Abstract
Grain size and weight are the key traits determining rice quality and yield and are mainly controlled by quantitative trait loci (QTL). In this study, one minor QTL that was previously mapped in the marker interval of JD1009-JD1019 using the Huanghuazhan/Jizi1560 (HHZ/JZ1560) recombinant inbred line (RIL) population, qTGW1-2, was validated to regulate grain size and weight across four rice-growing seasons using twenty-one near isogenic line (NIL)-F2 populations. The twenty-one populations were in two types of genetic background that were derived from the same parents HHZ and JZ1560. Twelve F9, F10 or F11 NIL-F2 populations with the sequential residual heterozygous regions covering JD1009-RM6840 were developed from one residual heterozygote (RH) in the HHZ/JZ1560 RIL population, and the remaining nine BC3F3, BC3F4 or BC3F5 NIL-F2 populations with the sequential residual heterozygous regions covering JD1009-RM6840 were constructed through consecutive backcrosses to the recurrent parent HHZ followed with marker assistant selection in each generation. Based on the QTL analysis of these genetic populations, qTGW1-2 was successfully confirmed to control grain length, width and weight and further dissected into two QTLs, qTGW1-2a and qTGW1-2b, which were respectively narrowed down to the marker intervals of JD1139-JD1127 (~ 978.2-kb) and JD1121-JD1102 (~ 54.8-kb). Furthermore, the two types of NIL-F2 populations were proved to be able to decrease the genetic background noise and increase the detection power of minor QTL. These results provided an important basis for further map-based cloning and molecular design breeding with the two QTLs in rice.
Subject terms: Genetics, Plant sciences
Rice (Oryza sativa L.) is one of the three major food crops, and feeds more than half of the consumers as their staple food in the world. Number of panicles per plant, number of grains per panicle, and grain weight are the three determined components of grain yield, among which grain weight is the direct factor to determine grain yield. Grain weight is generally evaluated by 1000-grain weight (TGW) and is closely related to grain length (GL), grain width (GW), grain thickness (GT) and length width ratio (LWR), which are comprehensively controlled by a large number of quantitative trait loci (QTL)1,2. GL and GW are stably inheritable traits that are less affected by environments, whereas GT and grain plumpness are easily affected by environmental factors and dependent on the filling process. Therefore, most QTLs for grain size and weight are map-based cloned through identifying GL, GW and TGW3.
In the past two decades, fine-mapping and cloning for yield traits, especially for grain weight, have achieved considerable progress. Up to now, 20 QTLs for grain weight and grain size have been cloned. Among them, both GL74/ GW75 and GS96 have opposite allelic directions of additive effects on GL and GW, regulating grain size but hardly affecting grain weight. GSA17 and GW6a8 have similar effects on GL and GW with same directions so that they have great influence on grain weight. The remaining 16 QTLs affect grain size and grain weight at the same time. Six of the 16 QTLs mainly control grain width and weight, including GW29, TGW210, GS511, qSW512/GW513, GW614 and GW815, and the other ten QTLs mainly control grain length and weight, including GS216/GL217, OsLG318, qLGY319/OsLG3b20, GS32, GL3.121/qGL322, TGW323/GL3.324, GL425, TGW626, GL627 and GLW728. Isolation and functional characterization of these QTLs have greatly enhanced our understanding of genetic control of grain size and weight in rice, but more investigations are needed to enrich the regulatory framework for these key agronomic traits29.
Although 20 QTLs with major effects have been successfully isolated, majority of these QTLs for grain size and weight have minor effects and are difficult to be repeatedly identified in different trials. Therefore, increasing the detection power of minor QTLs is the key for fine mapping or map-based cloning of them. For detecting minor QTLs, near isogenic line F2 (NIL-F2) populations are the ideal materials to eliminate the genetic noise of background and increase the detection power largely. In the NIL-F2 population, the region of target QTL is segregated with fixed genetic background, which will enable to eliminate the background noise and precisely estimate the effect of minor QTLs. NIL-F2 population can be produced by consecutive backcrossing strategy or residual heterozygous line method30. Consecutive backcrossing strategy includes continuous backcrosses to the recurrent parent and marker assistant selection in each generation. This type of NIL-F2 population has been used in cloning many QTLs7,19,28,31. NIL-F2 populations generated by self-pollinating residual heterozygotes (RH) in the recombinant inbred line (RIL) population have validated and dissected many QTLs for grain size and weight32–34.
In our previous study, we identified one stably expressed QTL regulating GL and grain weight, qTGW1-2/qGL1-2, using the HHZ/JZ1560 RIL population across two years23. To validate and fine map qTGW1-2/qGL1-2, two types of NIL-F2 populations including RH-derived population and advanced backcross population were constructed using HHZ and JZ1560 (Fig. 1). HHZ is an indica rice variety widely cultivated in China with small grains, and JZ1560 is a japonica material with super large grains.
Figure 1.
Development of two types of near isogenic line F2 populations.
Results
Validation and dissection of qTGW1-2 using ten RH-derived NIL-F2 populations
Descriptive statistics of TGW, GL and GW in the ten RH-derived populations in F9 and F10 are shown in Table 1. Three traits in all the ten populations showed continuous distributions with low skewness and kurtosis, which suggested the typical phenotypic distributions of quantitative traits.
Table 1.
Phenotypic performance of TGW, GL and GW in ten RH-derived populations in F9 or F10. TGW, 1000-grain weight (g); GL, grain length (mm); GW, grain width (mm).
| Population | Trait | Mean | SD | CV | Range | Skew | Kurt |
|---|---|---|---|---|---|---|---|
| YC1-1 | TGW | 36.90 | 2.37 | 0.06 | 32.13–43.59 | 0.41 | 0.20 |
| GL | 11.34 | 0.15 | 0.01 | 11.08–11.67 | 0.07 | − 0.88 | |
| GW | 3.27 | 0.06 | 0.02 | 3.13–3.42 | − 0.07 | − 0.11 | |
| YC1-2 | TGW | 37.71 | 2.00 | 0.05 | 33.56–44.15 | 0.64 | 0.86 |
| GL | 11.33 | 0.15 | 0.01 | 10.98–11.66 | − 0.02 | − 0.47 | |
| GW | 3.27 | 0.06 | 0.02 | 3.11–3.46 | 0.04 | 1.17 | |
| YC1-3 | TGW | 44.98 | 1.62 | 0.04 | 41.55–49.62 | 0.39 | 0.05 |
| GL | 11.03 | 0.16 | 0.01 | 10.63–11.51 | 0.45 | 0.17 | |
| GW | 3.42 | 0.07 | 0.02 | 3.29–3.59 | 0.35 | − 0.52 | |
| YC2-1 | TGW | 36.72 | 1.20 | 0.03 | 32.14–40.53 | 0.49 | 0.23 |
| GL | 10.61 | 0.15 | 0.01 | 10.20–11.13 | 0.63 | 0.62 | |
| GW | 2.99 | 0.05 | 0.02 | 2.88–3.10 | − 0.07 | − 0.35 | |
| YC2-2 | TGW | 37.02 | 1.50 | 0.04 | 33.53–40.63 | 0.14 | − 0.29 |
| GL | 11.04 | 0.16 | 0.01 | 10.71–11.47 | 0.47 | − 0.42 | |
| GW | 3.06 | 0.07 | 0.02 | 2.84–3.24 | − 0.06 | 0.46 | |
| YC2-3 | TGW | 37.57 | 1.13 | 0.03 | 33.53–40.82 | 0.79 | 0.55 |
| GL | 10.67 | 0.17 | 0.02 | 10.23–11.19 | 0.43 | 0.95 | |
| GW | 3.10 | 0.04 | 0.01 | 3.00–3.21 | 0.24 | − 0.22 | |
| YC2-4 | TGW | 39.38 | 1.55 | 0.04 | 35.09–43.21 | 0.11 | − 0.33 |
| GL | 10.62 | 0.18 | 0.02 | 10.25–11.06 | 0.29 | − 0.51 | |
| GW | 3.13 | 0.06 | 0.02 | 2.90–3.30 | − 0.16 | 0.96 | |
| YC2-5 | TGW | 36.58 | 1.05 | 0.03 | 33.83–38.88 | 0.14 | − 0.35 |
| GL | 10.44 | 0.13 | 0.01 | 10.09–10.80 | 0.17 | − 0.21 | |
| GW | 3.05 | 0.04 | 0.01 | 2.92–3.16 | 0.08 | 0.08 | |
| YC2-6 | TGW | 36.81 | 1.19 | 0.03 | 33.98–39.99 | 0.27 | − 0.32 |
| GL | 10.56 | 0.14 | 0.01 | 10.26–10.96 | 0.43 | 0.04 | |
| GW | 3.04 | 0.04 | 0.01 | 2.94–3.16 | 0.08 | − 0.34 | |
| YC2-7 | TGW | 37.02 | 1.16 | 0.03 | 33.94–39.69 | 0.14 | − 0.48 |
| GL | 10.55 | 0.13 | 0.01 | 10.27–10.96 | 0.34 | 0.09 | |
| GW | 3.03 | 0.04 | 0.01 | 2.94–3.11 | − 0.11 | − 0.50 |
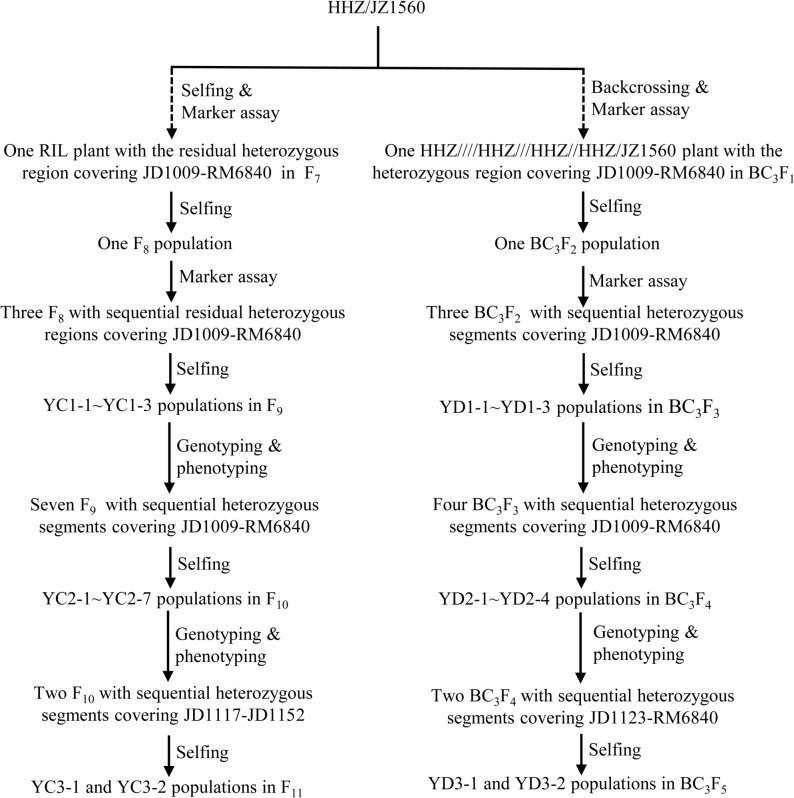
To confirm the location and genetic effect of qTGW1-2, three plants with sequential heterozygous regions covering JD1009-RM6840 were selected from one population in F8 and were self-crossed to generate three NIL-F2 populations named as YC1-1, YC1-2 and YC1-3 (Table 2; Fig. 1). Three segmental linkage maps were constructed for YC1-1, YC1-2 and YC1-3, respectively (Fig. 2a).
Table 2.
Rice populations and field experiments. HZ, Hangzhou, Zhejiang Province; LS, Lingshui, Hainan Province.
| Population | Segregating region | Number of plants | Location and growing season | ||
|---|---|---|---|---|---|
| Name | Generation | ||||
| RH-derived populations | |||||
| YC1-1 | F9 | JD1009-RM6840 | 120 | HZ: May–September 2018 | |
| YC1-2 | F9 | JD1009-JD1090 | 128 | HZ: May–September 2018 | |
| YC1-3 | F9 | JD1018-RM6840 | 224 | HZ: May–September 2018 | |
| YC2-1 | F10 | JD1009-JD1022 | 190 | HZ: May–September 2019 | |
| YC2-2 | F10 | JD1009-JD1062 | 190 | HZ: May–September 2019 | |
| YC2-3 | F10 | JD1140-JD1049 | 104 | HZ: May–September 2019 | |
| YC2-4 | F10 | JD1062-JD1052 | 190 | HZ: May–September 2019 | |
| YC2-5 | F10 | JD1018-JD1019 | 190 | HZ: May–September 2019 | |
| YC2-6 | F10 | JD1052-JD1090 | 190 | HZ: May–September 2019 | |
| YC2-7 | F10 | JD1062-RM6840 | 190 | HZ: May–September 2019 | |
| YC3-1 | F11 | JD1152-JD1136 | 190 | HZ: May–September 2020 | |
| YC3-2 | F11 | JD1136-JD1121 | 190 | HZ: May–September 2020 | |
| Backcross populations | |||||
| YD1-1 | BC3F3 | JD1009-JD1052 | 190 | HZ: May–September 2019 | |
| YD1-2 | BC3F3 | JD1009-JD1090 | 190 | HZ: May–September 2019 | |
| YD1-3 | BC3F3 | JD1018-RM6840 | 190 | HZ: May–September 2019 | |
| YD2-1 | BC3F4 | JD1009-JD1133 | 190 | LS: December 2019–April 2020 | |
| YD2-2 | BC3F4 | JD1127-JD1052 | 190 | LS: December 2019–April 2020 | |
| YD2-3 | BC3F4 | JD1022-JD1090 | 190 | LS: December 2019–April 2020 | |
| YD2-4 | BC3F4 | JD1134-RM6840 | 190 | LS: December 2019–April 2020 | |
| YD3-1 | BC3F5 | JD1134-JD1159 | 190 | HZ: May–September 2020 | |
| YD3-2 | BC3F5 | JD1152-RM6840 | 190 | HZ: May–September 2020 | |
Figure 2.
Genotypic compositions of the ten residual heterozygote-derived F2 populations in the segregating regions.
Based on the genotype and phenotype data, qTGW1-2 for TGW, GL and GW was identified in all the three populations, and the enhancing alleles for the three traits in all the three populations were all derived from JZ1560 (Table 1). In YC1-1, qTGW1-2 showed the additive effects of 1.66 g, 0.11 mm and 0.06 mm for TGW, GL and GW, and explained 19.3%, 24.4% and 33.9% of phenotypic variations (R2). In YC1-2, the additive effects for TGW, GL and GW were 1.59 g, 0.11 mm and 0.04 mm, explaining 23.8%, 26.3% and 18.9% of phenotypic variations. Similar additive effects in the same direction and similar R2 values confirmed the existence of qTGW1-2 in the common segregating region of YC1-1 and YC1-2. Compared with the additive effects for TGW in YC1-1 and YC1-2, a relatively smaller additive effect of 0.92 g was detected in the remaining population YC1-3. We further compared the segregating regions of JD1009-RM6840, JD1009-JD1090 and JD1018-RM6840 in YC1-1, YC1-2 and YC1-3 populations. The segregating regions of YC1-1 and YC1-2 covered all or part of the segregating region of YC1-3. These results suggested that there might be two or more QTLs with the same direction of additive effects in the primary interval of qTGW1-2.
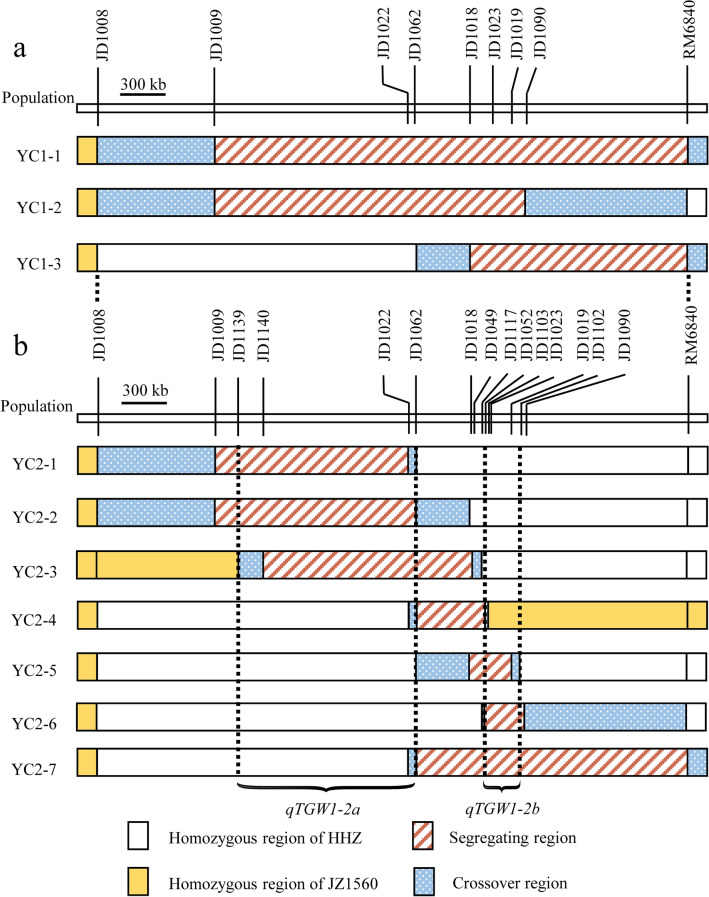
To confirm our conjecture, the segregating region responsible for qTGW1-2 should be subdivided into smaller heterozygous segments. Seven recombinant plants with sequential segregating regions covering the interval JD1009-RM6840 were selected from YC1-1, YC1-2 and YC1-3 populations and selfed to develop F10 RH-derived F2 populations, namely YC2-1, YC2-2, YC2-3, YC2-4, YC2-5, YC2-6 and YC2-7 (Table 2; Fig. 1). The seven F10 populations carried smaller segregating regions. TGW, GL and GW of each plant in the seven populations were measured and showed continuous segregations in each population (Table 1). Combined the genotype and phenotype information of each plant in the seven populations, seven segmental linkage maps were constructed (Fig. 2b). QTL analysis results for TGW, GL and GW using these populations are shown in Table 3. Except for YC2-4, QTLs responsible for TGW, GL and GW were detected in the remaining six populations. Since YC2-1 and YC2-2 showed the common segregating region (JD1139-JD1022) and no overlapping segregating region with YC2-5 and YC2-6, these results indicated that there should be two QTLs in the target interval of qTGW1-2. Furthermore, the segregating region of YC2-3 overlapped with the common segregating region (JD1139-JD1022), and similar additive effects for TGW, GL and GW were observed in YC1-1, YC1-2 and YC1-3. The result indicated that one QTL controlling TGW, GL and GW was located in the common segregating region of YC2-1, YC2-2 and YC2-3 flanked by JD1139 and JD1062, corresponding to a 1.2-Mb region in the Nipponbare genome. We designated this QTL as qTGW1-2a. Due to the fact that no QTL for all the three traits detected in YC2-4, the other QTL for TGW, GL and GW should be located in the common segregating region of YC2-5, YC2-6 and YC2-7 but outside the segregating region of YC2-4, with the interval flanked by JD1052 and JD1102, corresponding to a 235.1-kb region in the Nipponbare genome. We named this QTL as qTGW1-2b.
Table 3.
QTLs detected for TGW, GL, and GW in ten RH-derived populations in F9 and F10. TGW, 1000-grain weight (g); GL, grain length (mm); GW, grain width (mm); A, additive effect of replacing a HHZ allele with a JZ1560 allele; D, dominance effect; R2, proportion of phenotypic variance explained by the QTL effect; ns, no significance.
| Population | Segregating region | Trait | LOD | A | D | R2 (%) |
|---|---|---|---|---|---|---|
| YC1-1 | JD1009-RM6840 | TGW | 5.18 | 1.66 | − 0.77 | 19.3 |
| GL | 7.07 | 0.11 | 0.04 | 24.4 | ||
| GW | 9.80 | 0.06 | − 0.01 | 33.9 | ||
| YC1-2 | JD1009-JD1090 | TGW | 6.78 | 1.59 | − 0.38 | 23.8 |
| GL | 7.98 | 0.11 | − 0.04 | 26.3 | ||
| GW | 5.68 | 0.04 | − 0.02 | 18.9 | ||
| YC1-3 | JD1018-RM6840 | TGW | 8.34 | 0.92 | 0.03 | 17.0 |
| GL | 12.47 | 0.11 | − 0.02 | 24.4 | ||
| GW | 3.70 | 0.03 | 0.00 | 7.9 | ||
| YC2-1 | JD1009-JD1022 | TGW | 6.71 | 0.67 | − 0.01 | 15.4 |
| GL | 8.24 | 0.10 | − 0.03 | 18.5 | ||
| GW | 7.20 | 0.03 | 0.00 | 16.4 | ||
| YC2-2 | JD1009-JD1062 | TGW | 6.78 | 0.85 | 0.00 | 15.8 |
| GL | 4.13 | 0.07 | 0.01 | 9.9 | ||
| GW | 5.28 | 0.04 | 0.01 | 12.5 | ||
| YC2-3 | JD1140-JD1049 | TGW | 4.81 | 0.57 | − 0.66 | 19.7 |
| GL | 3.57 | 0.08 | − 0.09 | 15.0 | ||
| GW | 3.33 | 0.03 | − 0.01 | 14.1 | ||
| YC2-4 | JD1062-JD1052 | TGW | ns | ns | ns | ns |
| GL | ns | ns | ns | ns | ||
| GW | ns | ns | ns | ns | ||
| YC2-5 | JD1018-JD1019 | TGW | 6.36 | 0.59 | 0.09 | 14.9 |
| GL | 3.45 | 0.06 | 0.00 | 8.4 | ||
| GW | 3.01 | 0.02 | 0.00 | 7.4 | ||
| YC2-6 | JD1052-JD1090 | TGW | 13.11 | 0.87 | − 0.01 | 28.2 |
| GL | 9.39 | 0.09 | 0.01 | 21.2 | ||
| GW | 7.66 | 0.03 | 0.00 | 17.6 | ||
| YC2-7 | JD1062-RM6840 | TGW | 14.70 | 0.97 | − 0.06 | 32.1 |
| GL | 12.51 | 0.11 | − 0.03 | 27.4 | ||
| GW | 10.49 | 0.03 | 0.00 | 23.8 |
Validation and dissection of qTGW1-2 using seven NIL-F2 populations in BC3F3 and BC3F4
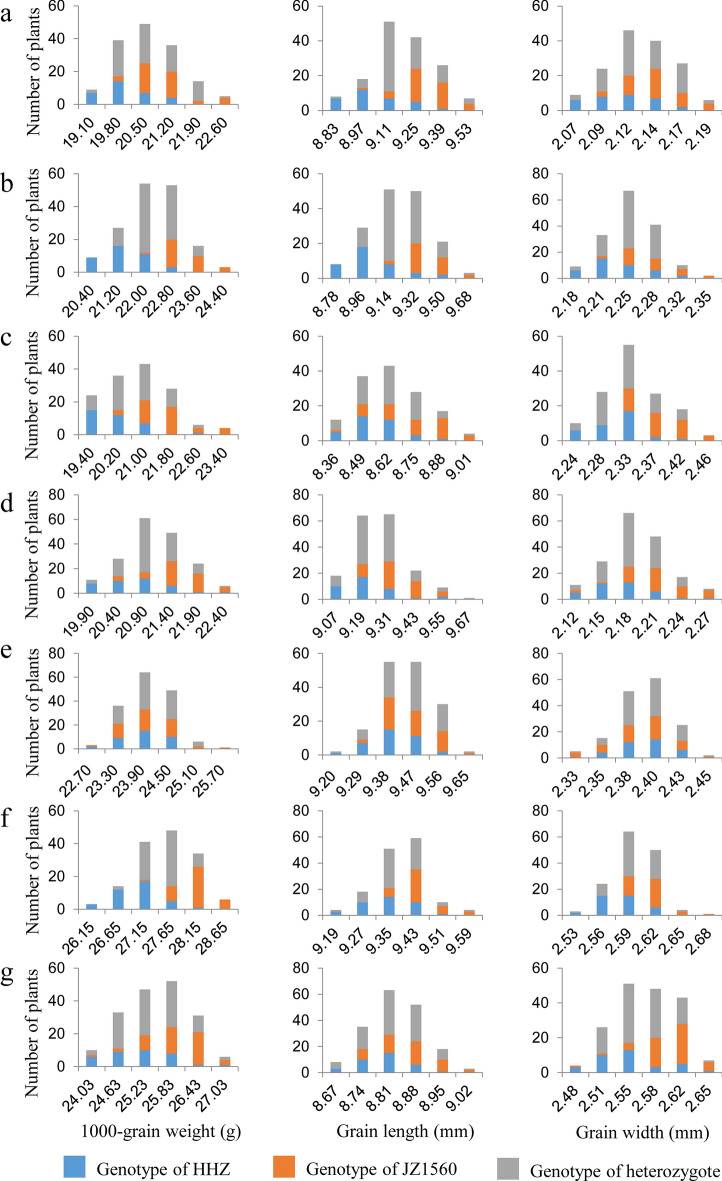
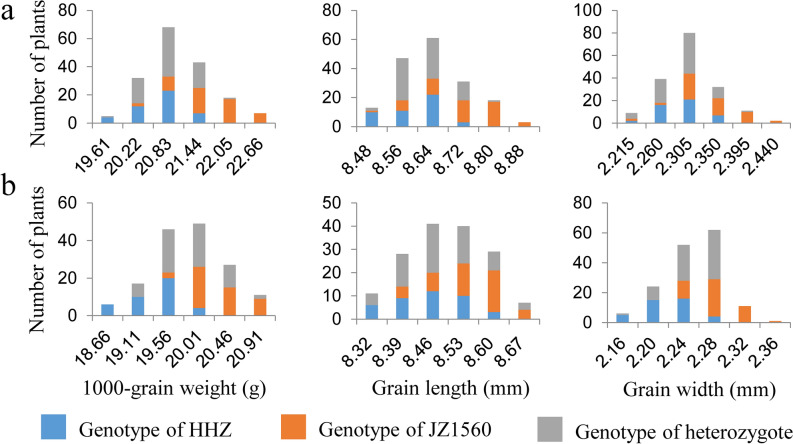
For further validation and dissection of qTGW1-2, we conducted another experiment using advanced backcross populations with the genetic background of HHZ. Three BC3F3 and four BC3F4 populations with the sequential segregating regions covering the interval JD1009-RM6840 were established and planted in three rice-growing seasons (Table 2; Fig. 1). For each plant of the seven populations, TGW, GL and GW were measured, and the distribution tendencies of the three traits are shown in Fig. 3. TGW, GL and GW showed continuous distributions in all the populations, but differentiation between the HHZ and JZ1560 homozygous genotypes was also observed. Except for in YD2-2 population, concentrations of the HHZ and JZ1560 homozygous plants were obviously distributed in the low- and high-value areas for TGW, GL and GW in YD1-1, YD1-2, YD1-3, YD2-1, YD2-3 and YD2-4 populations, which suggested qTGW1-2 was segregated in these populations (Fig. 3). Similar frequency distribution of the three traits of the HHZ and JZ1560 homozygous plants was observed in YD2-2 population.
Figure 3.
Distributions of 1000-grain weight, grain length, and grain width in the three BC3F3 and four BC3F4 populations. (a) YD1-1. (b) YD1-2. (c) YD1-3. (d) YD2-1. (e) YD2-2. (f) YD2-3. (g) YD2-4.
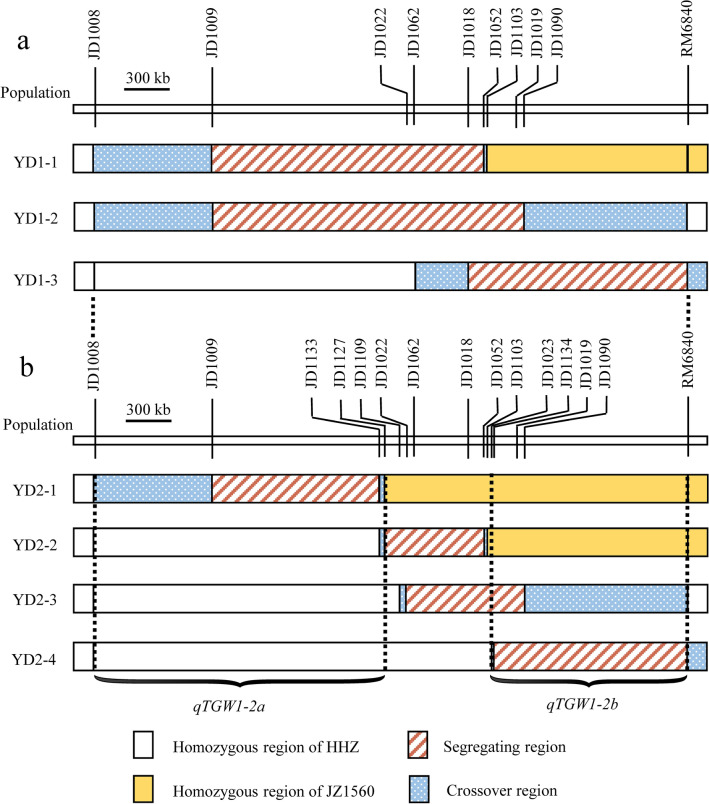
Combined the genotype and phenotype information of each plant in the seven populations, seven segmental linkage maps were constructed (Fig. 4). Results of QTL analysis for TGW, GL and GW using YD1-1, YD1-2 and YD1-3 populations were presented in Table 4. Significant QTL effects for the three traits were observed in all the three BC3F3 populations. For TGW, GL and GW, the enhancing alleles were derived from JZ1560 in all the three populations. In YD1-1, the additive effects were 0.54 g for TGW, 0.15 mm for GL and 0.02 mm for GW, explaining 22.8%, 38.6% and 16.9% of the phenotypic variances, respectively. In YD1-2, the additive effects were 0.87 g for TGW, 0.18 mm for GL and 0.02 mm for GW, explaining 45.6%, 39.6% and 13.6% of the phenotypic variations, respectively. In YD1-3, the additive effects were 0.72 g for TGW, 0.08 mm for GL and 0.03 mm for GW, explaining 31.4%, 13.4% and 22.7% of the phenotypic variances, respectively. The segregating region of YD1-2 overlapped the segregating regions of YD1-1 and YD1-3, and larger additive effects for TGW and GL were detected in YD1-2 than that in YD1-1 and YD1-3 populations (Fig. 4a). The results suggested there might be more than one QTL in the whole segregating region, which is consistent with the results of QTL analysis in the three F9 RH-derived populations.
Figure 4.
Genotypic compositions of the three BC3F3 and four BC3F4 populations in the segregating regions.
Table 4.
QTL detected for TGW, GL, and GW in seven backcross populations in BC3F3 or BC3F4. TGW, 1000-grain weight (g); GL, grain length (mm); GW, grain width (mm); A, additive effect of replacing a HHZ allele with a JZ1560 allele; D, dominance effect; R2, proportion of phenotypic variance explained by the QTL effect; ns, no significance.
| Population | Segregating region | Trait | LOD | A | D | R2 (%) |
|---|---|---|---|---|---|---|
| YD1-1 | JD1009-JD1052 | TGW | 9.84 | 0.54 | 0.15 | 22.8 |
| GL | 18.55 | 0.15 | 0.02 | 38.6 | ||
| GW | 7.08 | 0.02 | 0.01 | 16.9 | ||
| YD1-2 | JD1009-JD1090 | TGW | 25.03 | 0.87 | 0.07 | 45.6 |
| GL | 20.80 | 0.18 | 0.01 | 39.6 | ||
| GW | 6.01 | 0.02 | 0.00 | 13.6 | ||
| YD1-3 | JD1018-RM6840 | TGW | 15.13 | 0.72 | − 0.09 | 31.4 |
| GL | 5.81 | 0.08 | 0.00 | 13.4 | ||
| GW | 9.82 | 0.03 | − 0.01 | 22.7 | ||
| YD2-1 | JD1009-JD1133 | TGW | 10.82 | 0.40 | − 0.06 | 24.2 |
| GL | 6.37 | 0.07 | − 0.01 | 15.0 | ||
| GW | 7.44 | 0.02 | − 0.01 | 12.3 | ||
| YD2-2 | JD1127-JD1052 | TGW | ns | ns | ns | ns |
| GL | ns | ns | ns | ns | ||
| GW | ns | ns | ns | ns | ||
| YD2-3 | JD1022-JD1090 | TGW | 24.66 | 0.52 | − 0.05 | 51.8 |
| GL | 11.23 | 0.06 | − 0.01 | 28.0 | ||
| GW | 11.20 | 0.02 | 0.00 | 27.9 | ||
| YD2-4 | JD1134-RM6840 | TGW | 9.90 | 0.47 | − 0.15 | 22.0 |
| GL | 4.24 | 0.04 | 0.00 | 10.1 | ||
| GW | 9.85 | 0.03 | − 0.01 | 21.7 |
For further delimitation of qTGW1-2, we conducted QTL analysis of four BC3F4 populations with sequential segregating regions covering the interval JD1009-RM6840, which were developed from four recombinant plants in the YD1-1, YD1-2 and YD1-3 populations (Table 4). Significant QTL effects were detected in YD2-1, YD2-3 and YD2-4 but not in YD2-2. In YD2-1, the additive effects were 0.40 g for TGW, 0.07 mm for GL and 0.02 mm for GW, explaining 24.2%, 15.0% and 12.3% of the phenotypic variances, respectively. In view of non-significant QTL effects detected in YD2-2, the YD2-1 population was segregated for qTGW1-2a only. Thus, for the region of qTGW1-2a, the segregating region of YD2-2 was excluded and the cross-over region on the left of the InDel marker JD1127 should be included. In YD2-3 and YD2-4, significant QTL effects were identified for all the three traits. The enhancing alleles were all derived from JZ1560 with the additive effects of 0.52 g and 0.47 g for TGW, 0.06 mm and 0.04 mm for GL, 0.02 mm and 0.03 mm for GW in YD2-3 and YD2-4 populations, respectively. Considering that the similar additive effects of the three traits were detected in YD2-3 and YD2-4, we confirmed that the QTL qTGW1-2b should be located in the common segregating region and two cross-over regions flanked by JD1023 and RM6840 (Fig. 4b), which is in corresponding to the results of QTL analysis in the seven F10 RH-derived populations.
Fine-mapping of qTGW1-2b using four newly developed NIL-F2 populations
For further delimitation of qTGW1-2b, two RH-derived populations in F11 and two backcross populations in BC3F5 with sequential segregating regions covering the interval JD1117-RM6840 were developed from the last generation (Table 2; Fig. 1). Descriptive statistics of TGW, GL and GW in the two RH-derived populations in F11 are shown in Table 5. Three traits showed the same continuous distribution with low skewness and kurtosis as the previous RH-derived populations. Similarly, the distribution tendencies of the three traits are descripted in Fig. 5. The plants with HHZ and JZ1560 homozygous genotypes concentrated to low- and high-value areas, which indicated that qTGW1-2b was segregated in the two BC3F5 populations.
Table 5.
Phenotypic performance of TGW, GL and GW in two RH-derived populations in F11. TGW, 1000-grain weight (g); GL, grain length (mm); GW, grain width (mm).
| Population | Trait | Mean | SD | CV | Range | Skew | Kurt |
|---|---|---|---|---|---|---|---|
| YC3-1 | TGW | 32.48 | 1.64 | 0.05 | 28.41–36.15 | − 0.16 | − 0.20 |
| GL | 10.89 | 0.17 | 0.02 | 10.46–11.36 | 0.28 | − 0.32 | |
| GW | 3.28 | 0.07 | 0.02 | 3.13–3.45 | 0.22 | − 0.23 | |
| YC3-2 | TGW | 33.44 | 1.56 | 0.05 | 28.81–36.80 | − 0.26 | − 0.30 |
| GL | 10.78 | 0.12 | 0.01 | 10.49–11.21 | 0.13 | 0.83 | |
| GW | 3.28 | 0.06 | 0.02 | 3.06–3.41 | − 0.61 | 0.53 |
Figure 5.
Distributions of 1000-grain weight, grain length, and grain width in the two BC3F5 populations. (a) YD3-1. (b) YD3-2.
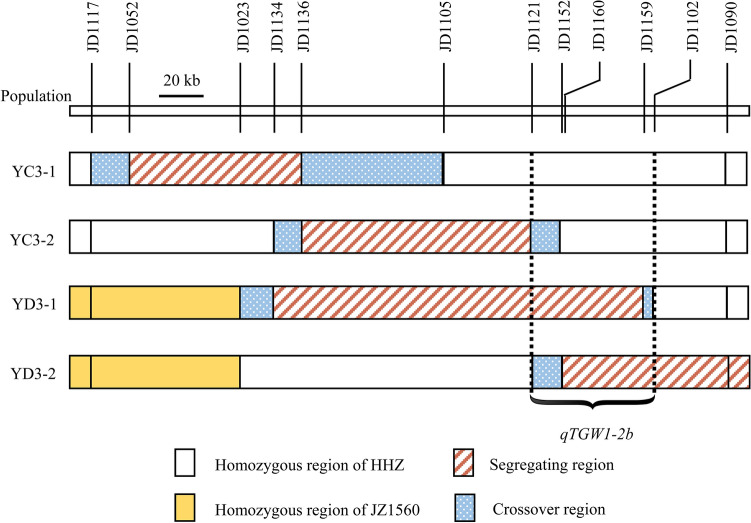
Genotypic compositions of the two F11 and two BC3F5 populations in the segregating regions are shown in Fig. 6. The segregating region covered the whole original interval of qTGW1-2b. QTL analysis results for TGW, GL and GW using these populations are shown in Table 6. Significant QTL effects were detected in YD3-1 and YD3-2 but not in YC3-1 and YC3-2. In YD3-1, the additive effects were 0.46 g for TGW, 0.06 mm for GL and 0.02 mm for GW, explaining 36.7%, 29.1% and 20.3% of the phenotypic variances, respectively. In YD3-2, the additive effects for TGW, GL and GW were 0.46 g, 0.05 mm and 0.03 mm, explaining 44.1%, 16.3% and 30.6% of phenotypic variations, respectively. Similar additive effects with the enhancing alleles derived from JZ1560 in the same direction comfirmed the existence of qTGW1-2b. In view of non-significant QTL effects detected in YC3-1 and YC3-2, qTGW1-2b was delimited to the common segregating region of YD3-1 and YD3-2 flanked by JD1121 and JD1102 which corresponded to a 54.8-kb region in the Nipponbare genome.
Figure 6.
Genotypic compositions of the two F11 and two BC3F5 populations in the segregating regions.
Table 6.
QTLs detected for TGW, GL, and GW in two RH-derived populations in F11 and two backcross populations in BC3F5. TGW, 1000-grain weight (g); GL, grain length (mm); GW, grain width (mm); A, additive effect of replacing a HHZ allele with a JZ1560 allele; D, dominance effect; R2, proportion of phenotypic variance explained by the QTL effect; ns, no significance.
| Population | Segregating region | Trait | LOD | A | D | R2 (%) |
|---|---|---|---|---|---|---|
| YC3-1 | JD1052-JD1136 | TGW | ns | ns | ns | ns |
| GL | ns | ns | ns | ns | ||
| GW | ns | ns | ns | ns | ||
| YC3-2 | JD1136-JD1121 | TGW | ns | ns | ns | ns |
| GL | ns | ns | ns | ns | ||
| GW | ns | ns | ns | ns | ||
| YD3-1 | JD1134-JD1159 | TGW | 17.30 | 0.46 | − 0.33 | 36.7 |
| GL | 13.02 | 0.06 | − 0.02 | 29.1 | ||
| GW | 8.59 | 0.02 | − 0.02 | 20.3 | ||
| YD3-2 | JD1152-RM6840 | TGW | 21.42 | 0.46 | 0.01 | 44.1 |
| GL | 6.78 | 0.05 | − 0.02 | 16.3 | ||
| GW | 15.34 | 0.03 | 0.00 | 30.6 |
Candidate gene analysis of qTGW1-2b
According to Rice Genome Annotation Project (http://rice.plantbiology.msu.edu), there are nine candidate genes in the 54.8-kb region for qTGW1-2b. Gene ID and products are listed in Table 7. Based on 100× whole genome re-sequencing of HHZ and JZ156023, we found five genes showing polymorphic sites between the two parents in the target 54.8-kb region. Among them, two genes showed 3-bp or 5-bp deletion in the UTR of the genes whereas one gene showed a 5-bp deletion in the intron. Only two genes showed polymorphic sites in the coding region between the two parents. LOC_Os01g72480 showed a 9-bp insertion in the first exon in JZ1560, which encodes a C3HC4-type RING finger E3 ubiquitin ligase of the RING/U-box superfamily, whereas LOC_Os01g72500 encoding a retrotransposon protein showed a 3-bp deletion in the first exon in JZ1560. According to the 9-bp sequence difference in LOC_Os01g72480, one InDel marker JD1160 was designed to genotype YC3-1, YC3-2, YD3-1 and YD3-2 segregating populations. As shown in Fig. 6, genotypes of YD3-1 and YD3-2 that showed significant additive effects were heterozygous at the polymorphic site of JD1160. In the segregating populations, the JZ1560 homozygous plants with 9-bp insertion in the first exon of LOC_Os01g72480 were obviously distributed in the high-value areas for all the three grain traits, whereas the HHZ homozygous plants were distributed in the low-value areas (Fig. 5), indicating that the 9-bp insertion might result in the phenotypic viriations of TGW, GL and GW.
Table 7.
Candidate genes in the target region of qTGW1-2b.
| Candidate gene | Gene product | Polymorphic information |
|---|---|---|
| LOC_Os01g72440 | Hypothetical protein | None |
| LOC_Os01g72450 | DNA binding protein | None |
| LOC_Os01g72460 | NADPH quinone oxidoreductase | None |
| LOC_Os01g72470 | Expressed protein | 3-bp deletion in 3’UTR |
| LOC_Os01g72480 | C3HC4-type RING finger E3 ubiquitin ligase | 9-bp insertion in the first exon |
| LOC_Os01g72490 | LRP1 | 5-bp deletion in 5’UTR |
| LOC_Os01g72500 | Retrotransposon protein | 3-bp deletion in the first exon |
| LOC_Os01g72510 | Eukaryotic aspartyl protease domain containing protein | None |
| LOC_Os01g72520 | Phosphoesterase family protein | 5-bp deletion in the first intron |
Discussion
In the past decade, significant progress has been achieved in the isolation and functional characterization of QTLs for yield traits in rice, especially for grain size and weight30,31,35. To date, more than 500 QTLs for rice grain weight distributing throughout the 12 chromosomes (http://www.gramene.org) have been identified in the primary mapping. Only 20 of them with major effects for grain size and weight have been cloned, which distributed on the eight chromosomes except for chromosomes 1, 10, 11 and 12. Majority of QTLs controlling grain size and weight show minor effects and are difficult to be cloned. In the present study, one minor QTL for grain weight and GL, qTGW1-2, was validated and dissected. No QTL for grain size and weight has been validated or cloned in the target region of qTGW1-2.
qTGW1-2 is a stably expressed QTL with minor effects for TGW, GL and GW. In this study, the minor effects of qTGW1-2 on TGW, GL and GW were stably observed across different populations, generations, rice-growing seasons and locations. Although the values of additive effects appeared to be changed in different experiments, the enhancing allele was derived from JZ1560 and the direction of additive effects remained unchanged for all the three grain traits. qTGW1-2 was further dissected into qTGW1-2a and qTGW1-2b. Due to the clustering distribution of QTLs controlling grain size and weight, genetic dissection of QTL regions into different QTLs has been frequently reported36–40. Based on the results of QTL analysis using seven RH-derived populations in F10 and four BC3F4 populations, qTGW1-2 was dissected into two QTLs (qTGW1-2a and qTGW1-2b), which were responsible for all the three grain traits. Compared the target intervals of these two QTLs in F10 RH-derived populations and BC3F4 populations, qTGW1-2a was delimited to the marker interval flanked by JD1139 and JD1127 (~ 978.2-kb), and qTGW1-2b was narrowed down into a ~ 186.0 kb region flanked by JD1023 and JD1102. Subsequently, two RH-derived populations in F11 and two BC3F5 populations were constructed, and qTGW1-2b was successfully fine-mapped to an accurate region flanked by JD1121 and JD1102, corresponding to a 54.8-kb region in the Nipponbare genome.
According to 100 × whole genome re-sequencing of HHZ and JZ1560, only two genes showed insertion/deletion polymorphic sites in the exons. LOC_Os01g72480 encoding a C3HC4-type RING finger E3 ubiquitin ligase of the RING/U-box superfamily, showed a 9-bp insertion in the first exon of JZ1560 compared to that of HHZ. It was reported that ubiquitin pathway is one of the most important regulatory pathways for seed development35. LOC_Os01g72500 showed a 3-bp deletion in the first exon and encodes a retrotransposon protein. No related evidence was reported that LOC_Os01g72500 involves in seed development. Therefore, LOC_Os01g72480 might be the possible candidate gene to regulate grain size and weight.
NIL-F2 populations developed from RHs of the RIL population and advanced backcross individuals are ideal materials for identifying minor QTLs. In this study, these two types of populations were both constructed to validate and dissect the minor QTL, qTGW1-2. In our previous primary mapping, qTGW1-2 was detected to be a pleiotropic QTL for TGW and GL because qTGW1-2 for TGW and qGL1-2 for GL were mapped in the same marker interval JD1009-JD1019 across two years in the HHZ/JZ1560 RIL population. No significant QTL effect for GW was observed in the target interval of qTGW1-2. Compared with the QTL effects of qTGW1-2 in the primary mapping RIL population, qTGW1-2 showed small but significant additive effects on GW besides TGW and GL in the NIL-F2 populations derived from both RHs and advanced backcross individuals. Furthermore, qTGW1-2 explained much higher phenotypic variation in NIL-F2 populations. For TGW, qTGW1-2 explained 9.20%-10.72% of phenotypic variations in the HHZ/JZ1560 RIL population. In the present study, qTGW1-2 accounted for 17.0%-23.8% and 22.8%-31.4% of the total phenotypic variations in RH-derived F9 populations and BC3F3 populations, respectively. These results indicated that elimination of genetic background noise in the NIL-F2 population increased the detection power of minor QTLs.
Compared to the cloned QTLs, both qTGW1-2a and qTGW1-2b showed relatively smaller effects, which might increase the difficulty in map-based cloning of them. The most likely candidate gene of qTGW1-2b, LOC_Os01g72480 encoding an E3 ubiquitin ligase might control grain size and weight through the ubiquitin-proteasome pathway. In rice, GW2 and WTG1 encoding an E3 ubiquitin ligase and a deubiquitinase respectively also regulate grain size by ubiquitin regulatory pathway9,41. In this study, we successfully delimited qTGW1-2a and qTGW1-2b to the marker intervals of JD1139-JD1127 (~ 978.2-kb) and JD1121-JD1102 (~ 54.8-kb) at the end of chromosome 1, respectively. The closely linked InDel markers, such as JD1160, JD1140 and JD1102, could be used in the marker assistant selection of qTGW1-2a and qTGW1-2b to improve rice varieties. Compared with the major QTLs, the breeders are hard to select the varieties which carry minor QTLs by the phenotypic identification. However, benefited from the development of gene markers or tightly linked markers, molecular marker assistant selection of the minor QTLs can be performed with high accuracy and efficiency in the early stage of breeding.
Materials and methods
Development of F9, F10 or F11 RH-derived NIL-F2 populations
Based on the previous genotyping of the HHZ/JZ1560 RIL population with 208 DNA markers23, one RIL plant with the residual heterozygous region covering JD1009-RM6840 was identified and selfed to produce one F8 population. To validate the genetic effect of qTGW1-2, three recombinant plants with sequential heterozygous regions covering JD1009-RM6840 were selected and selfed to construct three RH-derived F2 populations in F9 named as YC1-1, YC1-2 and YC1-3 (Fig. 1). These populations contained 120, 128 and 224 individuals, respectively. To further narrow down the marker interval of qTGW1-2, seven recombinant plants with sequential heterozygous regions covering JD1009-RM6840 were further selected from the three F9 NIL-F2 populations and selfed to generate YC2-1, YC2-2, YC2-3, YC2-4, YC2-5, YC2-6 and YC2-7 RH-derived NIL-F2 populations in F10 (Fig. 1). For further delimitation of qTGW1-2b, two F11 NIL-F2 populations with sequential heterozygous segments covering JD1117-JD1152 named as YC3-1 and YC3-2 were constructed (Fig. 1). Except for the YC2-3 population with 104 individuals, each of the remaining six populations contained 190 individuals.
Construction of NIL-F2 populations by consecutive backcrossing with the recurrent parent HHZ
The F1 plants derived from the cross between HHZ and JZ1560 were backcrossed with the recurrent parent HHZ for three consecutive generations. After screened by 208 DNA markers throughout the whole genome23, one BC3F1 (HHZ////HHZ///HHZ//HHZ/JZ1560) plant with the heterozygous region covering JD1009-RM6840 was selected and selfed to generate one BC3F2 population. To validate the genetic effect of qTGW1-2, three recombinant plants were selected from the BC3F2 population to construct three BC3F3 populations named as YD1-1, YD1-2 and YD1-3 (Fig. 1). These populations carried sequential heterozygous regions covering JD1009-RM6840. To further dissect qTGW1-2, four BC3F3 recombinant plants with sequential heterozygous regions covering JD1009-RM6840 were selected and selfed to produce YD2-1, YD2-2, YD2-3 and YD2-4 BC3F4 populations with sequential heterozygous regions covering JD1009-RM6840 (Fig. 1). Furthermore, two BC3F5 NIL-F2 populations named as YD3-1 and YD3-2 with sequential heterozygous segments covering JD1023-RM6840 were developed to narrow down the target region of qTGW1-2b (Fig. 1). Each of the populations mentioned above contained 190 individuals.
Field trails and phenotypic evaluation
The twenty-one populations were planted in the experimental stations of the China National Rice Research Institute, which were located at Hangzhou (120.2° E, 30.3° N) in Zhejiang Province or Lingshui (110.0° E, 18.5° N) in Hainan Province, China (Table 2). All the populations were planted with 16.7 cm between plants and 26.7 cm between rows. Field management followed local agricultural practice. At maturity, plants were harvested individually and sun-dried. Approximately 200 fully filled grains were selected by 3.5 mol/L NaCl solution. TGW, GL and GW were measured by the methods of Zhang et al32.
DNA marker analysis
DNA of each plant in the populations was extracted from the tender leaves following the method of Zheng et al42. PCR amplification was performed according to Chen et al43. The PCR profile was as follows: pre-denaturation of 3 min at 94°C, 30 cycles of 30 s at 94°C, 30 s at 55°C and 45 s at 72°C, followed by a final incubation at 72°C for 8 min. For genotyping the populations, a total of 23 insertion and deletion markers were designed with Primer3.0 (http://primer3.ut.ee/) according to the sequence differences between HHZ and JZ1560 detected by 100× whole genome re-sequencing. Primer sequences, physical position and PCR product sizes of HHZ and JZ1560 are listed in Table 8.
Table 8.
InDel markers developed in our study.
| Name | Forward primer (5′–3′) | Reverse primer (5′–3′) | Physical position (bp) | Product size (bp) | |
|---|---|---|---|---|---|
| HHZ | JZ1560 | ||||
| JD1018 | ATCGGTGCTGAGTGCTGAC | GAGGTGATGCGATTGGGAC | 41,727,488–41,727,915 | 417 | 427 |
| JD1022 | CCTGGAAACGGAGCGTATTT | GCAGTCGGTGGTGTAGTGGA | 41,318,246–41,318,669 | 403 | 423 |
| JD1023 | GAAATGTGAGCGTCAGAAGT | GTCGTCCGTTATGTTCAAGTA | 41,879,696–41,880,129 | 413 | 433 |
| JD1049 | AATGCAAACGTGAGAAATTG | AGGTAGAGAAAAACAGGCGA | 41,749,752–41,749,987 | 211 | 235 |
| JD1052 | TGAATTGGGTCATATCCTTGT | ATTGCTGGAATCGTATCGTAG | 41,830,564–41,830,867 | 283 | 303 |
| JD1062 | TGGAGAGTAAACAGAAAAGC | GGTGACAAGTAAGAAACGAG | 41,372,303–41,372,597 | 284 | 294 |
| JD1088 | AAACATGAATCGTGAAAGCA | ATCGGATCAACCACAGTAGC | 40,966,716–40,966,939 | 243 | 223 |
| JD1090 | GATGGATTGATGATAGCGCA | AACTCGTACAACCCAAGTGG | 42,098,975–42,099,272 | 277 | 297 |
| JD1102 | GTGATGCTCCTTTTCAATG | AGAATCGGGATACCACCT | 42,065,654–42,065,834 | 169 | 180 |
| JD1103 | TCCTTTCACCAATCACGG | AAGGGTTGGGAAGAGGCT | 41,850,660–41,850,861 | 182 | 201 |
| JD1105 | ATTCTGAATATCTGGTTGGATC | GGGGTTGACTTTGGAAAA | 41,971,742–41,971,957 | 225 | 215 |
| JD1113 | GCTTCGGTTTATTAGGGC | GGTCACTGGTCAGGGTCA | 41,109,254–41,109,638 | 349 | 376 |
| JD1117 | TGCCCATTGCTATGTAGA | CAACCTCACTGCTTACGG | 41,812,497–41,812,749 | 221 | 252 |
| JD1121 | CCAGTCACCGAAGGAAGT | CAGAGCAGATGAGCAGGA | 42,010,780–42,011,269 | 522 | 489 |
| JD1127 | GTAGCGTGCCTCCTGTTT | GGTCCAACTCTGGTTTCTTC | 41,180,095–41,173,703 | 155 | 168 |
| JD1133 | ACCTGATATTATTCGGGACA | GCAGCAACTTCAACTTCACT | 41,139,023–41,139,367 | 351 | 364 |
| JD1134 | GGCGTATGCTTATTGGAT | AAATAGACTTTTCTCACCCT | 41,894,706–41,895,072 | 385 | 366 |
| JD1136 | ATTAAGCATACATGAAGCC | AAGCCAACTATCACAAACTA | 41,907,772–41,908,134 | 339 | 362 |
| JD1139 | ACATTTTGTCCGCTACTG | CCACAACCATTCTTTCGT | 40,195,295–40,195,588 | 327 | 293 |
| JD1140 | GAAAATGGGTGCTCAAAA | GCTTACTAAAACGGCAGAA | 40,363,254–40,363,645 | 423 | 391 |
| JD1152 | CATAACTCGCCTGGAAAC | TCAACTTAGACCCCGTTT | 42,024,093–40,324,345 | 252 | 232 |
| JD1159 | ATGCTTCAAGTTACTCCCT | ACTCTTCCGCCTAATCTC | 42,060,810–42,061,277 | 486 | 497 |
| JD1160 | CGAACCCCTCGCCTCCTT | CGTGGTCGCATCCCTTGA | 42,025,727–42,026,161 | 425 | 434 |
Data analysis
Genetic maps of each population were constructed using Mapmaker/Exp 3.0, in which genetic distances between markers were presented in centiMorgans (cM) derived with Kosambi function. QTL analysis was performed with Windows QTL Cartographer 2.5, and the LOD value of 3.0 was taken as the threshold value.
Permission statement
Experimental research and field studies on plants, including the collection of plant materials, comply with relevant institutional, national, and international guidelines and legislation.
Conclusions
A total of 21 NIL-F2 populations were constructed to validate and dissect a minor and stably expressed QTL, qTGW1-2. Based on the QTL analysis of these genetic populations, qTGW1-2 was successfully validated to control grain length, width and weight with the enhancing allele derived from JZ1560. Furthermore, qTGW1-2 was further dissected into two QTLs, qTGW1-2a and qTGW1-2b, which were respectively narrowed down to the marker intervals of JD1139-JD1127 (~978.2-kb) and JD1121-JD1102 (~54.8-kb). These results supplied important basis for further map-based cloning and molecular design breeding in rice.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (32072050), State Key Laboratory of Subtropical Silviculture (KF2017-4), Chinese High-yielding Transgenic Program (2016ZX08001-004) and Zhejiang Provincial Natural Science Foundation of China under Grant No. LQ21C130003.
Author contributions
J.-Z.Y. and J.Z. designed the experiments. J.-Z.Y. and Y.-C.C. constructed the populations. Y.-C.C., G.L. and M.Y. performed the marker assay. Y.-C.C., T.-V.A., Y.-F.W. and X.-H. T. conducted the field trials. J.Z., Y.-C.C. and G.L. analyzed the data. J.-Z.Y. and Y.-C.C. wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yi-chen Cheng and Guan Li.
Contributor Information
Jian Zhang, Email: zhangjian@caas.cn.
Jie-zheng Ying, Email: yingjiezheng@caas.cn.
References
- 1.Huang R, et al. Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 2013;18:218–226. doi: 10.1016/j.tplants.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Fan C, et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 3.Zuo J, Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 2014;48:99–118. doi: 10.1146/annurev-genet-120213-092138. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015;47:944–948. doi: 10.1038/ng.3346. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015;47:949–954. doi: 10.1038/ng.3352. [DOI] [PubMed] [Google Scholar]
- 6.Zhao DS, et al. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 2015;9:1240–1253. doi: 10.1038/s41467-018-03616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong NQ, et al. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2015;11:2629–2644. doi: 10.1038/s41467-020-16403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 9.Song XJ, et al. Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. U S A. 2015;112:76–81. doi: 10.1073/pnas.1421127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruan B, et al. Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 2020;227:629–640. doi: 10.1111/nph.16540. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 12.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 13.Weng J, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 14.Shi CL, et al. A quantitative trait locus GW6 controls rice grain size and yield through the gibberellin pathway. Plant J. 2020;103:1174–1188. doi: 10.1111/tpj.14793. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012;44:950–954. doi: 10.1038/ng.2327. [DOI] [PubMed] [Google Scholar]
- 16.Hu J, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Che R, et al. Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants. 2015;2:15195–151201. doi: 10.1038/nplants.2015.195. [DOI] [PubMed] [Google Scholar]
- 18.Yu J, et al. OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 2017;15:28–45. doi: 10.1186/s12915-017-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q, et al. G-protein betagamma subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 2018;9:852–863. doi: 10.1038/s41467-018-03047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu, J. et al. Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol J. (2018). [DOI] [PMC free article] [PubMed]
- 21.Qi P, et al. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2018;22:1666–1680. doi: 10.1038/cr.2012.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U S A. 2012;109:21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying JZ, et al. TGW3, A major QTL that negatively modulates grain length and weight in rice. Mol. Plant. 2018;11:750–753. doi: 10.1016/j.molp.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Xia D, et al. GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol/ Plant. 2018;11:754–756. doi: 10.1016/j.molp.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Wu W, et al. A single-nucleotide polymorphism causes smaller grain size and loss of seed shattering during African rice domestication. Nat. Plants. 2017;3:17064–17070. doi: 10.1038/nplants.2017.64. [DOI] [PubMed] [Google Scholar]
- 26.Ishimaru K, et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet. 2013;45:707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- 27.Wang A, et al. The PLATZ transcription factor GL6 affects grain length and number in rice. Plant Physiol. 2019;180:2077–2090. doi: 10.1104/pp.18.01574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Si L, et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- 29.Li N, Li Y. Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 2016;33:23–32. doi: 10.1016/j.pbi.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Bai X, Wu B, Xing Y. Yield-related QTLs and their applications in rice genetic improvement. J. Integr. Plant Biol. 2012;54:300–311. doi: 10.1111/j.1744-7909.2012.01117.x. [DOI] [PubMed] [Google Scholar]
- 31.Chen JY, et al. Fine mapping of qHd1, a minor heading date QTL with pleiotropism for yield traits in rice (Oryza sativa L.) Theor. Appl. Genet. 2014;127:2515–2524. doi: 10.1007/s00122-014-2395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu XJ, et al. Identification and validation of quantitative trait loci for grain number in rice (Oryza sativa L.) Agronomy. 2020;10:180–194. doi: 10.3390/agronomy10020180. [DOI] [Google Scholar]
- 33.Zhang H, et al. Identification and verification of quantitative trait loci affecting milling yield of rice. Agronomy. 2020;10:75–87. doi: 10.3390/agronomy10010075. [DOI] [Google Scholar]
- 34.Zhu Y, et al. Fine mapping of qTGW10–20.8, a QTL having important contribution to grain weight variation in rice. Crop J. 2019;7:587–597. doi: 10.1016/j.cj.2019.01.006. [DOI] [Google Scholar]
- 35.Li N, Xu R, Duan P, Li Y. Control of grain size in rice. Plant Reprod. 2018;31(3):237–251. doi: 10.1007/s00497-018-0333-6. [DOI] [PubMed] [Google Scholar]
- 36.Guo L, et al. Dissection of two quantitative trait loci for grain weight linked in repulsion on the long arm of chromosome 1 of rice (Oryza sativa L.) The Crop Journal. 2013;1:70–76. doi: 10.1016/j.cj.2013.07.008. [DOI] [Google Scholar]
- 37.Wang LL, et al. Dissection of qTGW1.2 to three QTLs for grain weight and grain size in rice (Oryza sativa L.) Euphytica. 2014;202:119–127. doi: 10.1007/s10681-014-1237-7. [DOI] [Google Scholar]
- 38.Dong Q, et al. Dissection and fine-mapping of two QTL for grain size linked in a 460-kb region on chromosome 1 of rice. Rice (N Y). 2018;11:44–54. doi: 10.1186/s12284-018-0236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan AN, et al. Identification through fine mapping and verification using CRISPR/Cas9-targeted mutagenesis for a minor QTL controlling grain weight in rice. Theor. Appl. Genet. 2021;134:327–337. doi: 10.1007/s00122-020-03699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang HW, et al. Dissection of the qTGW1.1 region into two tightly-linked minor QTLs having stable effects for grain weight in rice. BMC Genet. 2016;17:98–107. doi: 10.1186/s12863-016-0410-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang K, et al. WIDE AND THICK GRAIN 1, which encodes an otubain-like protease with deubiquitination activity, influences grain size and shape in rice. Plant J. 2017;91:849–860. doi: 10.1111/tpj.13613. [DOI] [PubMed] [Google Scholar]
- 42.Zheng, K., Huang, N., Bennett, J. & Khush, G. S. PCR-based marker-assisted selection in rice breeding. In IRRI Discussion Paper Series No.12; International Rice Research Institute: Los Banos, Philippines (1995).
- 43.Chen X, Temnykh S, Xu Y, Cho YG, McCouch SR. Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L) Theor. Appl. Genet. 1997;95:553–567. doi: 10.1007/s001220050596. [DOI] [Google Scholar]