Abstract
Strains of Lactobacillus acidophilus WFA1 (KU877440), WFA2 (KU877441), and WFA3 (KU877442) were isolated from indigenous Dahi (yogurt), screened, and selected based on acid and bile tolerance along with the antimicrobial activity. These selected strains were further assessed for their probiotic and functional attributes. Results for simulated gastric and intestinal tolerance/ resistance revealed that all three strains can resist and survive under the following mentioned conditions. To access cell surface hydrophobicity, bacterial adhesion to hydrocarbons (BATH), cellular auto‐aggregation, and salt aggregation were performed. In BATH, adhesion of strains against three hydrocarbons namely xylene, dichloromethane, and hexadecane was conducted. The results show that strains showed the least adhesion to xylene (54.25%) as compared to dichloromethane (55.25%) and hexadecane (56.65%). WFA1 showed maximum adherence percentage (55.48%) followed WFA2 (55.48%) and WFA3 (51.38%). Cellular auto‐aggregation varied from 21.72% to 30.73% for WFA3 and WFA1, respectively. In the salt aggregation test (SAT), WFA1, WFA2, and WFA3 aggregated at 0.6, 1.0, and 2.0 molar concentrations of ammonium sulfate, respectively. PCR amplification of bile salt hydrolase gene (bsh) was performed and sequences were submitted to the public database of NCBI and Gene bank under accession numbers, KY689139, KY689140, and KY689141. Additionally, a cholesterol‐lowering assay was conducted and up to 26% reduction in cholesterol was observed by the strains. Regarding functional properties, exopolysaccharide (EPS) production, and antioxidant potential, strain WFA1 showed promising results EPS (1.027mg/ml), DPPH (80.66%), ABTS (81.97%), and reducing power (1.787). It can be concluded from the present study that the mentioned strains of L. acidophilus (WFA1, WFA2, and WFA3) are strongly hydrophobic; thus having an ability to survive and colonize under the gastrointestinal tract which confirms their probiotic nature. Regarding their functional properties, L. acidophilus WFA1 (KU877440) showed excellent properties of antioxidants and EPS production.
Keywords: antioxidants, bsh gene amplification, cell surface hydrophobicity, cellular auto‐aggregation, exopolysaccharide (EPS), gastrointestinal transit tolerance, hypocholesterolemic activity
Strains of L. acidophilus (WFA1, WFA2 and WFA3) have resisted and survived under GI tract conditions, furthermore; they have the capability to adhere and colonize in the intestinal cavity, thereby confirming their probiotic potential. These strains also have cholesterol‐lowering property along with high EPS production and antioxidant activity. Among the three strains, WFA1 was selected further due to its potential functional properties and was supplemented to produce probiotic fermented milk.

1. INTRODUCTION
The presence of L. acidophilus is reported in indigenously produced fermented milk products locally named as “dahi” of Pakistan, along with other members of lactic acid bacteria (LAB), L. acidophilus is promising, completely characterized, well‐documented probiotic member of LAB (Behbahani et al., 2019) and is already included in the generally recommended as safe (GRAS) category. According to FAO (Food and Agriculture Organization, 2006), “Probiotics are the live microorganisms which when administered in an adequate amount, confers health benefits to the host.” They are also categorized into a category of nutraceutical (Hill et al., 2014).
The most important criterion for probiotics is to remain viable under harsh environmental conditions of the gastrointestinal tract. Low pH in the stomach and gastric enzymes along with the presence of bile and intestinal enzymes in the intestine is considered fatal for bacterial strains. So for the selection of probiotics, strains have to access acid, bile, and digestive enzyme tolerance and gastrointestinal (GI) transit tolerance. Furthermore, they must possess the quality of intestinal colonization. This characteristic makes it the most suitable to not only remove/decrease the adherence of enteropathogens (Monteagudo‐Mera et al., 2019), but also need to exert their health benefits on their respective host.
Cell surface hydrophobicity of a probiotic strain is a measure of its intestinal colonization, that is, adhesion and persistence once they have entered the intestinal cavity. The higher the hydrophobicity, higher colonization was observed (de Souza et al., 2019). This was further resolved if there is better bacterial adhesion to hydrocarbons (BATH), cellular auto‐aggregation, and salt aggregation. BATH deals with the adhesion property of strain and both cellular auto‐aggregation and salt aggregation are clumping or self‐aggregation capacity of cells related to its persistence (Saito et al., 2019),
Probiotic has also been reported for their hypocholesterolemic activity which can be attributed to bile salt hydrolase (bsh) activity (Huang et al., 2019; Kumar et al., 2012). It is an enzymatic breakdown or deconjugation of bile salts or bile acids by probiotic LAB, increases its excretion or decrease its reabsorption (Ishimwe et al., 2015), and can be attributed to in vivo lowering of serum cholesterol levels (Patel et al., 2010).
Production of EPS by LAB is one of the most promising functional properties. EPS are naturally produced sugar polymers having a distinct and significant role, one of which is to enhance the taste and texture of food (Badel et al., 2011). Its bio‐functional property increases its importance as food‐grade hydrocolloids, emulsifiers, or bio stabilizers in fermented foods (Rühmann et al., 2015). Apart from this, EPS have also strongly antioxidant, anticancer, and anti‐inflammatory activity (Deepak, Ramachandran, et al., 2016). Along with other functional properties, yogurt bacteria were reported to have a role in reducing oxidative stress because they have a strong potential of scavenging free radicals such as reactive oxygen species (ROS), peroxide, and superoxide (Ji et al., 2015; Xing et al., 2015). Antioxidants prevent oxidative damage, thus capable of slowing down the process of aging and likewise protects the human body from the progression of different diseases.
Previously studies have confirmed the presence of L. acidophilus and other LAB in indigenous dahi collected from the area of Rawalpindi, Pakistan (Soomro & Masud, 2012). But their probiotic nature and functional properties were not assessed. Studies also lacked the molecular characterization. Thus, the present study was designed to assess the potential of local strains of L. acidophilus. Molecular characterization along with initial isolation and screening of L. acidophilus on the basis of acid and bile tolerance and antimicrobial activity against pathogens has already been published (Farid et al., 2016). Hence, the current paper discusses the GI transit tolerance, adhesion properties, and hypocholesterolemic activity of L. acidophilus strains (WFA1, WFA2, and WFA3). Furthermore, these strains were screened for exopolysaccharide (EPS) production and antioxidant activity to further uses in probiotic product development.
2. MATERIALS AND METHODS
In a preliminary isolation study, screening and molecular characterization of L. acidophilus were performed. Briefly, isolation was conducted on MRS media supplemented with 0.7% bile salts. Out of 57 strains of LAB, 18 were confirmed as L. acidophilus through API CHL 50. These 18 strains were further screened for acid tolerance and antimicrobial activity against different pathogens namely Escherichia coli ATCC 25922, Salmonella paratyphi ATCC 5702, Staphylococcus epidermis ATCC 12228, Acinetobacter baumannii ATCC 17978, and Staphylococcus aureus ATCC 29213. Strains showing higher antimicrobial activity were selected and characterized at the molecular level through polymerase chain reaction (PCR) using universal primers 9F and 1510R and sequencing, and then, their sequence was submitted to the NCBI Gene Bank as Lactobacillus acidophilus WFA1 (KU877440), WFA2 (KU877441) and WFA3 (KU877442) (Farid et al., 2016).
In the present study, the probiotic potential of these characterized strains was assessed including GI transit tolerance and cell surface hydrophobicity which ensures their survival and colonization in the GI tract. Furthermore, their functional properties, that is, bsh activity, cholesterol‐lowering ability, EPS quantification, and antioxidant properties, were also analyzed.
2.1. GI transit tolerance
For GI transit tolerance, the method of Zhou, Pan, Wang, and Li (2007) was followed. Firstly gastric (pH 2, 3, and 4) and small intestinal (pH 8 and 0.3% bile salts) juices were prepared with pepsin (3 g/L) and pancreatin (1 g/L) in saline solution (0.5%) respectively.
The bacterial cell pellet was harvested, washed twice, and resuspended in PBS buffer. A volume of 2 ml of gastric juice with pH 2 and 3 was taken in a sterilized Eppendorf tube containing 0.4 ml of bacterial cell suspension already mixed with 0.6 ml of saline and vortex gently before incubating at 30℃ for 5 hr. The same procedure was followed for intestinal tolerance but incubation was done for 12 and 24 hr. The plate count method of the colony‐forming unit (log CFU/ml) was followed to calculate the survival of bacterial strains.
2.2. Cell surface hydrophobicity
2.2.1. Bacterial Adhesion to Hydrocarbons (BATH)
The BATH was adopted from the method of (Kaushik et al., 2009). Overnight the cell pellet grown culture was washed and resuspended in a phosphate urea magnesium buffer (PUM) to an absorbance of 0.7 at 600 nm. A volume of 3.0 ml of Lactobacilli cell was mixed with 1.0 ml of xylene or dichloromethane or n‐hexadecane and incubated at 37℃ for 10 min. After vortexing, incubation for phase separation was carried out at 37℃ for 1 hr, where the absorbance of the aqueous phase was recorded at 600 nm. The surface hydrophobicity (percent) was calculated by using the following equation:
2.2.2. Cell aggregation
Cellular aggregation was conducted by Zuo et al. (2016). Bacterial cells were collected, washed, resuspended in sterilized PBS (step 1), and the concentration was adjusted to an absorbance of 0.5 at 600 nm using the respective buffer. The cell pellets were taken out and mixed with an equal volume of broth obtained at step 1. This mixture was incubated at 37℃ for 2 hr. The upper suspension was then taken to measure the absorbance (Abs final), using the broth as reference.
2.2.3. Salt aggregation test (SAT)
Salt aggregation test assay was conducted (Nwanyanwu & Abu, 2013) in which different molarities (M) of (NH4)2SO4 solution (ranging from 0.2 to 4.0 M) were used and further aided by 0.1% (w/v) methylene blue solution to improve visualization. Equal volumes (100 ul) of cell suspension and (NH4)2SO4 were mixed and checked for the concentration on which bacterial cells were clumped or salted out (lower the concentration, higher will be the hydrophobicity). The classification was expressed as follows:
Strongly hydrophobic = < 1.0 M, Hydrophobic = 1.0–2.0 M, Hydrophilic = > 2.0 M.
2.3. Cholesterol lowering assay
2.3.1. Bile salt hydrolase (bsh) activity
For bsh activity, overnight grown bacterial culture was streaked on MRS agar‐containing bile salts (0.5%) along with 0.37 g/L CaCl2 and incubated anaerobically at 37℃ for 48–72 hr. White opaque bacterial growth or presence of the precipitated hydrolyzed product of bile salts in or around the colony growth were recorded, it exhibited the indication of bsh activity (Pereira et al., 2003; Tsai et al., 2014).
2.3.2. Molecular confirmation for bsh gene
For PCR amplification already submitted sequence of bsh gene of L. acidophilus (Accession # AF091248.3 Lactobacillus acidophilus putative bile salt hydrolase operon) was taken as reference and NCBI primer blast designing software was used for primer designing (Table 1).
TABLE 1.
Sequence of forward and reverse primer for bsh gene amplification
| Primer sequences | ||
|---|---|---|
| Forward primer | Reverse primer | This study |
| 5´CAGGATTATGGCGAAGGCGT3´ | 5´TCGAGCGGTTGACGGTATTG3´ | |
Deoxyribonucleic acid (DNA) template was prepared by the method described by (Plourde‐Owobi et al., 2005) with some modification. Freshly grown colonies of L. acidophilus were picked and transferred to 10 ul Tris‐EDTA (ethylene diamine tetra‐acetic acid) buffer (10 ml of 1.0 M Tris‐HCl buffer (pH 7.5) and 2 ml of 0.5 M EDTA solution (pH 8) make the volume up to 1 L) by using sterilized toothpicks and incubated at 80℃ for 15 min, followed by centrifugation at 12,000 rpm for 15–20 min and then the supernatant was taken as the template. The reaction mixture, its concentration, and details of the PCR cycle are given in Tables 2 and 3, respectively.
TABLE 2.
Quantity of chemicals used in PCR amplification
| Reaction mixture | Quantity (ul) |
|---|---|
| Reaction buffer | 1.0 |
| MgCl2 | 0.6 |
| Primers | |
| (Reverse) | 0.5 |
| (Forward) | 0.5 |
| Taq DNA polymerase | 0.4 |
| dNTPs | 0.2 |
| DNA template | 2.0 |
| Water | 4.8 |
| Total | 10.0 |
TABLE 3.
Cycle conditions (temperature and time) for PCR amplification
| Temperature (oC) | Time (mins) | No. of cycles | |
|---|---|---|---|
| Initial heating | 94 | 2 | 1 |
| Denaturation | 94 | 1 | 33 |
| Annealing | 61.5 | 1 | |
| Extension | 72 | 1.5 | |
| Final extension | 72 | 20 | 1 |
Gel electrophoresis was performed by running agarose gel (1%) in TAE (Tris‐Acetate EDTA) buffer for about 20–30 min on 70 volts and 110 mA followed by staining in ethidium bromide solution and visualization conducted under UV transilluminator.
2.3.3. Sequencing, sequence analysis, and phylogenetic tree
The amplified products were sequenced from macrogen (Korea) in sense and antisense directions. The obtained sequences were analyzed by using Bio Edit sequence alignment software and BLAST tool of the National Centre for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Final sequences were submitted in the public database of NCBI under accession numbers, KY689139, KY689140, and KY689141. The phylogenetic tree was constructed using 15 closely related sequences (99%–100% genetic homologies) of bsh gene which were taken from NCBI after BLAST.
2.3.4. Hypocholesterolemic activity of L. acidophilus strains
MRS broth containing 0.2% bile salt (w/v) and 0.01% (w/v) cholesterol (polyoxyethanyl‐cholesteryl Sebacate; Sigma) was prepared, inoculated with overnight grown culture, and were incubated at 37℃ for 24 hr. Bacterial cells were removed after 6, 12, 18, and 24 hr of inoculation, and cholesterol contents were measured (Rudel & Morris, 1973).
2.4. EPS extraction and quantification
Exopolysaccharide extraction and quantification (6, 12, 18, and 24 hr of inoculation) was performed by the method described by Feldmane et al. (2013) with some modification. Overnight‐grown bacterial culture was boiled (20–30 min), cooled, and centrifuged (8,000 rpm for 10 min), followed by the addition of 17 ml of 85% trichloro‐acetic acid. Samples were cooled at 4℃ and centrifuged again. Cold ethanol (1:3) was finally used to precipitate EPS.
For quantification of EPS, an equal amount of the sample and phenol red solution (5%) were taken in a test tube (400 µl). For control, distilled water replaced the sample. Then 2 ml of concentrated sulfuric acid was added and left for 10 min, followed by shaking and incubation (30℃ for 10 min). Absorbance was recorded at 490 nm. The same procedure was followed for the preparation of the glucose concentration curve (0.2–1.0 mg/ml), taken as a standard for quantification of EPS.
2.5. Antioxidative assay
2.5.1. Preparation of cell free supernatant (CFS)
For CFS preparation, the pH of the overnight grown culture was maintained at 5.5 (1 M NaOH) then centrifuged (8,000 rpm for 10 min). The supernatant was separated and passed through a 0.2 um syringe filter to remove bacterial cells completely.
2.5.2. Antioxidant potential
Antioxidant activity or radical scavenging ability of CFS of L. acidophilus strains was conducted after 6, 12, 18, and 24 hr of inoculation and incubation. The maximum antioxidant activity was observed after 24 hr of incubation which were further compared with ascorbic acid (0.1%) (Water‐soluble antioxidant).
Determination of 2, 2‐diphenyl‐1‐picrylhydrazyl (DPPH) scavenging ability
The DPPH scavenging activity was measured according to the method of Arora and Chandra (2010). DPPH (0.1 mM) solution was prepared freshly in ethanol, 1 ml of this solution was mixed with 0.5 ml of CFS and thereafter shaken vigorously and was left to stand for 30 min in the dark. Change in color at 517 nm determines the DPPH radical scavenging activity, which was calculated according to the following equation:
whereas,
A0 = Absorbance of control, A1 = Absorbance of CFS.
A2 = Absorbance without DPPH and broth was taken as reference.
Determination of 2, 2'‐azino‐bis (3 ethylbenzothiazoline 6‐sulphonic acid) ABTS cation scavenging activity
ABTS cation scavenging assay was performed according to the procedure described by Ji et al. (2015). Firstly ABTS stock solution was prepared (7 mM ABTS and 2.45 mM potassium per sulfate) and left for –416 hr and diluted to an absorbance of 0.7 at 734 nm with ethanol. 0.3 ml of CFS and 2.7 ml of ABTS solution were also mixed for 45 s and after 1 min spectrophotometric absorbance was measured at 734 nm. The following equation was used to calculate scavenging activity: .
Abs (s) = Absorbance of sample Abs (c) = Absorbance of control.
Reducing power
The CFS reducing the power of CFS was determined as described by Xing et al. (2015) with slight modification. A sample (CFS, MRS broth, 0.5 ml) was briefly mixed with potassium ferricyanide (1%, 0.5 ml) and PBS (pH 6.6, 0.5 ml). Subsequently, the mixture was heated at 50℃ for 20 min and allowed to cool. Upon cooling, 0.5 ml of 10% trichloroacetic acid (TCA) was added to the mixture and then centrifuged at 3,000 g for 5 min. The supernatant (1 ml) was mixed with ferric chloride (0.1%, 1 ml) and was allowed to react for 10 min. The absorbance of the mixture was determined at 700 nm. Higher absorbance of the mixture indicated higher reducing activity.
2.6. Statistical analysis
Data were analyzed statistically by software Statistix 8.1 (for ANOVA), for percentages/ comparisons and standard deviation MS‐excel was used. Likewise, for the alignment of bsh gene sequences, BioEdit sequence alignment software was used.
3. RESULTS
3.1. GI Transit tolerance
Gastrointestinal transit tolerance and survival of LAB under these conditions is preliminary for the selection of probiotics. Results for simulated gastric and intestinal conditions of all the three strains (WFA1, WFA2, and WFA3) are presented in Table 4. These results show their ability to resist and survive under these harsh environmental conditions. Although a decrease in log CFU/ml has been observed in both simulated gastric and intestinal conditions but to a lesser extent.
TABLE 4.
Survival percentage of Lactobacillus acidophilus strains under simulated gastric (pH 2, 3, and 4) and intestinal conditions (incubation for 12 and 24 hr)
| Strains | Survival % under gastric conditions | Survival % under intestinal conditions | |||
|---|---|---|---|---|---|
| pH2 | pH3 | pH4 | 12 hr | 24 hr | |
| WFA1 | 47.35 | 63.25 | 80.5 | 78.87 | 64.26 |
| WFA2 | 50.45 | 62.5 | 78.7 | 69.95 | 57.45 |
| WFA3 | 43.12 | 57.9 | 79.85 | 84.65 | 71.36 |
Under simulated gastric conditions, pH 2 has a more severe impact on the survival percentage of strains as compared to pH 3 where a sharp decrease was observed in log CFU/ml at this pH. At pH 2, survival of strains ranged from 43.19% to 50.45% (almost about 49.55%–56.81% reduction). At pH 4, least impact of pH was observed where the survival rate was about 78.7%–80.5% (21.3%–19.5% reduction). Similarly in simulated intestinal conditions of incubation for 24 hr showed more decrease in log CFU/ml compared to 12 hr. Survival ranges from 57.45% to 71.36% (42.55%–28.64% reduction) after 24 hr of incubation. This decrease was obvious because bacterial strains were not acclimatized to these harsh conditions but their survival ability was not much affected.
3.2. Cell surface hydrophobicity
3.2.1. Bacterial adhesion to hydrocarbons
Bacterial adhesion to hydrocarbons measures the adhesion property or ability of LAB to adhere to the organic compounds (nonpolar). Higher the nonpolarity of surfaces, higher will be adhesion to hydrocarbons and more will be hydrophobicity.
The percent adhesion of strains to different hydrocarbons has been presented graphically in Figure 1. Statistical analysis revealed that both the adhesion ability of strains and adherence percentage of different hydrocarbons are different significantly. L. acidophilus WFA1 exhibited maximum adherence (59.28%), followed by L. acidophilus WFA2 (55.48%). While L. acidophilus WFA3 (51.38%) had the lowest adhering property. Among the three tested hydrocarbons, hexadecane (56.65%) showed the maximum score of adhesion followed by dichloromethane (55.25%) and xylene (54.25%).
FIGURE 1.
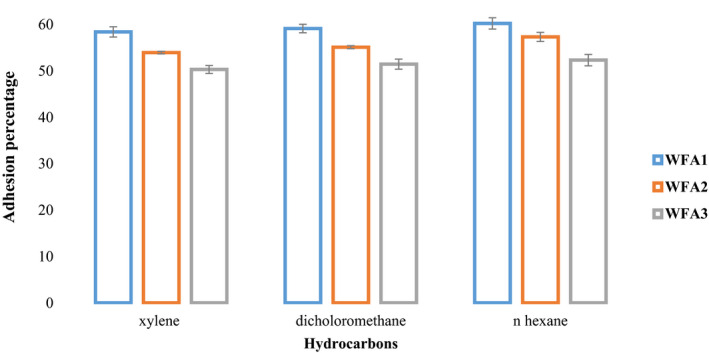
Graphical representation of the percentage of bacterial adherence to hydrocarbons of selected strains of Lactobacillus acidophilus by using xylene, dichloromethane, and hexadecane
3.2.2. Cellular auto‐aggregation
It is the measure of self‐clumping or the aggregating ability of cells of bacteria which is an important criterion for its persistence and colonization in the intestine. The percent cell auto‐aggregation of different strains of L. acidophilus presented in Table 5 shows that there is a significant difference in the aggregation property of all three probiotic strains. The mean value of aggregation among different strains ranged from 21.72% to 30.73%. L. acidophilus WFA1 was found to have maximum aggregating % (30.7%) followed by L. acidophilus WFA2 (27.34%). L. acidophilus WFA3 (21.72%) has the lowest adhering property among all tested strains.
TABLE 5.
Comparison of aggregation percentage, salt aggregation, and cholesterol‐lowering ability among selected strains of Lactobacillus acidophilus
| Strains | Aggregation (%) | SAT | Cholesterol‐lowering (%) |
|---|---|---|---|
| WFA1 | 30.73 ± 1.16a | 0.6 M | 26.55 ± 0.79a |
| WFA2 | 27.34 ± 0.93b | 1 M | 26.22 ± 1.36a |
| WFA3 | 21.72 ± 0.83c | 2 M | 26.43 ± 0.63a |
The values are means ± SE; each value is expressed as a mean of three experiments, different letters in a column indicated the significant difference.
3.3. Salt aggregation test (SAT)
Salt aggregation test is the measurement of aggregation or precipitation of probiotic L. acidophilus strains in different concentrations or molecular strength of ammonium sulfate solution ranging from 0.2 to 4 M (Tyfa et al., 2015). Results for the salt aggregation test have been presented in Table 5. According to the criterion, one of L. acidophilus strains falls in the category of being strongly hydrophobic by agglutinating at 0.6 M (WFA1) and the other two were hydrophobic as they clumped at 1 M (WFA2) and 2 M (WFA3) of ammonium sulfate. It was observed that lower the concentration of ammonium sulfate on which aggregation was recorded, the higher will be the hydrophobicity of strain.
3.4. Hypocholesterolemic activity
3.4.1. Bile salt hydrolase activity (Bsh gene amplification, sequencing, and phylogenetic tree)
All three strains showed thick white opaque colonies on differential media which confirmed their Bsh activity as shown in Figure 2 and were further subjected to PCR amplification of the bile salt hydrolase gene. A product of 975 bps was obtained and sequenced. Sequences were then submitted to the public database of Gene bank and NCBI under accession numbers, KY689139, KY689140, and KY689141. A Phylogenetic (Figure 3) tree of closely similar sequences of the bsh gene was constructed; sequences were obtained from BLAST conducted on NCBI.
FIGURE 2.
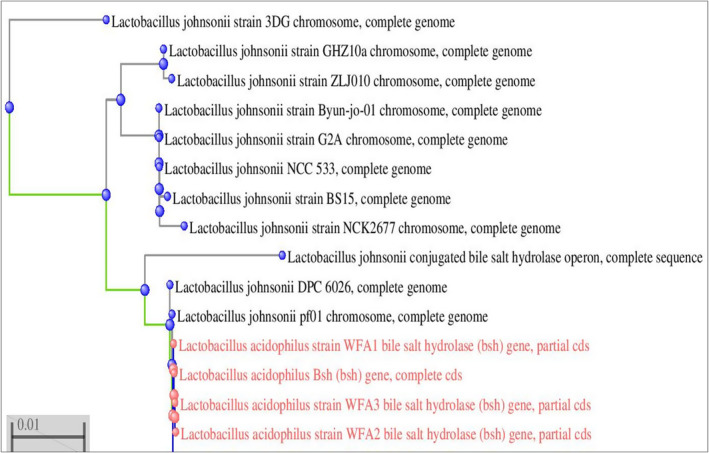
Phylogenetic tree of Bsh gene sequence of Lactobacillus acidophilus strains with the most similar/resembled sequences
FIGURE 3.
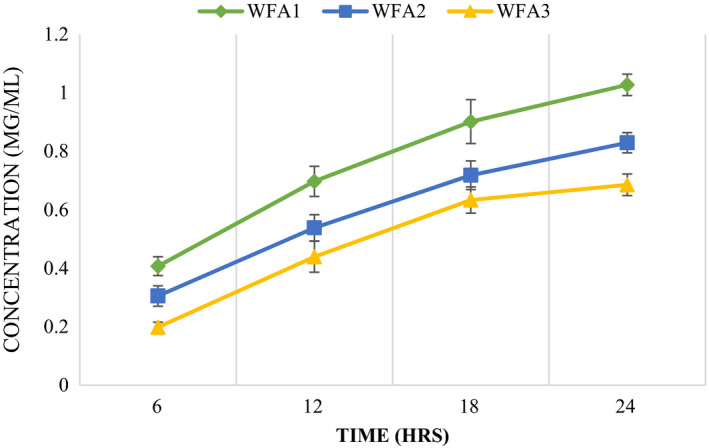
Comparative analysis of EPS quantification of Lactobacillus acidophilus strains at a different time interval at 37℃
3.4.2. Cholesterol‐lowering assay
Cholesterol‐lowering activity or cholesterol reduction percentage of different strains of L. acidophilus has been presented in Table 5. All three strains possess the ability to reduce cholesterol from bile salts (0.2%) supplemented media with a maximum reduction of about 26% was observed after 24 hr of inoculation.
3.5. EPS extraction and quantification
Results of EPS production by strains as well as treatments (with time increase in EPS production) differed statistically as presented in Figure 3. Maximum EPS was produced after 24 hr of inoculation and incubation at 37℃ by L. acidophilus WFA1 (1.027 mg/ml) followed by WFA2 and WFA3 (0.83 and 0.685 mg/ml), respectively. Among all treatments (with time), WFA1 produced the highest amount of EPS at 6 hr (0.407 mg/ml), 12 hr (0.697 mg/ml), 18 hr (0.901 mg/ml), and 24 hr (1.027 mg/ml).
3.6. Comparison of antioxidant activity (DPPH, ABTS, and reducing power) of CFS of L. acidophilus strains with ascorbic acid
Antioxidant forms an irreversible or a stable complex by donating electrons or hydrogen atoms to the free radical (DPPH and ABTS accept electrons or hydrogen atoms from antioxidant substances and are converted into irreversible stable molecules). Comparison of DPPH, ABTS, and reducing power with ascorbic acid has been represented graphically in Figures 4 and 5, respectively. It was evident from data that DPPH scavenging activity of WFA1 (80.66%) has a nonsignificant difference from that of ascorbic acid (85.49%). ABTS scavenging activity (81.97%) and reducing power (1.78) of WFA1 have significantly differed from ascorbic acid (89.94%) and (2.05), respectively. Both of the other strains, that is, WFA2 and WFA3 have also high ABTS, DPPH, and reducing power but less when compared to the potential of WFA1.
FIGURE 4.
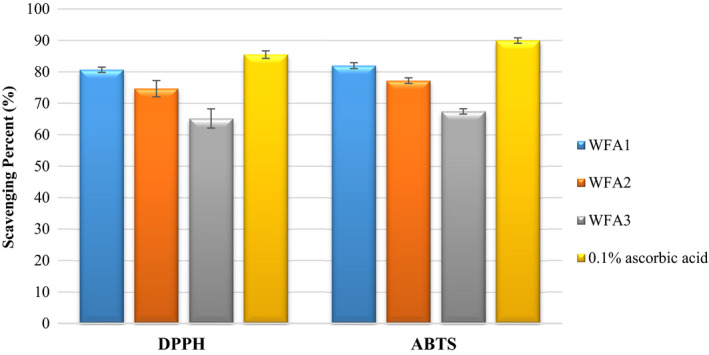
Comparison of DPPH and ABTS scavenging activity of Lactobacillus acidophilus strains after 24 hr of incubation with ascorbic acid
FIGURE 5.
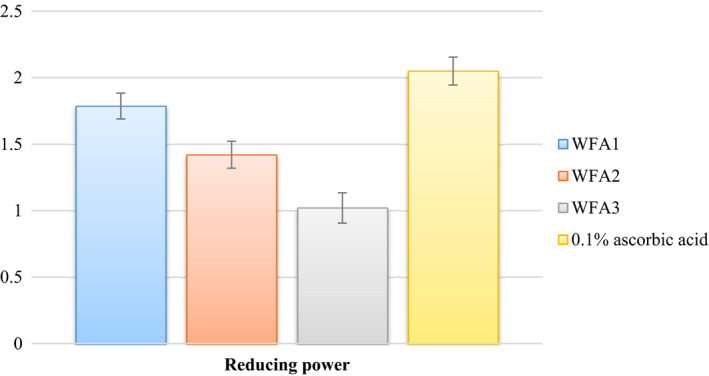
Comparison of reducing the power of Lactobacillus acidophilus strains after 24hrs of incubation with ascorbic acid
4. DISCUSSION
To colonize in an intestinal portion of the alimentary tract, probiotic strain must tolerate and remain viable in harsh environmental conditions of the stomach (pH ranges from 1.5 to 3 or 4 and gastric enzymes) and intestine (bile and enzymes). It is obvious from earlier studies that this condition decreases bacterial viability by inhibiting their metabolism and growth (Liu et al., 2013). Lactobacilli have been reported previously for their resistance and survival under acidic (pH 2 and 3) (Sahadeva et al., 2011) and intestinal conditions (Pisano et al., 2014). L. acidophilus is also well documented for its resistance and survival in the stomach and intestine (Khaleghi et al., 2010).
In vitro cell surface hydrophobicity deals with the adhesion of probiotics to epithelial cell lining of the intestine, higher the hydrophobicity enhanced adhesion will be observed. On the basis of the degree of adhesion to hydrocarbons, Tyfa et al. (2015) categorized and divided bacterial strains into three categories including strongly hydrophobic (>50%), moderately hydrophobic (20%–50%), and hydrophilic (<20%). According to this criterion, all three strains of L. acidophilus falls in the category of strongly hydrophobic. In vitro bacterial adhesion to hydrocarbons was conducted against n‐hexadecane, xylene, and dichloromethane (Jose et al., 2015). Different studies reported different adherence percentages of Lactobacilli. Kaushik et al. (2009) studied the adherence percentage of probiotic L. johnsonni LA1 (47%), L. acidophilus LA7 (57%–58%), and L. plantarum (37.1%–37.7%). Similarly, Schillinger, Guigas, and Holzapfel, (2005) also reported bacterial cell surface hydrophobicity ranging from 74% to 95% in L. acidophilus. Among hydrocarbons used, xylene has shown a low mean value compared to dichloromethane and hexadecane that might be owing to its toxic or destructive behavior on microbial cells.
Bacterial auto‐aggregation capacity is considered as an aggregation among the cells of the same strains which further assessed their persistence in the intestine. Moreover, this aggregation eliminated or reduced the adhesion of pathogenic bacteria. Kaushik et al. (2009) reported cellular auto aggregation by L. acidophilus LA7 and L. plantarum (Lp9), which was about 46.5% and 31%, respectively. The results confirmed the self‐aggregating ability of L. acidophilus.
Cellular auto‐aggregation and SAT along with BATH collectively contributed to the cell surface hydrophobicity, which bacterial strains must possess to adhere to the intestinal mucosal or epithelial lining. Overall, adhesion of bacteria is mainly a strain‐specific property depending on its origin, variation in cell surface protein expression level along with the influence of environment on the expression of certain proteins which are responsible for cell surface hydrophobicity or cell adhesion (Chaffanel et al., 2018; De Vries et al., 2006). As bacterial adhesion to the human cell is a very complex process involving many components including charges on bacterial cell and human cell, hydrophobicity, the presence of extracellular polysaccharide and cell surface proteins (Pan et al., 2017), therefore for proper, strong or irreversible adhesion, the bacterial cells have to overcome all of these hurdles. Adhesion property increases as the nonpolarity of both or one of the surfaces (microbial and host cell) increases.
Strains of LAB have been widely studied previously for their hypocholesterolemic activity both in vivo and in vitro. One of the probable mechanisms for this was the deconjugation of bile by Bsh. Bsh activity involves enzymatic breakdown or deconjugation of bile salts or bile acids by probiotic Lactobacilli and Bifidobacterium, increasing its excretion or a decrease in its reabsorption. Cholesterol, being a precursor of bile acids, converts its molecules to bile acids as compensation or to replace lost ones, thus decreasing the cholesterol level. In the control of serum cholesterol levels, this mechanism could be operated by conversion of deconjugated bile acids by colonic microbes (Patel et al., 2010). Bsh activity was supposed to be the important criteria as nondeconjugating organisms do not appear to be able to remove cholesterol from the culture medium to any significant extent (Kumar et al., 2012; Tahri et al., 1997).
Moreover, Gilliland et al. (1985) reported cholesterol removal by L. acidophilus strains from culture medium when grown under simulated intestinal conditions. As this is a strain‐dependent property, therefore in different studies the cholesterol‐lowering percentage differs significantly (Lye et al., 2010). Lim et al. (2004) reported about 31.5%–58.5% reduction, and Sirilun et al. (2010) observed 25.41 to 81.46% reduction in cholesterol. Similarly, about 26.74%–85.41% reduction of cholesterol from the growth medium after 20 hr of inoculation of different Lactobacilli strains was reported by Ramasamy et al. (2010).
Our results also confirm that there is a relation between bile salt hydrolase activity and the cholesterol‐lowering effect. Strains having Bsh activity also can reduce cholesterol levels from serum (Öner et al., 2014). Lactobacilli have also been reported previously for their cholesterol‐lowering attributes. Similar to our study, Jones et al. (2004) evaluated the cholesterol‐lowering potential of LAB and attributed this to bsh activity, due to deconjugation of conjugated glycodeoxycholic acid and taurodeoxycholic acid. Reduction of cholesterol by LAB and Bifidobacteria in the presence of bile salts were also been reported in previous researches (Huang et al., 2019; Tsai et al., 2014).
Quantification of EPS was conducted in many studies where Deepak et al. (2016a) reported about 400–597 mg/L of EPS produced by L. acidophilus after 24 hr of incubation under optimized conditions. Similarly, Polak‐Berecka et al. (2014) reported about 210.28 mg/L of EPS by L. rhamnosus E/N strain. EPS are polymers of sugars, arranged as long‐chained polysaccharides, while their functional properties may be due to the arrangement of monosaccharides, as well as the type and conformation of glycosidic linkages of the polysaccharide (Suresh Kumar et al., 2007). EPS of LAB are used in food to enhance its taste and texture, ability to operate as an emulsifier, and maintaining viscosity (bio stabilizer) of fermented foods. Apart from this, EPS also have strong antioxidant, anticancer, and anti‐inflammatory activities (Deepak, Ramachandran, et al., 2016; Liu et al., 2011).
Similarly, numerous researches were conducted on DPPH scavenging ability, ABTS cation radical scavenging potential, and reducing the power of CFS of LAB, respectively, and they also reported its high antioxidant potential (Afify et al., 2012; Gao, 2012; Xing et al., 2015). Mostly 0.05% of the ascorbic acid concentration was used previously in different studies. In the present study, we used an almost double percentage of ascorbic acid, that is, 0.1%. Ji et al. (2015) reported high DPPH and ABTS scavenging activity of CFS of Lactobacilli when compared to 0.5% ascorbic acid (that was considered as a high concentration of ascorbic acid). Furthermore, (Liang et al., 2016) also used ascorbic acid as a positive control or standard for comparison and reported high antioxidant potential. The antioxidant potential of different microbial or bacterial strains can be attributed to the production of their cell surface compounds, for example, the production of exo‐polysaccharide (Oh & Jung, 2015) and lipoteichoic acid (Yi et al., 2009).
5. CONCLUSION
Strains of L. acidophilus (WFA1, WFA2, and WFA3) have resisted and survived under GI tract conditions, furthermore; they can adhere and colonize in the intestinal cavity, thereby confirming their probiotic potential. These strains also have cholesterol‐lowering properties along with high EPS production and antioxidant activity. Among the three strains, WFA1 was selected further due to its potential functional properties and was supplemented to produce probiotic fermented milk.
CONFLICT OF INTEREST
The authors declared that there is no conflict of interest.
AUTHOR CONTRIBUTIONS
Wajiha Farid : Conceptualization (equal); Data curation (equal). Tariq Masud: Supervision (equal). Asma Sohail: Methodology (equal). Nazir Ahmad: Data curation (equal). S.M. Saqlan Naqvi: Visualization (equal). Sipper Khan : Software (equal). Amjad Ali: Supervision (equal). Salah Khalifa: Writing‐original draft (equal). ABID HUSSAIN: Resources (equal). Sartaj Ali: Validation (equal). Maryum Saghir: Formal analysis (equal). Azhari Siddeeg: Resources (equal). Muhammad Faisal Manzoor: Formal analysis (equal); Writing‐review & editing (equal).
ACKNOWLEDGMENT
The research was funded by the higher education of commission (HEC) of Pakistan.
Farid, W., Masud, T., Sohail, A., Ahmad, N., Naqvi, S. M. S., Khan, S., Ali, A., Khalifa, S. A., Hussain, A., Ali, S., Saghir, M., Siddeeg, A., & Manzoor, M. F. (2021). Gastrointestinal transit tolerance, cell surface hydrophobicity, and functional attributes of Lactobacillus Acidophilus strains isolated from Indigenous Dahi. Food Science & Nutrition, 9, 5092–5102. 10.1002/fsn3.2468
Contributor Information
Tariq Masud, Email: drmasud_tariq@hotmail.com.
Azhari Siddeeg, Email: azhari_siddeeg@uofg.edu.sd.
Muhammad Faisal Manzoor, Email: faisaluos26@gmail.com.
DATA AVAILABILITY STATEMENT
The dataset supporting the conclusions of this article is included within the article.
REFERENCES
- Afify, A.‐ E.‐M.‐ M., Romeilah, R. M., Sultan, S. I., & Hussein, M. M. (2012). Antioxidant activity and biological evaluations of probiotic bacteria strains. International Journal of Academic Research, 4(6), 131–139. [Google Scholar]
- Arora, D. S., & Chandra, P. (2010). Assay of antioxidant potential of two Aspergillus isolates by different methods under various physio‐chemical conditions. Brazilian Journal of Microbiology, 41(3), 765–777. 10.1590/S1517-83822010000300029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel, S., Bernardi, T., & Michaud, P. (2011). New perspectives for Lactobacilli exopolysaccharides. Biotechnology Advances, 29(1), 54–66. 10.1016/j.biotechadv.2010.08.011 [DOI] [PubMed] [Google Scholar]
- Behbahani, B. A., Noshad, M., & Falah, F. (2019). Inhibition of Escherichia coli adhesion to human intestinal Caco‐2 cells by probiotic candidate Lactobacillus plantarum strain L15. Microbial Pathogenesis, 136, 103677. [DOI] [PubMed] [Google Scholar]
- Chaffanel, F., Charron‐Bourgoin, F., Soligot, C., Kebouchi, M., Bertin, S., Payot, S., Le Roux, Y., & Leblond‐Bourget, N. (2018). Surface proteins involved in the adhesion of Streptococcus salivarius to human intestinal epithelial cells. Applied Microbiology and Biotechnology, 102(6), 2851–2865. 10.1007/s00253-018-8794-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza, B. M. S., Borgonovi, T. F., Casarotti, S. N., Todorov, S. D., & Penna, A. L. B. (2019). Lactobacillus casei and Lactobacillus fermentum strains isolated from mozzarella cheese: Probiotic potential, safety, acidifying kinetic parameters and viability under gastrointestinal tract conditions. Probiotics and Antimicrobial Proteins, 11(2), 382–396. 10.1007/s12602-018-9406-y [DOI] [PubMed] [Google Scholar]
- de Vries, M. C., Vaughan, E. E., Kleerebezem, M., & de Vos, W. M. (2006). Lactobacillus plantarum—Survival, functional and potential probiotic properties in the human intestinal tract. International Dairy Journal, 16(9), 1018–1028. 10.1016/j.idairyj.2005.09.003 [DOI] [Google Scholar]
- Deepak, V.Ram Kumar Pandian, S., Sivasubramaniam, S. D., Nellaiah, H., & Sundar, K. (2016). Optimization of anticancer exopolysaccharide production from probiotic Lactobacillus acidophilus by response surface methodology. Preparative Biochemistry and Biotechnology, 46(3), 288–297. [DOI] [PubMed] [Google Scholar]
- Deepak, V., Ramachandran, S., Balahmar, R. M., Pandian, S. R. K., Sivasubramaniam, S. D., Nellaiah, H., & Sundar, K. (2016). In vitro evaluation of anticancer properties of exopolysaccharides from Lactobacillus acidophilus in colon cancer cell lines. Vitro Cellular & Developmental Biology‐Animal, 52(2), 163–173. 10.1007/s11626-015-9970-3 [DOI] [PubMed] [Google Scholar]
- Farid, W., Masud, T., Sohail, A., Naqvi, S., & Qazalbash, M. A. (2016). Molecular characterization and 16S rRNA sequence analysis of probiotic lactobacillus acidophilus isolated from indigenous Dahi (Yoghurt). International Journal of Bioscience, 9(5), 19–27. [Google Scholar]
- Feldmane, J., Semjonovs, P., & Ciprovica, I. (2013). Potential of exopolysaccharides in yoghurt production. International Journal of Nutrition and Food Engineering, 7(8), 767–770. [Google Scholar]
- Food and Agriculture Organization . (2006). Probiotics in food: Health and nutritional properties and guidelines for evaluation. FAO. [Google Scholar]
- Gao, J. (2012). Antioxidant activity of Lacbacillus rhamnosus strains. M. Sc. Thesis, China. [Google Scholar]
- Gilliland, S., Nelson, C., & Maxwell, C. (1985). Assimilation of cholesterol by Lactobacillus acidophilus. Applied and Environmental Microbiology, 49(2), 377–381. 10.1128/aem.49.2.377-381.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., Morelli, L., Canani, R. B., Flint, H. J., Salminen, S., Calder, P. C., & Sanders, M. E. (2014). The International scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Reviews Gastroenterology & Hepatology, 11(8), 506–514. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- Huang, C.‐H., Ho, C.‐Y., Chen, C.‐T., Hsu, H.‐F., & Lin, Y.‐H. (2019). Probiotic BSH activity and anti‐obesity potential of Lactobacillus plantarum strain TCI378 isolated from Korean Kimchi. Preventive Nutrition and Food Science, 24(4), 434–441. 10.3746/pnf.2019.24.4.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimwe, N., Daliri, E. B., Lee, B. H., Fang, F., & Du, G. (2015). The perspective on cholesterol‐lowering mechanisms of probiotics. Molecular Nutrition & Food Research, 59(1), 94–105. 10.1002/mnfr.201400548 [DOI] [PubMed] [Google Scholar]
- Ji, K., Jang, N. Y., & Kim, Y. T. (2015). Isolation of lactic acid bacteria showing antioxidative and probiotic activities from kimchi and infant feces. Journal of Microbiology and Biotechnology, 25(9), 1568–1577. 10.4014/jmb.1501.01077 [DOI] [PubMed] [Google Scholar]
- Jones, M. L., Chen, H., Ouyang, W., Metz, T., & Prakash, S. (2004). Microencapsulated genetically engineered Lactobacillus plantarum 80 (pCBH1) for bile acid deconjugation and its implication in lowering cholesterol. Journal of Biomedicine and Biotechnology, 2004(1), 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose, N. M., Bunt, C. R., & Hussain, M. A. (2015). Comparison of microbiological and probiotic characteristics of lactobacilli isolates from dairy food products and animal rumen contents. Microorganisms, 3(2), 198–212. 10.3390/microorganisms3020198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik, J. K., Kumar, A., Duary, R. K., Mohanty, A. K., Grover, S., & Batish, V. K. (2009). Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS One, 4(12), e8099. 10.1371/journal.pone.0008099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghi, M., Kermanshahi, R. K., Yaghoobi, M. M., Zarkesh‐Esfahani, S., & Baghizadeh, A. (2010). Assessment of bile salt effects on S‐layer production, slp gene expression and some physicochemical properties of Lactobacillus acidophilus ATCC 4356. Journal of Microbiology and Biotechnology, 20(4), 749–756. [PubMed] [Google Scholar]
- Kumar, M., Nagpal, R., Kumar, R., Hemalatha, R., Verma, V., Kumar, A., Chakraborty, C., Singh, B., Marotta, F., Jain, S., & Yadav, H. (2012). Cholesterol‐lowering probiotics as potential biotherapeutics for metabolic diseases. Experimental Diabetes Research, 2012, 1–14. 10.1155/2012/902917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, T.‐W., Tseng, S.‐C., & Wang, S.‐L. (2016). Production and characterization of antioxidant properties of exopolysaccharide (s) from Peanibacillus mucilaginosus TKU032. Marine Drugs, 14(2), 40. 10.3390/md14020040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, H.‐J., Kim, S.‐Y., & Lee, W.‐K. (2004). Isolation of cholesterol‐lowering lactic acid bacteria from human intestine for probiotic use. Journal of Veterinary Science, 5(4), 391–395. 10.4142/jvs.2004.5.4.391 [DOI] [PubMed] [Google Scholar]
- Liu, C. F., Tseng, K. C., Chiang, S. S., Lee, B. H., Hsu, W. H., & Pan, T. M. (2011). Immunomodulatory and antioxidant potential of Lactobacillus exopolysaccharides. Journal of the Science of Food and Agriculture, 91(12), 2284–2291. 10.1002/jsfa.4456 [DOI] [PubMed] [Google Scholar]
- Liu, X., Liu, W., Zhang, Q., Tian, F., Wang, G., Zhang, H., & Chen, W. (2013). Screening of lactobacilli with antagonistic activity against enteroinvasive Escherichia coli. Food Control, 30(2), 563–568. 10.1016/j.foodcont.2012.09.002 [DOI] [Google Scholar]
- Lye, H.‐S., Rahmat‐Ali, G. R., & Liong, M.‐T. (2010). Mechanisms of cholesterol removal by lactobacilli under conditions that mimic the human gastrointestinal tract. International Dairy Journal, 20(3), 169–175. 10.1016/j.idairyj.2009.10.003 [DOI] [Google Scholar]
- Monteagudo‐Mera, A., Rastall, R. A., Gibson, G. R., Charalampopoulos, D., & Chatzifragkou, A. (2019). Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Applied Microbiology and Biotechnology, 103(16), 6463–6472. 10.1007/s00253-019-09978-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwanyanwu, C., & Abu, G. (2013). Influence of growth media on hydrophobicity of phenol‐utilizing bacteria found in petroleum refinery effluent. International Research Journal of Biological Sciences, 2, 6–11. [Google Scholar]
- Oh, Y. J., & Jung, D. S. (2015). Evaluation of probiotic properties of Lactobacillus and Pediococcus strains isolated from Omegisool, a traditionally fermented millet alcoholic beverage in Korea. LWT‐Food Science and Technology, 63(1), 437–444. [Google Scholar]
- Öner, Ö., Aslim, B., & Aydaş, S. B. (2014). Mechanisms of cholesterol‐lowering effects of lactobacilli and bifidobacteria strains as potential probiotics with their bsh gene analysis. Journal of Molecular Microbiology and Biotechnology, 24(1), 12–18. [DOI] [PubMed] [Google Scholar]
- Pan, M., Kumaree, K. K., & Shah, N. P. (2017). Physiological changes of surface membrane in Lactobacillus with prebiotics. Journal of Food Science, 82(3), 744–750. [DOI] [PubMed] [Google Scholar]
- Patel, A. K., Singhania, R. R., Pandey, A., & Chincholkar, S. B. (2010). Probiotic bile salt hydrolase: Current developments and perspectives. Applied Biochemistry and Biotechnology, 162(1), 166–180. 10.1007/s12010-009-8738-1 [DOI] [PubMed] [Google Scholar]
- Pereira, D. I., McCartney, A. L., & Gibson, G. R. (2003). An in vitro study of the probiotic potential of a bile‐salt‐hydrolyzing Lactobacillus fermentum strain, and determination of its cholesterol‐lowering properties. Applied and Environmental Microbiology, 69(8), 4743–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano, M. B., Viale, S., Conti, S., Fadda, M. E., Deplano, M., Melis, M. P., Deiana, M., & Cosentino, S. (2014). Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from Sardinian dairy products. BioMed Research International, 2014, 286390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plourde‐Owobi, L., Seguin, D., Baudin, M.‐A., Moste, C., & Rokbi, B. (2005). Molecular characterization of Clostridium tetani strains by pulsed‐field gel electrophoresis and colony PCR. Applied and Environmental Microbiology, 71(9), 5604–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak‐Berecka, M., Waśko, A., & Kubik‐Komar, A. (2014). Optimization of culture conditions for exopolysaccharide production by a probiotic strain of Lactobacillus rhamnosus E/N. Polish Journal of Microbiology, 63(2), 253–257. 10.33073/pjm-2014-034 [DOI] [PubMed] [Google Scholar]
- Ramasamy, K., Abdullah, N., Wong, M. C., Karuthan, C., & Ho, Y. W. (2010). Bile salt deconjugation and cholesterol removal from media by Lactobacillus strains used as probiotics in chickens. Journal of the Science of Food and Agriculture, 90(1), 65–69. [DOI] [PubMed] [Google Scholar]
- Rudel, L. L., & Morris, M. (1973). Determination of cholesterol using o‐phthalaldehyde. Journal of Lipid Research, 14(3), 364–366. 10.1016/S0022-2275(20)36896-6 [DOI] [PubMed] [Google Scholar]
- Rühmann, B., Schmid, J., & Sieber, V. (2015). High throughput exopolysaccharide screening platform: From strain cultivation to monosaccharide composition and carbohydrate fingerprinting in one day. Carbohydrate Polymers, 122, 212–220. 10.1016/j.carbpol.2014.12.021 [DOI] [PubMed] [Google Scholar]
- Sahadeva, R., Leong, S., Chua, K., Tan, C., Chan, H., Tong, E., Wong, S., & Chan, H. (2011). Survival of commercial probiotic strains to pH and bile. International Food Research Journal, 18(4), 1515–1522. [Google Scholar]
- Saito, K., Tomita, S., & Nakamura, T. (2019). Aggregation of Lactobacillus brevis associated with decrease in pH by glucose fermentation. Bioscience, Biotechnology, and Biochemistry, 83(8), 1523–1529. [DOI] [PubMed] [Google Scholar]
- Schillinger, U., Guigas, C., & Holzapfel, W. H. (2005). In vitro adherence and other properties of lactobacilli used in probiotic yoghurt‐like products. International Dairy Journal, 15(12), 1289–1297. [Google Scholar]
- Sirilun, S., Chaiyasut, C., Kantachote, D., & Luxananil, P. (2010). Characterisation of non human origin probiotic Lactobacillus plantarum with cholesterol‐lowering property. African Journal of Microbiology Research, 4(10), 994–1000. [Google Scholar]
- Soomro, A. H., & Masud, T. (2012). Probiotic characteristics of Lactobacillus spp. isolated from fermented milk product dahi. Food Science and Technology Research, 18(1), 91–98. [Google Scholar]
- Suresh Kumar, A., Mody, K., & Jha, B. (2007). Bacterial exopolysaccharides–A perception. Journal of Basic Microbiology, 47(2), 103–117. 10.1002/jobm.200610203 [DOI] [PubMed] [Google Scholar]
- Tahri, K., Grill, J. P., & Schneider, F. (1997). Involvement of trihydroxyconjugated bile salts in cholesterol assimilation by bifidobacteria. Current Microbiology, 34(2), 79–84. 10.1007/s002849900148 [DOI] [PubMed] [Google Scholar]
- Tsai, C.‐C., Lin, P.‐P., Hsieh, Y.‐M., Zhang, Z.‐Y., Wu, H.‐C., & Huang, C.‐C. (2014). Cholesterol‐lowering potentials of lactic acid bacteria based on bile‐salt hydrolase activity and effect of potent strains on cholesterol metabolism in vitro and in vivo. The Scientific World Journal, 2014, 690752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyfa, A., Kunicka‐Styczyńska, A., & Zabielska, J. (2015). Evaluation of hydrophobicity and quantitative analysis of biofilm formation by Alicyclobacillus sp. Acta Biochimica Polonica, 62(4), 785–790. 10.18388/abp.2015_1133 [DOI] [PubMed] [Google Scholar]
- Xing, J., Wang, G., Zhang, Q., Liu, X., Gu, Z., Zhang, H., Chen, Y. Q., & Chen, W. (2015). Determining antioxidant activities of lactobacilli cell‐free supernatants by cellular antioxidant assay: A comparison with traditional methods. PLoS One, 10(3), e0119058. 10.1371/journal.pone.0119058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, Z.‐J., Fu, Y.‐R., Li, M., Gao, K.‐S., & Zhang, X.‐G. (2009). Effect of LTA isolated from bifidobacteria on D‐galactose‐induced aging. Experimental Gerontology, 44(12), 760–765. 10.1016/j.exger.2009.08.011 [DOI] [PubMed] [Google Scholar]
- Zhou, X.‐X., Pan, Y.‐J., Wang, Y.‐B., & Li, W.‐F. (2007). In vitro assessment of gastrointestinal viability of two photosynthetic bacteria, Rhodopseudomonas palustris and Rhodobacter sphaeroides. Journal of Zhejiang University Science B, 8(9), 686–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, F., Yu, R., Feng, X., Chen, L., Zeng, Z., Khaskheli, G. B., Ma, H., & Chen, S. (2016). Characterization and in vitro properties of potential probiotic Bifidobacterium strains isolated from breast‐fed infant feces. Annals of Microbiology, 66(3), 1027–1037. 10.1007/s13213-015-1187-x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is included within the article.


