Abstract
Monitoring biodiversity is of increasing importance in natural ecosystems. Metabarcoding can be used as a powerful molecular tool to complement traditional biodiversity monitoring, as total environmental DNA can be analyzed from complex samples containing DNA of different origin. The aim of this research was to demonstrate the potential of pollen DNA metabarcoding using the chloroplast trnL partial gene sequencing to characterize plant biodiversity. Collecting airborne biological particles with gravimetric Tauber traps in four Natura 2000 habitats within the Natural Park of Paneveggio Pale di San Martino (Italian Alps), at three-time intervals in 1 year, metabarcoding identified 68 taxa belonging to 32 local plant families. Metabarcoding could identify with finer taxonomic resolution almost all non-rare families found by conventional light microscopy concurrently applied. However, compared to microscopy quantitative results, Poaceae, Betulaceae, and Oleaceae were found to contribute to a lesser extent to the plant biodiversity and Pinaceae were more represented. Temporal changes detected by metabarcoding matched the features of each pollen season, as defined by aerobiological studies running in parallel, and spatial heterogeneity was revealed between sites. Our results showcase that pollen metabarcoding is a promising approach in detecting plant species composition which could provide support to continuous monitoring required in Natura 2000 habitats for biodiversity conservation.
Subject terms: Biodiversity, Molecular ecology
Introduction
Monitoring biodiversity in natural ecosystems, and particularly in protected areas (e.g. the European network Natura 2000), is becoming of increasing importance in ecological research to document the consequences of the recognized biodiversity loss of the last decades1,2. Metabarcoding provides a powerful tool for rapid surveys of plant and animal communities to acquire high-resolution information on biodiversity and its changes over long timescales and large regions3. Environmental DNA (eDNA) analysis, in particular, that can identify multiple species in bulk samples4 has the potential to provide a swift improvement in estimating community biodiversity, fostering new insights on diversity components (alpha, beta and gamma diversity) for organisms and ecosystems5–9.
As regards plant biodiversity, pollen richness is often utilized as a proxy to estimate floristic richness in the catchment area10. Traditionally, pollen assemblages are analyzed via light microscopy based on pollen morphology11. Specific caveats are linked to peculiar morphological features of pollen grains, which can be shared among different species, genera (e.g. genera in Poaceae family) or even families (e.g. Cupressaceae and Taxaceae 12,13. This limitation to taxonomic precision can be tackled by the exploitation of the DNA-based approaches which recently revolutionized environmental monitoring14. Pollen DNA from environmental samples has been used in recent years as a matrix in different fields, including the study of airborne allergenic pollen12,15,16, plant–pollinator interactions17–19 and provenance of honey20,21.
High Throughput Sequencing (HTS) enables the recovery of DNA sequence data directly from environmental samples, increasing the scalability of biodiversity surveys22. HTS can quickly produce a massive amount of DNA sequences with ease23, and increasingly accessible DNA reference databases facilitate taxonomic assignment to single (barcoding) or multiple (metabarcoding) species24,25. Environmental samples, such as air samples, can easily undergo degradation26 and therefore short markers are used 3. Between different short fragment markers, the chloroplast trnL partial gene sequencing has been previously used with airborne pollen showing to be sufficiently variable and appropriate for amplification12,15. Previous surveys have compared different matrices (air samples, plant material, pollen traps) to detail overlapping results between pollen morphology and DNA analyses finding a good agreement between the methods15,16,18,27. However, a debate is still ongoing about the effectiveness of plant metabarcoding used for the relative quantification of plant communities21,28, with studies supporting the quantitative potential of a multi-locus approach29,30.
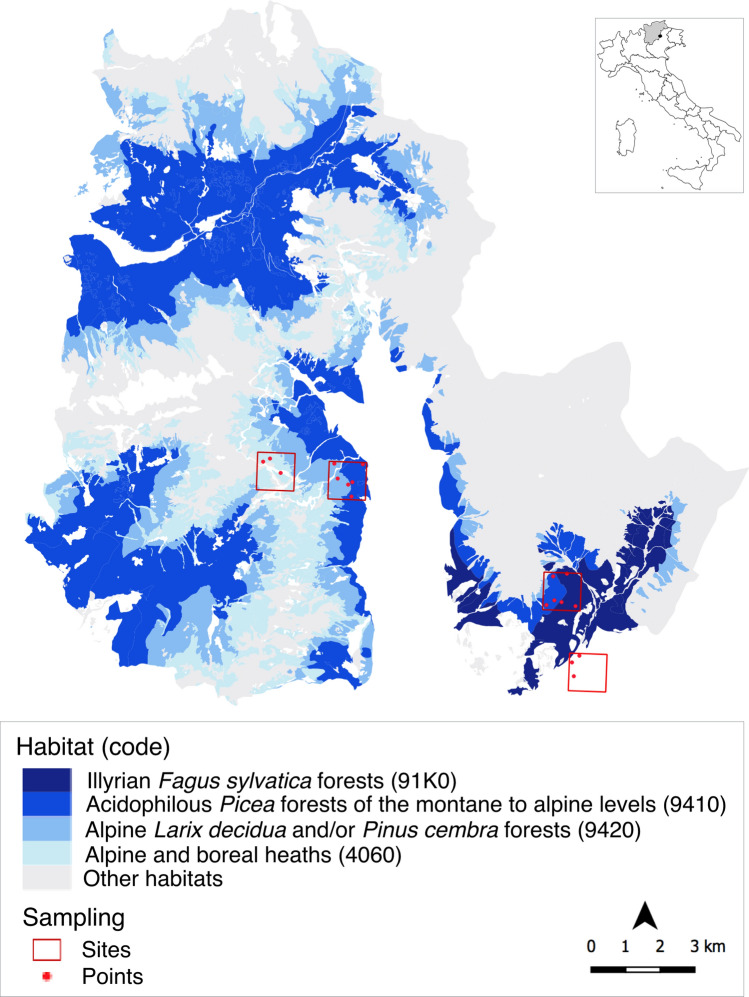
This study, following recently optimized protocols12, aimed to demonstrate the potential of pollen DNA metabarcoding, using trnL partial gene high throughput sequencing, to characterize and monitor plant biodiversity in protected areas, (e.g. the European ecological network Natura 2000) and to be used as a tool to inform efforts related to biodiversity conservation of species and habitats in a long-term perspective. A direct comparison with the traditional methods of pollen analysis was performed by light microscopy on a subset of samples to evaluate data overlapping and complementarity. We also investigated how the DNA-based trnL partial gene sequencing of pollen analysis can capture spatio-temporal patterns of airborne pollen, and, hence, its potential to semi-quantitatively track vegetation changes. The study encompassed the four most prevalent EU Natura 2000 habitats within the Natural Park of Paneveggio Pale di San Martino (eastern Italian Alps), a study site selected by several conservation and monitoring projects over the last decades. In particular, each habitat (i.e. Beech forest; Spruce forest, in two quadrates; Larch forest and Alpine heath) was monitored in three sampling points (within-habitat replicates) and three non-EU sampling points adjacent to the park were also monitored to account for low-altitude vegetation (Fig. 1). Overall, the study included 18 sampling points along an altitudinal gradient (Table 1) and the sampling covered a 1-year time span: October 2014–March 2015, March–July 2015 and July–October 2015.
Figure 1.
The sampling area in the National Park of Paneveggio–Pale di San Martino, Italy. The habitats in quadrats (sites) and sampling points (points) that were selected for the study are reported, as generated in QGIS3 (version 3.10.13, https://www.qgis.org).
Table 1.
The habitat types (according to the EU coding system) selected for the study are described.
| Habitat type | Habitat code | Altitudinal range (m) | Geographical location | % cover |
|---|---|---|---|---|
| Land principally occupied by agriculture, with significant areas of natural vegetation/coniferous forest (Lowland) | CLC 243/CLC 312 | 1050–1080 | Val Canali | – |
| Illyrian Fagus sylvatica forests (Beech) | 91K0 | 1290–1460 | Val Canali | 3.62 |
| Acidophilous Picea forests of the montane to alpine levels (Spruce low) | 9410 | 1530–1590 | Val Canali | 24.31 |
| Acidophilous Picea forests of the montane to alpine levels (Spruce high) | 9410 | 1620–1760 | Tognola | |
| Alpine Larix decidua and/or Pinus cembra forests (Larch) | 9420 | 1780–1860 | Tognola | 12.82 |
| Alpine and Boreal heaths (Alpine) | 4060 | 2040–2180 | Tognola | 10.92 |
A contracted description of the habitats is reported in brackets.
Results
Biodiversity assessment through DNA metabarcoding
Our analysis detected 160 Operational Taxonomic Units (OTUs) with 12,007,712 sequenced reads, 222,370 ± 41,954 (sd) reads per sample, for a total of 54 sequenced samples. The rarefaction curves showed good sequencing effort for the samples (Supplementary Figure S1) which were rarefied to the least count among samples corresponding to 135,443 reads. Twenty OTUs, (7.2% of the total), were assigned to taxa not relevant to our work (mainly to mosses and ferns during the periods October 2014–March 2015 and July–October 2015). From the remaining OTUs, 108 (88% of the reads) were taxonomically assigned to 32 families of vascular plants (68 identified taxa) (Table 2, Supplementary Table S1) and 32 OTUs (4.8% of the reads) remained unidentified either because of low sequence identity and/or query coverage percentage or the absence of any sequence classification result, even when compared to the complete ‘Nucleotide’ Genbank database. The results of the taxonomic assignment to vascular plants are presented in Supplementary Table S1. The OTU sequences were assigned to plant taxa with at least 95% identity and coverage, from which 70% of the OTUs had ≥ 98% sequence identity with the assigned taxa. The positive control of the DNA extraction, Corylus avellana pollen, was correctly identified after HTS. From the 19 negative controls included in the extraction plate, one negative control was selected for sequencing, the only one with sufficient amplicon concentration (2 ng μl−1). In this sample two OTUs were detected (263,649 reads), both assigned to Quercus spp. and contributing < 0.005% to the counts of the rest of the samples. The average amplicon concentration of the three pooled PCR products (before library preparation) was 8.7 ± 4.6 (sd) ng μl−1.
Table 2.
Quantitative data for the sequenced reads and the vascular plant taxa that were identified at the different sampling periods and habitats.
| No. of OTUs (sequenced reads) | No. of identified taxa | No. of identified families | |
|---|---|---|---|
| Sampling period | |||
| October 2014–March 2015 | 142 (3,744,854) | 64 | 30 |
| March–July 2015 | 135 (4,182,915) | 64 | 31 |
| July–October 2015 | 109 (4,079,943) | 46 | 21 |
| Habitat type | |||
| Lowland | 128 (2,225,089) | 63 | 29 |
| Beech forest | 129 (1,744,374) | 61 | 30 |
| Spruce forest (low) | 124 (2,195,166) | 58 | 28 |
| Spruce forest (high) | 129 (2,109,992) | 55 | 24 |
| Larch forest | 113 (1,851,947) | 52 | 22 |
| Alpine heath | 130 (1,881,144) | 53 | 24 |
| Total | 160 (12,007,712) | 68 | 32 |
The full names of the corresponding habitats are given in this table.
The sampling period July–October 2015 had a relatively lower number of OTUs and identified vascular plant taxa compared to the other periods (March–July 2015 and October 2014–March 2015). When grouped by habitat, the lowest number of OTUs and assigned taxa were found in Larch forests (Table 2). The alpha diversity of the habitats in each sampling period estimated as the number of OTUs per sample, is presented in Supplementary Figure S2. The interaction between sampling period and habitat category significantly influenced the alpha diversity (ANOVA, p < 0.01). Specifically, OTU richness decreased in July–October 2015 for all habitats, while in October 2014–March 2015 Larch forests and Alpine heath showed significantly lower plant biodiversity than Spruce, Beech and Lowland habitats.
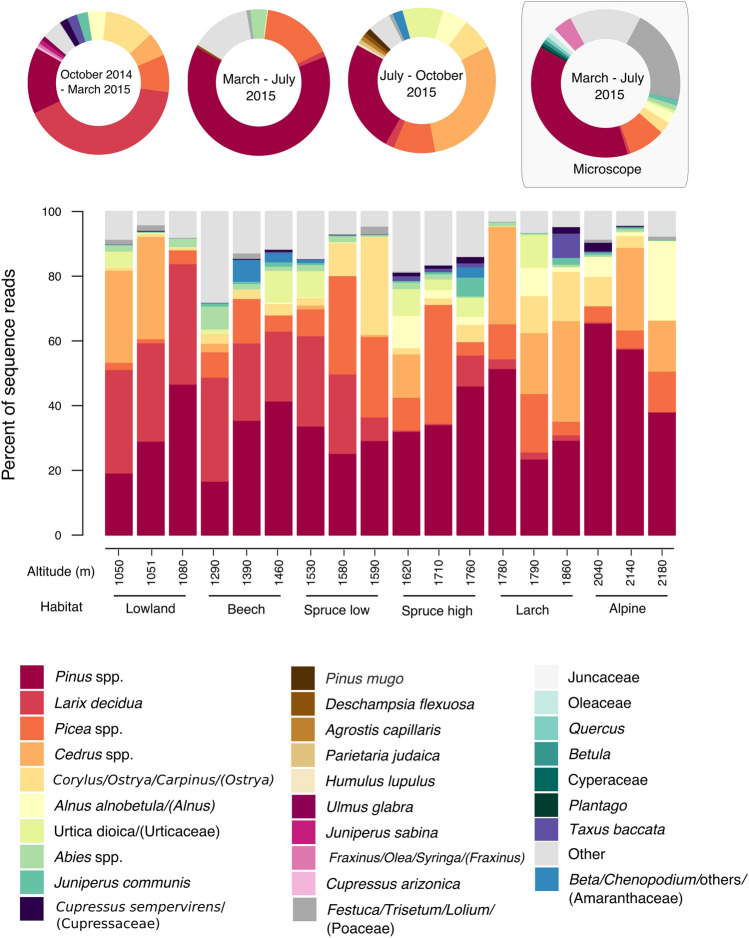
Of the 68 identified taxa from the HTS dataset, 42 were woody (trees and shrubs) and 26 herbaceous, 10 of which were graminoids (Supplementary Table S1). Fifteen of these taxa were not present in the plant checklist of the park31 (Supplementary Table S1). There were 13 main pollen taxa contributing at least 0.5% to the total number of the sequence reads (Supplementary Table S1). These were Pinus spp. (36.8%), Larix decidua (14.4%), Cedrus spp. (12.4%), Picea spp. (11.6%), Corylus/Ostrya/Carpinus (5%), Alnus alnobetula (2.9%), Urtica dioica (2.8%), Abies spp. (1.5%), Juniperus communis (0.7%), Beta/Chenopodium/others (0.6%), Taxus baccata (0.6%), Festuca/Trisetum/Lolium (0.5%) and Cupressus sempervirens (0.5%). The relative abundance of the most represented taxa (with a relative contribution at least 0.5% to the sequenced reads (in each period and in total) is presented in Fig. 2. Pinaceae taxa (Pinus, Picea, Cedrus, Abies) were dominant in all three periods, Betulaceae (Corylus/Ostrya/Carpinus, Alnus, Betula), Cupressaceae (Juniperus, Cupressus) and Taxaceae (Taxus) were also abundant in the period October 2014–March 2015 as well as Amaranthaceae (Beta/Chenopodium/others), Betulaceae (Corylus/Ostrya/Carpinus, Alnus, Betula), and Urticaceae (Urtica/Parietaria) in the period July–October 2015. Pinus spp. were the most represented plant taxa in all habitats for the entire 1-year dataset. As regards the other taxa, each habitat showed a distinct plant biodiversity spectrum, while Picea spp. were mostly found in the samples of the Spruce forests (Fig. 2).
Figure 2.
Taxa summary plots: Doughnut pie chart for all three sampling periods as derived by HTS and for the period March–July 2015, as derived by microscopy (up) and barplot for each sampling point (ordered in the x-axis from low to high altitudes) as derived by HTS. The charts represent relative abundance of sequences reads and microscope counts (≥ 0.5% of each period’s total for the doughnut plots and ≥ 0.5% of the annual total for the barplot). All the rest of the taxa are grouped under ‘Other' taxa. If the level of taxonomic identification differed between methods, given in parenthesis is the level after the microscopic method. For the full plant taxa assignment data see Supplementary Table S1. The full names of the corresponding habitats are given in Table 1.
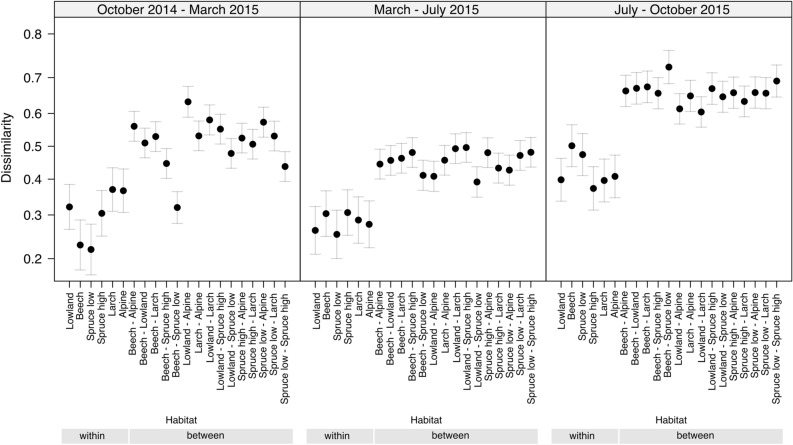
Beta diversity as calculated by Jaccard dissimilarity between samples is presented within and between habitats (Table S4, Fig. 3). Jaccard dissimilarity values were lower within habitats than between habitats for all three periods. For the within-habitat dissimilarity the period March–July 2015 had the highest values. Beta diversity was affected by the sampling period (PERMANOVA, p < 0.001, pseudo-F = 11.1, R2 = 0.30) and the habitat (PERMANOVA, p < 0.01, pseudo-F = 1.5, R2 = 0.39). According to the Generalized Linear Model predicting the Jaccard dissimilarity values as function of sampling period, habitat and their interaction (categorical variables), significant differences in beta diversity of different periods and habitats were detected, while also the interaction term of the latter variables had a significant effect on the variation of species composition (Supplementary Table S3). The effect of habitat (categorical variable) to Jaccard dissimilarity values is displayed in Fig. 3 showing higher dissimilarity between habitats than within habitats.
Figure 3.
Effect plots showing the differences between the habitat and period categories included in the Generalized Linear Model (quasi-binomial error distribution and logit link function) formulated with pairwise Jaccard dissimilarity index values (mean ± standard error) The full names of the corresponding habitats are given in Table 1.
Comparisons between DNA metabarcoding and microscopic analysis
The period March–July 2015, which is the main pollen season for most of the vascular plants of the study, was selected for microscopic analysis. The number of identified taxa is presented in Supplementary Table S4 for each habitat. The mean number in the habitats was 23 ± 4 (sd) taxa, while in total we could identify 50 taxa. The lowest number of identified taxa was recorded in Beech and Spruce high habitats, while for the same period, HTS recorded the lowest numbers in Spruce high and Larch forests. The most represented ones were Pinus spp. (37.9%), Poaceae (20.9%) and Picea spp. (8%) and 19 taxa contributed > 0.5% to the counts (Fig. 2, Supplementary Table S1). Beta diversity, as estimated by Jaccard distance (Supplementary Figure S4), showed to be affected by the habitat type (PERMANOVA, p < 0.05, pseudo-F = 1.45, R2 = 0.37).
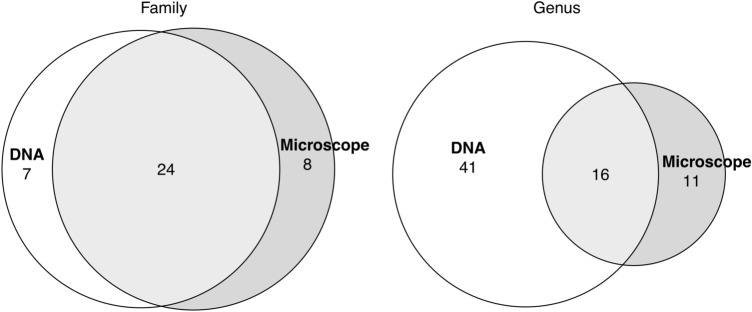
In total, DNA metabarcoding detected 68 plant taxa compared to 50 revealed by microscope (Supplementary Table S1). We identified a total of 39 families, 24 of which were identified by both methods (Fig. 4). Within these common families, DNA metabarcoding could distinguish 57 genera or groups of genera compared to 27 identified by the microscopic method (Fig. 4). In general, families not shown in the outputs of one of the two methods (eight and seven absent from the DNA metabarcoding and microscope outputs, respectively) were scarcely represented also by the other. Cyperaceae and Polygonaceae, although found with considerable abundance with conventional light microscopy analysis (0.4% and 0.6%, respectively), did not appear in the HTS results (Supplementary Table S1).
Figure 4.
Venn diagrams with the number of families found by DNA metabarcoding and microscope and the common genera from those families as detected during March–July 2015.
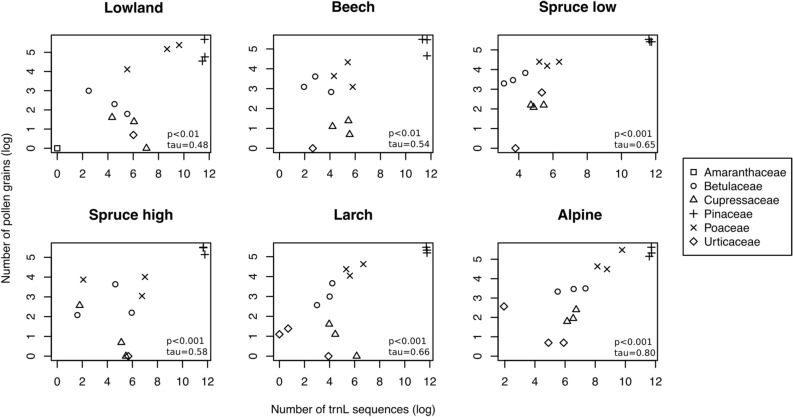
Log-scaled quantification results of pollen grains (microscope) and trnL sequence reads (metabarcoding) were compared between the two methods for the samples of March–July 2015 for each habitat within the altitudinal gradient (Fig. 5). For all habitats, there was significant correlation, i.e. p < 0.01 for Lowland (tau = 0.48) and Beech forest (tau = 0.54); p < 0.001 for Spruce low (tau = 0.59), Spruce high (tau = 0.67), Larch forest (tau = 0.67) and Alpine heath (tau = 0.8), which showed the highest correlation coefficient. Comparing the contribution of each family to the pollen spectra of all habitats, as derived by the two methods, we observe that the microscopic method systematically detected a higher frequency of Poaceae, Betulaceae and Oleaceae pollen than metabarcoding, whereas the latter showed a higher frequency of Pinaceae pollen (Fig. 2, Supplementary Table S1). For both DNA metabarcoding and pollen counts, Pinus was the most abundantly represented taxon (Supplementary Table S1). Notably, Fagus was not present in the HTS results and scarcely represented by the microscopic method. 11.7% of pollen grains were unidentified by the microscopic analysis due to unclear morphology, resulting mainly from the degradation of the pollen cell wall structure. 10.4% of sequence reads were unidentified (no significant match with reference sequences) by metabarcoding in the same period.
Figure 5.
Log-scaled quantification results as estimated by trnL metabarcoding counts (x-axis) and pollen microscope counts (y-axis). The results are summarized for the top five families identified by metabarcoding by habitat group for the period March–July 2015. P-values and the correlation coefficient from Kendall tau rank correlation tests are provided in the plot of each habitat. The full names of the corresponding habitats are given in Table 1.
Procrustes analysis revealed that there was no significant accordance between distance matrices generated by HTS and microscopic analysis for the period March–July 2015, based on the permutational test (no statistical significance of the Procrustean fit, correlation 0.4, 999 permutations) (Supplementary Figure S4).
Spatio-temporal patterns of pollen taxa
For each sampling period, the results of the HTS data for the identified plant taxa are presented in Supplementary Table S1. The main HTS pollen taxa contributing with at least 0.5% in the total reads of each period are Larix decidua, Pinus spp., Corylus/Ostrya/Carpinus, Picea spp., Cedrus spp., Alnus alnobetula, Cupressaceae (Juniperus communis, Cupressus sempervirens, Juniperus sabina), Taxus baccata, Ulmus glabra and Fraxinus/Olea/Syringa for the period October 2014–March 2015; Pinus spp., Picea spp., Abies spp., Larix decidua and Festuca/Trisetum/Lolium for March–July 2015; and Cedrus spp., Pinus spp., Picea spp., Urticaceae (Urtica dioica and Parietaria judaica), Corylus/Ostrya/Carpinus, Alnus alnobetula, Larix decidua, Beta/Chenopodium/others and Poaceae (Agrostis capillaris, Festuca/Trisetum/Lolium, Deschampsia flexuosa) for July–October 2015. Significantly higher occurrence according to ANOVA was recorded for Cupressus sempervirens (p < 0.001), Juniperus communis (p < 0.01) and Larix decidua (p < 0.001) in the period October 2014–March 2015, for Pinus spp. (p < 0.001) and Abies spp. (p < 0.001) in the period March–July 2015 and for Urtica dioica (p < 0.001), Cedrus spp. (p < 0.01) and Beta/Chenopodium/others (p < 0.05) in the period July–October 2015.
For the 13 main plant taxa, detailed quantitative HTS and microscope data are presented for each habitat during the period March–July 2015 (Supplementary Table S2). Significant spatial patterns were revealed according to ANOVA for Larix decidua with the highest occurrence recorded in the Larch forest (p < 0.05), Alnus alnobetula with the highest occurrence in the Alpine heath (p < 0.05) and Festuca/Trisetum/Lolium with the highest occurrence in the Lowland and Alpine heath (p < 0.05) (Supplementary Table S2). Similarly, for the microscope data spatial patterns were statistically significant for Larix, with the highest occurrence recorded in the Larch forest (p < 0.001), for Alnus, with the highest occurrence in the Alpine heath (p < 0.05), and for Cedrus with the highest occurrence in the low Spruce forest (p < 0.05). Occurrence of Picea was higher in the Spruce and Larch forests and for Poaceae in the Lowland and Alpine habitats, but in both cases differences were not significant, (p < 0.05) (Supplementary Table S2).
According to metabarcoding results, high read numbers of the main plant taxa were recorded when high pollen counts were also detected in airborne pollen data from the aerobiological station of the Park (Supplementary Figure S3). Exceptions are Corylus/Ostrya/Carpinus, Alnus alnobetula and Pinus spp., for which eDNA showed that their pollen was at relatively high concentrations both during their pollen seasons and at different times. In contrast, for Beta/Chenopodium/others and Urtica dioica, plant DNA was detected only in the period July–October 2015, although their pollen season was longer, starting within the period March–July 2015 (Supplementary Figure S3).
The majority of the main pollen taxa (11 of 13 taxa) have representatives in at least one of the habitat types examined31, except for Cupressus sempervirens and Cedrus spp. (Supplementary Figure S3) that do not occur within the Park and are probably transported from surrounding areas; notably, Cedrus is found at all altitudes of all periods. Also Pinus spp., Larix decidua and Picea spp. were retrieved from all samples (Supplementary Table S1).
Discussion
In this study, we characterized airborne pollen patterns by DNA metabarcoding within the most represented Natura 2000 habitats in Paneveggio Pale di San Martino Natural Park (North Italy), using eDNA from air samples. Chloroplast trnL partial gene sequencing provided an informative list of plant taxa growing along an altitudinal gradient of the Italian Alps with a finer taxonomic resolution when compared to the conventional light microscopy. It also uncovered spatio-temporal pollen patterns. Most of the taxa detected with at least 0.5% of total reads by DNA metabarcoding are represented in the vegetation of the park, and they were mostly recorded at the sampling times corresponding to their pollen season, as confirmed by the aerobiological sampling. This result is particularly useful for biodiversity assessments and monitoring in the present study area and also suggests that the method has the potential to be applied successfully in other areas.
Due to various factors that can influence quantitative results of metabarcoding, we applied semi-quantitative indices for our community analysis with HTS data: OTUs richness to estimate alpha diversity and the presence/absence-based Jaccard index as a measure of beta diversity32. Overall, alpha diversity for the habitats of lower altitude (1050–1760 m), i.e. Beech and Spruce forest was higher and revealed more vascular plant taxa with a total of 30 families when compared to habitats at higher altitudes (1780–2180 m), i.e. the Larch forest and Alpine Heath, with a total of 24 families (Supplementary Figure S2, Table 2). This suggests that a pollen metabarcoding approach can detect known differences in taxa richness along elevational gradients in mountainous areas33. In addition, the Jaccard dissimilarity of taxa within samples of the same period was significantly lower than between periods (PERMANOVA, p < 0.001, pseudo-F = 11.1, R2 = 0.30), showing that the sampling period significantly influences beta diversity. This is an important finding because it shows that pollen metabarcoding can detect changes with time and, hence, that it can be used for monitoring purposes. Beta diversity, estimated with Jaccard index, showed that the habitat replicates efficiently represented the different habitats by both conventional (PERMANOVA, p < 0.05, pseudo-F = 1.45, R2 = 0.37) and metabarcoding (PERMANOVA, p < 0.05, pseudo-F = 1.45, R2 = 0.37), since the variation in species composition was lower within than between habitats. Additionally, the Generalized Linear Model (quasi-binomial error distribution and logit link function) predicting the Jaccard dissimilarity values as function of sampling period, habitat and of their interaction term showed significant differences in the pairwise comparisons between habitats (Fig. 3, Supplementary Table S3).
When comparing qualitatively, in terms of identified taxa, conventional light microscopy and trnL partial gene sequencing, results showed good agreement, since 61% of the families were represented by both methods. For plant families detected by just one of the two methods, pollen abundance based on read counts or microscope counts was very low (Supplementary Table S1). Two families, Cyperaceae and Polygonaceae, were not detected by DNA metabarcoding, although considerably abundant in the light microscopy results. However, Cyperaceae is a family known to have low detection with the trnL barcode34. The result of not detecting specific taxa by this procedure could be due to primer mismatching during PCR and lack of amplification with a consequent exclusion from the sequencing results15,35 Other PCR biases such as differences in GC content36 or differences in cpDNA copy numbers12 may also affect the detection of specific taxa. To reduce PCR bias resulting from primer mismatching, an optimization of the PCR protocol or the use of a combination of primer pairs30 could be applied to improve sensitivity towards the plant species of interest.
Within the common families, the taxonomic information resulting from the metabarcoding method was richer (27% more taxa) and provided a finer resolution (53% more genera) when compared to the light microscopy results. The observed improvement in taxonomic information is in agreement with previous studies15,19,20,27. In particular, for some taxa that are usually identified only collectively (at family level or higher) with conventional light microscopy (e.g. grasses, for which pollen grains have common morphological features), the targeted HTS led to the identification of different genera13. This is highly relevant for biodiversity estimates, as well as for health issues linked to pollen allergy, e.g. more precise diagnosis and treatment of human sensitivity to aeroallergens37.
Nevertheless, quantitative estimates (abundance) of the plant biodiversity showed some notable differences between the DNA and microscope-based methods. When comparing the results of the two methods by habitat, Alpine heaths showed better correlation of relative abundance of reads and pollen grains (tau = 0.8, p < 0.001) compared to forested zones. This might also have led to overall low correlation generated by Procrustes analysis when comparing the ordinations generated by the two methods. In addition, when focusing in the most abundant taxa (at least 0.5% of contribution to the HTS counts), there were taxa that showed similar peaks in abundance compared to microscope results, such as Larix in Larch forest, Picea in Spruce forest, Poaceae and Alnus in the Alpine heaths.
For the main pollen season that we used for comparisons, although Pinus was the most abundantly represented taxon by both methods, Pinaceae were much more represented by DNA metabarcoding. In contrast, light microscopy identified more Poaceae, Betulaceae and Oleaceae. Similar differences were reported by other authors when either nuclear or chloroplast markers were used to analyze mixed pollen samples15,20,21,27. This quantitative issue could be linked to the amplification biases inherent in the PCR methodological step that can possibly lead to overamplification of some templates38,39. Other studies have applied capture enrichment which led to more accurate results avoiding PCR biases40. In addition, pollen structure can lead to over and under-representation of some taxa as demonstrated by previous studies41. Studies investigating predefined mock communities have attempted to give insight to quantitative issues42–44, but more controlled experiments with detailed knowledge of the eDNA ecology and simulations in all ecosystems would be needed to correlate the observed with the true abundance42.
In several cases, plant taxa were found in samples collected from sites where the producing plants were absent. This was the case for Taxus baccata, Beta/Chenopodium/others, Corylus/Ostrya/Carpinus, Abies spp. and Pinus spp. that grow in other sites inside the Park, but also for Cedrus spp. and Cupressus sempervirens that grow outside it. All Pinaceae taxa (Pinus spp., Larix spp., Cedrus spp., Picea spp., Abies spp.) were present in all seasons and almost all habitats. Since those taxa were in high quantity and they were not amplified in negative controls, we do not consider them as contaminants of the process. This finding can be partly explained by transfer from elsewhere45, but not only, since these plant taxa were detected at periods that do not correspond to their pollen seasons. For Pinaceae, this could be explained as the result of the huge amount of pollen produced locally, which may remain on the ground for a long time, gets resuspended by the wind and can be found in near ground-level samples even in periods when it is not produced and released by the plants. Such relic DNA has been detected and quantified in water and soil studies46–48; this study provides initial evidence that it can also be detected in air samples. Pinus was the most abundant plant/pollen taxon after either method, thanks to its very light pollen, easily transported by the wind49, more than that of other Pinaceae taxa. Van der Knaap50, using Tauber traps, found higher amounts of Pinus pollen above the treeline than in lower mountainous areas, indicating upslope transport by wind currents. Notably, the absence of Fagus (a prevalent tree in the Park) from the DNA metabarcoding results agreed with the aerobiological monitoring data of the Paneveggio Pale di San Martino Natural Park. Given the natural variability in the reproductive output of beech in different years51, this may be due to a much lower pollen production of beech in the year of study.
Overall, pollen metabarcoding increased considerably our ability to identify pollen52. Within the ecological network Natura 2000, eDNA analysis could be used as an efficient tool to characterize plant biodiversity and monitor diversity changes over space and time. Thanks to the fine taxonomic assignment, the method could be used in plant management, for instance to detect potential invasion of non-native species; this could be the case for ragweed pollen which can affect human health with its high allergenicity53. To resolve quantitative issues, more effective techniques could be applied in the future, such as complementing with other or new target barcodes, which would also give better resolution for some plant families29. At the same time, more extensive field experiments in different natural ecosystems could be planned at different time scales to examine the accuracy of the quantitative information42. DNA metabarcoding has therefore the potential to be applied successfully in other areas for biodiversity management and/or conservation purposes to ensure the protection of plant biodiversity and habitats in a long-term perspective.
Materials and methods
Sampling design
The sampling areas in the Natural Park of Paneveggio Pale di San Martino belong to the San Martino chain (Tognola mountain) in the west and the Val Canali chain in the southeast. The four most prevalent habitats54, covering 52% of the Park’s surface, were selected (among a total of 12 Natura 2000 habitats that cover > 90% of the area): Spruce forests (code 9410: Acidophilous Picea forests of the montane to alpine levels; 24.3%) in low and high altitudes, Larch forests (code 9420: Alpine Larix decidua and/or Pinus cembra forests; 12.8%), Alpine habitat (code 4060: Alpine and boreal heaths; 10.9%) and Beech forests (code 91K0: Illyrian Fagus sylvatica forests; 3.6%). An area outside of the Park was selected to account for lower altitude biodiversity, classified according to Corine Land Cover classification system (CLC) as Land principally occupied by agriculture, with significant areas of Natural vegetation (CLC 243; two points) and coniferous forest (CLC 312; one point). The study included 18 sampling points along the altitudinal gradient of the park. Three points were selected as replicates to account for within-habitat variability of the plant biodiversity. The sampling points were located randomly in quadrats (1 km × 1 km) of east exposure, with each quadrat spanning one or two different habitats. Spruce forest was replicated in the two different quadrats, to account for the variability related to geographic location (Table 1, Fig. 1).
A total of 18 sampling points were equipped with gravimetric Tauber traps, and pollen samples were collected over three different periods covering a 1-year time span: October 2014–March 2015, March–July 2015 and July–October 2015. At each sampling point two Tauber traps were positioned at the same pole, further used as site replicates for HTS and microscope analysis.
Tauber traps, sample processing and microscopic analysis
Each Tauber trap had 700 mL of a preservative solution (1:1:1 water, alcohol, glycerol, plus 2 g L−1 phenol)12. All traps were installed at the different sampling points close to the ground (half meter), tied to poles. A collar was placed on the apertures of the traps that was covered with a 5 mm mesh net to prevent collection of larger particles49. Airborne particles were collected by gravitational settling, and sampling solutions were filtered and processed following the protocol of Leontidou et al.12. The obtained pellets were the starting material for microscopic and molecular analysis and they are considered biological replicates, since they derived from exactly the same area after following the same sampling and processing methodology.
The period of March–July 2015 was selected for comparisons between metabarcoding and microscopic analysis, since it is the main pollen season for most of the plant families in the study area according to aerobiological data. Samples for the microscopic analysis (18 out of a total of 54) were processed by smearing a small aliquot of the pellet on a slide and coloring with basic fuchsin. Morphological pollen analysis was performed using an optical microscope (Leitz Diaplan, Ernst Leitz Wetzlar, Wetzlar, Germany) under 400× magnification. Approximately 400 pollen grains were analyzed per each sample. A local pollen reference collection was used as a quality check for pollen identification by light microscopy55,56.
Vegetation and aerobiological data
For vegetation data, we used the plant checklist of the Park31. Aerobiological data including the concentrations of pollen taxa per cubic meter of air were retrieved from the archive of the monitoring station in Villa Welsperg, the visitor’s centre of the Park (Val Canali, 46°11′57.3″N 11°52′06.3″E, 1050 m). The station is equipped with a Lanzoni volumetric sampler (VPSS, Lanzoni, Bologna, Italy) and pollen analysis is performed by light microscopy, following the European standard (UNI CEN/TS 16868:2015).
DNA extraction and sequencing
Pollen pellets originating from 54 Tauber traps were used as starting material for DNA extraction using Nucleomag kit (Macherey–Nagel, Düren, Germany) and the Kingfisher (Thermo Fisher Scientific, Waltham, MA, USA) automated DNA extraction system. In particular, with the use of distilled water, 200 μl were transferred from the initial pollen pellet sample to the extraction tube. To decrease lab contaminants, the tubes were transferred in a non-invasive DNA extraction room (separated from PCR or post-PCR rooms), where the lysis step and the preparation of the extraction plates were performed in a specialized safety cabinet. For the lysis, a step that is prone to lead to differences in DNA release between species15, we followed optimized protocols to increase DNA yield. In particular, one sterilized steel bead, lysis buffer and RNase were added in each tube, according to the manufacturer’s protocol and a high-energy agitation (Retsch MM200 mixer mill, Germany) was applied (30 Hz for 1 min, two steps) to mechanically disrupt cell walls. The final product was transferred in an extraction plate, where the rest of the reagents were added. We included a sample of a known species (Corylus avellana) as positive control and we had 19 negative controls in different stages of the extraction (14 with lysis buffer and post-lysis reagents and five wells of the plate filled only with the post-lysis reagents). The extracted DNA was eluted in 100 μl.
The eDNA was amplified with the chloroplast trnL primers c-A49325 and h-B49466 [5′-CGAAATCGGTAGACGCTACG-3′ and 5′-CCATTGAGTCTCTGCACCTATC-3′] (Sigma-Aldrich, Milan, Italy), with expected amplicon size 141 bp57. We followed the GoTaq protocol (Promega, Madison, WI, USA) in three separate reactions for each sample, to maximize the chance of detecting all species in our pollen pellets58. The DNA amplification was carried out in a final volume of 50 μl using 5 μl of extracted DNA. The amplification mixture contained 1.25 U GoTaq Polymerase, 1× of GoTaq Flexi Buffer, 3 mM of MgCl2, 0.2 mM of dNTPs and 0.2 μM of each primer. All PCR amplifications were carried out on a Veriti 96 well thermal cycler (Applied Biosystems, Foster City, CA, USA) with the following program: 2 min at 95 °C and 40 cycles of 15 s at 95 °C, 15 s at 52 °C and 30 s at 72 °C, followed by 5 min at 72 °C. The PCR products were checked using electrophoresis in Qiaxcel (Qiagen, GmbH, Hilden, Germany). Three PCR reaction products were pooled together in equal amounts. They were further gel purified using the MinElute Gel Purification kit (Qiagen) and eluted in 53 μl. The final PCR product was quantified with Qubit 2.0 Fluorometer (Life Technologies, Thermo Fisher Scientific).
Around 100 ng of the purified PCR product were used for library preparation. A total of 56 independent libraries were generated using TruSeq DNA sample preparation kit V2 (Illumina Inc., San Diego, CA, USA), pooled in equimolar ratio and then sequenced using MiSeq Reagent Kit v3 on an Illumina MiSeq platform.
Bioinformatics processing and taxonomic assignment
Raw Illumina reads were paired and pre-processed using VSEARCH v2.5.0 ‘merge pairs’ algorithm59. Reads were filtered out if ambiguous bases were detected and if the read lengths were less than 100 bp. Furthermore, an expected error (= 1) was used as an indicator of read accuracy. Pollen OTUs were obtained using the ‘cluster_fast’ algorithm with a 97% sequence identity and a depth of at least 100× reads for each cluster. The centroid of the resulting cluster was chosen as the representative sequence for the taxonomic assignment step. To decrease the false positive rate in the sequence population, a chimera detection analysis was performed on the obtained reference sequences. Since there is no reference database for the trnL gene for chimera detection, we used the ‘uchime_denovo' algorithm that carries out a de novo analysis without a reference.
Taxonomic assignments were performed by BLASTN+ software of ncbi-blast-2.2.31+60. Reference OTU sequences were aligned against our local reference database, constructed to include the most widespread anemophilous taxa of the study area (Trentino)12. Briefly, 1470 species (46 families) known to occur in the study area were searched in the database ‘Nucleotide’ of Genbank and 1188 sequences (403 species of 46 families) were retrieved and stored in a local database as described in Leontidou et al.12. The e-value parameter was set to a maximum value of 1e−40, above which the assignment was not considered valid. We also asked for sequence similarity and query coverage > 95%. An additional comparison with the non-redundant nucleotide NCBI database was performed with the same parameters to check the OTUs that were not assigned by our local reference database and search for contaminants or species not related to our study (e.g. ferns).
Statistical analysis
Diversity was analyzed with the R packages ‘vegan’61 and ‘phyloseq’62. First, a normalization of the read counts was performed to achieve equal sequencing depth among samples (rarefaction). In particular, the OTU table was rarefied (R vegan function ‘rarefy’) to the least number of sequence reads among samples and rarefaction curves were constructed (R vegan function ‘rarecurve’) to estimate the sufficiency of sequencing depth per sample. The normalized counts were used for further analysis.
Alpha diversity was estimated by calculating OTUs number in each sample (Richness). We also examined whether alpha diversity was affected by the sampling period and habitat using ANOVA tests.
To assess the effectiveness of trnL metabarcoding for pollen quantification we investigated the correlation between the number of trnL sequence reads and the number of pollen grains (microscope) for the period March–July 2015. Correlation coefficients and p-values were calculated using Kendall tau rank correlation tests.
As a measure of beta diversity, i.e. variation in species composition between samples, we used the presence/absence-based Jaccard pairwise dissimilarity index. Permutational multivariate analysis of variance (PERMANOVA) was used with a balanced design to estimate the significance of the categorical variables (habitat, sampling period) to the dissimilarity (R vegan function ‘adonis’). Then, we formulated a Generalized Linear Model (quasi-binomial error distribution and logit link function) predicting the Jaccard dissimilarity values as function of sampling period, habitat and of their interaction term to further elucidate the effect of the aforementioned variables including difference within and between groups. To display the effects of the independent variables to the dissimilarity values (dependent variable) we used the R package ‘effects’63.
To compare the communities identified by microscope and metabarcoding we performed Non-metric Multidimensional Scaling (NMDS) using the beta diversity (Jaccard) matrix and further applied Procrustes analysis on these ordinations.
Finally, we applied ANOVA tests to detect significant differences in the main contributing pollen taxa in different sampling periods and habitats.
Supplementary Information
Acknowledgements
The project was funded by Fondazione Edmund Mach (FEM) and FIRST (FEM International Research School in Trentino). We wish to thank Maria Cristina Viola (FEM Research and Innovation Centre), for microscopic analyses, field work and lab technical support; Maurizio Salvadori (Paneveggio Pale di San Martino Natural Park) for his help in field work and technical support, Cristiano Vernesi (FEM Research and Innovation Centre) for his help in field work and scientific support, Fabiana Cristofolini (FEM Research and Innovation Centre) for botanical identifications and Marco Ferretti for sampling design suggestions. We would also thank Elena Gottardini (FEM Research and Innovation Centre) for sharing the aerobiological monitoring data of Villa Welsperg (Paneveggio Pale di San Martino Natural Park).
Author contributions
K.L. carried out the sampling, lab work, data analysis and wrote the manuscript. M.Li. carried out the library preparation. A.B. and M.G. contributed to the experiments of the lab work. C.V. contributed to the design of lab experiments. A.S. performed the bioinformatics analysis and contributed to the statistical analysis. M.L. performed the statistical models and contributed to the statistical analysis. J.D.G. contributed to the data visualization and data management. M.C. coordinated the sequencing and bioinformatics analysis. D.V. participated in the experimental design, supervision, analytical approaches and in writing of the manuscript. A.C. was in charge of the project design, supervision and fund-raising, participated and coordinated sampling, lab work, experimental planning and writing of the manuscript. All authors revised the manuscript.
Data availability
The datasets generated and/or analysed during the current study are shared in figshare [10.6084/m9.figshare.12789950]. Raw reads were submitted to the European Nucleotide Archive (ΕΝΑ) under the Accession Number PRJEB40729.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-97619-3.
References
- 1.Assessment M. E. Ecosystems and Human Well-being. Island Press; 2005. p. 563. [Google Scholar]
- 2.Willis KJ, Birks HJB. What is natural? The need for a long-term perspective in biodiversity conservation. Science. 2006;314:1261–1265. doi: 10.1126/science.1122667. [DOI] [PubMed] [Google Scholar]
- 3.Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012;21:2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x. [DOI] [PubMed] [Google Scholar]
- 4.Pawlowski J, Apotheloz-Perret-Gentil L, Altermatt F. Environmental DNA: What’s behind the term? Clarifying the terminology and recommendations for its future use in biomonitoring. Mol Ecol. 2020;29:4258–4264. doi: 10.1111/mec.15643. [DOI] [PubMed] [Google Scholar]
- 5.Kelly RP, Port JA, Yamahara KM, Crowder LB. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE. 2014;9:e86175. doi: 10.1371/journal.pone.0086175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creer S, et al. The ecologist's field guide to sequence-based identification of biodiversity. Methods Ecol. Evol. 2016;7:1008–1018. doi: 10.1111/2041-210X.12574. [DOI] [Google Scholar]
- 7.Li Y, et al. Estimating fish alpha-and beta-diversity along a small stream with environmental DNA metabarcoding. Metabarcoding Metagenom. 2018;2:e24262. doi: 10.3897/mbmg.2.24262. [DOI] [Google Scholar]
- 8.Olds BP, et al. Estimating species richness using environmental DNA. Ecol. Evol. 2016;6:4214–4226. doi: 10.1002/ece3.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruppert KM, Kline RJ, Rahman MS. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019;17:e00547. doi: 10.1016/j.gecco.2019.e00547. [DOI] [Google Scholar]
- 10.Birks HJB, et al. Does pollen-assemblage richness reflect floristic richness? A review of recent developments and future challenges. Rev. Palaeobot. Palyno. 2016;228:1–25. doi: 10.1016/j.revpalbo.2015.12.011. [DOI] [Google Scholar]
- 11.Dell’Anna R, Cristofori A, Gottardini E, Monti F. A critical presentation of innovative techniques for automated pollen identification in aerobiological monitoring networks. In: Kaiser BJ, editor. Pollen: Structure, types and effects. Nova Science; 2010. pp. 273–288. [Google Scholar]
- 12.Leontidou K, et al. DNA metabarcoding of airborne pollen: New protocols for improved taxonomic identification of environmental samples. Aerobiologia. 2018;34:63–74. doi: 10.1007/s10453-017-9497-z. [DOI] [Google Scholar]
- 13.Brennan GL, et al. Temperate airborne grass pollen defined by spatio-temporal shifts in community composition. Nature Ecol. Evol. 2019;3:750–754. doi: 10.1038/s41559-019-0849-7. [DOI] [PubMed] [Google Scholar]
- 14.Longhi S, et al. Biomolecular identification of allergenic pollen: A new perspective for aerobiological monitoring? Ann. Allergy Asthma Immun. 2009;103:508–514. doi: 10.1016/S1081-1206(10)60268-2. [DOI] [PubMed] [Google Scholar]
- 15.Kraaijeveld K, et al. Efficient and sensitive identification and quantification of airborne pollen using next-generation DNA sequencing. Mol. Ecol. Resour. 2015;15:8–16. doi: 10.1111/1755-0998.12288. [DOI] [PubMed] [Google Scholar]
- 16.Núñez A, et al. Validation of the Hirst-type spore trap for simultaneous monitoring of prokaryotic and eukaryotic biodiversities in urban air samples by next-generation sequencing. Appl. Environ. Microbiol. 2017;83:e00472–e517. doi: 10.1128/AEM.00472-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell KL, et al. Applying pollen DNA metabarcoding to the study of plant–pollinator interactions. Appl. Plant Sci. 2017;5:1600124. doi: 10.3732/apps.1600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pornon A, et al. Using metabarcoding to reveal and quantify plant–pollinator interactions. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep27282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pornon A, Baksay S, Escaravage N, Burrus M, Andalo C. Pollinator specialization increases with a decrease in a mass-flowering plant in networks inferred from DNA metabarcoding. Ecol. Evol. 2019;9:13650–13662. doi: 10.1002/ece3.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawkins J, et al. Using DNA metabarcoding to identify the floral composition of honey: A new tool for investigating honey bee foraging preferences. PLoS ONE. 2015;10:e0134735. doi: 10.1371/journal.pone.0134735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richardson RT, et al. Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Appl. Plant Sci. 2015;3:1400066. doi: 10.3732/apps.1400066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Porter TM, Hajibabaei M. Scaling up: A guide to high-throughput genomic approaches for biodiversity analysis. Mol. Ecol. 2018;27:313–338. doi: 10.1111/mec.14478. [DOI] [PubMed] [Google Scholar]
- 23.Shokralla S, Spall JL, Gibson JF, Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 2012;21:1794–1805. doi: 10.1111/j.1365-294X.2012.05538.x. [DOI] [PubMed] [Google Scholar]
- 24.Sandionigi Α, et al. Analytical approaches for DNA barcoding data—How to find a way for plants? Plant Biosyst. 2012;146:805–813. doi: 10.1080/11263504.2012.740084. [DOI] [Google Scholar]
- 25.Yoccoz NG, et al. DNA from soil mirrors plant taxonomic and growth form diversity. Mol. Ecol. 2012;21:3647–3655. doi: 10.1111/j.1365-294X.2012.05545.x. [DOI] [PubMed] [Google Scholar]
- 26.Harrison JB, Sunday JM, Rogers SM. Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B. 2019;286:20191409. doi: 10.1098/rspb.2019.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller A, et al. Evaluating multiplexed next-generation sequencing as a method in palynology for mixed pollen samples. Plant Biol. 2015;17:558–566. doi: 10.1111/plb.12251. [DOI] [PubMed] [Google Scholar]
- 28.Bell KL, et al. Quantitative and qualitative assessment of pollen DNA metabarcoding using constructed species mixtures. Mol. Ecol. 2019;28:431–455. doi: 10.1111/mec.14840. [DOI] [PubMed] [Google Scholar]
- 29.Richardson RT, et al. Rank-based characterization of pollen assemblages collected by honey bees using a multi-locus metabarcoding approach. Appl. Plant Sci. 2015;3:1500043. doi: 10.3732/apps.1500043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson RT, et al. Quantitative multi-locus metabarcoding and waggle dance interpretation reveal honey bee spring foraging patterns in Midwest agroecosystems. Mol. Ecol. 2019;28:686–697. doi: 10.1111/mec.14975. [DOI] [PubMed] [Google Scholar]
- 31.Festi, F. & Prosser, F. La flora del Parco Naturale Paneveggio Pale di San Martino (Atlante corologico e repertorio delle segnalazioni, 2000).
- 32.Zinger L, Gobet A, Pommier T. Two decades of describing the unseen majority of aquatic microbial diversity. Mol. Ecol. 2012;21:1878–1896. doi: 10.1111/j.1365-294X.2011.05362.x. [DOI] [PubMed] [Google Scholar]
- 33.Charalampopoulos A, Damialis A, Tsiripidis I, Mavrommatis T, Halley JM, Vokou D. Pollen production and circulation patterns along an elevation gradient in Mt Olympos (Greece) National Park. Aerobiologia. 2013;29:455–472. doi: 10.1007/s10453-013-9296-0. [DOI] [Google Scholar]
- 34.De Barba M, Miquel C, Boyer F, Mercier C, Rioux D, Coissac E, Taberlet P. DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: Application to omnivorous diet. Mol. Ecol. Resour. 2014;14:306–323. doi: 10.1111/1755-0998.12188. [DOI] [PubMed] [Google Scholar]
- 35.Ishii K, Fukui M. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microb. 2001;67:3753–3755. doi: 10.1128/AEM.67.8.3753-3755.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laursen MF, Dalgaard MD, Bahl MI. Genomic GC-content affects the accuracy of 16S rRNA gene sequencing based microbial profiling due to PCR bias. Front. Microbiol. 2017;8:1934. doi: 10.3389/fmicb.2017.01934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghitarrini S, Galán C, Frenguelli G, Tedeschini E. Phenological analysis of grasses (Poaceae) as a support for the dissection of their pollen season in Perugia (Central Italy) Aerobiologia. 2017;33:339–349. doi: 10.1007/s10453-017-9473-7. [DOI] [Google Scholar]
- 38.Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microb. 1998;64:3724–3730. doi: 10.1128/AEM.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microb. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dowle EJ, Pochon X, C Banks J, Shearer K, Wood SA. Targeted gene enrichment and high-throughput sequencing for environmental biomonitoring: A case study using freshwater macroinvertebrates. Mol. Ecol. Resour. 2016;16:1240–1254. doi: 10.1111/1755-0998.12488. [DOI] [PubMed] [Google Scholar]
- 41.Swenson SJ, Gemeinholzer B. Testing the effect of pollen exine rupture on metabarcoding with Illumina sequencing. PLoS ONE. 2021;16:e0245611. doi: 10.1371/journal.pone.0245611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deiner K, et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017;26:5872–5895. doi: 10.1111/mec.14350. [DOI] [PubMed] [Google Scholar]
- 43.Elbrecht V, Leese F. Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass—sequence relationships with an innovative metabarcoding protocol. PLoS ONE. 2015;10:e0130324. doi: 10.1371/journal.pone.0130324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baksay S, et al. Experimental quantification of pollen with DNA metabarcoding using ITS1 and trnL. Sci. Rep. 2020;10:1–9. doi: 10.1038/s41598-020-61198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohanty RP, Buchheim MA, Anderson J, Levetin E. Molecular analysis confirms the long-distance transport of Juniperus ashei pollen. PLoS ONE. 2017;12:e0173465. doi: 10.1371/journal.pone.0173465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carini P, et al. Effects of spatial variability and relic DNA removal on the detection of temporal dynamics in soil microbial communities. MBio. 2020;11:e02776–-19. doi: 10.1128/mBio.02776-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paul JH, Jeffrey WH, David AW, DeFlaun MF, Cazares LH. Turnover of extracellular DNA in eutrophic and oligotrophic freshwater environments of southwest Florida. Appl. Environ. Microb. 1989;55:1823–1828. doi: 10.1128/aem.55.7.1823-1828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levy-Booth DJ, et al. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 2007;39:2977–2991. doi: 10.1016/j.soilbio.2007.06.020. [DOI] [Google Scholar]
- 49.Pidek AI, Piotrowska K, Kasprzyk I. Pollen–vegetation relationships for pine and spruce in southeast Poland on the basis of volumetric and Tauber trap records. Grana. 2010;49:215–226. doi: 10.1080/00173134.2010.514006. [DOI] [Google Scholar]
- 50.van der Knaap WO, van Leeuwen JF, Ammann B. Seven years of annual pollen influx at the forest limit in the Swiss Alps studied by pollen traps: Relations to vegetation and climate. Rev. Palaeobot. Palyno. 2001;117:31–52. doi: 10.1016/S0034-6667(01)00075-6. [DOI] [Google Scholar]
- 51.Pidek IA, et al. Variation in annual pollen accumulation rates of Fagus along a N-S transect in Europe based on pollen traps. Veg. Hist. Archaeobot. 2010;19:259–270. doi: 10.1007/s00334-010-0248-0. [DOI] [Google Scholar]
- 52.Kelly RP, et al. Genetic and manual survey methods yield different and complementary views of an ecosystem. Front. Marine Sci. 2017;3:283. doi: 10.3389/fmars.2016.00283. [DOI] [Google Scholar]
- 53.Damialis, A., Traidl-Hoffmann, C., & Treudler, R. Climate change and pollen allergies. In Biodiversity and Health in the Face of Climate Change, 47–66. (Springer, 2019).
- 54.Amadei, M., et al. Carta della Natura e Biodiversità nelle Aree Naturali Protette: il Parco Naturale Paneveggio-Pale di San Martino, Rapporti 56/2005 (2005).
- 55.Bucher, E., Kofler V., Vorwohl, G. & Zieger, E. Lo spettro pollinico dei mieli dell’Alto Adige (Laboratorio biologico dell'Agenzia per la protezione dell'ambiente e la tutela del lavoro, 2004).
- 56.Faegri K, Iversen J. Textbook of Pollen Analysis. Wiley; 1989. [Google Scholar]
- 57.Taberlet P, et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007;35:e14. doi: 10.1093/nar/gkl938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sickel W, et al. Increased efficiency in identifying mixed pollen samples by meta-barcoding with a dual-indexing approach. BMC Ecol. 2015;15:20. doi: 10.1186/s12898-015-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oksanen, J., et al. Vegan: community ecology package. R package version 1.17-4. http://CRAN.R-project.org/package=vegan (2010).
- 62.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fox J, Weisberg S. An R Companion to Applied Regression. 3. Sage Publications; 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are shared in figshare [10.6084/m9.figshare.12789950]. Raw reads were submitted to the European Nucleotide Archive (ΕΝΑ) under the Accession Number PRJEB40729.