Abstract
Inhibin βA (INHBA) serves a prognostic and tumor-promoting role in numerous types of cancer. The present study aimed to determine the clinical significance of INHBA in non-small cell lung cancer (NSCLC) and the mechanisms underlying its potential tumor-promoting effect. INHBA expression was detected in clinical NSCLC samples using immunohistochemistry. In vivo loss- and gain-of-function studies were performed to determine the effects of INHBA on NSCLC invasion. In addition, protein and mRNA expression levels of INHBA, yes-associated protein (YAP), large tumor suppressor 1/2 kinase (LATS1/2), connective tissue growth factor, cysteine rich angiogenic inducer 61 and Merlin were assessed using western blotting and reverse transcription-quantitative PCR, respectively, to investigate the mechanism by which INHBA may affect the invasion of NSCLC. The present study revealed that INHBA was significantly upregulated in 238 clinical NSCLC samples compared with its expression levels in paired adjacent non-cancerous tissues, and in metastatic nodules compared with in primary tumors. Notably, high INHBA expression was statistically associated with clinicopathological features, including poor differentiation and advanced tumor stage. INHBA positivity was statistically related to decreased 5-year overall survival, for which INHBA was an independent prognostic factor. Furthermore, INHBA promoted NSCLC invasion in vitro. In NSCLC, INHBA expression was associated with the nuclear levels of YAP and INHBA overexpression enhanced the invasive abilities of NSCLC cells via inhibiting the Hippo pathway. Mechanistically, INHBA inhibited l LATS1/2 phosphorylation and induced YAP nuclear translocation by downregulating the protein expression levels of Merlin. In conclusion, INHBA may negatively regulate the Hippo pathway to act as a tumor promotor, and could represent a marker of prognosis in NSCLC.
Keywords: Hippo pathway, inhibin βA, invasion, non-small cell lung cancer, yes-associated protein
Introduction
NSCLC is the most commonly diagnosed cancer and is also the main cause of cancer-related death worldwide. NSCLC has three major types: Adenocarcinoma (ADC), squamous cell carcinoma (SCC) and large cell carcinoma (LCC) (1,2). Despite advances in surgery, radiotherapy and chemotherapy, the 5-year survival rate of lung cancer is <15%, mainly because numerous patients are diagnosed at the late stage of the disease and due to the frequent occurrence of metastasis (3,4). In recent years, to treat lung cancer, certain genes (e.g., epidermal growth factor receptor, human epidermal growth factor receptor 2 and ALK receptor tyrosine kinase) have been successfully targeted and there have been ongoing advances in NSCLC care (5). In addition, the treatment of lung cancer with immunotherapy has made significant progress; however, only a few patients can benefit from these treatments (6). Therefore, it has become clear that additional therapeutic strategies are required to provide an improved survival benefit for patients with NSCLC. Inhibin βA (INHBA) is a disulfide-linked homodimer of activin A and is a member of the transforming growth factor (TGF)-β superfamily (7). Previous research has focused on the controversial function of activin A in cancer (8), and both tumor-promoting and tumor-suppressing roles of activin A have been identified (9–13). Notably, previous studies have demonstrated the tumor-promoting effects of INHBA on gastric cancer, urothelial carcinoma and nasopharyngeal carcinoma, and INHBA expression has been shown to be correlated with enhanced invasion, proliferation and poor prognosis (14–16). However, the role of INHBA in NSCLC pathogenesis remains unclear. Seder et al (17) revealed that in >70% of cases of lung ADC, INHBA was overexpressed, and a significantly worse prognosis was observed in patients with high INHBA transcription compared with those with low INHBA transcription (17). Wamsley et al (18) further demonstrated increased activin expression in primary LCC, SCC and ADC tumors. These studies suggested that metastasis and invasion of NSCLC may be regulated by INHBA; however, the potential molecular mechanism remains unknown.
Previous research has investigated the cross-talk between TGF-β and Hippo signaling. Varelas et al (19) and Narimatsu et al (20) suggested that tafazzin (TAZ) and yes-associated protein (YAP) may mediate nuclear Smad signals. Moreover, Fujii et al (21) suggested that YAP was critical for TGF-β-mediated induction of a small subset of target genes dependent on YAP (e.g., Smad2 and matrix metalloproteinase 7) (21). Therefore, we hypothesized that INHBA, the TGF-β superfamily member, may be associated with the Hippo pathway in NSCLC. When the Hippo signaling pathway is activated, its core components, mammalian sterile 20-like 1/MOB kinase activator and large tumor suppressor kinase (LATS)1/2 can regulate the transcriptional co-activator YAP/TAZ, forming a kinase cascade that culminates in phosphorylation and inactivation of YAP/TAZ, thereby inhibiting the development of lung cancer (22–24). Conversely, certain upstream regulators of the Hippo pathway, such as Merlin [encoded by the neurofibromin 2 (NF2) gene], FERM domain-containing 6 (FRMD6) and KIBRA [also known as WW and C2 domain-containing 1 (WWC1)], have also been shown to regulate NSCLC invasion and metastasis (25).
In view of the potential association of INHBA with the invasion and metastasis of NSCLC, the present study aimed to investigate the molecular mechanism underlying the INHBA-mediated regulation of invasion and metastasis of NSCLC. In addition, the clinical significance of INHBA in patients with NSCLC was assessed and the present study aimed to reveal effects of INHBA on regulation of the Hippo signaling pathway in NSCLC.
Materials and methods
Ethics statement
The Institute Research Medical Ethics Committee of Sun Yat-sen University Cancer Center (Guangzhou, China) approved the present study, and the patients involved in this study provided informed consent (verbal or written) for the retrospective analysis of tissue samples. All samples were subjected to anonymization.
Clinical samples
Tissue samples were collected from 238 cases of NSCLC diagnosed at the Sun Yat-sen University Cancer Center between January 2010 and December 2015. The samples were from 172 men and 66 women (median age, 58 years). The lung tumor histological criteria (2015) from the World Health Organization (26), were used to classify the samples as ADC (138 cases), SCC (82 cases) and LCC (18 cases). Differentiation was assessed as being high in 32 cases, moderate in 94 cases and poor in 112 cases. Lymph node metastases were present in 133 of the patients. Cases with lymph node metastases were selected for a comparison of metastatic nodules with the primary tumor. The tumor-node-metastasis (TNM) staging system of the International Union against Cancer (27) was used for tumor staging. A total of 100 patients had stage I disease and 138 patients had stage II–IV disease. Before surgery, none of the patients had been treated with radiotherapy or chemotherapy; however, after surgery, these two treatments were provided as standard care. For pathological analysis and diagnosis, all collected samples were fixed with 10% formalin at room temperature for 24 h, paraffin-embedded, cut into 4-µm thick sections and stained with hematoxylin and eosin for pathological analysis. Furthermore, an additional 24 fresh NSCLC and adjacent non-tumor tissues samples were obtained from 24 patients who underwent surgical resection for NSCLC at Sun Yat-sen University Cancer Center between January 2018 and December 2020. The 24 NSCLC samples were obtained from 17 (70.8%) men and 7 (29.2%) women (mean age, 59.0 years). In total, 14 cases were ADC, 9 cases were SCC and 1 case was LCC. Differentiation was high in 8 cases, moderate in 10 cases and poor in 6 cases. In addition, 10 cases showed lymph node metastases. All 24 samples were randomly selected from the fresh sample library diagnosed with NSCLC, which were frozen in liquid nitrogen immediately after resection and stored at −80°C until use.
Cell culture
The human lung cancer cell lines A549, H1299 and H460 were obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences in 2019. H157, H520 and normal human bronchial epithelial (HBE) cells were obtained from the American Type Culture Collection. RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Biological Industries), penicillin (100 U/ml), streptomycin (100 µg/ml), 2 mM glutamine and 10 mM HEPES buffer was used for cell culture at 37°C.
Immunohistochemistry (IHC)
For IHC, a tissue microarray (TMA) consisting of 238 primary NSCLC tissues, 30 paired metastatic nodule samples and 30 adjacent normal lung tissues was constructed. The paraffin-embedded samples were serially cut into 4-µm sections, de-paraffinized in xylene, rehydrated through a graded alcohol series, immersed in 3% hydrogen peroxide at room temperature for 10 min to block endogenous peroxidase activity and subjected to antigen retrieval by pressure cooking for 3 min in citrate buffer (pH 6.0). Subsequently, the slides were incubated with 10% normal goat serum (Beyotime Institute of Biotechnology) at room temperature for 30 min to reduce non-specific reactivity and then incubated with anti-YAP antibodies (1:400; cat. no. 14074; Cell Signaling Technology, Inc.) and anti-INHBA antibodies (1:200; cat. no. ab97705; Abcam) overnight at 4°C. Slides were then washed twice with PBS for 5 min, incubated with a secondary antibody (ready to use; cat. no. GK500705; Envision; Dako; Agilent Technologies, Inc.) for 1 h at room temperature, washed twice with PBS for 5 min and developed using 3,3-diaminobenzidine. Finally, the sections were counterstained with Mayer's hematoxylin for 3 min at room temperature, dehydrated and mounted with neutral gum. Negative controls were prepared by replacing the primary antibody with normal murine IgG (1:200; cat. no. A7028; Beyotime Institute of Biotechnology). Two experienced pathologists scored all the sections independently. As described previously (24), the IHC score was obtained by multiplying the score for the percentage of positively stained cells (0, <5%; 1, 6–25%; 2, 26–50%; 3, 51–75%; 4, 76–100%) by the score of the staining intensity (0, negative staining; 1, weak staining; 2, moderate staining and 3, strong staining) to give a final score for each section. Sections with a total score of ≥3 were defined as exhibiting positive staining. Nuclear YAP positivity was scored by determining the percentage of positive nuclei, regardless of cytoplasmic YAP expression and staining intensity. All tissues were examined using an Olympus BX53 microscope (Olympus Corporation; magnification, ×400).
Western blotting
Western blotting was performed according to a previously described method (28). Cells were lysed using sonication at 20 KHz or RIPA lysis buffer (Pierce; Thermo Fisher Scientific, Inc.) at 4°C for 20 min, and proteins were quantified using the Bradford protein assay. A total of 50 µg protein/lane was separated by SDS-PAGE on a 10% polyacrylamide gel. The separated proteins were transferred to polyvinylidene fluoride membranes (MilliporeSigma), which were blocked with 5% bovine serum albumin (Merck KGaA) at 37°C for 2 h. The membranes were then incubated overnight at 4°C with the following antibodies: YAP (1:500; cat. no. 14074), phosphorylated (p)YAP-S127 (1:500; cat. no. 57706), vimentin (1:1,000; cat. no. 12826), Snail (1:500; cat. no. 3895), GAPDH (1:1,000; cat. no. 5174s) (all from Cell Signaling Technology, Inc.); INHBA (1:500; cat. no. ab113489), LATS1/2 (1:500; cat. no. ab70565), pLATS1/2 (T1079 + T1041; 1:500; cat. no. ab111344), tubulin (1:1,000; cat. no. ab6160), Lamin B (1:1,000; cat. no. ab16048), connective tissue growth factor (CTGF; 1:500; cat. no. ab94939), cysteine-rich angiogenic inducer 61 (CYR61; 1:500; cat. no. ab228592), WWC1 (1:1,000; cat. no. ab216508), FRMD6 (1:1,000; cat. no. ab110675) (all from Abcam); E-cadherin (1:500; cat. no. 610181), N-cadherin (1:1,000; cat. no. 610920) (both from BD Biosciences) and Merlin (1:100; sc-331; Santa Cruz Biotechnology, Inc.). After incubation with anti-mouse (1:2,000; cat. no. E030110-01) or anti-rabbit (1:2,000; cat. no. E030120-01) IgG antibodies (EarthOx Life Sciences) at 37°C for 2 h, the protein bands were visualized using enhanced chemiluminescence (Thermo Fisher Scientific, Inc.) and semi-quantified using a BioImaging Systems platform. Relative protein expression levels were calculated using GAPDH as the loading control.
Construction of plasmids and transfection
Plasmids expressing full-length human INHBA and Merlin constructs, and empty vectors were all purchased from Shanghai GeneChem Co., Ltd. Specifically, the coding sequences of INHBA and Merlin were inserted between the BamHI and AgeI restriction enzyme sites of the Ubi-3FLAG-CBh-gcGFP-IRES-puromycin vector to generate the following overexpression plasmids of these two genes: Ubi-INHBA-3FLAG-CBh-gcGFP-IRES-puromycin and Ubi-Merlin-3FLAG-CBh-gcGFP-IRES-puromycin. The small interfering RNA (siRNA) sequences targeting INHBA and a negative control (NC) were all purchased from Guangzhou Ruibo Co., Ltd.; the sequences are shown in Table SI. The NC siRNA targets no known genes. Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform all transfections of lung cancer cells in 6-well plates. The concentration of plasmids transfected into cells was 1 µg/µl and that of siRNA was 20 pmol/µl. Briefly, A549 and H1299 cells were seeded 24 h prior to transfection in 6-well plates (5×105 cells/well) and were transfected using Lipofectamine 3000 according to the manufacturer's protocol. After transfection, cells were cultured at 37°C for 48 h, and then collected for reverse transcription-quantitative PCR (RT-qPCR) or western blotting.
RNA extraction and RT-qPCR
RT-qPCR was performed as described previously (28). TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from tissues and cells according to the manufacturer's protocol. Subsequently, 1 µg total RNA was used to produce cDNA using a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. The SYBR-Green PCR Master Mix and StepOne-Plus Real-Time PCR system (both Applied Biosystems; Thermo Fisher Scientific, Inc.) were used to perform qPCR. The thermocycling conditions were as follows: 94°C for 2 min, followed by 40 cycles at 94°C for 15 sec and 60°C for 1 min according to the manufacturer's protocol. The primers used for qPCR are shown in Table SII. The mRNA ΔCq values from each sample were calculated by normalizing to the reference gene 18S and using the 2−∆∆Cq method (29). Experiments were performed in triplicate.
In vitro migration and invasion
In vitro invasion and migration assays were performed as described previously (30). For the migration assay, cells (3–5×104 cells; 100 µl) were seeded into the upper chamber of a Transwell plate (8-µm pore size; Corning, Inc.) in 200 µl serum-free media, whereas the lower chamber was filled with 600 µl media supplemented with 20% FBS. For the invasion assay, cells (3–5×104 cells; 100 µl) in serum-free medium were seeded into the upper chamber of a Transwell plate, which had been pre-coated with Matrigel (BD Biosciences) at 4°C overnight and then 37°C for 30 min, and the lower chamber was filled with 600 µl media supplemented with 20% FBS. After incubation at 37°C for 24 h, migrated/invaded cells in the lower compartment were washed with PBS, fixed with 4% paraformaldehyde for 30 min and stained with 1% crystal violet for 20 min. Both the fixing and staining steps were conducted at room temperature. For each filter, the numbers of migrated/invaded cells on the lower surface of the membrane were counted in five randomly selected fields at ×200 magnification under a Nikon E200 light microscope (Nikon Corporation). The experiment was performed three times to calculate the mean number of cells.
A rescue experiment was performed using Verteporfin (VP; Selleck Chemicals), a small molecule Hippo pathway inhibitor that can inhibit the YAP-TEA domain transcription factor interaction. DMSO was used to dissolve VP, which was added to the H1299 cells with culture medium at a final concentration of 10 µM at 37°C for 24 h. Next, H1299 cells were collected for western blotting, and migration and invasion assays.
Subcellular fractionation
A Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Institute of Biotechnology) was used to separate the nuclear and cytoplasmic fractions of lung cancer cell extracts, according to the manufacturer's protocol. The proteins in the extracts were then analyzed using western blotting.
Co-immunoprecipitation
Proteins were extracted from the cells prior to and following transfection. The cells were collected and incubated with 300 µl RIPA lysis buffer (Beyotime Institute of Biotechnology) with protease inhibitors for 40 min on ice. After protein concentration was quantified using a Bradford assay, 6,000 µg each lysate was added to 2 µg INHBA, Merlin or IgG (cat. no. B900620; ProteinTech Group, Inc.) antibodies and incubated at 4°C overnight. Subsequently, 20 µl protein A/G-agarose beads (Santa Cruz Biotechnology, Inc.) were added and the samples were gently agitated for 3 h at 4°C. The samples were then centrifuged at 2,500 × g for 5 min at 4°C and washed three times with lysis buffer to collect the precipitate. Finally, the precipitate was boiled with 40 µl loading buffer at 100°C for 5 min and analyzed using western blotting as aforementioned.
Statistical analysis
All experiments were repeated more than three times, and the data are presented as the mean ± SD. All data were analyzed using SPSS statistical software package version 16.0 (SPSS, Inc.) and GraphPad Prism 5 software (GraphPad Software, Inc.). χ2 test was used to analyze the association between INHBA expression and the clinical characteristics of patients with NSCLC. Wilcoxon test and Mann-Whitney U tests were used for non-parametric data as appropriate. The Kaplan-Meier method and log-rank tests and Cox regression analysis were used to analyze the association between INHBA expression and OS and DFS in patients with NSCLC. An unpaired Student's t-test was used to compare the inter-group difference between two groups. Comparative data among multiple groups were analyzed using one-way ANOVA followed by Tukey's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical relevance of INHBA positivity in samples of NSCLC
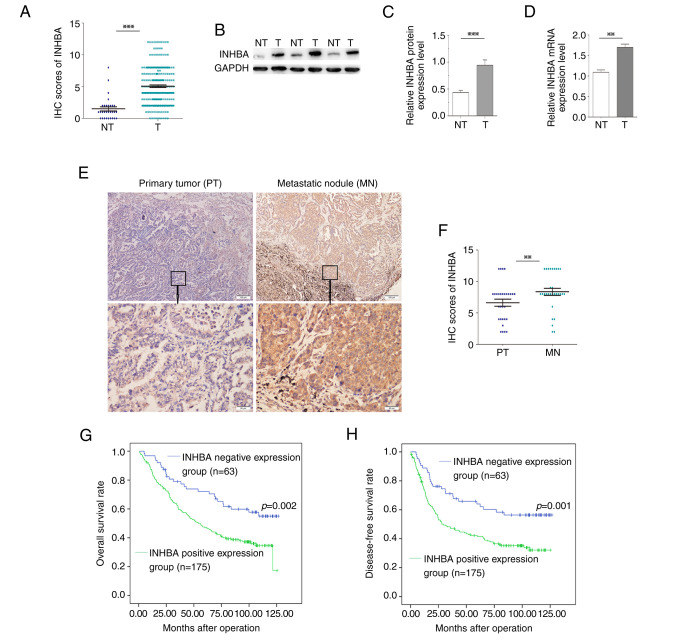
To determine the clinical relevance of INHBA in NSCLC, the expression levels of INHBA were detected using immunostaining of a TMA containing 238 NSCLC and 30 normal lung samples. The IHC scores of INHBA in 238 NSCLC tissues were significantly higher compared with those in the 30 cases of normal lung tissues (Fig. 1A). Moreover, INHBA immunoreactivity was graded as negative or positive. Out of the 238 cases, INHBA expression was positive in 73.5% (175/238) of NSCLC tissues, whereas only 13.3% (4/30) of normal lung samples demonstrated a positive INHBA signal (Table I). Furthermore, 24 NSCLC tissues and paired adjacent non-tumor tissues were processed for western blot analysis of INHBA. The protein expression levels of INHBA in NSCLC samples were significantly higher compared with those in non-tumor tissues (P<0.001; Fig. 1B and C). Similar results were observed with regard to the mRNA expression levels (P<0.01; Fig. 1D). In addition, INHBA expression was detected in 30 pairs of NSCLC primary tumors and corresponding metastatic nodules using IHC. The expression levels of INHBA were significantly increased in metastatic nodules compared with those in the primary tumors (P=0.003; Fig. 1E and F). Pearson χ2 tests for clinical association revealed that positive INHBA expression in NSCLC samples was significantly associated with advanced tumor stage (TNM stage II–IV; P=0.017) and poor differentiation (P=0.016) (Table I).
Figure 1.
Clinical significance of INHBA in non-small cell lung cancer. (A) IHC scores of INHBA in 238 NSCLC tissues were significantly higher compared with those in 30 cases of normal lung tissues. (B) Western blotting of INHBA expression levels in T tissues and in matched, adjacent NT tissues. (C) Semi-quantification of western blotting of 24 paired tissue samples. INHBA protein expression levels were significantly higher in T samples compared with those in NT samples. (D) mRNA expression levels of INHBA were significantly higher in T samples compared with those in NT samples. (E) Representative cases showed that INHBA expression was increased in MN tissues compared with in paired PT tissues (original magnification, ×40 or ×200). (F) Based on the scores from immunohistochemistry, the abundance of INHBA was elevated in MN tissues compared with in PT tissues (n=30). (G) Kaplan-Meier curve for overall survival in 238 patients. Higher INHBA expression levels in NSCLC, based on immunohistochemistry scores, were associated with poorer overall survival. (H) Kaplan-Meier curve for disease-free survival. Higher INHBA expression levels in NSCLC were associated with poorer disease-free survival. **P<0.01, ***P<0.001. INHBA, inhibin βA; IHC, immunohistochemistry; NSCLC, non-small cell lung cancer; T, tumor; NT, non-tumor; MN, metastatic nodule; PT, primary tumor.
Table I.
Clinical characteristics of patients (n=238) with non-small cell lung cancer who were positive or negative for INHBA expression.
| INHBA | ||||
|---|---|---|---|---|
|
|
||||
| Characteristic | Number of patients | Negative | Positive | P-value |
| Group | 0.001a | |||
| Non-tumor | 30 | 26 | 4 | |
| Tumor | 238 | 63 | 175 | |
| Age, years | 0.660 | |||
| <60 | 124 | 31 | 93 | |
| ≥60 | 114 | 32 | 82 | |
| Sex | 0.871 | |||
| Female | 66 | 18 | 48 | |
| Male | 172 | 45 | 127 | |
| Histology | 0.082 | |||
| ADC | 138 | 44 | 94 | |
| SCC | 82 | 16 | 66 | |
| LCC | 18 | 3 | 15 | |
| Carcinoma differentiation | 0.016a | |||
| Well | 32 | 10 | 22 | |
| Moderate | 94 | 33 | 61 | |
| Poor | 112 | 20 | 92 | |
| Venous infiltration | 0.431 | |||
| Absent | 76 | 23 | 53 | |
| Present | 162 | 40 | 122 | |
| TNM stage | 0.017a | |||
| I | 100 | 35 | 65 | |
| II–IV | 138 | 28 | 110 | |
ADC, adenocarcinoma; SCC, squamous cell carcinomas; LCC, large cell carcinoma; TNM, tumor-node-metastasis.
Statistically significant.
INHBA positivity is associated with poor prognosis for patients with NSCLC
To assess the ability of INHBA to predict patient prognosis in NSCLC, the present study examined whether INHBA expression was associated with the overall survival (OS) and disease-free survival (DFS) of patients with NSCLC. OS-based Kaplan-Meier survival curves were constructed to analyze cases with positive and negative staining of INHBA among patients with NSCLC. As shown in Fig. 1G, high INHBA expression in NSCLC was significantly associated with a shorter OS (log-rank=9.875; P=0.002); the OS rate decreased from 58.7% in patients with INHBA-negative NSCLC to 36.6% in patients with INHBA-positive tumors. Multivariate Cox regression revealed that in addition to tumor stage (TNM stage II–IV; P=0.001), INHBA expression was an independent prognostic marker for OS of patients with NSCLC (P=0.019) (Table II).
Table II.
Analysis of overall survival and disease-free survival using multivariate Cox regression.
| OS | DFS | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age, years (≥60 vs. <60) | 1.258 | 0.888–1.782 | 0.197 | 1.196 | 0.843–1.698 | 0.315 |
| Sex (male vs. female) | 0.881 | 0.598–1.297 | 0.521 | 0.873 | 0.591–1.288 | 0.493 |
| Histology | ||||||
| ADC vs. SCC | 0.502 | 0.278–1.136 | 0.221 | 1.253 | 0.478–3.525 | 0.680 |
| ADC vs. LCC | 1.601 | 0.889–2.569 | 0.086 | 0.917 | 0.293–2.990 | 0.821 |
| Carcinoma differentiation | ||||||
| Well vs. moderate | 1.698 | 0.601–4.112 | 0.396 | 0.478 | 0.259–1.632 | 0.102 |
| Well vs. poor | 0.852 | 0.421–1.698 | 0.395 | 2.459 | 1.125–4.963 | 0.061 |
| Venous infiltration (present vs. absent) | 1.040 | 0.714–1.515 | 0.836 | 1.035 | 0.711–1.505 | 0.859 |
| TNM stage (I vs. II–IV) | 2.773 | 1.887–4.074 | <0.0001a | 2.852 | 1.935–4.203 | 0.001a |
| INHBA (positive vs. negative) | 1.698 | 1.092–2.639 | 0.019a | 1.196 | 1.098–2.638 | 0.017a |
Statistically significant. OS, overall survival; DFS, disease-free survival; TNM, tumor-node-metastasis; HR, hazard ratio; CI, confidence interval.
Kaplan-Meier analysis also demonstrated that INHBA positivity was associated with shorter DFS (log-rank=10.511; P=0.001; Fig. 1H). Using multivariate Cox regression analysis, INHBA was revealed to be an independent indicator of DFS (P=0.017), in addition to the tumor stage (TNM stage II–IV; P=0.001) (Table II). These data suggested that INHBA might be a potential prognostic marker in NSCLC.
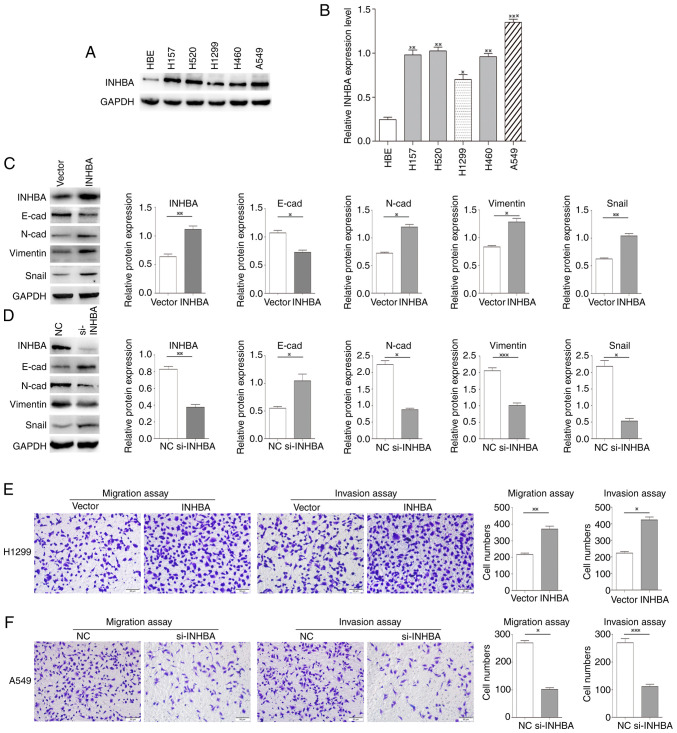
Overexpression of INHBA promotes the invasion of lung cancer cells
The expression of INHBA was significantly higher in all five NSCLC cell lines compared with HBE cells. To assess the effects of INHBA on lung cancer invasion, cells expressing low levels (H1299 cells) and high levels of INHBA (A549 cells) were selected for further study (Fig. 2A and B). Western blotting revealed that in H1299 cells, INHBA overexpression reduced the expression levels of E-cadherin, and increased the expression levels of N-cadherin, vimentin and Snail (P=0.02, P=0.01, P=0.03, P=0.001, respectively; Fig. 2C). By contrast, in A549 cells, siRNA-mediated INHBA knockdown increased E-cadherin expression levels, and decreased the expression levels of N-cadherin, vimentin and Snail (P=0.03, P=0.07, P=0.0001, P=0.02, respectively; Fig. 2D). Furthermore, the invasive and migratory abilities of H1299 cells were increased by INHBA overexpression (P=0.0025 for invasion; P=0.012 for migration; Fig. 2E), whereas in A549 cells, INHBA knockdown decreased invasive and migratory abilities (P=0.0123 for invasion; P=0.0004 for migration; Fig. 2F).
Figure 2.
INHBA overexpression promotes the invasion of lung cancer cells. (A) Western blotting and (B) semi-quantification of INHBA expression levels in five lung cancer cell lines and HBE cells. (C) Transient transfection of H1299 cells with the INHBA plasmid resulted in decreased expression levels of E-cadherin, and increased expression levels of N-cadherin, vimentin and Snail. (D) INHBA knockdown increased the expression levels of E-cadherin, and decreased the expression levels of N-cadherin, Snail and vimentin in A549 cells. (E) Cell invasion and migration were enhanced in H1299 cells transiently transfected with the INHBA plasmid. (F) Knockdown of INHBA decreased the invasive and migratory abilities of A549 cells. Original magnification, ×200. *P<0.05, **P<0.01, ***P<0.001. INHBA, inhibin βA; NC, negative control; si, small interfering; HBE, human bronchial epithelial.
Nuclear YAP protein levels are associated with INHBA expression in NSCLC tissues
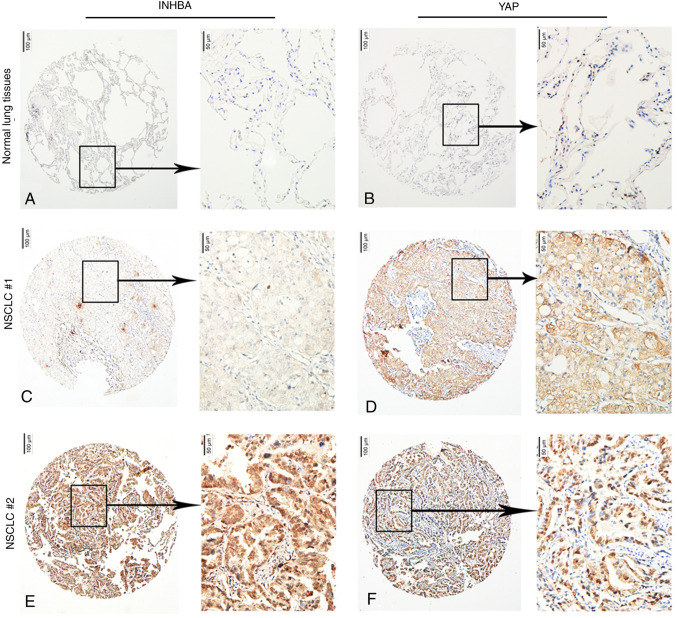
In NSCLC cases, YAP can be found in either the nucleus or the cytoplasm (31,32); therefore, the association between INHBA levels and nuclear and cytoplasmic YAP levels was examined. For the quantitative analysis of INHBA and YAP levels in 238 NSCLC samples, IHC scores were used. The results revealed INHBA levels negatively associated with cytoplasmic YAP levels (P=0.014) and positively associated with nuclear YAP levels (P=0.045; Fig. 3A-F; Table III).
Figure 3.
Representative photomicrographs of the results of a human tissue microarray for INHBA and YAP in 238 tumor tissues and 30 normal lung tissues. In normal lung tissues, INHBA expression was (A) negative and (B) positive YAP nuclear expression was limited to type II pneumocytes. Under conditions of (C) low INHBA protein expression levels, (D) nuclear YAP protein expression levels were low and the expression levels of cytoplasmic YAP were high. Under conditions of (E) high INHBA protein expression levels, (F) cytoplasmic YAP protein expression levels were low and nuclear YAP protein expression levels were high. Original magnification, ×40 or ×200. INHBA, inhibin βA; YAP, yes-associated protein; NSCLC, non-small cell lung cancer.
Table III.
Association between INHBA expression and the cytoplasmic and nuclear expression of YAP in NSCLC.
| INHBA expression | |||||
|---|---|---|---|---|---|
|
|
|||||
| Expression | Number of patients | Negative | Positive | χ2 | P-value |
| Cytoplasmic YAP expression | |||||
| Negative | 84 | 14 | 70 | 6.411 | 0.014a |
| Positive | 154 | 49 | 105 | ||
| Nuclear YAP expression | |||||
| Negative | 81 | 28 | 53 | 4.136 | 0.045a |
| Positive | 157 | 35 | 122 | ||
Statistically significant.
INHBA induces YAP nuclear translocation and activation in lung cancer cells
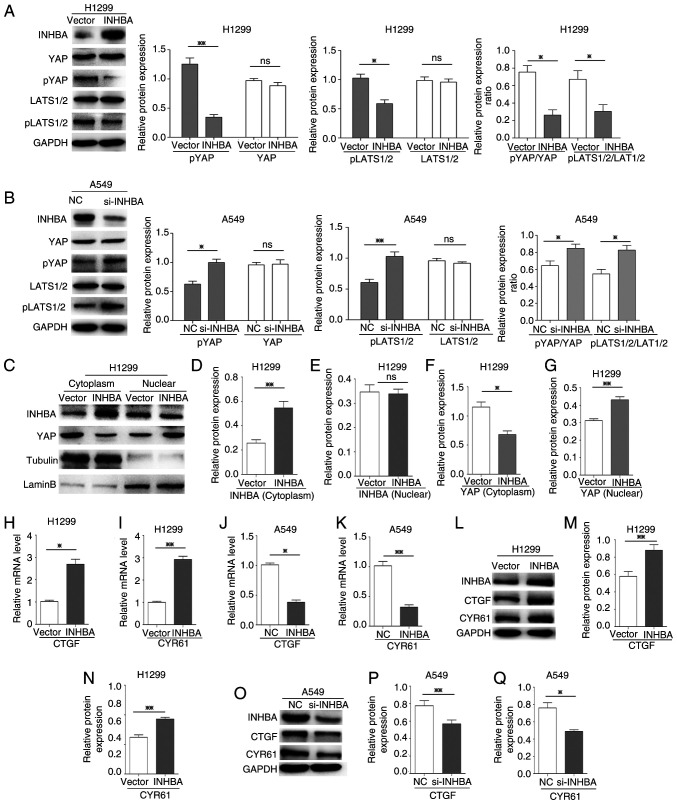
To assess if YAP was activated by INHBA, the present study analyzed the phosphorylation status of YAP and LATS1/2, which are major components of the Hippo pathway. Western blotting revealed that the protein expression levels of p-YAP (Ser127) and p-LATS1/2 (T1079 + T1041), and the ratio of p-YAP/YAP and p-LATS1/2/LATS1/2 were all significantly decreased (P=0.0045, P=0.0313, P=0.0378 and P=0.0145, respectively) at 48 h in H1299 cells transfected with the INHBA overexpression plasmid, whereas total YAP and LATS1/2 expression levels were not affected compared with those in cells transfected with the vector control plasmid (Fig. 4A). By contrast, in A549 cells where INHBA expression was knocked down using siRNA, the protein expression levels of p-YAP and p-LATS1/2, and the ratio of p-YAP/YAP and p-LATS1/2/LATS1/2 were higher compared with those in the NC-siRNA-transfected cells (P=0.0306, P=0.0028, P=0.0217 and P=0.0352, respectively; Fig. 4B). However, total YAP and LATS1/2 levels were unaffected by INHBA siRNA. These results indicated that INHBA may inhibit Hippo signaling. For YAP activation, its nuclear localization is the key regulatory mechanism (22); therefore, subcellular fractionation was performed to assess the cellular localization of YAP. Compared with the vector groups, YAP accumulated in the nucleus to a greater extent than in the cytoplasm in cells overexpressing INHBA (P=0.014 for cytoplasmic YAP; P=0.0041 for nuclear YAP; Fig. 4C-G). In addition, overexpression of INHBA increased transcription of the canonical YAP target genes CTGF and cysteine rich angiogenic inducer 61 (CYR61) (P=0.0173 for CTGF; P=0.0094 for CYR61; Fig. 4H and I). Conversely, knockdown of INHBA decreased the mRNA expression levels of the two target genes in A549 cells (P=0.0104 for CTGF; P=0.0057 for CYR61; Fig. 4J and K). Similar results were observed at the protein expression levels (P=0.0031 and 0.0046 for CTGF in the INHBA overexpression and knockdown groups, respectively; P=0.0015 and 0.0313 for CYR61, in the INHBA overexpression and knockdown groups, respectively; Fig. 4L-Q). These results suggested that in lung cancer cells, INHBA induced YAP nuclear translocation and activation by regulating LATS1/2 and then inhibiting Hippo signaling.
Figure 4.
INHBA activates YAP and induces its nuclear translocation in lung cancer cells. (A and B) Western blotting revealed that the expression levels of pYAP, pLATS1/2, and the ratio of pYAP/YAP and pLATS1/2/LATS1/2, were decreased in H1299 cells transfected with the INHBA overexpression plasmid and increased in A549 cells transfected with si-INHBA; total YAP and LATS1/2 levels were not affected. (C-G) Subcellular fractionation indicated that, after INHBA overexpression in cytoplasm, YAP accumulation in the cytoplasmic fraction decreased and YAP accumulation in the nuclear fraction increased. RT-qPCR indicated that H1299 cells transiently transfected with the INHBA plasmid exhibited increased (H) CTGF and (I) CYR61 mRNA expression levels. RT-qPCR demonstrated that A549 cells transiently transfected with si-INHBA exhibited decreased (J) CTGF and (K) CYR61 mRNA expression levels. (L-N) Western blotting indicated that H1299 cells transiently transfected with the INHBA plasmid exhibited increased CTGF and CYR61 protein expression levels. (O-Q) Western blotting demonstrated that A549 cells transiently transfected with si-INHBA exhibited decreased CTGF and CYR61 protein expression levels. *P<0.05, **P<0.01. INHBA, inhibin βA; YAP, yes-associated protein; si, small interfering; NC, negative control; LATS, large tumor suppressor kinase; p, phosphorylated; CTGF, connective tissue growth factor; CYR61, cysteine rich angiogenic inducer 61; RT-qPCR, reverse transcription-quantitative PCR.
INHBA promotes lung cancer cell invasion by negatively regulating Hippo signaling
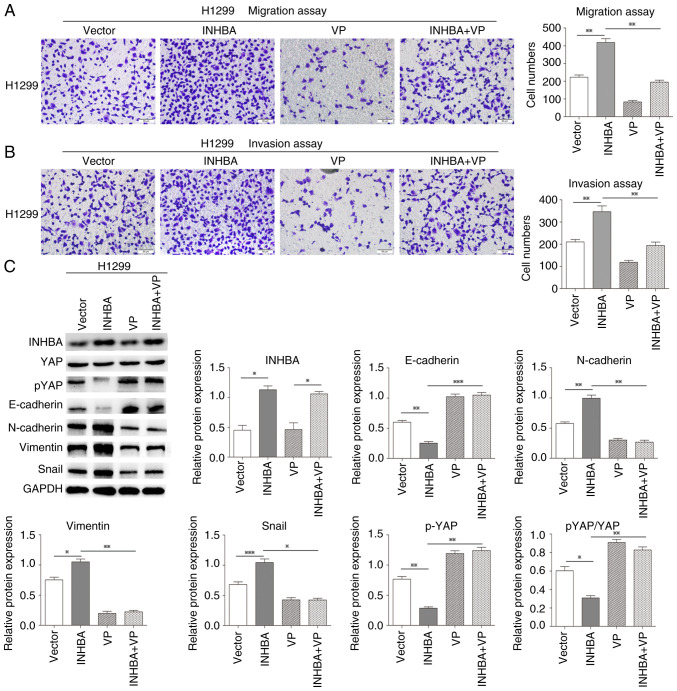
The Hippo-YAP pathway is involved in lung cancer (25). The present study revealed that in lung cancer cells INHBA overexpression induced YAP nuclear translocation and inhibited LATS1/2 phosphorylation. Thus, it was hypothesized that INHBA might function as an upstream regulator in tumor metastasis via the Hippo-YAP pathway. The present study further assessed whether INHBA overexpression could promote NSCLC invasion via activation of Hippo signaling and attenuating non-phosphorylated YAP nuclear translocation. Therefore, a rescue experiment was performed using VP, a small molecule inhibitor that can inhibit the YAP-TEA domain transcription factor interaction, in H1299 cells to determine its influence on the restoration of migration and invasion caused by INHBA overexpression. As expected, after VP treatment, the effects of INHBA overexpression on the promotion of NSCLC invasion and migration were significantly reversed in vitro (P=0.0026 for migration; P=0.0056 for invasion; Fig. 5A and B). Moreover, the increases in the protein expression levels of N-cadherin, vimentin and Snail, and the decreases in E-cadherin and pYAP expression, and pYAP/YAP ratio were also reversed after VP treatment in H1299 cells (P=0.0125 for N-cadherin, P=0.0024 for vimentin, P=0.0161 for Snail, P=0.0004 for E-cadherin, P=0.0058 for pYAP and P=0.0028 for pYAP/YAP, respectively; Fig. 5C). These findings indicated that increased INHBA expression may promote NSCLC invasion and epithelial-mesenchymal transition via inactivation of the Hippo pathway.
Figure 5.
INHBA promotes lung cancer cell invasion by negatively regulating Hippo signaling. Rescue experiments showed that INHBA overexpression-induced promotion of NSCLC (A) invasion and (B) migration were reversed following VP treatment in H1299 cells. Original magnification, ×200. (C) Rescue experiments showed that INHBA overexpression-induced increases in N-cadherin, vimentin and Snail expression levels, and decreases in E-cadherin, pYAP expression levels and the pYAP/YAP ratio were also reversed after VP treatment in H1299 cells. *P<0.05, **P<0.01, ***P<0.001. INHBA, inhibin βA; YAP, yes-associated protein; p, phosphorylated; VP, Verteporfin.
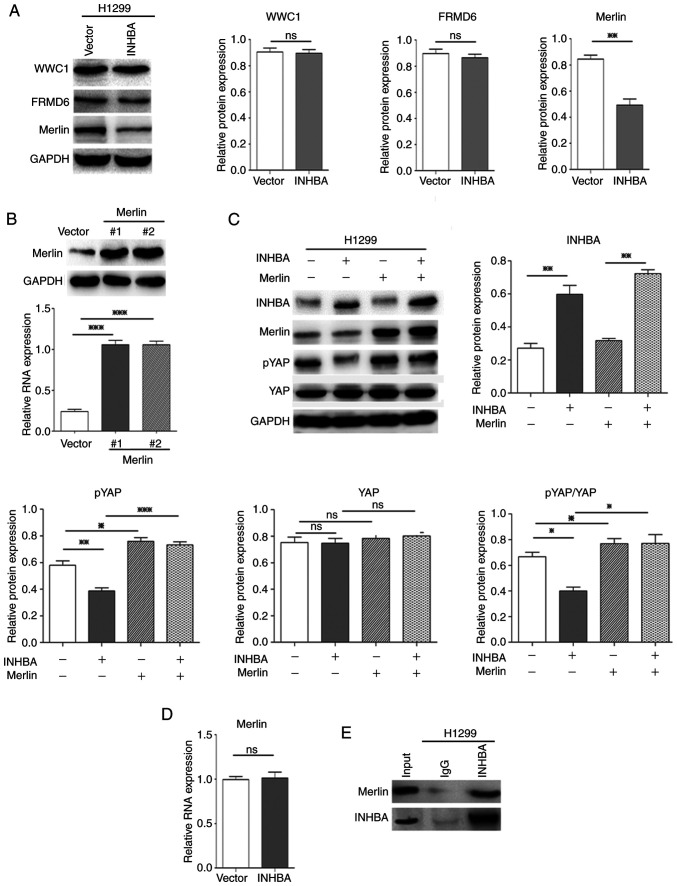
INHBA negatively regulates Hippo signaling by downregulating Merlin protein expression levels in lung cancer cells
To determine how INHBA enhances nuclear translocation of oncogenic YAP, the present study assessed the upstream regulators of the Hippo-YAP pathway. The present study had already verified that INHBA could affect the phosphorylation status of YAP and LATS1/2; therefore, it was further determined whether the other three important upstream regulators of Hippo-YAP signaling, Willin/FRMD6, Merlin/NF2 and KIBRA/WWC1, were regulated by INHBA. Western blotting revealed that in H1299 cells overexpressing INHBA, at 48 h, only the protein expression levels of Merlin were significantly decreased (P=0.0049, compared with cells transfected with the vector control plasmid; Fig. 6A). Merlin suppresses tumorigenesis by regulating certain pathways, including the mammalian target of rapamycin and Hippo signaling pathways. Merlin activates the Hippo pathway, which induces oncogenic YAP1 cytoplasmic retention (33). To further determine whether overexpression of INHBA inhibited Hippo signaling and increased YAP nuclear translocation via the downregulation of Merlin, a rescue experiment was performed using plasmid-mediated exogenous overexpression of Merlin. The effectiveness of Merlin overexpression plasmids was confirmed by western blotting (Fig. 6B). As expected, overexpression of Merlin significantly reversed the effect of INHBA on inhibiting the Hippo pathway and activating YAP in vitro in H1299 cells (P=0.0007 for pYAP, P=0.0124 for pYAP/YAP; Fig. 6C). Notably, INHBA overexpression only downregulated the protein expression levels of Merlin, whereas no significant alterations were noted in the mRNA expression levels (Fig. 6D). Moreover, INHBA could bind to the Merlin protein (Fig. 6E). These results suggested that INHBA may downregulate Merlin levels via post-translational modification. Overall, these results demonstrated that increased expression of INHBA induced NSCLC invasion via Hippo pathway inactivation by downregulating Merlin levels, which inhibits phosphorylation and promotes nuclear translocation of YAP, which then upregulates CTGF and CYR61 levels (Fig. 7A and B).
Figure 6.
INHBA negatively regulates Hippo signaling by downregulating Merlin protein expression levels in lung cancer cells. (A) Western blotting revealed that only Merlin protein expression levels were decreased in H1299 cells transfected with the INHBA overexpression plasmid. (B) Western blotting revealed the efficacy of Merlin overexpression plasmids in H1299 cells. (C) Rescue experiments showed that overexpression of Merlin reversed the effect of INHBA on activating YAP in H1299 cells. (D) Reverse transcription-quantitative PCR indicated no change in NF2 (encoding Merlin) mRNA expression levels after H1299 cells were transiently transfected with the INHBA plasmid. (E) Immunoprecipitation analysis revealed that INHBA could bind to Merlin. *P<0.05, **P<0.01, ***P<0.001. INHBA, inhibin βA; YAP, yes-associated protein; WWC1, WW and C2 domain-containing 1; FRMD6, FERM domain-containing 6; NF2, neurofibromin 2.
Figure 7.
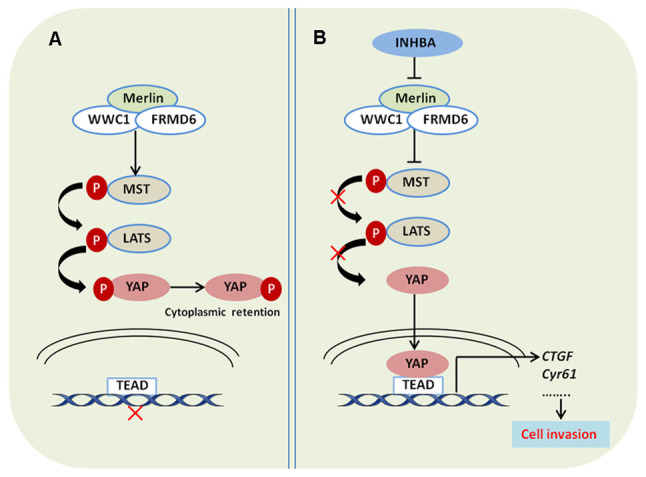
INHBA promotes lung cancer cell invasion via inhibiting the Hippo pathway. (A) Protein complex of Merlin, FRMD6 and WWC1 activates the Hippo pathway, which increases the phosphorylation of YAP and prevents its translocation and binding to TEAD in the nucleus. (B) INHBA inhibits the Hippo pathway by downregulating Merlin expression, which decreases the phosphorylation of YAP, and promotes its translocation and binding to TEAD in the nucleus. INHBA, inhibin βA; YAP, yes-associated protein; WWC1, WW and C2 domain-containing 1; FRMD6, FERM domain-containing 6; MST, mammalian sterile 20-like 1/MOB kinase; LATS, large tumor suppressor kinase; p, phosphorylated; TEAD, TEA domain; CTGF, connective tissue growth factor; Cyr61, cysteine rich angiogenic inducer 61.
Discussion
Investigations into the role of INHBA have demonstrated that positive INHBA expression is significantly associated with increasing pathological tumor stage, lymph node metastasis, higher histological grade and vascular invasion in urothelial carcinoma and gastric cancer (14,15). Moreover, Seder et al (17) suggested that in stage I lung ADC, INHBA may encourage cell proliferation, and that a worse outcome may be associated with upregulated INHBA expression (17). However, it remains unclear as to whether INHBA participates in the regulation of metastasis and invasion of NSCLC.
Previous studies have suggested that the subcellular localization of YAP implies its functional diversity in lung cancer. For example, Wang et al (31) revealed that the majority of YAP was accumulated in the nucleus, and that YAP overexpression contributed to the progression and poor prognosis of NSCLC (31). Conversely, in lung ADC, high levels of YAP in the cytoplasm were associated with better survival (32). The results of the present study demonstrated that, in NSCLC, INHBA expression was inversely associated with YAP expression in the cytoplasm and was positively associated with nuclear YAP levels. Moreover, it was revealed that, in NSCLC, the expression of INHBA was significantly increased, and it was positively associated with worse clinicopathological features, such as poor carcinoma differentiation and advanced TNM stage. These findings support the two of previously reported studies, which indicated that INHBA and activin expression levels were elevated in NSCLC compared with those in non-malignant lungs, and were associated with advanced stages of primary human NSCLC (17,18). Furthermore, the present study demonstrated that patients with INHBA positivity had an unfavorable prognosis and that INHBA expression was an independent predictive factor of prognosis. These results prompted the hypothesis that INHBA affects the invasion and metastasis of NSCLC. It is worth noting that there was a limitation in comparing INHBA expression between NSCLC and adjacent normal lung tissues. Due to insufficient materials, most of the NSCLC samples were not associated with adjacent normal lung tissue; thus, 30 unpaired adjacent tissues were used as the control. Previous studies have suggested that YAP may be vital for TGF-β-mediated induction of certain target genes, thus indicating that the expression of INHBA, as a member of the TGF-β superfamily, may be associated with YAP expression (21). The present study determined the association between the nuclear and cytoplasmic levels of YAP and INHBA. The expressions levels of INHBA were negatively associated with the expression levels of YAP in the cytoplasm and were positively associated with the expression levels of YAP in the nucleus. These findings suggested that INHBA may regulate the subcellular localization of YAP and could be involved in lung cancer development; this hypothesis warranted further exploration. Therefore, the present study assessed the underlying molecular mechanism by which INHBA affects NSCLC invasion and metastasis.
As expected, overexpression of INHBA promoted NSCLC cell invasion via inhibiting the Hippo pathway. Mechanistically, INHBA inhibited LATS1/2 phosphorylation and induced YAP nuclear translocation by downregulating Merlin levels. Notably, in the Hippo pathway, Merlin is an upstream regulator and a core component of the Merlin/KIBRA/FRMD6 complex, which promotes the phosphorylation of YAP and activates the Hippo pathway (33). Additionally, Merlin is encoded by the NF2 gene. Alcantara and Garcia (34) revealed that downregulation of NF2 promoted the migration and proliferation of lung cancer cells. Sánchez et al (35) also reported that invasive and metastatic lung adenocarcinoma exhibited lower Merlin protein levels compared with noninvasive tumors, and suggested that Merlin could be a promising therapeutic target to inhibit the progression of lung adenocarcinoma. Therefore, it may be hypothesized that Merlin has a cancer suppressive role in NSCLC, whereas INHBA may abrogate the cancer-suppressing effect of Merlin, and promote the invasion and metastasis of NSCLC.
Notably, in the present study, INHBA only downregulated Merlin at the protein level, but not at the mRNA level. Moreover, INHBA could bind to Merlin. These results suggested that INHBA may downregulate Merlin levels via post-translational modification. Previous studies have confirmed that the intracellular regulatory function of Merlin is lost via ubiquitination-mediated degradation (36,37); however, as a member of the TGF-β superfamily, INHBA itself has no ubiquitination function. Mota et al (38) confirmed that Merlin expression was associated with Smad7. Moreover, previous studies revealed that Smad7 is the ligation factor of E3 ubiquitin ligase of Smurf2, and the PPXY domain of Smad7 combines with the WW domain of Smurf2 to form an E3 ligase complex that regulates the ubiquitination-mediated degradation of related proteins (39,40). Therefore, it was hypothesized that INHBA may mediate the ubiquitination-mediated degradation of Merlin via the Smad7/Smurf2 E3 ligase complex. However, these speculations require further experimental verification.
In conclusion, the present study demonstrated that the invasive and metastatic potential of lung cancer cells was promoted by INHBA via the downregulation of Merlin expression, which may cause negative regulation of Hippo signaling, and then inhibit phosphorylation and promote nuclear translocation of YAP. These results suggested that INHBA may represent a potential therapeutic target for NSCLC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (grant nos. 82002668 and 81772489).
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 82002668 and 81772489).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YZ and EW conceived the research. YZ, SY and YaL designed the methodology. YZ, SY, YaY, JZ, YuL, PL, YoL, and YH performed the experiments. YZ wrote the original draft of the manuscript. EW reviewed and edited the manuscript, and supervised the study. YZ and EW confirm the authenticity of all the raw data All authors read and approved the final manuscript.
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee, and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The present study was approved by the both the Institute Research Medical Ethics Committee of Sun Yat-sen University and China Medical University, and informed consent (written or verbal) was obtained from the patients in this study for retrospective analysis of tissue samples. All samples were anonymized.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 2.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL, Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: Analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 3.Sun S, Schiller JH, Spinola M, Minna JD. New molecularly targeted therapies for lung cancer. J Clin Invest. 2007;117:2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 5.de Mello RA, Madureira P, Carvalho LS, Araújo A, O'Brien M, Popat S. EGFR and KRAS mutations, and ALK fusions: Current developments and personalized therapies for patients with advanced non-small-cell lung cancer. Pharmacogenomics. 2013;14:1765–1777. doi: 10.2217/pgs.13.177. [DOI] [PubMed] [Google Scholar]
- 6.Wagner G, Stollenwerk HK, Klerings I, Pecherstorfer M, Gartlehner G, Singer J. Efficacy and safety of immune checkpoint inhibitors in patients with advanced non-small cell lung cancer (NSCLC): A systematic literature review. Oncoimmunology. 2020;9:1774314. doi: 10.1080/2162402X.2020.1774314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaddy-Kurten D, Tsuchida K, Vale W. Activins and the receptor serine kinase superfamily. Recent Prog Horm Res. 1995;50:109–129. doi: 10.1016/b978-0-12-571150-0.50010-x. [DOI] [PubMed] [Google Scholar]
- 8.Loomans HA, Andl CD. Intertwining of activin A and TGFβ signaling: Dual roles in cancer progression and cancer cell invasion. Cancers (Basel) 2014;7:70–91. doi: 10.3390/cancers7010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green JB, New HV, Smith JC. Responses of embryonic xenopus cells to activin and FGF are separated by multiple dose thresholds and correspond to distinct axes of the mesoderm. Cell. 1992;71:731–739. doi: 10.1016/0092-8674(92)90550-V. [DOI] [PubMed] [Google Scholar]
- 10.Vale W, Rivier C, Hsueh A, Campen C, Meunier H, Bicsak T, Vaughan J, Corrigan A, Bardin W, Sawchenko P, et al. Chemical and biological characterization of the inhibin family of protein hormones. Recent Prog Horm Res. 1988;44:1–34. doi: 10.1016/b978-0-12-571144-9.50005-3. [DOI] [PubMed] [Google Scholar]
- 11.Kelner N, Rodrigues PC, Bufalino A, Fonseca FP, Santos-Silva AR, Miguel MC, Pinto CA, Leme AF, Graner E, Salo T, et al. Activin A immunoexpression as predictor of occult lymph node metastasis and overall survival in oral tongue squamous cell carcinoma. Head Neck. 2015;37:479–486. doi: 10.1002/hed.23627. [DOI] [PubMed] [Google Scholar]
- 12.Fu S, Zhang N, Yopp AC, Chen D, Mao M, Chen D, Zhang H, Ding Y, Bromberg JS. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25-precursors. Am J Transplant. 2004;4:1614–1627. doi: 10.1111/j.1600-6143.2004.00566.x. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa K, Funaba M. Activin in humoral immune responses. Vitam Horm. 2011;85:235–253. doi: 10.1016/B978-0-12-385961-7.00012-3. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 15.Lee HY, Li CC, Huang CN, Li WM, Yeh HC, Ke HL, Yang KF, Liang PI, Li CF, Wu WJ. INHBA overexpression indicates poor prognosis in urothelial carcinoma of urinary bladder and upper tract. J Surg Oncol. 2015;111:414–422. doi: 10.1002/jso.23836. [DOI] [PubMed] [Google Scholar]
- 16.Peng S, Wang J, Hu P, Zhang W, Li H, Xu L. INHBA knockdown inhibits proliferation and invasion of nasopharyngeal carcinoma SUNE1 cells in vitro. Int J Clin Exp Pathol. 2020;13:854–868. [PMC free article] [PubMed] [Google Scholar]
- 17.Seder CW, Hartojo W, Lin L, Silvers AL, Wang Z, Thomas DG, Giordano TJ, Chen G, Chang AC, Orringer MB, Beer DG. Upregulated INHBA expression may promote cell proliferation and is associated with poor survival in lung adenocarcinoma. Neoplasia. 2009;11:388–396. doi: 10.1593/neo.81582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wamsley JJ, Kumar M, Allison DF, Clift SH, Holzknecht CM, Szymura SJ, Hoang SA, Xu X, Moskaluk CA, Jones DR, et al. Activin upregulation by NF-κB is required to maintain mesenchymal features of cancer stem-like cells in non-small cell lung cancer. Cancer Res. 2015;75:426–435. doi: 10.1158/0008-5472.CAN-13-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Narimatsu M, Samavarchi-Tehrani P, Varelas X, Wrana JL. Distinct polarity cues direct Taz/Yap and TGFβ receptor localization to differentially control TGFβ-induced smad signaling. Dev Cell. 2015;32:652–656. doi: 10.1016/j.devcel.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Fujii M, Toyoda T, Nakanishi H, Yatabe Y, Sato A, Matsudaira Y, Ito H, Murakami H, Kondo Y, Kondo E, et al. TGF-β synergizes with defects in the Hippo pathway to stimulate human malignant mesothelioma growth. J Exp Med. 2012;209:479–494. doi: 10.1084/jem.20111653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating yorkie, the drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Pan D. The Hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Q, Lin X, Zhang X, Jiang G, Zhang Y, Miao Y, Rong X, Zheng X, Han Y, Han X, et al. WWC3 regulates the Wnt and Hippo pathways via dishevelled proteins and large tumour suppressor 1, to suppress lung cancer invasion and metastasis. J Pathol. 2017;242:435–447. doi: 10.1002/path.4919. [DOI] [PubMed] [Google Scholar]
- 26.Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol. 2015;10:1240–1242. doi: 10.1097/JTO.0000000000000663. [DOI] [PubMed] [Google Scholar]
- 27.Sobin LH, Gospodarowicz MK, Christian Wittekind C. 8th edition. Oxford: Wiley-Blackwell; 2017. International Union Against Cancer (UICC): TNM classification of malignant tumours. [Google Scholar]
- 28.Zhang Y, Zhao Y, Jiang G, Zhang X, Zhao H, Wu J, Xu K, Wang E. Impact of p120-catenin isoforms 1A and 3A on epithelial mesenchymal transition of lung cancer cells expressing E-cadherin in different subcellular locations. PLoS One. 2014;9:e88064. doi: 10.1371/journal.pone.0088064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Yan S, Chen J, Gan C, Chen D, Li Y, Wen J, Kremerskothen J, Chen S, Zhang J, Cao Y. WWC2 is an independent prognostic factor and prevents invasion via Hippo signalling in hepatocellular carcinoma. J Cell Mol Med. 2017;21:3718–3729. doi: 10.1111/jcmm.13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Dong Q, Zhang Q, Li Z, Wang E, Qiu X. Overexpression of yes-associated protein contributes to progression and poor prognosis of non-small-cell lung cancer. Cancer Sci. 2010;101:1279–1285. doi: 10.1111/j.1349-7006.2010.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun PL, Kim JE, Yoo SB, Kim H, Jin Y, Jheon S, Kim K, Lee CT, Chung JH. Cytoplasmic YAP expression is associated with prolonged survival in patients with lung adenocarcinomas and epidermal growth factor receptor tyrosine kinase inhibitor treatment. Ann Surg Oncol. 2014;21(Suppl 4):S610–S618. doi: 10.1245/s10434-014-3715-5. [DOI] [PubMed] [Google Scholar]
- 33.Su T, Ludwig MZ, Xu J, Fehon RG. Kibra and Merlin activate the Hippo pathway spatially distinct from and independent of expanded. Dev Cell. 2017;40:478–490.e3. doi: 10.1016/j.devcel.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcantara KMM, Garcia RL. MicroRNA-92a promotes cell proliferation, migration and survival by directly targeting the tumor suppressor gene NF2 in colorectal and lung cancer cells. Oncol Rep. 2019;41:2103–2116. doi: 10.3892/or.2019.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez NC, Medrano-Jiménez E, Aguilar-León D, Pérez-Martínez L, Pedraza-Alva G. Tumor necrosis factor-induced miR-146a upregulation promotes human lung adenocarcinoma metastasis by targeting Merlin. DNA Cell Biol. 2020;39:484–497. doi: 10.1089/dna.2019.4620. [DOI] [PubMed] [Google Scholar]
- 36.Wei Y, Yee PP, Liu Z, Zhang L, Guo H, Zheng H, Anderson B, Gulley M, Li W. NEDD4L-mediated Merlin ubiquitination facilitates Hippo pathway activation. EMBO Rep. 2020;21:e50642. doi: 10.15252/embr.202050642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang X, Jang SW, Wang X, Liu Z, Bahr SM, Sun SY, Brat D, Gutmann DH, Ye K. Akt phosphorylation regulates the tumour-suppressor Merlin through ubiquitination and degradation. Nat Cell Biol. 2007;9:1199–1207. doi: 10.1038/ncb1641. [DOI] [PubMed] [Google Scholar]
- 38.Mota MSV, Jackson WP, Bailey SK, Vayalil P, Landar A, Rostas JW, III, Mulekar MS, Samant RS, Shevde LA. Deficiency of tumor suppressor Merlin facilitates metabolic adaptation by co-operative engagement of SMAD-Hippo signaling in breast cancer. Carcinogenesis. 2018;39:1165–1175. doi: 10.1093/carcin/bgy078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/S1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Z, Liu C, Chen B, Tang W, Liu Z, Cao W, Li X. Smad7 down-regulation via ubiquitin degradation mediated by Smurf2 in fibroblasts of hypertrophic scars in burned patients. Burns. 2021;47:1333–1341. doi: 10.1016/j.burns.2020.12.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.