Abstract
The Rad25 protein in yeast is a DNA helicase and a subunit of the general transcription factor TFIIH. While in vitro studies have led to the hypothesis that TFIIH helicase activity plays a role in promoter melting, in vivo tests are lacking. Using potassium permanganate, which preferentially modifies single-stranded DNA, we show that a temperature-sensitive rad25ts mutant severely reduces the normally extensive promoter melting observed in vivo on the highly expressed genes TDH2 and PDC1 and on the induced heat shock gene HSP82. Loss of promoter melting can be observed in as little as 30 s after a shift to the nonpermissive temperature and is accompanied by a dramatic reduction in transcription. These effects on the promoter are specific, since the mutation does not affect TATA box occupancy or, in the case of HSP82, recruitment of TATA-binding protein to the TATA element or that of heat shock factor to heat shock elements. Additionally, using the technique of formaldehyde cross-linking coupled with restriction endonuclease cleavage and ligation-mediated PCR, we were able to map the polymerase density on the promoter of HSP82. This high-resolution mapping allowed us to determine that the polymerase II (Pol II) density on the promoter is also dramatically reduced after inactivation of TFIIH. These data provide strong support for the hypothesis that TFIIH functions with Pol II in the transcriptionally required step of promoter melting and show, surprisingly, that the extent of TFIIH-dependent promoter melting observed in vivo is several times larger than that seen in vitro.
Transcription of a eukaryotic gene is a complex process that requires completion of an ordered series of steps: opening of chromatin (33), TFIID binding (5, 27, 38, 40), general transcription factor and RNA polymerase II (Pol II) recruitment (5, 22, 34), open complex formation (18, 21, 25), initiation of transcription (19), promoter escape, and early elongation (14, 15). Binding of upstream regulatory factors to specific regulatory DNA elements can influence, in principle, the rates of one or more of these steps (4, 39). An essential component of the transcription machinery is the general transcription factor TFIIH, a complex composed of nine different subunits whose activities include DNA helicases (7, 18, 32, 35) and a CTD kinase (1, 9, 24, 30). These activities may act at one or several distinct steps in the transcription process.
One of the helicase subunits of TFIIH, Rad25p (an ERCC3 homologue), has a critical role in the early steps of the transcription cycle. This subunit of TFIIH has 3′→5′ helicase activity and ATP-binding motifs (17, 35). Qiu et al. (28) have shown previously that transcription of several mRNAs in yeast is rapidly and dramatically reduced when a conditional mutant, rad25ts, is raised to the nonpermissive temperature. In vitro, TFIIH helicases appear to unwind promoter DNA at the start of transcription (20, 26); however, it is not established what consequences a mutation in TFIIH might have on the broad promoter melting observed in vivo or on other interactions in the core promoter or upstream regulatory region.
Here we have evaluated the specific effects on gene expression and promoter architecture in vivo by using a rapidly acting, temperature-sensitive mutation in RAD25. We have examined the promoters of three genes—TDH2, PDC1, and HSP82—for RNA expression, TATA-binding protein (TBP) occupancy, and promoter melting. In the case of HSP82, heat shock induces several molecular changes in the yeast HSP82 promoter; normally, heat shock stimulates increased binding of heat shock factor (HSF) to upstream heat shock elements (HSEs) and increased TBP occupancy of the TATA box, triggers extensive Pol II-dependent melting of DNA at the promoter (about 50 bases of melted DNA), and gives rise to high levels of HSP82 transcription (11). We have examined these changes in vivo and compared the wild-type and mutant cells. Finally, to investigate the requirement of TFIIH for promoter associations of Pol II, we have employed a high-resolution modification of the formaldehyde cross-linking technique to map the density of polymerase at the promoter of HSP82.
MATERIALS AND METHODS
Primer extension.
Total yeast RNA was extracted with hot acid phenol as described in Current Protocols in Molecular Biology (3a). The total amount of RNA was quantified by absorbance readings at 260 nm (A260), and 30 μg of total RNA was used for primer extension reactions. Approximately 300 fmol of each end-labeled primer was used for primer extension. The primers used were as follows: for ACT1, GCTGATGTAGTAGAAGATCCTATTC; for TDH2, GCAATTCTCATGACCAATCTACCG; for PDC1, CCGAAAACGGTGTTAACGTTGACTTGC; for the 5′ end of HSP82, CAGCTTGAAATTCAAAAGTTTCACT; for the 3′ end of HSP82, GTTTTGTTTATAACCTATTCAAGGCC.
In vivo KMnO4 footprinting.
For HSP82, 4 ml of cells from a culture grown in synthetic complete medium and dextrose supplemented with all amino acids except tryptophan were treated with 25 μl of a 0.35 M solution of KMnO4. For heat shock samples, the culture was spun down and resuspended in half of the original volume of medium. An equal volume of fresh medium that had been prewarmed to 53°C was then added to the culture to instantaneously raise the temperature to 39°C. Four-milliliter samples were then taken and treated with KMnO4; the reaction was quenched by adding 45 ml of sorbitol stop solution. The DNA was purified and run through ligation-mediated PCR (LMPCR), as previously described (11). For TDH2 and PDC1, cells were grown in rich medium and treated with 300 μl of a 0.35 M solution of KMnO4. Cells were grown at 25°C to an optical density at 600 nm of 0.5. Half of the culture was transferred to a flask in a shaking water bath set at 37°C, while the remaining culture was allowed to continue growing at 25°C. After 2 h, a 4-ml sample was taken and treated with KMnO4; the reaction was quenched by adding 45 ml of sorbitol stop solution. The remainder of the procedure is the same as described above. The primers used for HSP82 were HSP82 UP1 (TCTCATCTTAATACCAACCAGGTCC) and HSP82 UP2 (GGTCCTTCCGCCACCCCCTAAAAC). The primers used for TDH2 were TDH2 UP1 (GCTAATATGTGTTTTGATAGTACCC) and TDH UP2 (GATAGTACCCAGTGATCGCAGACCTG). The primers used for PDC1 were PDC1 UP1 (GATGGCACATTTTTGCATAAACCTAGC) and PDC UP2 (CCTAGCTGTCCTCGTTGAACATAGG).
Formaldehyde cross-linking.
The protocol used for cross-linking RNA polymerase to the promoter of HSP82 was based on the methods of Aparicio et al. (2) except for the following modifications. Yeast cells were lysed in ice-cold lysis buffer containing 3% Sarkosyl and layered on top of a CsCl block gradient consisting of 1.5 ml of CsCl at 1.75 g/ml, 1 ml of CsCl at 1.5 g/ml, and 0.9 ml of CsCl at 1.3 g/ml. Each layer also contained 1.0% Sarkosyl and 1 mM EDTA (pH 8.0). The samples were centrifuged for 20 h at 30,000 rpm in an SW60 rotor at 20°C. Half-milliliter fractions were collected from the bottom of the tube, and samples with a refractive index of between 1.4 and 1.38 were pooled and dialyzed into 0.2% Sarkosyl, 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA. Restriction enzyme buffer NEB2 (supplied by New England Biolabs) was added to the DNA and supplemented with 100 μg of bovine serum albumin/ml and 0.8% Nonidet P-40 (NP-40). The DNA was cut with 100 units of HinfI and 50 units of MboII at 37°C for 4 h. Reactions were stopped by adding EDTA to a final concentration of 20 mM. A portion of the sample was set aside as a total DNA control. Immunocomplexes were collected with protein A-Sepharose beads (Sigma). Cross-linked protein-DNA samples were pretreated with 30 μl of a 50% solution (in lysis buffer) of protein A-Sepharose beads for 1 h at 4°C. The beads were pelleted by centrifugation for 1 min at 1,000 × g. The supernatant was treated with 1 μl of anti-RNA Pol II antiserum for 12 h at 4°C to bind polymerase-DNA adducts and then with 30 μl of a 50% solution of protein A-Sepharose beads for 3 h at 4°C. Bound complexes were collected by centrifugation for 1 min at 1,000 × g. The beads were washed at room temperature for 5 min in the following solutions: seven times in 1 ml of lysis buffer; once with 1 ml of lysis buffer containing 300 mM NaCl; once with 1 ml of a solution of 10 mM Tris-HCl (pH 8.0), 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, and 1 mM EDTA; and once with 1 ml of Tris-EDTA (TE). The cross-links were reversed at 65°C overnight and samples were treated with proteinase K for 2 h at 37°C. The DNA was extracted twice with phenol and once with chloroform. The DNA was precipitated and resuspended in 50 μl of TE. For mapping of the polymerase density on the DNA, we used LMPCR. For the cross-linked samples, we used 9 μl of our DNA and 9 μl of our total DNA sample that had been diluted 1:10,000 for ligation. The protocol and primers used subsequently are the same as those used for permanganate LMPCR (above). Bands were quantified with a Molecular Dynamics Storm PhosphorImager and the accompanying software, ImageQuant (IQMac) version 1.2.
RESULTS
rad25ts reduces the extensive promoter melting of constitutively expressed genes.
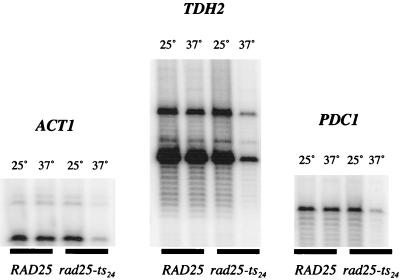
Previous experiments by Qiu et al. (28) have shown that the rad25ts mutation causes a striking reduction in the total amount of yeast mRNA and a number of specific mRNAs. We begin here by examining how this rad25ts mutation affects the expression and promoter architecture of two highly expressed genes (37), TDH2 and PDC1. In Fig. 1, primer extension assays show that the levels of TDH2 and PDC1 mRNAs decreased dramatically in rad25ts cells (but not the wild-type control) when raised to the nonpermissive temperature. Therefore, these and previous results (28) show that Pol II transcription of a variety of genes is severely compromised in the rad25ts mutant.
FIG. 1.
Primer extension of ACT1, TDH2, and PDC1 mRNAs from total RNA. Total RNA was isolated from exponentially growing yeast cultures at 25°C or after the culture had been shifted to 37°C for 2 h. The amount of total RNA was quantified, and equal amounts (30 μg) of RNA were used for primer extension with 32P end-labeled primers for ACT1, TDH2, and PDC1.
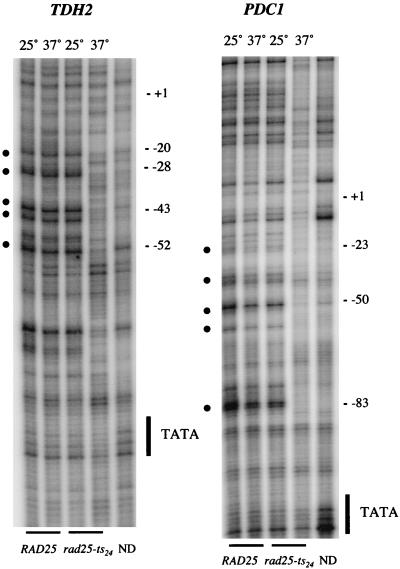
Because TFIIH plays a critical role in melting of DNA at the start site in vitro (18), we examined the broad in vivo promoter melting of TDH2 and PDC1 in yeast containing the rad25ts mutation. Wild-type cells and cells containing the rad25ts mutation were grown at the permissive temperature (25°C) or shifted to the nonpermissive temperature (37°C) for 2 h and treated in culture with potassium permanganate, which modifies thymine bases in melted or distorted DNA. The permanganate treatment was quenched after 1 min, and the DNA was isolated and cleaved at the modified bases with piperidine. The sites of DNA modification were mapped with LMPCR and electrophoresis of products on a denaturing gel. At the permissive temperature, both wild-type and rad25ts mutant cells show strong hypersensitivity (which we interpret as promoter melting [10]) that extends from about 30 to 40 bp downstream of the TATA box to about 20 bp upstream of the transcription start site (Fig. 2). This extensive promoter melting, which is in contrast to the tight DNA melting observed in vitro, covers approximately 30 to 50 bp and is similar to that reported for GAL1, GAL10, and HSP82 by Giardina and Lis (10, 11). However, shifting of the rad25ts mutant cells to the nonpermissive temperature severely reduced both expression and promoter melting of the TDH2 and PDC1 genes.
FIG. 2.
Potassium permanganate (KMnO4) reactivity of the TDH2 and PDC1 promoters in vivo. In vivo KMnO4 patterns at permissive (25°C) and nonpermissive (37°C) temperatures for both wild-type and rad25ts mutant cells are shown. The sites of cleavage were viewed by LMPCR with primers to display the bottom (transcribed) strand. The TATA sequence is labeled on the side of the gel, and the numbers indicate positions relative to the transcription start site. Permanganate-sensitive bands are labeled with bullets (•); naked DNA samples are labeled ND.
rad25ts rapidly affects transcription and extensive melting at the HSP82 promoter.
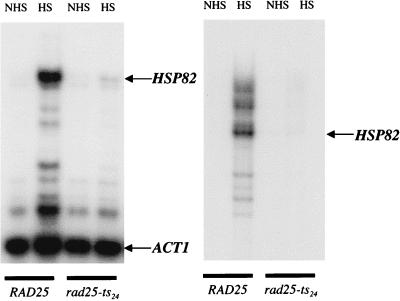
To evaluate the effects of the rad25ts mutation on activated transcription, we examined the HSP82 gene, which is highly and rapidly induced by a temperature shift. The optimal heat shock temperature is also a condition that leads to inactivation of the TFIIH helicase in the rad25ts strain. In Fig. 3, primer extension assays show that the level of HSP82 mRNA isolated in yeast cells containing the helicase mutant is dramatically reduced relative to the wild-type control at the nonpermissive (heat shock) temperature. Because the heat shock response is known to be extremely rapid, the inactivation of TFIIH must likewise be extremely rapid following the shift of rad25ts cells to the nonpermissive temperature. We note that the effect appears to be at the early steps in the transcription cycle, as both a 5′ primer and a primer complementary to the 3′ end of HSP82 show the same reduction in the heat-shocked mutant cells.
FIG. 3.
Primer extension of HSP82 mRNA from total RNA. Total RNA was isolated from exponentially growing yeast cultures at 25°C or after the culture had been shifted to 39°C for 30 min. The amount of total RNA was quantified, and equal amounts (30 μg) of RNA were used for primer extension with 32P end-labeled primers for both ACT1 and HSP82. The left panel shows primer extension reactions with an actin-specific primer and a primer specific for the 5′ end of HSP82. Actin was used as an internal control due to its long half-life in vivo. The right panel shows a primer extension reaction with a primer specific for the 3′ end of HSP82. NHS, RNA extracted from yeast under non-heat shock conditions; HS, RNA extracted from yeast under heat shock conditions.
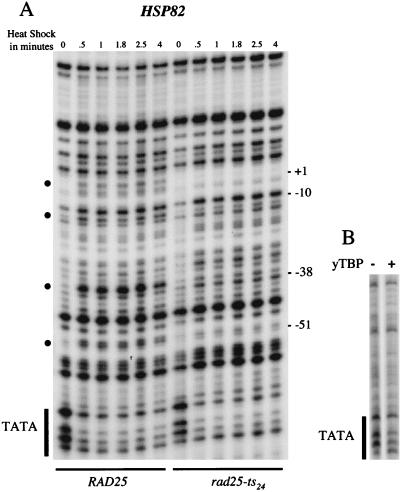
Like transcription, broad HSP82 promoter melting is also affected in the rad25ts mutant. In the case of wild-type cells, the HSP82 gene shows an obvious increase in sensitivity to permanganate at several sites that can be seen in as little as 30 s after induction and remains relatively constant up to 4 min (Fig. 4A). In contrast, the HSP82 promoter of the rad25ts mutant shows substantially reduced melting in as little as 30 s after induction. Interestingly, there is a band at −10 which becomes hypersensitive to permanganate in both wild-type and rad25ts mutant cells. This band may be a result of a change in the promoter architecture due to the recruitment of upstream factors in response to heat shock. Thus, the TFIIH mutation rapidly affects the extensive melting of promoter DNA.
FIG. 4.
Potassium permanganate (KMnO4) reactivity of the HSP82 promoter in vivo and in vitro. (A) In vivo KMnO4 patterns before and after heat shock for both wild-type and rad25ts mutant cells. A logarithmically growing culture was treated for 1 min with 2.2 mM KMnO4 at either 25 or 39°C. The DNA was then purified and cleaved at the modified bases. The sites of cleavage were viewed by LMPCR with primers to display the bottom (transcribed) strand. The TATA sequence is labeled on the side of the gel, and the numbers indicate positions relative to the transcription start site. Permanganate-sensitive bands are labeled with bullets (•). (B) In vitro KMnO4 patterns of naked DNA with and without added yeast TBP. The promoter region of the HSP82 gene was amplified from plasmid pMF13 by PCR, and 12 fmol of this fragment, containing the TATA sequence, was treated with 25 mM KMnO4 at 25°C for 30 s in either the presence or absence of 3 pmol of yeast TBP. The primers used to amplify the fragment were UP0.1 (GAACAGGAATAAAGCTTAATCGGAT) and LARRY82 (CAGCTTGAAATTCAAAAGTTTCACT).
The rad25ts mutation does not affect binding of TBP to the HSP82 promoter.
Giardina et al. (11, 12) have shown that the TATA sequence in naked DNA is sensitive to permanganate treatment, presumably due to the non-B-form nature of the TATA sequence (8). This TATA sensitivity is also apparent in genomic HSP82 promoter DNA under noninduced conditions. As shown in Fig. 4A, the TATA element is relatively sensitive to permanganate modification when cells are in the noninduced state. However, when either wild-type or rad25ts cells are heat shock induced, the TATA sequence is protected from modification, presumably due to the binding of TBP. To evaluate whether this pattern seen in vivo indeed represents TBP binding, we examined the effects of purified, recombinant yeast TBP on the permanganate sensitivity of a PCR-generated fragment (−85 to +90) of the HSP82 promoter. Figure 4B shows that the reduction in permanganate sensitivity can be reproduced in vitro with purified recombinant yeast TBP and naked DNA. In cells, the TFIIH mutation has a severe effect on promoter melting; however, occupancy of the adjacent TATA element by TBP is not detectably reduced. We conclude that, as in wild-type cells, TBP binding to the HSP82 TATA box is rapidly induced in the rad25ts mutant at the nonpermissive temperature.
Binding of HSF to upstream HSEs on the HSP82 promoter is not affected in the rad25ts mutant.
Additional protein interactions with the core promoter and upstream regulatory elements could potentially be influenced by TFIIH either directly or indirectly. For example, depletion of TFIIH activity might adversely affect the ability of upstream factors to bind to their DNA elements if the protein components of a fully active core promoter interact and stabilize upstream factor binding to DNA. To determine if the key heat shock regulatory factor HSF is able to gain access to HSEs in both wild-type and rad25ts mutant cells, we used in vivo DMS footprinting to identify HSF interaction with HSEs. HSF binding to HSEs creates a characteristic pattern of DMS protection and hypersensitivity that is enhanced upon heat shock. This enhanced pattern is indicative of a 20-fold increase in HSF binding to DNA, as was demonstrated with in vitro reconstruction experiments with purified HSF and promoter DNA (11, 16). We treated yeast cells with DMS under noninducing and inducing conditions and compared the pattern hypersensitivity and protection. As seen in Fig. 5, bands near the HSE borders become hypersensitive to DMS modification upon heat shock, while bands within HSE1 become protected (relative to hypersensitive bands in naked DNA). These changes in hypersensitivity and protection can be reproduced in vitro with purified DNA and yeast HSF (11). Importantly, these changes are seen in both wild-type and mutant cells at the nonpermissive temperature. Thus, it appears that for wild-type and mutant cells, HSF is able to access the HSE in the HSP82 promoter.
FIG. 5.
DMS footprinting of HSP82 HSEs in vivo before and after heat shock in both wild-type and rad25ts mutant cells. Four milliliters of yeast culture in exponential phase was treated with 4 μl of DMS at either 25 or 42°C for 1 min. Subsequent processing of the samples to map DMS modifications, as well as primers used to display both the top strand (left panel) and bottom strand (right panel), was performed as previously described (11). Open circles mark sites of protection, while filled circles mark sites of hypersensitivity. NHS, non-heat shock samples; HS, heat shock samples; ND, naked DNA samples.
Pol II binding to the HSP82 promoter appears to be compromised in rad25ts mutant cells.
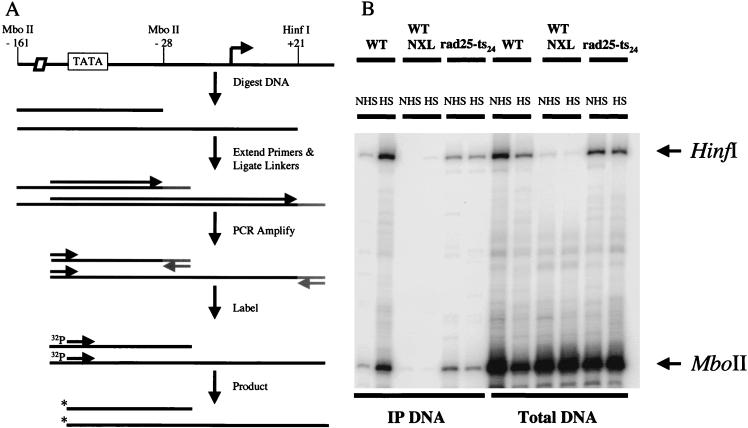
Protein-DNA cross-linking procedures allow examination of the interaction of Pol II with a specific promoter in vivo (13). Existing protocols for formaldehyde cross-linking in yeast call for sonication of the genomic DNA, giving an average size of about 500 bp (2, 31). This fails to provide sufficient resolution to distinguish Pol II that is associated with the melted promoter from Pol II that is transcribing downstream of the start site. To increase the resolution of the cross-linking, we added steps of restriction endonuclease cleavage and LMPCR to the standard protocol (14). After formaldehyde cross-linking, chromatin samples were layered onto a CsCl step gradient and centrifuged to separate DNA and DNA-protein complexes from free protein. The DNA containing fraction was then dialyzed and cleaved with MboII and HinfI, which cut the HSP82 promoter at −28 and +21, respectively. The cross-linked protein-DNA complexes were immunoprecipitated with an antibody raised against whole yeast Pol II. Cross-links were reversed, and a portion of the immunoprecipitated DNA was used for LMPCR to quantify the Pol II density on specific restriction fragments (Fig. 6A). Using this method, we were able to separate polymerase molecules cross-linked to the promoter DNA from polymerase molecules cross-linked not only to the promoter but also to the DNA within the transcribed portion of the gene. The ligation step in LMPCR ensures that the fragments assayed are actually cut by the restriction enzyme, as a partial digest will lead to fragments of different size. These additional larger fragments can also be quantified, as they provide information on the relative Pol II density on these sequences.
FIG. 6.
(A) Schematic representation of LMPCR. Primers specific for the HSP82 promoter are used to make blunt ends of restriction enzyme-cut DNA. Linkers are then annealed to the blunt-ended DNAs. The DNA is amplified with the original primer used to make the blunt end and one of the primers used to make the linker. The DNA is labeled with a 32P end-labeled primer which is internal to the first HSP82-specific primer. The products are then run on an 8.5% denaturing gel. (B) In vivo Pol II cross-linking to the HSP82 promoter in yeast cells before and after heat shock in both wild-type and rad25ts mutant cells. Yeast cells were treated with formaldehyde for 7 min either at 25°C or after a 5-min incubation at 39°C. The DNA was cut with the restriction enzymes MboII and HinfI to separate the promoter from the transcribed portion of the HSP82 gene. In addition to cross-linked samples, there are also samples that were not subjected to formaldehyde cross-linking that act as a background control (NXL). After restriction cutting and immunoprecipitation steps, isolated DNA was amplified by LMPCR. The two signals amplified were judged to be from DNA that had been cut by MboII (lower band) and HinfI (upper band). The percentages of total DNA cross-linked to RNA Pol II for each restriction fragment and each condition in the experiment shown are as follows: wild type non-heat shock, MboII 0.007, HinfI 0.024; wild type heat shock, MboII 0.276, HinfI 2.248; rad25ts non-heat shock, MboII 0.039, HinfI 0.134; rad25ts heat shock, MboII 0.018, HinfI 0.138.
Figure 6B shows that when wild-type yeast cells were raised to the heat shock temperature of 39°C, the amount of cross-linking to the promoter and downstream DNA sequences increased dramatically while the amount of cross-linking in rad25ts cells did not. Quantification revealed that in the wild-type control, polymerase cross-linking to the HSP82 promoter increased during heat shock approximately 40-fold for the MboII fragment and 90-fold for the HinfI fragment (in other experiments, the increase was about 50-fold [data not shown]). In contrast, cross-linking to the MboII fragment decreased during heat shock by 45% in the rad25ts mutant (in other experiments, the cross-linking increased by only 1.6-fold [data not shown]) and cross-linking to the HinfI fragment showed no increase. Therefore, the TFIIH mutation not only causes a lack of promoter melting but also may interfere with the ability of polymerase to gain access to the promoter.
DISCUSSION
Understanding the molecular mechanism of complex biological processes like transcription requires a combination of biochemical and genetic approaches. Over the past 15 years, development of highly specific cross-linking and footprinting assays has provided detailed views of promoter structures and at least some of the molecular rearrangements that occur at these promoters in living cells. Here, we have used these methods of probing protein-nucleic acid complexes to examine in vivo the changes in promoter structure and function that accompany rapid inactivation of a transcription factor. The conditional mutant rad25ts permits rapid inactivation of an activity of the critical general transcription factor TFIIH.
Inactivation of TFIIH in the rad25ts strain by a shift to the nonpermissive temperature leads to a shut-down in transcription and a specific change in promoter architecture. All three of the genes that we chose to examine show a reduction in transcription and a loss of the extensive promoter melting associated with highly active transcription units (10). These in vivo results, and previous in vitro studies of transcription of other genes (18, 26), support the general requirement of TFIIH in transcription and promoter melting.
The in vivo effects of a conditional mutation in TFIIH on the architecture and function of the HSP82 promoter are both rapid and specific. The HSP82 gene can be induced by an instantaneous heat shock (by mixing cells with prewarmed medium), and the effects on promoter melting in vivo can be detected with a 1-min potassium permanganate treatment applied 30 s after heat shock. This melting is blocked in the rad25ts mutant by this temperature shift. These results demonstrate that TFIIH is rapidly inactivated at nonpermissive temperatures and that the consequence of this inactivation on promoter melting is a primary effect and is unlikely to be caused indirectly through a defect in another process that then in turn affects promoter melting.
The specificity of the effect of TFIIH on promoter architecture is also clear. The extensive promoter melting is lost; however, other features of the active and activated promoter appear normal. In all three genes examined, the TATA box, which ranges from 20 to 40 bp from the beginning of the normally melted region of the promoter, appears to be occupied by TBP. Additionally, in the case of HSP82, the heat shock-induced recruitment of TBP to the TATA box and of the specific transcription activator HSF to its upstream regulatory sites remains efficient in the mutant.
To specifically examine polymerase density in the upstream regions of the HSP82 promoter, we needed to distinguish between the signal generated by Pol II cross-linked to the promoter and the signal generated by an actively transcribing polymerase downstream of the start site. To achieve this, we introduced a new addition to the procedure that allows resolution of cross-linked complexes at the level of a restriction map. In short, the cross-linked chromatin is cleaved with restriction enzymes prior to immunoprecipitation. Additionally the DNA is amplified by LMPCR, which places a linker on the ends that have been cleaved, ensuring that the fragments assayed by amplification have been cut. This cross-linking analysis indicates that Pol II is not only recruited to the transcription unit following heat shock but also exists on the activated HSP82 promoter and can cross-link to a DNA fragment that is 20 bp upstream of the transcription start site. In the rad25ts mutant, cross-linking of Pol II to the transcription unit and to the upstream fragment are both dramatically reduced. The simplest interpretation of this data is that the density of Pol II on these DNA sequences in vivo is correspondingly reduced due to an effect in the temperature-sensitive rad25ts subunit of TFIIH. However, we cannot rule out the possibility that the existence of melted DNA at the promoter potentially influences the efficiency with which formaldehyde cross-links protein to DNA, as formaldehyde modifies single-stranded DNA more readily than double-stranded DNA (36), and this difference could contribute to some of the difference in Pol II density between wild-type and mutant cells.
Since Rad25 has an essential DNA helicase activity that is required for transcription (17), it is tempting to speculate that this helicase activity itself creates the extensive promoter melting associated with highly active genes. However, Pol II levels in the promoter region of HSP82 also appear to be reduced in the rad25ts mutant. Moreover, the essential role of Pol II itself in promoter melting has been shown by using a temperature-sensitive mutation in the largest Pol II subunit, RBP1-1 (10). Taken together, these two studies suggest that Pol II and TFIIH cooperate in this activity, since disruption of either causes a reduction in the extensive promoter melting of active genes. While Pol II and TFIIH are both required for promoter melting and Pol II recruitment, recruitment of HSF and TBP is independent of the Rad25 activity of TFIIH. These results are consistent with a study from the Hahn laboratory that shows that TBP recruitment and recruitment of holoenzyme can occur at distinct steps in preinitiation complex formation (29).
The extensive melting observed on the promoters of active yeast genes in vivo (10) is much greater than expected from in vitro experiments that track TFIIH-dependent Pol II initiation with mammalian transcription components (18, 20, 26). This extensive melting begins approximately 20 bp downstream of the TATA box and extends nearly to the start of transcription. The location of the upstream edge of the melted region relative to the TATA box is similar to that seen on genes of higher eukaryotes (12) and may represent a common mechanism of Pol II entry relative to the TBP-TATA complex. Indeed, two-dimensional crystal structure studies with yeast Pol II show that the distance between the end of Pol II that interacts with TFIIB (and, by inference, TBP and the TATA box) and the Pol II active site is the equivalent of 30 bp (3, 6, 23). This is the length between the TATA box and the transcription start site in higher eukaryotes, whereas yeast differs from higher eukaryotes in that the apparent transcription start sites are further downstream from the TATA box. Perhaps the promoter entry site of Pol II in yeast is similar to that of higher eukaryotes but is followed by a Pol II tracking step to more distal start sites. This tracking may melt the DNA between the Pol II entry and start sites. Alternatively, Pol II may initiate at a site similar to that in higher eukaryotes and then reinitiate at the distal start sites, which then become the observed 5′ ends of yeast mRNAs. While these and other models of extensive melting remain to be tested, this promoter melting is clearly dependent in vivo on the function of TFIIH.
ACKNOWLEDGMENTS
We thank L. Prakash and S. Prakash for providing the yeast strains used in this study, C. Roberts for the yeast Pol II antibody, P. Mason for the contribution of purified yeast TBP and advice on in vitro permanganate mapping, C. Giardina for advice on in vivo permanganate mapping and HSF footprinting, and D. K. Lee for excellent suggestions on in vivo cross-linking and polymerase mapping. We also thank J. Roberts, T. Huffacker, P. Mason, and D. K. Lee for critical reading of the manuscript as well as the rest of the members of the Lis laboratory for input and encouragement.
This work was supported by National Institutes of Health grant GM25232 to J.T.L.
REFERENCES
- 1.Akoulitchev S, Makela T P, Weinberg R A, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 3.Asturias F J, Meredith G D, Poglitsch C L, Kornberg R D. Two conformations of RNA polymerase II revealed by electron crystallography. J Mol Biol. 1997;272:536–540. doi: 10.1006/jmbi.1997.1273. [DOI] [PubMed] [Google Scholar]
- 3a.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1997. [Google Scholar]
- 4.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–562. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 6.Darst S A, Edwards A M, Kubalek E W, Kornberg R D. Three-dimensional structure of yeast RNA polymerase II at 16 Å resolution. Cell. 1991;66:121–128. doi: 10.1016/0092-8674(91)90144-n. [DOI] [PubMed] [Google Scholar]
- 7.Drapkin R, Reinberg D. The multifunctional TFIIH complex and transcriptional control. Trends Biochem Sci. 1994;19:504–508. doi: 10.1016/0968-0004(94)90139-2. [DOI] [PubMed] [Google Scholar]
- 8.Elgin S C R, editor. Chromatin structure and gene expression. Vol. 9. Oxford, England: Oxford University Press; 1995. [Google Scholar]
- 9.Feaver W J, Gileadi O, Li Y, Kornberg R D. CTD kinase associated with yeast RNA polymerase II initiation factor B. Cell. 1991;67:1223–1230. doi: 10.1016/0092-8674(91)90298-d. [DOI] [PubMed] [Google Scholar]
- 10.Giardina C, Lis J T. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 11.Giardina C, Lis J T. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol. 1995;15:2737–2744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giardina C, Perez Riba M, Lis J T. Promoter melting and TFIID complexes on Drosophila genes in-vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- 13.Gilmour D S, Lis J T. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster. Mol Cell Biol. 1985;5:2009–2018. doi: 10.1128/mcb.5.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmour D S, Lis J T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 16.Gross D S, English K E, Collins K W, Lee S. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J Mol Biol. 1990;216:611–631. doi: 10.1016/0022-2836(90)90387-2. [DOI] [PubMed] [Google Scholar]
- 17.Guzder S N, Sung P, Bally V, Prakash L, Prakash S. RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature. 1994;369:578–581. doi: 10.1038/369578a0. [DOI] [PubMed] [Google Scholar]
- 18.Holstege F C, Van Der Vliet P C, Timmers H T M. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 1996;15:1666–1677. [PMC free article] [PubMed] [Google Scholar]
- 19.Holstege F C P, Fiedler U, Timmers H T M. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holstege F C P, Tantin D, Carey M, Van Der Vliet P C, Timmers H T M. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Yan M, Gralla J D. A three-step pathway of transcription initiation leading to promoter clearance at an activated RNA polymerase II promoter. Mol Cell Biol. 1996;16:1614–1621. doi: 10.1128/mcb.16.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 23.Leuther K K, Bushnell D A, Kornberg R D. Two-dimensional crystallography of TFIIB- and IIE-RNA polymerase II complexes: implications for start site selection and initiation complex formation. Cell. 1996;85:773–779. doi: 10.1016/s0092-8674(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 24.Lu H, Zawel L, Fisher L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 25.Luse D S, Kochel T, Kuempel E D, Coppolas J A, Cai H. Transcription initiation by RNA polymerase II in vitro. J Biol Chem. 1987;262:289–297. [PubMed] [Google Scholar]
- 26.Parvin J D, Sharp P A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 27.Purnell B A, Emanuel P A, Gilmour D S. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 28.Qiu H, Park E, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD25 is required for transcription by RNA polymerase II. Genes Dev. 1993;7:2161–2171. doi: 10.1101/gad.7.11.2161. [DOI] [PubMed] [Google Scholar]
- 29.Ranish J A, Yudkovski N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serizawa H, Makela T P, Conaway J W, Conaway R C, Weinberg R A, Young R A. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–287. doi: 10.1038/374280a0. [DOI] [PubMed] [Google Scholar]
- 31.Strahl Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 32.Svejstrup J Q, Vichi P, Egly J M. The multiple roles of transcription-repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 33.Taylor I C A, Workman J L, Schuetz T J, Kingston R E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 34.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 35.Tirode F, Busso D, Coin F, Egly J M. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 36.Utiyama H, Doty P. Kinetic studies of denaturation and reaction with formaldehyde on polydeoxyribonucleotides. Biochemistry. 1971;10:1254–1264. doi: 10.1021/bi00783a024. [DOI] [PubMed] [Google Scholar]
- 37.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 38.Workman J L, Roeder R G. Binding of transcription factor TFIID to the major late promoter during in-vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987;51:613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 39.Xiao H, Pearson A, Coulombe B, Truant R, Zhang S, Regier J L, Triezenberg S J, Reinberg D, Flores O, Ingles C J. Binding of basal transcription factor TFIIH to the acidic activation domains of VP16 and p53. Mol Cell Biol. 1994;14:7013–7024. doi: 10.1128/mcb.14.10.7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]