G. The CRISPR toolbox in medical mycology: State of the art and perspectives. PLoS Pathog. 2020;16:e1008201. doi: 10.1371/journal.ppat.1008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P. Genetic transformation in Cryptococcus species. J. Fungi (Basel) 2021;7:56. doi: 10.3390/jof7010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumoto Y, et al. Induction of signal transduction pathways related to the pathogenicity of Cryptococcus neoformans in the host environment. Drug Discov. Ther. 2019;13:177–182. doi: 10.5582/ddt.2019.01047. [DOI] [PubMed] [Google Scholar]
- 40.Cruz MC, Sia RA, Olson M, Cox GM, Heitman J. Comparison of the roles of calcineurin in physiology and virulence in serotype D and serotype A strains of Cryptococcus neoformans. Infect. Immun. 2000;68:982–985. doi: 10.1128/IAI.68.2.982-985.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fox DS, et al. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol. Microbiol. 2001;39:835–849. doi: 10.1046/j.1365-2958.2001.02295.x. [DOI] [PubMed] [Google Scholar]
- 42.Alshahni MM, Makimura K, Yamada T, Takatori K, Sawada T. Nourseothricin acetyltransferase: A new dominant selectable marker for the dermatophyte Trichophyton mentagrophytes. Med. Mycol. 2010;48:665–668. doi: 10.3109/13693780903330555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.