Abstract
β-Catenin stabilizes the cadherin cell adhesion complex but, as a component of the Wnt/Wg signaling pathway, also controls gene expression by forming a heterodimer with a transcription factor of the LEF-TCF family. We demonstrate that the substrate adhesion molecule fibronectin is a direct target of Wnt/Wg signaling. Nuclear depletion of β-catenin following cadherin transfection in Xenopus fibroblasts resulted in downregulation of fibronectin expression which was restored by activating the Wnt/Wg signaling cascade via LiCl treatment or transfection of either Xwnt-8 or β-catenin. We isolated the Xenopus fibronectin gene (FN) promoter and found four putative LEF-TCF binding sites. By comparing the activities of different fibronectin gene reporter constructs in fibroblasts and cadherin transfectants, the LEF-TCF site at position −368 was identified as a Wnt/Wg response element. LEF-1-related proteins were found in nuclei of the fibroblasts but were absent in a kidney epithelial cell line. Consistent with the lack of these transcription factors, the FN promoter was silent in the epithelial cells but was activated upon transfection of LEF-1. Wild-type Xenopus Tcf-3 (XTcf-3) was unable to activate FN promoter reporter constructs, while a mutant lacking the groucho binding region behaved like LEF-1. In contrast to XTcf-3, LEF-1 does not interact with groucho proteins, which turn TCFs into activators or repressors (J. Roose, M. Molenaar, J. Hurenkamp, J. Peterson, H. Brantjes, P. Moerer, M. van de Wetering, O. Destreé, and H. Clevers, Nature 395:608–612, 1998). Together these data provide evidence that expressing LEF-1 enables fibroblasts, in contrast to epithelial cells, to respond to the Wnt/Wg signal via β-catenin in stimulating fibronectin gene transcription. Our findings further promote the idea that due to its dual function, β-catenin regulates the balance between cell-cell and cell-substrate adhesion.
The β-catenin proto-oncogene, the vertebrate homolog to armadillo, links cell adhesion and cell differentiation; it stabilizes cell-cell adhesion by anchoring cadherins via α-catenin to the cytosekeleton (28, 55) and transduces the Wnt/Wg signal to target genes by interacting with transcription factors of the LEF-TCF family (1, 24, 39). The evolutionarily highly conserved Wnt/Wg signaling cascade includes a membrane-integrated receptor of the frizzled (fz) family, which activates the phosphoprotein dishevelled (dsh), which leads to inhibition of glycogen synthase kinase 3β. Because β-catenin is a substrate of this serine/threonine kinase, it remains hypophosphorylated upon Wnt/Wg signaling and accumulates in the cytoplasm. This promotes its binding to LEF-TCF transcription factors. The β-catenin–LEF-TCF heterodimer enters the nucleus and is able to activate or repress gene transcription (for detailed reviews, see references 4, 15, and 32). Important developmental target genes, such as siamois, twin, and nodal-related 3 in Xenopus as well as Ubx (Ultrabithorax) in Drosophila, were found to be controlled by direct binding of β-catenin–LEF-TCF to their promoters (3, 34, 36, 46).
Abnormally high concentrations of β-catenin have been reported in several tumor and carcinoma cell lines caused by mutations in the adenomatous polyposis coli gene or β-catenin gene. These mutations prevent the degradation of β-catenin (41), which then contributes to the formation of a constitutively active β-catenin–LEF-TCF transcription complex (25, 27, 31, 38, 40, 48, 54). Most recently, He et al. (18) have identified the proto-oncogene c-myc as a direct target gene of the β-catenin–Tcf-4 complex in a human colorectal cancer cell line. This links upregulation of β-catenin to loss of proliferation control in tumorigenesis.
The different functions of β-catenin, strengthening of cadherin-mediated cell adhesion and regulation of target genes of the Wnt/Wg signaling pathway, can compete with each other. When cytosolic or nuclear β-catenin is tethered to the plasma membrane by cadherin overexpression in Xenopus embryos, severe developmental defects are observed due to inhibition of β-catenin’s nuclear function (9, 19). Using a quite similar approach, we have previously shown that ectopic expression of cadherins in Xenopus XTC fibroblasts shifted β-catenin to the plasma membrane and led to downregulation of fibronectin and α3β1 integrin synthesis. Additionally, the transfectants altered their adhesive properties, losing their ability to adhere to substrate molecules (10). These findings give evidence for a cross talk between cell-cell and cell-substrate adhesion regulated by β-catenin. Other examples of mutual interference between the two adhesion systems have been reported for different cell types. When keratinocytes were treated with cadherin antibodies, expression of α6β1 integrin persisted during terminal differentiation (22), while dominant-negative expression of E-cadherin resulted in disappearance of α2β1 and α3β1 integrins (58). Recently, Novak et al. (42) have shown that overexpression of integrin-linked kinase repressed E-cadherin synthesis in epithelial cells. These reports emphazise a cross talk between the cell-cell and the cell-substrate adhesion systems; however, the underlying regulatory mechanisms remain to be clarified.
In a first step toward gaining insight into a possible regulatory principle, we extended our previous studies on fibroblasts stably transfected with cadherins (10). We analyzed the molecular mechanism by which cadherin expression leads to fibronectin gene repression. Here, we present evidence that β-catenin in its function as a Wnt/Wg signal transducer induces the expression of the cell-substrate adhesion molecule fibronectin. Downregulation of fibronectin gene expression in cadherin-transfected Xenopus fibroblasts was abolished when the Wnt/Wg signal pathway was activated. Consistent with these findings, we demonstrate that the isolated Xenopus fibronectin gene promoter was upregulated by β-catenin via a LEF-TCF target site. Furthermore, in epithelial cells the promoter was silent due to lack of LEF-1 expression, but it was active upon LEF-1 transfection. These studies assign β-catenin a key role in coordinating cell-cell and cell-substrate adhesion.
MATERIALS AND METHODS
Cell culture and transfection experiments.
The renal epithelial cell line A6 (ATCC CCL 102; American Type Culture Collection), the fibroblastic cell line XTC, and the cadherin transfectants, XTC-XB (10), were routinely grown in 70% Dulbecco modified Eagle medium supplemented with 10% fetal calf serum in the presence of 7% CO2. For transfection experiments, 1 to 5 μg of plasmid DNA was applied either by calcium phosphate precipitation (13) for XTC and XTC-XB cells or by the use of Lipofectin (Life Technologies, Karlsruhe, Germany) for A6 cells. Xwnt-5A- and Xwnt-8-containing plasmids, kindly provided by R. T. Moon (Seattle, Wash.), were subcloned by using the HindIII and XbaI restriction sites of pRc/CMV. pBAT–β-catenin and LEF-1 constructs were gifts from J. Behrens (1) (MDC, Berlin, Germany). Xenopus Tcf-3 (XTcf-3) was isolated in our hands from a Xenopus gastrula stage cDNA library and inserted into pCS2 by using XhoI sites. The mutant XTcf-3Δgrg was kindly provided by H. Clevers (47). For each experiment, cells were seeded 1 day prior to transfection at 60 to 70% confluency and harvested 2 days after transfection. Lithium treatment was carried out by applying 60 mM LiCl in complete medium for 10 min. Two days after lithium treatment, cells were harvested.
Immunoblotting.
For fibronectin detection, cells were lysed in the presence of 2 M urea. Twenty micrograms of total protein was separated by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis, transferred onto nitrocellulose, and incubated with monoclonal antibody 6D9 as described elsewhere (6, 10). For the detection of LEF-1-related proteins, cells were lysed in radioimmunoprecipitation assay buffer containing 10 mM HEPES (pH 7.5), 150 mM NaCl, 2 mM EGTA, 2 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, and 1 μg of leupeptin per ml. Ten micrograms of total protein was separated by SDS–7.5% PAGE, transferred onto nitrocellulose, and stained with a polyclonal antiserum against LEF-1 (a gift from J. P. von Kries, MDC, Berlin, Germany). As a secondary antibody, either goat anti-mouse or goat anti-rabbit immunoglobulin G coupled to horseradish peroxidase (Dianova, Hamburg, Germany) was used. Visualization was performed with the ECL detection system (Amersham, Braunschweig, Germany).
Immunostaining.
For visualizing the localization of β-catenin or LEF-1 homologs, cells were seeded on coverslips. At 48 h after transfection or LiCl treatment cells were fixed with 3% paraformaldehyde in amphibian buffered saline (APBS) (103 mM NaCl, 2.7 mM KCl, 0.15 mM KH2PO4, 0.7 mM NaH2PO4), incubated for 8 min with 0.1% Triton X-100 in APBS, and blocked with 1% bovine serum albumin. Cells were incubated overnight at 4°C with an affinity-purified polyclonal antiserum against β-catenin (provided by K. Herrenknecht, Eisai London Research, London, United Kingdom) or a polyclonal antiserum against LEF-1. Cells were rinsed in APBS before incubation with goat anti-rabbit immunoglobulin G–Cy3 conjugate (Dianova) at room temperature for 1 h. After extensive washing in APBS, nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole) (Merck, Darmstadt, Germany). Cells were mounted in elvanol and analyzed by confocal laser scanning microscopy (TCD Leica).
RT-PCR.
RNA was isolated as described by Chirgwin et al. (5). One microgram of total RNA was reverse transcribed by using random hexamer primers and Moloney murine leukemia virus (MMLV) reverse transcriptase (Life Technologies). For PCR, 1/25 of the reverse transcription (RT) reaction mixture was amplified with 28 cycles for fibronectin and histone 4 or 32 cycles for XLEF-1, XTcfs, and XGrgs. The following primer pairs were designed for detection: fibronectin, Fn1 (5′-TTCCTGCACAAGTGAGGGTC-3′) and Fn2 (5′-AGCATCGCACTTCCATCTGC-3′); histone 4, H4/1 (5′-CGGGATAACATTCAGGGTATCACT-3′) and H4/2 (5′-ATCCATGGCGGTAACTGTCTTCCT-3′); XTcf-3, XTcf-31 (5′-CAGCGCTCGCGCGTACTTAC-3′) and XTcf-32 (5′-GCCACGTNCGCCCAAGGATCTGGT-3′); XTcf-4, XTcf-41 (5′-AACCCCCCACCGCACTTTAC-3′) and XTcf-42 (5′-GGCTGCGCTTTCTTTTAACG-3′); XLEF-1, XLEF-11 (5′-GATCTTCGCCGAGATCAGTC-3′) and XLEF-12 (5′-GTGGGATCCCGGAGAAAAGT-3′); XGrg4, XGrg41 (5′-GTCATTCCTT TCTTGTCCCA-3′) and XGrg42 (5′-CCGAATGGGG TTGGATAAGG-3′); and XGrg5, XgGrg51 (5′-GATGAATTCC AACTGCTGCA-3′) and XGrg52 (5′-CTCGTTCAAT CGCTCCCAGG-3′). The XTcf-4-specific primers were designed based on the sequence of a 600-bp PCR fragment showing homology to human and murine Tcf-4 unpublished data.
Isolation of the fibronectin gene promoter.
A Xenopus genomic λ-FIX library was screened with probes representing the 5′ regions of Xenopus cDNA clones (8). Four independent clones were isolated. Southern blot analysis indicated that a 7.8-kb fragment, named XFN30.1, included the 5′ end of the cDNA probe. This fragment was subcloned into pUC19 and pBluescript. Sequence analysis was performed on both DNA strands by using either standard primers binding within the vectors or synthetic fibronectin oligonucleotides. The transcription start site was determined by primer extension studies with the primer sequences FN-ext.3 (5′-GGATACAAAAGCGGAGGCGAAAATAACCAG-3′, corresponding to positions +92 to +63) and FN-ext.4 (5′-GAAAGGAAAAGAAAGCGCAAAAGCTGCGAG-3′, corresponding to positions +53 to +24). Gel-purified deoxyoligonucleotides were labeled with 150 μCi of [γ-32P]ATP by using T4 polynucleotide kinase. Each deoxyoligonucleotide (105 cpm) was annealed to 50 μg of total cellular RNA isolated from Xenopus XTC fibroblasts (5). Oligonucleotides were extended by using MMLV reverse transcriptase (Life Technologies), and the extension products were analyzed following RNase A digestion on 8% acrylamide gels with 7 M urea.
Construction of fibronectin reporter gene constructs.
Two constructs for transient cell transfections (−3403/+20 and −1334/+20) were designed by ligating restriction fragments of XFN30.1 into pGL3-basic (Promega). This plasmid does not contain any promoter in front of the luciferase gene. Correct orientation of the fragments was verified by sequencing. Deletion mutants were constructed by PCR techniques with the following upstream primer sequences: −908, 5′-GGGCCCAAGCTTATGTGCTGGAAAAATATGTT-3′; −499, 5′-GGGCCCAAGCTTGCCTGTTTTTATATGGTCAT-3′; −309, 5′-GGGCCCAAGCTTAGAGCAAAGTGAACTAATAA-3′; and −199, 5′-GGGCCCAAGCTTAACGCTATAAAGACGAACCA-3′. Downstream primers were as follows: for all +20 constructs, 5′-CGCGCGAAGCTTCGCTAAGACAGAGG-3′; and for the −149 construct, 5′-CGCGCGAAGCTTGATTTGGTGGGGATGTGGGG-3′. The amplified fragments were inserted into pGL3-basic by using the primer-inserted HindIII site and sequenced. To prepare the −499/+20 mt construct, two PCR fragments were amplified independently by using the following primer sets: (i) the −499 upstream primer in combination with 5′-AATAAACAAGGGAGAGGCGCTGTTAATG-3′ and (ii) the +20 downstream primer in combination with 5′-CATTAACAGCGCCTCTCCCTTGTTTATT-3′. Both fragments were gel purified, combined, and again amplified by using the flanking primers. The resulting fragment was ligated into pGL3-basic and sequenced.
Reporter gene assays.
XTC fibroblasts or epithelial A6 cells at 70% confluency were transfected with 10 μg of fibronectin-luciferase constructs and 10 μg of pCMV-βgal (a gift of H. Steinbeisser, MPI, Tübingen, Germany). For cotransfection experiments, 5 μg of pRc/Xwnt-8, pRc/Xwnt-5A, pBAT-β-catenin, LEF-1, or pRc/CMV was transfected. After 48 h, cells were harvested. Luciferase activity was measured with a commercially available kit (Promega), and β-galactosidase activity was determined as described previously (49). As a negative control, cells were transfected with pGL3-basic. For positive controls, a cytomegalovirus (CMV)-luciferase construct, kindly provided by W. Knöchel (Ulm, Germany), was used. The luciferase activity was subsequently normalized to β-galactosidase activity to eliminate differences in transfection efficiency.
Fusion proteins, nuclear extracts, and bandshift studies.
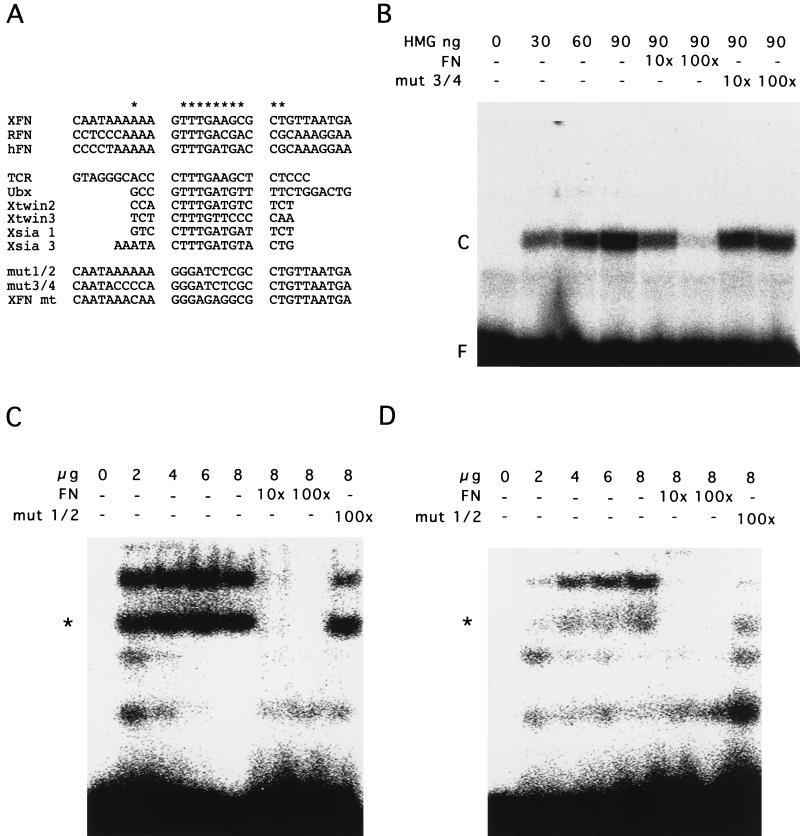
His-tagged fusion protein containing the LEF-1 HMG box (a gift of J. P. von Kries, MDC, Berlin, Germany) was expressed in BL21(DE3) cells and purified under native conditions by using nitrilotriacetic acid-agarose. Proteins were eluted by using an imidazole step gradient and monitored for homogeneity by SDS-PAGE and Coomassie blue staining procedures. Nuclear extracts were prepared as described previously (50). Prior to binding to labeled oligonucleotides, all protein solutions were dialyzed against binding buffer (20 mM Tris HCl [pH 8.0], 50 mM NaCl, 5% glycerol, 0.1 mg of bovine serum albumin per ml, 1 mM dithiothreitol, 1 mM MgCl2). Protein concentrations were measured by the Bradford assay. Binding reactions were carried out at room temperature for 20 min in a final volume of 15 μl containing 50 ng of poly[d(I-C)] and 0.1 pmol of labeled double-stranded deoxyoligonucleotide, and products were separated on 7% nondenaturing polyacrylamide gels. All oligonucleotides were gel purified prior to labeling with T4 polynucleotide kinase and [γ-32P]ATP. The sequences of the oligonucleotides and competitors used are indicated in Fig. 5A.
FIG. 5.
Interaction of the identified Wnt/Wg response element with HMG box fusion protein and nuclear extracts. (A) Comparison of putative Wnt/Wg response elements with the LEF-TCF consensus binding sequence in different promoters. XFN, Xenopus fibronectin gene promoter; RFN, rat fibronectin gene promoter (43); HFN, human fibronectin gene promoter (7); TCRα, T-cell receptor α (12), Ubx, ultrabithorax (46); Xtwin2 and -3, Xenopus twin (34); and Xsia 1 and 3, Xenopus siamois (3). mut 1/2 and mut 3/4, two mutated XFN sequences used for competition experiments; mt, mutated LEF-TCF target site in the −499/+20 mt construct. (B) Electrophoretic mobility shift assay with the XFN oligonucleotide and a fusion protein consisting of the LEF-1 HMG box. Amounts of HMG box protein are indicated. For competition studies, oligonucleotide mut 3/4 was used. F, free oligonucleotides; C, complex of oligonucleotide and protein. (C) Electrophoretic mobility shift assay with the XFN oligonucleotide and nuclear extracts of Xenopus fibroblasts (XTC). Amounts of nuclear extracts are indicated. For competition studies, oligonucleotide mut 1/2 was used. Two slower-migrating bands were identified. (D) Electrophoretic mobility shift assay with the XFN oligonucleotide and nuclear extracts of a Xenopus kidney epithelial cell line (A6). Amounts of nuclear extracts are indicated. For competition studies, oligonucleotide mut 1/2 was used. The band marked with an asterisk was specific in both XTC and A6 cells, whereas the slower-migrating band was specific only in XTC cells.
RESULTS
Fibronectin expression in cadherin-transfected fibroblasts is restored by activation of the Wnt/Wg signaling cascade.
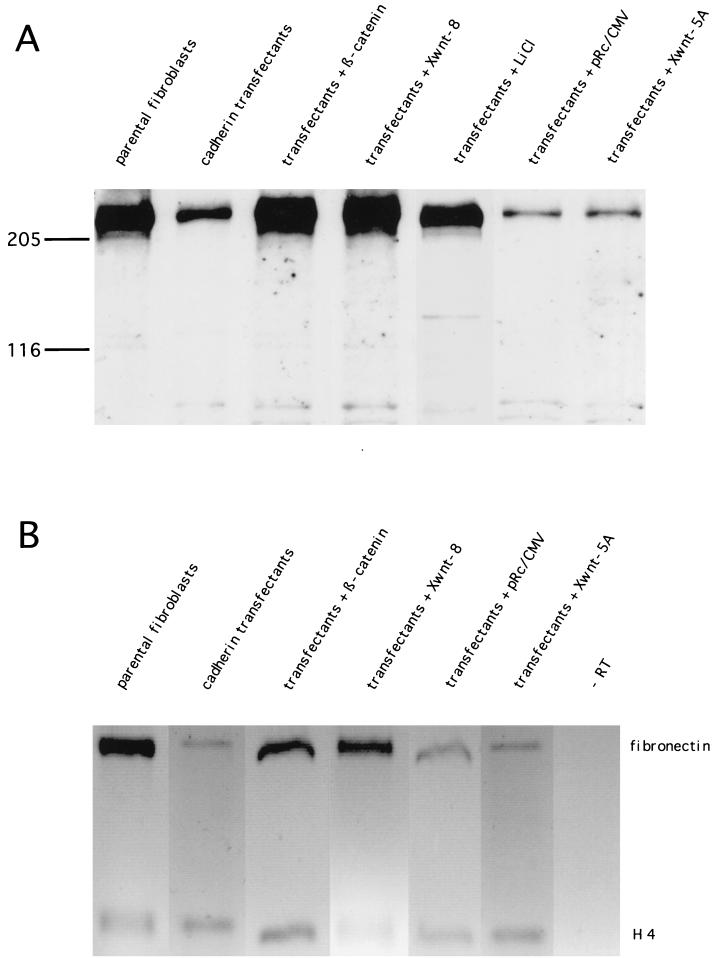
We have shown previously that expression of cadherins in Xenopus fibroblasts downregulates the synthesis of fibronectin and α3β1 integrin (10). Cadherins bind cytoplasmic β-catenin and could thus interfere with β-catenin’s function in the Wnt/Wg pathway (9). We therefore wanted to examine whether fibronectin gene expression is controlled by the Wnt/Wg pathway. Several clones of stably XB/U-cadherin-transfected Xenopus fibroblasts that show downregulated fibronectin synthesis were transiently transfected with Xwnt-8 or β-catenin cDNA or were treated with LiCl, which inhibits glycogen synthase kinase 3β (20, 30). Remarkably, we found by immunoblot and RT-PCR analyses that in all clones strong fibronectin protein and mRNA syntheses were restored (Fig. 1). Transfection of Xwnt-5A, which belongs to a different Wnt subfamily (53), did not reactivate fibronectin synthesis.
FIG. 1.
Fibronectin expression is restored in cadherin-transfected cells by transient cotransfection of β-catenin or Xwnt-8 or by lithium treatment. (A) Immunoblot analyses of fibronectin expression with monoclonal antibody 6D9, specific for Xenopus fibronectin. (B) RT-PCR analyses showing the 550-bp fragment of fibronectin and, as an internal standard, the 220-bp fragment of histone H4.
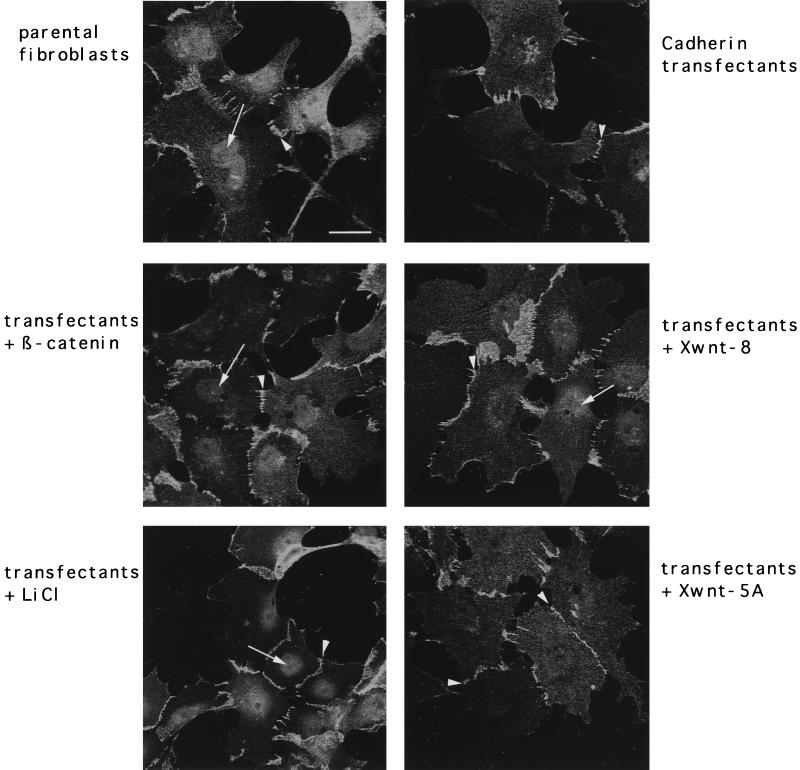
Nuclear localization of β-catenin indicates that the Wnt/Wg pathway is active (4). We were therefore interested in examining the cellular distribution of endogenous β-catenin in our cell lines. Cadherin transfection of Xenopus fibroblasts and stimulation of the Wnt/Wg signaling pathway in these transfectants resulted in a marked alteration of β-catenin distribution. In the parental cells (Fig. 2), β-catenin was localized at the cell membrane, in the cytoplasm, and also significantly in the nucleus. Membrane localization of β-catenin in these fibroblasts may be explained by the presence of endogenous N-cadherin and Xcadherin-11, which was detected by RT-PCR (data not shown). As expected, the cadherin transfectants showed enhanced membrane staining of β-catenin and some cytoplasmic staining surrounding the nucleus, while their nuclei were depleted (Fig. 2). However, additional transfection of either β-catenin or Xwnt-8 as well as LiCl treatment resulted in a reappearance of the nuclear β-catenin staining. Xwnt-5A expression did not alter the distribution of β-catenin (Fig. 2). As previously reported by others (20, 33), we also observed an increase in the amount of endogenous β-catenin upon Xwnt-8 transfection or LiCl treatment (data not shown). Thus, concomitant with the reactivation of fibronectin expression in the cadherin transfectants following cotransfection of β-catenin or Xwnt-8 or LiCl treatment, β-catenin was translocated to the nuclei.
FIG. 2.
Subcellular localization of endogenous β-catenin in fibroblasts and cadherin transfectants, prior to and after stimulation of the Wnt/Wg signaling cascade. In the parental fibroblastic cell line, β-catenin is found in nuclei (arrow) and the cell membrane (arrowhead). In the cadherin transfectants, the β-catenin signal is restricted to the cell membrane (arrowheads). Nuclear staining of β-catenin in the cadherin transfectants (arrows) was observed only following cotransfection with β-catenin or Xwnt-8 or LiCl treatment and not after cotransfection with Xwnt-5A. Bar, 10 μm.
Identification of a LEF-TCF target site and Wnt/Wg response element in the Xenopus fibronectin promoter.
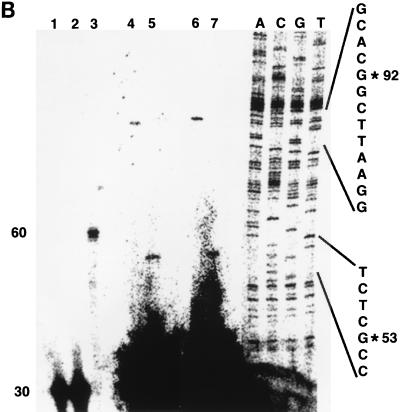
To investigate whether the fibronectin gene is a direct target gene of the Wnt/Wg signal, we isolated the Xenopus fibronectin gene (FN) promoter. We screened a Xenopus genomic library by using as a probe the 5′ region of the Xenopus fibronectin cDNA. We obtained a 3,400-bp genomic fragment that represents the 5′ flanking region of the fibronectin gene (EMBL accession no. Y13284). The transcription start site was mapped by primer extension with RNA isolated from XTC cells. The primer sequences used (FN-ext.3 and FN-ext.4) are described in Materials and Methods. While primer FN-ext.3 was extended to 92 nucleotides, the FN-ext.4 extension product was 53 nucleotides in length after the reverse transcriptase reaction (Fig. 3B). From this data one transcription start can be defined (Fig. 3A). Also in other organisms (rat and human) only one transcription start was found in the 5′ flanking region of the fibronectin gene. The distance between the transcription and translation start sites is in the same range as was found for the rat or human homolog (7, 43).
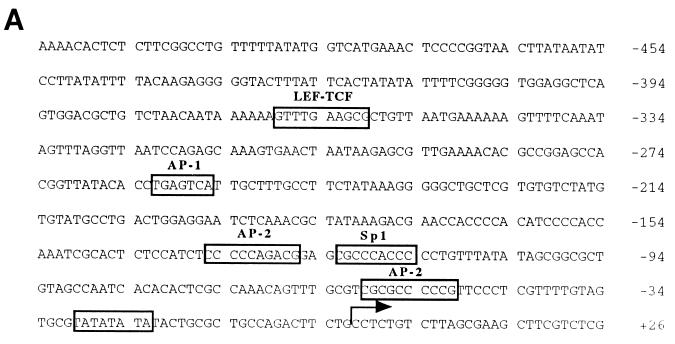
FIG. 3.
Determination of the transcription start site of the FN promoter. (A) Partial nucleotide sequence (−533/+27) of the genomic fibronectin clone XFN30.1. Numbering is with respect to the transcription start site (arrow). Conserved transcription factor binding sites are boxed. (B) Primer extension mapping of the 5′ termini of fibronectin transcripts. For primer extension, oligonucleotide FN-ext.3 (lane 1) or FN-ext.4 (lane 2), each 30 nucleotides in size, was labeled with T4 polynucleotide kinase. For size standards, either an unrelated oligonucleotide of 60 nucleotides was labeled (lane 3) or Sanger sequencing of an unrelated sequence of known composition was performed (lanes A, C, G, and T). Oligonucleotides were annealed to total CsCl-purified RNA from XTC cells and extended with MMLV reverse transcriptase. Two independent experiments are shown in lanes 4 and 5 and in lanes 6 and 7. Extension products of FN-ext.3 are shown in lanes 4 and 6, and those of FN-ext.4 are shown in lanes 5 and 7. RNA from lanes 6 and 7 was digested with DNase I prior to hybridization to exclude contamination with genomic DNA. No differences between the two experiments were detectable. In both cases, the extension product of FN-ext.3 was 92 nucleotides long, and that of FN-ext.4 was 53 nucleotides. None of the oligonucleotides annealed with tRNA (not shown).
Several putative binding motifs for transcription factors were found in the −3403/+20 fragment: two canonical TATA boxes at positions −29 and −27, a CCAAT box at −89, a potential SP-1 binding site at −122 (16), and two AP-1 sites at −268 and −628 (35) (Fig. 3A). Two binding sites for AP-2 surround the CCAAT and Sp1 boxes at positions −59 and −135 (26), and an element that strongly resembles CRE is located at −647. In addition, four putative LEF-TCF binding sites were found, at positions −2454, −1122, −1054, and −368.
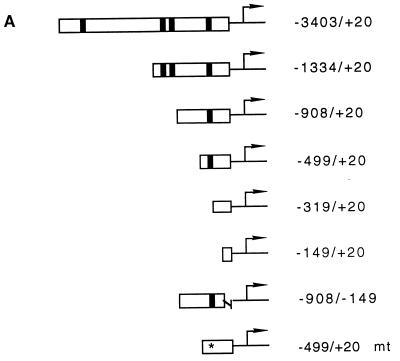
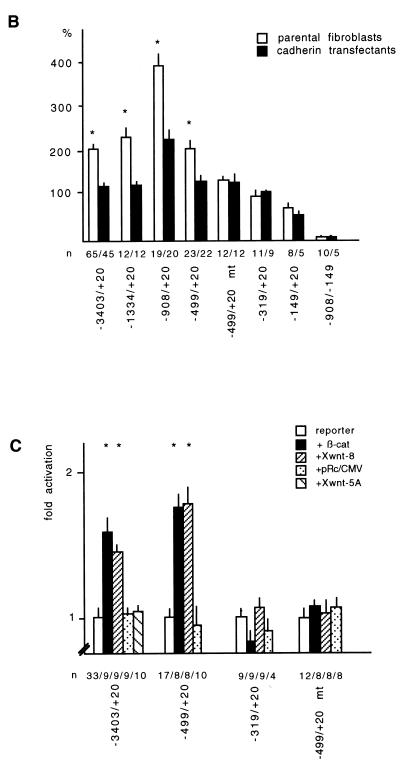
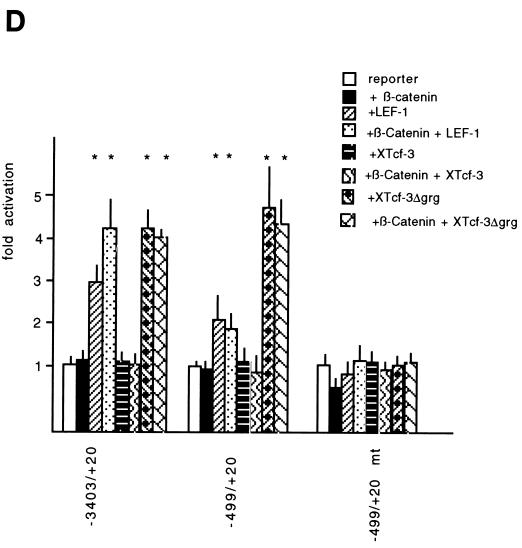
In order to test the effect of the Wnt/Wg signal on the fibronectin gene promoter, various deletion constructs in front of the luciferase gene (Fig. 4A) were examined in different cells and in the presence of several Wnt/Wg effectors. We found that the −3403/+20 promoter construct had a higher activity in parental Xenopus fibroblasts than in cadherin transfectants (Fig. 4B) (significant by the Student t test; P < 0.005). This difference persisted in the constructs that were truncated from their 5′ end and that lacked the putative LEF-TCF binding sites at positions −2454, −1122, and −1054 (Fig. 4A). These findings make it unlikely that the three distal LEF-TCF target sites are regulated by Wnt/Wg signaling in XTC cells. However, the difference in FN promoter activity between the cell lines was abolished when the promoter was deleted from position −499 to −319. This deletion resulted in loss of the putative LEF-TCF binding site at position −368, suggesting that this binding site may be responsible for activation by Wnt/Wg. We then mutated this LEF-TCF binding site in the −499/+20 mt construct by exchange of conserved nucleotides (Fig. 5A). This mutant construct had the same activity in both cell lines (Fig. 4B). Thus, our results demonstrate that the difference in the activity of the FN promoter in parental and cadherin-transfected fibroblasts is due to the proximal LEF-TCF binding site. To determine whether the isolated FN promoter indeed responds to the Wnt/Wg signaling pathway, we measured promoter activity in cadherin-expressing fibroblasts after transfection of different members of the Wnt/Wg signal cascade. Transfection of β-catenin or Xwnt-8, but not Xwnt-5A, together with the −3403/+20 reporter gene construct resulted in a significant increase of promoter activity (P < 0.005 by the Student t test) (Fig. 4C). Similar results were obtained when the promoter construct −499/+20 containing the proximal-most LEF-TCF binding site was used. Neither the −319/+20 construct lacking this site nor the mutated promoter (−499/+20 mt) was sensitive to Wnt/Wg signaling. These data show that the Xenopus fibronectin gene promoter is regulated through the most proximal LEF-TCF binding site, which we hereafter refer to as the Wnt/Wg response element. In all assays we found that the single LEF-TCF binding site at position −368 conferred an approximately twofold increase in activity to the FN promoter. A similar responsiveness has been reported for a single Wnt/Wg response element on the siamois and c-myc promoters, although in these cases the presence of several inducible LEF-TCF target sites results in stronger total activation (3, 18).
FIG. 4.
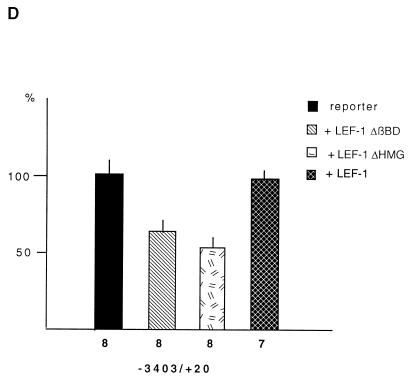
Identification of a Wnt/Wg response element in the Xenopus fibronectin promoter. (A) Luciferase reporter gene constructs used in this study. Putative LEF-TCF binding sites are indicated as filled boxes. The asterisk marks a mutated LEF-TCF site (for sequence comparison, see Fig. 5A). (B) Promoter activities of different constructs in parental fibroblasts (open bars) and cadherin-transfected fibroblasts (filled bars). Promoter activities were normalized to account for differences in transfection efficiency. The activities of the different constructs are shown as percentages relative to that of the CMV promoter. n, number of independent transfections; asterisks, significant differences by the Student t test (P < 0.005). (C) Influence of β-catenin (β-cat), Xwnt-8, and Xwnt-5A on fibronectin promoter activity in cadherin-transfected fibroblasts. n, number of experiments; asterisks, significant differences by the Student t test (P < 0.005). The activity of the corresponding reporter gene construct was set to 100%. (D) Influence of LEF-1 and LEF-1 deletion mutants, lacking either the β-catenin binding site (LEF-1 ΔβBD) or the DNA binding site (LEF-1 ΔHMG), on the fibronectin promoter in parental fibroblasts. The activity of the reporter was set as 100%. The number of experiments is shown below each bar.
To show that the Wnt/Wg response element is used in the parental cell line, we measured the activities of the FN promoter constructs in Xenopus fibroblasts upon transfection of LEF-1 mutants. Two different dominant-negative mutants, lacking either the β-catenin binding site (LEF-1 ΔßBD) (1) or the HMG box responsible for DNA binding (LEF-1 ΔHMG) (1), reduced the activity of the −3403/+20 FN promoter fragment (Fig. 4D), while transfection of wild-type LEF-1 had no influence. Similar results were obtained when the activity of the −499/+20 FN reporter construct was tested (data not shown). Constructs lacking the LEF-TCF target site (−319/+20) or containing the mutated Wnt/Wg response element (−499/+20 mt) were insensitive to the expression of either LEF-1 mutant (data not shown).
We next asked whether the identified Wnt/Wg response element of the Xenopus fibronectin gene promoter binds proteins of the LEF-TCF family. We used the bacterially expressed HMG box of LEF-1 in electrophoretic mobility shift assays with double-stranded 32P-labeled deoxyoligonucleotides containing the Wnt/Wg response element (XFN in Fig. 5A). A specifically retarded band was observed in the presence of the HMG box (Fig. 5B). Unlabeled oligonucleotide in 100-fold excess served as a competitor, whereas two control deoxyoligonucleotides (mut 1/2 and mut 3/4 [Fig. 5A]) did not compete. Crude nuclear extracts of Xenopus fibroblasts also interacted with the specific XFN deoxyoligonucleotide (Fig. 5C). Two complexes of different sizes, probably formed by different members of the LEF-TCF family, were found. Both protein-DNA complexes were competed by specific unlabeled oligonucleotides and not by mutant sequence motifs. In contrast, nuclear extracts isolated from a kidney epithelial cell line (A6) that does not express fibronectin showed only a weak binding with the specific oligonucleotide (Fig. 5D). In addition, we observed only one band (Fig. 5D) specifically interacting with the XFN oligonucleotide, while the upper band was also competed by the mutant sequence motifs. This prompted us to examine whether fibroblasts and epithelial cells differ in their content of LEF-TCF homologs.
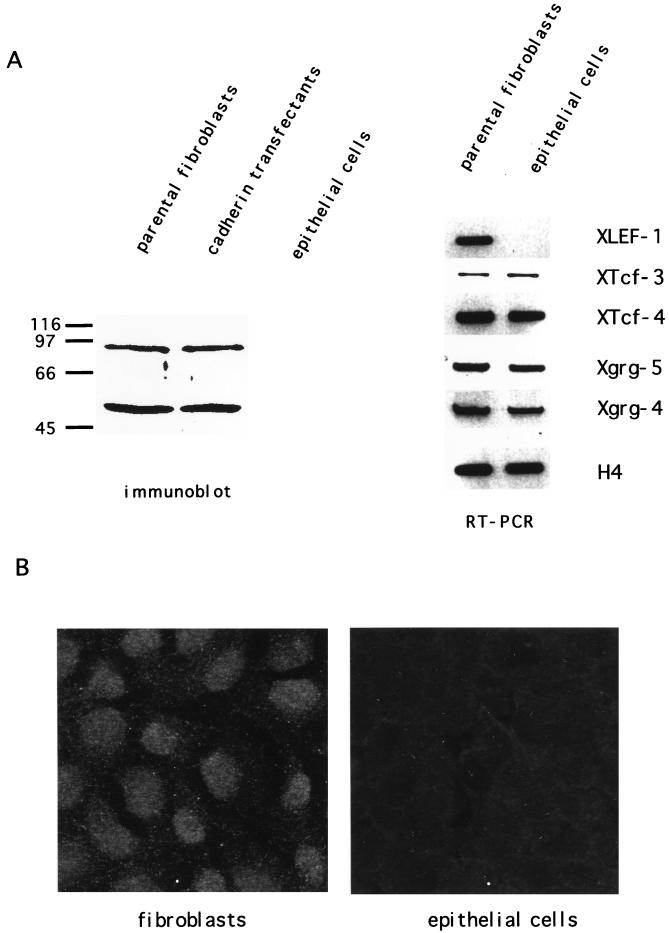
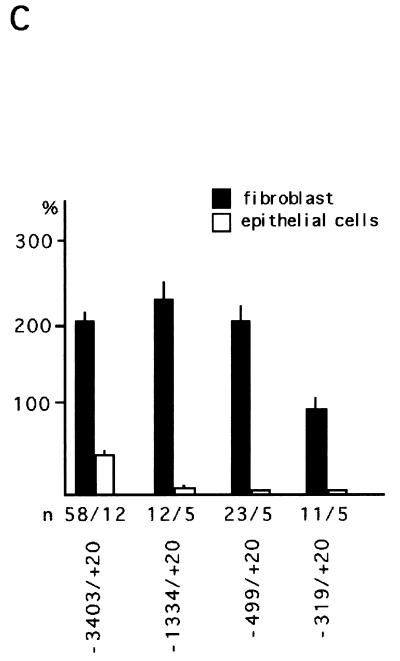
LEF-1-related proteins were identified in the parental and the cadherin-transfected fibroblasts but not in the kidney epithelial cell line (Fig. 6A). Only extracts from fibroblasts showed a distinct band at approximately 55 kDa, the size of murine LEF-1, in immunoblots stained with LEF-1 antiserum. The additional larger protein band may represent another LEF-1-related protein or an alternative splice product. The amounts of both proteins did not change in fibroblasts upon transfection with XB-cadherin. Immunostaining confirmed our immunoblot data, showing nuclear staining by LEF-1 antiserum in fibroblasts but not in epithelial cells (Fig. 6B). The protein results were verified by RT-PCR studies with primers specific for XLEF-1, XTcf-3, and XTcf-4. Remarkably, XTcf-3 and XTcf-4 are expressed in equal amounts in the fibroblasts and epithelial cells. In contrast, XLEF-1 expression was restricted to the fibroblasts (Fig. 6A). Taken together, these data suggest that the FN promoter contains a Wnt/Wg response element that binds to the HMG box of the LEF-TCF family. As demonstrated by the LEF-1 mutants, this LEF-TCF binding site is used to enhance FN promoter activity in fibroblasts.
FIG. 6.
Correlation between fibronectin promoter activity and presence of LEF-1-related proteins in kidney epithelial cells (A6). (A) LEF-1 immunoblot and RT-PCR of fibroblasts, cadherin-transfected fibroblasts, and epithelial cells. In transfected and untransfected fibroblasts a protein band of approximately 55 kDa which corresponds to the size of murine LEF-1 was detectable, while lysates of epithelial cells gave no signal. The protein of 90 kDa might represent a LEF-1-related HMG box-containing protein. The RT-PCR showed XTcf-3 and XTcf-4 expression in both fibroblasts and epithelial cells, whereas XLEF-1 was detected only in the fibroblasts. Both cell lines also express XGrg4 and XGrg5. As an internal standard, histone H4 was used. (B) Subcellular localization of LEF-TCF-related proteins in immunostaining with LEF-1 antiserum. In the fibroblasts the signal was found concentrated in the nuclei, while epithelial cells were negative in staining. (C) The fibronectin promoter was inactive in epithelial cells compared to fibroblasts. The activity of the promoter is shown in relation to that of the CMV promoter. The reporter gene constructs used are indicated. n, number of independent transfections. (D) The fibronectin promoter containing the Wnt/Wg response element was activated by LEF-1 alone or by the combination of LEF-1 and β-catenin, while β-catenin alone was not able to enhance promoter activity. Wild-type XTcf-3 does not influence FN promoter activity, either alone or in combination with β-catenin, while the mutant XTcf-3Δgrg upregulates the promoter activity. n, number of independent transfections; asterisks, significant differences by the Student t test (P < 0.005). Activity of the corresponding promoter construct was set at 100%.
The FN promoter is inducible by LEF-1 in the kidney epithelial cell line.
We next asked whether the lack of LEF-1-related proteins is responsible for the inactivity of the fibronectin gene in the epithelial cell line. Reporter gene assays revealed that none of the tested FN promoter fragments showed significant transcriptional activity in these cells (Fig. 6C). However, some differences in promoter activity between fibroblasts and epithelial cells persisted when the construct lacking the Wnt/Wg response element (−319/+20) was tested in both cell lines. This indicates that within the −319/+20 fragment, a further cell-type-specific regulatory element may exist.
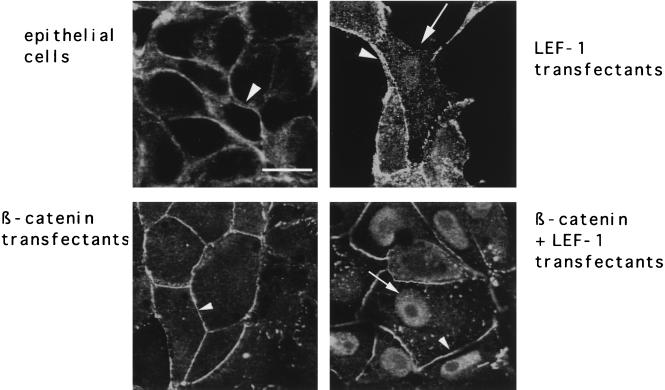
To prove that a shortage of LEF-1 homologs indeed contributes to the failure to express the fibronectin gene in epithelial cells, promoter activity was measured upon transfection of LEF-1 or β-catenin. Transfection of LEF-1 activated the −3403/+20 promoter fragment threefold (Fig. 6D), while β-catenin had no effect. However, transfection of LEF-1 combined with β-catenin led to a further increase in activity (Fig. 6D). The activating effect of LEF-1 was restricted to those constructs containing the Wnt/Wg response element. Constructs lacking the LEF-TCF binding site or containing the mutated motif were unable to respond to LEF-1 expression (Fig. 6D).
LEF-1 was also essential for nuclear localization of endogenous β-catenin in kidney epithelial cells. In mock-transfected or parental cells, β-catenin distribution was restricted to the plasma membrane (Fig. 7). This localization was not altered following transfection of β-catenin (Fig. 7). In contrast, β-catenin was found in the nuclei when either LEF-1 or LEF-1 together with β-catenin was transfected (Fig. 7). XTcf-3 behaved differently. Although it is endogenously expressed in the epithelial cell line, we proved its ability to influence FN promoter activity by exogenous overexpression. None of the tested constructs revealed an increase in promoter activity upon transfection (Fig. 6D). In contrast to LEF-1, XTcf-3 can bind groucho proteins (XGrg4 and XGrg5) (47), which we found expressed in the fibroblasts and the epithelial cells (Fig. 6A). Association with XGrg4 alters the role of XTcf-3 from an activator to a repressor, while XGrg5 enhances the XTcf-3 mediated gene activation (47). Therefore, we measured the FN promoter activity in the epithelial cell line after transfection of a mutant (XTcf-3Δgrg) lacking the Grg binding region. As shown in Fig. 6D, this mutant, in contrast to the wild-type XTcf-3, strongly activates the FN promoter, indicating that the inhibitory effect of XGrg4 dominates the putative enhancer XGrg5. Based on these data, we assume that the presence of XGrg4 prevents XTcf-3 from activating the FN promoter, giving this role to LEF-1. Thus, the observed difference in XLEF-1 expression between fibroblasts and epithelial cells, but not the lack of β-catenin, contributes to the low activity of the FN promoter in epithelial cells, indicating that β-catenin in a context with LEF-1 promotes fibroblast-specific fibronectin gene expression.
FIG. 7.
Nuclear localization of β-catenin in epithelial cells (A6) upon transfection with LEF-1 alone or LEF-1 combined with β-catenin. In epithelial cells β-catenin was detected exclusively at the plasma membrane (arrowheads). After introduction of LEF-1, either alone or in combination with β-catenin, a subpopulation of the cells revealed nuclear staining (arrows), whereas transfection with β-catenin gave no additional signal in the nuclei. Bar, 10 μm.
DISCUSSION
Here we report that β-catenin, which was initially characterized as a component of the cadherin adhesion complex, controls the expression of the cell-substrate adhesion molecule fibronectin. This regulation occurs at the level of gene transcription and requires the presence of transcription factors of the LEF-TCF family. We obtained the following findings: (i) cadherin transfection of fibroblasts downregulates fibronectin expression on the protein and mRNA levels and diminishes fibronectin promoter activity, (ii) promoter activity and fibronectin expression are restored following upregulation of nuclear β-catenin by stimulation of the Wnt/Wg signaling pathway, (iii) the activity of the fibronectin promoter depends on the presence of a functional LEF-TCF binding site, and (iv) the fibronectin promoter can be activated in epithelial cells by exogenous expression of LEF-1 but not of XTcf-3. The last effect seems to be blocked by binding to groucho proteins, because the XTcf-3Δgrg mutant, lacking the binding site for groucho proteins, strongly activates the FN promoter.
Most importantly, our findings suggest a novel mechanism by which the inverse relationship between cell-cell and cell-substrate adhesion is coordinated: the dual function of β-catenin. As a component of the cadherin adhesion complex, β-catenin is required to link cadherins to the cytoskeleton confering adhesiveness to the cells (28, 55). In association with E-cadherin, β-catenin is also responsible for basolateral polarity of epithelial cells, which was shown by E-cadherin transfections in murine fibroblasts (37). Not surprisingly, the loss of E-cadherin or presence of mutants in carcinoma cells and tumors correlates with increased invasive potential, which is abolished by introducing wild-type E-cadherin (11, 44, 56). With our observation that β-catenin as a component of the Wnt/Wg signaling cascade controls the expression of the fibronectin gene, a mesenchymal gene, a new aspect of tumorigenesis is brought up, i.e., the enforcement of cell transformation and cell motility caused by induction of mesenchymal genes.
The first identified target genes of the Wnt/Wg signaling cascade were developmental genes important for the formation of the dorsoventral axis in Xenopus (siamois [3] and twin [34]) and the differentiation of visceral mesoderm in Drosophila (ultrabithorax [46]). Their promoters possess a typical LEF-TCF binding site which was initially characterized in lymphocytes (12, 14). We found in the Xenopus fibronectin gene promoter a LEF-TCF binding motif and confirmed by gel retardation assays that it interacts with the HMG box of murine LEF-1. Analyzing the activities of FN promoter reporter constructs in Xenopus fibroblasts and their derivatives, the cadherin transfectants, we demonstrated that this promoter responds to the Wnt/Wg signal via a LEF-TCF binding site at position −368 (Fig. 3A). Consistent with the promoter data are our previous findings (10) that fibronectin expression is downregulated in fibroblasts when β-catenin is shifted to the plasma membrane following overexpression of cadherins. In this study, we present the direct molecular link between subcellular localization of β-catenin and regulation of fibronectin expression, as follows. Depletion of the Wnt/Wg signal transducer β-catenin from the cytosolic-nuclear pool reduces FN promoter activity and fibronectin expression. Refilling of the β-catenin pool by activation of the Wnt/Wg signaling cascade restores promoter activity and fibronectin synthesis (Fig. 1). LEF-TCF binding sites were also found in fibronectin promoters of other species (Fig. 5A), suggesting that the regulation of this extracellular matrix protein by Wnt/Wg signaling is a general phenomenon. In addition to fibronectin, some integrins and cadherins may also be direct targets of Wnt/Wg signaling. We found that α3β1 integrin expression was downregulated in our cadherin transfectants (10), and we also observed restored β1 integrin expression after activation of the Wnt/Wg signaling cascade (data not shown). Moreover, Xcadherin-11 is inducible by Xwnt-8 but not by siamois expression (17), which argues for a LEF-TCF binding motif in the promoter of this cadherin, which is expressed in mesenchymal tissues (23, 29). In the murine E-cadherin promoter a LEF-TCF binding site that is able to bind the heterodimer LEF-1–β-catenin has been identified (24). However, only an inverse relationship between LEF-1 and E-cadherin expression (24) or E-cadherin upregulation and Wnt-1 signaling (52) was observed in mice, leading to the speculation that E-cadherin might be repressed by LEF-TCF family members, which has to be verified.
Strengthening of calcium-dependent cell-cell adhesion by increased cadherin-catenin complex formation following Wnt expression was shown for different cell lines (2, 21). Accumulation of β-catenin due to its stabilization via Wnt/Wg signaling resulted in elevated amounts of N- or E-cadherin. Transcription of these cadherins remained unaltered; instead, their turnover decreased (2, 21). We also did not observe an induction of fibronectin expression or an activation of the FN promoter in a kidney epithelial cell line after β-catenin transfection (Fig. 6). However, we found that LEF-1-related proteins were lacking in these cells. Introduction of LEF-1 in the epithelial cell line resulted in translocalization of β-catenin into the nucleus and activation of the FN promoter (Fig. 6 and 7). XTcf-Δgrg also enhanced promoter activity, indicating that the presence of XGrg4 prevents XTcf-3 from activating the FN promoter. Although we cannot confirm that XTcf-4 behaves like XTcf-3, because XTcf-4 has not been cloned yet, it seems obvious that a lack of XLef is the limiting factor for nuclear function of β-catenin in the A6 epithelial cell line. In contrast, we did not detect a further increase in FN promoter activity or fibronectin synthesis upon activation of the Wnt/Wg pathway in the XTC fibroblasts. However, we observed a reduction in activity upon transfection of LEF-1 mutants (Fig. 4D), indicating that the signaling cascade is active in Xenopus fibroblasts. Therefore, decreasing the amount of cytosolic or nuclear β-catenin via exogenous expression of epithelial cadherins affects gene expression in these cells. These findings emphasize the idea that the transactivating function of β-catenin is specified by the subtype of the HMG box transcription factor. This might explain why rat fibroblasts (57) and PC12 cells (51) alter morphology and gene expression upon Wnt-1 transfection, whereas epithelial cells respond only with increased cell-cell adhesion (21).
The function of β-catenin described here, i.e., driving, together with transcription factors of the LEF-TCF family, expression of fibronectin and probably that of other mesenchymal adhesion molecules, addresses the question of whether accumulation of β-catenin in presence of LEF-TCF transcription factors is sufficient to induce the epithelial-mesenchymal transition in epithelial tumors. Recently, mutations in β-catenin have been observed in several carcinoma and melanoma cell lines, revealing a constitutive active β-catenin–LEF-TCF complex demonstrated by the activity of synthethic LEF-TCF reporter constructs (TOPFLASH and TOPGAL) (31, 40, 48). In a first attempt, we therefore investigated the activity of the FN promoter in SW480 colocarcinoma cells, which are characterized by stable β-catenin and Tcf-4 expression and which show high activity of the synthetic LEF-TCF promoters (31). However, these cells are unable to activate the FN promoter. As in the case of Xenopus epithelial cells (Fig. 6 and 7), additional LEF-1 transfection was required (data not shown). Importantly, an FN promoter fragment lacking the Wnt/Wg response element (−319/+20) had different activities in Xenopus fibroblasts and epithelial cells (Fig. 6C). This indicates that additional elements regulating the cell-type-specific activity may be present in the −319/+20 fragment. For example, this fragment contains a binding site for AP-1, which was shown to trigger loss of polarity in mammary epithelial cell lines (45). Independently of this observation, our results show that the FN promoter is indeed regulated by the Wnt/Wg signaling cascade and that its activity is restricted to cells endowed with all components of this signal pathway as known from Xenopus fibroblasts. In epithelial cells, the lack of LEF-1 transcription factor and not β-catenin limits FN promoter activity. Thus, overexpression of β-catenin is not sufficient to induce the epithelial-mesenchymal transition. However, in cells that express LEF-1 homologs, Wnt/Wg signaling may promote this process.
ACKNOWLEDGMENTS
We thank P. Dietmann, M. Hess, and V. Bührmann for excellent technical assistance. We also acknowledge J. Behrens, W. Birchmeier, D. W. DeSimone, J. von Kries, R. Moon, H. Steinbeisser, and M. Torres for providing cDNA clones and antibodies. We are grateful to W. Birchmeier and A. Starzinski-Powitz for helpful suggestions on the manuscript.
This research was supported by DFG grant Kn 200/4-1 to D.W. and by financial research support by the Land Baden-Württemberg to M.K.
The first two authors contributed equally to this study.
REFERENCES
- 1.Behrens J, von der Kries J P, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 2.Bradley R S, Cowin P, Brown A M C. Expression of Wnt-1 in PC12 cells results in modulation of plakoglobin and E-cadherin and increased cellular adhesion. J Cell Biol. 1993;123:1857–1865. doi: 10.1083/jcb.123.6.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brannon M, Gomperts M, Sumoy L, Moon R T, Kimelman D. A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997;11:2359–2370. doi: 10.1101/gad.11.18.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavallo R, Rubinstein D, Peifer M. Armadillo and dTCF: a marriage made in the nucleus. Curr Opin Genet Dev. 1997;7:459–466. doi: 10.1016/s0959-437x(97)80071-8. [DOI] [PubMed] [Google Scholar]
- 5.Chirgwin J M, Przybyia A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 6.Danker K, Hacke H, Ramos J, DeSimone D, Wedlich D. V+-fibronectin expression and localization prior to gastrulation in Xenopus laevis embryos. Mech Dev. 1993;44:155–165. doi: 10.1016/0925-4773(93)90064-5. [DOI] [PubMed] [Google Scholar]
- 7.Dean D C, Bowlus C L, Bourgeois S. Cloning and analysis of the promoter region of the human fibronectin gene. Proc Natl Acad Sci USA. 1987;84:1876–1880. doi: 10.1073/pnas.84.7.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeSimone D W, Norton P A, Hynes R O. Identification and characterization of alternatively spliced fibronectin mRNAs expressed in early Xenopus embryos. Dev Biol. 1992;149:357–369. doi: 10.1016/0012-1606(92)90291-n. [DOI] [PubMed] [Google Scholar]
- 9.Fagotto F, Gumbiner B M. Cell contact-dependent signaling. Dev Biol. 1996;180:445–454. doi: 10.1006/dbio.1996.0318. [DOI] [PubMed] [Google Scholar]
- 10.Finnemann S, Kühl M, Otto G, Wedlich D. Cadherin transfection of Xenopus XTC cells downregulates expression of substrate adhesion molecules. Mol Cell Biol. 1995;15:5082–5091. doi: 10.1128/mcb.15.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frixen U H, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Löchner D, Birchmeier W. E-Cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giese K, Cox J, Grosschedl R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structure. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 13.Gorman C. High efficiency gene transfer into mammalian cells. In: Glover D M, editor. DNA cloning: a practical approach. Vol. 2. Oxford, United Kingdom: IRL Press; 1985. pp. 143–190. [Google Scholar]
- 14.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 15.Gumbiner B M. Signal transduction by β-catenin. J Cell Biol. 1995;7:634–640. doi: 10.1016/0955-0674(95)80104-9. [DOI] [PubMed] [Google Scholar]
- 16.Gustafson T A, Kedes L. Identification of multiple proteins that interact with functional regions of the human cardiac α-actin promoter. Mol Cell Biol. 1989;9:3269–3283. doi: 10.1128/mcb.9.8.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadeball B, Borchers A, Wedlich D. Xenopus cadherin-11 (Xcadherin-11) expression requires the Wnt/Wg signal. Mech Dev. 1998;72:101–113. doi: 10.1016/s0925-4773(98)00022-7. [DOI] [PubMed] [Google Scholar]
- 18.He T-C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Identification of c-myc as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 19.Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, Yoshida-Noro C, Wylie C. Overexpression of cadherins and underexpression of β-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 20.Hedgepeth C M, Conrad L J, Zhang J, Huang H-C, Lee V M Y, Klein P S. Activation of the Wnt signaling pathway: a molecular mechanism for lithium action. Dev Biol. 1997;185:82–91. doi: 10.1006/dbio.1997.8552. [DOI] [PubMed] [Google Scholar]
- 21.Hinck L, Nelson W J, Papkoff J. Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing β-catenin binding to the cell adhesion protein cadherin. J Cell Biol. 1994;124:729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodivala K J, Watt F M. Evidence that cadherins play a role in the downregulation of integrin expression that occurs during keratinocyte terminal differentiation. J Cell Biol. 1994;124:589–600. doi: 10.1083/jcb.124.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann I, Balling R. Cloning and expression analysis of a novel mesodermally expressed cadherin. Dev Biol. 1995;169:337–346. doi: 10.1006/dbio.1995.1148. [DOI] [PubMed] [Google Scholar]
- 24.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Nuclear localization of β-catenin by interaction with the transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 25.Ilyas M, Tomlinson I P M, Rowan A, Pignatelli M, Bodmer W F. β-Catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imagawa M, Chiu R, Karin M. Transcription factor AP-2 mediates induction by two different signal-transduction pathways: protein kinase C and cAMP. Cell. 1987;51:251–260. doi: 10.1016/0092-8674(87)90152-8. [DOI] [PubMed] [Google Scholar]
- 27.Iwao K, Nakamori S, Kameyama M, Imaoka S, Kinoshita M, Fukui T, Ishiguro S, Nakamura Y, Miyoshi Y. Activation of the beta-catenin gene by interstitial deletions involving exon 3 in primary colorectal carcinomas without adenomatous polyposis coli mutations. Cancer Res. 1998;58:1021–1026. [PubMed] [Google Scholar]
- 28.Kemler R. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 1993;9:317–321. doi: 10.1016/0168-9525(93)90250-l. [DOI] [PubMed] [Google Scholar]
- 29.Kimura Y, Matsunami H, Takeichi M. Expression of cadherin-11 delineates boundaries, neuromeres, and nuclei in the developing mouse brain. Dev Dynamics. 1997;206:455–462. doi: 10.1002/(SICI)1097-0177(199608)206:4<455::AID-AJA11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 30.Klein P S, Melton D A. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1996;93:8455–9459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 32.Kühl M, Wedlich D. Wnt signalling goes nuclear. Bioessays. 1997;19:101–104. doi: 10.1002/bies.950190204. [DOI] [PubMed] [Google Scholar]
- 33.Larabell C A, Torres M, Rowning B A, Yost C, Miller J R, Wu M, Kimelman D, Moon R T. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in β-catenin that are modulated by the Wnt signaling pathway. J Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurent M N, Blitz I L, Hashimoto C, Rothbächer U, Cho K W-Y. The Xenopus homeobox gene Twin mediates Wnt induction of goosecoid in establishment of Spemann’s organizer. Development. 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- 35.Lee W, Mitchell P, Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1997;49:741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 36.McKendry R, Hsu S-C, Harland R, Grosschedl R. Lef-1/TCF proteins mediate Wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Dev Biol. 1997;192:420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]
- 37.McNeill H, Ozawa M, Kemler R, Nelson W J. Novel function of the cell adhesion molecule uvomorulin as an inducer of cell surface polarity. Cell. 1990;62:309–316. doi: 10.1016/0092-8674(90)90368-o. [DOI] [PubMed] [Google Scholar]
- 38.Miyoshi Y, Iwao K, Nagasawa Y, Aihara T, Sasaki Y, Imaoka S, Murata M, Shimamo T, Nakamura Y. Activation of the beta-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res. 1998;58:2524–2527. [PubMed] [Google Scholar]
- 39.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 40.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 41.Munemitsu S, Alert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular β-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novak A, Hso S-C, Leung-Hegesteijn C, Radeva G, Papkoff J, Montesano R, Roskelcly C, Grosschedl R, Dedhar S. Cell adhesion and the integrin-linked kinase regulate LEF-1 and β-catenin signaling pathways. Proc Natl Acad Sci USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel R, Odermatt E, Schwarzbauer J E, Hynes R O. Organization of the fibronectin gene provides evidence for exons huffling during evolution. EMBO J. 1987;6:2565–2572. doi: 10.1002/j.1460-2075.1987.tb02545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perl A-K, Wilgenbus P, Dahl U, Semb H, Christofori G. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190–193. doi: 10.1038/32433. [DOI] [PubMed] [Google Scholar]
- 45.Reichmann E, Schwarz H, Deiner E M, Leitner I, Eilers M, Berger J, Busslinger M, Beug H. Activation of an inducible c-FosER fusion protein causes loss of epithelial polarity and triggers epithelial-fibroblastoid cell conversion. Cell. 1992;71:1103–1116. doi: 10.1016/s0092-8674(05)80060-1. [DOI] [PubMed] [Google Scholar]
- 46.Riese J, Yu X, Munnerlyn A, Hsu S-C, Grosschedl R, Bienz M. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]
- 47.Roose J, Molenaar M, Hurenkamp J, Peterson J, Brantjes H, Moerer P, van de Wetering M, Destrée O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 48.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Schreiber E, Matthias P, Müller M, Schaffner W. Rapid detection of octamer binding proteins with ’mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shackleford G M, Willert K, Wang J, Vamus H. The wnt-1 proto-oncogene induces changes in morphology, gene expression, and growth factor responsiveness in PC12 cells. Neuron. 1993;11:865–875. doi: 10.1016/0896-6273(93)90116-9. [DOI] [PubMed] [Google Scholar]
- 52.Shimamura K, Hirano S, McMahon A P, Takeichi M. Wnt-1-dependent regulation of local E-cadherin and αN-catenin expression in the embryonic mouse brain. Development. 1994;120:2225–2234. doi: 10.1242/dev.120.8.2225. [DOI] [PubMed] [Google Scholar]
- 53.Slusarski D C, Yang-Snyder J, Busa W B, Moon R T. Modulation of embryonic intracellular Ca++ signaling by Wnt-5A. Dev Biol. 1997;182:114–120. doi: 10.1006/dbio.1996.8463. [DOI] [PubMed] [Google Scholar]
- 54.Sparks A B, Morin P J, Vogelstein B, Kinzler K W. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 55.Takeichi M. Morphogenetic role of classic cadherins. Curr Opin Cell Biol. 1995;7:619–627. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 56.Vleminckx K, Vakaet L, Mereel M, Fiers W, van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-m. [DOI] [PubMed] [Google Scholar]
- 57.Young C S, Kitamura M, Hardy S, Kitajewski J. Wnt-1 induces growth, cytosolic β-catenin, and Tcf/Lef transcriptional activation in rat-1 fibroblasts. Mol Cell Biol. 1998;18:2474–2485. doi: 10.1128/mcb.18.5.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu A J, Watt F M. Expression of a dominant negative cadherin mutant inhibits proliferation and stimulates terminal differentiation of human epithelial keratinocytes. J Cell Sci. 1996;109:3013–3023. doi: 10.1242/jcs.109.13.3013. [DOI] [PubMed] [Google Scholar]