Abstract
Progress has been made in understanding coeliac disease, a relatively frequent and underappreciated immune-mediated condition that occurs in genetically predisposed individuals. However, several gaps remain in knowledge related to diagnosis and management. The gluten-free diet, currently the only available management, is not curative or universally effective (some adherent patients have ongoing duodenal injury). Unprecedented numbers of emerging therapies, including some with novel tolerogenic mechanisms, are currently being investigated in clinical trials. In March 2020, the Celiac Disease Foundation and the Society for the Study of Celiac Disease convened a consensus workshop to identify high-yield areas of research that should be prioritized. Workshop participants included leading experts in clinical practice, academia, government and pharmaceutical development, as well as representatives from patient support groups in North America. This Roadmap summarizes key advances in the field of coeliac disease and provides information on important discussions from the consensus approach to address gaps and opportunities related to the pathogenesis, diagnosis and management of coeliac disease. The morbidity of coeliac disease is often underestimated, which has led to an unmet need to improve the management of these patients. Expanded research funding is needed as coeliac disease is a potentially curable disease.
Subject terms: Coeliac disease, Coeliac disease
Coeliac disease is a serious condition and a model disease that can shed light into the mechanisms that underlie autoimmunity and chronic inflammation. This Roadmap summarizes key advances in coeliac disease and provides recommendations from a consensus workshop to address the gaps and opportunities in the pathogenesis, diagnosis and management of coeliac disease, providing a path forward.
Key points
Coeliac disease is a common and serious medical condition that is under-recognized among the health-care provider community, government and the public.
This Roadmap summarizes consensus recommendations to address gaps and opportunities in pathogenesis, diagnosis and management of coeliac disease.
Various animal models are available to translate hypotheses generated from human studies, and progress is being made in the development of physiological coeliac epithelial models based on organoid technology.
Coeliac-specific serology is highly reliable for the diagnosis of coeliac disease; however, there is disagreement between experts as to the necessity of intestinal biopsy to confirm the diagnosis.
There is increasing need for development of programmes for proper clinical management of coeliac disease, and the number of potential therapeutic targets and clinical trials has grown exponentially over the past 15 years.
Increased funding for coeliac disease research is crucial to improve clinical management and facilitate development of therapies for this condition.
Introduction
Coeliac disease, an immune-mediated enteropathy triggered by gluten, is a relatively common condition affecting ~1% of the world population that can develop across the lifespan1,2. The incidence has been increasing rapidly over the past four decades worldwide3. A diagnosis of coeliac disease is associated with increased overall mortality4. Currently, the primary management for coeliac disease is a strict gluten-free diet, which is practically challenging and socially difficult to follow5. Thus, patients often have inadequate symptom control on a gluten-free diet, yet the few alternative therapies available are repurposed medications, such as budesonide, that are not disease specific.
To establish priorities for the support of relevant and high-yield areas of research, the Celiac Disease Foundation (CDF) and the Society for the Study of Celiac Disease (SSCD) convened the first SSCD consensus workshop on ‘Research Opportunities in Celiac Disease’ in March 2020 (ref.6). The consensus workshop was attended by leaders from academia and pharmaceutical companies, patient support group members and scientists from government institutions (NIH) that support research in coeliac disease. The consensus workshop was organized under the umbrella of SSCD and CDF. Workshop participants were identified and invited based on their area of expertise. A workshop outline was determined a priori, and each expert was asked to address specific topics. A round-table discussion followed each presentation. After the workshop, participants ranked the initiatives in a formal voting process (25 participants submitted votes for each topic), to determine prioritization. The complete list of attendants of the workshop in addition to the authors is detailed in the Acknowledgments. The authors of this article were selected by the SSCD to report on the discussion and recommendations arising from the workshop.
This Roadmap summarizes the latest advances and controversies in the field (Table 1) and provides information on selected milestones and consensus recommendations to address gaps and opportunities in pathogenesis and genetics (Table 2), and diagnosis, management and prevention of coeliac disease (Table 3). Recognized gaps in epidemiology are related to the unknown reason for the increasing incidence of coeliac disease and the prevalence of coeliac disease outside North America and Europe. Major questions have emerged about who to screen for coeliac disease, when and how to confirm the diagnosis. In terms of pathogenesis, although research has yielded various animal models of coeliac disease, there remain substantial gaps in understanding how gluten tolerance is lost and tissue destruction ensues. Gaps in disease management are also identified, as research has shown that patients are not able to maintain a strict gluten-free diet owing to the ubiquitous presence of gluten in the food supply. There is also a concern regarding persistent low diagnostic rates and poor understanding of the complications related to coeliac disease within the medical community and broader population that can be addressed through further education and development of stronger partnerships between patients, clinicians and academic researchers.
Table 1.
Selected milestones and gaps in coeliac disease
| Area of interest | Milestones | Challenges | Opportunities | Readiness |
|---|---|---|---|---|
| Pathogenesis | Key pathways revealed by animal models (MHC class II HLA-DQ2 or HLA-DQ8 and transgenic for IL-15) | Limited understanding of pathways of disease tolerance and tissue destruction, as well as different phenotypes of the disease |
Current knowledge of mechanisms generates opportunities in translational research Animal and organoid models are useful to investigate mechanisms for biomarker and drug discovery |
Tools available |
| Diagnosis | Once thought to afflict primarily children of Irish descent, coeliac disease is a common condition affecting nearly 1% of the worldwide population, and the incidence is increasing3 |
The reason for increasing incidence over time is unclear There are insufficient data to support mass screening There is disagreement on the need for duodenal biopsy to confirm coeliac disease |
Clinical trials on high-risk population using novel approaches such as HLA-DQ–gluten tetramer assays or IL-2 release to measure immune response | Tools available |
| Disease management | Novel tools to improve detection of gluten to improve disease management |
GFD is difficult to follow and a great proportion of those with coeliac disease remain symptomatic Histological recovery is inconsistent with clinical presentation and coeliac disease-specific serology Lack of coeliac disease academic research centres and experts No approved disease-specific therapy |
Identification of markers of preclinical disease and development of more accurate tools to assess disease activity, which could be tested in preclinical models Continuing education of community in coeliac disease Development of pharmacological therapies |
Needs development |
| Funding | Research efforts in coeliac disease have proved highly efficient, leading to a better understanding of the disease | Funding for research is lower than for other less prevalent conditions, such as Crohn’s disease55 | Increasing funding would catalyse and sustain coeliac disease research centres, supporting patients and generating new knowledge | Insufficient |
GFD, gluten-free diet.
Table 2.
Selected milestones and current and future research objectives in pathogenesis and genetics
| Mechanism or feature (time frame) | Milestones | Current and future research objectives |
|---|---|---|
| Cereal protein chemistry and genetics (1900s–) | Osborne66 characterized cereal grain proteins as ‘prolamins’. Starting with wheat α-gliadin, protein and gene sequencing provided key information for defining candidate toxic and/or antigenic gluten peptides67,68 | Refining understanding of a safe threshold for dietary gluten; improving food testing for gluten; breeding cereals with reduced immunogenicity; defining other cereal components that trigger symptoms; understanding gluten digestion, absorption and systemic handling in health and disease |
| Environmental antigens (1950s–) | Protease-resistant peptides from prolamins in wheat, rye, barley and, possibly, oats69 drive coeliac disease | More comprehensive molecular characterization of targets for gluten-driven immunity in all genetic subtypes of coeliac disease; triggers for disease onset; microbiome and infections as modifiers of gluten immunity and tissue injury |
| Genetic susceptibility (1970s–) | Starting with HLA associations70 and defining HLA-DQ2.5 as the primary genetic susceptibility, subsequent genome-wide studies identify multiple non-HLA linkages and genes in common with other HLA-linked autoimmune diseases such as type 1 diabetes mellitus71 | Precise localization and functional characterization of germline non-MHC genes and any additional MHC genes implicated in coeliac disease and gluten immunity; understanding gene–gene interactions, DNA modification and acquired T cell and B cell receptor mutations facilitating gluten immunity |
| Animal models, ex vivo tissue and in vitro cell culture (1970s–) | Intestinal biopsies, cell lines and clones developed as bioassays for gluten; gluten immunity tested in humanized HLA-transgenic mice from 2000s72 | Refinement of bioengineering, organoid and microfluidic technologies to develop organ-on-a-chip models to complement humanized mouse models |
| Autoimmunity in coeliac disease (1980s–) | Endomysial antibody later determined to recognize tissue transglutaminase 2, the main autoantigen for gluten-dependent autoantibodies in active coeliac disease45 | Molecular characterization of targets for autoimmunity accounting for extraintestinal manifestations; more detailed understanding of humoral and cellular autoimmunity, and its relationship to gluten immunity |
| Host receptor-mediated recognition of gluten (1990s–) | The molecular and cellular basis for recognition of gluten was determined by cloning intestinal gluten-specific CD4+ T cells exclusively from patients with coeliac disease73, which were complemented by HLA-DQ–peptide binding assays, determining epitope restriction elements and, more recently, structural biology studies9,74 | Refining understanding of gluten recognition and contributions of innate and adaptive immunity or other pathways facilitating gluten-mediated intestinal and extraintestinal manifestations of coeliac disease; expanding understanding of antigen-presenting cells including B cells specific for gluten and transglutaminase |
| Immune–intestinal epithelial cell interactions (1990s–) | Identification of crucial innate immune pathways involving IEC–NK cell receptor interactions supported by IL-15 that are conducive to atrophy75 | Characterization of the role of IEC–NK cell receptor interactions across coeliac disease states and further elucidation of signals leading to licensing of cytotoxicity |
| Extraintestinal immune response to dietary gluten (2000s–) | Gluten ingestion drives expansion of peripheral blood gut-homing gluten-reactive CD4+ T cells and CD8+ T cells days later29. Serum cytokines, especially IL-2, are elevated within hours76 | Improved characterization of the sources and effects of gluten-stimulated systemic cytokine release; understanding of the cause and effects of intestinal CD8+ T cell expansion in blood and gut tissue; establishing phenotypic and functional changes in gluten-specific CD4+ T cells |
| Additional environmental triggers (2010s–) | Resurgence of the microbial hypothesis supported by sequencing technology and longitudinal at-risk cohorts. Used in combination with humanized models, microorganisms could influence key mechanisms in coeliac disease (e.g. gluten metabolism, loss of tolerance, molecular mimicry)77 | Continued identification of microorganisms and mechanisms that protect or incite breakdown of tolerance to gluten; identify and validate new microbial therapeutics that modulate pathogenic targets in coeliac disease |
| Gluten peptide-specific activation and modulation of gluten immunity (2010s–) | Systemically administered immunogenic gluten peptides cause acute digestive symptoms and immune activation with subsequent tachyphylaxis11 | Enhanced understanding of natural mechanisms that regulate gluten immunity and disease manifestations; understanding of the basis for potential coeliac disease versus highly symptomatic disease; more complete understanding of molecular events enabling development of neoplasia associated with coeliac disease |
IEC, intestinal epithelial cell; MHC, major histocompatibility complex; NK cell, natural killer cell.
Table 3.
Selected milestones and current and future research objectives in diagnosis, management, therapeutics and prevention of coeliac disease
| Feature (time frame) | Milestone | Research objectives |
|---|---|---|
| Diagnosis | ||
| Established diagnostic markers of coeliac disease (1950s–) |
First descriptions of histological features of gluten enteropathy and subsequent development of capsule biopsy78 for histological diagnosis Advances in serodiagnostics and HLA-DQ genotyping have led to serology-based diagnosis without biopsy36 |
Future studies need to address the need for diagnosis confirmation by biopsy, particularly in adult populations; regular updates of evidence-based diagnostic guidelines informed by large, well-designed clinical trials |
| Novel diagnostic markers of gluten immunity and coeliac disease (2000s–) | Detection of peripheral blood CD4+ T cells enable redefinition of coeliac disease and diagnosis without requiring intestinal biopsy while regularly consuming gluten41 | Well-designed clinical trials of novel diagnostics; development of accurate blood diagnostics that minimize invasive procedures; biomarkers indicative of gluten immunity that do not require sustained re-introduction of gluten |
| Management and therapeutics | ||
| Centres of clinical excellence and research (1950s–) |
Paediatric centres established early, but adult centres for transition of care and the bulk of new diagnoses resulting from increased awareness since the ‘coeliac disease iceberg’ was identified have lagged Misdiagnosis and underdiagnosis of coeliac disease are commonplace, unnecessary use of GFD is widespread and follow-up of patients after diagnosis is inconsistent79 |
More centres for training health-care professionals in all aspects of medical care of coeliac disease are warranted to enhance current standard of care for diagnosis, management and follow-up of patients; centres integrating patient care and translational research in key areas such as immunology, dietetics and therapeutics development will drive future advances in clinical care |
| Monitoring on GFD (1960s) |
Reversal of intestinal injury with strict GFD reported in children having follow-up capsule biopsy80 Follow-up serology poorly sensitive for mucosal injury on GFD81. Debate continues regarding the utility of follow-up biopsy Quantitative histology studies suggest villous atrophy is under-reported in treated, seronegative coeliac disease82,83 Tools that objectively detect gluten immunogenic peptides84 developed in 2000s |
Enhance standard practice for handling and assessing small bowel biopsies to accurately assess villous atrophy; development of molecular markers that quantify mucosal injury; clinical guidelines for integration of new tests such as gluten immunogenic peptides in clinical use and follow-up; development of models of care for specialized dietitian and medical assessment during lifelong GFD |
| Repurposing drugs for coeliac disease (1970s–) | Many immunosuppressive drugs starting with prednisolone shown to reduce gluten-mediated intestinal injury, but undesirable long-term adverse effects deter their widespread use85 | Assessment of pharmaceuticals approved for other indications that target key mediators of gluten-dependent disease and immunity and have acceptable safety profile |
| Drug development specifically for coeliac disease (2000s–) |
‘Druggable’ targets identified: gluten, microorganism–gluten–immune interactions, transglutaminase, epithelial gluten transport, HLA-DQ and gluten-specific immunity53,86 Drug development evolves to include symptom and histological end points87,88 Regulatory approval for treatment of coeliac disease not achieved |
Comprehensive assessment of immune and non-immune drug targets to augment or replace GFD; understand mechanisms and interventions to restore gluten-specific immune tolerance; identify cytokines responsible for pathology; advance understanding of transglutaminase effects on gluten and normal tissues; understand gluten metabolism in vivo |
| Prevention | ||
| Prevention of coeliac disease (2000s–) | Prospective gluten feeding studies in at-risk infants failed to identify a window of opportunity to decrease coeliac disease risk89. Gluten dose and infections and/or microbial changes might affect risk | Development of strategies to reduce the age of diagnosis and overall incidence of coeliac disease; understand potential coeliac disease and progression from health to development of active disease |
GFD, gluten-free diet.
Milestones and gaps in pathogenesis
Coeliac disease is triggered by the ingestion of gluten in a subgroup of genetically predisposed individuals2. Specifically, coeliac disease confirmed by recall response to gluten challenge is currently limited to individuals positive for HLA-DQ2, HLA-DQ8 or, rarely, HLA-DQ7 (ref.7). Gluten-specific CD4+ T cells in coeliac disease are activated by particular peptide fragments derived from gluten proteins and related prolamins present in wheat, barley and rye when they are bound to HLA-DQ2 or HLA-DQ8 (currently no HLA-DQ7 gluten epitopes are defined)8. Deamidation of wild-type gluten in the lamina propria by tissue transglutaminase 2 (TG2) increases binding efficiency to HLA-DQ2 (ref.9) and HLA-DQ8 (ref.10), which preferentially accommodate peptides with negatively charged residues at anchor sites. HLA-DQ–gluten peptide complexes activate CD4+ T cells, leading to the release of pro-inflammatory cytokines and chemokines that favour inflammatory cell infiltration and CD8+ T cell cytotoxic activity that contributes to mucosal damage in coeliac disease11. Patients with coeliac disease can develop typical upper gastrointestinal symptoms, such as nausea and abdominal pain and systemic markers of immune activation within hours of oral or intradermal gluten exposure11. It is clear that both gluten and permissive HLA-DQ genotypes are required for disease development; however, other genetic12 and environmental13 factors (such as enteric infections and high gluten load) also influence disease risk. As such, coeliac disease represents a model condition in gastroenterology and autoimmunity with exquisite pathophysiological insight.
Although studies performed in biopsy samples and human tissue pioneered important discoveries in coeliac disease and continue to be key in the study of its pathogenesis, the development of animal models in coeliac disease has long been an unmet need, in particular to promote and accelerate development and testing of preclinical therapies. Various animal models have been developed that reveal key relevant pathways amenable to diagnostic and therapeutic modulation, as well as additional risk factors that constitute future targets for primary prevention. Most models are based on MHC class II transgenic mice expressing HLA-DQ2 or HLA-DQ8 (refs14,15), as well as transgenic expression of IL-15 (ref.16), a key cytokine in cytotoxic transformation of intraepithelial lymphocytes. Animal models have also revealed the role of microbial factors in the metabolism of the main dietary trigger, gluten, as well as other dietary components (such as the essential amino acid tryptophan) that participate in the activation of immune pathways that can affect coeliac disease risk17–19. Models that combine genetic susceptibility and transgenic expression of IL-15 have been essential to unravel epithelial-cytotoxicity mechanisms and the role of transient viral infection in the breakdown of oral tolerance to gluten20. Although no single mouse model recapitulates all features of human disease, well-characterized humanized mouse models expressing different key features of coeliac disease pathogenesis and disease stages are now available and represent an invaluable research tool.
In vitro models based on organoid technology have now gained attention. As in other intestinal inflammatory disorders, 3D organoids, as well as 2D organoid-derived monolayers from patients with coeliac disease, expressed phenotypic differences compared with those derived from healthy individuals as controls21–23. Advantages of these tools reside on the physiological complexity of the intestinal epithelium compared with classic transformed or primary cell lines. The limitations are common to organoid modelling, including lack of vasculature and immune cells, high cost, technical difficulties and the closed nature of the 3D structure. Chip technology, microfluidics and co-culturing systems could address some of these limitations24,25. However, collectively, the cellular and mouse models developed in the past decade provide a long-awaited milestone that should stimulate translational and preclinical research in coeliac disease.
Challenges
Despite the key milestones achieved, there is limited understanding of pathways of disease tolerance and tissue destruction. This gap in knowledge limits the development of preventive agents, pharmaceutical adjuncts to diet or curative therapies for coeliac disease. Furthermore, one consequence of the lack of prescription therapies and alternatives to a gluten-free diet is that patients might not consistently receive follow-up care26, so the medical community and, to a lesser extent, the research community, is disengaged from patient needs and the burden of disease. In addition, as suspected by many in the research community, several different coeliac disease phenotypes exist that should be better defined, as differing treatments could be required. In particular, improved quantitative assays that enable clinical and immune phenotype to be accurately defined will play a crucial part in future efforts to develop therapeutics, and might also enable coeliac disease to be redefined and diagnosed on the basis of a recall immune response to gluten.
Opportunities
In contrast to other T cell-mediated organ-specific immune diseases, many of which are genetically linked or associated with coeliac disease, researchers can undertake patient-based so-called proof-of-concept studies with outcomes broadly relevant to understanding and treating other conditions. For example, antigen-specific adaptive immunity has a central role in many serious autoimmune diseases27, but coeliac disease is the only one for which molecular determinants of cellular and humoral immunity are confidently known, and tissue from the affected organ can be sampled while exposure to the causative antigen is safely manipulated. Blood and tissue biomarkers of humoral immunity including gliadin and autoantigen (TG2)-specific antibodies, B cells and plasma cells can be assessed, and the gluten-specific CD4+ T cell population can be studied in gut tissue, repeatedly in blood for single-cell analyses or in fresh samples for functional assessments28. A newly characterized human serum cytokine release profile dominated by pronounced, early elevations in the T cell-associated cytokine IL-2 following acute gluten exposure provides an additional biomarker for coeliac disease research and promises to inform diagnostics and therapeutics development11,29. Relevant animal models are available to reverse-translate hypotheses generated from human studies, and progress is being made in the development of physiological coeliac epithelial models based on organoid technology to study variations in gene expression21, microbiota–host interactions22 and immune response to gliadin23. Altogether in vitro systems and the latest mouse models enable understanding of specific pathogenic mechanisms that are difficult to study clinically and for preclinical drug testing. Thus, researchers have developed a unique toolbox for understanding and developing treatment of antigen-specific cell-mediated human disease by focusing their efforts on coeliac disease. In fact, patients with coeliac disease are often affected by other common autoimmune diseases such as type 1 diabetes mellitus30. Future research implementing mechanistic models, including organoids, to selectively modify gluten immunity is likely to be broadly applicable to antigen-specific immunotherapy of autoimmune disease.
Milestones and gaps in diagnosis
By epidemiological standards, coeliac disease is common (affecting nearly 1% of the worldwide population) and has shown increasing incidence over the past several decades3. On the basis of birth cohort studies in Finland and Colorado, USA, there is evidence that disease penetration is deeper than commonly assumed31.
Challenges
Researchers do not know why disease incidence is increasing, although environmental factors including viral infections during childhood or changes in gut microbiota (composition or metabolite production)32, which have led to an increase in many autoimmune and allergic conditions, have been implicated. Despite the combination of high prevalence and low diagnosis, major questions have emerged about who to test for coeliac disease and when. Although screening of first-degree relatives might be feasible, there is insufficient data to determine whether there is a benefit to screening asymptomatic individuals, particularly adults33. The alternative to screening is a case-finding approach, but the optimal set of clinical criteria that should trigger testing has not been established, and there is not a well-defined set of symptoms that increases the probability of a diagnosis of coeliac disease34,35. The primary limitation of case finding, even when well implemented, is that subclinical cases will be missed and, owing to the many protean manifestations of coeliac disease and symptom overlap, diagnosis will be delayed as the condition is overlooked or not considered in the differential diagnosis. The correct balance between the costs and unintended consequences of mass screening and missed opportunities to improve health outcomes by addressing perpetually low coeliac disease diagnosis rates has yet to be found.
There is also disagreement regarding diagnostic standards as to the necessity of intestinal biopsy. A biopsy-free approach for those with markedly elevated serum TG2 IgA levels (>10 times the upper limit of normal of the assay) was delineated by the European Society for Paediatric Gastroenterology, Hepatology and Nutrition (ESGPHAN) in 2020 (ref.36), but this approach has not been formally adopted by paediatric counterparts in the USA, and the more rigid non-biopsy pathway outlined in the ESPGHAN 2012 guideline was not necessarily implemented, even in central Europe37. Guidelines for diagnosis of coeliac disease in adults have continued to advocate for intestinal biopsy5, although a biopsy-avoidant approach was offered by the British Society of Gastroenterology in 2020 in the context of the COVID-19 pandemic38. There is also debate surrounding best practices for gluten challenge in patients already on a gluten-free diet before testing for coeliac disease, how to evaluate patients whose diagnosis of coeliac disease did not include a small intestinal biopsy for eligibility in clinical trials and how to manage these patients during clinical evaluation of non-responsive coeliac disease5,39,40.
Opportunities
The uncertainty regarding whether and how to screen for coeliac disease in average-risk and high-risk individuals provides the clinical equipoise necessary to justify a randomized trial. For feasibility, testing screening strategies should first be planned in high-risk groups such as those with a family history of coeliac disease. Although a non-biopsy approach will remain the subject of ongoing debate, it might be rendered moot if a non-invasive diagnostic method is validated to have high-performance characteristics, such as a simple, HLA-DQ-diagnostic blood-based assay identifying gluten-specific T cells or biomarkers released by T cells in response to gluten in vivo or ex vivo11,41,42. These modern assays are less dependent on gluten avoidance and/or exposure and might ultimately prove to obviate a gluten challenge. Such a test could foreshadow redefinition of coeliac disease as an acquired antigen-specific immune disorder diagnosed by the presence of gluten-specific T cells. With improved alignment of international and paediatric and adult coeliac disease diagnostic approaches, resources would be freed to focus on crucial questions related to treatment outcomes.
Milestones and gaps in management
Major advances in coeliac disease management to date have been identification of the role of gluten in perpetuating the disease43, identification of HLA-DQ2.5 and HLA-DQ8 as the main haplotypes associated with coeliac disease44, and confirmation of TG2 as an auto-antigen and TG2 IgA antibodies as a biomarker of untreated disease45. The capacity to identify gluten peptide sequences in foods and the consensus for 20 ppm as the upper threshold of gluten for a foodstuff to be considered gluten free46 have led to more unified regulations in food processing and labelling worldwide.
Challenges
Studies47,48 show that patients are not able to maintain a strict gluten-free diet owing to the great difficulty of adherence, but, as we are all aware, no alternatives exist as yet. In practice, many patients are lost to follow-up, particularly those who do not adopt or who discontinue a gluten-free diet, so the long-term population outcomes are not well characterized26,49. Nevertheless, it is known that histological recovery (villi and/or gut healing) is inconsistent, and it seems that younger patients recover better than older patients50. Researchers acknowledge that the long-term implications of undiagnosed or undertreated coeliac disease are poorly understood51. There was broad acknowledgement among the workshop attendees that poor understanding of the disease in the medical community and broader population, coupled with misconceptions about indications for a gluten-free diet, has confused many about the seriousness of coeliac disease. On the other hand, the higher cost of gluten-free products, as well as difficulties in accessing an expert coeliac dietitian, are major challenges for the patient community. There was also consensus that the scarcity of coeliac disease academic research centres, particularly in North America (in contrast to the many for inflammatory bowel disease (IBD) and type 1 diabetes mellitus) and the lack of young researchers studying coeliac disease, were major impediments. Initiatives to partner with primary or secondary centres to facilitate the diagnosis and management of patients with coeliac disease, for instance, through more timely and efficient access to endoscopy and dietetic services, would improve diagnosis and clinical management, providing that health-care professionals are educated in the current standards. Quality certification programmes for clinics or health-care professionals involved in the care of patients with coeliac disease, recognized by scientific societies, can help to achieve this milestone (coeliac disease centres). The dearth of pharmaco-economic data52 has led to an underappreciation of disease burden by employers, insurers and government, and has also severely limited the development of treatments.
Opportunities
There is increasing interest and support from industry for development programmes in coeliac disease, and the number of potential therapeutic targets and clinical trials has grown exponentially over the past 15 years53,54. In addition to the much-needed therapies in coeliac disease, there is a need to improve non-pharmacological management of coeliac disease through optimization of the only currently available management — the gluten-free diet. This progress can be achieved by providing financial support to patients; intensifying education efforts in the patient and medical community and the food industry; improving access to a coeliac dietitian, which requires training programmes and patient access through coverage of these services; implementation of new monitoring tools in food, urine and stool gluten immunogenic peptide detection tests in standard of care. New biomarkers (such as IL-2 and circulating gluten-specific T cells11,29,41) could both simplify the diagnosis, especially when the individual is already on a gluten-free diet, and reduce the burden of study procedures in drug trials in coeliac disease.
Milestones and gaps in funding
Research in coeliac disease has long been a case of accomplishing much with little. Consistent with the authors’ anecdotal experience, systematic review of NIH funding has shown that coeliac disease receives proportionally less funding than a range of comparable conditions (US$3 million annually compared with US$16 million for Crohn’s disease, US$13 million for Barrett oesophagus and US$7 million for nonalcoholic fatty liver disease (NAFLD)55, a situation that likely is the cause rather than the effect of a relatively small pool of dedicated investigators. Research efforts in coeliac disease have proved highly efficient, leading to better understanding of its pathophysiology than that of many better-funded gastrointestinal conditions, such as IBD and Barrett oesophagus. Understanding of the key steps in pathophysiology has led to excellent diagnostic tests, which have in turn enabled large and highly accurate epidemiology studies that have uncovered very large populations of coeliac disease outside Europe and North America, which has previously not been recognized, as well as new insights into age and risk factors for coeliac disease onset. With some high-profile exceptions56,57, many of these studies were funded by sources outside the USA. The 2020 federal budget passed by the United States Congress explicitly encourages the NIH to devote sufficient, focused research to the study of coeliac disease; this new milestone might mark an inflexion point in the dedication of public funds to support coeliac disease research in the USA. We are unaware of similar mandates outside the USA.
Challenges
The coeliac disease research and dedicated clinical community is barely at replacement levels. There is a desperate need in many areas for new expert coeliac disease centres, and in some areas coeliac groups have disappeared with the retirement or move of a single dedicated clinician or researcher. Along with providing high-quality care for patients with coeliac disease, which is currently not routine in non-specialized practices, these clinics are synergistic with local research efforts providing access to patients and patient materials for research. New and increased funding would catalyse and sustain a new generation of coeliac disease centres, supporting patients and research in the coming years. Efforts should be made to improve training of community physicians and other health-care providers through symposia, webinars and development of clinical guidelines, as well as increasing opportunities for research funding coming from industry or government (Table 4).
Table 4.
Models for innovation and potential application in coeliac disease
| Model | Examples outside coeliac disease | Dividends | Potential applications in coeliac disease |
|---|---|---|---|
| Clinical trials consortium |
Children’s Oncology Group TrialNet (type 1 diabetes); FARE (Food Allergy Research Education) |
Successful multicentre treatment and prevention trials in difficult-to-recruit populations (e.g. double-blind placebo with oral peanut challenge in anaphylaxis); overall survival for childhood cancer >80%62; patients outside major academic centres participate in research | More rapid evaluation of increasing number of emerging therapies; increased generalizability of findings and increased rate of discovery and innovation; more efficient use of existing resources; identification of well-defined patients |
| Inception cohort study | Epic-IBD | Identification of prognostic features | Risk stratification at diagnosis; personalized dietary prescription |
| Screening study | Colorectal cancer screening trials | Justification for population-based cancer screening | Evidence-based strategies regarding who to screen and why |
| Collaborative care network | Cystic Fibrosis Foundation Accredited Care Centers; ImproveCareNow (IBD) | Improved quality of care and life expectancy of patients with cystic fibrosis; increase in number of children with IBD in remission off steroids | Personalized treatment for different disease phenotypes; improved understanding of natural history of treated coeliac disease |
IBD, inflammatory bowel disease.
Opportunities
As with many areas of research today, laboratories and clinical groups working individually will only be able to make incremental gains. Larger problems will only be able to be solved by larger teams. For both clinical and basic research, multinational networks, as exist for other conditions such as type 1 diabetes mellitus and IBD will be needed to address some of the most fundamental questions in coeliac disease. Relevant examples include TrialNet (in the diabetes research community) and ImproveCareNow for IBD, both of which have accelerated research in their respective fields. Researchers, physicians and stakeholders are working collaboratively with the community to accelerate innovation, discovery and the application of new knowledge to disease management, such as adoption of novel tools to detect gluten excretion in urine and stool in clinical practice58,59. Such networks will enable efficient clinical trials providing sufficient numbers of heterogeneous patients to ensure that findings are generalizable and reproduceable. In addition, networks of laboratories will enable distribution of the cost and expertise needed to implement the latest technologies, including immune cell phenotyping, microbiome characterization, genome sequencing and reverse translation into improved animal models. In turn, this network would serve as an incubator for new clinicians and researchers, thereby forming a virtuous cycle.
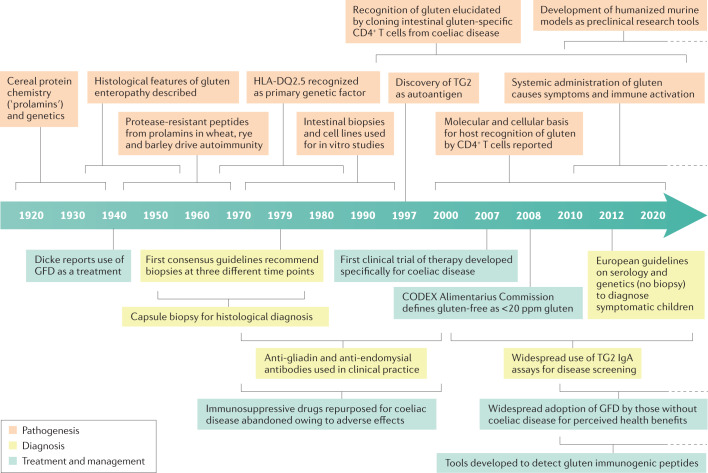
As discussed in the next section, creation of an international coeliac disease consortium should be considered a key priority for the next decade. A summary of the milestones and gaps identified in coeliac disease is provided in Table 1, and a timeline of milestones in pathophysiology, diagnosis, treatment and management of coeliac disease is provided in Fig. 1. These discoveries include examples of how coeliac disease researchers have made substantial contributions through partnership with type 1 diabetes mellitus researchers to include secondary aims related to coeliac disease in multi-site multinational collaboratives (for example, the TEDDY study60) and of how new therapeutic concepts in the autoimmunity field have been applied to coeliac disease61.
Fig. 1. Timeline of major advances and discoveries in coeliac disease.
Graphical timeline description of the major milestones and discoveries in coeliac disease diagnosis, pathogenesis and clinical management since the discovery of its main environmental trigger. Detailed information and references related to each milestone are provided in Tables 2 and 3. GFD, gluten-free diet; TG2, tissue transglutaminase 2.
Final comments and reflections
Over the past decades, the field of coeliac disease has had many research accomplishments to celebrate. We have gained tremendous understanding of the physiopathology of the disease, which is one of very few autoimmune diseases for which the inciting antigen can be administered safely, with the ability to track antigen-specific T cell responses. Thus, coeliac disease is a very attractive model for antigen-specific immune tolerance and other therapies. There are great opportunities for major research in coeliac disease with a potential for high returns on investment. However, as described already, the most important challenges and opportunities require the coordination of efforts in the laboratory and in the clinic and systematic data collection that exceeds the abilities of research groups working in isolation. As coeliac disease is common and heterogeneous with both individual and regional variations, a multinational consortium will be essential to solving the current gaps in diagnosis and management. At the same time, the strong current foundation of knowledge regarding coeliac disease pathophysiology and clinical impact, along with the advanced assortment of powerful clinically available tests such as highly accurate serology and laboratory technologies including isoloation of circulating gluten-specific T cells, suggests that with targeted investment, there is the opportunity to make substantial progress in coeliac disease that would benefit autoimmune disorders broadly.
Specifically, this multidisciplinary working group makes the following recommendations for coeliac disease.
Increase government funding, as philanthropic and industry funding is insufficient to achieve common goals. This approach would require lobbying government to establish priority to allocate research funding in coeliac disease. Focusing on early detection and improvement in treatment and management of coeliac disease would reduce health-care costs, which will ultimately benefit the community.
Improve understanding of coeliac disease as a serious medical condition among the health-care provider community, government and the public.
Develop the infrastructure to support an international consortium that links clinical, microbiome, genetic and immunological data, alongside adult and paediatric patient data, into one accessible community resource that can promote mechanistic study of human disease, evidence-based management approaches and clinical trials.
Innovative models that have been successful in managing other conditions could potentially be implemented in coeliac disease. For instance, successful multicentre trials for treatment and prevention in populations with known difficulties in recruitment have been implemented by various groups, such as paediatric oncology62 and food allergy63. Moreover, population databases to establish risk factors as well as screening programmes have been implemented for IBD64 and cancer (such as the SEER database, which is supported by the Surveillance Research Program in the NCI’s Division of Cancer Control and Population Sciences. Collaborative multidisciplinary networks have improved the quality of care of patients with cystic fibrosis65 and IBD (such as the Promoting Access and Care through Centres of Excellence (PACE) network in Canada). More detailed explanation of various models for innovation that can be applied in coeliac disease are shown in Table 4.
Conclusions
Overall, we remain confident that research breakthroughs in the near term have the potential to greatly improve clinical care in coeliac disease and facilitate innovations across autoimmunity. However, if funding opportunities do not increase for coeliac disease in the coming years, then the breakthroughs needed to continue to advance and possibly cure this morbid condition will be delayed or lost. We hope that this Roadmap will provide guidance to private organizations and governmental agencies justifying and prioritizing funding for coeliac disease, a common and serious yet underestimated condition.
Acknowledgements
M.I.P.-S. received an Innovation Grant from CCC, Medicine Internal Career Research Award, Division of Gastroenterology AFP grant, and Farncombe Family Digestive Health Research Institute Award. J.A.S. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH under award number K23DK119584. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Canadian Institutes of Health Research. E.F.V is funded by a CIHR grant 168840 and holds a Canada Research Chair. R.P.A. declares no funding. The authors thank the hospitality of the Faculty House at Columbia University and Implementation Committee, C. Beckman and K. Cervenka. The authors thank the organizing committee, P. H. R. Green and J. A. Murray, the scientific committee P. H. R. Green (Society for the Study of Celiac disease (SSCD) Past President), C.P.K. (SSCD President), B.L. (SSCD President Elect), J. A. Murray. (SSCD Past President) and E.F.V. (SSCD Past President). Special thanks for the contribution of other Consensus Workshop Faculty: D. Adams (Vanderbilt Center Celiac Clinic, USA); A. Alaedini, G. Bhagat, S. Krishnareddy, N. R. Reilly, A. R. Lee (Columbia University Celiac Disease Center, USA); S. Moleski (Jefferson Celiac Center, USA); B. Tycko (John Threurer Cancer Center, USA); M. J. Blaser (Rutgers University, USA); S. P. Burke, C. Elson; R. S. Chuong and L. Harris (Mayo Clinic. USA); B. Jabri, C. Semrad and R. Verma (University of Chicago, USA); M. Leonard and A. Fasano (Center for Celiac Research and Treatment at Massachussets General Hospital, USA); E. Liu (Children’s Hospital Colorado, USA); A. Cartee (University of Michigan, USA); E. Charles (University of Alabama, USA); A. Rothermel (NIAID, NIH, DHHS); M. Geller, D. Ceizler and R. English (Celiac Disease Foundation); F. Leon (Provention Bio) and A. Sapone (Takeda Pharmaceuticals). Only participants of the workshop included as authors were involved in the writing of this Roadmap Review.
Author contributions
All authors participated in the conception, writing and editing of the article.
Competing interests
B.L. has consulted for Takeda and Anokion. D.A.L. receives salary support from Takeda Pharmaceuticals unrelated to this manuscript. M.I.P.-S. has consulted for Takeda and Lupin unrelated to this manuscript. J.A.S. has consulted for Takeda Pharmaceuticals and received grant funding from Glutenostics, Milky Way Life Sciences, the Celiac Disease Foundation and BEYOND Celiac. E.F.V. received grant funding from Biocodex Foundation and Gilead unrelated to this manuscript. R.P.A. has served as consultant for Takeda, GSK, Anokion, Allero Therapeutics, TregTherapeutics and EVOQ Therapeutics. R.P.A. is founder and shareholder of Novoviah Pharmaceuticals and is the inventor of patents relating to the diagnosis and treatment of coeliac disease. The remaining authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks Stefano Guandalini, Knut Lundin, David Sanders and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Celiac Disease Foundation: https://celiac.org/
Coeliac disease centres: https://celiac.org/celiacdiseasecentersandprograms/
ImproveCareNow: https://www.improvecarenow.org/
SEER database: https://seer.cancer.gov/
Surveillance Research Program: https://surveillance.cancer.gov/
Society for the Study of Celiac Disease: https://www.theceliacsociety.org/
TrialNet: https://www.trialnet.org/
These authors contributed equally: M. Ines Pinto-Sanchez, Jocelyn A. Silvester.
References
- 1.Singh P, et al. Global prevalence of celiac disease: systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2018;16:823–836. doi: 10.1016/j.cgh.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 2.Lebwohl B, Sanders DS, Green PHR. Coeliac disease. Lancet. 2018;391:70–81. doi: 10.1016/S0140-6736(17)31796-8. [DOI] [PubMed] [Google Scholar]
- 3.King JA, et al. Incidence of celiac disease is increasing over time: a systematic review and meta-analysis. Am. J. Gastroenterol. 2020;115:507–525. doi: 10.14309/ajg.0000000000000523. [DOI] [PubMed] [Google Scholar]
- 4.Lebwohl B, Green PHR, Söderling J, Roelstraete B, Ludvigsson JF. Association between celiac disease and mortality risk in a swedish population. JAMA. 2020;323:1277–1285. doi: 10.1001/jama.2020.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubio-Tapia A, Hill ID, Kelly CP, Calderwood AH, Murray JA. ACG clinical guidelines: diagnosis and management of celiac disease. Am. J. Gastroenterol. 2013;108:656–676. doi: 10.1038/ajg.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geller, M. G. SSCD consensus workshop on celiac disease research – Marilyn’s Message March 2020. Celiac Disease Foundationhttps://celiac.org/about-the-foundation/featured-news/2020/03/marilyns-message-march-2020/ (2020).
- 7.Tye-Din JA, et al. Patient factors influencing acute gluten reactions and cytokine release in treated coeliac disease. BMC Med. 2020;18:362. doi: 10.1186/s12916-020-01828-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sollid LM, et al. Update 2020: nomenclature and listing of celiac disease–relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics. 2020;72:85–88. doi: 10.1007/s00251-019-01141-w. [DOI] [PubMed] [Google Scholar]
- 9.Kim C-Y, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA-DQ2-mediated presentation of gluten epitopes in celiac disease. Proc. Natl Acad. Sci. USA. 2004;101:4175–4179. doi: 10.1073/pnas.0306885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson KN, et al. A structural and immunological basis for the role of human leukocyte antigen DQ8 in celiac disease. Immunity. 2007;27:23–34. doi: 10.1016/j.immuni.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Goel G, et al. Cytokine release and gastrointestinal symptoms after gluten challenge in celiac disease. Sci. Adv. 2019;5:eaaw7756. doi: 10.1126/sciadv.aaw7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutierrez-Achury J, et al. Fine mapping in the MHC region accounts for 18% additional genetic risk for celiac disease. Nat. Genet. 2015;47:577–578. doi: 10.1038/ng.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuja-Halkola R, et al. Heritability of non-HLA genetics in coeliac disease: a population-based study in 107 000 twins. Gut. 2016;65:1793–1798. doi: 10.1136/gutjnl-2016-311713. [DOI] [PubMed] [Google Scholar]
- 14.Marietta EV, Murray JA. Animal models to study gluten sensitivity. Semin. Immunopathol. 2012;34:497–511. doi: 10.1007/s00281-012-0315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Kauwe AL, et al. Resistance to celiac disease in humanized HLA-DR3-DQ2-Transgenic mice expressing specific anti-gliadin CD4+ T cells. J. Immunol. 2009;182:7440–7450. doi: 10.4049/jimmunol.0900233. [DOI] [PubMed] [Google Scholar]
- 16.Abadie V, et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature. 2020;578:600–604. doi: 10.1038/s41586-020-2003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caminero A, et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2. Nat. Commun. 2019;10:1198. doi: 10.1038/s41467-019-09037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caminero A, et al. Lactobacilli degrade wheat amylase trypsin inhibitors to reduce intestinal dysfunction induced by immunogenic wheat products. Gastroenterology. 2019;156:2266–2280. doi: 10.1053/j.gastro.2019.02.028. [DOI] [PubMed] [Google Scholar]
- 19.Lamas B, et al. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci. Transl. Med. 2020;12:eaba0624. doi: 10.1126/scitranslmed.aba0624. [DOI] [PubMed] [Google Scholar]
- 20.Bouziat R, et al. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science. 2017;356:44–50. doi: 10.1126/science.aah5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dieterich W, Neurath MF, Zopf Y. Intestinal ex vivo organoid culture reveals altered programmed crypt stem cells in patients with celiac disease. Sci. Rep. 2020;10:3535. doi: 10.1038/s41598-020-60521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freire R, et al. Human gut derived-organoids provide model to study gluten response and effects of microbiota-derived molecules in celiac disease. Sci. Rep. 2019;9:7029. doi: 10.1038/s41598-019-43426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serena G, et al. Intestinal epithelium modulates macrophage response to gliadin in celiac disease. Front. Nutr. 2019;6:167. doi: 10.3389/fnut.2019.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujii M, Clevers H, Sato T. Modeling human digestive diseases with CRISPR–Cas9-modified organoids. Gastroenterology. 2019;156:562–576. doi: 10.1053/j.gastro.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 25.Sarvestani SK, et al. Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat. Commun. 2021;12:262. doi: 10.1038/s41467-020-20351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blansky BA, et al. Lack of follow-up of pediatric patients with celiac disease. Clin. Gastroenterol. Hepatol. 2019;17:2603–2604. doi: 10.1016/j.cgh.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayter SM, Cook MC. Updated assessment of the prevalence, spectrum and case definition of autoimmune disease. Autoimmun. Rev. 2012;11:754–765. doi: 10.1016/j.autrev.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 28.Christophersen A, et al. Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat. Med. 2019;25:734–737. doi: 10.1038/s41591-019-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leonard MM, et al. Evaluating responses to gluten challenge: a randomized, double-blind, 2-dose gluten challenge trial. Gastroenterology. 2021;160:720–733.e8. doi: 10.1053/j.gastro.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdu EF, Danska JS. Common ground: shared risk factors for type 1 diabetes and celiac disease. Nat. Immunol. 2018;19:685–695. doi: 10.1038/s41590-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 31.Andrén Aronsson C, et al. Association of gluten intake during the first 5 years of life with incidence of celiac disease autoimmunity and celiac disease among children at increased risk. JAMA. 2019;322:514–523. doi: 10.1001/jama.2019.10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindfors K, et al. Metagenomics of the faecal virome indicate a cumulative effect of enterovirus and gluten amount on the risk of coeliac disease autoimmunity in genetically at risk children: the TEDDY study. Gut. 2020;69:1416–1422. doi: 10.1136/gutjnl-2019-319809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bibbins-Domingo K, et al. Screening for celiac disease: US preventive services task force recommendation statement. JAMA. 2017;317:1252–1257. doi: 10.1001/jama.2017.1462. [DOI] [PubMed] [Google Scholar]
- 34.Hujoel IA, et al. Natural history and clinical detection of undiagnosed coeliac disease in a North American community. Aliment. Pharmacol. Ther. 2018;47:1358–1366. doi: 10.1111/apt.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl MG, et al. Mass screening for celiac disease: the autoimmunity screening for kids study. Am. J. Gastroenterol. 2021;116:180–187. doi: 10.14309/ajg.0000000000000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Husby S, et al. European society paediatric gastroenterology, hepatology and nutrition guidelines for diagnosing coeliac disease 2020. J. Pediatr. Gastroenterol. Nutr. 2020;70:141–156. doi: 10.1097/MPG.0000000000002497. [DOI] [PubMed] [Google Scholar]
- 37.Riznik P, et al. The use of biopsy and ‘no-biopsy’ approach for diagnosing paediatric coeliac disease in the central european region. Gastroenterol. Res. Pract. 2019;2019:9370397. doi: 10.1155/2019/9370397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rees CJ, et al. Restarting gastrointestinal endoscopy in the deceleration and early recovery phases of COVID-19 pandemic: guidance from the British Society of Gastroenterology. Clin. Med. 2020;20:352–358. doi: 10.7861/clinmed.2020-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leffler D, et al. Kinetics of the histological, serological and symptomatic responses to gluten challenge in adults with coeliac disease. Gut. 2013;62:996–1004. doi: 10.1136/gutjnl-2012-302196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penny HA, Baggus EMR, Rej A, Snowden JA, Sanders DS. Non-responsive coeliac disease: a comprehensive review from the NHS england national centre for refractory coeliac disease. Nutrients. 2020;12:216. doi: 10.3390/nu12010216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarna VK, et al. HLA-DQ–gluten tetramer blood test accurately identifies patients with and without celiac disease in absence of gluten consumption. Gastroenterology. 2018;154:886–896.e6. doi: 10.1053/j.gastro.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Anderson RP, et al. Whole blood interleukin-2 release test to detect and characterize rare circulating gluten-specific T cell responses in coeliac disease. Clin. Exp. Immunol. 2021;204:321–334. doi: 10.1111/cei.13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Kamer JH, Weijers HA, Dicke WK. An investigation inthe the injurious constituents of wheat in connection with their action on patients with coeliac disease. Acta Paediatr. 1953;42:223–231. doi: 10.1111/j.1651-2227.1953.tb05586.x. [DOI] [PubMed] [Google Scholar]
- 44.Sollid L, et al. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J. Exp. Med. 1989;169:345–350. doi: 10.1084/jem.169.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dieterich W, et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 1997;3:797–801. doi: 10.1038/nm0797-797. [DOI] [PubMed] [Google Scholar]
- 46.Codex Alimentarius Commission. Codex standard for foods for special dietary use for persons intolerant to gluten. CSX 118-1979 (WHO, 2008).
- 47.Abu-Janb N, Jaana M. Facilitators and barriers to adherence to gluten-free diet among adults with celiac disease: a systematic review. J. Hum. Nutr. Diet. 2020;33:786–810. doi: 10.1111/jhn.12754. [DOI] [PubMed] [Google Scholar]
- 48.Silvester JA, et al. Most patients with celiac disease on gluten-free diets consume measurable amounts of gluten. Gastroenterology. 2020;158:1497–1499.e1. doi: 10.1053/j.gastro.2019.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Leary C, et al. Celiac disease and the transition from childhood to adulthood: a 28-year follow-up. Am. J. Gastroenterol. 2004;99:2437–2441. doi: 10.1111/j.1572-0241.2004.40182.x. [DOI] [PubMed] [Google Scholar]
- 50.Lebwohl B, Murray JA, Rubio-Tapia A, Green PHR, Ludvigsson JF. Predictors of persistent villous atrophy in coeliac disease: a population-based study. Aliment. Pharmacol. Ther. 2014;39:488–495. doi: 10.1111/apt.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choung RS, et al. Prevalence and morbidity of undiagnosed celiac disease from a community-based study. Gastroenterology. 2017;152:830–839.e5. doi: 10.1053/j.gastro.2016.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mearns ES, et al. Systematic literature review of the economic burden of celiac disease. Pharmacoeconomics. 2019;37:45–61. doi: 10.1007/s40273-018-0707-5. [DOI] [PubMed] [Google Scholar]
- 53.Kivelä L, et al. Current and emerging therapies for coeliac disease. Nat. Rev. Gastroenterol. Hepatol. 2021;18:181–195. doi: 10.1038/s41575-020-00378-1. [DOI] [PubMed] [Google Scholar]
- 54.Schuppan D, et al. A randomized trial of a transglutaminase 2 inhibitor for celiac disease. N. Engl. J. Med. 2021;385:35–45. doi: 10.1056/NEJMoa2032441. [DOI] [PubMed] [Google Scholar]
- 55.Clerx E, Kupfer SS, Leffler DA. Disparities among gastrointestinal disorders in research funding from the national institutes of health. Gastroenterology. 2017;153:877–880. doi: 10.1053/j.gastro.2017.08.051. [DOI] [PubMed] [Google Scholar]
- 56.Liu E, et al. Risk of pediatric celiac disease according to HLA haplotype and country. N. Engl. J. Med. 2014;371:42–49. doi: 10.1056/NEJMoa1313977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubio-Tapia A, Ludvigsson JF, Brantner TL, Murray JA, Everhart JE. The prevalence of celiac disease in the United States. Am. J. Gastroenterol. 2012;107:1538–1544. doi: 10.1038/ajg.2012.219. [DOI] [PubMed] [Google Scholar]
- 58.Comino I, et al. Monitoring of gluten-free diet compliance in celiac patients by assessment of gliadin 33-mer equivalent epitopes in feces. Am. J. Clin. Nutr. 2012;95:670–677. doi: 10.3945/ajcn.111.026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silvester JA, et al. Exposure sources, amounts and time course of gluten ingestion and excretion in patients with coeliac disease on a gluten-free diet. Aliment. Pharmacol. Ther. 2020;52:1469–1479. doi: 10.1111/apt.16075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.TEDDY Study Group The environmental determinants of diabetes in the young (TEDDY) study: study design. Pediatr. Diabetes. 2007;8:286–298. doi: 10.1111/j.1399-5448.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- 61.Getts DR, Shea LD, Miller SD, King NJC. Harnessing nanoparticles for immune modulation. Trends Immunol. 2015;36:419–427. doi: 10.1016/j.it.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Leary M, Krailo M, Anderson JR, Reaman GH. Progress in childhood cancer: 50 years of research collaboration, a report from the Children’s Oncology Group. Semin. Oncol. 2008;35:484–493. doi: 10.1053/j.seminoncol.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.PALISADE Group of Clinical Investigators. AR101 oral immunotherapy for peanut allergy. N. Engl. J. Med. 2018;379:1991–2001. doi: 10.1056/NEJMoa1812856. [DOI] [PubMed] [Google Scholar]
- 64.Egberg MD, Kappelman MD, Gulati AS. Improving care in pediatric inflammatory bowel disease. Gastroenterol. Clin. North. Am. 2018;47:909–919. doi: 10.1016/j.gtc.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 65.Mogayzel PJ, Dunitz J, Marrow LC, Hazle LA. Improving chronic care delivery and outcomes: the impact of the cystic fibrosis Care Center Network. BMJ Qual. Saf. 2014;23:3–8. doi: 10.1136/bmjqs-2013-002363. [DOI] [PubMed] [Google Scholar]
- 66.Osborne TB. Our present knowledge of plant proteins. Science. 1908;28:417–427. doi: 10.1126/science.28.718.417. [DOI] [PubMed] [Google Scholar]
- 67.Rej A, Aziz I, Sanders DS. Breaking bread! Proc. Nutr. Soc. 2019;78:118–125. doi: 10.1017/S0029665118002549. [DOI] [PubMed] [Google Scholar]
- 68.Asri N, Rostami-Nejad M, Anderson RP, Rostami K. The gluten gene: unlocking the understanding of gluten sensitivity and intolerance. Appl. Clin. Genet. 2021;14:37–50. doi: 10.2147/TACG.S276596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hardy MY, et al. Ingestion of oats and barley in patients with celiac disease mobilizes cross-reactive T cells activated by avenin peptides and immuno-dominant hordein peptides. J. Autoimmun. 2015;56:56–65. doi: 10.1016/j.jaut.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 70.Solheim BG, et al. HLA antigens in dermatitis herpetiformis and coeliac disease. Tissue Antigens. 1976;7:57–59. doi: 10.1111/j.1399-0039.1976.tb01031.x. [DOI] [PubMed] [Google Scholar]
- 71.Sallese M, Lopetuso LR, Efthymakis K, Neri M. Beyond the HLA genes in gluten-related disorders. Front. Nutr. 2020;7:1–7. doi: 10.3389/fnut.2020.575844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ju JM, Marietta EV, Murray JA. Generating transgenic mouse models for studying celiac disease. Methods Mol. Biol. 2015;1326:23–33. doi: 10.1007/978-1-4939-2839-2_3. [DOI] [PubMed] [Google Scholar]
- 73.Lundin KE, et al. Gliadin-specific, HLA-DQ(alpha 1*0501,beta 1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J. Exp. Med. 1993;178:187–196. doi: 10.1084/jem.178.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen X, et al. Structural basis for antigen recognition by transglutaminase 2-specific autoantibodies in celiac disease. J. Biol. Chem. 2015;290:21365–21375. doi: 10.1074/jbc.M115.669895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mayassi T, Jabri B. Human intraepithelial lymphocytes. Mucosal Immunol. 2018;11:1281–1289. doi: 10.1038/s41385-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tye-Din JA, et al. Elevated serum interleukin-2 after gluten correlates with symptoms and is a potential diagnostic biomarker for coeliac disease. Aliment. Pharmacol. Ther. 2019;50:901–910. doi: 10.1111/apt.15477. [DOI] [PubMed] [Google Scholar]
- 77.Caminero A, Verdu EF. Celiac disease: should we care about microbes? Am. J. Physiol. - Gastrointest. Liver Physiol. 2019;317:G161–G170. doi: 10.1152/ajpgi.00099.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shiner M. Duodenal biopsy. Lancet. 1956;270:17–19. doi: 10.1016/S0140-6736(56)91854-2. [DOI] [PubMed] [Google Scholar]
- 79.Cichewicz AB, et al. Diagnosis and treatment patterns in celiac disease. Dig. Dis. Sci. 2019;64:2095–2106. doi: 10.1007/s10620-019-05528-3. [DOI] [PubMed] [Google Scholar]
- 80.Anderson CM. Histological changes in the duodenal mucosa in coeliac disease: Reversibility during treatment with a wheat gluten free diet. Arch. Dis. Child. 1960;35:419–427. doi: 10.1136/adc.35.183.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Silvester JA, et al. Tests for serum transglutaminase and endomysial antibodies do not detect most patients with celiac disease and persistent villous atrophy on gluten-free diets: a meta-analysis. Gastroenterology. 2017;153:689–701.e1. doi: 10.1053/j.gastro.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lähdeaho ML, et al. Safety and efficacy of AMG 714 in adults with coeliac disease exposed to gluten challenge: a phase 2a, randomised, double-blind, placebo-controlled study. Lancet Gastroenterol. Hepatol. 2019;4:948–959. doi: 10.1016/S2468-1253(19)30264-X. [DOI] [PubMed] [Google Scholar]
- 83.Daveson AJM, et al. Baseline quantitative histology in therapeutics trials reveals villus atrophy in most patients with coeliac disease who appear well controlled on gluten-free diet. GastroHep. 2020;2:22–30. doi: 10.1002/ygh2.380. [DOI] [Google Scholar]
- 84.Morón B, et al. Toward the assessment of food toxicity for celiac patients: characterization of monoclonal antibodies to a main immunogenic gluten peptide. PLoS ONE. 2008;3:e2294. doi: 10.1371/journal.pone.0002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shalimar, et al. Effect of addition of short course of prednisolone to gluten-free diet on mucosal epithelial cell regeneration and apoptosis in celiac disease: a pilot randomized controlled trial. Dig. Dis. Sci. 2012;57:3116–3125. doi: 10.1007/s10620-012-2294-1. [DOI] [PubMed] [Google Scholar]
- 86.Lionetti E, et al. Introduction of gluten, HLA status, and the risk of celiac disease in children. N. Engl. J. Med. 2014;371:1295–1303. doi: 10.1056/NEJMoa1400697. [DOI] [PubMed] [Google Scholar]
- 87.Gottlieb K, Dawson J, Hussain F, Murray JA. Development of drugs for celiac disease: review of endpoints for phase 2 and 3 trials. Gastroenterol. Rep. 2015;3:91–102. doi: 10.1093/gastro/gov006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leffler D, et al. Development of celiac disease therapeutics: report of the third gastroenterology regulatory endpoints and advancement of therapeutics workshop. Gastroenterology. 2016;151:407–411. doi: 10.1053/j.gastro.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 89.Vriezinga SL, et al. Randomized feeding intervention in infants at high risk for celiac disease. N. Engl. J. Med. 2014;371:1304–1315. doi: 10.1056/NEJMoa1404172. [DOI] [PubMed] [Google Scholar]