Abstract
TRIP4 is one of the subunits of the transcriptional coregulator ASC-1, a ribonucleoprotein complex that participates in transcriptional coactivation and RNA processing events. Recessive variants in the TRIP4 gene have been associated with spinal muscular atrophy with bone fractures as well as a severe form of congenital muscular dystrophy. Here we present the diagnostic journey of a patient with cerebellar hypoplasia and spinal muscular atrophy (PCH1) and congenital bone fractures. Initial exome sequencing analysis revealed no candidate variants. Reanalysis of the exome data by inclusion in the Solve-RD project resulted in the identification of a homozygous stop-gain variant in the TRIP4 gene, previously reported as disease-causing. This highlights the importance of analysis reiteration and improved and updated bioinformatic pipelines. Proteomic profile of the patient’s fibroblasts showed altered RNA-processing and impaired exosome activity supporting the pathogenicity of the detected variant. In addition, we identified a novel genetic form of PCH1, further strengthening the link of this characteristic phenotype with altered RNA metabolism.
Subject terms: Genetics research, Next-generation sequencing
Introduction
The thyroid receptor interacting protein 4, encoded by the TRIP4 gene, is one of the four subunits of the transcriptional coregulator ASC-1. TRIP4, together with ASCC1, ASCC2 and ASCC3, form a ribonucleoprotein complex that participates in transcriptional coactivation and RNA processing events [1]. TRIP4 binds transcription factors, such as AP-1 and NF-kappa-B through its conserved cysteine rich Zn-chelating domain [2], whilst the C-terminal RNA-binding PUA domain is involved in RNA processing [3].
Recessive variants in TRIP4 have been associated with two distinct phenotypes: spinal muscular atrophy with bone fractures (OMIM #616866; ref. [1]) and a severe form of congenital muscular dystrophy (CMD)(OMIM #617066; ref. [4, 5]). Interestingly, changes in another subunit of the transcriptional coregulator ASC-1, ASCC1, also result in a prenatal muscle weakness with arthrogryposis and congenital bone fractures [1, 6]. Since the first reports in 2016 describing the two associated phenotypes, only five additional CMD families have been reported [5]. Here we present the diagnostic journey of a patient with cerebellar hypoplasia, spinal muscular atrophy (PCH1-like) and congenital bone fractures, where we identified a homozygous stop-gain variant in the TRIP4 gene; the ninth case reported world-wide.
Subjects and methods
This study was approved by the Ethics Committee of University of Duisburg-Essen (19-9011-BO). All the involved subjects gave their written informed consent.
Exome sequencing
DNA from the index case and his parents was subjected to whole exome sequencing (WES) using Nextera Rapid Exome Capture (Illumina)(Supplementary material). Exonic and splice site variants with a minor allele frequency of <1% in external databases (i.e. ExAC, 1000 genomes) were prioritized. In silico prediction tools such as Polyphen2 (http://genetics.bwh.harvard.edu/pph2/), CADD (https://cadd.gs.washington.edu/) and SIFT (http://sift.jcvi.org/) were used to assess pathogenicity.
WES reanalysis
The fastq files for the index case and his parents were submitted to Solve-RD [7] through the Genome-Phenome Analysis Platform (GPAP, https://platform.rd-connect.eu/) as part of the European Reference Network for Neuromuscular Disease (ERN-EURO-NMD, https://ern-euro-nmd.eu/), and processed using the RD-Connect bioinformatics pipeline [8]. The reanalysis strategy carried out by the SNV-Indels Working Group [7, 9] implemented automated data filtration to specifically identify previously reported disease-causing variants (as annotated in ClinVar v.13-01-2020) from a candidate gene list of 594 genes known to be associated with NMD [9]. The identified variant has been submitted to ClinVar (SUB8878662; https://www.ncbi.nlm.nih.gov/clinvar).
Mass spectrometry based proteomic analysis
Control and patient-derived fibroblasts were grown to a 70% confluency and mechanically harvested. Label free shotgun proteomic analysis was performed as previously described [10]. To interrogate protein-dysregulation pathways a Gene Ontology (GO) term analysis was performed. Proteomaps (www.proteomaps.net) was used to visualise affected cellular functions. The mass spectrometry proteomics data was submitted to the ProteomeXchange Consortium via the PRIDE (PXD023584; https://www.ebi.ac.uk/pride).
Results
Clinical findings
The patient was the first child of consanguineous parents, born at 38 weeks of gestational age by Caesarean section (Apgar score: 5/5/5). Pregnancy was complicated by oligohydramnios, placental insufficiency and growth retardation. The newborn presented small for gestational age at birth (weight 2610 g [6th percentile], length 42 cm [<3th percentile], head circumference 34.5 cm [40th percentile]). Multiple contractures and intrauterine-originated fractures of both femurs and the right humerus were present at birth. He also showed respiratory insufficiency complicated by absence of spontaneous movements so that invasive ventilation was initiated. Clinical course was further characterised by pleural effusion and chylothorax, biliary calculi and soft tissue emphysema. Cranial and thoracic MRI performed at the age of 6 weeks revealed a hypoplastic cerebellum but normal pons, necrosis and haemorrhage of the occipital cortex, pronounced atrophy of skeletal musculature and confirmed bilateral pleural effusion (Fig. 1). After excluding 5q-SMA by genetic analysis of the SMN1 gene, a vastus lateralis muscle biopsy was performed for further diagnostic evaluation, yet due to the massive atrophy, the number of intact fibres was too small to allow a valid pathological examination. Given the unfavourable prognosis, after consultation with the parents and the clinical ethical committee, invasive ventilation was terminated at the age of 8 weeks when the patient died. The combination of SMA and cerebellar hypoplasia resembled patients with disease-causing variants in components of the human RNA exosome [11].
Fig. 1. Magnetic resonance imaging (MRI).
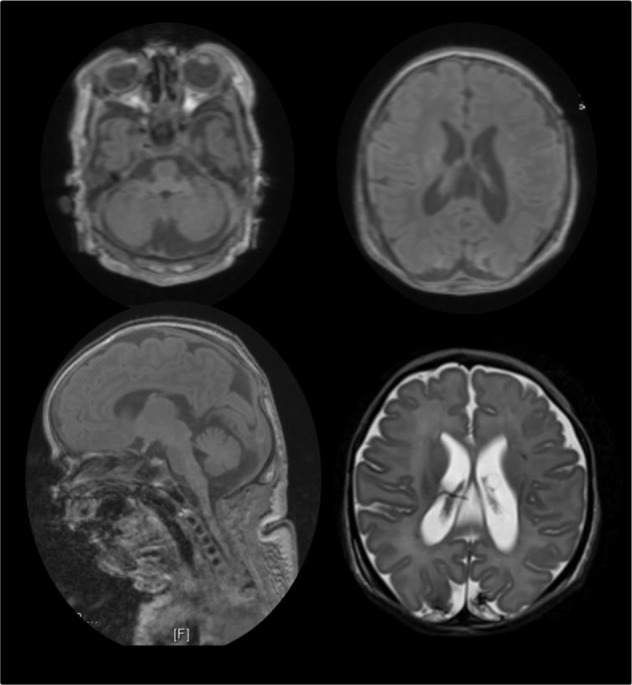
Cranial and thoracic MRI performed at the age of 6 weeks revealed a hypoplastic cerebellum, necrosis and haemorrhage of the occipital cortex, pronounced atrophy of skeletal musculature and confirmed bilateral pleural effusion.
Initial genetic analysis
SMN1 and additional single-gene screening of ACTA1, BIN1, DNM2, MTM1, RYR1, SEPN1, TNNT1, TPM2 and TPM3 was negative. Trio exome sequencing was undertaken, with the first round of analysis taking place in 2017. Stringent filtering criteria retrieved no variants in known disease-causing or strong candidate genes.
Reanalysis of exome data
The ‘low hanging fruit’ strategy implemented in 2020 by the SNV-Indels Working Group of Solve-RD [7, 9] meant only variants already known to be associated with disease were selected. Thus, a homozygous stop-gain variant [hg19:chr15:64698591 C > T; NM_016213.5:c.760 C > T; p.(Arg254*)] in the TRIP4 gene was identified. This variant had been previously reported in three patients (from two families) with Prenatal Spinal Muscular Atrophy and Congenital Bone Fractures [1], proving to be a phenotypic match and final diagnosis to our patient.
Proteomic findings
To further validate the pathogenicity of the detected variant and to obtain biochemical insights into the underlying pathophysiology, a proteomic profile on patient-derived fibroblasts was carried out. Applying a data independent acquisition (DIA) approach, a total of 3558 proteins were quantified. Among these, 323 were significantly dysregulated (p-ANOVA ≤ 0.05): 143 (4.2%) and 180 (5.6%) proteins showed increased and decreased abundance, respectively. An intersection of dysregulated proteins with genetic variants revealed no significant correlations. Proteomaps and a GO term analysis revealed a dysregulation of diverse signaling cascades including the NF-kappa-B pathway. We also detected an increased abundance in proteins of the RNA-processing machinery, while decreased proteins suggested impaired exosomal activity (Fig. 2).
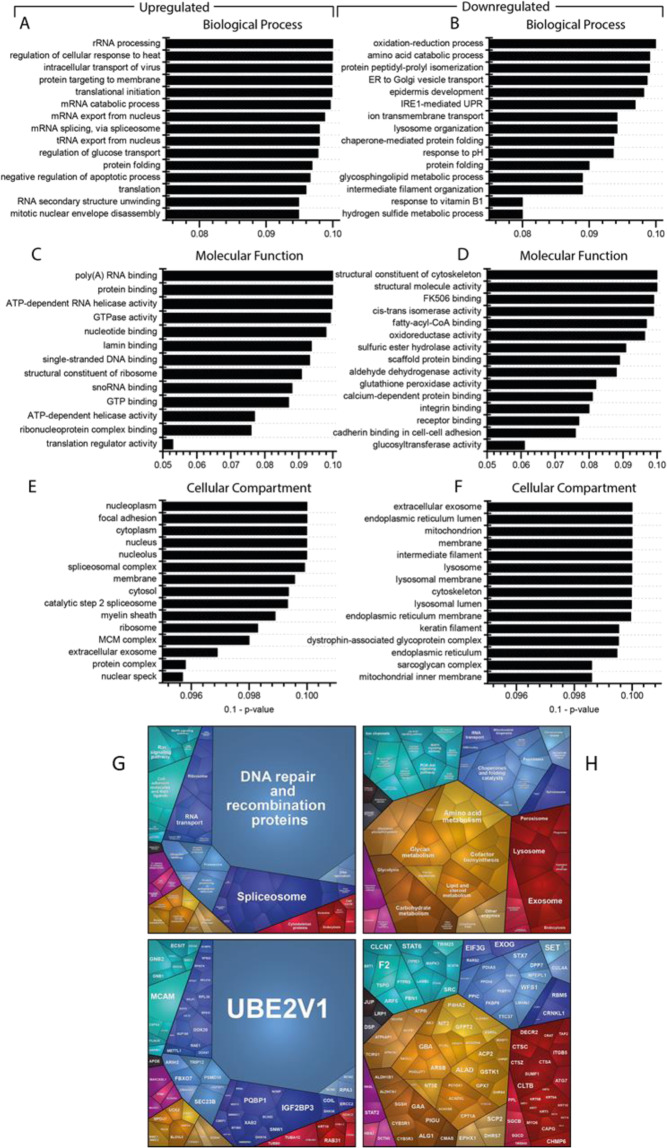
Fig. 2. In silico studies of proteomic data.
Gene Ontology (GO) term analysis was performed focussing on the up- and downregulated proteins separately: (A) “Biological Processes”revealed that the majority of increased proteins is involved in RNA processing, whereas most of decreased proteins (B) control the response to oxidative stress or are involved in protein folding. “Molecular Function” revealed that proteins involved in RNA-binding and modulating nucleotide binding are increased (C), and those involved in the cytoskeleton structure and the modulation of oxidative stress predominate amongst the downregulated (D). “Cellular Component” showed that the proteins upregulated in the patient-derived fibroblasts mostly belong to the nucleus (E) and the ER, the lysosome, mitochondria, and the cytoskeleton are downregulated (F). The proteomaps-based pathway analysis also focussed on increased and decreased proteins separately. G For increased proteins, it confirmed changes in RNA-transport and RNA-processing, and indicated an activation of the protein clearance machinery (Ubiquitin-mediated proteolysis via the proteasome) as well as altered signalling processes (Ras, ErbB and MAPK pathways). H For decreased proteins, indicated a vulnerability of cellular metabolism accompanied by perturbed signalling cascades (Jak-STAT, ErbB, MAPK, PI3K-AKT & NF-kappa-B pathways) and reduced lysosomal as well as exosome function.
Discussion
We describe the diagnostic journey of a patient presenting with a cerebellar hypoplasia and SMA (PCH1-like) phenotype with bone fractures, in whom the exome reanalysis using the RD-Connect/Solve-RD pipeline [8, 9] identified a homozygous stop-gain variant in the TRIP4 gene. Intriguingly, the disease-causing variant was not found during the first round of analysis (circa 2017), even though coverage at this position was >40x. TRIP4 was associated with disease only in 2016 [1, 4], however the initial WES analysis searched for candidate variants in the whole exome, and not just a gene list of known associated genes. Since the TRIP4 variant is a highly deleterious homozygous variant, it seems unlikely that it would have not been picked as a strong candidate. In fact, by interrogating filtered VCF files we speculate that the variant must have been filtered out during the bioinformatics steps. This emphasises that reevaluation of legacy exome data should be performed not only by periodic reiteration of the analysis [12], but also by improved and updated bioinformatic pipelines, such as those implemented in Solve-RD [7–9].
TRIP4 encodes a transcription coactivator which facilitates nuclear receptors-mediated transcription and ribosome-associated quality control in mammalian cells [13]. It has been also shown to play a role in transactivation of NF-kappa-B [2].
The combination of genomics, transcriptomics and proteomics is increasingly being used to correlate protein abundances with genetic variants, as well as to delineate pathways affected by the loss of a functional protein [14, 15]. Such approaches build on the assumption that protein interaction networks can be viewed as maps in which diseases can be identified with characteristic proteomic signatures [14, 16]. Tissues such as blood, skin or muscle biopsies provide a source of DNA, RNA and protein [14] suitable to study the molecular etiology of these diseases [10]. Here, we showed how proteomic profiling was carried out in a patient with PCH1-like disease with bone fractures to evaluate the pathogenicity of the detected variant and to obtain insights into the biochemical etiology. Our pathway-analyses of proteomic findings highlighted perturbations in RNA-processing and signaling processes including the NF-kappa-B pathway. We detected decrease in abundance of the RNA exosome complexes, in line with the clinical presentation of PCH1 [17], however no variant was detected in any of the known genes encoding subunits or accessory proteins of the human exosome complex. Although the TRIP4 protein was not detectable on the proteomics profile, alteration of other proteins resulted in a “proteomic signature”, highlighting the molecular consequences of the loss of TRIP4. The similar clinical presentation of patients with variants in TRIP4 and exosomal proteins further highlights that PCH1 is genetically heterogeneous, and linked with ribosome dysfunction and abnormal RNA processing [18].
Clinical and biochemical synopsis
A neurological syndrome combining cerebellar hypoplasia and spinal motor muscular atrophy is already linked to exosomal protein variants expanding the list of human exosomopathies [11]. The RNA exosome represents a conserved multi-protein complex essential for gene expression via processing and degradation of mRNA [19]. Proteomic pathway analysis in a patient with a novel TRIP4 variant revealed a dysregulation of RNA-processing and down-regulation of exosomal proteins. These findings suggest a common pathomechanism leading to the clinical manifestation of this neurological syndrome. However, comparative biochemical studies in in vitro or in vivo models are crucial to elucidate the precise molecular nature of the common pathophysiology.
Supplementary information
Acknowledgments
Solve-RD SNV-indel working group
Leslie Matalonga4, Enzo Cohen12, Isabel Cuesta13, Daniel Danis14, Anne-Sophie Denommé-Pichon15, Yannis Duffourd15,16, Christian Gilissen17,18, Mridul Johari19, Steven Laurie4, Shuang Li20, Isabelle Nelson12, Ida Paramonov4, Sophia Peters21, Sivakumar Prasanth22, Peter Robinson4, Karolis Sablauskas17,18, Marco Savarese19, Wouter Steyaert17,18, Ana Töpf1, Joeri K. van der Velde20, Antonio Vitobello15
Solve-RD-DITF-euroNMD
Ana Töpf1, Jonathan Baets23,24,25, Danique Beijer24,25, Gisèle Bonne12, Enzo Cohen12, Judith Cossins26, Teresinha Evangelista12, Alessandra Ferlini27, Peter Hackman19, Michael G. Hanna22, Rita Horvath5, Henry Houlden22, Mridul Johari19, Jarred Lau28, Hanns Lochmüller4,28,29,30,31, William L. Macken22, Francesco Musacchia32,33, Andres Nascimento34, Daniel Natera-de Benito34, Vincenzo Nigro33, Giulio Piluso32, Veronica Pini35, Robert D. S. Pitceathly22, Kiran Polavarapu28,31, Pedro M. Rodriguez Cruz26,36, Anna Sarkozy35, Marco Savarese19, Rita Selvatici27, Rachel Thompson28, Annalaura Torella32, Bjarne Udd19, Liedewei Van de Vondel24,25, Jana Vandrovcova22, Irina Zaharieva35
Funding
The Solve-RD project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 779257. Several authors of this study are members of the European Reference Network for Neuromuscular disease, Project ID No 739543. This study was supported by the AFM (grant 21644 to AR), the framework of the NME-GPS project by the European Regional Development Fund (AR/US/AS), the support by the Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen, the Regierende Bürgermeister von Berlin-inkl. Wissenschaft und Forschung, and the Bundesministerium für Bildung und Forschung (AH and AS), the European Research Council (RH), the Wellcome Investigator Award (109915/Z/15/Z to RH), the Medical Research Council UK (MR/N025431/1 to RH), the Newton Fund (MR/N027302/1 to RH), the Lily Foundation (RH) and the Evelyn Trust (RH). This research was supported by an MRC strategic award to establish an International Centre for Genomic Medicine in Neuromuscular Diseases (ICGNMD) MR/S005021/1 and the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Members of the Solve-RD SNV-indel working group and Solve-RD DITF-euroNMD are listed above Funding.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Andreas Roos, Rita Horvath
Contributor Information
Andreas Roos, Email: Andreas.Roos@uk-essen.de.
Rita Horvath, Email: rh732@medschl.cam.ac.uk.
Solve-RD SNV-indel working group:
Enzo Cohen, Isabel Cuesta, Daniel Danis, Anne-Sophie Denommé-Pichon, Yannis Duffourd, Christian Gilissen, Mridul Johari, Steven Laurie, Shuang Li, Isabelle Nelson, Ida Paramonov, Sophia Peters, Sivakumar Prasanth, Peter Robinson, Karolis Sablauskas, Marco Savarese, Wouter Steyaert, Joeri K. van der Velde, and Antonio Vitobello
Solve-RD DITF-euroNMD:
Jonathan Baets, Danique Beijer, Gisèle Bonne, Judith Cossins, Teresinha Evangelista, Alessandra Ferlini, Peter Hackman, Michael G. Hanna, Henry Houlden, Jarred Lau, Hanns Lochmüller, William L. Macken, Francesco Musacchia, Andres Nascimento, Daniel Natera-de Benito, Vincenzo Nigro, Giulio Piluso, Veronica Pini, Robert D. S. Pitceathly, Kiran Polavarapu, Pedro M. Rodriguez Cruz, Anna Sarkozy, Rita Selvatici, Rachel Thompson, Annalaura Torella, Bjarne Udd, Liedewei Van de Vondel, Jana Vandrovcova, and Irina Zaharieva
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00851-8.
References
- 1.Knierim E, Hirata H, Wolf NI, Morales-Gonzalez S, Schottmann G, Tanaka Y, et al. M. Mutations in Subunits of the Activating Signal Cointegrator 1 Complex Are Associated with Prenatal Spinal Muscular Atrophy and Congenital Bone Fractures. Am J Hum Genet. 2016;98(Mar):473–89. doi: 10.1016/j.ajhg.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung D-J, Sung H-S, Goo Y-W, Lee HM, Park OK, Jung S-Y, et al. Novel transcription coactivator complex containing activating signal cointegrator 1. Mol Cell Biol. 2002;22:5203–11. doi: 10.1128/MCB.22.14.5203-5211.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iyer LM, Burroughs AM, Aravind L. The ASCH superfamily: novel domains with a fold related to the PUA domain and a potential role in RNA metabolism. Bioinformatics. 2006;2:257–63. doi: 10.1093/bioinformatics/bti767. [DOI] [PubMed] [Google Scholar]
- 4.Davignon L, Chauveau C, Julien C, Dill C, Duband-Goulet I, Cabet E, et al. The transcription coactivator ASC-1 is a regulator of skeletal myogenesis, and its deficiency causes a novel form of congenital muscle disease. Hum Mol Genet. 2016;25(Apr):1559–73. doi: 10.1093/hmg/ddw033. [DOI] [PubMed] [Google Scholar]
- 5.Villar-Quiles RN, Catervi F, Cabet E, Juntas-Morales R, Genetti CA, Gidaro T, et al. ASC-1 Is a Cell Cycle Regulator Associated with Severe and Mild Forms of Myopathy. Ann Neurol. 2020;87(Feb):217–32. doi: 10.1002/ana.25660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böhm J, Malfatti E, Oates E, Jones K, Brochier G, Boland A, et al. Novel ASCC1 mutations causing prenatal-onset muscle weakness with arthrogryposis and congenital bone fractures. J Med Genet. 2019;56:617–21. doi: 10.1136/jmedgenet-2018-105390. [DOI] [PubMed] [Google Scholar]
- 7.Zurek B, Ellwanger K, Vissers LELM, Schüle R, Synofzik M, Töpf A, et al. Solve-RD: systematic Pan-European data sharing and collaborative analysis to solve Rare Diseases. Submitted to EJHG, reference 698-20-EJHG. [DOI] [PMC free article] [PubMed]
- 8.Laurie S, Fernandez-Callejo M, Marco-Sola S, Trotta JR, Camps J, Chacón A, et al. From Wet-Lab to Variations: Concordance and Speed of Bioinformatics Pipelines for Whole Genome and Whole Exome Sequencing. Hum Mutat. 2016;37(Dec):1263–1271. doi: 10.1002/humu.23114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matalonga L, Hernández-Ferrer C, Piscia D, Solve-RD SNV-indel working group, et al. Solving patients with rare diseases through programmatic reanalysis of genome-phenome data. Submitted to EJHG, reference 703-20-EJHG. [DOI] [PMC free article] [PubMed]
- 10.Hentschel A, Czech A, Münchberg U, Freier E, Schara-Schmidt U, Sickmann A, et al. Protein Signature of Human Skin Fibroblasts Allows the Study of the Molecular Etiology of Rare Neurological Diseases. Research Square, Jul 2020. [DOI] [PMC free article] [PubMed]
- 11.Burns DT, Donkervoort S, Müller JS, Knierim E, Bharucha-Goebel D, Faqeih EA, et al. Variants in EXOSC9 Disrupt the RNA Exosome and Result in Cerebellar Atrophy with Spinal Motor Neuronopathy. Am J Hum Genet. 2018;102:858–73. doi: 10.1016/j.ajhg.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker SW, Murrell JR, Nesbitt AI, Pechter KB, Balciuniene J, Zhao X, et al. Automated Clinical Exome Reanalysis Reveals Novel Diagnoses. J Mol Diagn. 2019;21(Jan):38–48. doi: 10.1016/j.jmoldx.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto S, Sugiyama T, Yamazaki R, Nobuta R, Inada T. Identification of a novel trigger complex that facilitates ribosome-associated quality control in mammalian cells. Sci Rep. 2020;10:3422. doi: 10.1038/s41598-020-60241-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roos A, Thompson R, Horvath R, Lochmüller H, Sickmann A. Intersection of Proteomics and Genomics to “Solve the Unsolved” in Rare Disorders such as Neurodegenerative and Neuromuscular Diseases. Proteomics Clin Appl. 2017;1700073. [DOI] [PubMed]
- 15.Thompson R, Spendiff S, Roos A, Bourque PR, Warman Chardon J, Kirschner J, et al. Advances in the diagnosis of inherited neuromuscular diseases and implications for therapy development. Lancet Neurol. 2020;19:522–32. doi: 10.1016/S1474-4422(20)30028-4. [DOI] [PubMed] [Google Scholar]
- 16.Ghiassian SD, Menche J, Barabási AL. A DIseAse MOdule Detection (DIAMOnD) algorithm derived from a systematic analysis of connectivity patterns of disease proteins in the human interactome. PLoS Comput Biol. 2015;11:e1004120. doi: 10.1371/journal.pcbi.1004120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boczonadi V, Müller JS, Pyle A, Munkley J, Dor T, Quartararo J, et al. EXOSC8 mutations alter mRNA metabolism and cause hypomyelination with spinal muscular atrophy and cerebellar hypoplasia. Nat Commun. 2014;5:4287. doi: 10.1038/ncomms5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fabre A, Badens C. Human Mendelian diseases related to abnormalities of the RNA exosome or its cofactors. Intractable Rare Dis Res. 2014;3(Feb):8–11. doi: 10.5582/irdr.3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Comparative Study Genome Res. 2004;14(Jul):1315–23. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.