Abstract
Glioblastoma (GBM) is one of the most common and malignant types of primary cancer in the central nervous system; however, the clinical outcomes of patients with GBM remain poor. Circular RNAs (circRNAs) have been revealed to serve important roles in diverse biological processes, such as regulating cell proliferation, epithelial-mesenchymal transition and tumor development. However, the underlying biological function of circRNA filamin A (circFLNA) and its potential role in GBM remain to be determined. The present study aimed to identify differentially expressed circRNAs in GBM. Reverse transcription-quantitative PCR was used to analyze the expression levels of circFLNA. The results demonstrated that the expression levels of circFLNA were significantly upregulated in clinical GBM samples and GBM cells compared with adjacent healthy brain tissues and normal human astrocytes, respectively. The results of the Cell Counting Kit-8 and Transwell assays revealed that circFLNA knockdown significantly inhibited the proliferative and invasive abilities of GBM cell lines. Moreover, high circFLNA expression levels were associated with a worse prognosis in GBM. MicroRNA (miR)-199-3p was subsequently predicted to be target of circFLNA. The inhibitory effect of miR-199-3p on cell proliferation and invasion was partially reversed following circFLNA knockdown. In conclusion, the findings of the present study identified novel roles for circFLNA in GBM and indicated that the circFLNA/miR-199-3p signaling axis may serve an important role in GBM progression. Therefore, circFLNA may represent a novel target for the diagnosis and treatment of GBM.
Keywords: glioblastoma, circular RNA, circular RNA filamin A, microRNA-199-3p, proliferation, invasion
Introduction
Glioblastoma (GBM) is one of the most aggressive and malignant types of primary cancer in the central nervous system, accounting for ~3% of all cancers diagnosed worldwide (1) and 90,000 patient deaths per year (2). Currently, the overall survival of patients with GBM is <2 years (3,4). The low survival rate is, at least in part, due to traditional therapies, such as surgical resection, chemotherapy and radiotherapy, being unsatisfactory for the treatment of GBM (5). For example, the recurrence rate of patients following surgery is high and the 2-year survival rate after surgery was found to be <35% (6,7). Therefore, there remains an urgent requirement to determine more efficient GBM diagnostic and prognostic biomarkers, and to develop novel strategies for the treatment of GBM.
Circular RNAs (circRNAs) are a class of non-coding RNAs, which, similar to microRNAs (miRNAs/miRs), lack 5′ or 3′ ends, and form a circular structure with covalent bonds (8). Accumulating evidence has indicated that the aberrant expression of circRNAs may exert important biological functions in the progression of numerous types of cancer, including hepatocellular carcinoma, gastric carcinoma, colorectal cancer and glioma (9,10). In fact, previous studies have demonstrated that dysregulated circRNAs served as novel regulators of cancer progression, including in GBM (11,12). The tumor invasion-associated biomarkers, MMP-2 and MMP-9, belong to the MMP family, and are known to promote extracellular matrix degradation and invasion of cancer cells into adjacent healthy tissues during tumor development (13).
circRNA filamin A (circFLNA), also known as hsa_circ_0092012, is a newly discovered oncogene, that originates from exon 9 to 15 of the FLNA gene and has a spliced mature sequence length of 543 base pairs (14). Aberrant expression levels of circFLNA have been reported in a variety of human cancer types, including gastric cancer, oral squamous cell carcinoma and laryngeal squamous cell carcinoma (14–17). However, to the best of our knowledge, the role of circFLNA in GBM remains to be determined.
miRNAs are small, non-coding RNAs that negatively regulate gene expression levels by binding to the 3′-untranslated region (UTR) of target mRNAs (18). The aberrant expression of miRNAs was found to be associated with the occurrence and progression of a number of human diseases, including breast cancer, glioblastoma, thyroid papillary carcinoma, hepatocellular carcinoma, lung cancer, colon cancer and endocrine pancreatic tumors (18,19). In particular, the expression levels of miR-199-3p have been reported to be downregulated in multiple types of cancer, including GBM (20,21). However, the role of miR-199-3p in GBM progression remains elusive.
Thus, the present study aimed to detect the role and underlying mechanism of circFLNA in glioblastoma and screened the circRNAs-miRNA network that exist during the progression of human.
Materials and methods
Cell lines
Human GBM cell lines (U251, LN229, T98G, A172 and SHG44) were obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. Normal human astrocytes (NHAs) were purchased from American Type Culture Collection. Cells were cultured in DMEM (cat. no. 670087; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (cat. no. 16140071; Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere with 5% CO2 at 37°C.
Patient studies
A total of 50 human GBM and paired adjacent healthy brain tissues were collected from patients with GBM (25 male patients and 25 female patients; median age, 40.5 years; age range, 18–69 years) at Harbin Medical University Cancer Hospital (Harbin, China) between January 2015 and January 2019. Inclusion criteria: i) Newly diagnosed GBM; ii) patients older than 18 years; iii) GBM cases with confirmed pathology; and iv) patients with GBM were treated by surgery. Exclusion criteria: i) GBM cases with unconfirmed pathology; ii) GBM cases with spinal involvement; ii) GBM cases with incomplete data records; and iv) patients receiving chemotherapy and radiotherapy prior to the surgery. All GBM tissues were histopathologically confirmed by two senior pathologists. The patient were divided into the high or low circFLNA and high or low miR-199-3p expression group according to the median expression level of clinical patients with GBM or patients from The Cancer Genome Atlas (TCGA)-GBM database. Informed written consent was obtained from all patients prior to participation. The study was approved by The Institutional Review Board of Harbin Medical University (approval no. 2019HMUIRB0171).
Gene expression profiles of circFLNA expression
The edgeR Bioconductor software package (RStudio, Inc. version.4.1) was used to identify differentially expressed circRNAs in two Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) datasets, GSE92322 (22) and GSE86202 (23). Overall survival data based on the expression levels of miRNAs were obtained from patients with GBM from TCGA database (http://cancergenome.nih.gov). The DEseq2 package (version 3.11; http://bioconductor.org/packages/release/bioc/html/DESeq2.html) was used to detect the differentially expressed circRNAs using the following criteria as significant cut-off values: Log2 fold-change (FC)>2 and false-discovery rate (FDR)<0.01. Clinicopathological data were obtained from 50 patients with GBM who underwent surgical resection. Gene Ontology (GO) functional term enrichment analysis was performed using The Database for Annotation, Visualization and Integrated Discovery version 6.8 (https://david.ncifcrf.gov). Circos plots were constructed using http://www.bioinformatics.com.cn, an online platform for data analysis and visualization.
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was used to analyze the expression levels of circFLNA and miR-199-3p. Briefly, total RNA was extracted from clinical tissues or cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using a PrimeScript™ RT kit (cat. no. RR014A; Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. qPCR for mRNA detection was subsequently performed using a SYBR-Green PCR Master Mix (cat. no. DRR820A; Takara Biotechnology Co., Ltd.). RNase-R was applied to detect the presence of circFLNA and eliminate the influence of linear RNAs. The expression levels of miRNAs were analyzed using a miScript PCR system (cat. no. 339306; Qiagen GmbH). The following primers were used for the qPCR: circFLNA forward, 5′-CCAGCTGAGGCTCTACCGTGCC-3′ and reverse, 5′-GAGGCGTCAGCATCCCCAACAG-3′; miR-199-3p forward, 5′-ACACTCCAGCTGGGTCCCTGAGACCCTTTA-3′ and reverse, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCA-3′; miR-296-5p forward, 5′-ATGGCGGACGAGGAGAAGCTGC-3′ and reverse, 5′-TCACTCAGTGCGGAGGATGATG-3′; miR-515-3p forward, 5-CGGGTTCTCCAAAAGAAAGCA-3′ and reverse, 5-CAGCCACAAAAGAGCACAAT-3′ MMP-2 forward, 5′-CAGGACATTGTCTTTGATGGCATCGC-3′ and reverse, 5′-TGAAGAAGTAGCTATGACCACCGCC-3′; MMP-9 forward, 5′-ATCCCCCACCTTTACCA-3′ and reverse, 5′-TCAGAACCGACCCTACAA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; U1 forward, 5′-GGACTCATCAAGACTCATCA-3′; and reverse, 5′-GTGAGGACGAAACTGCCTTG-3′; and GAPDH forward, 5′-AGGCTGTTGGGAAAGTTCTTC-3′ and reverse, 5′-ACTGTTGGAACTCGGAATGC-3′. The following thermocycling conditions were used for qPCR: Initial denaturation at 94°C for 10 min; followed by 40 cycles of denaturation at 94°C for 5 sec, annealing at 60°C for 30 sec and extension at 72°C for 45 sec. GAPDH and U6 were used as the endogenous controls. The relative gene expression levels were calculated using the 2−ΔΔCq method (24) and normalized to the expression levels of the endogenous controls, GAPDH (for mRNA) and U6 (for miRNA).
Isolation of cytoplasmic and nuclear RNA
Cellular cytoplasmic and nuclear RNA of glioblastoma cells was extracted and purified using a PARIS kit (cat. no. AM1921; Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Cell transfection
Small interfering RNA (siRNA/si)-circFLNA, si-negative control (NC), miR-199-3p mimic, miR-199-3p inhibitor, NC mimic, NC inhibitor, pcDNA3.1-circFLNA and pcDNA3.1 (empty vector) were purchased from Shanghai GenePharma Co., Ltd. The sequences of the constructs were as follows: si-circFLNA forward, 5′-AGCCCCTTCAGGGAGCTGGCA-3′ and reverse, 5′-CAACAGCCCCTTCAGGGAGCT-3′; pcDNA3.1-circFLNA forward, 5′-GUGCCAGCUCCCUGAAGGGTT-3′ and reverse, 5′-GCCAGCUCCCUGAAGGGGCTT-3′; si-NC forward, 5′-GGTAAGCAGTGGCTCCTCTAA-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′; miR-199-3p mimic forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-AGGGCCCCCCCUCAAUCCUGU-3′; miR-199-3p inhibitor forward, 5′-ACAGGAUUGAGGGGGGGCCCU-3′; NC mimic forward, 5′-CAGUACUUUUGUGUAGUACAA-3′ and reverse, 5′-CAGUACUUUUGUGUAGUACAA-3′; and NC inhibitor forward, 5′-GGUAAGCAGUGGCUCCUCUAA-3′ and reverse, 5′-ACGUGACACGUUCGGAGAAUU-3′. Cells were added in 6-well plates at 1×105 cells/well and were transfected with 20 µM miR-199-3p mimic, miR-199-3p inhibitor, si-circFLNA, pcDNA3.1-circFLNA or respective NCs using Lipofectamine® 2000 (cat. no. 11668030; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following transfection at 37°C for 6 h, the culture medium was replaced and cells were subsequently obtained at 24 h post-transfection for further experiments.
Dual luciferase reporter assay
RNA22 (https://cm.jefferson.edu/rna22/Interactive) and starBase (http://starbase.sysu.edu.cn) databases were used to predict the potential target miRNAs of circFLNA. Among the candidate miRNAs, the top three miRNAs, namely miR-199-3p, miR-296-5p and miR-515-5p, were selected according to their prediction score. Wild-type (WT) and mutant (MUT) putative miR-199-3p binding sites of the 3′-untranslated region (UTR) in circFLNA (Shanghai GenePharma Co., Ltd.), were cloned into psiCHECK2 luciferase reporter vectors (Shanghai GenePharma Co., Ltd.). LN229 cells were seeded at the density of 3×105 cells/well into 6-well plates and co-transfected with 20 µl psiCHECK2-circFLNA-WT or -MUT (108 TU/ml) and 20 µM miR-199-3p mimic or NC mimic using Lipofectamine 2000. The medium was removed with fresh medium at 4 h post-transfection. The relative luciferase activity was detected at 48 h post-transfection using a Dual Luciferase Reporter assay system (cat. no. E1910; Promega Corporation) according to the manufacturer's instructions. Firefly luciferase activity was normalized to Renilla luciferase activity.
Cell proliferation assay
Cell proliferation was measured using a Cell Counting Kit-8 (CCK-8) assay (cat. no. C0037; Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Briefly, following transfection, GBM cells were seeded (1×104 cells/well) into 96-well plates and incubated for 24, 48 or 72 h. The rescue experiment was performed at 72 h. Following the incubation, 10 µl CCK-8 reagent was added to each well and incubated for a further 4 h at 37°C. The absorbance was measured using a microplate reader (Infinite F50; Tecan Group, Ltd.) at a wavelength of 450 nm.
Transwell invasion assay
A total of 5×104 LN229 and A172 cells were plated in serum-free DMEM into the upper chambers of Transwell plates (Corning, Inc.), which were precoated with Matrigel (37°C for 30 min), and incubated at 37°C for 24 h. The lower chambers were filled with DMEM supplemented with 20% FBS before the incubation. Following incubation, the invasive cells in the lower chamber were fixed with 4% paraformaldehyde for 15 min at room temperature and stained with 1 mg/ml crystal violet for 20 min at room temperature. The invasive cells were counted in five randomly selected fields using a light microscope (Nikon Corporation; magnification, ×100).
Western blotting
Total protein was extracted from LN229 and A172 cells using RIPA lysis buffer (cat. no. P0013B; Beyotime Institute of Biotechnology) supplemented with complete Protease Inhibitor Cocktail (cat. no. 04693124001; Roche Applied Science). Total protein was quantified using a BCA protein assay kit (cat. no. P0012; Beyotime Institute of Biotechnology). The absorbance was measured using a microplate reader (Infinite F50; Tecan Group, Ltd.) at a wavelength of 562 nm and 50 µg protein/lane was separated via 10% SDS-PAGE. The separated proteins were subsequently transferred onto PVDF membranes (cat. no. FFP24; Beyotime Institute of Biotechnology) and blocked with 5% BSA (cat. no. AR0004; Wuhan Boster Biological Technology, Ltd.) at 4°C for 1 h. The membranes were then incubated with the following primary antibodies at 4°C overnight: Rabbit anti-MMP-2 (1:1,000; cat. no. 10373-2-AP; ProteinTech Group, Inc.), rabbit anti-MMP-9 (1:1,000; cat. no. 10375-2-AP; ProteinTech Group, Inc.) and mouse anti-GAPDH (1:1,000; cat. no. SC-47724; Santa Cruz Biotechnology, Inc.). Following primary antibody incubation, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h (anti-mouse or anti-rabbit; cat. nos. ab6721 and ab6728; 1:2,000; Abcam). Protein bands were visualized using ECL reagent (cat. no. P0018S; Beyotime Institute of Biotechnology) on a ChemiDoc™ MP Imaging system (cat. no. 12003154; Bio-Rad Laboratories, Inc.). GAPDH was used as the loading control.
Statistical analysis
Statistical analyses were performed using SPSS 20.0 software (IBM Corp.) and data are presented as the mean ± SD. Statistical differences between groups were determined using one-way or two-way ANOVA followed by Tukey's post hoc test or an unpaired Student's t-test. The expression levels of circFLNA and miR-199-3p in clinical GBM tissues were analyzed using a Wilcoxon signed-rank test. Kaplan-Meier curves were used to determine the overall survival and a log-rank test was conducted to analyze the significant differences in survival using GraphPad Prism 5.0 software (GraphPad Software, Inc.). The association between circFLNA expression levels and the clinicopathological features of patients with GBM was determined using a χ2 test. The correlation between the expression of circFLNA and miR-199-3p was analyzed using Pearson's correlation analysis. Experiments were independently performed in triplicate. P<0.05 was considered to indicate a statistically significant difference.
Results
circRNA expression profile analysis
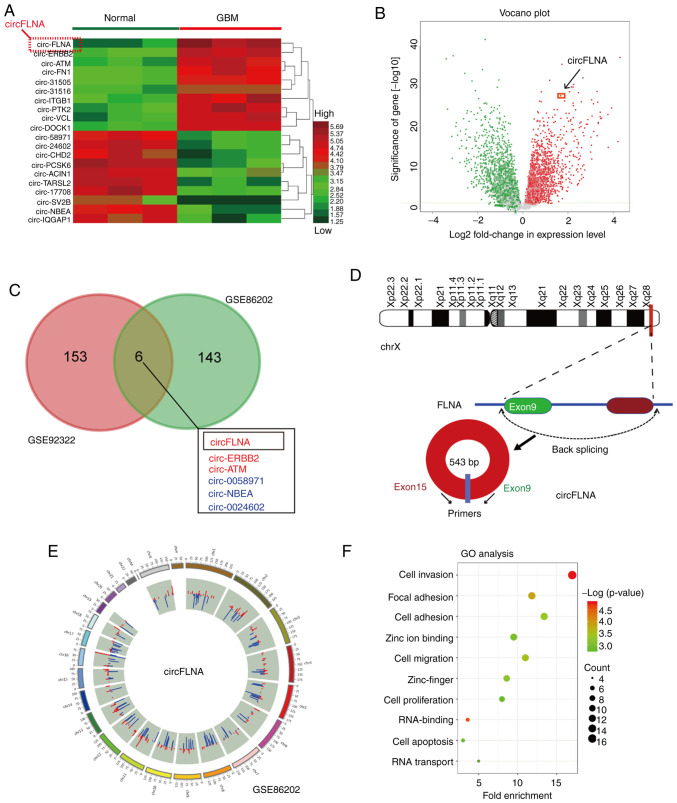
The present study investigated the expression levels of the top 20 differentially expressed circRNAs in the GSE86202 dataset obtained from the GEO database; the cut-off values were determined based on the Benjamini-Hochberg method (log2FC>2 and FDR<0.01) (25) and the circRNAs were identified according to the log2FC level (Fig. 1A). Among the differentially expressed circRNAs, circFLNA expression levels were the highest in GBM tissues, demonstrating significantly upregulated expression levels compared with adjacent healthy brain tissues (Fig. 1A and B). To further determine whether circFLNA served a role in the progression of GBM, the expression levels of circRNAs were evaluated using microarray data downloaded from two GEO datasets and GSE86202. As shown in Fig. 1C, six intersecting circRNAs (three upregulated, circ-FLNA, circ-ERBB2, circ-ATM; and three downregulated, circ-0058971, circ-NBEA, circ-0024602) were identified and investigated according to their log2FC level. As the expression levels of circFLNA were upregulated in both GEO databases, circFLNA was further analyzed in subsequent experiments. circFLNA was discovered to originate from exon 9 to 15 of the FLNA gene, with a spliced mature sequence length of 543 base pairs (Fig. 1D). The differentially expressed circRNAs identified between GBM and healthy tissues from the GEO database are displayed in Fig. 1E. circFLNA was subsequently subjected to GO functional term enrichment analysis to predict its function in GBM progression. The results revealed that circFLNA was associated with the biological process of glioblastoma, and ‘Cell invasion’, ‘Cell migration’ and ‘Focal adhesion’ were identified as the most enriched malignant biological processes (Fig. 1F). These results suggested that circFLNA may participate in GBM development.
Figure 1.
circFLNA expression levels are upregulated in GBM. (A) Heatmap demonstrating the differentially expressed circRNAs (fold-change) in the GEO dataset, GSE86202. (B) Volcano plot indicated that circFLNA expression levels were increased in the two groups of circRNAs from the GEO database. (C) A total of six significantly differentially expressed circRNAs were identified in the GSE86202 and GSE92322 GEO datasets. (D) Genomic loci of the FLNA gene and formation of circFLNA. The spliced mature sequence length of circFLNA is 543 base pairs in length. (E) Overview of differentially expressed circRNAs and their chromosomal location (using the GSE86202 dataset). The blue bar (downregulated circRNAs) and red bar (upregulated circRNAs) formed the inner circle. (F) Genes positively associated with circFLNA in the GEO datasets (GSE86202) were identified using GO analysis. circRNA, circular RNA; circFLNA, circRNA filamin A; GBM, glioblastoma; GEO, Gene Expression Omnibus; TPM, trans per kilobase of exon model per million; GO, Gene Ontology; Chr, chromosome.
circFLNA expression levels are upregulated in GBM tissues
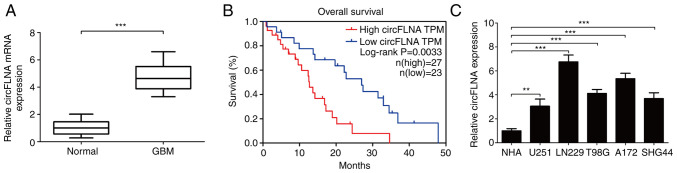
circFLNA expression levels were subsequently analyzed in 50 clinical GBM cases using RT-qPCR. The results demonstrated that circFLNA expression levels were significantly upregulated in GBM tissues compared with healthy tissues (Fig. 2A). Furthermore, analysis of the clinical data of the patients with GBM revealed that high expression levels of circFLNA in GBM were associated with a worse overall survival according to the median survival time of patients with GBM (Fig. 2B). The expression levels of circFLNA were also significantly associated with the presence of necrosis in MRI scans (n=50; P=0.001; Table I). To determine the function of circFLNA in GBM, the expression levels of circFLNA in GBM cell lines (U251, T98G, LN229, SHG44 and A172) were compared with NHAs (Fig. 2C). The expression levels of circFLNA were upregulated in GBM cell lines compared with NHAs and the expression level was highest in LN229 and A172 cells. Thus, these cells were selected for use in subsequent experiment.
Figure 2.
Expression levels of circFLNA are upregulated in human GBM tissues and cell lines. (A) Expression levels of circFLNA were upregulated in GBM tissues (n=50) compared with the adjacent healthy brain tissues. (B) Kaplan-Meier analysis identified that high expression levels of circFLNA were associated with a worse prognosis for patients with GBM (n=50). (C) circFLNA expression levels in GBM cell lines (U251, LN229, T98G, A172 and SHG44) and NHAs were analyzed using reverse transcription-quantitative PCR. **P<0.01, ***P<0.001. Data are presented as the mean ± SD of three independent experiments. circFLNA, circular RNA filamin A; GBM, glioblastoma; NHA, normal human astrocytes; TPM, transcripts per kilobase of exon model per million mapped reads.
Table I.
Association between circFLNA expression levels and the clinical characteristics of patients with glioblastoma (n=50).
| circFLNA expression | ||||
|---|---|---|---|---|
|
|
||||
| Variable | n | Low | High | P-value |
| Age, years | 0.845 | |||
| <60 | 21 | 10 | 11 | |
| ≥60 | 29 | 13 | 16 | |
| Sex | 0.777 | |||
| Male | 25 | 12 | 13 | |
| Female | 25 | 11 | 14 | |
| Karnofsky performance status scale | 0.260 | |||
| <60 | 16 | 6 | 10 | |
| ≥60 | 34 | 17 | 17 | |
| Mean tumor diameter, cm | 0.951 | |||
| <5 | 15 | 7 | 8 | |
| ≥5 | 35 | 16 | 19 | |
| Presence of necrosis on MRI | 0.001a | |||
| Yes | 26 | 6 | 20 | |
| No | 24 | 17 | 7 | |
| Seizure | 0.586 | |||
| Yes | 24 | 12 | 12 | |
| No | 26 | 11 | 15 | |
P<0.05. circFLNA, circular RNA filamin A.
circFLNA knockdown inhibits GBM cell proliferation and invasion
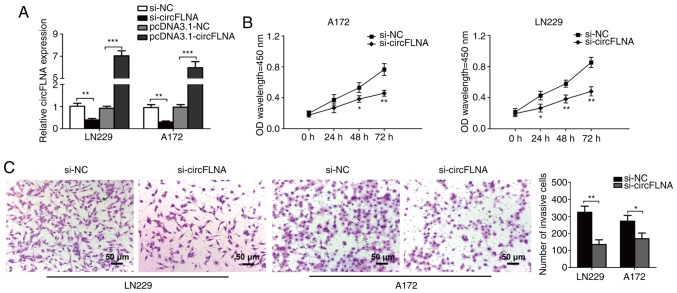
To investigate the effect of circFLNA on GBM proliferation and invasion, circFLNA expression levels were knocked down in LN229 and A172 cells. To identify the biological function of circFLNA in the progression of glioblastoma, LN229 and A172 cells were transfected with si-circFLNA/si-NC or pcDNA3.1-circFLNA/pcDNA3.1-NC, and the proliferative and invasive abilities were determined. The transfection efficiencies were confirmed via RT-qPCR, where circFLNA expression was significantly decreased or increased compared with corresponding NC groups (Fig. 3A). Furthermore, the results of the CCK-8 assay discovered that circFLNA knockdown significantly inhibited the proliferative ability of GBM cells following 48–72 h of incubation compared with GBM cells transfected with si-NC (Fig. 3B). In addition, the findings of the Transwell invasion assay demonstrated that the invasive ability of GBM cells was significantly inhibited by si-circFLNA compared with si-NC (Fig. 3C). These results suggested that the proliferative and invasive abilities of GBM cells may be suppressed by circFLNA knockdown in vitro.
Figure 3.
circFLNA knockdown inhibits the proliferative and invasive abilities of GBM cells in vitro. (A) circFLNA expression levels were detected in LN229 and A172 cells following the transfection of si-circFLNA or pcDNA3.1-circFLNA and respective NCs. (B) GBM cell proliferation was analyzed using a Cell Counting Kit-8 assay (si-circFLNA vs. si-NC). *P<0.05, **P<0.01 vs. si-NC. (C) Transwell invasion assays were performed to determine the invasive ability of GBM cells. Magnification, ×100; scale bar, 50-µm. *P<0.05, **P<0.01, ***P<0.001. Data are presented as the mean ± SD of three independent experiments. circFLNA, circular RNA filamin A; NC, negative control; GBM, glioblastoma; si, small interfering RNA; OD, optical density.
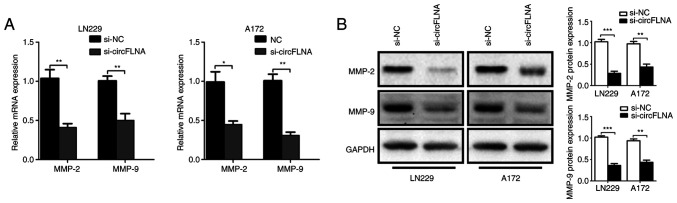
To further explore the effect of circFLNA on GBM cell invasion, the mRNA and protein expression levels of tumor invasion-related biomarkers, MMP-2 and MMP-9, in GBM cells were analyzed using RT-qPCR and western blotting, respectively. The results revealed that circFLNA knockdown significantly downregulated the expression levels of MMP-2 and MMP-9 in GBM cells compared with the si-NC group (Fig. 4A and B). Based on these results, it was suggested that circFLNA may regulate the invasive ability of GBM cells.
Figure 4.
Expression levels of MMP-2 and MMP-9 in GBM cells. (A) Reverse transcription-quantitative PCR analysis of MMP-2 and MMP-9 expression levels. (B) Western blotting was used to analyze the expression levels of MMP-2 and MMP-9 in si-circFLNA-transfected cells. Data are presented as the mean ± SD of three independent experiments. *P<0.05, **P<0.01, ***P<0.001. NC, negative control; si, small interfering RNA; circFLNA, circular RNA filamin A.
Association between circFLNA and miR-199-3p expression levels
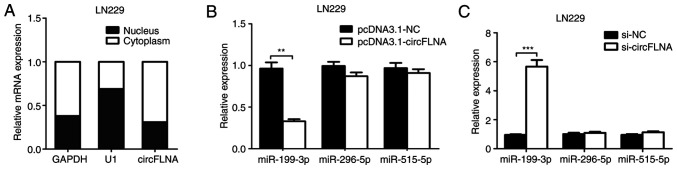
RT-qPCR analysis was used to determine the subcellular localization of circFLNA in LN229 cells. As shown in Fig. 5A, circFLNA was found to be primarily localized in the cytoplasm of the GBM cells, indicating that circFLNA may exert both transcriptional and post-transcriptional regulatory effects in GBM. Using StarBase and RNA22 database blast prediction, three potential miRNA targets of circFLNA (miR-199-3p, miR-296-5p and miR-515-5p) were identified. To investigate the regulatory relationship between circFLNA and the miRNAs, RT-qPCR was performed to analyze miR-199-3p, miR-296-5p and miR-515-5p expression levels in LN229 cells transfected with pcDNA3.1-circFLNA plasmids, pcDNA3.1-NC plasmids, si-circFLNA or si-NC. The data revealed that transfection with pcDNA3.1-circFLNA significantly downregulated the expression levels of miR-199-3p compared with the pcDNA3.1-NC plasmid, whereas circFLNA knockdown significantly upregulated the expression levels of miR-199-3p compared with si-NC-transfected cells; however, the expression levels of miR-296-5p and miR-515-5p were unaffected by the overexpression or knockdown of circFLNA (Fig. 5B and C).
Figure 5.
Identification of potential binding miRNAs of circFLNA in glioblastoma. (A) Subcellular localization of circFLNA was determined using RT-qPCR in LN229 cells. (B) Expression levels of miR-199-3p, miR-296-5p and miR-515-5p in LN229 cells transfected with pcDNA3.1-circFLNA were analyzed using RT-qPCR. Expression levels of miR-199-3p were downregulated following transfection with pcDNA3.1-circFLNA. (C) RT-qPCR was used to determine that the expression levels of miR-199-3p were upregulated following the transfection with si-circFLNA. Data are presented as the mean ± SD of three independent experiments. **P<0.01, ***P<0.001. miR, microRNA; NC, negative control; si, small interfering RNA; circFLNA, circular RNA filamin A; RT-qPCR, reverse transcription-quantitative PCR.
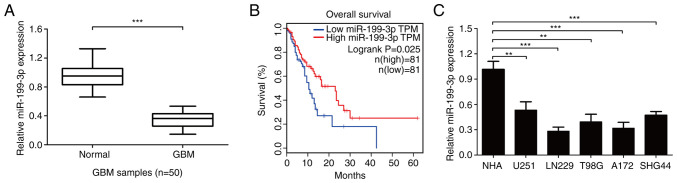
The expression levels of miR-199-3p in 50 GBM and adjacent healthy brain tissues were subsequently determined. The expression levels were significantly downregulated in GBM tissues compared with adjacent healthy tissues (Fig. 6A). Kaplan-Meier analysis showed that miR-199-3p expression was positively associated with the prognosis of patients with GBM from TCGA-GBM database according to the median overall survival time of patients with GBM (Fig. 6B). In addition, miR-199-3p expression levels were significantly downregulated in GBM cell lines compared with NHAs (Fig. 6C).
Figure 6.
Expression levels of miR-199-3p in GBM. (A) Expression levels of miR-199-3p were analyzed in clinical GBM tissues. (B) Patients with GBM with high expression levels of miR-199-3p had an improved overall survival compared with patients with low miR-505-3p expression levels, as determined from data from The Cancer Genome Atlas database (log-rank test, P=0.025). (C) miR-199-3p expression levels were analyzed in GBM cells using reverse transcription-quantitative PCR. Data are presented as the mean ± SD of three independent experiments. **P<0.01, ***P<0.001. miR, microRNA; GBM, glioblastoma.
miR-199-3p targets the 3′-UTR of circFLNA
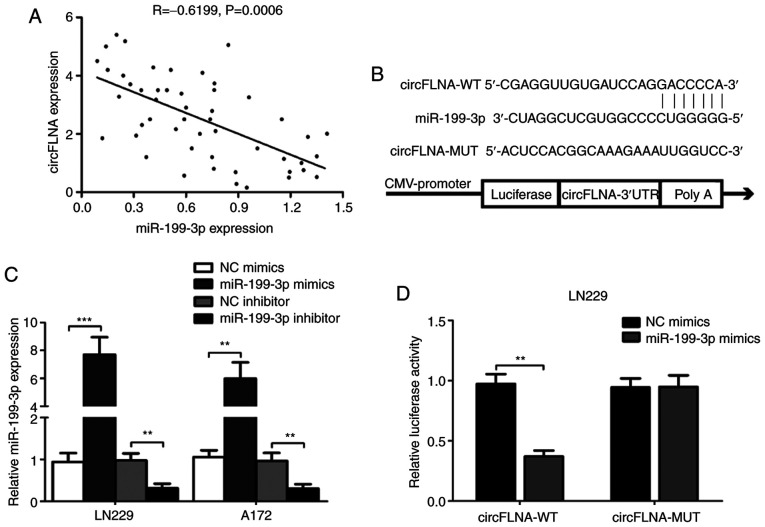
miR-199-3p and circFLNA expression levels in tissues from patients with GBM (n=50) were negatively correlated (n=50; R=−0.6199; P=0.0006; Fig. 7A). Using the RNA22 and StarBase databases, the complementary sequence between circFLNA and miR-199-3p was identified (Fig. 7B). miR-199-3p mimic (miR-199-3p), NC mimic or inhibitors were transfected into LN229 and A172 cells and the transfection efficiencies were detected. The results demonstrated that miR-199-3p was significantly increased or decreased in transfected cells compared with NC groups (Fig. 7C). Subsequently, the circFLNA-WT or circFLNA-MUT 3′-UTRs, which contained the predicted miR-199-3p binding site, were cloned into psiCHECK2 luciferase reporter vectors. The relative luciferase activity was significantly decreased following the co-transfection of the circFLNA-WT vector and miR-199-3p mimic compared with the circFLNA-WT and miR-NC, whereas no significant differences were observed following the co-transfection with the miR-199-3p mimic or miR-NC and circFLNA-MUT vector in LN229 cells (Fig. 7D). These results indicated that circFLNA may interact with miR-199-3p in GBM cells.
Figure 7.
miR-199-3p targets the 3′-UTR of circFLNA. (A) Correlation between circFLNA and miR-199-3p expression levels was determined. (B) Predicted binding site of miR-199-3p on circFLNA is shown. (C) miR-199-3p expression levels were analyzed in LN229 and A172 cells following transfection with miR-199-3p mimic, NC mimic, miR-199-3p inhibitor or NC inhibitor using reverse transcription-quantitative PCR. (D) Relative luciferase activity was determined following co-transfection of LN229 cells with miR-199-3p mimics or NC mimics and circFLNA-WT or circFLNA-MUT. Data are presented as the mean ± SD of three independent experiments. **P<0.01, ***P<0.001. circFLNA, circular RNA filamin A; miR, microRNA; WT, wild-type; MUT, mutant; NC, negative control; si, small interfering RNA; UTR, untranslated region.
miR-199-3p reverses the effect of circFLNA in GBM cells
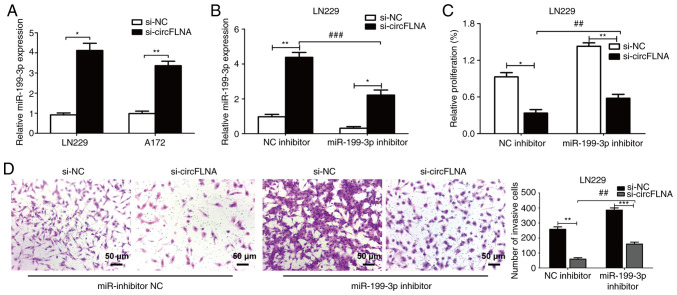
As aforementioned, miR-199-3p expression levels were found to be negatively correlated with circFLNA expression levels in GBM cells. LN229 cells were subsequently used to perform rescue experiments. The expression levels of miR-199-3p were significantly upregulated following transfection with si-circFLNA compared with si-NC in GBM cells (Fig. 8A). The co-transfection of miR-199-3p inhibitor+si-circFLNA significantly downregulated the expression levels of miR-199-3p vs. the NC inhibitor + si-circFLNA group (Fig. 8B). Conversely, the expression levels of miR-199-3p were significantly upregulated following the co-transfection of cells with si-circFLNA vs. the si-NC + miR-199-3p inhibitor group. Transfection with miR-199-3p inhibitor also partially attenuated the suppressive effect of circFLNA knockdown on the viability and invasion of LN229 cells (Fig. 8C and D). These data indicated that miR-199-3p may be a crucial mediator of circFLNA-regulated tumor proliferation and invasion processes.
Figure 8.
miR-199-3p reverses the effect of circFLNA on glioblastoma cells. (A) miR-199-3p expression levels were upregulated following circFLNA knockdown in LN229 and A172 cells. (B) Expression levels of miR-199-3p in LN229 cells co-transfected with combinations of si-circFLNA, si-NC, miR-199-3p inhibitor and NC inhibitor. (C) Cell viability was detected using a Cell Counting Kit-8 assay. (D) Invasive ability was analyzed using a Transwell assay. Magnification, ×100; scale bar, 50 µm. Data are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001; ##P<0.01, ###P<0.001. miR, microRNA; circFLNA, circular RNA filamin A; NC, negative control; si, small interfering RNA.
Discussion
GBM is an aggressive and malignant type of primary brain cancer with a >90% 5-year mortality (1,2). Although significant progress has been achieved in research into treatments for GBM, the therapeutic strategies available (resection techniques, chemotherapy strategies and radiation therapy) for GBM remain unsatisfactory (5). Accumulating evidence has reported that circRNAs were associated with the occurrence of numerous types of cancer and the aberrant expression of circRNAs was found to be associated with promoting the tumorigenesis of cancer (26–29). however, its underlying mechanism of action requires further investigation.
The mechanism via which circFLNA acts as an oncogene in GBM remains unknown. In the present study, circFLNA expression was significantly increased in GBM tissues compared with adjacent normal tissues. In addition, miR-199-3p expression was decreased in GBM tissues, and miR-199-3p expression was negatively correlated with circFLNA expression. Using bioinformatics analysis, miR-199-3p was predicted as the potential target of circFLNA. Notably, circFLNA knockdown suppressed the proliferative and invasive abilities of GBM cells, whereas co-transfection with miR-199-3p inhibitor partially reversed these trends. Therefore, circFLNA knockdown may suppress the proliferation and invasion of GBM, indicating a potential therapeutic strategy for GBM.
The competing endogenous RNA (ceRNA) theory is considered to be an important mechanism for circRNAs, in which circRNAs have been demonstrated to act as sponges to regulate the expression and function of miRNAs (11,30). A previous study reported that circRNA_001783 expression levels were downregulated in breast cancer, which regulated cancer cell proliferation via miR-200c-3p (31). circRNA activin A receptor type 2A was also demonstrated to function as a ceRNA for miR-626, which suppressed cell proliferation and invasion in bladder cancer (32). In gastric cancer, circRNA Ran GTPase activating protein 1 regulated invasion and metastasis by upregulating VEGFA expression levels via interacting with miR-877-3p (33). It was hypothesized that circFLNA may act as a ceRNA towards miR-199-3p. Accumulating evidence has revealed that the aberrant expression of miRNAs serves important roles in the occurrence and development of numerous types of cancer (34–38). For example, Zhang et al (39) reported that miR-199-3p directly regulated the expression levels of snail family transcriptional repressor 1 in hepatoma cells. Koshizuka et al (40) demonstrated that miR-199-3p expression levels were downregulated in head and neck cancer, which suppressed malignant biological behaviors. The present study detected that circFLNA knockdown may suppress the proliferation and invasion of GBM, these results highlighted the potential role of the circFLNA/miR-199-3p axis in GBM development.
In conclusion, the findings of the present study suggested that circFLNA may serve as an oncogenic circRNA by acting as a ceRNA and sponging miR-199-3p in GBM. In a future study, the patient-derived xenograft model will be used to follow the effects of circRNA on tumorigenesis, which is a more clinically predictive model of human glioblastoma. The results identified a novel role of circFLNA in GBM and elucidated the underlying mechanisms of circFLNA in the progression of GBM. Therefore, circFLNA may be regarded as a novel approach for the treatment of GBM.
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the Scientific Research Project of Health Commission of Heilongjiang Province (grant no. 2019-368).
Funding
The present study was supported by the Scientific Research Project of Health Commission of Heilongjiang Province (grant no. 2019-368).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YS, GM and HX designed the study, performed the experiments, analyzed the data and wrote the manuscript. XW, HX and HW performed the in vitro experiments. FQ, YS and CL analyzed the data and drafted the manuscript. CL, YZ and GM designed and supervised the study, and edited the manuscript. YS, YZ and CL confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Written informed consent was obtained from all patients and the study protocol was approved by the Ethics Committee of Harbin Medical University (Harbin, China; approval no. 2019HMUIRB0171). All procedures were performed in accordance with national (D.L.n.26, March 4th, 2014) and international laws and policies (directive 2010/63/EU) (41).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Bernstock JD, Mooney JH, Ilyas A, Chagoya G, Estevez-Ordonez D, Ibrahim A, Nakano I. Molecular and cellular intratumoral heterogeneity in primary glioblastoma: Clinical and translational implications. J Neurosurg. 2019;23:1–9. doi: 10.3171/2019.5.JNS19364. [DOI] [PubMed] [Google Scholar]
- 4.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 5.Hara A, Kanayama T, Noguchi K, Niwa A, Miyai M, Kawaguchi M, Ishida K, Hatano Y, Niwa M, Tomita H. Treatment strategies based on histological targets against invasive and resistant glioblastoma. J Oncol. 2019;2019:2964783. doi: 10.1155/2019/2964783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garton ALA, Kinslow CJ, Rae AI, Mehta A, Pannullo SC, Magge RS, Ramakrishna R, McKhann GM, Sisti MB, Bruce JN, et al. Extent of resection, molecular signature, and survival in 1p19q-codeleted gliomas. J Neurosurg. 2020;134:1357–1367. doi: 10.3171/2020.2.JNS192767. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Zhang W, Mao XG, Cao WD, Zhen HN, Hu SJ. Malignant intracranial high grade glioma and current treatment strategy. Curr Cancer Drug Targets. 2019;19:101–108. doi: 10.2174/1568009618666180530090922. [DOI] [PubMed] [Google Scholar]
- 8.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Y, Long J, He Q, Li Y, Liao Y, He P, Zhu W. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10:5015–5021. doi: 10.7150/jca.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao Z, Hu S, Liu Z, Song W, Zhao Y, Li M. Circular RNAs: Functions and prospects in glioma. J Mol Neurosci. 2019;67:72–81. doi: 10.1007/s12031-018-1211-2. [DOI] [PubMed] [Google Scholar]
- 12.Cheng J, Meng J, Zhu L, Peng Y. Exosomal noncoding RNAs in Glioma: Biological functions and potential clinical applications. Mol Cancer. 2020;19:66. doi: 10.1186/s12943-020-01189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hingorani DV, Lippert CN, Crisp JL, Savariar EN, Hasselmann JPC, Kuo C, Nguyen QT, Tsien RY, Whitney MA, Ellies LG. Impact of MMP-2 and MMP-9 enzyme activity on wound healing, tumor growth and RACPP cleavage. PLoS One. 2018;13:e0198464. doi: 10.1371/journal.pone.0198464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu J, Yang J, Chen M, Wei R, Tian J. CircFLNA Acts as a sponge of miR-646 to facilitate the proliferation, metastasis, glycolysis, and apoptosis inhibition of gastric cancer by targeting PFKFB2. Cancer Manag Res. 2020;12:8093–8103. doi: 10.2147/CMAR.S264674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang N, Gao L, Ren W, Li S, Zhang D, Song X, Zhao C, Zhi K. Fucoidan affects oral squamous cell carcinoma cell functions in vitro by regulating FLNA derived circular RNA. Ann N Y Acad Sci. 2020;1462:65–78. doi: 10.1111/nyas.14190. [DOI] [PubMed] [Google Scholar]
- 16.Lu C, Shi X, Wang AY, Tao Y, Wang Z, Huang C, Qiao Y, Hu H, Liu L. RNA-Seq profiling of circular RNAs in human laryngeal squamous cell carcinomas. Mol Cancer. 2018;17:86. doi: 10.1186/s12943-018-0833-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JX, Liu Y, Jia XJ, Liu SX, Dong JH, Ren XM, Xu O, Zhang HZ, Duan HJ, Shan CG. Upregulation of circFLNA contributes to laryngeal squamous cell carcinoma migration by circFLNA-miR-486-3p-FLNA axis. Cancer Cell Int. 2019;19:196. doi: 10.1186/s12935-019-0924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;11:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 19.Chen Z, Li J, Tian L, Zhou C, Gao Y, Zhou F, Shi S, Feng X, Sun N, Yao R, et al. MiRNA expression profile reveals a prognostic signature for esophageal squamous cell carcinoma. Cancer Lett. 2014;350:34–42. doi: 10.1016/j.canlet.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Ye B, Wang P, Yao F, Zhang C, Yu G. Overview of microRNA-199a regulation in cancer. Cancer Manag Res. 2019;11:10327–10335. doi: 10.2147/CMAR.S231971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chi GN, Yang FW, Xu DH, Liu WM. Silencing hsa_circ_PVT1 (circPVT1) suppresses the growth and metastasis of glioblastoma multiforme cells by up-regulation of miR-199a-5p. Artif Cells Nanomed Biotechnol. 2020;48:188–196. doi: 10.1080/21691401.2019.1699825. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Ye J, Zhang L, Xia L, Hu H, Jiang H, Wan Z, Sheng F, Ma Y, Li W, et al. Differential expression of circular RNAs in glioblastoma multiforme and its correlation with prognosis. Transl Oncol. 2017;10:271–279. doi: 10.1016/j.tranon.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Y, Jiaoming L, Xiang W, Yanhui L, Shu J, Maling G, Qing M. Analyzing the interactions of mRNAs, miRNAs, lncRNAs and circRNAs to predict competing endogenous RNA networks in glioblastoma. J Neurooncol. 2018;137:493–502. doi: 10.1007/s11060-018-2757-0. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate-A practical and powerful approach to multiple testing. J R Stat Soc. 1995;1:289–300. [Google Scholar]
- 26.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: A novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 27.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: Opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao W, Dong M, Pan J, Wang Y, Zhou J, Ma J, Liu S. Circular RNAs: A novel target among non-coding RNAs with potential roles in malignant tumors (Review) Mol Med Rep. 2019;20:3463–3474. doi: 10.3892/mmr.2019.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15:995–1005. doi: 10.1080/15476286.2018.1486659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, Andrews R, Zhong W, Zhang X, Song E, Gong C. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10:55. doi: 10.1038/s41419-018-1287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q, Wang Q, Xie R, Su Y, Yang M, et al. Circular RNA ACVR2A suppresses bladder cancer cells proliferation and metastasis through miR-626/EYA4 axis. Mol Cancer. 2019;18:95. doi: 10.1186/s12943-019-1025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 34.Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017;8:45–56. doi: 10.4331/wjbc.v8.i1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu M, Wang G, Tian W, Deng Y, Xu Y. miRNA-based therapeutics for lung cancer. Curr Pharm Des. 2018;23:5989–5996. doi: 10.2174/1381612823666170714151715. [DOI] [PubMed] [Google Scholar]
- 36.Shin VY, Chu KM. miRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432–10439. doi: 10.3748/wjg.v20.i30.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fridrichova I, Zmetakova I. MicroRNAs contribute to breast cancer invasiveness. Cells. 2019;8:1361. doi: 10.3390/cells8111361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng JH, Deng Q, Kuo CH, Delaney SW, Ying SY. miRNA targets of prostate cancer. Methods Mol Biol. 2013;936:357–369. doi: 10.1007/978-1-62703-083-0_27. [DOI] [PubMed] [Google Scholar]
- 39.Zhang HY, Li CH, Wang XC, Luo YQ, Cao XD, Chen JJ. miR-199 inhibits EMT and invasion of hepatoma cells through inhibition of Snail expression. Eur Rev Med Pharmacol Sci. 2019;23:7884–7891. doi: 10.26355/eurrev_201909_18998. [DOI] [PubMed] [Google Scholar]
- 40.Koshizuka K, Hanazawa T, Kikkawa N, Arai T, Okato A, Kurozumi A, Kato M, Katada K, Okamoto Y, Seki N. Regulation of ITGA3 by the anti-tumor miR-199 family inhibits cancer cell migration and invasion in head and neck cancer. Cancer Sci. 2017;108:1681–1692. doi: 10.1111/cas.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLOS Biol. 2020;18:e3000410. doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.