Abstract
PANX1, one of the members of the pannexin family, is a highly glycosylated channel-forming protein. Recently, we identified heterozygous variants in PANX1 that follow an autosomal dominant inheritance pattern and cause female infertility characterized by oocyte death. In this study, we screened for novel PANX1 variants in patients with the phenotype of oocyte death and discovered a new type of inheritance pattern accompanying PANX1 variants. We identified two novel homozygous missense variants in PANX1 [NM_015368.4 c.712T>C (p.(Ser238Pro) and c.899G>A (p.(Arg300Gln))] associated with the oocyte death phenotype in two families. Both of the homozygous variants altered the PANX1 glycosylation pattern in cultured cells, led to aberrant PANX1 channel activation, and resulted in mouse oocyte death after fertilization in vitro. It is worth noting that the destructive effect of the two homozygous variants on PANX1 function was weaker than that caused by the recently reported heterozygous variants. Our findings enrich the variational spectrum of PANX1 and expand the inheritance pattern of PANX1 variants to an autosomal recessive mode. This highlights the critical role of PANX1 in human oocyte development and helps us to better understand the genetic basis of female infertility due to oocyte death.
Subject terms: Infertility, Genetics research
Introduction
Infertility affects ~10–15% of couples worldwide [1], and the application of assisted reproductive technology, including in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI), has helped a large number of infertile couples to successfully give birth [2]. However, there are still many couples who undergo recurrent failure of IVF/ICSI attempts. Normal oocyte maturation, fertilization, and embryonic development are necessary for successful IVF/ICSI, and abnormalities in any of these processes will lead to female infertility [3–5]. Genetic variants account for many patients with abnormalities in these processes, and several variant genes have been found to be responsible for oocyte maturation abnormalities (TUBB8 [6], PATL2 [7]), fertilization failure (TLE6 [8], WEE2 [9]) and early embryonic developmental arrest (PADI6 [10], NLRP2, and NLRP5 [11]), potently demonstrating the contributions of genetic factors to female infertility.
In a recent study, we identified four families with a new kind of phenotype termed oocyte death that manifested as oocyte cytoplasmic shrinkage, blackening, and death before or after fertilization. These phenotypes caused recurrent IVF/ICSI failure and female infertility, and we identified different heterozygous variants in PANX1 that were responsible for the phenotype [12]. PANX1, which encodes a highly glycosylated channel protein, is widely expressed in multiple human tissues and organs, especially in the brain and oocytes [12, 13]. The main function of PANX1 is to form large-pore channels that release ATP and other small metabolites, and thus it plays a critical role in information exchange between cells [14, 15]. Although PANX1 is involved in multiple physiological and pathological functions [16–20], Panx1 knockout mice are viable, fertile, and have no obvious phenotype, which weakens the importance of its physiological role in vivo [21–24]. However, we previously found that heterozygous variants in PANX1 altered the protein’s glycosylation pattern, influenced its subcellular localization, and led to aberrant PANX1 channel activity and ATP release in oocytes, and mice that overexpressed a patient-derived variant were infertile due to oocyte death [12]. Our findings thus demonstrated the critical role of PANX1 in human oocyte development. With the publication of our study, Six articles on the structure of PANX1 were published, showing that it is a heptameric channel protein [25–30], which further suggests that PANX1 might play a significant role in cell communication.
In this study, we identified two homozygous variants in two families with the phenotype of oocyte death after fertilization, indicating that in addition to the dominant inheritance pattern reported before, PANX1 variants can also be inherited in a recessive pattern. We investigated the effects of the variants in cultured cells, in Xenopus laevis oocytes, and in mouse oocytes. The results showed that the effect of homozygous missense variants on PANX1 function was weaker than that of previously reported heterozygous variants.
Materials and methods
Human subjects and study design
Infertility patients with the oocyte death phenotype were recruited from the Second Affiliated Hospital of Zhengzhou University and Affiliated Hospital of Chifeng University. DNA was extracted from the peripheral blood based on a previously reported protocol [6]. Then, sanger sequencing was performed to identify variants in PANX1. Functional impairment of identified variants was evaluated by glycosylation assay, electrophysiological assay, and mouse oocyte cRNA microinjection (Fig. S1).
Screening of PANX1 variants
Peripheral blood was collected from the patients, their family members, and controls after obtaining informed consent. Genomic DNA samples were extracted from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). All exons and splicing sites of PANX1 were amplified, and the corresponding primers are shown in Table S1. Amplified fragments were directly sequenced using an ABI 3100 DNA analyzer (Applied Biosystems, Foster City, CA, USA). The PANX1 variants were submitted to LOVD at https://www.LOVD.nl/PANX1.
Expression vector construction and mutagenesis
The full-length sequence encoding human PANX1 (NM_015368.4) was amplified and cloned into the pCMV6-Entry vector containing an engineered stop codon to express untagged proteins. Site-directed mutagenesis was performed to introduce the identified variants c.712T>C (p.(Ser238Pro)) and c.899G>A (p.(Arg300Gln)) into the wild-type (WT) vector using the site-directed KOD-Plus-Mutagenesis Kit (Toyobo, TKY, Japan) according to the manufacturer’s instructions. WT and mutant clones were confirmed by Sanger sequencing.
Cell culture and transfection
HeLa cells obtained from the Cell Bank of Shanghai Institute for Biological Sciences were cultured in high-glucose Dulbecco’s minimum essential medium (Gibco, Waltham, MA, USA) supplemented with 1% penicillin/streptomycin and 10% (v/v) fetal bovine serum (FBS; Gibco, Waltham, MA, USA), and maintained at 37 °C in a humidified 5% CO2 incubator. PANX1 WT and mutant constructs were transfected into HeLa cells using the PolyJet In Vitro DNA Transfection Reagent (Signagen, Frederick, MD, USA) according to the manufacturer’s instructions.
Western blotting
HeLa cells were harvested 36 h after transfection and washed three times with cold phosphate-buffered saline (PBS). Cells were lysed in RIPA lysis buffer (Shanghai Wei AO Biological, SH, China) with 1% protease inhibitor cocktail (Bimake, Houston, TX, USA) and centrifuged at 12,000 × g for 30 min at 4 °C. Supernatants were collected, mixed with 5× sodium dodecyl sulfate (SDS) loading buffer, and heated at 100 °C for 10 min. Equal amounts of protein were separated using SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose filter membranes (Pall Corporation, NYC, USA). The membranes were blocked with 5% nonfat milk diluted in PBS with 0.1% Tween 20 (PBST) for 1 h and then incubated at 4 °C overnight with rabbit anti-PANX1 (1:1000 dilution, Cell Signaling Technology, Danvers, MA, USA) and rabbit anti-vinculin (1:1000 dilution, Cell Signaling Technology, Danvers, MA, USA). The membranes were washed with PBST three times and incubated with goat anti-rabbit IgG secondary antibodies (1:5000 dilution, Abmart, SH, China) for 1 h at room temperature followed by washing again with PBST three times. Finally, the membranes were incubated with ECL Western Blotting Substrate (Tanon, SH, China) and imaged on a chemiluminescent imaging system (5200, Tanon, SH, China). Quantitation of western blotting results was performed with the ImageJ software.
cRNA transcription
WT and mutant constructs were linearized by digestion with the AgeI restriction enzyme (New England BioLabs, 240 County Road Ipswich, MA, USA) at 37 °C for 3 h. Purified linearized DNA was used as a template to transcribe PANX1 cRNA, followed by DNase I treatment and poly(A) polymerase tailing using the HiScribe T7 ARCA mRNA Kit (New England BioLabs, 240 County Road Ipswich, MA, USA). Finally, the cRNAs were purified and dissolved in nuclease-free water using the RNeasy MinElute Cleanup Kit (Qiagen, 240 County Road Ipswich, MA, USA).
Mouse oocyte collection, microinjection, and fertilization in vitro
Ovaries were isolated from 6 to 8-week-old female ICR (Institute of Cancer Research) mice (Beijing Vital River Laboratory Animal Technology Co, Changping County, BJ, China). The ovaries were chopped up with a razor blade, and GV oocytes with diameters of about 80 µm were collected by mouth pipetting on the stage of a dissecting microscope. The GV oocytes were cultured in M2 medium (Sigma-Aldrich, NSW, Australia) with 10% FBS under mineral oil (Sigma-Aldrich, NSW, Australia) at 37 °C in an atmosphere of 5% CO2.
The mouse GV oocytes were microinjected with WT or mutant cRNAs using a Leica Hoffman microscope (DMi8; leica, Wetzlar, Germany) equipped with a TransferMan 4r micromanipulator, InjectMan 4, and FemtoJet 4i (Eppendorf, Saxony, Germany). About 5–10 pl of cRNA solution (200–1000 ng/µl) was microinjected into the cytoplasm of each mouse GV oocyte. Injected GV oocytes were matured in vitro in M2 medium (Sigma-Aldrich, NSW, Australia) containing 10% FBS and penicillin-streptomycin (Gibco) for 12 h. Mature oocytes were then collected and mixed with sperm in human tubal fluid medium (Millipore, USA) for fertilization. All oocytes were cultured at 37 °C in an atmosphere of 5% CO2. All experimental mouse protocols were reviewed and approved by the Shanghai Medical College of Fudan University.
Two-electrode voltage-clamp electrophysiology
X. laevis oocytes were injected with 150 ng/μl WT or mutant PANX1 cRNAs. At 12–18 h after injection, a two-electrode voltage-clamp experiment was performed in the standard external solution containing 2 mM CaCl2, 2 mM KCl, 1 mM MgCl2, 90 mM NaCl, and 5 mM HEPES with or without 10 mM carbenoxolone (CBX) (Sigma-Aldrich, NSW, Australia). The pH was adjusted to 7.4 using KOH. Initially, the membrane potential was held at −60 mV for 100 ms, then changed from −100 to +60 mV in 2 s ramps with a 20 mV increase per step. Data were captured and analyzed using the pClamp10 software (Molecular Devices, San Jose, CA, USA).
Statistical analyses
All data are representative of at least three independent experiments. GraphPad Prism was used to perform the statistical analysis. Values were analyzed by Student’s t tests when comparing experimental groups, and P values <0.05 were considered significant.
Results
Clinical characteristics of the probands
The two probands from the two independent families had been diagnosed with primary infertility for several years (Fig. 1). The proband (II-1) in family 1 was 29 years old at examination and had regular menstrual cycles and normal sex hormone concentrations. She had undergone a failed IVF and a failed ICSI attempt. In the IVF attempt, as noted in her medical records, a total of 52 oocytes were retrieved (Table 1), and 43 of them were successfully fertilized. However, all fertilized oocytes gradually degenerated and died within 48 h, accompanied by cytoplasmic shrinkage and darkening as previously described [12]. In the ICSI attempt, 17 first polar body (pb1) oocytes were successfully fertilized. However, all fertilized oocytes died within 24 h. Individual II-1 had one older sister (II-2) who had given birth normally.
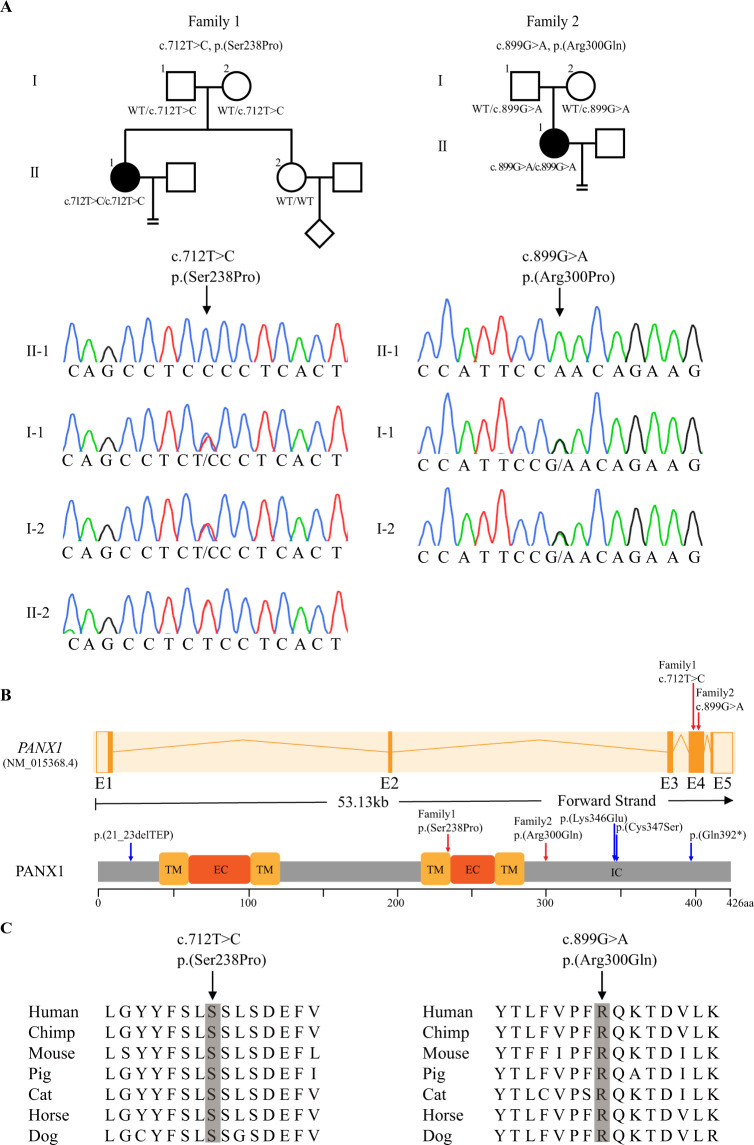
Fig. 1. Identification of pathogenic variants in PANX1.
A Two pedigrees carrying PANX1 variants that lead to infertility with the oocyte death phenotype. Sanger sequencing confirmation is shown below the pedigrees. Squares denote male family members, circles denote female members, the diamond denotes unknown gender, black solid circles denote the probands, and the equal sign denotes infertility. B Locations of the newly identified homozygous variants in PANX1 exons and the protein structure of PANX1. Red arrows indicate the newly identified variants and the blue arrow indicates the variants reported previously. TM transmembrane region, EC extracellular region, IC intracellular region. C The affected amino acids were compared among seven mammalian species in a conservation analysis.
Table 1.
Clinical characteristics of the patients with PANX1 variants.
| Individual | Age (years) | Duration of infertility (years) | IVF/ICSI attempts | Total oocytes | Fertilized oocytes | Oocytes that died or degenerated after fertilization | Outcomes |
|---|---|---|---|---|---|---|---|
| II-1 in family 1 | 28 | 6 | First IVF | 52 | 43 | 43 | – |
| Second ICSI | 31 | 17 | 17 | – | |||
| II-1 in family 2 | 36 | 7 | First IVF | 12 | 11 | 11 | – |
| Second IVF | 6 | 5 | 4 | One viable embryo (grade II, 8-cell) that failed to establish pregnancy |
IVF in vitro fertilization, ICSI intracytoplasmic sperm injection.
The proband in family 2 was 34 years old at examination and had undergone two failed IVF attempts. In the first attempt, a total of 12 oocytes were retrieved (Table 1), of which 11 morphologically normal pb1 oocytes were successfully fertilized with two pronuclei, but all of them gradually died within 48 h in the same manner as seen in patient II-1 in family 1. In her second IVF attempt, six morphologically normal pb1 oocytes were retrieved, and five of them were successfully fertilized. Likewise, four fertilized oocytes died within 48 h and only one zygote was viable and developed to a grade II 8-cell embryo, but it failed to establish pregnancy (Table 1).
Identification of homozygous variants in PANX1
Pathogenic variants in PANX1 were recently shown to cause the oocyte death phenotype [12]. Because of the oocyte death phenotype observed in the probands of the two families, screening of PANX1 variants was performed. All members in families 1 and 2 underwent Sanger sequencing of the PANX1 exons. As expected, both probands had likely pathogenic variants in PANX1. However, instead of the dominant inheritance pattern that was seen in the previous study, the two probands here possessed homozygous variants in PANX1 that showed a recessive inheritance pattern. The proband in family 1 had a homozygous missense variant c.712T>C (p.(Ser238Pro)), while the proband in family 2 had a homozygous missense variant c.899G>A (p.(Arg300Gln)). The parents of the two probands were heterozygous carriers, and the sister of the proband in family 1, who was fertile, had WT alleles (Fig. 1A). Specific information on the genomic position of variants, their frequency and their in silico analysis is provided in Table 2. The variant c.712T>C (p.(Ser238Pro)) is located at the end of the third transmembrane region, while the variant c.899G>A (p.(Arg300Gln)) is located in the intracellular region at the C-terminus of PANX1 (Fig. 1B). The residue Ser238 and Arg300 are highly conserved across species (Fig. 1C).
Table 2.
Overview of the PANX1 variants observed in two families.
| Probands in families | Genomic position on Chr11 (bp) | cDNA change | Protein change | Variant type | Inheritance | gnomADa | SIFTb | PPH2b |
|---|---|---|---|---|---|---|---|---|
| Family 1 | 93912934 | c.712T>C | p.(Ser238Pro) | Missense | AR | Absent | T | B |
| Family 2 | 93913121 | c.899G>A | p.(Arg300Gln) | Missense | AR | Absent | D | P |
The genomic reference sequence is NM_015368.4 (hg19).
AR autosomal recessive, T tolerated, D damaging, B benign, P probably damaging.
aFrequency of corresponding variants in the gnomAD browser.
bVariant assessment by SIFT and PolyPhen-2 (PPH2).
Effects of homozygous variants on PANX1 glycosylation in vitro
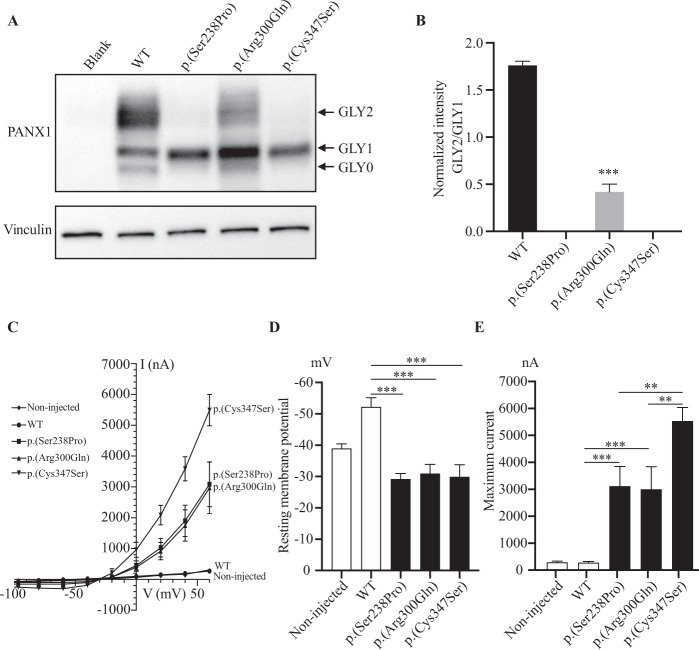
PANX1 is a highly glycosylated membrane protein that exists as three species, including GLY0 (the non-glycosylated protein), GLY1 (the high mannose-type glycoprotein), and GLY2 (the fully processed glycoprotein) [31, 32]. To evaluate the effects of the homozygous variants on PANX1 glycosylation in vitro, WT and mutant PANX1 constructs were transfected into HeLa cells. The variant p.(Cys347Ser), which resulted in oocyte death after fertilization in our previous study, was used as the positive control [12]. Compared with WT PANX1, the variant p.(Ser238Pro) resulted in the complete absence of the GLY2 band, which was consistent with the effect of the variant p.(Cys347Ser) (Fig. 2A). As for the variant p.(Arg300Gln), although the GLY2 band was retained, the GLY2/GLY1 intensity ratio was significantly reduced compared with WT (Fig. 2A, B). Taken together, these results indicated that the two homozygous variants in PANX1 resulted in an altered glycosylation pattern in HeLa cells in vitro.
Fig. 2. Effects of PANX1 pathogenic variants on glycosylation and channel properties.
A Western blot analysis of HeLa cell extracts after transfection with WT or mutant PANX1 constructs. The positions of full-length bands are shown on the right. Vinculin was used as the loading control (bottom). B The ratio of GLY2 to GLY1 of PANX1. The GLY2/GLY1 of the variant p.(Arg300Gln) was significantly reduced compared with WT. Three independent experiments were performed. ***P < 0.001. C Average I–V (current voltage) curves from X. laevis oocytes expressing WT or mutant PANX1 (p.(Ser238Pro), p.(Arg300Gln), and p.(Cys347Ser). The voltage steps ranged from −100 mV to +60 mV in 20 mV increments. Data are shown as the means ± SD of four to five oocytes. D Resting membrane potentials in X. laevis oocytes after expression of WT or mutant PANX1. Data are shown as the means ± SD of four to five oocytes. ***P < 0.001. E Maximum currents in X. laevis oocytes after expression of WT or mutant PANX1. Data are shown as the means ± SD of four to five oocytes. ***P < 0.001.
The homozygous variants influenced PANX1 channel properties but had less of an effect compared to the heterozygous variant p.(Cys347Ser)
To determine whether homozygous variants altered the activity of the PANX1 channel, we used X. laevis oocytes to analyze the effect of variant PANX1 on the biophysical properties of channel activity in a two-electrode voltage-clamp electrophysiology experiment. As shown in Fig. 2C, the homozygous variant groups [c.712T>C (p.(Ser238Pro) and c.899G>A (p.(Arg300Gln))] had a significant increase in channel activity compared with the non-injected and WT groups. The homozygous variant groups also had reduced resting membrane potentials and much higher maximum current compared with the non-injected and WT groups (Fig. 2D, E). In addition, we used the channel inhibitor CBX in a rescue experiment. The application of CBX decreased the current amplitudes in the injected group (Fig. S2a–e), indicating the impairment of channel activity as a result of the homozygous variants p.(Ser238Pro) and p.(Arg300Gln). It should be noted that the effect of the two homozygous variants on channel activity was much lower than that of the heterozygous variant p.(Cys347Ser) (Fig. 2C, E).
The homozygous variants caused mouse oocyte death but required a higher dosage of cRNA than the heterozygous variant p.(Cys347Ser)
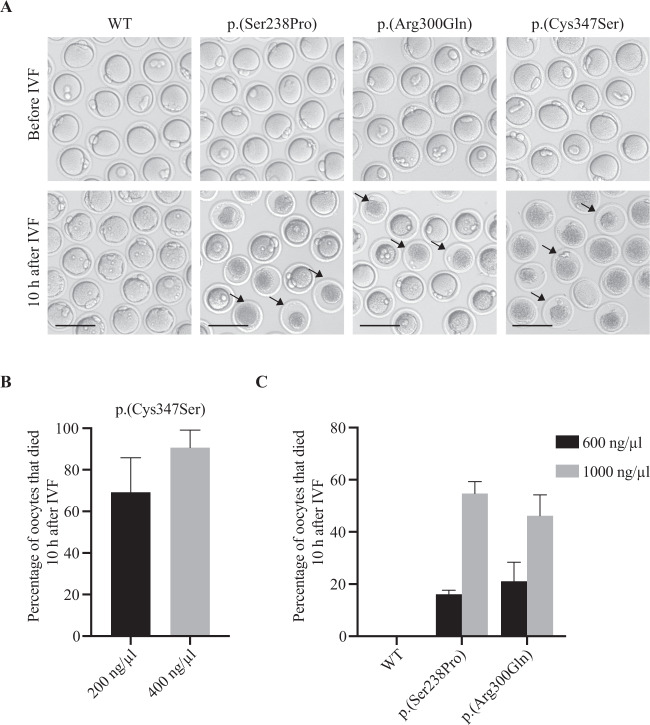
Finally, to mimic the phenotype of oocyte death, we directly injected WT or mutant PANX1 cRNAs into mouse GV oocytes at different concentrations. GV oocytes were allowed to mature in vitro for 12 h and then used for IVF. After cultivation, oocytes injected with mutant cRNAs matured normally by extruding the first polar body (Fig. 3A). However, these oocytes gradually died within 10 h after IVF in a dose-dependent manner. For oocytes injected with p.(Cys347Ser) cRNA, about 70% died within 10 h after IVF at a concentration of 200 ng/μl, and more than 90% died when the concentration was increased to 400 ng/μl (Fig. 3A, B). Injection of homozygous p.(Ser238Pro) and p.(Arg300Gln) cRNAs also resulted in mouse oocyte death, but required a much higher concentration than p.(Cys347Ser) cRNA. Even when injecting p.(Ser238Pro) and p.(Arg300Gln) cRNAs at 1000 ng/μl, the rate of oocyte death was only about 50%, which was significantly lower than the 90% death rate seen for the heterozygous variant p.(Cys347Ser) at a concentration of 400 ng/μl (Fig. 3A, C). These results together with the changes in channel activity (Fig. 2D, E) lead us conclude that the newly identified homozygous variants are disease causing but have milder effects on PANX1 function compared with previously identified heterozygous variants. This might explain why the probands’ mothers, who had heterozygous variants c.712T>C (p.(Ser238Pro)) or c.899G>A (p.(Arg300Gln)), were fertile.
Fig. 3. Mimicking the oocyte death phenotype in mouse oocytes in vitro.
A Images of mouse pb1 oocytes before and after fertilization. GV oocytes injected with WT (600 or 1000 ng/μl), p.(Ser238Pro) (600 or 1000 ng/μl), p.(Arg300Gln) (600 or 1000 ng/μl) or p.(Cys347Ser) (200 or 400 ng/μl) PANX1 cRNAs were allowed to mature in vitro for 10 h and before using for IVF. Arrows indicate dead oocytes. Scale bars, 100 μm. B, C The percentage of oocytes that died after fertilization at different concentrations of cRNAs injection. Three independent experiments were performed, each group includes 60–80 oocytes, and the data are shown as the means ± SEM.
Discussion
In the present study, we identified two homozygous variants [c.712T>C (p.(Ser238Pro)) and c.899G>A (p.(Arg300Gln))] in PANX1 from two independent families with the phenotype of oocyte death after fertilization. Unlike the previously reported dominant inheritance pattern [12], the newly identified variants showed a recessive inheritance pattern. The homozygous variants altered the PANX1 glycosylation pattern, affected membrane electrophysiological properties, and resulted in mouse oocyte death in vitro.
Recently we reported that four heterozygous variants [c.1174C>T (p.(Gln392*)), c.1036A>G (p.(Lys346Glu)), c.1040G>C (p.(Cys347Ser)), and c.61_69delACGGAGCCC (p.(21_23delTEP))] in PANX1 are responsible for oocyte death [12]. In this study, homozygous variants c.712T>C (p.(Ser238Pro)) and c.899G>A (p.(Arg300Gln)) in PANX1 were also shown to cause oocyte death. It is not uncommon that different inheritance patterns of pathogenic variants in the same gene result in similar diseases or phenotypes. For example, we showed that both dominant and recessive variants in TUBB8 are responsible for oocyte maturation arrest and its range of phenotypes [6, 33, 34]. This can be explained by the fact that variants in different locations in a gene might have different effects on the protein, and different inheritance patterns might thereby arise due the different effects of the variants. In addition, our previous study showed that patients with heterozygous variants c.1174C>T (p.(Gln392*)) and c.1036A>G (p.(Lys346Glu)) had a more severe phenotype with oocyte death occurring before fertilization, while oocytes from patients carrying the heterozygous variants c.1040G>C (p.(Cys347Ser)) and c.61_69delACGGAGCCC (p.(21_23delTEP)) did not die until after fertilization. In this study, the two patients also had the phenotype of oocyte death after fertilization. Oocytes expressing p.(Ser238Pro) and p.(Arg300Gln) cRNAs had a lower degree of channel activation than oocytes expressing p.(Cys347Ser) cRNA (Fig. 2C), and the rate of oocyte death after injection with the homozygous mutant cRNAs was significantly lower than for oocytes injected with p.(Cys347Ser) cRNA (Fig. 3B, C). These results suggest that compared to the effects of heterozygous variants, the impairment of PANX1 function due to the homozygous variants was mild. This might explain why the variants c.712T>C (p.(Ser238Pro)) and c.899G>A (p.(Arg300Gln)) resulted in oocyte death only in a homozygous state, while heterozygous c.712T>C (p.(Ser238Pro)) or c.899G>A (p.(Arg300Gln)) did not affect oocyte development and thus the mothers of the probands, as heterozygous carriers, were still able to give birth normally.
There was also a phenotypic difference between patients with homozygous variants. The patient with the variant c.899G>A (p.(Arg300Gln)) could produce a viable embryo (grade II, 8-cell) using IVF, while in the patient with the variant c.712T>C (p.(Ser238Pro)) all of the oocytes died after fertilization (Table 1). PANX1 is a glycoprotein that exists in different glycosylated forms in the endoplasmic reticulum and Golgi apparatus, and the level of glycosylation is critical for the cellular localization and the function of the channel [35]. We found that the PANX1 GLY2 species was completely absent in the patient with the variant c.712T>C (p.(Ser238Pro)), while the patient with the variant c.899G>A (p.(Arg300Gln)) still had the PANX1 GLY2 species, but at a significantly lower level compared to WT (Fig. 2A). It is likely that the degree of phenotype severity is dependent on the impairment of PANX1 glycosylation resulting from the different variants.
Previous studies have found that Panx1 knockout mice are viable, fertile, and have no obvious reproductive phenotype [21]. In addition, the engineered OE-PANX1Q392* female mice were completely infertile with the oocyte death phenotype [12], suggesting that the oocyte death phenotype was caused by gain-of-function effect. Therefore, the oocyte death phenotype can be mimicked by injecting mutant cRNAs in mouse oocyte, even in the presence of WT allele.
The two newly identified homozygous variants c.712T>C (p.(Ser238Pro)) c.899G>A (p.(Arg300Gln)) as well as two previously reported heterozygous variants c.1040G>C (p.(Cys347Ser)) and c.61_69delACGGAGCCC (p.(21_23delTEP)) resulted in oocyte death only after fertilization, but the molecular mechanism for this phenotype remains unknown. Upon fertilization, oocyte activation consists of a coordinated series of events, including repeated increases in cytoplasmic Ca2+ concentration (Ca2+ oscillations) [36], cortical granule exocytosis [37], the resumption of the second phase of meiosis, and extrusion of the second polar body [38]. We speculate that oocyte death after fertilization might be related to these physiological changes and relevant pathways, and this requires further exploration in transgenic mice.
In conclusion, we have identified the homozygous variants c.712T>C (p.(Ser238Pro)) and c.899G>A (p.(Arg300Gln)) in PANX1 as responsible for oocyte death. Our findings confirm the vital role of PANX1 in oocyte development and female fertility and provide additional genetic markers for infertility patients.
Supplementary information
Acknowledgements
We thank the patients, their families, and the healthy volunteers for participating in this study. This work was supported by the National Key Research and Development Program of China (2018YFC1003800, 2017YFC1001500, and 2016YFC1000600), the National Natural Science Foundation of China (81725006, 81822019, 81771581, 81971450, and 81971382), the project supported by Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), Project of Shanghai Municipal Science and Technology Commission (19JC1411001), the Natural Science Foundation of Shanghai (19ZR1444500), Shuguang Program of Shanghai Education Development Foundation and Shanghai Municipal Education Commission (18SG03), the Foundation of Shanghai Health and Family Planning Commission (20154Y0162), the Capacity Building Planning Program for Shanghai Women and Children’s Health Service, the collaborative innovation center project construction for Shanghai Women and Children’s Health.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Our study was approved by the Ethics Committee of the Medical College of Fudan University and the Reproductive Study Ethics Committee of the hospital (No. 148).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Weijie Wang, Ronggui Qu, Qian Dou, Fengyan Wu
Contributor Information
Lei Wang, Email: wangleiwanglei@fudan.edu.cn.
Qing Sang, Email: sangqing@fudan.edu.cn.
Supplementary information
The online version of this article (10.1038/s41431-020-00807-4) contains supplementary material, which is available to authorized users.
References
- 1.Tamrakar SR, Bastakoti R. Determinants of infertility in couples. J Nepal Health Res Counc. 2019;17:85–9. doi: 10.33314/jnhrc.v17i01.1827. [DOI] [PubMed] [Google Scholar]
- 2.DeAngelis AM, Martini AE, Owen CM. Assisted reproductive technology and epigenetics. Semin Reprod Med. 2018;36:221–32. doi: 10.1055/s-0038-1675780. [DOI] [PubMed] [Google Scholar]
- 3.Levran D, Farhi J, Nahum H, Glezerman M, Weissman A. Maturation arrest of human oocytes as a cause of infertility: case report. Hum Reprod. 2002;17:1604–9. doi: 10.1093/humrep/17.6.1604. [DOI] [PubMed] [Google Scholar]
- 4.Edwards RG, Bavister BD, Steptoe PC. Early stages of fertilization in vitro of human oocytes matured in vitro. Nature. 1969;221:632–5. doi: 10.1038/221632a0. [DOI] [PubMed] [Google Scholar]
- 5.Gardner DK, Lane M. Culture and selection of viable blastocysts: a feasible proposition for human IVF? Hum Reprod Update. 1997;3:367–82. doi: 10.1093/humupd/3.4.367. [DOI] [PubMed] [Google Scholar]
- 6.Feng R, Sang Q, Kuang Y, Sun X, Yan Z, Zhang S, et al. Mutations in TUBB8 and human oocyte meiotic arrest. N Engl J Med. 2016;374:223–32. doi: 10.1056/NEJMoa1510791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen B, Zhang Z, Sun X, Kuang Y, Mao X, Wang X, et al. Biallelic mutations in PATL2 cause female infertility characterized by oocyte maturation arrest. Am J Hum Genet. 2017;101:609–15. doi: 10.1016/j.ajhg.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alazami AM, Awad SM, Coskun S, Al-Hassan S, Hijazi H, Abdulwahab FM, et al. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16:240. doi: 10.1186/s13059-015-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sang Q, Li B, Kuang Y, Wang X, Zhang Z, Chen B, et al. Homozygous mutations in WEE2 cause fertilization failure and female infertility. Am J Hum Genet. 2018;102:649–57. doi: 10.1016/j.ajhg.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Shi Y, Fu J, Yu M, Feng R, Sang Q, et al. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am J Hum Genet. 2016;99:744–52. doi: 10.1016/j.ajhg.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mu J, Wang W, Chen B, Wu L, Li B, Mao X, et al. Mutations in NLRP2 and NLRP5 cause female infertility characterised by early embryonic arrest. J Med Genet. 2019;56:471–80. doi: 10.1136/jmedgenet-2018-105936. [DOI] [PubMed] [Google Scholar]
- 12.Sang Q, Zhang Z, Shi J, Sun X, Li B, Yan Z, et al. A pannexin 1 channelopathy causes human oocyte death. Sci Transl Med. 2019;11. [DOI] [PubMed]
- 13.Baranova A, Ivanov D, Petrash N, Pestova A, Skoblov M, Kelmanson I, et al. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics. 2004;83:706–16. doi: 10.1016/j.ygeno.2003.09.025. [DOI] [PubMed] [Google Scholar]
- 14.Bao L, Locovei S, Dahl G. Pannexin membrane channels are mechanosensitive conduits for ATP. Febs Lett. 2004;572:65–8. doi: 10.1016/j.febslet.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. Embo J. 2006;25:5071–82. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi G, Liu C, Yang Y, Song L, Liu X, Wang C, et al. Panx1 promotes invasion-metastasis cascade in hepatocellular carcinoma. J Cancer. 2019;10:5681–8. doi: 10.7150/jca.32986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SP, Qin T, Seidel JL, Zheng Y, Eikermann M, Ferrari MD, et al. Inhibition of the P2X7-PANX1 complex suppresses spreading depolarization and neuroinflammation. Brain. 2017;140:1643–56. doi: 10.1093/brain/awx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma AK, Charles EJ, Zhao Y, Narahari AK, Baderdinni PK, Good ME, et al. Pannexin-1 channels on endothelial cells mediate vascular inflammation during lung ischemia-reperfusion injury. Am J Physiol Lung Cell Mol Physiol. 2018;315:L301–12. doi: 10.1152/ajplung.00004.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meens MJ, Kwak BR, Duffy HS. Role of connexins and pannexins in cardiovascular physiology. Cell Mol Life Sci. 2015;72:2779–92. doi: 10.1007/s00018-015-1959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molica F, Figueroa XF, Kwak BR, Isakson BE, Gibbins JM. Connexins and pannexins in vascular function and disease. Int J Mol Sci. 2018;19:1663. doi: 10.3390/ijms19061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao HB, Zhu Y, Liang C, Chen J. Pannexin 1 deficiency can induce hearing loss. Biochem Biophys Res Commun. 2015;463:143–7. doi: 10.1016/j.bbrc.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Zhu Y, Liang C, Chen J, Zhao HB. Pannexin1 channels dominate ATP release in the cochlea ensuring endocochlear potential and auditory receptor potential generation and hearing. Sci Rep. 2015;5:10762. doi: 10.1038/srep10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bargiotas P, Krenz A, Hormuzdi SG, Ridder DA, Herb A, Barakat W, et al. Pannexins in ischemia-induced neurodegeneration. Proc Natl Acad Sci USA. 2011;108:20772–7. doi: 10.1073/pnas.1018262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seminario-Vidal L, Okada SF, Sesma JI, Kreda SM, van Heusden CA, Zhu Y, et al. Rho signaling regulates pannexin 1-mediated ATP release from airway epithelia. J Biol Chem. 2011;286:26277–86. doi: 10.1074/jbc.M111.260562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Q, Zhang B, Zheng X, Li N, Xu L, Xie Y, et al. Cryo-EM structures of human pannexin 1 channel. Cell Res. 2020;30:449–51. doi: 10.1038/s41422-020-0310-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Z, He Z, Maksaev G, Bitter RM, Rau M, Fitzpatrick JAJ, et al. Cryo-EM structures of the ATP release channel pannexin 1. Nat Struct Mol Biol. 2020;27:373–81. doi: 10.1038/s41594-020-0401-0. [DOI] [PubMed] [Google Scholar]
- 27.Mou L, Ke M, Song M, Shan Y, Xiao Q, Liu Q, et al. Structural basis for gating mechanism of Pannexin 1 channel. Cell Res. 2020;30:452–4. doi: 10.1038/s41422-020-0313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalski K, Syrjanen JL, Henze E, Kumpf J, Furukawa H, Kawate T. The Cryo-EM structure of pannexin 1 reveals unique motifs for ion selection and inhibition. Elife. 2020;9. [DOI] [PMC free article] [PubMed]
- 29.Qu R, Dong L, Zhang J, Yu X, Wang L, Zhu S. Cryo-EM structure of human heptameric Pannexin 1 channel. Cell Res. 2020;30:446–8. doi: 10.1038/s41422-020-0298-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan Z, Orozco IJ, Du J, Lu W. Structures of human pannexin 1 reveal ion pathways and mechanism of gating. Nature. 2020;584:646–51. doi: 10.1038/s41586-020-2357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penuela S, Bhalla R, Gong XQ, Cowan KN, Celetti SJ, Cowan BJ, et al. Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J Cell Sci. 2007;120:3772–83. doi: 10.1242/jcs.009514. [DOI] [PubMed] [Google Scholar]
- 32.Boassa D, Ambrosi C, Qiu F, Dahl G, Gaietta G, Sosinsky G. Pannexin1 channels contain a glycosylation site that targets the hexamer to the plasma membrane. J Biol Chem. 2007;282:31733–43. doi: 10.1074/jbc.M702422200. [DOI] [PubMed] [Google Scholar]
- 33.Feng R, Yan Z, Li B, Yu M, Sang Q, Tian G, et al. Mutations in TUBB8 cause a multiplicity of phenotypes in human oocytes and early embryos. J Med Genet. 2016;53:662–71. doi: 10.1136/jmedgenet-2016-103891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B, Li B, Li D, Yan Z, Mao X, Xu Y, et al. Novel mutations and structural deletions in TUBB8: expanding mutational and phenotypic spectrum of patients with arrest in oocyte maturation, fertilization or early embryonic development. Hum Reprod. 2017;32:457–64. doi: 10.1093/humrep/dew322. [DOI] [PubMed] [Google Scholar]
- 35.Penuela S, Simek J, Thompson RJ. Regulation of pannexin channels by post-translational modifications. Febs Lett. 2014;588:1411–5. doi: 10.1016/j.febslet.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki S, Shirakawa H, Nakada K, Honda Y. Essential role of the inositol 1,4,5-trisphosphate receptor/Ca2+ release channel in Ca2+ waves and Ca2+ oscillations at fertilization of mammalian eggs. Dev Biol. 1993;158:62–78. doi: 10.1006/dbio.1993.1168. [DOI] [PubMed] [Google Scholar]
- 37.Vogt EJ, Tokuhiro K, Guo M, Dale R, Yang G, Shin SW, et al. Anchoring cortical granules in the cortex ensures trafficking to the plasma membrane for post-fertilization exocytosis. Nat Commun. 2019;10:2271. doi: 10.1038/s41467-019-10171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horner VL, Wolfner MF. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev Dyn. 2008;237:527–44. doi: 10.1002/dvdy.21454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.