Summary
Turning non-inflamed (cold) tumors into inflamed (hot) tumors is important for maximizing the effect of immune checkpoint inhibitors (ICIs) against malignancies. We showed that lactate, a product of the Warburg effect, inhibited the efficacy of ICIs and suppressed IL-12 p40 expression in dendritic cells (DCs) through reducing NF-κB p65, p50, and c-Rel DNA-binding activity to the IL-12 p40 promoter. Additionally, lactate promoted the expression of early growth response protein 1 (EGR1), whose expression was increased in human invasive melanoma compared with non-invasive melanoma. We also found that EGR1 interacts with serum response factor (SRF) and represses the expression of CD80 in DCs. These findings suggest that lactate and its induced EGR1 are key factors that turn hot tumors into cold tumors and may represent targets in cancer treatment with ICIs.
Subject areas: immunology, immune response, cancer
Graphical abstract

Highlights
-
•
Lactate suppressed IL-12 p40 expression in dendritic cells
-
•
Lactate promoted the expression of early growth response protein 1 (EGR1)
-
•
EGR1 interacts with serum response factor (SRF) and represses the expression of CD80
-
•
Lactate and EGR1 switch inflamed (hot) tumors to non-inflamed (cold) tumors
Immunology; Immune response; Cancer
Introduction
Malignant neoplasms are life-threatening diseases for which effective treatments are urgently needed. Immune checkpoint inhibitors (ICIs) have recently proven to be effective against tumors, demonstrating that the immune system plays a critical role in suppressing malignancies. However, the therapeutic responses to ICIs vary considerably among individual patients; therefore, elucidation of their mechanisms is required to design strategies for improving their therapeutic efficacy (Chen and Mellman, 2017). One important factor that determines the effectiveness of ICIs may be inflammation of the tumor environment. ICIs are effective against inflamed tumors, so-called “hot tumors,” in which many pro-inflammatory cytokines are detected (Herbst et al., 2014). Conversely, non-inflamed tumors, so-called “cold tumors,” have no tumor-suppressive cytokines or co-stimulators of antigen-presenting cells and rarely respond to ICIs (Herbst et al., 2014). Because dendritic cells (DCs) secrete various cytokines and also have antigen-presenting ability, they may be key regulators in ICI treatment and greatly affect treatment effectiveness (Chen and Mellman, 2017).
Recently, tumor-derived metabolites were reported to have an important effect on immune cells (O'Neill et al., 2016). Lactate accumulation in the tumor environment occurs as a result of the accelerated glucose metabolism in tumor cells, the so-called Warburg effect (Brand et al., 2016). Accumulation of lactate in solid tumors is a pivotal step in the development of malignancies (Hirschhaeuser et al., 2011). In human primary tumors, high levels of lactate correlate with the incidence of distant metastases (Walenta et al., 2000). Recent studies have suggested that the tumor-promoting effect of lactate is related to its immunosuppressive effect within the tumor environment (Brand et al., 2016). For example, one study reported that tumor-derived lactate inhibits the activation and differentiation of T cells and monocytes in vitro (Dietl et al., 2010; Fischer et al., 2007) and promotes the development of immunosuppressive M2-like macrophages (Colegio et al., 2014). Yang et al. (Yang et al., 2020) reported that lactate suppresses the macrophage pro-inflammatory response to LPS stimulation by inhibition of YAP and NF-κB activation via GPR81-mediated signaling. Gottfried et al. (Gottfried et al., 2006) reported that lactate inhibits DC activation during antigen-specific T cell stimulation. Other studies reported that lactate skews DC differentiation into a tolerogenic phenotype, as exemplified by increased production of IL-10 and loss of IL-12 (Dong and Bullock, 2014; Nasi et al., 2013). Another report showed that lactate attenuates IFNα induction and induces pro-tumor reprogramming in plasmacytoid DCs (Raychaudhuri et al., 2019). These results suggest the possible inhibitory effect of tumor-derived lactate on DCs and the therapeutic activity of ICIs, which may represent key factors to determine the hot or cold status of a tumor. Here, we focused on lactate and examined its effect on tumor immunity and particularly its effect on DCs.
Results
Lactate suppresses IL-12 p40 expression in DCs and promotes tumor growth in a mouse model
To investigate the in vitro effects of lactate on cytotoxicity of cytotoxic T lymphocyte (CTLs) against tumor cells, we performed a CTL killing assay using the xCELLigence system (Figure S1). We found that lactate treatment attenuated the cytotoxicity of CTLs against B16-F1 cells (Figure 1A). We also performed antigen-presentation assays with OT-I transgenic mouse cells and observed markedly reduced proliferation in OT-I CD8+ cells cultured with bone marrow-derived DCs (BMDCs) stimulated by lactate compared with control cells (Figure 1B), which suggests the attenuation of the antigen-presenting ability of lactate-treated DCs. To examine whether lactate induced apoptosis of effector cells and BMDCs, we evaluated apoptosis using the Annexin V/Propidium Iodide apoptosis assay. The ratio of Annexin V+ PI+ apoptotic cells was similar in effector cells and BMDCs stimulated with 0–20 mM lactate (Figure S2), which is consistent with the previous report (Yang et al., 2020).
Figure 1.
Lactate (Lac) attenuates the antigen-presenting ability of dendritic cells (DCs) and suppresses IL-12 p40 expression in DCs
(A) Cytotoxicity of Lac-treated (20 mM) cytotoxic T lymphocytes (CTLs) and control CTLs against B16-F1 target cells at the indicated effector:target (E:T, CTL:B16-F1 cell) ratios. %Cytolysis was determined by the xCELLigence system.
(B) Carboxyfluorescein succinimidyl ester (CFSE)-labeled CD8+ T cells from OT-I transgenic mice and Lac-treated (20 mM) bone marrow-derived DCs (BMDCs) or control BMDCs from WT mice pulsed with Ova 257–264 peptide were co-cultured at decreasing dilutions (CD8+ T cell:BMDC = 1:1–1:1/16). Proliferation of CD8+ T cells was assessed after 84 hr by flow cytometry (top graph). Representative plots are shown (bottom panels).
(C) Tumor growth kinetics in diphtheria toxin (DTX)-treated CD11c-DTR bone marrow chimeric mice with subcutaneous injection with 1 × 106 B16-F1 melanoma cells and treated intratumorally with Lac (5 mM) or PBS-treated BMDCs (1×106) as indicated. Data are shown as mean ± SD (n = 4 mice per group).
(D) BMDCs from WT mice were stimulated with LPS (100 ng/mL) and/or Lac (20 mM) for 4 hr, and the total RNA from these cells was subjected to microarray analysis. The log2 ratio for each of the corresponding genes in BMDCs stimulated with LPS was subtracted from that of BMDCs stimulated with LPS plus Lac, and the results were arranged in descending order. Genes categorized in the indicated Kyoto Encyclopedia of Genes and Genomes pathways were analyzed. The top 20 most highly differentially expressed genes are indicated in the box.
(E) Levels of IL-12 p40 secreted by BMDCs following stimulation by LPS and/or indicated concentrations of Lac or sodium Lac for 24 hr were measured by ELISA (left and center). pH levels at the indicated Lac concentrations are also shown. Levels of IL-12 p40 secreted by BMDCs under the indicated pH (adjusted by HCl) are also shown (right).
(F) Levels of IL-12 p40 secreted by BMDCs following stimulation by LPS (100 ng/mL) Lac (20 mM), and/or NaHCO3 for 24 hr were measured by ELISA.
Data are expressed as mean ± SD. Significance was analyzed using a two-tailed Student's t-test, Welch's t test, or Mann-Whitney's U test. ∗p < 0.05; ∗∗p < 0.01; n.s., not significant (p > 0.05). Ctrl: control. See also Figures S1–S6.
These results prompted us to investigate the effects of lactate on DCs. To investigate the in vivo effects of lactate on DCs, we established a tumor mouse model using DC-depleted mice by subcutaneously injecting mice with B16-F1 melanoma cells. After CD11c-DTR bone marrow chimeric mice were treated with diphtheria toxin and DCs were depleted, mice were subcutaneously injected with lactate-treated (5 mM) BMDCs or non-treated control BMDCs. Increased tumor growth was observed in the mice injected with lactate-treated BMDCs compared with mice injected with control BMDCs (Figure 1C). In addition, increased tumor growth was observed in the mice treated with lactate (5 mM) compared with controls, and the anti-tumor efficacy of anti-PD-L1 was reduced in the lactate treatment group compared with the mice treated with PBS (Figure S3).
Because TLR4 agonists are one of the damage-associated molecular patterns, which are important stimulators of DCs in the tumor microenvironment, we assessed the effects of lactate on the response to LPS, the TLR4 ligand. We performed microarray analyses using BMDCs from WT mice that had been stimulated with LPS, lactate, or LPS together with lactate. Within several Kyoto Encyclopedia of Genes and Genomes (KEGG) categories, such as the cytokine-cytokine receptor interaction, pathways in cancer, Th1 and Th2 cell differentiation, and Toll-like receptor signaling pathway, the Il12b gene was the most differentially expressed gene in BMDCs stimulated with LPS and lactate compared those stimulated with LPS (Figures 1D and S4). We further found that lactate attenuated the LPS-mediated induction of IL-12 p40 in BMDCs and bone marrow-derived macrophages (BMDMs) in a dose-dependent manner (Figures 1E and S5), which is consistent with previous reports (Gottfried et al., 2006; Nasi et al., 2013). Furthermore, we confirmed that acidity (pH 6–7) mimicked the effects of lactate on IL-12 p40, and the significant inhibitory effect of lactate on LPS-mediated induction of IL-12 p40 levels in BMDCs was eliminated upon neutralization of lactate using NaHCO3 (Figure 1F). This result suggests that acidity decreased the expression of IL-12 p40 in BMDCs. We further examined the actual pH values in human melanoma and mouse tumor tissues. We found that the pH values of both tumor issues were approximately 6–7, and the tumor weights correlated with lactate levels in B16-F1 cell-derived tumors (Figure S6). Taken together, these results suggest that lactate suppressed the LPS-mediated induction of IL-12 p40, likely depending on its acidity.
Lactate suppresses NF-κB DNA-binding activity to the Il12b promoter
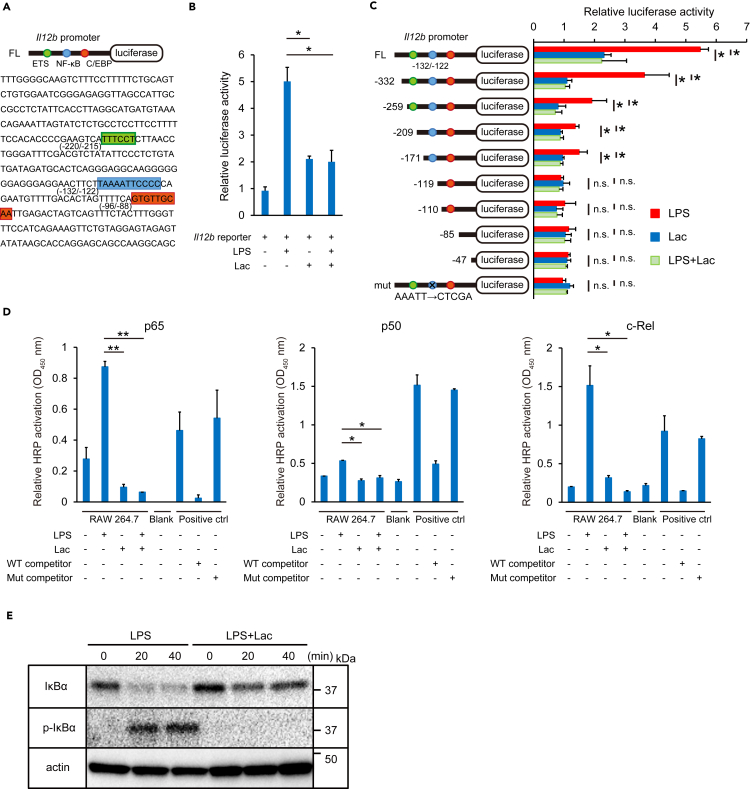
The inhibition of LPS-mediated induction of IL-12 p40 levels by lactate led us to hypothesize that lactate might affect IL-12 p40 gene levels by regulating transcription factors on the Il12b promoter. We first performed luciferase reporter assays using a construct with the Il12b promoter upstream of a luciferase reporter gene (Figure 2A). Luciferase activities were suppressed by the addition of lactate to cells treated with LPS compared with only LPS stimulation (Figure 2B). To identify the Il12b promoter elements that are important for transcriptional regulation by lactate, we next performed serial deletion and substitution mutant analyses. Deletion of the NF-κB binding site, but not the E26 transformation-specific (ETS) binding site, eliminated the inhibitory effect of lactate on LPS-mediated luciferase reporter activity (Figure 2C). The construct containing only the CCAAT/enhancer-binding protein (C/EBP) binding element did not show significant activation by LPS or lactate. In addition, mutation of the NF-κB binding site in the reporter construct eliminated the inhibition of luciferase reporter activity induced by lactate (Figure 2C). These results indicate that the NF-κB binding sequence in the Il12b promoter is important for the transcriptional suppression of Il12b by lactate.
Figure 2.
Lactate (Lac) suppresses NF-κB DNA-binding activity to the Il12b promoter
(A) A schematic of the luciferase reporter construct with the mouse Il12b promoter (top) and the mouse Il12b promoter sequence (bottom). Potential binding sites and consensus sequences for the transcription factors ETS (green), NF-κB (blue), and C/EBP (red) are shown.
(B) Luciferase assays. The mouse Il12b promoter luciferase reporter construct was transfected into RAW 264.7 cells. Cells were activated with LPS (100 ng/mL) and/or Lac (20 mM) at 24 hr post-transfection, and luciferase activity was measured 24 hr after stimulation. Relative luciferase activity was determined compared with non-activated control cells.
(C) Luciferase assays with deletion mutants and a substitution mutant (mut) of the Il12b promoter. Full-length (FL) or mutant constructs were transfected into RAW 264.7 cells. After 24 hr, cells were activated with LPS (100 ng/mL) and/or Lac (20 mM) for 24 hr. Relative luciferase activity was determined compared with non-activated control cells.
(D) The DNA-binding activity of NF-κB p65, p50, and c-Rel was measured with a TransAM NF-κB Transcription Factor Assay Kit.
(E) The IκBα and p-IκBα expressions in cytosolic extracts of RAW 264.7 activated with LPS (100 ng/mL) and/or Lac (20 mM) for 0, 20 or 40 min were analyzed by Western blotting.
Data are expressed as mean ± SD. Significance was analyzed using a two-tailed Student's t-test, Welch's t test, or Mann-Whitney's U test. ∗p < 0.05; ∗∗p < 0.01; n.s., not significant (p > 0.05). See also Figure S7.
We next evaluated the influence of lactate on the DNA-binding activity of NF-κB. Downregulation of the DNA-binding activity of NF-κB p65, p50, and c-Rel subunits was observed in nuclear extracts from cells stimulated with lactate compared with nuclear extracts from LPS-stimulated cells (Figure 2D). Similarly, LPS-induced NF-κB p65 DNA-binding activity was decreased in acidic conditions (pH 6.0), which suggests that the effect of lactate on NF-κB transcriptional activity may depend on acidity (Figure S7). Additionally, silencing of GPR81, a receptor for lactate, attenuated the suppressive effect of lactate on the NF-κB p65 DNA-binding activity, which is consistent with the previous report (Yang et al., 2020). These results suggest that both the acidity of lactate and the signaling pathway through GPR81 may be important in the suppressive effect of lactate on NF-κB p65 DNA-binding activity.
We also considered how lactate suppressed p65 DNA-binding activity. We found that IκBα expression levels were higher in LPS plus lactate-stimulated BMDCs than in LPS-stimulated BMDCs (Figure 2E). In addition, phospho-IκBα expression levels were decreased in LPS and lactate-stimulated BMDCs compared with levels in cells stimulated with LPS only. Taken together, these results suggest that lactate causes suppressed phosphorylation of IκBα, which leads to IκBα accumulation and subsequent inhibition of NF-κB. This eliminates the ability of NF-κB to interact with the NF-κB binding site in the Il12b promoter and leads to reduction of IL-12 p40 expression.
Lactate promotes the expression of transcription factor EGR1 in DCs
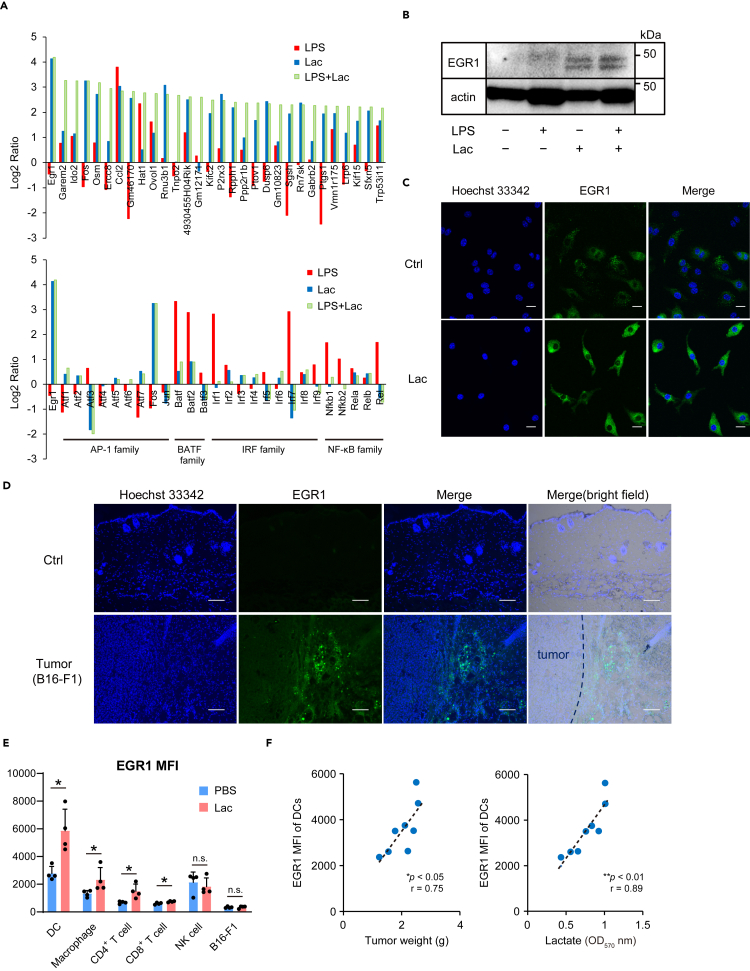
We performed a comprehensive gene expression analysis, including transcription factor genes, in BMDCs treated with LPS, lactate, and LPS together with lactate. Microarray analyses showed that the early growth response protein 1 (EGR1) transcription factor gene was the most highly expressed gene in BMDCs stimulated by lactate, as well as LPS plus lactate (Figure 3A). Notably, the gene expression of EGR1 was induced by lactate to higher levels compared with other transcription factor genes including the NF-κB family. The induction of EGR1 by lactate was confirmed by Western blotting (Figure 3B). Immunohistochemistry showed that lactate-induced EGR1 localized in the cytoplasm and partially in the nucleus (Figure 3C).
Figure 3.
Lactate (Lac) induces the expression of early growth response protein 1 (EGR1)
(A) BMDCs from WT mice were stimulated with LPS (100 ng/mL) and/or Lac (20 mM) for 4 hr, and the total RNA from these cells was subjected to microarray analysis. The log2 ratios for the indicated genes in BMDCs stimulated with LPS plus Lac are arranged in descending order.
(B) Western blot analysis of EGR1 expression in BMDCs stimulated with LPS (100 ng/mL) and/or Lac (20 mM) for 24 hr.
(C) EGR1 expression (green) in RAW 264.7 cells stimulated with Lac (20 mM) or control medium (Ctrl) for 24 hr. Nuclei were stained with Hoechst 33,342 (blue). Scale bars = 10 μm.
(D) Tumor tissues or control normal skin (Ctrl) from WT mice at two weeks after B16-F1 cell implantation were examined for EGR1 expression (green). Scale bars = 100 μm.
(E) Median fluorescence intensity (MFI) of EGR1 in leukocyte subpopulations in tumor tissues of WT mice 3 weeks post-B16-F1 cell implantation was analyzed using flow cytometry. On days 6, 9 and 12 after injection of B16-F1 cells, Lac (5 mM, 200 μL) or PBS control (200 μL) was intratumorally administered. Data are expressed as mean ± SD.
(F) The relationship between EGR1 MFI of tumor-infiltrating DCs and tumor weight (left) or lactate level (right) in the tumor tissues of WT mice 3–4 weeks post-B16-F1 cell implantation. Individual findings are plotted and each data point represents one mouse.
Significance was analyzed using a two-tailed Student's t-test, Welch's t test, or Mann–Whitney's U test. Pearson's correlation coefficient was used to evaluate the correlations. ∗p < 0.05; ∗∗p < 0.01; n.s., not significant (p > 0.05). See also Figures S8 and S9.
Because EGR1 has been reported to be induced by stress signals by the MAPK signaling pathway (Lim et al., 1998), we speculated that stress signals related to acidity from lactate may lead to EGR1 expression. We confirmed that EGR1 was induced by acidity (pH 6.0), and increased expressions of phospho-p38 and phospho-JNK were observed in lactate-treated BMDCs compared with control BMDCs (Figure S8). We examined other cell types and found that lactate slightly induced the expression of EGR1 in CD4+ T cells and CD8+ T cells but not in NK cells and B16-F1 melanoma cells (Figure S9).
Based on the results of the increased expression of EGR1 induced by lactate, we hypothesized that the expression levels of EGR1 in tumor tissues may be increased by tumor-derived lactate induced by the Warburg effect. We first examined the expression levels of mouse EGR1 in tumors from the mouse model established by subcutaneous injection with B16-F1 melanoma cells. We found that EGR1 was expressed in tumor tissues but not in normal skin (Figure 3D). Flow cytometric analyses revealed that the EGR1 mean fluorescence intensity (MFI) was higher in DCs in B16-F1 cell-derived tumors compared with other immune cells or tumor cells (Figure 3E). Additionally, the EGR1 MFI in DCs, macrophages, CD4+ T cells, and CD8+ T cells was increased in mice treated with lactate compared with those with PBS. The EGR1 MFI of DCs correlated with tumor weight and lactate levels in tumor tissues (Figure 3F).
Next, we investigated the expression of EGR1 in 19 human melanoma tissue samples (Table S1). We found a significant increase in EGR1-positive expression in tumor tissues of patients with invasive melanoma compared with primary tissues of non-invasive melanoma in situ (Figures 4A and 4B). Additionally, we found a correlation of the EGR1-positive ratio with tumor thickness as well as serum 5-S-cysteinyl-DOPA level, which is a common tumor marker for melanoma (Figure 4C). CD11c+ DCs highly expressed EGR1, while the expression of EGR1 was moderate in other cell types (Figure 4D). Finally, database analyses using TCGA revealed a correlation between EGR1 expression and the expression of lactate dehydrogenase (LDHA), a glycolytic marker, in primary invasive melanoma (Figure 4E). Collectively, these data show that EGR1 was expressed in DCs and its expression was increased in primary invasive melanoma tissues and correlated with LDHA, suggesting that the expression of EGR1 induced by lactate may have some role in DC function within the tumor microenvironment.
Figure 4.
EGR1 is expressed in DCs and correlates with LDHA in human melanoma tissues
(A) Tumor tissues from patients with melanoma in situ and invasive melanoma were examined by fluorescent staining (Hoechst 33,342, blue), as well as by immunofluorescence analysis for EGR1 (green). Nuclei (enclosed within light blue lines) and EGR1-positive cells (enclosed within magenta lines) were counted automatically using the In Cell Analyzer software. Scale bars = 50 μm.
(B) EGR1-positive ratios in tumor tissues of melanoma in situ and invasive melanoma are plotted (see also Table S1). Each data point represents one patient. Bars indicate the mean.
(C) Relationship between EGR1-positive ratio and tumor thickness (left) or serum 5-S-cysteinyl-DOPA (5-S-CD) levels (right). Individual findings are plotted and each data point represents one patient.
(D) Primary tumor tissues from patients with invasive melanoma were examined by fluorescent staining as indicated. Scale bars = 50 μm.
(E) Correlation analysis of EGR1 and LDHA expression levels using RNA-seq data set acquired from primary tumors from 45 skin cutaneous melanoma patients in TCGA (TCGA-SKCM).
Significance was analyzed using a two-tailed Student's t-test, Welch's t test, or Mann–Whitney's U test. Pearson's correlation coefficient was used to evaluate the correlations. ∗p < 0.05; ∗∗p < 0.01.
CD80 expression is increased EGR1-deficient DCs
Our data suggest that EGR1 induced by lactate may be important for DC function. Therefore, we performed analyses using BMDCs from Egr1−/− mice (Figure S10) compared with DCs from WT mice. To investigate the effect of EGR1 on cytokine expression, we performed ELISAs to examine the levels of IL-12 p40, TNF-α, and IL-6 secreted by Egr1−/− and WT BMDCs following stimulation by LPS. However, there were no significant differences in IL-12 p40, TNF-α, and IL-6 levels in culture medium of Egr1−/− BMDCs and WT BMDCs (Figure 5A). We also did not detect significant differences in the number of splenic macrophages, DCs, or T cells between Egr1−/− and WT controls (Figure S11).
Figure 5.
CD80 expression level is increased EGR1-deficient DCs
(A) Levels of IL-12 p40, TNF-α, and IL-6 secreted by BMDCs stimulated with LPS (100 ng/mL) for 24 hr were measured by ELISA. Data are expressed as mean ± SD from two independent experiments (n = 3–5).
(B) RNA from BMDCs from WT and Egr1−/− mice was subjected to microarray analysis. The log2 ratio was determined for the corresponding genes categorized in cell adhesion molecules within KEGG pathways and the results were arranged in descending order. Genes that overlapped with TLR signaling pathways are in red boxes.
(C) Flow cytometric analyses of CD80 and CD86 in CD11c+ DCs within splenocytes from WT and Egr1−/− mice. Representative plots are shown (left). Data are expressed as mean ± SD from two independent experiments (right, n = 3). MFI, median fluorescence intensity.
(D) BMDCs from WT or Egr1−/− mice were stimulated with LPS (100 ng/mL) and/or lactate (Lac, 20 mM) for 24 hr and the MFI of CD80 was assessed (n = 3). Data are expressed as mean ± SD.
(E) CFSE-labeled CD8+ T cells from OT-I transgenic mice were co-cultured with BMDCs from WT or Egr1−/− mice pulsed with Ova 257–264 peptide. Proliferation of OT-I CD8+ T cells was assessed after 72 hr by flow cytometry.
(F and G) Tumor growth kinetics in WT or Egr1−/− mice subcutaneously injected with 1 × 106 B16-F1 melanoma cells and treated with anti-PD-L1 antibody or control antibody (Ctrl), Lac and/or PBS as indicated. On days 6, 9, and 12 after injection of B16-F1 cells, 200 μg of control IgG Ab or anti-PD-L1 Ab were injected intraperitoneally (F), and Lac (5 mM, 200 μL) or PBS control (200 μL) was intratumorally administered (G). Data are shown as mean ± SD of 3–4 mice per group.
(H) Tumor growth kinetics in diphtheria toxin (DTX)-treated CD11c-DTR bone marrow chimeric mice with subcutaneous injection with 1 × 106 B16-F1 melanoma cells and treated intratumorally with 1×106 WT or Egr1−/− BMDCs. Data are shown as mean ± SD (n = 4 mice per group).
Significance was analyzed using a two-tailed Student's t-test, Welch's t test, or Mann–Whitney's U test. ∗p < 0.05; ∗∗p < 0.01; n.s., not significant (p > 0.05). See also Figures S10 and S11.
We next performed microarray analysis using Egr1−/− and WT BMDCs (Figure 5B). Within the KEGG category of the cell adhesion molecules and TLR signaling pathways, Cd80, Cd86, and Cd40 overlapped. Among the three genes, Cd80 showed the greatest increase in expression in Egr1−/− DCs compared with WT DCs, and a significant increase of CD80 expression in Egr1−/− DCs was also confirmed using flow cytometric analysis (Figure 5C). Furthermore, lactate reduced LPS-induced CD80 MFI in BMDCs, and the MFI of CD80 was higher in Egr1−/− BMDCs compared with WT BMDCs treated with LPS and lactate (Figure 5D). Antigen presentation assays revealed increased proliferation of OT-I CD8+ cells cultured with Egr1−/− BMDCs compared with those cultured with WT BMDCs (Figure 5E). However, there were no significant differences in tumor growth between Egr1−/− and WT mice, and the anti-tumor efficacy of anti-PD-L1 was similar between Egr1−/− and WT mice (Figure 5F). Similarly, there were no significant differences in tumor growth between Egr1−/− and WT mice treated with 5 mM lactate (Figure 5G).
We speculated these results may be because whole body EGR1-knockout mice were used, and thus other immune cells were affected by EGR1 depletion. Therefore, we performed similar experiments using CD11c-DTR bone marrow chimeric mice and found that tumor growth was suppressed in DC-depleted mice transferred with Egr1−/− BMDCs compared those transferred with WT BMDCs (Figure 5H). These results suggest that EGR1-expressing DCs have pro-tumor activities.
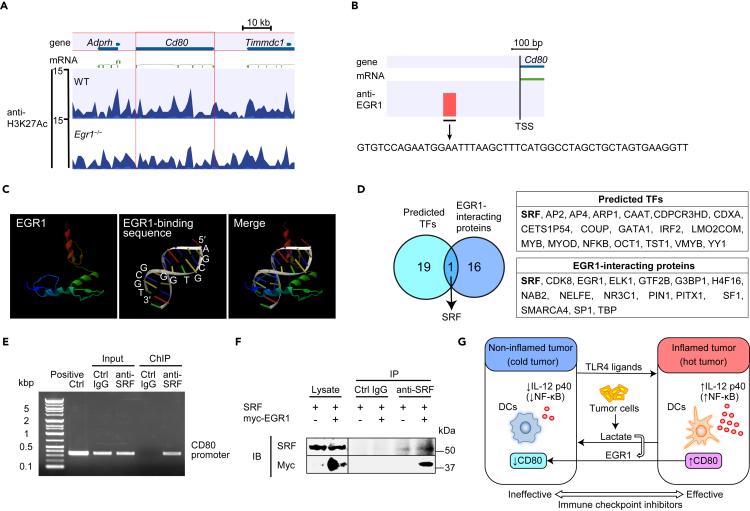
Based on the result of increased expression of CD80 in Egr1−/− BMDCs, we analyzed the status of H3K27Ac in LPS and lactate-stimulated WT and Egr1−/− BMDCs using ChIP-sequencing (ChIP-seq). Contrary to our expectations, the H3K27Ac status on the Cd80 gene region was comparable between WT and Egr1−/− BMDCs (Figure 6A). The H3K27Ac status on Il12b, Il6, Tnf, and Cd86 gene regions were also similar between WT and Egr1−/− BMDCs (Figure S12). Next, we analyzed the proximal TSS region on Cd80 promoter by ChIP-seq using anti-EGR1 antibody and identified a potential EGR1-interacting region (Figure 6B). We searched for potential transcription factors that bind to the identified region using CLC Genomics Workbench and TFBIND software (Tsunoda and Takagi, 1999). Analysis of the 3D protein structure from Protein DataBank (Elrod-Erickson et al., 1996) and TFBIND analysis did not identify any EGR1-binding sequence in the CD80 promoter (Figure 6C). NF-κB was predicted as a potential transcription factor that binds to this region; however, the NF-κB p65 DNA-binding activities were similar between nuclear extracts overexpressing EGR1 and controls (Figure S13). Serum response factor (SRF) was also predicted as a potential transcription factor that binds to this region, and SRF was previously reported to interact with EGR1 (Wang et al., 2005) (Figure 6D). Another study also reported that Cd80 was downregulated in Srf−/− neutrophils compared with WT neutrophils in RNA deep sequencing (Taylor et al., 2014). Our ChIP-PCR analysis using SRF antibody also demonstrated that SRF interacted with the CD80 promoter (Figure 6E). Therefore, we hypothesized that EGR1 might interact with the CD80 promoter through binding to SRF. Co-immunoprecipitation experiments revealed that EGR1 interacts with SRF (Figure 6F), which is consistent with the previous report (Wang et al., 2005). Additionally, ChIP-PCR analysis in cells knocked down for SRF by siRNA demonstrated that EGR1 binding on the CD80 promoter was lost when SRF was silenced (Figure S14). Taken together, these results suggest that EGR1 interacts with SRF and may regulate its activation on the Cd80 promoter.
Figure 6.
EGR1 interacts with SRF and may regulate its activation on the Cd80 promoter
(A) ChIP-seq enrichment profiles for H3K27Ac at the Cd80 locus were generated using WT or Egr1−/− BMDCs stimulated with LPS (100 ng/mL) and lactate (Lac, 20 mM) for 24 hr.
(B) ChIP-seq enrichment profiles for EGR1 at the transcription start site (TSS) at the Cd80 locus were generated using WT BMDCs stimulated by LPS (100 ng/mL) and Lac (20 mM) for 24 hr.
(C) The 3D protein structure of EGR1 and the EGR1 binding sequence from Protein DataBank were analyzed by CLC Genomics Workbench.
(D) SRF was predicted as a potential transcription factor that binds to the region identified in (B) using TFBIND software. SRF was previously reported to interact with EGR1. TF, transcription factor.
(E) ChIP-PCR analysis was performed using RAW 264.7 cells overexpressing Srf. ChIP was performed using control IgG (Ctrl) or anti-SRF antibody. PCR was performed using primers that detect sequences in the Cd80 promoter.
(F) Co-immunoprecipitation experiments assessing EGR1 binding to SRF. Cell extracts from RAW 264.7 cells overexpressing both Srf and Myc-tagged Egr1 were immunoprecipitated with anti-SRF antibody or control IgG (Ctrl) and analyzed with anti-c-Myc and anti-SRF antibody by Western blotting.
(G) Proposed schematic model of how lactate and its induction of EGR1 switches a hot tumor to a cold tumor. Lactate induces a shift from a hot tumor (presence of inflammation) to a cold (non-inflamed) tumor, while TLR4 ligands cause the opposite shift. Lactate upregulates the expression of EGR1, which may downregulate the expression of CD80.
See also Figures S12–S14.
Discussion
Since the demonstrated clinical success of ICIs, one of the most active research focuses today is augmenting the responses to ICIs by targeting the tumor microenvironment (Chen and Mellman, 2017). Tumors with an inflamed tumor microenvironment are considered “hot tumors” and show good responses to ICIs. Our results show that lactate inhibited the effects of ICI in a mouse tumor model, indicating that lactate is possibly associated with one of the mechanisms of the cooling switch. LPS-induced anti-tumor cytokines and molecules, such as IL-12 p40 and CD80, were decreased by lactate in DCs, which suggests that DCs can be directed to non-inflammatory conditions by lactate. Luciferase assays showed that suppression of the DNA-binding activity of NF-κB by lactate is important for the regulation of IL-12 p40. Our results suggested that lactate causes suppressed phosphorylation of IκBα, which leads to IκBα accumulation and subsequent inhibition of NF-κB. These results were consistent with the recent report showing that lactate attenuated LPS-stimulated macrophage TNF-α and IL-6 production and suppressed LPS-stimulated NF-κB (Yang et al., 2020). Acidity also mimicked the effects of lactate on IL-12 p40, EGR1 and NF-κB activation; therefore, we speculate the effects of lactate on DCs may be from the acidity. Our results also indicate that lactate mediates the suppressive effect on NF-κB activation via GPR81, which was consistent with a previous report (Yang et al., 2020). Together, our findings suggest that both the acidity of lactate and the signaling pathways through GPR81 may be important in the suppressive effect of lactate on NF-κB p65 DNA-binding activity.
Despite the remarkable suppression of NF-κB activity by lactate, microarray analysis showed that the decrease of NF-κB family mRNAs by lactate was mild. Conversely, the EGR1 gene was the most highly induced transcription factor in DCs stimulated with lactate. EGR1 is an inflammatory transcriptional factor (Sukhatme, 1990) that regulates a large set of inflammatory genes, such as IL-2 and TNF-α genes (Krämer et al., 1994; Skerka et al., 1995). Aberrant EGR1 expression is linked to various human diseases including cancers (Bhattacharyya et al., 2013). With database analysis using TCGA, we identified a correlation between EGR1 and LDHA expression in primary melanoma. As LDHA is a glycolytic marker, this result suggests the association of EGR1 with glycolysis. Additionally, histological analysis showed that the expression of EGR1 was increased in human primary invasive melanoma tissues, and CD11c+ DCs expressed EGR1 in human melanoma. Studies have shown that as tumor thickness increases, there is a significant decrease in survival rates (Balch et al., 2009). Therefore, increased EGR1 expression might be a marker of invasive melanoma. Consistent with these observations, the expression levels of EGR1 were correlated with tumor thickness and 5-S-cysteinyl-DOPA, which is correlated with prognosis in melanoma (Bánfalvi et al., 2003). Notably, these results are preliminary findings, as the number of human samples was small; therefore, further studies with a larger sample number are needed.
We hypothesized that EGR1 might be a key transcription factor to regulate cytokines and co-stimulatory factors in DCs. However, there were no significant differences in the expression levels of IL-12 p40, TNF-α, and IL-6 between Egr1−/− and WT BMDCs. However, CD80 was increased in Egr1−/− BMDCs compared with WT BMDCs. These results were consistent with the recent report that Egr1 silencing led to elevated expression of CD80 in toxoplasma-infected DCs and LPS-challenged immature DCs (Ten Hoeve et al., 2019). Despite the significant difference of the expression of CD80 and antigen presentation assay in vitro, the B16-F1 cell–derived tumor growth rate was similar between WT and Egr1−/− mice. Consistent with these findings, Caso et al. reported reduced growth of Lewis lung carcinoma but not B16-F10 cell-derived tumors in Egr1−/− mice. Fahmy et al. also reported similar results, which suggest other EGR1 mechanisms, including FGF-dependent angiogenesis during neovascularization and tumor growth by EGR1 (Fahmy et al., 2003). Therefore, to eliminate the possible effect of EGR1 in other cells except DCs, we used a DC-specific knockout model and found that tumor growth was suppressed in DC-depleted mice transferred with Egr1−/− BMDCs compared those transferred with WT BMDCs. These results suggest that EGR1 in DCs has a pro-tumor activity.
ChIP-Seq results identified a region with a potential interaction site for EGR1 in the Cd80 promoter. The region was not identical to the EGR1-binding sequence, according to the analysis by TFBIND and data from Protein DataBank. However, our data show that EGR1 interacts with the SRF transcription factor on the Cd80 promoter. A previous study reported that Cd80 was downregulated in Srf−/− neutrophils compared with WT neutrophils (Taylor et al., 2014), and thus EGR1 may regulate SRF activation on the Cd80 promoter. Taken together, these results suggest that the expression of IL-12 p40 can be suppressed by lactate but this is independent of EGR1, while the expression of CD80 may be dependent on both lactate and EGR1, which itself is induced by lactate. Although increased expression of CD80 was observed in Egr1−/− BMDCs compared with WT BMDCs under stimulation of LPS and lactate, the difference was mild. Therefore, our results suggest that lactate suppresses LPS-induced CD80 expression but its suppressive effect via upregulation of EGR1 may be partial.
In conclusion, our results suggest that lactate and its induced EGR1 may be key factors that determine the inflamed or non-inflamed tumor status (Figure 6G). Lactate and EGR1 may be targets for stimulating antitumor immunity and may also be important in cancer treatment with ICIs.
Limitations of the study
Our study has several limitations. Our methods have some artificial aspects as experimental models that may not completely represent real life conditions, such as the possible effect of buffering systems existing in vivo. Additionally, lactate has been reported to differentially affect not only DCs but also other tumor infiltrating immune cells, such as CD8+ T cells and Tregs (Haas et al., 2015; Speiser et al., 2016). Therefore, our results may just reflect part of the mechanisms underlying the immunomodulating effect of lactate on immune cells. Therefore, further studies using experimental models that more closely reflect real life tumors are needed to elucidate the full mechanisms of lactate on tumors including other tumor-infiltrating immune cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse EGR1 | Abcam | ab133695 |
| anti-mouse EGR1 | Abcam | ab174509 |
| anti-human EGR1 | Abcam | ab194357 |
| anti-mouse PD-L1 | Abcam | ab213480 |
| anti-c-Myc antibody (Y69) | Abcam | ab32072 |
| anti-β-actin (C4) | Santa Cruz Biotechnology | sc-47778 |
| goat anti-rabbit IgG (H+L)-HRP conjugate | Bio-Rad | 170-6515 |
| anti-β-actin-HRP (AC-15) | Abcam | ab49900 |
| FITC anti-mouse CD3 | BioLegend | 17A2 |
| PE anti-mouse CD80 | BioLegend | 16-10A1 |
| PE anti-mouse CD86 | BioLegend | GL-1 |
| PerCP/Cy5.5 anti-mouse CD11b | BioLegend | M1/70 |
| PerCP/Cy5.5 anti-mouse CD45RA | BD Pharmingen | 14.8 |
| PerCP/Cy5.5 anti-mouse CD4 | BioLegend | GK1.5 |
| APC anti-mouse F4/80 | BioLegend | BM8 |
| APC anti-mouse CD11c | BioLegend | N418 |
| APC anti-mouse CD8a | BioLegend | 53-6.7 |
| InVivoMAb anti-mouse PD-L1 (B7-H1) | Bio X Cell | 10F.9G2 |
| InVivoMAb rat IgG2b isotype control, anti-keyhole limpet hemocyanin | Bio X Cell | LTF-2 |
| Alexa Fluor 647-goat anti-rabbit IgG (H+L) | Thermo Fisher Scientific | A-21245 |
| anti-EGR1 (used for ChIP) | Cell Signaling Technology | 15F7 |
| anti-histone H3 (acetyl K27) | Abcam | ab4729 |
| normal rat IgG | Santa Cruz Biotechnology | sc-2026 |
| rat anti-mouse SRF antibody | Active Motif | 2C5 |
| rabbit anti-mouse SRF antibody | Abcam | ab53147 |
| goat anti-rabbit IgG H&L (Alexa Fluor 405) | Abcam | ab175652 |
| APC anti-human CD20 Antibody | BioLegend | 2H7 |
| Alexa Fluor 594 anti-human MART-1 antibody | BioLegend | M2-7C10 |
| APC anti-human CD56 (NCAM) antibody | BioLegend | MEM-188 |
| APC anti-human CD68 antibody | BioLegend | Y1/82A |
| APC anti-human CD11c antibody | BioLegend | 3.9 |
| anti-p-p38 antibody (Thr 180/Tyr 182) | Santa Cruz Biotechnology | sc-17852-R |
| anti-phospho-SAPK/JNK (Thr183/Tyr185) | Cell Signaling Technology | 81E11 |
| anti-IκBα mouse antibody | Cell Signaling Technology | L35A5 |
| anti-phospho-IκBα (Ser32/36) | Cell Signaling Technology | 5A5 |
| anti-melanoma gp100 antibody | Abcam | ab137078 |
| donkey anti-goat IgG H&L (HRP) | Abcam | ab97110 |
| human/mouse c-Rel antibody | R&D | AF2699 |
| PE anti-EGR1 rabbit antibody | Cell Signaling Technology | 44D5 |
| anti-GPR81 antibody | Thermo Fisher Scientific | PA5-67873 |
| Rabbit IgG Isotype Control | Cell Signaling Technology | DA1E |
| Chemicals, peptides, and recombinant proteins | ||
| mouse GM-CSF | Prospec | CYT-222 |
| mouse M-CSF | PeproTech | 315-02 |
| LPS (LPS-SM ultrapure) | InvivoGen | tlrl-smlps |
| L-(+)-Lactic acid | Sigma-Aldrich | L6402 |
| Ova 257–264 peptide (SIINFEKL) | MBL | OF1060-B |
| recombinant mouse IFN-γ (carrier-free) | BioLegend | 575306 |
| Hoechst 33342 solution | Dojindo | H342 |
| protein G-Sepharose | Abcam | ab193259 |
| sodium L-lactate | Sigma-Aldrich | 71718 |
| diphtheria toxin | Sigma-Aldrich | D0564 |
| collagenase type II | Sigma-Aldrich | C6885 |
| Critical commercial assays | ||
| High Pure RNA Isolation Kit | Roche | 11828665001 |
| ReverTra Ace | Toyobo | TRT-101 |
| Real-time PCR Master Mix | Toyobo | QPK-101 |
| mouse IL-12 p40 DuoSet ELISA | R&D | DY-499-05 |
| mouse TNF-α DuoSet ELISA | R&D | DY410-05 |
| mouse IL-6 DuoSet ELISA | R&D | DY406-05 |
| mouse CD4+ T Cell Isolation Kit | Miltenyi Biotec | 130-104-454 |
| mouse CD8+ T Cell Isolation Kit | Miltenyi Biotec | 130-104-075 |
| mouse NK Cell Isolation Kit II | Miltenyi Biotec | 130-096-892 |
| CFSE Cell Division Tracker Kit | BioLegend | 423801 |
| Dual-Luciferase Reporter Assay System | Promega | E1910 |
| PrimeSTAR Mutagenesis Basal Kit | Takara | R046A |
| TransAM NF-κB p65 ELISA kit | Active Motif | 40096 |
| TransAM Flexi NF-κB p50 | Active Motif | 41098 |
| TransAM Flexi NF-κB Family | Active Motif | 43298 |
| ChIP-IT Express Enzymatic | Active Motif | 53009 |
| Chromatin IP DNA Purification Kit | Active Motif | 58002 |
| Foxp3/Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | 00-5523-00 |
| FITC Annexin V Apoptosis Detection Kit I | BD Pharmingen | 556547 |
| Lactate Assay Kit (Colorimetric) | Cell Biolabs | MET-5012 |
| Deposited data | ||
| BMDC microarray data | This paper | GEO: GSE163978 |
| ChIP-seq data | This paper | GEO: GSE164027 |
| Experimental models: Cell lines | ||
| B16-F1 cells | Riken BRC | RCB2649 |
| RAW 264.7 cells | Riken BRC | RCB0535 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Charles River Laboratories Japan | https://www.crj.co.jp/product/rm/detail/b6j |
| Mouse: Egr1−/− | Jackson laboratory | JAX stock #012924, Lee et al., 1995 |
| Mouse: C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I) | Jackson laboratory | JAX stock #003831, Hogquist et al., 1994 |
| Mouse: CD11c-DTR | Jackson laboratory | JAX stock #004509, Jung et al., 2002 |
| Oligonucleotides | ||
| TaqMan probes: 18S rRNA | Thermo Fisher Scientific | Mm03928990_g1 |
| TaqMan probes: Cd274 | Thermo Fisher Scientific | Mm00452054_m1 |
| TaqMan probes: Egr1 | Thermo Fisher Scientific | Mm00656724_m1 |
| Genotyping primers for Egr1−/− mice, Common: 5′-GGGCACAGGGGATGGGA ATG-3′ |
Jackson Laboratory | Lee et al., 1995 |
| Genotyping primers for Egr1−/− mice, WT: 5′-AACCGGCCCAGCAAGACACC-3′ | Jackson Laboratory | Lee et al., 1995 |
| Genotyping primers for Egr1−/− mice, Mut: 5′-CTCGTGCTTTACGGTATCGC-3′ | Jackson Laboratory | Lee et al., 1995 |
| Primers used for CD80 promoter ChIP analysis Fwd, 5′-TGGCAGTCACTCTGTGTCCAG-3′ | This paper | N/A |
| Primers used for CD80 promoter ChIP analysis Rvs, 5′-CTAGTAAGAGTCTATTGAGG-3′ | This paper | N/A |
| GPR81 siRNAs | Thermo Fisher Scientific | s211558 |
| SRF siRNAs | Thermo Fisher Scientific | s74390 |
| Silencer negative control No. 1 siRNA | Thermo Fisher Scientific | AM4611 |
| Recombinant DNA | ||
| Plasmid: Egr1 | OriGene Technologies | MC217560 |
| Plasmid: Myc-tagged Egr1 | OriGene Technologies | MR227136 |
| Plasmid: Srf | OriGene Technologies | MC217127 |
| Plasmid: Il12b 400bp pro-Luc | Addgene | #20020 |
| Software and algorithms | ||
| FlowJo analysis software V10 | Ashland | https://www.flowjo.com/ |
| Developer Toolbox of IN Cell Analyzer | GE Healthcare | https://www.cytivalifesciences.co.jp/catalog/1705.html |
| CLC Genomics Workbench 10 | Qiagen | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench/ |
| TFBIND software | RIKEN | Tsunoda and Takagi, 1999 |
| GraphPad Prism v8 | GraphPad | https://www.graphpad.com/ |
| Other | ||
| QuantStudio 12K Flex | Applied Biosystems | N/A |
| Multiskan FC | Thermo Fisher Scientific | N/A |
| ChemiDoc XRS+ System | Bio-Rad | N/A |
| NovoCyte | ACEA Biosciences | N/A |
| xCELLigence S16 system | Agilent Technologies | N/A |
| Neon Transfection System | Thermo Fisher Scientific | N/A |
| EnSpire | PerkinElmer | N/A |
| LB 941 | Berthold Technologies | N/A |
| ABI 3130xl | Applied Biosystems | N/A |
| BZ-9000 | Keyence | N/A |
| FV1200 | Olympus | N/A |
| IN Cell Analyzer 2200 | GE Healthcare | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hisashi Kanemaru (hisashikanemaru@gmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
Mice were maintained in our specific pathogen-free animal facility according to the institutional guidelines. C57BL/6J mice were obtained from Charles River Laboratories Japan. OT-I transgenic mice, Egr1−/− mice and CD11c-DTR transgenic mice were obtained from the Jackson Laboratory (Hogquist et al., 1994; Jung et al., 2002; Lee et al., 1995). Experiments were generally performed with female and male mice at 6–12 weeks of age. All animal experiments were performed with approval from the Animal Research Committee of Kumamoto University (Kumamoto, Japan).
Cells
B16-F1 (Riken BRC, Ibaraki, Japan) cells were grown in D-MEM (High Glucose, 043-30085, FUJIFILM Wako, Osaka, Japan) containing 10% fetal bovine serum (FBS). RAW 264.7 (Riken BRC) cells were grown in MEM (Minimum Essential Medium Eagle, M4655, Sigma-Aldrich, St. Louis, MO, USA) containing 10% FBS. BMDCs were generated as described previously (Kanemaru et al., 2017). Briefly, mouse bone marrow cells were obtained from femurs and differentiated into DCs in MEM with 10% FBS containing 10 ng/ml granulocyte macrophage colony-stimulating factor (GM-CSF; Prospec, East Brunswick, NJ, USA). Medium was replaced with fresh medium containing GM-CSF every two days. On day 6, cells were collected. BMDMs were also generated as described previously (Kanemaru et al., 2017). Bone marrow cells were obtained and incubated in MEM with 10% FBS containing 3 ng/ml macrophage colony-stimulating factor (M-CSF, PeproTech, Rocky Hill, NJ, USA). Medium was changed on day 3. On day 5, macrophages were harvested. BMDCs and BMDMs were stimulated with LPS (100 ng/ml; LPS-SM ultrapure, InvivoGen, San Diego, CA, USA) and/or L-(+)-Lactic acid (Sigma-Aldrich), and/or sodium L-lactate (Sigma-Aldrich).
Plasmids
Plasmids encoding Egr1, Myc-tagged Egr1, and Srf were purchased from OriGene Technologies (MC217560, MR227136, and MC217127). Il12b 400bp pro-Luc plasmid was a gift from Dr. Stephen Smale (Addgene plasmid #20020; http://n2t.net/addgene:20020; RRID:Addgene_20020) (Plevy et al., 1997).
Tumor model
B16-F1 cells were grown in DMEM culture media with 10% FBS. Cells were washed with PBS, dispersed in a 0.25% solution of trypsin, and resuspended in fresh culture media. After centrifugation, the cell pellet was resuspended in PBS and the concentration was adjusted to 107 cells/ml. Mice were then injected with 0.1 ml of the suspension (1 × 106 B16-F1 cells). Tumors were measured with a dial-caliper every 3 days, and tumor volumes were determined using the formula (width)2 × length × 0.52, as previously described (Colegio et al., 2014; O'Reilly et al., 1997). When the mean tumor diameter reached 1.2 cm, the experiments were terminated, and mice were sacrificed and autopsied.
Method details
In vivo anti-PD-L1 antibody and lactate treatment
In vivo treatment of the tumor mouse model with anti-PD-L1 antibody was performed as previously described (Tsukamoto et al., 2018). On days 6, 9 and 12 after injection of B16-F1 cells, 200 μg of control IgG Ab (LTF-2, Bio X Cell, Lebanon, NH, USA) or anti-PD-L1 Ab (10F.9G2, Bio X Cell) were injected intraperitoneally. In addition, lactate (5 mM, 200 μl) or PBS control (200 μl) was subcutaneously transferred into the backs of mice on days 6, 9 and 12. The tumors were measured with a dial caliper every 3 days, and the tumor volumes were determined as described above.
Bone marrow transfer experiments were performed as previously described (Kanemaru et al., 2017). Recipient C57BL/6J mice (6 weeks old) were lethally irradiated with 9 Gy and intravenously injected with 2×107 bone marrow cells obtained from CD11c-DTR (Itgax-DTR+/−) mice. For the depletion of CD11c+ cells, CD11c-DTR bone marrow chimeric mice were intraperitoneally treated with 0.5 μg diphtheria toxin (Sigma-Aldrich) twice per week for a total of four treatments, and mice were treated intratumorally with 1×106 BMDCs on days 5, 6, 7, 12, 13, and 14, as previously described (Tsuchiya et al., 2019).
CTL killing assay
The CTL killing assays were performed using the xCELLigence S16 system (Agilent Technologies, Santa Clara, CA, USA) as previously described (Delconte et al., 2016; Fasbender and Watzl, 2018; Hildner et al., 2008; Lecoeur et al., 2001; Zaritskaya et al., 2010). Whole splenocytes (4×107) obtained from B16-F1 cell–tumor-bearing WT mice 14 days after implantation were cultured with 2×106 IFN-γ-treated (100 U/ml, BioLegend, San Diego, CA, USA) B16-F1 cells. After 5 days, the cells were harvested and used as effector cells in a cytotoxicity assay. As lactate-treated CTLs, effector cells were stimulated with lactate (20 mM) for 24 h. To generate target cells, B16-F1 tumor cells were treated with 100 U/ml IFN-γ for 48 h before use. Target cells were washed and seeded at 20,000 per well into the wells of E-Plates in 150 μl of media. Cell growth was dynamically monitored with the impedance-based xCELLigence S16 system until cells reached log growth phase and formed a monolayer (approximately 18 h). Effector cells were then added directly to individual wells containing the target cells at different ratios (E:T=12.5:1, 25:1, 50:1). For background controls, effector cells were added to wells that contained no target cells, and target cells were added to wells without the addition of effector cells. After addition of effector cells, the system continued to take measurements every 15 min.
Antigen-presentation assays
Antigen-presentation assays were performed as previously described (Hildner et al., 2008; Thevenot et al., 2014). Ovalbumin-specific CD8+ OT-I cells were obtained from mouse spleens and lymph nodes using a CD8+ T Cell Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany). For OT-I T cell proliferation, CD8+ T cells from the splenocytes and lymph nodes of OT-I transgenic mice were labeled with carboxyfluorescein succinimidyl ester (CFSE). BMDCs were pulsed with 1 μM Ova 257-264 peptide (SIINFEKL, MBL, Nagano, Japan) for 3 h, and then cells were washed twice with PBS and co-cultured with 5×104 CFSE-labeled naïve OT-I transgenic CD8+ T cells at decreasing dilutions (1:1–1:1/16, OT-I:BMDCs). Proliferation of OT-I cells was measured after 84 h by flow cytometry.
Microarray analysis
BMDCs from WT and Egr1−/− mice were stimulated with LPS (100 ng/mL) and/or lactate (20 mM) for 4 h. Total RNA was then isolated using a High Pure RNA Isolation Kit (Roche, Basel, Switzerland) and subjected to Agilent Expression microarray analysis. The microarray was performed by Takara Bio, and the resulting data were analyzed by Aqua (Takara, Shiga, Japan).
Quantitative PCR
RNA was extracted from cells using a High Pure RNA Isolation kit (Roche) according to the manufacturer’s instructions. Reverse transcription was performed with 4 μl of ReverTra Ace (Toyobo, Osaka, Japan) in a total volume of 20 μl. Subsequently, 1 μl of cDNA fragments was amplified using 10 μl of real-time PCR Master Mix (Toyobo) with 1 μl of TaqMan probes (Thermo Fisher Scientific, Waltham, MA, USA) in a total volume of 20 μl. Fluorescence from the TaqMan probe for each gene was detected using a QuantStudio 12K Flex (Applied Biosystems, Foster City, CA, USA). The mRNA expression level of each gene was normalized to the 18S rRNA expression level (Mm03928990_g1, Thermo Fisher Scientific). The following TaqMan probes (Thermo Fisher Scientific) were used for quantitative PCR: Cd274 (Mm00452054_m1) and Egr1 (Mm00656724_m1).
ELISA.
IL-12 p40, TNF-α, and IL-6 levels were determined by ELISAs (DuoSet ELISA, R&D, Minneapolis, MN, USA) performed according to the manufacturer’s instructions. The absorbance of each ELISA plate was measured using a microplate reader (Multiskan FC, Thermo Fisher Scientific).
Immunoblot analysis
Cells were lyzed with lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP40) containing Complete Protease Inhibitor Cocktail (Roche). The cell lysates were separated by standard SDS-PAGE (e-PAGEL, ATTO, Tokyo, Japan) and analyzed by immunoblotting. The following antibodies were used for immunoblotting: anti-PD-L1 (ab213480, Abcam, Cambridge, UK), anti-EGR1 (ab133695, Abcam), anti-c-Myc antibody (Y69) (ab32072, Abcam), goat anti-rabbit IgG (H+L)-HRP Conjugate (Bio-Rad), anti-phospho-p38 antibody (Thr 180/Tyr 182; sc-17852-R, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-phospho-SAPK/JNK (Thr183/Tyr185; 81E11, Cell Signaling Technology, Danvers, MA, USA), anti-IκBα mouse antibody (L35A5, Cell Signaling Technology), anti-phospho-IκBα (Ser32/36; 5A5, Cell Signaling Technology), anti-β-actin (C4) (sc-47778, Santa Cruz Biotechnology), and anti-β-actin-HRP (AC-15) (ab49900, Abcam). The Western HRP Substrate (Pierce ECL Western Blotting Substrate, Thermo Fisher Scientific) was used for the development of positive signals, and chemiluminescence was detected using a ChemiDoc XRS+ System (Bio-Rad, Hercules, CA, USA).
Flow cytometry
Cells were washed with magnetic-activated cell sorting buffer, incubated with antibodies for 15 min, and then washed twice. The antibodies for flow cytometry were purchased from commercial sources as follows: anti-CD3-FITC (17A2; BioLegend), anti-CD80-PE (16-10A1; BioLegend), anti-CD86-PE (GL-1; BioLegend), anti-CD11b-PerCP/Cy5.5 (M1/70; BioLegend), anti-CD45RA-PerCP/Cy5.5 (14.8; BD Pharmingen, San Diego, CA, USA), anti-CD4-PerCP/Cy5.5 (GK1.5; BioLegend), anti-F4/80-APC (BM8; BioLegend), anti-CD11c-APC (N418; BioLegend), anti-CD8a-APC (53-6.7; BioLegend), and PE anti-EGR1 rabbit antibody (Cell Signaling Technology). The dilution ratio was 1:100–200 for each antibody. For EGR1 staining, a Foxp3/Transcription Factor Staining Buffer Set (eBioscience, San Diego, CA, USA) was used. Data were collected using a NovoCyte (ACEA Biosciences, San Diego, CA, USA) and analyzed using FlowJo analysis software (Ashland, OR, USA).
Luciferase reporter assays
Luciferase reporter assays were performed according to the manufacturer’s instructions (Dual-Luciferase Reporter Assay System, Promega, Madison, WI, USA). For transfections, 1×106 RAW 264.7 cells were suspended in 100 μl of a solution (buffer R) containing 8 μg of Il12b 400-bp pro-Luc plasmid, and 1 μg of pRL-TK. Cells were electroporated using the Neon Transfection System (Thermo Fisher Scientific) at 1680 V for 20 msec and then plated in a 24-well plate at 1×105 cells/well. After 24 h, cells were activated by LPS (100 ng/ml) or lactate (20 mM) for 24 h. Following activation, cell extracts were prepared using Passive Lysis Buffer (Promega). Luciferase activity was determined from a 20-μl cell extract and measured on the EnSpire (PerkinElmer, Waltham, MA, USA) or LB 941 (Berthold Technologies, Bad Wildbad, Germany) microplate reader.
To generate deletion mutants for luciferase reporter assays, mutant DNA fragments were produced by PCR and cloned into the KpnI-BglII sites of the Il12b pro-Luc construct as previously described (Murphy et al., 1995). To generate substitution mutants, the mutant constructs were produced using a PrimeSTAR Mutagenesis Basal Kit (Takara) according to the manufacturer’s recommendations. All plasmids were verified by restriction mapping and by sequencing using ABI 3130xl (Applied Biosystems).
NF-κB DNA-binding assay
The NF-κB p65 DNA-binding activity was measured with the TransAM NF-κB p65, TransAM Flexi NF-κB p50, and TransAM Flexi NF-κB Family ELISA kit and according to the manufacturer’s recommendations (Active Motif, Carlsbad, CA, USA). Nuclear extracts were prepared from 5 × 106 RAW 264.7 cells. After the cells were activated with LPS (100 ng/mL) or lactate (20 mM) for 1 h, cell lysates were prepared in 200 μl lysis buffer [10 mM Hepes-KOH, pH 7.8, 10 mM KCl, 0.1 mM EDTA, pH 8.0, protease inhibitor mixture (Roche), and 0.1% Nonidet P-40], and nuclear extracts were obtained in 60 μl buffer [50 mM Hepes-KOH, pH 7.8, 420 mM KCl, 0.1 mM EDTA, pH 8.0, 5 mM MgCl2, protease inhibitor mixture (Roche), and 20% glycerol] as previously described (Kanemaru et al., 2017). Nuclear extracts (5 μl) in microwells were used to evaluate NF-κB DNA-binding activity.
Histological analysis
Frozen 10-μm-thick mouse tumor tissue sections in Tissue-Tek OCT compound (Sakura Finetek, Torrance, CA, USA) or cells plated in chamber slides (Lab-Tek II Chamber Slide; Thermo Fisher Scientific) were fixed in 4% paraformaldehyde and blocked with 3% BSA in PBS. Formalin-fixed paraffin-embedded sections of primary tumor tissues (4 μm thick) from patients with no history of systemic treatment, including treatment with ICIs, were used for immunofluorescence analyses.
Immunofluorescent staining was performed using the following antibodies and reagents: anti-mouse EGR1 (ab174509, Abcam), anti-mouse EGR1 (ab133695, Abcam), anti-human EGR1 (ab194357, Abcam), Alexa Fluor 647-goat anti-rabbit IgG (H+L) (A-21245; Thermo Fisher Scientific), Alexa Fluor 405-goat anti-rabbit IgG H&L (Abcam), Hoechst 33342 solution (Dojindo, Kumamoto, Japan), APC anti-human CD20 Antibody (BioLegend), Alexa Fluor 594 anti-MART-1 antibody (BioLegend), APC anti-human CD56 (NCAM) antibody (BioLegend), APC anti-human CD68 antibody (BioLegend), and APC anti-human CD11c antibody (BioLegend). The dilution ratios for anti-EGR1 and Hoechst 33342 solution were 1:500 and 1:5000, respectively. The dilution ratios for other antibodies were 1:100. Images were obtained using a BZ-9000 (Keyence, Osaka, Japan), FV1200 (Olympus, Tokyo, Japan) or IN Cell Analyzer 2200 (GE Healthcare, Little Chalfont, UK). The cell-counting software Developer Toolbox of IN Cell Analyzer was used to determine the ratio of EGR1-positive cells to the total number of cells in whole tumor sections by counting EGR1-positive cells and cell nuclei (Tagawa et al., 2020).
Analysis of human specimens
Informed consent was obtained from the participant involved in the study. All procedures performed in studies were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Tissue digestion for cell population analyses
Tissues were isolated from mice and placed in 10 mL of MEM containing 1 mg/mL collagenase type II (C6885; Sigma-Aldrich). Tissues were disrupted with surgical scissors and incubated at 37 °C for 30 min. Samples were filtered through 40-μm filters, and erythrocytes were removed by incubation for 3 min with ACK Lysing Buffer (Gibco, Life Technologies). After two washes with magnetic-activated cell sorting buffer (0.5% BSA and 2 mM EDTA in PBS, pH 7.2), samples were stained for analysis by flow cytometry, as previously described (Kanemaru et al., 2017). Leukocyte subpopulations were generally defined as follows: DCs, CD45+ CD11c+ cells; macrophages, CD45+ CD11b+ F4/80+ cells; CD4+ T cells, CD45+ CD3+ CD4+ cells; CD8+ T cells, CD45+ CD3+ CD8+ cells; natural killer cells, CD45+ NK1.1+ cells; and B16-F1 tumor cells, CD45− melanoma gp100+ cells.
Measurement of lactate concentration in tumors
The concentrations of lactate in tumors were measured as previously described (Maeda-Otsuka et al., 2019; Ohashi et al., 2017). Tumor specimens from mice were homogenized in PBS (1 μl per 1 mg tumor) and centrifuged. The concentrations of lactate in the obtained supernatants were measured using a Colorimetric Lactate Assay Kit (Cell Biolabs, San Diego, CA).
ChIP-seq analysis
ChIP analysis was performed using ChIP-IT Express Enzymatic (Active Motif) (Kanemaru et al., 2018). BMDCs (2×107) were stimulated with LPS (100 ng/ml) and lactate (20 mM). After 24 h, the cells were fixed with 20 ml of 1% formaldehyde for 10 min, followed by a wash with 10 ml of PBS, and the fixation reaction was stopped by adding 10 ml of glycine stop fix solution for 5 min. Cell extracts were prepared in lysis buffer and zhomogenized using a Dounce homogenizer (BioMasherII; Nippi, Tokyo, Japan) on ice. Enzymatic shearing to produce DNA fragments was performed for 30 min. Immunoprecipitation was then performed using 3 μg of anti-EGR1 (15F7) (Cell Signaling Technology) or anti-histone H3 (acetyl K27) (ab4729; Abcam). The samples were incubated overnight with 25 μl of protein G magnetic beads, and then the beads were washed. DNA was eluted in 50 μl of elution buffer, and 50 μl of reverse cross-linking buffer was added. Samples were centrifuged, and the resulting supernatants were collected and incubated at 95 °C for 15 min. After samples were returned to room temperature, RNase and proteinase K were added, and the samples were incubated for 1 h at 37 °C. DNA was then purified using the Chromatin IP DNA Purification Kit (58002; Active Motif) according to the manufacturer’s instructions. ChIP-seq was performed by Filgen (Aichi, Japan), and the resulting data were analyzed by CLC Genomics Workbench 10 (Qiagen, Hilden, Germany). The potential transcription factor–binding regions were estimated using TFBIND software (RIKEN, Yokohama, Japan) (Tsunoda and Takagi, 1999).
ChIP-PCR analysis
ChIP analysis was performed using ChIP-IT Express Enzymatic (Active Motif). RAW 264.7 cells (1.5×107) were transfected with 60 μg of plasmid encoding Srf using the Neon Transfection System (Thermo Fisher Scientific) at 1680 V for 20 msec. After 12 h, the cells were fixed with 1% formaldehyde for 10 min followed by a wash with PBS; the fixation reaction was stopped by adding glycine stop fix solution for 5 min. Cell extracts were prepared in lysis buffer and zhomogenized using a Dounce homogenizer (BioMasherII, Nippi) on ice. Enzymatic shearing was performed for 30 min, and DNA fragments were obtained (Input). Immunoprecipitation was performed using 3 μg of normal rat IgG (sc-2026; Santa Cruz Biotechnology) or rat anti-SRF antibody (2C5; Active Motif). Samples were incubated overnight with 25 μl of protein G magnetic beads, and then the beads were washed. DNA was eluted in 50 μl of elution buffer and 50 μl of reverse cross-linking buffer was added. The supernatants were then collected and incubated at 95 °C for 15 min. After samples were returned to room temperature, RNase and proteinase K was added, and the samples were incubated for 1 h at 37 °C. Input samples and precipitated DNA were analyzed by PCR using primers that detect sequences containing the SRF binding site of the CD80 promoter: 5′-TGGCAGTCACTCTGTGTCCAG-3′ and 5′- CTAGTAAGAGTCTATTGAGG-3′.
Co-immunoprecipitation
Cells (1 × 106) were suspended in 100 μL of a solution (buffer R) containing 8 μg of Myc-tagged Egr1 plasmid and 8 μg of Srf plasmid. Cells were electroporated using the Neon Transfection System (Thermo Fisher Scientific) at 1,680 V for 20 ms. Transfected cells were plated at 5 × 106 cells per dish. After 12 h, cell extracts were prepared using lysis buffer [10 mM Hepes-KOH, pH 7.8, 10 mM KCl, 0.1 mM EDTA, pH 8.0, protease inhibitor mixture (Roche), and 0.1% Nonidet P-40]. Lysates (50 μg) were incubated with 4 μg of normal rat IgG (sc-2026; Santa Cruz Biotechnology) or rat anti-SRF (2C5; Active Motif) in a total volume of 110 μL. The samples were incubated at 4 °C for 1 h on a tube rotator. Protein G-Sepharose (25 μL of 50% slurry; ab193259, Abcam) was added, and the mixture was agitated at 4 °C for 1 h. After centrifugation, the pellet was washed four times with TBS (50 mM Tris and 150 mM NaCl, pH 7.5). Samples were analyzed by standard SDS-PAGE immunoblotting using anti-c-Myc antibody (ab32072, Abcam) and anti-SRF antibody (ab53147, Abcam) from a different host species.
siRNA transfection
RAW 264.7 cells (1 × 106) were suspended in 100 μL of a solution (buffer R) containing 100 nM of Gpr81 siRNAs, Srf siRNAs, or negative control siRNAs (Thermo Fisher Scientific). Cells were electroporated using the Neon Transfection System (Thermo Fisher Scientific) at 1,680 V for 20 ms. Transfected cells were plated at 5 × 106 cells per dish. After 24 h, cells were collected for experiments. Knockdown of the genes were confirmed by immunoblotting.
Quantification and statistical analysis
Database analysis
The results of database analysis shown here are in whole or part based upon data generated by TCGA Research Network: https://www.cancer.gov/tcga. Gene expression analysis was determined with the RNA-seq dataset acquired from skin cutaneous melanoma patients in TCGA data (TCGA-SKCM). Properties of patients selected here were filtered based on the following: sample type; primary tumor, site of resection; skin, stage; II and III, primary diagnosis; nodular melanoma, amelanotic melanoma, lentigo maligna melanoma, acral lentiginous melanoma, malignant melanoma (NOS).
Statistics
All statistical analyses were performed using GraphPad Prism software v8 (GraphPad, San Diego, CA, USA). A two-tailed Student’s t-test or Welch’s t-test was performed for the comparison of two independent normally distributed groups. Mann–Whitney’s U test was used to compare two sets of non-normally distributed data. Pearson’s correlation coefficient was used to evaluate the correlations. A p-value of less than 0.05 was considered statistically significant for all tests.
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (JSPS, Grant Numbers 18H06228, 19K21328 and 20K17353) and Center for Animal Resources and Development of Kumamoto University. We thank Prof. Seiji Okada and Dr. Ryusho Kariya for technical assistance in bone marrow transfer experiments. We thank Gabrielle White Wolf, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Author contributions
H.Kanemaru, Y.M., S.F. and H.I. developed the concept. H.Kanemaru, A.K. and H.T. designed and performed the experiments and analyzed the data. H.Kanemaru, A.K., T.K., H. Kuriyama, S.S., I.K., K.M., A.M., J.A., T.M., S.M., S.F., and H.I. provided patient specimens and/or clinical data. H.Kanemaru wrote the paper.
Declaration of interests
The authors declare no potential conflicts of interest.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103067.
Supplemental information
Data and code availability
The microarray and ChIP-seq datasets have been deposited in the NCBI Gene Expression Omnibus (GEO) database (accession no. GSE163978 and GSE164027). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bánfalvi T., Gilde K., Gergye M., Boldizsár M., Kremmer T., Ottó S. Use of serum 5-S-CD and S-100B protein levels to monitor the clinical course of malignant melanoma. Eur. J. Cancer. 2003;39:164–169. doi: 10.1016/s0959-8049(02)00487-2. [DOI] [PubMed] [Google Scholar]
- Balch C., Gershenwald J., Soong S., Thompson J., Atkins M., Byrd D., Buzaid A., Cochran A., Coit D., Ding S. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S., Fang F., Tourtellotte W., Varga J. Egr-1: new conductor for the tissue repair orchestra directs harmony (regeneration) or cacophony (fibrosis) J. Pathol. 2013;229:286–297. doi: 10.1002/path.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A., Singer K., Koehl G.E., Kolitzus M., Schoenhammer G., Thiel A., Matos C., Bruss C., Klobuch S., Peter K. LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24:657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- Colegio O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., Cyrus N., Brokowski C.E., Eisenbarth S.C., Phillips G.M. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delconte R.B., Kolesnik T.B., Dagley L.F., Rautela J., Shi W., Putz E.M., Stannard K., Zhang J.G., Teh C., Firth M. CIS is a potent checkpoint in NK cell-mediated tumor immunity. Nat. Immunol. 2016;17:816–824. doi: 10.1038/ni.3470. [DOI] [PubMed] [Google Scholar]
- Dietl K., Renner K., Dettmer K., Timischl B., Eberhart K., Dorn C., Hellerbrand C., Kastenberger M., Kunz-Schughart L.A., Oefner P.J. Lactic acid and acidification inhibit TNF secretion and glycolysis of human monocytes. J. Immunol. 2010;184:1200–1209. doi: 10.4049/jimmunol.0902584. [DOI] [PubMed] [Google Scholar]
- Dong H., Bullock T.N. Metabolic influences that regulate dendritic cell function in tumors. Front. Immunol. 2014;5:24. doi: 10.3389/fimmu.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson M., Rould M.A., Nekludova L., Pabo C.O. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- Fahmy R.G., Dass C.R., Sun L.Q., Chesterman C.N., Khachigian L.M. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nat. Med. 2003;9:1026–1032. doi: 10.1038/nm905. [DOI] [PubMed] [Google Scholar]
- Fasbender F., Watzl C. Impedance-based analysis of Natural Killer cell stimulation. Sci. Rep. 2018;8:4938. doi: 10.1038/s41598-018-23368-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K., Hoffmann P., Voelkl S., Meidenbauer N., Ammer J., Edinger M., Gottfried E., Schwarz S., Rothe G., Hoves S. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood. 2007;109:3812–3819. doi: 10.1182/blood-2006-07-035972. [DOI] [PubMed] [Google Scholar]
- Gottfried E., Kunz-Schughart L.A., Ebner S., Mueller-Klieser W., Hoves S., Andreesen R., Mackensen A., Kreutz M. Tumor-derived lactic acid modulates dendritic cell activation and antigen expression. Blood. 2006;107:2013–2021. doi: 10.1182/blood-2005-05-1795. [DOI] [PubMed] [Google Scholar]
- Haas R., Smith J., Rocher-Ros V., Nadkarni S., Montero-Melendez T., D'Acquisto F., Bland E.J., Bombardieri M., Pitzalis C., Perretti M. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. Plos Biol. 2015;13:e1002202. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R.S., Soria J.C., Kowanetz M., Fine G.D., Hamid O., Gordon M.S., Sosman J.A., McDermott D.F., Powderly J.D., Gettinger S.N. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhaeuser F., Sattler U.G.A., Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71:6921–6925. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., Carbone F.R. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Jung S., Unutmaz D., Wong P., Sano G., De los Santos K., Sparwasser T., Wu S., Vuthoori S., Ko K., Zavala F. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaru H., Yamane F., Fukushima K., Matsuki T., Kawasaki T., Ebina I., Kuniyoshi K., Tanaka H., Maruyama K., Maeda K. Antitumor effect of Batf2 through IL-12 p40 up-regulation in tumor-associated macrophages. Proc. Natl. Acad. Sci. U S A. 2017;114:E7331–E7340. doi: 10.1073/pnas.1708598114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaru H., Yamane F., Tanaka H., Maeda K., Satoh T., Akira S. BATF2 activates DUSP2 gene expression and up-regulates NF-κB activity via phospho-STAT3 dephosphorylation. Int. Immunol. 2018;30:255–265. doi: 10.1093/intimm/dxy023. [DOI] [PubMed] [Google Scholar]
- Krämer B., Meichle A., Hensel G., Charnay P., Krönke M. Characterization of an Krox-24/Egr-1-responsive element in the human tumor necrosis factor promoter. Biochim. Biophys. Acta. 1994;1219:413–421. doi: 10.1016/0167-4781(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Lecoeur H., Fevrier M., Garcia S., Riviere Y., Gougeon M.L. A novel flow cytometric assay for quantitation and multiparametric characterization of cell-mediated cytotoxicity. J. Immunol. Methods. 2001;253:177–187. doi: 10.1016/s0022-1759(01)00359-3. [DOI] [PubMed] [Google Scholar]
- Lee S.L., Tourtellotte L.C., Wesselschmidt R.L., Milbrandt J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J. Biol. Chem. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]
- Lim C.P., Jain N., Cao X. Stress-induced immediate-early gene, egr-1, involves activation of p38/JNK1. Oncogene. 1998;16:2915–2926. doi: 10.1038/sj.onc.1201834. [DOI] [PubMed] [Google Scholar]
- Maeda-Otsuka S., Kajihara I., Tasaki Y., Yamada-Kanazawa S., Sakamoto R., Sawamura S., Masuzawa M., Masuzawa M., Amoh Y., Hoshina D. Hypoxia accelerates the progression of angiosarcoma through the regulation of angiosarcoma cells and tumor microenvironment. J. Dermatol. Sci. 2019;93:123–132. doi: 10.1016/j.jdermsci.2019.01.005. [DOI] [PubMed] [Google Scholar]
- Murphy T.L., Cleveland M.G., Kulesza P., Magram J., Murphy K.M. Regulation of interleukin 12 p40 expression through an NF-κB half-site. Mol. Cell. Biol. 1995;15:5258–5267. doi: 10.1128/mcb.15.10.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasi A., Fekete T., Krishnamurthy A., Snowden S., Rajnavölgyi E., Catrina A.I., Wheelock C.E., Vivar N., Rethi B. Dendritic cell reprogramming by endogenously produced lactic acid. J. Immunol. 2013;191:3090–3099. doi: 10.4049/jimmunol.1300772. [DOI] [PubMed] [Google Scholar]
- O'Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly M.S., Boehm T., Shing Y., Fukai N., Vasios G., Lane W.S., Flynn E., Birkhead J.R., Olsen B.R., Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- Ohashi T., Aoki M., Tomita H., Akazawa T., Sato K., Kuze B., Mizuta K., Hara A., Nagaoka H., Inoue N. M2-like macrophage polarization in high lactic acid-producing head and neck cancer. Cancer Sci. 2017;108:1128–1134. doi: 10.1111/cas.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevy S.E., Gemberling J.H., Hsu S., Dorner A.J., Smale S.T. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell. Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri D., Bhattacharya R., Sinha B.P., Liu C.S.C., Ghosh A.R., Rahaman O., Bandopadhyay P., Sarif J., D'Rozario R., Paul S. Lactate induces pro-tumor reprogramming in intratumoral plasmacytoid dendritic cells. Front. Immunol. 2019;10:1878. doi: 10.3389/fimmu.2019.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerka C., Decker E.L., Zipfel P.F. A regulatory element in the human interleukin 2 gene promoter is a binding site for the zinc finger proteins Sp1 and EGR-1. J. Biol. Chem. 1995;270:22500–22506. doi: 10.1074/jbc.270.38.22500. [DOI] [PubMed] [Google Scholar]
- Speiser D.E., Ho P.C., Verdeil G. Regulatory circuits of T cell function in cancer. Nat. Rev. Immunol. 2016;16:599–611. doi: 10.1038/nri.2016.80. [DOI] [PubMed] [Google Scholar]
- Sukhatme V.P. Early transcriptional events in cell growth: the Egr family. J. Am. Soc. Nephrol. 1990;1:859–866. doi: 10.1681/ASN.V16859. [DOI] [PubMed] [Google Scholar]
- Tagawa H., Kanemaru H., Kimura T., Kuriyama H., Sawamura S., Kajihara I., Miyashita A., Aoi J., Fukushima S., Ihn H. BATF2 expression as a novel marker for invasive phenotype in malignant melanoma. J. Dermatol. 2020;47:e372–e373. doi: 10.1111/1346-8138.15495. [DOI] [PubMed] [Google Scholar]
- Taylor A., Tang W., Bruscia E.M., Zhang P.X., Lin A., Gaines P., Wu D., Halene S. SRF is required for neutrophil migration in response to inflammation. Blood. 2014;123:3027–3036. doi: 10.1182/blood-2013-06-507582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hoeve A.L., Hakimi M.A., Barragan A. Sustained Egr-1 response via p38 MAP kinase signaling modulates early immune responses of dendritic cells parasitized by Toxoplasma gondii. Front. Cell. Infect. Microbiol. 2019;9:349. doi: 10.3389/fcimb.2019.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenot P.T., Sierra R.A., Raber P.L., Al-Khami A.A., Trillo-Tinoco J., Zarreii P., Ochoa A.C., Cui Y., Del Valle L., Rodriguez P.C. The stress-response sensor chop regulates the function and accumulation of myeloid-derived suppressor cells in tumors. Immunity. 2014;41:389–401. doi: 10.1016/j.immuni.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya N., Zhang R., Iwama T., Ueda N., Liu T., Tatsumi M., Sasaki Y., Shimoda R., Osako Y., Sawada Y. Type I interferon delivery by iPSC-derived myeloid cells elicits antitumor immunity via XCR1+ dendritic cells. Cell Rep. 2019;29:162–175.e9. doi: 10.1016/j.celrep.2019.08.086. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H., Fujieda K., Miyashita A., Fukushima S., Ikeda T., Kubo Y., Senju S., Ihn H., Nishimura Y., Oshiumi H. Combined blockade of IL6 and PD-1/PD-L1 signaling abrogates mutual regulation of their immunosuppressive effects in the tumor microenvironment. Cancer Res. 2018;78:5011–5022. doi: 10.1158/0008-5472.CAN-18-0118. [DOI] [PubMed] [Google Scholar]
- Tsunoda T., Takagi T. Estimating transcription factor bindability on DNA. Bioinformatics. 1999;15:622–630. doi: 10.1093/bioinformatics/15.7.622. [DOI] [PubMed] [Google Scholar]
- Walenta S., Wetterling M., Lehrke M., Schwickert G., Sundfør K., Rofstad E.K., Mueller-Klieser W. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000;60:916–921. [PubMed] [Google Scholar]
- Wang G., Balamotis M.A., Stevens J.L., Yamaguchi Y., Handa H., Berk A.J. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Yang K., Xu J., Fan M., Tu F., Wang X., Ha T., Williams D.L., Li C. Lactate suppresses macrophage Pro-inflammatory response to LPS stimulation by inhibition of YAP and NF-κB activation via GPR81-mediated signaling. Front. Immunol. 2020;11:587913. doi: 10.3389/fimmu.2020.587913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaritskaya L., Shurin M.R., Sayers T.J., Malyguine A.M. New flow cytometric assays for monitoring cell-mediated cytotoxicity. Expert Rev. Vaccin. 2010;9:601–616. doi: 10.1586/erv.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray and ChIP-seq datasets have been deposited in the NCBI Gene Expression Omnibus (GEO) database (accession no. GSE163978 and GSE164027). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.