Abstract
Porcine deltacoronavirus (PDCoV) is one of the most important enteropathogenic pathogens, and it causes enormous economic losses to the global commercial pork industry. PDCoV was initially reported in Hong Kong (China) in 2012 and subsequently emerged in swine herds with diarrhea in Ohio (USA) in 2014. Since then, it has spread to Canada, South Korea, mainland China, and several Southeast Asian countries. Information about the epidemiology, evolution, prevention, and control of PDCoV and its prevalence in China has not been comprehensively reported, especially in the last five years. This review is an update of current information on the general characteristics, epidemiology, geographical distribution, and evolutionary relationships, and the status of PDCoV vaccine development, focusing on the prevalence of PDCoV in China and vaccine research in particular. Together, this information will provide us with a greater understanding of PDCoV infection and will be helpful for establishing new strategies for controlling this virus worldwide.
Introduction
Coronaviruses (CoVs) are a family of enveloped viruses with a positive-stranded RNA genome, and they can be genetically divided into four genera (Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus) that belong to the family Coronaviridae of the order Nidovirales [1].
CoVs are distributed widely among mammals and birds [2–5]. Individual CoVs usually infect their hosts in a species-specific manner, with alpha- and betacoronaviruses mainly infecting mammals, gammacoronaviruses normally infecting avian species, and deltacoronaviruses infecting both mammals and avian species. Attention to coronaviruses has increased in recent years because of the emergence of severe acute respiratory syndrome (SARS), Middle-East respiratory syndrome (MERS), and the newly emerging coronavirus disease 2019 (COVID-19), all of which cause acute respiratory illness, with mortality rates up to 9.5% in SARS and 35% in MERS, and with an inferred infection fatality rate varying from 0.00% to 1.63%, with corrected values varying from 0.00% to 1.54% for COVID-19 [6–10].
Porcine deltacoronavirus (PDCoV), a member of the genus Deltacoronavirus, is a novel swine enteropathogenic coronavirus that causes acute diarrhea, vomiting, and dehydration in neonatal piglets [11–15]. PDCoV was initially identified in 2012 during a molecular surveillance study in Hong Kong and emerged later in swine herds with diarrhea in Ohio (USA) in 2014 [2, 16]. Subsequently, the outbreak exhibited a global spread, and the virus has been detected in fecal samples from piglets in South Korea, mainland China, Thailand, Vietnam, and Laos [17–20].
The epidemiological, clinical, and pathological features of PDCoV are similar to those of transmissible gastroenteritis virus (TGEV) and porcine epidemic diarrhea virus (PEDV), but PDCoV has a lower clinical impact and disease severity than TGEV and PEDV [21, 22]. Pathogenicity experiments have confirmed that PDCoV exhibits enteropathogenicity, causing severe diarrhea and vomiting in suckling piglets. Histological examination has revealed lesions characteristic of atrophic enteritis, primarily in the jejunum and ileum, which were characterized by intestinal villi atrophy and shortening [12, 13, 23–26].
PDCoV infections have caused significant economic losses in the swine industry worldwide. Therefore, the development of effective vaccines is essential for the prevention and control of PDCoV. However, there are currently no effective vaccines available for PDCoV. This review summarizes the latest discoveries in the study of PDCoV, with particular emphasis on its epidemiology, genetic evolution, and prevalence in China as well as vaccine research.
Genomic organization and general characteristics
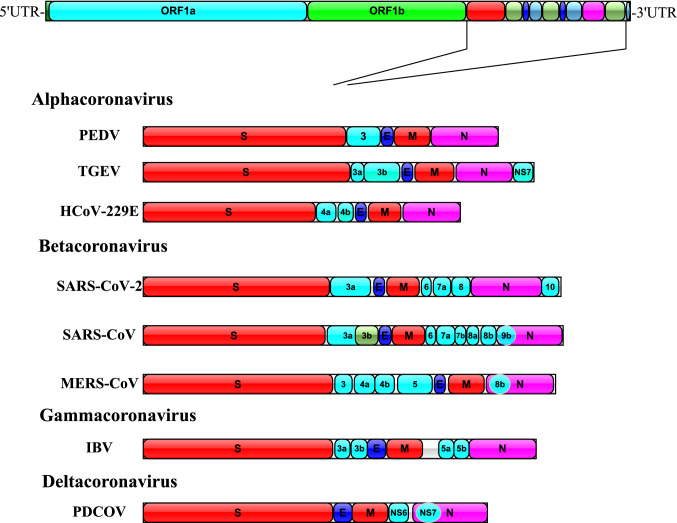
The genome of PDCoV is approximately 25.4 kb in length [16, 27], making it the smallest genome known among CoVs. The genome organization of PDCoV is typical of CoVs and contains six common coronaviral genes in the following conserved order: 5ʹUTR-ORF1a-ORF1b-S-E-M-NS6-N-NS7-3ʹUTR [2, 27] (Fig. 1).
Fig. 1.
Schematic diagram showing the genome organization of eight known CoVs of the genera Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. The expanded regions below show the structural and accessory proteins in the 3′ regions of PEDV, TGEV, HCoV-229E, SARS-CoV-2, SARS-CoV, MERS-CoV, IBV, and PDCoV.
ORF1a and ORF1b occupy nearly three-fourths of the viral genome and encode two overlapping replicase polyproteins [28, 29]. Downstream of ORF1a and ORF1b, there are several additional ORFs that code for the structural proteins: spike (S), envelope (E), membrane (M), nonstructural protein 6 (NS6), nucleocapsid (N), and nonstructural protein 7 (NS7) [17, 30]. The S protein forms peplomers on the virion surface and plays an important role in receptor attachment and viral and host cell membrane fusion [31–33]. It is a major target for virus neutralizing antibodies, and its structure is divided into S1 and S2 domains [34, 35]. The E and M proteins are transmembrane proteins that function in viral envelope formation and release [28, 36]. The N protein is the most abundant and multifunctional viral component. The primary role of the CoV N protein is to package the genomic viral genome into long, flexible, helical ribonucleoprotein (RNP) complexes called capsids, and the capsid protects the genome and ensures its timely replication and reliable transmission [37]. In addition, the PDCoV N protein associates with viral pathogenesis by interfering with the early activation of pRIG-I in the host antiviral response [38, 39]. NS6 and NS7 encode an accessory protein [40, 41], and NS7 of PDCoV overlaps with the N protein [42].
Survival and inactivation
Many factors can influence virus survival outside of the host, including temperature, relative humidity (RH), desiccation, irradiation, the milieu in which the virus is suspended, and the physicochemical properties of the virus. CoVs survive for extended periods of time in feces, feed, and feed ingredients [43]. This prolonged survival can be reduced by feed additives [44] and heat [45].
PEDV can survive for 1–7 days at 70% RH at 60 °C, TGEV can exist for up to 14 days at 60 °C, and PEDV can survive for more than 4 weeks in wet complete feed but for only 1 week in dry complete feed [46, 47].
There is a lack of data on the presence and survival of PDCoV. Theoretically, the physical and chemical properties of PDCoV are comparable to those of other intestinal coronaviruses, such as TGEV and PEDV, but there are some differences.
A survey conducted by a Chinese team showed that PDCoV is resistant to acid and organic solvents but is sensitive to heat. PDCoV is stable at pH 3.0 and is tolerant to ether and chloroform treatment. Most PDCoV strains are absolutely inactivated at 50 °C after 60 min of exposure [48]. The Weibull distribution model was used to analyze the inactivation kinetics of PDCoV in various feed ingredient matrices, and the results showed that the virus survived the longest in soybean meal and that the first log reduction of virus titer of PDCoV and TGEV in soybean meal was observed at 42.04 days and 42 days, respectively [43].
Further study is needed to measure the comparative survival of coronaviruses in the environment so that effective control strategies can be designed and implemented.
Host range and susceptibility
CoVs have remarkable potential for crossing species barriers, which plays a major role in virus evolution and diversification [49, 50]. CoVs have been detected in a wide range of hosts, covering avian and mammalian species, including humans, poultry, bats, dogs, swine, and cattle [3, 8, 51–55], causing respiratory, neurological and gastrointestinal diseases, but coronaviruses of the genera gamma- and deltacoronavirus have been isolated primarily in birds.
Deltacoronaviruses were previously identified primarily in multiple songbird species, Asian leopard cats, and Chinese ferret badgers [2]. Pigs are the main hosts of PDCoV, and pigs of all ages are susceptible to PDCoV [15], with suckling piglets being the most susceptible [14]. PDCoV in piglets is mainly characterized by varying degrees of diarrhea, vomiting, dehydration, and histopathological lesions typical of atrophic enteritis [12].
In addition to swine, calves and chickens have been shown by experimental testing to be susceptible to PDCoV infection [3, 5, 56]. Jung et al. demonstrated that gnotobiotic calves inoculated orally with PDCoV develop an acute infection with persistent fecal PDCoV RNA shedding and PDCoV-specific serum IgG antibody responses but show no signs of significant intestinal lesions or clinical disease [3]. Liang et al. found that PDCoV can infect and be continuously passaged in chicken embryos, and chickens showed mild diarrhea symptoms, with viral RNA being detected in multiple organs and intestinal contents after inoculation [56]. A recent report of PDCoV infection in poultry in the United States showed similar features of diarrhea, persistent viral RNA shedding, and PDCoV-specific IgY antibody responses [5].
Although there have been no reports of PDCoV infecting humans, some researchers have observed that PDCoV efficiently infects human cells [57, 58]. Cruz-Pulido et al. analyzed the transcriptomes of human and pig intestinal epithelial cells after PDCoV infection and found that human cells exhibited a more pronounced and upregulated response to PDCoV infection in comparison to pig cells and speculated that humans could be a new host for PDCoV [57].
Epidemiology and geographical distribution in China
PDCoV was first reported in Hong Kong in 2012 and was subsequently detected in pigs from the United States in February 2014 [2, 16]. Shortly thereafter, PDCoV was introduced into South Korea, China, Thailand, Vietnam, Canada, and Mexico, exhibiting a global distribution trend [18, 20, 59–61].
In China, PDCoV was first reported in 2015 and quickly spread across the country. However, it remains unclear when PDCoV was introduced into China. A retrospective study on clinical samples collected during 2004–2014 from Hubei, Anhui, Jiangsu, and Guangxi provinces revealed that PDCoV was present in the Chinese mainland as early as 2004 [18]. After the PDCoV outbreaks in 2015, detailed studies of the epidemiology and evolution of PDCoV were conducted in China, which led to the accumulation of a large amount of data (Table 1). To date, PDCoV infection has been reported in 26 provinces of China, including all major traditional breeding areas [15, 27, 62–75].
Table 1.
Studies of PDCoV infection of pigs in China
| Reference | Provinces | No. of samples | Positive rate % (n) | Year | Sample types | ||
|---|---|---|---|---|---|---|---|
| PDCOV | PEDV | PDCOV + PEDV | |||||
| [59] | Gansu, Qinghai, Sichuan | 189 | 3.70 (7) | 27.51 (52) | 2.11 (4) | 2016, 2017 | Faecal samples |
| [60] | 18 provinces (Heilongjiang, Liaoning, Beijing, Hebei, Henan, Shanxi, Shandong, Hubei, Anhui, Hunan, Jiangxi, Zhejiang, Jiangsu, Guangxi, Yunnan, Fujian, Sichuan, Gansu) | 719 | 13.07 (94) | 36.72 (267) | 4.73 (34) | 2016–2018 | Faeces, faecal swabs, small intestines |
| [61] | Hebei | 871 | 11.02 (96) | – | – | 2015, 2016 | Serum |
| [62] | Heilongjiang | 319 | 11.59 (37) | – | – | 2014, 2015 | Serum |
| [63] | Guangdong | 84 | 41.7 (35) | – | – | – | Faeces, faecal swabs, small intestines |
| [64] | 9 Provinces (Henan, Shaanxi, Liaoning, Gansu, Ningxia, Chongqing, Hainan, Jiangxi, Qinghai) | 398 | 36.18 (144) | 19.60 (78) | – | 2015–2017 | Faeces |
| [65] | Heilongjiang, Jilin, Liaoning | 672 | 3.87 (26) | 19.05 (128) | – | 2017, 2018 | Faeces |
| [27] | Jiangxi | 356 | 33.71 (120) | 64.89 (231) | 19.66 (120) | 2012–2015 | Intestinal and fecal samples |
| [66] | Shaanxi, Henan, Hubei | 70 | 2.9 (2) | 84.2 (59) | 2.9 (2) | 2015, 2016 | Intestinal and fecal samples |
| [67] | Guangdong, Guangxi, Hainan | 390 | 1.28 (5) | 22.56 (88) | 1.28 (5) | 2012–2015 | Faeces |
| [15] | Henan | 177 | 69.49 (123) | 0 | 0 | 2017–2019 | Fecal samples, small intestinal content |
| [68] | Jiangxi, Zhejiang, Fujian, Guangdong, Hunan | 2987 | 27.22 (813) | 57.32 (1712) | 12.72 (380) | 2012–2018 | Faeces, intestine, milk |
| [69] | Henan | 430 | 23.49 (101) | – | 14.18 (61) | 2015–2018 | Faecal and intestinal samples |
| [70] | Guangdong | 252 | 21.8 (55) | 65.5 (165) | 0.79 (2) | 2015, 2016 | Faecal and intestinal samples |
| [118] | Guangdong | 420 | 13.33 (56) | 31.9 (134) | 5.95 (25) | 2012–2016 | Fecal samples, small intestinal content |
| [119] | Guangxi | 1547 | 4.52 (70) | 54.94 (843) | 1.1 (17) | 2013–2018 | Faecal samples |
| [71] | Jiangxi | 249 | 31.33 (78) | – | – | 2012–2015 | Faeces, faecal swabs, small intestines |
| [72] | Liaoning, Shaanxi, Shandong, Chongqing, Ningxia, Gansu | 354 | 34.2 (121) | 9.6 (34) | 1.4 (5) | 2015–2018 | Faecal samples |
From March 2016 to June 2018, a survey covering 18 provinces revealed that the infection rate of PDCoV and PEDV among Chinese pig populations was 13.07% and 36.72%, respectively, and the coinfection rate of PDCoV and PEDV was 4.73% [63]. In another study, a total of 2987 field diarrheal samples collected from 168 pig farms in five provinces (Jiangxi, Zhejiang, Fujian, Guangdong, and Hunan) in southern China during 2012–2018 were tested, the positive rates of PDCoV infection varied from 19.62% to 31.47% among samples from different years, and the positive rates of different provinces varied from 19.57% (Guangdong) to 33.32% (Fujian). Of 2987 samples, 813 (27.22%) were PDCoV positive, and of the 813 PDCoV-positive samples, 380 (46.74%) were coinfected with PEDV [71]. Interestingly, we also found that PDCoV was detectable in pigs of all ages (sows, suckling piglets, nursery pigs, finishing pigs), and the positive rates were 27.79% (187/673), 29.52% (501/1697), 20.45% (73/357), and 16.74% (36/215), respectively, indicating that suckling piglets are more susceptible to PDCoV infection [71]. These results are consistent with those of another study conducted in 2019 [15].
In general, the infection rates of PEDV and PDCoV and their coinfection rates in China vary from region to region, and the infection rate of PEDV is higher than that of PDCoV, indicating that although there is a commercial vaccine, PEDV prevention and control in China are still inadequate. These findings also reveal the possibility that PDCoV is one of the major pathogens, in addition to PEDV, causing outbreaks of diarrhea in swine farms on China.
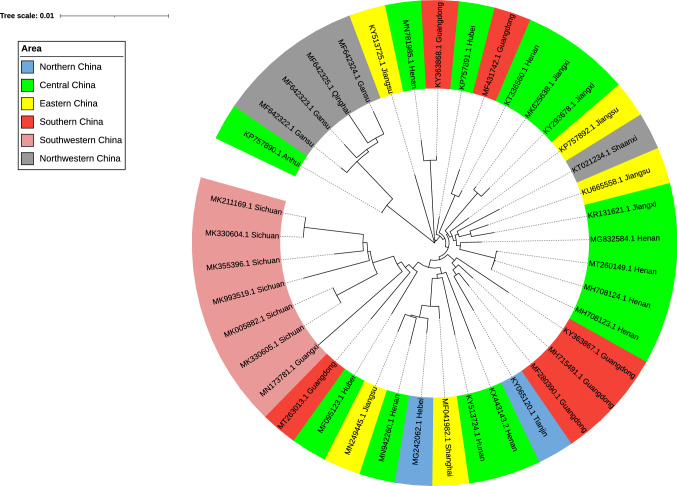
In terms of the geographical distribution in China (Fig. 2), the infection rate of PDCoV in the northeastern (Heilongjaing, Jinlin, Liaoning) [68] and northwestern regions (Gansu, Qinghai) [62] is comparatively low, but the infection rate of PDCoV in central China (Henan, Hubei) [15, 64] and southern China (Guangdong, Jiangxi) [66, 73, 74] is higher, which may be related to the greater amount of pig production and the higher frequency of pig transport in these provinces.
Fig. 2.
Geographical distribution of PDCoV strains in different regions of China. The genome sequences of 40 strains from 15 different provinces of China were downloaded from the NCBI database, and phylogenetic trees were constructed using the neighbor-joining method in MEGA7. Green represents the area of central China (Henan, Anhui, Hubei, Hunan, Jiangxi), gray represents the area of northwestern China (Shaanxi, Gansu, Qinghai), yellow represents the area of eastern China (Jiangsu, Shanghai), blue represents the area of northern China (Hebei, Tianjin), pink represents the area of southwestern China (Sichuan), and red represents the area of southern China (Guangdong, Guangxi).
These data provided us with a basic reference for understanding the epidemiology of PDCoV in China, and statistics show that PDCoV infection occurs extensively in the pig population in China.
However, because China is a country with a vast territory and a large number of breeding stock, these epidemiological studies are limited by sample size, sampling location, and season. Therefore, the epidemic features and incidence trends of PDCoV infection are not completely understood. More epidemiological data should be monitored dynamically in future studies in China.
Transmission
CoVs cause respiratory infections in humans and intestinal infections in pigs, and viruses with different phenotypes spread in different ways. Respiratory pathogenic coronaviruses (e.g., SARS-CoV, MERS-CoV, SARS-CoV-2) spread primarily through respiratory droplets and close contact [76–78], while enteropathogenic coronaviruses (TGEV, PEDV, PDCoV) are spread mainly by contact with feces, vomitus, and contaminated feed and trucks [79, 80]. Although some possibilities have been considered for how PDCoV was introduced and rapidly disseminated throughout pig farms around the world, the exact mechanism by which PDCoV entered and disseminated is not completely understood.
Direct transmission
PDCoV is transmitted horizontally (pig-to-pig infection) through both direct and indirect contact. Direct transmission occurs by contact with various secretions (feces, vomitus, saliva, milk, nasal secretions, oral fluids, semen) from infected pigs.
A total of 293 porcine samples from the United States and Canada were tested, and 52% (27/52) of intestinal samples, 40% (30/75) of fecal samples, 32% (6/19) of feed samples, and 19% (10/54) of saliva samples were positive for PDCoV [81]. The contamination of samples from different sources is indicative of the complexity of potential transmission.
The fecal-oral route is considered to be the most important mode of PDCoV transmission. Feces and vomitus from infected pigs contain high levels of the virus [81, 82]. Fecal shedding of PDCoV initiates even before the presentation of clinical signs and normally lasts for several weeks [12, 14, 25]. In some cases, fecal shedding may continue even after clinical signs have completely disappeared [25].
Indirect transmission
Indirect transmission occurs through virus-contaminated feed, feed trucks, transport trailers, and breeders. Contaminated feed and transport trailers may play an important role in the spread of the virus in and among farms [80].
Lowe et al. investigated the role of transport in the spread of porcine epidemic diarrhea virus infection in the United States and found that 6.6% (38/575) of the trailers were contaminated prior to unloading, and of the trailers not contaminated at arrival, 5.2% (28/537) were contaminated during the unloading process [80]. Greiner surveyed feed mills from various regions in the United States and found that 5% of the truck foot pedals and 1% of the bulk-ingredient pits tested suspect for PEDV, 3.4% of the foot pedals of the trucks tested positive, and 2.2% of the office floors tested suspect for PDCoV [83].
Previous research has suggested that PDCoV can survive in feed and feed ingredients at room temperature for up to 56 days [43], indicating that feed may be another route for PDCoV transmission. However, the spray drying process of feed production is hot enough to inactivate the infectious virus; therefore, the spread of the virus caused by transport should be given more attention.
Airborne
Studies have demonstrated the possibility of virus transmission through aerosols. In a study performed by Alonso and colleagues, it was shown that aerosolized PEDV can be transported up to 10 miles downwind through the air [79]. Interestingly, PEDV was not detected in air samples under field conditions, but pigs infected with experimental air samples experienced PEDV symptoms of moderate to severe diarrhea. Another study showed that despite obstacles to farrowing stalls and physical barriers, PDCoV infection would likely spread rapidly throughout modern farrowing and gestation barns [13]. The above case shows that airborne transmission should be considered as a potential route of PDCoV dissemination.
Origin and evolution
Origin
In the past 20 years, the world witnessed three serious zoonotic events in humans, SARS-CoV in 2003, MERS-CoV in 2012, and the SARS-CoV-2 that is circulating this year.
SARS-CoV was confirmed to have been transmitted to humans from Paguma larvata in 2002, causing outbreaks in 37 countries, with more than 8000 cases and 774 deaths [6]. MERS-CoV is reported to have spread from dromedary camels to humans in 2012 and has caused lethal respiratory infections in humans and 712 deaths [8]. In the ongoing COVID-19 epidemic, it has been suggested that SARS-CoV-2 was transmitted from pangolins to humans [84], and this virus has spread to more than 180 countries around the world. All three epidemics are thought to have originated from bat CoVs, which are transmitted to humans directly or indirectly through an intermediate host [85, 86].
There are four major known enteric coronaviruses that affect the pig industry: TGEV, PEDV, PDCoV, and the newly emerging swine acute diarrhea syndrome coronavirus (SADS-CoV). PEDV shows a close phylogenetic relationship to a CoV detected in T. brasiliensis bats in Brazil, indicating a possibility of bat origin [87]. SADS-CoV is the first bat-origin coronavirus shown to cause severe disease in domestic animals, and it shares 98.48% sequence identity with Rhinolophus bat coronavirus HKU2 [88].
To date, the origin of the novel PDCoV is still unknown. It is generally believed that the deltacoronavirus originated in birds [2, 89], but the partial genome sequences of PDCoV isolated from birds are too short to reflect evolutionary characteristics. It has been speculated that avian coronavirus may be the genetic source of mammalian deltacoronaviruses, and there may be cross-species transmission from birds to mammals [24, 58].
Compared to many other species, pigs are in frequent contact with humans, birds, and other animals and theoretically have a greater chance to be involved in cross-species transmission of viruses [90].
A study by Woo et al. showed that PorCoV HKU15 and SpCoV HKU17 are members of the same species, which implies that interspecies jumping from birds to pigs may have occurred [2]. Li et al. observed that PDCoV can efficiently infect cells with an unusually broad species range, including human and chicken cells [58]. Similar results were also reported in the comparative transcriptome profiling of human and pig intestinal epithelial cells after PDCoV infection [57]. Therefore, it appears inevitable that similar zoonotic events will occur again in the future, so more epidemiological studies need to be conducted to clarify the origin, epidemiology, and interspecific transmission mechanisms of coronaviruses.
Evolution
To date, 104 complete genome sequences of PDCoV isolates from the United States, China, Korea, Laos, Vietnam, Thailand, and Japan are available in the GenBank database. To investigate the molecular origin and evolution of PDCoV in mainland China, we performed a detailed phylogenetic analysis at the genome level using strains that have been reported in countries for the first time or for which epidemiological data show a high prevalence.
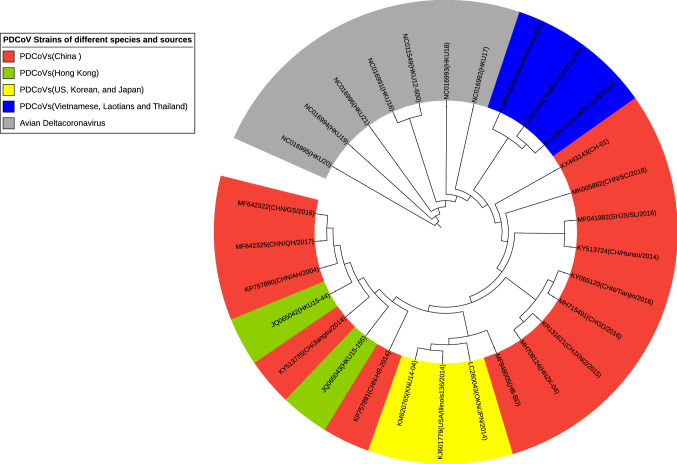
A total of 22 strains of porcine deltacoronavirus and seven avian isolates from seven countries were selected for phylogenetic analysis. Of these, 14 PDCoV isolates were from 13 different provinces of mainland China (Fig. 3).
Fig. 3.
Phylogenetic analysis of the complete genome sequences of 29 members of the genus Deltacoronavirus. The tree was constructed using the distance-based neighbor-joining method in MEGA7.0. Bootstrap analysis was carried out on 1000 replicate data sets, and values are shown adjacent to the branching points. Red represents the Chinese PDCoVs, blue represents the Vietnamese, Laotian, and Thai PDCoVs, yellow represents the US, Korean, and Japanese PDCoVs, green represents the Hong Kong (China) PDCoVs, and grey represents the avian deltacoronaviruses.
The phylogenetic analysis showed that the PDCoV isolates clustered together, while the avian deltacoronaviruses formed a separate cluster, and the evolutionary relationship between the two is distant. All porcine deltacoronavirus strains are closely related in the genetic evolution and have a high degree of sequence similarity, indicating that they might have originated from a common ancestor. The US, Korean, and Japanese PDCoV isolates grouped in the same branch with up to 99.9% nucleotide sequence identity and therefore might represent the same strain. The isolates from Vietnam, Laos, and Thailand belong to another branch with 98.4–99.8% whole-genome nucleotide sequence identity. It is clear that the Southeast Asian PDCoV isolates are much more closely related to the Chinese strains.
The nucleotide sequence identity of the 14 strains from mainland China ranged from 97.7% to 99.7%, with the isolates from Qinghai and Gansu and the Anhui strains belonging to the same branch with 99.1–99.5% nucleotide sequence identity. The isolates CHN/AH/2004, CHN/GS/2016, and CHN/QH/2017 were found to be more closely related to HKU15-44, while the Jiangsu isolate (CHN/Jiangsu/2014) was more closely related to HKU15-155.
Compared with the US, Korean, and Japanese PDCoV strains, most Chinese strains (except HKU15-44, CHN/AH/2004, CHN/GS/2016, and CHN/QH/2017) have a continuous deletion mutation of three nucleotides (AAT) at one site in the S gene. Previous reports have shown that the mutation rate of the S gene is relatively high, which may lead to altered tissue tropism, virulence, and even host specificity [27]. Whether the deletion of ATT has an effect on the virulence of the virus needs to be studied further.
In general, analysis of genetic evolution based on whole-genome sequencing shows that PDCoV has undergone extensive variation in different regions, and the mutations occur mainly in the S gene. Thus, it is important to monitor genetic variations occurring in the PDCoV S gene as well as to evaluate the impact of these variations on pathogenicity in order to develop an effective vaccine to control the disease.
Virulence and pathogenicity
Coronavirus infection has been documented previously in livestock and companion animals [55, 91]. TGEV and PEDV and the newly reported SADS-CoV mainly cause severe intestinal infections in piglets, leading to high morbidity and mortality and vast economic losses [88, 92, 93]. Bovine CoV, rat CoV, and infectious bronchitis virus (IBV) cause mild to severe respiratory tract infections in cattle, rats, and chickens, respectively [94–96]. Feline infectious peritonitis virus (FIPV) causes highly lethal disease in domestic cats [97].
Clinically, PDCoV can cause infection in pigs of various ages but mainly causes infection in newborn piglets, characterized by mild to severe diarrhea, vomiting, dehydration, anorexia, and growth retardation [13, 15, 98]. Inoculation experiments have suggested that although PDCoV exhibits enteropathogenicity in both gnotobiotic and conventional piglets, infected pigs display milder signs of clinical impact and disease severity than those infected with PEDV and TGEV [12, 24, 82].
Due to their underdeveloped immune system, neonatal piglets are highly susceptible to viral infection during their first few weeks. Mortality rates are highest in neonatal piglets, often reaching nearly 100%. Commercial fattening pigs and sows can also exhibit typical clinical features, such as diarrhea, inappetence, and persistent viral shedding in feces, but the symptoms of fattening pigs and sows are relatively mild, and the mortality rate is lower, with the animals gradually recovering [15].
Within 20–48 h postinfection, diarrhea is observed. Diarrhea typically lasts for at least 1 week. Vomiting symptoms were inconsistent in different experiments, which may be due to differences in virulence between strains. Core body temperatures remained within normal limits, and no respiratory signs were observed. Viral shedding peaked on day 7 postinfection, and virus was still detectable in feces and in the ileum at day 21 postinfection, which may enhance the risk for viral transmission [12–14, 21, 24, 52].
Pathological changes are characterized by intestinal villous atrophy and shortening, and villous changes are associated with extensive intestinal epithelial degeneration and necrosis. Gross lesions are observed in the small intestines, and no significant lesions are observed in extraintestinal tissues except the lung. PDCoV infection can cause mild interstitial pneumonia in gnotobiotic piglets [12], which has not been reported for PEDV or TGEV.
Vaccine and control strategies
Vaccines remain the most effective means to control coronavirus infections. However, there are no effective vaccines available for PDCoV. Strategies for PDCoV vaccine development include inactivated virus vaccines, subunit vaccines, viral vector vaccines, and live-attenuated virus vaccines, each of which has both advantages and disadvantages. Multiple routes of vaccine research are being evaluated for PDCoV and other enteric coronaviruses.
Inactivated virus vaccines
Inactivated virus vaccines use chemicals or radiation to render the virus noninfectious while preserving its antigenicity. The most recent PDCoV vaccine was developed by the State Key Laboratory of Veterinary Biotechnology in China. The vaccine is based on inactivated virus formulated with an adjuvant. When administered to seronegative sows using a prime/boost strategy 20 and 40 days before delivery, high levels of spike (S)-specific IgG and neutralizing antibody against PDCoV were found in colostrum and milk, as well as in the serum of piglets born to vaccinated sows [12]. Piglets were infected orally at 5 days of life with 105 TCID50 of PDCoV. The experiment showed that 87.1% of all piglets (n = 31) born to immunized sows were protected against lethal infection, and the infected piglets showed milder diarrhea, less viral shedding, and only minor damage to intestinal villi. In contrast, piglets from unimmunized sows had moderate diarrhea, which quickly worsened at 2 days postinfection and remained severe until the end of the experiment.
Live-attenuated vaccines
Nonreplicating vaccines (inactivated vaccines, subunit vaccines) usually generate short-lived neutralizing antibody responses with comparatively low titers. In contrast, live-attenuated vaccines are generally more immunogenic than nonreplicating vaccines; they can induce long-lasting immunity, produce a comprehensive spectrum of native viral antigens, and present antigens to the immune system in the same manner as in natural infections.
Zhang et al. [99] generated a full-length infectious cDNA clone of PDCoV, which they manipulated by replacing the NS6 gene with a green fluorescent protein (GFP) to generate rPDCoV-ΔNS6-GFP. Growth kinetics studies suggested that rPDCoV-ΔNS6-GFP showed a substantial reduction in viral replication in cell cultures and was highly attenuated in neonatal piglets, indicating that PDCoV lacking NS6 might be an ideal live-attenuated vaccine candidate.
Generally, live-attenuated virus vaccines are promising candidates for use against coronavirus infections, but they also have decreased safety and stability compared to inactivated vaccines, and some live-attenuated virus vaccines have the potential to spontaneously revert to virulence post-vaccination.
Vectored vaccines
Vectored vaccines function as viral gene delivery systems that rely on a host viral genome from a different virus, such as adenovirus, poxvirus, measles virus, parainfluenza virus, rabies virus, or vesicular stomatitis virus, and they have been used in the development of vaccines for CoVs [100–106].
Porcine adenovirus was used to deliver the core neutralizing epitope of PEDV, and this resulted in robust humoral and mucosal immune responses in piglets [100]. A recombinant vesicular stomatitis virus expressing the PEDV spike protein was developed, and sows immunized with this recombinant vaccine provided protective lactogenic immunity against a virulent G2b PEDV challenge to their piglets [104]. In addition, Yuan et al. used swinepox virus to express an epitope of the S protein of TGEV and a truncated spike protein of PEDV [101, 102].
Virus-like particles (VLPs)
Virus-like particles (VLPs) have drawn increasing attention in recent years. VLPs containing one or more viral structural proteins structurally resemble the native virus, can be easily recognized by antigen-presenting cells and B cells, and are capable of eliciting robust humoral and cell-mediated immune responses that are comparable to those achieved with inactivated or live-attenuated virus vaccines. There have been few reports about PDCoV VLPs, but studies of other animal coronavirus VLPs can provide a reference for PDCoV vaccine research.
Wang et al. [107] produced PEDV virus-like particles (VLPs) composed of S, M, and E proteins using a baculovirus expression system and showed that they induced a high level of anti-PEDV-neutralizing antibodies in mice. Xu et al. [108] developed chimeric IBV VLPs expressing M, E, and a recombinant S protein in baculoviruses. These induced a high level of IBV-specific antibodies and neutralizing antibodies that were comparable to those induced by an inactivated M41 virus via subcutaneous inoculation.
Moreover, other vaccine approaches for expressing the S, E, M, and N genes of two or more coronaviruses as well as other viral genes in bacteria, yeast, plants, and nanoparticles have been assessed [109–116]. However, the efficacy of these VLPs against lethal infection has not been tested in piglets, and further studies need to be performed.
Currently, most commercial vaccines for enteric coronaviruses are designed to induce lactogenic immunity by vaccinating the sow during pregnancy, and antibodies are passively transferred from sows to neonatal piglets via colostrum and milk.
Coronavirus infections are generally initiated at mucosal surfaces, and it is critical to induce localized intestinal sIgA and T cell immune responses to mucosal infections. For maternal immunity, oral vaccines or intentional infection of the sow may initiate the gut-mammary sIgA axis [117, 118].
A previous study showed that oral inoculation of sows with attenuated TGEV, followed by intramuscular injection with a recombinant subunit vaccine expressing the S protein of TGEV as a booster generated high titers of sIgA antibodies and neutralizing antibodies in colostrum and milk [119]. Similar prime/boost strategies can be applied to PDCoV vaccines to induce active immunity in newborn piglets.
Coronaviruses are an important group of pathogens that can have a devastating impact on humans and animals. New zoonotic coronaviruses are continually emerging or reemerging. In addition to good production management and strict biosecurity measures, the most effective way to control PDCoV is vaccination. Consequently, new vaccine development platforms and technologies are highly desirable, and further research will provide a better understanding of PDCoV replication and pathogenesis, a prerequisite for the development of new and promising vaccines to prevent, control, and ultimately eliminate the virus.
Acknowledgments
This work was supported by the State National Key Laboratory of Animal Genetic Engineering Vaccine (grant no. AGVSKL-ZD-202007) and Leading Talents in Science and Technology Innovation of Zhongyuan Thousand Talents Project in Henan Province (grant no. 204200510012).
Author contributions
All authors contributed to the study conception and design. The idea and original draft preparation came from Pan Tang and Enhui Cui. Shujuan Wang and Congcong Lei contributed to the literature search and data analysis. Ruoqian Yan and Jingyu Wang provided critical review and substantially revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the State National Key Laboratory of Animal Genetic Engineering Vaccine (grant no. AGVSKL-ZD-202007), and Leading Talents in Science and Technology Innovation of Zhongyuan Thousand Talents Project in Henan Province (grant no. 204200510012).
Availability of data and material
All data generated or analysed during this review are included in this published article.
Declarations
Conflict of interest
No conflict of interest exits in the submission of this manuscript, and the manuscript was approved by all authors for publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ruoqian Yan, Email: yrq1688@126.com.
Jingyu Wang, Email: nwsuaf4409@126.com.
References
- 1.De Groot R, Ziebuhr J, Poon L (2008) Revision of the family Coronaviridae
- 2.Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung K, Hu H, Saif LJ. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch Virol. 2017;162(8):2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vlasova AN, Wang Q, Jung K, Langel SN, Malik YS, Saif LJ. Porcine coronaviruses. In: Malik YS, Singh RK, Yadav MP, editors. Emerging and transboundary animal viruses livestock diseases and management. Singapore: Springer; 2020. pp. 79–110. [Google Scholar]
- 5.Boley PA, Alhamo MA, Lossie G, Yadav KK, Vasquez-Lee M, Saif LJ, Kenney SP. Porcine deltacoronavirus infection and transmission in poultry, United States. Emerg Infect Dis. 2020;26(2):255–265. doi: 10.3201/eid2602.190346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilgenfeld R, Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 2013;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chafekar A, Fielding BC (2018) MERS-CoV: understanding the latest human coronavirus threat. Viruses 10(2). 10.3390/v10020093 [DOI] [PMC free article] [PubMed]
- 9.Giwa AL, Desai A, Duca A. Novel 2019 coronavirus SARS-CoV-2 (COVID-19): an updated overview for emergency clinicians. Emerg Med Pract. 2020;22(5):1–28. [PubMed] [Google Scholar]
- 10.Ioannidis JPA (2021) Infection fatality rate of COVID-19 inferred from seroprevalence data. 99:19-33F. WHO, Bulletin of the World Health Organization. 10.2471/BLT.20.265892 [DOI] [PMC free article] [PubMed]
- 11.Xu Z, Zhong H, Zhou Q, Du Y, Chen L, Zhang Y, Xue C, Cao Y. A highly pathogenic strain of porcine deltacoronavirus caused watery diarrhea in newborn piglets. Virol Sin. 2018;33(2):131–141. doi: 10.1007/s12250-018-0003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Chen J, Liu Y, Da S, Shi H, Zhang X, Liu J, Cao L, Zhu X, Wang X, Ji Z, Feng L. Pathogenicity of porcine deltacoronavirus (PDCoV) strain NH and immunization of pregnant sows with an inactivated PDCoV vaccine protects 5-day-old neonatal piglets from virulent challenge. Transbound Emerg Dis. 2020;67(2):572–583. doi: 10.1111/tbed.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitosh-Sillman S, Loy JD, Brodersen B, Kelling C, Doster A, Topliff C, Nelson E, Bai J, Schirtzinger E, Poulsen E, Meadors B, Anderson J, Hause B, Anderson G, Hesse R. Experimental infection of conventional nursing pigs and their dams with porcine deltacoronavirus. J Vet Diagn Invest. 2016;28(5):486–497. doi: 10.1177/1040638716654200. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Gauger P, Stafne M, Thomas J, Arruda P, Burrough E, Madson D, Brodie J, Magstadt D, Derscheid R, Welch M, Zhang J. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology. 2015;482:51–59. doi: 10.1016/j.virol.2015.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, Zheng L, Li H, Ding Q, Wang Y, Wei Z (2019) Porcine deltacoronavirus causes diarrhea in various ages of field-infected pigs in China. Biosci Rep 39(9). 10.1042/BSR20190676 [DOI] [PMC free article] [PubMed]
- 16.Wang L, Byrum B. Zhang Y (2014) Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA. Emerg Infect Dis. 2014;20(7):1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S, Lee C (2014) Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc 2(6). 10.1128/genomeA.01191-14 [DOI] [PMC free article] [PubMed]
- 18.Dong N, Fang L, Zeng S, Sun Q, Chen H, Xiao S. Porcine deltacoronavirus in mainland China. Emerg Infect Dis. 2015;21(12):2254–2255. doi: 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janetanakit T, Lumyai M, Bunpapong N, Boonyapisitsopa S, Chaiyawong S, Nonthabenjawan N, Kesdaengsakonwut S. Amonsin A (2016) porcine deltacoronavirus, Thailand. Emerg Infect Dis. 2015;22(4):757–759. doi: 10.3201/eid2204.151852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, Tantituvanont A, Kaewprommal P, Piriyapongsa J, Nilubol D. Different lineage of porcine deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis. 2017;64(1):3–10. doi: 10.1111/tbed.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Qu H, Hu J, Fu J, Chen R, Li C, Cao S, Wen Y, Wu R, Zhao Q, Yan Q, Wen X, Huang X. Characterization and pathogenicity of the porcine deltacoronavirus isolated in Southwest China. Viruses. 2019;11(11):1074. doi: 10.3390/v11111074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia L, Yang Y, Wang J, Jing Y, Yang Q. Impact of TGEV infection on the pig small intestine. Virol J. 2018;15(1):102. doi: 10.1186/s12985-018-1012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, Xiao S. Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol. 2016;196:98–106. doi: 10.1016/j.vetmic.2016.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J (2015) Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 6(2):e00064. 10.1128/mBio.00064-15 [DOI] [PMC free article] [PubMed]
- 25.Suzuki T, Shibahara T, Imai N, Yamamoto T, Ohashi S. Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect Genet Evol. 2018;61:176–182. doi: 10.1016/j.meegid.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saeng-Chuto K, Jermsutjarit P, Stott CJ, Vui DT, Tantituvanont A, Nilubol D. Retrospective study, full-length genome characterization and evaluation of viral infectivity and pathogenicity of chimeric porcine deltacoronavirus detected in Vietnam. Transbound Emerg Dis. 2020;67(1):183–198. doi: 10.1111/tbed.13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis. 2015;62(6):575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masters PS (2006) the molecular biology of coronaviruses. In: Advances in virus research, pp 193–292. 10.1016/s0065-3527(06)66005-3 [DOI] [PMC free article] [PubMed]
- 29.Woo PC, Huang Y, Lau SK, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen F, Zhu Y, Wu M, Ku X, Yao L, He Q (2015) Full-length genome characterization of chinese porcine deltacoronavirus strain CH/SXD1/2015. Genome Announc 3(5). 10.1128/genomeA.01284-15 [DOI] [PMC free article] [PubMed]
- 31.Zhang J, Chen J, Shi D, Shi H, Zhang X, Liu J, Cao L, Zhu X, Liu Y, Wang X, Ji Z, Feng L. Porcine deltacoronavirus enters cells via two pathways: a protease-mediated one at the cell surface and another facilitated by cathepsins in the endosome. J Biol Chem. 2019;294(25):9830–9843. doi: 10.1074/jbc.RA119.007779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X, Liu S, Wang X, Luo Z, Shi Y, Wang D, Peng G, Chen H, Fang L, Xiao S. Contribution of porcine aminopeptidase N to porcine deltacoronavirus infection. Emerg Microbes Infect. 2018;7(1):65. doi: 10.1038/s41426-018-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen R, Fu J, Hu J, Li C, Zhao Y, Qu H, Wen X, Cao S, Wen Y, Wu R, Zhao Q, Yan Q, Huang Y, Ma X, Han X, Huang X. Identification of the immunodominant neutralizing regions in the spike glycoprotein of porcine deltacoronavirus. Virus Res. 2020;276:197834. doi: 10.1016/j.virusres.2019.197834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang J, Zheng Y, Yang Y, Liu C, Geng Q, Tai W, Du L, Zhou Y, Zhang W, Li F (2018) Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J Virol 92(4). 10.1128/JVI.01556-17 [DOI] [PMC free article] [PubMed]
- 35.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. 2019;16(1):69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015;208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Likai J, Shasha L, Wenxian Z, Jingjiao M, Jianhe S, Hengan W, Yaxian Y. Porcine deltacoronavirus nucleocapsid protein suppressed IFN-beta production by interfering porcine RIG-I dsRNA-binding and K63-linked polyubiquitination. Front Immunol. 2019;10:1024. doi: 10.3389/fimmu.2019.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang P, Fang L, Liu X, Hong Y, Wang Y, Dong N, Ma P, Bi J, Wang D, Xiao S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology. 2016;499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi S, Lee C. Functional characterization and proteomic analysis of porcine deltacoronavirus accessory protein NS7. J Microbiol Biotechnol. 2019;29(11):1817–1829. doi: 10.4014/jmb.1908.08013. [DOI] [PubMed] [Google Scholar]
- 42.Fang P, Fang L, Hong Y, Liu X, Dong N, Ma P, Bi J, Wang D, Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J Gen Virol. 2017;98(2):173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trudeau MP, Verma H, Sampedro F, Urriola PE, Shurson GC, Goyal SM. Environmental persistence of porcine coronaviruses in feed and feed ingredients. PLoS One. 2017;12(5):e0178094. doi: 10.1371/journal.pone.0178094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cottingim KM, Verma H, Urriola PE, Sampedro F, Shurson GC, Goyal SM. Feed additives decrease survival of delta coronavirus in nursery pig diets. Porcine Health Manage. 2017;3:5. doi: 10.1186/s40813-016-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trudeau MP, Verma H, Sampedro F, Urriola PE, Shurson GC, McKelvey J, Pillai SD, Goyal SM. Comparison of thermal and non-thermal processing of swine feed and the use of selected feed additives on inactivation of porcine epidemic diarrhea virus (PEDV) PLoS One. 2016;11(6):e0158128. doi: 10.1371/journal.pone.0158128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Environmental stability of PEDv (2014)
- 47.Quist-Rybachuk GV, Nauwynck HJ, Kalmar ID. Sensitivity of porcine epidemic diarrhea virus (PEDV) to pH and heat treatment in the presence or absence of porcine plasma. Vet Microbiol. 2015;181(3–4):283–288. doi: 10.1016/j.vetmic.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Zheng L, Li X, Yan M, Ren W, Zhang L, Lu C, Tian X, Han W. Isolation, identification and biological characteristics analysis of porcine deltacoronavirus TJ1, China. Anim Husbandry Vet Med. 2017;45(1):219–224. [Google Scholar]
- 49.Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84(7):3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skowronski DM, Astell C, Brunham RC, Low DE, Petric M, Roper RL, Talbot PJ, Tam T, Babiuk L. Severe acute respiratory syndrome (SARS): a year in review. Annu Rev Med. 2005;56:357–381. doi: 10.1146/annurev.med.56.091103.134135. [DOI] [PubMed] [Google Scholar]
- 52.Jung K, Hu H, Eyerly B, Lu Z, Chepngeno J, Saif LJ. Pathogenicity of 2 porcine deltacoronavirus strains in gnotobiotic pigs. Emerg Infect Dis. 2015;21(4):650–654. doi: 10.3201/eid2104.141859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torres CA, Hora AS, Tonietti PO, Taniwaki SA, Cecchinato M, Villarreal LY, Brandao PE. Gammacoronavirus and deltacoronavirus in quail. Avian Dis. 2016;60(3):656–661. doi: 10.1637/11412-032316-Reg.1. [DOI] [PubMed] [Google Scholar]
- 54.Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K (2019) bats and coronaviruses. Viruses 11(1). 10.3390/v11010041 [DOI] [PMC free article] [PubMed]
- 55.Sowman HR, Cave NJ, Dunowska M. A survey of canine respiratory pathogens in New Zealand dogs. N Z Vet J. 2018;66(5):236–242. doi: 10.1080/00480169.2018.1490214. [DOI] [PubMed] [Google Scholar]
- 56.Liang Q, Zhang H, Li B, Ding Q, Wang Y, Gao W, Guo D, Wei Z, Hu H (2019) Susceptibility of chickens to porcine deltacoronavirus infection. Viruses 11(6). 10.3390/v11060573 [DOI] [PMC free article] [PubMed]
- 57.Cruz-Pulido D, Boley PA, Ouma WZ, Alhamo MA, Saif LJ, Kenney SP (2021) Comparative transcriptome profiling of human and pig intestinal epithelial cells after porcine deltacoronavirus infection. Viruses 13(2). 10.3390/v13020292 [DOI] [PMC free article] [PubMed]
- 58.Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA. 2018;115(22):E5135–E5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JH, Chung HC, Nguyen VG, Moon HJ, Kim HK, Park SJ, Lee CH, Lee GE, Park BK. Detection and phylogenetic analysis of porcine deltacoronavirus in Korean swine farms, 2015. Transbound Emerg Dis. 2016;63(3):248–252. doi: 10.1111/tbed.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Rivera C, Ramirez-Mendoza H, Mendoza-Elvira S, Segura-Velazquez R, Sanchez-Betancourt JI. First report and phylogenetic analysis of porcine deltacoronavirus in Mexico. Transbound Emerg Dis. 2019;66(4):1436–1441. doi: 10.1111/tbed.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ajayi T, Dara R, Misener M, Pasma T, Moser L, Poljak Z. Herd-level prevalence and incidence of porcine epidemic diarrhoea virus (PEDV) and porcine deltacoronavirus (PDCoV) in swine herds in Ontario, Canada. Transbound Emerg Dis. 2018;65(5):1197–1207. doi: 10.1111/tbed.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang M, Wang Y, Baloch AR, Pan Y, Tian L, Xu F, Shivaramu S, Chen S, Zeng Q. Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai-Tibet Plateau of China. Transbound Emerg Dis. 2018;65(2):363–369. doi: 10.1111/tbed.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Cheng Y, Xing G, Yu J, Liao A, Du L, Lei J, Lian X, Zhou J, Gu J. Detection and spike gene characterization in porcine deltacoronavirus in China during 2016–2018. Infect Genet Evol. 2019;73:151–158. doi: 10.1016/j.meegid.2019.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo SX, Fan JH, Opriessnig T, Di JM, Liu BJ, Zuo YZ. Development and application of a recombinant M protein-based indirect ELISA for the detection of porcine deltacoronavirus IgG antibodies. J Virol Methods. 2017;249:76–78. doi: 10.1016/j.jviromet.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su M, Li C, Guo D, Wei S, Wang X, Geng Y, Yao S, Gao J, Wang E, Zhao X, Wang Z, Wang J, Wu R, Feng L, Sun D. A recombinant nucleocapsid protein-based indirect enzyme-linked immunosorbent assay to detect antibodies against porcine deltacoronavirus. J Vet Med Sci. 2016;78(4):601–606. doi: 10.1292/jvms.15-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ma L, Zeng F, Huang B, Cong F, Huang R, Ma J, Guo P. Development of a conventional RT-PCR assay for rapid detection of porcine deltacoronavirus with the same detection limit as a SYBR green-based real-time RT-PCR assay. Biomed Res Int. 2018;2018:5035139. doi: 10.1155/2018/5035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding G, Fu Y, Li B, Chen J, Wang J, Yin B, Sha W, Liu G. Development of a multiplex RT-PCR for the detection of major diarrhoeal viruses in pig herds in China. Transbound Emerg Dis. 2020;67(2):678–685. doi: 10.1111/tbed.13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jia S, Feng B, Wang Z, Ma Y, Gao X, Jiang Y, Cui W, Qiao X, Tang L, Li Y, Wang L, Xu Y. Dual priming oligonucleotide (DPO)-based real-time RT-PCR assay for accurate differentiation of four major viruses causing porcine viral diarrhea. Mol Cell Probes. 2019;47:101435. doi: 10.1016/j.mcp.2019.101435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li D, Feng H, Liu Y, Chen Y, Wei Q, Wang J, Liu D, Huang H, Su Y, Wang D, Cui Y, Zhang G. Molecular evolution of porcine epidemic diarrhea virus and porcine deltacoronavirus strains in Central China. Res Vet Sci. 2018;120:63–69. doi: 10.1016/j.rvsc.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhai SL, Wei WK, Li XP, Wen XH, Zhou X, Zhang H, Lv DH, Li F, Wang D. Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol J. 2016;13:136. doi: 10.1186/s12985-016-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang F, Luo S, Gu J, Li Z, Li K, Yuan W, Ye Y, Li H, Ding Z, Song D, Tang Y. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet Res. 2019;15(1):470. doi: 10.1186/s12917-019-2212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang H, Liang Q, Li B, Cui X, Wei X, Ding Q, Wang Y, Hu H. Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan province, China. Prev Vet Med. 2019;166:8–15. doi: 10.1016/j.prevetmed.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mai K, Feng J, Chen G, Li D, Zhou L, Bai Y, Wu Q, Ma J. The detection and phylogenetic analysis of porcine deltacoronavirus from Guangdong Province in Southern China. Transbound Emerg Dis. 2018;65(1):166–173. doi: 10.1111/tbed.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang F, Song D, Zhou X, Huang D, Li A, Peng Q, Chen Y, Wu Q, He H, Tang Y. Establishment and application of a RT-PCR assay for detection of newly emerged porcine deltacoronavirus. Sci Agric Sin. 2016;49(7):1406–1416. [Google Scholar]
- 75.Huang X, Chen J, Yao G, Guo Q, Wang J, Liu G. A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses. Appl Microbiol Biotechnol. 2019;103(12):4943–4952. doi: 10.1007/s00253-019-09835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Otter JA, Donskey C, Yezli S, Douthwaite S, Goldenberg SD, Weber DJ. Transmission of SARS and MERS coronaviruses and influenza virus in healthcare settings: the possible role of dry surface contamination. J Hosp Infect. 2016;92(3):235–250. doi: 10.1016/j.jhin.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ding Y, He L, Zhang Q, Huang Z, Che X, Hou J, Wang H, Shen H, Qiu L, Li Z, Geng J, Cai J, Han H, Li X, Kang W, Weng D, Liang P, Jiang S. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J Pathol. 2004;203(2):622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fan C, Lei D, Fang C, Li C, Wang M, Liu Y, Bao Y, Sun Y, Huang J, Guo Y, Yu Y, Wang S. Perinatal transmission of COVID-19 associated SARS-CoV-2: should we worry? Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alonso C, Goede DP, Morrison RB, Davies PR, Rovira A, Marthaler DG, Torremorell M. Evidence of infectivity of airborne porcine epidemic diarrhea virus and detection of airborne viral RNA at long distances from infected herds. Vet Res. 2014;45:73. doi: 10.1186/s13567-014-0073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lowe J, Gauger P, Harmon K, Zhang J, Connor J, Yeske P, Loula T, Levis I, Dufresne L, Main R. Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerg Infect Dis. 2014;20(5):872–874. doi: 10.3201/eid2005.131628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marthaler D, Raymond L, Jiang Y, Collins J, Rossow K, Rovira A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg Infect Dis. 2014;20(8):1347–1350. doi: 10.3201/eid2008.140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu H, Jung K, Vlasova AN, Saif LJ. Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine deltacoronavirus strain OH-FD22. Arch Virol. 2016;161(12):3421–3434. doi: 10.1007/s00705-016-3056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greiner L. Evaluation of the likelihood of detection of porcine epidemic diarrhea virus or porcine delta coronavirus ribonucleic acid in areas within feed mills. J Swine Health Prod. 2016;24(4):198–204. [Google Scholar]
- 84.Lam TT, Shum MH, Zhu HC, Tong YG, Ni XB, Liao YS, Wei W, Cheung WY, Li WJ, Li LF, Leung GM, Holmes EC, Hu YL, Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 85.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, Xie JZ, Shen XR, Zhang YZ, Wang N, Luo DS, Zheng XS, Wang MN, Daszak P, Wang LF, Cui J, Shi ZL. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13(11):e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Simas PV, Barnabe AC, Duraes-Carvalho R, Neto DF, Caserta LC, Artacho L, Jacomassa FA, Martini MC, Bianchi Dos Santos MM, Felippe PA, Ferreira HL, Arns CW. Bat coronavirus in Brazil related to appalachian ridge and porcine epidemic diarrhea viruses. Emerg Infect Dis. 2015;21(4):729–731. doi: 10.3201/eid2104.141783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 2018;556(7700):255–258. doi: 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woo PC, Lau SK, Lam CS, Lai KK, Huang Y, Lee P, Luk GS, Dyrting KC, Chan KH, Yuen KY. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J Virol. 2009;83(2):908–917. doi: 10.1128/JVI.01977-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang L, Su S, Bi Y, Wong G, Gao GF. Bat-origin coronaviruses expand their host range to pigs. Trends Microbiol. 2018;26(6):466–470. doi: 10.1016/j.tim.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao S, Li W, Schuurman N, van Kuppeveld F, Bosch BJ, Egberink H (2019) Serological screening for coronavirus infections in cats. Viruses 11(8). 10.3390/v11080743 [DOI] [PMC free article] [PubMed]
- 92.Wang D, Fang L, Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo R, Fan B, Chang X, Zhou J, Zhao Y, Shi D, Yu Z, He K, Li B. Characterization and evaluation of the pathogenicity of a natural recombinant transmissible gastroenteritis virus in China. Virology. 2020;545:24–32. doi: 10.1016/j.virol.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38(2):281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 95.Gomez DE, Arroyo LG, Poljak Z, Viel L, Weese JS. Detection of bovine coronavirus in healthy and diarrheic dairy calves. J Vet Intern Med. 2017;31(6):1884–1891. doi: 10.1111/jvim.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haick AK, Rzepka JP, Brandon E, Balemba OB, Miura TA. Neutrophils are needed for an effective immune response against pulmonary rat coronavirus infection, but also contribute to pathology. J Gen Virol. 2014;95(Pt 3):578–590. doi: 10.1099/vir.0.061986-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tekes G, Thiel HJ. Feline coronaviruses: pathogenesis of feline infectious peritonitis. Adv Virus Res. 2016;96:193–218. doi: 10.1016/bs.aivir.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Curry SM, Gibson KA, Burrough ER, Schwartz KJ, Yoon KJ, Gabler NK. Nursery pig growth performance and tissue accretion modulation due to porcine epidemic diarrhea virus or porcine deltacoronavirus challenge. J Anim Sci. 2017;95(1):173–181. doi: 10.2527/jas.2016.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang M, Li W, Zhou P, Liu D, Luo R, Jongkaewwattana A, He Q. Genetic manipulation of porcine deltacoronavirus reveals insights into NS6 and NS7 functions: a novel strategy for vaccine design. Emerg Microbes Infect. 2020;9(1):20–31. doi: 10.1080/22221751.2019.1701391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Do VT, Jang J, Park J, Dao HT, Kim K, Hahn TW. Recombinant adenovirus carrying a core neutralizing epitope of porcine epidemic diarrhea virus and heat-labile enterotoxin B of Escherichia coli as a mucosal vaccine. Arch Virol. 2020;165(3):609–618. doi: 10.1007/s00705-019-04492-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yuan X, Lin H, Li B, He K, Fan H. Efficacy and immunogenicity of recombinant swinepox virus expressing the truncated S protein of a novel isolate of porcine epidemic diarrhea virus. Arch Virol. 2017;162(12):3779–3789. doi: 10.1007/s00705-017-3548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yuan X, Lin H, Fan H. Efficacy and immunogenicity of recombinant swinepox virus expressing the A epitope of the TGEV S protein. Vaccine. 2015;33(32):3900–3906. doi: 10.1016/j.vaccine.2015.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liniger M, Zuniga A, Tamin A, Azzouz-Morin TN, Knuchel M, Marty RR, Wiegand M, Weibel S, Kelvin D, Rota PA, Naim HY. Induction of neutralising antibodies and cellular immune responses against SARS coronavirus by recombinant measles viruses. Vaccine. 2008;26(17):2164–2174. doi: 10.1016/j.vaccine.2008.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ke Y, Yu D, Zhang F, Gao J, Wang X, Fang X, Wang H, Sun T. Recombinant vesicular stomatitis virus expressing the spike protein of genotype 2b porcine epidemic diarrhea virus: a platform for vaccine development against emerging epidemic isolates. Virology. 2019;533:77–85. doi: 10.1016/j.virol.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bukreyev A, Lamirande EW, Buchholz UJ, Vogel LN, Elkins WR, St Claire M, Murphy BR, Subbarao K, Collins PL. Mucosal immunisation of African green monkeys (Cercopithecus aethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363(9427):2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kato H, Takayama-Ito M, Iizuka-Shiota I, Fukushi S, Posadas-Herrera G, Horiya M, Satoh M, Yoshikawa T, Yamada S, Harada S, Fujii H, Shibamura M, Inagaki T, Morimoto K, Saijo M, Lim CK. Development of a recombinant replication-deficient rabies virus-based bivalent-vaccine against MERS-CoV and rabies virus and its humoral immunogenicity in mice. PLoS One. 2019;14(10):e0223684. doi: 10.1371/journal.pone.0223684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang C, Yan F, Zheng X, Wang H, Jin H, Wang C, Zhao Y, Feng N, Wang T, Gao Y, Yang S, Xia X. Porcine epidemic diarrhea virus virus-like particles produced in insect cells induce specific immune responses in mice. Virus Genes. 2017;53(4):548–554. doi: 10.1007/s11262-017-1450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu PW, Wu X, Wang HN, Ma BC, Ding MD, Yang X. Assembly and immunogenicity of baculovirus-derived infectious bronchitis virus-like particles carrying membrane, envelope and the recombinant spike proteins. Biotechnol Lett. 2016;38(2):299–304. doi: 10.1007/s10529-015-1973-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen WH, Chag SM, Poongavanam MV, Biter AB, Ewere EA, Rezende W, Seid CA, Hudspeth EM, Pollet J, McAtee CP, Strych U, Bottazzi ME, Hotez PJ. Optimization of the production process and characterization of the yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1), a SARS vaccine candidate. J Pharm Sci. 2017;106(8):1961–1970. doi: 10.1016/j.xphs.2017.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jiang X, Hou X, Tang L, Jiang Y, Ma G, Li Y. A phase trial of the oral Lactobacillus casei vaccine polarizes Th2 cell immunity against transmissible gastroenteritis coronavirus infection. Appl Microbiol Biotechnol. 2016;100(17):7457–7469. doi: 10.1007/s00253-016-7424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.LeCureux JS, Dean GA (2018) Lactobacillus mucosal vaccine vectors: immune responses against bacterial and viral antigens. mSphere 3(3). 10.1128/mSphere.00061-18 [DOI] [PMC free article] [PubMed]
- 112.Ma F, Zhang E, Li Q, Xu Q, Ou J, Yin H, Li K, Wang L, Zhao X, Niu X, Li X, Zhang S, Wang Y, Deng R, Zhou E, Zhang G (2020) A plant-produced recombinant fusion protein-based newcastle disease subunit vaccine and rapid differential diagnosis platform. Vaccines (Basel) 8(1). 10.3390/vaccines8010122 [DOI] [PMC free article] [PubMed]
- 113.Michon C, Langella P, Eijsink VG, Mathiesen G, Chatel JM. Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb Cell Fact. 2016;15:70. doi: 10.1186/s12934-016-0468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sirichokchatchawan W, Temeeyasen G, Nilubol D, Prapasarakul N. Protective effects of cell-free supernatant and live lactic acid bacteria isolated from Thai pigs against a pandemic strain of porcine epidemic diarrhea virus. Probiotics Antimicrob Proteins. 2018;10(2):383–390. doi: 10.1007/s12602-017-9281-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Szatraj K, Szczepankowska AK, Chmielewska-Jeznach M. Lactic acid bacteria—promising vaccine vectors: possibilities, limitations, doubts. J Appl Microbiol. 2017;123(2):325–339. doi: 10.1111/jam.13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang X, Wang Z, Xu H, Xiang B, Dang R, Yang Z. Orally administrated whole yeast vaccine against porcine epidemic diarrhea virus induced high levels of IgA response in mice and piglets. Viral Immunol. 2016;29(9):526–531. doi: 10.1089/vim.2016.0067. [DOI] [PubMed] [Google Scholar]
- 117.Chattha KS, Roth JA, Saif LJ. Strategies for design and application of enteric viral vaccines. Annu Rev Anim Biosci. 2015;3:375–395. doi: 10.1146/annurev-animal-022114-111038. [DOI] [PubMed] [Google Scholar]
- 118.Gerdts V, Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet Microbiol. 2017;206:45–51. doi: 10.1016/j.vetmic.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Park S, Sestak K, Hodgins DC, Shoup DI, Ward LA, Jackwood DJ, Saif LJ. Immune response of sows vaccinated with attenuated transmissible gastroenteritis virus (TGEV) and recombinant TGEV spike protein vaccines and protection of their suckling pigs against virulent TGEV challenge exposure. Am J Vet Res. 1998;59(8):1002–1008. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this review are included in this published article.