Abstract
Tumor necrosis factor receptor (TNFR)-associated factor 3 (TRAF3) plays context-specific roles in multiple receptor-mediated signaling pathways in different cell types. Mice lacking TRAF3 in T cells display defective T-cell-mediated immune responses to immunization and infection and demonstrate defective early signaling via the TCR complex. However, the role of TRAF3 in the function of GITR/TNFRSF18, an important costimulatory member of the TNFR superfamily, is unclear. Here we investigated the impact of T cell TRAF3 status on both GITR expression and activation of specific kinases in the GITR signaling pathway in T cells. Our results indicate that TRAF3 negatively regulates GITR functions in several ways. First, expression of GITR protein was elevated in TRAF3-deficient T cells, resulting from both transcriptional and posttranslational regulation that led to greater GITR transcript levels, as well as enhanced GITR protein stability. TRAF3 associated with T cell GITR in a manner dependent upon GITR ligation. TRAF3 also inhibited several events of the GITR mediated early signaling cascade, in a manner independent of recruitment of phosphatases, a mechanism by which TRAF3 inhibits signaling through several other cytokine receptors. These results add new information to our understanding of GITR signaling and function in T cells, which is relevant to the potential use of GITR to enhance immune therapies.
Keywords: adaptor protein, T-cell, TRAF, receptor, signal transduction, GITR
Abbreviations: CAR-T, chimeric antigen receptor T cell; JAK1, Janus-activated kinase 1; LMC, littermate control; MAPK, mitogen-activated protein kinase; NKT, natural killer T cell; NLS, nuclear localization sequence; TNFR, tumor necrosis factor receptor; TRAF3, TNFR-associated factor 3; Treg, T regulatory cell
Glucocorticoid-induced TNFR-related protein (GITR, CD357, TNFRSF18) is a type 1 transmembrane cell surface protein expressed by most immune cell subsets, including T regulatory cells (Treg) (1), conventional CD4+ and CD8+ T cells (2), invariant natural killer T cells (NKT) (3, 4), and at low levels by B cells, NK cells, macrophages, and dendritic cells (5, 6). GITR-derived signals are indispensable for both CD4+ and CD8+ T cells to mount maximal immune responses against both viral infections and tumors (1, 7, 8, 9, 10). GITR ligand (GITRL, TNFSF18) is found on antigen-presenting cells (11, 12, 13) and endothelial cells (14, 15, 16, 17). When combined with suboptimal CD3 stimulation, GITR engagement by GITRL triggers enhanced expansion of CD4+ and CD8+ T cells and augments production of IFN-γ, IL2, and CD25 expression (2, 18, 19). GITR induces enhanced expression of Bcl-XL, a prosurvival factor, in TCR-activated CD8+ cells (15). In Treg, which have high levels of GITR expression, GITR signaling participates in regulation of proliferation and modulation of suppressive function (5, 20, 21). Targeting GITR via anti-GITR agonistic antibody (22, 23, 24, 25), recombinant Fc-GITRL proteins (26, 27, 28), or engineered Chimeric Antigen Receptor T cells (CAR-T) expressing recombinant receptors with the GITR cytosolic domain (29, 30, 31) can exert potent therapeutic activities. However, GITR-mediated intracellular signaling remains only partially characterized. GITR has a relatively short intracellular domain that does not possess intrinsic enzymatic activity (18, 32). As is typical of TNFR superfamily (TNFRSF) members, GITR relies upon recruitment of signaling adaptors, primarily the TNFR-associated factors (TRAFs) to mediate downstream signaling (18). GITR-induced activation of NFκB, mitogen-activated protein kinases (MAPKs), and the ribosomal protein S6 pathway require TRAF2 and TRAF5 (10, 18, 33). However, little is known about other regulatory components of the GITR signaling complex, such as inhibitory factors that may restrain GITR signaling. TRAF3 interacts with the cytoplasmic domain of GITR (14, 34), and we speculated that it may serve as a positive or negative regulator. Here we report that the adapter TRAF3 inhibits T cell GITR function, via several mechanisms.
TRAF3 regulates multiple signaling pathways via various molecular pathways (35). One canonical function of TRAF3 is regulation of NFκB2 activation. NFκB2 (p52 and its precursor p100) plays a central role in the immune system by regulating processes ranging from the development and survival of lymphocytes and lymphoid organs to the control of immune responses and malignant transformation, especially in B cells. Roles of this pathway in survival, activation, and differentiation of diverse subtypes of immune cells under many pathological settings have been demonstrated (36). The proteolytic generation of p52 from its p100 precursor is triggered by the upstream kinase NIK, which is tightly regulated by the TRAF2/TRAF3/cIAP ubiquitination complex (37). TRAF3 is also essential to promote T-cell-mediated immune responses, evident from the impaired immunity of mice with a T cell conditional Traf3 deficiency (38). We previously showed TRAF3 is required for enhancing early TCR signaling, which primes many other T cell effector functions (38, 39, 40). TRAF3 also regulates the differentiation and function of Treg (41) by recruiting the phosphatase PTPN2 to the IL-2 receptor. There, PTPN2 inhibits IL-2R signaling by dephosphorylating JAK/STAT pathway components (42). Similarly, TRAF3 facilitates association of the phosphatase PTPN22 with Janus-activated kinase 1 (JAK1) and thus functions as an inhibitor of IL-6R signaling in B cells (43). In contrast, TRAF3 promotes TCR and IL-15R-mediated iNKT cell differentiation (44). The impaired TCR signaling in TRAF3-deficient T cells is due in part to increased plasma membrane localization of the phosphatase PTPN22 and the inhibitory kinase Csk (40). These precedents indicate that relocalization of cellular phosphatases is one strategy utilized by TRAF3 to exert its regulatory functions (39). A remaining knowledge gap in understanding the essential role of TRAF3 in regulating T lymphocyte functions is how TCR costimulatory signals are impacted by TRAF3. GITR is constitutively expressed by T cells, not requiring prior TCR signals to induce its expression (2, 45). Here we show that in contrast to its promotion of TCR signaling, TRAF3 inhibits both GITR expression and GITR-mediated early signaling events, highlighting the diverse roles of TRAF3 in signaling pathways in T lymphocytes.
Results
TRAF3 restricts GITR expression by T lymphocytes
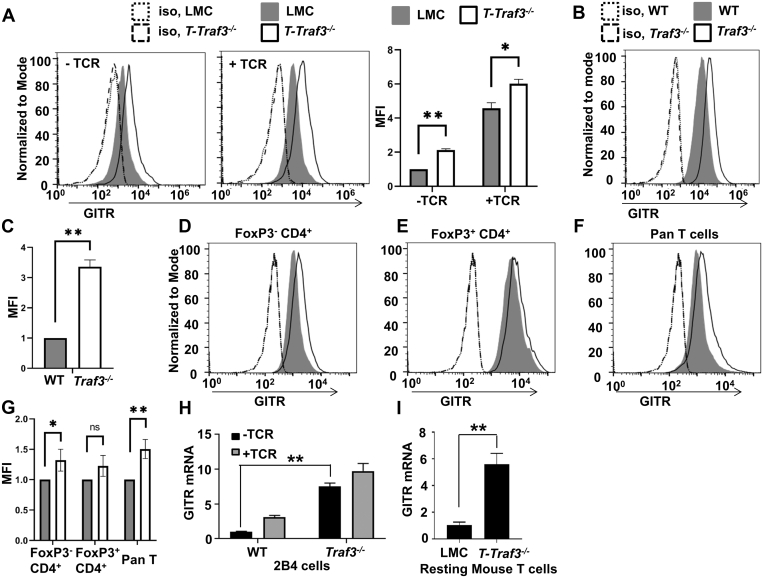
TRAF3 is required for optimal TCR-mediated signals to T cells (38, 40). Expression of TCR costimulatory TNFRSF receptors often requires TCR stimulation (46). An exception is GITR, which is constitutively expressed by resting T cells (47). We thus wondered whether GITR expression is impacted by T cell TRAF3. We first examined the amount of GITR on the surface of Traf3−/− primary mouse T cells compared with those from wild-type littermate control (LMC) mice, either unstimulated or following engagement of CD3 and CD28. T cells lacking TRAF3 displayed enhanced expression of GITR surface protein, either before or after CD3+CD28 stimulation (Fig. 1A). As a complementary model, we deleted Traf3 in the mouse T cell line 2B4 as described in Experimental procedures and observed the same trend of increased membrane GITR expression in the absence of TRAF3 (Fig. 1, B and C).
Figure 1.
Regulation of T cell GITR expression by TRAF3.A, splenic T cells isolated from LMC or T-Traf3−/− mice as indicated above the histograms were unstimulated (-TCR) or stimulated with plate bound Ab to CD3 (0.5 μg/ml) and soluble Ab to CD28 (5 μg/ml) (+TCR) for 16 h. Washed cells were then stained with anti-GITR or an isotype control antibody (Iso) and analyzed by flow cytometry. Mean fluorescence intensity (MFI) ± SEM of three experiments are shown to the right of a representative experiment. B and C, unstimulated WT or Traf3−/− subclones of the mouse T cell line 2B4 were analyzed as in (A). A representative result (B) and MFI ± SEM (n = 6) (C) are shown. D–G, unstimulated primary splenic T cells (D, conventional T cells; E, regulatory T cells; F, pan T cells) were analyzed for GITR surface expression as in A–C. One representative of three replicate experiments is shown. Quantification of MFI ± SEM (n = 4) is shown in G. LMC: filled; T-Traf3−/−: open. For A, C, and G, ∗∗p < 0.01, ∗p < 0.05, and n.s indicates no significant difference by unpaired t test. H–I, GITR relative mRNA levels, 2−ΔΔCt normalized to GAPDH (y axis), were determined for RNA isolated from 2B4 (H) or primary splenic T cells (I) by quantitative RT-PCR, as described in Experimental procedures. Cells in H were unstimulated (−TCR) or stimulated for 4 h with Abs to CD3 and CD28, as in (A) (+TCR). Error bars in F and G represent mean values ± SEM; ∗∗p < 0.01 by unpaired t test. N = 12.
T-Traf3−/− mice have a 2- to 3-fold greater frequency of Foxp3+ regulatory T cells (Treg), resulting from enhanced differentiation of Treg precursors to mature Treg cells (42). This occurs because T cell TRAF3 normally recruits the phosphatase PTPN2/TCPTP to the IL-2R, to inhibit early signaling events (42). Treg expresses higher levels of GITR than conventional T cells (1), so we asked if the increased frequency of Treg in the T-Traf3−/− mouse accounts for the overall elevated expression of GITR seen in Figure 1A, although this could not account for the increased GITR expression by TRAF3-deficient 2B4 cell subclones shown in Figure 1, B and C. As expected, we isolated approximately 2-fold more Treg from T-Traf3−/− than LMC mice (Fig. S1). To examine the contribution of this expanded Treg population to the observed enhancement in GITR expression in T-Traf3−/− T cells, we measured relative GITR levels on total T cells compared with those separated on the basis of FoxP3 expression. We found that the FoxP3+ population did not display a notable difference in GITR levels between TRAF3-sufficient and TRAF3-deficient T cells; GITR was increased in both total (1F) and FoxP3− (1D) T cells (Fig. 1, D–G). Therefore, elevated GITR expression in T-Traf3−/− mice was not due to the increased Treg subpopulation.
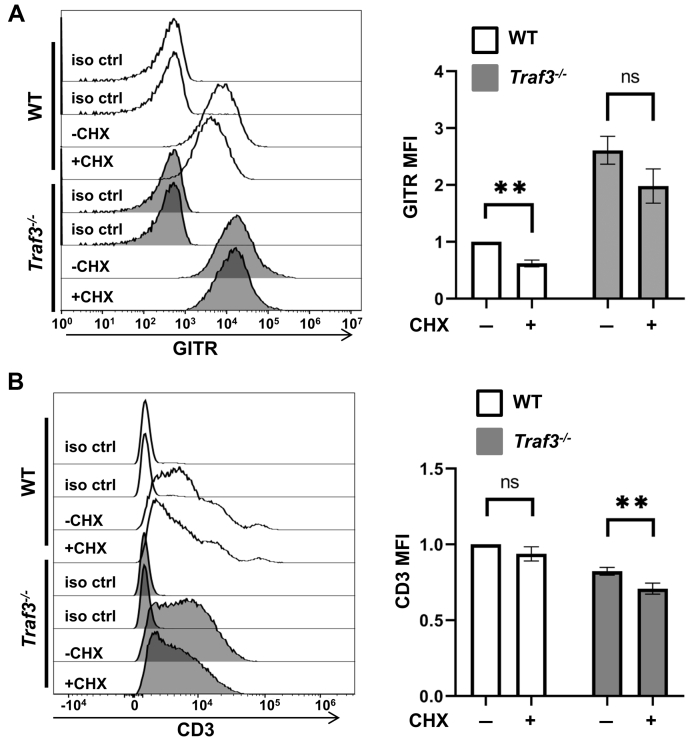
To determine how TRAF3 restrains GITR expression, we investigated two stages impacting protein levels—transcription and posttranslational degradation (protein turnover). TRAF3 deficiency resulted in a ≥ 5-fold increase in GITR mRNA in both unstimulated and CD3+CD28-stimulated 2B4 (Fig. 1H) and mouse primary total T cells. (Fig. 1I). We also monitored GITR protein turnover rates by inhibiting protein synthesis with cycloheximide. In 2B4 T cells lacking TRAF3, the rate of GITR degradation was markedly decreased compared with WT T cells (Fig. 2). Thus, reduced GITR expression by TRAF3-sufficient T cells correlates with reduced GITR gene expression, as well as potential effects on protein stability.
Figure 2.
GITR protein turnover. WT or Traf3−/− 2B4 cells were treated with 30 μg/ml cycloheximide (CHX) for 4 h, then stained for GITR (A) or for CD3 (B) prior to flow cytometry analysis. “Iso ctrl” indicates staining using isotype control Abs. Quantification of MFI ± SEM (n = 4) is shown to the right of a representative result in both B and C. ∗∗p < 0.01, and n.s indicates no significant difference by unpaired t test.
Role of noncanonical NFκB2 activation in enhanced expression of GITR by TRAF3-deficient T cells
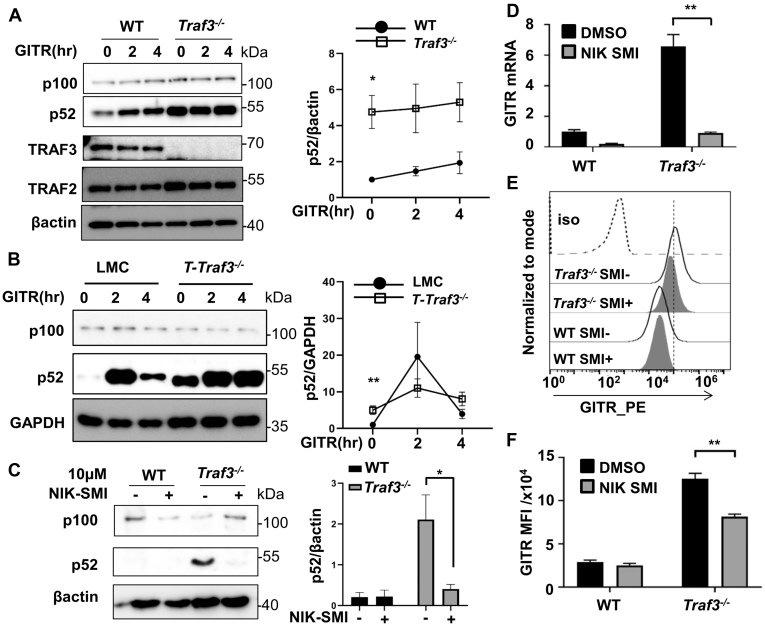
The transcription factors NFκB and FoxP3 regulate GITR transcription (48, 49). Constitutive activation of NFκB2 is seen in all TRAF3-deficient cell types examined to date, including T lymphocytes (50). We thus investigated the role of this elevated NFκB2 in the elevated GITR expression seen in T cells lacking TRAF3. Processing of the precursor protein p100 to p52 was induced following GITR stimulation in TRAF3-sufficient 2B4 cells and primary mouse splenic T cells. Both 2B4 cells and primary T cells lacking TRAF3 exhibited constitutively elevated p52, as previously reported (38, 51) (Fig. 3, A and B). NFκB2 activation occurs when receptors such as CD40 recruit cytoplasmic TRAF3, together with associated TRAF2 and cIAP, to the receptor. TRAFs 2 and 3 are then themselves polyubiquitinated and degraded (37). This releases the NFκB inducing kinase (NIK), normally constitutively targeted for degradation by this complex, to activate the NFκB2 pathway (37). Interestingly, long-term GITR stimulation also led to degradation of TRAF3 in T cells (Fig. S2, A and B). We considered the possibility that elevated NFκB2 activation mediated by NIK contributed to enhanced GITR expression in T cells lacking TRAF3. To test this, we used the NIK-specific kinase inhibitor NIK-SMI, an ATP competitive SMI. NIK-SMI has IC50 values of ∼40 nM for mouse NIK and ∼23 nM for human NIK and has limited off-target activities (52). Treatment of 2B4 T cells with NIK-SMI markedly inhibited the processing of p100 to p52 in both TRAF3-sufficent and TRAF3-deficient subclones (Fig. 3C). Following treatment with NIK-SMI, GITR mRNA was markedly decreased in both cell types as well (Fig. 3D). In the presence of NIK-SMI, GITR protein on the surface of TRAF3-deficient 2B4 cells was also reduced significantly (Fig. 3, E and F). Interestingly, NIK inhibition reduced the mRNA levels of GITR in Traf3−/− 2B4 cells to a level comparable to TRAF3-sufficient T cells, but only partially reduced surface protein levels of GITR in Traf3−/− T cells, which remained well above those in WT cells (Fig. 3, E and F). Collectively, these results suggest that constitutive basal activation of NFκB2 associated with TRAF3 deficiency in T cells contributes to enhanced GITR expression, but other factors including reduced GITR turnover also contribute to the observed enhancement of GITR protein in the absence of TRAF3.
Figure 3.
Role of noncanonical NFκB2 activation in enhanced GITR expression by TRAF3-deficient T cells.A, WT or Traf3−/− 2B4 T cells were stimulated via GITR as described in Experimental procedures, for the indicated times. Whole cell lysates were subjected to Western blot analysis for the indicated proteins, as described in Experimental procedures. Quantification (n = 3) of band intensity of p52 normalized to loading control (βactin) is shown on the right. B, total splenic T cells from the indicated mouse strains were stimulated via GITR as in (A), for the indicated times. Protein levels of p100 and p52 in whole cell lysates were determined by Western blot. Quantification (n = 5) of band intensity of p52 that is normalized to loading control (GAPDH) is shown on the right. C, inhibition of constitutive NFκB2 p100 processing. WT or Traf3−/− 2B4 cells were incubated for 2 h with 10 μM NIK-SMI, a small-molecule inhibitor of NIK kinase, prior to processing for Western blotting as described above. Quantification (n = 3) of band intensity of p52 normalized to loading control (GAPDH) is shown on the right. For A–C, ∗∗p < 0.01, and ∗p < 0.05 by unpaired t test. D, GITR mRNA levels of cells treated as in (C). In D, error bars represent mean ± SEM of values of one representative of three experiments. y-axis shows 2−ΔΔCt value of GITR mRNA as normalized to GAPDH. ∗∗p < 0.01 by unpaired t test. N = 12. E and F, inhibition of enhanced cell surface expression of GITR in TRAF3-deficient 2B4 T cells by NIK-SMI. WT or TRAF3-deficient 2B4 cells were treated with 10 μM NIK SMI for 2 h, prior to staining for GITR and analysis by flow cytometry. MFI ± SEM (n = 3) for GITR is shown in (F). A representative experiment is shown in (E). ∗∗p < 0.01 by unpaired t test.
Role of nuclear TRAF3 in T cell GITR expression
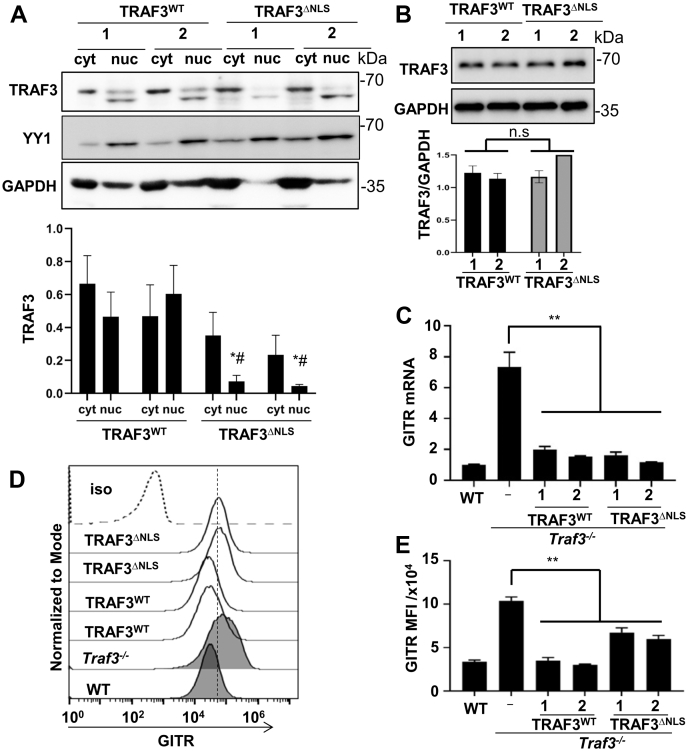
We previously reported that TRAF3 is constitutively present in the B cell nucleus, and nuclear TRAF3 specifically inhibits CREB-mediated transcription of prosurvival genes in B cells (53). We thus examined the potential role played by nuclear TRAF3 in regulating T cell GITR expression. As in B cells, 2B4 T cells have TRAF3 constitutively localized in their nuclei (Fig. 4A). To determine the importance of this nuclear TRAF3 in GITR expression, we transfected Traf3−/− 2B4 cells with either WT TRAF3 (TRAFWT) or TRAF3 with a defective nuclear localization sequence (NLS) that we previously produced (TRAF3ΔNLS) (53). Figure 4B shows that expressions of these two forms of TRAF3 were similar in all subclones tested. TRAF3ΔNLS had reduced nuclear localization in transfected T cells, compared with WT TRAF3 (Fig. 4A). However, TRAF3WT and TRAF3ΔNLS both restored normal levels of GITR mRNA in TRAF3-deficient T cells (Fig. 4C). Thus, robust localization of TRAF3 to the nucleus is not required for its inhibition of GITR mRNA levels. Interestingly, surface expression of GITR in T cells expressing TRAF3ΔNLS was intermediate between the levels seen in WT T cells and TRAF3−/− T cells (Fig. 4, D and E). The NLS mutation changes the sequence RDYKRRKQ to RDYARRAQ in the TRAF domain of TRAF3 (53). It may be that this mutation, in addition to altering the localization of TRAF3, alters the ability of TRAF3 to mediate turnover of GITR protein.
Figure 4.
Role of nuclear TRAF3 in suppression of GITR expression.A, subcellular distribution of WT TRAF3 and TRAF3 ΔNLS in 2B4 T cells. Whole-cell lysates from subclones of Traf3−/− 2B4 cells stably expressing either WT TRAF3 (TRAF3WT) or ΔNLS TRAF3 (TRAF3ΔNLS) were separated into nuclear (marker YY1), and cytosolic (marker GAPDH) fractions and subjected to Western blot analysis. For A–E, results using two individual subclones from the indicated stable transfections are shown. Quantification (n = 5) of normalized band intensity of TRAF3 variants is shown at the bottom of a representative Western blot. ∗p < 0.05 versus stable transfectant line 1 of TRAF3WT; #p < 0.05 versus stable transfectant line 2 of TRAF3WT. B, TRAF3 protein expression in whole cell lysates of two TRAFWT and two TRAF3ΔNLS 2B4 subclones, analyzed by Western blot. Quantification (n = 4) of normalized band intensity of TRAF3 variants is shown at the bottom of a representative Western blot. n.s indicates no significant difference by unpaired t test. C, relative GITR mRNA levels, 2−ΔΔCt value of GITR mRNA as normalized to GAPDH, were determined in TRAF3WT and TRAF3ΔNLS 2B4 subclones by quantitative RT-PCR. Graph depicts mean ± SEM, ∗∗p < 0.01 by unpaired t test. N = 10. One representative result of three similar experiments is shown. D and E, cell surface expression of GITR in TRAF3WT and TRAF3ΔNLS 2B4 subclones (two of each type, as in A–C), was determined by GITR staining and flow cytometry analysis. Iso = isotype control Ab staining of the parent Traf3−/− 2B4 strain. Parental 2B4 is indicated as “WT,” untransfected Traf3−/− 2B4 cells as “−,” and the two subclones of each type of transfected Traf3−/− 2B4 cells are shown for comparison. Results in (D) are representative of three similar experiments. The mean ± SEM of mean fluorescence intensity (MFI) values from (D) are presented in (E) (three experiments. ∗∗p < 0.01 by unpaired t test).
Impact of TRAF3 on GITR-mediated signaling in T cells
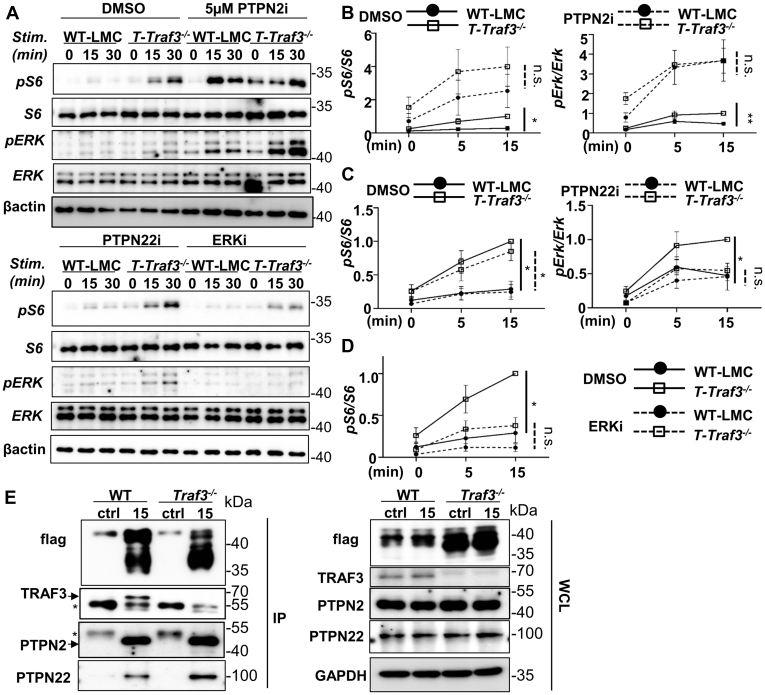
GITR signaling is essential to maintain normal T cell responses during adaptive immune responses (46). Several major signaling pathways, including the NFκB pathway, MAPK cascades, and the mTOR-AKT-S6 kinase axis are activated upon GITR stimulation (2, 10, 54, 55). We found that GITR-induced activation of ERK p44/42 at T202/Y204, phosphorylation of the NFκB1 inhibitory protein IκBα, and phosphorylation of ribosomal protein S6 at S235/236, the terminal event of mTOR-AKT pathway (10, 56) were markedly elevated in TRAF3-deficient total T cells (Fig. 5, A–C). We observed a trend in increased phosphorylation of AKTT308 at later time points, although this did not reach statistical significance (Fig. 5A). Because phosphorylation of S6 can also be activated through the MAPK cascade (57), and because GITR-mediated ERK activation was markedly enhanced in TRAF3-deficient cells, we asked if ERK was responsible for the increased phosphorylation of S6 in TRAF3-deficient T cells. We thus treated splenic-T cells with a small molecule drug targeting MEK1/2 (U0126, labeled as ERKi) (58). ERKi completely abolished phosphorylation of Erk and partially reduced the phosphorylation of S6 to a level not significantly different from that observed in TRAF3-sufficient T cells (Fig. 6, A and D). This suggests that GITR-activated S6 phosphorylation is in part mediated by ERK and regulated by TRAF3 in T cells.
Figure 5.
Impact of TRAF3 on GITR signaling to T cells.A, splenic T cells isolated from littermate control (LMC) or T-Traf3−/− mice were stimulated via GITR for the indicated times. Whole cell lysates were subjected to Western blot analysis as described in Experimental procedures. Results are representative of three similar experiments. B and C, quantification of three independent experiments with three pairs of animals of each of the indicated genotypes. Graphs represent change in pS6 (pS235/236) normalized to total S6 (B) and pERK (pT202/Y204) normalized to total ERK (C) over the course of stimulation via GITR. Curves represent mean ± SEM of relative band intensities, where statistical significance is determined by two-way ANOVA. ∗∗p < 0.01.
Figure 6.
The roles of phosphatases PTPN2 and PTPN22 in restraining GITR signaling.A, splenic T cells isolated from littermate control (LMC) or T-Traf3−/− mice were pretreated with DMSO (upper-left), inhibitors (5 μM) of PTPN2 (SF1670, upper-right), PTPN22 (LTV-1, bottom-left) or ERK (U0126) (bottom-right) before being stimulated via GITR for the indicated times. Whole cell lysates were subjected to SDS-PAGE and Western blot analysis. Representative blots of four similar experiments are shown. B–D, quantification of four independent experiments including representative blots shown in A. Curves represent mean ± SEM of relative band intensities, where statistical significance was determined by two-way ANOVA. ∗p < 0.05; ∗∗p < 0.01; n.s indicates no significant difference. E, subclones of HuT28.11 or Traf3−/− HuT28.11 T cells expressing hCD40-GITR (FLAG-tagged) were stimulated via anti-hCD40 Ab-conjugated protein G beads (see Experimental procedures) for times indicated and the hCD40-GITR signaling complex was immunoprecipitated. IPs (left) were subjected to SDS-PAGE and Western blotting for TRAF3, PTPN2 and PTPN22. Blots of whole cell lysates are shown on the right. Ctrl = IP with protein G beads conjugated with isotype control mAb. Positions of TRAF3 and PTPN2 bands are indicated by arrows. ∗ indicates the heavy chain of the IP Ab remaining in the IP sample. Blots in E are representative of ≥ 3 similar experiments. Quantification of band intensity is summarized in Fig. S4.
We also used a complementary method to measure early GITR-mediated signaling events in the human CD4+ T cell line HuT28.11, which does not express endogenous GITR. As described in Experimental procedures, we stably transfected WT and TRAF3-deficient subclones of HuT28.11 with a hybrid human (h)CD40-GITR receptor, which can be engaged with agonistic anti-hCD40 mAb. Figure S3, A and B demonstrates that the TRAF3-deficient subclones had markedly enhanced GITR-mediated ERK activation, as well as a trend toward enhanced phosphorylation of S6 kinase. These data support an important role for T cell TRAF3 in restraining GITR-mediated early signaling events.
Involvement of phosphatases PTPN2 and PTPN22 in regulation of GITR signaling
TRAF3 recruits phosphates PTPN2 (TCPTP) and PTPN22 to inhibit IL-2R and IL-6R-mediated signaling, respectively (42, 43). We thus considered the possibility that TRAF3 normally restrains early GITR signaling events via recruitment of one or both phosphatases. To test the role of phosphatase activity, primary splenic T cells were pretreated with the small-molecule inhibitors LTV-1, targeting PTPN22 (PTPN22i), and SF1670, targeting PTPN2 (PTPN2i) prior to stimulation through GITR. Inhibition of PTPN2 increased phosphorylation of ERK and S6 kinase in both WT and TRAF3-deficienct T cells (Fig. 6, A and B). This suggests that while PTPN2 acts to restrain GITR-mediated ERK phosphorylation and activation of ribosomal protein S6, it does not account for the specifically enhanced GITR activated signal seen in TRAF3-deficient T cells. In contrast, inhibition of PTPN22 in splenic T cells resulted in no obvious changes in phosphorylation of S6 and ERK (Fig. 6, A and C). Previously, we showed both PTPN2 and PTPN22 directly interact with TRAF3 in T cells (40, 42). However, association of PTPN2 and PTPN22 with GITR was not disrupted in the absence of TRAF3 (Fig. 6E; Fig. S4). These data indicate that GITR associates with PTPN2 and PTPN22 in T cells, independently of TRAF3.
Impact of TRAF3 upon GITR association with signaling molecules
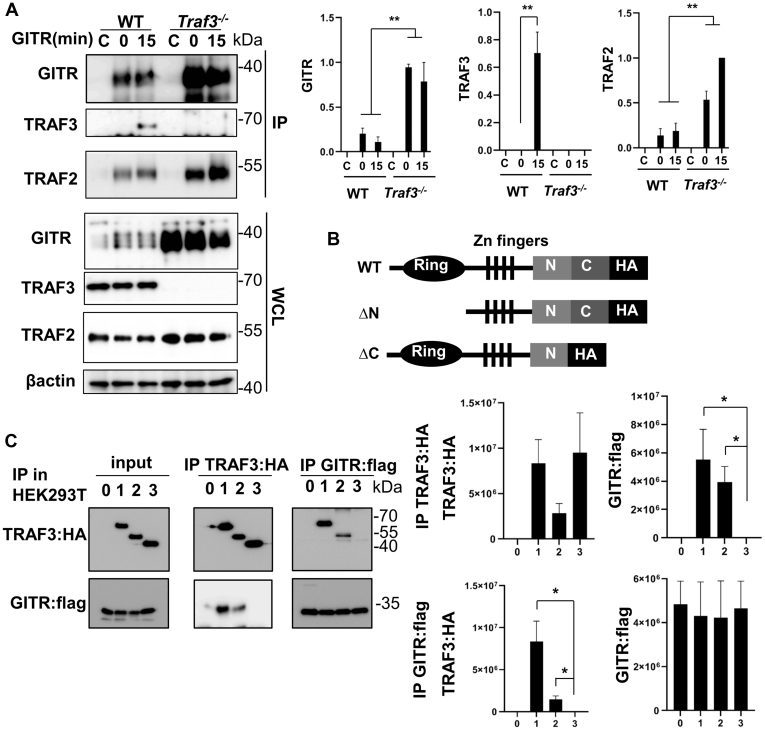
Overexpression models in nonimmune cells showed that TRAFs 2/3/5 interact with GITR (14, 34). Additionally, TRAF2 and TRAF5 induce GITR-mediated NFκB activation in CD8+ T cells (59). To determine how TRAF3 impacts composition of the T cell GITR signaling complex, we immunoprecipitated the receptor complex in 2B4 T cells, finding that TRAF3 interacts with GITR upon its stimulation (Fig. 7A). In contrast, TRAF3, unlike TRAF2, does not associate with GITR in resting 2B4 T cells. As shown in earlier figures, TRAF3-deficient T cells also express more GITR protein, reflected here in the immunoprecipitated complex; this increased total GITR likely accounts for the increased TRAF2 seen in the immunoprecipitates in Traf3−/− T cells. The ratios of TRAF2:GITR calculated from GITR immunoprecipitates at 15 min after GITR stimulation are not statistically different between WT and TRAF3-deficient T cells (Fig. 7A). We also expressed WT or structural mutants of TRAF3 shown in Figure 7B, together with GITR, by transient transfection of HEK293 T cells. Results showed that TRAF3 and GITR associated in the receptor complex (Fig. 7C). This interaction required the TRAFC domain, but not the RING domain of TRAF3 (Fig. 7, B and C). Taken together, these results clearly indicate that TRAF3 interacts with the GITR intracellular domain upon GITR engagement.
Figure 7.
Association of GITR, TRAF3, and TRAF2 in T cells.A, WT or Traf3−/− 2B4 cells were stimulated via anti-GITR Ab-conjugated protein G beads for times indicated, after which GITR signaling complexes were immunoprecipitated. C = control IP, where protein G beads conjugated with only the secondary anti-rat IgG Ab were used to IP GITR in unstimulated 2B4 cells, as described in Experimental procedures. IP samples were subjected to SDS-PAGE, and association of TRAF2 and TRAF3 with GITR was assessed by Western blot. Quantification (n = 3) of normalized band intensity of each protein is shown to the right of a representative figure. B, HA-tagged TRAF3 constructs used to transfect HEK293 cells for structure–function analysis. Zn Ring, Zn fingers, TRAF-N (N), and TRAFC (C) domains are shown. C, HEK293 T cells were transiently transfected with plasmids of the following combinations: Flag-tagged GITR plasmid alone (0) or + (1) WT TRAF3 (2), ΔN TRAF3, or (3) ΔC TRAF3. Cell lysates were immunoprecipitated with Abs to HA (TRAF3) or Flag (GITR), and association of GITR with TRAF3 mutants was assessed by Western blot. Protein expression in input lysates (each is 6% of total lysate) is also shown. Quantification of band intensity of indicated proteins is shown to the right of a representative figure. N = 3 for TRAF3:HA IP; N = 4 for GITR:flag IP. For both A and C, ∗∗p < 0.01, and ∗p < 0.05 by unpaired t test.
Discussion
GITR is an important TNFRSF member critical for full T cell activation subsequent to initial TCR stimulation (46). GITR is expressed constitutively on conventional naïve and memory T cells and is rapidly upregulated upon TCR activation (2, 15). High levels of GITR are found on Treg, likely as a result of transcriptional activation by Foxp3 (48). GITR is costimulatory to both CD4+ and CD8+ T cells (10, 59) and important to effective T cell control of chronic LCMV infection (10). GITR signaling stimulates IFNγ production, enhanced expression of IL-2R and CD69, and promotes hyperproliferation and survival of CD4+ Th1 cells, CD8+ effector cells, and Treg (2, 10, 12, 19, 54, 55, 60). Similar to other TNFRSF members, GITR lacks intrinsic enzymatic activity and utilizes TRAF2 and 5 to promote signaling (18, 32). However, the role of TRAF3 in GITR signaling was previously undefined.
The multiple functions of TRAF3 are often context-dependent; TRAF3 has both cell-type-specific roles, as well as distinct roles in regulating the function of different types of receptors, even in the same cell type (35, 61). For example, TRAF3 inhibits CD40 signals in B cells (62, 63), but is a positive regulator of signaling mediated in B cells by the Epstein–Barr virus-encoded CD40 mimic Latent Membrane Protein 1 (64). In T cells, TRAF3 enhances TCR complex signaling and IL-15R signals, while restraining IL-2R signaling (38, 42, 44). Therefore, knowledge about the regulatory mechanisms of TRAF3 cannot be completely extrapolated from one cell type or receptor to another. In this study we reveal that TRAF3 clearly inhibits the function of the TNFR superfamily costimulator GITR, via multiple mechanisms.
This study introduces a new role for T cell TRAF3: control of the surface levels of a TNFRSF member (Fig. 8). TRAF3 deficiency led to abnormal elevation of GITR expression at both mRNA and protein levels in CD4 and CD8 T cells. The 5′ region upstream of the coding regions of the Tnfrsf18 locus contains an NFκB consensus binding site (49, 65). Our results demonstrate that enhanced expression of GITR protein is at least partially due to the constitutive activation of NFκB2 in TRAF3-deficient T cells. Unlike the many context-dependent alterations in cell phenotype and functions caused by TRAF3 deficiency, constitutive NFκB2 activation is common to all TRAF3-deficient cell types (50). However, GITR upregulation in TRAF3−/− T cells was not completely suppressed by NIK inhibition, suggesting that other unknown TRAF3-regulated factors may contribute to this phenotype. An additional mechanism contributing to enhanced GITR levels in TRAF3-deficient T cells is suggested by reduced GITR protein turnover rates in Traf3−/− 2B4 T cells. To date, TRAF3 per se was not reported to have validated E3 ubiquitin ligase activity in lymphocytes under physiological circumstances (66, 67). Future study is needed to examine if polyubiquitination or other posttranslational modifications of GITR are regulated by TRAF3 in T cells. Although nuclear TRAF3 plays an important role in regulating certain survival pathways in B cells (53), and we observed here that TRAF3 can also localize to the T cell nucleus, GITR expression was normalized by restoration of TRAF3, even mutant TRAF3 that is mostly excluded from the nucleus.
Figure 8.
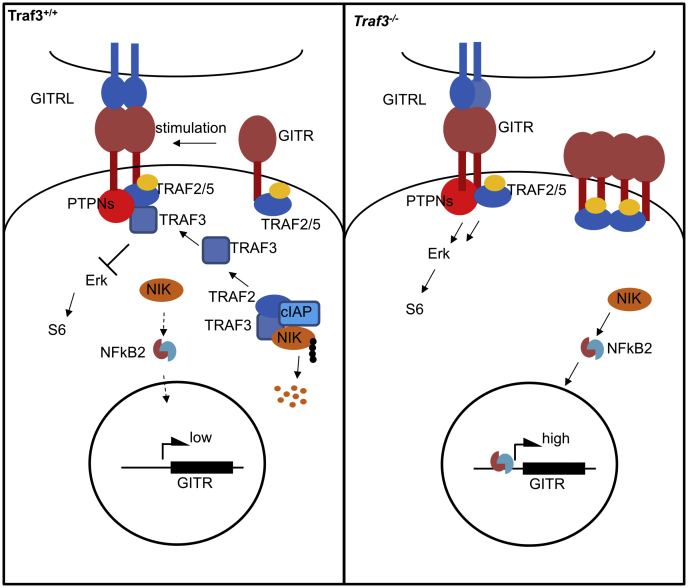
Model: TRAF3 suppresses GITR expression and signaling in T cells. GITR transcription is stringently regulated by activity of NFκB2. In normal T cells (left panel), the major upstream kinase for activation of NFκB2, NIK is constitutively polyubiquitinated (represented as chain of black dots attached to NIK) and targeted for degradation by an E3 ubiquitination complex containing TRAF2, TRAF3, and cIAP. Engagement of GITR with GITRL leads to recruitment of TRAF3 and disassembly of the TRAF2/TRAF3/cIAP E3 complex, which releases NIK and activates downstream NFκB2. In addition, phosphatases PTPN2 and PTPN22 attenuate the signaling strength of the GITR complex independent of TRAF3. Negative regulation of GITR signaling by TRAF3 may also involve competition with TRAF2 and TRAF5 (yellow and blue ovals), both of which can promote GITR signaling (59). In the absence of TRAF3 (right panel), NFκB2 is constitutively active and upregulates GITR transcription. Increased levels of the GITR receptor and lack of TRAF3 regulation of the GITR signaling cascade lead to elevated GITR signaling in TRAF3-deficient T cells.
We also analyzed responsiveness of TRAF3-deficient T cells to GITR engagement independent of TCR stimulation and observed that TRAF3 attenuates GITR signaling, in particular, activation of MAP kinases and the ribosomal protein S6 signaling axis. Results presented here reveal TRAF3 as the first TRAF family member shown to restrain GITR signaling (Fig. 8). In contrast, TRAF2 and TRAF5 are reported to be required for maximal GITR signaling in CD8+ T cells (59). In 2B4 T cells, TRAF2 constitutively associates with GITR independent of GITR ligation status. In contrast, TRAF3 is recruited to the receptor upon stimulation. A similar constitutive association is observed for TRAF5 in CD4+ HuT28.11 T cells. In 2B4 T cells, it is hard to assess possible competition between TRAF3 and TRAF2/5 to associate with the TRAF-binding motif of GITR, because the increased total GITR likely contributes to the increased TRAF2 seen in coimmunoprecipitates in TRAF3-deficient T cells. We introduced ectopic expression of human GITR in HuT28.11 cells, in which no endogenous GITR is expressed. While comparable GITR amounts are observed in TRAF3 sufficient and deficient HuT28.11 transfectants, no obvious increased association of TRAF2/5 with GITR is seen in the absence of TRAF3. This tends to argue against simple displacement of other TRAFs by TRAF3 at the receptor. The specific TRAF3-regulated inhibitory mechanism of the early GITR signaling cascade thus remains to be identified in future studies.
TRAF3 inhibits IL-2R signaling in T cells, promoting Treg differentiation (42), and suppresses signaling mediated by the IL-6R and the B cell antigen receptor in B cells (43, 68). Signaling through IL-2 and IL-6 receptors is tightly regulated by TRAF3 recruited phosphatases, PTPN2 and PTPN22, respectively (39). Additionally, TRAF3 inhibits association of PTPN22 with the TCR complex (40). We observed that PTPN2 but not PTPN22 restrained GITR-mediated phosphorylation of MAP kinase and ribosomal protein S6. However, these phosphatases associated with GITR independent of the TRAF3 status of the T cells.
A potential important future question is how the increased GITR levels of TRAF3-deficient T cells might impact reverse signaling through GITRL in cells that form immune synapses with T cells. It has been shown that GITRL reverse signals enhance primary NK cell cytotoxicity and IFNγ production (69) and modulate activation and migration of monocytes/macrophages (70). Future efforts will be directed to address this question. Immunotherapeutic designs based on GITR are currently gaining interest, due to the versatile functions of GITR in various subsets of T cells and in macrophages, NK, and B cells (71, 72, 73). Our characterization of TRAF3's effects of restraining GITR expression and signaling is relevant to understanding how GITR itself is regulated in participating in T cell functions, including design of therapeutic approaches that are based upon GITR functions.
Experimental procedures
Mice
The T-Traf3−/− mice were created by crossing Traf3flox/flox mice (74), backcrossed >10 generations to C57B1/6 mice, to CD4-Cre mice (40, 42). Their LMCs are generated from the same cross. Adult mice (2–4 months) were used for all experiments, with similar numbers of male and female mice. Mice were housed in specific pathogen-free conditions and used in accordance with NIH guidelines, under a protocol approved by the Animal Care and Use Committee, the University of Iowa, Iowa City, IA.
DNA constructs
To construct the human GITR (hGITR) expressing plasmid, hGITR cDNA was initially amplified from a cDNA library constructed from human PBMC. The primers used were: hGITR_sense: 5′ ATG GCA CAG CAC GGG GCG ATG 3′, and hGITR_antisense: 5′ TCA CAC CCA CAG GTC TCC CAG 3′. We added a Kozak sequence and restriction enzyme sites by PCR, using the primer pair hGITR_HindIII_kozak_S: 5′ AAA AAA GCT TGC CGC CAC CAT GGC ACA GCA CGG GGC GAT G 3′ and hGITR_XbaI_CAS: TAT ATC TAG ATT CAC TTG TCG TCA TCG TCT TTG TAG TCC ACC CAC AGG TCT CCC AGC CG. The PCR product was then digested with restriction enzymes HindIII and XbaI and ligated to linearized pcDNA3.1 vector digested with HindIII and XbaI. To construct the hCD40-GITR hybrid receptor expressing plasmid, one fragment containing the extracellular domain of hCD40 was amplified by PCR with the primer pair hCD40_HindIII_kozak_S: 5′ AAA AAA GCT TGC CGC CAC CAT GGT TCG TCT GCC TCT GCA G 3′, and hCD40_ex_antisense: 5′ TCT CAG CCG ATC CTG GGG AC 3′. A fragment containing the sequence encoding the transmembrane and cytosolic domains of human GITR was amplified with a sense primer that partially overlaps with hCD40_ex_antisense at its 5′ end, hGITR_TM_CD40linker_S: 5′ GTC CCC AGG ATC GGC TGA GAC TTG GGT GGC TGA CCG TCG TC 3′, and the anti-sense primer contains a 5′ sequence that encodes FLAG tag (DYKDDDDK): hGITR_XbaI_CAS: 5′ TAT ATC TAG ATT CAC TTG TCG TCA TCG TCT TTG TAG TCC ACC CAC AGG TCT CCC AGC CG 3′. These two fragments were then joined by a second round of PCR, using the primer pair hCD40_HindIII_Kozak_S and hGITR_XbaI_CAS. The final PCR product was digested with HindIII and XbaI restriction enzymes and ligated to linearized pcDNA3.1 vector as above. To produce the hCD40-GITR expressing plasmid in which expression is driven by the RSV promoter, the hCD40-GITR insert was subcloned, via addition of HindIII and XbaI restriction enzyme sites, into pRSV.5(neo) (75) or pRSV.5∗.puro (a modified version of pRSV.5(neo), in which the neomycin resistance cassette was replaced with a puromycin resistance gene). The plasmid pOPRSV.mTRAF3 encodes full-length mouse TRAF3, while the variant pOPRSV.mTRAF3ΔNLS encodes mouse TRAF3 with a mutant NLS sequence; both contain a 3xFLAG epitope tag and have been previously described (53). pcDNA.TRAF3.HA encodes full-length TRAF3 with an HA tag, pcDNA.ΔN_TRAF3 encodes TRAF3 lacking the N-terminal RING domain, and pcDNA.ΔC_TRAF3 encodes TRAF3 lacking the C-terminal TRAFC domain; these constructs and PTPN22_R-flag and PTPN22-W-flag expressing plasmids were described in (40). PTPN2-TC45 and PTPN2-TC48 were previously described in (42).
Cell lines
2B4 is a mouse hybridoma T cell line, originally generated by fusing splenocytes from a C57B10.A mouse with the mouse AKR strain-derived thymoma BW5147 (76). HuT28.11, a subclone of the human CD4+ T cell line HuT78 transfected to stably express CD28 (77), was the gift of Dr Arthur Weiss, University of California, San Francisco. Both 2B4 and HuT28.11 cells were cultured in RPMI 1640 medium supplemented with 100 U/ml penicillin, 100 U/ml streptomycin, 2 mM L-glutamine, 10 μM β-mercaptoethanol, and 10% fetal calf serum (FCS) (BCM10). Subclones of Traf3−/− 2B4 cells (see below) with stably transfected full-length TRAF3 or ΔNLS TRAF3 were cultured in BCM10 with 400 μg/ml G418. HuT28.11 was transfected with hCD40-GITR in pRSV5∗.puro. Clones were selected by puromycin resistance and cultured in BCM10 with 2 μg/ml puromycin.
Production of Traf3−/− 2B4 subclones using CRISPR/cas9
Guide RNA/cas9 vector constructs for disruption of the Traf3 gene (NCBI gene ID:22031) were designed according to (78), using the CRISPR design tool (crispr.mit.edu) maintained by Dr Feng Zhang (MIT, Cambridge, MA). Two pX330 (Addgene ID 42230) based constructs were made targeting exon 3 and exon 6, respectively. In total, 2.5 μg of each of these targeting constructs was then transfected into 1 × 106 2B4 cells, together with the plasmid pEGFP-C1 that constitutively expresses EGFP, via electroporation (225V for 30 ms, BTX square wave electroporator). GFP-expressing cells were sorted at 1 cell/well into 96-well plates 5 days after transfection. Clones containing the desired deletion were identified by PCR, and the loss of TRAF3 protein was confirmed by Western blot.
HEK293T cell transfection
HEK293T cells (1 × 106/ml) were grown overnight in DMEM supplemented with 100 U/ml penicillin, 100 U/ml streptomycin, 2 mM L-glutamine, 10 μM β-mercaptoethanol, and 10% FCS (DMEM10). Prior to transfection, DMEM was replaced with Opti-MEM medium (Thermo Fisher) for 30 min. Cells were transfected with plasmids indicated in the figure legends, using a lipofectamine 2000 transfection kit (Thermo Fisher), according to the manufacturer's protocol. In total, 6–8 h later, medium was removed and fresh DMEM10 was added. Two days later, cells were washed with PBS and lysed with lysis buffer (0.5% TritonX-100, 100 mM NaCl, 40 mM Tris[pH7.5], 1 mM MgCl2, 1 mM CaCl2, 0.05 mg/ml DNase added prior to use, and one EDTA-free mini-complete protease inhibitor tablet [Roche]/10 ml) for 10 min on ice. Cell lysates were collected, and an immunoprecipitation was performed on some samples, as detailed below.
Antibodies and reagents
Antibodies (Abs) used in immunoblotting included anti-HA (HA-7, #128M4846), anti-flag (M2), and the anti-mouse actin purchased from Sigma. The anti-ERK (T202/204, 9101), anti-ERK (9102), anti-S6 (S235/236, 91B2), anti-S6 (5610), anti-pIkBα (S32, 14D4), anti-IkBα (9242), anti-pAKT (S473, D9E), anti-pAKT (T308, 244F9), anti-AKT (9272), anti-PTEN (138G6), anti-NFκB2 (4882), anti-PTPN22 (D6D1H) and anti-human TRAF5 (D3E2R) Abs were purchased from Cell Signaling Technology. The anti-TRAF3 (H122), anti-YY1 (H-10), and anti-GAPDH (D-6) Abs were from Santa Cruz Biotechnology, and the anti-TRAF2 (MBL592) Ab was purchased from MBL International. The anti-mouse GITR (AF524) and anti-PTPN2 (MAB1930) Abs were purchased from R&D Systems. HRP-conjugated goat-anti-mouse IgG and goat-anti-rabbit Ig Abs were from Jackson ImmunoResearch Laboratories. Abs used for stimulation include anti-mouse CD3 (145-2C11), anti-CD28 (37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51) (Thermo Fisher), and anti-mouse GITR (DTA-1) (BioXCell). Anti-human CD40 (G28.5, mouse IgG1) was produced in our lab from a hybridoma obtained from the ATCC. The isotype control mAb (iso) for G28.5, MOPC31c, was purchased from Sigma. For immunoprecipitations (IPs), 10 μg of anti-GITR Ab (DTA-1, rat) or 10 μg of anti-CD40 Ab (G28.5, mouse) was conjugated to 10 μl of protein G beads via 10 μg of goat-anti-rat or goat-anti-mouse IgG antibodies, respectively. All fluorescently labeled conjugates used for flow cytometry were purchased from eBioscience or BioLegend. Cycloheximide and the MAPK pathway inhibitor (U0126) were purchased from Sigma. The small-molecule inhibitors for PTPN2 (SF1670) and PTPN22 (LTV-1) were purchased from Tocris Bioscience. The NIK inhibitor, SMI, was graciously provided by Genentech.
Immunoprecipitation
Ten million cells per time point were stimulated with anti-GITR Ab-conjugated magnetic protein G beads (Dynabeads, Invitrogen) for times indicated in the figures, before being pelleted by centrifugation at 700g for 2 min. Cell pellets were then lysed by resuspending in 800 μl lysis buffer, and the lysate was incubated on ice for 10 min. Fifty microliter of each cell lysate was saved as an input control. Using a magnet, beads were washed five times with lysis buffer. For unstimulated samples (labeled as 0 min), cells were lysed in 800 μl lysis buffer before adding the same amount of agonistic Ab-conjugated protein G beads. The IP negative control (labeled as C or ctrl) was performed with unstimulated cells using protein G beads conjugated with isotype control antibodies. Samples were then subjected to SDS-PAGE and Western blot analysis, as previously described (40).
GITR stimulation
Cells (0.5 million/time point) were aliquoted and prewarmed in a 37 °C water bath, before being stimulated with soluble anti-GITR (DTA-1) agonistic Ab at a final concentration of 10 μg/ml, as previously described (79). After stimulation, all samples were put on ice for 5 min and pelleted by centrifugation at 1000g for 2 min. Supernatants were then removed, and cells were resuspended in 100 μl of SDS-PAGE sample buffer. All samples were sonicated to fragment genomic DNA before analysis by SDS-PAGE, followed by Western blot analysis. Imaging of Western blots was performed with a chemiluminescent substrate (Supersignal Pico) and a low-light imaging system (LAS-4000, Fujifilm). Quantitation of band intensity was done with MultiGauge (Fujifilm Software Co)
Flow cytometry
Primary mouse splenic T cells were isolated using a Pan T cell negative selection kit or a CD4+ T cell isolation kit (Stem Cell Technologies). Primary T cells or 2B4 cells were stained with fluorescently labeled Abs against GITR, CD4, CD8 (53–6.7), or FoxP3 (FJK-16S). For intracellular staining, cells were fixed and permeabilized with a fixation/permeabilization kit (BD Cytofix/Cytoperm, BD Biosciences). All flow cytometry and cell sorting were performed with Accuri C6 (BD Bioscience) in our own lab, or LSRII with UV (BD Bioscience), and Aria Fusion cell sorter (BD Bioscience) at the University of Iowa Flow Cytometry Facility, which is a Carver College of Medicine/Holden Comprehensive Cancer Center core research facility.
GITR turnover rates measurement
Analysis of protein stability was measured by the cycloheximide chase assay, slightly modified from (80). Parental WT or Traf3−/− 2B4 cells were treated with a DMSO solvent control or 30 μg/ml cycloheximide for 4 h, before being stained for surface GITR expression with PE conjugated anti-GITR mAb. Relative levels of cell surface GITR were measured by flow cytometry.
RT-qPCR
RNA was extracted from 2 million cells/sample using the RNeasy Mini Kit (Qiagen). One microgram of total RNA per sample was used in the reverse transcription reaction using the iScript reverse transcription kit (Bio-Rad). For real-time PCR, the primer set for GITR was as previously described (81). For GAPDH, the sense primer was 5′ TCCACCACCCTGTTGCTGTA 3′, and the antisense primer was 5′ ACCAGAGTCCATGCCATCAC 3′. qPCR was carried out in the Genomics Core at the University of Iowa, Iowa City, IA, using a SYBR green qPCR kit (Applied Biosystems). Gene expression was quantitated relative to the expression of GAPDH.
Subcellular fractionation
To isolate nuclear and cytoplasmic fractions (53), ten million T cells were incubated in 200 μl of hypotonic buffer (10 mM Hepes pH 7.9, 10 mM KCl, 0.1 mM EDTA, 5 mM MgCl2) supplemented by 1 mM DTT and a protease inhibitor tablet (Roche) for 30 min. Swollen cells were lysed with 0.1% Triton X-100 and vortexed for 20s, before being centrifuged at 13,000g for 10 min at 4 °C. The cytoplasmic fraction was carefully removed to another tube, and an equal volume of 2× SDS sampling buffer was added. The nuclear pellet was resuspended in 200 μl of hypertonic buffer (20 mM Hepes pH7.9, 0.4 M NaCl, 1 mM EDTA) supplemented with 1 mM DTT, and a protease inhibitor tablet (Roche), and incubated for 30 min. An equal volume of 2× SDS sampling buffer was then added to the resuspended nuclear fraction.
Statistical analysis
All statistical analysis was performed using GraphPad Prism software. Unpaired t test was used when comparing two groups. Two-way ANOVA was used when comparing quantification of bands in Western blots. Data were considered statistically significantly different with p < 0.05.
Data availability
All data needed to evaluate conclusions of this study are contained within the manuscript or the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
G. A. B. is a senior research career scientist of the VAMC. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors acknowledge the services of the Flow Cytometry and Genomics Cores, which receive support from NIH P30 CA086862. This work was also supported by the Holden Comprehensive Cancer Center at The University of Iowa and its National Cancer Institute Award P30CA086862. This material is based upon work supported in part by facilities and equipment provided by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development.
Author contributions
B. S. H., T. A., and G. A. B., conceptualization; H. L., data curation; H. L., B. S. H., T. A., and G. A. B., formal analysis; G. A. B., funding acquisition; H. L. and T. A., investigation; B. S. H., methodology; G. A. B., project administration; T. A., resources; G. A. B., supervision; G. A. B., validation; H. L., writing–original draft; B. S. H. and G. A. B., writing–review and editing.
Funding and additional information
This work was supported by NIH R01 AI123107 to G. A. B. T. A. received support from NIH T32 awards AI007485 and GM007337. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Peter Cresswell
Supporting information
References
- 1.Ronchetti S., Ricci E., Petrillo M.G., Cari L., Migliorati G., Nocentini G., Riccardi C. Glucocorticoid-induced TNFR-related protein: A key marker of functional regulatory T cells. J. Immunol. Res. 2015;2015:171520. doi: 10.1155/2015/171520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanamaru F., Youngnak P., Hashiguchi M., Nishioka T., Takahashi T., Sakaguchi S., Ishikawa I., Azuma M. Costimulation via glucocorticoid-induced TNFR in both conventional and CD25+ regulatory CD4+ T cells. J. Immunol. 2004;172:7306–7314. doi: 10.4049/jimmunol.172.12.7306. [DOI] [PubMed] [Google Scholar]
- 3.Schaer D.A., Murphy J.T., Wolchok J.D. Modulation of GITR for cancer immunotherapy. Curr. Opin. Immunol. 2012;24:217–224. doi: 10.1016/j.coi.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nocentini G., Ronchetti S., Petrillo M.G., Riccardi C. Pharmacological modulation of GITRL/GITR system: Therapeutic perspectives. Br. J. Pharmacol. 2012;165:2089–2099. doi: 10.1111/j.1476-5381.2011.01753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu J., Yamazaki S., Takahashi T., Ishida Y., Sakaguchi S. Stimulation of CD25+ CD4+ regulatory T cells through GITR breaks immunological self-tolerance. Nat. Immunol. 2002;3:135–142. doi: 10.1038/ni759. [DOI] [PubMed] [Google Scholar]
- 6.Liu B., Li Z., Mahesh S.P., Pantanelli S., Hwang F.S., Siu W.O., Nussenblatt R.B. Glucocorticoid-induced TNFR negatively regulates activation of human primary natural killer (NK) cells by blocking proliferative signals and increasing NK cell apoptosis. J. Biol. Chem. 2008;283:8202–8210. doi: 10.1074/jbc.M708944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Batista N.V., Chang Y.-H., Chu K.-L., Wang K.C., Girard M., Watts T.H. T Cell–intrinsic CX3CR1 marks the most differentiated effector CD4+ T cells, but is largely dispensable for CD4+ T cell responses during chronic viral infection. ImmunoHorizons. 2020;4:701–712. doi: 10.4049/immunohorizons.2000059. [DOI] [PubMed] [Google Scholar]
- 8.Fabian K.P., Malamas A.S., Padget M.R., Solocinski K., Wolfson B., Fujii R., Sater H.A., Schlom J., Hodge J.W. Therapy of established tumors with rationally designed multiple agents targeting diverse immune-tumor interactions: Engage, expand, enable. Cancer Immunol. Res. 2020;9:239–252. doi: 10.1158/2326-6066.CIR-20-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim H.J., Kim H.Y., Kim B.K., Kim S., Chung D.H. Engagement of glucocorticoid-induced TNF receptor costimulates NKT cell activation in vitro and in vivo. J. Immunol. 2006;176:3507–3515. doi: 10.4049/jimmunol.176.6.3507. [DOI] [PubMed] [Google Scholar]
- 10.Clouthier D.L., Zhou A.C., Wortzman M.E., Luft O., Levy G.A., Watts T.H. GITR intrinsically sustains early type 1 and late follicular helper CD4 T cell accumulation to control a chronic viral infection. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nocentini G., Riccardi C. GITR: A modulator of immune response and inflammation. Ther. Targets TNF Superfamily. 2009;647:156–173. doi: 10.1007/978-0-387-89520-8_11. [DOI] [PubMed] [Google Scholar]
- 12.Tone M., Tone Y., Adams E., Yates S.F., Frewin M.R., Cobbold S.P., Waldmann H. Mouse glucocorticoid-induced tumor necrosis factor receptor ligand is costimulatory for T cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15059–15064. doi: 10.1073/pnas.2334901100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shevach E.M., Stephens G.L. The GITR–GITRL interaction: Co-stimulation or contrasuppression of regulatory activity? Nat. Rev. Immunol. 2006;6:613–618. doi: 10.1038/nri1867. [DOI] [PubMed] [Google Scholar]
- 14.Kwon B., Yu K.-Y., Ni J., Yu G.-L., Jang I.-K., Kim Y.-J., Xing L., Liu D., Wang S.-X., Kwon B.S. Identification of a novel activation-inducible protein of the TNFR superfamily and its ligand. J. Biol. Chem. 1999;274:6056–6061. doi: 10.1074/jbc.274.10.6056. [DOI] [PubMed] [Google Scholar]
- 15.Gurney A., Marsters S., Huang A., Pitti R., Mark M., Baldwin D., Gray A., Dowd P., Brush J., Heldens S. Identification of a new member of the TNF family and its receptor, a human ortholog of mouse GITR. Curr. Biol. 1999;9:215–218. doi: 10.1016/s0960-9822(99)80093-1. [DOI] [PubMed] [Google Scholar]
- 16.Kim J., Choi B., Bae J., Lee U., Han I., Lee H., Youn B., Vinay D., Kwon B. Cloning and characterization of GITR ligand. Genes Immun. 2003;4:564–569. doi: 10.1038/sj.gene.6364026. [DOI] [PubMed] [Google Scholar]
- 17.Kwon B., Youn B.-S., Kwon B.S. Functions of newly identified members of the TNFR/ligand superfamilies in lymphocytes. Curr. Opin. Immunol. 1999;11:340–345. doi: 10.1016/s0952-7915(99)80054-5. [DOI] [PubMed] [Google Scholar]
- 18.Snell L.M., Lin G.H., McPherson A.J., Moraes T.J., Watts T.H. T-cell intrinsic effects of GITR and 4-1BB during viral infection and cancer immunotherapy. Immunol. Rev. 2011;244:197–217. doi: 10.1111/j.1600-065X.2011.01063.x. [DOI] [PubMed] [Google Scholar]
- 19.Ronchetti S., Nocentini G., Bianchini R., Krausz L.T., Migliorati G., Riccardi C. Glucocorticoid-induced TNFR-related protein lowers the threshold of CD28 costimulation in CD8+ T cells. J. Immunol. 2007;179:5916–5926. doi: 10.4049/jimmunol.179.9.5916. [DOI] [PubMed] [Google Scholar]
- 20.Ephrem A., Epstein A.L., Stephens G.L., Thornton A.M., Glass D., Shevach E.M. Modulation of T reg cells/T effector function by GITR signaling is context–dependent. Eur. J. Immunol. 2013;43:2421–2429. doi: 10.1002/eji.201343451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacHugh R., Piccirilo C., Young D., Shevach E., Collins M., Byrne C. CD4CD25 immunoregulatory T cells: Gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity. 2002;16:311–323. doi: 10.1016/s1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 22.Rosenzweig M., Ponte J., Apostolou I., Doty D., Guild J., Slavonic M., Ponath P., Vaickus L. Development of TRX518, an aglycosyl humanized Mab agonist of huGITR. J. Clin. Oncol. 2010;28 [Google Scholar]
- 23.Sukumar S., Wilson D.C., Yu Y., Wong J., Naravula S., Ermakov G., Riener R., Bhagwat B., Necheva A.S., Grein J. Characterization of MK-4166, a clinical agonistic antibody that targets human GITR and inhibits the generation and suppressive effects of T regulatory cells. Cancer Res. 2017;77:4378–4388. doi: 10.1158/0008-5472.CAN-16-1439. [DOI] [PubMed] [Google Scholar]
- 24.Tigue N.J., Bamber L., Andrews J., Ireland S., Hair J., Carter E., Sridharan S., Jovanović J., Rees D.G., Springall J.S. MEDI1873, a potent, stabilized hexameric agonist of human GITR with regulatory T-cell targeting potential. Oncoimmunology. 2017;6 doi: 10.1080/2162402X.2017.1280645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez A.M., Manrique M., Swiech L., Horn T., Waight J., Liu Y., Lin S., Underwood D., Breous E., Leger O. American Association for Cancer Research Annual Meeting. 2017. INCAGN1876, a unique GITR agonist antibody that facilitates GITR oligomerization. [Google Scholar]
- 26.Ko K., Yamazaki S., Nakamura K., Nishioka T., Hirota K., Yamaguchi T., Shimizu J., Nomura T., Chiba T., Sakaguchi S. Treatment of advanced tumors with agonistic anti-GITR mAb and its effects on tumor-infiltrating Foxp3+ CD25+ CD4+ regulatory T cells. J. Exp. Med. 2005;202:885–891. doi: 10.1084/jem.20050940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen A.D., Schaer D.A., Liu C., Li Y., Hirschhorn-Cymmerman D., Kim S.C., Diab A., Rizzuto G., Duan F., Perales M.A. Agonist anti-GITR mAb induces melanoma tumor immunity in mice by altering regulatory T cell stability and intra-tumor accumulation. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turk M.J., Guevara-Patiño J.A., Rizzuto G.A., Engelhorn M.E., Houghton A.N. Concomitant tumor immunity to a poorly immunogenic melanoma is prevented by regulatory T cells. J. Exp. Med. 2004;200:771–782. doi: 10.1084/jem.20041130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golubovskaya V., Berahovich R., Xu Q., Zhou H., Xu S., Guan J., Harto H., Li L., Wu L. GITR domain inside CAR co-stimulates activity of CAR-T cells against cancer. Front. Biosci. 2018;23:2245–2254. doi: 10.2741/4703. [DOI] [PubMed] [Google Scholar]
- 30.Kintz H., Nylen E., Barber A. Inclusion of Dap10 or 4-1BB costimulation domains in the chPD1 receptor enhances anti-tumor efficacy of T cells in murine models of lymphoma and melanoma. Cell. Immunol. 2020;351:104069. doi: 10.1016/j.cellimm.2020.104069. [DOI] [PubMed] [Google Scholar]
- 31.Xi B., Berahovich R., Zhou H., Xu S., Wei Y., Guan J., Harto H., Guan J., Wu L., Santa Ana D. A real-time potency assay for chimeric antigen receptor T cells targeting solid and hematological cancer cells. JoVE. 2019;153 doi: 10.3791/59033. [DOI] [PubMed] [Google Scholar]
- 32.Chattopadhyay K., Lazar-Molnar E., Yan Q., Rubinstein R., Zhan C., Vigdorovich V., Ramagopal U.A., Bonanno J., Nathenson S.G., Almo S.C. Sequence, structure, function, immunity: Structural genomics of costimulation. Immunol. Rev. 2009;229:356–386. doi: 10.1111/j.1600-065X.2009.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esparza E., Lindsten T., Stockhausen J., Arch R. TRAF5 is a critical intermediate of costimulatory signaling pathways triggered by GITR in T cells. J. Biol. Chem. 2006;281:8559–8564. doi: 10.1074/jbc.M512915200. [DOI] [PubMed] [Google Scholar]
- 34.Hauer J., Püschner S., Ramakrishnan P., Simon U., Bongers M., Federle C., Engelmann H. TRAF3 serves as an inhibitor of TRAF2/5-mediated activation of the noncanonical NF-κB pathway by TRAF-binding TNFRs. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2874–2879. doi: 10.1073/pnas.0500187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bishop G.A. TRAF3 as a powerful and multitalented regulator of lymphocyte functions. J. Leuk. Biol. 2016;100:919–926. doi: 10.1189/jlb.2MR0216-063R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallabhapurapu S., Karin M. Regulation and function of NF-κB transcription factors in the immune system. Ann. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 37.Sun S.-C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie P., Kraus Z.J., Stunz L.L., Liu Y., Bishop G.A. TRAF3 is required for T cell-mediated immunity and TCR/CD28 signaling. J. Immunol. 2011;186:143–155. doi: 10.4049/jimmunol.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wallis A.M., Bishop G.A. TRAF3 regulation of inhibitory signaling pathways in B and T lymphocytes by kinase and phosphatase localization. J. Leuk. Biol. 2018;103:1089–1098. doi: 10.1002/JLB.2MIR0817-339RR. [DOI] [PubMed] [Google Scholar]
- 40.Wallis A.M., Wallace E.C., Hostager B.S., Yi Z., Houtman J.C., Bishop G.A. TRAF3 enhances TCR signaling by regulating the inhibitors Csk and PTPN22. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-02280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang J.-H., Hu H., Jin J., Puebla-Osorio N., Xiao Y., Gilbert B.E., Brink R., Ullrich S.E., Sun S.-C. TRAF3 regulates the effector function of regulatory T cells and humoral immune responses. J. Exp. Med. 2014;211:137–151. doi: 10.1084/jem.20131019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yi Z., Lin W.W., Stunz L.L., Bishop G.A. The adaptor TRAF3 restrains the lineage determination of thymic regulatory T cells by modulating signaling via the receptor for IL-2. Nat. Immunol. 2014;15:866–874. doi: 10.1038/ni.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin W.W., Yi Z., Stunz L.L., Maine C.J., Sherman L.A., Bishop G.A. The adaptor protein TRAF3 inhibits IL-6 receptor signaling in B cells to limit plasma cell development. Sci. Signaling. 2015;8 doi: 10.1126/scisignal.aaa5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yi Z., Stunz L.L., Bishop G.A. TRAF3 plays a key role in development and function of invariant NK T cells. J. Exp. Med. 2013;210:1079–1086. doi: 10.1084/jem.20122135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McHugh R.S., Whitters M.J., Piccirillo C.A., Young D.A., Shevach E.M., Collins M., Byrne M.C. CD4+ CD25+ immunoregulatory T cells: Gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity. 2002;16:311–323. doi: 10.1016/s1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 46.Watts T.H. TNF/TNFR family members in costimulation of T cell responses. Annu. Rev. Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 47.Vinay D.S., Kwon B.S. TNF superfamily: Costimulation and clinical applications. Cell Biol. Int. 2009;33:453–465. doi: 10.1016/j.cellbi.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tone Y., Kidani Y., Ogawa C., Yamamoto K., Tsuda M., Peter C., Waldmann H., Tone M. Gene expression in the Gitr locus is regulated by NF-κB and Foxp3 through an enhancer. J. Immunol. 2014;192:3915–3924. doi: 10.4049/jimmunol.1302174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhan Y., Gerondakis S., Coghill E., Bourges D., Xu Y., Brady J.L., Lew A.M. Glucocorticoid-induced TNFR expression by T cells is reciprocally regulated by NF-κB and NFAT. J. Immunol. 2008;181:5405–5413. doi: 10.4049/jimmunol.181.8.5405. [DOI] [PubMed] [Google Scholar]
- 50.Lin W.W., Hildebrand J.M., Bishop G.A. A complex relationship between TRAF3 and non-canonical NF-κB2 activation in B lymphocytes. Front. Immunol. 2013;4:477. doi: 10.3389/fimmu.2013.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardam S., Sierro F., Basten A., Mackay F., Brink R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 2008;28:391–401. doi: 10.1016/j.immuni.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Brightbill H.D., Suto E., Blaquiere N., Ramamoorthi N., Sujatha-Bhaskar S., Gogol E.B., Castanedo G.M., Jackson B.T., Kwon Y.C., Haller S. NF-κB inducing kinase is a therapeutic target for systemic lupus erythematosus. Nat. Comm. 2018;9:1–14. doi: 10.1038/s41467-017-02672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mambetsariev N., Lin W.W., Stunz L.L., Hanson B.M., Hildebrand J.M., Bishop G.A. Nuclear TRAF3 is a negative regulator of CREB in B cells. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1032–1037. doi: 10.1073/pnas.1514586113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ronchetti S., Zollo O., Bruscoli S., Agostini M., Bianchini R., Nocentini G., Ayroldi E., Riccardi C. Frontline: GITR, a member of the TNFR superfamily, is costimulatory to mouse T lymphocyte subpopulations. Eur. J. Immunol. 2004;34:613–622. doi: 10.1002/eji.200324804. [DOI] [PubMed] [Google Scholar]
- 55.Esparza E.M., Arch R.H. Glucocorticoid-induced TNFR, a costimulatory receptor on naive and activated T cells, uses TRAF2 in a novel fashion as an inhibitor of NF-κB activation. J. Immunol. 2005;174:7875–7882. doi: 10.4049/jimmunol.174.12.7875. [DOI] [PubMed] [Google Scholar]
- 56.Owonikoko T.K., Khuri F.R. Targeting the PI3K/AKT/mTOR pathway: Biomarkers of success and tribulation. Am. Soc. Clin. Oncol. Education. 2013;33:e395–e401. doi: 10.1200/EdBook_AM.2013.33.e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frödin M., Gammeltoft S. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell. Endocrinol. 1999;151:65–77. doi: 10.1016/s0303-7207(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 58.Favata M.F., Horiuchi K.Y., Manos E.J., Daulerio A.J., Stradley D.A., Feeser W.S., Van Dyk D.E., Pitts W.J., Earl R.A., Hobbs F. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 59.Snell L.M., McPherson A.J., Lin G.H., Sakaguchi S., Pandolfi P.P., Riccardi C., Watts T.H. CD8 T cell-intrinsic GITR is required for T cell clonal expansion and mouse survival following severe influenza infection. J. Immunol. 2010;185:7223–7234. doi: 10.4049/jimmunol.1001912. [DOI] [PubMed] [Google Scholar]
- 60.Stephens G.L., McHugh R.S., Whitters M.J., Young D.A., Luxenberg D., Carreno B.M., Collins M., Shevach E.M. Engagement of glucocorticoid-induced TNFR family-related receptor on effector T cells by its ligand mediates resistance to suppression by CD4+ CD25+ T cells. J. Immunol. 2004;173:5008–5020. doi: 10.4049/jimmunol.173.8.5008. [DOI] [PubMed] [Google Scholar]
- 61.Bishop G.A., Stunz L.L., Hostager B.S. TRAF3 as a multifaceted regulator of B lymphocyte survival and activation. Front. Immunol. 2018;9:2161. doi: 10.3389/fimmu.2018.02161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hostager B.S., Bishop G.A. Cutting edge: Contrasting roles of TRAF2 and TRAF3 in CD40-activated B lymphocyte differentiation. J. Immunol. 1999;162:6307–6311. [PubMed] [Google Scholar]
- 63.Haxhinasto S.A., Hostager B.S., Bishop G.A. Cutting edge: Molecular mechanisms of synergy between CD40 and the B cell antigen receptor: Role for TRAF2 in receptor interaction. J. Immunol. 2002;169:1145–1149. doi: 10.4049/jimmunol.169.3.1145. [DOI] [PubMed] [Google Scholar]
- 64.Xie P., Hostager B.S., Bishop G.A. Requirement for TRAF3 in signaling by LMP1 but not CD40 in B lymphocytes. J. Exp. Med. 2004;199:661–671. doi: 10.1084/jem.20031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nocentini G., Ronchetti S., Bartoli A., Spinicelli S., Delfino D., Brunetti L., Migliorati G., Riccardi C. Identification of three novel mRNA splice variants of GITR. Cell Death Diff. 2000;7:408–410. doi: 10.1038/sj.cdd.4400670. [DOI] [PubMed] [Google Scholar]
- 66.Moore C.R., Bishop G.A. Differential regulation of CD40-mediated TRAF degradation in B lymphocytes. J. Immunol. 2005;175:3780–3789. doi: 10.4049/jimmunol.175.6.3780. [DOI] [PubMed] [Google Scholar]
- 67.Lin W.W., Hostager B.S., Bishop G.A. TRAF3, ubiquitination, and B-lymphocyte regulation. Immunol. Rev. 2015;266:46–55. doi: 10.1111/imr.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whillock A.L., Ybarra T.K., Bishop G.A. TRAF3 restrains B cell receptor signaling in normal and malignant B cells. J. Biol. Chem. 2021;296:100465. doi: 10.1016/j.jbc.2021.100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hanabuchi S., Watanabe N., Wang Y.-H., Wang Y.-H., Ito T., Shaw J., Cao W., Qin F.X.-F., Liu Y.-J. Human plasmacytoid predendritic cells activate NK cells through GITRL. Blood. 2006;107:3617–3623. doi: 10.1182/blood-2005-08-3419. [DOI] [PubMed] [Google Scholar]
- 70.Bae E.M., Kim W.-J., Suk K., Kang Y.-M., Park J.-E., Kim W.Y., Choi E.M., Choi B.K., Kwon B.S., Lee W.-H. Reverse signaling initiated from GITRL induces NF-κB activation through ERK in the inflammatory activation of macrophages. Mol. Immunol. 2008;45:523–533. doi: 10.1016/j.molimm.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 71.Bae E., Kim W.J., Kang Y.M., Suk K., Koh E.M., Cha H.S., Ahn K.S., Huh T.L., Lee W.H. Glucocorticoid-induced TNFR-related protein-mediated macrophage stimulation may induce cellular adhesion and cytokine expression in rheumatoid arthritis. Clin. Exp. Immunol. 2007;148:410–418. doi: 10.1111/j.1365-2249.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim W.J., Bae E.M., Kang Y.J., Bae H.U., Hong S.H., Lee J.Y., Park J.E., Kwon B.S., Suk K., Lee W.H. GITR mediates inflammatory activation of macrophages that can destabilize atherosclerotic plaques. Immunology. 2006;119:421–429. doi: 10.1111/j.1365-2567.2006.02453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teodorovic L.S., Riccardi C., Torres R.M., Pelanda R. Murine B cell development and antibody responses to model antigens are not impaired in the absence of the TNF receptor GITR. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie P., Stunz L.L., Larison K.D., Yang B., Bishop G.A. TRAF3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity. 2007;27:253–267. doi: 10.1016/j.immuni.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Long E., Rosen-Bronson S., Karp D., Malnati M., Sekaly R., Jaraquemada D. Efficient cDNA expression vectors for stable and transient expression of HLA-DR in transfected fibroblast and lymphoid cells. Hum. Immunol. 1991;31:229–235. doi: 10.1016/0198-8859(91)90092-n. [DOI] [PubMed] [Google Scholar]
- 76.Ashwell J.D., Cunningham R.E., Noguchi P.D., Hernandez D. Cell growth cycle block of T cell hybridomas upon activation with antigen. J. Exp. Med. 1987;165:173–194. doi: 10.1084/jem.165.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stein P.H., Fraser J.D., Weiss A. The cytoplasmic domain of CD28 is both necessary and sufficient for costimulation of IL-2 secretion and association with phosphatidylinositol 3'-kinase. Mol. Cell. Biol. 1994;14:3392–3402. doi: 10.1128/mcb.14.5.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bauer D.E., Canver M.C., Orkin S.H. Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J. Vis. Exp. 2015 doi: 10.3791/52118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nowakowska D.J., Kissler S. Ptpn22 modifies regulatory T cell homeostasis via GITR upregulation. J. Immunol. 2016;196:2145–2152. doi: 10.4049/jimmunol.1501877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kao S.-H., Wang W.-L., Chen C.-Y., Chang Y.-L., Wu Y.-Y., Wang Y.-T., Wang S.-P., Nesvizhskii A.I., Chen Y.-J., Hong T.-M. Analysis of protein stability by the cycloheximide chase assay. Bio-protocol. 2015;5:e1374. doi: 10.21769/BioProtoc.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santucci L., Agostini M., Bruscoli S., Mencarelli A., Ronchetti S., Ayroldi E., Morelli A., Baldoni M., Riccardi C. GITR modulates innate and adaptive mucosal immunity during the development of experimental colitis in mice. Gut. 2007;56:52–60. doi: 10.1136/gut.2006.091181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate conclusions of this study are contained within the manuscript or the supporting information.