Summary
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is an emerging respiratory virus responsible for the ongoing coronavirus disease 19 (COVID‐19) pandemic. More than a year into this pandemic, the COVID‐19 fatigue is still escalating and takes hold of the entire world population. Driven by the ongoing geographical expansion and upcoming mutations, the COVID‐19 pandemic has taken a new shape in the form of emerging SARS‐CoV‐2 variants. These mutations in the viral spike (S) protein enhance the virulence of SARS‐CoV‐2 variants by improving viral infectivity, transmissibility and immune evasion abilities. Such variants have resulted in cluster outbreaks and fresh infection waves in various parts of the world with increased disease severity and poor clinical outcomes. Hence, the variants of SARS‐CoV‐2 pose a threat to human health and public safety. This review enlists the most recent updates regarding the presently characterized variants of SARS‐CoV‐2 recognized by the global regulatory health authorities (WHO, CDC). Based on the slender literature on SARS‐CoV‐2 variants, we collate information on the biological implications of these mutations on virus pathology. We also shed light on the efficacy of therapeutics and COVID‐19 vaccines against the emerging SARS‐CoV‐2 variants.
Introduction
The past year has witnessed a severe collapse of the global healthcare system and downturned leading economies, disrupting livelihoods, impacting all trade sectors, every individual in every part of the world (Kaye et al., 2020). All these are repercussions following the emergence and widespread dissemination of the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), resulting from the outbreak of coronavirus disease 19 (COVID‐19) pandemic. This enigmatic pathogen has been regarded as an emerging respiratory virus incurring a high risk to human life (Çelik et al., 2020). The SARS‐CoV‐2 is an enveloped virus displaying characteristic crown‐like projections on its surface. Structurally, the virus codes for four structural proteins, namely the envelope (E), membrane (M), spike (S) and nucleocapsid (N) proteins (Kaur and Gupta, 2020). SARS‐CoV‐2 has surpassed other coronaviruses like SARS‐CoV and the Middle East respiratory syndrome coronavirus (MERS‐CoV) in terms of transmission rate, reproduction number (R0) and global mortalities (Abdelrahman et al., 2020). In contrast, its case fatality rate remains to be low (1%–3%) in comparison to SARS‐CoV (9.6%) and MERS‐CoV (34.4%) (Hu et al., 2020b). Ever since the first reports of a pneumonia‐like outbreak in Wuhan, China, in mid‐December 2019, mysteries surrounding the origin of SARS‐CoV‐2 have intensified, raising speculations over the theories of laboratory escapes, or natural selection in an animal host before zoonotic transfer, or natural selection in humans and subsequent zoonotic transfer (Andersen et al., 2020). In chronology, the World Health Organization (WHO) was informed about the pneumonia‐like outbreak on 31st December 2019 (Fig. 1), following which the etiological agent was identified as a ‘novel coronavirus’ on 9th January 2020 (Hu et al., 2020a). On 20th January 2020, the situation became dire as reports confirmed the incidence of human‐to‐human transmission (Hu et al., 2020a). These events coincided with the Chinese Lunar New Year facilitating human travel alongside transmission of SARS‐CoV‐2 in disguise throughout the country. In the coming months, virus transmission accelerated multi‐folds, and large clusters of SARS‐CoV‐2 infections were reported across several countries. These urged the WHO to officially declare the global COVID‐19 outbreak as a pandemic (WHO, 2020). Since then, COVID‐19 has spread across 220 countries and territories spanning all six continents, with more than 180 million confirmed cases and over 3.9 million deaths (Worldometers, 2021).
Fig. 1.
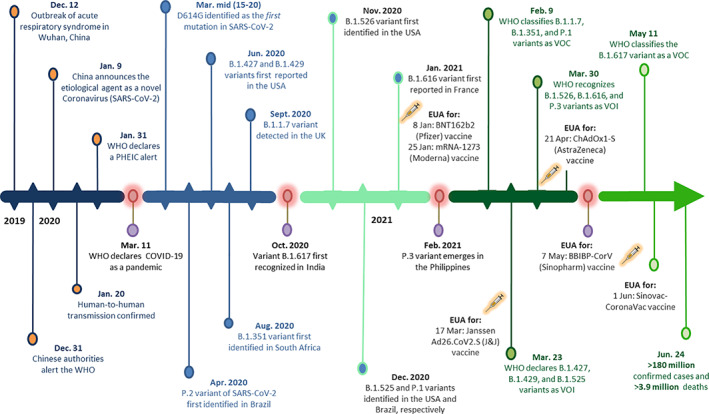
Timeline of key events unfolding the emergence of SARS‐CoV‐2 variants during the COVID‐19 pandemic. The first confirmed cases of SARS‐CoV‐2 were reported in December 2019 in Wuhan, China. Following confirmation of human‐to‐human transmission and its global dissemination, the WHO announced COVID‐19 as a pandemic. In the successive 12 months, hundreds of SARS‐CoV‐2 variants have emerged, out of which four have been designated as variant of concern (VOC) and seven as variant of interest (VOI) by the WHO. Five COVID‐19 vaccines have also been approved for emergency use authorization (EUA) by the WHO. Since then, more than 180 million cases of SARS‐CoV‐2 have been confirmed, with nearly 3.9 million deaths worldwide.
The Centre for Diseases Control and Prevention (CDC) suggests preventive measures to manage the spread of COVID‐19 by maintaining social distancing, use of face masks, sanitization of probable fomites, regular washing of hands, or use of alcohol‐based hand sanitizers and practicing personal hygiene (CDC, 2021a). However, its high transmissibility owing to dissemination by direct human contact, fomites and even respiratory aerosols has exerted a snowballing effect over COVID‐19 transmission. Nevertheless, numerous scientific milestones have also been achieved in unravelling the complex biology of SARS‐CoV‐2, repurposing therapeutic drugs and convalescent plasma therapy for COVID‐19 patients, and developing efficacious COVID‐19 vaccines (Nature news, 2021). In such a short span, ground‐breaking efforts by scientists in collaboration with pharmaceutical giants have yielded six potent WHO's Strategic Advisory Group of Experts on Immunization (SAGE) approved COVID‐19 vaccines like the mRNA‐1273 (Moderna), BNT162b2 mRNA (Pfizer), AZD1222 (Oxford/AstraZeneca), Janssen Ad26.CoV2.S (Johnson & Johnson), BBIBP‐CorV (Sinopharm) and CoronaVac (Sinovac Biotech) (WHO, 2021a). Concomitantly, mutants of SARS‐CoV‐2 have surfaced across the globe as the virus's geographical range and infection cycle has been continuously expanding (Fig. 2). Since SARS‐CoV‐2 contains an unstable positive‐sense ssRNA genome, it poses an invariable threat towards the emergence of mutant strains that may outcompete the existing ones by natural selection (Wang et al., 2020). These SARS‐CoV‐2 mutants can exhibit accelerated viral replication and transmission, with the ability to slip past the host immunosurveillance system and elude virus neutralization by antibodies, thereby driving the resurgence of COVID‐19 infections (Lauring and Hodcroft, 2021). During the early months of the COVID‐19 pandemic, the first global SARS‐CoV‐2 variant predominantly circulating in Europe was detected with a missense mutation (D614G) in its S protein (Isabel et al., 2020). Since then, various mutants of SARS‐CoV‐2 have been related to a fresh spike in the COVID‐19 infections across different countries. These mutations or amino acid substitutions have been predominantly identified in the S protein of SARS‐CoV‐2, primarily the N‐terminal domain and receptor‐binding domain (RBD) of the S1 subunit (Guruprasad, 2021). Considering the functional implications of viral S protein in seizing the host cells by targeting the human angiotensin‐converting enzyme 2 (hACE2) receptor, the prevalence of mutations in the S protein can confer immune evasion and increased infectivity to the mutant types. Very recently, mutations in the S protein have been strongly associated with different infection waves worldwide, resulting in mutant virus strains with a high R0 (transmissibility), infection rates, disease severity and mortality (Korber et al., 2020). In India, nearly 7000 mutations were recently detected in over 5000 SARS‐CoV‐2 genomes reportedly circulating in the country (Srivastava et al., 2021). Interestingly, variants of SARS‐CoV‐2 have evolved worldwide regardless of the environmental conditions and geographical locations. However, a community with high population density is speculated to offer prolonged viral infection and transmission cycles (Bhadra et al., 2021; Kadi and Khelfaoui, 2020), permitting high viral replication rates and the subsequent selection of a variant by natural selection (Lauring and Hodcroft, 2021). The phylogenetic diversity of SARS‐CoV‐2 variants with respect to the S protein has been depicted in Fig. 3. The emergence of such worrisome mutants has made the epidemiologists and researchers reiterate their line of action before the mutants unleash a wave of violent infections. Hence, in this review, we collate the literature concerning the presently characterized SARS‐CoV‐2 mutants and provide insights into the functional implications, mutational properties, disease transmission and severity and the current knowledge on WHO‐approved vaccines reported to generate immune responses against the variants of SARS‐CoV‐2.
Fig. 2.
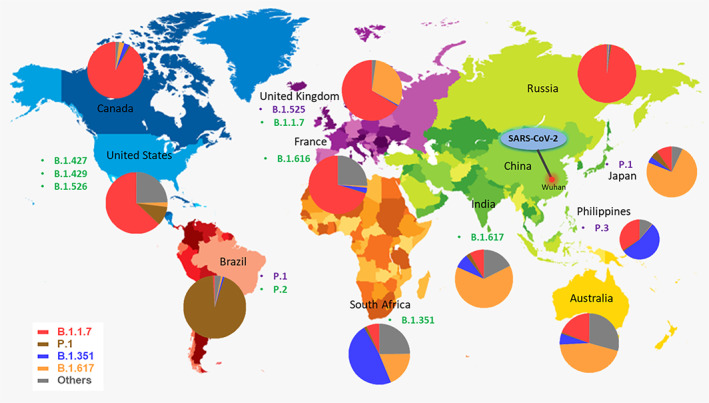
World map depicting the geographical distribution of SARS‐CoV‐2 variants (VOI and VOC) till 29th May 2021. The epicentre of the COVID‐19 pandemic (Wuhan, China) has been shown in a red marking. The VOI and VOC have been listed alongside the countries they were first reported in with green and violet colours respectively. The pie charts depict the relative frequencies of SARS‐CoV‐2 VOC in different countries where the variants originated. The variants are colour‐coded as follows B.1.1.7 ( ), B.1.351 (
), B.1.351 ( ), B.1.617 (
), B.1.617 ( ), P.1 (
), P.1 ( ) and others (
) and others ( ). Others represent VOI.
Source: https://covariants.org/.
). Others represent VOI.
Source: https://covariants.org/.
Fig. 3.
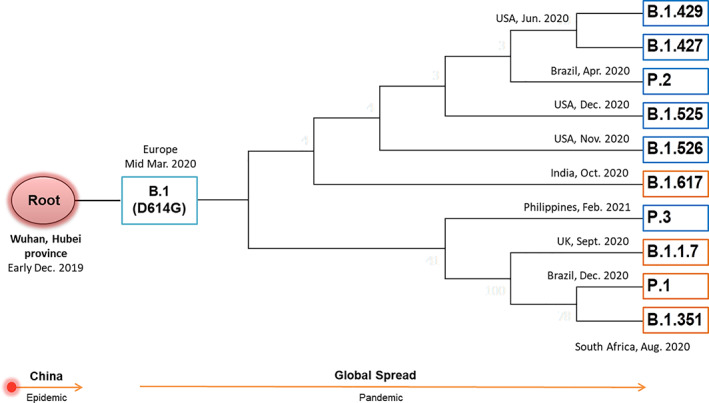
A simplified phylogenetic tree illustrating the diversity of SARS‐CoV‐2 variants concerning the S protein. The variants diverge from their ancestral root (Wuhan), accumulating mutations in the S protein as they spread globally. The boxes represent various lineages of the SARS‐CoV‐2 variants, and the branches indicate the inferred month of emergence of different variants. VOC and VOI are shown in orange and blue boxes respectively.
Variants of SARS‐CoV‐2 and their characterized mutations: classification and functional implications
Numerous variants of the SARS‐CoV‐2 have emerged across the world during the COVID‐19 pandemic. These mutants have been circulating across countries and held responsible for abrupt infectious waves. SARS‐CoV‐2 variants and their mutations have been precisely monitored using epidemiological studies, bench‐based experiments and sequence‐based surveillance systems. The WHO classifies variants of SARS‐CoV‐2 into two categories on the grounds of threat levels, transmissibility, disease severity and their ability to evade diagnostic detection, vaccines and treatment strategies (WHO, 2021b). These comprise the variant of interest (VOI) and variant of concern (VOC). In addition, the US CDC defines another category called the variant of high consequence (VOHC; CDC, 2021b). Each class attributes virulence hallmarks of the SARS‐CoV‐2 variants based on epidemiological investigations and scientific evidence. Each category's status is dynamic and subject to escalation or de‐escalation as per the emerging reports and incoming scientific literature. This section describes each class of SARS‐CoV‐2 variants with specific examples of mutant viral types and their characteristics in terms of virulence.
Variant of concern
SARS‐CoV‐2 variants for which confirmatory evidence suggest their association with increased virulence traits in terms of virus transmissibility, infectivity, disease severity accompanied by a notable reduction in the efficacy of present‐day therapeutic drugs, monoclonal antibodies (mAbs), neutralization by antibodies generated from previous exposure or post‐vaccination and even failure of available diagnostic techniques are classified as VOC (WHO, 2021b). These attributes sharpen viral virulence that ultimately alters the clinical disease presentation, exerting unpredictable yet adverse changes in COVID‐19 epidemiology, thereby incapacitating the public health and disease control measures or available diagnostics and viable therapies. These variants harbour mutations in vital genetic markers, primarily the S protein, driving severe viral pathology and disease outcomes (Table 1). Therefore, VOC increases the gravity of this COVID‐19 pandemic requiring immediate public health actions like controlling the transmission of SARS‐CoV‐2 mutants, increasing COVID‐19 testing, fast‐tracking sequencing and data analysis, stringent screening and surveillance of mutants, epidemiological research and prompt investigations to assess the effectiveness of available or novel therapeutic drugs and vaccines against the variants. We also augment data from clinical studies that illustrate the efficacies of the WHO‐approved COVID‐19 vaccines against each VOC in Table 2.
Table 1.
Characteristic mutations and virulence attributes of WHO‐designated SARS‐CoV‐2 VOC.
| S. No. | PANGO lineage | Synonymous name | Mutations reported in spike protein | Impact on virulence and virus phenotype |
|---|---|---|---|---|
| 1 | B.1.1.7 | 20I/501Y.V1 | ∆69–70, ∆144, E484K, S494P, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H, K1191N | Increased transmissibility by 43%–90% (Davies et al., 2021a). Potential increase in risk of hospitalization (40%–64%) and risk of death (30%; Davies et al., 2021a). Diagnostic failure in RT‐PCR tests (Srivastava et al., 2021). No significant effect on vaccine efficacy (Raddad et al., 2021). |
| 2 | B.1.351 | 20H/501Y.V2 | D80A, D215G, ∆241–243, K417N, E484K, N501Y, D614G, A701V | Increased transmissibility by ~50% (Zhou et al., 2021a). Potential increase in in‐hospital mortality by 20% (Williams et al., 2021). Ability to evade neutralizing antibodies after infection and after vaccination (Madhi et al., 2021). |
| 3 | B.1.617 | 21A/S:154 K | G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H |
Increased transmissibility by ~50% (PHE, 2021). Resistant to mAbs (Bamlanivimab) and evades antibodies generated from infection and immunization (Hoffmann et al., 2021). Vaccine breakthrough cases observed (Ferreira et al., 2021). |
| 4 | P.1 | 20 J/501Y.V3 | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I | Increased transmissibility by 40% (Gan et al., 2021). Three‐fold increase in death rate in people between 20 and 39 years (Freitas et al., 2021). Evades protective immunity after viral infection and post‐vaccination (Beltran et al., 2021). |
Table 2.
A brief account on vaccine trial efficacy and/or virus neutralization of WHO‐approved vaccines against SARS‐CoV‐2 VOC.
| Variant of concern (VOC) | Notable findings from COVID‐19 vaccine trials in terms of efficacy and/or virus neutralization | |||||
|---|---|---|---|---|---|---|
| AZD1222 (AstraZeneca) | Ad26.COV2.S (Johnson & Johnson) | BNT162b2 (Pfizer) | mRNA‐1273 (Moderna) | BBIBP‐CorV (Sinopharm) | CoronaVac (Sinovac Biotech) | |
| Prototype B.1 strain (D614G) | 66.7% Vaccine efficacy (Voysey et al., 2021). | 66% Vaccine efficacy (Karim and Oliveira, 2021). | 95% Vaccine efficacy (Karim and Oliveira, 2021). | 94% Vaccine efficacy (Karim and Oliveira, 2021). | 79% Vaccine efficacy (Karim and Oliveira, 2021). | 50.7% Vaccine efficacy (Shapiro et al., 2021) |
| B.1.1.7 | 70.4% Vaccine efficacy (Emary et al., 2021). | Not available | 93.4% Vaccine efficacy (Bernal et al., 2021). Reduction in serum neutralizing activity by 2 folds (Karim and Oliveira, 2021). | Effectively susceptible to post‐vaccination sera, with no significant reduction in neutralizing activity (Wu et al., 2021). | Slight decrease (1.4 folds) in neutralizing activity of vaccinee sera (Wang et al., 2021). | Not available |
| B.1.351 | No protection, extremely low vaccine efficacy of 10.4% (Madhi et al., 2021). | 64% Vaccine efficacy (Sadoff et al., 2021). | 75% Vaccine efficacy (Raddad et al., 2021). Reduction in serum neutralizing activity by ~6.5 folds (Karim and Oliveira, 2021). | Significant reduction (~5–10 folds) in serum neutralizing activity post‐immunization (Wu et al., 2021). | Resistance to post‐vaccination sera (2.5–3.3 folds) with complete or partial loss in neutralizing activity (Wang et al., 2021). | Not available |
| B.1.617 | 59.8% Vaccine efficacy (Bernal et al., 2021). | Not available | 87.9% Vaccine efficacy (Bernal et al., 2021). Reduction in serum neutralizing activity of vaccinees by approximately seven folds (Edara et al., 2021). | Reduction in serum neutralizing activity of vaccinees by sevenfolds (Edara et al., 2021). | Not available. | Not available |
| P.1 | Not available. | Not available | Effectively susceptible to vaccine‐elicited serum (Liu et al., 2021). But with reduction in serum neutralizing activity of vaccinees by ~6.7 folds (Karim and Oliveira, 2021). | Reduction in serum neutralizing activity of vaccinees by ~4.5 folds (Karim and Oliveira, 2021). | Not available. | 49.6% vaccine efficacy (Shapiro et al., 2021). |
The B.1.351 variant
The B.1.351 variant (also called 501Y.V2) of SARS‐CoV‐2 is a lineage of concern first detected in August 2020 in South Africa. At present, the B.1.351 variant spans 85 countries worldwide with 3 circulating sub‐lineages B.1.351.1 (Botswana), B.1.351.2 (Mayotte) and B.1.351.3 (Bangladesh and Singapore) (O'Toole et al., 2021). This variant has been commonly detected in South Africa, Germany, France, USA and Sweden. The B.1.351 variant possesses 10 mutations in its S protein, including the D80A, D215G, ∆241–243, K417N, E484K, N501Y, D614G and A701V (CDC, 2021c). The RBD of B.1.351 variant harbours both the E484K mutation that aids in immune evasion and the N501Y mutation, which expedites viral transmission (Zhou et al., 2021a). Besides, the K417N mutation, along with the latter, confers increased binding affinity of the variant with the hACE2 receptor due to more electrostatic interactions (Khan et al., 2021). These mutations have accounted for a ~ 50% increase in the transmissibility of this B.1.352 variant (Pearson et al., 2021). This variant displays a massive reduction (215 folds) on the clinical front in susceptibility to the combination of bamlanivimab and etesevimab (FDA, 2021a). But other EUA mAbs like casirivimab and imdevimab have proved effective in neutralizing this variant (FDA, 2021b). Although convalescent‐ and post‐vaccination (mRNA‐1273, AZD1222) sera have demonstrated a significant neutralizing effect against the B.1.351 variant, their efficacy remains reduced (Wu et al., 2021; Wang et al., 2021b). The BNT162b2 mRNA vaccine (Pfizer) has shown 72.1% effectiveness against severe forms of infection with the B.1.351 variant (Raddad et al., 2021). However, even after administering the second vaccine shot, neutralizing antibody titers remained 14‐folds lower against this variant (Planas et al., 2021). Also, the AZD1222 vaccine (Covishield) and Ad26.CoV2.S (Johnson & Johnson) demonstrated low efficacies of 10.4% and 57% against the B.1.351 variant (Madhi et al., 2021). These findings pitch concerns over the long‐term repercussions that such resilient SARS‐CoV‐2 variants may impose.
The B.1.1.7 variant
The B.1.1.7 variant of SARS‐CoV‐2 is a VOC first reported in the UK on 20th September 2020. It has become a global variant and has spread to 122 countries as of May 2021 (O'Toole et al., 2021). Countries most affected by the B.1.1.7 variant include the UK, USA, Germany, Denmark and Sweden (PANGO lineages: http://github.com/cov-lineages/pangolin). About 13 mutations in the S protein have been identified in this variant. These include the ∆69–70, ∆144, E484K, S494P, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H and K1191N (CDC, 2021c). Evidence suggests that the N501Y mutation within the RBD of S protein enhances the binding affinity of the SARS‐CoV‐2 virus to the hACE2 receptor, increasing transmissibility by 43% to 90% (Davies et al., 2021a). Interestingly, the unique P681H mutation of this variant has been linked with increased proteolytic activation of SARS‐CoV‐2 by furin‐like proteases that may impact viral entry or dissemination within the host (Lubinski et al., 2021). The N501Y and P681H mutations have also been related to an exponential increase in the worldwide frequency of this variant (Maison et al., 2021). Moreover, the ∆69–70 in this variant leads to diagnostic failure in RT‐PCR tests targeting the S gene (Srivastava et al., 2021). The B.1.1.7 variant also exhibits heightened disease severity in hospitalizations with increased death hazard (~61%) and case fatality rates (Davies et al., 2021b). Nonetheless, the EUA mAbs in treatment have been shown to retain their neutralizing capabilities against the B.1.1.7 variant (FDA, 2021a; FDA, 2021b). Reports suggest that this variant extends a two‐ to five‐fold reduction in the neutralizing antibody titre, but the vaccines elicit an effective immune response with high efficacy (70.4% to 89.5%) against this variant (Emary et al., 2021; Raddad et al., 2021; Shen et al., 2021). Despite its increased transmissibility, cohort studies suggest that the timely administration of COVID‐19 vaccines can easily control this variant.
The P.1 variant
The P.1 variant (alias B.1.1.28.1) of SARS‐CoV‐2 is a descendant of the B.1.1.28 lineage, first identified in Brazil in December 2020. Recently, Japan reported mutations in several strains of SARS‐CoV‐2 variants derived from the P.1 lineage (Hirotsu and Omata, 2021). The variant has been detected maximally in the USA, Brazil, Italy and commonly in 45 other countries (PANGO lineages: http://github.com/cov-lineages/pangolin). The P.1 variant harbours 11 noteworthy mutations in its S protein, including L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y and T1027I (CDC, 2021c). The trio of K417T, E484K and N501Y mutations have previously been of great concern among virologists. Their presence is associated with increased viral transmissibility due to enhanced binding affinity for the hACE2 receptor (Gan et al., 2021). These mutations in the P.1 variant have been demonstrated to boost viral transmissibility by 1.7–2.4 folds (Faria et al., 2021). Moreover, the P.1 variant has been shown to infect young adults causing severe complications (Taylor, 2021). The variant not only exhibits increased transmissibility but is also more lethal and capable of dodging the host immune system. Reports have pointed towards a 2.7 fold increase in the variant‐inflicted death toll among people aged between 20 and 39 years (Freitas et al., 2021). This variant has also been shown to evade 25%–61% of the protective immunity generated from previous viral exposure compared with the non‐P.1 lineages (Faria et al., 2021). Besides, the P.1 variant exhibits reduced susceptibility to the combination of bamlanivimab and etesevimab mAbs by over 46 folds (FDA, 2021a). However, casirivimab demonstrates effective neutralizing capabilities against the P.1 variant (FDA, 2021b). The variant is also refractory to neutralization by convalescent plasma (by 3.4–6.5 folds) and post‐vaccination sera derived from the mRNA‐1273 (Moderna) and BNT162b2 mRNA (Pfizer) vaccinees by 4.8‐ and 3.8‐folds respectively (Wang et al., 2021a). Considering the global spread and heightened virulence of the P.1 variant, it demands immediate attention from the global task forces on COVID‐19 for implementing countermeasures, including mass vaccinations, therapeutics and rapid diagnostics.
The B.1.617 variant
The B.1.617 lineage is a variant of the SARS‐CoV‐2 belonging to the Indian lineage. It was first detected in October 2020 in the state of Maharashtra, India. On 11th May 2021, the WHO classified the B.1.617 variant as a VOC owing to its high transmission rate, frequent cluster outbreaks in several countries and reduced effectiveness to mAbs and present‐day COVID‐19 therapeutics (WHO, 2021c). The variant has spread across 50 countries, but the caseload has predominantly been reported from India (PANGO lineages: http://github.com/cov-lineages/pangolin). The B.1.617 and B.1.1.7 variants circulating in the Indian subcontinent have been linked to the most recent surge in COVID‐19 infections (Ranjan et al., 2021). It is characterized by seven amino acid substitutions in the viral S protein, including G142D, E154K, L452R, E484Q, D614G, P681R and Q1071H (Yadav et al., 2021b). Of these, three mutations are essential, namely the L452R, E484Q and D614G substitutions. Studies on the RBD of B.1.617 variant suggest that mutations result in an increased hACE2 binding along with the rate of S1‐S2 cleavage, enhancing virus transmissibility and its ability to escape neutralizing antibodies (Cherian et al., 2021). The mutations reported in the B.1.1.7 variant have previously been shown to boost viral infectivity and to evade the host immune responses (Tchesnokova et al., 2021). Reports on modelled growth estimates suggest that the B.1.617 variant is as contagious as the B.1.1.7 variant with ~50% increased transmissibility (PHE, 2021).
Moreover, this variant also resists neutralization by EUA mAbs (FDA, 2021a). Infection with B.1.617 variants in hamster models resulted in severe clinical manifestations in the form of increased weight loss, high viral load and notable lesions in the lungs (Yadav et al., 2021a). Also, the B.1.617 variant demonstrated resistance against bamlanivimab along with increased entry into the Caco‐2 (intestinal) and Calu‐3 (lung‐derived) cell lines (Hoffmann et al., 2021). Studies assessing the impact of the B.1.617 variant on vaccine efficacy, therapeutics, or reinfection risks are in their infancy. Nevertheless, preliminary data revealed a significant reduction in the neutralization effectiveness against the B.1.617 variant among BNT162b2 mRNA (Pfizer) and mRNA‐1273 (Moderna) vaccinees by seven folds (Edara et al., 2021). Nevertheless, studies conducted with sera of BBV152 (Covaxin) and AZD1222 (Covishield) vaccinees showed promising neutralization abilities against the B.1.617 variant, but with a twofold reduction in the antibody titre (Yadav et al., 2021b; Yadav et al., 2021c). In this view, three sub‐lineages of the B.1.617 variant have evolved, which differ by a few, but potential mutations in the S protein and their global prevalence (WHO, 2021c). Since the B.1.617 variant and its sub‐lineages are on the rise globally, because of its high transmission rate, infectivity and receptivity to COVID‐19 therapies, effective administrative intervention to arrest the rapid spread of this variant is the need of the hour.
Variant of interest
Mutants of SARS‐CoV‐2 are designated to be VOI if their phenotypic characteristics have evolved compared with a reference (pre‐existing) strain due to genomic alterations resulting in significant amino acid substitutions contributing to increased virulence (WHO, 2021b). In other words, variants with peculiar genetic markers that are directly linked to enhanced receptor binding, lowered virus neutralization by antibodies generated from previous exposure or vaccination (immune evasion), ability to impact diagnostics and reduced treatment efficacy with an anticipated increase in contagiousness and disease severity. VOI may be attributed to increased caseload or unique outbreak clusters. Hence, these demand sturdy public health actions, including laboratory‐based characterization, watchful sequence surveillance, epidemiological scrutiny and therapeutic evaluation of in‐line drugs and vaccines. The implications of each VOI have been highlighted in Table 3.
Table 3.
Characteristic mutations and biological implications reported for WHO‐designated SARS‐CoV‐2 VOI.
| S. No. | PANGO lineage | Synonymous name | Mutations reported in spike protein | Impact on virulence and virus phenotype |
|---|---|---|---|---|
| 1 | B.1.427 | 20C/S:452R | S13I, W152C, L452R, D614G | Increased transmissibility by over 20%. Two‐fold increase in viral shedding (Deng et al., 2021). Decreased susceptibility to neutralizing antibodies generated from viral infection and post‐vaccination (FDA, 2021a). |
| 2 | B.1.429 | 20C/S:452R | S13I, W152C, L452R, D614G | Increased transmissibility by over 20% (Deng et al., 2021). Decreased susceptibility to neutralizing antibodies generated from viral infection and post‐vaccination (FDA, 2021a) |
| 3 | B.1.525 | 20A/S:484 K | A67V, ∆69–70, ∆144, E484K, D614G, Q677H, F888L | Clinical implication yet to be described. |
| 4 | B.1.526 | 20C/S:484 K | L5F, T95I, D253G, S477N/G, E484K, A701V | Increased binding to hACE2 receptor. No impact on public health reported (Schrörs et al., 2021). |
| 5 | B.1.616 | 20C | H66D, G142V, ∆144, D215G, V483A, D614G, H655Y, G669S, Q949R, N1187D | Poor detection in upper respiratory tract samples by RT‐PCR. Increased lethality by ~44% (Fillatre et al., 2021). |
| 6 | P.2 | 20B/S.484 K | F565L, E484K, D614G, V1176F | Evades CD8+ T cell‐mediated host immune responses (Pretti et al., 2021). |
| 7 | P.3 | – | ∆141–143, E484K, N501Y, D614G, P681H, E1092K, H1101Y, V1176F | No accounts on virulence and imposed clinical implications. |
The P.2 variant
The lineage P.2 (alias B.1.1.28.2) of SARS‐CoV‐2 has evolved from the Brazilian parent lineage B.1.1.28 and was identified on 15th April 2020 in Rio de Janeiro, Brazil. This variant has developed independently of the P.1 lineage (VOC) and currently manifests 33 countries, of which the significant case proportion has been reported from the USA and Brazil (PANGO lineages: http://github.com/cov-lineages/pangolin). It contains four prime mutations in the S protein, namely F565L, D614G, V1176F and the already characterized E484K mutation (CDC, 2021c). The P.2 strain has been spreading rapidly and recently detected in cases of SARS‐CoV‐2 reinfection (Nonaka et al., 2021). A recent study indicated the ability of the P.2 variant to evade the CD8+ T cell‐mediated host immune responses following virus exposure or immunization as a consequence of poorly presented viral peptides by HLA‐I molecules (Pretti et al., 2021). In addition, the P.2 variant also exhibited reduced neutralization with the BNT162b2 mRNA (Pfizer) and mRNA‐1273 (Moderna) vaccines by 5.8‐ and 2.9‐folds respectively, in fully vaccinated individuals (Beltran et al., 2021). This virulence feature was attributed to the E484K mutation of the variant. Although the P.2 variant of SARS‐CoV‐2 exhibits increased virulence by immune evasion, the effect of reported mutations on transmissibility and disease severity needs to be elucidated. Till then, this variant remains as a VOI with active monitoring and global surveillance.
The B.1.427 variant
The B.1.427 variant of SARS‐CoV‐2 belongs to the USA lineage (CA) and was first identified in June 2020 in California, USA. The variant is regarded as a VOC by the US CDC and has spread across 28 countries worldwide (PANGO lineages: http://github.com/cov-lineages/pangolin). It contains four mutations in the S protein, of which the L452R and D614G amino acid substitutions contribute to virulence. Reports suggest the role of L452R mutation in accelerating viral shedding and transmissibility of the B.1.427 variant by two‐folds and over 20% respectively (Deng et al., 2021). Viral infectivity with L452R mutation was enhanced by ~15 folds in 293T cells and by ~10 folds in human airway organoids, relative to the D614G mutation alone (Deng et al., 2021). Also, plasma from convalescent patients and fully‐vaccinated recipients (BNT162b2 mRNA or mRNA‐1273) showed a significant reduction in the susceptibility profile of the B.1.427 variant to neutralizing antibodies. Other studies have reported similar findings relating the L452R mutation of the B.1.427 variant abolishing antibody‐mediated neutralization and immune evasion (McCallum et al., 2021). Nevertheless, few EUA mAbs exhibit no significant changes in their neutralizing ability against the B.1.427 variant (FDA, 2021b). Since the B.1.427 variant is a more transmissible type with immune escape abilities but with a comparatively low impact on public health, further investigations are warranted to unearth its biological implications.
The B.1.429 variant
The lineage B.1.429 is a variant of SARS‐CoV‐2 predominantly circulating in California, USA, where its incidence was first reported in June 2020. The B.1.429 variant has been recognized as a VOC by the US CDC and currently spans 31 countries, majorly affecting the USA (PANGO lineages: http://github.com/cov-lineages/pangolin). The variant displays four amino acid substitutions in the viral S protein, including S13I, W152C, L452R and D614G (CDC, 2021c). The B.1.429 variant is quite similar to the B.1.427 variant of SARS‐CoV‐2 in terms of mutations, transmissibility and response to present‐day therapeutics. Studies have associated the L452R substitution of the B.1.429 variant with a 20% increase in transmissibility, two folds surge in the viral shedding in vivo and cell infectivity (Deng et al., 2021). Consequently, the antibodies from convalescent‐ and post‐vaccination sera revealed a significant reduction in the B.1.429 neutralizing titres. Similar investigations have also disclosed the ability of the B.1.429 variant to overcome antibody‐mediated neutralization, eliciting immune escape (McCallum et al., 2021). The variant also exhibits a nine‐fold reduction in the neutralizing capabilities of bamlanivimab and etesevimab in combination (FDA, 2021a). However, the mAbs casirivimab and imdevimab have effectively neutralized the B.1.429 variant (FDA, 2021b). Although the B.1.429 remains a VOC, its incidence rate in the USA has dropped suddenly from February 2021 (PANGO lineages: http://github.com/cov-lineages/pangolin). The variant is now being outcompeted by the highly contagious B.1.1.7 variant of SARS‐CoV‐2. This fact draws attention as it showcases the potential of mutations in shaping disease epidemiology by dictating viral virulence (transmissibility), thereby competing with one another in nature.
The B.1.526 variant
Lineage B.1.526 is a variant of SARS‐CoV‐2 first detected on 23rd November 2020 in New York City, USA. This lineage predominantly circulated in the American subcontinent and accounted for 25% of all sequenced SARS‐CoV‐2 viruses in the USA till February 2021 (Thompson et al., 2021). As of May 2021, the B.1.526 variant has been detected in 48 states of the USA and 35 other countries (PANGO lineages: http://github.com/cov-lineages/pangolin). Two sorts of this variant have been identified, bearing the common D614G mutation in the S protein and four novel mutations (L5F, S477N/G, E484K, A701V) and two notable mutations: T95I and D253G in the RBD (Zhou et al., 2021b). The S477N/G mutation is present in smaller fractions among this variant, while E484K has been reported in more than half of the lineage (Annavajhala et al., 2021). Recently, the S477N/G mutation has been shown to strengthen the binding of S protein with the hACE2 receptor (Schrörs et al., 2021). The clinical implications of this B.1.526 variant are widely undescribed. However, the E484K spike mutation has been attributed to a 31‐fold reduced susceptibility to bamlanivimab and etesevimab mAb cocktail (FDA, 2021a). In a similar context, the E484K mutation in B.1.351 variant (VOC) has been shown to lower the protective efficacy of the BNT162b2 mRNA vaccine (Pfizer) in populations with a prevalence of B.1.351 variant (Tada et al., 2021). Moreover, the E484K mutation is also known to reduce the neutralization efficiency of convalescent and post‐vaccination sera (Jangra et al., 2021). These findings suggest the combative role of E484K mutation in SARS‐CoV‐2 variants resulting in evasion of vaccine‐elicited antibodies. Nevertheless, casivirimab and imdevimab, independently and in combination, have been reported to neutralize this variant effectively (FDA, 2021b). At present, sub‐lineages of this variant have emerged, but their global impact remains unexplored.
The sub‐lineage of this variant, B.1.526.1, was first detected in New York City. This variant has been characterized with several unique mutations and remains majorly restricted to the USA. The US CDC lists this variant as a VOI (CDC, 2021c). The peculiar modifications of the S protein include D80G, F157S, L452R, D614G, T791I, T859N and D950H (CDC, 2021c). Among these mutations, D614G has been shown to enhance the transmissibility of the variants (Korber et al., 2020), while L452R increases viral infectivity with a reinforced affinity for the hACE2 receptor, boosting viral replication capacity and simultaneously evading cellular immunity (Motozono et al., 2021). Regarding the functional implications, L452R mutation in B.1.427 and B.1.429 lineages has been shown to lower virus neutralization by nine folds with a cocktail of bamlanivimab and etesevimab, thereby reducing antibody efficacy (FDA, 2021a). Also, this mutant exhibits reduced neutralization by vaccine sera (CDC, 2021b). However, casivirimab and imdevimab have been shown to neutralize this mutant strain effectively (FDA, 2021b). Its prevalence among the circulating variants is extremely low; hence, this strain may eventually be inconsequential.
B.1.525 variant
The Lineage B.1.525 variant of SARS‐CoV‐2 was first reported in the United Kingdom (UK) and Nigeria on 11th December 2020. It is now regarded as an international variant of SARS‐CoV‐2 spanning 51 countries, with maximum cases reported in the USA (PANGO lineages: http://github.com/cov-lineages/pangolin). The defining mutations of the B.1.525 variant comprise A67V, ∆69–70, ∆144, D614G, Q677H, F888L and the spike mutation E484K (CDC, 2021c). This variant is similar to the highly transmissible VOC B.1.1.7 lineage with the common E484K mutation. The latter has been previously described to aid virus escape from neutralizing antibodies in the variants that emerged in South Africa (B.1.351 lineage) and Brazil (P.1 and P.2 lineages). The biological characteristics of this new variant remain widely unexplored. However, this mutant directs the reduction of antibody‐mediated virus neutralization against convalescent‐ and post‐vaccination sera (Jangra et al., 2021) and emergency use authorization (EUA) mAbs (FDA, 2021a). As mentioned above, the clinical implications of the mutations on virus transmissibility and infectivity are yet to be described as most investigations are underway.
The B.1.616 variant
The B.1.616 variant of SARS‐CoV‐2 has evolved independently and given rise to the French lineage of COVID‐19. It was first detected on 29th January 2021 in France and remains restricted to the country (PANGO lineages: http://github.com/cov-lineages/pangolin). The S protein of the B.1.616 variant has been characterized with a unique constellation of mutations like H66D, G142V, ∆144, D215G, V483A, D614G, H655Y, G669S, Q949R and N1187D (WHO, 2021c). V483A. Few of these amino acid substitutions have been of pronounced concern, particularly V483A that imparts resistance to mAbs (Li et al., 2020), D614G, which increases viral transmissibility (Korber et al., 2020) and H655Y in conferring escape from human mAbs (Braun et al., 2021). Recent reports also propound that RT‐PCR poorly detects the B.1.616 variant on nasopharyngeal samples, increased disease severity and lethality with a high 28‐day mortality of ~44% (Fillatre et al., 2021). While this variant is on the surge, it is closely monitored by the WHO and European Centre for Disease Prevention and Control (ECDC). More investigations are warranted to delineate the clinical implications of this variant on the efficacy of therapeutic drugs and COVID‐19 vaccines.
The P.3 variant
The P.3 variant (alias B.1.1.28.3) of SARS‐CoV‐2 is listed as a VOI by the WHO. This lineage was first reported in the Philippines in February 2021 and presently spans 12 countries (PANGO lineages: http://github.com/cov-lineages/pangolin). However, the caseload from this variant predominantly comes from the Philippines. The P.3 variant displays several characteristic mutations within the viral S protein, such as ∆141–143, E484K, N501Y, D614G, P681H, E1092K, H1101Y and V1176F (CDC, 2021c). The presence of worrisome E484K, N501Y, D614G and P681H mutations in the P.3 variant suggest that it may exhibit increased infectivity, transmissibility and immune evasion (Bascos et al., 2021). However, no studies to date describe the effect of such mutations on the virulence of the P.3 variant. Moreover, there is no accounted evidence to comment on the transmissibility or severity imposed by this variant as most investigations are underway. The biological significance of the P.3 variant will be unravelled only when conclusive findings are reported in the literature.
Based on cited literature, we have summarized the biological consequences of the characterized S protein substitutions in the emerging SARS‐CoV‐2 variants in Fig. 4. The overall effect of such amino acid substitutions (mutations) on viral pathology and virulence has been depicted.
Fig. 4.
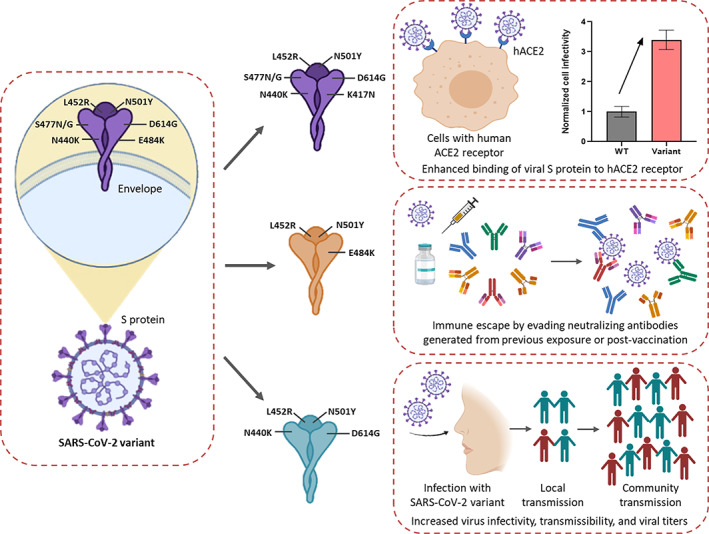
Mechanistic insight into the biological implications of the mutations reported in SARS‐CoV‐2 VOC and VOI. Individual or a combination of mutations in the spike (S) protein drives the virulence attributes of SARS‐CoV‐2 variants. Amino acid substitutions in the S protein have been shown to boost viral infectivity by enhancing binding to the hACE2 receptor. Mutations increase the immune evasion abilities of SARS‐CoV‐2 variants by evading antibodies and protective immune responses generated from previous viral infection and vaccination. SARS‐CoV‐2 variants also exhibit increased transmissibility leading to fresh cluster outbreaks and community transmission.
Variant of high consequence
Apart from VOI and VOC, the US CDC defines another class of SARS‐CoV‐2 variants, called the VOHC. Any SARS‐CoV‐2 variant that exhibits pinpoint evidence regarding reduced effectiveness of preventive public health and social measures, diagnostic techniques and medical intervention strategies relative to the circulating variants is designated as a VOHC (CDC, 2021b). Their possible attributes include those of VOC, in addition to detection failure by diagnostic procedures, increased viral transmissibility or infectivity, increased hospitalizations and extreme disease conditions, a significant reduction in COVID‐19 vaccine efficacy, or a massive increase in vaccine breakthrough cases, or weakened vaccine‐driven protection and resistance against EUA prophylaxis and approved therapeutics. If a VOHC is identified, it demands immediate communication to the WHO and global task forces on COVID‐19 and a declaration stating containment strategies and suggestions to upgrade the line of treatment and vaccine formulations (CDC, 2021c). Fortunately, there are no variants of the SARS‐CoV‐2 that fall into the category of VOHC.
Conclusion
The COVID‐19 pandemic has unleashed a wave of unprecedented devastation across the world with nationwide lockdowns, hitting hard on all trade sectors and economic activities. The global dissemination of the SARS‐CoV‐2 has given rise to innumerable lineages bearing diverse variants with characteristic genome‐wide mutations. Natural selection processes drive the evolution of SARS‐CoV‐2 variants in various populations and geographical locations. A constellation of unique amino acid substitutions in the viral S protein has been shown to boost the virulence of SARS‐CoV‐2 variants by enhancing transmissibility, infectivity and host immune evasion. These variants have been classified into different categories based on their threat levels: VOI, VOC and VOHC. The VOI exhibits alterations in specific viral genetic markers predicted to enhance virulence and remain under strict surveillance. A VOC is attributed to a tremendous increase in virulence and resistance to existing therapies. On the other hand, VOHC is associated with escalated virulence, disease severity, diagnostics failure and resistance to present therapeutics. Although intervention and treatment strategies are in place, these variants are co‐evolving at an unprecedented rate. SARS‐CoV‐2 variants have also shown reduced effectiveness to the current lot of COVID‐19 vaccines with unexpected vaccine breakthrough cases. Hence, such rapidly evolving variants have acquired mutations that confer resistance to neutralizing antibodies generated from previous viral exposure or immunization. In summary, the longevity of this COVID‐19 pandemic decides the fate of forthcoming variants. As this resilient virus keeps expanding in diverse populations, there are high chances of accumulating different genome‐wide mutations that may exacerbate virulence. Therefore, variants of SARS‐CoV‐2 require immediate attention from the scientific community to develop novel COVID‐19 therapeutics and upgraded vaccines. Until then, COVID‐19 vaccines are the only resort at our disposal. Thus, vaccination programs must be driven at full gears with close monitoring of SARS‐CoV‐2 variants by global surveillance initiatives to enable necessary countermeasures.
Abbreviations
- CDC
Centres for Disease Control and Prevention
- COVID‐19
coronavirus disease‐19
- ECDC
European Centre for Disease Prevention and Control
- EUA
emergency use authorization
- hACE2
human angiotensin‐converting enzyme 2
- mAb
monoclonal antibody
- MERS‐CoV
Middle East respiratory syndrome coronavirus
- NTD
N‐terminal domain
- R0
reproduction number
- RBD
receptor‐binding domain
- RT‐PCR
reverse transcriptase polymerase chain reaction
- S protein
spike protein
- SARS‐CoV
severe acute respiratory syndrome coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- VOC
variant of concern
- VOHC
Variant of high consequence
- VOI
variant of interest
- WHO
World Health Organization
Acknowledgements
We want to thank the Indian Council of Medical Research (ICMR) and Prime Minister's Research Fellow (PMRF) Scheme, Govt. of India for providing fellowship to JC and NM respectively. We also thank Prof. Kusum Harjai, Panjab University and Prof. Sanjay Chhibber, ICMR Emeritus Scientist for their continuous support and motivation.
Data availability statement
The datasets generated and analysed during the current study are available at NCBI SARS‐CoV‐2 Resources (https://www.ncbi.nlm.nih.gov/sars-cov-2/), CoVariants (enabled by data from GISAID; https://covariants.org/) and PANGO lineages (http://github.com/cov-lineages/pangolin).
References
- Abdelrahman, Z. , Li, M. , and Wang, X. (2020) Comparative review of SARS‐CoV‐2, SARS‐CoV, MERS‐CoV, and influenza a respiratory viruses. Front Immunol 11: 552909. 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, K.G. , Rambaut, A. , Lipkin, W.I. , Holmes, E.C. , and Garry, R.F. (2020) The proximal origin of SARS‐CoV‐2. Nat Med 26: 450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annavajhala, M.K. , Mohri, H. , Wang, P. , Zucker, J.E. , Sheng, Z. , Gomez‐Simmonds, A. , et al. (2021) A novel SARS‐CoV‐2 variant of concern, B.1.526, identified in New York. medRxiv preprint. doi: 10.1101/2021.02.23.21252259. [DOI]
- Bascos, N.A.D. , Mirano‐Bascos, D. , and Saloma, C.P. (2021) Structural analysis of spike protein mutations in an emergent SARS‐CoV‐2 variant from The Philippines. bioRxiv preprint. doi: 10.1101/2021.03.06.434059. [DOI]
- Beltran, G.W.F. , Lam, E.C. , St. Denis, K. , Nitido, A.D. , Garcia, Z.H. , Hauser, B.M. , et al. (2021) Multiple SARS‐CoV‐2 variants escape neutralization by vaccine‐induced humoral immunity. Cell 184: 2372–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal, L.J. , Andrews, N. , Gower, N. , Gallagher, E. , Simmons, R. , Thelwall, S. , et al. (2021) Effectiveness of COVID‐19 vaccines against the B.1.617.2 variant. medRxiv preprint. doi: 10.1101/2021.05.22.21257658. [DOI]
- Bhadra, A. , Mukherjee, A. , and Sarkar, K. (2021) Impact of population density on Covid‐19 infected and mortality rate in India. Model Earth Syst Environ 7: 623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, K.M. , Moreno, G.K. , Halfmann, P.J. , Hodcroft, E.B. , Baker, D.A. , Boehm, E.C. , et al. (2021) Transmission of SARS‐CoV‐2 in domestic cats imposes a narrow bottleneck. PLoS Pathog 17: e1009373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (2021a) How to protect yourself and others. 2021. Centers for Disease Control and Prevention. URL https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/prevention.html.
- CDC (2021b) Update on emerging SARS‐CoV‐2 variants and vaccine considerations. 2021. Centers for Disease Control and Prevention. URL https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html. [PubMed]
- CDC (2021c) Coronavirus disease 2019 (COVID‐19). 2021. Centers for Disease Control and Prevention. URL https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html.
- Çelik, İ. , Saatçi, E. , and Eyüboğlu, A.F. (2020) Emerging and reemerging respiratory viral infections up to Covid‐19. Turk J Med Sci 50: 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian, S. , Potdar, V. , Jadhav, S. , Yadav, P. , Gupta, N. , Das, M. , et al. (2021) Convergent evolution of SARS‐CoV‐2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID‐19 in Maharashtra, India bioRxiv preprint. doi: 10.1101/2021.04.22.440932. [DOI] [PMC free article] [PubMed]
- Davies, N.G. , Abbott, S. , Barnard, R.C. , Jarvis, C.I. , Kucharski, A.J. , Munday, J.D. , et al. (2021a) Estimated transmissibility and impact of SARS‐CoV‐2 lineage B.1.1.7 in England. Science 9: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, N.G. , Jarvis, C.I. , Edmunds, W.J. , Jewell, N.P. , Diaz‐Ordaz, K. , and Keogh, R.H. (2021b) Increased mortality in community‐tested cases of SARS‐CoV‐2 lineage B.1.1.7. Nature 593: 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Garcia‐Knight, M.A. , Khalid, M.M. , Servellita, V. , Wang, C. , Morris, M.K. , et al. (2021) Transmission, infectivity, and neutralization of a spike L452R SARS‐CoV‐2 variant. Cell S0092‐8674: 505–505. 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara, V.V. , Lai, L. , Sahoo, M.K. , Floyd, K. , Sibai, M. , Solis, D. , et al. (2021) Infection and vaccine‐induced neutralizing antibody responses to the SARS‐CoV‐2 B.1.617.1 variant. bioRxiv preprint. doi: 10.1101/2021.05.09.443299. [DOI] [PMC free article] [PubMed]
- Emary, K.R.W. , Golubchik, T. , Aley, P.K. , Ariani, C.V. , Angus, B. , Bibi, S. , et al. (2021) Efficacy of ChAdOx1 nCoV‐19 (AZD1222) vaccine against SARS‐CoV‐2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. Lancet 397: 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria, N.R. , Mellan, T.A. , Whittaker, C. , Claro, I.M. , Candido, D.D.S. , Mishra, S. , et al. (2021) Genomics and epidemiology of the P.1 SARS‐CoV‐2 lineage in Manaus, Brazil. Science 372: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA (2021a) Fact sheet for health care providers emergency use authorization (EUA) of Bamlanivimab and Etesevimab. 2021. Food and Drug Administration. URL https://www.fda.gov/media/145802.
- FDA (2021b) Fact sheet for health care providers emergency use authorization (EUA) of REGEN‐COV™ (Casirivimab with Imdevimab). 2021. Food and Drug Administration. URL https://www.fda.gov/media/145611.
- Ferreira, I. , Datir, R. , Papa, G. , Kemp, S. , Meng, B. , Rakshit, P. , et al. (2021) SARS‐CoV‐2 B.1.617 emergence and sensitivity to vaccine‐elicited antibodies. bioRxiv preprint. doi: 10.1101/2021.05.08.443253. [DOI]
- Fillatre, P. , Dufour, M.J. , Behillil, S. , Vatan, R. , Reusse, P. , Gabellec, A. , et al. (2021) A new SARS‐CoV‐2 variant poorly detected by RT‐PCR on nasopharyngeal samples, with high lethality. medRxiv preprint. doi: 10.1101/2021.05.05.21256690. [DOI] [PMC free article] [PubMed]
- Freitas, A.R.R. , Beckedorff, O.A. , Cavalcanti, L.G. , Siqueira, A.M. , Castro, D.B. , Costa, C.F. , et al. (2021) The emergence of novel SARS‐CoV‐2 variant P.1 in Amazonas (Brazil) was temporally associated with a change in the age and gender profile of COVID‐19 mortality. SciELOPreprints . doi: 10.1590/SciELOPreprints.2030. [DOI] [PMC free article] [PubMed]
- Gan, H.H. , Twaddle, A. , Marchand, B. , and Gunsalus, K.C. (2021) Structural modeling of the SARS‐CoV‐2 spike/human ACE2 complex Interface can identify high‐affinity variants associated with increased transmissibility. J Mol Biol 433: 167051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruprasad, L. (2021) Human SARS CoV‐2 spike protein mutations. Proteins 89: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu, Y. , and Omata, M. (2021) Discovery of a SARS‐CoV‐2 variant from the P.1 lineage harboring K417T/E484K/N501Y mutations in Kofu, Japan. J Infect 82: 276–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M. , Hofmann‐Winkler, H. , Krüger, N. , Kempf, A. , Nehlmeier, I. , Graichen, L. , et al. (2021) SARS‐CoV‐2 variant B.1.617 is resistant to Bamlanivimab and evades antibodies induced by infection and vaccination. bioRxiv preprint. doi: 10.1101/2021.05.04.442663. [DOI] [PMC free article] [PubMed]
- Hu, B. , Guo, H. , Zhou, P. , and Shi, Z.L. (2020a) Characteristics of SARS‐CoV‐2 and COVID‐19. Nat Rev Microbiol 19: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, T. , Liu, Y. , Zhao, M. , Zhuang, Q. , Xu, L. , and He, Q. (2020b) A comparison of COVID‐19, SARS and MERS. PeerJ 8: e9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel, S. , Graña‐Miraglia, L. , Gutierrez, J.M. , Bundalovic‐Torma, C. , Groves, H.E. , Isabel, M.R. , et al. (2020) Evolutionary and structural analyses of SARS‐CoV‐2 D614G spike protein mutation now documented worldwide. Sci Rep 10: 14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra, S. , Ye, C. , Rathnasinghe, R. , Stadlbauer, D. , Krammer, F. , Simon, V. , et al. (2021) SARS‐CoV‐2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2: 283–284. 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadi, N. , and Khelfaoui, M. (2020) Population density, a factor in the spread of COVID‐19 in Algeria: statistic study. Bull Natl Res Cent 44: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim, A.S.S. , and Oliveira, T. (2021) New SARS‐CoV‐2 variants – clinical, public health, and vaccine implications. N Engl J Med 384: 1866–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S.P. , and Gupta, V. (2020) COVID‐19 Vaccine: A comprehensive status report. Virus Res 288: 198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye, A.D. , Okeagu, C.N. , Pham, A.D. , Silva, R.A. , Hurley, J.J. , Arron, B.L. , et al. (2020) Economic impact of COVID‐19 pandemic on healthcare facilities and systems: international perspectives. Best Pract Res Clin Anaesthesiol. 10.1016/j.bpa.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. , Zia, T. , Suleman, M. , Khan, T. , Ali, S.S. , Abbasi, A.A. , et al. (2021) Higher infectivity of the SARS‐CoV‐2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J Cell Physiol. 10.1002/jcp.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, B. , Fischer, W.M. , Gnanakaran, S. , Yoon, H. , Theiler, J. , Abfalterer, W. , et al. (2020) Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell 182: 812–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring, A.S. , and Hodcroft, E.B. (2021) Genetic variants of SARS‐CoV‐2—what do they mean? JAMA 325: 529–531. [DOI] [PubMed] [Google Scholar]
- Li, Q. , Wu, J. , Nie, J. , Zhang, L. , Hao, H. , Liu, S. , et al. (2020) The impact of mutations in SARS‐CoV‐2 spike on viral infectivity and antigenicity. Cell 182: 1284–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Liu, J. , Xia, H. , Zhang, X. , Fontes‐Garfias, C.R. , Swanson, K.A. , et al. (2021) Neutralizing activity of BNT162b2‐elicited serum. N Engl J Med 384: 1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski, B. , Tang, T. , Daniel, S. , Jaimes, J.A. , Whittaker, G.R. (2021) Functional evaluation of proteolytic activation for the SARS‐CoV‐2 variant B.1.1.7: role of the P681H mutation. bioRxiv preprint. doi: 10.1101/2021.04.06.438731. [DOI] [PMC free article] [PubMed]
- Madhi, S.A. , Baillie, V. , Cutland, C.L. , Voysey, M. , Koen, A.L. , Fairlie, L. , et al. (2021) Efficacy of the ChAdOx1 nCoV‐19 Covid‐19 vaccine against the B.1.351 variant. N Engl J Med 384: 1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison, D.P. , Ching, L.L. , Shikuma, C.M. , and Nerurkar, V.R. (2021) Genetic characteristics and phylogeny of 969‐bp S gene sequence of SARS‐CoV‐2 from Hawai'i reveals the worldwide emerging P681H mutation. Hawaii J Health Soc Welf 80: 52–61. [PMC free article] [PubMed] [Google Scholar]
- McCallum, M. , Bassi, J. , Marco, A.D. , Chen, A. , Walls, A.C. , Iulio, J.D. , et al. (2021) SARS‐CoV‐2 immune evasion by variant B.1.427/B.1.429. bioRxiv preprint. doi: 10.1101/2021.03.31.437925. [DOI]
- Motozono, C. , Toyoda, M. , Zahradnik, J. , Ikeda, T. , Saito, A. , Tan, T.S. , et al. (2021) An emerging SARS‐CoV‐2 mutant evading cellular immunity and increasing viral infectivity. bioRxiv preprint. doi: 10.1101/2021.04.02.438288. [DOI]
- Nature news . (2021) COVID research: a year of scientific milestones. Nature. 10.1038/d41586-020-00502-w. [DOI] [PubMed] [Google Scholar]
- Nonaka, V.C.K. , Miranda Franco M., Gräf, T. , Almeida Mendes, A.V. , Santana de Aguiar, R. , Giovanetti, M. , et al. (2021) Genomic evidence of a Sars‐Cov‐2 reinfection case with E484K spike mutation in Brazil. Preprints . doi: 10.20944/preprints202101.0132.v1. [DOI]
- O'Toole, Á. , Hill, V. , and Pybus, O.G. (2021) Tracking the international spread of SARS‐CoV‐2 lineages B.1.1.7 and B.1.351/501Y‐V2. Wellcome Open Res. 6: 121–127. 10.12688/wellcomeopenres.16661.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C.A.B. , Russell, T.W. , Davies, N.G. , Kucharski, A.J. , CMMID COVID‐19 Working Group , Edmunds, J. , et al. (2021) Estimates of severity and transmissibility of novel SARS‐CoV‐2 variant 501Y.V2 in South Africa. 2021. URL https://cmmid.github.io/topics/covid19/reports/sa‐novelvariant/2021_01_11_Transmissibility_and_severity_of_501Y_V2_in_SA.
- PHE (2021). SARS‐CoV‐2 variants of concern and variants under investigation in England (Technical briefing 10). 2021. Public Health England. URL https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/984274/Variants_of_Concern_VOC_Technical_Briefing_10_England.pdf.
- Planas, D. , Bruel, T. , Grzelak, L. , Guivel‐Benhassine, F. , Staropoli, I. , Porrot, F. , et al. (2021) Sensitivity of infectious SARS‐CoV‐2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 27: 917–924. [DOI] [PubMed] [Google Scholar]
- Pretti, M.A.M. , Galvani, R.G. , Farias, A.S. , and Boroni, M. (2021) New SARS‐CoV‐2 lineages could evade CD8+ T‐cells response. bioRxiv preprint. doi: 10.1101/2021.03.09.434584. [DOI]
- Raddad, A.L.J. , Chemaitelly, H. , and Butt, A.A. (2021) Effectiveness of the BNT162b2 Covid‐19 vaccine against the B.1.1.7 and B.1.351 variants. N Engl J Med 385: 187–189. 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan, R. , Sharma, A. , and Verma, M.K. (2021) Characterization of the second wave of COVID‐19 in India. medRxiv preprint. doi: 10.1101/2021.04.17.21255665. [DOI]
- Sadoff, J. , Gray, G. , Vandebosch, A. , Cárdenas, V. , Shukarev, G. , Grinsztejn, B. , et al. (2021) Safety and efficacy of single‐dose Ad26.COV2.S vaccine against Covid‐19. N Engl J Med. 384: 2187–2201. 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrörs, B. , Gudimella, R. , Bukur, T. , Rösler, T. , Löwer, M. , and Sahin, U. (2021) Large‐scale analysis of SARS‐CoV‐2 spike‐glycoprotein mutants demonstrates the need for continuous screening of virus isolates. bioRxiv preprint. doi: 10.1101/2021.02.04.429765. [DOI] [PMC free article] [PubMed]
- Shapiro, J. , Dean, N.E. , Madewell, Z.J. , Yang, Y. , Halloran, M.E. , Longini, I. (2021) Efficacy estimates for various COVID‐19 vaccines: what we know from the literature and reports. medRxiv preprint. doi: 10.1101/2021.05.20.21257461. [DOI]
- Shen, X. , Tang, H. , McDanal, C. , Wagh, K. , Fischer, W. , Theiler, J. , et al. (2021) SARS‐CoV‐2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe 29: 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, S. , Banu, S. , Singh, P. , Sowpati, D.T. , and Mishra, R.K. (2021) SARS‐CoV‐2 genomics: an Indian perspective on sequencing viral variants. J Biosci 46: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, T. , Dcosta, B.M. , Samanovic‐Golden, M. , Herati, R.S. , Cornelius, A. , Mulligan, M.J. , et al. (2021) Neutralization of viruses with European, south African, and United States SARS‐CoV‐2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine‐elicited antibodies. bioRxiv preprint. doi: 10.1101/2021.02.05.430003. [DOI]
- Taylor, L. (2021) Covid‐19: Brazil's spiralling crisis is increasingly affecting young people. BMJ 373: 879. 10.1136/bmj.n879. [DOI] [PubMed] [Google Scholar]
- Tchesnokova, V. , Kulakesara, H. , Larson, L. , Bowers, V. , Rechkina, E. , Kisiela, D. , et al. (2021) Acquisition of the L452R mutation in the ACE2‐binding interface of spike protein triggers recent massive expansion of SARS‐Cov‐2 variants. bioRxiv preprint. doi: 10.1101/2021.02.22.432189. [DOI] [PMC free article] [PubMed]
- Thompson, C.N. , Hughes, S. , Ngai, S. , Baumgartner, J. , Wang, J.C. , McGibbon, E. , et al. (2021) Rapid emergence and epidemiologic characteristics of the SARS‐CoV‐2 B.1.526 variant – New York City, New York, January 1–April 5, 2021. MMWR Morb Mortal Wkly Rep 70: 712–716. 10.15585/mmwr.mm7019e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey, M. , Costa Clemens, S.A. , Madhi, S.A. , Weckx, L.Y. , Folegatti, P.M. , Aley, P.K. , et al. (2021) Single‐dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV‐19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet 397: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G.L. , Wang, Z.Y. , Duan, L.J. , Meng, Q.C. , Jiang, M.D. , Cao, J. , et al. (2021) Susceptibility of circulating SARS‐CoV‐2 variants to neutralization. N Engl J Med. 384: 2354–2356. 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Li, X. , Li, T. , Zhang, S. , Wang, L. , Wu, X. , et al. (2020) The genetic sequence, origin, and diagnosis of SARS‐CoV‐2. Eur J Clin Microbiol Infect Dis 39: 1629–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Casner, R.G. , Nair, M.S. , Wang, M. , Yu, J. , Cerutti, G. , et al. (2021a) Increased resistance of SARS‐CoV‐2 variant P.1 to antibody neutralization. Cell Host Microbe 29: 747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Nair, M.S. , Liu, L. , Iketani, S. , Luo, Y. , Guo, Y. , et al. (2021b) Increased resistance of SARS‐CoV‐2 variants B.1.351 and B.1.1.7 to antibody neutralization. Nature 593: 130–135. [DOI] [PubMed] [Google Scholar]
- WHO (2020) Coronavirus disease 2019 (COVID‐19): situation report, 51. 2020. World Health Organization. URL https://apps.who.int/iris/handle/10665/331475.
- WHO (2021a) WHO lists additional COVID‐19 vaccine for emergency use and issues interim policy recommendations. 2021. World Health Organization. URL https://www.who.int/news/item/07‐05‐2021‐who‐lists‐additional‐covid‐19‐vaccine‐for‐emergency‐use‐and‐issues‐interim‐policy‐recommendations.
- WHO (2021b) COVID‐19 Weekly Epidemiological Update. Special edition: Proposed working definitions of SARS‐CoV‐2 Variants of Interest and Variants of Concern. 2021. World Health Organization. URL https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update.
- WHO (2021c) COVID‐19 Weekly Epidemiological Update. 2021. World Health Organization. URL https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19-11-may-2021
- Williams, H. , Hutchinson, D. , and Stone, H. (2021) Watching brief: the evolution and impact of COVID‐19 variants B.1.1.7, B.1.351, P.1 and B.1.617. Global Biosecurity 3: 1–27. 10.31646/gbio.112. [DOI] [Google Scholar]
- Worldometers (2021) [24 June, 2021]. URL https://wwwworldometersinfo/coronavirus/.
- Wu, K. , Werner, A.P. , Moliva, J.I. , Koch, M. , Choi, A. , Stewart Jones, G.B.E. , et al. (2021) mRNA‐1273 vaccine induces neutralizing antibodies against spike mutants from global SARS‐CoV‐2 variants. bioRxiv preprint. doi: 10.1101/2021.01.25.427948. [DOI]
- Yadav, P.D. , Mohandas, S. , Shete, A.M. , Nyayanit, D.A. , Gupta, N. , Patil, D.Y. , et al. (2021a) SARS CoV‐2 variant B.1.617.1 is highly pathogenic in hamsters than B.1 variant. bioRxiv preprint. doi: 10.1101/2021.05.05.442760. [DOI]
- Yadav, P.D. , Sapkal, G.N. , Abraham, P. , Ella, R. , Deshpande, G. , Patil, D.Y. , et al. (2021b) Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. bioRxiv preprint. doi: 10.1101/2021.04.23.441101. [DOI] [PubMed]
- Yadav, P.D. , Sapkal, G.N. , Abraham, P. , Ella, R. , Deshpande, G. , Patil, D.Y. , et al. (2021c) Neutralization potential of Covishield vaccinated individuals against B.1.617.1t. bioRxiv preprint. doi: 10.1101/2021.05.12.443645. [DOI] [PubMed]
- Zhou, D. , Dejnirattisai, W. , Supasa, P. , Liu, C. , Mentzer, A.J. , Ginn, H.M. , et al. (2021a) Evidence of escape of SARS‐CoV‐2 variant B.1.351 from natural and vaccine‐induced sera. Cell 184: 2348–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Dcosta, B.M. , Samanovic, M.I. , Mulligan, M.J. , Landau, N.R. , and Tada, T. (2021b) B.1.526 SARS‐CoV‐2 variants identified in new York City are neutralized by vaccine‐elicited and therapeutic monoclonal antibodies. bioRxiv preprint. doi: 10.1101/2021.03.24.436620. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analysed during the current study are available at NCBI SARS‐CoV‐2 Resources (https://www.ncbi.nlm.nih.gov/sars-cov-2/), CoVariants (enabled by data from GISAID; https://covariants.org/) and PANGO lineages (http://github.com/cov-lineages/pangolin).


