Abstract
DNA replication starts at multiple discrete sites across the human chromosomal c-myc region, including two or more sites within 2.4 kb upstream of the c-myc gene. The corresponding 2.4-kb c-myc origin fragment confers autonomously replicating sequence (ARS) activity on plasmids, which specifically initiate replication in the origin fragment in vitro and in vivo. To test whether the region that displays plasmid replicator activity also acts as a chromosomal replicator, HeLa cell sublines that each contain a single copy of the Saccharomyces cerevisiae FLP recombinase target (FRT) sequence flanked by selectable markers were constructed. A clonal line containing a single unrearranged copy of the transduced c-myc origin was produced by cotransfecting a donor plasmid containing the 2.4-kb c-myc origin fragment and FRT, along with a plasmid expressing the yeast FLP recombinase, into cells containing a chromosomal FRT acceptor site. The amount of short nascent DNA strands at the chromosomal acceptor site was quantitated before and after targeted integration of the origin fragment. Competitive PCR quantitation showed that the c-myc origin construct substantially increased the amount of nascent DNA relative to that at the unoccupied acceptor site and to that after the insertion of non-myc DNA. The abundance of nascent strands was greatest close to the c-myc insert of the integrated donor plasmid, and significant increases in nascent strand abundance were observed at sites flanking the insertion. These results provide biochemical and genetic evidence for the existence of chromosomal replicators in metazoan cells and are consistent with the presence of chromosomal replicator activity in the 2.4-kb region of c-myc origin DNA.
The bacterial replicon model of Jacob et al. (17) proposed that a trans-acting initiator protein binds to a cis-acting replicator element, defined as “a specific element of recognition upon which the corresponding initiator would act, allowing the replication of the DNA attached to the replicator.” Consistent with this model, replication initiation in the simple eucaryote Saccharomyces cerevisiae is regulated by the interaction of cell cycle-dependent trans-acting factors with cis-acting DNA elements to establish replication bubbles and cause the initiation of DNA synthesis (6, 36). In metazoans, however, putative cis-acting replicator elements may be distributed over larger distances than the compact replicators of S. cerevisiae (4, 37), effect initiation at multiple sites, and comprise features of nuclear, chromatin, or DNA structure as well as DNA sequence (6, 8, 14, 15). Biochemical assays suggest that DNA synthesis can initiate at multiple sites over regions as large as 55 kb in the hamster dihydrofolate reductase (DHFR) locus (9), the human c-myc locus (39, 41), and elsewhere (2, 7, 24, 35, 42). Replication does not initiate randomly across these large zones, although the degree to which replication initiates at preferred sites varies between replicons (13, 18, 20, 32). Nevertheless, the start sites for DNA synthesis may be more numerous than the elements that control replicon firing.
Our laboratory was the first to report that DNA replication begins 5′ to the human c-myc gene, within a 2.4-kb HindIII-XhoI restriction fragment (23, 28). Subsequent work has confirmed that replication starts in this location and at additional sites over a 12-kb region encompassing the c-myc gene (27, 29, 33, 38–41). The 2.4-kb c-myc origin fragment contains a DNA unwinding element (DUE), which is a common feature of eucaryotic origins (11), a unique nucleosome organization, and three matches to the S. cerevisiae autonomously replicating sequence (ARS) consensus (39). Plasmids containing the 2.4-kb fragment of the c-myc origin replicate autonomously in vitro and when transfected into HeLa cells (3, 16, 27). As in the chromosome, c-myc plasmid replication does not initiate at random but begins in a zone centered over the 2.4-kb origin fragment (39, 41). These observations suggest that the 2.4-kb fragment of c-myc 5′ flanking DNA contains cis-acting replicator sequences that control replication initiation.
To test whether the 2.4-kb 5′ flanking sequence contains chromosomal replicator activity as well as start sites for DNA synthesis, we used the yeast FLP recombinase to integrate the 2.4-kb c-myc origin fragment at a single FLP recombinase target (FRT) site in the HeLa genome. Competitive PCR was used to quantitate the amount of short nascent DNAs at the acceptor site before and after the introduction of the c-myc origin sequences, or after the introduction of non-myc control sequences. The results show that a significant increase in the amount of nascent DNA occurred only after insertion of the c-myc origin construct. The greatest quantity of nascent DNA was observed at the sites closest to the c-myc sequences, but the amount of nascent DNA was also substantially increased at sites flanking the FRT acceptor. PCR mapping of short nascent strands indicated that the c-myc origin construct induced replication at new flanking sites in the chromosome. These data suggest that the 2.4-kb c-myc origin fragment can activate replication in cis at a novel chromosomal location and support the chromosomal replicator function of the c-myc origin DNA.
MATERIALS AND METHODS
Cell culture and DNA isolation.
HeLa cells were maintained in Dulbecco’s modified Eagle’s minimal medium (DMEM) with 10% newborn calf serum (Gibco-BRL) and 50 μg of gentamicin/ml in a humidified 5% CO2 atmosphere at 37°C. For diagnostic PCR, genomic DNA was obtained from single colonies growing in 24-well tissue culture plates (22). DNA for Southern blot analysis was isolated from ∼4.8 × 107 cells (six 10-cm plates) by standard methods (34).
Constructs and transfections.
Overlap extension PCR was used to place the 48-bp FRT sequence GAAGTTCCTATTCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTC in reading frame into a fusion gene encoding hygromycin phosphotransferase and herpes simplex virus (HSV) thymidine kinase (TK) in the plasmid pHyg.TK.Fus (25) to generate the acceptor site plasmid pHyg.FRT.TK. The plasmid was linearized with BglI and transfected at a low concentration into HeLa cells by electroporation. Stably transfected colonies were selected in hygromycin (Sigma) at 100, 200, or 400 μg/ml. The origin donor construct pFRT.Myc was derived from the plasmid pNeo.Myc-2.4 (27, 28), which contains the 2.4-kb HindIII-XhoI fragment of c-myc 5′ flanking sequences. The mouse metallothionein I promoter was removed from the neomycin phosphotransferase gene and replaced by a modified 47-bp FRT. The 47-bp FRT contained a single-base deletion that did not compromise the recombination function of the FRT and allowed expression of neomycin resistance after recombination at the acceptor site cytomegalovirus (CMV) promoter but shifted the HSV TK reading frame, making accurately targeted recombinants resistant to ganciclovir. A third plasmid, pHyg.FRT.Neo, was constructed to mimic the structure of the recombinant between the Hyg.FRT.TK acceptor and the pFRT.Myc donor; it was transfected into HeLa cells to confirm the activity of the Hyg-FRT-Neo fusion protein and to provide control DNA for PCR with primers specific for chromosomal recombination events. The structures of all plasmid constructs were confirmed by DNA sequencing. The control donor construct pOG45, containing one 48-bp FRT, bacterial vector sequences, and a neomycin phosphotransferase expression cassette, and construct pOG44, expressing FLP recombinase, were purchased from Stratagene. 406 cells were cotransfected with pOG44 and pFRT.Myc as described elsewhere (31). Colonies resistant to G418 and ganciclovir were obtained at a frequency of approximately 0.02%.
In vitro FLP reactions.
Plasmids were prepared by alkaline lysis and CsCl centrifugation. Plasmids were linearized, and 100 ng of a single plasmid, or a mixture of two plasmids digested to give chimeric recombination products of a distinct size, was added to FLP reaction buffer (25 mM 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid [TAPS] [pH 8.0], 1 mM EDTA, 2.5 mg of bovine serum albumin [BSA]/ml, 10% polyethylene glycol [wt/vol], 20% glycerol [vol/vol] and preheated to 30°C. Purified FLP protein (a gift from M. Cox) in FLP dilution buffer (25 mM TAPS [pH 8.0], 2.5 mg of BSA/ml, 5% glycerol, 1 M NaCl) was added to a concentration of 136 to 200 nM, and the reaction mixtures were incubated for 30 min at 30°C. Reactions were stopped by the addition of GED (50% glycerol, 50 mM EDTA, 0.1% bromophenol blue)–10% sodium dodecyl sulfate (SDS) in a 3:2 ratio, and reaction products were electrophoresed through 1.0% agarose gels in TAE (40 mM Tris-acetate [pH 8.3]–1 mM EDTA). DNA was stained in ethidium bromide, photographed, and subjected to Southern blot hybridization.
MTT assay.
Cells were plated in 96-well tissue culture plates (500 cells/well) in 200 μl of DMEM 24 h before the addition of ganciclovir (15 μM). Cells were grown with or without drug for 6 days, and on the 7th day after plating, the medium was removed and replaced with fresh DMEM containing 0.5 mg of MTT (thiazolyl blue tetrazolium bromide; Amresco). Plates were incubated in the dark at 37°C for 4 h, the medium was removed, and the converted dye was solubilized by the addition of 50 μl of dimethyl sulfoxide per well. One hundred fifty microliters of 0.1 M glycine (pH 10)–0.16 M NaCl buffer was added, and the A570 of each well was measured in a plate reader (Spectromax 250; Molecular Devices Inc.).
Diagnostic PCR analysis.
Fifty nanograms of genomic DNA was used for PCR with primer sets specific for either the chromosomal acceptor construct or the acceptor construct after integration of pFRT.Myc or pOG45. Reaction mixtures contained 10 mM Tris-Cl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM (each) dGTP, dCTP, dATP, and dTTP, 1.25 U of Taq polymerase (Gibco-BRL), and 25 pmol of each primer for the following amplification conditions: 30 to 35 cycles of 94°C for 30 s, 50 to 57°C for 40 s, and 72°C for 30 s. Products were electrophoresed on 2% agarose gels and visualized by ethidium bromide staining.
Southern blot hybridization.
Ten micrograms of genomic DNA was digested with 200 U of BamHI (Gibco-BRL) overnight, electrophoresed on 0.8% agarose gels in TAE, and blot transferred to Hybond N+ membranes (34). After blotting, the filters were soaked in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 20 min, UV cross-linked (Stratalinker), and prehybridized (in 6× SSC, 0.2% Ficoll 400, 0.2% polyvinylpyrrolidone, 0.2% BSA, 1% SDS, and 50 μg of denatured salmon sperm DNA/ml) for 3 h at 42.5°C. Hybridization was performed overnight in 50% formamide–6× SSC–1% SDS–50 μg of denatured salmon sperm DNA/ml at 42.5°C with an [α-32P]dCTP labeled, randomly primed probe (Pharmacia), which was a gel-purified 465-bp PCR product of the TK gene in pHyg.FRT.TK. Filters were exposed to Kodak XR-5 film at −80°C with intensifying screens.
Isolation of nascent DNA.
Nascent DNA was isolated essentially as described previously (39). Approximately 3 × 108 exponentially growing cells were labeled with 33 μM 5-bromodeoxyuridine (BrdUrd; Sigma) for 15 min at 37°C. All procedures after this step were carried out under yellow lights to prevent breakage of the labeled DNA. Plates were rinsed with ice-cold phosphate-buffered saline, and the cells were lysed with 3 ml of lysis buffer (50 mM Tris-Cl [pH 8.0]–10 mM EDTA–0.5% SDS) and gentle rocking. Lysates were first incubated with 10 μg of RNase A/ml for 30 min at 37°C, then incubated overnight at 37°C after the addition of proteinase K (to 800 μg/ml), and dialyzed for 24 h against 10 mM Tris-Cl–1 mM EDTA (pH 8.0) at 4°C. The DNA was concentrated by extraction with sec-butanol, denatured by the addition of NaOH to a final concentration of 0.2 N, and adjusted to a density of 1.81 g/ml (refractive index [RI] = 1.4095) by the addition of a saturated alkaline CsCl solution (in 50 mM NaOH–1 mM EDTA). Isopycnic gradients were centrifuged at 35 krpm for 72 h at 25°C (in a Beckman 75 Ti rotor) and fractionated from below. Fractions containing heavy DNA (RI, 1.4105 to 1.413; density, 1.82 to 1.84 g/ml) were pooled, the RI was adjusted to 1.4107 with an alkaline CsCl solution, and the DNA was recentrifuged under the same conditions. Control experiments using high-molecular-weight BrdUrd-labeled nascent DNA or lambda DNA showed no detectable decrease in DNA size as a result of these procedures. The BrdUrd-labeled DNA was precipitated with isopropanol and pelleted at 12 krpm in a Sorvall SS-34 rotor for 30 min, then resuspended in sterile water. Approximately 0.03 pg of BrdUrd DNA/cell (range, 0.015 to 0.05 pg/cell) was typically isolated from each of the pulse-labeled cell lines. Nascent DNA was size fractionated by alkaline agarose electrophoresis and eluted by using a QIAquick Gel Extraction Kit (Qiagen).
Construction of PCR competitors.
Competitors were designed to be identical to the genomic sequence-tagged sites (STSs) with the exception of a 20- to 30-nucleotide (nt) deletion. Competitors were constructed by amplifying genomic DNA with a wild-type primer and a mutagenic primer. Mutagenic primers (32 to 38 nt) spanned a 20- to 30-nt deletion of wild-type sequence 14 to 16 nt from the 3′ end. The 5′ 18 to 22 nt were identical to the corresponding wild-type primer sequence, so that the mutagenized PCR product could be amplified by the wild-type primers. The competitor product was purified by 8% acrylamide gel electrophoresis after amplification with wild-type primers. The effective concentration of each competitor was quantitated by titration against a known amount of genomic template (18). Competitor DNA was diluted from concentrated stocks and quantitated before each set of PCRs. The amounts of nascent DNA at a given STS in different cell lines were quantitated in parallel.
Competitive PCR analysis of nascent DNA.
A constant volume of size-fractionated (0.85 to 1.5 kb) nascent DNA was added to amplification reactions containing increasing amounts of competitor template. The PCR conditions were the same as those described earlier except that 40 to 45 cycles were performed (10). PCR products were resolved on 8% polyacrylamide gels run at 140 V for 3 h in TBE (9 mM Tris-borate [pH 8.0]–0.4 mM EDTA). Gels were stained in ethidium bromide for 20 min and photographed by using a Fotodyne charge-coupled device camera imaging system. The log of the ratio of the signal intensities for the competitor and nascent template PCR products was calculated by using Collage 4.0 software (Fotodyne, Inc.) and plotted against the log of the amount of competitor added to each reaction mixture. GraphPad Prism was used to fit a straight line to the points, and the equation of this line was used to quantitate the amounts of nascent DNA template (5, 10). Reaction conditions were selected to avoid heteroduplex formation between the competitor and nascent PCR products, as evidenced by the r2 values near unity (18). At least four to six titrations were performed for each competitor with nascent DNA from two or three independent preparations, and the results were averaged. To compare DNA from different cell cultures, the amount of nascent DNA in each preparation was normalized by PhosphorImager quantitation of slot blots (as shown in Fig. 5) hybridized to a genomic DNA probe.
FIG. 5.
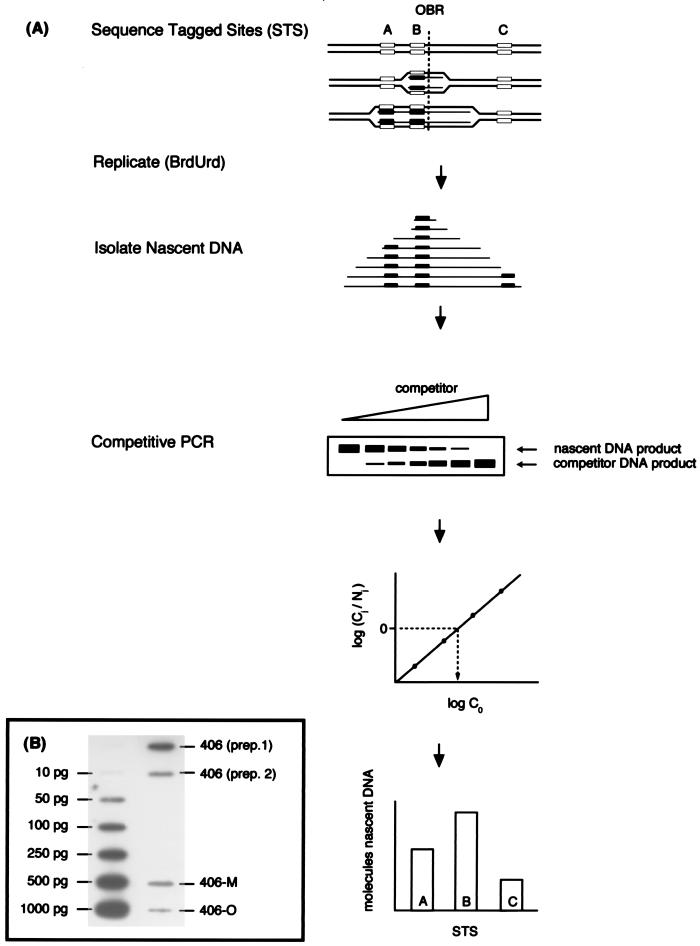
Competitive PCR assay of nascent DNA abundance. (A) Unsynchronized cells were pulse labeled with BrdUrd to allow the synthesis of dense nascent fragments of various lengths. Isolated nascent DNA of 0.85 to 1.5 kb was quantitated at STS-A, -B, and -C by coamplification against known amounts of competitor template specific for each STS. Competitor templates were designed to be ca. 20 bp smaller so that the competitor and nascent PCR products were resolvable by polyacrylamide gel electrophoresis. The logarithm of the relative intensities of the nascent (Nj) and competitor (Cj) products produced in each reaction was plotted against the logarithm of the amount of competitor added (C0). The initial quantity of nascent DNA for each STS was determined by the amount of competitor template required to produce equal amounts of competitor and nascent products after amplification. The STS where the abundance of nascent DNA was greatest would be located closest to a replication origin. (B) Representative slot blot autoradiogram of filter used for PhosphorImager quantitation of nascent DNA, by interpolation on a straight-line plot (r2 > 0.99) of DNA amount (10 to 1,000 pg) versus PhosphorImager signal intensity.
RESULTS
Efficient FLP-mediated recombination in vitro.
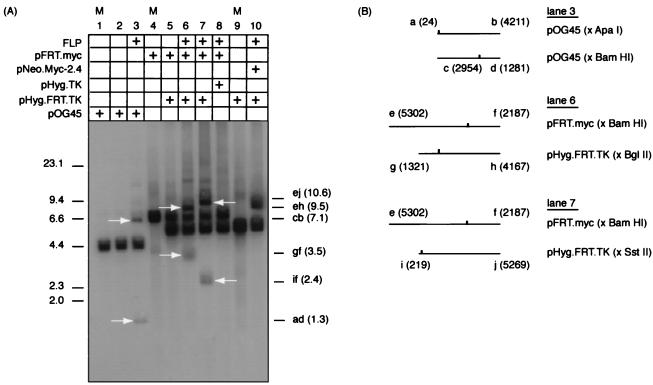
To examine the pattern of replicative intermediates at the same chromosomal locus before and after the integration of c-myc origin sequences, a site-specific recombination system based on yeast FLP recombinase was adapted for use in HeLa cells (Fig. 1). O’Gorman et al. (31) showed that plasmids containing an FRT could be targeted to recombine at a simian cell chromosomal FRT site when cotransfected with a plasmid expressing the FLP recombinase. In the present work the fidelity and orientation of recombination of the FRT plasmids constructed for use in cell culture were confirmed in vitro prior to in vivo recombination. Plasmids containing an FRT (pFRT.Myc, pHyg.FRT.TK, and pOG45) and plasmids without an FRT (pNeo.Myc-2.4 and pHyg.TK) were combined as substrates in the presence of purified FLP recombinase. Plasmids were linearized with restriction enzymes such that the resulting recombination products would be of different sizes than the input substrates. Recombination products were obtained only when an FRT was present in both substrate molecules (Fig. 2, lanes 3, 6, and 7). In reactions where only one of two substrate plasmids contained an FRT, no chimeric recombination products were observed (Fig. 2, lanes 8 and 10). Similarly, no chimeric recombination products were observed when FLP recombinase was omitted from reactions in which both substrate molecules contained an FRT (Fig. 2, lane 5). The intensities of the recombination products produced with the commercially obtained FRT plasmid pOG45 (Fig. 2, lane 3) were similar to the intensities of products resulting from recombination of the donor and acceptor constructs, suggesting that these plasmids recombined at comparable efficiencies in the presence of FLP recombinase.
FIG. 1.
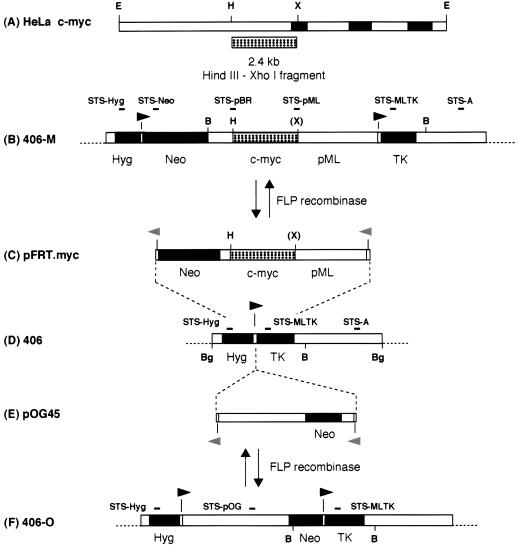
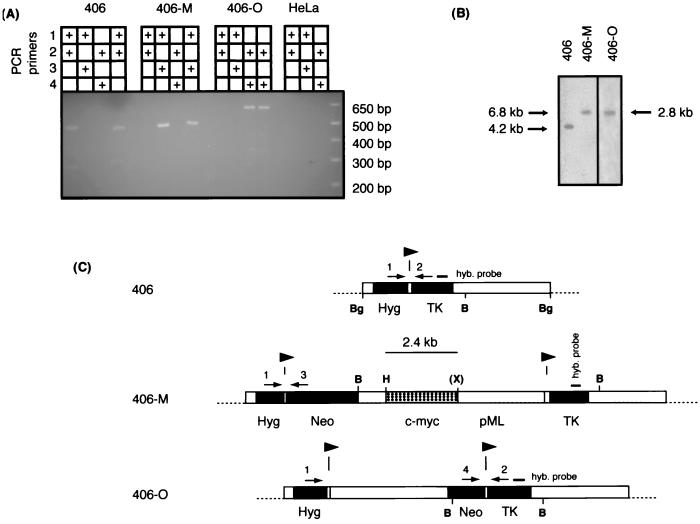
DNA maps. (A) HeLa endogenous c-myc locus. Solid boxes, exons; open boxes, introns or flanking DNA; stippled box, the 2.4-kb c-myc HindIII-XhoI origin fragment. (B) Acceptor site in 406-M cells after FLP-mediated recombination of pFRT.Myc into the chromosomal FRT of 406 cells. Solid arrowheads indicate locations and polarity of FRT sites. Hyg, hygromycin resistance gene; Neo, neomycin (G418) resistance gene; pML and open boxes, vector sequences; TK, HSV TK gene. (C) The FRT donor construct pFRT.Myc (shown linearized at the FRT XbaI site). c-myc, 2.4-kb HindIII-XhoI c-myc origin fragment; Neo, promoterless neomycin phosphotransferase gene. Dashed lines indicate FLP-mediated integration of the donor construct pFRT.Myc at the pHyg.FRT.TK chromosomal acceptor locus in 406 cells. Shaded (left-pointing) arrowheads indicate locations and polarity of FRT half-sites. (D) Chromosomal acceptor site in 406 cells after the introduction of the FRT construct pHyg.FRT.TK. (Note that the DNA is shown linearized at the BglI site, although the exact ends of the integrated construct have not been mapped.) (E) Control non-myc DNA FRT donor construct (shown linearized at the FRT XbaI site). Note that the Neo cassette is a different size in pOG45 than in pFRT.Myc. (F) Acceptor site in 406-O cells after FLP-mediated recombination of pOG45 into the chromosomal FRT of 406 cells. B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; X, XhoI; (X), XhoI site destroyed in cloning. STSs used in the experiments of Fig. 6 and 8 are indicated above the maps.
FIG. 2.
In vitro FLP recombinase reactions. (A) Acceptor and donor plasmid constructs, with or without an FRT, were linearized and incubated in the presence or absence of purified FLP recombinase. Lanes marked M are size markers generated from control DNA. Lanes 1 and 2, pOG45 linearized with ApaI. No chimeric products are detectable. Lane 3, a mixture of pOG45 linearized with ApaI or BamHI. Lane 4, pFRT.Myc (cut with BamHI). Lanes 5 and 6, pFRT.Myc (BamHI cut) and pHyg.FRT.TK (BglII cut). Lane 7, pFRT.Myc (BamHI cut) and pHyg.FRT.TK (SstII cut). Lane 8, pFRT.Myc (BamHI cut) and pHyg.TK (BglII cut). Lane 9, pHyg.FRT.TK (SstII cut). Lane 10, pNeo.Myc-2.4 (BamHI cut) and pHyg.FRT.TK (HindIII cut). The products were resolved by gel electrophoresis and identified by hybridization to a probe with sequences common to all of the constructs. Chimeric fragments produced by recombination were of the expected sizes and are indicated by arrows. (B) Maps of FRT plasmid digests used in panel A. FRT position is indicated by an upward tick mark.
FLP-mediated recombination in HeLa cells.
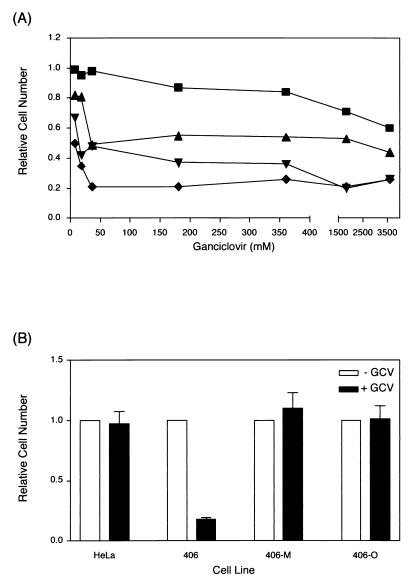
HeLa cells were transfected with linearized pHyg.FRT.TK and selected at one of three concentrations of hygromycin (100, 200, or 400 μg/ml). All three populations of transfected cells showed an increased sensitivity to ganciclovir compared to untransfected HeLa cells (Fig. 3) due to expression of the HSV TK gene. In cells selected at the highest concentration of hygromycin (400 μg/ml), the 50% lethal concentration (LC50) of ganciclovir was lowest (∼10 μM). An acceptor cell line, line 406, resulting from selection at 400 μg of hygromycin/ml, was characterized as having a single, unrearranged copy of the Hyg.FRT.TK acceptor construct. Southern blot analysis with a probe complementary to sequences in the TK gene demonstrated that digestion of 406 genomic DNA with BamHI produced a single band of approximately 4.2 kb (see Fig. 4B). This band resulted from digestion at the single BamHI site in the acceptor construct and a BamHI site in the 5′ flanking chromosomal DNA; it indicated that only one copy of the acceptor construct was present in this cell line. That the acceptor site construct was not rearranged during integration in 406 cells was confirmed by additional restriction enzyme digestions (data not shown) and by the resistance of the cells to hygromycin and their sensitivity to ganciclovir.
FIG. 3.
Ganciclovir sensitivity determined by MTT assay. (A) HeLa cells transfected with the linearized acceptor construct pHyg.FRT.TK and selected at different concentrations of hygromycin were exposed to increasing amounts of ganciclovir. The number of cells surviving drug selection after 7 days is expressed as a fraction of the number of untransfected HeLa cells surviving drug selection after the same period of time. ■, untransfected HeLa cells; ▴, 100 μg of hygromycin/ml; ▾, 200 μg of hygromycin/ml; ⧫, 400 μg of hygromycin/ml. (B) Ganciclovir resistance was measured before and after FRT-mediated integration of donor constructs. HeLa, untransfected HeLa cells; 406, chromosomal FRT acceptor cell line; 406-M, 406 cells with pFRT.Myc integrated at the chromosomal FRT; 406-O, 406 cells with pOG45 integrated at the 406 chromosomal FRT. Cells were grown in the absence (− GCV) or presence (+ GCV) of 15 μM ganciclovir for 7 days. The number of cells surviving drug selection is expressed as the fraction of the number of cells present without drug selection after the same period (plus the standard error of the mean).
FIG. 4.
Site-specific recombination in vivo. (A) Genomic DNA was isolated from the cell lines indicated and used in PCR amplifications with primers 1 through 4 as shown. HeLa DNA was used as a control for nonspecific amplification. The last lane for each cell line is an amplification of control DNA to produce PCR products specific for accurate FLP recombination. (B) Genomic DNA was isolated from 406, 406-M, and 406-O cells and digested with BamHI. Cells containing a single, unrearranged copy of the acceptor construct (406) and those that had integrated either the pFRT.Myc or the pOG45 donor plasmid specifically at the chromosomal FRT locus were identified by using a probe that was complementary to sequences common to all three cell lines. (C) Maps of the chromosomal acceptor sites in the 406, 406-M, and 406-O cell lines. Primer oligonucleotides 1 through 4 and their positions in acceptor (406), pFRT.Myc-integrated (406-M), and pOG45-integrated (406-O) cells are indicated above the linear maps by arrows. Primers 1 and 2 are specific to sequences found only in the acceptor construct, while primers 3 and 4 are complementary to Neo sequences found in both the pFRT.Myc and pOG45 donor constructs. Probes used in the Southern hybridization analysis are indicated above the maps. Symbols and abbreviations are as defined for Fig. 1.
Integration of pFRT.Myc and pOG45 at the chromosomal FRT.
Cotransfection of the pFRT.Myc origin donor plasmid and an FLP recombinase expression plasmid (pOG44) into the acceptor cell line 406 resulted in G418-resistant colonies due to the expression of the promoterless neomycin resistance cassette from the acceptor site CMV immediate-early promoter. Culture in G418 selected strongly for appropriately integrated donor plasmids, since expression of the promoterless neo gene due to random integration is extremely rare (31). Because there was no selective pressure for retention of the donor plasmid as an episome, it was lost during clonal expansion of transfected cells and was not detected by PCR or DNA hybridization using a Neo probe. Nor were donor plasmid sequences detected at any non-FRT site. Integration of pFRT.Myc introduced simian virus 40 (SV40) polyadenylation sequences, multiple translational stop codons in the pFRT.Myc vector, and a single-base deletion in the downstream FRT that disrupted the HSV TK reading frame, so that the accurately targeted acceptor site no longer expressed the HSV TK activity. FLP-recombined cells could therefore be selected by their resistance to both G418 and ganciclovir (Fig. 3B). Similarly, FLP-mediated integration of pOG45 resulted in cell lines that were resistant to G418 due to expression of the neomycin phosphotransferase gene from the SV40 early promoter and resistant to ganciclovir due to disruption of the HSV TK reading frame (Fig. 3B).
Cell lines containing pFRT.Myc (406-M) or pOG45 (406-O) were characterized further. Diagnostic PCR with primers specific to junction regions formed by integration of either pFRT.Myc or pOG45 showed the presence of 502- or 635-bp PCR products, respectively, confirming that these cell lines contained pFRT.Myc or pOG45 constructs integrated appropriately at the chromosomal FRT in 406 cells (Fig. 4A, lanes 406-M and 406-O). Acceptor 406 cells prior to donor plasmid integration produced a 484-bp PCR product when amplified with primers complementary to regions flanking the chromosomal FRT (Fig. 4A). This product was absent in 406-M, 406-O, and HeLa cells (Fig. 4A).
BamHI digestion produced a 4.2-kb fragment at the acceptor site in 406 cells (Fig. 4B). Site-specific integration of unrearranged pFRT.Myc at the chromosomal FRT site in 406 cells was predicted to yield a 6.8-kb BamHI restriction fragment. Digestion of 406-M genomic DNA with BamHI produced one band, of the expected size, when the DNA was hybridized with a probe specific for TK sequences (Fig. 4B). Site-specific integration of unrearranged pOG45 at the chromosomal FRT site in 406 cells was predicted to yield a 2.8-kb BamHI restriction fragment. Digestion of 406-O genomic DNA with BamHI also produced one band, of the expected size, when the DNA was hybridized with the same probe (Fig. 4B). In both cases, multiple probes and restriction digestion with other enzymes confirmed that only one pFRT.Myc integration or one pOG45 integration had occurred, respectively, in the 406-M or 406-O cell line (data not shown).
Quantitation of acceptor site nascent DNAs.
The competitive PCR assay for quantitating nascent DNA synthesized in vivo is outlined in Fig. 5. An asynchronous population of cells was labeled with BrdUrd for 15 min, and the DNA was extracted. Low-molecular-weight, uniformly BrdUrd-pulse-labeled (nascent) DNA was separated from unreplicated, parental DNA (unlabeled) by two rounds of alkaline CsCl centrifugation. The nascent DNA was size fractionated on an alkaline agarose gel, and the DNA was extracted from the size fraction containing fragments of ∼850 to 1,500 nt. Nascent strand abundance at selected STSs was then quantitated by PCR amplifications of a fixed amount of nascent DNA in the presence of several different quantities of competitor template and by use of the equation for PCR amplification given by Connolly et al. (5), log (Cj/Nj) = log C0 − log N0, where Cj is the amount of competitor template after j cycles of amplification, C0 is the input amount of competitor template, Nj is the amount of nascent template after j cycles of amplification, and N0 is the input amount of nascent template. For each STS, the quantity of competitor template required to produce equal amounts of competitor and nascent DNA PCR products in a single reaction was calculated by plotting the logarithm of the amount of competitor template against the logarithm of the signal ratio of the products from the competitor and nascent templates. The amount of competitor which yielded log (Cj/Nj) = 0 was interpolated from the plot, at which point log C0 = log N0 and C0 = N0 (12, 20, 32, 38). To allow comparison between the abundances of nascent strands at STSs in different cell lines, the competitive PCR data were normalized to equivalent amounts of total nascent DNA, determined by PhosphorImager quantitation (Fig. 5B).
Quantitation of 406, 406-M, and 406-O nascent DNA by competitive PCR.
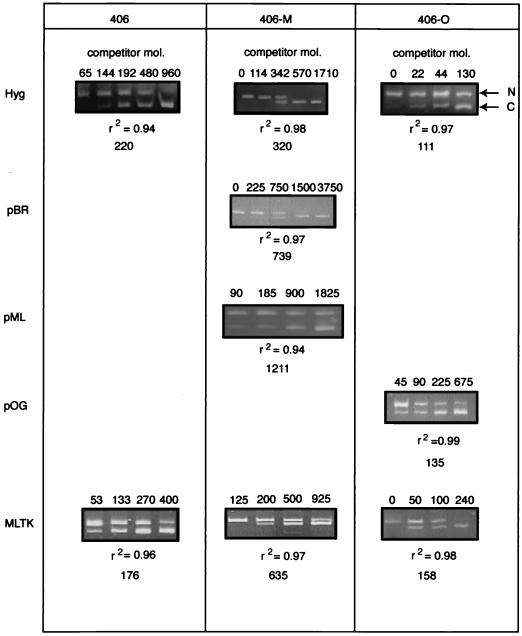
Nascent DNA was isolated from 406, 406-M, and 406-O cells. Competitive PCR was used to quantitate the amount of nascent strands in the 0.85- to 1.5-kb fraction from five STSs: STS-Hyg and STS-MLTK (present in all three cell lines), STS-pBR and STS-pML (present in 406-M cells only), and STS-pOG (present in 406-O cells only). Figure 6 shows representative data produced by competitive PCR of 406, 406-M, and 406-O nascent DNA. Each competitive PCR experiment was repeated at least four to six times for each STS. The logarithm of the signal ratio of competitor template to nascent DNA template was plotted against the logarithm of the amount of competitor added, and the total number of nascent DNA molecules (copies) was derived from this plot, as indicated below each gel. The coefficient of determination, r2, was calculated for each plot as an indicator of the validity of the titration.
FIG. 6.
Competitive PCR analysis of 406, 406-M, and 406-O nascent DNA. Representative gels of competitive PCRs with nascent DNA from the 406, 406-M, and 406-O cell lines are shown for the following STSs: Hyg, pBR, pML, pOG, and MLTK. pBR and pML are specific for the integrated pFRT.Myc sequences, while pOG is specific for the pOG45 sequences. Arrows indicate the positions of the nascent (N) and competitor (C) PCR products. The number of competitor molecules (mol.) added to each reaction is indicated at the top of each lane. The number of nascent molecules, calculated from a plot of the logarithm of the signal ratio versus the logarithm of the amount of competitor added, and the r2 value are shown below each gel.
Figure 7 summarizes the results of quantitation of the short nascent DNAs at each STS in 406, 406-M, and 406-O cells. Before the integration of c-myc origin sequences, roughly 500 to 600 copies of nascent DNA were calculated to be present at STS-Hyg or STS-MLTK in a standard aliquot (2%) of the 0.85- to 1.5-kb nascent DNA fraction. After integration of the c-myc origin fragment in 406-M cells, more than twice as many nascent strands were detected at STS-Hyg (P < 0.001) and STS-MLTK (P < 0.001) in equivalent amounts of nascent DNA. The greatest abundance of nascent DNA in 406-M cells was detected close to the c-myc origin sequences, at STS-pML. The amount of nascent DNA at this STS was enriched approximately 6- to 8-fold over the quantities observed at the acceptor site (STS-Hyg and STS-MLTK) before integration of the c-myc sequences in 406 cells and was 8- to 10-fold greater than the quantities observed at these STSs in 406-O cells. In addition, there was no statistically significant difference in the amounts of short nascent DNA at STS-Hyg, STS-pOG, and STS-MLTK in 406-O cells.
FIG. 7.
Nascent DNA abundance at the chromosomal acceptor site. (A) The relative quantity of nascent strands as determined by competitive PCR is shown at each STS assayed in 406, 406-M, and 406-O cells. The data are means (plus standard deviations) of at least four to six different competitive PCR titrations of two or three independent nascent DNA preparations for each cell line. To allow comparison between cell lines, the quantities shown have been normalized to adjust for the different amounts of nascent DNA in each preparation (based on PhosphorImager quantitation) and are expressed relative to the abundance of nascent strands at STS-Hyg in 406 cells (defined as 1.0). (B) Acceptor site maps. Symbols and abbreviations are as defined for Fig. 1.
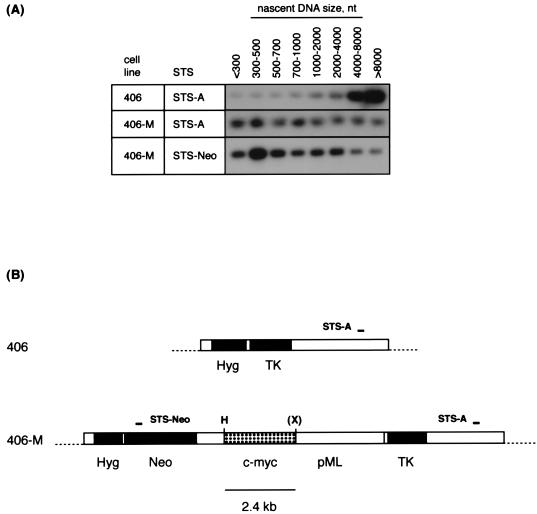
These data indicate that the c-myc origin construct induces replication in cis near the acceptor FRT. To confirm this conclusion, PCR mapping was performed on size-fractionated nascent DNA from 406 cells and 406-M cells by using a primer set (STS-A) located in bacterial vector sequences at the acceptor site. With nascent DNA from 406 cells, amplification occurs primarily in the highest-molecular-weight fraction, containing strands of >8 to 23 kb (Fig. 8). This suggests that if replication of STS-A is from a bidirectional origin, then replication initiates at least 4 to 11 kb away from STS-A in these cells. If replication is unidirectional, the origin is >8 to 23 kb away. The strong preferential amplification of relatively large nascent strands in this assay also indicates that nascent DNA is not degraded during the isolation procedure used in these experiments.
FIG. 8.
PCR mapping of nascent DNA near the FRT acceptor site. (A) Nascent DNA was isolated from 406 and 406-M cells and size fractionated by alkaline gel electrophoresis. A 5% aliquot of each size fraction was subjected to PCR in the linear range of amplification as described previously (39). Products were detected after gel electrophoresis by hybridization to probes specific to the internal regions of the products. Indicated above the lanes are the approximate sizes of the nascent DNA in each fraction. Note that the STS-Neo and STS-A PCR products are ∼230 bp and would not amplify smaller Okazaki fragments. (B) Acceptor site maps showing the locations of STS-A and STS-Neo. Symbols and abbreviations are as defined for Fig. 1.
In contrast, when the c-myc donor plasmid is integrated at the acceptor site in 406-M cells, short nascent strands of less than 300 to 500 nt are readily amplified in equivalent preparations of nascent DNA from equal numbers of cells. Hence, integration of the c-myc donor plasmid causes replication to initiate efficiently in the bacterial sequences at the acceptor site, within 300 to 500 nt of STS-A.
We have recently reported that replication initiates at multiple sites in the region of the c-myc origin (39, 41). Here, PCR mapping shows that replication initiates efficiently within 300 to 500 nt of a second bacterial sequence, STS-Neo, located in the donor plasmid. STS-Neo is located more than 10 kb from STS-A, yet replication initiates within 1 kb of each of these sites in 406-M cells. Thus, as at the endogenous locus, replication initiates at multiple sites at the transduced c-myc origin region. Taken together, these data provide biochemical and genetic evidence that the c-myc origin plasmid displays replicator activity that influences the pattern of replicative intermediates in cis in the chromosome.
DISCUSSION
Data obtained in vitro and in vivo suggest a model of c-myc replication in which DNA unwinding begins within 2.4 kb 5′ of the c-myc gene and exposes potential start sites for DNA synthesis on each template strand. The frequency of initiation at any of the potential start sites depends on the likelihood of its exposure as single-stranded DNA. Hence, replication may begin at several possible sites over an extended region, but sites nearest the initial DUE will be used more often. In this model a critical regulatory event is DNA unwinding, and the sequences or structural elements that control unwinding comprise an essential part of the genetically defined replicator. To test this model, the 2.4-kb upstream region of the human c-myc gene has been moved to a new chromosomal position and tested for its ability to initiate replication and influence the pattern of replication intermediates in its vicinity.
The 2.4-kb 5′ flanking sequences contain several structural elements common to other eucaryotic replication origins, including positioned nucleosomes, constitutive nuclease hypersensitive sites, regions of bent DNA, a DUE, and transcription factor binding sites (3, 11, 26, 30). We have shown previously that the chromatin structure of the 2.4-kb c-myc upstream region, as revealed by the positioning of nucleosomes and DNase-hypersensitive sites, is not altered when this segment is moved to new chromosomal locations by adeno-associated virus transduction (21).
In the present experiments the S. cerevisiae FLP recombinase allowed the site-specific targeting of the c-myc origin fragment to a specific acceptor location and analysis of the pattern of replicative intermediates before and after integration of the origin construct. The data indicate not only that the amounts of nascent DNA at the acceptor site increase after the introduction of the c-myc construct but also that there is a strong peak of short nascent fragments near the c-myc origin sequences compared to their abundance at two flanking locations in the acceptor site. This distribution of nascent strands is similar to that found by using competitive PCR to analyze the hamster DHFR origin (18, 32). In contrast, the introduction of non-myc sequences did not result in any detectable increase in the amounts of nascent DNA at either the flanking acceptor sequences or the introduced sequences.
The c-myc origin sequences increased the abundance of short nascent strands at both STS-Hyg and STS-MLTK. Since STS-Hyg and STS-MLTK are 3 to 4 kb away from the c-myc origin sequences after integration, these data suggest that the presence of an origin can alter the pattern of replicative intermediates in the surrounding area over large (6- to 8-kb) distances. Additionally, since the size of the nascent DNA assayed was 0.85 to 1.5 kb, the short nascent strands found at STS-Hyg and STS-MLTK did not initiate at the same sites. Similarly, STS-Neo and STS-A are more than 10 kb apart, and both appear to initiate replication at sites within 1 kb after introduction of the c-myc origin plasmid. Therefore, the introduction of the c-myc replicator exposes new sites for replication initiation in the flanking DNA. In contrast, the levels of nascent DNA observed in 406-O cells remained constant across the acceptor locus, suggesting that the addition of non-myc sequences did not influence the pattern of replicative intermediates in the same manner as did the c-myc donor plasmid sequences.
The non-myc DNA used as a control in 406-O cells comprises a bacterial transcription unit, not mammalian DNA. Hence it could be argued that bacterial DNA is less capable of supporting initiation in a mammalian background than is DNA of mammalian origin. That this is not the case was clearly shown by Calos and coworkers, who directly compared the abilities of mammalian and bacterial sequences to initiate replication and stated explicitly that there is no significant bias in the initiation of replication in bacterial versus mammalian sequences in mammalian cells (19). Moreover, our PCR mapping data show that the pattern of replication at the bacterial site STS-A is changed completely upon integration of the c-myc plasmid at the acceptor site in 406-M cells. The results at STS-A and STS-Neo suggest that replication can initiate in bacterial sequences at the FRT acceptor site, although the abundance of nascent strands is lower than that near the neighboring c-myc origin DNA.
Although the pFRT.Myc origin construct and the negative-control (pOG) construct differ primarily in the presence of the c-myc origin fragment, their bacterial sequences are not completely identical. While we have not excluded the possibility that the bacterial sequences possess replicator activity, this is unlikely for several reasons. First, in the absence of c-myc sequences, the pFRT.Myc vector does not display replicator activity in more permissive ARS assays in vitro or in vivo (3, 27, 29). Second, the abundance of replicative intermediates peaks near the c-myc insert of the integrated pFRT.Myc, suggesting that replication initiation activity is concentrated in this area. Finally, constructs that are identical to pFRT.Myc except for deletions of 100 to 1,400 bp of c-myc DNA, removing specific transcription factor consensus binding sites, show 50 to 100% loss of replication initiation activity when integrated at the acceptor site in 406 cells (unpublished data). Therefore, in cells that are isogenic except for small deletions in the acceptor site c-myc origin DNA, replicator activity depends on the c-myc DNA sequences.
According to the model above, the increase in abundance of short nascent strands at the flanking STSs can be explained by the introduction of the early-firing c-myc DUE (41). If replication initiates at the c-myc origin before synthesis from the endogenous origin for the acceptor site, initiations from the c-myc origin will appear as a peak in the quantities of nascent DNA at the acceptor site. Experiments are under way using the FLP recombinase system and site-specific mutants of the c-myc core origin to test the hypothesis that early-S-phase activation of the c-myc DUE is responsible for exposing flanking initiation sites for replication.
During the preparation of this paper, Aladjem et al. reported that short nascent DNAs were synthesized preferentially in a fragment of the human beta globin replicon integrated by the FLP recombinase at an ectopic location in monkey cells (1).
ACKNOWLEDGMENTS
We thank J. Bode for the pHyg.TK.fus plasmid, M. Cox for purified FLP recombinase and extensive technical advice, and Poonam Khaira for technical input and helpful discussions.
M. Malott was supported by the Wright State University Biomedical Sciences Ph.D. program. This work was supported by PHS grant GM 53819 from the NIGMS to M.L.
REFERENCES
- 1.Aladjem M I, Rodewald L W, Kolman J L, Wahl G M. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 2.Benard M, Lagnel C, Pierron G. Site-specific initiation of DNA replication within the non-transcribed spacer of Physarum rDNA. Nucleic Acids Res. 1995;23:1447–1453. doi: 10.1093/nar/23.9.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berberich S, Trivedi A, Daniel D C, Johnson E M, Leffak M. In vitro replication of plasmids containing human c-myc DNA. J Mol Biol. 1995;245:92–109. doi: 10.1006/jmbi.1994.0010. [DOI] [PubMed] [Google Scholar]
- 4.Bielinsky A K, Gerbi S A. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- 5.Connolly A R, Cleland L G, Kirkham B W. Mathematical considerations of competitive polymerase chain reaction. J Immunol Methods. 1995;187:201–211. doi: 10.1016/0022-1759(95)00185-2. [DOI] [PubMed] [Google Scholar]
- 6.Coverley D, Laskey R A. Regulation of eukaryotic DNA replication. Annu Rev Biochem. 1994;63:745–776. doi: 10.1146/annurev.bi.63.070194.003525. [DOI] [PubMed] [Google Scholar]
- 7.Delidakis C, Kafatos F C. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 1989;8:891–901. doi: 10.1002/j.1460-2075.1989.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DePamphilis M. Origins of DNA replication. In: DePamphilis M, editor. DNA replication in eucaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 45–86. [Google Scholar]
- 9.Dijkwel P A, Hamlin J L. Initiation of DNA replication in the dihydrofolate reductase locus is confined to the early S period in CHO cells synchronized with the plant amino acid mimosine. Mol Cell Biol. 1992;12:3715–3722. doi: 10.1128/mcb.12.9.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diviacco S, Norio P, Zentilin L, Menzo S, Clementi M, Biamonti G, Riva S, Falaschi A, Giacca M. A novel procedure for quantitative polymerase chain reaction by coamplification of competitive templates. Gene. 1992;122:313–320. doi: 10.1016/0378-1119(92)90220-j. [DOI] [PubMed] [Google Scholar]
- 11.Dobbs D L, Shaiu W L, Benbow R M. Modular sequence elements associated with origin regions in eukaryotic chromosomal DNA. Nucleic Acids Res. 1994;22:2479–2489. doi: 10.1093/nar/22.13.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giacca M, Pelizon C, Falaschi A. Mapping replication origins by quantifying relative abundance of nascent DNA strands using competitive polymerase chain reaction. Methods (Orlando) 1997;13:301–312. doi: 10.1006/meth.1997.0529. [DOI] [PubMed] [Google Scholar]
- 13.Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S, et al. Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci USA. 1994;91:7119–7123. doi: 10.1073/pnas.91.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert D M. Replication origins in yeast versus metazoa: separation of the haves and the have nots. Curr Opin Genet Dev. 1998;8:194–199. doi: 10.1016/s0959-437x(98)80141-x. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert D M, Miyazawa H, DePamphilis M L. Site-specific initiation of DNA replication in Xenopus egg extract requires nuclear structure. Mol Cell Biol. 1995;15:2942–2954. doi: 10.1128/mcb.15.6.2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishimi Y, Matsumoto K, Ohba R. DNA replication from initiation zones of mammalian cells in a model system. Mol Cell Biol. 1994;14:6489–6496. doi: 10.1128/mcb.14.10.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp Quant Biol. 1963;28:329–348. [Google Scholar]
- 18.Kobayashi T, Rein T, DePamphilis M L. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol Cell Biol. 1998;18:3266–3277. doi: 10.1128/mcb.18.6.3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krysan P J, Smith J G, Calos M P. Autonomous replication in human cells of multimers of specific human and bacterial DNA sequences. Mol Cell Biol. 1993;13:2688–2696. doi: 10.1128/mcb.13.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Giacca M, Norio P, Biamonti G, Riva S, Falaschi A. Utilization of the same DNA replication origin by human cells of different derivation. Nucleic Acids Res. 1996;24:3289–3294. doi: 10.1093/nar/24.17.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Leffak M. Conserved chromatin structure in c-myc 5′ flanking DNA after viral transduction. J Mol Biol. 1991;222:45–57. doi: 10.1016/0022-2836(91)90736-p. [DOI] [PubMed] [Google Scholar]
- 22.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leffak M, James C D. Opposite replication polarity of the germ line c-myc gene in HeLa cells compared with that of two Burkitt lymphoma cell lines. Mol Cell Biol. 1989;9:586–593. doi: 10.1128/mcb.9.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little R D, Platt T H, Schildkraut C L. Initiation and termination of DNA replication in human rRNA genes. Mol Cell Biol. 1993;13:6600–6613. doi: 10.1128/mcb.13.10.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupton S D, Brunton L L, Kalberg V A, Overell R W. Dominant positive and negative selection using a hygromycin phosphotransferase-thymidine kinase fusion gene. Mol Cell Biol. 1991;11:3374–3378. doi: 10.1128/mcb.11.6.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcu K B, Bossone S A, Patel A. MYC function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- 27.McWhinney C, Leffak M. Autonomous replication of a DNA fragment containing the chromosomal replication origin of the human c-myc gene. Nucleic Acids Res. 1990;18:1233–1242. doi: 10.1093/nar/18.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McWhinney C, Leffak M. Episomal persistence of a plasmid containing human c-myc DNA. Cancer Cells. 1988;6:467–471. [Google Scholar]
- 29.McWhinney C, Waltz S E, Leffak M. Cis-acting effects of sequences within 2.4-kb upstream of the human c-myc gene on autonomous plasmid replication in HeLa cells. DNA Cell Biol. 1995;14:565–579. doi: 10.1089/dna.1995.14.565. [DOI] [PubMed] [Google Scholar]
- 30.Michelotti G A, Michelotti E F, Pullner A, Duncan R C, Eick D, Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Gorman S, Fox D T, Wahl G M. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 32.Pelizon C, Diviacco S, Falaschi A, Giacca M. High-resolution mapping of the origin of DNA replication in the hamster dihydrofolate reductase gene domain by competitive PCR. Mol Cell Biol. 1996;16:5358–5364. doi: 10.1128/mcb.16.10.5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phi-van L, Sellke C, von Bodenhausen A, Stratling W H. An initiation zone of chromosomal DNA replication at the chicken lysozyme gene locus. J Biol Chem. 1998;273:18300–18307. doi: 10.1074/jbc.273.29.18300. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Shinomiya T, Ina S. Mapping an initiation region of DNA replication at a single-copy chromosomal locus in Drosophila melanogaster cells by two-dimensional gel methods and PCR-mediated nascent-strand analysis: multiple replication origins in a broad zone. Mol Cell Biol. 1994;14:7394–7403. doi: 10.1128/mcb.14.11.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 37.Stinchcomb D T, Struhl K, Davis R W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 38.Tao L, Nielsen T, Friedlander P, Zannis-Hadjopoulos M, Price G. Differential DNA replication origin activities in human normal skin fibroblast and HeLa cell lines. J Mol Biol. 1997;273:509–518. doi: 10.1006/jmbi.1997.1352. [DOI] [PubMed] [Google Scholar]
- 39.Trivedi A, Waltz S E, Kamath S, Leffak M. Multiple initiations in the c-myc replication origin independent of chromosomal location. DNA Cell Biol. 1998;17:885–896. doi: 10.1089/dna.1998.17.885. [DOI] [PubMed] [Google Scholar]
- 40.Vassilev L, Johnson E M. An initiation zone of chromosomal DNA replication located upstream of the c-myc gene in proliferating HeLa cells. Mol Cell Biol. 1990;10:4899–4904. doi: 10.1128/mcb.10.9.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waltz S E, Trivedi A A, Leffak M. DNA replication initiates non-randomly at multiple sites near the c-myc gene in HeLa cells. Nucleic Acids Res. 1996;24:1887–1894. doi: 10.1093/nar/24.10.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Dijkwel P A, Hamlin J L. Lagging-strand, early-labelling, and two-dimensional gel assays suggest multiple potential initiation sites in the Chinese hamster dihydrofolate reductase origin. Mol Cell Biol. 1998;18:39–50. doi: 10.1128/mcb.18.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]