Abstract
The initiation of translation in eukaryotes requires several multisubunit complexes, including eukaryotic translation initiation factor 4F (eIF4F). In higher eukaryotes eIF4F is composed of the cap binding protein eIF4E, the adapter protein eIF4G, and the RNA-stimulated ATPase eIF4A. The association of eIF4A with Saccharomyces cerevisiae eIF4F has not yet been demonstrated, and therefore the degree to which eIF4A’s conserved function relies upon this association has remained unclear. Here we report an interaction between yeast eIF4G and eIF4A. Specifically, we found that the growth arrest phenotype associated with three temperature-sensitive alleles of yeast eIF4G2 was suppressed by excess eIF4A and that this suppression was allele specific. In addition, in vitro translation extracts derived from an eIF4G2 mutant strain could be heat inactivated, and this inactivation could be reversed upon the addition of recombinant eIF4A. Finally, in vitro binding between yeast eIF4G and eIF4A was demonstrated, as was diminished binding between mutant eIF4G2 proteins and eIF4A. In total, these data indicate that yeast eIF4G and eIF4A physically associate and that this association performs an essential function.
The recruitment of the 40S ribosomal subunit to the initiator codon of a eukaryotic mRNA is mediated by the interplay of several different translation initiation complexes (reviewed in reference 18). A key complex in this process is eukaryotic translation initiation factor 4F (eIF4F). The eIF4F complex in higher eukaryotes is composed of three proteins, eIF4G, eIF4A, and eIF4E (7). The eIF4E protein binds to the m7G cap structure at the 5′ end of the mRNA, as well as to eIF4G. The eIF4G protein acts as a bridging factor in that it binds to eIF4E, to the poly(A) tail binding protein Pab1p, and to eIF3, a complex of proteins which is tightly associated with the 40S ribosomal subunit (11, 13, 19, 20, 23). The RNA-stimulated ATPase eIF4A is also associated with eIF4G in higher eukaryotes and is thought to use its ATPase activity to unwind RNA secondary structure in the 5′ leader of each message (21). This presumably allows for the unimpeded scanning of the 40S ribosomal subunit along the 5′ leader as it searches for the initiator codon.
The mechanism by which the eIF4F complex mediates translation initiation in the yeast Saccharomyces cerevisiae is slightly more obscure. This organism encodes the eIF4E protein with the CDC33 gene (3), the eIF4A protein with two different genes (TIF1 and TIF2) (17), and the two eIF4G proteins (eIF4G1 and eIF4G2) with the TIF4631 and TIF4632 genes, respectively (8). Although S. cerevisiae expresses all of these putative components of eIF4F, the yeast eIF4A protein has not previously been shown to bind to eIF4G. For example, the purification of eIF4E from higher eukaryotes by cap analog chromatography results in the copurification of eIF4A and eIF4G (reviewed in reference 7), whereas purification of yeast eIF4E through a similar procedure results in the copurification of eIF4G but not eIF4A (9, 26). Furthermore, the initial mapping of the eIF4A binding domain in rabbit eIF4G placed it within the C-terminal third of the protein (16). The yeast eIF4G protein, although homologous to much of mammalian eIF4G, lacks homology to this entire C-terminal region (8). Each of these observations has brought into question whether eIF4A and eIF4G associate in yeast and, more generally, whether the mechanism by which translation initiation occurs in yeast involves the early recruitment of eIF4A to the 5′ end of the mRNA.
During the course of our study it was reported that mammalian eIF4G contains a second eIF4A binding site that is distinct from its previously identified C-terminal binding site (14). This site lies in the middle of the protein, which is conserved in all eukaryotic eIF4G proteins. It was shown in that study that the presence of both eIF4A binding sites on mammalian eIF4G was required for eIF4G to stimulate translation in vitro. Based on extensive sequence homology between yeast and mammalian eIF4G proteins, it was also proposed that yeast eIF4A binds to eIF4G within this region.
Here we present evidence that yeast eIF4A and eIF4G functionally and physically interact. Overexpression of eIF4A in several different tif4632 mutants led to suppression of their temperature-sensitive phenotype. This suppression was shown to be allele specific. A heat-inactivated translation extract from a tif4632 mutant could have its translational activity significantly stimulated by the addition of excess eIF4A, and this stimulation was also shown to be tif4632 allele specific. Finally, the binding of recombinant yeast eIF4A to eIF4G1 and eIF4G2 in vitro was demonstrated for the first time, as was a decreased ability of the several mutant eIF4G2 proteins to bind to eIF4A. These data show that yeast eIF4G contains an eIF4A binding site, and they indicate that an interaction between yeast eIF4A and eIF4G is required for translation and is essential for yeast cell viability.
MATERIALS AND METHODS
Yeast techniques.
Yeast strains (Table 1) were propagated on standard YPD or YM medium containing either 2% glucose or 2% galactose as the carbon source and all of the amino acids and nucleotides necessary for growth (10). All plasmids indicated in Table 2 and elsewhere in this paper were introduced into yeast by lithium acetate transformation and plating onto YM medium containing the appropriate additives.
TABLE 1.
Yeast strains used in this study
| Yeast strain (YAS no.a) | Mutationb/plasmid (BAS no.c) |
|---|---|
| 1856 | tif4631::LEU2 |
| 1888 | cdc33-1 |
| 1948 | tif4631::LEU2 tif4632::ura3/pTIF4632URA3CEN (2004) |
| 1951 | tif4631::LEU2 tif4632::ura3/pTIF4632TRP1CENI (2068) |
| 1993 | tif4631::LEU2 tif4632::ura3/ptif4632-1HIS3CEN (3013) |
| 1995 | tif4631::LEU2 tif4632::ura3/ptif4632-6HIS3CEN (3015) |
| 1996 | tif4631::LEU2 tif4632::ura3/ptif4632-8HIS3CEN (3016) |
| 2000 | tif4631::LEU2 tif4632::ura3/ptif4632-8TRP1CEN (3029) |
| 2002 | tif4631::LEU2 tif4632::ura3/ptif4632-430TRP1CEN (3037) |
| 2096 | 1993 + pTRP12μ (3437) |
| 2099 | 1993 + pTIF1TRP12μ (3137) |
| 2100 | 1995 + pTRP12μ (3437) |
| 2103 | 1995 + pTIF1TRP12μ (3137) |
| 2106 | 1948 + pTIF1TRP12μ (3137) |
| 2429 | 1996 + pTRP12μ (3437) |
| 2430 | 1996 + pTIF1TRP12μ (3137) |
| 2431 | 1996 + pTIF1TRP1CEN (3431)d |
| 2434 | 2002 + pTIF1URA32μ (3432) |
| 2435 | 1888 + pTIF1TRP12μ (3137) |
| 2436 | 1993 + pTIF2TRP12μ (3433) |
| 2437 | 1995 + pTIF2TRP12μ (3433) |
| 2438 | 1996 + pTIF2URA32μ (3434) |
| 2441 | 2002 + pTIF2URA32μ (3434) |
| 2442 | 1993 + pGPF::CDC33URA3CEN (3163)e |
| 2443 | 1995 + pGPF::CDC33URA3CEN (3163) |
| 2444 | 1996 + pGPF::CDC33URA3CEN (3163) |
| 2447 | 2002 + pGPF::CDC33URA3CEN (3163) |
| 2448 | 1993 + pDED1TRP12μ (3435) |
| 2449 | 1995 + pDED1TRP12μ (3435) |
| 2450 | 1996 + pDED1TRP12μ (3435) |
| 2453 | 2002 + pDED1URA32μ (3436)f |
| 2454 | 1888 + pDED1TRP12μ (3435) |
| 2455 | 1888 + pDED1URA32μ (3436) |
| 2456 | 1856 + p(HA)TIF4632TRP1CEN (2077) + pURA3CEN (989) |
| 2457 | 1856 + p(HA)TIF4632TRP1CEN (2077) + pTIF1URA32μ (3432) |
| 2458 | 1856 + p(HA)tif4632-1TRP1CEN (3040) + pURA3CEN (989) |
| 2459 | 1856 + p(HA)tif4632-1TRP1CEN (3040) + pTIF1URA32μ (3432) |
| 2460 | 1856 + p(HA)tif4632-6TRP1CEN (3041) + pURA3CEN (989) |
| 2461 | 1856 + p(HA)tif4632-6TRP1CEN (3041) + pTIF1URA32μ (3432) |
| 2462 | 1856 + p(HA)tif4632-8TRP1CEN (3042) + pURA3CEN (989) |
| 2463 | 1856 + p(HA)tif4632-8TRP1CEN (3042) + pTIF1URA32μ (3432) |
The YAS number is to be used for requesting strains.
All strains derived from YAS1993, -1995, and -1996 have the following genotype: MATα ade2-1 his3-11,15 leu2-3 trp1-1 ura3-1. All other strains have the following genotype: MATa ade2-1 his3-11,15 leu2-3 trp1-1 ura3-1 pep4::HIS3.
The number of the bacterial strain that contains the indicated plasmid.
This plasmid was provided by P. Linder (24).
This plasmid was provided by J. McCarthy (29).
This plasmid was provided by T.-H. Chang (4).
TABLE 2.
Genetic suppression of temperature-sensitive alleles of TIF4632
| Allele (YAS no.a) | Temperature sensitivity suppressed by excessb:
|
|||
|---|---|---|---|---|
| TIF1 | TIF2 | CDC33 | DED1 | |
| tif4632-1 (1993) | Yes | Yes | No | No |
| tif4632-6 (1995) | Yes | Yes | No | No |
| tif4632-8 (1996) | Yes | Yes | No | No |
| tif4632-430 (2002) | No | No | Yes | No |
| cdc33-1 (1888) | No | NDc | Yes | Yes |
Strain number to be used when making requests.
The indicated genes were expressed in the listed strains on multicopy plasmids (TIF1, TIF2, and DED1) or as a promoter fusion (GPF::CDC33) on a CEN plasmid (29).
ND, not determined.
TIF4632 mutagenesis and dosage suppression.
Yeast strain YAS1948 (Table 1), which contains TIF4632 on a URA3CEN plasmid as its sole source of eIF4G, was transformed with a mixture of DNA containing (i) BamHI- and EcoRI-digested pAS481 (26), which yields a TRP1CEN vector containing the entire TIF4632 gene but lacking the open reading frame, and (ii) linear fragments of the TIF4632 gene containing its open reading frame and approximately 200 nucleotides on either side. These fragments contained random mutations at approximately 400-bp intervals as a result of their amplification by PCR with Taq DNA polymerase (22) and recombined with the gapped plasmid by homologous recombination within the yeast to yield circular TRP1CEN plasmids with an intact TIF4632 gene. Following selection for tryptophan prototrophs at 30°C, strains were replica plated onto YM medium containing 5-fluoro-orotic acid (5-FOA) (2) at 30°C. Viable colonies on this plate were replica plated onto YM medium lacking tryptophan at 30 and 37°C. Plasmid DNAs from the temperature-sensitive colonies were rescued by standard techniques (10) and retested in YAS1948 for their ability to confer temperature sensitivity to this strain after growth on 5-FOA at 30°C. Those plasmids containing conditional tif4632 alleles had both strands of their DNA sequenced in the region encoding amino acids 400 to 800 of eIF4G2. Confirmation that the identified mutations within this region were responsible for the conditional phenotype was obtained by subcloning this region of DNA into an otherwise wild-type TIF4632 gene and showing that the resulting recombinant gene conferred upon cells a temperature-sensitive phenotype that was suppressed by multicopy TIF1 (data not shown).
For the isolation of multicopy suppressor plasmids, rapidly growing cultures (optical density at 600 nm [OD600] = 0.5 to 1.0) of yeast strains YAS1993 (tif4632-1) and YAS1996 (tif4632-8) were transformed with a URA3/2μ genomic yeast library and allowed to recover on YM plates that selected for uracil prototrophs for 8 hours at 30°C. The plates were then shifted to 37°C for 7 days, and temperature-resistant colonies were identified. To test that the suppression of the temperature sensitivity depended upon the URA3 library plasmid, the temperature-resistant isolates were plated onto YM medium containing 5-FOA and then retested for their ability to grow at 37°C on YM plates. For those strains which were unable to grow at 37°C after growth on 5-FOA, their plasmids were isolated by standard techniques and transformed into the bacterial strain MC1061 by electroporation. Bacterial plasmid DNA preparation and DNA sequencing were performed by standard methodology.
Detection of mutant proteins at the nonpermissive temperature.
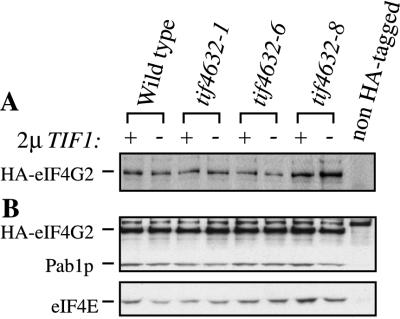
The yeast strains indicated in Fig. 3 were grown in YM medium with selection for uracil and tryptophan at 37°C and harvested at an OD600 of 1.8 to 2.0. The cells were washed, resuspended in an equal volume of translation buffer A (25), lysed by vortexing with glass beads as described previously for a small-scale extract preparation (25), and clarified by microcentrifugation for 10 min at 4°C. The extracts were normalized by their OD260 readings, and approximately 10 μg of total protein for each sample was boiled in sodium dodecyl sulfate (SDS) sample buffer and resolved by SDS–10% polyacrylamide gel electrophoresis (SDS–10% PAGE). Hemagglutinin (HA)-tagged eIF4G2 was identified by Western analysis as described previously (26) with the monoclonal antibody 12CA5. For the coimmunoprecipitation experiments shown in Fig. 3B, 90-μl portions of equivalent amounts of extract in buffer A were rocked for 2 h at 4°C with a 10-μl bed volume of protein A-Sepharose (Santa Cruz Biotechnology), which had been preincubated with 5 μl of anti-HA monoclonal antibodies (12CA5) and washed in phosphate-buffered saline (140 mM NaCl, 10 mM Na2HPO4, 3 mM KCl, 2 mM KH2PO4) plus 0.1% Triton X-100 and 0.01% SDS (PBSTS). The beads were then washed four times in 500 μl of cold PBSTS and boiled in SDS sample buffer, and the bound proteins were resolved by SDS–10% PAGE. HA-eIF4G, Pab1p, and eIF4E were detected by Western analysis.
FIG. 3.
Expression and initiation factor association of mutant eIF4G2 proteins in the presence and absence of excess eIF4A. (A) Excess eIF4A does not lead to overproduction of eIF4G2. Crude lysates prepared from yeast strains YAS2456 to -2463 grown at 37°C were analyzed by Western analysis for their content of the indicated epitope-tagged HA-eIF4G2. Each strain either lacked (−) or contained (+) the multicopy plasmid containing TIF1. All strains contained non-epitope-tagged eIF4G2 as well and therefore were viable at 37°C. Extracts from a strain (YAS2106) lacking an HA-tagged eIF4G (non HA-tagged) were used as a negative control. (B) Association of mutant forms of eIF4G2 with eIF4E and Pab1p. The epitope-tagged HA-eIF4G2 proteins in the crude lysates described for panel A were immunoprecipitated, resolved by SDS-PAGE, and visualized by Western analysis. The degree of coimmunoprecipitation of Pab1p and eIF4E with eIF4G2 was determined by Western analysis of the immunoprecipitate with Pab1p- or eIF4E-specific antibodies.
Quantification of eIF4A levels.
YAS2429, -2430, and -2431 were grown in YM medium with selection for tryptophan at 30°C and harvested at an OD600 of 1.8 to 2.0. The cells were washed and resuspended in an equal volume of translation buffer A. Extracts were made by vortexing the cells with glass beads as described previously for a small-scale extract preparation (26) and clarified by microcentrifugation for 10 min at 4°C. The total protein concentration in each extract was determined with the Bio-Rad protein reagent, and an equal amount of total protein for each extract was boiled in SDS sample buffer and loaded on a 10% SDS gel in twofold serial dilutions (beginning with 3 μg). The eIF4A in each dilution of extract was detected by Western analysis, and the levels of overexpression of eIF4A in YAS2430 and YAS2431 were determined by noting at which dilution the Western signal disappeared compared with that for an extract containing no excess eIF4A (YAS2429).
The amount of eIF4A in the translation extracts was determined by loading twofold serial dilutions of the extract (beginning with 3 μg of total protein) on an SDS-polyacrylamide gel next to serial dilutions of recombinant eIF4A (beginning with 0.03 μg) plus 2 μg of bovine serum albumin (to prevent protein adsorption). The gels were subjected to Western analysis, and the amount of eIF4A was determined by noting at which dilution the Western signal disappeared when comparing the translation extract lanes to the lanes containing recombinant eIF4A. The anti-eIF4A antibody was used at a 1:10,000 dilution.
Recombinant protein production and purification.
The glutathione S-transferase (GST)-eIF4G(His)6 fusion proteins were expressed from the pGEX2T vector (Pharmacia) in the BL21 bacterial strain. The different eIF4G genes were subcloned as BamHI/EcoRI fragments (26) into the vector’s BamHI and EcoRI sites (30) to yield BAS3214 (eIF4G2), BAS3438 (eIF4G2-1), BAS3439 (eIF4G2-6), BAS3440 (eIF4G2-8), and BAS3470 (eIF4G2-430). Protein expression was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) (26), and cells were washed in buffer B (150 mM NaCl, 10 mM Na2HPO4, 4 mM NaH2PO4, pH 7.3). Following resuspension of the cell pellet in 3 ml of buffer B per g of cells and freezing, the cells were quick-thawed and lysed by sonication after the addition of lysozyme to 250 μg/ml. The extract was clarified by microcentrifugation and brought to 0.1% Triton X-100, 10 mM imidazole, and 1 M NaCl. The extract was then adsorbed to 250 μl of buffer B-equilibrated ProBond nickel agarose (Invitrogen) per 600 ml of culture. After 1.5 h of gentle rocking at 4°C, the Ni beads were washed twice in 5 ml of buffer BT (buffer B plus 0.1% Triton X-100) plus 1 M NaCl and 20 mM imidazole and twice in 5 ml of buffer BT plus 1 M NaCl and 80 mM imidazole. Proteins were eluted once with 200 μl of buffer BT plus 250 mM imidazole and twice with 200 μl of buffer BT plus 500 mM imidazole, and these eluates were pooled.
Recombinant GST-eIF4G2(His)6 proteins that were to be added back to translation extracts were batch adsorbed to 100 μl of glutathione-Sepharose resin (Pharmacia) preequilibrated in buffer B by gentle rocking for 1 h at 4°C. They were eluted with 100 μl of 20 mM glutathione–100 mM Tris-HCl by gentle rocking at room temperature for 20 min and dialyzed against translation buffer A in 50% glycerol.
Recombinant GST-eIF4G2(His)6 proteins that were used in the binding studies had their concentrations on the resin normalized as follows. Eighty microliters of the Ni-agarose eluate was incubated with 10 μl of glutathione-Sepharose resin, the resin was washed four times with 1 ml of buffer BT at 4°C, and each sample was boiled in SDS sample buffer. SDS-PAGE and Coomassie brilliant blue staining were used to determine the amounts of each GST-eIF4G2 protein on the resin. The amounts of nickel-agarose eluate added to the glutathione-Sepharose resin were then adjusted accordingly so that each GST-eIF4G2 protein was equally concentrated on the resin prior to the start of the binding assays. To decrease background eIF4A binding, the resin-bound proteins were washed once in buffer BT at room temperature for 20 min before use.
The production and purification of recombinant eIF4E from BAS2024 were as described previously (6, 29). Recombinant eIF4A was a kind gift from Jon Lorsch and Dan Herschlag (Stanford University). This protein was also the source of material used to raise rabbit polyclonal anti-eIF4A antibodies by standard procedures.
In vitro binding assays.
Approximately 400 ng of each GST-eIF4G2 fusion protein immobilized on a 10-μl bed volume of glutathione-Sepharose resin was incubated with 2.5 μg of recombinant eIF4A or 500 ng of recombinant eIF4E at 26°C for 20 min with occasional mixing. The beads were then washed four times with 500 μl of cold buffer BT and boiled in SDS sample buffer, and the bound proteins were resolved by SDS–10% PAGE and detected by either Coomassie brilliant blue staining (eIF4G) or Western analysis (eIF4A and eIF4E). Each binding reaction was done in a 20-μl final volume.
In vitro translation assays.
Yeast extracts were prepared from strains YAS1951, YAS2000, and YAS2002 as described previously (12, 25). All experiments used non-nuclease-treated extracts to which 2 mM EGTA was added just before use. The addition of EGTA led to greater reproducibility of the extracts over time. Heated extracts were put at 30°C for the indicated amount of time and immediately quick-frozen in liquid nitrogen. Nonheated extracts were quick-frozen at time zero. After thawing, approximately 90 ng of the indicated mRNA was added in a 7.5-μl mix of translational components to 7.5 μl of extract as described previously (25). Each reaction mixture was incubated at 26°C for the indicated amount of time and stopped by quick-freezing in liquid nitrogen. Luciferase (LUC) production was assayed with 10 μl of each reaction mixture and 50 μl of luciferin reagent (Promega). Luminescence was measured for 15 s with a Turner TD-20e luminometer.
For recombinant protein addition experiments, protein was added to each extract and incubated on ice for 20 min prior to heating. Each protein was added in a 2-μl volume. To compensate for the added volume, 2 μl of H2O was left out of the translational component mix. Reaction mixtures without added protein had 2 μl of buffer A added to them prior to heating. Cap analog inhibitor studies used m7GpppG at between 0.25 and 1.0 mM with nuclease-treated extracts, as previously described (27). Each assay was performed at least three times with at least two different extract preparations. Reported values are representative of the results from these experiments.
RESULTS
Excess eIF4A suppresses the temperature sensitivity of several tif4632 mutants.
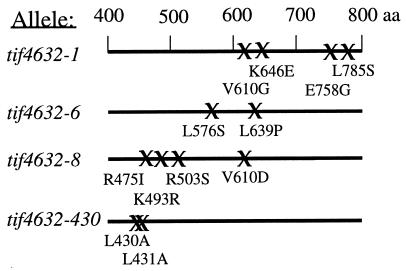
Three different temperature-sensitive alleles of the yeast tif4632 gene were identified by a plasmid-shuffling scheme with randomly mutagenized plasmid DNA (see Materials and Methods). These are referred to as tif4632-1, -6, and -8. Each of these mutants exhibited growth arrest on YPD plates at 37°C after three to five generations. Sequencing of these mutated genes revealed they contained several different mutations within the region of eIF4G2 known to be essential for cell viability (28) (Fig. 1). Interestingly, the tif4632-1 and tif4632-8 mutants both contained a mutation in residue 610. This residue lies within the RNP2 consensus sequence of the putative RNA recognition motif of eIF4G, and therefore it is possible that these mutations alter the structure of that domain (8, 15). We also note that a mutated residue within the tif4632-6 product (L576S) is conserved in mammalian eIF4G. When this residue is mutated in combination with other residues in that protein, a loss of eIF4A affinity has been observed (14).
FIG. 1.
Alleles of TIF4632 used in this study. The position and type of amino acid substitution corresponding to each of the alleles are shown. The numbering of amino acids shown here includes the extra two amino acids introduced at the N terminus of the recombinant protein (26).
We chose to isolate multicopy suppressors of two of these mutations (tif4632-1 and tif4632-8) as a means to identify partners interacting with eIF4G2. Yeast strains containing either of these genes as the sole source of eIF4G within the cell were grown at the permissive temperature, transformed with a multicopy library of yeast genomic DNA, and then selected for growth at 37°C. Library plasmids that allowed for temperature-resistant growth were isolated, and their inserts were identified by sequencing (see Materials and Methods).
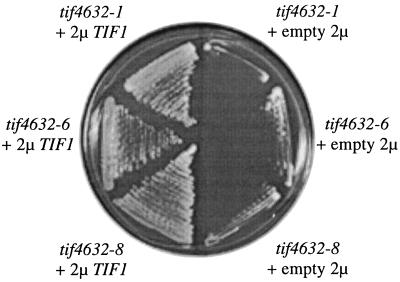
The inserts within these plasmids fell into three categories. One category consisted of inserts containing TIF4631 (7 isolates), another consisted of inserts containing TIF4632 (2 isolates), and the last consisted of inserts containing the yeast eIF4A1 gene TIF1 (23 isolates). The TIF1 gene was shown to be responsible for the suppression phenotype by the finding that a multicopy yeast plasmid containing only the TIF1 gene was able to rescue the temperature sensitivity of the tif4632-1, -6, and -8 mutants (Fig. 2). Excess TIF1 did not allow for cell growth in the absence of both the TIF4631 and TIF4632 genes, thereby indicating that overexpression of eIF4A did not lead to bypass suppression (data not shown). The eIF4A2 gene TIF2, which encodes a protein with a sequence identical to that of eIF4A1 (17), was not isolated in the original experiment. However, it was able to suppress the temperature sensitivity of each of these three mutant strains when it was expressed on a multicopy plasmid (Table 2). These data indicate that excess eIF4A within the three tif4632 mutant strains can reverse their growth arrest at 37°C.
FIG. 2.
Overexpression of eIF4A suppresses the temperature sensitivity of the tif4632-1, -6, and -8 mutants. The yeast eIF4A gene TIF1 in a multicopy (2μ) plasmid or the plasmid with no insert (empty 2μ) were transformed into the indicated tif4632 mutants to yield strains YAS2096, YAS2099, YAS2100, YAS2103, YAS2429, and YAS2430. The ability of the transformants to grow on minimal medium plates after 5 days at 37°C is shown.
Introduction of TIF1 on a centromeric (i.e., low-copy) plasmid into the tif4632-1, -6, and -8 strains also allowed for their temperature-resistant growth (data not shown). Comparative Western analysis of extracts derived from one of these strains (YAS2431) with antibodies directed against eIF4A revealed that it contained approximately twice the amount of eIF4A as the parental strain (YAS2429) (see Materials and Methods). This result suggests that approximately twice the normal level of eIF4A within the mutant cell is sufficient for suppression to be observed.
The temperature sensitivity of the conditional allele tif4632-430 was not suppressed by TIF1 on a multicopy plasmid (Table 2). This allele was previously characterized as being defective in eIF4E binding (27). Excess TIF1 also did not suppress the temperature sensitivity of the cdc33-1 strain, which contains a mutation within the eIF4E gene (1). Overexpression of the yeast cap binding protein eIF4E, which can suppress the temperature-sensitive phenotype of a tif4632-430 mutant (27), or the putative RNA helicase Ded1p, which can suppress the temperature sensitivity of various cdc33 mutants (5), did not suppress the growth phenotypes of the tif4632-1, -6, and -8 strains (Table 2). These data suggest that the TIF1-mediated suppression of the phenotypes of the tif4632-1, -6, and -8 strains is allele specific and not a general phenomenon of overexpression of an initiation factor.
Biochemical characterization of the eIF4G2-1, -6, and -8 proteins.
The expression of the mutant eIF4G2 proteins was examined at 37°C in order to test whether overexpression of eIF4A led to changes in their levels. Each of the mutant eIF4G2 proteins was epitope tagged with the influenza virus HA peptide at its N terminus and then expressed in cells also containing an untagged version of eIF4G2 in the genome and either TIF1 or no insert on a multicopy plasmid. The presence of the wild-type eIF4G2 protein allowed these cells to remain viable at 37°C. Comparative Western analysis of the amount of epitope-tagged eIF4G2 protein within crude extracts revealed nearly equal expression of the wild-type and the mutant eIF4G2 proteins in the presence or absence of excess TIF1 (Fig. 3A). This suggests that the TIF1-mediated suppression of the temperature-sensitive growth phenotype is not the result of increasing the levels of the mutant eIF4G2 proteins within the cell.
The association of the mutant eIF4G2 proteins with the cap binding protein eIF4E and the poly(A) binding protein Pab1p was also measured by coimmunoprecipitation of the epitope-tagged eIF4G2 proteins within crude lysates. As shown in Fig. 3B, the wild-type and mutant eIF4G2 proteins exhibited nearly equal abilities to associate with eIF4E and Pab1p within the extracts in the presence or absence of excess TIF1. This implies that the TIF1-mediated suppression of the tif4632-1, -6, and -8 phenotypes is not the result of enhanced complex formation between eIF4G2 and Pab1p or eIF4E. Western analysis of these samples with various antisera failed to reveal yeast eIF3 subunits or eIF4A within them (data not shown).
Heat-inactivated tif4632-8 translation extracts are stimulated by the addition of eIF4G2 or eIF4A.
One likely explanation for the above in vivo results is that translation is compromised in the eIF4G2 mutant strains. This possibility was tested by asking whether in vitro translation extracts from one of these strains, the tif4632-8 strain, exhibited a defect in translation and, if so, whether excess eIF4A could rescue this defect. The tif4632-8 strain was chosen as the source of mutant extract since it exhibited the most rapid arrest of growth at 37°C (data not shown).
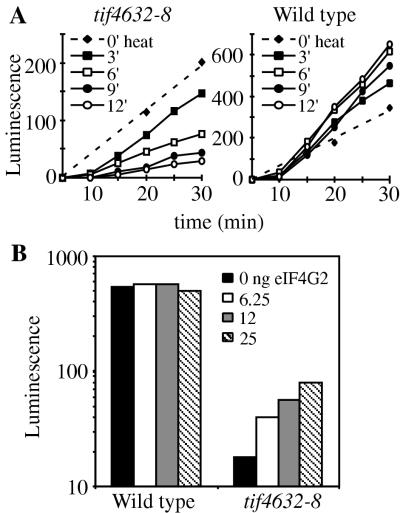
The translational capacities of the extracts were determined by measuring their ability to produce LUC protein when programmed with LUC mRNA. LUC protein expression was measured with a luminescence assay. As shown in Fig. 4A (dashed lines), the ability of the tif4632-8 extract to translate a polyadenylated LUC mRNA (LUCpA) was not significantly different from that of the TIF4632 extract. However, when the tif4632-8 extract was preheated at 30°C for increasing lengths of time, its translational activity at 26°C decreased nearly 10-fold after 12 min of heating. Similar results were obtained when the tif4632-8 extract was programmed with a capped and polyadenylated (capLUCpA) reporter mRNA, although the decrease in its activity after 12 min of heating was only fivefold (data not shown). In contrast, the wild-type extract gained activity (Fig. 4A). Extracts from the tif4632-430 mutant also gained activity, although not to the same degree (data not shown). We believe that this increase in activity is a result of nonspecific nucleases within the extract liberating translation factors, since pretreatment of the extracts with micrococcal nuclease has a similar stimulatory effect (data not shown). Western analysis of heated extracts indicated that heating did not change their amount of intact eIF4G2 (data not shown).
FIG. 4.
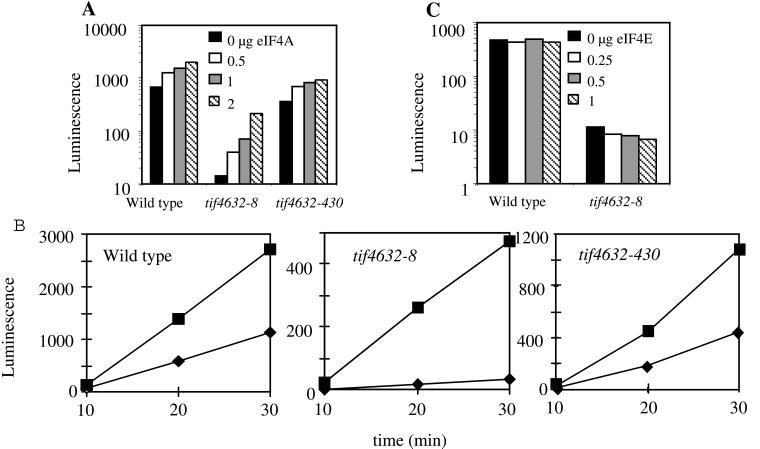
Characterization of eIF4G2 mutant and wild-type translation extracts. (A) Heat inactivation of tif4632-8 mutant extracts. TIF4632- or tif4632-8-derived translation extracts were heated at 30°C for the indicated times, frozen, thawed, and then mixed with LUCpA mRNA and incubated at 26°C for the indicated times under conditions permissive for translation. The amount of LUC protein produced in each of these mixtures was measured by using a luminescence assay. (B) Restoration of translation in heat-inactivated tif4632-8 extracts by the addition of recombinant GST-eIF4G2. The indicated translation extract was heated at 30°C for 12 min in the presence of the indicated amount of GST-eIF4G2 protein. After freezing, thawing, and mixing of the extracts with capLUCpA mRNA, they were incubated at 26°C for 30 min under conditions permissive for translation. The amount of LUC protein produced in each of these mixtures was measured by using a luminescence assay.
The heat inactivation of the tif4632-8 extract could be reversed upon the addition of recombinant eIF4G2 (Fig. 4B). Increasing amounts of highly purified GST-eIF4G2 were added to either wild-type or mutant extracts, which were then heated at 30°C for 12 min. The translational activity of each mixture was determined by programming it with capLUCpA mRNA and measuring the production of LUC protein at 26°C for 30 min. While the wild-type extract was not stimulated by the addition of GST-eIF4G2, this protein significantly stimulated the tif4632-8 extract, with 25 ng of protein returning it to its original level of activity before heating. Addition of GST-eIF4G2 in excess of 25 ng did not lead to any greater stimulation of the tif4632-8 extract and was inhibitory to the wild-type extract (data not shown). Stimulation of the tif4632-430 extract was also achieved upon the addition of GST-eIF4G2 (data not shown). The addition of GST protein alone did not stimulate the activity of any extract (data not shown). These data indicate that the tif4632-8 and tif4632-430 extracts are defective in translation as a result of inactive eIF4G2 proteins.
The translational activities of the heated wild-type and mutant extracts in the presence or absence of excess eIF4A were also assayed. Increasing amounts of recombinant eIF4A were added to the extracts, which were then heated for 12 min at 30°C. The translational activity of each mixture was then determined by programming it with capLUCpA mRNA and measuring the production of LUC protein at 26°C for 30 min. The maximal amount of eIF4A added in these experiments (2 μg) results in approximately a fourfold increase over the amount of endogenous eIF4A in the extract (see Materials and Methods). Therefore, the amount of eIF4A used in vitro is similar to the amount found within the yeast strains overexpressing eIF4A on a multicopy (eightfold) or centromeric (twofold) plasmid (see Materials and Methods).
In these experiments we were surprised to find that the wild-type extract was stimulated threefold by addition of 2 μg of recombinant eIF4A (Fig. 5A). A similar degree of stimulation was seen in the tif4632-430 extract (Fig. 5A). eIF4A also stimulated the tif4632-8 extract significantly, with 2 μg of eIF4A leading to an almost 15-fold increase in activity, which exceeds the extract’s original levels before inactivation. This level of activity was approximately one-third of the wild-type extract’s activity in the absence of excess eIF4A. It is important to note that the total amount of stimulation for the mutant extract (∼200 U) was less than that of the wild-type extract (∼1,350 U). While we cannot exclude the possibility that the mutant extract is limited by another factor, this finding is consistent with a less efficient association between eIF4G2-8 and eIF4A.
FIG. 5.
Stimulation of translation in tif4632-8 extracts by the addition of eIF4A. (A) Stimulation by eIF4A. The indicated extracts were heated at 30°C for 12 min in the presence of the indicated amounts of eIF4A and then assayed for translation of the capLUCpA mRNA as described in the legend to Fig. 4. (B) Rates of LUC production in extracts lacking and containing excess eIF4A. The indicated extracts were heated as described for panel A in the presence (squares) or absence (diamonds) of 2 μg of eIF4A. Aliquots of translation reaction mixtures incubated at 26°C and programmed with capLUCpA mRNA were withdrawn at the indicated times and assayed for LUC protein with a luminescence assay. (C) eIF4E does not stimulate translation in tif4632-8 extracts. Extracts were heated and assayed for LUC protein production from capLUCpA mRNA as described above in the presence of the indicated amounts of eIF4E.
These measurements were performed during a time period in which the amount of LUC protein within the extract was increasing linearly (Fig. 5B). This stimulatory effect of eIF4A can thus be attributed to an effect on the rates of protein synthesis. The translational activity in eIF4A-stimulated extracts was inhibited by cap analog, indicating that this translation was cap dependent and not a result of increased internal initiation. The stimulatory effect on the extract caused by excess eIF4A was not a general phenomenon of addition of a translation initiation factor, since the addition of eIF4E to the tif4632-8 extract had no stimulatory effect (Fig. 5C). In summary, these data show that the tif4632-8 extract is limiting for eIF4G2 (Fig. 4B) and that it is highly responsive to the addition of eIF4A (Fig. 5A).
Association of yeast eIF4A with eIF4G in vitro.
The experiments described above showed that overexpression of eIF4A led to the ability of the tif4632-1, -6, and -8 strains to grow at 37°C, that this dosage suppression was not the result of indirect effects on the mutant eIF4G2 expression levels, and that excess eIF4A had less of a stimulatory effect on extracts derived from the tif4632-8 strain than on those derived from a wild-type strain. Although these data are all consistent with the hypothesis that eIF4G and eIF4A associate in yeast and that the eIF4G2-8 protein associates poorly with eIF4A, it also was possible that the eIF4A effects were nonspecific.
In order to test directly whether the eIF4G2-8 protein bound less well to eIF4A than its wild-type counterpart, we first needed to investigate whether yeast eIF4G can bind eIF4A. Attempts to detect eIF4A in immunoprecipitates of eIF4G2 from crude extracts by using several conditions similar to those described in Fig. 3B failed to reveal significant eIF4A association. Because these failures could have been due to the intrinsically low affinity of eIF4A for eIF4G, we decided to further pursue these binding studies with recombinant proteins under conditions that maintained their high concentrations during most of the assay procedure.
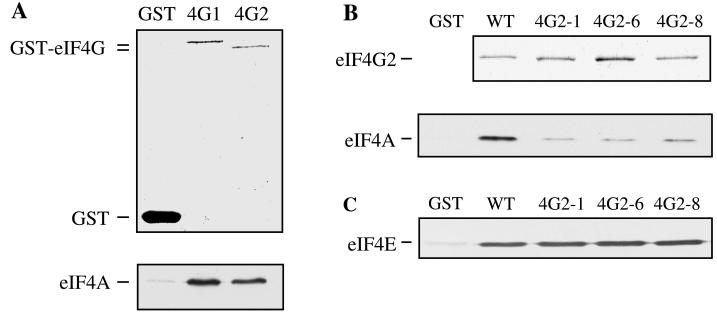
Highly purified recombinant eIF4G1 or eIF4G2 proteins fused at their N termini to GST were immobilized on glutathione-Sepharose resin and incubated with recombinant yeast eIF4A. After extensive washing of the resin, bound proteins were eluted with SDS, resolved by SDS-PAGE, and detected by either Coomassie brilliant blue staining (eIF4G) or Western analysis (eIF4A). As shown in Fig. 6A, binding of eIF4A to GST-eIF4G1 and to GST-eIF4G2, but not to GST alone, was observed. This interaction was not sensitive to prior treatment of the GST-fusion proteins with RNase, indicating that this association was not the result of a nonspecific tethering phenomenon (26) (data not shown). The amount of eIF4A bound to the eIF4G fusion proteins was substoichiometric in these assays, since eIF4A in the eluted fractions was not visible by Coomassie brilliant blue staining (see Discussion). These data show that yeast eIF4A and eIF4G can bind in the absence of other proteins.
FIG. 6.
In vitro binding of eIF4A to eIF4G. (A) Yeast eIF4A binds to eIF4G1 and eIF4G2 in vitro. Purified GST or GST-eIF4G1 or GST-eIF4G2 fusion protein immobilized on glutathione-Sepharose resin was incubated with recombinant eIF4A and then washed extensively. Bound proteins were eluted with SDS, resolved on an SDS–10% polyacrylamide gel, and detected by either Coomassie brilliant blue staining (GST-eIF4G) or Western analysis (eIF4A). (B) Differential in vitro binding of eIF4A to wild-type and mutant eIF4G2 proteins. The indicated GST-eIF4G2 fusion proteins were immobilized on glutathione-Sepharose resin and analyzed for eIF4A binding as described for panel A. The eIF4G protein was detected by Coomassie brilliant blue staining, and the eIF4A protein was detected by Western analysis. WT, wild type. (C) Association with eIF4E. The procedure was the same as for panel A, except eIF4E was incubated with the indicated GST-eIF4G fusion proteins and was detected by Western analysis with anti-eIF4E antibodies.
Highly purified GST–eIF4G2-1, -6, and -8 proteins were also immobilized on glutathione resin and incubated with recombinant eIF4A. The amount of eIF4A that remained associated with these forms of eIF4G2 after washing of the resin was significantly lower than the amount bound to the wild-type GST-eIF4G2 protein (Fig. 6B). In contrast, the GST–eIF4G2-430 protein bound eIF4A as well as the wild-type protein (data not shown). As a positive control for the ability of the mutant fusion proteins to bind an associated factor, we also analyzed their binding to eIF4E. As shown in Fig. 6C, the mutant and wild-type GST-eIF4G2 proteins associated with eIF4E equally well. Therefore, the differential ability of wild-type and mutant eIF4G2 proteins to bind to eIF4A in these assays suggests that the mutant eIF4G2 proteins are defective in their eIF4A binding capabilities.
DISCUSSION
This work presents evidence for the association between yeast eIF4G and eIF4A. Specifically, we found that excess eIF4A suppresses the temperature sensitivity of specific tif4632 mutants. Also, heat-inactivated translation extracts from the tif4632-8 mutant could be significantly stimulated by recombinant eIF4G2 and by the addition of excess eIF4A. Finally, we found that eIF4G fusion proteins bound to eIF4A in vitro and that mutant eIF4G2 proteins bound less well. Taken together, these data provide evidence to support the conclusion that yeast eIF4G and eIF4A interact within the translational apparatus.
Several observations suggest that the in vitro translation data accurately reflect what is seen in vivo. First, both the tif4632-8 and -430 extracts were limited for eIF4G, an expected result since they both contain mutated versions of eIF4G2. Second, the fold stimulation upon addition of eIF4A was much lower in the tif4632-430 extract than in the tif4632-8 extract. This parallels the in vivo result that multicopy TIF1 was unable to rescue the temperature sensitivity of the tif4632-430 strain. Third, the significant translational stimulation of the tif4632-8 extract upon addition of eIF4A mimics the stimulation of growth of the mutants expressing excess eIF4A. Finally, the amount of eIF4A needed to restore near-normal rates of protein synthesis in heat-inactivated extracts was comparable to the amount of excess eIF4A that was needed to support growth of the tif4632 mutants at 37°C.
Our observation that wild-type extracts are also stimulated by excess eIF4A raises at least two important points. First, it suggests that the amount of eIF4A in extracts limits their translational capacity. This is a somewhat unexpected finding, since eIF4A is one of the most abundant translation initiation factors. The second point is that the stimulation of the tif4632-8 extract by eIF4A, as well as the genetic suppression by excess eIF4A, could be due to general stimulatory effects of eIF4A on translation. While we cannot completely exclude this possibility, the allele specificity of TIF1-mediated suppression, the inability of eIF4A to stimulate in the tif4632-8 extract the same amount of total translation as in the wild-type extract, and the in vitro binding data more fully support the alternative hypothesis that eIF4G and eIF4A do interact and that all of our observations reflect a loss of affinity between these proteins.
Why has the association of yeast eIF4G and eIF4A not been seen previously? One possibility is that the affinity of yeast eIF4G for eIF4A is too low for the complex to have withstood previous experimental measures. This possibility is supported by several of our observations. Recombinant eIF4A bound to recombinant eIF4G at much less than a 1:1 stoichiometry in our in vitro binding studies. We could not successfully coimmunoprecipitate eIF4A with eIF4G2. Finally, we were unable to detect significant amounts of eIF4A by Western analysis in the mixture of proteins that copurify with yeast eIF4E by cap analog chromatography (data not shown).
This lowered affinity between yeast eIF4G and eIF4A could result from the fact that yeast eIF4G, unlike mammalian eIF4G, does not contain the second, C-terminally localized eIF4A binding site. It is possible that the presence of two eIF4A binding sites within mammalian eIF4G stabilizes eIF4A binding. Perhaps the second binding site for eIF4A is found on another yeast initiation factor, and eIF4A is retained with affinities comparable to those of mammalian eIF4A when that protein becomes part of the initiation complex. Future work will need to address this possibility in more detail.
In summary, this work has provided biochemical and genetic evidence to support the conclusion that yeast eIF4G and eIF4A functionally and physically interact. The recruitment of eIF4A to the 5′ end of mRNA via its association with eIF4G is thought to be an essential step in the translation initiation process. Our identification of this interaction within the yeast system lends support to the generality of this model. Future goals are now to understand the details of how eIF4G and eIF4A interact and how this interaction leads to the stimulation of translation.
ACKNOWLEDGMENTS
We thank Jon Lorsch for the generous supply of recombinant eIF4A and members of our group for critical reading of the manuscript.
This work was supported by NIH grant 50308 to A.B.S.
REFERENCES
- 1.Altmann M, Trachsel H. Altered mRNA cap recognition activity of initiation factor 4E in the yeast cell cycle division mutant cdc33. Nucleic Acids Res. 1989;17:5923–5931. doi: 10.1093/nar/17.15.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeke J D, Trueheart J, Natsoulis G, Fink G. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 3.Brenner C, Nakayama N, Goebl M, Tanaka K, Toh-e A, Matsumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chuang R Y, Weaver P L, Liu Z, Chang T H. Requirement of the DEAD-box protein Ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 5.Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edery I, Altmann M, Sonenberg N. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene. 1988;74:517–525. doi: 10.1016/0378-1119(88)90184-9. [DOI] [PubMed] [Google Scholar]
- 7.Gingras, A.-C., B. Raught, and N. Sonenberg. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem, in press. [DOI] [PubMed]
- 8.Goyer C, Altmann M, Lee H S, Blanc A, Deshmukh M, Woolford J L, Trachsel H, Sonenberg N. TIF4631 and TIF4632: two genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif sequence and carry out an essential function. Mol Cell Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goyer C, Altmann M, Trachsel H, Sonenberg N. Identification and characterization of cap-binding proteins from yeast. J Biol Chem. 1989;264:7603–7610. [PubMed] [Google Scholar]
- 10.Guthrie C, Fink G. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:14–15. [PubMed] [Google Scholar]
- 11.Hentze M W. eIF4G: a multipurpose ribosome adaptor? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 12.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 16.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 17.Linder P, Slonimski P P. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc Natl Acad Sci USA. 1989;86:2286–2290. doi: 10.1073/pnas.86.7.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrick W C, Hershey J W B. The pathway and mechanism of eukaryotic protein synthesis. In: Hershey J W B, Matthews M, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 31–69. [Google Scholar]
- 19.Morley S J, Curtis P S, Pain V M. eIF4G: translation’s mystery factor begins to yield its secrets. RNA. 1997;3:1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 20.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4G1 and evicts poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors eIF4A and eIF4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachs A B, Deardorff J A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 23.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eucaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 24.Schmid S R, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarun S, Sachs A B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 26.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 27.Tarun S Z, Sachs A B. Binding of eIF4E to eIF4G represses translation of uncapped mRNA. Mol Cell Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarun S Z, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF-4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasilescu S, Ptushkina M, Linz B, Muller P P, McCarthy J E G. Mutants of eukaryotic initiation factor eIF-4E with altered mRNA cap binding specificity reprogram mRNA selection by ribosomes in Saccharomyces cerevisiae. J Biol Chem. 1996;271:7030–7037. doi: 10.1074/jbc.271.12.7030. [DOI] [PubMed] [Google Scholar]
- 30.Wells S E, Hillner P, Vale R, Sachs A B. Circularization of mRNA by eucaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]