Abstract
Here we present evidence that CIF150 (hTAFII150), the human homolog of Drosophila TAFII150, plays an important and selective role in establishing gene expression patterns necessary for progression through the cell cycle. Gel filtration experiments demonstrate that CIF150 (hTAFII150) seems to be less tightly associated with human transcription factor IID than hTAFII130 is associated with hTAFII250. The transient functional knockout of CIF150 (hTAFII150) protein led to cell cycle arrest at the G2/M transition in mammalian cell lines. PCR display analysis with the RNA derived from CIF150-depleted cells indicated that CIF150 (hTAFII150) is required for the transcription of only a subset of RNA polymerase II genes. CIF150 (hTAFII150) directly stimulated cyclin B1 and cyclin A transcription in cotransfection assays and in vitro assays, suggesting that the expression of these genes is dependent on CIF150 (hTAFII150) function. We defined a CIF150 (hTAFII150) consensus binding site and demonstrated that a CIF150-responsive cis element is present in the cyclin B1 core promoter. These results suggest that one function of CIF150 (hTAFII150) is to select specific RNA polymerase II core promoter elements involved in cell cycle progression.
The regulation of cell growth at the molecular level is a central problem in cancer biology research. Tumor cells have established different mechanisms to bypass the normal regulation of cellular proliferation including defects in cell cycle checkpoints. The eukaryotic cell cycle is composed of four distinct stages: G1, S, G2, and M (28). The transitions between cell cycle stages are partly controlled by cyclins, regulatory subunits of the cyclin-dependent kinases (34). The gene expression of cyclins itself is cell cycle regulated (21, 33) and is crucial for maintaining normal cellular growth; the deregulation of cyclin A and cyclin D1 has been correlated with different human tumors (3, 13). Despite our extensive knowledge of cyclin-dependent signaling pathways, the molecular mechanisms for the periodic expression of the cyclins are not well understood.
The binding of general transcription factor IID (TFIID) to core promoter elements is considered to initiate the formation of the RNA polymerase II preinitiation complex (reviewed in references 20, 32, 37, and 49). The TATA box, the initiator (Inr) sequence, a downstream promoter element (DPE), and a TFIIB-specific sequence have been identified as core promoter cis elements (4, 5, 19; for a review, see reference 43). For promoters that contain a TATA box the core promoter recognition is carried out by the TATA-binding protein (TBP). However, many of the cyclin core promoters do not contain a functional TATA box and are lacking defined transcriptional start sites. In vitro binding studies have suggested that TBP-associated factors (TAFIIs), which are subunits of the TFIID complex, play a critical role in selecting core promoters (4, 5, 16, 22, 35, 47, 48). Recently, a new TAFII-containing complex lacking TBP has been described, supporting the view that TAFs play a role in promoter recognition (53), especially on TATA-less promoters.
TAFIIs were originally described as required coactivators for activated transcription. Activated transcription can not be reconstituted by TBP alone in in vitro transcription assays but can be observed in the presence of TFIID (reviewed in references 37 and 49). These results led to the hypothesis that TAFIIs directly interact with activator molecules bound to upstream sequences and thereby recruit general transcription factors, including RNA polymerase II, to the promoter. This view has been challenged by functional knockout experiments in yeast, where activated transcription seems not to be globally affected in the absence of certain TAFIIs. These results suggested that some TAFIIs are dispensable for the activated transcription of specific genes in yeast (1, 12, 26, 50, 51). More recently it has been shown that other histone-like TAFIIs are more broadly required for transcription in yeast (2, 25, 27). According to these reports, a critical role of some yeast TAFIIs seems to be core promoter recognition rather than the mediation of activated transcription in general (for a review, see references 9, 20, and 44). Recent in vitro studies using crude HeLa nuclear extracts have also suggested that TAFIIs are not universally required for activated transcription (29). It should be noted that the two models for TAFII function are not mutually exclusive and might only illustrate the functional redundancy of TAFIIs and other coactivators in promoting activated transcription. One unifying explanation might be that only a specific set of TAFIIs is required for mediating transcriptional activation of a particular activator and that distinct TFIID complexes might exist. For example Zhou and collaborators demonstrated the in vivo importance of two TAFIIs in Drosophila and showed that this TAFII dependence was most severe in a subset of cells expressing the target gene snail (55). Even more surprising is the discovery that one subset of TAFIIs in the TFIID complex is also an integral component of histone acetylase complexes connecting the polymerase II machinery with chromatin-modifying activities (8, 27, 30). A better understanding of TAFII function in core promoter recognition and activation requires additional comprehensive in vivo analysis.
We reported the identification of an essential cofactor for TFIID-dependent Inr function, CIF150 (cofactor of Inr function) (17, 18). CIF150 protein shows striking homology to the Drosophila TAFII150 protein (dTAFII150) and some similarity to the essential yeast protein TSM-1 (18, 36, 47). More recently, essentially the same cDNA was cloned independently by Martinez and colleagues and named human TAFII150 (hTAFII150) (23). In contrast to what we published initially for CIF150, their fractionation and immunopurification data suggest that hTAFII150 (CIF150) is a tightly associated component of the human TFIID complex. Interestingly, dTAFII250 and dTAFII150 have been implicated in core promoter recognition (47, 48), suggesting that hTAFII150 (CIF150) might also have sequence-specific DNA binding activity. In fact, a temperature-sensitive mutant of the yeast homolog of hTAFII150 (CIF150), TSM-1, has been reported to block cell cycle progression through the G2 or M phase, revealing a degree of promoter selectivity (36, 50).
In this study, we investigated the function of hTAFII150 (CIF150) in mammalian cells. Using HeLa nuclear extracts in gel filtration experiments we demonstrated that in contrast to hTAFII250 or hTAFII130, hTAFII150 (CIF150) readily dissociates from human TFIID complex. We demonstrated that transient functional in vivo knockout experiments with hTAFII150 (CIF150) led to cell cycle arrest in G2 or M, similar to the phenotype of the temperature-sensitive mutant of TSM-1 in yeast. The absence of functional hTAFII150 (CIF150) protein affected only a subset of class II genes, indicating that hTAFII150 (CIF150) seemed not to be required for RNA polymerase II transcription in general. Furthermore, we demonstrated that hTAFII150 (CIF150) has sequence-specific binding activity which might explain the observed specificity. Finally, we demonstrated that hTAFII150 (CIF150) is a positive regulator of cyclin B1 transcription by characterizing a functional CIF150 binding element (CBE) in the cyclin B1 promoter. Taken together, our results indicate that hTAFII150 (CIF150) functions in the recognition and selection of core promoters including cell cycle-specific genes like cyclins A and B1.
MATERIALS AND METHODS
Purification of proteins and gel filtration of HeLa nuclear extracts.
HeLa nuclear extracts and hTAFII150 (CIF150)-depleted nuclear extract were prepared as described previously (7, 17, 18) and concentrated by ammonium sulfate precipitation (30%). The depletion of hTAFII150 (CIF150) protein was confirmed by Western blotting (data not shown; see reference 18). One hundred microliters (10 mg/ml) of ammonium sulfate-precipitated nuclear extracts, dialyzed against buffer A (20 mM HEPES [pH 7.9], 1 mM EDTA, 3 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 20% glycerol) containing 0.1 or 1 M KCl, was applied to a Superose 6 column (Pharmacia). The proteins were eluted with buffer A (10% glycerol) containing 0.1 or 1 M KCl (flow rate, 0.5 ml/min) and 500-μl fractions were collected. Every second protein-containing fraction was analyzed by sodium dodecyl sulfate (SDS)–6% polyacrylamide gel electrophoresis (PAGE), followed by immunoblot analysis with an hTAFII150 (CIF150)-specific antibody generated against the N-terminal peptide MNRKKGDKGFESPRP or monoclonal antibodies specific for hTAFII250 and hTAFII130 (Santa Cruz Biotechnology, Inc.). Immunoblot analyses were performed as described before (17). A control gel filtration with three appropriate molecular weight standards was performed under the same conditions. Recombinant hTAFII150 (CIF150) protein was purified from SF9 cells under native conditions by using the baculovirus expression system (pBlueBacHis2; Invitrogen). Whole-cell extract from baculovirus-infected SF9 cells was prepared by sonication and applied to TALON metal affinity resin (Clontech) according to the manufacturer’s protocol applying imidazole step elution. Recombinant hTAFII150 (CIF150) protein was tested for CIF150 activity as described previously (17, 18) and analyzed by SDS–8% PAGE, followed by silver staining.
Cell culture and cell cycle analysis.
IMR90 (normal human lung fibroblasts) and HeLa cells were transfected with the phosphothioate oligomers (100 to 400 nM) according to the protocol of the manufacturer (Sequitur, Inc.) by using lipofection and OptiMEM (Gibco BRL). The efficiency of transfection (around 90%) was determined on the basis of green fluorescent protein (GFP) fluorescence by using GFP-tagged control oligomers. The transfected cells were harvested at different time points postlipofection for fluorescence-activated cell sorting (FACS) and Western and Northern analysis. Initially, four single-strand antisense oligomers were designed to target distinct regions of hTAFII150 (CIF150) mRNA. The functional antisense oligomer designated B (5′TGCTCATGGAAGCATAAGCAGCCAC3′) was used in combination with a control oligonucleotide Bx (5′CACCGACGAATACGAAGGTACTCGT3′) containing the reverse sequence (3′→5′) of oligonucleotide B, to ensure identical nucleotide content. To monitor cell synchrony, 106 cells from each sample were fixed in ethanol, treated with RNase A (0.5 mg/ml for 1 h at 37°C), stained with propidium iodide (40 μg/ml) for 2 h at 4°C, and then analyzed by flow cytometry on a Becton Dickinson FACScan apparatus. Differential display was performed as described by the manufacturer (Genomyx Corp.).
Transfection and reporter assays.
Cyclin A (nucleotides −887 to +136) (10) and cyclin B1 (nucleotides −893 to +110) (6) promoter fragments were generated by PCR with HeLa DNA and cloned into the promoterless pGL3 luciferase reporter vector (Promega). All PCR-amplified fragments were verified by DNA sequencing. Cotransfections were performed in HeLa cells by using different amounts of a pEVRF1 (24)-based CIF150 expression vector (pEVRF-CIF150) or a pEVRF1-Ob expression plasmid in combination with the indicated reporter constructs. The expression of CIF150 (hTAFII150) protein was confirmed by Western blotting (data not shown). The pEVRF1-Ob, cytomegalovirus (CMV), and minimal c-Fos–luciferase constructs were a kind gift from K. Giese. Luciferase activity was determined 36 h after transfection according to the manufacturer’s protocol (Promega).
In vitro transcription and in vitro DNA binding assays.
In vitro transcription reactions were performed with the templates containing the G-less cassette as described before (reference 17 and references therein). Plasmid DNAs containing cyclin A, cyclin B1, and CMV promoter fragments were cloned upstream of a 180-bp G-less cassette by using a PCR protocol (17). The immunoglobulin H (IgH) promoter construct fused to a G-less cassette was a kind gift from J. D. Parvin and P. A. Sharp. For the complementation assay, 8 μl of the CIF150-depleted nuclear extract (4 mg/ml) were preincubated for 30 min at 4°C in the presence of a DNA template with 1, 2, and 4 μl of recombinant CIF150 protein (see Fig. 3A; 10 μl loaded), followed by the addition of ribonucleoside triphosphates to yield the following final concentrations: 500 μM ATP, 500 μM CTP, 500 μM GTP, and 30 μM [α-32P]UTP. 32P-labeled RNA products were digested for 15 min with RNase H1, resolved on an 8% polyacrylamide-urea gel, and visualized by autoradiography. Electrophoretic mobility shift assays were performed by using 3 and 6 μl of CIF150 in 40 μl of GL buffer as previously described (18), with the exception that the reaction mixture contained 50 ng of dG-dC oligomer as competitor. After 30 min of incubation at 4°C the binding mixtures were loaded on a Tris-borate-EDTA (0.5×)–6% polyacrylamide gel, and the signals were visualized by autoradiography. The signals were quantitated by PhosphorImager analysis (Bio-Rad, Inc.).
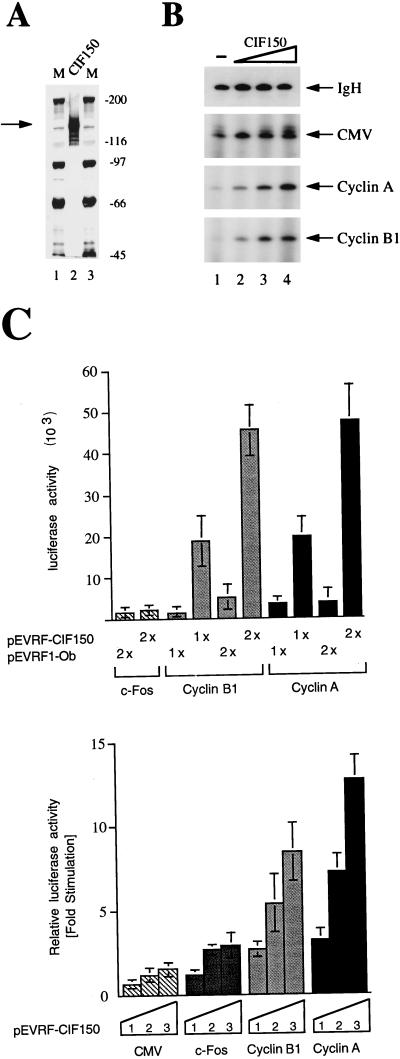
FIG. 3.
CIF150 (hTAFII150) is a positive regulator of cyclin B1 and cyclin A transcription. (A) Purified CIF150 was visualized after SDS-PAGE by silver staining (20 μl loaded in lane 2). M, molecular size standards (in kilodaltons). (B) CIF150 is required for cyclin B1 and cyclin A transcription but not for IgH and CMV transcription. In vitro transcription was performed by using CIF150-depleted nuclear extracts (lane 1) alone or in combination with increasing amounts of recombinant CIF150 protein (lanes 2 to 4). (C) Cotransfection of increasing amounts of CIF150 expression plasmid with cyclin B1, cyclin A, CMV, and Fos promoter-driven luciferase reporter constructs. Relative luciferase units are shown in the upper panel, and fold stimulation is shown in the lower panel. The amount of CMV-derived vector in each transfection assay was kept constant by using an unrelated expression plasmid (pEVRF1-Ob). The values shown are the averages from at least four independent transfection experiments ± standard deviations (error bars).
RESULTS
hTAFII150 (CIF150) is not as tightly associated with hTAFII250 as hTAFII130.
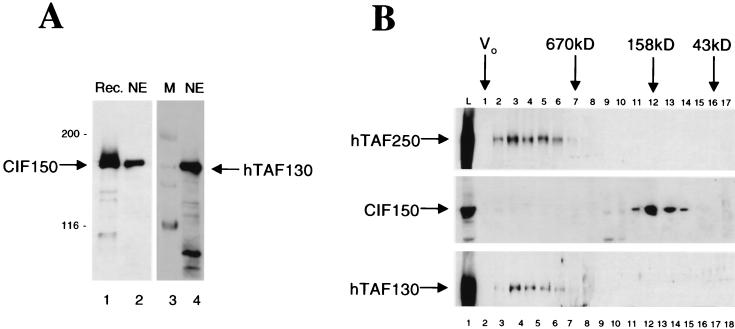
At least eight TAFIIs are associated with TBP in a complex called TFIID (for reviews, see references 9, 10, 44, and 46). Although the definition of a bona fide TAFII (TATA-binding protein-associated factor) is controversial, one biochemical criterion seems to be that a TAFII copurifies with TBP when immunoaffinity methods are employed under high salt conditions (1 M KCl in a phosphocellulose D fraction). However, there is increasing evidence for distinct TAFII-containing complexes in vivo, including a TBP-free TAF complex (53) and the recently discovered SAGA-PCAF complex (8, 30; for a review see reference 44). In the Drosophila system dTAFII150 is tightly associated with the TFIID complex, whereas in the mammalian system it is still controversial whether CIF150 (hTAFII150) is an integral component of human TFIID. Martinez et al. recently described a TAFII150-containing TFIID complex in which TAFII150 (CIF150) is apparently associated with human TFIID (23). These results contradict our earlier published results that CIF150 is only substoichiometrically present in the TFIID complex. Here, we performed side-by-side immunoblot experiments comparing CIF150 electrophoretic mobility to that of hTAFII130 by SDS-PAGE (Fig. 1A). We used a polyclonal antibody highly specific for recombinant CIF150 protein purified from bacculo-virus extracts (Fig. 1A, lane 1; see also Fig. 3A). Recombinant CIF150, native CIF150, and hTAFII130 have similar electrophoretic mobilities (Fig. 1A). This result explains, as suggested by Martinez et al., the difficulties to detect the 150-kDa protein in the immunopurified TFIID complex (23). However, it was still puzzling that we had originally succeeded in purifying hTAFII150 (CIF150) in a complementation assay with TFIID-containing fractions (17). To test the hypothesis that our nuclear extract preparation method disrupts the association of TFIID and CIF150, we performed gel filtration experiments. hTAFII250 and hTAFII130 coeluted from the gel filtration column with an apparent molecular weight expected for TFIID (Fig. 1B, fraction 3). This result clearly indicates that these two TAFIIs are stably associated in the TFIID complex in our nuclear extracts. However, CIF150 (hTAFII150) eluted at a later fraction, suggesting that the majority of CIF150 protein is not associated with the TFIID complex (Fig. 1B, middle panel). We performed these experiments under high-salt (1 M KCl) as well as low-salt (0.1 M KCl) conditions with the same result, suggesting that the disruption of hTAFII150 (CIF150) and TFIID occurs during or before our nuclear extract preparation.
FIG. 1.
CIF150 (hTAFII150) seems to be less tightly associated with holo TFIID than hTAFII250 and hTAFII130. (A) Recombinant and native CIF150 have an electrophoretic mobility in SDS–6% PAGE very similar to that of hTAFII130. Shown is a side-by-side immunoblot analysis of recombinant CIF150 (Rec.) (lane 1; 3 μl), native CIF150 (lane 2; 10 μl of HeLA nuclear extract [NE]) in comparison to hTAFII130 (lane 4; 10 μl of HeLA nuclear extract [NE]). Lane 3 contains molecular mass (M) standards (in kilodaltons). (B) Gel filtration experiments with HeLa nuclear extracts (buffer A [10% glycerol, 0.1 M KCl]) indicate a dissociation of CIF150 and human TFIID. Immunoblot assays were performed with 4 μl of HeLa nuclear extract (lane 1) (L) and gel filtration fractions 1 to 17 (lane 2 to 18, 20 μl loaded). The elutions of molecular mass standards are indicated. V0, voided volume.
It is worth noting that we were able to detect a very weak hTAFII150 (CIF150)-specific signal in fraction 3 (Fig. 1B, lane 4) when we overexposed the immunoblot (data not shown), indicating that a small portion of hTAFII150 (CIF150) protein might be present in the TFIID complex. As mentioned before we previously demonstrated by Far-Western analysis that hTAFII150 (CIF150) interacts directly with TAFII130, suggesting that hTAFII150 (CIF150) is able to associate with TFIID under certain conditions. One possible difference in the nuclear extract preparations might be that our extracts contained less DNA, since we used ammonium sulfate precipitation to concentrate our nuclear extracts. Contaminating DNA might lead to coimmunoprecipitation of DNA binding proteins, explaining the tight association of hTAFII150 (CIF150) and TFIID in the above-mentioned experiments by Martinez et al. (23). Taken together, however, our data demonstrate that under conditions where TAFII250 and TAFII130 form a highly stable complex, the majority of hTAFII150 (CIF150) was not associated with the TFIID complex.
Functional knockout of hTAFII150 (CIF150) is leading to a cell cycle arrest in G2 or M.
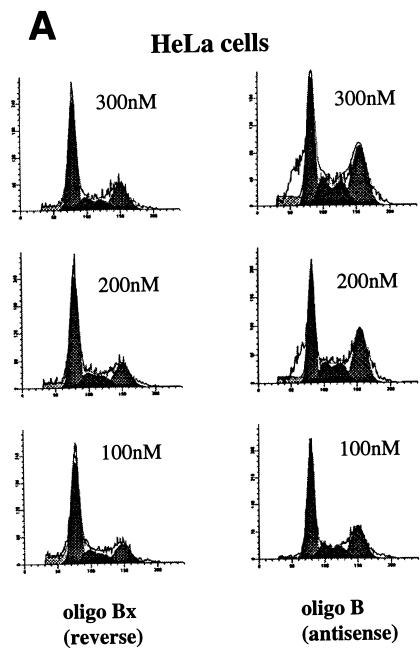
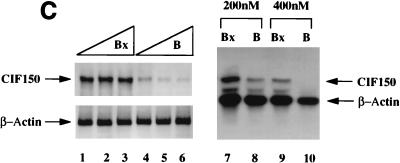
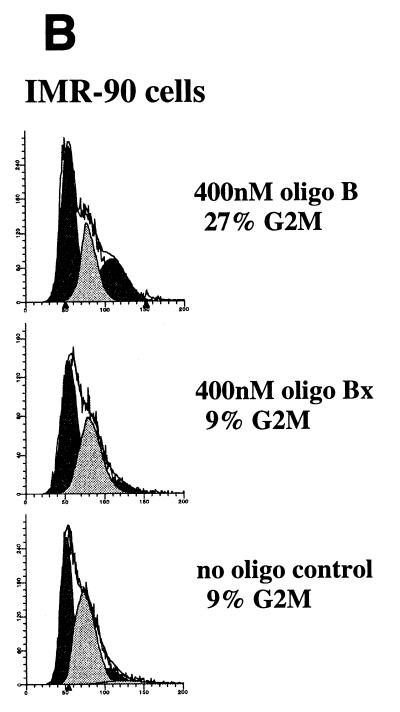
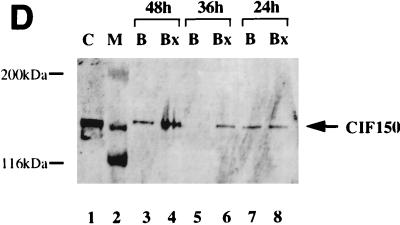
The above results suggested that CIF150 (hTAFII150) might function independently of the holo TFIID complex. To investigate the potential role of CIF150 protein in vivo, we attempted to knock out the function of CIF150 transiently in human cells. Initially, we tested four different CIF150-specific antisense phosphorothioate oligonucleotides for their ability to modulate CIF150 function. The transfection of one antisense oligonucleotide (B) into HeLa cells led to a concentration-dependent increase of cells in the G2/M phase and a decrease of cells in the G1 phase of the cell cycle (Fig. 2A, right panel; data not shown), whereas the other three oligomers had no effect on cell cycle progression (data not shown). Since HeLa cells are highly transformed and bear defects in cell cycle checkpoints (p53− and RB−), we confirmed the antisense effect by using the primary lung fibroblast cell line IMR90 (Fig. 2B). A control oligonucleotide (Bx [reverse sequence of B]) did not affect the cell cycle progression in either cell line (Fig. 2A and B). In Fig. 2C, we analyzed the RNA derived from HeLa cells treated with antisense oligonucleotide B and control oligonucleotide Bx. Quantitative reverse transcription (RT)-PCR (Fig. 1C, lanes 1 to 6) as well as Northern hybridization (lanes 7 to 10) revealed dramatically reduced CIF150 mRNA levels 24 h after antisense oligonucleotide treatment. In order to demonstrate the antisense oligonucleotide effect on the CIF150 protein level we performed an immunoblot analysis with CIF150-specific antiserum on cell lysates prepared from different time points after antisense oligonucleotide treatment (Fig. 2D and E). The level of CIF150 protein decreased 36 h after antisense oligonucleotide treatment but remained unchanged after 24 h or in control oligonucleotide-treated cells. Since the antisense oligomer was transiently transfected, the CIF150 protein level increased again after 48 h (Fig. 2D, lane 3). At this point we do not know the exact level of hTAFII150 (CIF150) depletion after our antisense oligonucleotide treatment. It is possible that the remaining hTAFII150 (CIF150) protein, which is undetectable by immunoblotting, is sufficient for some hTAFII150 (CIF150) function. One technical problem is the immanent toxicity and instability of the oligonucleotides, preventing us from achieving a more complete depletion for a longer period of time (Fig. 2D). Nevertheless, the demonstrated dose-dependent effect with different concentrations of antisense oligonucleotide B (Fig. 2E and A) suggests that we at least partially deplete hTAFII150 (CIF150) activity. Taken together our data indicate that hTAFII150 (CIF150) is likely to be involved in cell cycle progression through G2/M, which is in agreement with one of the described functions of the yeast homolog TSM-1 (50).
FIG. 2.
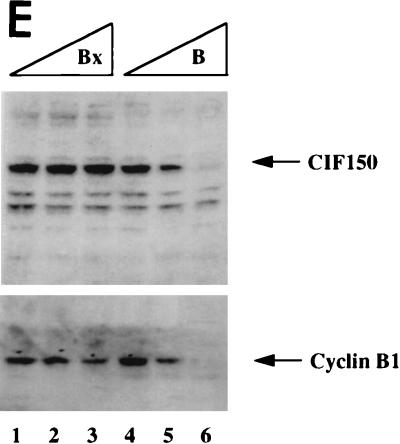
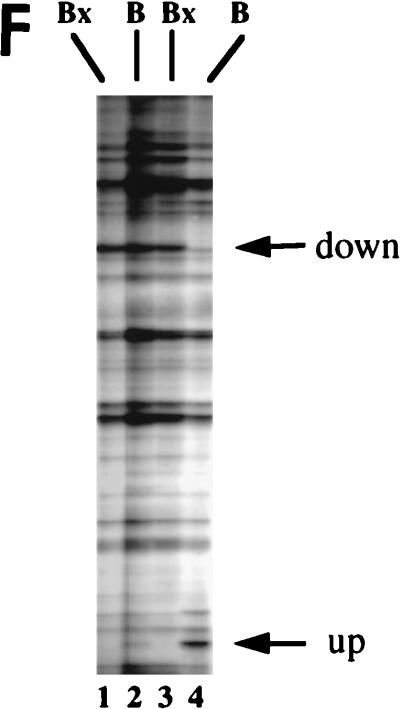
Functional knockout of CIF150 (hTAFII150) protein leads to cell cycle arrest in G2 or M and to reduced gene expression of cyclin B1. (A) Analyses of the cell cycle of HeLa cells 36 h after transfection with the CIF150 specific antisense oligomer B or oligomer Bx (reverse sequence). The different concentrations of the oligomers are indicated. (B) Cell cycle analyses of IMR90 cells after 36 h of oligomer treatment. (C) Total RNA, derived from HeLa cells treated with oligomer B or Bx, analyzed by quantitative RT-PCR (lanes 1 to 6, 100, 200, and 300 nM concentrations of oligomer B and Bx) and Northern blot analysis (lanes 7 to 10) with CIF150- and β-actin-specific primers or 32P-labeled cDNA probes. (D) HeLa cell extracts from different time points, after antisense oligomer (400 nM) treatment, analyzed by immunoblotting for the decrease of CIF150 protein. Control (C) nuclear extract (lane 1) and molecular size markers (M) (lane 2) were loaded. (E) Cyclin B1 protein expression decreases after CIF150 antisense treatment (100, 200, and 300 nM) of HeLa cells. (F) Differential PCR display identifies genes that are transcriptionally dependent on CIF150. A representative nondenaturing gel of differential display products is shown. Total RNA was prepared 24 h (lane 1 and 2) or 36 h (lane 3 and 4) after oligomer transfection. Arrows indicate cDNAs which are specifically upregulated or downregulated after antisense treatment.
Expression of a specific set of class II genes, including cyclin B1, seems to be dependent on hTAFII150 (CIF150) activity.
To confirm the results of our FACS analysis, we analyzed cyclin B1 expression by using extracts from antisense and control oligomer-treated HeLa cells. Again, CIF150 protein levels were concentration-dependently reduced by using antisense oligonucleotide B (Fig. 2E). The level of cyclin B1 expression seemed to be affected in the same way by the antisense treatment, indicating a cell cycle arrest and confirming the results of our FACS analysis (Fig. 2E). In order to address the question of whether the decrease of cyclin B1 was the result of a more general effect on transcription in the absence of CIF150, we set out to determine the expression levels of other mRNAs. As a first approach we analyzed RNA derived from cells treated with the antisense or control oligomer (at 36 h [Fig. 2D]) by differential PCR display. In this assay each band represents the RT-PCR product of a specific mRNA. Only a minority of transcripts seemed to be affected by the loss of hTAFII150 (CIF150) function (Fig. 2F), suggesting that hTAFII150 (CIF150) mediates the selective transcription of a specific set of class II genes. In order to identify potential hTAFII150 (CIF150)-dependent cDNAs, we analyzed a limited set of the affected PCR fragments. In our initial screen (with 40 PCR products analyzed) we identified the following potential target genes of hTAFII150 (CIF150): ribosomal proteins L44, S10, and L7a, cyclin B1, ISGF-3, metallothionein II, lipid kinase (68 kDa; type I), DNA methyltransferase, and 10 novel human cDNAs including a human homolog of a Xenopus mitotic phosphoprotein (data not shown). Similar gene-specific effects have been reported for a temperature-sensitive mutant of mammalian TAFII250 (11, 38, 40, 41, 45, 52) and after functional knockout experiments using yeast TAFII90 (1) and TAFII145 (42, 50, 51). A more comprehensive analysis has demonstrated more recently that about 15% of all yeast genes show a significant decrease in expression upon heat shock of a temperature-sensitive allele of yeast TAFII145, establishing that not all RNA polymerase II transcription is dependent on the yeast homolog of human TAFII250 (12). For the later part of this article we decided to concentrate on the hypothesis that hTAFII150 (CIF150) might be directly involved in cyclin B1 and cyclin A (G2/M cyclins) transcription (Fig. 2E).
hTAFII150 (CIF150) directly stimulates the TATA-less cyclin B1 and cyclin A promoters.
Our differential PCR display analysis, as well as the CIF150 or cyclin B1 immunoblot data after antisense oligonucleotide treatment (Fig. 2E), indicated a correlation between CIF150 activity and cyclin B1 transcription. These results suggested that hTAFII150 (CIF150) might be a positive regulator of this TATA-less promoter. In order to test this hypothesis more directly we analyzed the effect of CIF150 on in vitro transcription and cotransfection assays. For in vitro transcription we used highly purified recombinant CIF150 protein (Fig. 3A) in combination with nuclear extracts chromatographically depleted of CIF150 activity as described previously (17, 18). In the absence of CIF150 activity we did not observe cyclin B1 and A promoter-dependent transcription. The titration of CIF150 protein stimulated the TATA-less cyclin A and B1 promoters, indicating that CIF150 is required for their transcription (Fig. 3B; compare lane 1 with lanes 2, 3, and 4). The TATA-containing control promoters (IgH and CMV) were not affected by the absence of CIF150. To demonstrate the TATA-less promoter-specific stimulation effect of CIF150 in vivo, we performed cotransfection experiments with CIF150 expression vectors in combination with a luciferase reporter gene fused to cyclin B1 and A promoter fragments. The overexpression of CIF150 had only a minor effect on TATA-containing CMV and on minimal Fos promoter activity but was able to stimulate cyclin B1 and cyclin A transcription in HeLa cells (Fig. 3C). The upper panel of Fig. 3C shows the absolute luciferase activities by using the c-Fos, cyclin B1, and cyclin A promoters in cotransfection experiments with an unrelated expression plasmid (pEVRF1-Ob) and CIF150 expression plasmid (pEVRF-CIF150). Both the cyclin A and B1 promoters are preferentially stimulated by CIF150, compared to the TATA-containing promoters c-Fos and CMV (Fig. 3C). These findings support our in vitro data and suggest that hTAFII150 (CIF150) is required for cyclin B1 and A transcription but is more dispensable for the transcription of TATA-containing promoters, including the CMV, IgH, and minimal Fos promoters. It has been shown previously that a 90-bp upstream region of the cyclin B1 promoter is sufficient to mediate cell cycle-dependent transcription, supporting the idea of an involvement of the core promoter in regulation (14). In addition, the preferential stimulation of the TATA-less cyclin promoters by the overexpression of hTAFII150 (CIF150) seems to confirm our biochemical data that hTAFII150 (CIF150) is only substoichiometrically present in the human TFIID complex (Fig. 1B). This indicates that hTAFII150 (CIF150) activity might be limiting on specific sets of promoters.
hTAFII150 (CIF150) binds DNA sequence specifically.
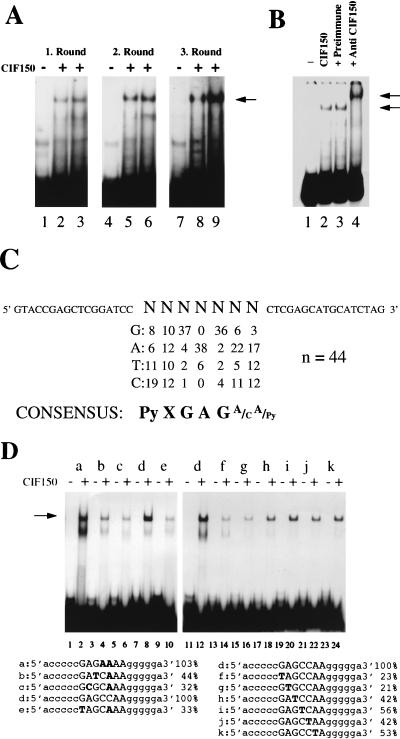
Since dTAFII150 has been reported to recognize specific core promoter elements (47, 48), we have begun to identify a cis-acting CIF150-responsive element. As a first step, we performed binding-site selection (31) by using highly purified recombinant CIF150 (Fig. 3A). A pool of DNA fragments with 7 bp of randomized nucleotide pairs were 32P labeled and used in three successive rounds of gel shift experiments (Fig. 4A). The data in Fig. 4B show that the selected protein-DNA complex was specific for the CIF150 protein as demonstrated by a supershift induced by CIF150-specific antiserum. The DNA sequences of 44 selected PCR fragments revealed a statistically significant enrichment of fragments with the core sequence 5′GAG3′ after the alignment of the sequences (Fig. 4C; data not shown). We estimate that we improved the binding affinity of CIF150 only fivefold after four rounds of selection (Fig. 4A; data not shown). This might be due to the presence of cryptic binding sites within the flanking sequences of the PCR primers (Fig. 4C) and/or the ability of CIF150 to form relatively stable complexes nonspecifically with DNA. It is therefore possible that the consensus sequence we identified depended partly on some initial nonrandomized flanking sequence. To validate the putative sequence element we performed electrophoretic mobility shift experiments by using DNA fragments with defined base pair substitutions (Fig. 4D). Substitutions of the GAG core sequence reduced CIF150 binding dramatically, compared to flanking substitutions (Fig. 4D; compare oligonucleotides a and d with oligonucleotides b, c, e, f, and g). These experiments demonstrate that CIF150 binds the GAG core sequence with higher affinity than randomized DNA, indicating a potential role of CIF150 (hTAFII150) in promoter recognition and selection. The final selected consensus sequence is very short (Py X GAG [A/C] [A/Py]), and we do not know yet whether this sequence is part of a larger high-affinity site. Therefore, using longer randomized sequences for the initial selection experiment might lead to a stronger hTAFII150 (CIF150) consensus binding site. Taken together the demonstrated preference for hTAFII150 (CIF150) binding to a GAG-containing core sequence might only be a starting point for a more detailed analysis. However, a loosely defined consensus sequence has also been reported for the Inr (Py Py A +1 N [T/A] Py Py) and DPE (G [A/T] C G) elements, which are both considered to be recognized by TAFIIs (4, 5, 43).
FIG. 4.
CIF150 (hTAFII150) has sequence-specific binding activity. (A) CIF150 binding-site selection. The arrow indicates the specific protein-DNA complex. (B) The addition of anti-CIF150 (lane 4) but not that of preimmune serum (lane 3) supershifted the retarded band. (C) Consensus CBE derived after four rounds of binding-site selection. (D) Point mutational analysis of the CBE by gel shift experiments. The sequences of the oligonucleotides used are shown at the bottom, with the nucleotides (in bold) that differ from oligonucleotide d (100%). The results are representative of three independent experiments.
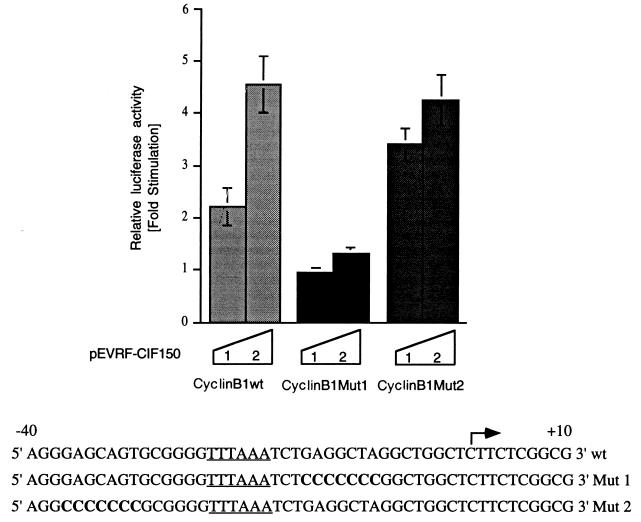
Mutation in one putative CIF50 binding element (CBE) in the cyclin B1 core promoter abolishes stimulation by CIF150 (hTAFII150).
The cyclin B1 core promoter contains two putative CBEs (GAG core sequences) flanking a noncanonical TATA box (Fig. 5; see reference 6). In order to test the role of CIF150 in promoter recognition directly, we mutated both putative CBEs in the cyclin B1 core promoter and tested these constructs in cotransfection experiments. The mutation of the upstream site (Fig. 5, Mut 2) did not affect the stimulation of the cyclin B1 promoter by CIF150. However, the substitution of a putative binding site (5′GAGGCTA3′) just 4 bp downstream of a noncanonical TATA box abolished the stimulation of reporter activity by CIF150 (Fig. 5). These data suggest that CIF150 interacts with the cyclin B1 promoter in a sequence-specific manner to stimulate transcription. Moreover, these results provide more evidence to support the view that one of the regulatory functions of TAFIIs, including CIF150, is to select core promoters through the sequence-specific recognition of DNA. Although we originally identified CIF150 (hTAFII150) as a cofactor for Inr function, the DNA-binding analyses as well as our mutational analyses of the cyclin B1 core promoter suggest that hTAFII150 (CIF150) may not be involved in Inr recognition itself. Our current working model is that hTAFII150 (CIF150) contacts DNA downstream of the −25 region but upstream of the start site and thereby stabilizes TFIID binding preferentially on TATA-less promoters, whereas another protein in the TFIID complex (most likely, TAFII250) binds directly to the Inr element.
FIG. 5.
Identification of a CBE in the cyclin B1 core promoter. Shown is the cotransfection of increasing amounts of CIF150 expression plasmid (1 and 2 μg) with cyclin B1 wild-type (wt) and cyclin B1 promoter mutant constructs fused to a luciferase reporter plasmid. The amount of CMV-derived vector in each transfection assay was kept constant by using an unrelated expression plasmid (pEVRF1-Ob). The values shown are the averages of four independent experiments ± standard deviations (error bars). An arrow indicates the transcription start site, and the −25 region of the promoter is underlined. Bold nucleotides indicate the changes in the mutated promoter constructs.
DISCUSSION
It is evident from in vitro studies that TAFIIs serve multiple functions as coactivators for activated transcription and also play an important role as promoter selectivity factors (reviewed in references 9, 20, 37, 49). Although TAFIIs are integral components of the general transcription machinery, individual TAFIIs might be required by only a distinct subset of activators or a subset of RNA polymerase II core promoters. We employed transient functional knockout experiments with these essential genes as one approach to elucidate the function of individual TAFIIs in vivo. Our experiments demonstrated for the first time that the transient depletion of CIF150 (hTAFII150) in mammalian cells leads to a cell cycle arrest phenotype. These results are in agreement with experiments obtained with temperature-sensitive mutants of the yeast homolog TSM-1 (50). Analysis of the RNA derived from HeLa cells depleted of hTAFII150 (CIF150) protein suggests that hTAFII150 (CIF150) is not an absolute requirement for all RNA polymerase II-dependent transcription in mammalian cells. Furthermore, we present evidence that hTAFII150 (CIF150) exhibited specificity for TATA-less promoters, including the cell cycle-specific cyclin A and B1 promoters. However, at this point we do not know whether the hTAFII150 (CIF150)-dependent transcription of the TATA-less cyclin B1 and cyclin A promoters is TBP dependent.
It is worth noting that the TBP-free TFTC complex described by Wieczorek et al. (53) did contain additional polypeptides, including one of the same size as hTAFII150 (CIF150). An attractive hypothesis is that hTAFII150 (CIF150) binding to the CBE might compensate for the absence of TBP binding to the TATA box. Similar models have been suggested for the Inr-, TAF-, and DPE-dTAFII60 interactions (4, 5, 48, 49), which are most likely functionally analogous to the CIF150-CBE interaction in compensating for a weak or missing TATA box interaction. However, hTAFII150 (CIF150)-dependent transcription seems to be different in that the overexpression of hTAFII150 (CIF150) directly stimulates transcription from TATA-less cyclin promoters, suggesting that CIF150 activity, but not TFIID, is limiting on these promoters. Furthermore, we were able to demonstrate that hTAFII150 (CIF150) seemed to be more dispensable for the transcription of TATA-containing promoters by using in vitro complementation assays and cotransfection assays. These results together with the functional knockout experiments presented suggest that hTAFII150 (CIF150) might be initiating gene-specific transcription without necessarily being stably integrated into the human TFIID complex. Our gel filtration results demonstrated that at least a portion of the hTAFII150 (CIF150) protein can dissociate from TFIID under low-salt conditions. As mentioned before, the absence of a tight interaction between TFIID and hTAFII150 (CIF150) is reminiscent of the TFIID-TFIIA interaction. TFIIA and dTAFII150 are tightly associated with the TFIID complex in Drosophila cells but not in human cells (54). A hypothesis that would include our results is that hTAFII150 (CIF150) may have a function related to TFIIA in stabilizing the TFIID-DNA interaction (18).
Whether hTAFII150 (CIF150) also plays a role in mediating activated transcription, as a direct target of activation domains is currently unknown and warrants further study. Addressing this question is important in light of studies demonstrating that TAFII250 function in cyclin A promoter-dependent transcription is apparently associated with both enhancer and core promoter elements (52).
The implication of hTAFII150 (CIF150) in cell cycle progression is not unprecedented. It has been demonstrated that other TAFIIs play an important role in the regulation of cell growth and cell cycle progression (26, 38, 39, 40, 42, 45, 52). Further experiments are needed to elucidate the cell cycle regulation of hTAFII150 (CIF150) itself. hTAFII150 (CIF150) might be phosphorylated by cyclin–cyclin-dependent kinase complexes to modulate CIF150 activity or degradation of CIF150 protein during cell cycle progression. We wish to emphasize that the positive regulation of cyclin B1 and cyclin A transcription by hTAFII150 (CIF150) does not exclude other activating and repressing mechanisms of cell cycle-regulated transcription. Several mechanisms of transcriptional repression have been proposed, including chromatin structure changes of the promoter region, inhibition of general transcription factors, and DNA binding competition (for a review, see reference 15).
In summary, our data suggest that the mammalian TAF-like protein CIF150 (hTAFII150) is a necessary positive transcriptional regulator of cell cycle progression through G2/M. The depletion of CIF150 (hTAFII150) activity in vivo seems to selectively affect a subset of class II promoters, and CIF150 activity appears to be one rate-limiting step for cyclin B1 transcription. Future studies of the regulatory role of hTAFII150 (CIF150) in signal transduction pathways should provide more insights into the regulation of the eukaryotic cell cycle.
ACKNOWLEDGMENTS
We thank T. Shi and T. Brown for FACS analysis, L. Cornroy and S. Widger for baculovirus expression of CIF150, T. Wolf for selection and synthesis of the antisense oligomers, and M. Innis, N. Marini, A. Klippel, C. Reinhardt, K. Giese, and K. Ahrens for their helpful discussions and comments on the manuscript. We also thank J. Escobedo and L. T. Williams for their support.
REFERENCES
- 1.Apone L M, Virbasius C A, Reese J C, Green M R. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 2.Apone L M, Virbasius C A, Holstege F C, Wang J, Young R A, Green M R. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol Cell. 1998;2:653–661. doi: 10.1016/s1097-2765(00)80163-x. [DOI] [PubMed] [Google Scholar]
- 3.Brechot C. Oncogenic activation of cyclin A. Curr Opin Genet Dev. 1993;3:11–18. doi: 10.1016/s0959-437x(05)80335-1. [DOI] [PubMed] [Google Scholar]
- 4.Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 5.Burke T W, Kadonaga J T. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogswell J P, Godlevski M M, Bonham M, Bisi J, Babiss L. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol Cell Biol. 1995;15:2782–2790. doi: 10.1128/mcb.15.5.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates III J R, Workman J L. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 9.Hahn S. The role of TAFs in RNA polymerase II transcription. Cell. 1998;95:579–582. doi: 10.1016/s0092-8674(00)81625-6. [DOI] [PubMed] [Google Scholar]
- 10.Henglein B, Chenivesse X, Wang J, Eick D, Brechot C. Structure and cell cycle-regulated transcription of the human cyclin A gene. Proc Natl Acad Sci USA. 1994;91:5490–5494. doi: 10.1073/pnas.91.12.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hisatake K, Hasegawa S, Takada R, Nakatani Y, Horikoshi M, Roeder R G. The p250 subunit of native TATA-binding factor TFIID is the cell-cycle regulatory protein CCG1. Nature. 1993;362:179–181. doi: 10.1038/362179a0. [DOI] [PubMed] [Google Scholar]
- 12.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 13.Hunter T, Pines J. Cyclins and cancer II: cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 14.Hwang A, Maity A, McKenna W G, Muschel R J. Cell cycle-dependent regulation of the cyclin B1 promoter. J Biol Chem. 1995;270:28419–28424. doi: 10.1074/jbc.270.47.28419. [DOI] [PubMed] [Google Scholar]
- 15.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 16.Kaufmann J, Smale S T. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann J, Verrijzer C P, Shao J, Smale S T. CIF, an essential cofactor for TFIID-dependent initiator function. Genes Dev. 1996;10:873–886. doi: 10.1101/gad.10.7.873. [DOI] [PubMed] [Google Scholar]
- 18.Kaufmann J, Ahrens K, Koop R, Smale S T, Mueller R. CIF150, a human cofactor for transcription factor IID-dependent initiator function. Mol Cell Biol. 1998;18:233–239. doi: 10.1128/mcb.18.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagrange T, Kapanidis A N, Tang H, Reinberg D, Ebright R H. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee T I, Young R A. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 21.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 22.Martinez E, Chiang C M, Ge H, Roeder R G. TAFs in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez E, Ge H, Tao Y, Yuan C X, Palhan V, Roeder R G. Novel cofactors and TFIIA mediate functional core promoter selectivity by the human TAFII150-containing TFIID complex. Mol Cell Biol. 1998;18:6571–6583. doi: 10.1128/mcb.18.11.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthias P, Mueller M M, Schreiber E, Rusconi S, Schaffner W. Eukaryotic expression vectors for analysis of mutant proteins. Nucleic Acids Res. 1989;17:6418. doi: 10.1093/nar/17.15.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michel B, Komarnitsky P, Buratowski S. Histone-like TAFs are essential for transcription in vivo. Mol Cell. 1998;2:663–673. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- 26.Moqtaderi Z, Bai Y, Poon D, Weil A P, Struhl K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature. 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 27.Moqtaderi Z, Keaveney M, Struhl K. The histone H3-like TAF is broadly required for transcription in yeast. Mol Cell. 1998;2:675–682. doi: 10.1016/s1097-2765(00)80165-3. [DOI] [PubMed] [Google Scholar]
- 28.Murray A, Hunt T. The cell cycle. W. H. New York, N.Y: Freeman and Company; 1993. [Google Scholar]
- 29.Oelgeschlager T, Tao Y, Kang Y K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol Cell. 1998;1:925–931. doi: 10.1016/s1097-2765(00)80092-1. [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko V V, Kotani T, Zhang X, Schiltz R L, Howard T, Yang X-J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 31.Oliphant A R, Brandl C J, Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 33.Pines J, Hunter T. Human cyclin A is adenovirus E1A-associated protein p60 and behaves differently from cyclin B. Nature. 1990;346:760–763. doi: 10.1038/346760a0. [DOI] [PubMed] [Google Scholar]
- 34.Pines J. Cyclins and cyclin-dependent kinases: take your partners. Trends Biochem Sci. 1993;18:195–197. doi: 10.1016/0968-0004(93)90185-p. [DOI] [PubMed] [Google Scholar]
- 35.Purnell B A, Emanuel P A, Gilmour D S. TFIID sequence recognition of the initiator and sequence farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 36.Ray B L, White C I, Haber J E. The TSM1 gene of Saccharomyces cerevisiae overlaps the MAT locus. Curr Genet. 1991;20:25–31. doi: 10.1007/BF00312761. [DOI] [PubMed] [Google Scholar]
- 37.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 38.Ruppert S, Wang E H, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993;362:175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- 39.Segil N, Guermah M, Hoffmann A, Roeder R G, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- 40.Sekiguchi T, Nohiro Y, Nakamura Y, Hisamoto N, Nishimoto T. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol Cell Biol. 1991;11:3317–3325. doi: 10.1128/mcb.11.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sekiguchi T, Noguchi E, Hatashida T, Nakshima T, Toyoshima H, Nichimoto T, Hunter T. D-type cyclin expression is decreased and p21 and p27 CDK inhibitor expression is increased when tsBN462 CCG1/TAFII250 mutant cells arrest in G1 at the restrictive temperature. Genes Cell. 1996;1:687–705. doi: 10.1046/j.1365-2443.1996.00259.x. [DOI] [PubMed] [Google Scholar]
- 42.Shen W-C, Green M R. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 43.Smale S T. Architecture of core promoters for eukaryotic protein-coding genes. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 63–81. [Google Scholar]
- 44.Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki-Yagawa Y, Guermah M, Roeder R G. The ts13 mutation in the TAFII250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tansey W P, Herr W. TAFs: guilt by association? Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- 47.Verrijzer C P, Yokomori K, Chen J-L, Tjian R. Drosophila TAFII150: similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- 48.Verrijzer C P, Chen J-L, Yokomori K, Tjian R. Binding of TAF’s to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 49.Verrijzer C P, Tjian R. TAF’s mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 50.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 51.Walker S S, Shen W-C, Reese J C, Apone L M, Green M R. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 52.Wang E H, Zou S, Tjian R. TAFII250-dependent transcription of cyclin A is directed by ATF activator proteins. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wieczorek E, Brand M, Jacq X, Tora L. Function of TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- 54.Yokomori K, Admon A, Goodrich J A, Chen J-L, Tjian R. Drosophila TFIIA-L is processed into two subunits that are associated with the TBP/TAF complex. Genes Dev. 1993;7:2235–2245. doi: 10.1101/gad.7.11.2235. [DOI] [PubMed] [Google Scholar]
- 55.Zhou J, Zwicker J, Szymanski P, Levine M, Tjian R. TAFII mutations disrupt Dorsal activation in the Drosophila embryo. Proc Natl Acad Sci USA. 1998;95:13483–13488. doi: 10.1073/pnas.95.23.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]