SUMMARY
We have identified and validated a spaceflight-associated microRNA (miRNA) signature that is shared by rodents and humans in response to simulated, short-duration and long-duration spaceflight. Previous studies have identified miRNAs that regulate rodent responses to spaceflight in low-Earth orbit, and we have confirmed the expression of these proposed spaceflight-associated miRNAs in rodents reacting to simulated spaceflight conditions. Moreover, astronaut samples from the NASA Twins Study confirmed these expression signatures in miRNA sequencing, single-cell RNA sequencing (scRNA-seq), and single-cell assay for transposase accessible chromatin (scATAC-seq) data. Additionally, a subset of these miRNAs (miR-125, miR-16, and let-7a) was found to regulate vascular damage caused by simulated deep space radiation. To demonstrate the physiological relevance of key spaceflight-associated miRNAs, we utilized antagomirs to inhibit their expression and successfully rescue simulated deep-space-radiation-mediated damage in human 3D vascular constructs.
Graphical Abstract
In Brief
Malkani et al. uncover the role of circulating microRNAs as both a potential biomarker for health risks associated with spaceflight and a countermeasure to mitigate the damage caused to the body by the space environment.
INTRODUCTION
Humans and vertebrate organisms exhibit a unique set of biological responses to the space environment, including bone and muscle atrophy (Burkhart et al., 2019; Gerbaix et al., 2017), immune dysfunction (Baqai et al., 2009), neurobehavioral changes (Ronca et al., 2019), vision impairment (Mader et al., 2011), and cardiovascular damage (Elgart et al., 2018). Ionizing radiation (IR) and microgravity are the main biological stressors in low-Earth orbit (LEO), which is the region encompassed by the Earth’s magnetosphere where all crewed missions currently occur. Although exposure to deep space radiation, which is primarily composed of galactic cosmic rays (GCRs), is reduced within LEO due to the magnetosphere, IR still poses a risk to astronauts in LEO, and GCRs are a critical health risk in the upcoming lunar and Mars missions (Townsend, 2005). Thus, combining LEO experiments with ground-based analogs of spaceflight stressors, including simulated GCRs, allows investigations into the mechanisms behind biological responses to spaceflight.
MicroRNAs (miRNAs) are short (~22 nucleotides long) sequences of non-coding RNA that can regulate gene expression at the transcriptional and translational level and interact directly with proteins (Vanderburg and Beheshti, 2020). miRNAs have been implicated as predictors and regulators for several diseases, including cancer (Macfarlane and Murphy, 2010), neurodegenerative diseases (Konovalova et al., 2019), and autoimmune disorders (Chen et al., 2016). miRNAs have also been found to be involved in biological responses to both microgravity (Girardi et al., 2014) and IR (Simone et al., 2009). Therefore, identifying miRNAs (and their networks) that are shared between humans and model organisms in response to spaceflight stressors could reveal targets for biomarker and countermeasure development to reduce the associated health risks.
Previously, we used spaceflown rodent RNA sequencing (RNA-seq) datasets from the NASA GeneLab database to make unbiased predictions about miRNAs potentially involved in regulating the transcriptional response to spaceflight (Beheshti et al., 2018). The datasets analyzed in that study are described in the STAR Methods. Our analysis identified a set of 13 spaceflight-associated miRNAs that were predicted to drive a systemic response and suggested transforming growth factor (TGF)-β1 as an overarching regulator of their expression (Beheshti et al., 2018). TGF-β1 is a major regulatory cytokine mediating the progression of cancer (Rooke and Crosier, 2001), autoimmunity (Marie et al., 2006), and neuroinflammation (Cekanaviciute et al., 2014). However, it is important to note that this spaceflight-associated miRNA signature was defined computationally, based on transcriptomic analysis, and not measured experimentally.
Here, we experimentally validated the predicted spaceflight-associated miRNA signature by quantifying circulating miRNAs in serum from rodents exposed to simulated spaceflight conditions: microgravity, partial gravity, LEO-relevant IR, simulated deep space radiation, and combined exposures to reduced gravity and simulated space radiation, as well as true spaceflight in LEO. We supported spaceflight-stressor-mediated changes in the expression of the predicted miRNAs by performing agnostic miRNA-seq in selected datasets, and our experimental validation also incorporated simulated deep space radiation conditions to help evaluate the extent to which the miRNA signature may be shared by different space environments. Finally, we addressed the human relevance of our findings by analyzing miRNAs released by irradiated human immune cells ex vivo and miRNA expression changes in astronauts from the NASA Twins Study in vivo (Garrett-Bakelman et al., 2019).
We expanded our initial panel of 13 candidate miRNAs (Beheshti et al., 2018) to include 15 target miRNAs with the following justifications: (1) miRNA let-7 was subdivided into two more specific family members (let-7a and let-7c) identified in recent space biology studies (Režen et al., 2014; Wei et al., 2018), and (2) an additional miRNA (miR-92a) was added after analyzing astronaut data from the NASA Twins Study. Furthermore, we investigated the potential downstream effects of spaceflight-associated human miRNAs using single-cell RNA (scRNA) sequencing of a targeted gene panel of leukocyte-depleted fractions in the NASA Twins Study (Garrett-Bakelman et al., 2019). Specifically, we compared gene expression in cells collected within 3 days upon spaceflown twin’s (TW) return to his preflight and postflight samples, as well as control samples taken from the ground control twin (HR). Single-cell transcriptomics also let us determine the individual cell types that display the strongest perturbations in gene expression consistent with miRNA signatures identified in bulk transcriptomics, and single-cell assay for transposase accessible chromatin (scATAC) sequencing data were used to delineate chromatin changes that may have altered the accessibility of the spaceflight-associated miRNAs.
RESULTS
Predicted Spaceflight miRNA Signature in Rodents Was Validated Using Simulated Microgravity and Deep Space Radiation
To validate the predicted spaceflight-associated miRNA panel, we quantified their changes in response to ground analogs of individual spaceflight stressors in rodent models: simulated microgravity using hindlimb unloading (HU) (Morey-Holton and Globus, 1998) as well as IR using either gamma rays or major components of galactic cosmic rays—protons and/or high mass and charge 56Fe particles (deep space relevant exposure) (Mewaldt, 1994). We exposed male 16-week-old C57BL/6J mice to HU, IR, or their combination and measured the set of selected 15 miRNAs in their serum at 1 or 11 days post-exposure using droplet digital PCR (ddPCR) (Hindson et al., 2013).
Euclidean clustering of serum miRNA concentrations separated the miRNAs into two main clusters: (1) deep-space-radiation-associated clusters, which are significantly regulated by 56Fe IR and 56Fe IR in combination with HU; and (2) microgravity-associated clusters, primarily altered in HU-only conditions (Figures 1A–1C). An additional subset of miRNAs was significantly increased in mouse serum only, early (1 day) after exposure to the combination of mixed proton/56Fe irradiation and HU, but was not affected by individual stressors (Figure 1A). Of note, we have provided a discussion regarding potential hemolysis that might occur with the serum/plasma samples utilized throughout this manuscript in the STAR Methods.
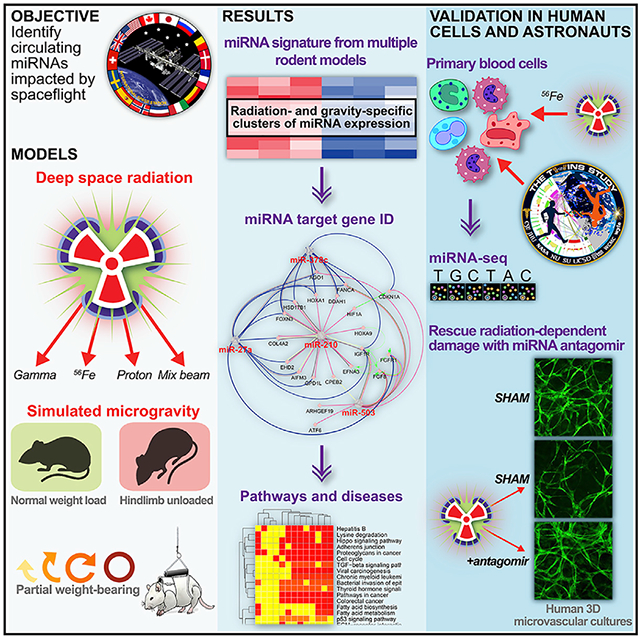
Figure 1. Quantified miRNAs in Serum from Rodents Exposed to Ground Analogs of Spaceflight.
Microgravity and radiation conditions of spaceflight were simulated alone and in combination.
(A) Quantification of miRNAs in serum from rodents exposed to hindlimb unloading (HU) or normal loading (NL) for 3 days before 2 Gy gamma, 1 Gy proton, 1 Gy 600MeV/n 56Fe, 2 Gy 600MeV/n56Fe, or sham irradiation. HU or NL was continued for another 1 or 11 days after irradiation.
(B) Fold changes (log2) of miRNAs compared to NL sham mice. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student’s t test. The error bars represent SEM.
(C) t-Distributed Stochastic Neighbor Embedding (t-SNE) plot showing how the significantly regulated miRNAs are associated with each experimental group. Statistical significance is based on comparing each group with the sham irradiated NL mice.
(D) Top 50 predicted gene targets for all statistically significant miRNAs determined by Cytoscape plugin ClueGo.
(E) The predicted diseases regulated by the miRNA signature determined through miRNet. The blue nodes represent cancer; green, cardiovascular disease; yellow, neurological diseases; orange, muscle degeneration; purple, digestive issues.
(F) Gene Ontology (GO) pathways predicted to be regulated by all statistically significant spaceflight-associated miRNAs determined by the DIANA-microT-CDS (v5.0) algorithm.
(G) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways predicted to be regulated by all statistically significant spaceflight-associated miRNAs.
Most miRNAs (with the exception of miR-217) were observed to increase with radiation dose and linear energy transfer (LET; gamma < 150 MeV proton < 600 MeV/n 56Fe) in mouse serum both at 1 day and 11 days post-IR. At 11 days post-IR, the only significant miRNA increases were observed in IR-only (no HU) mice irradiated with either 1 Gy or 2 Gy 600 MeV/n 56Fe or 1 Gy protons (Figures 1A and 1B). In combination, these results indicate that the miRNA signature is mainly driven by the particle radiation component, which is proportional to dose and LET.
Based on all miRNAs that were significantly different in spaceflight-associated conditions, we predicted the top 50 targeted genes (Figure 1D), key diseases regulated by the miRNAs (Figure 1E), and significant pathways (Figures 1F, 1G, and S1). The top 50 genes targeted by these miRNAs (Figure 1D) primarily regulate cell-cycle pathways and protein kinase activity, in addition to hematopoiesis and blood cell development (Figures 1F and S1). The majority of diseases regulated by these miRNAs are related to cancer (Figure 1E), though we also uncovered other diseases associated with spaceflight, including cardiovascular (green circles), neurological (yellow circles) and digestive disorders (purple circles), and muscular degeneration (orange circles) (Figure 1E). Similarly, key significantly regulated pathways include carcinogenesis as well as TGF-β signaling, which supports our previous prediction of TGF-β as a key regulator of spaceflight-associated miRNA functions and implies downstream carcinogenic and immune effects. Finally, the pathways also include adherens junctions, which, combined with cardiovascular disease associations, suggests vascular impacts (Figures 1F and 1G).
To examine the contribution of reduced gravity to the miRNA signature at higher resolution, a partial weight bearing (PWB) Wistar male rat model (Mortreux et al., 2018) was used to simulate varying degrees of gravitational loading. We observed that decreasing amounts of gravitational loading increased the quantities of miRNAs of interest, suggesting a gravity-dependent dose response (Figures 2A–2C). These results also validated a reduced-gravity-specific miRNA signature in both mice and rats and separated the miRNAs associated with reduced gravity from those associated with exposure to simulated deep space radiation in mice. The only observed exception was miR-217, which did not change. A similar miRNA dose response was observed based on correlation with muscle mass (Figure 2D): the miRNAs of interest increased with reduced muscle mass (which is associated with lower simulated gravity both in this rat model and in astronauts).
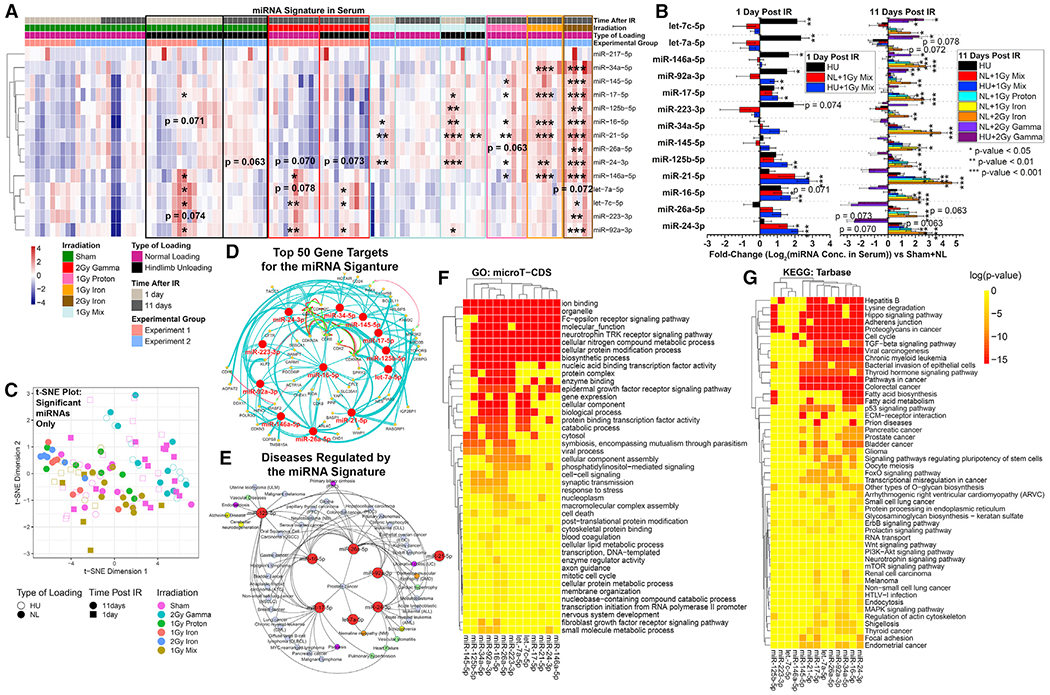
Figure 2. Serum miRNA Quantification from PWB Rats Modeled under Four Different Gravity Conditions.
(A) Global comparisons of miRNAs from 20%, 40%, 70%, and 100% gravitational loading conditions. The 100% loaded group is further split into FH or NH.
(B) t-SNE plot reveals distinct separation of the miRNAs between different experimental groups for each gravity condition, as well as a separation between the NH and FH 100% gravity groups.
(C) Fold changes (log2) for miRNA increases compared to 100% gravity with FH group.
(D) Correlation between serum miRNA increases and physiological muscle mass over different gravity conditions (PWB). Significance indicated in circles. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student’s t test. Circle size is proportional to the correlation coefficient, also indicated by color (legend on right-hand side).
(E) Global comparison of miRNAs in serum from mice flown on the International Space Station (ISS) for 30 days, sacrificed 4 days after returning to Earth.
(F) Serum miRNA comparisons for only the 16-week-old mice.
(G) Serum miRNA comparisons for only the 37-week-old mice.
(H) t-SNE plot of all mice flown on the ISS shows subtle separation between young and old groups.
(I and J) t-SNE plot of old mice shows separation between spaceflight and ground mice, while t-SNE plot of young mice fails to show separation.
(K) Correlation of miRNA quantities and organ weights for all mice flown on the ISS. The significance of each correlation is indicated in the circles. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student’s t test. Circle size is proportional to the correlation coefficient.
Notably, this model included two control groups of animals exposed to full gravitational loading: no harness (NH) and full harness (FH) (Figures 2A and 2B). Both conditions had been included in prior studies, and no functional or phenotypical differences were recorded, although there was a difference in miRNA expression between NH and FH 100% gravity groups (Figures 2A and 2B), possibly due to a trend in reduction in muscle mass in FH controls (not significant). Therefore, further comparison between groups included only the FH control (Figure 2C).
Predicted Spaceflight miRNA Signature Was Partially Observed in Serum from Mice Exposed to LEO
Next, we investigated whether the spaceflight-environment-linked miRNA signature is distinguishable in true spaceflight samples, using serum from young and old female BALB/cAnNTac mice flown to the International Space Station (ISS) on Rodent Research Mission 8. In contrast with the simulated spaceflight cohorts, these mice experienced short-lasting hypergravity during takeoff and landing in addition to microgravity while in orbit, spent 4 days reacclimating on ground after the flight before sample collection, and received a significantly lower dose of IR than simulation experiments: 0.01128 Gy over approximately 30 days on the ISS.
Although the combined mouse cohort did not exhibit any significantly increased miRNAs compared to basal (no spaceflight, normal housing conditions) controls (Figure 2E), when the samples were separated by age (Figures 2F and 2G), we observed clustering between spaceflight and basal controls in the older group (31–37 weeks at sacrifice) corresponding to middle-age in humans, which is more representative of typical astronaut cohorts, and no detectable clustering in the younger group (10–16 weeks at sacrifice) (Figures 2H–2J). Considering that these mice had 4 days to reacclimate to ground conditions before the samples were collected, these results may suggest that it takes longer for serum miRNA levels to return to basal levels in older mice, while younger mice recover faster. This finding is consistent with our prior results from simulated spaceflight studies, in which miRNA levels in irradiated mice start to return to control by 11 days following an over-100-fold-higher dose of IR than received by the spaceflown mice (Figure 1A).
Another notable result from this spaceflight experiment is the correlation between organ weights and miRNA concentration in spaceflight mice and basal controls. Normalized to total body weight, adrenal gland weight increases with concentration of the majority of quantified miRNAs, and, conversely, brain weight decreases with increasing miRNA concentrations. While these correlations are not statistically significant, they suggest the possibility of lingering impacts on specific organs (Figure 2K). Increases in adrenal gland weight might be related to chronic stress, since the adrenal gland regulates stress responses (Ulrich-Lai et al., 2006) that have been previously associated with increased systemic corticosterone levels during spaceflight (Pecaut et al., 2017). Meanwhile, a volumetric reduction of brain features has been observed in cosmonauts (Pechenkova et al., 2019), though the physiological relevance of these findings remains to be elucidated. Since systemic increases in corticosterone levels also lead to reduced brain weight (Coburn-Litvak et al., 2004), chronic stress might be an underlying factor behind both adrenal gland and brain results, and future studies are needed to examine miRNA as its potential biomarker.
Spaceflight-Relevant miRNA Signature Was Also Identified in Agnostic Sequencing Study of Simulated-Spaceflight-Exposed Mice
To understand the specificity and variation of spaceflight-relevant miRNA levels within the global miRNA population, miRNA-seq was performed on tissue samples from 16-week-old female C57BL/6J mice after exposure to simulated spaceflight conditions: normal loading (NL) or HU for 2 weeks prior to irradiation with 5 Gy gamma, 1 Gy simulated solar particle event (simSPE), 0.5 Gy simulated simplified galactic cosmic rays (SimGCRSim), or sham control. SimGCRSim consists of five ions: protons at 1000 MeV, 28Si at 600 MeV/n, 4He at 250 MeV/n, 16O at 350 MeV/n, 56Fe at 600 MeV/n, and protons at 250 MeV (Simonsen et al., 2020). This dose of radiation is equivalent to what an astronaut is predicted to receive in deep space during a Mars mission, though it is modeled as a single exposure over 25 min instead of the actual chronic exposures over 1.5 years. Simulated solar particle event (SimSPE) irradiation consists of protons from 50 MeV to 150 MeV. Twenty-four hours after irradiation, the mice were sacrificed, and miRNA-seq was performed on tissue samples from the plasma, heart, liver, and soleus muscle.
We observed highly variable miRNA expression profiles between organs, with the exception of heart and soleus muscle (Figures 3A and 3B). This variation underscores the importance of establishing a circulating systemic miRNA signature that can be measured in plasma, which is the most accessible source for biomarker evaluation in spaceflight conditions. Additionally, the high correlation between miRNA expression changes in heart and soleus muscle unsurprisingly suggests that the damage in these muscle tissues may be driven by similar changes in miRNAs (Figures 3A and 3B). Notably, miRNA-seq profiles from plasma also are more heterogeneous than those from individual organs, suggesting that plasma miRNA quantification might allow a more finely tuned spaceflight signature that could differentiate between different types of exposures or provide personalized risk assessment (Figures 3A and 3B).
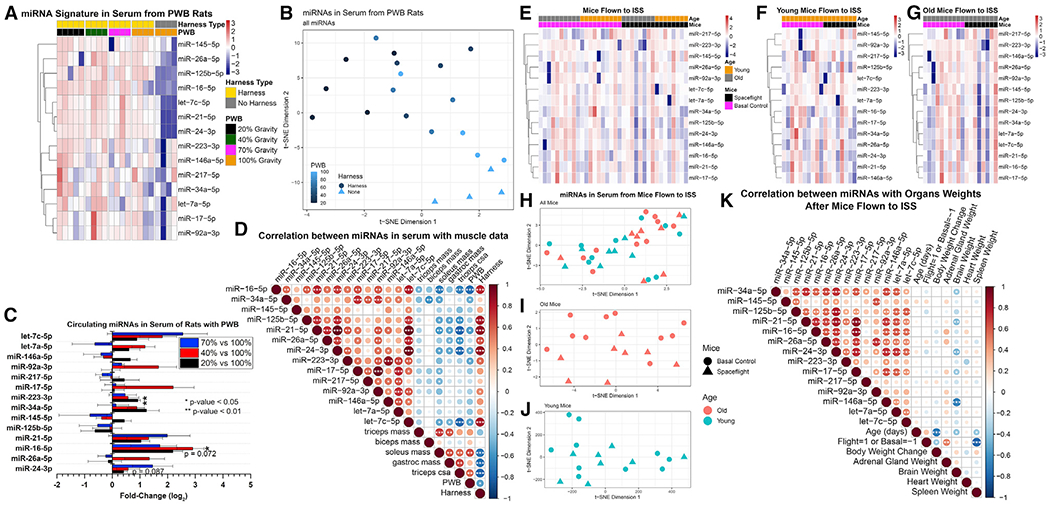
Figure 3. miRNA-seq of Heart, Liver, Plasma, and Soleus Muscle from Mice Exposed to Simulated Spaceflight Stressors.
(A) Comparison of miRNA expression profiles between organs.
(B) Combined and individual t-SNE plots for significantly expressed miRNAs in each organ.
(C) Overlap of significant differentially expressed miRNAs between organs for GCR HU and sham NL and for SPE HU versus sham NL. The center, dark gray region indicates significant miRNAs shared by all organs.
(D) Top 20 genes regulated by the four common miRNAs differentially expressed in GCR HU versus sham NL.
(E) Pathways that are significantly regulated by the top 20 genes shown in (D).
(F) miRNA fold changes between HU and NL groups for each irradiation condition. miRNAs from our proposed signature are included, as well as the four additional miRNAs implicated by miRNA-seq in (C).
(G) C2 pathways analysis from miRNA-seq data shows pathways that are being suppressed. The larger the circle, the more significant the effect, and darker purple indicates a larger magnitude of suppression. miRNAs highlighted in yellow or blue were included in our predicted spaceflight signature. miRNAs highlighted in yellow are known to be involved in spaceflight-induced cardiac damage.
Comparison of significantly regulated miRNAs identified four miRNAs that are increased in all organs in the GCR+HU (“combined spaceflight stressors”) group compared to sham NL (“no spaceflight stressors” control) (Figure 3C). Therefore, we decided to focus on the GCR+HU irradiated group for further analysis. We computed the top 20 genes regulated by the four common differentially expressed miRNAs (Figure 3D) as well as the pathways that were significantly regulated by these genes (Figure 3E) and found potential cardiovascular as well as systemic circulatory targets (Figure 3E).
We next quantified miRNA fold changes between GCR/SPE/Gamma+HU and sham NL conditions, focusing on the panel of spaceflight-associated miRNAs (top 15) as well as the four additional miRNAs that were found to be shared among all tissues for GCR+HU versus NL by miRNA-seq (bottom 4) (Figure 3F). Additional data is available in a companion paper (Wuu et al., 2020). We observed trends or significance for multiple spaceflight miRNAs, and the same miRNA family members differentially expressed across multiple tissues. For example, for miRNA-503-5p, expression was decreased in the liver, soleus, and heart but strongly increased in plasma (Figure 3F), indicating potential organ-dependent miRNA changes with the key systemic stressor response stemming from the circulating miRNAs. MiR-503 belongs to the miR-16 extended family (Caporali and Emanueli, 2011; Wang et al., 2019), which is one of the miRNAs predicted to be involved in the spaceflight response (Figure 1A) and impacts cardiovascular diseases through the inhibition of angiogenesis (Besnier et al., 2019; Caporali and Emanueli, 2011).
Finally, we performed pathway analysis utilizing the miRNA-seq data with C2 curated gene sets from MSigDB on the miRNA-seq data and displayed all significantly regulated pathways (Figure 3G) for the liver and soleus muscle. Data for the plasma and heart tissues are in companion papers and show similar results (Paul et al., 2020; Wuu et al., 2020), and the pathways that showed suppression (false discovery rate [FDR]-adjusted p value < 0.25; Figure 3G) have been previously implicated in cardiovascular adaptations to spaceflight (Beheshti et al., 2018, 2019). These specific pathways were selected for the countermeasure experiments (below). This experiment demonstrates that although the individual response of the miRNAs might not be significantly regulated, the global response of miRNAs on downstream gene targets reveals regulation by miRNAs belonging to the spaceflight signature.
Spaceflight-Associated Rodent miRNA Signature Was Partially Shared by Human Responses Ex Vivo and In Vivo
We next show that spaceflight-associated miRNA signature is partially conserved in human samples that had been exposed to real or simulated space environment conditions. First, we analyzed human peripheral blood mononuclear cell (PBMC) responses to 0.3 Gy 600 MeV/n 56Fe particles that form a biologically significant component of GCRs, using PBMCs isolated from 12 healthy human (non-astronaut) donors. Thus, this experiment simulated deep-space-relevant radiation, without simulating microgravity. We observed an increase in spaceflight-associated miRNAs in the supernatant of irradiated PBMCs 4 hours, post-irradiation (Figures 4A–4D), as well as a positive correlation with increasing donor BMI and cell number (Figures 4B and 4D; Table S3) .
Figure 4. Quantification of miRNAs in Human Samples Exposed to Spaceflight or IR.
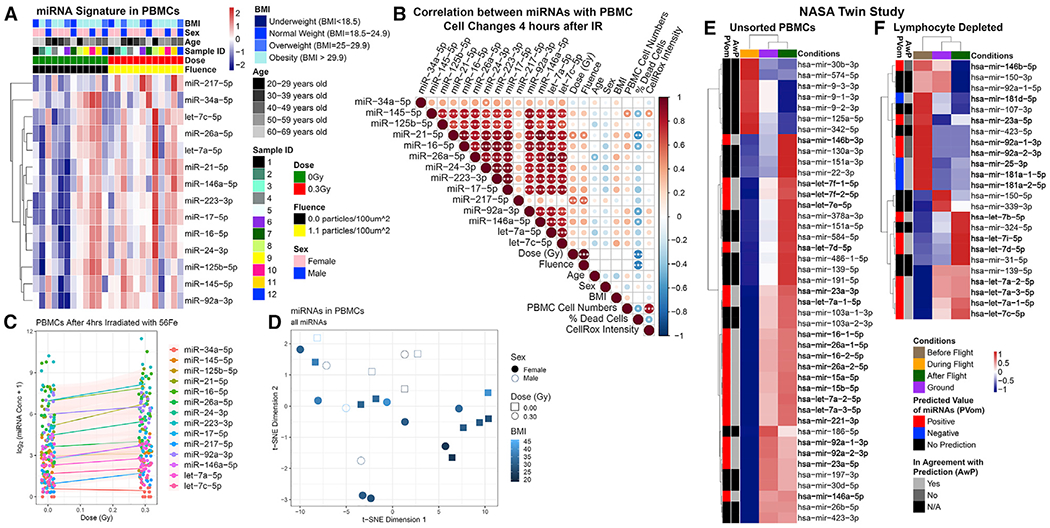
(A) Comparison of human peripheral blood mononuclear cell (PBMC) samples from 12 subjects 4 h post-irradiation with 0.3 Gy or sham (0 Gy) 56Fe irradiation.
(B) Correlation between miRNA quantities and cell phenotypes 4 h post-irradiation. The significance of each correlation is indicated in the circles. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed Student’s t test. Circle size is proportional to the correlation coefficient.
(C) General additive model fit of the miRNA quantities as a function of dose.
(D) t-SNE plot of the samples clustering as a function of BMI based on miRNA concentration.
(E and F) NASA Twins Study data for (E) unsorted PBMCs and (F) LD cells in comparison to the spaceflight miRNA signature predictions.
Finally, in order to assess the impact of the real spaceflight environment on human cells, we analyzed miRNA-sequence data on total PBMC and lymphocyte-depleted (LD) samples from the NASA Twins Study. This study involved actual LEO (low-LET radiation and microgravity) exposure for 340 days for a total of 146.34 mSv (or 0.1463 Gy). Unsorted PBMCs isolated during and after LEO spaceflight showed persistent changes in miRNA expression for the astronaut in space, which partially follows the predicted spaceflight signature (Figure 4E). Similarly, LD cells sampled before and after flight also indicated persistent changes in miRNA expression, though not all miRNAs behaved as predicted by the spaceflight signature (Figure 4F). Notably, the spaceflown twin (TW) had consistently higher expression of the miRNA target genes compared to the HR even at baseline/preflight, despite having higher levels of miRNAs as well (Figures 4E and 4F); thus, TW pre-flight levels were considered a better control than ground control twin (HR) pre-flight levels. This difference in miRNA expression is not surprising, due to known discordant miRNA expression profiles in twins (Xiao et al., 2019).
Of the miRNAs found in the irradiated PBMC panel, only miR-92a-3p was significantly downregulated and let-7c-5p was upregulated in the TW miRNA-sequence data (Figure 4F). However, the downstream targets of the irradiation-associated miRNAs were significantly decreased between preflight and postflight samples, except the targets of hsa-miR-92a-3p and hsa-miR-26a-5p. This suggests that there was an increase in some irradiation-associated miRNA functions in TW, and the effects of miR-92a-3p and miR-26a-5p were more acute than the other miRNAs. Similar to the lack of response of miR-217 in animal models based on ddPCR results, miR-217 only had three targets within the targeted panel, none of which were significantly changed by spaceflight, indicating that miR-217, although implicated by original unbiased in silico predictions, does not respond to the stress of the space environment.
We next analyzed the changes in miRNA expression across different blood cell populations that were associated with spaceflight-mediated miRNA differences in the same NASA Twins Study. Specifically, we conducted targeted scRNA-seq and scAb-seq (epitope mapping) of LD Twins Study samples across multiple time points. In total, 23,408 cells were sequenced across 12 samples: 6 TW and 6 HR (Figure S2 and Table S4). Cell types were identified on a per cluster basis by both RNA and epitope levels (Figure 5A). Module scores for the targets of the subset of spaceflight-associated miRNAs that were found to be upregulated in irradiated human PBMCs were calculated using Seurat and compared among TW preflight, return, and postflight samples (Figures 5B–5N).
Figure 5. Spaceflight-Associated miRNA Target Genes Are Downregulated upon TW Return and Postflight Compared to Preflight, with Individual Cell Types Being Differentially Affected.
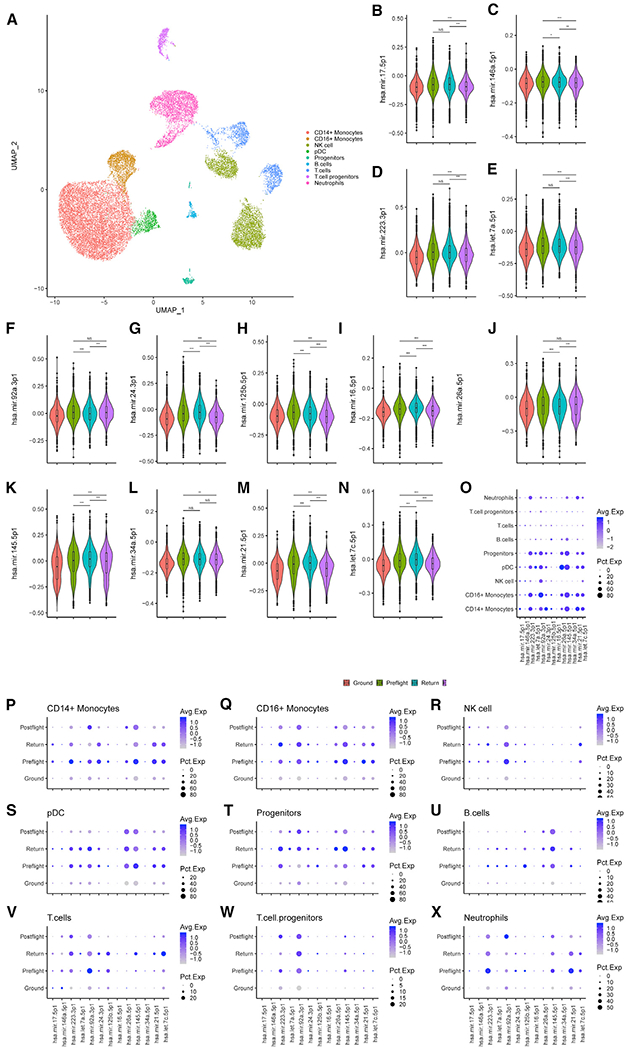
(A) Uniform manifold approximation and projection (UMAP) plot of 23,408 cells grouped by cell-type-identified clusters.
(B–N) Violin plots with mean and first and third quartiles of the indicated miRNA target gene module scores. Cells were separated into four groups: cells from HR GD114, GD138, GD182, GD3, GD-66, and GD66 as Ground; TW L-162 and L-148 days as Preflight; TW R0 as Return; and TW R36 and R191 as Postflight. Differences in expression of miRNA target gene modules scores was tested by Wilcoxon rank sum test. *p < 0.05, **p < 0.01, ***p < 0.001.
(O–W) Dot plots colored by average expression and size based on percent expression of the spaceflight-associated mRNA target gene module scores of cell types grouped by Ground, Preflight, Return, and Postflight.
When the cell population was split by cell type, we observed that the changes in miRNA-mediated gene expression were highly variable across cell types (Figure 5O). Dividing each cell type into ground, preflight, return, and postflight groups demonstrated that individual cell types were differentially affected by spaceflight-associated miRNAs. For example, CD14+ and CD16+ monocytes responded differently to hsa-miR-92a-3p: CD14+ monocytes showed decreased expression of hsa-miR-92a-3p targets, while CD16+ monocytes expressed higher levels of hsa-miR-92a-3p target genes upon return compared to either preflight or postflight. Progenitor (circulating hematopoietic stem) cells stood out in this analysis, as expression of the spaceflight-associated miRNA target genes increased specifically at the return time point. These differences among cell types indicate that immune cell composition may play a role in the downstream effects of spaceflight-associated miRNA increases and suggest return to Earth as a major stressor affecting miRNA expression.
The Increase in Expression of Spaceflight-Associated miRNAs Is Not Driven by Epigenetic Changes
To determine whether the changes in miRNA expression were due to spaceflight-induced chromatin changes, we used scATAC-seq on 11,344 cells (Figure S3 and Table S5). SnapTools and SnapATAC were used to create 5-kb bins across the genome and determine genome accessibility based on ATAC activity per cell, and then genes and miRNAs were assigned the ATAC activity based on the overlapping bin. ATAC activity is associated with chromatin accessibility, which can be used to determine whether the miRNAs upregulated in irradiated PBMCs were differently accessible in TW during spaceflight (Figure 6B). Of note, open chromatin peaks were observed at let-7c-5p, miR-34a-5p, miR-125b-5p, let-7a-5p, and miR-146a-5p. Furthermore, both control PBMCs and HR ground PMBCs displayed consistent ATAC peaks across the majority of the miRNA loci. Together, this suggests that the chromatin around these miRNA loci is open but not strongly affected by spaceflight. In addition, we assessed ATAC activity in different cell types (Figures 6C and 6D). The module score and dot plots of the miRNAs upregulated in irradiated PBMCs demonstrate that CD14+ monocytes and pre-dendritic cells (pDCs) show more miRNA accessibility that other cell populations (Figures 6C and 6D), indicating that chromatin states depend more on immune cell type than spaceflight.
Figure 6. Spaceflight Does Not Induce Epigenetic Changes around Spaceflight-Associated miRNA.
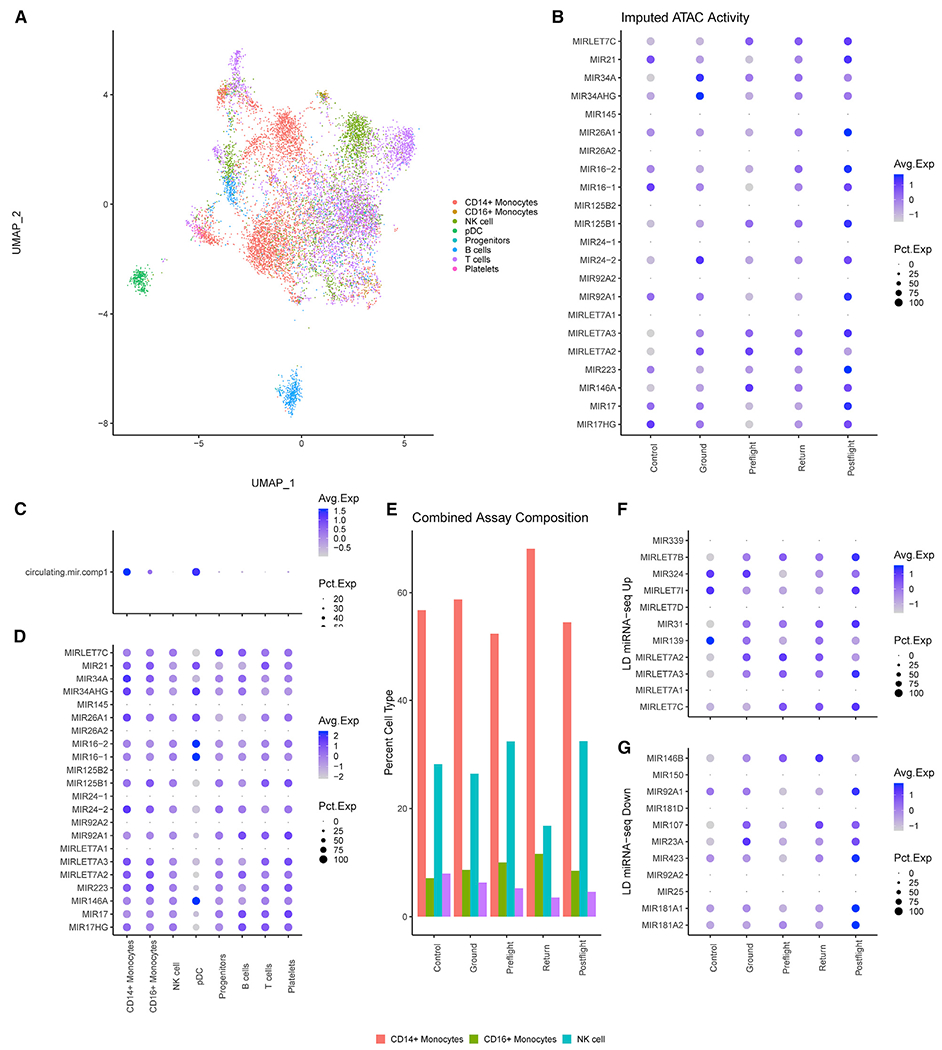
(A) UMAP plot of 11,344 cells grouped by cell type predicted by Seurat’s transfer anchor function from the 10× PBMC dataset (see Method Details).
(B) Dot plot colored by ATAC activity and sized based on percentage ATAC activity at the 5-kb bins associated with spaceflight-associated miRNA. Cells were separated into five groups: cells from the control (non-LD) PBMCs as Control, HR GD-66 and GD125 as Ground, TW L-71 as Preflight, TW R3 as Return, and TW R36 as Postflight.
(C) Dot plot of spaceflight-associated miRNA module scores with cells grouped by cell type across all samples.
(D) Dot plot of the individual spaceflight-associated miRNA ATAC activity with cells grouped by cell type.
(E) Composition analysis per group based after removal of B and T cells across both scATAC and scRNA analysis.
(F and G) Dot plots of ATAC activity at miRNA upregulated (F) or downregulated (G) in the LD miRNA-seq.
We next investigated immune cell composition at different time points during flight, specifically the proportion of CD14+ monocytes. Due to a low number of cells in the TW preflight sample, data from ATAC-seq and scRNA-seq experiments were combined to determine the immune cell distribution in LD fractions. (Figure 6E). Notably, the TW return samples had the highest proportion of CD14+ monocytes. This result, combined with the scRNA-seq of downstream targets, suggests that miRNA expression during spaceflight is regulated not by chromatin accessibility, but by immune cell distribution, and is particularly high in CD14+ monocytes. Finally, we evaluated whether specific ATAC peak changes correlated with expression levels from the Twins Study miRNA-seq (Figures 6F and 6G). However, ATAC activity was observed in the HR samples for upregulated miRNAs, indicating that the chromatin was already open prior to spaceflight.
Selective Inhibition of Spaceflight-Associated miRNAs Reduces Cardiovascular Damage in a Human Tissue Model after Exposure to Simulated Deep Space Radiation
Finally, we developed and utilized a mature human 3D microvascular tissue model (Grabham et al., 2011; Templin et al., 2016; Wuu et al., 2020) seeded with human umbilical vein endothelial cells (HUVECs) to examine the physiological effects of the candidate miRNAs on structural cardiovascular damage caused by heavy ion irradiation (Grabham et al., 2011; unpublished data). Forty-eight hours after irradiation with 0.5 Gy SimGCRSim, several of the candidate miRNAs showed marked increases compared to controls (Figure 7A), though the increases were not significant, possibly due to low sample numbers.
Figure 7. miRNA Changes and Impacts in a 3D Microvascular Tissue Model.
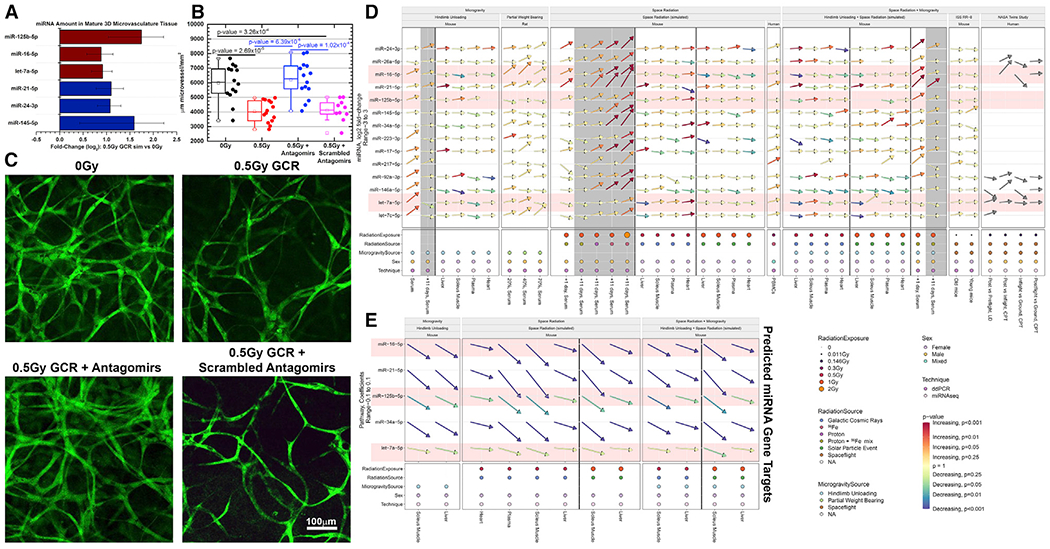
(A) Quantification of miRNAs from a 3D culture of mature human microvessels with human umbilical vein endothelial cells (HUVECs) irradiated with 0.5 Gy of simplified simulated galactic cosmic rays (SimGCRSim) compared to sham irradiated samples 48 h after irradiation. All candidate miRNAs were examined, but only the miRNAs that seemed to respond differently for each group are shown. The error bars represent SEM.
(B and C) Mature microvessels fixed and fluorescently stained with 5-(4,6-dichlorotriazinyl) aminofluorescein (DTAF) (Method Details) 48 h after SimGCRSim irradiation with or without antagomir-induced inhibition of miR-125b, miR-16, and let-7a starting at 24 h prior to irradiation. Scrambled version of the antagomir was used a vehicle control.
(D) Summary of all the fold-change values for all experiments utilized in this manuscript (including both ddPCR and miRNA-seq data).
(E) The summary of the overall impact of the miRNAs on the gene targets determined in Figure 3D through pathway analysis on the miRNA-seq data. The arrows indicate the degree of up- or downregulation, and the colors of the arrows indicate the significance (i.e., p values). The gray-shaded regions represent the 11-day time points after irradiation. The red-shaded miRNAs represent the miRNAs used for the antagomir experiments in (B) and (C).
Three of the miRNAs (miR-125, miR-16, and let-7a) that were increased by SimGCRSim irradiation were also previously predicted to be associated with space-radiation-induced cardiovascular damage (Beheshti et al., 2019) and were chosen to be inhibited using antagomirs (Krützfeldt et al., 2005). Further data showing that these miRNAs are associated with disrupted angiogenesis following IR have been reported elsewhere (Wuu, et al., 2020). In addition, these miRNAs show consistent presence and impact on downstream functions in the space environment (i.e., Twins Study data) compared to all other miRNAs. Antagomirs were applied 24 h before irradiation, and morphology of the tissue constructs was assessed 48 h after irradiation with 0.5 Gy SimGCRSim. We observed irradiation-mediated loss of vascularization because the microvessels had shrunk or collapsed, indicating that 0.5 Gy SimGCRSim is sufficient to disrupt microvessels. This impact of SimGCRSim exposure was reversed by application of the three antagomirs 24 h before irradiation, while a scrambled antagomir control did not induce the rescue phenotype (Figures 7B and 7C). Based on the summary of the of spaceflight-stressor-induced changes across all miRNAs (Figure 7D) and the miRNAs that have global significant impact on the downstream gene targets determined in Figure 3D (Figure 7E), it is apparent that these three miRNAs have a uniform trend across all different conditions (red shaded miRNAs in Figures 7D and 7E). Thus, microvessel structure was protected by selectively inhibiting three miRNAs that are the most universally associated with spaceflight responses, proving that the vascular impairments caused by simulated deep space radiation are at least partially mediated through miRNAs and in the future might be rescued using antagomirs in human tissue constructs. Future steps to further explore the impact of this potential countermeasure will be performed in murine models and are currently ongoing. Although the results with the antagomirs are promising in the in vitro model, we still cannot account for the complexity in an in vivo model and the efficacy of this treatment.
DISCUSSION
In summary, we have validated the previously predicted panel of candidate spaceflight-relevant miRNAs using a combination of spaceflight, simulated microgravity, partial gravity, and simulated deep space radiation conditions in male and female mouse and rat in vivo, astronaut, and human ex vivo models. The majority of rodent serum miRNAs followed the pattern of dose-dependent increase in response to IR, with the notable exception of miR-217 (Figures 7D and 7E). We evaluated the applicability of this miRNA signature to humans by examining miRNA content in the supernatant of primary human immune cell samples exposed to simulated deep space radiation, as well as immune cells from the NASA Twins Study, and observed partial conservation across species. The functional significance of key miRNAs was predicted by analyzing miRNA-dependent changes in gene expression at the single-cell level in the NASA Twins Study and experimentally evaluated utilizing antagomirs to mitigate vascular damage caused by simulated deep space radiation in a 3D human microvascular tissue model. When viewing these results independently, it may appear that some level of variability occurs for the miRNA between the different conditions (Figures 1, 2, 3, and 4), but when observing all conditions together, this variability is in fact accounted for, based on which component of the space environment is being observed. A summary of how all the miRNA patterns and downstream targets demonstrate the consistency over all the models utilized for this study is shown in Figures 7D and 7E. For example, certain miRNAs (e.g., let-7a-5p, let-7c-5p, miR-223-3p) are more significantly regulated with microgravity experiments as opposed space radiation experiments, while other miRNAs (e.g., miR-16-5p) are consistently being expressed in all conditions (Figure 7D).
Collectively, our samples identify two spaceflight miRNA signatures, the first associated with reduced gravity and LEO and the second representing the responses to simulated deep space radiation. Specifically, gamma-irradiated mice, sham HU mice, PWB rats, ISS-flown mice, and the Twins Study data collectively display a LEO/microgravity miRNA signature, while 600 MeV/n 56Fe and proton-irradiated mice, human PBMCs, and human 3D microvascular tissue models exposed to 600 MeV/n 56Fe ions and simplified simulated GCRs show a deep-space-radiation-dependent signature (Figure 7D). Notably, significant overlaps exist between both signatures, indicating a miRNA panel that might be particularly relevant to future lunar and Mars missions that will combine both stressors. The absence of significant differences in miRNA-217 expression in these datasets serves as a negative control and demonstrates that the responses that had been observed in the remaining candidate miRNAs are unlikely to be caused by universal miRNA increases induced by an arbitrary stressor.
A miRNA signature unique to the spaceflight response has potential uses as a biomarker of systemic biological damage as well as a source for targets for mitigating the health risks caused by spaceflight and deep space radiation. While specific miRNAs have previously been implicated in spaceflight-associated biological damage (Girardi et al., 2014; Simone et al., 2009; Teodori et al., 2019), recent systems biology studies indicate that using an entire miRNA network as a biomarker can provide a more robust association with biological changes (Beheshti et al., 2017). In addition, we have observed that individual components of the space environment have unique miRNA signatures: some miRNAs are associated with the microgravity component of real or simulated spaceflight, others are associated with exposure to deep space radiation, and still others are only significantly increased after exposure to both, suggesting that different miRNA-based biomarkers would be applicable to different conditions, with the latter being particularly important in deep space exploration. The sensitivity of miRNA responses is underscored by the fact that the magnitude of miRNA increases appears to be dose dependent for both radiation and microgravity, as observed in the mouse simulated space environment model and the PWB rat model, respectively. Similarly, for the IR component, the signature is also LET dependent, as we report more significant miRNA changes after 600 MeV/n 56Fe compared to proton or gamma irradiation, which suggests its potential relevance for future deep space missions. Finally, conserved increases of key miRNAs across species, sexes, and experiments emphasizes the reliability of the miRNA signature.
Since the majority of the data in this paper are from males, there can be sex-specific differences that are not captured here. It has been shown that certain miRNAs and family members of miRNAs are significantly expressed in one sex over the other, in normal and cancerous (e.g., The Cancer Genome Atlas) tissues (Guo et al., 2017). For example, miR-503 is linked to female-related cardiovascular disease (Florijn et al., 2018), indicating that miRNAs in the same family have similar behaviors in different sexes while still linked to the original family of miRNAs. Future research will need to explore the sex differences and the spaceflight signatures for specific miRNAs as well as related miRNA family members.
Of note, miRNA signatures may be applied for improving astronaut health by providing the first step toward identifying a network of circulating regulatory molecules that are activated by spaceflight. Identifying, validating and targeting such systemic regulatory networks, as opposed to addressing symptomatic damage in isolated organs and tissues, may accelerate our ability to mitigate the damage to several organ systems at once. Additionally, this approach may be used for preemptive risk reduction.
Indeed, we experimentally demonstrated the countermeasure potential of miRNAs by using antagomirs to inhibit three miRNAs associated with spaceflight cardiovascular health risks in human 3D tissue models. In general, antagomirs are a powerful tool for preventing the downstream effects of miRNA, which can be extensive due to the ability of a single miRNA to interact with hundreds of genes at the transcriptional, translational, and protein levels (Krützfeldt et al., 2005). Antagomirs have been shown to effectively inhibit their miRNA targets in vivo in mouse models of disease; however, more research is required for understanding their off-target effects and tissue specificity (Ishida and Selaru, 2013). In our model, antagomirs were applied as a countermeasure prior to deep-space-relevant irradiation and successfully prevented microvascular damage. Thus, our suggested network of miRNAs and their implied targets not only provides mechanistic insight into spaceflight-induced organ damage, but also may be a valuable platform for preventing the adverse health effects of spaceflight. However, in our experiments, the antagomir therapy was only applied against the cardiovascular component of the miRNA signature due to the model only recapitulating human microvasculature. It would be important to measure the effects of miRNA inhibition of the rest of the miRNA signature in a model that more fully captures spaceflight-mediated immune dysfunction and cancer progression as well as in vivo.
The panel of spaceflight-associated miRNAs has been predicted to disrupt cell signaling and metabolic pathways, at least in part via their interactions with TGF-β1 (Beheshti et al., 2018). TGF-β1 is a pleiotropic cytokine that has been shown previously to be significantly modulated during spaceflight conditions (Jessup et al., 2000; Westerlind and Turner, 1995), and its expression has been successfully altered as a therapeutic approach in cancer and inflammation. However, TGF-β1 is involved in a wide variety of endothelial and immune cell processes, which makes it a risky target (Otten et al., 2010). Furthermore, given that TGF-β1 has been found to exert both tumor-suppressive and tumor-promoting effects at different stages of tumorigenesis (Ikushima and Miyazono, 2010), the ability of miRNAs to target the TGF-β1 pathway at multiple specific sites may suggest a more effective and dynamic modulation of TGF-β1 effects than simple overexpression or inhibition (Butz et al., 2012). Thus, antagomir-mediated miRNA inhibition may allow more context-specific TGF-β1 pathway modulation and may be a promising countermeasure for spaceflight applications as well.
The interactions between the proposed miRNA candidates and TGF-β1 have important consequences for inflammation during different stages of spaceflight, particularly during landing. miR-145 and miR-146a, which are both associated with a pro-inflammatory response (Tahamtan et al., 2018), have been found to be increased during landing (Capri et al., 2019). miR-21, which is involved in the resolution of inflammation (Tahamtan et al., 2018), has been found to be decreased during spaceflight (Hughes-Fulford et al., 2015; Makedonas et al., 2018) and increased during landing (Capri et al., 2019). Changes in these miRNAs during different stages of spaceflight likely contribute to the overall dysregulation of inflammation, which may persist long after astronauts return to Earth (Garrett-Bakelman et al., 2019)
In general, miRNAs have yet to be studied extensively in the context of space biology. Previous rodent and human spaceflight (LEO) experiments that specifically examined miRNAs have been limited to in vitro cellular models (human fibroblasts and leukocytes), though some miRNAs (miR-34, miR-21, let-7a, and let-7c) identified in those studies do overlap with our proposed spaceflight signature (Hughes-Fulford et al., 2015; Zhang et al., 2016). Simulated microgravity studies in humans undergoing 21-day bedrest have also reported let-7a, let-7c, miR-25, miR-125b, and miR-92a from our panel (Rullman et al., 2016). Finally, simulated deep space radiation studies in mice have implicated miR-34 and miR-125 (Templin et al., 2012), which we have also observed in irradiated mouse serum.
Further studies will be necessary to compare spaceflight-associated miRNA effects at the individual organ, tissue, and cellular levels and to explore potential sex differences. Although we have observed strong similarities in miRNA expression in cardiac and soleus muscles (Figures 7D and 7E), the miRNA content in these organs was not sufficient to explain the systemic changes in plasma miRNAs, indicating that other organs and tissues not quantified in this study may play a major role in miRNA expression. Therefore, there is a need for a more comprehensive and unbiased assessment of all miRNA through miRNA-seq in a wide range of tissues from each of the experimental models investigated here. In addition, scRNA-seq has indicated major changes in miRNA-associated gene expression among different blood immune cell types in the human blood from the NASA Twins Study (Gertz et al., 2020). CD14+ monocytes are particularly likely to express and respond to miRNAs, suggesting a potential regulatory role of miRNAs on the innate immune response that is known to be dysregulated in spaceflight (Crucian et al., 2011). Despite the demonstrated contribution of CD14+ monocytes to increases in the circulating miRNAs, it is also possible that the miRNAs could be coming from another organ where they are more abundantly expressed. Thus, further studies will be necessary to determine the exact origin of the increased miRNAs and tissue response, such as with cell-free DNA profiling (Bezdan et al., 2020), which will be beneficial for mapping and developing effective countermeasures. Finally, the successful prevention of vascular damage through antagomir therapy in our proof-of-concept human tissue model demonstrates the need for further experiments to fully characterize antagomir therapies in vivo to understand their clinical safety and efficacy and to examine which components of the spaceflight miRNA signature make effective targets for antagomir therapies.
Collectively, our results provide experimental confirmation of a previously predicted spaceflight-associated miRNA signature (Beheshti et al., 2018) in multiple rodent and human models in true spaceflight, simulated gravitational changes, and simulated deep space radiation. We also demonstrate the potential for preventing biological damage by strategic inhibition of specific miRNAs with antagomirs. The immediate impact of this study is the first step toward developing a systemic miRNA signature as a novel space biology research metric for assessing spaceflight-induced damage in astronauts and animal models of true spaceflight as well as ground analogs, while its expected long-term outcome is revealing new avenues for spaceflight countermeasure development that modulates systemic effects rather than isolated organ damage.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Afshin Beheshti (afshin.beheshti@nasa.gov).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets generated and analyzed during this study. All miRNA-sequence raw data were deposited on the Database: NASA’s GeneLab data repository/platform. The following identifiers are used for the data in this paper: for all heart tissue related data: GLDS-334, https://doi.org/10.26030/cg2g-as49; for all liver tissue related data: GLDS-335, https://doi.org/10.26030/72ke-1k67 for all soleus muscle related data: GLDS-337, https://doi.org/10.26030/m73g-2477 and for all plasma related data: GLDS-336, https://doi.org/10.26030/qasa-rr29.
Deposited data from the sequencing data from the NASA Twin Study can be found on the NASA Life Sciences Data Archive (LSDA) and the accession code is not available due to privacy concerns. LSDA is the repository for all human and animal research data, including that associated with this study. LSDA has a public facing portal where data requests can be initiated (https://lsda.jsc.nasa.gov/Request/dataRequestFAQ). The LSDA team provides the appropriate processes, tools, and secure infrastructure for archival of experimental data and dissemination while complying with applicable rules, regulations, policies, and procedures governing the management and archival of sensitive data and information. The LSDA team enables data and information dissemination to the public or to authorized personnel either by providing public access to information or via an approved request process for information and data from the LSDA in accordance with NASA Human Research Program and JSC Institutional Review Board direction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rodent models
Simulated space environment studies
Mouse serum was obtained from Dr. Ann-Sofie Schreurs from a simulated microgravity and radiation study (Table S1). Forty 16-week old male C57BL/6J (Jackson Labs, Sacramento, CA) mice were divided into four equal groups: sham, irradiated (IR), hindlimb unloaded (HU), or HU + IR. Mice were given sham or 2Gy of gamma (137Cs) irradiation, on day 3 of normal loading or HU, and were sacrificed on day 14.
Animals assigned to the HU group were suspended with traction tape based on previously established procedures (Morey-Holton and Globus, 1998). The room was maintained at an average 73.3°F, 31% humidity, 12-hour light/dark cycle, with food and water provided ad libitum. Animal weights were monitored throughout the duration of the experiment to assess health of the mice. The NASA Ames Research Center Institutional Animal Care and Use Committee (IACUC) approved all procedures. All experiments were performed in accordance with relevant guidelines and regulations. All animals were individually housed.
At 16 weeks of age, mice were exposed to 2 Gy gamma total body irradiation (137Cs at 0.83 Gy/min, JL Shepherd Mark I, San Gabriel, CA). For this treatment, mice were housed individually in a custom animal holding cage, maintaining HU mice in a head-down position. Mice were placed in the irradiator chamber and exposed to total body irradiation. The unloading of mice was maintained during irradiation by custom small HU cages. Sham-irradiated controls were handled similarly and placed in the irradiation chamber with the radiation source turned off.
Additional mouse serum was obtained from Dr. Yasaman Shirazi from a simulated microgravity and radiation study (Table S1). Eighty 16-week old C57BL/6J male mice (Jackson Laboratory Maine, BNL protocol Globus 2015 #481) were divided into seven groups: sham (n = 14), 1Gy proton irradiation (1H, 150 MeV/n, LET ~0.52 KeV/um) (n = 9), 1Gy iron (56Fe, 600 MeV/n, LET ~175 KeV/um) irradiation (n = 9), 2Gy iron irradiation (n = 6), 1Gy mixed iron and proton irradiation (1H/56Fe/1H at 25/50/25cGy) (n = 14), hindlimb unloaded (HU) (n = 14), and HU+1Gy mixed iron and proton irradiation (n = 14). Total body irradiations were carried out at NASA Space Radiation Laboratory (NSRL) at Brookhaven National Laboratory (BNL) on conscious mice with 600 MeV/n 56Fe (1 Gy), 150 MeV/n 1H (1Gy), or mixed beam (1H/56Fe/1H at 25/50/25cGy), maintaining respective hindlimb unloading (HU) or normal loading (NL) conditions. Mice were irradiated after 3 days of HU or NL, and maintained HU or NL for 1 or 11 days before being sacrificed.
For both the above simulated microgravity and radiation studies, serum was isolated from blood draws from the vena cava of the mice. In order to prevent hemolysis from interfering in the results, the following protocol was uniformly conducted for all serum isolations: the blood was placed in a 4mL EDTA coated blood collection tube (BD Vacutainer Lavender K2-EDTA Cat# 367861) and kept at room temperature for 30 mins to allow for clotting. Then, the tubes were centrifuged at 2000 g for 15 minutes at 4°C followed by immediate freezing of the aliquots at −80°C. Samples were always mixed gently to prevent hemolysis.
Partial weight bearing rat model
Partial weight bearing rat serum was donated by Dr. Marie Mortreux and Dr. Seward Rutkove from a previous study. All experimental protocols were approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. 40 adult male Wistar rats (14 weeks of age at baseline, Charles River Laboratories, Wilmington, MA) were assigned to 1 of 5 groups to ensure equal distribution of the body weights at baseline (428.3 ± 5.2g). Animals were assigned to either normal loading without apparatus (PWB100 N), normal loading with full harness (PWB100 FH), 70% of normal loading (PWB70), 40% of normal loading (PWB40), or 20% of normal loading (PWB20) for 28 consecutive days. Partial weight-bearing (PWB) was performed in custom cages using a quadrupedal unloading composed of a forelimb jacket and a pelvic harness and extensively described elsewhere (Mortreux et al., 2018).
Across our 28-day study, the only significant difference between the NH and FH control groups was observed in weight gain where the 100% gravity NH group gained significantly more weight than the 100% gravity FH group. The weight gain was also associated with a slightly (but not significant) increased daily food intake that did not result in a different cumulative food intake throughout our study. The device did not exert any pressure nor prevented the animals from moving freely in their cage.
Rats were singly housed in a room maintained at 22 ± 2°C with a 12h:12h light-dark cycle starting at 7:00 am, and allowed 48 hours to acclimate before baseline. On day 28, animals were euthanized using CO2 inhalation per IACUC guidelines and blood was collected via cardiac puncture. Blood was allowed to clot at room temperature and was then centrifuged for 10 minutes at 10,000g. Serum was collected and frozen upon further analysis. 4 animals per group were selected randomly to perform miRNA analysis, and are outlined in Table S1.
ISS-flown mice
Serum was isolated from 36 space-flown female BALB/cAnNTac mice (Taconic) (Table S1). These mice were launched to the ISS on Rodent Research Reference Mission 1 (RRRM-1)/Rodent Research Mission 8 (RR-8) on 12/05/2018, docked to the ISS on 12/08/2018 and were returned to Earth as live animals on 01/13/2019, a total of 39 days after launch. The total absorbed radiation dose the mice received was 11.28mGy. The mice were sacrificed 4 days after returning to Earth. There were two different age groups, young (n = 18, 10-16 weeks) and old (n = 18, 31-37 weeks) as outlined in Table S1.
Space irradiation and hindlimb unloading of mice for miRNA-sequencing
15-week ± 3-day old, C57BL/6J wild-type (Wt) female mice were purchased from Jackson Laboratories and housed at Brookhaven National Laboratory (BNL, Upton, NY). Upon arrival to BNL, mice were quarantined and acclimated to a standard 12:12h light:dark cycle, with controlled temperature/humidity for 1-week prior to cage acclimation. Food and water were given ad libitum, and standard bedding was changed once per week. Mice were cage acclimated (n = 10 mice per group; 2 mice per cage to maintain social interaction) 3-days prior to HU, followed by 14-days either normally loaded (NL) or hindlimb unloaded (HU, see details below). Irradiation was administered on day 13 and blood tissues were collected at 24-hours post-irradiation and post-euthanasia by CO2 overdose, followed by cervical dislocation. Blood was collected via the abdominal aorta in EDTA-coated tubes (0.5 M) and plasma was separated by centrifugation at 2,000xg for 15 minutes. Plasma was collected and flash frozen in liquid nitrogen for −80°C storage. A 100 μL aliquot of cellular fraction was flash frozen and stored at −80°C for RNA analyses, while the remaining cellular fraction was lysed with 1x RBC lysis buffer (Thermo Fisher Scientific) followed by flow cytometric preparation and analyses, as describe below. The heart, liver, and soleus muscle were flash frozen and collected at the time of dissection. All tissues were stored at −80°C. Body weight tracking was performed on days −3, 0, 7 and 14.
Hindlimb unloading (HU) was performed using the adjusted Morey-Holton method for social housing (n = 2 per cage) (Tahimic et al., 2019). Briefly, mice were suspended from the tail using non-invasive traction tape attached to an adjustable pulley mounted on the top of a standard rodent cage. The adjustable nature of the device allows the user to position the animal in a head-down position (approximately 30° to the horizon) once attached. The crossbar height, lateral crossbar position, and chain length are adjustable. The pulley is free to move along the crossbar that spans the length of the top of the cage enabling the mouse to navigate freely on one half of the cage using its forepaws and to interact with the second mouse in the cage, without enabling ambulation of the limbs. Individual food and water sources are provided to each mouse and replaced daily. Control mice were housed in identical cages with normal ambulation.
On day 13, mice were transported on BNL base to the NASA Space Radiation Laboratory (NSRL) facility by animal care staff and were transferred to individual HU boxes. The following doses of irradiation were administered; simplified GCR sim (0.5 Gy), SPE (1 Gy), Gamma (5 Gy) and Sham control (0 Gy). Mice were positioned in the plateau region of the Bragg curve and irradiated at room temperature in individual HU boxes to enable whole body irradiation while maintaining hindlimb suspension. The NSRL physics staff performed the dosimetry. To simulate GCR, we used the simplified GCR simulation of ions, energy, and doses determined by a NASA consensus formula that consists of 5 ions: protons at 1000 MeV, 28Si at 600 MeV/n, 4He at 250 MeV/n, 16O at 350 MeV/n, 56Fe at 600 MeV/n, and protons at 250 MeV. This dose of radiation is equivalent to what an astronaut is predicted to receive in deep space during a Mars mission, though it is modeled as a single exposure over 25 minutes instead of the actual chronic exposures over 1.5 years. Further, GCR sim is a mixture of high and low LET ions in a ratio of 15% to 85%, respectfully. To simulate SPE, we used a total dose of 1Gy protons with energy ranges from 50MeV to 150MeV. For all irradiations a 60x60 beam was utilized at NSRL. For radiation dose equivalence and biological reference, we used 5 Gy gamma irradiation with the cesium resource available at BNL, in the absence of HU due to resource limitations. Sham controls were treated similar to GCR/SPE irradiated mice, including HU cage boxes and beam line (without irradiation) for the same duration as GCR simualtion, i.e., 25 minutes.
In vitro models
Irradiated human PBMCs
Peripheral blood mononuclear cells (PBMCs) were obtained from Dr. Sylvain Costes from a human irradiation sensitivity study (NASA HRP #NNJ16HP24I, NASA Ames Research Center Human Research IRB #HRI-357) (Table S1). Briefly, PBMCs were isolated from blood donated by 12 healthy adults: 8 females and 4 males. PBMC isolation was performed using Ficoll-Paque (VWR Cat# 95038-168) density gradient centrifugation followed by red blood cell lysis (VWR Cat# 420301-BL), and cells were frozen at 1x107 cells/mL until the day before irradiation. All subjects were of European descent and 20 - 64 years old.
Cells were plated in 12-well plates (VWR Cat# 29442-040) 24 hours before irradiation in RPMI 1640 (Sigma Cat# R8758) supplemented with 10% fetal bovine serum (VWR Cat# 1300-500H) and 1% Penicillin/Streptomycin/Glutamine (Fisher Cat# 10-378-016). Each well received approximately 1.6x106 cells and 2mL media. 24 hours later, the cells were irradiated with 0Gy or 0.3Gy 600MeV/n 56Fe. Cell supernatants were collected 4 hours after irradiation and stored at −80°C until miRNA extractions.
3D human vascular model
Mature human 3D microvascular tissue models are grown by seeding-human umbilical vein endothelial cells (HUVECs) (Lonza Cat# CC-2519) into a collagen/matrigel mixture as described (Grabham et al., 2011). Briefly, HUVECs were first grown in EGM Endothelial Cell Growth Medium (Cat# CC-3124), then cells (1 × 106/ml) were embedded into a collagen/matrigel mixture (Cat# 354236 and Cat# 356230). Microvessels are grown in EGM-2 Endothelial Cell Growth Medium (Cat# CC-3162) plus 50 nM Phorbol-12-myristate-13-acetate: (PMA) (Cat# 524400) for 7 days to form tubular microvessels before irradiations.
These tissue models were used to study vascular changes associated with specific miRNAs (Table S1). Irradiations were carried out at Brookhaven National Lab using 50cGy of SimGCRSim (5-ion simplified galactic cosmic ray simulation: protons at 1000MeV, 28Si at 600 MeV/n, 4He at 250 MeV/n, 16O at 350 MeV/n, 56Fe at 600 MeV/n, and protons at 250 MeV). A dose of 0.5 μm of each antagomir was administered to the appropriate groups 24 hours prior to irradiation. Prior work has shown that lentivirus particles efficiently incorporate into these constructs, therefore it was assumed that antagomirs would have no trouble either. The tissue constructs used for fluorescence analysis were fixed with 4% paraformaldehyde (Sigma Cat# 158127100G) for 5 minutes at 37°C followed by 0.5% Triton X-100 (Sigma Cat# X100) to solubilize the fats. Cell structures were stained with 5-(4,6-Dichlorotriazinyl) Aminofluorescein (DTAF) (Cat# D 16) and Images captured on a Nikon TE 200 confocal C1 microscope with EZ-C1 software. The tissue constructs used for miRNA analysis were dissolved by submerging in TRIzol (ThermoFisher Cat# 15596026) 48 hours post irradiation, without any fixation or homogenization.
Simulated GCR irradiation
To simulate GCR, we used the simplified GCR simulation of ions, energy, and doses determined by a NASA consensus formula that consists of 5 ions: protons at 1000 MeV, 28Si at 600 MeV/n, 4He at 250 MeV/n, 16O at 350 MeV/n, 56Fe at 600 MeV/n, and protons at 250 MeV, in the following proportions; Protons 100MeV at 34.8%, Si at 1.1%, He at 18%, O at 5.8%, 56Fe at 1%, and Protons 250 MeV at 39.3%. This mixture is the simplified version of the full GCR simulation and represents the proportions of ions found in space and thus translates to exploratory class missions (Simonsen et al., 2020). The low LET particles (Protons and Helium) make up the majority of radiation although high LET ions generally have a greater RBE.
NASA Twins study samples
The NASA Twins Study involved two male twin subjects, aged 50 years old at the time of launch. The flight twin spent 340 days aboard the ISS while his identical twin stayed on Earth as the ground control.
Blood samples were collected into 4mL CPT tubes (BD Biosciences Cat # 362760) per manufacturer’s recommendations. Cell separation was performed by centrifugation in at 1800 X g for 20 minutes at room temperature, both on the ISS and for the ground-based samples. Ambient blood collected samples slated for immediate return on Soyuz capsule were stored at 4°C until processing (average of 35-37 hours after collection, including repatriation time). Samples collected on Earth and the ISS and planned for long-term storage were mixed by inversion and immediately frozen at −80°C.
Fresh processing of CPT tubes was performed using the following steps: 1. plasma was retrieved from the CPT tubes and flash frozen prior to long term storage at −80°C. 2. The peripheral blood mononuclear cells (PBMCs) were recovered and washed in PBS. 0.5 million PBMCs were retrieved from one pre-flight and one post-flight samples, pelleted, flash frozen and stored at −80°C until use for RNA extractions. The flow through from cell sorting steps was recovered as the lymphocyte depleted (LD) fraction; LD and PBMC cell specimens were lysed into RLT+ buffer (QIAGEN Cat# 1053393), flash frozen and stored at −80°C until use.
METHOD DETAILS
Accounting for Hemolysis in Samples
Although hemolysis is unavoidable for blood/serum samples obtained from in vivo studies similar to what is described throughout this manuscript, we believe that we have accounted for this to the best to our ability for our analysis. There are many papers discussing this topic and impact on circulating miRNAs with the conclusions from these papers stating that for in vivo samples hemolysis can’t be avoided but procedures can be done to reduce the effects dramatically (Blondal et al., 2013; Pizzamiglio et al., 2017; Poel et al., 2018). Due the constraints for space biology experiments, the main step to reduce hemolysis variations for miRNAs is by handling all the samples identically. Both the controls and all other samples were handled under the exact same conditions throughout the manuscript. So if miRNAs levels would have been altered due to hemolyzed blood, then all miRNA levels would have been altered the same way between our comparisons. In fact in the negative controls all the miRNAs from our spaceflight signature are either nonexistent or at extremely low levels compared to samples under experimental conditions. This indicated that although it has been reported that hemolysis will alter miRNAs levels for all samples (Pizzamiglio et al., 2017), the levels of the miRNAs should have been altered identically in our controls and experimental conditions, allowing for reduced variability between results.
miRNA extraction
MiRNA extractions from serum, plasma, all tissues, and cell supernatants were carried out using the QIAGEN miRNeasy serum/plasma kit (#217184). MiRNA extractions from dissolved microvessel constructs were carried out using the QIAGEN miRNeasy Mini Kit (#217004). Quantitation of miRNA samples was done using a NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific).
Total RNA extraction from the NASA Twin Study LD and PBMC lysates was carried out using the QIAGEN Allprep kit (Cat# 80204). Quantitation of RNA was assessed using Qubit 2.0 (Invitrogen) per manufacturer’s recommendations. RNA quality control was performed using RNA Nano kit (Agilent) on a Bioanalyzer 2100 (Agilent) per manufacturer’s recommendations.
cDNA generation and Droplet Digital PCR
First, cDNA was synthesized from miRNA samples using the QIAGEN miRCURY LNA RT Kit (Cat# 339340) using a concentration of 5ng/μl for the miRNA per sample. Next, samples were mixed with a 1:20 dilution of the generated cDNA with the BioRad QX200 ddPCR Evagreen Supermix (Cat# 1864034) and the appropriate miRNA primers from miRCURY LNA miRNA PCR Assays (QIAGEN). BioRad QX200 Automated Droplet Generator (Cat# 1864101) was used to create emulsion droplets. With the C1000 Touch Thermal Cycler with 96–Deep Well Reaction Module (Bio-Rad) the following PCR reaction was used for all the primers: 1 cycle 95°C for 5 min, 40 cycles of 95°C for 30 s and 58°C for 1 min (the annealing temperature can change depending on the primer), 1 cycle of 4°C for 5 min, and 1 cycle of 90°C for 5 min. Not all miRNA primers sets for ddPCR will have the same annealing temperature, so optimizing the annealing temperature is required for each primer set. Their respective annealing temperatures are found in Table S2. Finally, the QX200 Droplet Digital PCR System (Bio-Rad) quantified the amount of miRNA for each primer set per sample. QuantaSoft software (Bio-Rad) generated the data for each primer set and sample. The same threshold setting was used for all samples per primer set. The concentration (miRNA copies/μl) value generated by QuantaSoft was converted to miRNA copies/ng of serum. These values were used for all miRNA analysis. For all analysis the miRNA concentrations were log2(x+1) transformed to allow for easy comparison between miRNAs and samples.
miRNA sequencing on murine samples
Library construction and sequencing was performed from miRNAs isolated from plasma, liver, heart, and soleus muscle from the mouse experiments described above. The miRNA extraction was carried out using the QIAgen miRNeasy kit (#217004). The total RNA quality and quantity were analyzed using a Bioanalyzer 2100 (Agilent, CA, USA) with RIN number > 7. Approximately 1 μg of total RNA was used to prepare small RNA library according to protocol of TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA). Single-end sequencing was performed using 50 bp on an Illumina Hiseq 2500 at the LC Sciences (Hangzhou, China) following the vendor’s recommended protocol. All miRNA-sequence raw data was deposited on NASA’s GeneLab data repository/platform with the following identifiers: for all heart tissue related data: GLDS-334, https://doi.org/10.26030/cg2g-as49 for all liver tissue related data: GLDS-335, https://doi.org/10.26030/72ke-1k67: for all soleus muscle related data: GLDS-337, https://doi.org/10.26030/m73g-2477 and for all plasma related data: GLDS-336, https://doi.org/10.26030/qasa-rr29.
miRNA sequencing on NASA Twins Study samples
Small RNA libraries were prepared from 50 ng total RNA using the NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (NEB #E7560) per manufacturer’s recommendations with the following modifications: adaptors and RT primer were diluted four fold, 17 cycles of PCR were used for library amplification, and no size selection was performed. The i7 primers in the NEBNext® Multiplex Oligos for Illumina® Dual Index Primers (NEB# E7600, NEB#E7780) were used to supplement the index primers in NEB #E7560. The libraries were sequenced in an Illumina NextSeq instrument (1x50bp).
Single cell RNA and epitope sequencing on NASA Twin Study samples
Cells were isolated prepared using the BD Rhapsody platform according to the BD Rhapsody Express Single-Cell Analysis System Instrument User Guide, with a custom-designed RNA and epitope panel. Briefly, cells from each sample were labeled with sample tags, then pooled after being washed twice with FACS buffer. Combined samples were then washed an additional time before being stained with the BD AbSeq Ab-Oligo reagents. After staining, the cells were washed twice before resuspension at approximately 20,000 cells in 620 μL. These cells were then isolated using the Single-Cell Analysis System and cDNA Synthesis with the BD Rhapsody Express Single-Cell Analysis System using the manufacturers’ protocol (BD Biosciences). Cells were loaded onto 3 BD Rhapsody nanowell cartridges. Cartridges were loaded with Cell Capture Beads (BD Biosciences) before shaking for 15 s at 1,000 rpm. Cells were lysed and cell capture beads were retrieved and washed prior to Exonuclease I treatment and reverse transcription. Targeted amplification of cDNA with the Human Immune Response Panel primers and custom supplemental panel (all targets listed in Table S3) was done through 10 PCR cycles. PCR products were purified, and mRNA and AbSeq products were separated by SPRIselect beads with double-sided selection. mRNA products were amplified further with 15 PCR cycles. Final libraries were indexed with 8 PCR cycles. Library quality was assessed by Bioanalyzer (D1000 HS, Agilent). Library DNA concentration was quantified using a Qubit dsDNA HS Kit (ThermoFisher, #Q32854) on a Qubit Fluorometer. Libraries were diluted to 2nM and multiplexed before being sequenced on three lanes of a Novaseq-6000. Mean read depth per cell was 13,244.31 mRNA reads per cell and 11,506.98 AbSeq reads per cell for a combined 243751.30 reads per cell.
Single cell ATAC sequencing on NASA Twins Study samples
Cells were isolated and prepared according to the SureCell ATAC-Seq Library Prep Kit User Guide (17004620, Bio-Rad) by the Weill Cornell Epigenomics Core. Briefly, lysis and tagmentation was performed simultaneously on the leukocyte depleted fraction of PBMC samples. Cells were washed before resuspension in Whole Cell Tagmentation Mix. Cells were tagmented using a thermal protocol before being mixed and agitated for 30 minutes at 37°C. Prior to encapsulation, tagmented cells were kept on ice. Tagmented cells were loaded onto the ddSEQ Single-Cell Isolator (12004336, Bio-Rad). The SureCell ATAC-Seq Library Prep Kit (17004620, Bio-Rad) and SureCell ddSEQ Index Kit (12009360, Bio-Rad) were used to prepare scATAC libraries. Ampure XP beads (A63880, Beckman Coulter) were used to clean up the libraries. Barcoded amplicons were further amplified through 7 PCR cycles. These PCR products were cleaned up using Ampure XP beads and quality was assessed on both a TapeStation (D5000, Agilent) and Bioanalyzer. Library concentrations were quantified by on a Qubit Fluorometer by Qubit dsDNA HS Kit. Experiments were then pooled and sequenced by Bio-Rad on four lanes of a Novaseq-6000.
QUANTIFICATION AND STATISTICAL ANALYSIS
miRNA quantification analysis from ddPCR data
We previously utilized NASA GeneLab database to make unbiased predictions about miRNAs potentially involved in regulating the spaceflight response (Beheshti et al., 2018). The transcriptomic datasets analyzed in this study include GLDS-25 (livers from female C57BL/6J mice, n = 15 per group), GLDS-21 (gastrocnemius from female C57BL/6J mice, n = 4 per group), GLDS-63 (mammary gland, liver, and adipose tissue from pregnant female Sprague-Dawley rats, n = 4 per group), GLDS-111 (soleus and extensor digitorum longus from male C57/BL6 mice, n = 3 per group), GLDS-4 (thymus from C57BL/6NTac mice, n = 4 per group), GLDS-61 (skin from male C57Bl/J10 mice, n = 3 per group), and GLDS-48 (liver from female C57BL/6J mice, n = 6 per group). Details for this work and analysis is available in Beheshti et al. (Beheshti et al., 2018) and is the basis of the analysis we are describing in these methods.
Experimental groups and the number of replicates (n) within each group are summarized in Table S1. For analysis on the archived serum from the simulated murine experiments, we grouped all miRNA copies/ng values for NL Sham controls together and all other experimental groups were compared to the NL Sham conditions. For all analysis the p values reported are two-sided and adjusted with the significance indicated by * p value < 0.05, ** p value < 0.01, and *** p value < 0.001. Hierarchical clustering of the miRNA values were displayed as heatmaps with pheamap and t-Distributed Stochastic Neighbor Embedding (t-SNE) analysis (van der Maaten and Hinton, 2008) was done to determine how the samples cluster together based on the miRNA values. The miRNA values were also plotted using R-program ggplot2 (v3.2.1) (Wickham, 2016.) as scatterplots and data points were then fit using a generalized additive model (GAM) (Hastie and Tibshirani, 1986; Wood et al., 2015). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway predictions from the miRNAs were determined by the DIANA-TarBase v7.0 algorithm. Correlation plots between miRNAs and different conditions were plotted using R program corrplot v0.84. Lastly, OriginPro 2015 was used to plot bar plots and of the fold-change values between conditions and also boxplots.
In silico predictions of genes from miRNAs
Through the use of a Cytoscape (Shannon et al., 2003) plug-in called ClueGo/CluePedia (Bindea et al., 2013), we were able to predict genes targeted by the miRNAs determined. This involved entering all miRNAs in ClueGo and allowing the software to determine the top 50 genes that were significantly regulated and connected to the miRNAs. The predictions only reflect the functions that will be regulated by the miRNAs and do not show whether the function will be activated or inhibited. We also predicted the impact of the miRNAs on overall pathways by utilizing DIANA microT-CDS tool which allows for pathway discovery from targeted miRNAs (Paraskevopoulou et al., 2013; Reczko et al., 2012). Lastly, miRNet 2.0 was utilized to predict the diseases that are associated with the miRNAs (Chang et al., 2020).
Analysis of miRNA sequencing from murine samples
Raw reads were subjected to an in-house software program, ACGT101-miR (LC Sciences, Houston, Texas, USA) to remove adaptor dimers, junk, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA) and repeats. Subsequently, unique sequences with length in 18~26 nucleotide were mapped to specific species precursors in miRBase 22.0 (Kozomara et al., 2019) by BLAST search to identify known miRNAs and novel 3p- and 5p- derived miRNAs. Length variation at both 3′ and 5′ ends and one mismatch inside of the sequence were allowed in the alignment. The unique sequences mapping to specific species mature miRNAs in hairpin arms were identified as known miRNAs. The unique sequences mapping to the other arm of known specific species precursor hairpin opposite to the annotated mature miRNA-containing arm were considered to be novel 5p- or 3p- derived miRNA candidates. The remaining sequences were mapped to other selected species precursors (with the exclusion of specific species) in miRBase 22.0 by BLAST search, and the mapped pre-miRNAs were further BLASTed against the specific species genomes to determine their genomic locations. The above two we defined as known miRNAs. The unmapped sequences were BLASTed against the specific genomes, and the hairpin RNA structures containing sequences were predicated from the flank 80 nt sequences using RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The criteria for secondary structure prediction were: (1) number of nucleotides in one bulge in stem (≤12); (2) number of base pairs in the stem region of the predicted hairpin (≥16); (3) cutoff of free energy (kCal/mol ≤ −15); (4) length of hairpin (up and down stems + terminal loop ≥ 50); (5) length of hairpin loop (≤20); (6) number of nucleotides in one bulge in mature region (≤8); (7) number of biased errors in one bulge in mature region (≤4); (8) number of biased bulges in mature region (≤2); (9) number of errors in mature region (≤7); (10) number of base pairs in the mature region of the predicted hairpin (≥12); and (11) percent of mature in stem (≥80). Differential expression of miRNAs based on normalized deep-sequencing counts was analyzed by selectively using Fisher exact test, Chi-square 2X2 test, Chi-square nXn test, Student t test, or ANOVA based on the experimental design. The significance threshold was set to be 0.01 and 0.05 in each test.
To determine gene ontology (GO) (Smith and Eppig, 2009) and C2 pathways being regulated by the miRNAs we performed miRNA gene set analysis utilizing the RBiomirGS v0.2.12 R package (Zhang and Storey, 2018) from the processed miRNA analysis for all conditions in the plasma and heart tissues. From the pathways we chose an FDR < 0.25 cutoff for significantly regulated pathways. We plotted the specific pathways with R packages ggplot2.
Analysis of miRNA sequencing from NASA Twin Study samples
Standard libraries were preprocessed and quality controlled using miRTrace (Kang et al., 2018). Subsequently, reads were mapped against MirGeneDB sequences (Fromm et al., 2020) using the miRDeep2 (Friedländer et al., 2012) quantifier module. Expression values were normalized to reads per million (RPM) considering only miRNA counts. The normalized RPM value were utilized for all analysis. If the value for the miRNA was zero for all samples that miRNA was excluded from the analysis. To determine statistically significant miRNAs, ANOVA analysis with p value < 0.2 was independently performed for each cell type. For flight only comparisons, the same statistical significance was applied for all time points excluding the ground samples and all ambient return samples. All statistics were run independently for each cell condition/type. To determine the overlap of the miRNA signature in this paper with the Twin Study miRNA-seq data, the proposed miRNAs were used as well as all mature components of the miRNAs included in the miRNA family.
Analysis of scRNA and abseq from NASA Twins Study samples
Fastq files were uploaded to Seven Bridges Genomics. Data was demultiplexed and sequences analyzed with BD’s Rhapsody pipeline (BD Rhapsody Analysis Pipeline 1.4 Beta) on Seven Bridges (https://www.sevenbridges.com). Data was then loaded into Seurat (version 3.2.0) for analysis (Stuart et al., 2019). This generated a sparse matrix file of features by barcodes. This sparse matrix data was then read into R using the R package Seurat 3.2.0, and standard quality control was run to remove cells with few genes. Data was then scaled and normalized. MAST (Finak et al., 2015) was used to determine DEGs between all combinations of preflight, return and postflight for both TW and HR samples. Linear dimensional reduction was performed by calculation of PCA from the most variable genes. Cells were then clustered using a resolution value of 0.5 and visualized by UMAP. Marker genes were checked for RNA and epitope expression for each cluster and assigned to the indicated cell types, with cell subtypes being grouped to match the scATAC data (Figures S2E and S2F). multiMiR was used to extract target genes for spaceflight-associated miRNA (Ru et al., 2014). Module scores for each spaceflight-associated miRNA target signature was calculated by AddModuleScore with a control value of 5. Violin plots were created with ggplot2 (version 3.3.2, (Wickham, 2016.) and significance tested by Wilcoxon rank sum using the geom_signif function. Cells were subsetted by cell type and dot plots were generated using the Seurat DotPlot function. Full list of RNA and AbSeq target genes are found in Table S3.
Analysis of scATAC from NASA Twins Study samples
Fastq files from each sample were processed individually through BioRad’s scATAC processing pipeline according to the Bio-Rad ATAC-seq Analysis Toolkit Tutorial using the Bio-Rad docker images to run FastQC, debarcode, align reads, filter and deconvolute beads, and cell filtration. The aligned reads for each experiment were then processed using SnapTools (version 1.2.3) to create 5kb bins and a bin by cell matrix (Fang et al., 2020). The matrices for each sample were combined and then processed using SnapATAC (version 1.0.0) using the suggested parameters to filter for high-quality barcodes and remove bins that overlapped with ENCODE hg38 blacklisted regions or ChrM (Fang et al., 2020). Diffusion maps were computed for dimensionality reduction, and significant components were determined. Genes were assigned to each bin based off of GRCh38.p13. The resulting Snap object was then converted to a Seurat object and exported to Seurat (version 3.2.0). Samples were scaled and normalized, and anchors were transferred from the 10X PBMC dataset to the experiment from date 1/30/2019, as it contained the control, non-leukocyte depleted PBMCs. This data was then integrated using the IntegrateData function with the experiment from date 4/18/2019, and anchors were transferred from the 1/30/2019 experiment to the integrated object. Cells were assigned cell types based off of the predicted score, with cell subtypes being grouped to match the scRNA and epitope data (Figure S3I). Due to the sparsity of the data, MAGIC (Rmagic 2.0.3) was then used to impute activity using the default parameters (van Dijk et al., 2018). The imputed activity was then used to generate dot plots and violin plots, with significance being tested by Wilcoxon rank sum using the geom_signif function. Cell composition percentages of cells after T and B cell removal was calculated by pooling the scATAC and scRNA/epitope cell identities per condition. Table S4 has all the determined DEGs between all combinations of preflight, return and postflight for both TW and HR samples.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Human PBMCs | Consented Donors | N/A |
| NASA Twin Study Unsorted PBMCs | Consented Donors | N/A |
| NASA Twin Study Lymphocyte Depleted Cells | Consented Donors | N/A |
| NASA Twin Study CD8 CD4, CD14 and CD19 Cells | Consented Donors | N/A |
| Mouse Blood | C57BL/6J Wt female | N/A |
| Mouse Plasma | C57BL/6J Wt female | N/A |
| Mouse Liver | C57BL/6J Wt female | N/A |
| Mouse Heart | C57BL/6J Wt female | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RLT+ Buffer | QIAGEN | Cat# 1053393 |
| Triton X-100 | Sigma | Cat# X100 |
| TRIzol | ThermoFisher | Cat# 15596026 |
| Ficoll-Paque | VWR | Cat# 95038-168 |
| 1x Phosphate Buffered Saline (pH 7.4) | Thermofisher Scientific | Cat# 10010023 |
| EGM-2 Endothelial Cell Growth Medium-2 Bullet-Kit | Lonza | Cat# CC-3162 |
| EGM Endothelial Cell Growth Medium Bullet-Kit | Lonza | Cat# CC-3124 |
| Rat tail Collagen Type 1 | BD Biosciences | Cat# 354236 |
| Matrigel Matrix | BD Biosciences | Cat# 356230 |
| Phorbol-12-myristate-13-acetate: PMA | Millipore | Cat# 524400 |
| 5-(4,6-Dichlorotriazinyl) Aminofluorescein (DTAF) | Thermofisher Scientific | Cat# D-16 |
| MP Biomedicals Trypsin-EDTA Solution | Thermofisher Scientific | Cat# ICN1689149 |
| Critical Commercial Assays | ||
| miRNeasy serum/plasma kit | QIAGEN | Cat# 217184 |
| miRNeasy mini kit | QIAGEN | Cat# 217004 |
| Allprep kit | QIAGEN | Cat# 80204 |
| miRCURY LNA RT kit | QIAGEN | Cat# 339340 |
| QX200 ddPCR Evagreen Supermix | BioRad | Cat# 1864034 |
| miRNA primers | QIAGEN | Listed under “Oligonucleotides” |
| NEBNext® Multiplex Small RNA Library Prep Set for Illumina® | NEB | NEB# E7560 |
| NEBNext® Multiplex Oligos for Illumina® Dual Index Primers | NEB | NEB# E7600, NEB# E7780 |
| Index primers | NEB | NEB# E7560 |
| iSCRIPT™ cDNA synthesis kit | Bio-Rad | Cat# 1708890 |
| iQ™ SYBR® Green Supermix | Bio-Rad | Cat# 170-8880 |
| TruSeq Small RNA Sample Prep Kits | Illumina | RS-200-0012 |
| Rhapsody AbSeq reagent pack | BD Biosciences | Cat# 633771 |
| Rhapsody Human Immune Response panel | BD Biosciences | Cat# 633750 |
| Rhapsody custom gene panel (See Table S3) | BD Biosciences | Cat# 633743 |
| Human Single cell multiplexing kit | BD Biosciences | Cat# 633781 |
| SureCell ATAC-seq Library Prep Kit | Bio-Rad | Cat# 17004620 |
| SureCell ddSEQ Index Kit | Bio-Rad | Cat# 12009360 |
| Deposited Data | ||
| MirGeneDB sequences | (Fromm et al., 2020) | https://mirgenedb.org/ |
| miRNA-seq for mice data | NASA GeneLab (https://genelab.nasa.gov/) | GLDS-334, GLDS-335, GLDS-336, GLDS-337 |
| NASA Twin Study miRNA-seq results | This paper | N/A |
| miRNA ddPCR raw data | This paper | LSDA (NASA) |
| NASA Twin Study BD Rhapsody Data | This paper and Getz et al. (2020) | N/A |
| NASA Twin Study scATAC Data | This paper | N/A |
| Experimental Models: Cell Lines | ||
| Human: HUVECs | Lonza | Cat# CC-2519 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Labs, Sacramento, CA | (IMSR Cat# JAX:000664, RRID:IMSR_JAX:000664) |
| Mouse: C57BL/6J | Jackson Laboratory Maine, | (IMSR Cat# JAX:000664, RRID:IMSR_JAX:000664) |
| Rat: Wistar | Charles River Laboratories, Wilmington, MA | (RGD Cat# 2312511, RRID:RGD_2312511) |
| Mouse: BALB/cAnNTac | Taconic | (IMSR Cat# TAC:balb, RRID:IMSR_TAC:balb) |
| Oligonucleotides | ||
| hsa-miR-17-5p miRCURY LNA miRNA PCR Assay: MIMAT0000070: 5′CAAAGUGCUUACAGUGCAGGUAG | QIAGEN | Cat# YP02119304 |
| hsa-miR-146a-5p miRCURY LNA miRNA PCR Assay: MIMAT0000449: 5′UGAGAACUGAAUUCCAUGGGUU | QIAGEN | Cat# YP00204688 |
| hsa-miR-223-3p miRCURY LNA miRNA PCR Assay: MIMAT0000280: 5′UGUCAGUUUGUCAAAUACCCCA | QIAGEN | Cat# YP00205986 |
| hsa-let-7a-5p miRCURY LNA miRNA PCR Assay: MIMAT0000062: 5′UGAGGUAGUAGGUUGUAUAGUU | QIAGEN | Cat# YP00205727 |
| hsa-mir-92a-3p miRCURY LNA miRNA PCR Assay: MIMAT0000092: 5′UAUUGCACUUGUCCCGGCCUGU | QIAGEN | Cat# YP00204258 |
| hsa-miR-24-3p miRCURY LNA miRNA PCR Assay: MIMAT0000080: 5′UGGCUCAGUUCAGCAGGAACAG | QIAGEN | Cat# YP00204260 |
| hsa-miR-125b-5p miRCURY LNA miRNA PCR Assay: MIMAT0000423: 5′UCCCUGAGACCCUAACUUGUGA | QIAGEN | Cat# YP00205713 |
| hsa-miR-16-5p miRCURY LNA miRNA PCR Assay: MIMAT0000069: 5′UAGCAGCACGUAAAUAUUGGCG | QIAGEN | Cat# YP00205702 |
| hsa-miR-26a-5p miRCURY LNA miRNA PCR Assay: MIMAT0000082: 5′UUCAAGUAAUCCAGGAUAGGCU | QIAGEN | Cat# YP00206023 |
| hsa-miR-145-5p miRCURY LNA miRNA PCR Assay: MIMAT0000437: 5′GUCCAGUUUUCCCAGGAAUCCCU | QIAGEN | Cat# YP00204483 |
| mmu-miR-217-5p miRCURY LNA miRNA PCR Assay: MIMAT0000679: 5′UACUGCAUCAGGAACUGACUGGA | QIAGEN | Cat# YP00205070 |
| hsa-miR-34a-5p miRCURY LNA miRNA PCR Assay: MIMAT0000255: 5′UGGCAGUGUCUUAGCUGGUUGU | QIAGEN | Cat# YP00204486 |
| hsa-miR-21-5p miRCURY LNA miRNA PCR Assay: MIMAT0000076: 5′UAGCUUAUCAGACUGAUGUUGA | QIAGEN | Cat# YP00204230 |
| mmu-miR-25-5p miRCURY LNA miRNA PCR Assay: MIMAT0017049: 5′AGGCGGAGACUUGGGCAAUUGC | QIAGEN | Cat# YP02118413 |
| hsa-let-7c-5p miRCURY LNA miRNA PCR Assay: MIMAT0000064: 5′UGAGGUAGUAGGUUGUAUGGUU | QIAGEN | Cat# YP00204767 |
| Software and Algorithms | ||
| miRTrace | (Kang et al., 2018) | https://github.com/friedlanderlab/mirtrace |
| miRDeep2 | (Friedländer et al., 2012) | https://www.mdc-berlin.de/content/mirdeep2-documentation; RRID:SCR_010829 |
| Cytoscape | (Shannon et al., 2003) | https://cytoscape.org; RRID:SCR_003032 |
| ClueGo/CluePedia | (Bindea et al., 2013) | http://www.ici.upmc.fr/cluego/; RRID:SCR_005748 |
| DIANA microT-CDS tool | (Paraskevopoulou et al., 2013; Reczko et al., 2012) | http://carolina.imis.athena-innovation.gr/dianatools/web/index.php; RRID:SCR_016510 |
| miRNet 2.0 | (Chang et al., 2020) | https://www.mirnet.ca/ |
| Rtsne v0.15 | (van der Maaten and Hinton, 2008) | https://github.com/jkrijthe/Rtsne; RRID:SCR_016342 |
| corrplot v0.84 | n/a | https://github.com/taiyun/corrplot |
| ggplot2 v3.3.1 | (Wickham, 2016) | https://ggplot2.tidyverse.org; RRID:SCR_014601 |
| pheatmap V1.0.12 | Kolde Rpheatmap: Pretty heatmaps software; 2015 | https://cran.r-project.org/web/packages/pheatmap/index.html; RRID:SCR_016418 |
| ACGT101-miR | LC Sciences | N/A |
| miRBase 22.0 | (Kozomara et al., 2019) | http://www.mirbase.org/; RRID:SCR_003152 |
| RBiomirGS V0.2.12 R package | (Zhang and Storey, 2018) | https://github.com/jzhangc/git_RBiomirGS |
| GO Mouse Genome Informatics (MGI) | (Smith and Eppig, 2009) | http://www.informatics.jax.org/; RRID:SCR_006489 |
| R package version 3.6.1 | https://www.r-project.org/; RRID:SCR_001905 | |
| QuantaSoft V1.7.4.0917 | Bio-Rad Laboratories | https://www.bio-rad.com/webroot/web/pdf/lsr/literature/10047467.pdf |
| OriginPro 2015 | OriginLab Corporation | https://www.originlab.com; RRID:SCR_014212 |
| R package version 4.0.2 (for single cell analysis) | https://www.r-project.org/; RRID:SCR_001905 | |
| Seurat 3.2.0 | (Stuart et al., 2019) | https://github.com/satijalab/seurat; RRID:SCR_007322 |
| MAST | (Finak et al., 2015) | https://www.bioconductor.org/packages/release/bioc/html/MAST.html; RRID:SCR_016340 |
| multiMiR | (Ru et al., 2014) | https://www.bioconductor.org/packages/release/bioc/html/multiMiR.html |
| SnapTools 1.4.1 | (Fang et al., 2020) | https://github.com/r3fang/SnapTools; RRID:SCR_018097 |
| SnapATAC | (Fang et al., 2020) | https://github.com/r3fang/SnapATAC |
| Rmagic 2.0.3 | (van Dijk et al., 2018) | https://github.com/KrishnaswamyLab/MAGIC |
| Patchwork 1.0.1 | https://github.com/thomasp85/patchwork; RRID:SCR_000072 | |
| Other | ||
| QX200 Automated Droplet Generator | BioRad | Cat# 1864101 |
| QX200 Droplet Reader | BioRad | Cat# 1864003 |
| Illumina Hiseq 2500 | LC Sciences | N/A |
| ABI 7500 Real-Time PCR | Applied Biosystems | N/A |
| NovaSeq 6000 | Illumina | N/A |
| BD Rhapsody | BD Biosciences | Cat# 633701 |
| ddSEQ Single-Cell Isolator | Bio-Rad | Cat# 12004366 |
| BioAnalyzer | Agilent | Cat# D1000 HS |
| Simplified 5-ion GCR Simulation: Protons at 1000 MeV, 28Si at 600 MeV/n, 4He at 250 MeV/n, 16O at 350 MeV/n, 56Fe at 600 MeV/n, and protons at 250 MeV | Brookhaven National Lab - NSRL | N/A |
| Simulated SPE: Protons at 50MeV to 150MeV | Brookhaven National Lab - NSRL | N/A |
| Gamma: 137Cs source (5 Gy) | Brookhaven National Lab – NSRL | N/A |
| 56Fe Irradiation: 600 MeV/n | Brookhaven National Lab – NSRL | N/A |
Highlights.
Spaceflight miRNA signature validated in multiple organism models
Components of miRNA signature related to space radiation and microgravity
Downstream targets and circulating dependence of miRNAs in NASA Twins Study
Inhibition of key microvasculature miRNAs mitigates space radiation impact
ACKNOWLEDGMENTS
Special thanks to Dr. Ruth Globus for her input on the manuscript and to Carolyn Reifsnyder, Zach Burkett, and Kelly Kaihara from Bio-Rad for early access to scATAC. We would also like to thank Drs. Peter Guida, Adam Rusek, Michael Sivertz, and the NSRL team at BNL for their assistance and expertise with irradiation studies. C.E.M. would like to thank the Epigenomics Core Facility at Weill Cornell Medicine, the Scientific Computing Unit (SCU), XSEDE Supercomputing Resources, STARR I13-0052, the Vallee Foundation, the WorldQuant Foundation, The Pershing Square Sohn Cancer Research Alliance, NASA (NNX14AH50G, NNX17AB26G, 19-19OMNI_2-0109), and the National Institutes for Health (R01MH117406, R01CA249054, TRISH [NNX16AO69A:0107, NNX16AO69A:0061], LLS 9238-16, Mak, LLS-MCL7001-18). This work was also supported by NASA grant 16-ROSBFP_GL-0005: NNH16ZTT001N-FG Appendix G: Solicitation of Proposals for Flight and Ground Space Biology Research (award 80NSSC19K0883) and The Translational Research Institute for Space Health through NASA Cooperative Agreement NNX16AO69A (T-0404) awarded to A.B. ISS-related mouse tissue research reported in this publication was supported by the International Space Station U.S. National Laboratory under award UA-2019-018 given to A.B.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108448.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Baqai FP, Gridley DS, Slater JM, Luo-Owen X, Stodieck LS, Ferguson V, Chapes SK, and Pecaut MJ (2009). Effects of spaceflight on innate immune function and antioxidant gene expression. J. Appl. Physiol 106, 1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheshti A, Vanderburg C, McDonald JT, Ramkumar C, Kadungure T, Zhang H, Gartenhaus RB, and Evens AM (2017). A Circulating microRNA Signature Predicts Age-Based Development of Lymphoma. PLoS ONE 12, e0170521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheshti A, Ray S, Fogle H, Berrios D, and Costes SV (2018). A microRNA signature and TGF-β1 response were identified as the key master regulators for spaceflight response. PLoS ONE 13, e0199621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheshti A, McDonald JT, Miller J, Grabham P, and Costes SV (2019). GeneLab Database Analyses Suggest Long-Term Impact of Space Radiation on the Cardiovascular System by the Activation of FYN Through Reactive Oxygen Species. Int. J. Mol. Sci 20, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnier M, Shantikumar S, Anwar M, Dixit P, Chamorro-Jorganes A, Sweaad W, Sala-Newby G, Madeddu P, Thomas AC, Howard L, et al. (2019). miR-15a/-16 Inhibit Angiogenesis by Targeting the Tie2 Coding Sequence: Therapeutic Potential of a miR-15a/16 Decoy System in Limb Ischemia. Mol. Ther. Nucleic Acids 17, 49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezdan D, et al. (2020). Cell-free DNA (cfDNA) and exosome profiling from a year-long human spaceflight reveals circulating biomarkers. iScience. 10.1016/j.isci.2020.101844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Galon J, and Mlecnik B (2013). CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 29, 661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, and Dahlsveen IK (2013). Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59, S1–S6. [DOI] [PubMed] [Google Scholar]
- Burkhart K, Allaire B, and Bouxsein ML (2019). Negative Effects of Long-duration Spaceflight on Paraspinal Muscle Morphology. Spine 44, 879–886. [DOI] [PubMed] [Google Scholar]
- Butz H, Rácz K, Hunyady L, and Patócs A (2012). Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol. Sci 33, 382–393. [DOI] [PubMed] [Google Scholar]
- Caporali A, and Emanueli C (2011). MicroRNA-503 and the extended microRNA-16 family in angiogenesis. Trends Cardiovasc. Med 21, 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capri M, Morsiani C, Santoro A, Moriggi M, Conte M, Martucci M, Bellavista E, Fabbri C, Giampieri E, Albracht K, et al. (2019). Recovery from 6-month spaceflight at the International Space Station: muscle-related stress into a proinflammatory setting. FASEB J. 33, 5168–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekanaviciute E, Fathali N, Doyle KP, Williams AM, Han J, and Buckwalter MS (2014). Astrocytic transforming growth factor-beta signaling reduces subacute neuroinflammation after stroke in mice. Glia 62, 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Zhou G, Soufan O, and Xia J (2020). miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 48, W244–W251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JQ, Papp G, Szodoray P, and Zeher M (2016). The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun. Rev 15, 1171–1180. [DOI] [PubMed] [Google Scholar]
- Coburn-Litvak PS, Tata DA, Gorby HE, McCloskey DP, Richardson G, and Anderson BJ (2004). Chronic corticosterone affects brain weight, and mitochondrial, but not glial volume fraction in hippocampal area CA3. Neuroscience 124, 429–438. [DOI] [PubMed] [Google Scholar]
- Crucian B, Stowe R, Quiriarte H, Pierson D, and Sams C (2011). Monocyte phenotype and cytokine production profiles are dysregulated by short-duration spaceflight. Aviat. Space Environ. Med 82, 857–862. [DOI] [PubMed] [Google Scholar]
- Elgart SR, Little MP, Chappell LJ, Milder CM, Shavers MR, Huff JL, and Patel ZS (2018). Radiation Exposure and Mortality from Cardiovascular Disease and Cancer in Early NASA Astronauts. Sci. Rep 8, 8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R, Preissl S, Li Y, Hou X, Lucero J, Wang X, Motamedi A, Shiau AK, Zhou X, Xie F, et al. (2020). SnapATAC: A Comprehensive Analysis Package for Single Cell ATAC-seq. bioRxiv. 10.1101/615179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florijn BW, Bijkerk R, van der Veer EP, and van Zonneveld AJ (2018). Gender and cardiovascular disease: are sex-biased microRNA networks a driving force behind heart failure with preserved ejection fraction in women? Cardiovasc. Res 114, 210–225. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Mackowiak SD, Li N, Chen W, and Rajewsky N (2012). miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Domanska D, Høye E, Ovchinnikov V, Kang W, Aparicio-Puerta E, Johansen M, Flatmark K, Mathelier A, Hovig E, et al. (2020). MirGeneDB 2.0: the metazoan microRNA complement. Nucleic Acids Res. 48, D132–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Bakelman FE, Darshi M, Green SJ, Gur RC, Lin L, Macias BR, McKenna MJ, Meydan C, Mishra T, Nasrini J, et al. (2019). The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science 364, eaau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbaix M, Gnyubkin V, Farlay D, Olivier C, Ammann P, Courbon G, Laroche N, Genthial R, Follet H, Peyrin F, et al. (2017). One-month spaceflight compromises the bone microstructure, tissue-level mechanical properties, osteocyte survival and lacunae volume in mature mice skeletons. Sci. Rep 7, 2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz ML, Chin CR, Tomoiaga D, MacKay M, Butler D, Afshinnekoo E, Bezdan D, Schmidt MA, Mozsary C, Melnick AM, et al. (2020). Multiomic, Single-cell, and Biochemical Profiles of Astronauts Guide Pharmacological Strategies for Returning to Gravity. Cell Rep. Published online November 25, 2020. 10.1016/j.celrep.2020.108429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardi C, De Pittà C, Casara S, Calura E, Romualdi C, Celotti L, and Mognato M (2014). Integration analysis of microRNA and mRNA expression profiles in human peripheral blood lymphocytes cultured in modeled microgravity. BioMed Res. Int 2014, 296747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabham P, Hu B, Sharma P, and Geard C (2011). Effects of ionizing radiation on three-dimensional human vessel models: differential effects according to radiation quality and cellular development. Radiat. Res 175, 21–28. [DOI] [PubMed] [Google Scholar]
- Guo L, Zhang Q, Ma X, Wang J, and Liang T (2017). miRNA and mRNA expression analysis reveals potential sex-biased miRNA expression. Sci. Rep 7, 39812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, and Tibshirani R (1986). Generalized Additive Models. Stat. Sci 1, 297–310. [DOI] [PubMed] [Google Scholar]
- Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, and Tewari M (2013). Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10, 1003–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes-Fulford M, Chang TT, Martinez EM, and Li CF (2015). Spaceflight alters expression of microRNA during T-cell activation. FASEB J. 29, 4893–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, and Miyazono K (2010). Cellular context-dependent “colors” of transforming growth factor-beta signaling. Cancer Sci. 101, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M, and Selaru FM (2013). miRNA-Based Therapeutic Strategies. Curr. Anesthesiol. Rep 1, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup JM, Frantz M, Sonmez-Alpan E, Locker J, Skena K, Waller H, Battle P, Nachman A, Bhatti, Weber ME, et al. (2000). Microgravity culture reduces apoptosis and increases the differentiation of a human colorectal carcinoma cell line. In Vitro Cell Dev. Biol. Anim 36, 367–373. [DOI] [PubMed] [Google Scholar]
- Kang W, Eldfjell Y, Fromm B, Estivill X, Biryukova I, and Friedländer MR (2018). miRTrace reveals the organismal origins of microRNA sequencing data. Genome Biol. 19, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova J, Gerasymchuk D, Parkkinen I, Chmielarz P, and Domanskyi A (2019). Interplay between MicroRNAs and Oxidative Stress in Neurodegenerative Diseases. Int. J. Mol. Sci 20, 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, and Griffiths-Jones S (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, and Stoffel M (2005). Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689. [DOI] [PubMed] [Google Scholar]
- Macfarlane LA, and Murphy PR (2010). MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genomics 11, 537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader TH, Gibson CR, Pass AF, Kramer LA, Lee AG, Fogarty J, Tarver WJ, Dervay JP, Hamilton DR, Sargsyan A, et al. (2011). Optic disc edema, globe flattening, choroidal folds, and hyperopic shifts observed in astronauts after long-duration spaceflight. Ophthalmology 118, 2058–2069. [DOI] [PubMed] [Google Scholar]
- Makedonas G, Chouker A, Mehta S, Simpson R, Stowe R, Sams C, Pierson D, and Crucian B (2018). Mechanistic Clues to Overcome Spaceflight-Induced Immune Dysregulation. Curr. Pathobiol. Rep 6, 185–192. [Google Scholar]
- Marie JC, Liggitt D, and Rudensky AY (2006). Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity 25, 441–454. [DOI] [PubMed] [Google Scholar]
- Mewaldt RA (1994). Galactic cosmic ray composition and energy spectra. Adv. Space Res 14, 737–747. [DOI] [PubMed] [Google Scholar]
- Morey-Holton ER, and Globus RK (1998). Hindlimb unloading of growing rats: a model for predicting skeletal changes during space flight. Bone 22, 83S–88S. [DOI] [PubMed] [Google Scholar]
- Mortreux M, Nagy JA, Ko FC, Bouxsein ML, and Rutkove SB (2018). A novel partial gravity ground-based analog for rats via quadrupedal unloading. J. Appl. Physiol 125, 175–182. [DOI] [PubMed] [Google Scholar]
- Otten J, Bokemeyer C, and Fiedler W (2010). Tgf-Beta superfamily receptors-targets for antiangiogenic therapy? J. Oncol 2010, 317068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, and Hatzigeorgiou AG (2013). DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 41, W169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AM, Cheng-Campbell M, Blaber EA, Anand S, Bhattacharya S, Zwart S, Crucian B, Smith SM, Meller R, Grabham P, et al. (2020). Beyond Low-Earth Orbit: Characterizing the Immune Profile Following Simulated Spaceflight Conditions for Deep Space Missions. iScience. Published online November 25, 2020. 10.1016/j.isci.2020.101747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecaut MJ, Mao XW, Bellinger DL, Jonscher KR, Stodieck LS, Ferguson VL, Bateman TA, Mohney RP, and Gridley DS (2017). Is spaceflight-induced immune dysfunction linked to systemic changes in metabolism? PLoS ONE 12, e0174174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechenkova E, Nosikova I, Rumshiskaya A, Litvinova L, Rukavishnikov I, Mershina E, Sinitsyn V, Van Ombergen A, Jeurissen B, Jillings S, et al. (2019). Alterations of Functional Brain Connectivity After Long-Duration Spaceflight as Revealed by fMRI. Front. Physiol 10, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzamiglio S, Zanutto S, Ciniselli CM, Belfiore A, Bottelli S, Gariboldi M, and Verderio P (2017). A methodological procedure for evaluating the impact of hemolysis on circulating microRNAs. Oncol. Lett 13, 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poel D, Buffart TE, Oosterling-Jansen J, Verheul HMW, and Voortman J (2018). Evaluation of several methodological challenges in circulating miRNA qPCR studies in patients with head and neck cancer. Exp. Mol. Med 50, e454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczko M, Maragkakis M, Alexiou P, Grosse I, and Hatzigeorgiou AG (2012). Functional microRNA targets in protein coding sequences. Bioinformatics 28, 771–776. [DOI] [PubMed] [Google Scholar]
- Režen T, Kovanda A, Eiken O, Mekjavic IB, and Rogelj B (2014). Expression changes in human skeletal muscle miRNAs following 10 days of bed rest in young healthy males. Acta Physiol. (Oxf.) 210, 655–666. [DOI] [PubMed] [Google Scholar]
- Ronca AE, Moyer EL, Talyansky Y, Lowe M, Padmanabhan S, Choi S, Gong C, Cadena SM, Stodieck L, and Globus RK (2019). Behavior of mice aboard the International Space Station. Sci. Rep 9, 4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooke HM, and Crosier KE (2001). The smad proteins and TGFbeta signalling: uncovering a pathway critical in cancer. Pathology 33, 73–84. [PubMed] [Google Scholar]
- Ru Y, Kechris KJ, Tabakoff B, Hoffman P, Radcliffe RA, Bowler R, Mahaffey S, Rossi S, Calin GA, Bemis L, and Theodorescu D (2014). The multiMiR R package and database: integration of microRNA-target interactions along with their disease and drug associations. Nucleic Acids Res. 42, e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rullman E, Mekjavic IB, Fischer H, and Eiken O (2016). PlanHab (Planetary Habitat Simulation): the combined and separate effects of 21 days bed rest and hypoxic confinement on human skeletal muscle miRNA expression. Physiol. Rep 4, e12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Gius D, and Mitchell JB (2009). Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS ONE 4, e6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen LC, Slaba TC, Guida P, and Rusek A (2020). NASA’s first ground-based Galactic Cosmic Ray Simulator: Enabling a new era in space radiobiology research. PLoS Biol. 18, e3000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, and Eppig JT (2009). The mammalian phenotype ontology: enabling robust annotation and comparative analysis. Wiley Interdiscip. Rev. Syst. Biol. Med 1, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A, Teymoori-Rad M, Nakstad B, and Salimi V (2018). Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol 9, 1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahimic CGT, Paul AM, Schreurs AS, Torres SM, Rubinstein L, Steczina S, Lowe M, Bhattacharya S, Alwood JS, Ronca AE, and Globus RK (2019). Influence of Social Isolation During Prolonged Simulated Weightlessness by Hindlimb Unloading. Front. Physiol 10, 1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templin T, Young EF, and Smilenov LB (2012). Proton radiation-induced miRNA signatures in mouse blood: characterization and comparison with 56Fe-ion and gamma radiation. Int. J. Radiat. Biol 88, 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templin T, Sharma P, Guida P, and Grabham P (2016). Short-Term Effects of Low-LET Radiation on the Endothelial Barrier: Uncoupling of PECAM-1 and the Production of Endothelial Microparticles. Radiat. Res 186, 602–613. [DOI] [PubMed] [Google Scholar]
- Teodori L, Costa A, Campanella L, and Albertini MC (2019). Skeletal Muscle Atrophy in Simulated Microgravity Might Be Triggered by Immune-Related microRNAs. Front. Physiol 9, 1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend LW (2005). Implications of the space radiation environment for human exploration in deep space. Radiat. Prot. Dosimetry 115, 44–50. [DOI] [PubMed] [Google Scholar]
- Ulrich-Lai YM, Figueiredo HF, Ostrander MM, Choi DC, Engeland WC, and Herman JP (2006). Chronic stress induces adrenal hyperplasia and hypertrophy in a subregion-specific manner. Am. J. Physiol. Endocrinol. Metab 291, E965–E973. [DOI] [PubMed] [Google Scholar]
- van der Maaten LJP, and Hinton GE (2008). Visualizing High-Dimensional Data Using t-SNE. J. Mach. Learn. Res 9, 2579–2605. [Google Scholar]
- van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, et al. (2018). Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 174, 716–729.e727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderburg C, and Beheshti A (2020). MicroRNAs (miRNAs), the Final Frontier: The Hidden Master Regulators Impacting Biological Response in All Organisms Due to Spaceflight (THREE). [Google Scholar]
- Wang S, Zhu W, Xu J, Guo Y, Yan J, Meng L, Jiang C, and Lu S (2019). Interpreting the MicroRNA-15/107 family: interaction identification by combining network based and experiment supported approach. BMC Med. Genet 20, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Wang J, He J, and Xie X (2018). Serum microRNA as noninvasive indicator for space radiation. Acta Astronaut. 152, 101–104. [Google Scholar]
- Westerlind KC, and Turner RT (1995). The skeletal effects of spaceflight in growing rats: tissue-specific alterations in mRNA levels for TGF-beta. J. Bone Miner. Res 10, 843–848. [DOI] [PubMed] [Google Scholar]
- Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag). [Google Scholar]
- Wood SN, Pya N, and Säfken B (2015). Smoothing Parameter and Model Selection for General Smooth Models. J. Am. Stat. Assoc 111, 1548–1563. [Google Scholar]
- Wuu YR, Hu B, Okunola H, Paul AM, Blaber EA, Cheng-Campbell M, Beheshti A, and Grabham P (2020). LET Dependent Low Dose and Synergistic Inhibition of Human Angiogenesis by Charged Particles: Validation of Mi RNAs that Drive Inhibition. iScience. Published online November 25, 2020. 10.1016/j.isci.2020.101771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Pan C, Liu E, He H, Liu C, Huang Y, Yi S, and Huang D (2019). Differences of microRNA expression profiles between monozygotic twins’ blood samples. Forensic Sci. Int. Genet 41, 152–158. [DOI] [PubMed] [Google Scholar]
- Zhang J, and Storey KB (2018). RBiomirGS: an all-in-one miRNA gene set analysis solution featuring target mRNA mapping and expression profile integration. PeerJ 6, e4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu T, Wong M, Wang X, Stodieck L, Karouia F, Story M, and Wu H (2016). Transient gene and microRNA expression profile changes of confluent human fibroblast cells in spaceflight. FASEB J. 30, 2211–2224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated and analyzed during this study. All miRNA-sequence raw data were deposited on the Database: NASA’s GeneLab data repository/platform. The following identifiers are used for the data in this paper: for all heart tissue related data: GLDS-334, https://doi.org/10.26030/cg2g-as49; for all liver tissue related data: GLDS-335, https://doi.org/10.26030/72ke-1k67 for all soleus muscle related data: GLDS-337, https://doi.org/10.26030/m73g-2477 and for all plasma related data: GLDS-336, https://doi.org/10.26030/qasa-rr29.
Deposited data from the sequencing data from the NASA Twin Study can be found on the NASA Life Sciences Data Archive (LSDA) and the accession code is not available due to privacy concerns. LSDA is the repository for all human and animal research data, including that associated with this study. LSDA has a public facing portal where data requests can be initiated (https://lsda.jsc.nasa.gov/Request/dataRequestFAQ). The LSDA team provides the appropriate processes, tools, and secure infrastructure for archival of experimental data and dissemination while complying with applicable rules, regulations, policies, and procedures governing the management and archival of sensitive data and information. The LSDA team enables data and information dissemination to the public or to authorized personnel either by providing public access to information or via an approved request process for information and data from the LSDA in accordance with NASA Human Research Program and JSC Institutional Review Board direction.


