Abstract
Coronavirus disease‐19 (COVID‐19) is a complex disorder caused by the pandemic diffusion of a novel coronavirus named SARS‐CoV‐2. Clinical manifestations vary from silent infection to severe pneumonia, disseminated thrombosis, multi‐organ failure, and death. COVID‐19 pathogenesis is still not fully elucidated, while increasing evidence suggests that disease phenotypes are strongly related to the virus‐induced immune system's dysregulation. Indeed, when the virus‐host cross talk is out of control, the occurrence of an aberrant systemic inflammatory reaction, named “cytokine storm,” leads to a detrimental impairment of the adaptive immune response. Dendritic cells (DCs) are the most potent antigen‐presenting cells able to support innate immune and promote adaptive responses. Besides, DCs play a key role in the anti‐viral defense. The aim of this review is to focus on DC involvement in SARS‐CoV‐2 infection to better understand pathogenesis and clinical behavior of COVID‐19 and explore potential implications for immune‐based therapy strategies.
Keywords: COVID‐19, dendritic cells, immune‐therapies, SARS‐CoV‐2, vaccines
1. INTRODUCTION
At the end of 2019, several human cases of infection by a novel coronavirus named SARS‐CoV‐2 were related to the Huanan Seafood Wholesale Market, Wuhan, China. Since then, the wide dissemination of the infection and of its related disease, called COVID‐19, to every continent is forcing us to live with this virus for a probably very long time. COVID‐19 is a complex disorder, with the lung being the most frequently affected site. Manifestations of COVID‐19 range from mild upper respiratory illness to severe bilateral pneumonia, acute respiratory distress syndrome (ARDS), disseminated thrombosis, multi‐organ failure, and death. 1 , 2 , 3 However, not all infected individuals develop a clinically significant disease. Early identification of healthy virus carriers has been the cornerstone of campaigns to prevent infection spread since Germany's first report which demonstrated their potential contagiousness. 4
SARS‐CoV‐2 is a lipid‐enveloped positive‐sense RNA virus that uses a heavily glycosylated spike (S) protein to attach to the cell membrane and penetrate the host cell. 5 , 6 , 7 The S protein consists of two functional subunits: S1 contains the receptor‐binding domain that allows the engagement of the angiotensin‐converting enzyme (ACE)‐2 on the target cell surface, while S2 is involved in the virus‐cell fusion process. 7 , 8 As ACE‐2 is highly expressed in trachea and bronchial epithelia, 9 in type II alveolar cells, 10 and in nasal epithelial cells, 11 these locations have a key role in the initial viral infection and spread. Nevertheless, ACE‐2 expression in additional tissue/organ‐specific cells makes them at potentially high risk for infection. 12 Likewise, infection by SARS‐CoV‐2 can occur regardless of ACE‐2 expression, 13 through alternative receptors including C‐type lectin receptors (CLR), toll‐like receptors (TLR), neuropilin (NRP)‐1, dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing non‐integrin (DC‐SIGN), homolog dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing non‐integrin related (L‐SIGN), and macrophage galactose‐type lectin (MGL). 13 , 14
Pathogenesis of COVID‐19 is a puzzle not fully elucidated. 15 , 16 , 17 , 18 However, based on clinical and experimental data, there is growing evidence that disease phenotypes are strongly related to the degree of the immune system's dysregulation in response to the virus, as depicted in detail in the review by Sokolowska et al 19 Briefly, innate immune cells act as the first line of defense through the recognition of pathogen‐associated molecular patterns (PAMPs), which allow the activation 20 , 21 of signaling pathways leading to the expression of anti‐viral molecules, including interferons (IFNs), interferon‐stimulated genes (ISGs), and inflammatory chemokines and cytokines. 20 , 22 However, when the virus‐immune system cross talk is out of control, a detrimental systemic inflammatory reaction, named “cytokine storm,” can occur in infected individuals along with a simultaneous impairment of the adaptive immune response. 15 , 20 , 23 Unfortunately, this scenario is not an infrequent occurrence in the COVID‐19 patient and is burdened by devastating effects because the host cannot efficiently counter the virus while harming itself. 24 , 25 In line with these considerations, a first Chinese report showed that 99 subjects investigated in Wuhan had increased total neutrophils (38%), reduced total lymphocytes (35%), increased serum levels of interleukin (IL)‐6 (52%), and increased c‐reactive protein (CRP) (84%). 25 , 26 Also, high amounts of IL‐1β, interferon (IFN)‐γ, IFN induced protein‐10 (IP‐10), and monocyte chemoattractant protein (MCP)‐1 were observed in infected patients, resulting in activated T‐helper (Th)‐1 immune responses. Interestingly, higher levels of IL‐6, granulocyte‐colony stimulating factor (G‐CSF), IP‐10, MCP‐1, macrophage inflammatory protein (MIP)‐1α, and tumor necrosis factor (TNF)‐α were associated with disease severity. 25 , 26 A retrospective study performed in 452 SARS‐CoV‐2‐infected patients has demonstrated that depletion of CD4+ T cells was more prominent in advanced disease. Conversely, there were no significant changes in the number of CD8+T cells and B lymphocytes. 27 An additional report on 140 COVID‐19 patients further suggested low blood counts of eosinophils and lymphocytes as potential markers for diagnosis purposes. 19 , 28 However, the pathophysiological mechanism of these observations merits further study. In particular, when looking at lymphopenia (i.e., apoptosis, virus‐mediated cytopathic effect) it seems that it occurs along with functional T‐cell defects. 19 There is evidence that T‐helper 17 cells actively participate to the cytokine storm response, 29 while T regulatory cells are decreased. 30 Also, highly activated lymphocytes exhibit an exhausted phenotype with high PD‐1 expression, which decreases with virus clearance along with cell function restoration. 31 Finally, a further explanation of impairment of T‐cell functionality and anti‐viral activities may rely on a defective cell priming upon specific antigen (Ag) presentation. In this issue, regulation of the immune response through the conversion of type‐1 to type‐2, activation of M2 macrophages, and maturation of dendritic cells (DCs) by mesenchymal stem cells has been proposed as an alternative therapeutic strategy in COVID‐19. 32
Notably, DCs and macrophages are the main components of the mononuclear phagocytic system. In particular, DCs are the most specialized and potent Ag‐presenting cells (APCs) of the immune system. They promote the activation of naive and memory T cells, thus polarizing the immune response. Broadly studied in different experimental and internal medicine areas, including transplantation, allergy, autoimmunity, infectious diseases, and cancer, 33 , 34 , 35 , 36 DCs also play a crucial role in the host defense against viruses and exert tolerogenic activities. 37
Given the multiple and pleiotropic properties of DCs and their “bridge” position between innate and adaptive immunity, the purpose of this review is to put together the available data concerning the involvement of these cells in SARS‐CoV‐2 infection. This effort aims to better understand the pathogenesis of COVID‐19 and make considerations on the potential use of DCs in immune‐based treatments to counter it.
2. OVERVIEW ON DENDRITIC CELL CLASSIFICATION AND FUNCTION
DCs encompass a heterogeneous family of bone marrow‐derived cells and are found in peripheral blood, lymphoid organs, and tissues. According to the last DC classification, conventional DCs (cDCs) and plasmacytoid DCs (pDCs) are the two main functional DC subtypes. 38 , 39 Circulating DCs include CD11C+ cDCs, that are cDC2 CD1C+ and cDC1 CD141+ cells, and CD11C− pDCs, that are CD123+ and CD303+ cells. 38 , 39 Conventional DCs play a fundamental role in the Ag processing and presentation to lymphocytes. On the other, pDCs display high anti‐viral activities due to their ability to produce type I interferon (IFN) and are thought to be involved in immune tolerance. 40 , 41 Conventional DCs encompass a new subset with monocyte‐like characteristics, named monocyte‐derived (mo)‐DCs or DC3. 42 This DC subtype expresses CD11c, CD14, CD1c, CD163, receptor of the Fc region of immunoglobulins E (FcεR)‐1, IFN regulatory factor (IRF)‐4, and Zbtb64. Mo‐DCs express high levels of tumor necrosis factor (TNF) and C‐C motif chemokine ligand 2 (CCL2), prompt the activation of tissue‐resident memory cells, are engaged in the inflammatory site and endowed with a strong potential to regulate tumor immunity. 43 , 44 , 45 , 46
In reality, human DC subsets are quite heterogeneous with still little agreement on the identification and naming of new subtypes. This makes their classification a dynamic process with the need for continuous updating. About that, the finer characterization of at least two subtypes of cDCs2 (A and B) seems to have gained consensus more recently. Conventional DCs2‐A express higher levels of CD11c, CD1c, and major histocompatibility complex (MHC) class II genes along with CD32+ and CD5. Previously named CD3, cDCs2‐B are CD14+/CD36+/CD163+ and express low levels of CD5 (CD5low) and high amounts of inflammatory cytokines. 39 , 47 , 48 High‐dimensional cytometry has permitted to redraw the blood cDC2 phenotype in CD1c+ BTLA+ with low (cDC2‐B) to high expression of CD5 (previously cDC2‐A), and DC3 phenotype in CD88−CD1c+CD163+ with low to high expression of CD14, improving functional description of DC3 as a lineage of inflammatory DCs holding a strong potentiality to regulate anti‐tumor cytotoxic T‐cell immune responses. 43 , 45 , 46 Additionally, single‐cell RNAseq studies have identified a further subtype of CD16+/CD141−/CD1c− DCs that cluster separately from monocytes. 42 The expression of a broad range of toll‐like receptors (TLR) and inflammatory cytokines upon lipopolysaccharide/IFN‐γ stimulation makes these cells as ideal immune sentinels for detecting invading pathogens. 49 Finally, the existence in the human system of non‐canonical DC subtypes with merged characteristics of cDCs and pDCs has also been proposed. This is the case of blood CD123+ cells expressing the distinctive markers AXL and SIGLEC6. 42 , 50 While their identity is still unclear, AXL+SIGLEC6+ CD123low CD11chigh DCs look closely related to cDCs2. By the other, AXL+SIGLEC6+ CD123high CD11clow DCs are phenotypically similar to pDCs but functionally assimilable to cDCs2. 42 , 45 , 50
With reference to tissue‐resident DCs, they are subdivided into lymphoid and non‐lymphoid. Lymphoid‐resident DCs are usually found in the thymus, spleen, and lymph nodes and consist of phenotypically different subtypes able to process and present antigens entering these sites. 51 Specific tissue localization discriminates non‐lymphoid DCs, into distinct cell subsets such as Langerhans cells (LC), dermal DCs in the skin, and interstitial DCs in various tissues. Mucosal surface‐associated DCs are present in the mucosa of the oral cavity, intestinal and respiratory tracts. These cells develop from blood precursors and continuously re‐circulate from tissues to lymph nodes where they present tissue‐derived Ags to T cells. 51 , 52
Figure 1 summarizes the current classification of DCs.
FIGURE 1.
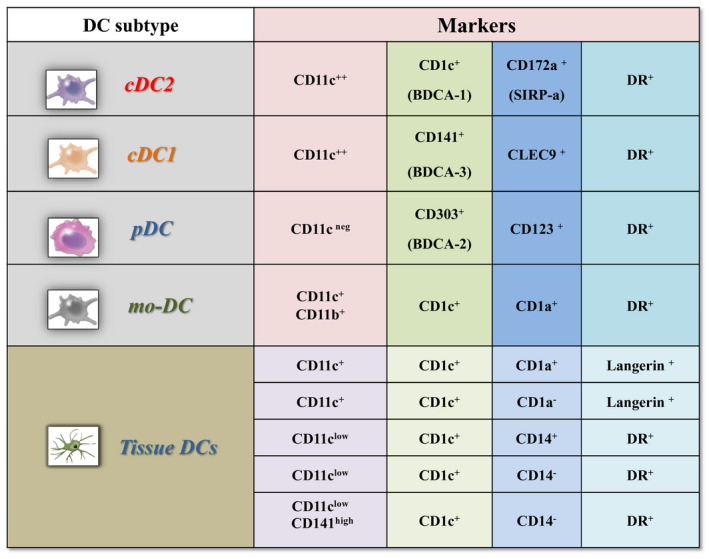
Dendritic cell classification. A systematic summary of dendritic cell classification according to the different expressions of surface markers. Abbreviations: DC, dendritic cell; mo‐DC, monocyte‐derived‐DC; BDCA, Blood Dendritic Cells Antigens; SIRPα, Signal regulatory protein α
Phenotypic profiles and functional pathways are different in naïve (immature) and mature DCs. Immature DCs display low levels of MHC and co‐stimulatory molecules. They are mainly located in barrier tissues, such as the skin, the mucosal surfaces, and lymphoid organs, where they act as sentinels committed to Ag uptake and processing. Tissue damage, inflammatory processes, microorganisms, and tumor‐derived products may promote the maturation of DCs. After that, DCs lose their endocytic activity and acquire the ability to produce a wide array of immune‐stimulating cytokines, such as IL‐12 and type I IFN, which in turn regulate the activity of effector innate immune cells. Moreover, mature DCs possess an increased migratory potential to lymph nodes and lymphoid organs and up‐regulate the expression of CD80‐CD86 co‐stimulatory molecules to drive adaptive immune responses through an efficient Ag presentation to CD4 and CD8 T lymphocytes. 53 , 54
3. DENDRITIC CELLS ARE TARGETS OF SARS‐CoV‐2
A large number of studies have amply demonstrated the crucial role that DCs play in infectious diseases. Accordingly, these cells are highly likely to also contribute to the pathogenesis of SARS‐CoV‐2 infection. 55 , 56 , 57 As previously mentioned, ACE‐2 is an essential receptor for SARS‐CoV‐2 entry into host cells through the interaction with the spike protein. ACE‐2 expression by interstitial lung DCs suggests that these cells can be infected by SARS‐CoV‐2. 8 Even though DCs express intermediate ACE‐2 levels, the infection could also occur due to micropinocytosis or interaction with the dipeptidyl peptidase (DPP) 4 receptor. 8 , 13 More recently, Wang et al have shown that CD147, a transmembrane glycoprotein, can act as a new receptor allowing SARS‐CoV‐2 entry in DCs. 58 CD147 and ACE‐2 expressions are exclusively independent on the cell surface, suggesting that they could mediate virus infection with complementary functions. In particular, loss of CD147 or blocking CD147 can inhibit SARS‐CoV‐2 replication, while CD147 over‐expression promotes virus entry. 58 Dendritic cells also express DC‐SIGN, which is specialized in the recognition of viral proteins with high mannose glycans. 59 Cumulative evidence has shown that both DC‐SIGN and L‐SIGN were involved in the enhancement of SARS‐CoV infection. However, similarly to human immunodeficiency virus (HIV) infection, 60 binding to DC‐SIGN promoted SARS‐CoV transmission from DCs to susceptible targets. 61 , 62 , 63 , 64 In line with these findings, it has been recently reported that SARS‐CoV‐2 upregulates the expression of DC‐SIGN in mo‐DCs thus allowing the trans‐infection of other cell types. 65
4. CURRENT EVIDENCE OF DC INVOLVEMENT IN SARS‐CoV‐2 INFECTION
The subtle interplay between sentinel DCs and SARS‐CoV‐2 remains elusive, while some similarities are likely with the SARS‐CoV infection model. DC maturation was impaired by SARS‐CoV, hampering an appropriate activation of anti‐viral adaptive immunity through a decreased expression of class I and II MHC and co‐stimulatory molecules (CD40 and CD86). 66 Also, SARS‐CoV failed to trigger a significant production of anti‐viral cytokines such as IFN‐α, IFN‐β, or IFN‐γ by infected DCs, promoting a moderate increase of pro‐inflammatory cytokines including TNF‐α and IL‐6. Similarly, SARS‐CoV‐infected DCs displayed a significant increment of inflammatory chemokines such as MIP‐1α, regulated on activation normal T cell expressed and secreted (RANTES), IP‐10, and MCP‐1. Finally, DCs were suggested to serve as a SARS‐CoV reservoir, providing more continuous exposure to the virus and contributing to the infection's persistence and chronicity. 62 , 63
In line with these observations, viral proteins released during SARS‐CoV‐2 infection are not able to induce the secretion of anti‐viral type I (IFN‐α and IFN‐β), II (IFN‐γ), or III (IFN‐λ1) IFNs in mo‐DCs and macrophages. 65 Likewise, SARS‐CoV‐2‐infected lung DCs and macrophages are leading sources of pro‐inflammatory cytokines that worsen the clinical manifestation of COVID‐19. 65 The attenuation of IFN‐related host responses is consistent with previous findings in SARS‐CoV‐2‐infected patients. 67 An effective innate immune response against viral infection consists of an efficient secretion of type I IFN, preventing viral replication pushing toward an effective adaptive immune response. 68 , 69 It has been estimated that about the 15% of patients affected by life‐threatening COVID‐19 pneumonia displayed a defective type I IFN immunity. This defect is characterized by inborn errors of TLR‐3 and IFN regulatory factor (IRF) 7‐dependent type I IFN production 70 and preexisting neutralizing autoantibodies to type I IFN. 71 All nucleated cells can produce type I IFN upon viral infection. However, pDCs are the only ones that constitutively express high levels of IRF‐7 and produce more type I IFN than other cell types. 72 A recent study has shown that pDCs are resistant to SARS‐CoV‐2 infection while are efficiently activated by the virus regardless of ACE‐2 expression. In particular, SARS‐CoV‐2 particles promote pDCs diversification in three activated subsets (P1, P2, and P3) that may contribute to type I IFN‐dependent immunity against SARS‐CoV‐2 infection. 73
Therefore, acute COVID‐19 patients could display DC defects acquired during the disease's progression that are not directly related to the virus itself. Proportions of conventional and plasmacytoid DC subsets have been found lower in severe COVID‐19 patients than in healthy individuals, 74 , 75 with restoration of DC impairment during convalescence. 75 Similarly, the recovery of pre‐DC2 and DC3 absolute numbers was reported in severe COVID‐19 patients following virus clearance and seroconversion. 76 Single‐cell RNA sequencing analysis has furtherly shown that decreased proportions of pDCs and cDCs were detectable in the peripheral blood of eight patients with COVID‐19. Notably, there was a significant depletion of cDCs in samples from patients with adult respiratory distress syndrome (ARDS). 77 Accordingly, the proportions and activation profiles of DC subtypes have also been evaluated in 64 COVID‐19 patients stratified according to different clinical severity levels. Data showed a cDC2 depletion in peripheral blood related to specific recruitment to the lungs in patients with severe COVID‐19. On the contrary, blood circulating cDC1 and pDCs were dramatically reduced in all COVID‐19 patients regardless of their clinical status and were undetectable in the lungs of cases developing severe ARDS. 78 Frequencies of circulating pDCs were also reduced in additional 50 SARS‐CoV‐2‐infected patients regardless of disease severity. 79 Interestingly, new findings of multi‐process defects affecting the pDCs displayed both increased pro‐apoptotic pathways and decreased viral innate sensing, through loss of TLR7 and DEAH‐Box Helicase (DHX)36. Also, cDC2 effector functions could be impaired through a decrease of MHC class II‐related genes and of MHC class II trans‐activator activity, suggesting viral inhibition of Ag presentation in severe COVID‐19. These mechanisms uncovered previously unknown defects related to both innate and adaptive immunity in SARS‐Cov‐2 infection. 80 In this regard, it has been demonstrated that low frequencies of CD8+ T lymphocytes predict high mortality and clinical severity of COVID‐19 pneumonia. 81 , 82 In analogy with these findings, it may be hypothesized that any progressive immune‐associated injury and inadequacy could have detrimental effects on acute COVID‐19 outcomes. Based on this assumption, a recent study performed in 41 COVID‐19 patients, including 17 acute and 24 convalescent cases, revealed that the frequency and functionality of blood circulating DC subsets were significantly reduced in patients compared to healthy controls. Also, patients with acute disease displayed proportions of cDCs significantly lower than pDCs. Reduction of DCs was associated with functional impairment for maturation and CD8+T‐cell activation, resulting in an imbalance between the increase of specific antibodies against SARS‐CoV‐2 and the loss of T‐cell responses during the acute phase of the infection. 83 Similarly, early (first week) reduction of mature CD80+/CD86+ cDCs and total pDCs has been correlated with disease severity. 84 Also, high‐dimensional flow cytometry revealed an affected developmental phenotype in the cDC2 lineage and expansion of monocyte‐myeloid derived suppressor cells (mo‐MDSC)‐like which allowed the discrimination of different patient clusters, with an IFN‐imprint in cDCs1 in all patients and a decreased CD200R expression in pre‐DCs, cDCs2, and DC3. Interestingly, changes in circulating monocyte and DC lineages were mirrored in the lung tissue. 76
The abundant expression of ACE‐2 in the lung and small intestine epithelia makes these organs the gateway to the virus. However, additional targets include the liver, heart, kidney, skin, muscle, vessels, and central nervous system. 85 , 86 To the best of our knowledge, data on tissue DCs in SARS‐CoV‐2 infection/COVID‐19 are scare. Activated mast cells and DCs along with neutrophils were increased in the broncho‐alveolar lavage of COVID‐19 patients than in healthy controls. 87 Conversely, depletion of DC subsets and reduced expression of pro‐inflammatory cytokines were found in the gastro‐intestinal (GI) tract of good prognosis COVID‐19 patients presenting only GI symptoms. 88 Unlike natural killer cells and B lymphocytes, CD11c+ DCs and CD11b+ macrophages specifically infiltrated the lung and intestine and not the liver and kidney of three COVID‐19 patients. Both cell types did not express the leukocyte activation marker DR with the local milieu being characterized by increased TNF‐α and IL‐10 expression. 86
In conclusion, through a synoptic evaluation of the presented data, it could be speculated that the reduced numbers of DCs and the dysregulated IFN signaling may be involved in the impaired progression from innate to adaptive immunity as evasion strategies adopted by SARS‐CoV‐2 with significant impact on disease outcomes. It is possible to assume that this scenario may contribute to the failure of the so‐called trained immunity to keep viral infection under control and avoid hyper‐inflammation, in analogy with other models of infection of DCs. 89 , 90 Interestingly, the weakness of this process, which enables “memory” innate immune cells to quickly respond to unrelated recalling stimuli through the epigenetic reprogramming of IFN and immune signaling, has been linked to recognized risk factors for a worst COVID‐19 prognosis, including older age, comorbidities, and male gender. 91 With respect to this last issue, epidemiological data have clearly shown sex‐based differences in COVID‐19 outcome, with men accounting for about 70% of deaths. 92 The sex‐based difference in antibody response is a deeply characterized immunological process that may account for this observation as well. Interestingly, it has been hypothesized that X‐linked immune genes involved in pDC‐mediated type I IFN signaling occur more efficiently in females leading to the efficient production of SARS‐CoV‐2 neutralizing antibodies. 71 The ability of estrogen to stimulate DCs is an additional protective factor that may help explain why women are affected but less severe forms of COVID‐19 with reduced mortality. 93
An overview of the DC involvement in SARS‐CoV‐2 infection is illustrated in Figure 2.
FIGURE 2.
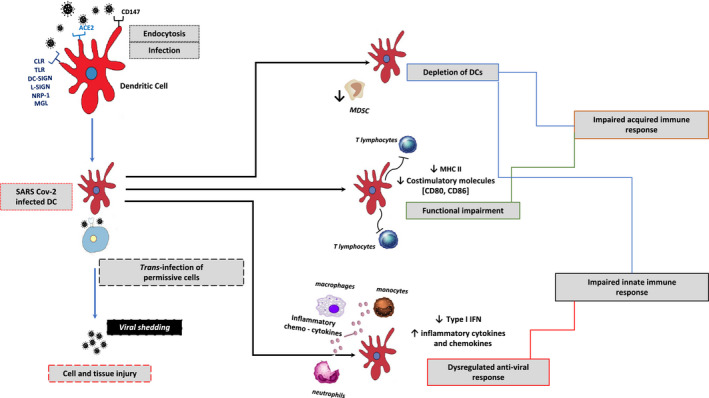
Overview of dendritic cell involvement in SARS‐CoV‐2 infection. SARS‐CoV‐2 can infect human DCs through the engagement of different receptors apart from ACE‐2. The virus binding to DC‐SIGN and L‐SIGN promotes the infection spreading from DCs to trans‐infected permissive cells. As a result of SARS‐CoV‐2 infection, quantitative reduction and functional impairment of DCs lead to a dysregulation of both innate and adaptive immune responses. Overall, these alterations are likely to contribute to the cytokine storm and a reduced host ability to counteract the virus dissemination, which may ultimately account for an uncontrolled disease progression. Abbreviations: DC, dendritic cell; ACE, angiotensin‐converting enzyme; CLR, C‐type lectin receptors; TLR, toll‐like receptor; NRP, neuropilin; MLG, macrophage galactose‐type lectin; DC‐SIGN, dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing non‐integrin; L‐SIGN, homolog dendritic cell‐specific intercellular adhesion molecule‐3‐grabbing non‐integrin related; IFN, interferon; MHC, major histocompatibility complex; mo‐MDSC, monocyte‐myeloid derived suppressor cells
5. DENDRITIC CELLS AND COVID‐19 IN AGED PATIENTS
The SARS‐CoV‐2‐related pandemic has demonstrated a striking demographic bias in the number of cases and deaths affecting the elderly subjects and especially those with comorbidities with an increased risk for worst prognosis and mortality. Older patients for these characteristics also represent a sensitive target in the case of vaccination against COVID‐19 for the greater risk of anaphylactic reactions, as recently reported by Bousquet et al 2 , 94
Immune‐senescence results from the imbalance between inflammatory and anti‐inflammatory mechanisms that lead to chronic inflammation with a significant impact on aged individuals’ survival and fragility. Interestingly, immune‐senescence is part of the so‐called "inflamm‐aging," which is characterized by reduced ability to tolerate an inflammatory milieu and increased production of inflammatory cytokines, acute‐phase proteins, and oxidative stress. 95 , 96 It has been shown that immune‐senescence can influence the function of cDCs upon stimulation with a TLR‐7 ligand, affecting the activation of naive CD8+ T lymphocytes and the generation of effector cytotoxic T cells. 97 Furthermore, DC dysfunction occurring in aged individuals promotes the expression of pro‐inflammatory cytokines and reduces that of anti‐inflammatory and immune‐regulatory cytokines. In particular, impairment of type I and III IFNs secretion increases the aged subjects’ susceptibility to viral and bacterial infections. 95 Given these findings, it is likely that any functional alteration of DCs may be contributing to an increase in the susceptibility to severe COVID‐19 clinical phenotypes and poor outcomes in the elderly. Of course, these aspects deserve further study for a better characterization of the hypothetical phenomena.
6. DENDRITIC CELLS AND COVID‐19 IN CHILDREN
To date, prevalence of SARS‐CoV‐2 infection is lower in children than in adults. Most infected children are asymptomatic or have mild symptoms not requiring hospitalization. 98 , 99 Several hypotheses have postulated children's lower risk of being infected by SARS‐CoV‐2, including the different nature of the innate immune response, 100 but a comprehensive explanation is still lacking. It is likely that children have less inflammation driven by PAMP activation along with a type‐2 skewed immunity. These features while decrease the host protection against the virus are, however, beneficial as they guarantee an hypo‐inflammatory milieu. 101
Interestingly, some studies have reported small percentages of children developing a clinical picture named multisystem‐inflammatory syndrome in children (MIS‐C), emerging as a late manifestation of SARS‐CoV‐2 infection. 102 MIS‐C is characterized by persistent fever and systemic hyper‐inflammation, with clinical features resembling the Kawasaki disease. Recent evidence suggests that decreased levels of innate immune cells including pDCs and low expression levels of HLA‐DR and CD86 on monocytes and DCs may account for an impairment of Ag presentation in these patients. 102 , 103 A comparison study between MIS‐C and COVID‐19 children has revealed that circulating pDCs were more severely decreased in the former than in the latter. 104 Interestingly, COVID‐19 children also showed higher IFN‐α plasma levels and increased expression of type I IFNs inducible genes than MIS‐C patients, clearly strengthening the key role that pDCs have in anti‐viral immunity. 69
7. DENDRITIC CELLS AND COVID‐19 IN PATIENTS SUFFERING FROM ALLERGIC DISEASES
Current evidence does not suggest a higher risk for severe COVID‐19 in allergic subjects. 105 One explanation may rely on the finding that asthma and allergic respiratory disorders have been associated with a significant reduction of ACE‐2 expression in nasal and bronchial cells, both in adults and children. 19 , 106 However, dysfunction of epithelial barriers in allergic diseases may predispose patients to viral infections. 105 , 107 , 108 This because the allergic environment itself can affect the innate immune system, including DCs. Data show that pDCs from atopic subjects and asthmatics have an impaired IFN‐α secretion upon ex vivo exposure to influenza viruses or rhinoviruses. 109 , 110 This defect is inversely related to IgE serum levels which in turn are correlated with the strinking decrease of TLR expression on pDCs in asthmatics, clearly explaining their increased susceptibility to respiratory viral infections triggering exacerbation events. 107 , 110 , 111 Treatments targeting type‐2 immune responses, that is, omalizumab, by blocking free IgE allow to recover the ability of pDCs to produce IFN‐α thus improving anti‐viral activity and reducing disease exacerbation rates. 111 , 112 , 113 , 114 Taken together, these observations suggest that asthmatics may have greater fragility in countering SARS‐CoV‐2 infection due to a predisposing DC dysfunction. However, to the best of our knowledge, no focused studies in this patient population are available. Certainly, additional considerations are of interest with regard to the type‐2 immune response which characterizes these patients. Indeed, it is likely that it may have beneficial effects on the cytokine storm in acute COVID‐19, 15 , 20 , 23 through the antagonization of pro‐inflammatory cytokines, while contrasting the virus spreading through the inhibition of ACE‐2 expression. 115 As a consequence, inhibition of type‐2 immune responses in severe and critical COVID‐19 cases may cause an aggravation of the disease. In line with this feeling, there is indication that all biological therapies should be discontinued in allergic patients with COVID‐19 until viral clearance is achieved. 105 Conversely, there are no concerns about a decreased tolerability of allergen immunotherapy in the COVID‐19 setting. 116
8. STRATEGIES FOR DC‐BASED THERAPIES AGAINST SARS‐CoV‐2 INFECTION
As of January 2021, 68 anti‐COVID‐19 vaccines have been tested in clinical trials around the world. Four of them have already been approved, while 20 and 24 are still in phase III and phase II clinical trials, respectively. 117 Anti‐COVID‐19 vaccines are mainly using viral vectors, nucleic acids (RNA), protein subunits, and inactivated or killed viruses. Overall, vaccines based on these platforms require adjuvants to improve their ability to induce an immune response since they are less immunogenic than live attenuated virus vaccines. 118
DCs may represent an alternative vaccine candidate thanks to their ability to process and present Ags to immune cells. This option has proven effective in countering cancer and viral infections. 119 In particular, several data have established the clinical safety and potency of DC‐based vaccines to activate natural killer (NK) cells, CD8+ and CD4+ T lymphocytes. 119 Additionally, the faculty of DCs to move between lymphoid and non‐lymphoid tissues and to modulate cytokine and chemokine secretion, thus regulating lymphocyte homing and inflammation, represents an intrinsic essential feature for effective immunotherapy. 120 Personalized vaccines employing ex vivo generated DCs, mainly obtained from peripheral blood mononuclear cells (PBMCs), mo‐DCs, or CD34+ hematopoietic stem cell progenitors loaded with different Ags, have been extensively investigated in numerous preclinical and clinical studies. 119 Although several of these efforts have clearly demonstrated the safety and immunogenicity of these DC‐based vaccines, the clinical benefit was, however, poor. 121 , 122 Next‐generation DC‐based vaccines using naturally circulating DCs have successfully overcame this limitation as blood‐derived specific DC subtypes are superior to their in vitro generated counterparts for therapeutic vaccination purposes. 123 More recently, alternative approaches employing the direct in vivo Ag delivery to DCs by using chimeric proteins and adjuvants to boost the immune response are under investigation in cancer therapy with promising results. 124 , 125 , 126
Some preclinical trials are actually testing DCs in the generation of anti‐COVID‐19 vaccines. The first attempt has adopted the chimeric antigen receptor (CAR) T‐cell strategy which is an immune‐oncology approach. It is under testing in phase I‐II multicenter trials including healthy subjects and infected individuals aged between 6 months and 80 years. In detail, a lentiviral vector system carrying minigenes expressing SARS‐CoV‐2‐related proteins (S, M, E, and N) in combination with the P polyprotein protease gene is used to modulate DCs (LV‐SMENP DCs, NCT04276896). The trial objectives are first to check the vaccine safety, and then to assess the cytotoxic T‐cell responses and clinical improvement of SARS‐CoV‐2‐infected individuals in the same age range.
The second strategy is to test autologous DCs previously loaded ex vivo with the SARS‐CoV‐2 S protein (even pulsed or not with GM‐CSF) in healthy adults >18 years old. The trial aims (enrollment not yet initiated) will be to evaluate the vaccine safety and its efficacy by measuring the levels of circulating anti‐S specific IgG (NCT04386252).
Finally, a novel approach may consist of combining DCs with nanotechnology by using extraordinary functional molecules such as nanoparticles (NP). NPs can specifically activate DCs in vivo to induce anti‐virus immune responses. NP‐based vaccines employ self‐assembling viruses (virus‐like particles), lipids (liposomes), proteins, metals, and polymers which also act as their adjuvant. 127 Numerous pieces of evidence have reported that NP‐based vaccines have a good safety profile and high immunogenic potential, with the ability to target DCs for efficient Ag processing and presentation. 128 In particular, the simultaneous targeting of DC subsets (i.e., DC‐SIGN+ and BDCA3+) by NPs can significantly improve vaccine efficacy against tumors and pathogens through the synergic triggering of T cell–mediated immune responses. 129
It is of interest that some of the most hopeful anti‐COVID‐19 vaccines use nanocarriers for packaging to improve stability and ensure efficient Ag processing. In particular, NVX‐CoV2373, a self‐assembled nanoparticle vaccine derived from the recombinant expression of full‐length S protein by a Baculovirus expression system in moth cells (Novarax), has an efficacy of 89.3% in a phase III clinical trial performed in the UK (NCT04611802). Accordingly, the NP‐based messenger RNA (mRNA) BNT162b2 (BioNtech and Pfizer) and mRNA‐1273 (Moderna) anti‐COVID‐19 vaccines have been shown to be effective in preventing COVID‐19 in randomized placebo‐controlled phase III clinical trials. In this regard, two doses of BNT162b2 have a protective effect of 94.8%. 130 Likewise, the mRNA‐1273 vaccine has an estimated 94.1% effectiveness in preventing COVID‐19, including severe illness. 131 , 132 The prophylactic effectiveness of these vaccines goes together with a favorable safety profile. Indeed, serious allergic reactions to anti‐COVID‐19 vaccines are infrequent events that are likely to primarily occur in subjects with history of allergic reaction to vaccine components. 133
Table 1 schematically illustrates clinical trials adopting anti‐SARS‐CoV‐2 vaccines using DC‐based or DC‐boosting strategies.
TABLE 1.
Clinical trials adopting anti‐SARS‐CoV‐2 vaccines using DC‐based or DC‐boosting strategies
| Vaccine type (description) | Administration route | Phase | Reference | Status |
|---|---|---|---|---|
| DC‐based vaccines | ||||
| Injection and infusion of LV‐SMENP DC vaccine and antigen‐specific CTLs | IM and IN | I‐II | NCT04276896 | Recruiting |
| Autologous DCs loaded ex vivo with SARS‐CoV−2 S protein (AV‐COVID−19), even pulsed or not with GM‐CSF | SC | I‐II | NCT04386252 | Not yet recruiting |
| NP‐based vaccines boosting DC functions | ||||
| Injections of SARS‐CoV−2 recombinant S protein nanoparticle vaccine (SARS‐CoV−2rS) with matrix‐M1 adjuvant or placebo | IM | III | NCT04611802 | Active, not recruiting |
| Injections of nanoparticle‐formulated, nucleoside‐modified RNA vaccineencoding the SARS‐CoV−2 full‐length spike a | IM | III | NCT04368728 | Recruiting |
| Injections of nanoparticle‐formulated, nucleoside‐modified RNA vaccineencoding the SARS‐CoV−2 full‐length spike a | IM | III | NCT04470427 | Active, not recruiting |
Abbreviations: CTLs, cytotoxic T lymphocytes; DC, dendritic cell; EMA, European medicines agency; FDA, food drug administration; GM‐CSF, granulocyte macrophage‐colony stimulating factor; NP, nanoparticle.
Following the approval of regulatory agencies (FDA, EMA, etc.) both BNT162b2 (BioNtech and Pfizer) and mRNA‐1273 (Moderna), are already in use worldwide.
9. CONCLUSIONS
Dendritic cells are key players tailoring innate and adaptive immune responses against viral infections. An increasing number of information begins to emerge about SARS‐CoV‐2 infection. To date, available data show that acute SARS‐CoV‐2 infection leads to a rapid depletion of host DCs along with T‐cell function abnormalities. The immune response's dysregulation could have significant implications in disease pathogenesis, clinical severity, contagiousness, and vulnerability to future virus re‐exposure. Also, these alterations can harm the induction and persistence of immunological memory and the preparation and efficacy of vaccines.
Future efforts should be to analyze in greater detail the interactions between SARS‐CoV‐2 and DC subsets during the different phases of the natural history of the infection/disease. Indeed, a more comprehensive understanding will help to decipher still unclear aspects of pathogenesis while offering the opportunity to identify sensitive targets for new therapies.
CONFLICT OF INTEREST
All authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
All authors contributed to the writing and editing of the review. All authors approved the final version.
ETHICAL APPROVAL AND ETHICAL STANDARDS
Not applicable. This is a review and not an original paper.
INFORMED CONSENT
Not applicable. This is a review and not an original paper.
Galati D, Zanotta S, Capitelli L, Bocchino M. A bird's eye view on the role of dendritic cells in SARS‐CoV‐2 infection: Perspectives for immune‐based vaccines. Allergy. 2021;77:100–110. 10.1111/all.15004
Domenico Galati and Serena Zanotta contributed equally to this study.
Funding information
No funding.
Contributor Information
Domenico Galati, Email: d.galati@istitutotumori.na.it.
Marialuisa Bocchino, Email: marialuisa.bocchino@unina.it.
REFERENCES
- 1. Zhang Q, Wang Z, Lv Y, et al. Clinical features and prognostic factors of patients with COVID‐19 in Henan Province, China. Hum Cell. 2021;34(2):419‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239‐1242. [DOI] [PubMed] [Google Scholar]
- 3. Osuchowski MF, Winkler MS, Skirecki T, et al. The COVID‐19 puzzle: deciphering pathophysiology and phenotypes of a new disease entity. Lancet Respir Med. 2021;9(6):622‐642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019‐nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970‐971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Casalino L, Gaieb Z, Goldsmith JA, et al. Beyond shielding: the roles of glycans in the SARS‐CoV‐2 spike protein. ACS Cent Sci. 2020;6(10):1722‐1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gadanec LK, McSweeney KR, Qaradakhi T, Ali B, Zulli A, Apostolopoulos V. Can SARS‐CoV‐2 virus use multiple receptors to enter host cells? Int J Mol Sci. 2021;22(3):992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell. 2020;183(6):1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215‐220. [DOI] [PubMed] [Google Scholar]
- 9. Bertram S, Heurich A, Lavender H, et al. Influenza and SARS‐coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One. 2012;7(4):e35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hirano T, Murakami M. COVID‐19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52(5):731‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sungnak W, Huang N, Becavin C, Berg M; Network HCALB . SARS‐CoV‐2 entry genes are most highly expressed in nasal goblet and ciliated cells within human airways. ArXiv. 2020:681–687. 10.1038/s41591-020-0868-6 [DOI] [Google Scholar]
- 12. Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single‐cell RNA‐seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019‐nCoV infection. Front Med. 2020;14(2):185‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao C, Zeng J, Jia N, et al. SARS‐CoV‐2 spike protein interacts with multiple innate immune receptors. bioRxiv. 2020:1–26. 10.1101/2020.07.29.227462 [DOI] [Google Scholar]
- 14. Amraie R, Napoleon MA, Yin W, et al. CD209L/L‐SIGN and CD209/DC‐SIGN act as receptors for SARS‐CoV‐2 and are differentially expressed in lung and kidney epithelial and endothelial cells. bioRxiv. 2020:1–27. 10.1101/2020.06.22.165803 [DOI] [Google Scholar]
- 15. Pedersen SF, Ho YC. SARS‐CoV‐2: a storm is raging. J Clin Invest. 2020;130(5):2202‐2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi Y, Wang Y, Shao C, et al. COVID‐19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27(5):1451‐1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vellingiri B, Jayaramayya K, Iyer M, et al. COVID‐19: a promising cure for the global panic. Sci Total Environ. 2020;725:138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sokolowska M, Lukasik ZM, Agache I, et al. Immunology of COVID‐19: mechanisms, clinical outcome, diagnostics, and perspectives‐a report of the European academy of allergy and clinical immunology (EAACI). Allergy. 2020;75(10):2445‐2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID‐19 cytokine storm; what we know so far. Front Immunol. 2020;11:1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thompson MR, Kaminski JJ, Kurt‐Jones EA, Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3(6):920‐940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39(5):529‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parackova Z, Zentsova I, Bloomfield M, et al. Disharmonic inflammatory signatures in COVID‐19: augmented neutrophils’ but impaired monocytes’ and dendritic cells’ responsiveness. Cells. 2020;9(10):2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen G, Wu DI, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620‐2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID‐19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1‐9. [DOI] [PubMed] [Google Scholar]
- 27. Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with COVID‐19 in Wuhan, China. Clin Infect Dis. 2020;71(5):762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy. 2020;75(7):1730‐1741. [DOI] [PubMed] [Google Scholar]
- 29. Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420‐422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gursel M, Gursel I. Is global BCG vaccination‐induced trained immunity relevant to the progression of SARS‐CoV‐2 pandemic? Allergy. 2020;75(7):1815‐1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Diao BO, Wang C, Tan Y, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID‐19). Front Immunol. 2020;11:827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mallis P, Michalopoulos E, Chatzistamatiou T, Stavropoulos‐Giokas C. Mesenchymal stromal cells as potential immunomodulatory players in severe acute respiratory distress syndrome induced by SARS‐CoV‐2 infection. World J Stem Cells. 2020;12(8):731‐751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galati D, Zanotta S, Canora A, et al. Severe depletion of peripheral blood dendritic cell subsets in obstructive sleep apnea patients: a new link with cancer? Cytokine. 2020;125:154831. [DOI] [PubMed] [Google Scholar]
- 34. Galati D, Zanotta S, Polistina GE, Coppola A, Capitelli L, Bocchino M. Circulating dendritic cells are severely decreased in idiopathic pulmonary fibrosis with a potential value for prognosis prediction. Clin Immunol. 2020;215:108454. [DOI] [PubMed] [Google Scholar]
- 35. Galati D, Zanotta S, Corazzelli G, et al. Circulating dendritic cells deficiencies as a new biomarker in classical Hodgkin lymphoma. Br J Haematol. 2019;184(4):594‐604. [DOI] [PubMed] [Google Scholar]
- 36. Galgani M, Fabozzi I, Perna F, et al. Imbalance of circulating dendritic cell subsets in chronic obstructive pulmonary disease. Clin Immunol. 2010;137(1):102‐110. [DOI] [PubMed] [Google Scholar]
- 37. Steinman RM. Decisions about dendritic cells: past, present, and future. Annu Rev Immunol. 2012;30:1‐22. [DOI] [PubMed] [Google Scholar]
- 38. Ziegler‐Heitbrock L, Ancuta P, Crowe S, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116(16):e74‐e80. [DOI] [PubMed] [Google Scholar]
- 39. Mildner A, Jung S. Development and function of dendritic cell subsets. Immunity. 2014;40(5):642‐656. [DOI] [PubMed] [Google Scholar]
- 40. Haniffa M, Collin M, Ginhoux F. Ontogeny and functional specialization of dendritic cells in human and mouse. Adv Immunol. 2013;120:1‐49. [DOI] [PubMed] [Google Scholar]
- 41. Dai H, Thomson AW, Rogers NM. Dendritic cells as sensors, mediators, and regulators of ischemic injury. Front Immunol. 2019;10:2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Villani AC, Satija R, Reynolds G, et al. Single‐cell RNA‐seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356(6335):eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dutertre CA, Becht E, Irac SE, et al. Single‐cell analysis of human mononuclear phagocytes reveals subset‐defining markers and identifies circulating inflammatory dendritic cells. Immunity. 2019;51(3):573‐589.e8. [DOI] [PubMed] [Google Scholar]
- 44. Collin M, Bigley V. Human dendritic cell subsets: an update. Immunology. 2018;154(1):3‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bourdely P, Anselmi G, Vaivode K, et al. Transcriptional and functional analysis of CD1c(+) human dendritic cells identifies a CD163(+) subset priming CD8(+)CD103(+) T cells. Immunity. 2020;53(2):335‐352.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cytlak U, Resteu A, Pagan S, et al. Differential IRF8 transcription factor requirement defines two pathways of dendritic cell development in humans. Immunity. 2020;53(2):353‐370.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yin X, Yu H, Jin X, et al. Human blood CD1c+ dendritic cells encompass CD5high and CD5low subsets that differ significantly in phenotype, gene expression, and functions. J Immunol. 2017;198(4):1553‐1564. [DOI] [PubMed] [Google Scholar]
- 48. Korenfeld D, Gorvel L, Munk A, et al. A type of human skin dendritic cell marked by CD5 is associated with the development of inflammatory skin disease. JCI Insight. 2017;2(18):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rhodes JW, Tong O, Harman AN, Turville SG. Human dendritic cell subsets, ontogeny, and impact on HIV infection. Front Immunol. 2019;10:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. See P, Dutertre CA, Chen J, et al. Mapping the human DC lineage through the integration of high‐dimensional techniques. Science. 2017;356(6342):eaag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Galati D, Corazzelli G, De Filippi R, Pinto A. Dendritic cells in hematological malignancies. Crit Rev Oncol Hematol. 2016;108:86‐96. [DOI] [PubMed] [Google Scholar]
- 52. Sato K, Fujita S. Dendritic cells: nature and classification. Allergol Int. 2007;56(3):183‐191. [DOI] [PubMed] [Google Scholar]
- 53. Bertho N, Adamski H, Toujas L, Debove M, Davoust J, Quillien V. Efficient migration of dendritic cells toward lymph node chemokines and induction of T(H)1 responses require maturation stimulus and apoptotic cell interaction. Blood. 2005;106(5):1734‐1741. [DOI] [PubMed] [Google Scholar]
- 54. Ohl L, Mohaupt M, Czeloth N, et al. CCR7 governs skin dendritic cell migration under inflammatory and steady‐state conditions. Immunity. 2004;21(2):279‐288. [DOI] [PubMed] [Google Scholar]
- 55. Alamri A, Fisk D, Upreti D, Kung SKP. A missing link: engagements of dendritic cells in the pathogenesis of SARS‐CoV‐2 infections. Int J Mol Sci. 2021;22(3):1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Borges RC, Hohmann MS, Borghi SM. Dendritic cells in COVID‐19 immunopathogenesis: insights for a possible role in determining disease outcome. Int Rev Immunol. 2020;1‐18. [DOI] [PubMed] [Google Scholar]
- 57. Campana P, Parisi V, Leosco D, Bencivenga D, Della Ragione F, Borriello A. Dendritic cells and SARS‐CoV‐2 infection: still an unclarified connection. Cells. 2020;9(9):2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang KE, Chen W, Zhang Z, et al. CD147‐spike protein is a novel route for SARS‐CoV‐2 infection to host cells. Signal Transduct Target Ther. 2020;5(1):283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soilleux EJ, Morris LS, Leslie G, et al. Constitutive and induced expression of DC‐SIGN on dendritic cell and macrophage subpopulations in situ and in vitro. J Leukoc Biol. 2002;71(3):445‐457. [PubMed] [Google Scholar]
- 60. de Witte L, Nabatov A, Geijtenbeek TB. Distinct roles for DC‐SIGN+‐dendritic cells and Langerhans cells in HIV‐1 transmission. Trends Mol Med. 2008;14(1):12‐19. [DOI] [PubMed] [Google Scholar]
- 61. Jeffers SA, Tusell SM, Gillim‐Ross L, et al. CD209L (L‐SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci U S A. 2004;101(44):15748‐15753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marzi A, Gramberg T, Simmons G, et al. DC‐SIGN and DC‐SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J Virol. 2004;78(21):12090‐12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang ZY, Huang Y, Ganesh L, et al. pH‐dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC‐SIGN. J Virol. 2004;78(11):5642‐5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hofmann H, Pohlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12(10):466‐472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang D, Chu H, Hou Y, et al. Attenuated interferon and proinflammatory response in SARS‐CoV‐2‐infected human dendritic cells is associated with viral antagonism of STAT1 phosphorylation. J Infect Dis. 2020;222(5):734‐745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Spiegel M, Schneider K, Weber F, Weidmann M, Hufert FT. Interaction of severe acute respiratory syndrome‐associated coronavirus with dendritic cells. J Gen Virol. 2006;87(7):1953‐1960. [DOI] [PubMed] [Google Scholar]
- 67. Mantlo E, Bukreyeva N, Maruyama J, Paessler S, Huang C. Antiviral activities of type I interferons to SARS‐CoV‐2 infection. Antiviral Res. 2020;179:104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chu H, Chan JFW, Wang Y, et al. Comparative replication and immune activation profiles of SARS‐CoV‐2 and SARS‐CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID‐19. Clin Infect Dis. 2020;71(6):1400‐1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Blanco‐Melo D, Nilsson‐Payant BE, Liu W‐C, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181(5):1036‐1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life‐threatening COVID‐19. Science. 2020;370(6515):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bastard P, Rosen LB, Zhang Q, et al. Autoantibodies against type I IFNs in patients with life‐threatening COVID‐19. Science. 2020;370(6515):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu YJ. IPC: professional type 1 interferon‐producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275‐306. [DOI] [PubMed] [Google Scholar]
- 73. Onodi F, Bonnet‐Madin L, Meertens L, et al. SARS‐CoV‐2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4. J Exp Med. 2021;218(4):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Matic S, Popovic S, Djurdjevic P, et al. SARS‐CoV‐2 infection induces mixed M1/M2 phenotype in circulating monocytes and alterations in both dendritic cell and monocyte subsets. PLoS One. 2020;15(12):e0241097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen Q, Yu B, Yang Y, et al. Immunological and inflammatory profiles during acute and convalescent phases of severe/ critically ill COVID‐19 patients. Int Immunopharmacol 2021;97:107685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kvedaraite E, Hertwig L, Sinha I, et al. Major alterations in the mononuclear phagocyte landscape associated with COVID‐19 severity. Proc Natl Acad Sci U S A. 2021;118(6):e2018587118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wilk AJ, Rustagi A, Zhao NQ, et al. A single‐cell atlas of the peripheral immune response in patients with severe COVID‐19. Nat Med. 2020;26(7):1070‐1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sanchez‐Cerrillo I, Landete P, Aldave B, et al. Differential redistribution of activated monocyte and dendritic cell subsets to the lung associates with severity of COVID‐19. J Clin Invest. 2020;130(12):6290–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID‐19 patients. Science. 2020;369(6504):718‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Saichi M, Ladjemi MZ, Korniotis S, et al. Single‐cell RNA sequencing of blood antigen‐presenting cells in severe COVID‐19 reveals multi‐process defects in antiviral immunity. Nat Cell Biol. 2021;23(5):538‐551. [DOI] [PubMed] [Google Scholar]
- 81. Du RH, Liang LR, Yang CQ, et al. Predictors of mortality for patients with COVID‐19 pneumonia caused by SARS‐CoV‐2: a prospective cohort study. Eur Respir J. 2020;55(5):2000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu BO, Fan CY, Wang AL, et al. Suppressed T cell‐mediated immunity in patients with COVID‐19: a clinical retrospective study in Wuhan, China. J Infect. 2020;81(1):e51‐e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhou R, To KKW, Wong YC, et al. Acute SARS‐CoV‐2 infection impairs dendritic cell and T cell responses. Immunity 2020;53(4):864‐877.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kulkarni‐Munje A, Palkar S, Shrivastava S, Lalwani S, Mishra AC, Arankalle VA. Disease‐duration based comparison of subsets of immune cells in SARS CoV‐2 infected patients presenting with mild or severe symptoms identifies prognostic markers for severity. Immun Inflamm Dis. 2021;9(2):419‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Criado PR, Pagliari C, Carneiro FRO, Quaresma JAS. Lessons from dermatology about inflammatory responses in Covid‐19. Rev Med Virol. 2020;30(5):e2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang C, Xu J, Wang S, et al. Imaging mass cytometric analysis of postmortem tissues reveals dysregulated immune cell and cytokine responses in multiple organs of COVID‐19 patients. Front Microbiol. 2020;11:600989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhou Z, Ren L, Zhang LI, et al. Heightened innate immune responses in the respiratory tract of COVID‐19 patients. Cell Host Microbe. 2020;27(6):883‐890.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Livanos AE, Jha D, Cossarini F, et al. Intestinal host response to SARS‐CoV‐2 infection and COVID‐19 outcomes in patients with gastrointestinal symptoms. Gastroenterology. 2021;160(7):2435–2450.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Prakash S, Agrawal S, Cao JN, Gupta S, Agrawal A. Impaired secretion of interferons by dendritic cells from aged subjects to influenza: role of histone modifications. Age (Dordr). 2013;35(5):1785‐1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ma S, Wan X, Deng Z, et al. Epigenetic regulator CXXC5 recruits DNA demethylase Tet2 to regulate TLR7/9‐elicited IFN response in pDCs. J Exp Med. 2017;214(5):1471‐1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gabriele L, Fragale A, Romagnoli G, et al. Type I IFN‐dependent antibody response at the basis of sex dimorphism in the outcome of COVID‐19. Cytokine Growth Factor Rev. 2021;58:66‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL. Considering how biological sex impacts immune responses and COVID‐19 outcomes. Nat Rev Immunol. 2020;20(7):442‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Laffont S, Seillet C, Guery JC. Estrogen receptor‐dependent regulation of dendritic cell development and function. Front Immunol. 2017;8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bousquet J, Agache I, Blain H, et al. Management of anaphylaxis due to COVID‐19 vaccines in the elderly. Allergy. 2021:1–28. 10.1111/all.14838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Agrawal A, Agrawal S, Gupta S. Role of dendritic cells in inflammation and loss of tolerance in the elderly. Front Immunol. 2017;8:896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Pawelec G. Age and immunity: what is "immunosenescence"? Exp Gerontol. 2018;105:4‐9. [DOI] [PubMed] [Google Scholar]
- 97. Zacca ER, Crespo MI, Acland RP, et al. Aging impairs the ability of conventional dendritic cells to cross‐prime CD8+ T cells upon stimulation with a TLR7 ligand. PLoS One. 2015;10(10):e0140672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhou Y, Guo Y, Liu Y. Health, income and poverty: evidence from China's rural household survey. Int J Equity Health. 2020;19(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zimmermann P, Curtis N. COVID‐19 in children, pregnancy and neonates: a review of epidemiologic and clinical features. Pediatr Infect Dis J. 2020;39(6):469‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lauro R, Irrera N, Eid AH, Bitto A. Could antigen presenting cells represent a protective element during SARS‐CoV‐2 infection in children? Pathogens. 2021;10(4):476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Maddux AB, Douglas IS. Is the developmentally immature immune response in paediatric sepsis a recapitulation of immune tolerance? Immunology. 2015;145(1):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Carter MJ, Fish M, Jennings A, et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS‐CoV‐2 infection. Nat Med. 2020;26(11):1701‐1707. [DOI] [PubMed] [Google Scholar]
- 103. Gruber CN, Patel RS, Trachtman R, et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS‐C). Cell. 2020;183(4):982‐995.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Caldarale F, Giacomelli M, Garrafa E, et al. Plasmacytoid dendritic cells depletion and elevation of IFN‐gamma dependent chemokines CXCL9 and CXCL10 in children with multisystem inflammatory syndrome. Front Immunol. 2021;12:654587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Vultaggio A, Agache I, Akdis CA, et al. Considerations on biologicals for patients with allergic disease in times of the COVID‐19 pandemic: an EAACI statement. Allergy. 2020;75(11):2764‐2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Jackson DJ, Busse WW, Bacharier LB, et al. Association of respiratory allergy, asthma, and expression of the SARS‐CoV‐2 receptor ACE2. J Allergy Clin Immunol. 2020;146(1):203‐206.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Rowe RK, Gill MA. Targeting antiviral pathways for treatment of allergic diseases. J Pediatric Infect Dis Soc. 2018;7(suppl 2):S54‐S56. [DOI] [PubMed] [Google Scholar]
- 108. Kortekaas Krohn I, Seys SF, Lund G, et al. Nasal epithelial barrier dysfunction increases sensitization and mast cell degranulation in the absence of allergic inflammation. Allergy. 2020;75(5):1155‐1164. [DOI] [PubMed] [Google Scholar]
- 109. Gonzales‐van Horn SR, Farrar JD. Interferon at the crossroads of allergy and viral infections. J Leukoc Biol. 2015;98(2):185‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Gern JE. How rhinovirus infections cause exacerbations of asthma. Clin Exp Allergy. 2015;45(1):32‐42. [DOI] [PubMed] [Google Scholar]
- 111. Esquivel A, Busse WW, Calatroni A, et al. Effects of omalizumab on rhinovirus infections, illnesses, and exacerbations of asthma. Am J Respir Crit Care Med. 2017;196(8):985‐992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Palomares O, Untersmayr E, Gutermuth J, et al. Biologicals in allergic diseases and asthma: toward personalized medicine and precision health: highlights of the 3rd EAACI master class on biologicals, San Lorenzo de El Escorial, Madrid, 2019. Allergy. 2020;75(4):936‐940. [DOI] [PubMed] [Google Scholar]
- 113. Teach SJ, Gill MA, Togias A, et al. Preseasonal treatment with either omalizumab or an inhaled corticosteroid boost to prevent fall asthma exacerbations. J Allergy Clin Immunol. 2015;136(6):1476‐1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Holguin F, Cardet JC, Chung KF, et al. Management of severe asthma: a European respiratory society/American thoracic society guideline. Eur Respir J. 2020;55(1):1900588. [DOI] [PubMed] [Google Scholar]
- 115. Wong JJM, Leong JY, Lee JH, Albani S, Yeo JG. Insights into the immuno‐pathogenesis of acute respiratory distress syndrome. Ann Transl Med. 2019;7(19):504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Pfaar O, Agache I, Bonini M, et al. COVID‐19 pandemic and allergen immunotherapy ‐ an EAACI survey. Allergy. 2021;76:3504–3516. 10.1111/all.14793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kaur SP, Gupta V. COVID‐19 vaccine: a comprehensive status report. Virus Res. 2020;288:198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Pollard AJ, Bijker EM. A guide to vaccinology: from basic principles to new developments. Nat Rev Immunol. 2021;21(2):83‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sabado RL, Balan S, Bhardwaj N. Dendritic cell‐based immunotherapy. Cell Res. 2017;27(1):74‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mastelic‐Gavillet B, Balint K, Boudousquie C, Gannon PO, Kandalaft LE. Personalized dendritic cell vaccines‐recent breakthroughs and encouraging clinical results. Front Immunol. 2019;10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Ahmed MS, Bae YS. Dendritic cell‐based therapeutic cancer vaccines: past, present and future. Clin Exp Vaccine Res. 2014;3(2):113‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Anguille S, Smits EL, Bryant C, et al. Dendritic cells as pharmacological tools for cancer immunotherapy. Pharmacol Rev. 2015;67(4):731‐753. [DOI] [PubMed] [Google Scholar]
- 123. Tel J, Aarntzen EHJG, Baba T, et al. Natural human plasmacytoid dendritic cells induce antigen‐specific T‐cell responses in melanoma patients. Cancer Res. 2013;73(3):1063‐1075. [DOI] [PubMed] [Google Scholar]
- 124. Caminschi I, Maraskovsky E, Heath WR. Targeting dendritic cells in vivo for cancer therapy. Front Immunol. 2012;3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. van Dinther D, Stolk DA, van de Ven R, van Kooyk Y, de Gruijl TD, den Haan JMM. Targeting C‐type lectin receptors: a high‐carbohydrate diet for dendritic cells to improve cancer vaccines. J Leukoc Biol. 2017;102(4):1017‐1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Macri C, Dumont C, Johnston AP, Mintern JD. Targeting dendritic cells: a promising strategy to improve vaccine effectiveness. Clin Transl Immunology. 2016;5(3):e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Pati R, Shevtsov M, Sonawane A. Nanoparticle vaccines against infectious diseases. Front Immunol. 2018;9:2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhao L, Seth A, Wibowo N, et al. Nanoparticle vaccines. Vaccine. 2014;32(3):327‐337. [DOI] [PubMed] [Google Scholar]
- 129. Sehgal K, Ragheb R, Fahmy TM, Dhodapkar MV, Dhodapkar KM. Nanoparticle‐mediated combinatorial targeting of multiple human dendritic cell (DC) subsets leads to enhanced T cell activation via IL‐15‐dependent DC crosstalk. J Immunol. 2014;193(5):2297‐2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA covid‐19 vaccine. N Engl J Med. 2020;383(27):2603‐2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Baden LR, El Sahly HM, Essink B, et al. Efficacy and safety of the mRNA‐1273 SARS‐CoV‐2 vaccine. N Engl J Med. 2021;384(5):403‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sokolowska M, Eiwegger T, Ollert M, et al. EAACI statement on the diagnosis, management and prevention of severe allergic reactions to COVID‐19 vaccines. Allergy. 2021;76(6):1629‐1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sampath V, Rabinowitz G, Shah M, et al. Vaccines and allergic reactions: the past, the current COVID‐19 pandemic, and future perspectives. Allergy. 2021;76(6):1640–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]


