Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is responsible for coronavirus disease 2019 (COVID‐19), one of the most challenging global pandemics of the modern era. Potential treatment strategies against COVID‐19 are yet to be devised. It is crucial that antivirals that interfere with the SARS‐CoV‐2 life cycle be identified and developed. 3‐Chymotrypsin‐like protease (3CLpro) is an attractive antiviral drug target against SARS‐CoV‐2, and coronaviruses in general, because of its role in the processing of viral polyproteins. Inhibitors of 3CLpro activity are screened in enzyme assays before further development of the most promising leads. Dimethyl sulfoxide (DMSO) is a common additive used in such assays and enhances the solubility of assay components. However, it may also potentially affect the stability and efficiency of 3CLpro but, to date, this effect had not been analyzed in detail. Here, we investigated the effect of DMSO on 3CLpro‐catalyzed reaction. While DMSO (5%‐20%) decreased the optimum temperature of catalysis and thermodynamic stability of 3CLpro, it only marginally affected the kinetic stability of the enzyme. Increasing the DMSO concentration up to 20% improved the catalytic efficiency and peptide‐binding affinity of 3CLpro. At such high DMSO concentration, the solubility and stability of peptide substrate were improved because of reduced aggregation. In conclusion, we recommend 20% DMSO as the minimum concentration to be used in screens of 3CLpro inhibitors as lead compounds for the development of antiviral drugs against COVID‐19.
Keywords: 3‐chymotrypsin‐like protease (3CLpro), COVID‐19, DMSO, initial velocity, SARS‐CoV‐2, thermodynamic stability
Abbreviations
- 3CLpro
3‐chymotrypsin‐like protease
- COVID‐19
coronavirus disease 2019
- DMSO
dimethyl sulfoxide
- DSC
differential scanning calorimetry
- DSF
differential scanning fluorimetry
- FRET
fluorescence resonance energy transfer
- k cat
turnover number
- K m
Michaelis constant
- k U
rate of unfolding
- MD
molecular dynamics
- nsp
nonstructural protein
- PEDV
porcine epidemic diarrhea virus
- RdRp
RNA‐dependent RNA polymerase
- RMSD
root‐mean‐square deviation
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- T m
melting temperature
- T Opt
temperature of catalysis
- WT
wild type
- ΔH cal
calorimetric enthalpy
- ΔH ‡
enthalpy of activation
- ΔT 1/2
half‐peak height
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) belongs to a diverse group of coronaviruses that infect animals, and cause mild to severe upper and lower respiratory infections in human.1, 2, 3, 4, 5 In addition to the novel coronavirus disease 2019 (COVID‐19), which emerged in Wuhan (China) in 2019, coronaviruses have been responsible for other pandemics, including severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome, which emerged in 2002 and 2012, respectively. This renders coronaviruses a worldwide health concern.6, 7, 8
Discovered in the 1960s, coronaviruses are enveloped viruses containing a single‐stranded, positive‐sense RNA genome.9, 10, 11, 12 Upon entry into the cell, viral RNA‐dependent RNA polymerase (RdRp) replicates the viral genomic RNA. Then, the protein synthesis machinery of the host cell translates the viral RNA to produce two polyproteins, pp1a and pp1ab.13 Next, pp1a and pp1ab are processed by two coronavirus proteases, 3‐chymotrypsin‐like protease (3CLpro) and papain‐like protease, into 4 structural proteins and 16 nonstructural proteins (nsps). The latter assemble into the replication and transcription complex for viral replication and transcription.14, 15, 16, 17, 18 3CLpro (Nsp5) acts on 11 cleavage sites to generate 13 nsps (Nsp4‐Nsp16), including RdRp, which is critical for viral replication and maturation. The two SARS‐CoV‐2 proteases have been proposed as important targets for the development of antiviral therapies to prevent the processing and maturation of new viral particles.14, 16, 17, 19, 20
3CLpro from different coronaviruses share high structural fold similarity.14, 15, 16, 17, 19, 21, 22, 23, 24, 25 3CLpro is catalytically active in a dimeric state. The monomer contains three domains: I (residues 10‐96), II (residues 102‐180), and III (residues 200‐303).16, 19 The active site of 3CLpro is highly conserved and located at the interface between domains I and II. The Cys‐His catalytic dyad is required for catalysis. SARS‐CoV‐2 3CLpro cleaves the viral polypeptides at N‐Leu Gln↓Ser Ala Gly‐C sites, where ↓ marks the cleavage site.14, 15, 16, 19, 26, 27 Cys145 is the catalytic residue of SARS‐CoV‐2 3CLpro and interacts with the backbone carbonyl carbon of glutamine at P1 site of the peptide substrate.16, 19, 21, 28, 29 His41 of the catalytic dyad of SARS‐CoV‐2 3CLpro is required for catalysis, specifically, for the deprotonation of the thiol side chain of Cys145, enabling its nucleophilic attack on the carbonyl carbon of the P1 glutamine on the polyprotein backbone.16, 21, 28
A highly sensitive enzymatic assay based on fluorescence resonance energy transfer (FRET) is the most common assay used for monitoring the catalytic activity of 3CLpro. In the assay, the proteolytic rate is determined based on the cleavage of a fluorescent peptide substrate.14, 15, 16, 19, 26, 27, 30, 31, 32 As a caveat, the peptide substrate used in the assay is hydrophobic, with low water solubility. Consequently, the peptide substrate for 3CLpro activity determinations is usually dissolved in Dimethyl sulfoxide (DMSO). Different DMSO concentrations are used in the activity assays of 3CLpro from different coronaviruses, ranging from 0.1% to 10%.25, 32, 33, 34, 35, 36, 37, 38 The latter is one of the highest reported concentrations of DMSO used in a SARS‐CoV 3CLpro assay, and was shown to not significantly affect the enzymatic activity.35 In addition to the requirement for DMSO in the 3CLpro reaction, to enhance the solubility of the peptide substrate, DMSO is also incorporated in high‐throughput screens of 3CLpro inhibitors. For example, libraries of small molecules used in such high‐throughput screens are solubilized in DMSO, which necessitates the presence of DMSO in the 3CLpro reaction mixture.
It is well documented that DMSO induces major structural perturbations and thermodynamic destabilization of proteins, leading to a completely unfolded state at high DMSO concentrations.39 The binding of DMSO to hydrophobic and aromatic amino acids facilitates conformational changes of the protein and an exposure of protein core segments.40 However, at low DMSO concentrations, most proteins are relatively stable, with a reduced thermal stability and only a minimal change in the gross secondary protein structure.39 Considering these observations, determining the effect of DMSO on SARS‐CoV‐2 3CLpro stability and enzymatic activity is of critical importance, not only in terms of the development of an optimal enzymatic assay but also for a reliable identification of prospective 3CLpro inhibitors. Accordingly, we here performed a detailed biochemical and kinetic characterization of the effects of DMSO on the stability and catalytic efficiency of SARS‐CoV‐2 3CLpro. We show that increased DMSO concentrations decreased the thermodynamic stability and optimum temperature for 3CLpro catalysis. Surprisingly, increased DMSO concentration enhanced the enzymatic efficiency even though the protein thermodynamic stability was reduced. Fluorescence microscopy and molecular dynamic (MD) simulations revealed that the enhanced catalytic efficiency of 3CLpro was a consequence of an increased stability of the peptide substrate, whose rate of aggregation was reduced and optimum binding to the active site enhanced by DMSO. Furthermore, the kinetic stability of 3CLpro was not compromised even in the presence of 20% DMSO. We conclude that 20% DMSO is required for the optimum catalytic efficiency of SARS‐CoV‐2 3CLpro, to facilitate the identification and characterization of 3CLpro inhibitors to be used as anti‐COVID‐19 drugs.
2. MATERIALS AND METHODS
2.1. Expression and purification of recombinant 3CLpro
Gene encoding recombinant 3CLpro from SARS‐CoV‐2 was introduced into pET28b(+) bacterial expression vector by GenScript Inc (Piscataway, NJ), and the His × 6‐tagged 3CLpro protein was expressed and purified as described previously.41 Briefly, recombinant 3CLpro was expressed in Escherichia coli BL21(DE3)CodonPlus‐RIL cells (Agilent Technologies, Santa Clara, CA) grown in terrific broth. The cell lysate was loaded onto a ProBond nickel‐chelating resin (Thermo Fisher Scientific), followed by a HiLoad Superdex 200 size‐exclusion column (GE Healthcare), using an AKTA purifier core system (GE Healthcare). The final protein sample was purified in a buffer containing 20 mM Hepes, pH 7.5, 150 mM NaCl, and 0.5 mM tris(2‐chloroethyl) phosphate (TCEP). The final protein sample was collected and concentrated to approximately 150 µM, as determined by Bradford assay (Bio‐Rad Laboratories). The sample purity was assessed by SDS‐PAGE.
2.2. Thermodynamic stability of 3CLpro at different DMSO concentrations
Differential scanning fluorimetry measurements were performed using qPCR instrument (Mx3005P QPCR system, Agilent Technologies, La Jolla, CA). T m of 3CLpro was determined in the presence of SYPRO Orange, the fluorescent reporter dye, using a 96‐well thin‐walled PCR microplate (Bio‐Rad, Cat. No. 223 94444), with excitation and emission at 492 and 610 nm, respectively. 3CLpro concentration was 7.5 µM and 3× SYPRO Orange dye was used under all conditions tested. Protein unfolding signals were acquired from 25 to 85°C at 1°C/min temperature ramp rate. The qPCR instrument was equipped with a Peltier‐based thermal system for uniform temperature ramping and thermal accuracy to ensure reproducibility of the data. The data were fitted to Boltzmann sigmoidal function using the Excel add‐on package XLfit (IDBS limited, Bridgewater, NJ), and the T m was calculated at the middle of the denaturation transition as described previously.42 The T m value of 3CLpro was measured at different DMSO concentrations to determine its effect on the stability of the protease in a buffer containing 20 mM Hepes, pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP.
The thermodynamic stability of 3CLpro was measured using Nano‐differential scanning calorimetry (DSC) (TA Instruments) that had been calibrated using chicken egg white lysozyme as an external standard. The DSC thermograms were acquired in solutions containing 30 µM 3CLpro and different DMSO concentrations. The reference and 3CLpro sample cells contained the same buffer (20 mM Hepes, pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP). The samples were scanned from 15 to 75°C at a temperature ramp rate of 1°C/min at 3 atm pressure. Each sample was ramped two times to acquire two thermograms, and the second scan was used as the buffer background scan for each sample. The melting transitions of 3CLpro samples were irreversible, as indicated by a lack of signal from the second ramp temperature scan. The DSC scans were normalized for protein concentration, baseline‐corrected by subtracting the corresponding buffer baseline, and then converted to plots of excess heat capacity (C p) as a function of temperature. T m of 3CLpro was equal to the temperature at the apex of the thermal transition. Calorimetric enthalpy (ΔH cal) of the transitions was estimated from the area under the thermal transition using NanoAnalyze Software v3.11.0 from TA instruments.
2.3. Thermal kinetic stability of 3CLpro at different DMSO concentrations
The thermal kinetic stability of 3CLpro was determined by monitoring the rate of thermal unfolding after incubating 3CLpro (30 µM) at varying temperatures, from 42 to 55°C, in a buffer containing 20 mM Hepes, pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP. The unfolding rate was measured at different concentrations of DMSO; however, Circular dichroism (CD) scanning of 3CLpro reactions was not feasible because of the high absorbance background associated with the presence of DMSO, as described previously.41 As an alternative to CD measurements, DSF was used for measuring the kinetic stability of 3CLpro in the presence of DMSO, where the unfolding of 3CLpro was continuously monitoring (for 8 minutes) by measuring the increase in fluorescence signal upon binding of the SYPRO Orange reporter dye to hydrophobic patches on 3CLpro. The fluorescence signal was then normalized and the unfolded protein fraction (F U) size was calculated from the fluorescence intensity data using the equation:
where y is the fluorescence intensity signal of SYPRO Orange dye observed at a specific time point; and y N and y D are the fluorescence intensities of native and denatured states of 3CLpro, respectively, where they were obtained at low and high incubation temperatures of 3CLpro, respectively. The rate of protein unfolding (k U) was determined from the slope of the linear region of the unfolding time course after fitting the data to a straight line using the Excel add‐on package XLfit (IDBS limited, Bridgewater, NJ).
2.4. Initial velocity studies of 3CLpro at different DMSO concentrations
The catalytic activity of 3CLpro was determined using a FRET cleavage continuous assay, with (DABCYL)KTSAVLQ↓SGFRKME(EDANS)‐NH2 (GL Biochem; Shanghai, China) as the fluorogenic peptide substrate, as described previously.14, 15, 26, 30, 31 The enzymatic rate of 3CLpro was determined by monitoring the cleavage of the fluorogenic substrate, with an increase in the fluorescence signal upon the release of the EDANS group monitored using a Cytation 5 multi‐mode microplate reader (Biotek Instruments, Winooski, VT) in a 96‐well plate assay format. The reaction was initiated upon addition of 3CLpro enzyme to the peptide substrate in 20 mM Hepes, pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP. The enzyme concentration was fixed at 3 μM and the reaction rate was measured in the presence of different amounts of DMSO for 5 minutes at 30°C, in a thermostatically controlled cell compartment. The concentration of the peptide substrate was varied from 20 to 500 μM to determine the initial velocity pattern of 3CLpro. The cleavage rate data were fitted to Michaelis‐Menten equation using the global fitting analysis function in the kinetics module of SigmaPlot (Systat Software, Inc San Jose, CA). The kinetic parameters of 3CLpro in the presence of different concentrations of DMSO were determined from the initial velocity pattern of 3CLpro. Error bars were calculated from triplicate measurements of each reaction, and are presented as the mean ± standard deviation (SD).
The inner filter effect of FRET enzymatic assay was also considered. The effect depends on the path length, the concentration of the quencher (DABCYL), and the type of instrument and its configuration used for the measurement of the emission signal. The excitation coefficient of free EDANS was determined in the absence of the peptide substrate by varying the concentrations of free EDANS, , at λex 360 nm and λem 500 nm as the excitation and emission wavelengths, respectively. As shown previously, the correction factor (Corr%) of the inner filter effect is important for correcting the decrease in the emission signal of the fluorogenic substrate in the presence of the quencher (DABCYL).26, 31, 43, 44 The fluorescence measurements with various concentrations (from 20 to 500 µM) of the peptide substrate were performed in the absence, , and presence, , of a fixed concentration (50 µM) of free EDANS. Then, the fluorescence signal of free EDANS, , at a specific substrate concentration was determined from the equation:
Corr% was determined from the emission reduction of free EDANS at a specific substrate concentration, , compared with that of EDANS in the absence of peptide substrate, .
The calculated Corr% varied depending on the concentration of the peptide substrate. This was taken into consideration when measuring the cleavage rate of 3CLpro, with the corresponding Corr% applied at specific substrate concentrations.
2.5. Optimum temperature of catalysis of the 3CLpro reaction
To investigate the effect of DMSO on T Opt, the proteolytic activity of 3CLpro was continuously monitored using FRET‐based enzymatic assay, as described above. The assay reaction contained fixed concentrations of 3CLpro (3 μM) and peptide substrate (60 μM) in an assay buffer of 20 mM Hepes, pH 7.0, 150 mM NaCl, 1 mM EDTA, and 1 mM TCEP. T Opt was determined in the presence of different concentrations of DMSO, based on the 3CLpro enzymatic activity over a temperature range from 23 to 65°C. The reaction mixture was incubated for 5 minutes at the desired temperature before initiating the reaction by the addition of the enzyme. The proteolytic activity was monitored for 5 minutes and the rate was determined from the linear fit of the initial time course. Error bars were calculated from triplicate measurements of each reaction, and are presented as the mean ± SD.
2.6. Fluorescence microscopy measurements of the aggregation rate of the peptide substrate of 3CLpro
The aggregation of 3CLpro peptide substrate was followed by injecting 100 µL of 250 µM peptide into a 6‐channel microfluidic chamber, μ‐Slide VI 0.5 (Ibidi, Munich, Germany; 0.54 mm channel height), and imaged using an Olympus FluoView FV1000 confocal microscope (Olympus, Tokyo, Japan), in transmission mode, at 10× magnification. A time‐lapse series was acquired at 2‐minutes intervals. Images acquired after 15, 45 minutes, 1, and 3 hours are shown in Figure 5A. The total transmission signal at 488 nm was normalized and fitted to sigmoidal function.
FIGURE 5.
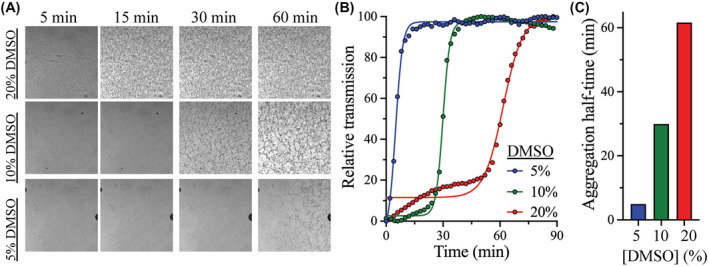
Effect of DMSO on the aggregation kinetics of 3CLpro peptide substrate. A, Bright field confocal images of the aggregation of 3CLpro peptide substrate 250 µM after incubation with different concentrations of DMSO. The aggregation of the peptide was monitored for 90 minutes, with images acquired every 2 minutes at room temperature. The condensed dark areas are peptide aggregates that formed dark‐colored particles. Regions with lesser aggregation remained clear. B, Effect of increasing DMSO concentration on the aggregation kinetics of 3CLpro peptide substrate. Peptide aggregation curves were determined from the total transmitted signal, where high transmission indicates faster aggregation rate of the peptide substrate. Increasing the DMSO concentration reduced peptide aggregation, with no aggregation observed at 20% DMSO. C, Bar plot of the halftime of aggregation of the peptide substrate at different concentrations of DMSO. The fastest aggregation rate of 5 minutes for the 3CLpro peptide substrate was observed at 5% DMSO and the slowest aggregation rate of 62 minutes was measured at 20% DMSO
2.7. MD simulation of the apo‐ and peptide‐bound 3CLpro in DMSO
For the modeling, monomeric‐state 3CLpro in complex with the peptide substrate was prepared using the crystal structure of the apo enzyme (PDB ID: 6Y2E) aligned with the crystal structure of porcine epidemic diarrhea virus (PEDV) 3CLpro (PDB ID: 4ZUH) containing the peptide substrate.16, 29 The homodimeric state of 3CLpro in complex with the peptide substrate was established using the BIOMT data in the PDB file of the monomer system using the Visual Molecular Dynamics (VMD) software.45 The protonation states of amino acid residues were set at pH 7.0 via the ProteinPrep tool in Schrödinger Maestro software.46 Subsequently, a coarse‐grained model of the homodimer of peptide‐bound 3CLpro was generated according to the elastic network model of Martini using the Martini Solution Maker of CHARMM‐GUI.47, 48 The model was solvated and neutralized with a 15 Å water layer and 0.15 M NaCl, respectively. In addition to the MD simulation in water, 3CLpro was modeled in 20% DMSO using the “gmx insert‐molecules” module of the GROningen MAchine for Chemical Simulations (GROMACS) package.49 The coarse‐grained representation of DMSO was generated using Auto_MARTINI, with the DMSO molecules inserted into the generated water box, with 20% of the total box consisting of DMSO molecules.50 The coarse‐grained simulations of peptide‐bound 3CLpro systems in water and 20% DMSO were performed using the GROMACS package.49 The systems were minimized via the steepest descent algorithm and equilibrated in the NPT ensemble. Subsequently, the systems were simulated in 10‐ps time‐steps, with the temperature of 303.15 K and pressure of 1 atm controlled by V‐rescale and Parrinello‐Rahman barostat respectively.51, 52 LINCS algorithm was exploited to constrain the stiff bonds.53 3CLpro in 20% DMSO was simulated for 2 μs in duplicate, whereas the coarse‐grained simulations in the absence of DMSO were performed for 3 μs in triplicate.
2.8. Coarse‐grained simulations of the aggregation of free peptide substrate
The effect of DMSO on the aggregation of peptide substrate was modeled using the peptide substrate of PEDV 3CLpro (PDB ID: 4ZUH).29 Thereafter, the peptide substrate was duplicated using the “gmx insert‐molecules” command of GROMACS.49 The simulation model of the peptide substrate was prepared as described above for the 3CLpro system, in water and in the presence of 20% DMSO. The coarse‐grained simulations of peptide substrate systems were performed for 1.2 μs under the same conditions as those used for the apo‐ and peptide‐bound 3CLpro systems. Analyses of the obtained trajectories were concatenated using the VMD software to demonstrate the impact of DMSO on the dynamic and structural properties of 3CLpro and its peptide substrate, as well as the effect of DMSO on the aggregation of free peptide substrate.45 root‐mean‐square deviation (RMSD) of peptide substrate in the 3CLpro active site was calculated over the simulated time by the “gmx rms” module of GROMACS using the following formula49:
where d n(t) and correspond to the current coordinate at time t and reference coordinates, respectively. The probability distribution of distances was measured over the simulation time using the “gmx distance” module of GROMACS, where the calculated timeline data were converted into probability plots.49 The minimum and maximum distance values within the systems were determined. The data were sampled at 2 Å from the minimum to the maximum distance values, and the frequencies of each interval were calculated to determine the probability of each sampled distance. Energy values for the binding interactions between the peptide substrate and 3CLpro active site were calculated using the “gmx energy” module of GROMACS.49
3. RESULTS
3.1. Experimental overview
SARS‐CoV‐2 3CLpro was expressed in E coli as a recombinant His‐tagged protein and purified using Ni‐NTA affinity column, followed by size‐exclusion gel filtration chromatography, as described previously.41 A highly sensitive FRET‐based assay was used to monitor the proteolytic activity of 3CLpro over time. Fluorescent peptide substrate labeled with a fluorophore and a quencher, (DABCYL)KTSAVLQ↓SGFRKME(EDANS)‐NH2, was utilized to monitor the cleavage rate of 3CLpro (↓ marks the cleavage site).14, 15, 16, 19, 26, 27, 30, 31, 32 The above peptide substrate is hydrophobic because of the large number of hydrophobic amino acid residues, rendering it water‐insoluble. Consequently, the 3CLpro enzymatic assay was conducted in the presence of DMSO to enhance the solubility of the peptide substrate.
3.2. Effect of DMSO on the thermodynamic stability of 3CLpro
The thermodynamic stability of 3CLpro in the presence of different concentrations of DMSO was explored using two thermoanalytical techniques: differential scanning fluorimetry (DSF) and DSC. The melting temperature (T m) of 3CLpro was determined by DSF in the presence of SYPRO Orange reporter dye at pH 7.0, at a temperature ramping rate of 1°C/min. An increase in the florescence signal was observed upon binding of the SYPRO Orange reporter dye to the hydrophobic surfaces of protein upon its unfolding and exposure of its hydrophobic core (Figure 1A). T m was calculated at the midpoint of the DSF thermal transition, as increasing the DMSO concentration reduced the stability of 3CLpro. It decreased from 52.5 ± 0.1°C in the absence of DMSO to 51.4 ± 0.1°C, 50.5 ± 0.1°C, and 47.7 ± 0.1°C at 5%, 10%, and 20% (v/v) DMSO, respectively (Figure 1B). All DMSO concentrations indicated in the manuscript are v/v. The addition of 20% DMSO decreased T m of 3CLpro by approximately 5°C, which was expected since DMSO destabilizes the structural fold of proteins by interacting with them and exposing their hydrophobic core.39, 40
FIGURE 1.
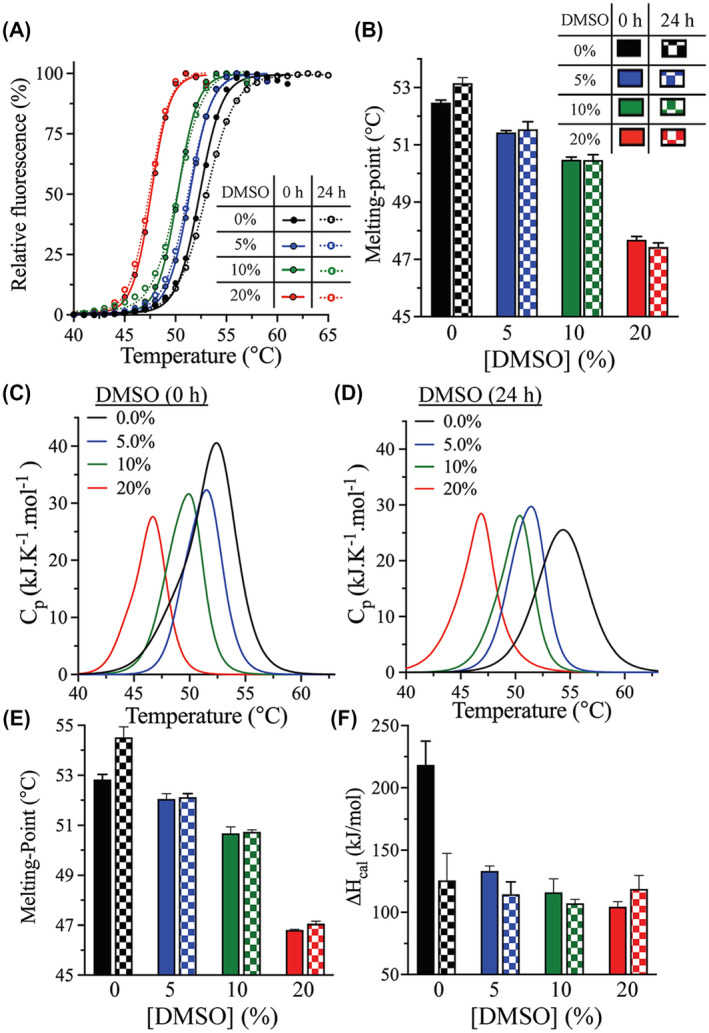
Effect of DMSO on the thermodynamic stability of 3CLpro. A, DSF thermal scans of 3CLpro in the presence of SYPRO Orange reporter dye. The scans were acquired at 0‐hour (solid line) and after 24‐hours incubation (dotted line) in the absence or presence of different concentrations of DMSO. B, Bar plot of T m calculated from the midpoint of DSF thermal scans of 3CLpro at 0‐hour (solid bars) and after 24‐hours (checkered bars) incubation in the presence of DMSO. C and D, DSC thermograms of WT 3CLpro at 0‐hour (C) and after 24‐hours (D) incubation in the presence of different amounts of DMSO. The sample was heated at a rate of 1.0 °C/min. E and F, Bar plots of T m and ΔH cal calculated from the DSC thermal scans of 3CLpro at 0‐hour and after 24‐hours incubation with DMSO. Symbols and colors are as in panel B. Data presented in bar plots are shown as the mean + SD from triplicate experiments
DSC confirmed the decrease in the thermodynamic stability of 3CLpro in the presence of DMSO. DSC thermograms of 3CLpro were acquired at 0%‐20% DMSO at pH 7.0, and exhibited single transitions, with the T m values calculated at the apex of the melting peak (Figure 1C). The T m values determined by DSC were in close agreement with those determined by DSF, with the 3CLpro stability decreasing with an increasing DMSO concentration. T m dropped from 52.8 ± 0.2°C in the absence of DMSO to 52.0 ± 0.2°C, 50.7 ± 0.3°C, and 46.8 ± 0.1°C in the presence of 5%, 10%, and 20% DMSO, respectively (Figure 1E). It decreased by approximately 5°C in the presence of 20% DMSO, which was in close agreement with what was observed using DSF.
Calorimetric enthalpy (ΔH cal) of unfolding was next determined from the area under the thermographic peak in 3CLpro DSC scans. The value notably decreased, from 218 ± 19 kJ/mol in the absence of DMSO to 133 ± 4 kJ/mol in the presence of 5% DMSO (Figure 1F). In comparison with the DSC scan data at 5% DMSO, the amplitude of DSC thermographic transitions (and the calculated ΔH cal) slightly decreased to 107 ± 3 and 104 ± 4 kJ/mol at 10% and 20% DMSO, respectively.
Another parameter that can be extracted from the DSC scan is the width at half‐peak height (ΔT 1/2) of the calorimetric transition. For 3CLpro, this parameter decreased in the presence of DMSO. Next, the ΔT 1/2 values were used to determine the cooperativity of the phase transition of DSC thermograms. The transition was narrower in the presence of DMSO, and ΔT 1/2 decreased from 2.3°C in the absence of DMSO to 1.5°C in the presence of 20% DMSO. On the other hand, 5% and 10% DMSO did not significantly affect the ΔT 1/2 value. The phase property of 3CLpro is dependent on DMSO, indicating less cooperativity of the phase transition in the absence of DMSO. Therefore, DMSO enhances the cooperativity of transition of 3CLpro.
We next investigated the effect of prolonged DMSO exposure, which was reflective of the timeframe of typical enzymatic rate measurements for kinetic characterization, and the screening and identification of inhibitors of 3CLpro. Accordingly, the enzyme's thermodynamic stability was assessed after its 24‐hours incubation in the presence of different concentrations of DMSO. Based on DSF analysis, T m of 3CLpro did not change significantly during that time; however, it slightly increased (by 1°C) in the absence of DMSO after 24 hours (Figure 1A,B). Similarly, the DSC thermographic scans of 3CLpro were not significantly affected by 24‐hours incubation with different DMSO concentrations, with similar T m values noted as those for 0‐hours incubation (Figure 1D,E). As for DSF analysis, the DSC scans revealed a small increase (1.7°C) in the T m value of 3CLpro after 24‐hours incubation in the absence of DMSO. The ΔH cal was also slightly altered, by less than 19 kJ/mol, upon 24‐hours incubation with DMSO (Figure 1F). However, 24‐hours incubation in the absence of DMSO resulted in the largest change of ΔH cal (a drop by 93 ± 21 kJ/mol). Overall, the DSF and DSC analyses revealed that the presence of DMSO decreased the thermodynamic stability of 3CLpro. However, 24‐hours incubation with DMSO did not further destabilize the protein's thermodynamic stability, with similar DSF and DSC scans at 0‐hour and after 24‐hours incubation with 5%‐20% DMSO.
3.3. Effect of DMSO on the optimum temperature for 3CLpro catalysis
The effect of temperature on the catalytic activity of 3CLpro was next assessed at different DMSO concentrations, to determine the effect of DMSO on the optimum temperature for 3CLpro catalysis (T Opt). Accordingly, 3CLpro activity at different temperatures (23‐ 65°C) and DMSO concentrations [5%‐20% (v/v)] was followed using a FRET‐based enzymatic assay. Because of the poor aqueous solubility of the peptide substrate, the proteolytic rate of 3CLpro could not be determined in the absence of DMSO. The T Opt plots of 3Clpro at different DMSO concentrations displayed classic bell‐shaped curves, with the cleavage activity recorded as a function of temperature (Figure 2A). T Opt was determined at the apex of the 3Clpro temperature profile, which corresponded to the highest proteolytic rate at a given DMSO concentration. Compared with 5% DMSO, the T Opt value of 43 °C for 3Clpro did not change at 10% DMSO; however, it decreased to 32 °C at 20% DMSO (Figure 2B). Similar to the thermodynamic stability analysis, increasing the DMSO concentration resulted in a decrease in the T Opt of 3Clpro. Interestingly, the cleavage rate of 3Clpro increased with an increasing DMSO concentration. At T Opt, the cleavage rate of 3Clpro increased from 64 ± 6 s−1 at 5% DMSO to 93 ± 6 s−1 and 186 ± 4 s−1 at 10% and 20% DMSO, respectively. The enhanced catalytic rate of 3Clpro at the higher DMSO concentrations tested was unexpected since the enzyme was thermodynamically less stable at these concentrations. We proceeded to investigate this phenomenon in more detail to reveal the absolute effect of DMSO on the kinetic properties and catalytic efficiency of 3Clpro reaction.
FIGURE 2.
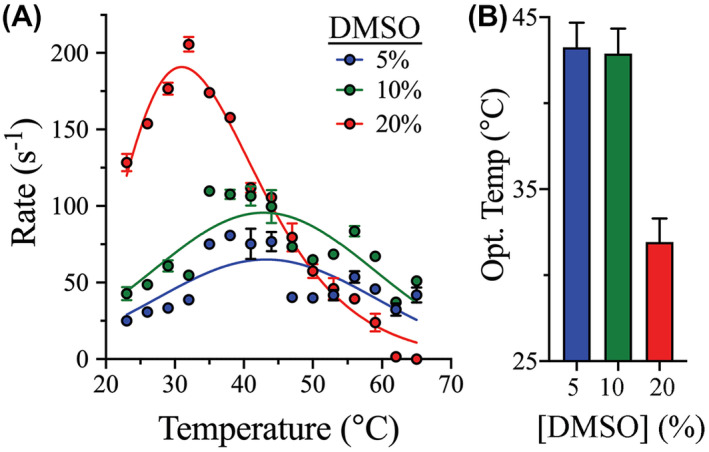
Effect of DMSO on the optimum temperature for 3CLpro catalysis. A, The optimum temperature for catalysis at different DMSO concentrations was determined by measuring the proteolytic cleavage rate of 3CLpro at different temperatures. Concentrations of the enzyme and its peptide substrate were fixed at 3 and 60 μM, respectively, for all measurements and the reactions were monitored in the same reaction buffer at pH 7.0. The bell‐shaped temperature profiles shifted to a lower temperature upon increasing the DMSO concentration to 20%. Despite this, the 3CLpro cleavage rate increased at higher DMSO concentrations. B, Bar plot of the optimum temperature for 3CLpro catalysis. T Opt was the same in the presence of 5% and 10% DMSO; however, it dropped from 43 to 32°C in the presence of 20% DMSO. Data in all panels are shown as the mean ± SD from triplicate experiments
3.4. Effect of DMSO on the kinetic parameters of 3Clpro
To investigate the effect of DMSO on the kinetic parameters of SARS‐CoV‐2 3Clpro, initial velocity studies were conducted, by measuring the 3Clpro cleavage rate at different concentrations of the peptide substrate, with fixed enzyme concentration, in the presence of 5%‐20% (v/v) DMSO (Figure 3A). Similar to the rate enhancement indicated by the T opt measurements, the turnover number (k cat) of 3Clpro increased by 1.5‐ and 2.5‐fold upon increasing the DMSO concentration from 5% to 10% and 20%, respectively (Figure 3C and Table 1). The affinity of 3Clpro for the peptide substrate also increased after increasing the DMSO concentration from 5% to 20%, as illustrated by an approximately twofold decrease in the Michaelis constant (K m) (Figure 3D and Table 1). The peptide affinity and K m value for 3Clpro at 5% and 10% DMSO were the same. Similarly, the catalytic efficiency (k cat/K m) increased by 1.5‐ and 4.7‐fold upon increasing the DMSO concentration from 5% to 10% and 20%, respectively (Figure 3E and Table 1). These observations confirm the enhancement of 3Clpro catalytic activity at increased DMSO concentrations despite its decreased thermodynamic stability.
FIGURE 3.
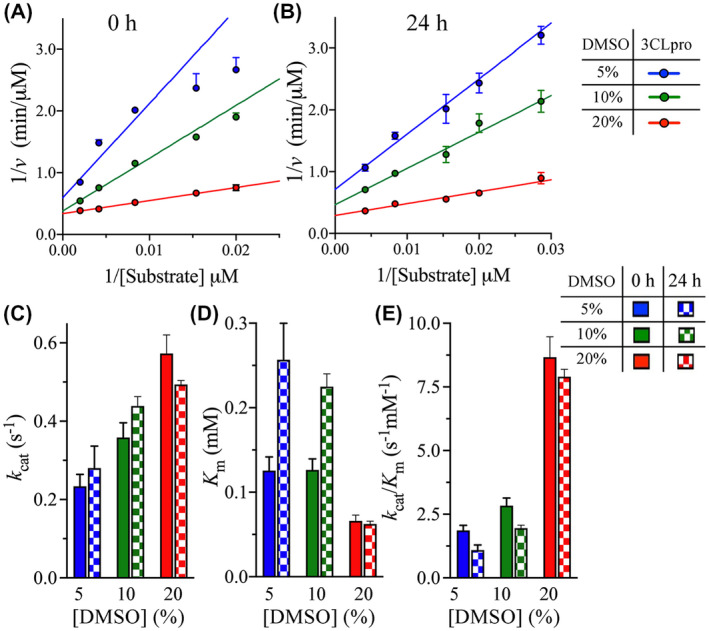
Effect of DMSO on the kinetic parameters of 3CLpro. A and B, Lineweaver‐Burk plots of the initial velocity of 3CLpro at different concentrations of DMSO. The cleavage rate was measured at 30°C and pH 7.0 at 0‐hour and after 24‐hours enzyme incubation with the indicated DMSO concentrations. The rate was measured at fixed 3CLpro concentration (3 μM), while varying the peptide substrate concentration (from 20 to 500 μM). C and E, Bar plots of k cat and K m, and the catalytic efficiency k cat/K m as a function of DMSO concentration at 0‐hour and after 24‐hours incubation with DMSO. Data in all panels are shown as the mean ± SD from triplicate experiments
TABLE 1.
Kinetic parameters of 3CLpro at different DMSO concentrations, determined at 30°C and pH 7.0
| DMSO (%) | kcat (s−1) Fold change | Km (mM) Fold change | kcat/Km (s−1 mM−1) Fold change | |
|---|---|---|---|---|
| 0‐hour incubation | 5 | 0.23 ± 0.03 | 0.13 ± 0.02 | 1.86 ± 0.24 |
| 10 | 0.36 ± 0.04 | 0.13 ± 0.01 | 2.84 ± 0.29 | |
| (+1.5) a | (1.0) a | (+1.5) a | ||
| 20 | 0.57 ± 0.05 | 0.066 ± 0.007 | 8.67 ± 0.80 | |
| (+2.5) a | (−1.9) a | (+4.7) a | ||
| 24‐hours incubation | 5 | 0.28 ± 0.06 | 0.26 ± 0.04 | 1.1 ± 0.2 |
| (+1.2) b | (+2.0) b | (−1.7) b | ||
| 10 | 0.44 ± 0.02 | 0.22 ± 0.02 | 2.0 ± 0.2 | |
| (+1.2) b | (+1.8) b | (−1.5) b | ||
| 20 | 0.49 ± 0.01 | 0.062 ± 0.003 | 7.9 ± 0.5 | |
| (−1.2) b | (−1.1) b | (−1.1) b |
Bold indicats discussion of the data.
Compared with the value at 5% DMSO after 0‐hour incubation.
Compared with the value at the same DMSO concentration after 0‐hour incubation.
An important concern during the measurements of 3Clpro enzymatic activity is the need for prolonged exposure to DMSO. To evaluate the effect of prolonged DMSO exposure on 3Clpro kinetic parameters, the enzyme was incubated for 24 hours with DMSO, and the kinetic parameters were then determined (Figure 3B). The 24‐hours incubation of 3Clpro with DMSO slightly altered the k cat value, by approximately 1.2‐fold, at the different DMSO concentrations tested (Figure 3C and Table 1). Interestingly, 24‐hours incubation of 3Clpro with 5% and 10% DMSO increased the K m value by approximately twofold, but with no change was observed at 20% DMSO (Figure 3D and Table 1). Therefore, prolonged incubation of 3Clpro with 5% and 10% DMSO reduced its affinity for the peptide substrate. The k cat/K m parameter slightly decreased by approximately 1.6‐fold after 24‐hours incubation with 5% and 10% DMSO, with no significant changes observed at 20% DMSO (Figure 3E and Table 1). Consequently, the prolonged 3Clpro exposure to 20% DMSO did not greatly affect the enzyme’s catalytic parameters. This confirms the importance of assaying 3Clpro activity at 20% DMSO, to ensure high enzymatic efficiency.
3.5. Effect of DMSO on the thermal kinetic stability of 3Clpro catalysis
Next, to determine the thermal kinetics of 3Clpro unfolding at different DMSO concentrations, the rate of isothermal protein denaturation was monitored by DSF using SYPRO Orange as the reporter dye. CD spectroscopy is often the preferred method for determining the thermal unfolding rates of protein; however, it was not possible to use CD here to monitor the 3Clpro unfolding signal in the presence of DMSO concentrations above 0.5% because of the high signal‐to‐noise ratio. Previously, CD was used to monitor the rate of unfolding of 3Clpro and determine its kinetic stability at different pH values.41 Instead, DSF was used here to monitor the rate of 3Clpro thermal unfolding at different incubation temperatures (42‐ 55°C) and in the presence of different concentrations of DMSO (0%‐20% [(v/v]) (Figure 4A‐D). The thermal unfolding rate (k U) of 3Clpro was calculated from the linear slope of the denaturation signal monitored by the increase in the fluorescence of SYPRO Orange reporter dye upon its binding to the exposed hydrophobic surfaces on the protein. The native (folded) and fully unfolded states of 3Clpro were then used to calculate the unfolded fraction size at different time points for up to 8 minutes.
FIGURE 4.
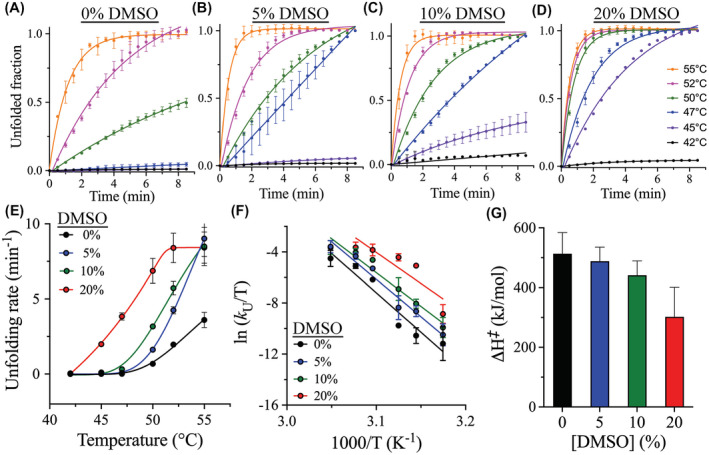
Effect of DMSO on the thermal kinetic stability of 3CLpro. A‐D, Time course of the thermal unfolding of 3CLpro at various temperatures from 42 to 55°C in the presence of 0%‐20% (v/v) DMSO, as monitored by DSF. The size of the unfolded fraction of 3CLpro was calculated from the increase in fluorescence signal of the SYPRO Orange reporter dye. Points on the graphs represent experimental data, and lines are the theoretical fit of the data to a single exponential function. Rate of unfolding (k U) was calculated from the linear portion of the time course. E, Plot of k U as a function of temperature at different DSMO concentrations. The fastest unfolding kinetics of 3CLpro were recorded at 20% DMSO, and the highest kinetic stability corresponding to the slowest rate of protein unfolding was observed in the absence of DMSO. F, Eyring plot of the temperature dependence of k U for 3CLpro at different DMSO concentrations. Points on the graph represent experimental data, and lines are the linear least square regressions of the data. A change in the slope is a result of the variation in ΔH ‡. G, Bar plot of ΔH ‡ at different DMSO concentrations with the stability and ΔH ‡ value decreasing with an increasing DMSO concentration. Data in all panels are shown as the mean ± SD values from triplicate experiments
The k U of 3Clpro increased with increasing the incubation temperature from 42 to 55°C at the different DMSO concentrations tested here (Figure 4E). Temperature dependence of the unfolding rate constant was represented by Eyring plot, with ln(k U/T) plotted against 1/T (Figure 4F). The slope of the resultant lines was used to calculate the enthalpy of activation (ΔH ‡). The temperature dependence of the second‐order rate constants of 3Clpro unfolding was fitted to Eyring equation, shown below, for the calculation of ΔH ‡:
where k B is Boltzmann’s constant; h is Planck’s constant; R is the gas constant; T is the absolute temperature; and ΔS ‡ is the entropy of activation. As determined, ΔH ‡ of 3Clpro did not change significantly upon the addition of DMSO, with a relatively small drop in ΔH ‡, from 513 kJ/mol in the absence of DMSO to 406 kJ/mol in the presence of 20% DMSO (Figure 4G). In addition, ΔH ‡ slightly changed, from 488 to 441 kJ/mol, upon increasing the DMSO concentration from 5% to 10%, respectively. Overall, the kinetic stability of 3Clpro was relatively similar in the absence or presence of DMSO.
3.6. Effect of DMSO on the aggregation of 3Clpro peptide substrate
As demonstrated above, with an increasing DMSO concentration, the thermodynamic stability of 3Clpro decreased with a slight change on its kinetic stability. However, the catalytic efficiency of 3Clpro increased with DMSO, where 20% DMSO represented the best concentration tested here. We asked whether this effect was associated with the behavior of the peptide substrate in the presence of DMSO: the 13 residue peptide substrate (KTSAVLQSGFRKM) contains six hydrophobic amino acids that contribute to its insolubility in water and might also reduce its stability in aqueous solutions. Since DMSO compromised the thermodynamic stability of 3Clpro, the higher rate of 3Clpro catalysis in the presence of increasing DMSO concentrations could be related to an enhanced stability of the peptide substrate. Accordingly, to determine the stability of the peptide substrate in solution, its aggregation rate was monitored using a confocal fluorescence microscope in the presence of 5%‐20% (v/v) DMSO (Figure 5A). The aggregation kinetics were assessed based on the total transmission signal curves fitted to a sigmoidal function (Figure 5B). At DMSO concentrations below 10%, an initial slow aggregation rate was observed, followed by fast aggregate formation. The peptide substrate did not aggregate at 20% DMSO, and was stable for up to 3 hours, the maximum time tested (Figure 5A,B). However, decreasing the DMSO concentration induced the formation of larger aggregates, with the aggregation halftime of 5, 30, and 62 minutes in the presence of 5%, 10%, and 20% DMSO, respectively (Figure 5C). The high aggregation rate of the peptide substrate at low DMSO concentrations was expected, because of its high hydrophobicity and low stability in aqueous solutions.
The DMSO‐dependent aggregation pattern observed by fluorescence microscopy was then investigated by coarse‐grained MD simulations. Free peptide substrate was extracted from the structure of PEDV 3CLpro (PDB ID: 4ZUH).29 Thereafter, the peptide substrate was duplicated, and two systems were setup to compare peptide stability in the absence and presence of 20% DMSO. The DMSO‐incorporating system was prepared by immersing the peptide in crystal water molecules in a solvent box with the dimensions of 200 Å × 200 Å × 200 Å, and containing 4419 DMSO molecules. The internal geometry of the DMSO molecules was kept rigid during the MD simulations, with a total number of 76 092 atoms in the simulated system; the calculations were performed at 298 K and 1 atm. The impact of DMSO on the stability, solubility, and availability of the free peptide substrate was consistent with the fluorescence microscopy analysis, with low peptide aggregation in the presence of 20% DMSO. DMSO decreased the formation of large aggregates of the peptide substrate, compared with peptide behavior in aqueous solutions. The coarse‐grained MD simulations in the presence of 20% DMSO revealed the formation of only small peptide substrate clusters, which confirms the importance of DMSO for the solubility and stability of the peptide substrate, by preventing its aggregation.
3.7. MD simulations of the impact of DMSO on the peptide substrate‐binding affinity to 3CLpro
Next, MD simulations were performed to investigate the impact of DMSO on the efficiency of peptide binding to the 3CLpro active site. The binding interaction and affinity of the peptide substrate to the 3CLpro active site were examined in the absence and presence of 20% DMSO. To prepare the peptide‐bound state of SARS‐CoV‐2 3CLpro, the crystal structure of the apo‐state of SARS‐CoV‐2 3CLpro (PDB ID: 6Y2E) was aligned with that of PEDV 3CLpro with a bound peptide substrate (PDB ID: 4ZUH).16, 29 The peptide‐bound state of SARS‐CoV‐2 3CLpro was then subjected to two separate MD simulations in water and in 20% DMSO, as described in the preceding paragraph, for 1.2 μs. The RMSD value of the backbone of the peptide substrate in the 3CLpro active site was reduced in the presence of 20% DMSO, which implied reduced fluctuation of the peptide substrate and increased binding stability in the active site of 3CLpro (Figure 6A). The presence of DMSO also reduced the distance between the peptide substrate and Cys145 of the catalytic dyad. Shorter distances were sampled between glutamine of P1 site in the peptide substrate and Cys145 in 20% DMSO than in its absence (Figure 6B). Presumably, the reduced distance between the backbone of P1 site of the peptide substrate and Cys145 in the presence of DMSO could contribute to an increased cleavage activity of 3CLpro. The interaction energy between the peptide substrate and 3CLpro was also calculated as, on average, −389 and −458 kJ/mol in the absence and presence of 20% DMSO, respectively. The energy of interaction between the peptide substrate and 3CLpro was higher in the presence of DMSO than in its absence, which indicated increased binding affinity. These observations are consistent with the initial velocity studies, in which a decrease in the K m value of the peptide substrate was observed in the presence of DMSO. MD simulations and the initial velocity studies confirmed that the peptide substrate‐binding affinity to 3CLpro increases in the presence of 20% DMSO.
FIGURE 6.
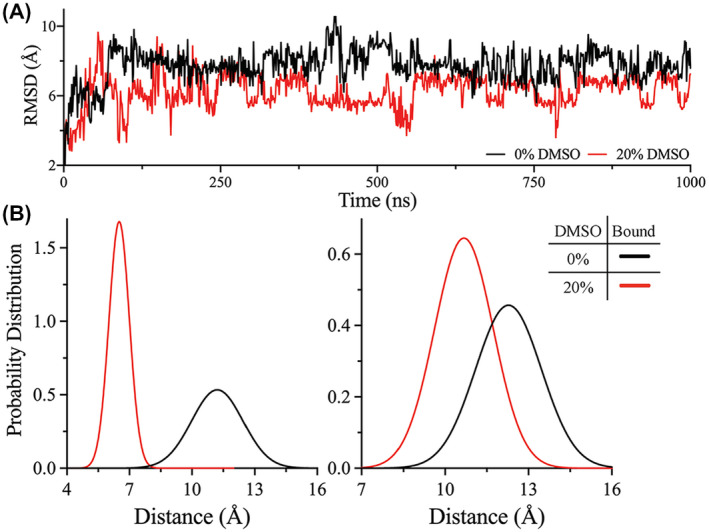
MD simulation of the DMSO effect on the peptide substrate bound to 3CLpro. A, RMSD of the peptide substrate obtained from the trajectories of peptide substrate bound to 3CLpro in the absence (black) or presence (red) of 20% DMSO. The peptide substrate demonstrated a reduced RMSD value in the presence of DMSO, indicating enhanced binding stability to the 3CLpro active site. B, Probability distribution of the distance between the catalytic residue, Cys145, and the backbone carbonyl carbon of glutamine at P1 site of the peptide substrate for the first (left) and second (right) protomer of the 3CLpro dimer. In the presence of 20% DMSO (red), the probability distribution of the distance between Cys145 and the peptide substrate was decreased, which might contribute for the enhanced catalytic activity of 3CLpro. The probability distribution of the distance was higher in the absence of DMSO (black) than in 20% DMSO
4. DISCUSSION
The 2019 SARS‐CoV‐2 disease outbreak has become one of the largest pandemics, with no treatment for coronavirus infections approved by the US Food and Drug Administration to date. Development of antiviral agents against SARS‐CoV‐2 is essential for the treatment of COVID‐19 and any future outbreaks of disease caused by novel coronaviruses. Coronavirus proteases are vital targets for the development of antiviral treatments and drug discovery studies, including those against COVID‐19. Identification of optimum assay conditions for the screening of highly effective SARS‐CoV‐2 protease inhibitors that could be developed into antiviral treatments against COVID‐19 is critical. While DMSO is a common additive in 3CLpro activity assays, most studies either fail to report the DMSO concentrations used or involve low DMSO concentrations, with 10% being the highest DMSO concentration reported for coronaviruses.32, 54 Here, we showed that 20% DMSO is the optimum concentration for assaying the enzymatic activity of 3CLpro, even though the 3CLpro thermodynamic stability and optimum temperature of catalysis are reduced at this concentration.
DMSO is a dipolar aprotic solvent that is commonly used in various enzymatic and biological assays, and also for the preparation of small molecule libraries for drug target screening. Likewise, DMSO is an important additive in the 3CLpro assay because of the low aqueous solubility of the 3CLpro peptide substrate. To date, the effect of DMSO on the stability and kinetic efficacy of 3CLpro from different coronaviruses has not been investigated in details nor its optimum concentration determined for 3CLpro activity assay. The thermal analytical determinations undertaken in the current study revealed that the thermodynamic stability of SARS‐CoV‐2 3CLpro was reduced in the presence of DMSO with a drop in the T m value that was DMSO concentration dependent. A decrease in the ΔH cal value was also observed with the decrease of thermodynamic stability of 3CLpro in the presence of DMSO. In general, the addition of DMSO decreases the thermodynamic stability of proteins, as it perturbs their secondary structure and structural fold.39 The thermodynamic destabilization of 3CLpro by DMSO observed herein can be a result of a partial exposure of the enzyme's hydrophobic core upon DMSO binding, and its interactions with the hydrophobic and aromatic amino acids in the core.40 This explanation is consistent with the decrease in ΔH cal of 3CLpro in the presence of DMSO, upon increased 3CLpro hydrophobicity. Indeed, protein ΔH cal reduction is associated with an increase in protein hydrophobicity linked to a partial exposure of the protein hydrophobic core.55, 56, 57, 58 However, increasing DMSO concentration from 5% to 20% did not alter ΔH cal of 3CLpro, which indicates that the enzyme structural fold was not significantly affected by the higher DMSO concentrations. DMSO also decreased ΔT 1/2 of 3CLpro, as determined based on the width of the DSC thermogram peak, which was indicative of the cooperativity of protein unfolding. Hence, the addition of DMSO increased the cooperativity of 3CLpro denaturation process.
TOpt was another thermal property of 3CLpro that decreased with an increasing DMSO concentration. In the current study, the lowest T Opt (32°C) was recorded at 20% DMSO. Even at 20%, the highest DMSO concentration tested here, T Opt of 3CLpro was higher than the room temperature (approximately 25°C), and represents the optimal temperature to be used in an assay of 3CLpro activity. Consequently, the subsequent 3CLpro assays measuring the protease cleavage rate were performed at 30°C. However, as noted during the T Opt measurements, the catalytic rate of 3CLpro increased by threefold upon increasing the DMSO concentration from 5% to 20% under the same reaction conditions. This was unexpected, as the thermodynamic stability and T Opt of 3CLpro decreased in the presence of increased DMSO concentrations. Therefore, we investigated the effect of DMSO on the catalytic efficiency of 3CLpro in more detail. The initial velocity studies revealed an enhancement of catalytic properties of 3CLpro in the presence of 20% DMSO. The catalytic efficiency of 3CLpro increased by approximately fivefold upon increasing the DMSO concentration from 5% to 20%, which was a result of an increased k cat value and an enhanced affinity of 3CLpro for the peptide substrate.
3CLpro exhibits enhanced catalytic efficiency at high DMSO concentrations of 20%, which can be explained by the data generated by different analyses in the current study. First, fluorescence imaging and MD simulations confirmed that increasing the DMSO concentration enhances the solubility and availability of the peptide substrate to 3CLpro. The peptide substrate was most stable at 20% DMSO: its stability was time‐sensitive, with aggregation and precipitation observed at low DMSO concentrations. In addition, MD simulations confirmed the stabilizing effect of DMSO on the binding interactions between the peptide substrate and residues at the 3CLpro active site. Another important observation was that the kinetic stability of 3CLpro did not change significantly upon the addition of up to 20% DMSO, with small variations in ΔH ‡ of 3CLpro observed. The kinetic stability of an enzyme is related to the free energy barrier between the native (functional) and unfolded (non‐functional) states.59 The energy barrier is related to the enthalpy of activation, where a higher free‐energy barrier preserves enzyme activity and catalytic function even if the enzyme is thermodynamically less stable in that scenario. In the current study, the value of ΔH ‡ decreased slightly in the presence of DMSO, which can explain the ability of 3CLpro to maintain a catalytically active state at 20% DMSO.
Overall, DMSO reduced the thermodynamic stability of 3CLpro and altered its structural fold from one dominated by polar hydrophilic bonding interactions to one with a more hydrophobic character. However, the kinetic stability was maintained in the presence of up to 20% DMSO, which is the minimum recommended DMSO concentration to be used in SARS‐CoV‐2 3CLpro activity assay. This DMSO concentration ensures stability of the peptide substrate, preventing its aggregation and allowing efficient binding to the 3CLpro active site. These conditions are especially important when screening small molecule inhibitors of 3CLpro, with the enzyme exposed to DMSO for an extended period of time. The addition of DMSO reduced the thermodynamic stability of 3CLpro; however, the prolonged (24 hours) incubation with DMSO did not affect the T m of 3CLpro and ΔH cal only slightly altered. It is clear that the addition of 20% DMSO to the catalytic assay of 3CLpro will reduce the stability of the enzyme, but it will enhance the catalytic rate measurement with a more stable and soluble peptide substrate. In the fight against COVID‐19, the identification of safe and effective antiviral therapeutics is of great importance, even in light of the development of effective SARS‐CoV‐2 vaccines.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest with the contents of this article.
AUTHOR CONTRIBUTIONS
J.C. Ferreira and S. Fadl expressed 3CLpro, conducted the biochemical analysis, and analyzed the data. M. Ilter and H. Pekel contributed equally to the MD simulations and computational modeling. R. Rezgui performed the fluorescence microscope imaging. O. Sensoy supervised the MD simulations and computational modeling, and assisted in writing the manuscript. W.M. Rabeh designed the biochemical experiments, supervised the project, and wrote the manuscript.
ACKNOWLEDGMENTS
This work was supported by New York University Abu Dhabi through research funds to the laboratory of WMR and the COVID‐19 Facilitator Research Fund (grant number: ADC05). This research was partially carried out using the Core Technology Platform resources at New York University Abu Dhabi. The authors thank Istanbul Medipol University, and Tubitak Ulakbim High Performance and Grid Computing Center (TRUBA resources) for providing access to the high‐performance computing system.
Ferreira JC, Fadl S, Ilter M, et al. Dimethyl sulfoxide reduces the stability but enhances catalytic activity of the main SARS‐CoV‐2 protease 3CLpro. The FASEB Journal. 2021;35:e21774. 10.1096/fj.202100994
This article was fast‐tracked under a recently instituted interim policy in which editors may, at their discretion, accept coronavirus‐related manuscripts submitted for the Methods, Review, Perspective, and Hypothesis categories without additional review.
DATA AVAILABILITY STATEMENT
All data, associated protocols, methods, and sources of materials are contained within the manuscript.
REFERENCES
- 1.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and coronavirus disease‐2019 (COVID‐19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acter T, Uddin N, Das J, Akhter A, Choudhury TR, Kim S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) as coronavirus disease 2019 (COVID‐19) pandemic: a global health emergency. Sci Total Environ. 2020;730:138996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn DG, Shin HJ, Kim MH, et al. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID‐19). J Microbiol Biotechn. 2020;30:313‐324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cascella M, Rajnik M, Cuomo A, Dulebohn SC, Di Napoli R. Features, Evaluation and Treatment Coronavirus (COVID‐19). Treasure Island, FL: StatPearls Publishing; 2020. [PubMed] [Google Scholar]
- 6.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohrabi C, Alsafi Z, O'Neill N, et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID‐19). Int J Surg. 2020;76:71‐76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS‐CoV‐2 and COVID‐19. Nat Rev Microbiol. 2020;19(3):141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahn JS, McIntosh K. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 2005;24:S223‐S227, discussion S226. [DOI] [PubMed] [Google Scholar]
- 10.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS‐CoV‐2. Nat Med. 2020;26:450‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liya G, Yuguang W, Jian L, et al. Studies on viral pneumonia related to novel coronavirus SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV: a literature review. APMIS. 2020;128(6):423‐432. [DOI] [PubMed] [Google Scholar]
- 13.Khan S, Siddique R, Shereen MA, et al. Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: biology and therapeutic options. J Clin Microbiol. 2020;58(5):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti‐SARS drugs. Science. 2003;300:1763‐1767. [DOI] [PubMed] [Google Scholar]
- 15.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085‐4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Lin D, Sun X, et al. Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved alpha‐ketoamide inhibitors. Science. 2020;368:409‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019‐nCoV). Nat Rev Drug Discov. 2020;19:149‐150. [DOI] [PubMed] [Google Scholar]
- 19.Jin Z, Du X, Xu Y, et al. Structure of M(pro) from SARS‐CoV‐2 and discovery of its inhibitors. Nature. 2020;582:289‐293. [DOI] [PubMed] [Google Scholar]
- 20.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C, Wei P, Fan K, Liu Y, Lai L. 3C‐like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568‐4574. [DOI] [PubMed] [Google Scholar]
- 22.Weiss SR, Navas‐Martin S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 2005;69:635‐664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan JF, Lau SK, Woo PC. The emerging novel Middle East respiratory syndrome coronavirus: the “knowns” and “unknowns”. J Formos Med Assoc. 2013;112:372‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Needle D, Lountos GT, Waugh DS. Structures of the Middle East respiratory syndrome coronavirus 3C‐like protease reveal insights into substrate specificity. Acta Crystallogr D Biol Crystallogr. 2015;71:1102‐1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomar S, Johnston ML, St John SE, et al. Ligand‐induced dimerization of middle east respiratory syndrome (MERS) coronavirus nsp5 protease (3CLpro): implications for nsp5 regulation and the development of antivirals. J Biol Chem. 2015;290:19403‐19422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue X, Yang H, Shen W, et al. Production of authentic SARS‐CoV M(pro) with enhanced activity: application as a novel tag‐cleavage endopeptidase for protein overproduction. J Mol Biol. 2007;366:965‐975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gioia M, Ciaccio C, Calligari P, et al. Role of proteolytic enzymes in the COVID‐19 infection and promising therapeutic approaches. Biochem Pharmacol. 2020;182:114225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratia K, Saikatendu KS, Santarsiero BD, et al. Severe acute respiratory syndrome coronavirus papain‐like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci U S A. 2006;103:5717‐5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye G, Deng F, Shen Z, et al. Structural basis for the dimerization and substrate recognition specificity of porcine epidemic diarrhea virus 3C‐like protease. Virology. 2016;494:225‐235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao RY, To AP, Ng LW, et al. Characterization of SARS‐CoV main protease and identification of biologically active small molecule inhibitors using a continuous fluorescence‐based assay. FEBS Lett. 2004;576:325‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuo CJ, Chi YH, Hsu JT, Liang PH. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem Biophys Res Commun. 2004;318:862‐867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grum‐Tokars V, Ratia K, Begaye A, Baker SC, Mesecar AD. Evaluating the 3C‐like protease activity of SARS‐Coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 2008;133:63‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratia K, Pegan S, Takayama J, et al. A noncovalent class of papain‐like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci U S A. 2008;105:16119‐16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacha U, Barrila J, Gabelli SB, Kiso Y, Mario Amzel L, Freire E. Development of broad‐spectrum halomethyl ketone inhibitors against coronavirus main protease 3CL(pro). Chem Biol Drug Des. 2008;72:34‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, Mittal A, Patel K, et al. Identification of novel drug scaffolds for inhibition of SARS‐CoV 3‐Chymotrypsin‐like protease using virtual and high‐throughput screenings. Bioorg Med Chem. 2014;22:167‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rathnayake AD, Zheng J, Kim Y, et al. 3C‐like protease inhibitors block coronavirus replication in vitro and improve survival in MERS‐CoV–infected mice. Sci Transl Med. 2020;12(557):eabc5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abian O, Ortega‐Alarcon D, Jimenez‐Alesanco A, et al. Structural stability of SARS‐CoV‐2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int J Biol Macromol. 2020;164:1693‐1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mody V, Ho J, Wills S, et al. Identification of 3‐chymotrypsin like protease (3CLPro) inhibitors as potential anti‐SARS‐CoV‐2 agents. Commun Biol. 2021;4:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson M, Mantsch HH. Beware of proteins in DMSO. Biochim Biophys Acta. 1991;1078:231‐235. [DOI] [PubMed] [Google Scholar]
- 40.Arakawa T, Kita Y, Timasheff SN. Protein precipitation and denaturation by dimethyl sulfoxide. Biophys Chem. 2007;131:62‐70. [DOI] [PubMed] [Google Scholar]
- 41.Ferreira JC, Rabeh WM. Biochemical and biophysical characterization of the main protease, 3‐chymotrypsin‐like protease (3CLpro) from the novel coronavirus SARS‐CoV 2. Sci Rep. 2020;10:22200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira JC, Khrbtli AR, Shetler CL, et al. Linker residues regulate the activity and stability of hexokinase 2, a promising anticancer target. J Biol Chem. 2020;296:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aggarwal M, Sharma R, Kumar P, Parida M, Tomar S. Kinetic characterization of trans‐proteolytic activity of Chikungunya virus capsid protease and development of a FRET‐based HTS assay. Sci Rep. 2015;5:14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCartney CE, Davies PL. FRET‐based assays to determine Calpain activity. Methods Mol Biol. 2019;1915:39‐55. [DOI] [PubMed] [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33‐38, 27‐38. [DOI] [PubMed] [Google Scholar]
- 46.Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221‐234. [DOI] [PubMed] [Google Scholar]
- 47.Jo S, Kim T, Iyer VG, Im W. CHARMM‐GUI: a web‐based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859‐1865. [DOI] [PubMed] [Google Scholar]
- 48.Qi Y, Ingolfsson HI, Cheng X, Lee J, Marrink SJ, Im W. CHARMM‐GUI Martini maker for coarse‐grained simulations with the martini force Field. J Chem Theory Comput. 2015;11:4486‐4494. [DOI] [PubMed] [Google Scholar]
- 49.Abraham MJ, Murtola T, Schulz R, et al. GROMACS: High performance molecular simulations through multi‐level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19‐25. [Google Scholar]
- 50.Bereau T, Kremer K. Automated parametrization of the coarse‐grained Martini force field for small organic molecules. J Chem Theory Comput. 2015;11:2783‐2791. [DOI] [PubMed] [Google Scholar]
- 51.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182‐7190. [Google Scholar]
- 52.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. [DOI] [PubMed] [Google Scholar]
- 53.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463‐1472. [Google Scholar]
- 54.Lee J, Worrall LJ, Vuckovic M, et al. Crystallographic structure of wild‐type SARS‐CoV‐2 main protease acyl‐enzyme intermediate with physiological C‐terminal autoprocessing site. Nat Commun. 2020;11:5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Privalov PL, Khechinashvili NN. A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J Mol Biol. 1974;86:665‐684. [DOI] [PubMed] [Google Scholar]
- 56.Arntfield SD, Murray ED. The influence of processing parameters on food protein functionality I. Differential scanning calorimetry as an indicator of protein denaturation. Can Inst Food Sci Technol. 1981;14:289‐294. [Google Scholar]
- 57.Ortiz SEM, Anon MC. Enzymatic hydrolysis of soy protein isolates ‐ DSC study. J Therm Anal Calorim. 2001;66:489‐499. [Google Scholar]
- 58.Nawaz MH, Ferreira JC, Nedyalkova L, et al. The catalytic inactivation of the N‐half of human hexokinase 2 and structural and biochemical characterization of its mitochondrial conformation. Biosci Rep. 2018;38(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanchez‐Ruiz JM. Protein kinetic stability. Biophys Chem. 2010;148:1‐15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data, associated protocols, methods, and sources of materials are contained within the manuscript.


