Abstract
We have devised a cis-antisense rescue assay of cleavage and polyadenylation to determine how long it takes the simian virus 40 (SV40) early poly(A) signal to commit itself to processing in vivo. An inverted copy of the poly(A) signal placed immediately downstream of the authentic one inhibited processing by means of sense-antisense duplex formation in the RNA. The antisense inhibition was gradually relieved when the inverted signal was moved increasing distances downstream, presumably because cleavage and polyadenylation occur before the polymerase reaches the antisense sequence. Antisense inhibition was unaffected when the inverted signal was moved upstream. Based on the known rate of transcription, we estimate that the cleavage-polyadenylation process takes between 10 and 20 s for the SV40 early poly(A) site to complete in vivo. Relief from inhibition occurred earlier for shorter antisense sequences than for longer ones. This indicates that a brief period of assembly is sufficient for the poly(A) signal to shield itself from a short (50- to 70-nucleotide) antisense sequence but that more assembly time is required for the signal to become immune to the longer ones (∼200 nucleotides). The simplest explanation for this target size effect is that the assembly process progressively sequesters more and more of the RNA surrounding the poly(A) signal up to a maximum of about 200 nucleotides, which we infer to be the domain of the mature apparatus. We compared strong and weak poly(A) sites. The SV40 late poly(A) site, one of the strongest, assembles several times faster than the weaker SV40 early or synthetic poly(A) site.
Compared to splicing, the cleavage-polyadenylation process is expected to be fairly straightforward. The basic reaction in vertebrates involves only one site on the RNA, not three as for splicing, and the core apparatus is comprised entirely of proteins—no small nuclear RNAs are required (14, 65). Spliceosome assembly occurs naturally in discrete steps (61) both in vivo (7) and in crude extracts in vitro (22). In contrast, no natural assembly intermediates have been observed for the cleavage-polyadenylation apparatus in vitro with crude extracts (38). Until now, however, there has been no information on the nature of the process in vivo. We report here that the assembly of the cleavage-polyadenylation apparatus in vivo is more complex than expected, being a gradual multistep process. Furthermore, we show that the strong simian virus 40 (SV40) late poly(A) site assembles considerably more rapidly in vivo than other sites known to be weaker.
The core poly(A) signal in vertebrates consists of two recognition elements flanking a cleavage-polyadenylation site. Typically, an almost invariant AAUAAA hexamer lies 20 to 50 nucleotides (nt) upstream of a more variable element rich in U or GU residues (11, 23, 48). Cleavage between these two elements is usually on the 3′ side of an A residue (11) and, in vitro, is mediated by a large, multicomponent protein complex that can be separated into five distinguishable factors (14, 65). Four of these factors are essential for cleavage in vitro at all poly(A) sites so far examined. They are the cleavage and polyadenylation specificity factor (CPSF), which binds the AAUAAA motif; the cleavage stimulation factor (CstF), which binds the downstream U-rich element; and two additional cleavage factors (CF I and CF II) that are less well characterized (14, 65). The fifth factor is the poly(A) polymerase, which in addition to its obvious role in the polyadenylation reaction must be present in most cases for the cleavage step as well (13, 64).
CPSF, CstF, CF I, and CF II cofractionate as a single large complex upon gel filtration of crude extracts (62). This has led to the suggestion that the 3′-end processing apparatus may assemble in vivo in a single step (46). However, multistep assembly is also an attractive possibility (27). New data indicate that assembly in some cases may even begin with the recruitment of CPSF and CstF to the transcription complex during initiation and elongation, with both factors then being snared by the poly(A) signal as it emerges from the polymerase (17, 47). By this scenario, the remaining factors would then be recruited to the growing apparatus on the nascent transcript. This order of assembly is consistent with in vitro studies using purified factors (65).
Once assembled, the 3′-end processing apparatus directs cleavage and polyadenylation. In vivo, the efficiency with which different poly(A) sites are processed varies considerably (10, 19, 21). The physical basis for this variation in poly(A) site “strength” is not known. In principle, a site could be strong because it directs faster assembly or because it assembles a more stable apparatus. Because poly(A) site strengths are correlated with the stabilities of the 3′-end processing complexes that form in vitro, it has been suggested that poly(A) site strength is based on complex stability (65, 66). However, it is easier to envision a kinetic explanation for poly(A) site strength when it is recognized that the complete cleavage and polyadenylation reaction takes place in seconds in vivo (6) while complex stabilities in vitro are measured in terms of minutes or hours. Preliminary experiments reported below comparing the rates of assembly of poly(A) sites of various strength are consistent with the kinetic view.
The fully mature 3′-end processing complex probably sequesters considerably more RNA than just the core poly(A) signal itself. There are numerous examples of poly(A) signals for which the flanking RNA immediately upstream (8, 10, 26, 33, 50, 53, 60) or downstream (5, 12, 26) of the core poly(A) signal is essential for full activity. In some cases, specific elements in these flanking sequences serve as binding sites for additional trans-acting factors (5, 44, 50). In other cases, the involvement of additional factors is not apparent though the flanking RNA is known to be important (12, 25, 26, 32, 33). For several poly(A) sites, direct interactions of the flanking RNA with CPSF and CstF have been demonstrated (25, 50) or are likely (26, 33). Thus, the emerging picture is of a core poly(A) signal at the center of a larger poly(A) signal domain.
The various elements in this domain are functionally intolerant of significant variations in position (2, 5, 9, 11, 24, 33, 59). For example, the U/GU-rich element occupies a well-defined position relative to the AAUAAA hexamer and the site of cleavage (11). Function is destroyed by separating these elements with inserted RNA but is restored by sequestering the extra RNA in secondary structure so as to bring the elements together again (2, 9). Similarly, elements in the upstream flanking RNA lose function if separated from the rest of the domain by unstructured RNA but regain function if the extra RNA is sequestered in secondary structure (24, 33). The weakness of the interactions comprising the core cleavage-polyadenylation structure and the role of cooperativity in holding them together have been emphasized previously (65). The similar dependence of function on proximity for the core and flanking elements suggests that both are part of a single, integrated structure held together cooperatively by multiple weak interactions. Presumably the weakness of the individual interactions within the structure accounts for the sensitivity to distance (34). Other elements, found further upstream (56) and downstream (42, 43) of the poly(A) site, are less distance dependent in their function and may belong to a distinct class of polyadenylation enhancers.
In the work presented below, we explored the assembly and function of the poly(A) signal domain in vivo by using cis-antisense sequences that antagonize poly(A) site utilization. By moving the antagonistic sequence gradually downstream of the poly(A) site, we were able to determine the distance downstream at which the antagonist lost its effect and poly(A) site function returned. An extensive series of controls established that the return of poly(A) site function, as the antisense element was moved downstream, corresponded to the time required for the RNA polymerase to reach and transcribe the antisense sequence. Using this approach, we determined for the SV40 early poly(A) site that assembly of the cleavage-polyadenylation apparatus in vivo begins immediately upon transcription of the poly(A) signal, covers an extensive region of RNA, and requires about 20 s to reach completion. The kinetics of assembly and the progressive nature of the process add to our understanding of the structure of the 3′-end processing apparatus and raise interesting questions regarding the meaning of poly(A) site strength in vivo and the mechanism by which poly(A)-dependent termination may be signaled to the elongating polymerase.
MATERIALS AND METHODS
Expression assays.
Transfections for chloramphenicol acetyltransferase (CAT) assays were performed by the calcium phosphate method as described previously (67). The transfected DNA included a cotransfected luciferase-expressing plasmid, pRSVfl, in which the HindIII-SacI fragment of Promega’s pGem-luc replaces the HindIII-BalI segment of pRSVcat. For each 100-mm-diameter plate, 3.3 μg of pRSVfl was used together with a threefold molar amount of experimental plasmid and carrier (pBR322) DNA to bring the total amount of DNA to 20 μg. Cells were rinsed twice with cold phosphate-buffered saline at 44 to 46 h after transfection, lysed in situ by 15 min of incubation at room temperature with 900 μl of Promega’s 1× reporter lysis buffer, transferred to a screw-cap microcentrifuge tube, freeze-thawed (using liquid nitrogen and room-temperature water, respectively), and centrifuged. After 200 μl of the supernatant was removed for the luciferase assay, the rest was heat inactivated, for the CAT assay, at 65°C for 10 min (16), followed by a 5-min spin. The supernatant was stored at −60°C. For the luciferase assay, 20 μl of extract was added to 80 μl of 2× luciferase assay buffer (0.2 M K2HPO4 [pH 7.8], 10 mM ATP, 2 mM dithiothreitol) and assayed in a luminometer (LUMAT model LB 9501) that injects 100 μl of luciferin solution (0.1 M K2HPO4 [pH 7.8], 1 mM dithiothreitol, 943 μM luciferin) into each reaction mixture and then captures the light emitted over a 10-s period. All samples were assayed in duplicate, and volumes of extract containing equivalent amounts of luciferase activity were then assayed in duplicate for CAT activity (31). Heat-inactivated extract was added to 0.67 mM (final concentration) acetyl coenzyme A, 0.01 μCi of [14C]chloramphenicol (54 mCi/mmol), 24.8 μM unlabeled chloramphenicol, and enough 0.25 M Tris (pH 8.0) for a final reaction volume of 120 μl. After 20 to 60 min at 37°C, the reaction was stopped by ethyl acetate extraction and the products were fractionated by thin-layer chromatography and then quantitated with a Molecular Dynamics PhosphorImager. Assay results were normalized to those for a positive control transfected in parallel.
For more recent work we have used Lipofectamine (Gibco BRL) or FuGENE 6 (Boehringer Mannheim) for transfection and a dual luciferase system (Promega) for expression, in accordance with the manufacturers’ instructions. Cells in six-well plates were transfected with a DNA mixture containing 35 ng of pRSVfl and 50 times this molar amount of the experimental plasmid. Cells were harvested by use of passive lysis buffer (Promega) 48 to 50 h after transfection, and the resulting lysates were cleared by microcentrifugation for 30 s at 4°C. All samples were assayed in duplicate, and the averaged Renilla luciferase values were then normalized to the averaged firefly values. A normalized average of duplicates was considered a single experimental point for purposes of further analysis and was additionally normalized to data for a positive control transfected in parallel.
RNase protection assays.
Standard procedures were used for the RNase protection assay (4). All probes were extracted with phenol or TRIzol (Gibco BRL) and then gel purified before use. Cells were transfected in 100-mm-diameter plates by using FuGENE 6. Cytoplasmic RNA was prepared by using RNeasy columns (Qiagen) in accordance with the manufacturer’s protocol. For nuclear RNA, the nuclear pellet was washed once with RLN (Qiagen) and then processed as recommended for obtaining total animal cell RNA. Target RNA was treated with DNase I (Ambion) (20), coprecipitated with ∼106 cpm of [α-32P]CTP-labeled probe, and then resuspended and hybridized at 56°C. RNase digestion was carried out with RNase T1 only. The final results were quantitated, with adjustment for C content, by using a PhosphorImager with ImageQuant software (Molecular Dynamics).
RESULTS
cis-antisense rescue, a method for measuring the rate of commitment to cleavage and polyadenylation in vivo.
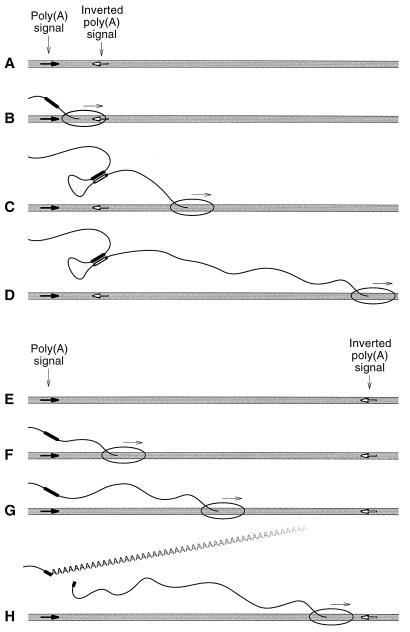
The rationale for cis-antisense rescue is based on cis-antisense-mediated inhibition of cleavage and polyadenylation (28). A copy of the poly(A) signal to be tested is inserted in the reverse orientation downstream of the parent poly(A) site (Fig. 1A). Although the poly(A) site is transcribed (Fig. 1B), before processing can occur the antisense transcript from the inverted poly(A) signal blocks cleavage and polyadenylation by forming a duplex with the transcribed poly(A) site (Fig. 1C and D). However, if the inverted poly(A) signal is separated from the authentic one by a sufficient length of spacer DNA (Fig. 1E), the additional time required for the polymerase to reach the inverted signal (Fig. 1F and G) allows processing (or commitment to processing) at the authentic site to proceed (Fig. 1H) and the poly(A) site is rescued. By measuring the degree of poly(A) site rescue as a function of DNA spacer length, the rate at which the poly(A) site becomes committed to cleavage and polyadenylation can be inferred from the known rate of transcription (63).
FIG. 1.
Rationale for measuring the rate of commitment to cleavage and polyadenylation in vivo by the cis-antisense rescue assay. See the text for details.
The above rationale assumes the following: (i) that an antisense sequence can target an upstream poly(A) site and block polyadenylation (cis-antisense inhibition), (ii) that moving the sense and antisense sequences apart will relieve this antisense effect (separation-dependent rescue) and (iii) that the rescue is attributable to a simple kinetic competition between the rate of commitment to cleavage and polyadenylation and the rate of transcription. Below we present experimental support for these three assumptions.
cis-antisense inhibition of polyadenylation occurs and is rescued by separation.
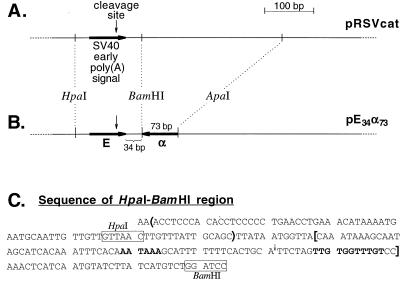
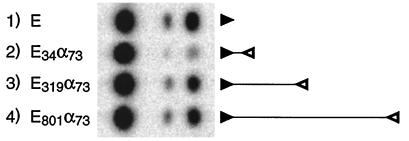
In this section we address the first two assumptions mentioned above. For our initial experiments we employed the expression vector pRSVcat, which contains the SV40 early poly(A) signal (Fig. 2A). We made a PCR copy of this poly(A) signal and used it, in the reverse orientation, to replace the BamHI-ApaI fragment of pRSVcat (Fig. 2B). In the resulting plasmid, pE34α73, the SV40 early poly(A) signal, E, is separated by 34 bp from its antisense-coding sequence, α, of 73 bp. To assess poly(A) site function, we then measured CAT expression in pE34α73-transfected COS cells. Other things being equal, gene expression is proportional to the amount of poly(A) site activity (21, 53). Figure 3 shows, in support of the first assumption (cis-antisense inhibition), that CAT expression for pE34α73 (lane 2) is drastically reduced compared to that of a control that simply lacks the BamHI-ApaI segment of pRSVcat (lane 1). Note that because the antisense sequence is located downstream of the 3′ end of the canonical pRSVcat mRNA, the only way that antisense activity can block expression is by interfering with mRNA 3′-end formation.
FIG. 2.
Construction of pE34α73. (A) The 3′ end of the CAT transcription unit of pRSVcat (30). (B) The 3′ end of the CAT transcription unit of pE34α73. To obtain pE34α73 the sequence bracketed in panel C was amplified with the primers 5′-CGGGCCCAAATAAAGCAATAGCATCACAAA and 5′-GGGATCCGGACAAACCACAACTAGAATGCAG. The PCR product was cut with ApaI and BamHI and then inserted by sticky-end ligation into BamHI- and ApaI-digested pRSVcat. (C) Sequence of the 3′ end of pRSVcat. The regions targeted by antisense sequence in pHα73 and in the pEα73 series of constructs are enclosed in parentheses and brackets, respectively. The sequence within the brackets actually encodes two interdigitated poly(A) signals. The upstream and downstream motifs for the dominant signal are shown in boldface and are used exclusively in vivo to direct cleavage as shown by the arrow (15). The sense-antisense separation for pE34α73 and its derivatives is arbitrarily defined as the number of base pairs separating the last C of the poly(A) signal region bracketed in the sequence and the first G of its inverted repeat downstream.
FIG. 3.
CAT assay of cis-antisense inhibition and rescue for the SV40 early poly(A) site. Diagrammed on the right for each construct are the poly(A) signal (solid arrowhead), the inverted poly(A) signal (open arrowhead), and the separation between them, drawn to scale.
Thus, a downstream antisense sequence can block processing, but is it by an antisense mechanism? To test this, we asked first whether the mere insertion of DNA into the BamHI site of pRSVcat (Fig. 2A) is deleterious to expression. Accordingly, we inserted various randomly chosen DNA sequences into pRSVcat to yield constructs pCat1 to pCat4 (Table 1), which we then assayed for the ability to direct CAT expression in transfected COS cells. These constructs did not differ appreciably from each other or from pRSVcat in their ability to express CAT (data not shown). Since pRSVcat depends on the presence of the SV40 early poly(A) site for significant levels of CAT expression (54), we conclude that the mere insertion of DNA into the pRSVcat BamHI site does not significantly affect cleavage and polyadenylation at the SV40 early poly(A) site.
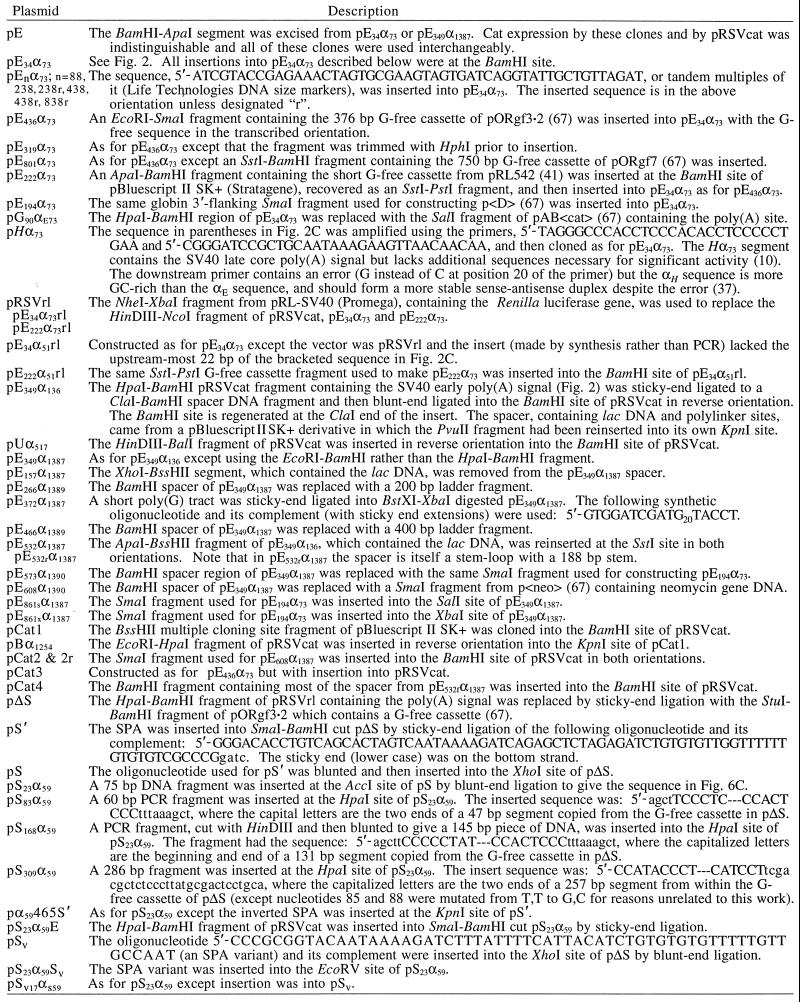
TABLE 1.
Plasmids constructed for these studies
To verify that it is the downstream insert that is responsible for extinguishing CAT expression in pE34α73 (Fig. 3, lane 2), rather than second-site damage to the plasmid, we removed the inverted poly(A) signal by BamHI-ApaI digestion. Full potential to express CAT was restored (data not shown). This showed that inhibition of CAT expression in pE34α73 is a specific effect of the inverted poly(A) signal.
Finally, we sought to confirm explicitly that this is an effect directed at the upstream poly(A) signal and that it is mediated by sequence complementarity. To check the role of sequence complementarity, we replaced the SV40 early poly(A) signal of pE34α73 with the chicken βH-globin poly(A) signal. The resulting construct, pG90αE73, recovered 91% ± 12% (mean ± standard deviation) of the original ability to express CAT, demonstrating that the globin poly(A) signal does not serve as a significant target for the inverted SV40 sequence, with which it shares little sequence homology. To confirm that sequence complementarity specifically to a poly(A) signal, rather than generally to the 3′ end of the message, is principally responsible for the inhibition of CAT expression in pE34α73, we prepared a final control construct, pHα73. This construct is like pE34α73 except that the antisense sequence is directed to a region surrounding the HpaI site just upstream of the poly(A) signal (Fig. 2C) rather than to the poly(A) signal itself. After transfection of pHα73 into COS cells, we found that it expressed substantial CAT activity (58% ± 22% of that of pRSVcat). This demonstrates that the highly effective inhibition of CAT activity by the inverted poly(A) signal in pE34α73 (13% ± 4% of that of pRSVcat) (Table 2) is a consequence of targeting the poly(A) signal specifically and is not simply a result of antisense activity generally.
TABLE 2.
Unadjusted CAT rescue values for Eα73 series constructs used for Fig. 8
| Eα73 spacer | CAT rescue valuea ± SD (avg/SD) | No. of transfections |
|---|---|---|
| 34 | 12.5 ± 3.9 (0.31) | 14 |
| 88 | 26.2 ± 3.7 (0.14) | 5 |
| 194 | 47.9 ± 9.5 (0.20) | 3 |
| 222 | 54.7 ± 10.6 (0.19) | 5 |
| 238 | 64.1 ± 19.6 (0.31) | 5 |
| 238r | 68.4 ± 21.2 (0.31) | 4 |
| 319 | 51.6 ± 15.2 (0.29) | 7 |
| 436 | 65.6 ± 13.1 (0.20) | 6 |
| 438 | 66.4 ± 21.1 (0.32) | 5 |
| 438r | 82.2 ± 23.4 (0.28) | 4 |
| 801 | 86.4 ± 10.8 (0.12) | 5 |
| 838r | 79.8 ± 14.9 (0.19) | 4 |
Expressed as the average percentage of activity versus that of pE, which was transfected in parallel. Prior to calculation of averages, outliers were removed, using the criterion that the standard deviation divided by the average should be less than 0.33. This resulted in the removal of four outliers from a total of 71 datum points.
The controls described above showed that the inverted poly(A) signal in pE34α73 blocks CAT activity by means of authentic cis-antisense inhibition, thereby satisfying the first assumption on which the rationale for measurement of the rate of commitment to cleavage and polyadenylation in vivo is based. The second assumption requires that the antisense inhibition be dependent on adequate proximity of the sense and antisense sequences so that moving them apart will progressively reduce the effect. For example, a nonstoichiometric, catalytic type of antisense interference (49) would not give rise to separation-dependent rescue. The data from lanes 3 and 4 of Fig. 3 show that when the poly(A) signal and its antisense sequence are moved apart, CAT expression is indeed rescued. This confirms the second assumption on which the rationale is based.
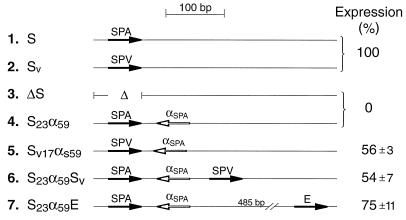
Do cis-antisense inhibition and separation-dependent rescue occur for poly(A) signals other than the SV40 early poly(A) signal? Looking first at cis-antisense inhibition, we repeated the above-described control experiments with a series of antisense constructs based on the synthetic poly(A) signal (SPA) of Levitt et al. (39) together with a variation of this signal (SPV) bearing 53% sequence identity to the original SPA. Figure 4 summarizes these additional controls. In the reference constructs (pS and pSv) (Fig. 4, lines 1 and 2) the SPA and SPV were placed so as to support expression of a gene encoding Renilla luciferase. A control plasmid with no inserted poly(A) site was used to set the baseline (pΔS) (Fig. 4, line 3). An inverted SPA downstream of the authentic SPA reduced luciferase expression to background levels (Fig. 4, line 4), in agreement with the results for pE34α73 (Fig. 3, lane 2). Expression was largely rescued when the sequence complementarity between sense and antisense elements was reduced to 59% (G-U pairs were counted as complementary) by replacing the upstream SPA with the related SPV (Fig. 4, line 5). This confirms that a high level of sequence complementarity and, presumably, duplex formation are required for antisense-mediated inhibition and that the antisense sequence is not a nonspecific “poison” to poly(A) site function. Expression was also rescued by placement of additional poly(A) sites downstream of a sense-antisense pair (Fig. 4, lines 6 and 7), showing that the mere presence of secondary structure in this region of the RNA, or pausing or termination of transcription, cannot be responsible for the poor expression that characterizes our cis-antisense constructs.
FIG. 4.
Dual luciferase assay of cis-antisense inhibition and rescue for the synthetic poly(A) site (SPA). All transfections and assays of the plasmids in lines 2 to 7 were performed in parallel with those of the positive control, pS, of line 1. Construct pΔS, which contains no added poly(A) site, served as the negative control. The background expression exhibited by pΔS (mean ± standard deviation, 34% ± 5% of that of pS) was high, due in part to cryptic poly(A) sites elsewhere in the plasmid (52). The basic antisense construct, pS23α59, exhibited a level of expression (30% ± 10% of that of pS) not significantly different from that for pΔS, so the average of these two constructs was taken as the baseline for calculating the expression levels of the rest. The expression levels of the remaining constructs (lines 5 to 7) were taken to be that which exceeds the pΔS:pS23α59 average, expressed as a percentage of the range between the negative (lines 3 and 4) and the positive (line 1 or 2) controls. Construct pS (line 1) was used as the 100% control for the SPA-based plasmids (lines 4, 6, and 7), and construct pSv (line 2), which expressed 43% more than pS, was used as the 100% control for the SPV-based plasmid (line 5).
The control experiments described above confirm that the simplest interpretation of the CAT rescue results in Fig. 3 is one based on the rationale presented in Fig. 1. Sense-antisense duplex formation prevents polyadenylation when a poly(A) signal is followed closely by an inverted copy of itself. As a result, only small amounts of mature mRNA reach the cytoplasm, and the level of expression is low (Fig. 3, lane 2). However, polyadenylation is rescued if the inverted copy is moved downstream, allowing the polyadenylation apparatus time to assemble. A clear prediction of this scenario is that cells transfected with cis-antisense-inhibited plasmids not only should lack mRNA in the cytoplasm but also should accumulate uncleaved pre-mRNA in their nuclei (15).
cis-antisense inhibition causes accumulation of uncleaved nuclear pre-mRNA.
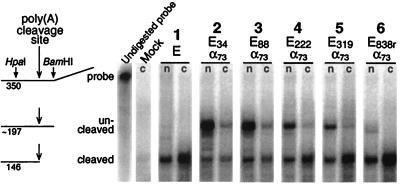
We used RNase protection to assay for uncleaved pre-mRNA. Figure 5, lane 2n, confirms that most nuclear SV40 early RNA produced by pE34α73 is uncleaved. Little of this RNA reaches the cytoplasm (lane 2c). The small amount of uncleaved RNA that does appear in the cytoplasm presumably results from cryptic poly(A) site activity downstream (52). Lanes 3 to 6 of Fig. 5 show the results of RNase protection assays for constructs in which the poly(A) site was separated from its antisense sequence by increasing lengths of spacer DNA. The results show that the proportion of uncleaved RNA in both the nucleus and the cytoplasm decreases as the spacing between sense and antisense sequences increases, as expected. Thus, at least for the SV40 early poly(A) site, cis-antisense-mediated inhibition of polyadenylation appears to operate as envisioned (Fig. 1), simply by occluding the upstream poly(A) signal and allowing uncleaved pre-mRNA to accumulate. The effect is relieved, and the poly(A) site is rescued, by increasing the separation between sense and antisense sequences.
FIG. 5.
RNase protection assay of cis-antisense inhibition and rescue for the SV40 early poly(A) site. The probe used was a T7 RNA polymerase transcript of DraI-digested pCat1. The RNA lengths given in the figure refer to T1 digestion at susceptible G’s. The probe lengths corresponding to uncleaved pre-mRNA varied from 196 to 198 nt depending on the construct. For the six numbered lanes, 24 to 31 μg of nuclear RNA (n) and 60 to 62 μg of cytoplasmic RNA (c) were analyzed. For the mock lane, 30 μg was used. The faint band at the cleaved position for RNA from mock-transfected cells reflects the low level of SV40 early mRNA produced by the COS cells themselves (29). After subtracting the estimated cellular contribution of cleaved RNA, the molar percentages of uncleaved nuclear RNA for lanes 2 to 6 were 88, 73, 49, 40, and 9, respectively.
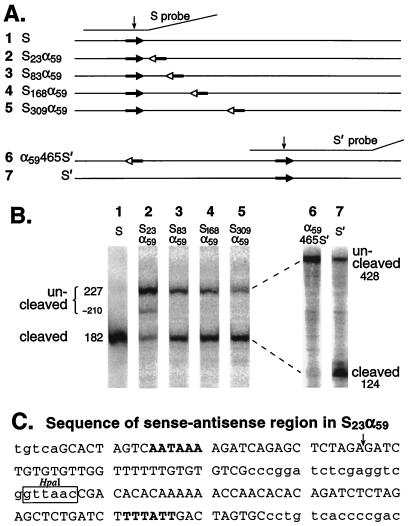
To determine whether the situation is similar for the unrelated SPA, RNase protection was used to assay nuclear RNA from cells transfected with a series of SPA-containing constructs. When the SPA is followed closely by an inverted SPA (Fig. 6A, construct 2), the nuclei of transfected cells accumulate mostly uncleaved RNA (Fig. 6B, lane 2). When the inverted SPA is moved progressively downstream (Fig. 6A, constructs 3 to 5), cleavage at the authentic SPA is progressively rescued (Fig. 6B, lanes 3 to 5). Thus, the SPA exhibits antisense inhibition and separation-dependent rescue just as does the SV40 early poly(A) site.
FIG. 6.
Distance-dependent rescue occurs for downstream but not upstream antisense sequences. (A) Diagrammatic representation, drawn to scale, of the plasmids and probes used for panel B. The downward arrows indicate the positions of poly(A) site cleavage. (B) RNase protection assays of nuclear RNA. The amounts of RNA used were as follows: 4 to 15 μg for lanes 2 to 5, 20 μg for lane 1, and 40 μg for lanes 6 and 7. The molar percentages of uncleaved RNA for lanes 2 to 7 were 73, 31, 25, 19, 73, and 5, respectively. The S probe was a T3 RNA polymerase transcript of a DraI-digested plasmid obtained by inserting the PvuII fragment of pBluescript SK(+) into the HpaI site of pS23α59. The S′ probe was a T7 RNA polymerase transcript of a plasmid obtained by inserting the ApaI fragment of pα59465S′ into the SacI site of pBluescript SK(+). An NdeI site was inserted into this plasmid by in vitro mutagenesis (QuikChange; Stratagene) to allow production of a 541-nt truncated transcript. The RNA lengths given in the figure refer to T1 digestion at susceptible G’s. The probe has a variable tendency to be cleaved at an internal position downstream of the poly(A) signal to yield a ∼210-nt fragment. (C) Sequence of the SPA–anti-SPA region in pS23α59. The SPA and its antisense sequence are capitalized, and the poly(A) hexamer and its antisense sequences are shown in boldface type. The probable poly(A) cleavage site (3) is indicated by an arrow.
cis-antisense rescue is attributable to 3′-end processing events that occur before the polymerase transcribes the antisense sequence.
In this section we address the third assumption on which the cis-antisense rescue rationale is based, namely, that rescue occurs because the time required for the polymerase to reach the inverted poly(A) signal exceeds the time required to commit to processing (Fig. 1). This assumption could fail in either of two ways. On the one hand, separation-dependent rescue of a poly(A) signal from cis-antisense inhibition could reflect a nonspecific transcript packaging process which precedes the recruitment of processing factors to the poly(A) signal. In this case, the rate of rescue would reflect the rate of the nonspecific event, not the rate of commitment to cleavage and polyadenylation. On the other hand, increasing the separation of the sense sequence from the antisense sequence could conceivably lead to rescue because of the increased time required for their mutual search through three-dimensional space.
To test the third assumption, we prepared construct 6 of Fig. 6A, in which the antisense sequence lies upstream rather than downstream of the poly(A) site. If separation-dependent rescue reflects a generic transcript packaging process, or the time during which sense and antisense sequences search for each other through three-dimensional space, then construct 6 (Fig. 6A), whose sense and antisense sequences are well separated from each other, should exhibit less inhibition of processing than construct 5, in which the sense and antisense sequences are closer together. The opposite is observed, however: the cleavage-polyadenylation process for construct 6 is mostly inhibited, whereas for construct 5 it is mostly rescued (Fig. 6B, lanes 5 and 6). A comparison of the results for construct 6 with those for construct 2 is even more revealing. After adjusting for the C contents of the two different probes, quantitation of the results reveals equal molar proportions (73%) of uncleaved RNA for both constructs. Thus, even when moved 465 nt upstream of the poly(A) site (construct 6), the antisense sequence remains just as effective as when it is almost immediately adjacent (construct 2). Clearly, for construct 6, the antisense sequence is neither shielded from the poly(A) site by packaging nor delayed in its assault by search time. Therefore, the rescue of cleavage and polyadenylation observed for construct 5 is attributable to the processing (or commitment to processing) that occurs during the time it takes the polymerase to reach the antisense sequence.
Careful attention was paid to the design of the constructs used for Fig. 6. First, the spacer inserts for constructs 3 to 5 were excerpts of the same sequence used for construct 6. Second, this sequence, shared by all of the constructs, is a G-free cassette (see below), a relatively monotonous sequence whose transcript is likely to have little or no secondary structure. Therefore, the differences between construct 6 on the one hand and constructs 3 to 5 on the other are not likely to reflect sequence-specific effects on RNA structure or transcription time. These data support the conclusion that the separation dependence of cis-antisense rescue is a measure of the rate of commitment to cleavage and polyadenylation.
Efficiency of rescue varies with transcript abundance.
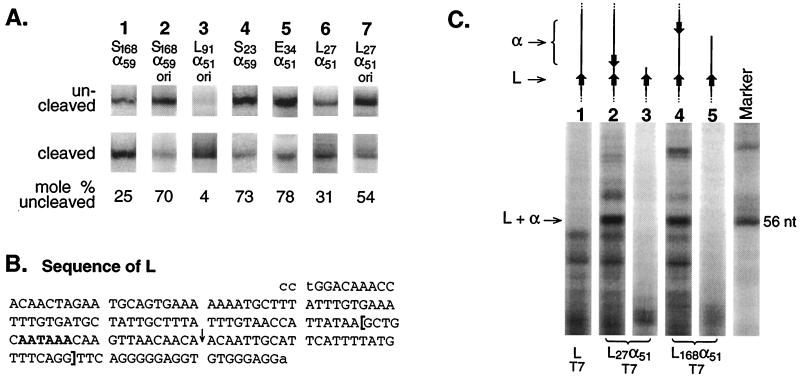
If the efficiency of cis-antisense rescue is related to the processing rate and not simply to the generic packaging properties of the RNA surrounding the poly(A) site, then any change in the rate of poly(A) site processing should alter the efficiency of cis-antisense rescue. For example, at any given separation of sense sequence from antisense sequence, a decrease in the processing rate is predicted to result in less rescue because less cleavage and polyadenylation will occur before the antisense sequence is transcribed. The efficiency of the cleavage-polyadenylation process is known to be affected by processing factor abundance in vivo (14). We therefore reasoned that the rate at which individual pre-mRNAs are processed would be lower, and rescue would be less, for a replicating plasmid whose many transcripts must compete for a fixed pool of processing factors. To test this, we replaced the Rous sarcoma virus promoter of pS168α59 (Fig. 6A, construct 4) with the SV40 early promoter, which contains the SV40 origin of replication (18). This generated a replicating version of the plasmid, which we called pS168α59ori. The RNase protection assay results of Fig. 7 confirm that there is less rescue (less processing) for the replicating plasmid (lane 2) than for its nonreplicating parent (lane 1). Rescue was similarly impaired by plasmid copy number for an entirely unrelated poly(A) site (Fig. 7, lanes 6 and 7). It is formally possible that the SV40 promoter, rather than the origin of replication, in these plasmids is responsible for the altered processing (17). However, the important point is that the replicating and nonreplicating plasmids are identical for several kilobases surrounding their poly(A) sites (e.g., more than 2 kb upstream and 1.5 kb downstream for pS168α59 and pS168α59ori), thus confirming that even remote changes that affect processing rates are reflected in the efficiency of cis-antisense rescue.
FIG. 7.
Effect of plasmid multiplicity and poly(A) site strength on rate of commitment to cleavage and polyadenylation. (A) RNase protection assays of nuclear RNA. The probe lengths for uncleaved S, L, and E RNAs are 227, 197, and 239 nt, respectively. The corresponding cleaved lengths are 182, 146, and 140 nt. For ease of comparison, the bands are arranged side by side in the figure. Lanes 1 and 4 are taken from lanes 4 and 2, respectively, of Fig. 6B. Minor bands corresponding to uncleaved RNA were produced in the assays of lanes 4, 6, and 7. These bands were included in the calculation of mole percent uncleaved RNA but are not shown in the figure. The L plasmids are based on a derivative of pCat1 in which the Renilla luciferase gene was inserted as for pRSVrl and an EcoRV site was introduced 100 bp upstream of the HpaI site by an in vitro mutagenic conversion of G to T. The EcoRV-HindIII fragment of this plasmid was then replaced with the DNA fragment shown in panel B by sticky-end ligation to give pL. The bracketed sequence in panel B, extended with AGCT at the 5′ end and with A at the 3′ end, was then synthesized, as was its complement, extended with AGCTT at its 5′ end. These two oligonucleotides were annealed and ligated into HindIII-cut pL to give pL27α51 (in which the insert is in the antisense orientation with a unique HindIII site between the sense and antisense sequences). The PCR fragment used for pS83α59 was then inserted into HindIII-cut pL27α51 to yield pL91α51. To replace the RSV promoter with the SV40 origin, the latter was obtained as an NdeI-BstBI fragment from pRL-SV40 (which had been modified to contain an NdeI site by converting a C to T 11 bp upstream of the BglII site) and then used to replace the NdeI-BstBI fragment of the appropriate RSV plasmid. For completeness we point out that each ori plasmid also possessed (for reasons unrelated to this work) a SmaI-EcoNI G-free cassette fragment from pORgf3·2 (67) inserted at the AatII site 1 to 2 kb downstream of the poly(A) site in a nonfunctional part of the plasmid. Also the L ori plasmids contained a HindIII-BamHI G-free cassette insert from pORgf3·2 at their KpnI sites downstream of the transcription unit, making them nearly identical to the S plasmids, all of which already have this fragment at a similar position. For RNase protection, the E and S probes of Fig. 5 and 6 were used. The L probe was a T7 RNA polymerase transcript of SspI-digested pL. Although this probe traverses the HindIII site into which insertions were made for the derivatives of pL, most uncleaved cellular RNA nevertheless protected the full length of the probe by looping out the inserted sequences. A small amount of probe was protected only up to the HindIII site. This band is not shown in the figure but is included as uncleaved RNA in the mole percent calculation. (B) Top strand of a StuI-HindIII-trimmed SV40 late poly(A) site PCR fragment. The bottom strand is 4 nt longer because of the HindIII sticky end. The poly(A) hexamer is shown in boldface, and the cleavage site (45) is indicated with an arrow. The sequence corresponding to the 51-nt antisense target in the RNA is enclosed in brackets. (C) L-α duplex formation in vitro. Linearized plasmid DNAs were transcribed with T7 RNA polymerase at 37°C for 1 h in a solution containing 6 mM MgCl2, 2 mM spermidine, 40 mM Tris (pH 7.9), and 10 mM dithiothreitol. Following transcription, DNaseI was added to 20 U/ml, KCl was added to a final concentration of 0.1 M, and RNase A and RNase T1 (RNase Cocktail; Ambion) were added to final concentrations of 6 and 250 U/ml, respectively. After digestion at 25°C for 30 min, the samples were analyzed as for RNase protection. Similar results were obtained using four times as much RNase Cocktail. The marker is a 56-nt piece of G-free RNA prepared by T1 RNase digestion. The plasmid templates were constructed as follows: pLT7, as for pL in panel A except with insertion into HpaI- and HindIII-cut pAP<cat> (67); pL27α51T7, as for pL27α51 in panel A except starting with pLT7; and pL168α51T7, as for pL91α51ori in panel A except that the PCR fragment used for pS168α59 was inserted into HindIII-cut pL27α51 by sticky-end ligation. The transcripts had the following lengths and were produced from templates linearized at the following restriction sites: lane 1, 1,384 nt, DraI; lane 2, 1,440 nt, DraI; lane 3, 808 nt, HindIII; lane 4, 2,299 nt, EcoNI; and lane 5, 943 nt, DraI. The central region surrounding L and α is shown drawn to scale in the figure. In all cases, the T7 promoter (not shown) was 731 nt upstream of L.
A strong poly(A) site commits to processing faster than weaker poly(A) sites.
It is thought that the strength of a poly(A) signal reflects the stability of its processing complex (65, 66). However, the data in Fig. 5 and 6 suggest that after the poly(A) signal is transcribed, commitment to cleavage and polyadenylation occurs during the time it takes the polymerase to travel the next few hundred base pairs. Since this time period is very short compared to the off rates measured in vitro (66), it is unlikely that complex stabilities measured in vitro are relevant to poly(A) site strengths expressed in vivo. It is more likely that, in vivo, kinetic parameters are determinative of poly(A) site strength. To determine whether poly(A) site strength is correlated with the rate of processing in vivo, we carried out some preliminary comparisons of processing rates for the SV40 late, the SV40 early, and the synthetic poly(A) sites (abbreviated L, E, and S, respectively, in our plasmid nomenclature). Carswell and Alwine have shown that L is five- to sixfold stronger than E in vivo (10), and we have found, in similar experiments, that E and S are of equivalent strength (data not shown).
Figure 7 shows, for a replicating plasmid, that S is slow to act, with most of its nuclear RNA remaining vulnerable to an antisense sequence located 168 nt downstream, which sequesters it and prevents cleavage (lane 2). In contrast, L acts rapidly and rescues itself almost completely from an antisense sequence which is only 91 nt downstream (lane 3). Similar, though less dramatic, results were obtained in a comparison of L to both S and E in a nonreplicating background (lanes 4 to 6). It is not surprising that L enjoys a greater advantage when factors are limiting (i.e., for a replicating plasmid). These results show that L, one of the strongest poly(A) sites known, commits to processing considerably faster than the weaker poly(A) sites, S and E.
Graveley et al. have shown that RNA secondary structure is an important determinant of poly(A) site strength in vitro (32). To determine whether the ability of L to rescue itself so rapidly from antisense attack is attributable to rapid folding into a secondary structure that resists duplex formation with the antisense sequence, we carried out nuclease digestions in vitro. RNA produced by T7 RNA polymerase was RNase digested in the presence of 0.1 M KCl and 6 mM Mg2+ immediately following transcription (Fig. 7C). Lane 1 of Fig. 7C shows that only a collection of short RNAs survived RNase digestion of transcripts that contained L but no antisense sequence. Lane 2 shows that transcripts of a plasmid with the inverted repeat 27 nt downstream of L yielded a prominent RNase-resistant band close to the length expected for RNase digestion of an L-α hairpin (53 to 54 nt). This band was still present for a plasmid in which α had been moved 141 bp farther downstream (lane 4). If either plasmid was cut between L and α before transcription, no protected band appeared (lanes 3 and 5), confirming that α was responsible for the RNase resistance. These results indicate that the SV40 late poly(A) site is not intrinsically resistant to formation of a duplex with an antisense sequence.
Commitment to cleavage and polyadenylation is a multistep process.
To estimate quantitatively the rate of commitment to cleavage and polyadenylation for the SV40 early poly(A) site, the spacer constructs of Fig. 5, and several additional constructs in the same series, were transfected into COS cells and assayed for CAT expression as described for Fig. 3. After normalization for transfection efficiency and adjustment for the actual fraction of cytoplasmic RNA polyadenylated at the SV40 early poly(A) site, the resulting CAT activity values were plotted in Fig. 8A as percentages of the activity of the positive control (i.e., percent rescue) versus the distance separating sense sequence from antisense sequence.
FIG. 8.
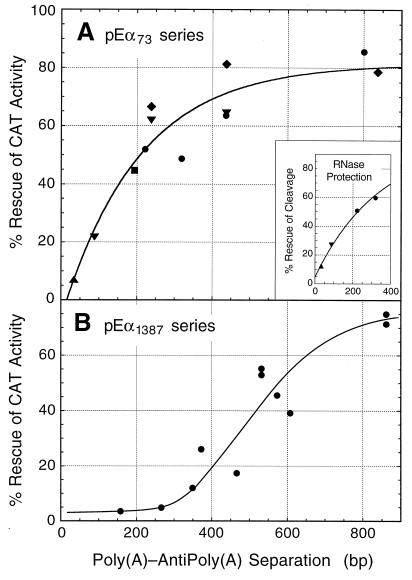
Distance dependence of cis-antisense rescue for short and long antisense sequences targeted to the SV40 early poly(A) site. (A) Rescue from the 73-nt antisense element. The CAT expression data in Table 2 were corrected for the presence of extended mRNAs polyadenylated at cryptic sites downstream (estimated from the cytoplasmic lanes of Fig. 5). The corrected data were then plotted and fitted to the following version of the first-order rate equation: (c0 − c)/c0 = A[1 − e−k(s + x)], where (c0 − c)/c0 is the fraction of poly(A) sites rescued as the polymerase travels a distance s + x down the template, c0 is the starting number of unprocessed poly(A) sites, c is the number of unprocessed poly(A) sites remaining after the polymerase has traveled a distance s + x, s is the separation between sense and antisense sequences as defined in the legend to Fig. 2C, x is a correction factor to be evaluated by curve fitting, and A is the maximum fraction of CAT activity that can be rescued in our experimental system. Notice that s + x is used here as a proxy for time t, which can be determined by dividing s + x by 40 nt/s, the known rate of RNA polymerase II elongation in mammalian cells (63). The curve in the figure returns the following values for the variables floated during the fit: A = 81%, k = 4.9 × 10−3 bp−1, and x = −18 bp. The inset summarizes the RNase protection data of Fig. 5 fit to the same first-order equation. The symbols for the sequence types inserted into the BamHI site of pE34α73 are as follows: ▴, no inserted sequence; ▾, 50 bp forward; ⧫, 50 bp reverse; ●, G free; and ■, chicken globin. (B) Rescue from the 1,387-nt antisense element, with separation defined exactly as for pE34α73. Data were obtained and corrected as for panel A, but curve fitting was by eye.
We wanted to be able to convert the separation distance into time based on the average rate of transcription (63). We therefore needed to ensure that the DNA spacers used were representative of average DNA sequences. In addition, at the RNA level, we needed to ensure that the spacers did not introduce unanticipated biases due to RNA folding. Accordingly, we chose carefully the types of DNA to be used as spacers in the construction of the various plasmids employed in the experiment shown in Fig. 8A. In one approach to generating spacers, we inserted tandem multiples of a 50-bp sequence to ensure that long spacers were simply longer and did not include new sequence variables. We made plasmids containing both forward and reverse orientations of the basic sequence, thus obtaining two spacer series with what are essentially different sequences. A second approach to minimizing the effects of sequence made use of spacers whose DNA sequence is relatively monotonous and whose transcripts are expected to be devoid of secondary structure. The inserts for these plasmids were all derived from the original G-free cassette of Sawadogo and Roeder (58). In the G-free orientation, these cassettes yield transcripts devoid of G’s, for which MulFold secondary-structure prediction (37) indicates that no significant secondary structure should exist. In addition to the carefully chosen sequences just described, we also included a plasmid whose spacer was taken from the 3′-flanking region of the chicken βH-globin gene.
The CAT rescue data in Fig. 8A show that as the antisense sequence is moved progressively downstream from the poly(A) signal, the CAT activities of the various constructs steadily recover from inhibition and are 50% rescued by about 200 bp. The relationship between separation and CAT rescue fits first-order kinetics, as shown by the curve in the figure (see the legend to Fig. 8). Candidate rate-limiting first-order (or pseudo-first-order) processes include factor binding and RNA folding steps that would sequester the poly(A) signal and make it inaccessible to the antisense sequence. The curve in Fig. 8A shows that all of the plasmid constructs used yielded data that scatter about the same first-order curve. Indeed, each of the three plasmid series (50-bp forward, 50-bp reverse, and G free) gave points that scattered on both sides of the curve. We therefore conclude that the results of this cis-antisense rescue assay are not significantly compromised by sequence effects and that they provide a reliable measure of commitment to 3′-end processing as a function of transcription time [assuming there is no poly(A)-dependent change in the transcription rate].
The asymptote for the curve in Fig. 8A is less than 100%, suggesting that complete rescue is not achieved. Perhaps this reflects a residual trans-antisense effect from neighboring transcripts. For the large sense-antisense separations at the asymptote, the antisense segment of each transcript has no cis target because the poly(A) site has already been sequestered by factors or processed (Fig. 1H). However, before the antisense segment is degraded, perhaps it can assault nearby nascent transcripts in trans as previously described by Liu et al. (40). Such a trans effect would presumably be minimal at short separations in which the antisense sequence is engaged in the cis interaction (Fig. 1C and D).
The rate of poly(A) site rescue (Fig. 8A) was analyzed not only by CAT expression but, as shown in the inset to Fig. 8A, also by RNase protection, using the data of Fig. 5. The percentage of cleaved pre-mRNA in the nucleus was determined and then analyzed by plotting and curve fitting as for the main part of Fig. 8A. The curve in the inset confirms that cleavage is 50% rescued by about 200 bp for the SV40 early poly(A) site. Thus, there is good agreement between conclusions based on measurements of cytoplasmic mRNA expression relative to a control (Fig. 8A) and measurements of cleaved-RNA levels in the nucleus as a percentage of the total (Fig. 8A, inset). Note, however, that the latter approach is affected by the unknown half-lives of the cleaved and uncleaved RNAs in the nucleus and by the unknown extent to which the cis-antisense sequences in the uncleaved RNA compete with the radioactive probe during hybridization.
During the course of these studies, when different lengths of antisense sequence were used, we noticed that the poly(A) site required more time to become immune to a longer antisense sequence than to a shorter one. Figure 8B shows, for example, that when a very long antisense sequence is used (1,387 nt), 50% rescue is not attained until the antisense sequence is moved 550 bp downstream of the sense element. Moreover, there is a distinct lag of about 300 bp before rescue itself actually begins, indicating a requirement for one or more preparatory steps prior to the event that finally achieves rescue. The simplest explanation for the antisense length-dependent lag is that the 3′-end processing complex is assembled progressively and that early stages of assembly are sufficient to protect against the shorter antisense sequences but only a more mature complex can resist the disruptive influence of a long antisense sequence.
The above explanation assumes that the rescue following the lag in Fig. 8B results from events taking place at the SV40 early poly(A) site. Although it is formally possible that transcription termination between sense and antisense sequences contributes to the rescue, this is not likely because poly(A) sites do not drive termination in the plasmid background we have used (67). We should also point out that although the 1,387-nt antisense sequence happens to contain the SV40 late poly(A) site (10), the presence of the complementary sense sequence upstream ensures that this downstream site never becomes accessible for processing (Fig. 6B, lane 6). We have used RNase protection assays to confirm this and to further establish that rescue is due exclusively to polyadenylation at the early site (data not shown).
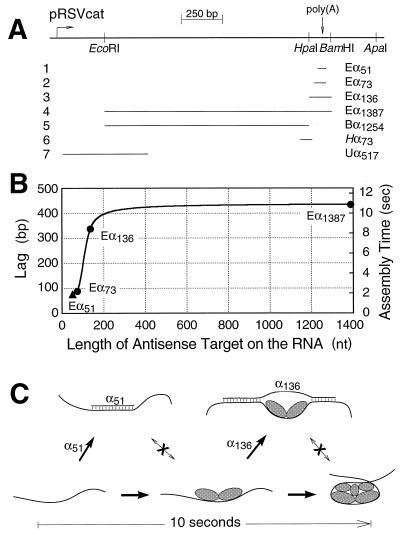
To estimate the amount of time required for complete assembly of the SV40 early 3′-end processing complex, we prepared additional constructs with various separations and with antisense sequences targeted to either 51 or 136 nt surrounding the poly(A) site (Fig. 9A, constructs 1 and 3). The rescue data for these constructs, together with the data of Fig. 8 (i.e., for constructs in series 2 and 4 of Fig. 9A), provided us with the relationship shown in Fig. 9B, in which the length of the antisense target on the RNA is plotted against the lag preceding rescue. Because the lag is difficult to estimate for the shorter antisense sequences, we have, for the purposes of Fig. 9B, operationally defined the lag as the separation required for 25% rescue. Thus, in Fig. 8B, for example, 435 bp are required for sufficient poly(A) site maturation to occur so that rescue can begin and proceed to the extent of 25%. Figure 9B shows that the lag increases with antisense target size until a maximum target of about 200 nt is reached, for which a lag corresponding to about 400 bp of transcription is required to reach 25% rescue. From this we infer an assembly time of about 10 s for the first poly(A) sites to become completely resistant to antisense sequences and an assembly domain of about 200 nt for the SV40 early poly(A) site. Because of the asynchrony of the process, it takes a further 10 s (i.e., 400 bp more) for all rescue to reach completion (Fig. 8B).
FIG. 9.
Progressive assembly of the cleavage-polyadenylation processing complex. (A) The antisense targets on pRSVcat of the four Eα series of plasmids used to obtain the data for panel B, as well as the targets of three control plasmids. For the control plasmids, the sense-antisense separation (172, 34, and 33 nt for plasmids 5 to 7, respectively) is defined as the number of base pairs separating the last C of the poly(A) signal region bracketed in the sequence of Fig. 2C and the beginning of the downstream antisense sequence. This definition maintains the poly(A) signal as the reference point even though the antisense sequences are targeted to non-poly(A) signal regions of the transcript. This is because rescue from a downstream antisense element for these constructs cannot begin until the tether between the transcript and its antisense sequence is broken by cleavage and polyadenylation. (B) The separation of sense from antisense sequences (the lag) required to achieve 25% rescue from antisense elements of increasing length. The circles represent measurements made on the pRSVcat-based antisense constructs indicated. The triangle is an estimated value based on the relationships between the pEα51rl and the pEα73rl classes of constructs, which are based on pRSVrl. The Eα51rl measurements could not be used directly, we discovered, because polyadenylation rates differ significantly for mRNAs containing different coding sequences. (C) Model depicting progressive assembly of the cleavage-polyadenylation apparatus. Short antisense elements (e.g., α51) interfere only with early stages of assembly. Longer antisense elements (e.g., α136) can interfere also with partially assembled complexes, but mature complexes are immune to antisense sequences. Although assembly is clearly a multistep process, we show here only a single step.
Since increasing the antisense target size beyond 200 nt requires little additional assembly time to effect rescue (Fig. 9B), it appears that most of the upstream target of the long antisense sequence in the Eα1387 series of constructs lies outside of the assembly domain of the 3′-end processing complex. Results obtained using clones 5 to 7 of Fig. 9A are consistent with the notion that the poly(A) site assembly domain does not cover much more than 200 nt. Clone 5 (Bα1254) expresses CAT at 42% ± 10% of control levels for a separation of 172 bp. This is over 10-fold more expression than for the clone 4 series (Eα1387) at a comparable separation (<4% expression compared to controls [Fig. 8B]). Yet the antisense sequence of clone 5 targets 90% of the same sequence as does the antisense sequence in the clones of series 4 (Fig. 9A). Thus, clone 5 lacks antisense sequence to 133 nt surrounding the poly(A) site, and the remaining 1,254 nt of antisense sequence are largely ineffective at inhibiting expression. Clone 6 of Fig. 9A, Hα73, is targeted to a region at the edge but still within the 200-nt assembly domain of the poly(A) site. As may be expected, and as described at the beginning of Results, it has a mild inhibitory effect on polyadenylation. In contrast, the much longer antisense sequence in clone 7 (Uα517), which is targeted to an upstream portion of the transcript, has no effect on expression (100% ± 25% of control). These results strengthen the interpretation that Fig. 9B identifies an assembly domain surrounding the poly(A) site that encompasses about 200 nt and has an assembly time of about 10 s (Fig. 9C).
DISCUSSION
cis-antisense rescue—measuring the kinetics of commitment to 3′-end processing in vivo.
We have described a method for using cis-antisense sequences to interrogate poly(A) signals following their synthesis in vivo. By progressively moving an antisense element downstream from its target poly(A) site in a series of constructs, we provided measured increases in potential assembly time to the poly(A) signal before the inhibitory antisense sequence was transcribed. When the time provided in this way became equal to the time required for commitment to cleavage and polyadenylation, we witnessed restored poly(A) site function (rescue).
We studied the SV40 early poly(A) site and an artificial poly(A) site, the SPA (39), using this assay. Numerous controls were carried out to confirm that a simple antisense mechanism was involved in inhibition and that relief of inhibition upon moving the antisense sequence downstream reflected simply the travel time required for the RNA polymerase to reach the antisense element. Thus, inhibition depended on the presence of adequate sequence complementarity between the antisense element and the upstream poly(A) signal to which it was targeted; inhibition gave rise to large amounts of uncleaved pre-mRNA in the nuclei of transfected cells, and inhibition was relieved only by moving the antisense sequence downstream of the poly(A) site so that its appearance was transcriptionally delayed—an antisense moved upstream was just as inhibitory as one immediately adjacent to the poly(A) site.
Cleavage-polyadenylation complex assembly and folding.
One of the principal conclusions from this study derives from the observation that the time required for poly(A) site assembly and rescue (for example, the lag evident in Fig. 8B) increased with the size of the antisense target region containing the poly(A) site (Fig. 9B). This indicates that the poly(A) site is progressively assembled into structures that are resistant to inhibition by ever-more-potent (i.e., longer) segments of antisense sequence. A model which can explain this result is presented in Fig. 9C. Our data do not suggest the nature of the progressive steps in maturation, but for the sake of discussion, we consider a scenario involving stepwise addition of factors to the growing complex (27). As shown in Fig. 9C (for simplicity, only a single intermediate step is illustrated), a short antisense sequence could inhibit processing by forming a duplex with the naked poly(A) signal, thereby interfering with the first step of factor binding. However, if the factors initiate binding before the poly(A) signal encounters the antisense element, the bound factors will rescue the poly(A) site from the short antisense sequence. Nevertheless, the partially assembled poly(A) site remains susceptible to inhibition by longer antisense sequences. Thus, a short antisense element is effective only if transcribed very soon after the poly(A) signal itself, before factor binding gets seriously under way. A longer antisense sequence, on the other hand, remains effective even when its appearance is delayed somewhat by the time required for transcription, because it can bind to and, according to this scenario, inactivate partially assembled complexes. A fully mature complex is resistant to antisense sequence of any length, possibly because cleavage immediately ensues, severing the cis relationship between the sense and the antisense sequences. Other scenarios are also possible, especially the simultaneous binding to the RNA of all factors in the form of a holocomplex followed by an ordered series of conformational transitions that increase the footprint of the complex on the RNA. Note, however, that the progressively greater effectiveness of longer and longer antisense sequences cannot be explained as trivial kinetic or stability effects related to length since even a short antisense element (59 nt) is capable of acting with undiminished effectiveness across at least 465 nt of upstream RNA (Fig. 6B, lane 6).
Assuming that the progressivity of complex maturation reflects stepwise factor binding, CPSF and CstF would presumably be the first to bind, perhaps by direct transfer from the polymerase (14, 17, 47, 65). Our data are consistent with an early occurrence of the initiating event, as implied by such a scenario. Lane 2 of Fig. 6B (same as Fig. 7A, lane 4) shows that rescue of 3′-end cleavage from the relatively short (59-nt) anti-SPA sequence is already under way by the time the antisense element, only 23 nt downstream, is extruded from the polymerase. Rescue is even faster (i.e., more cleavage) for the strong SV40 late poly(A) site (Fig. 7A, lane 6). Thus, the poly(A) signal begins recruiting factors while it is still in the vicinity of the transcription complex. Indeed, the converse may occur: it may be the polymerase-bound CPSF and CstF that recruit the poly(A) signal (35), with subsequent assembly steps then taking place in association with the transcriptional apparatus.
Figure 9C, which presents a physical interpretation of the present work, also accommodates our functional understanding of the 3′-end processing domain (see the introduction). Thus, RNA sequences immediately flanking the core poly(A) signal are important for function (5, 8, 10, 12, 26, 33, 50, 53, 60), and in at least one in vitro study, the presence of flanking RNA per se was important, regardless of the sequence (36). Also, all elements in the poly(A) signal domain (upstream flanking, AAUAAA hexamer, cleavage site, U/GU rich, and downstream flanking) occupy narrowly prescribed positions relative to each other (2, 5, 9, 11, 24, 33, 59), suggesting that they, and the factors that bind to them, comprise a single, well-defined structure. The fact that elements in the flanking RNA, when examined, are found directly associated with CPSF or CstF (25, 26, 33, 50) supports this interpretation. We suggest that it is this structure whose assembly we have observed in vivo and which is shown schematically in Fig. 9C.
Our estimate of 10 s for the minimum time to maturation of an SV40 early 3′-end processing complex (and 20 s for the completion of all processing) is consistent with previous estimates of the time required for cleavage and polyadenylation (1, 6, 51, 57). In the early work, time was measured directly, using pulse-labeling, and rough estimates of approximately 1 min for the completion of both cleavage and polyadenylation were obtained (1, 51, 57). Very recently, Baurén et al. (6) used reverse transcription-PCR to determine that cleavage of nascent Balbiani ring 1 transcripts in Chironomus occurs about 600 bp downstream of the poly(A) site. This result is remarkably similar to our results showing that assembly of the cleavage-polyadenylation complex is complete at about the same position on the template (Fig. 8B). Although it is unlikely that cleavage occurs at the same distance downstream for all poly(A) sites, this coincidence does strengthen the possibility that fully mature complexes, as defined by cis-antisense rescue, proceed immediately to cleavage.
Does assembly rate govern poly(A) site strength?
Assembly of the cleavage-polyadenylation apparatus is a multistep process that takes a significant length of time. One may therefore expect strong poly(A) sites to be those that can assemble quickly in vivo so as to minimize the opportunity for interference with the process. The results of Fig. 7 show that the rate of processing complex assembly in vivo is indeed correlated with poly(A) site strength, with the strong SV40 late poly(A) site committing to cleavage more rapidly than the weaker synthetic or SV40 early poly(A) sites placed in comparable plasmid backgrounds (compare lane 3 with lane 2 and lane 6 with lanes 4 and 5). Moreover, the cis-antisense rescue assay itself illustrates that rapid assembly leads to poly(A) site strength. The synthetic poly(A) site, with an antisense sequence located 168 nt downstream, behaves like an extremely weak poly(A) site (Fig. 7, lane 2) because it is unable to process an appreciable fraction of the pre-mRNA molecules before the downstream antisense sequence interferes. In contrast, the SV40 late site processes very quickly and is scarcely interfered with at all by an antisense sequence located a similar distance downstream (Fig. 7, lane 3). [The SV40 late antisense sequence is effective, however, if located sufficiently close to the poly(A) site (Fig. 7, lane 7).] Thus, at least with these experimental constructs, the strength of a poly(A) site is a direct consequence of its rate of assembly.
The realizations that weak sites assemble slowly and that the assembly process can be interfered with at any point in the pathway have regulatory implications. A particularly intriguing possibility in this regard concerns poly(A) sites that are subject to downstream regulation. These sites delay the decision to process until they encounter elements that may lie from several hundred base pairs (43) to more than 1.5 kb (3) downstream. Perhaps such sites have strategic impediments to assembly that maintain them in a partially assembled but arrested state while awaiting regulatory input.
The gradual nature of processing complex assembly may also be relevant to the mechanism by which the poly(A) signal instructs RNA polymerase II to terminate. Current models invoke events at the two extremes of the assembly continuum, either the initial binding of factors (47) or the final cleavage of the RNA (15, 55). To these we can now add the intermediate steps of the assembly process as possible triggers of termination, particularly if assembly occurs astride the polymerase.
ACKNOWLEDGMENTS
L.C.C. and A.J. contributed equally to this work.
We thank D. H. Liu for assistance with plasmid construction and E. Landaw for advice on data analysis.
This work was supported by NIH grant GM50863.
REFERENCES
- 1.Acheson N H. Kinetics and efficiency of polyadenylation of late polyomavirus nuclear RNA: generation of oligomeric polyadenylated RNAs and their processing into mRNA. Mol Cell Biol. 1984;4:722–729. doi: 10.1128/mcb.4.4.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed Y F, Gilmartin G M, Hanly S M, Nevins J R, Greene W C. The HTLV-I Rex response element mediates a novel form of mRNA polyadenylation. Cell. 1991;64:727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- 3.Ashe M P, Pearson L H, Proudfoot N J. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1988. [Google Scholar]
- 5.Bagga P S, Ford L P, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baurén G, Belikov S, Wieslander L. Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′-end formation and excision of the 3′-terminal intron. Genes Dev. 1998;12:2759–2769. doi: 10.1101/gad.12.17.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyer A L, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 8.Brackenridge S, Ashe H L, Giacca M, Proudfoot N J. Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res. 1997;25:2326–2336. doi: 10.1093/nar/25.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown P H, Tiley L S, Cullen B R. Effect of RNA secondary structure on polyadenylation site selection. Genes Dev. 1991;5:1277–1284. doi: 10.1101/gad.5.7.1277. [DOI] [PubMed] [Google Scholar]
- 10.Carswell S, Alwine J C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, MacDonald C C, Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen F, Wilusz J. Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res. 1998;26:2891–2898. doi: 10.1093/nar/26.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christofori G, Keller W. Poly(A) polymerase purified from HeLa cell nuclear extract is required for both cleavage and polyadenylation of pre-mRNA in vitro. Mol Cell Biol. 1989;9:193–203. doi: 10.1128/mcb.9.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 15.Connelly S, Manley J L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 16.Crabb D W, Dixon J E. A method for increasing the sensitivity of chloramphenicol acetyltransferase assays in extracts of transfected cultured cells. Anal Biochem. 1987;163:88–92. doi: 10.1016/0003-2697(87)90096-0. [DOI] [PubMed] [Google Scholar]
- 17.Dantonel J C, Murthy K G, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 18.DeLucia A L, Deb S, Partin K, Tegtmeyer P. Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J Virol. 1986;57:138–144. doi: 10.1128/jvi.57.1.138-144.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denome R M, Cole C N. Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol Cell Biol. 1988;8:4829–4839. doi: 10.1128/mcb.8.11.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon D A, Vaitkus D L, Prescott S M. DNase I treatment of total RNA improves the accuracy of ribonuclease protection assay. BioTechniques. 1998;24:732–734. doi: 10.2144/98245bm07. [DOI] [PubMed] [Google Scholar]
- 21.Edwalds-Gilbert G, Prescott J, Falck-Pedersen E. 3′ RNA processing efficiency plays a primary role in generating termination-competent RNA polymerase II elongation complexes. Mol Cell Biol. 1993;13:3472–3480. doi: 10.1128/mcb.13.6.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frendewey D, Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985;42:355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- 23.Gil A, Proudfoot N J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit β-globin mRNA 3′ end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 24.Gilmartin G M, Fleming E S, Oetjen J. Activation of HIV-1 pre-mRNA 3′ processing in vitro requires both an upstream element and TAR. EMBO J. 1992;11:4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 26.Gilmartin G M, Hung S L, DeZazzo J D, Fleming E S, Imperiale M J. Sequences regulating poly(A) site selection within the adenovirus major late transcription unit influence the interaction of constitutive processing factors with the pre-mRNA. J Virol. 1996;70:1775–1783. doi: 10.1128/jvi.70.3.1775-1783.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilmartin G M, Nevins J R. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3:2180–2190. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 28.Gimmi E R, Reff M E, Deckman I C. Alterations in the pre-mRNA topology of the bovine growth hormone polyadenylation region decrease poly(A) site efficiency. Nucleic Acids Res. 1989;17:6983–6998. doi: 10.1093/nar/17.17.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 30.Gorman C M, Merlino G T, Willingham M C, Pastan I, Howard B H. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci USA. 1982;79:6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graveley B R, Fleming E S, Gilmartin G M. RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol. 1996;16:4942–4951. doi: 10.1128/mcb.16.9.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graveley B R, Gilmartin G M. A common mechanism for the enhancement of mRNA 3′ processing by U3 sequences in two distantly related lentiviruses. J Virol. 1996;70:1612–1617. doi: 10.1128/jvi.70.3.1612-1617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graveley B R, Hertel K J, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17:6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 36.Hou W, Russnak R, Platt T. Poly(A) site selection in the yeast Ty retroelement requires an upstream region and sequence-specific titratable factor(s) in vitro. EMBO J. 1994;13:446–452. doi: 10.1002/j.1460-2075.1994.tb06279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller W. 3′-end cleavage and polyadenylation of nuclear messenger RNA precursors. In: Lamond A I, editor. Pre-mRNA processing. Austin, Tex: R. G. Landes Company; 1995. pp. 113–134. [Google Scholar]
- 39.Levitt N, Briggs D, Gil A, Proudfoot N J. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z, Batt D B, Carmichael G G. Targeted nuclear antisense RNA mimics natural antisense-induced degradation of polyoma virus early RNA. Proc Natl Acad Sci USA. 1994;91:4258–4262. doi: 10.1073/pnas.91.10.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.London L, Keene R G, Landick R. Analysis of premature termination in c-myc during transcription by RNA polymerase II in a HeLa nuclear extract. Mol Cell Biol. 1991;11:4599–4615. doi: 10.1128/mcb.11.9.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lou H, Helfman D M, Gagel R F, Berget S M. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lou H, Yang Y, Cote G J, Berget S M, Gagel R F. An intron enhancer containing a 5′ splice site sequence in the human calcitonin/calcitonin gene-related peptide gene. Mol Cell Biol. 1995;15:7135–7142. doi: 10.1128/mcb.15.12.7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lutz C S, Alwine J C. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 45.MacDonald C C, Wilusz J, Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol Cell Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manley J L. A complex protein assembly catalyzes polyadenylation of mRNA precursors. Curr Opin Genet Dev. 1995;5:222–228. doi: 10.1016/0959-437x(95)80012-3. [DOI] [PubMed] [Google Scholar]
- 47.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 48.McDevitt M A, Hart R P, Wong W W, Nevins J R. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986;5:2907–2913. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montgomery M K, Xu S-Q, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nevins J R, Darnell J E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978;15:1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- 52.Nordstrom J L, Westhafer M A. Splicing and polyadenylylation at cryptic sites in RNA transcribed from pSV2-neo. Biochim Biophys Acta. 1986;867:152–162. doi: 10.1016/0167-4781(86)90075-8. [DOI] [PubMed] [Google Scholar]
- 53.Prescott J, Falck-Pedersen E. Sequence elements upstream of the 3′ cleavage site confer substrate strength to the adenovirus L1 and L3 polyadenylation sites. Mol Cell Biol. 1994;14:4682–4693. doi: 10.1128/mcb.14.7.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pribyl T M, Martinson H G. Transcription termination at the chicken βH-globin gene. Mol Cell Biol. 1988;8:5369–5377. doi: 10.1128/mcb.8.12.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Proudfoot N J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989;14:105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- 56.Russnak R H. Regulation of polyadenylation in hepatitis B viruses: stimulation by the upstream activating signal PS1 is orientation-dependent, distance-independent, and additive. Nucleic Acids Res. 1991;19:6449–6456. doi: 10.1093/nar/19.23.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salditt-Georgieff M, Harpold M, Sawicki S, Nevins J, Darnell J E., Jr Addition of poly(A) to nuclear RNA occurs soon after RNA synthesis. J Cell Biol. 1980;86:844–848. doi: 10.1083/jcb.86.3.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schek N, Cooke C, Alwine J C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sittler A, Gallinaro H, Jacob M. Upstream and downstream cis-acting elements for cleavage at the L4 polyadenylation site of adenovirus-2. Nucleic Acids Res. 1994;22:222–231. doi: 10.1093/nar/22.2.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 62.Takagaki Y, Ryner L C, Manley J L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 63.Tennyson C N, Klamut H J, Worton R G. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- 64.Wahle E. 3′-end cleavage and polyadenylation of mRNA precursors. Biochim Biophys Acta. 1995;1261:183–194. doi: 10.1016/0167-4781(94)00248-2. [DOI] [PubMed] [Google Scholar]
- 65.Wahle E, Kuhn U. The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog Nucleic Acid Res Mol Biol. 1997;57:41–71. doi: 10.1016/s0079-6603(08)60277-9. [DOI] [PubMed] [Google Scholar]
- 66.Weiss E A, Gilmartin G M, Nevins J R. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991;10:215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeung G, Choi L M, Chao L C, Park N J, Liu D, Jamil A, Martinson H G. Poly(A)-driven and poly(A)-assisted termination: two different modes of poly(A)-dependent transcription termination. Mol Cell Biol. 1998;18:276–289. doi: 10.1128/mcb.18.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]