Abstract
Over the last few decades, a wealth of evidence has formed the basis for “the Old Friends hypothesis” suggesting that, in contrast to the past, increasingly people are living in environments with limited and less diverse microbial exposure, with potential consequences for their health. Hence, including safe live or heat‐killed microbes in the diet may be beneficial in promoting and maintaining human health. In order to assess the safety of microbes beyond the current use of standardized cultures and probiotic supplements, new approaches are being developed. Here, we present evidence for the safety of heat‐killed Mycolicibacterium aurum Aogashima as a novel food, utilizing the decision tree approach developed by Pariza and colleagues (2015). We provide evidence that the genome of M. aurum Aogashima is free of (1) genetic elements associated with pathogenicity or toxigenicity, (2) transferable antibiotic resistance gene DNA, and (3) genes coding for antibiotics used in human or veterinary medicine. Moreover, a 90‐day oral toxicity study in rats showed that (4) the no observed adverse effect level (NOAEL) was the highest concentration tested, namely 2000 μg/kg BW/day. We conclude that oral consumption of heat‐killed M. aurum Aogashima is safe and warrants further evaluation as a novel food ingredient.
Keywords: DSM 33539, Mycolicibacterium aurum Aogashima, novel food, safety
blurb for etocThere is growing realization of the health benefits of restoring human exposure to a more diverse range of microbes than we experience in increasingly urbanized lifestyles. In addition to probiotics, other environmental nonpathogenic organisms, such as environmental saprophytic nontuberculous (NTB) mycobacteria species, were once commonly present in the human diet. Here, we present evidence for the safety of heat‐killed Mycolicibacterium aurum Aogashima as a novel food, utilizing the decision tree approach developed by Pariza and colleagues (2015).
1. INTRODUCTION
The interaction between the human host and nonpathogenic ubiquitous environmental microorganisms, present throughout human evolution, recently emerged as an area of scientific interest and has evolved into “the Old Friends hypothesis” (Flandroy et al., 2018; Lowry et al., 2016; Rook et al., 2004, 2013). This awareness has reached consumers alike, who are increasingly willing to adjust their dietary habits to achieve improved well‐being (Marco et al., 2020). Indeed, intake of specific food and food supplements is one way to modulate exposure to what have been broadly considered “good” bacteria. The long list of “healthy” foods containing such bacteria includes fermented dairy products like yogurt and kefir, as well as fermented foods such as miso, kimchi, and sauerkraut and beverages such as kombucha tea. Members of the genera Bacillus, Bifidobacterium, Enterococcus, Lactobacillus, Saccharomyces, and Streptococcus are most commonly found in these foods, and together they are known as “probiotics” (Di Cerbo et al., 2016; Hori et al., 2020). According to the revised definition of the Food and Agriculture Organization (FAO)/World Health Organization (WHO), as well as in the public perception, probiotics are nonpathogenic live microorganisms that, when administered in adequate amounts, confer a health benefit to the host, such as improvement in metabolism and intestinal flora and modulation of immune functions (Aponte et al., 2020; FAO & WHO, 2002; Hill et al., 2014; Wilkins & Sequoia, 2017). The probiotic market is growing rapidly, buoyed by both foods and supplements intended to enhance wellness in healthy individuals, and by preparations for the dietary management of diseases (Grumet et al., 2020).
In addition to probiotics, other environmental nonpathogenic organisms are, or were at some point, commonly present in the human diet, such as environmental saprophytic nontuberculous (NTB) mycobacteria species. Based on recent comparative genomic studies, the genus Mycobacterium (Lehmann, 1896) was divided into an emended genus Mycobacterium, to which pathogenic species belong, and four novel genera: Mycolicibacter (type species: Mycolicibacter terrae), Mycolicibacillus (type species: Mycolicibacillus trivialis), Mycobacteroides (type species: Mycobacteroides abscessus), and Mycolicibacterium (type species: Mycolicibacterium fortuitum) (Gupta et al., 2018). The genus Mycolicibacterium encompasses rapidly growing NTB species, many of which have routinely been isolated from municipal water supplies (Falkinham et al., 2001; Falkinham, 2009; Fernandez‐Rendon et al., 2012; Imwidthaya et al., 1989; Kubalek & Mysak, 1996; Le Dantec et al., 2002a, 2002b; Martın et al., 2000; Moghim et al., 2012; Nasr‐Esfahani et al., 2012; Pontiroli et al., 2013; Scarlata et al., 1985; Vaerewijck et al., 2005). NTB mycobacteria, which include the mycolicibacteria, are not a permanent constituent of the microbiome, but because they have been regularly encountered in the diet and the environment, there is evidence for their evolutionary adaptedness (Rook, 2010). Interestingly, akin to the bacteria which make up the gut and skin microbiota, researchers have now identified communities of several of these operational mycobacterial taxonomic units in the oral cavity of healthy individuals (Macovei et al., 2015). This is presumably a reflection of the significant exposure to environmental NTB mycobacteria by this route. The extent to which NTB mycobacteria such as mycolicibacteria hold promise, like probiotics, for influencing human well‐being is the subject of ongoing research. Nevertheless, “the Old Friends hypothesis” makes a case for their benefit to human health as revealed by the drastic reduction of exposure to saprophytic environmental NTB mycobacteria in modern living conditions (Flandroy et al., 2018; Lowry et al., 2016; Rook et al., 2004, 2013).
Until recently, the assumption has been that probiotics should be viable to exert positive effects. Instead, there is now increasing evidence to show that nonviable probiotics maintain their health‐promoting benefits and a new term “postbiotic” has been coined to indicate preparations of inanimate microorganisms and/or their components that confer a health benefit to the host (Aguilar‐Toalá et al., 2018; Barros et al., 2020; Seminen et al., 2021; Taverniti & Guglielmetti, 2011). From a commercial standpoint, the use of nonviable bacteria has several advantages, including easing the challenges associated with product storage to maintain viability, reduction of safety concerns arising from horizontal virulence gene transfer from pathogenic bacteria, and the ability to deliver exact numbers of microorganisms per dose. In light of these issues, nonviable bacteria are now under consideration as novel food ingredients. In this report, we present evidence for the safety of heat‐killed Mycolicibacterium aurum Aogashima as a novel food ingredient. This is an environmental saprophytic organism which may not have the documented history of safe use that food‐associated probiotics have, but nonetheless, is likely to have been evolutionarily present in the diet, through exposure to untreated and even treated water supplies. The safety of this novel food was determined using the decision tree approach developed by Pariza and colleagues which relies on assessment of lack of allergenicity risk, confirmation that resistance to various antimicrobials is intrinsic and nontransmissible and that no harmful effects are detected in standard toxicology testing (Pariza et al., 2015). Our data support the conclusion that heat‐killed M. aurum Aogashima is safe as a novel food ingredient.
2. MATERIALS AND METHODS
2.1. Manufacture
M. aurum Aogashima has been deposited at the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Germany) under the accession number DSM 33539. M. aurum Aogashima is manufactured following Good Manufacturing Practice (GMP) and Good Laboratory Practice (GLP) principles. The organism is grown in a bioreactor of either five or twenty‐five liters. Once an appropriate biomass is reached, the bacteria are recovered by centrifugation and resuspension in water, before heat inactivation at 121ºC for ≥20min. The resulting M. aurum Aogashima biomass is then further diluted with water and stored prior to use.
2.2. Safety evaluation process
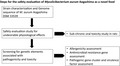
The safety of M. aurum Aogashima was assessed based on the decision tree approach developed by Pariza and colleagues (2015). A flow chart describing the steps is depicted in Figure 1.
FIGURE 1.
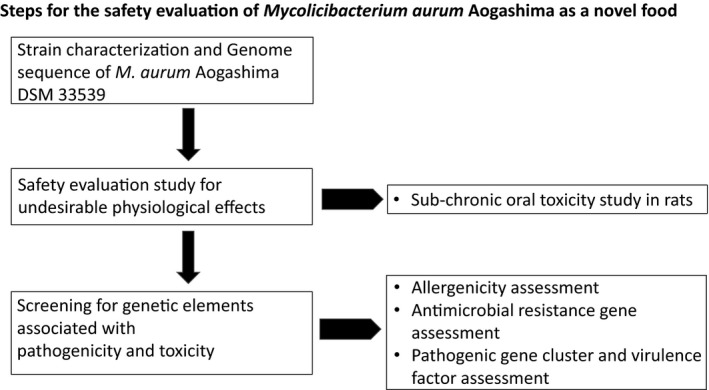
Flow chart describing the steps of the safety evaluation process based on Pariza et al., (2015)
2.3. Subchronic oral toxicity study
The safety of heat‐killed M. aurum Aogashima was investigated in a 90‐day toxicity study in rats. Doses tested were selected on the basis of a 14‐day oral dose range finding study. Studies were GLP compliant, performed at Sequani Ltd (UK) according to United Kingdom GLP Regulations 1999, SI 1999 No. 3,106, as amended by SI 2004 No. 994, in accordance with the Organization for Economic Cooperation and Development (OECD) Guidelines (OECD, 1998). Briefly, 6‐ to 8‐week‐old male (n = 40) and female (n = 40) Crl:WI(Han) rats (Charles River, UK) were divided in groups of 10 males and 10 females and were dosed at 0 (vehicle control), 20, 200, or 2000 μg/kg/day of heat‐killed M. aurum Aogashima, once daily by oral gavage, at a dose volume of 1 mL/kg body weight for at least 90 days, until the day before necropsy. Animals were housed in groups of 5, by sex and provided food and water ad libitum.
Animals were examined twice daily for mortality and morbidity. Any clinical signs of toxicity or changes in behavior or appearance were checked for daily. Body weights and food intake were recorded weekly until necropsy. Blood samples were taken for clinical pathology during week 13 according to OECD guidelines (1998). Hematological parameters investigated included changes in immune cell population counts. Blood chemistry parameters measured included markers associated with liver or kidney cellular toxicity, such as alanine and aspartate aminotransferase (ALT and AST, respectively), alkaline phosphatase (AP), urea, and creatinine. Animals were also subjected to a functional observational battery consisting of standard arena observations at predose and once weekly, together with an assessment at week 13 which included grip strength, motor activity, and sensorimotor responses to visual, acoustic, and proprioceptive stimuli according to OECD test guidelines (OECD, 1998). At the end of the treatment period, all animals were subjected to a gross necropsy, internal organs were weighed, and organ tissues from the control and high dose animals were examined microscopically.
2.4. Genome sequencing and analysis
DNA was extracted from a culture of M. aurum Aogashima as described in Amaro et al., (2008). Genome sequencing was performed using an Illumina MiSeq instrument, as previously described (Sangal et al., 2015). The genomes were assembled into contigs using SPAdes 3.9.0 with a kmer length of 127 and subsequently annotated using the Rapid Annotation of microbial genomes Subsystems Technology (RAST) server (Aziz et al., 2008; Bankevich et al., 2012).
2.5. Allergenicity assessment
Allergenicity potential of M. aurum Aogashima was assessed by AllerCatPro (https://allercatpro.bii.a‐star.edu.sg/), the most up‐to‐date database, comprising 4,180 unique allergenic protein sequences (Maurer‐Stroh et al., 2019). Briefly, linear sequences in the genome of M. aurum Aogashima were first compared to the allergen database to identify sequence windows of 80 residues with at least 35% of identity with proteins known to be allergenic as defined by FAO & WHO (2001). The amino acid sequences of M. aurum Aogashima genome were obtained after translation of nucleotide sequences using Prodigal software v2.6.3. The sequences with an identity above this threshold were then 3D‐modeled, and a B‐cell epitope‐like 3D surface was calculated and compared. Proteins with epitopes presenting an identity level of above 35% were considered allergens as outlined in Maurer‐Stroh et al., (2019).
2.6. Antimicrobial resistance gene assessment
The presence of genes coding for antibiotic resistance (AMR) was assessed in the genome of M. aurum Aogashima. The whole genomic sequence was compared against ResFinder databases and the Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al., 2020; Zankari et al., 2012). Briefly, in silico genome analysis for the AMR genes was carried out by ResFinder 3.2 webserver which encompasses 15‐drug classes in its database: aminoglycoside, beta‐lactam, colistin, a fluoroquinolone, fosfomycin, fusidic acid, glycopeptide, macrolide‐lincosamide‐streptogramin B, nitroimidazole, oxazolidinone, phenicol, rifampicin, sulphonamide, tetracycline, and trimethoprim (Zankari et al., 2012). The percent identity and perfect alignment were set at 70% and 60%, respectively. The minimum length or the number of nucleotides that must overlap a resistant gene to count as a hit was set at the default of 60%.
The genome sequence of strain M. aurum Aogashima was interrogated for the presence of AMR genes based on CARD and using Resistance Gene Identifier (RGI) software for resistome analysis and prediction (Alcock et al., 2020). Each predicted AMR gene was manually mapped and annotated using the SEED and the RAST server (Aziz et al., 2012). Protein domains of AMR genes were confirmed after comparison with those available in the Conserved Domains Database (CDD) of NCBI (Marchler‐Bauer et al., 2015). Any hits were reported and analyzed.
2.7. Pathogenic gene clusters and virulence factors assessment
The draft genome sequence of M. aurum Aogashima was screened for pathogenic island and virulence factors using the Virulence Factor database (VFDB) (Liu et al., 2019). Experimentally validated virulence factors of major medically important bacterial pathogens belonging to 24 genera were considered. In addition, predicted coding sequences were identified using the GLIMMER3 system (system for finding genes in microbial DNA) prior to using the VFanalyzer tool (Virulence Factor analyzer tool). Lastly, blastp and Conserved Domain tools of NCBI were used to identify the virulence factors associated amino acid sequences of M. aurum and determine their functional similarity with those of Mycobacterium tuberculosis H37Rv. The established threshold of 60% for functional protein similarity was adopted (Addou et al., 2009).
2.8. Statistical Analysis
In vivo data were analyzed using Graph Pad Prism to give group mean values and standard error. Where appropriate and within each sex, one‐way ANOVA followed by Sidak's multiple comparisons test was used to determine statistical differences upon comparison of groups receiving different doses of heat‐killed M. aurum Aogashima versus the control group.
3. RESULTS
3.1. Subchronic oral toxicity study
A 14‐day oral dose range finding study in Crl:WI(Han) rats was performed to determine doses to be tested further in a 90‐day oral toxicity study. This dose range study showed that administration of both 200 and 2000μg/kg/day was well tolerated and 2000 μg/kg/day was selected as the highest dose level in a subchronic oral toxicity study. Eighty Crl:WI(Han) rats (40 males and 40 females) were allocated into different dose groups receiving heat‐killed M. aurum Aogashima orally at 0 (vehicle control), 20, 200, and 2000 μg/kg/day (10 males and 10 females per group) for 90 days. Daily visual examinations from the start of treatment showed no deaths, no treatment‐related clinical signs of morbidity, toxicity, nor changes in behavior. Weekly measurements of body weight and food intake revealed no significant differences between groups. All groups gained a similar amount of weight (Figure 2) and ate a similar amount of food (data not shown) when compared to control groups. Animals showed no evidence for treatment‐related neurotoxicity based on functional observation battery assessments. Indeed, there were no effects on functional arena observations or on grip strength or motor activity and sensorimotor responses to visual, acoustic, and proprioceptive stimuli (data not shown). At the end of the treatment period, all animals were subjected to a gross necropsy where organs were weighed and examined macroscopically. We detected no effect on organ weights in the male groups regardless of treatment. In the female group, we observed only a significant decrease in the liver weight and only in the group receiving 20 μg/kg/day (Table 1). Nevertheless, there were no differences detected in the percentages of organ weight in relation to body weight in the 20 μg/kg/day dose group compared to control group (3.32 ± .067 versus 3.39 ± .037%, respectively). Moreover, no abnormal macroscopic observations were reported. Hence, because the observed difference in the weights of livers was not dose dependent, did not translate into changes in organ weight percentages or in macroscopic changes, and was found not to be associated with increases in liver enzymes indicative of hepatocellular toxicity such as AST, ALT, and AP (Table 2), it was deemed not to have biological significance. Furthermore, there were no macroscopic or when performed microscopic abnormalities related to oral consumption of heat‐killed M. aurum Aogashima in any organs of any groups.
FIGURE 2.
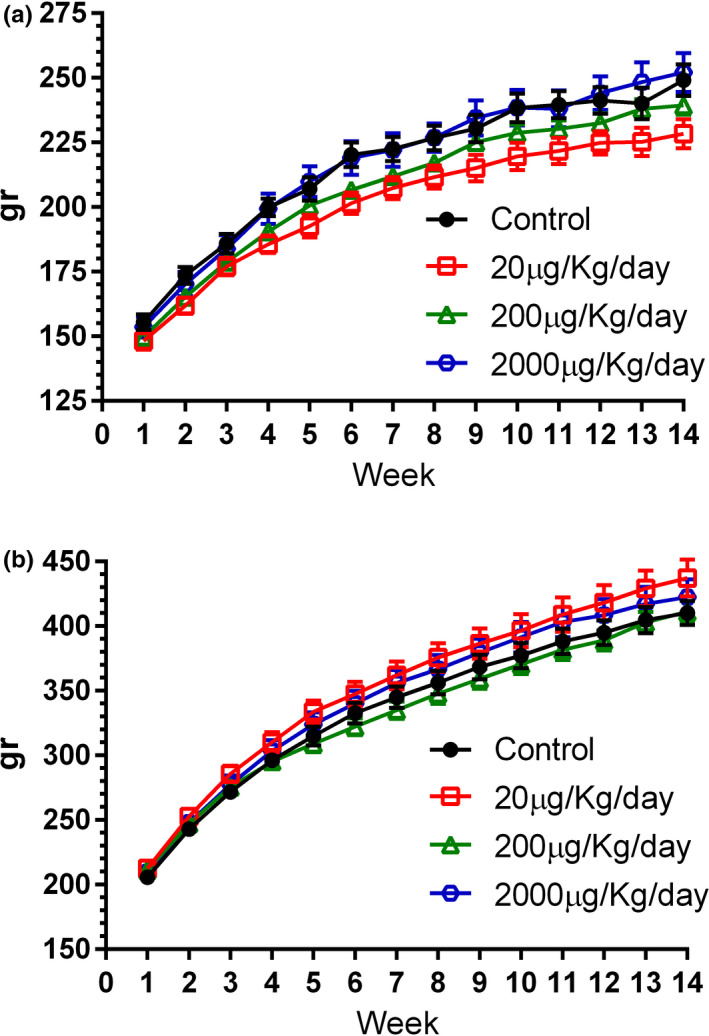
Comparison of body weight (gr) in female (A) and male (B) rats receiving vehicle control or 20, 200, and 2000 μg/kg BW/day (10 males and 10 females per group) M. aurum Aogashima for 90 days. There was no treatment‐related effect on body weight or overall body weight gain. Data are shown as mean ± SEM
TABLE 1.
Effects of feeding different doses of M. aurum Aogashima on selected tissue organs. Data are shown as mean (n = 10) ± SEM. No significant differences were observed compared to relevant vehicle control group in the males; * indicates significant differences in the female group
| Group |
Heart gr |
Kidneys gr |
Liver gr |
Spleen gr |
Thyroid mg |
Adrenal mg |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | |
|
0 μg/Kg/day Vehicle Control |
.85 ± .04 | 1.15 ± .04 | 1.64 ± .05 | 2.41 ± .05 | 8.47 ± .28 | 13.58 ± .44 | .57 ± .03 | .65 ± .02 | 18.80 ± 1.11 | 22.2 ± .44 | 73 ± 4.33 | 60 ± 3 |
| 20 μg/Kg/day | .77 ± .03 | 1.26 ± .04 | 1.57 ± .05 | 2.51 ± .09 |
7.54 ± .26 *p =.039 |
13.79 ± .43 | .56 ± .03 | .68 ± .03 | 16.70 ± 1.32 | 23.1 ± .82 | 80 ± 3.67 | 57 ± 1.67 |
| 200 μg/Kg/day | .82 ± .02 | 1.17 ± .04 | 1.61 ± .05 | 2.46 ± .09 | 8.02 ± .22 | 13.49 ± .45 | .56 ± .02 | .65 ± .02 | 17.90 ± .88 | 21.70 ± 1.11 | 75 ± 2.67 | 57 ± 3.33 |
| 2000 μg/Kg/day | .86 ± .02 | 1.22 ± .04 | 1.66 ± .05 | 2.56 ± .07 | 8.42 ± .26 | 13.82 ± .54 | .54 ± .04 | .69 ± .02 | 17.10 ± 1.42 | 22.20 ± 1.39 | 73 ± 2.33 | 59 ± 3.33 |
TABLE 2.
Effects of feeding different doses of M. aurum Aogashima on selected blood chemistry parameters. Data are shown as mean(n = 10) ± SEM. No significant differences were observed compared to relevant vehicle control group
| Group |
Alanine Aminotrasferase U/l |
Aspartate Aminotransferase U/l |
Alkaline Phosphatase U/l |
Urea mg/dl |
Creatine mg/dl |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | |
|
0 μg/Kg/day Vehicle Control |
29.40 ± 2.70 | 36.90 ± 2.08 | 71.70 ± 11.02 | 61.50 ± 2.69 | 49.90 ± 4.09 | 114.30 ± 8.31 | 34.77 ± 1.32 | 37.30 ± 1.61 | .26 ± .01 | .26 ± .01 |
| 20 μg/Kg/day | 28.20 ± 1.30 | 33.50 ± 2.28 | 64.30 ± 3.45 | 60.10 ± 2.50 | 57.80 ± 4.52 | 105.60 ± 6.06 | 34.38 ± .96 | 37.55 ± 1.16 | .25 ± .004 | .25 ± .01 |
| 200 μg/Kg/day | 28.10 ± .81 | 31.80 ± .62 | 59.00 ± 2.83 | 53.10 ± 1.24 | 51.60 ± 4.53 | 105.30 ± 7.89 | 34.5 ± 1.27 | 35.55 ± 1.1 | .26 ± .01 | .25 ± .01 |
| 2000 μg/Kg/day | 29.10 ± 1.37 | 31.80 ± 1.69 | 62.30 ± 5.77 | 60.80 ± 3.59 | 53.50 ± 4.26 | 104.30 ± 4.69 | 37.53 ± 2.03 | 34.63 ± .73 | .26 ± .003 | .22 ± .01 |
Prior to necropsy, blood samples were taken for clinical pathology analysis at week 13. Statistical analysis revealed no significant differences in the groups on any of the blood chemistry parameters which are routinely considered to be broadly indicative of hepatocellular or renal toxicity (Table 2). Among all blood chemistry parameters measured, we observed an out of “normal” range significant difference (*p <.05) in glucose levels compared to control animals. Levels (mg/dl) in control male rats (191.8 ± 7) were comparable to level in rats receiving 20 and 2000 μg/Kg/day (193.7 ± 6.5 and 196.3 ± 7, respectively) but significantly lower than levels in rats receiving 200 μg/Kg/day (221.1 ± 8.6). Similarly, levels in control female rats (163.7 ± 4.5) were comparable to levels in rats receiving 20 μg/Kg/day (179.8 ± 6.7) but significantly lower than those in rat receiving 200 and 2000 μg/Kg/day (191.7 ± 6 and 190.3 ± 5.2, respectively). As glucose levels were already higher than normal (106–184 in males and 89–163 mg/dl in females) in the control groups and rats were not fasted overnight prior to blood sampling, we consider these may be normal biological variations due to food consumption and circadian rhythms.
Hematological parameters were also assessed. There were no statistically significant changes in total white blood cell and immune cell population specific counts in the female groups, aside from a significant decrease in monocytes in the group receiving 200 μg/Kg/day (Table 3). However, when cell population percentages were calculated, no significant differences were detected in any cell populations, including monocytes, regardless of dose received (Table 4). In males, we observed a significant decrease in white blood cell, neutrophils, and lymphocytes absolute counts only in the group receiving 200 μg/Kg/day (Table 3). However, we found no evidence for changes in the percentages of these populations or in any other immune cell populations (Table 4). Moreover, values remained within normal range (white blood cells 1.98–11.06 x 103/μl; neutrophils .33–1.98 103/μl). Due to the lack of a dose relationship, given no differences were detected in the highest dose groups (which received concentrations 10 times of those where differences were observed), and because there were no changes in overall immune cell population percentages, we consider these observations part of normal biological variation rather than any effect of oral consumption of M. aurum Aogashima. Hence, the reported data concluded no observed adverse effect level (NOAEL) at all doses tested including the highest dose tested of 2000μg/Kg/day.
TABLE 3.
Effects of feeding different doses of M. aurum Aogashima on selected hematological parameters. Data are shown as mean (n = 10) ± SEM. * indicates significant differences in the male and female group
| Group |
White Blood Cell 103/μl |
Neutrophils 103/μl |
Lymphocytes 103/μl |
Monocytes 103/μl |
Eosinophils 103/μl |
Basophils 103/μl |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | |
|
0 μg/Kg/day Vehicle Control |
5.52 ± .47 | 7.79 ± .36 | .63 ± .10 | 1.± .09 | 4.56 ± .36 | 6.45 ± .29 | .16 ± .02 | .14 ± .02 | .13 ± .02 | 14 ± .01 | .01 ± .0 | .03 ± .0 |
| 20 μg/Kg/day | 5.45 ± .44 | 6.77 ± .26 | .70 ± .04 | .93 ± .05 | 4.45 ± .40 | 5.46 ± .25 | .13 ± .01 | .15 ± .03 | .12 ± .02 | .18 ± .03 | .01 ± .0 | .02 ± .0 |
| 200 μg/Kg/day | 4.39 ± .33 |
6.12 ± .39 *p =.009 |
.58 ± .07 |
.76 ± .05 *p =.045 |
3.57 ± .28 |
5.07 ± .4 *p =.026 |
.10 ± .01 *p =.016 |
.13 ± .01 | .11 ± .01 | .11 ± .01 | .01 ± .0 | .02 ± .0 |
| 2000 μg/Kg/day | 4.9 ± .32 | 6.83 ± .34 | .66 ± .08 | .87 ± .05 | 3.98 ± .27 | 5.66 ± .35 | .13 ± .02 | .12 ± .01 | .1 ± .01 | .13 ± .01 | .01 ± .0 | .02 ± .0 |
TABLE 4.
Effects of feeding different doses of M. aurum Aogashima on selected hematological parameters. Data are shown as mean (n = 10) ± SEM. No significant differences were observed compared to relevant vehicle control group
| Group |
% Neutrophils |
% Lymphocytes |
% Monocytes |
% Eosinophils |
% Basophils |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | ♀ | ♂ | |
|
0 μg/Kg/day Vehicle Control |
11.08 ± .97 | 12.79 ± .90 | 82.87 ± 1.14 | 82.81 ± .9 | 2.95 ± .43 | 1.72 ± .23 | 2.30 ± .22 | 1.79 ± .13 | .19 ± .03 | .33 ± .05 |
| 20 μg/Kg/day | 13.28 ± .87 | 13.82 ± .88 | 81.30 ± 1.08 | 80.6 ± 1.32 | 2.46 ± .19 | 2.25 ± .41 | 2.28 ± .33 | 2.58 ± .39 | .18 ± .03 | .26 ± .03 |
| 200 μg/Kg/day | 13.22 ± 1.31 | 12.89 ± 1.21 | 81.29 ± 1.34 | 82.31 ± 1.31 | 2.34 ± .15 | 2.08 ± .21 | 2.60 ± .18 | 1.87 ± .20 | .12 ± .03 | .29 ± .03 |
| 2000 μg/Kg/day | 13.45 ± 1.38 | 13.02 ± 1.12 | 81.23 ± 1.4 | 82.49 ± 1.33 | 2.60 ± .30 | 1.79 ± .15 | 2.04 ± .20 | 1.88 ± .18 | .16 ± .03 | .34 ± .07 |
3.2. Allergenicity
The allergenicity potential of M. aurum Aogashima was assessed by an innovative 3D‐modeling‐based analysis, using the AllerCatPro database. Only fifteen potentially allergenic protein sequences were detected with linear sequence window identity above the thresholds of 35% (Table 5). Most of the detected amino acids sequences in the genome of M. aurum Aogashima corresponded to allergenic proteins previously found in fungi (56%), while 26%, 15%, and 3% of the predicted proteins belong to foods, arthropods, and mammals (just one sequence), respectively. None of these proteins showed a 3D epitope identity, and therefore, we concluded that there was no evidence for allergenicity following consumption of heat‐killed M. aurum Aogashima (Table 5).
TABLE 5.
In silico genome analysis for allergenic proteins of M. aurum Aogashima using AllerCatPro server
| Known allergen hit name | % identity linear 88aa window | % identity 3D epitope | |
|---|---|---|---|
| Alcohol Dehydrogenase | Candida albicans | 41.1 – 67.5 | ‐ |
| ALF_CANAL fructose‐bisphosphate aldolase | Candida albicans | 55 | ‐ |
| Aldehyde dehydrogenase Cla h 10 | Cladosporium herbarum | 48.8 | ‐ |
| Mannitol dehydrogenase Cla h8 | Cladosporium herbarum | 37.5 – 38.2 | ‐ |
| NADP‐dependent mannitol dehydrogenase | Alternaria alternata | 37.7 – 40.8 | ‐ |
| Probable beta‐glucosidase ARB_05654 BGLA_ARTBC | Arthroderma benhamiae | 51.2 | ‐ |
| THIO_COPCM Thiredoxin | Coprinus comatus | 44.9 | ‐ |
| Asp f IAO | Aspergillus fumigatus | 43 | ‐ |
| Asp f FDH | Aspergillus fumigatus | 40 | ‐ |
| Pen C | Penicillum citrinum | 58.8 | ‐ |
| Seed maturation‐like protein precursor | Sesamum | 37.5 – 45 | ‐ |
| Tri a 34.0101 (glyceraldehyde−3‐phosphate dehydrogenase) | Triticum aestivum | 71.2 | ‐ |
| Aldehyde dehydrogenase‐like protein Tyrophagus | Tyrophagus putrescentiae | 56.2 | ‐ |
| Cul n 8 | Culicoides nubeculosus | 41.1 | ‐ |
| Cyclophilin, CyP | Mammals | 57.5 | ‐ |
3.3. Antimicrobial resistance gene assessment
We found no hits between the genome of M. aurum Aogashima and the AMR genes included in ResFinder databases. Instead, 3 hits above 70% identity: rbpA (RbpA bacterial RNA polymerase‐binding protein), mtrA (resistance‐nodulation‐cell division antibiotic efflux pump), and murA transferase (Mycobacterium tuberculosis intrinsic murA conferring resistance to fosfomycin) were reported using CARD webserver. These genes confer resistance to rifampicin, penam, and fosfomycin, respectively. However, as reported in Table 6, all hits were below the general reference value for gene homology (97%). Furthermore, these genes have been reported to be widely present in the Mycobacteriaceae and, therefore, not surprisingly also in the M. aurum type strain DSM 43999T (Table 6).
TABLE 6.
AMR genes detected in the genome sequence of M. aurum Aogashima and its relative M. aurum type strain DSM 43999T with an identity value ≥70%
| % Identity matching Region | % length or reference sequence | |||||
|---|---|---|---|---|---|---|
|
Genes AMR Gene Family |
Drug Class | Resistance Mechanisms | M. aurum Aogashima |
M. aurum Type strain |
M. aurum Aogashima |
M. aurum Type strain |
| rbpA (RbpA bacterial RNA polymerase‐binding protein) | Rifampicin | Antibiotic target protection | 96.4 | 96.4 | 97.4 | 100 |
| mtrA (resistance‐nodulation‐cell division antibiotic efflux pump) | Macrolide antibiotic, penam | Antibiotic efflux | 96.9 | 96.9 | 100 | 97.4 |
| murA transferase (Mycobacterium tuberculosis intrinsic murA conferring resistance to fosfomycin) | Fosfomycin | Antibiotic target alterations | 92.4 | 92.4 | 98.5 | 98.5 |
3.4. Pathogenic gene clusters and virulence factors assessment
The whole genome sequence of M. aurum Aogashima was screened to identify genetic element sequences that encode for virulence factors or protein toxins. We found no evidence for pathogenic islands. Screening of the genome of M. aurum Aogashima for all known virulence factors associated genes showed that most of the predicted genes were found in nonpathogenic or commensal bacteria and are involved in host interaction, survival, and maintenance of basic functions (Table 7). As shown in Table 7, several proteins with experimentally verified virulence factors were present in the genome of M. aurum Aogashima but their amino acid sequence similarity with that of the pathogenic M. tuberculosis is below the 60% cutoff value for functional homology (Table 7).
TABLE 7.
Virulence factors associated genes detected in the genome sequences of M. aurum Aogashima and M. tuberculosis H37Rv
| VFclass | Virulence factors | Related genes |
M. aurum Aogashima |
M. tuberculosis H37Rv |
Similarity (%) |
comments | e‐values |
|---|---|---|---|---|---|---|---|
| Amino acid and purine metabolism | Glutamine synthesis | glnA1 | orf04741 | Rv2220 | 84.3 | <95% | 0 |
| Leucine synthesis | leuD | orf04409 | Rv2987c | 84.2 | <95% | 6E−127 | |
| Lysine synthesis | lysA | orf04608 | Rv1293 | 79.3 | <95% | 0 | |
| Nitrate/nitrite transporter | narK2 | orf00718 | Rv1737c | 25.6 | <60% | 1E−14 | |
| Catabolism of cholesterol | Cyp125 | cyp125 | orf01099; orf01566; orf04038; orf04661 | Rv3545c | 54.4 | <95% | 4E−172 |
| FadE28 | fadE28 | orf04037 | Rv3544c | 68.5 | <95% | 1E−164 | |
| FadE29 | fadE29 | orf04036 | Rv3543c | 81.6 | <95% | 0 | |
| Cell surface components | Carboxylesterase | caeA | orf04747; orf04749 | Rv2224c | 53.6 | <60% | 3E−169 |
| Exported repetitive protein | erp | orf04235 | Rv3810 | 49.6 | <60% | 6E−58 | |
| fad23 | orf01422 | Rv1185c | 60.2 | <95% | 0 | ||
| fadE5 | orf01415; orf05079 | Rv0244c | 82.7 and 66.1 | <95% | 0 | ||
| gtf1 | orf01592 | Rv1526c | 48.8 | <95% | 1.3 | ||
| gtf 2 | orf01265; orf01577 | Rv1524 | 58.2 and 53.8 | <95% | 9E−162 | ||
| mmpL10 | orf01421 | Rv1183 | 57.2 | <95% | 0 | ||
| mmpS4 | orf00497; orf04089 | Rv0451c | 47.5 | <95% | 3E−47 | ||
| mps1 | orf02234 | Rv0101 | 49.3 | <95% | 0 | ||
| papA3 | orf01420 | Rv1182 | 54.5 | <95% | 0 | ||
| rmlA | orf01604; orf05016 | Rv0334 | 46.7 | <95% | 4E−161 | ||
| Heparin‐binding hemagglutinin | hbhA | orf03209 | Rv0475 | 67.9 | >60% | 5E−64 | |
| Lipoprotein | lprG | orf05150 | Rv1411c | 52.3 | <95 | 4E−78 | |
| Methyltransferase | mmaA4 | orf00819 | Rv0642c | 68.1 | <95 | 1.5 | |
| MymA operon | adhD | orf03671; orf04049; orf05070; orf05579 | Rv3086 | 34.1 | <60% | .016 | |
| fadD13 | orf01899 | Rv3089 | 36.4 | <60% | 2E−95 | ||
| mymA | orf05192 | Rv3083 | 50.0 | <60% | why not 95% | ||
| ddrA | orf00094; orf01270; orf02392 | Rv2936 | 46.4, 51.5, 67.8 | <60% | 1E−79 | ||
| ddrB | orf02393 | Rv2937 | 43.1 | <60% | 1.00E−76 | ||
| drrC | orf02394 | Rv2938 | 46.7 | <60% | 2E−90 | ||
| fadD26 | orf02385; orf02387 | Rv2930 | 60.2, 55.1 | <60% | 0 | ||
| fadD28 | orf01272; orf01571 | Rv2941 | 55.3, 63.4 | <60% | 0 | ||
| ppsA | orf02386 | Rv2931 | 56.0 | <60% | 0 | ||
| ppsB | orf02388; orf02389 | Rv2932 | 54.0, 55.1 | <60% | 0 | ||
| ppsD | orf02390 | Rv2934 | 58.8 | <60% | 0 | ||
| ppsE | orf02391 | Rv2935 | 34,7 | <60% | 1E−125 | ||
| Proximal cyclopropane synthase of alpha mycolates | pcaA | orf00241; orf00242; orf00820; orf01613 | Rv0470c | 60.0, 56.3, 68.9, 96.2 | <95 | 2E−123 | |
| Sulfolipid−1 biosynthesis and transport | mmpL8 | orf01266 | Rv3823c | 50.4 | <95 | 0 | |
| papA1 | orf01262 | Rv3824c | 51.0 | <95 | 2E−170 | ||
| lpqY | orf03429 | Rv1235 | 28.3 | <60% | 2E−53 | ||
| sugA | orf03428 | Rv1236 | 53.0 | <60% | 3E−91 | ||
| sugB | orf03427 | Rv1237 | 57.1 | <60% | 3E−109 | ||
| sugC | orf03426 | Rv1238 | 53.9 | <60% | 8E−142 | ||
| Copper uptake | Copper exporter | ctpV | orf03576 | Rv0969 | 54.3 | <60% | 0 |
| fadD33 | orf02653 | Rv1345 | 61.7 | <95% | 0 | ||
| MbtA | orf00510 | Rv2384 | 64.3 | <95% | 0 | ||
| MbtB | orf00509 | Rv2383c | 60.9 | <95% | 1E−23 | ||
| MbtC | orf00507 | Rv2382c | 75.0 | <95% | 0 | ||
| MbtD | orf00506 | Rv2381c | 51.6 | <95% | 0 | ||
| MbtE | orf00505 | Rv2380c | 67.4 | <95% | 0 | ||
| MbtF | orf00504 | Rv2379c | 55.6 | <95% | 0 | ||
| MbtG | orf00503 | Rv2378c | 76.9 | <95% | .16 | ||
| MbtH | orf00502; orf01576 | Rv2377c | 73.5 | <95% | 6E−41 | ||
| MbtJ | orf04363 | Rv2385 | 60.6 | <95% | 4E−131 | ||
| MbtK | orf05262 | Rv1347c | 62.7 | <95% | 4E−88 | ||
| Pantothenate synthesis | PanC | orf05603 | Rv3602c | 70.5 | <95% | 6E−138 | |
| PanD | orf00584 | Rv3601c | 63.1 | <95% | 5E−56 | ||
| Antiapoptosis factor | NuoG | NuoG | orf02508 | Rv3151 | 73.1 | >60% | 0 |
| Mammalian cell entry (mce) operons | Mce1 | mce1A | orf01133 | Rv0169 | 53.3 | ‐ | 1E−150 |
| mce 1B | ‐ | Rv0170 | ‐ | ‐ | |||
| mce 1C | ‐ | Rv0171 | ‐ | ‐ | |||
| mce1D | orf04143 | Rv0172 | 67.6 | 0 | |||
| mce1E | ‐ | Rv0173 | ‐ | ‐ | |||
| mce1F | orf04141 | Rv0174 | 68.1 | 0 | |||
| Mce2 | mce2A | orf04716 | Rv0589 | 66.5 | ‐ | 0 | |
| mce2B | orf04717 | Rv0590 | 72.4 | 3E−129 | |||
| mce2C | ‐ | Rv0591 | ‐ | ‐ | |||
| mce2D | ‐ | Rv0592 | ‐ | ‐ | |||
| mce2E | orf04142 | Rv0593 | 67.8 | 0 | |||
| mce2F | ‐ | Rv0594 | ‐ | ‐ | |||
| Mce3 | mce3A | orf03117 | Rv1966 | 65.3 | >60% | 6E−180 | |
| mce3B | orf03118; orf05701 | Rv1967 | 68.1 | >60% | 1E−163 | ||
| mce3C | orf03119 | Rv1968 | 61.5 | >60% | 4E−164 | ||
| mce3D | orf03120 | Rv1969 | 63.6 | >60% | 2E−04 | ||
| mce3E | orf03121; orf04341 | Rv1970 | >60% | 0 | |||
| mce3F | orf03122; orf04340 | Rv1971 | 58.9 | >60% | 1E−179 | ||
| Mce4 | mce4A | orf01047; orf01461 | Rv3499c | 66.7 | >60% | 0 | |
| mce4B | orf01048; orf01460 | Rv3498c | 69.1 | >60% | 1E−168 | ||
| mce4C | orf01049; orf01459 | Rv3497c | 67.7 | >60% | 2E−171 | ||
| mce4D | orf01050; orf01458 | Rv3496c | 63.8 | >60% | 0 | ||
| mce4E | orf01051; orf01457 | Rv3495c | 64.4 | >60% | 6E−179 | ||
| mce4F | orf01052; orf01456 | Rv3494c | 69.6 | >60% | 0 | ||
| Phagosome arresting | Nucleoside diphosphate kinase | ndk | orf05384 | Rv2445c | 80.7 | >60% | 2E−81 |
| PE family protein | PE_PGRS30 | ‐ | Rv1651c | ||||
| Tyrosine phosphatase | ptpA | orf00358 | Rv2234 | 70.0 | >60% | 3E−82 | |
| Secreted proteins | 19‐kD protein | lpqH | orf04515 | Rv3763 | 60.1 | <95% | 3E−59 |
| Alpha‐crystallin | hspX | orf00236; orf05393 | Rv2031c | 39.0 | <60% | 3E−18 | |
| Antigen 85 complex | eis | orf02486 | Rv3804c | 70.6 | >60% | 4E−150 | |
| fbpB | ‐ | Rv1886c | |||||
| fbpC | orf01653; orf02135; orf04229; orf04918 | Rv0129c | 76.1 | >60% | 5E−175 | ||
| Enhanced intracellular survival protein | eis | orf05310 | Rv2416c | 56.1 | <60% | 4E−150 | |
| ESX−1 (T7SS) | PE35 | orf05503 | Rv3872 | 47.7 | <60% | 3E−26 | |
| PPE68 | orf05504 | Rv3873 | 41.5 | <60% | 7E−63 | ||
| eccA1 | orf05499 | Rv3868 | 76.7 | >60% | 0 | ||
| eccB1 | orf05500 | Rv3869 | 64.2 | >60% | 0 | ||
| eccCa1 | orf05501 | Rv3870 | 80.0 | >60% | 0 | ||
| eccCb1 | orf05502 | Rv3871 | 73.3 | >60% | 0 | ||
| eccD1 | orf05508 | Rv3877 | 66.6 | >60% | 0 | ||
| eccE1 | orf04675 | Rv3882c | 67.5 | >60% | 0 | ||
| espI | orf03155; orf05507 | Rv3876 | 34.9 | <60% | 6E−49 | ||
| espJ | orf05509 | Rv3878 | 32.2 | <60% | 2E−12 | ||
| espK | orf05512 | Rv3879c | 55.0 | <60% | 5E−88 | ||
| espL | orf05699 | Rv3880c | 53.8 | <60% | 4E−31 | ||
| espR | orf05183 | Rv3849 | 80.1 | >60% | 4E−80 | ||
| esxA | orf05506 | Rv3875 | 54.4 | <60% | 5E−31 | ||
| esxB | orf05505 | Rv3874 | 43.4 | <60% | 2E−19 | ||
| mycP1 | orf04676 | Rv3883c | 71.8 | >60% | 0 | ||
| ESX−3 (T7SS) | PE5 | orf01528 | Rv0285 | 69.8 | >60% | 7E−31 | |
| PPE4 | orf01529 | Rv0286 | 58.4 | <60% | 4E−79 | ||
| eccA3 | orf01525 | Rv0282 | 73.1 | >60% | 0 | ||
| eccB3 | orf01526 | Rv0283 | 63.0 | >60% | 0 | ||
| eccC3 | orf01527 | Rv0284 | 74.7 | >60% | 0 | ||
| eccD3 | orf01533 | Rv0290 | 62.8 | >60% | 9E−176 | ||
| eccE3 | orf01535 | Rv0292 | 50.8 | <60% | 1E−80 | ||
| espG3 | orf01532 | Rv0289 | 55.3 | <60% | 3E−115 | ||
| esxG | orf01530 | Rv0287 | 76.6 | >60% | 7E−40 | ||
| esxH | orf01531 | Rv0288 | 72.6 | >60% | 6E−52 | ||
| mycP3 | orf01534 | Rv0291 | 60.9 | >60% | 0 | ||
| ESX−4 (T7SS) | eccB4 | orf00970 | Rv3450c | 48.5 | <60% | 5E−125 | |
| eccC4 | orf00973 | Rv3447c | 51.8 | <60% | 0 | ||
| eccD4 | orf00972 | Rv3448 | 30.0 | <60% | 4E−16 | ||
| esxT | orf00976 | Rv3444c | 64.8 | >60% | 3E−44 | ||
| esxU | orf00975 | Rv3445c | 64.3 | >60% | 3E−46 | ||
| mycP4 | orf00971 | Rv3449 | 54.4 | <60% | 2E−139 | ||
| cyp143 | orf00285 | Rv1785c | 60.0 | borderline of functional identity | 4E−168 | ||
| Catalase‐peroxidase | katG | orf02989; orf04948 | Rv1908c | 65.5 | >60% | 0 | |
| Cu | sodC | orf02589; orf04790 | Rv0432 | 33.3 | <60% | 2.4 |
4. DISCUSSION
Humans have evolved in a microbial world. The resulting evolutionary adaptedness is based on microbes’ colonization of human skin and mucosal surfaces as well as regular microbial contact in the air, surfaces, and in food and beverages (Rook, 2010). Until very recently, diet provided the most exposure through raw, minimally processed or fermented foods and beverages and through untreated water. A link between consumption of live microbes—such as those found in fermented food—and health has been reported in both intervention and associative studies as well as randomized controlled trials (Marco et al., 2017; Sanlier et al., 2019). As health evidence is mounting, there have been calls to include recommendations for the consumption of microbes in dietary guidelines, akin to the ones related to dietary fibers (Marco et al., 2020). For these reasons, microbes have gained increasing interest as potential novel food ingredients.
Numerous microbes are currently being investigated for their safety as novel food ingredients and for their potential benefit to human health. These include novel probiotics such as Clostridium butyricum CBM588, as well as postbiotics, defined as inanimate bacterial preparations which confer health benefit to the host. The latter would include Yarrowia lipolyticus, Mycobacterium setense strain Manresensis, and pasteurized Akkermansia municiphila among others (Aguilar‐Toalá et al., 2018; Akter et al., 2020; Barros et al., 2020; Cani & de Vos, 2017; EFSA Panel on Nutrition et al., 2019a; 2019b; Kanai & Mikani, 2015; Salminen et al., 2021; Taverniti & Guglielmetti, 2011). The food use of dead microbes has several advantages compared to live organisms: the difficulties of ensuring cell viability at the levels reported in the product description and for the duration of their shelf‐life are avoided, for example. Similarly, using heat‐killed organisms limits concerns arising from use of these products in vulnerable groups such as the very young and immunosuppressed individuals and allows for more widespread use (Piqué et al., 2019). Interestingly, the organism under study in this report, heat‐killed M. aurum Aogashima, may fall within the definition of postbiotic, should a health benefit for this preparation be shown in separately presented studies. The purpose of the work described here, however, is solely to present and assess the evidence for the safe use of heat‐killed M. aurum Aogashima as a novel food ingredient.
This environmental saprophytic organism is likely to have been long present in the diet as a harmless water contaminant (Falkinham et al., 2001; Le Dantec et al., 2002a, 2002b; Vaerewijck et al., 2005). Safety of heat‐killed M. aurum Aogashima as a novel food ingredient was assessed according to the decision tree approach developed by Pariza and colleagues (2015). The interest in expanding the number of microbes being considered as novel food, beyond the current standardized cultures and probiotics supplements, has driven a new approach to assess safety. This new framework is also pertinent to those cultures that are perceived to lack an established history of safe use for their intended application. We provide evidence that the genome of M. aurum Aogashima is free of (1) genetic elements associated with pathogenicity or toxigenicity, (2) transferable antibiotic resistance gene DNA, and (3) genes coding for antibiotics used in human or veterinary medicine. Moreover, our evidence shows that (4) the no observed adverse effect level (NOAEL) was the highest dose tested, 2000 μg/kg BW/day.
Genetic elements associated with pathogenicity or toxigenicity were investigated by extensive in silico analysis and showed no evidence of pathogen‐specific virulence factors in M. aurum Aogashima. Indeed, the virulence factors associated genes identified were common to both pathogenic and nonpathogenic and commensal bacteria and associated with highly conserved functions such as amino acid and purine metabolism and the catabolism of cholesterol and were not located on pathogenic island (Niu et al., 2013). Hence, these highly conserved coding sequences are not considered appropriate markers of pathogenicity of M. aurum Aogashima. In this context, the presence of secreted protein associated genes (e.g., fbpA) is expected because they play a fundamental role in cell envelope maintenance (Belisle et al., 1997). The same can be said with respect to the genes ptpA and ptpB which are widely distributed among pathogenic and nonpathogenic mycobacterial species and also found in the genomes of other prokaryotes, including Lactobacillus spp. (Altermann et al., 2005; Boekhorst et al., 2006). It should also be noted that following comprehensive phylogenomics and comparative genomic analysis on 150 genomes of Mycobacterium spp, Gupta and his colleagues have reclassified the Mycobacterium genus into five distinct monophyletic groups (Gupta et al., 2018). As a result, what was once known as Mycobacterium aurum has been reclassified into the novel genus Mycolicibacterium (“Fortuitum‐Vaccae” clade) which is comprised of rapidly growing environmental species that are divergent from the clinical pathogenic Mycobacterium species. Hence, the absence of true virulence genes and pathogenic island in the genome sequence of M. aurum Aogashima are in line with its assignment to the species M. aurum known for its nonpathogenic trait (Risk group 1).
Antimicrobial resistance gene assessment was made by screening the genome using both ResFinder and CARD webservers to ensure coverage of all AMR determinants (i.e., acquired resistance genes, resistant mutations of housekeeping genes, efflux overexpression, etc.), drug targets, antibiotic molecules and drug classes, and the molecular mechanisms of resistance (Alcock et al., 2020; Zankari et al., 2012). We found no evidence for any resistance genes associated with the most common antimicrobial compounds of concern in food (namely, Ampicillin, Chloramphenicol, Kanamycin, Streptomycin; Erythromycin, Gentamycin, Tetracyclin, Vanomycin, and Lincomycin). We did, however, detect similarities with rbpA, mtrA, and murA. While these genes are known to confer resistance to rifampicin, penam, and fosfomycin, respectively, their identity values were close to, but still below, the cutoff of 97% homology. Moreover, these genes are commonly present in mycobacteria as they are likely involved in essential cell functions (Maitra et al., 2019; Newell et al., 2006). Finally, there is no evidence for transferability. Hence, the absence of significant resistance genes in M. aurum Aogashima as well as the use of this organism as a heat‐killed preparation intended as novel food ingredient supports the conclusion that this is a safe product.
M. aurum Aogashima was also evaluated for allergenic potential using the traditional FAO/WHO issued guidelines as well as an innovative 3D‐modeling‐based analysis (AllerCatPro database). It was concluded that M. aurum Aogashima would not trigger any allergenic or hypersensitivity reactions in humans. Based on the low number of the predicted allergenic protein sequences detected in the genome by the traditional methodology, and the absence of 3D epitope similarity, it is highly probable that this organism does not produce any true allergenic proteins. Indeed, only fifteen protein sequences were deemed as potentially allergenic based on linear sequence window identity (80 residues) above the thresholds of 35% (traditional methodology). Of those, only two predicted allergenic proteins in M. aurum Aogashima related to food. None of those showed 3D epitope identity, strongly suggesting that the predicted protein sequence matches might be false positives.
Safety of heat‐killed M. aurum Aogashima was further assessed by toxicology testing, including a subchronic (90 day) oral challenge using male and female Crl:WI(Han) adult rats. All doses tested, including the highest doses of 2000 μg/Kg/day, had no treatment‐related adverse effects. No relevant abnormalities between groups receiving M. aurum Aogashima and the control group were detected upon statistical analysis in a variety of parameters evaluated. Indeed, statistical differences were limited to differences in specific immune cell counts, but did not apply to differences in percentages of the same cell population. Furthermore, all cellular values remained well within the natural healthy range for adult rats (Giknis & Clifford, 2008). In the case of the reported glucose levels, we consider these may be normal biological variations due to continuous access to food and the effects of circadian rhythms (Kohsaka & Bass, 2007). For these reasons, and because of the absence of a dose relationship (given no differences were detected in the highest dose groups which received doses 10 times of those where differences were observed), we consider these differences part of normal biological variation rather than any effect of consumption of heat‐killed M. aurum Aogashima.
Based on the findings of the work and analysis described here, our conclusion is that the use of heat‐killed M. aurum Aogashima in food products is safe and that it is suitable for being evaluated as a novel food ingredient.
5. ETHICAL APPROVAL
All animal work performed at Sequani Ltd was conducted conforming to the UK legislation under the Animal (Scientific Procedures) Act 1986 (ASPA) Amendment Regulations (SI 2012/3039). Sequani Ltd is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
CONFLICT OF INTEREST
TD is a senior executive and holds stock in Aurum Switzerland AG. IN has no conflict of interest to declare.
AUTHOR CONTRIBUTION
Imen Nouioui: Conceptualization (supporting); Data curation (lead); Formal analysis (lead); Investigation (lead); Methodology (lead); Project administration (supporting); Resources (lead); Supervision (lead); Validation (equal); Visualization (supporting); Writing‐original draft (equal); Writing‐review & editing (equal). Timothy Dye: Conceptualization (lead); Formal analysis (supporting); Funding acquisition (lead); Project administration (lead); Supervision (supporting); Validation (equal); Visualization (lead); Writing‐original draft (equal); Writing‐review & editing (equal).
ACKNOWLEDGMENT
We thank Professor Hans‐Peter Klenk and his colleagues for assistance with the work conducted at the University of Newcastle (UK). We also wish to thank Roberto Suarez of Pen & Tec Consulting for critically reviewing the manuscript.
Nouioui, I., & Dye, T. (2021). Heat‐killed Mycolicibacterium aurum Aogashima: An environmental nonpathogenic actinobacteria under development as a safe novel food ingredient. Food Science & Nutrition, 9, 4839–4854. 10.1002/fsn3.2413
Funding information
Work presented in this paper was funded by Aurum Switzerland AG
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- EFSA Panel on Nutrition , Novel Foods and Food Allergens (NDA) , Turck D., Castenmiller J., Henauw S. D., Hirsch‐Ernst K. I., Kearney J., Maciuk A., Mangelsdorf I., McArdle H. J., Naska A., Pelaez C., Pentieva K., Siani A., Thies F., Tsabouri S., Vinceti M., Cubadda F., Engel K. H., Frenzel T., Heinonen M., Marchelli R., Neuhäuser‐Berthold M., Pöting A., Poulsen M., Sanz Y., Schlatter J. R., van Loveren H., Sun Q., Gelbmann W., Knutsen H. K. (2019). Safety of Yarrowia lipolytica yeast biomass as a novel food pursuant to Regulation (EU) 2015/2283. EFSA Journal, 12, e05594. 10.2903/j.efsa.2019.5594 [DOI] [Google Scholar]
- EFSA Panel on Nutrition , Novel Foods and Food Allergens (NDA) , Turck D., Castenmiller J., Henauw S. D., Hirsch‐Ernst K. I., Kearney J., Maciuk A., Mangelsdorf I., McArdle H. J., Naska A., Pelaez C., Pentieva K., Siani A., Thies F., Tsabouri S., Vinceti M., Cubadda F., Engel K. H., Frenzel T., Heinonen M., Marchelli R., Neuhäuser‐Berthold M., Poulsen M., Sanz Y., Schlatter J. R., van Loveren H., Sun Q., Gelbmann W., Knutsen H. K. (2019). Safety of heat‐killed Mycobacterium setense manresensis as a novel food pursuant to Regulation (EU) 2015/2283. EFSA Journal, 17, e05824. 10.2903/j.efsa.2019.5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addou, S., Rentzsch, R., Lee, D., & Orengo, C. (2009). Domain‐based and family‐specific sequence identity thresholds increase the levels of reliable protein function transfer. Journal Molecular Biology, 387, 416–430. 10.1016/j.jmb.2008.12.045 [DOI] [PubMed] [Google Scholar]
- Aguilar‐Toalá, J., Garcia‐Varela, R., Garcia, H., Mata‐Haro, V., González‐Córdova, A., Vallejo‐Cordoba, B., & Hernández‐Mendoza, A. (2018). Postbiotics: An evolving term within the functional foods field. Trends Food Science & Technology, 75, 105–114. 10.1016/j.tifs.2018.03.009 [DOI] [Google Scholar]
- Akter, S., Park, J., & Jung, H. (2020). Potential Health‐Promoting Benefits of Paraprobiotics, Inactivated Probiotic Cells. Journal of Microbiology & Biotechnology, 30, 477–481. 10.4014/jmb.1911.11019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock, BP, Raphenya, AR, Lau, TTY, Tsang, KK, Bouchard, M, Edalatmand, A, Huynh, W, Nguyen, ALV, Cheng, AA, Liu, S, Min, SY, Miroshnichenko, A, Tran, HK, Werfalli, RE, Nasir, JA, Oloni, M, Speicher, DJ, Florescu, A, Singh, B, Faltyn, M, Hernandez‐Koutoucheva, A, Sharma, AN, Bordeleau, E, Pawlowski, AC, Zubyk, HL, Dooley, D, Griffiths, E, Maguire, F, Winsor, GL, Beiko, RG., Brinkman, FSL., Hsiao, WWL, Domselaar, GV, & McArthur, AG. (2020). CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research, 48, D517–D525. 10.1093/nar/gkz935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altermann, E., Russell, W. M., Azcarate‐Peril, M. A., Barrangou, R., Buck, B. L., McAuliffe, O., Souther, N., Dobson, A., Duong, T., Callanan, M., Lick, S., Hamrick, A., Cano, R., & Klaenhammer, T. R. (2005). Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proceeding National Academy of Science USA, 102, 3906–3912. 10.1073/pnas.0409188102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro, A., Duarte, E., Amado, A., Ferronha, H., & Botelho, A. (2008). Comparison of three DNA 179 extraction methods for Mycobacterium bovis, Mycobacterium tuberculosis and 180 Mycobacterium avium subsp avium. Letters Applied Microbiology, 47, 8–11. 10.1111/j.1472-765X.2008.02372.x [DOI] [PubMed] [Google Scholar]
- Aponte M., Murru N., & Shoukat, M. (2020). Therapeutic, Prophylactic, and Functional Use of Probiotics: A Current Perspective. Frontiers in Microbiology, 11, 10.3389/fmicb.2020.562048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., Formsma, K., Gerdes, S., Glass, E. M., Kubal, M., Meyer, F., Olsen, G. J., Olson, R., Osterman, A. L., Overbeek, R. A., McNeil, L. K., Paarmann, D., Paczian, T., Parrello, B., Pusch, G. D., Reich, C., Stevens, R., Vassieva, O., Vonstein, V., Wilke, A., & Zagnitko, O. (2008). The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics, 9(1), 75– 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, R., Devoid, S., Disz, T., Edwards, R., Henry, C., Olsen, G., Overbeek, R., Parrello, B., Pusch, G. D., Stevens, R. L., Vonstein V., & Xia, F. (2012). SEED servers: 227 high‐performance access to the SEED genomes, annotations and metabolic models. PLoS One, 7, e48053. 10.1371/journal.pone.0048053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., Lesin, V. M., Nikolenko, S. I., Pham, S., Prjibelski, A. D., Pyshkin, A. V., Sirotkin, A. V., Vyahhi, N., Tesler, G., Alekseyev, M. A., & Pevzner, P. A. (2012). SPAdes: A new genome assembly algorithm and its applications to single‐cell sequencing. Journal of Computation Biology, 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros, C. P., Guimaraes, J. T., Asmerino, R. A., Duarte, M. C. K. H., Silva, M. C., Silva, R., Ferreira B. M., Sant’Ana A. S., Freitas M. Q., & Cruz, A. G. (2020). Paraprobiotics and postbiotics:Concepts and potential application in diary products. Current Opinion Food Science, 32, 1–8. 10.1016/j.cofs.2019.12.003 [DOI] [Google Scholar]
- Belisle, J., Vissa, V., Sievert, T., Takayama, K., Brennan, P., & Besra, G. S. (1997). Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science, 276, 1420–1422. 10.1126/science.276.5317.1420 [DOI] [PubMed] [Google Scholar]
- Boekhorst, J., Wels, M., Kleerebezem, M., & Siezen, R. (2006). The predicted secretome of Lactobacillus plantarum WCFS1 sheds light on interactions with its environment. Microbiology, 152, 3175–3183. 10.1099/mic.0.29217-0 [DOI] [PubMed] [Google Scholar]
- Cani, P., & de Vos, W. (2017). Next‐Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Frontiers in Microbiology, 8, 1765. 10.3389/fmicb.2017.01765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cerbo, A., Palmieri, B., Aponte, M., Morales‐Medina, J., & Iannitti, T. (2016). Mechanisms and therapeutic effectiveness of lactobacilli. Journal Clinical Pathology, 69, 187–203. 10.1136/jclinpath-2015-202976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham 3rd, J. O., Norton, C. D., & LeChevallier, M. W. (2001). Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare, and other Mycobacteria in drinking water distribution systems. Applied Environmental Microbiology, 67, 1225–1231. 10.1128/AEM.67.3.1225-1231.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkinham, J. (2009). Surrounded by mycobacteria: Nontuberculous mycobacteria in the human environment. Journal Applied Microbiology, 107, 356–367. 10.1111/j.1365-2672.2009.04161.x [DOI] [PubMed] [Google Scholar]
- FAO , & WHO , (2002). Retrieved from http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf
- Fernandez‐Rendon, E., Cerna‐Cortes, J., Ramirez‐Medina, M., Helguera‐Repetto, A., Rivera‐Gutierrez, S., EstradaGarcia, T., & Gonzalez‐Y‐Merchand, J. (2012). Mycobacterium mucogenicum and other non‐tuberculous mycobacteria in potable water of a trauma hospital: A potential source for human infection. Journal Hospital Infection, 80, 74–76. 10.1016/j.jhin.2011.10.003 [DOI] [PubMed] [Google Scholar]
- Flandroy, L., Poutahidis, T., Berg, G., Clarke, G., Dao, M. C., Decaesteker, E., Furman E., Haahtela T., Massart S., Plovier H., Sanz Y., Rook, G. (2018). The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Science of the Total Environment, 627, 1018–1038. 10.1016/j.scitotenv.2018.01.288 [DOI] [PubMed] [Google Scholar]
- Grumet, L., Tromp, Y., & Stiegelbauer, V. (2020). The Development of High‐Quality Multispecies Probiotic Formulations. Nutrients, 12, 2453. 10.3390/nu12082453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R., Lo, B., & Son, J. (2018). Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Frontiers in Microbiology, 9, 67–77. 10.3389/fmicb.2018.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., Morelli L., Canani R. B., Flint H. J., Salminen S., Calder P. C., Sanders M. E., Sanders, M. (2014). The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nature Review Gastroenterology, 11, 506–514. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- Hori, T., Matsuda, K., & Oishi, K. (2020). Probiotics: A Dietary Factor to Modulate the Gut Microbiome, Host Immune System, and Gut‐Brain Interaction. Microorganisms, 10.3390/microorganisms8091401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwidthaya, P., Suthiravitayavaniz, K., & Phongpanich, S. (1989). Mycobacterium other than tubercle bacilli in various environments in Bangkok. Journal of the Medical Association of Thailand, 72, 317–320 [PubMed] [Google Scholar]
- Kanai, T., & Mikani, Y. H. (2015). A breakthrough in probiotics: Clostridium butyricum regulates gut homeostasis and anti‐inflammatory response in inflammatory bowel disease. Journal of Gastroenterology, 50, 928–939. 10.1007/s00535-015-1084-x [DOI] [PubMed] [Google Scholar]
- Kubalek, I., & Mysak, J. (1996). The prevalence of environmental mycobacteria in drinking water supply systems in a demarcated region in Czech Republic, in the period 1984–1989. European Journal of Epidemiology, 12, 471–474. 10.1007/BF00143998 [DOI] [PubMed] [Google Scholar]
- Le Dantec, C., Duguet, J., Montiel, A., Dumoutier, N., Dubrou, S., & Vincent, V. (2002). Occurrence of mycobacteria in water treatment lines and in water distribution systems. Applied Environmental Microbiology, 68, 5318–5325. 10.1128/aem.68.11.5318-5325.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Dantec, C., Duguet, J., Montiel, A., Dumoutier, N., Dubrou, S., & Vincent, V. (2002). Chlorine disinfection of atypical mycobacteria isolated from a water distribution system. Applied Environmental Microbiology, 68, 1025–1032. 10.1128/aem.68.3.1025-1032.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K. B., & Neumann R. O. (1896). Atlas und Grundriss der Bakteriologie und Lehrbuch der speziellen bakteriologischen Diagnostik. J.F. Lehmann, 1st ed [Google Scholar]
- Liu, B., Zheng, D., Jin, Q., Chen, L., & Yang, J. (2019). VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Research, 47(D1), D687–D692. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, C. A., Smith, D. G., Siebler, P. H., Schmidt, D., Stamper, C. E., Hassell Jr, J. E., Yamashita, P. S., Fox, J. H., Reber, S. O., Brenner, L. A., Hoisington, A. J., Postolache, T. T., Kinney, K. A., Marciani, D., Hernandez, M., Hemmings, S. M. J., Malan‐Muller, S., Wright, K. P., Knight, R., Raison, C. L., Rook, GAW. (2016). The microbiota, Immunoregulation and mental health: Implications for public health. Current Environmental Health Reports, 3, 270–286. 10.1007/s40572-016-0100-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei, L., McCafferty, J., Chen, T., Teles, F., Hasturk, H., Paster, B., & Campos‐Neto, A. (2015). The hidden mycobacteriome of the human healthy oral cavity and upper respiratory tract. Journal Oral Microbiology, 7, 26094–26105. 10.3402/jom.v7.26094. eCollection 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra, A., Munshi, T., Healy, J., Martin, L., Vollmer, W., Keep, N., & Bhakta, S. (2019). Cell wall peptidoglycan in Mycobacterium tuberculosis: An Achilles’ heel for the TB‐causing pathogen. FEMS Microbiology Reviews, 43, 548–575. 10.1093/femsre/fuz016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler‐Bauer, A., Derbyshire, M., Gonzales, N., Lu, S., Chitsaz, F., Geer, L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Marchler G. H., Song J. S., Thanki N., Wang Z., Yamashita R. A., Zhang D., Zheng C., Bryant, S. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Research, 43(Database, issue):D222–6. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco, M., Heeney, D., Binda, S., Cifelli, C., Cotter, P., Foligne, B., Gänzle M., Kort R., Pasin G., Pihlanto A., Smid E. J., Hutkins, R. (2017). Health benefits of fermented foods: Microbiota and beyond. Current Opinion Biotechnology, 44, 94–102. 10.1016/j.copbio.2016.11.010 [DOI] [PubMed] [Google Scholar]
- Marco, M., Hill, C., Hutkins, R., Slavin, J., Tancredi, D., Merenstein, D., & Sanders, M. (2020). Should There Be a Recommended Daily Intakeof Microbes? Journal of Nutrition, 150, 3061–3067. 10.1093/jn/nxaa323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martın Casabona, N., & Rossello Urgell, J. (2000). Environmental mycobacteria in Spain: Isolations in the 1976 1996 period. Medicina Clınica, 115, 663–670. 10.1016/s0025-7753(00)71655-1 [DOI] [PubMed] [Google Scholar]
- Maurer‐Stroh, S., Krutz, N., Kern, P., Gunalan, V., Nguyen, M., Limviphuvadh, V., Eisenhaber F., Gerberick, G. (2019). AllerCatPro—prediction of protein allergenicity potential from the protein sequence. Bioinformatics, 35, 3020–3027. 10.1093/bioinformatics/btz029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghim, S., Sarikhani, E., Nasr‐Esfahani, B., & Faghri, J. (2012). Identification of nontuberculous mycobacteria species isolated from water samples using phenotypic and molecular methods and determination of their antibiotic resistance patterns by E‐ test method in Isfahan. Iran. Iranian Journal of Basic Medical Science, 15, 1076–1082 [PMC free article] [PubMed] [Google Scholar]
- Nasr‐Esfahani, B., Sarikhani, E., Moghim, S., Faghri, J., Fazeli, H., Hoseini, N., & Rezaei‐Yazdi, H. (2012). Molecular characterization of environmental non‐tuberculous mycobacteria using PCR‐ RFLP analysis of 441 Bp heat shock protein 65 fragments. Iranian Journal of Public Health, 41, 108–114 [PMC free article] [PubMed] [Google Scholar]
- Newell, K., Thomas, D., Brekasis, D., & Paget, M. (2006). The RNA polymerase‐binding protein RbpA confers basal levels of rifampicin resistance on Streptomyces coelicolor. Molecular Microbiology, 60, 687–696. 10.1111/j.1365-2958.2006.05116.x [DOI] [PubMed] [Google Scholar]
- Niu, C., Yu, D., Wang, Y., Ren, H., Jin, Y., Zhou, W., Li B., Cheng Y., Yue J., Gao Z., Liang, L. (2013). Common and pathogen‐specific virulence factors are different in function and structure. Virulence, 4, 473–482. 10.4161/viru.25730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pariza, M., Gillies, K., Kraak‐Ripple, S., Leyer, G., & Smith, A. (2015). Determining the safety of microbiola cultures for the consumption by humans and animals. RegulatoryToxicology & Pharmacology, 73, 164–171. 10.1016/j.yrtph.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Pehrsson, E., Tsukayama, P., Patel, S., Mejia‐Bautista, M., Sosa‐Soto, G., Navarrete, K. M., Calderon M., Cabrera L., Hoyos‐Arango W., Bertoli M. T., Berg D. E., Gilman R. H., & Dantas, G. (2016). Interconnected microbiomes and resistomes in low‐income human habitats. Nature, 533, 212–216. 10.1038/nature17672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piqué, N., Berlanga, M., & Miñana‐Galbis, D. (2019). Health Benefits of Heat‐Killed (Tyndallized) Probiotics: An Overview. International Journal Molecular Science, 20, 2534. 10.3390/ijms20102534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontiroli, A., Khera, T. T., Oakley, B. B., Mason, S., Dowd, S. E., Travis, E. R., Erenso, G., Aseffa, A., Courtenay, O., Wellington, EMH. (2013). Prospecting environmental mycobacteria: Combined molecular approaches reveal unprecedented diversity. PLoS One, 8, e68648. 10.1371/journal.pone.0068648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, X, & Huang, Y. (2014). New approaches to prokaryotic systematics. In Goodfellow M., Sutcliffe I., & Chun J., Methods in Microbiology. 41:2‐327 [Google Scholar]
- Rook, G. (2010). 99th Dahlem conference on infection, inflammation and chronic inflammatory disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis. Clinical Experimental Immunology, 160, 70–79. 10.1111/j.1365-2249.2010.04133.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook, G., Adams, V., Hunt, J., Palmer, R., Martinlli, R., & Rosa Brunet, L. (2004). Mycobacterial and other environmental organisms as immunomodulators for immunoregulatory disorders. Springer Seminar in Immunopathology, 25, 237–255. 10.1007/s00281-003-0148-9 [DOI] [PubMed] [Google Scholar]
- Rook, G., Lowry, C., & Raison, C. (2013). Microbial “old Friends”, immunoregulation and stress resilience. Evolution, Medicine & Public Health, 1, 46–64. 10.1093/emph/eot004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangal, V., Jones, A., Goodfellow, M., Hoskinsson, P., Kaemper, P., & Sutcliffe, I. (2015). Genomic 220 analyses confirm close relatedness between Rhodococcus defluvii and Rhodococcus equi 221 (Rhodococcus hoagii). Archives in Microbiology, 197, 113–116. 10.1007/s00203-014-1060-5 [DOI] [PubMed] [Google Scholar]
- Sanlier, N., Gokcen, B., & Sezgin, A. (2019). Health benefits of fermented foods. Critical Review in Food Science & Nutrition, 59, 506–527. 10.1080/10408398.2017.1383355 [DOI] [PubMed] [Google Scholar]
- Scarlata, G., Pellerito, A., Di Benedetto, M., Massenti, M., & Nastasi, A. (1985). Isolation of Mycobacteria from drinking water in Palermo. Bollettino Istituto Sieroterapico Milanese, 64, 479–482 [PubMed] [Google Scholar]
- Seminen, S., Collado, M. C., Endo, A., Hill, C., Lebeer, S., Quigley, E. M. M., Mary E. S., Shamir R., Swann J. R., Szajewska H., & Vinderola, G. (2021). The international scientific association of probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nature Review Gastroenetrology Hepatology, 10.1038/s41575-021-00440-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverniti, V., & Guglielmetti, S. (2011). The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: Proposal of paraprobiotic concept). Genes & Nutrition, 6, 261–274. 10.1007/s12263-011-0218-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaerewijck, M., Huys, G., Palomino, J., Swings, J., & Portaels, F. (2005). Mycobacteria in drinking water distribution systems: Ecology and significance for human health. FEMS Microbiology Reviews, 29, 911–934. 10.1016/j.femsre.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Wilkins, T., & Sequoia, J. (2017). Probiotics for Gastrointestinal Conditions: A Summary of the Evidence. American Family Physician, 96, 170–178 [PubMed] [Google Scholar]
- Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., & Larsen, M. (2012). Identification of acquired antimicrobial resistance genes. Journal Antimicrobial Chemotherapy, 67, 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


