Abstract
CpG island methylation plays an important role in normal cellular processes, such as genomic imprinting and X-chromosome inactivation, as well as in abnormal processes, such as neoplasia. However, the dynamics of de novo CpG island methylation, during which a CpG island is converted from an unmethylated, active state to a densely methylated, inactive state, are largely unknown. It is unclear whether the development of de novo CpG island methylation is a progressive process, in which a subset of CpG sites are initially methylated with a subsequent increase in methylation density, or a single event, in which the initial methylation event encompasses the entire CpG island. The tumor suppressor gene p16/CDKN2a/INK4a (p16) is inactivated by CpG island methylation during neoplastic progression in a variety of human cancers. We investigated the development of methylation in the p16 CpG island in primary human mammary epithelial cell strains during escape from mortality stage 0 (M0) growth arrest. The methylation status of 47 CpG sites in the p16 CpG island on individual DNA molecules was determined by sequencing PCR clones of bisulfite-treated genomic DNA. The p16 CpG island was initially methylated at a subset of sites in three discrete regions in association with p16 transcriptional repression and escape from M0 growth arrest. With continued passage, methylation gradually increased in density and methylation expanded to sites in adjacent regions. Thus, de novo methylation in the p16 CpG island is a progressive process that is neither site specific nor completely random but instead is region specific. Our results suggest that early detection of methylation in the CpG island of the p16 gene will require methylation analysis of the three regions and that the identification of region-specific methylation patterns in other genes may be essential for an accurate assessment of methylation-mediated transcriptional silencing.
The methylation of CpG islands plays a critical role in heritable states of gene expression. De novo CpG island methylation is established during gametogenesis at imprinted loci as well as during early development in X-chromosome inactivation, resulting in the stable maintenance of monoallelic expression in somatic cells (46, 55). In addition, de novo methylation occurs aberrantly during neoplastic progression as well as in fragile X syndrome, resulting in the stable transcriptional silencing of the methylated genes (4, 39). The differential methylation patterns of the active and inactive states have been extensively studied by comparing the alleles on active and inactive X chromosomes, maternal and paternal alleles of imprinted genes, genes in normal and cancer tissue, and the FMR1 gene in males with normal X chromosomes and males with fragile X syndrome (26, 42, 53, 54, 57). Typically, the CpG island of a transcriptionally active allele is completely unmethylated, whereas the CpG island of a transcriptionally inactive allele is densely methylated. Although differential CpG island methylation has been extensively studied, the dynamics of de novo methylation in endogenous CpG islands that mediate the transition from the unmethylated, active state to the densely methylated, inactive state remain largely unknown. However, based on the differential CpG island methylation states, two models have been proposed for the de novo methylation process (53). The first model proposes that de novo CpG island methylation is a progressive process in which a subset of sites are initially methylated, followed by an increase in methylation density. The alternative model proposes that de novo CpG island methylation is a single event that encompasses the entire CpG island and is stably maintained. In addition, it remains unclear whether the addition of methyl groups to specific sites or regions in the CpG island plays an important role in the de novo methylation process (29, 47, 59).
In this study, we investigated the temporal development of methylation in the CpG island of the p16/CDKN2a/INK4a (p16) gene, one of the most commonly inactivated tumor suppressor genes in human cancer (50). p16 is a cyclin-dependent kinase inhibitor that regulates progression through the G1 phase of the cell cycle by binding and inhibiting cyclin-dependent kinases 4 and 6 (30, 49). p16 is also thought to be involved in senescence because its levels are induced in senescent cells but are either low or undetectable in immortalized cells (1, 35, 38, 45). p16 alleles can be inactivated during neoplastic progression by multiple mechanisms, including deletion, mutation, and CpG island methylation (10, 11, 37). The 5′ CpG island of the p16 gene spans the putative transcription start sites and exon 1α and has been found to be methylated at a high frequency in several types of human cancer but not in normal cells (44, 48, 61). p16 CpG island methylation correlates with the loss of expression in various cell lines and primary tumors, and p16 expression can be reactivated with the DNA methyltransferase inhibitor 5-azacytidine (20, 25, 37). Thus, methylation-mediated silencing of the p16 gene is an important epigenetic event during neoplastic progression in a variety of human cancers.
To investigate the temporal development of methylation in the p16 CpG island, we used primary human mammary epithelial cell strains (HMECs), which have previously been shown to undergo spontaneous selection for p16 transcriptional silencing by p16 CpG island methylation (9, 18, 28). HMECs, derived from normal breast tissue, undergo a proliferative block termed mortality stage 0 (M0), which is the first mortality stage of this cell type (17, 18). At M0, the cells enlarge and flatten, accumulate in G1 or G0, express senescence-associated β-galactosidase, and have increased p16 expression (9, 17, 28, 52). A small subpopulation of cells that can escape M0 is characterized by the methylation of the p16 CpG island and a marked decrease in p16 mRNA and protein levels. Human papillomavirus 16 (HPV16) E7, through its ability to disrupt retinoblastoma (Rb) function, allows HMECs to bypass M0 arrest in the presence of abundant p16 (17, 18). In contrast, HMECs expressing HPV16 E6 arrest at M0 in the presence of abundant p16 and undergo a selection for p16 methylation similar to that of normal cells, but unlike normal cells, the subpopulation that escapes M0 has an extended life span (18, 33). Thus, inactivation of the Rb/p16 pathway by either p16 methylation or E7 expression allows the continued proliferation of HMECs past M0.
Using bisulfite genomic sequencing, we investigated the changes in methylation profiles of the p16 CpG island over time as normal and E6-expressing HMECs escaped M0 arrest and lost p16 expression. In contrast to methylation-sensitive restriction enzymes that are limited to a subset of CpG cytosines, bisulfite genomic sequencing allows the analysis of the methylation status at each cytosine in a CpG island (19). In addition, sequencing individual PCR clones of bisulfite-treated DNA allows the determination of the methylation profiles of individual DNA molecules (epigenotypes). In this study, we demonstrated that the subpopulation of HMECs escaping M0 arrest underwent methylation preferentially in three distinct regions of the p16 CpG island with associated loss of p16 mRNA and protein expression. With continued passage, methylation in the HMECs expressing E6 increased in density and expanded to sites in adjacent regions of the p16 CpG island, demonstrating that de novo CpG island methylation is progressive and region specific.
MATERIALS AND METHODS
Cell culture.
HMEC cultures were derived from reduction mammoplasty specimens as described previously (18). HMECs 4, 6, and 9 (HMEC4, HMEC6, and HMEC9, respectively) were derived from tissues of three patients. Cells were cultured in DFCI-1 medium (3). LXSN-based retroviruses expressing HPV16 oncogenes (E6 and/or E7) (23) were used to infect HMECs after the establishment of the cultures, followed by selection with 100 μg of G418 per ml. E6-expressing HMEC6, E6-expressing HMEC9, and E6/E7-expressing HMEC9 were used in this study, and each achieved greater than 50 passages. HMEC4 was not infected with E6 or E7 and achieved 20 passages. H249 and H1618, lung cancer cell lines provided by J. Herman and S. Baylin, were used as controls for unmethylated and methylated p16 CpG islands, respectively (24).
Western analysis.
Whole-cell protein extracts were used to prepare Western blots as described previously (18). Antibodies against p16 (PharMingen clone G175-405) and p27 (Transduction Laboratories no. K25020) were used as probes. Proteins were visualized with horseradish peroxidase-conjugated anti-mouse immunoglobulin G (Jackson Immunoresearch Laboratories) and chemiluminescence (kit from Dupont NEN). Quantification was done on a scanned image with ImageQuant software (Molecular Dynamics).
Northern analysis.
Poly(A)+ RNA was isolated from 100 μg of total RNA and used to prepare Northern blots as described previously (18). Blots were probed with a p16 exon 1 probe generated by PCR as described previously (37) and labeled with [32P]dCTP by using the random-primed DNA labeling kit (Boehringer Mannheim). The Northern blots were stripped and reprobed with 36B4 as an RNA-loading control (34). Quantification was done on a phosphorimager with ImageQuant software (Molecular Dynamics).
Bisulfite conversion reactions.
Bisulfite converts unmethylated cytosines to uracils; methylated cytosines are resistant to conversion. Bisulfite sequence analysis of the 200 CpG cytosines from the H249 cell line (unmethylated control) and the >14,000 non-CpG cytosines from all the samples in the study indicated that the bisulfite conversion of unmethylated cytosines was at least 99.8% efficient. Analysis of the 185 CpG cytosines from the H1618 cell line (methylated control) indicated that methylation conferred at least 99.5% resistance to bisulfite conversion.
Total genomic DNA was extracted either with the QIAamp blood kit (Qiagen) or as described previously (16). Bisulfite reactions were performed as described previously (61). Each DNA sample (75 to 150 ng) was denatured in freshly prepared NaOH at a final concentration of 0.3 M for 20 min at 42°C. A freshly prepared mixture of 3.8 M sodium bisulfite (Sigma) and 1.0 mM hydroquinone (Sigma) (pH 5.0) was added to each sample, which was then incubated at 55°C for 6 to 8 h. The DNA samples were purified with QIAquick columns (Qiagen), desulfonated with NaOH at a final concentration of 0.3 M for 20 min at 37°C, and ethanol precipitated.
PCR amplification of bisulfite-treated DNA.
Each bisulfite-treated DNA sample was whole-genome amplified by using a degenerate 15-mer and the primer extension preamplification (PEP) protocol in a final volume of 60 μl, as described previously (63). p16-specific PCR amplifications were performed in a mixture containing 2 to 5 μl of PEP DNA, 200 μM deoxynucleoside triphosphates, 1.5 to 1.75 mM MgCl2, 20 pmol of each primer, GeneAmp PCR buffer (Perkin-Elmer Corp.), and 1.25 U of AmpliTaq Gold (Perkin-Elmer Corp.) in a final volume of 25 μl. Primer set A was described previously by Gonzalgo and Jones: 5′ GTA GGT GGG GAG GAG TTT AGT T 3′ (−355 to −334) and 5′ TCT AAT AAC CAA CCA ACC CCT CC 3′ (−95 to −73) (22). The nucleotide positions are numbered relative to the translation start site (+1). Reaction conditions were as described previously, except with a 64°C annealing temperature. PCR product A started just upstream of the transcription start sites and extended into the untranslated 5′ sequence, and it contained 15 CpG and 61 non-CpG cytosines internal to the primers. Primer set B was described previously by Herman et al.: 5′ TTT TTA GAG GAT TTG AGG GAT AGG 3′ (−159 to −136) and 5′ CTA CCT AAT TCC AAT TCC CCT ACA 3′ (+209 to +233) (24). PCR product B overlapped with the 3′ end of PCR product A and extended to within a few bases of the 3′ end of exon 1, and it contained 35 CpG and 48 non-CpG cytosines internal to the primers. Reaction conditions were as described previously, except with a 59°C annealing temperature. The overlapping regions of PCR products A and B share three CpG and five non-CpG cytosines. Both primer sets A and B allowed the determination of methylation on the coding strand of the p16 gene. All PCRs were performed with an MJ DNA Engine Tetrad thermal cycler (MJ Research, Inc.).
Cloning and sequencing PCR fragments amplified from bisulfite-treated DNA.
PCR products A and B were purified from a 2% agarose gel with the QIAquick gel extraction kit (Qiagen). The gel-purified PCR fragments were TA cloned with the pCR2.1 plasmid vector and INVαF′-competent cells (Invitrogen). Individual clones were sequenced with M13 forward and/or reverse primers on a PRISM 377 DNA sequencer (Applied Biosystems), using the PRISM dye primer or dye terminator cycle sequencing kit with AmpliTaq DNA polymerase (Applied Biosystems).
RESULTS
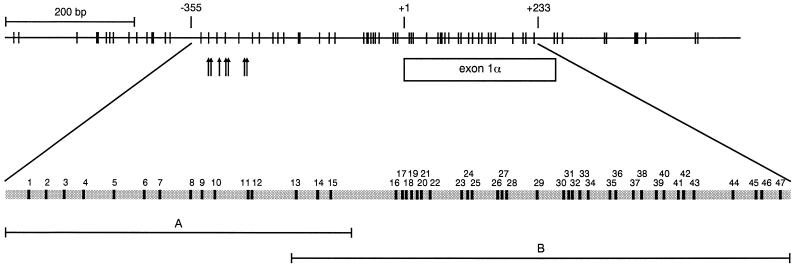
We used bisulfite sequencing to investigate the development of CpG island methylation in three independent HMECs: HMEC4, HMEC6, and HMEC9. Genomic DNA was treated with bisulfite, and then the core region of the p16 CpG island, spanning the putative transcription start sites and exon 1α, was amplified with primer sets A and B (Fig. 1). Cloned PCR products were sequenced, each clone representing an individual epigenotype of the cell population (Table 1). The H249 and H1618 cell lines, which were previously shown to have unmethylated and methylated p16 CpG islands, respectively, were used as controls for the bisulfite conversion reaction (24). Analysis of the H249 and H1618 cell lines and non-CpG cytosines showed that the bisulfite conversion was at least 99.8% efficient and at least 99.5% specific (Table 1; also see Materials and Methods).
FIG. 1.
Genomic map of the 5′ CpG island of the p16 gene. This CpG map is based on published genomic DNA sequences (GenBank accession no. AF022809, U12818, and AC000048). CpG sites and their genomic positions within 1.2 kb of the p16 CpG island (−621 to +521) are represented by vertical lines at the top. Nucleotide positions are numbered relative to the translation start site (+1). The genomic positions of the putative transcription start sites for the p16 gene are represented by arrows. The coding region of exon 1α is shown. The gray bar at the bottom represents a magnification of the region, from −355 to +233, that was analyzed for methylation in this study. The 47 CpG sites (black boxes within the gray bar) in this region are numbered according to their 5′-to-3′ order in the p16 genomic sequence and positioned based on their location within the genomic sequence. PCR products A (−355 to −73) and B (−159 to +233), amplified from bisulfite-treated DNA for sequencing, are shown.
TABLE 1.
Methylation in the p16 CpG island
| Cell culture, passage, and gene expressed | PCR product (CpG cytosinesa)
|
Mean no. of methylated sites per 47 total CpG sitesb | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A (1–15)
|
B (13–47)
|
||||||||
| No. of PCR clones sequenced | No. of methylated CpG sites/clone
|
No. of PCR clones sequenced | No. of methylated CpG sites/clone
|
||||||
| Meanc | Median | Range | Mean | Median | Range | ||||
| HMEC9 | |||||||||
| 3 | 10 | 0.1 (0.0, 0.3) | 0 | 0–1 | 10 | 0.5 (0.0, 1.1) | 0 | 0–2 | 0.6 |
| 8 | 9 | 0.2 (0.0, 0.5) | 0 | 0–1 | 9 | 0.2 (0.0, 0.5) | 0 | 0–1 | 0.4 |
| 16, E6 | 10 | 2.9 (1.5, 4.3) | 2 | 1–8 | 11 | 9.1 (7.0, 11.2) | 9 | 5–16 | 11.8 |
| 29, E6 | 7 | 2.9 (1.7, 4.1) | 3 | 1–5 | 10 | 13.6 (12.2, 15.0) | 14 | 10–16 | 16.2 |
| 42, E6 | 10 | 4.6 (2.5, 6.7) | 3.5 | 1–11 | 12 | 19.2 (15.4, 23.0) | 20 | 9–28 | 23.0 |
| 32, E6/E7 | 10 | 0.0 (0.0, 0.0) | 0 | 0 | 10 | 0.2 (0.0, 0.5) | 0 | 0–1 | 0.2 |
| HMEC6 | |||||||||
| 3 | 10 | 0.0 (0.0, 0.0) | 0 | 0 | 11 | 0.4 (0.0, 0.8) | 0 | 0–2 | 0.4 |
| 12, E6 | 10 | 2.0 (0.7, 3.3) | 1 | 0–5 | 12 | 9.3 (7.2, 11.4) | 8.5 | 6–17 | 10.9 |
| 25, E6 | 10 | 2.1 (1.1, 3.1) | 2.5 | 0–4 | 12 | 19.2 (16.4, 22.0) | 19 | 9–25 | 20.6 |
| HMEC4 | |||||||||
| 5 | 10 | 0.9 (0.0, 2.1) | 0 | 0–5 | 10 | 0.7 (0.0, 1.5) | 0 | 0–3 | 1.5 |
| 12 | 10 | 1.5 (0.4, 2.6) | 1.5 | 0–4 | 12 | 8.3 (4.7, 11.9) | 8.5 | 0–17 | 9.5 |
| 18 | 9 | 2.9 (2.3, 3.5) | 3 | 2–4 | 12 | 5.4 (3.2, 7.6) | 6 | 1–11 | 7.7 |
| H249 | 4 | 0.0 (0.0, 0.0) | 0 | 0 | 4 | 0.0 (0.0, 0.0) | 0 | 0 | 0.0 |
| H1618 | 3 | 14.7 (13.3, 15.0) | 15 | 14–15 | 4 | 35.0 (35.0, 35.0) | 35 | 35 | 46.9 |
Calculated by adding the methylation frequency per clone at each of the 47 sites (the methylation frequencies at CpG sites 13 to 15 were calculated with data from both PCR product A and B clones).
Ninety-five percent confidence intervals of the means are shown in parentheses.
Preferential methylation in three discrete regions of the p16 CpG island.
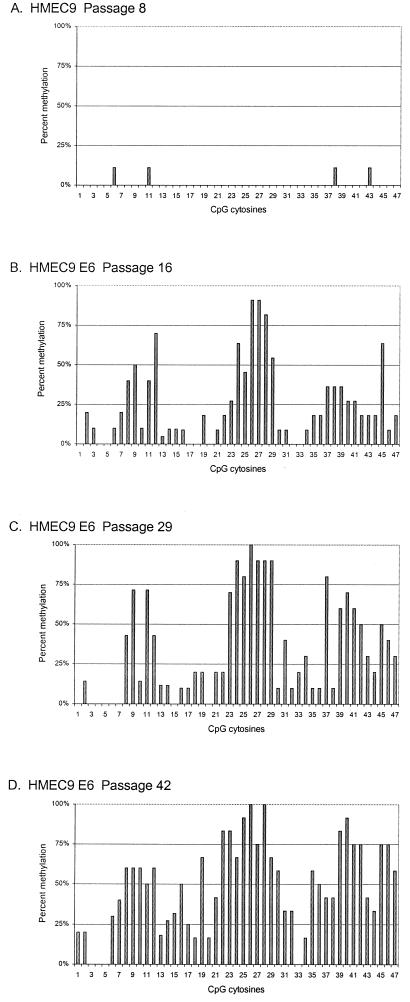
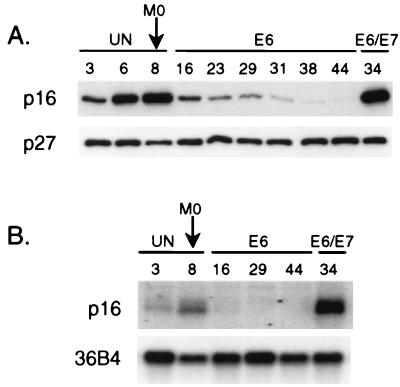
We investigated the development of methylation in the p16 CpG island as HMECs were selected for p16 transcriptional silencing and escape from proliferation arrest at M0. The p16 CpG island in HMEC9 was almost completely unmethylated at passages 3 and 8 (Table 1 and Fig. 2A). At passage 16, the subpopulation of E6-expressing HMEC9 that escaped M0 arrest underwent methylation (Fig. 2B). The p16 CpG island was partially methylated, with a mean of 11.8 methylated sites out of 47 CpGs (∼25%) per DNA strand (Table 1). Strikingly, methyl groups were not distributed uniformly across the 47 CpG sites but instead were clustered in three different regions of the p16 CpG island: CpGs 8 to 12 (−206 to −164) (region I), CpGs 23 to 29 (−18 to +35) (region II), and CpGs 37 to 45 (+101 to +186) (region III) (Fig. 2B). This region-specific methylation correlated with 80 and 90% decreases in mRNA and protein expression, respectively (Fig. 3). In contrast, HMEC9 expressing E6 and E7, which bypassed M0 arrest with abundant p16 mRNA and protein expression, remained unmethylated, similar to HMEC9 before M0 escape (Table 1 and Fig. 3). Therefore, the subpopulation of E6-expressing HMEC9 cells that escaped M0 arrest acquired a region-specific methylation pattern in the p16 CpG island with associated p16 transcriptional repression.
FIG. 2.
Development of methylation in the p16 CpG island of HMEC9. The 47 CpG sites are in numerical order according to their 5′-to-3′ order in the p16 genomic sequence (−355 to +233). CpG sites are not spaced out on the x axis according to their relative positions in the p16 genomic sequence. Percent methylation at each CpG site was calculated as the percentage of clones with a methylated cytosine at that site. Percent methylation for CpGs 13, 14, and 15 was calculated from data from clones of both PCR products A and B.
FIG. 3.
Expression of p16 in HMEC9. (A) Western analysis of p16 and p27. (B) Northern analysis of p16 with 36B4 loading control. Passage numbers are indicated above each lane. Cells were predominantly arrested at M0 by passage 8. A stable population of proliferating cells emerged by passage 14. UN, uninfected. E6 or E6/E7, infected with E6- or E6/E7-expressing retrovirus.
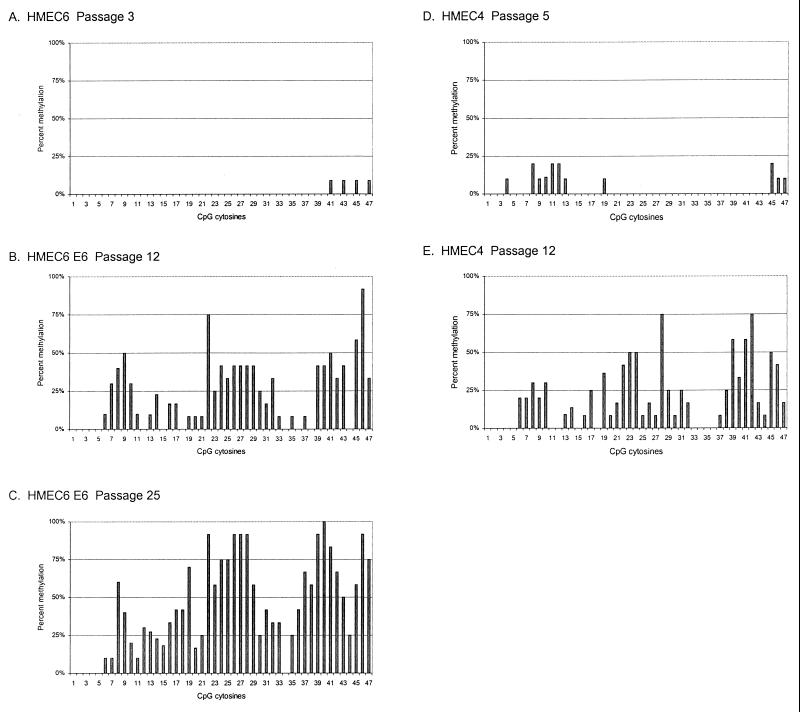
In HMEC6, a similar progression was observed. The p16 CpG island of pre-M0 HMEC6 was almost completely unmethylated at passage 3 (Fig. 4A), while post-M0 E6-expressing HMEC6 underwent p16 CpG island methylation at passage 12 (Fig. 4B), correlating with the loss of p16 expression (18). The p16 CpG island was again methylated in the region-specific pattern, with methyl groups clustered in three regions similar to those of HMEC9 (Fig. 2B and 4B).
FIG. 4.
Development of methylation in the p16 CpG island of HMEC4 and HMEC6. The CpG sites are numbered and spaced as in Fig. 3. Percent methylation at each CpG site was determined as for Fig. 3.
We also examined the development of p16 CpG island methylation in HMEC4, which, unlike the other two cell strains, did not express E6. Similar to those in HMEC6 and HMEC9, the p16 CpG island of pre-M0 HMEC4 was primarily unmethylated at passage 5 (Fig. 4D). Post-M0 uninfected HMEC4 acquired p16 CpG island methyl groups at passage 12 that also clustered in three regions similar to those of HMEC6 and HMEC9 (Fig. 4E) and correlated with the loss of p16 expression (18). Therefore, the region-specific patterns of p16 CpG island methylation and associated transcriptional silencing were similar among all three cell strains and independent of E6 expression.
Progressive increase in methylation density in the p16 CpG island with continued passages.
With increasing passage after M0 escape, the density of methylation in the p16 CpG island progressively increased. In HMEC9, the methylation density increased from a mean of 11.8 methylated sites (∼25%) at passage 16 to means of 16.2 (∼34%) at passage 29 and 23.0 (∼49%) at passage 42 (Fig. 2C and D and Table 1). The frequency of methylation increased at CpG sites across the p16 CpG island, but the region-specific pattern was maintained. p16 protein levels continued to decrease between passages 16 and 42, but p16 mRNA levels were barely detectable by passage 16, probably, in part, due to the limited sensitivity of the Northern blot analysis (Fig. 3). A similar progression was observed in HMEC6. The methylation density increased across the p16 CpG island from a mean of 10.9 sites (∼23%) at passage 12 to 20.6 (∼44%) at passage 25 (Table 1 and Fig. 4B and C). Thus, with continued passages, the methylation in the p16 CpG island increased in density. A statistically significant change in methylation density was not observed with continued passages of HMEC4, probably because of the short life span of this culture and the small sample of PCR clones (Table 1).
Despite an increase in methylation density, some CpG sites were methylated infrequently in all three HMECs. CpG sites 1 to 5, which are upstream of region I and encompass the putative transcription start sites of the p16 gene, were rarely methylated in all three cell strains (Fig. 2B to D, 4B to C, and 4E). In addition, CpG site 20 (between regions I and II), as well as CpG sites 33 and 34 (between regions II and III), were methylated infrequently. Interestingly, although CpG site 44, within region III, was often methylated, this occurred at a notably lower frequency than methylation at the surrounding CpG sites in region III at each passage examined in all three HMECs. Thus, the region-specific methylation pattern in the p16 CpG island of the post-M0 HMECs was characterized not only by the three preferentially methylated regions but also by the sites at which methylation seldom occurred even at later passages.
Methylation in the p16 CpG island is region specific, but not site specific.
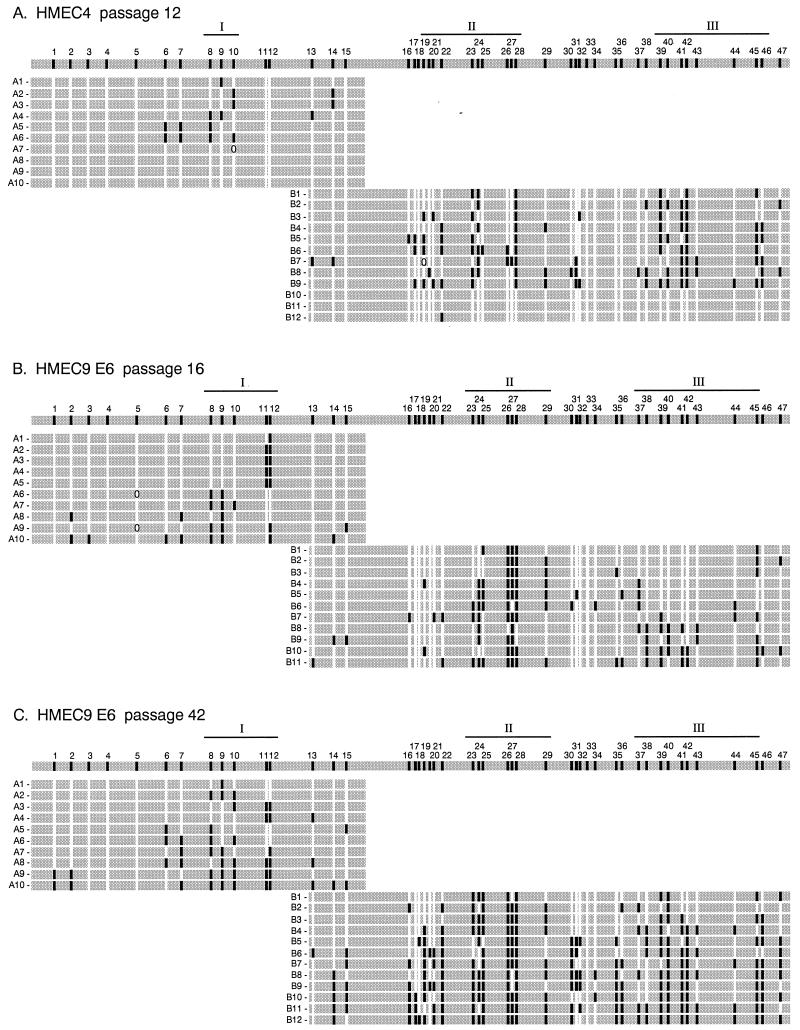
Each individual p16 CpG island epigenotype in post-M0 HMEC6 and HMEC9 had methyl groups clustered in three regions (Fig. 5B and data not shown). In contrast, post-M0 HMEC4 had two distinct populations of p16 CpG island epigenotypes, one that underwent methylation (Fig. 5A, clones B1 to B9) and one that was almost completely unmethylated (Fig. 5A, clones B10 to B12). Each methylated epigenotype from post-M0 HMEC4 had a methylation pattern and density similar to those of the epigenotypes from post-M0 HMEC6 and HMEC9 (Fig. 5A and B and data not shown). Therefore, the methylated p16 CpG island epigenotypes had a consistent region-specific methylation pattern and density within a single HMEC as well as among all three cell strains.
FIG. 5.
Individual epigenotypes at the p16 CpG island. The gray bar at the top of each panel represents the 588-bp region from −355 to +233 analyzed in this study. Individual epigenotypes are each represented by a gray bar with letters (A and B) and numbers (1 to 10, 11, and 12) on the left that represent the region and clone, respectively. CpG sites on the top gray bar are numbered from 1 to 47 according to their 5′-to-3′ order in the p16 genomic sequence and are spaced out according to their relative positions in the p16 genomic sequence. The methylation status of each CpG site is indicated at its relative position in the p16 genomic sequence by either a white (unmethylated) or black (methylated) box on the gray bar; an ellipse indicates ambiguous sequence information. The three preferentially methylated regions (I, II, and III) in each HMEC are based on the epigenotypes from the earliest passage after M0 (Fig. 3B and 4E). The lengths of the regions were slightly different among the different HMECs.
Although methyl groups in each epigenotype were clustered in three regions, the p16 CpG island epigenotypes were rarely identical, having a variable subset of methylated sites in each of the regions (Fig. 5). The specific CpG sites that were methylated within each region, as well as the number of methylated sites in the p16 CpG island, varied among the individual epigenotypes in each of the three post-M0 HMECs. Each HMEC strain had a few particular CpG sites that were methylated in almost every clone, but these sites differed among the three cell strains. For example, for HMEC6 at passage 12, CpG site 46 was methylated in 11 of 12 clones, whereas for HMEC9 at passage 16, the same site was methylated in only 1 of 11 clones (Fig. 2B and 4B). Thus, each post-M0 cell strain consisted of a heterogeneous population of p16 CpG island epigenotypes, suggesting that although methylation in the p16 CpG island is region specific, it is not site specific.
Progressive expansion of methylation in the p16 CpG island.
Analysis of individual p16 epigenotypes over time demonstrated that methylation expanded to encompass CpG sites outside of regions I, II, and III upon continued passage of HMEC6 and HMEC9. For example, in E6-expressing HMEC9, all epigenotypes were methylated within CpG sites 26 to 28 in region II at passages 16 and 42 (Fig. 2B and D and 5B and C). However, by passage 42, the majority of clones underwent methylation at adjacent sites, such as CpG sites 16 to 22, located 5′ of region II. Our data suggest that methylation in the p16 CpG island progressively expanded to sites outside the three regions during continued passages.
Non-CpG methylation in the p16 CpG island.
Although methylation is found primarily at CpG cytosines, a previous study using plasmid DNA transfected into mouse cell lines demonstrated that methylation at CpNpG cytosines can be heritable (15). We found that CpNpG methylation was rare in the p16 CpG island (only 19 of the >5,000 sites, not including CGG sites), consistent with previous studies of other CpG islands (53, 54). However, these CpNpG cytosines comprised the majority of nonconverted non-CpG cytosines, specifically 19 of 25, 12 of which were CCG cytosines. Methylation of the outer cytosine in a CCG trinucleotide located in region II (CpG site 26) was detected in all three post-M0 passages of HMEC9 (one clone at passage 16, three clones at passage 29, and three clones at passage 42), suggesting that methylation at a CpNpG site in an endogenous gene can be maintained.
DISCUSSION
Our results clearly distinguish between existing models of de novo methylation of CpG islands. We have demonstrated that de novo methylation of the p16 CpG island developed initially at a small subset of sites and gradually increased in density, rather than occurring at once throughout the entire CpG island. This initial methylation at a subset of sites was associated with transcriptional silencing of the p16 gene and escape from growth arrest at M0. Moreover, the de novo methylation was neither site specific nor completely random but instead developed preferentially in three discrete regions of the p16 CpG island and progressively expanded to sites in the adjacent regions.
In this study, we performed a detailed investigation of the evolution of CpG island methylation at the resolution of individual DNA molecules in three independent primary HMECs as the p16 CpG island was converted from an unmethylated, active state to a densely methylated, inactive state during escape from proliferative arrest at M0 over a time course of 40 passages. Previous studies have extensively investigated end stages of the de novo methylation process by comparing alleles on active and inactive X chromosomes, maternal and paternal alleles of imprinted genes, genes in normal and cancer tissue, and the FMR1 gene in males with normal X chromosomes and males with and fragile X syndrome (26, 42, 53, 54, 57). Demonstrations of the unmethylated, active and the densely methylated, inactive states by these studies have suggested two models for the development of CpG island methylation: initial methylation at a subset of sites followed by an increase in methylation density (progressive model) or a single methylation event encompassing the entire CpG island (single-event model) (53). We have demonstrated that the development of methylation in the CpG island of the p16 gene was initiated at a small subset of CpG sites clustered in three distinct regions, followed by an increase in methylation density and the expansion of methylation to neighboring regions. Thus, we have shown that de novo CpG island methylation is a progressive process, not a single event.
Previous studies using exogenously methylated genes, including p16, have shown that the methylation of only a subset of sites is sufficient for transcriptional repression of the transfected genes (7, 21, 27, 32). Consistent with these in vitro experiments, we have demonstrated in an endogenous system that the initial methylation of a subset of sites is associated with transcriptional downregulation and escape from proliferation arrest. Thus, our results suggest a progressive model for p16 CpG island methylation in which a cell undergoes methylation at a subset of sites in a region-specific pattern and consequently gains a selective proliferative advantage because of decreased p16 gene activity.
p16 CpG island methylation was preferentially clustered in three discrete regions in each of the individual epigenotypes. Previous studies demonstrated that the methylation of specific sites can directly disrupt transcription factor binding and that the resulting transcriptional repression is highly site specific, suggesting that site-specific methylation may be important in the regulation of gene expression (29, 59). However, the specific CpG sites that were methylated within the three regions varied among the epigenotypes in the three HMECs, suggesting that methylation in the p16 CpG island was region specific rather than site specific. Previous studies of the CpG islands of the O6-methylguanine-DNA methyltransferase gene in human tumor cell lines and the inactive-X-chromosome hypoxanthine phosphoribosyltransferase gene in mouse tumor cell lines similarly found regions of preferential methylation, suggesting that region-specific patterns may be a common feature of methylation in CpG island-containing genes (40, 43, 60). The basis for the regional differences in methylation that we have discovered is unclear and will require further investigation. They may be a result of regions having differential susceptibilities to DNA methylase(s) or DNA demethylase(s) due to differences in primary DNA sequence, secondary structures, local chromatin structure, and DNA-binding proteins (5, 6, 14, 62). For example, the three preferentially methylated regions in the p16 CpG island may be more favorable substrates for DNA methylase(s) or less favorable substrates for DNA demethylase(s). Alternatively, the extent of transcriptional repression may differ depending on the regional location of methylation within the p16 CpG island. For example, methylation at CpG sites within the three regions may result in a greater degree of transcriptional downregulation than methylation at sites outside those regions. Clonal selection of cells with lower p16 expression and higher proliferation rates would result in a population with preferentially methylated regions. Thus, region-specific p16 CpG island methylation suggests that the de novo methylation process is influenced by differences in the susceptibilities to DNA methylase(s) or DNA demethylase(s) and/or in the degree of methylation-mediated silencing.
Although methyl groups were preferentially clustered in the three regions, the methylation patterns of p16 CpG island epigenotypes were highly variable throughout the de novo methylation process. Molecule-to-molecule variation in methylation patterns have been similarly observed for densely methylated CpG islands in other tissue culture systems, in tumor tissue, and in leukocytes from males with fragile X syndrome, suggesting that the variability of methylation patterns is characteristic of methylated alleles (41, 51, 54). This complex heterogeneity of the p16 CpG island epigenotypes is consistent with a dynamic, stochastic methylation model proposed by Pfeifer et al. (41) which predicts that the de novo methylation process is an interplay of methyl group gain by a de novo methylase and a maintenance methylase and of methyl group loss by a demethylase and errors of the maintenance methylase (47). According to this model, the different frequencies of methylation at each CpG site in a cell population are the result of differential probabilities of methyl group gain and loss at each site.
The HMECs expressing E6 (HMEC6 and HMEC9) had a longer life span after M0 escape, enabling investigation of methylation at later passages. As the E6-expressing cells continued to divide after M0 escape, the methylation density in the p16 CpG island increased, expanding from sites in the three preferentially methylated regions to sites in adjacent regions. The progressive increase in the methylation density in the p16 CpG island correlated with further reduction in p16 protein levels and with p16 mRNA levels, which were barely detectable due to the limited sensitivity of the Northern blot analysis (Fig. 3) (18). Expansion of methylation may be important for the further reduction in gene expression or for increasing the stability of methylation-mediated transcriptional repression (7, 21, 27).
The basis for the progressive nature of the de novo methylation process that we have discovered remains unknown. This progressive process may be a result of increases in the levels of a DNA methylase or decreases in the levels of a DNA demethylase, both of which may increase the de novo methylation rate (58). Previous studies have demonstrated that cis-acting Sp1 elements protect CpG islands from de novo methylation, suggesting that the progressive methylation may be due to decreases in the levels of the trans-acting factors that interact with these Sp1 elements (8, 36). Alternatively, the methylation could be purely a result of a progressive accumulation of stochastic errors by a DNA methylase or a DNA demethylase combined with clonal selection (31). In addition, the progressive methylation process may be due to an initial methylation rendering other sites in the CpG island more susceptible to methylation (13, 56).
We and others have demonstrated that p16 expression is generally silenced by biallelic methylation in HMEC subpopulations that escape M0 (18, 28). Each p16 CpG island epigenotype in post-M0 HMEC6 and HMEC9 underwent methylation in the region-specific pattern, and these post-M0 populations remained heterozygous at the 9p21 locus (data not shown) (18). In contrast, post-M0 HMEC4 had two subpopulations of epigenotypes: a major one with a methylation pattern and density similar to those of post-M0 HMEC6 and HMEC9 and a minor one that was essentially unmethylated. The subpopulation of the HMEC4 cells with unmethylated epigenotypes may have been inactivated by an alternative mechanism. We previously reported that 3 of 10 clones of post-M0 HMEC4 had homozygous deletions at the c5.1 STS marker located just 3′ of p16 exon 2 which have also been frequently observed in cell lines and primary tumors (2, 11, 12, 18). Thus, p16 expression in HMECs is usually transcriptionally silenced by biallelic methylation and possibly inactivated at a lower frequency by homozygous deletion.
An important implication of our results lies in screening for p16 inactivation in cancer. Our data suggest that CpG sites within the three preferentially methylated regions may serve as better markers for methylation-mediated transcriptional silencing of the p16 gene. p16 CpG island methylation is currently screened for by methylation-specific PCR or methylation-sensitive restriction enzymes. We previously demonstrated that the original p16 methylation-specific PCR primers did not detect p16 methylation in the HMECs until many passages after p16 transcriptional repression (18). These methylation-specific PCR primers assay CpG sites 16 to 18 and 30 to 33 (24), which we show here are outside of the three preferentially methylated regions in the p16 CpG island. Similarly, almost all methylation-sensitive restriction enzymes, which have been used in other studies to screen for p16 methylation, are specific for sites outside of the three regions (20, 25, 37). Thus, our results suggest that screening by these methods may be subject to false negatives that would underestimate the frequency of p16 methylation in cancerous or precancerous conditions. Further investigation of primary tumor tissue will be necessary to determine if the region-specific methylation pattern and the subsequent expansion of methylation are characteristic of neoplastic progression in vivo. Characterization of the methylation patterns in tumor suppressor genes may be essential for the early detection of epigenetic lesions during premalignant stages of cancer.
In summary, our results demonstrate that de novo methylation of the p16 CpG island initially develops at a subset of sites within discrete regions and then gradually increases in density and expands to sites in adjacent regions. Thus, de novo methylation is a progressive process rather than a single event and is neither site specific nor completely random but instead is region specific.
ACKNOWLEDGMENTS
We thank the Biotechnology Center and the Image Analysis Center at the Fred Hutchinson Cancer Research Center (FHCRC). We thank Reinhard Stoger for his assistance with the graphical design of Fig. 5. We also thank Charles Laird, Michael Barrett, Thomas Paulson, and Reinhard Stoger for their advice and review of the manuscript.
This investigation was supported by the following: Poncin Scholarship Fund; National Cancer Institute RO1CA61202; American Cancer Society grant EDT80683; National Cancer Institute RO1CA64795; National Institute of General Medical Sciences, Medical Scientist Training Program grant 5T32GM07266; and the Molecular and Cellular Biology Program of the University of Washington and FHCRC, Seattle.
REFERENCES
- 1.Alcorta D A, Xiong Y, Phelps D, Hannon G, Beach D, Barrett J C. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci USA. 1996;93:13742–13747. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An H X, Niederacher D, Picard F, van Roeyen C, Bender H G, Beckmann M W. Frequent allele loss on 9p21-22 defines a smallest common region in the vicinity of the CDKN2 gene in sporadic breast cancer. Genes Chromosomes Cancer. 1996;17:14–20. doi: 10.1002/(SICI)1098-2264(199609)17:1<14::AID-GCC3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 3.Band V, Sager R. Distinctive traits of normal and tumor-derived human mammary epithelial cells expressed in a medium that supports long-term growth of both cell types. Proc Natl Acad Sci USA. 1989;86:1249–1253. doi: 10.1073/pnas.86.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 5.Bestor T. Supercoiling-dependent sequence specificity of mammalian DNA methyltransferase. Nucleic Acids Res. 1987;15:3835–3843. doi: 10.1093/nar/15.9.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolden A H, Nalin C M, Ward C A, Poonian M S, Weissbach A. Primary DNA sequence determines sites of maintenance and de novo methylation by mammalian DNA methyltransferases. Mol Cell Biol. 1986;6:1135–1140. doi: 10.1128/mcb.6.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–333. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 9.Brenner A J, Stampfer M R, Aldaz C M. Increased p16 expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene. 1998;17:199–205. doi: 10.1038/sj.onc.1201919. [DOI] [PubMed] [Google Scholar]
- 10.Cairns P, Mao L, Merlo A, Lee D J, Schwab D, Eby Y, Tokino K, van der Riet P, Blaugrund J E, Sidransky D. Rates of p16 (MTS1) mutations in primary tumors with 9p loss. Science. 1994;265:415–417. doi: 10.1126/science.8023167. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 11.Cairns P, Polascik T J, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- 12.Caldas C, Hahn S A, da Costa L T, Redston M S, Schutte M, Seymour A B, Weinstein C L, Hruban R H, Yeo C J, Kern S E. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. . (Erratum, 8:410.) [DOI] [PubMed] [Google Scholar]
- 13.Carotti D, Funiciello S, Palitti F, Strom R. Influence of pre-existing methylation on the de novo activity of eukaryotic DNA methyltransferase. Biochemistry. 1998;37:1101–1108. doi: 10.1021/bi971031i. [DOI] [PubMed] [Google Scholar]
- 14.Carotti D, Palitti F, Lavia P, Strom R. In vitro methylation of CpG-rich islands. Nucleic Acids Res. 1989;17:9219–9229. doi: 10.1093/nar/17.22.9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark S J, Harrison J, Frommer M. CpNpG methylation in mammalian cells. Nat Genet. 1995;10:20–27. doi: 10.1038/ng0595-20. [DOI] [PubMed] [Google Scholar]
- 16.Erlich H A. PCR technology: principles and applications for DNA amplification. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 17.Foster S A, Galloway D A. Human papillomavirus type 16 E7 alleviates a proliferation block in early passage human mammary epithelial cells. Oncogene. 1996;12:1773–1779. [PubMed] [Google Scholar]
- 18.Foster S A, Wong D J, Barrett M T, Galloway D A. Inactivation of p16 in human mammary epithelial cells by CpG island methylation. Mol Cell Biol. 1998;18:1793–1801. doi: 10.1128/mcb.18.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frommer M, McDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez-Zulueta M, Bender C M, Yang A S, Nguyen T, Beart R W, Van Tornout J M, Jones P A. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 21.Gonzalgo M L, Hayashida T, Bender C M, Pao M M, Tsai Y C, Gonzales F A, Nguyen H D, Nguyen T T, Jones P A. The role of DNA methylation in expression of the p19/p16 locus in human bladder cancer cell lines. Cancer Res. 1998;58:1245–1252. [PubMed] [Google Scholar]
- 22.Gonzalgo M L, Jones P A. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE) Nucleic Acids Res. 1997;25:2529–2531. doi: 10.1093/nar/25.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halbert C L, Demers G W, Galloway D A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S B. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 26.Hornstra I K, Yang T P. High-resolution methylation analysis of the human hypoxanthine phosphoribosyltransferase gene 5′ region on the active and inactive X chromosomes: correlation with binding sites for transcription factors. Mol Cell Biol. 1994;14:1419–1430. doi: 10.1128/mcb.14.2.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh C-L. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huschtscha L I, Noble J R, Neumann A A, Moy E L, Barry P, Melki J R, Clark S J, Reddel R R. Loss of p16INK4 expression by methylation is associated with lifespan extension of human mammary epithelial cells. Cancer Res. 1998;58:3508–3512. [PubMed] [Google Scholar]
- 29.Iguchi-Ariga S M, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 30.Jiang H, Chou H S, Zhu L. Requirement of cyclin E-Cdk2 inhibition in p16INK4a-mediated growth suppression. Mol Cell Biol. 1998;18:5284–5290. doi: 10.1128/mcb.18.9.5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones P A. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 32.Kass S U, Goddard J P, Adams R L P. Inactive chromatin spreads from a focus of methylation. Mol Cell Biol. 1993;13:7372–7379. doi: 10.1128/mcb.13.12.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 34.Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res. 1991;19:3998. doi: 10.1093/nar/19.14.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loughran O, Malliri A, Owens D, Gallimore P H, Stanley M A, Ozanne B, Frame M C, Parkinson E K. Association of CDKN2A/p16INK4A with human head and neck keratinocyte replicative senescence: relationship of dysfunction to immortality and neoplasia. Oncogene. 1996;13:561–568. [PubMed] [Google Scholar]
- 36.Macleod D, Charlton J, Mullins J, Bird A P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 37.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 38.Noble J R, Rogan E M, Neumann A A, Maclean K, Bryan T M, Reddel R R. Association of extended in vitro proliferative potential with loss of p16INK4 expression. Oncogene. 1996;13:1259–1268. [PubMed] [Google Scholar]
- 39.Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas M F, Mandel J L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 40.Park J-G, Chapman V M. CpG island promoter region methylation patterns of the inactive-X-chromosome hypoxanthine phosphoribosyltransferase (Hprt) gene. Mol Cell Biol. 1994;14:7975–7983. doi: 10.1128/mcb.14.12.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeifer G P, Steigerwald S D, Hansen R S, Gartler S M, Riggs A D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeifer G P, Tanguay R L, Steigerwald S D, Riggs A D. In vivo footprint and methylation analysis by PCR-aided genomic sequencing: comparison of active and inactive X chromosomal DNA at the CpG island and promoter of human PGK-1. Genes Dev. 1990;4:1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- 43.Qian X C, Brent T P. Methylation hot spots in the 5′ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997;57:3672–3677. [PubMed] [Google Scholar]
- 44.Reed A L, Califano J, Cairns P, Westra W H, Jones R M, Koch W, Ahrendt S, Eby Y, Sewell D, Nawroz H, Bartek J, Sidransky D. High frequency of p16 (CDKN2/MTS-1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56:3630–3633. [PubMed] [Google Scholar]
- 45.Reznikoff C A, Yeager T R, Belair C D, Savelieva E, Puthenveettil J A, Stadler W M. Elevated p16 at senescence and loss of p16 at immortalization in human papillomavirus 16 E6, but not E7, transformed human uroepithelial cells. Cancer Res. 1996;56:2886–2890. [PubMed] [Google Scholar]
- 46.Riggs A D, Pfeifer G P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 47.Riggs A D, Xiong Z, Wang L, LeBon J M. Methylation dynamics, epigenetic fidelity and X chromosome structure. Novartis Found Symp. 1998;214:214–225. doi: 10.1002/9780470515501.ch13. [DOI] [PubMed] [Google Scholar]
- 48.Schutte M, Hruban R H, Geradts J, Maynard R, Hilgers W, Rabindran S K, Moskaluk C A, Hahn S A, Schwarte-Waldhoff I, Schmiegel W, Baylin S B, Kern S E, Herman J G. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 49.Serrano M, Hannon G J, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 50.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 51.Silva A J, Ward K, White R. Mosaic methylation in clonal tissue. Dev Biol. 1993;156:391–398. doi: 10.1006/dbio.1993.1086. [DOI] [PubMed] [Google Scholar]
- 52.Stampfer M R. Isolation and growth of human mammary epithelial cells. J Tissue Cult Methods. 1985;9:107–115. [Google Scholar]
- 53.Stirzaker C, Millar D S, Paul C L, Warnecke P M, Harrison J, Vincent P C, Frommer M, Clark S J. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229–2237. [PubMed] [Google Scholar]
- 54.Stoger R, Kajimura T M, Brown W T, Laird C D. Epigenetic variation illustrated by DNA methylation patterns of the fragile-X gene FMR1. Hum Mol Genet. 1997;6:1791–1801. doi: 10.1093/hmg/6.11.1791. [DOI] [PubMed] [Google Scholar]
- 55.Surani M A. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 56.Toth M, Lichtenberg U, Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci USA. 1989;86:3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tremblay K D, Duran K L, Bartolomei M S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vertino P M, Yen R-W C, Gao J, Baylin S B. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996;16:4555–4565. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watt F, Molloy P L. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 60.Watts G S, Pieper R O, Costello J F, Peng Y-M, Dalton W S, Futscher B W. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17:5612–5619. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong D J, Barrett M T, Stoger R, Emond M J, Reid B J. p16INK4a promoter is hypermethylated at a high frequency in esophageal adenocarcinomas. Cancer Res. 1997;57:2619–2622. [PubMed] [Google Scholar]
- 62.Yoder J A, Soman N S, Verdine G L, Bestor T H. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270:385–395. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- 63.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci USA. 1992;89:5847–5851. doi: 10.1073/pnas.89.13.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]