Abstract
Interleukin-4 (IL-4) is a four-α-helical bundle type I cytokine with broad pleiotropic actions on multiple lineages. Major actions of IL-4 were initially discovered for B and T cells, but this cytokine acts on more than a dozen different target cells spanning the innate and adaptive immune systems and is produced by multiple different cellular sources. While IL-4 was discovered just under 40 years ago in 1982, the interest in and discoveries related to this cytokine continue to markedly expand. There are important new advances related to its biological actions and to its mechanisms of signaling, including critical genes and downstream targets in a range of cell types. IL-4 is critical not only for careful control of immunoglobulin production but also related to inflammation, fibrosis, allergic reactions, and antitumor activity, with actions of IL-4 occurring through two different types of receptors, one of which is also used by IL-13, a closely related cytokine with partially overlapping actions. In this review, we cover critical older information but also highlight newer advances. An area of evolving interest relates to the therapeutic blockade of IL-4 signaling pathway to treat atopic dermatitis and asthma. Thus, this cytokine is historically important, and research in this area has both elucidated major biological pathways and led to therapeutic advances for diseases that affect millions of individuals.
Keywords: IL-4, IL-13, cytokine, STAT6, receptor, IL-4Ralpha1, IL-13Ralpha1, common gamma chain
Overview
Interleukin 4 (IL-4) was first identified as a factor that was produced by T cells and could enhance the proliferative response of B cells to anti-IgM and also was capable of inducible immunoglobulin (Ig) isotype switch of B cells to produce IgG, particularly IgG11,2. Subsequently, this cytokine was demonstrated to be even more pleiotropic/polyfunctional. For example, IL-4 is also the key cytokine for the differentiation of T helper 2 (Th2) cells, which control infection by extracellular parasites and contribute to allergic responses3,4, and it can induce the differentiation of M2 macrophages, which control infection by the protozoan Trypanosoma cruzi5. IL-4 acts through two types of receptors—the type I and type II IL-4 receptor—which share the IL-4 receptor α chain but have a different secondary receptor, γc versus IL-13Rα1, respectively (discussed in detail in the text), in part depending on cell type, and a highly related cytokine, IL-13, uses only the type II IL-4 receptor. Not only have there been extraordinary advances in the biology of these cytokines and their signaling mechanisms, but because of the critical roles of IL-4 related to the mediation of allergic responses, controlling the actions of this cytokine has been of profound therapeutic interest. Indeed, we now are at a point of tremendous excitement where blockade of IL-4 and IL-13 is effective in the treatment of allergic inflammatory responses, including atopic dermatitis and moderate to severe asthma.
Biology of IL-4
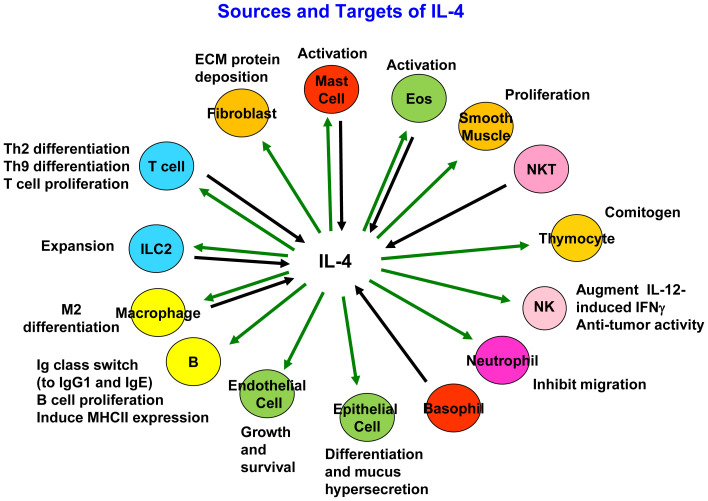
Although IL-4 is produced primarily by activated CD4+ T cells4, it also is produced by CD4+NK1.1+ “natural” T (NKT) cells, macrophages, eosinophils, basophils, mast cells, and type 2 innate lymphoid cells (ILC2s), although the amounts of IL-4 produced by these cell types are not uniform6–8 (Figure 1). IL-4 was initially described as a major B-cell growth factor and a promoter of Ig class switch that enhanced the production and secretion of mouse IgG1 (human IgG4)1,2. Subsequent work revealed that IL-4 is essential for the production of IgE, which is critical for allergen sensitization as well as the physiological response to parasites, including helminths. IL-4 induces elevated cell surface expression of CD23 (the low-affinity IgE receptor) on B cells and of class II major histocompatibility complex (MHC) molecules. In addition, IL-4 acts as a T-cell growth factor in both humans and mice and promotes the differentiation of Th2 cells. When combined with phorbol 2-myristate 3-acetate, IL-4 is also a potent co-mitogen for thymocytes. Furthermore, IL-4 derived from NKT cells is responsible for the expansion of memory-like CD8 T cells in BALB/c mice9. Overall, IL-4 can exert actions on a broad range of target cells, including macrophages, hematopoietic precursor cells, stromal cells, NK cells, and fibroblasts7, and can act as a potent anti-apoptotic factor.
Figure 1. Schematic of cellular sources of interleukin 4 (IL-4) and target cells for IL-4.
IL-4 potentially acts on other cells that are not listed. ECM, extracellular matrix; IFNγ, interferon gamma; MHC, major histocompatibility complex; NK, natural killer; NKT, natural killer T; Th, T helper.
IL-4 receptors
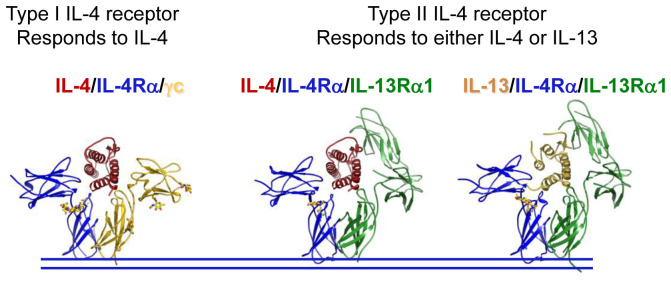
As noted above, IL-4 acts on both lympho-hematopoietic and non-lympho-hematopoietic cells10. Interestingly, there are two types of IL-4 receptors (Figure 2): type I IL-4 receptors are composed of the 140-kDa IL-4Rα and γc and are expressed on lympho-hematopoietic cells11,12; in contrast, type II IL-4 receptors comprising IL-4Rα and IL-13Rα1 are found on non-hematopoietic cells, and type II receptors also serve as the functional receptor for IL-1313, and both IL-4 and IL-13 induce similar actions via this type of receptor. Expression of IL-4Rα tends to be quite low, and cells that potently respond to IL-4 often express only a few hundred receptors per cell. For example, there are only about 300 IL-4 binding sites on resting lymphocytes, although receptor numbers can increase 5- to 10-fold upon cellular activation4. In reconstitution experiments, it was found that IL-4 binds to IL-4Rα with strong affinity (Kd ~ 266 pM), and in the presence of γc, the binding affinity is increased to “high” affinity (Kd ~79 pM)12, with the interaction of IL-4-IL-4Rα with γc being weak (Kd in > 500 nM)14. The type II IL-4 receptor, containing IL-4Rα and IL-13Rα1, is not expressed on mature T cells but is expressed on multiple other cell types and can transduce signals in these cells. Interestingly, type II receptors are expressed on immature T cells, including neonatal Th1 cells, and these receptors are implicated as mediating apoptosis of these cells15. The differential expression of γc and IL-13Rα1 determines whether IL-4 will signal via the type I or type II receptor, and IL-13 shares effects with IL-4 on the cells with type II IL-4 receptors. Consistent with this functional similarity, IL-4 and IL-13 are encoded by adjacent genes on mouse chromosome 11 and closely positioned on human chromosome 5q31; in both species, they are located between the RAD50 and KIF3A genes, consistent with a common ancestral gene. Interestingly, the genes encoding γc and IL-13Rα1 are both located on the X chromosome and share some general properties, suggesting that they may also have arisen by gene duplication.
Figure 2. Crystal structure of the type I interleukin 4 (IL-4) receptor bound to IL-4 (left) or the type II IL-4 receptor bound to either IL-4 (center) or IL-13 (right).
This figure was adapted using parts of Figure 3 from Elsevier, 132, LaPorte et al., Molecular and Structural Basis of Cytokine Receptor Pleiotropy in the Interleukin-4/13 System, 259–72, 200814, with permission from Elsevier.
Both Il4−/− and Il4ra−/− mice have normal numbers of T and B cells, indicating that IL-4 and IL-13 are not required for development of these lineages, but they have diminished IgG1 production and greatly decreased IgE production, after infection with Nippostrongylus brasiliensis, whereas levels of IgM and other IgG isotypes are produced at normal levels16,17. Interestingly, another γc family cytokine, IL-21, has some properties similar to those of IL-4 but also major differences. For example, the development of T and B cells is normal in Il21r−/− mice; however, IgG1 levels are greatly diminished, and IL-21 is also required for IgG3 production by CD40-activated naïve human B cells18,19. Whereas IL-4 favors the production of IgG1, IL-21 favors IgG3 production by CD40-activated naïve human B cells18, and whereas mice lacking IL-4 signaling have low IgE, IgE levels are elevated in Il21r−/− mice after immunization20, and this may be at least in part due to an inhibitory effect of IL-21 on IL-4-induced germ line Cε transcription21. IL-4 and IL-21 appear to cooperate for Ig production as indicated by a pan-hypogammaglobulinemia and poorly organized germinal centers in mice lacking both IL-4 and IL-21 signaling20. According to these observations, both IL-4 and IL-21 contribute to Ig production, Ig isotype switch, and the differentiation of B cells into plasma cells. Interestingly, cells that express only IL-4 are more efficient at promoting IgG1 class switch and plasma cell differentiation, whereas cells that secrete only IL-21 are more efficient at promoting somatic hypermutation and affinity maturation in B cells, and cells that express both IL-4 and IL-21 can serve both functions22. This illustrates the important but complex manner in which IL-4 and IL-21 functionally interact in B-cell biology.
IL-4 signaling
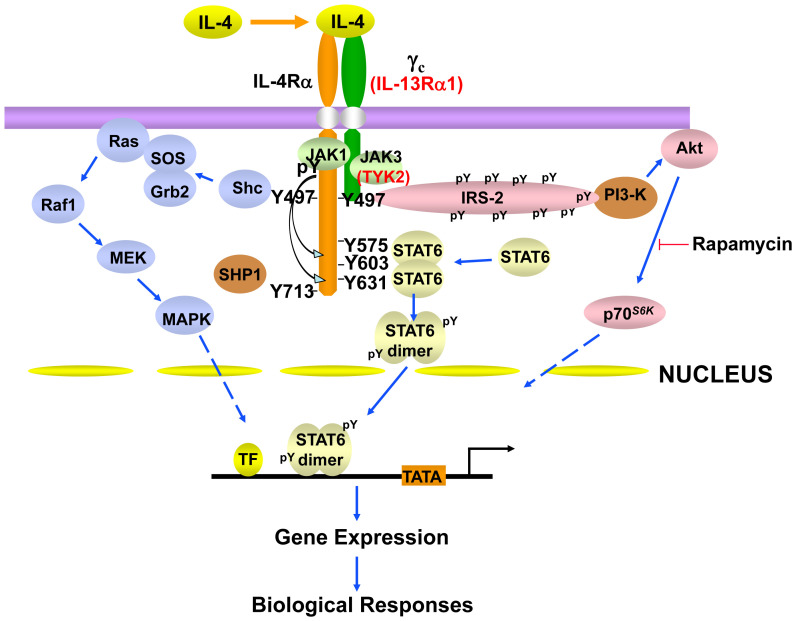
Like the other γc family cytokines, IL-4 activates multiple signaling pathways (Figure 3). IL-4 activates JAK1 and JAK3 via the type I IL-4 receptor; however, IL-4 activates JAK1 and either JAK2 or TYK2 (depending on the cell type) via type II IL-4 receptors. Although there are subtle signaling differences between the type I and type II receptors23, IL-4 and IL-13 are both distinctive in their potent activation of STAT6, which docks on key phosphotyrosines on IL-4Rα (Figure 3)24,25. STAT6 is so vital for the actions of IL-4 that Stat6-deficient mice phenocopy many of the defects observed with Il4- or Il4ra-deficient mice (e.g., related to Th2 differentiation and Ig class switch). However, in addition to JAK-STAT activation, like insulin, IL-4 can induce tyrosine phosphorylation of an IRS1-like molecule in hematopoietic cells, denoted IRS226,27. Studies in Irs2-deficient mice indicate that IL-4-activated IRS2 mediates negative regulatory feedback for the PI3K pathway in macrophages by targeting the phosphorylation of IRS1.
Figure 3. Schematic of some of the major signaling pathways and molecules involved in interleukin 4 (IL-4) signaling.
IL-4 signaling via the type I IL-4 receptor, consisting of IL-4Rα and γc, activates JAK1 and JAK3, whereas JAK1 and TYK2 are the tyrosine kinases involved in IL-4 signaling via the type II IL-4 receptor, which consists of IL-4Rα and IL-13Rα1. Type II IL-4 receptor–specific molecules are shown in red font. IRS2 and Shc interact with phosphorylated Y497, whereas Y575, Y603, and Y631, when phosphorylated, are docking sites for the SH2 domain of STAT6. Collectively, these signaling molecules mediate IL-4 signaling as well. IL-13 signals via the type II IL-4 receptor and activates JAK1 and either JAK2 or TYK2 but not JAK3. Nevertheless, the major signaling pathways are the same as what is shown given that IL-4Rα is the dominant molecule for the docking of signaling proteins. All of the positions of tyrosines indicated in the figure are derived from human IL-4Rα. STAT3 can also be recruited to the IL-13Rα1 in the type II receptor in human macrophages.
Recently, there has been considerable interest in IL4I1, which is encoded by an early IL-4-inducible gene in B cells28 that belongs to the L-amino-acid oxidase family and catalyzes the oxidation of amino acids, including L-phenylanine29. It inhibits T-cell proliferation by releasing its enzymatic catabolite, hydrogen peroxide, which results in lower expression of T-cell receptor zeta (TCRζ). However, it also contributes to macrophage programming and is a regulator of M2 macrophage polarization that inhibits T-cell activation via L-tryptophan and L-arginine depletion as well as increased IL-10 production30. Moreover, it activates the aryl hydrocarbon receptor and promotes tumor progression31. IL4I1 also was reported to be a prognostic biomarker affecting the local T-cell response in human cutaneous melanoma32 and to be capable of promoting central nervous system remyelination by modulating T cell–driven inflammation33.
Effects of IL-4 on T-cell differentiation
IL-4 is one of the key signature cytokines of Th2 cells3,34–37. Interestingly, IL-4 can also drive Th2 cell differentiation, which constitutes a powerful positive feedback mechanism during T-cell differentiation38. IL-4 may directly act on naïve CD4 T cells during T-cell activation to induce Th2 cell differentiation. STAT6 activation by IL-4 is necessary and sufficient during this process39. The initial source of IL-4 could be basophils, NKT cells, or naïve CD4 T cells themselves. Low dose of antigen stimulation may result in IL-2-dependent STAT5-driven early IL-4 production, which in turn further promotes Th2 cell differentiation40. Both IL-2- and IL-4-mediated signaling are also capable of inducing the expression of IL-4Rα, which can further enhance cellular responsiveness to IL-439,41,42. IL-4/STAT6 signaling in activated CD4 T cells induces the expression of the Th2 master transcription factor GATA3, which is critical for Th2 cell function and maintenance43,44. GATA3 directly activates Il5 transcription45; it also plays an important role in chromatin remodeling at the Il4/Il13 cytokine locus during Th2 cell differentiation46–48. In addition, the activation of STAT5 proteins (activated by IL-2 and potentially by other cytokines) is essential for initiating and maintaining Th2 differentiation44,49,50. IL-2 induces IL-4Rα expression early in Th2 differentiation, facilitating responsiveness of cells to IL-4, which promotes and sustains GATA3 expression. The induction of IL-4Rα by IL-2 is attributed to the binding of STAT5 proteins to the GAS motifs in the first intron of the Il4ra gene, and constitutive expression of Il4ra in Il2−/− CD4+ T cells restores Th2 differentiation in these cells51. Many Th2-specific genes, including those encoding IL-5, IL-13, and T1/ST2 (IL-33 receptor α chain), are GATA3 direct targets46; the clustering of some of these genes may help to explain their coordinated induction during Th2 differentiation. IL-4 can also expand GATA3-expressing Th2 cells through STAT6-mediated upregulation of transcription factor Gfi-152. IL-4 not only can drive Th2 cell polarization from naïve CD4 T cells but also induces a Th2 cell phenotype in already-differentiated Th cells, including Th17 cells53. Genome-wide CRISPR screens have also been performed and have revealed crosstalk between differentiation and activation in the Th2 differentiation process, underscoring the roles of factors, including Pparg and Bhlhe40, in controlling Th2 cell differentiation54.
IL-4 may also indirectly promote Th2 cell differentiation. It has been reported that IL-4 may induce TSLP55, which is a critical mediator of Th2 responses in vivo through its actions on dendritic cells (DCs) or T cells56–59. IL-4 and IL-13 can also act directly on intestinal epithelium to induce tuft cells to produce IL-2560, a potent activator of ILC2s, which are the counterparts of Th2 cells within the innate immune system8,61–63. ILC2s can mediate type 2 inflammation through their production of IL-5 and IL-13 even in the absence of Th2 cells, but under certain circumstances, ILC2s may also promote the differentiation of CD4+ T cells toward the Th2 cell fate64. Furthermore, ILC2s and Th2 cells may directly interact with each other through MHCII–TCR interaction65. Therefore, ILC2s and Th2 cells may exhibit crosstalk and collaborate in type 2 immune responses in vivo via multiple mechanisms8,61,66–68.
Although IL-4 seems to be required for Th2 cell differentiation in vitro, both IL-4-dependent and IL-4-independent Th2 cell differentiation have been reported in vivo3. Although GATA3 is absolutely required for Th2 cell differentiation both in vitro and in vivo, Th2 cell differentiation may occur even in CD4 T cells expressing low levels of GATA3 if these cells also receive a strong STAT5 signal44,50. Multiple cytokines, including IL-2, IL-7, IL-9, IL-15, and TSLP, can activate STAT569,70, and TSLP has been shown to program a pathogenic Th2 cell state71, which may provide a basis for IL-4-independent but GATA3-dependent Th2 cell differentiation in vivo. Indeed, our unpublished data indicate that high amounts of IL-7 or TSLP are capable of inducing Th2 cell differentiation to a certain degree in the absence of IL-4 signaling in vitro. This finding may partially explain the difference between in vitro and in vivo differentiation. However, it is possible that in vivo cell–cell interactions through co-stimulatory molecules, such as OX-4072,73 or Notch signaling74, are involved in IL-4-independent Th2 cell differentiation.
A recent study further assessed the requirement of IL-4 signaling for the differentiation of Th2 cell subsets in vivo75. Interestingly, in draining lymph nodes, while the development of IL-4-producing T follicular helper (Tfh) cells is not affected, non-Tfh IL-4-producing cells (Th2 effector cells) are reduced in the absence of IL-4 signaling. Furthermore, in dermal and lung tissues, whereas the generation of IL-13-only-producing Th2 cells is IL-4-independent, the generation of IL-4-producing Th2 cells (with or without IL-13 co-expression) depends on IL-4 signaling. Interestingly, TSLP drives the differentiation of IL-13-expressing Th2 cells76. These results indicate that IL-4 and TSLP can influence the differentiation or the expansion (or both) of distinct Th2 cell subsets. IL-4 can act in an autocrine fashion related to cell proliferation and survival, which may explain the preferential expansion of IL-4-producing Th2 cells as compared with other Th2 cells in vivo.
Together with transforming growth factor beta (TGFβ), IL-4 through activating STAT6 has been reported to induce IL-9-producing (Th9) cells from either naïve CD4 T cells or from Th2 cells77–79. Not only can IL-4-producing Th2 cells convert into IL-9-producing Th9 cells, IL-9-producing cells can—according to our unpublished data—also become IL-4-producing cells in the absence of TGFβ. Therefore, it is possible that a transcription factor whose expression is regulated by the balance between TGFβ and IL-4 serves as a switch for determining conversion from Th2 to Th9 cells or vice versa. Interestingly, together with IL-1β, IL-4 can induce the differentiation of anti-tumor Th9 cells in the absence of TGFβ signaling80, and these cells are phenotypically distinct from the Th9 cells induced by the combination of IL-4 and TGFβ.
IL-4 on ILC2s and other ILCs
Like Th2 cells, ILC2s can produce IL-5 and IL-13; although ILC2s also produce IL-4 under certain circumstances81–84, they produce much less IL-4 than Th2 cells85. However, the ability of ILC2s to produce IL-4 in vivo and the physiological importance of such production require further investigation. Nevertheless, ILC2s can be identified by an Il4 reporter construct in mice in steady state and after IL-25 treatment, indicating that the Il4 locus has an open chromatin configuration in these cells86. The reason why ILC2s produce much less IL-4 than Th2 cells do is unknown, but it seems likely that the chromatin structure at the Th2 cytokine loci containing Il4/Il13/Il5 is quite distinct87. Interestingly, lung Th2 cells produce more IL-13, but draining lymph node Th2 cells produce more IL-488,89; thus, it is also possible that preferential expression of IL-13 over IL-4 or vice versa is determined in a tissue-specific fashion90. GATA3 expression levels vary between ILC2s and Th2 cells in different locations. Thus, it is conceivable that the level of expression of GATA3, by regulating the formation of distinct chromatin structures at the Th2 cytokine loci, determines expression of IL-4 versus IL-13. Although many studies have demonstrated the physiological importance of IL-13 production by ILC2s, fewer have studied the importance of IL-4 production by these cells. Interestingly, however, it was reported that IL-4 produced by ILC2s drives Th2 cell differentiation84 and that IL-4 produced by ILC2s in IL-4Rα Y709F mutant mice (Y709 in mice corresponds to Y713 in humans) may promote food allergy by blocking the generation of allergen-specific regulatory T cells91.
ILC2s can be divided into IL-33-responsive nILC2s (natural ILC2s) and IL-25-responsive iILC2s (inflammatory ILC2s)92,93. iILC2s are located mainly in the gut but can migrate to the lung tissue after Nippostrongylus brasiliensis infection94 or IL-25 treatment95 and produce both IL-4 and IL-1394. Interestingly, deficiency of BATF, a Fos family member that is a component of AP1 complexes96 and that helps to form AP1-IRF4 composite elements97–100, selectively affects iILC2s but not nILC2s in the lung after infection94. iILC2s may turn into nILC2-like cells or ILC3-like cells92. c-Kit-expressing ILC2s in humans may resemble iILC2s in mice, and these cells can be converted into RORγt-expressing cells by IL-1 and IL-23 stimulation; this conversion is promoted by TGFβ but inhibited by IL-4101. By contrast, during ILC2 development, TGFβ was recently reported to induce T1/ST2 expression and thus to promote ILC2 development from its progenitors102. IL-1 together with IL-12 may convert ILC2s into interferon gamma (IFNγ)-producing ILCs, but IL-4 can inhibit this effect103–105. In addition to an important role of IL-4 in maintaining ILC2 identity, IL-4 or IL-13 (or both) made by Th2 cells may act directly on ILC2s to induce local expansion of ILC2s during type 2 immune responses106. Furthermore, basophil-derived IL-4 controls ILC2 actions, including the secretion of IL-13 during protease allergen–induced airway inflammation107.
IL-4 and macrophages
IL-4 is known to regulate the phenotypes of macrophages108–116. Since macrophages are mediators of host defense and play critical roles in a range of physiological processes, including tissue homeostasis and repair, recent studies have focused on understanding macrophage populations that mediate these specific functions in the context of diverse settings such as allergic inflammation, nematode infection, cancer, muscle regeneration, and thermogenesis117–125. Macrophages can adopt a wide spectrum of phenotypes controlled by tissue and environmental signals. A pro-inflammatory phenotype exists at one end of the spectrum (previously termed “M1” for lipopolysaccharide [LPS]/IFNγ-activated macrophages), and the pro-tissue repair phenotype at the other end (previously termed “M2,” “M2a,” or alternatively activated macrophages [AAMs] for the IL-4- or IL-13-activated macrophages). These strongly polarized phenotypes were initially characterized in vitro where addition of specific recombinant cytokines could push phenotypes to extremes in isolation108,109. M1 cells are typically found in abundance in tissues during infections with intracellular pathogens, exposure to components of bacterial cell walls, and high levels of IFNγ. In contrast, M2 cells are found in abundance in tissues high in IL-4 and IL-13, as is seen in parasitic worm infection or allergic inflammation. However, it is now appreciated that the heterogeneity of macrophage phenotypes is more complex in vivo, and traditional markers used for M1 versus M2 delineation, CD86 and CD206, respectively, are not sufficient to resolve the complexity of macrophage phenotypes in vivo113,115,116,126. Furthermore, the limited models of macrophage-specific or -selective deletion make it difficult to unambiguously assign roles for certain types of macrophages in disease processes127–129.
To better phenotype expression of markers by macrophage populations in tissue, Sommerfeld et al.130 recently used single-cell RNA sequencing (scRNA-Seq) to characterize macrophage phenotypes in mouse muscle tissue undergoing injury response to agents that drive either a pro-fibrotic (F) or a pro-regenerative (R) response. They identified two unique macrophage phenotypes in muscle macrophages in each condition (F1, F2 and R1, R2 respectively). Under fibrotic conditions, muscle macrophages (defined as CD45+CD64+F4/80hi) were characterized by gene expression as CD9−CD301b−MHCIIhi (F1) and CD9hiCD301b− MHCII-IL-36g+ (F2). Under regenerative conditions, muscle macrophages were characterized as CD9+CD301b+MHCIIhi (R1) and CD9−CD301b+CD206+ (R2). Interestingly, the R2 macrophages were sustained in the tissue for up to 6 weeks after injury and upregulated genes that are associated with classic M2 or M-IL-4 macrophages, including Chil3, Mrc1, Ccl24, and Il4ra. Because IL-4 plays an important role in muscle repair131, it is likely that the R2 macrophages differentiated in response to environmental IL-4 or IL-13 are present in the injured muscle. The R2 gene expression profile overlapped with that of human liver macrophages132, suggesting that M-IL-4 macrophages might participate in tissue repair and regeneration. Further analysis of macrophages found in other tissues undergoing type II inflammatory responses will be needed to determine whether the addition of cell surface markers such as CD301b and IL-4Rαhi to other “M2” marker panels, including CD206 and transglutaminase-2 (TG2)112, will provide consistent discrimination of macrophages activated by IL-4 or IL-13 in vivo. In this regard, Zhou et al.133 described an increase in CD206+CD103+ interstitial lung macrophages in response to endothelial cell–derived R-spondin 3 in vitro and during the repair phase of an LPS-induced lung injury model in vivo. The R-spondin 3-induced increase in anti-inflammatory M2-like macrophage phenotype (expressing Mrc1, Arg1, Retnla, ChiL3, and Il10) was necessary to limit endotoxemia-induced lung damage and death.
An enhanced understanding of the role for IL-4-activated M2 macrophages in tissue repair and host immune response will be needed to fully appreciate and take full advantage of the potential of these cells. For example, IL-4-coated gold nanoparticles were used to enhance recovery of injured skeletal muscle, resulting in a 40% increase in muscle force134. The IL-4 nanoparticles increased the percentage of muscle M2 macrophages twofold, and the beneficial effect was abrogated by monocyte/macrophage depletion.
A number of studies have implicated M2 macrophages as vital contributors to the anti-helminth immune response135–137, to the severity of allergic inflammation119,138,139, and to tissue repair134,140. However, truly understanding their function has been hampered by the lack of robust and specific mechanisms for their deletion. Methods used in the past include transiently poisoning macrophages with clodronate-loaded liposomes or conditional deletion using LysM-Cre deleter mice, but neither approach is specific for M2 cells127–129. However, a recent publication141 describes the generation of mice (RetnlaCre_R26tdTomato) reporting expression of RELMα, a protein induced by IL-4 in macrophages, and mice that support conditional deletion of RELMα-expressing cells by treatment with diphtheria toxin (RetnlaCre_R26iDTR). Using the reporter mice, the authors found that white adipose tissue, peritoneum, and gut were enriched with Relmα-tdTomato+ macrophages at a steady state. In addition, Relmα expression was detected in alveolar epithelial type 2 cells and in subsets of eosinophils and neutrophils. Interestingly, alveolar macrophages were largely negative for Relmα-dtTomato expression, even when stimulated with IL-4 in vitro, although a minor subset produced a low level of Relmα. These results are consistent with a study142 demonstrating that alveolar macrophages were much less responsive to IL-4 than peritoneal macrophages, potentially because of impaired glycolysis in the pulmonary niche. The alveolar macrophages regained responsiveness to IL-4 after their removal from the lungs in a glycolysis-dependent pathway. Surprisingly, the basal expression of Relmα-dtTomato in peritoneal macrophages was not dependent on IL-4/IL-13 or STAT6, in contrast to the low basal expression in alveolar macrophages, which was dependent on IL-4/IL-13/STAT6. Infection with the nematode N. brasiliensis resulted in an increase in lung Relmα-dtTomato+ interstitial macrophages and eosinophils in an IL-4/IL-13- and STAT6-dependent manner.
Using RetnlaCre_R26iDTR mice, the authors showed that deletion of RELMα-expressing cells with diptheria toxin (DT) during a primary infection with N. brasiliensis resulted in lethality but that control mice treated with DT cleared the infection and survived. Furthermore, removal of RELMα-expressing cells by DT during a secondary infection impaired protection, as assessed by larger parasite burdens in lung and small intestine and lower parasite counts in the skin, the site of parasite entry. Similar results were obtained using a strain with a Cre-inducible DTR in CSF1R (CD115)+ cells (RetnlaCre_CD115iDTR) that allowed for specific deletion of RELMα+ macrophages. Use of these RetnlaCre strains, in addition to the Csf1rLsL-DTR143 and CD206DTR144 mice, will enhance the ability to specifically delete M2 macrophages and specific genes they express. This will allow the investigation of their roles in diverse effector functions, including protection from helminthic parasites, eosinophilic inflammatory diseases, tissue repair, fat and glucose metabolism, and tumor growth.
IL-4 and chromatin remodeling in macrophages
Macrophage phenotypes are controlled by epigenetic mechanisms, including histone modifications145–147. Early studies demonstrated that the histone 3 Lys 27 (H3K27) demethylase jumonji-domain containing-3 (JMJD3) is essential for development of M2 cells in vitro and in vivo148,149. IL-4-induced activation of STAT6 resulted in increased abundance of JMJD3, which resulted in augmented expression of Irf4 and other downstream M2 genes by regulating histone modifications148. JMJD3 activity resulted in methylation of H3K27, thus relieving this negative histone mark. It was further reported that IL-4 induced an increase of H3K4 methylation and acetylation of H3 bound to M2 genes, marks of active chromatin. IL-4-induced polarization to the M2 phenotype is influenced by sex differences in humans and mice138,150. It was recently shown that female alveolar macrophages have increased IL-4Rα and estrogen receptor (ER) α expression compared with male counterparts, and greater epigenetically poised chromatin at M2 gene promoters, while the promoters were epigenetically silenced in male macrophages151. Furthermore, estradiol (E2) acting through ERα supported enhanced expression of IL-4-induced M2 genes. Exposure of female macrophages to E2 permanently changed the responsiveness of the cells to subsequent Th2 inflammatory stimulus even after macrophages were removed from all exogenous E2 up to 10 days later. These data have important implications for diseases in which E2 and macrophages play a key role, such as allergic asthma and breast cancer, and suggest that sex may influence the outcome of therapeutics targeting macrophages151.
Large-scale “omic” studies have recently shown that opposing macrophage polarization programs induced by IL-4 versus IFNγ show epigenomic and transcriptional cross-regulation152, and each cytokine is able to suppress a subset of genes regulated by the opposing cytokine. Upon IL-4 stimulation of macrophages, greater H3K27 acetylation was observed in genomic regions with high representation of canonical binding sites for STAT6. These regions were mainly near IL-4-inducible genes that were suppressed by IFNγ. Interestingly, a small set of IL-4-regulated genomic regions resistant to IFNγ suppression was enriched for Myc-binding E-box sequences. IL-4 increased the abundance of Myc protein and this was important for the induction of genes resistant to IFNγ suppression, including Ccl2, Ccl17, and Ccl12152. Tang et al. further showed that IL-4-induced changes in chromatin accessibility can differ in different types of macrophages153. The regions of chromatin altered by IL-4 were enriched for PU.1 binding motifs, notably in intronic regions capable of modulating higher-order structure of DNA (minor groove width and twist). Polymorphisms in C57BL/6 versus BALB/c mice in sequences flanking PU.1 binding motifs that altered DNA shape configurations were shown to contribute to differences in chromatin accessibility in response to IL-4. Moreover, global transcriptional profiling showed that macrophages from the different strains responded differently to IL-4. For example, IL-4-stimulated macrophages from BALB/c mice expressed lower levels of cell cycle–related genes, PDL-2, and MHC class II than macrophages from C57BL/6 mice. These results support the model that genetic polymorphisms that cause alterations in local DNA structure can influence changes in chromatin accessibility stimulated by IL-4. Whether this is directly correlated with differences in their transcriptional responses to IL-4 remains to be determined153. Furthermore, it has not yet been determined whether modulating epigenetic pathways will be useful in controlling macrophage polarization in disease states154. Indeed, in addition to positive effects, STAT6 has been shown to repress a set of inflammatory enhancers, thereby limiting the activation of AAMs155. Moreover, IL-4 via STAT6 can promote TET2-dependent demethylation to favor DC differentiation156; conversely, DCs can act to reinforce Th2 cytokine production in allergic disease157.
IL-4 and neutrophil function
Neutrophils are normally considered the first line of defense against bacterial infections and are the first immune cell type to enter a site of inflammation. Neutrophilic infiltration is most associated with Th1 and Th17 immune responses, but neutrophils also have been reported to play important roles in anti-helminth immunity158–160. Priming of macrophages to become M2 cells and engage in the killing and clearance of helminthic parasites was dependent on the presence of neutrophils159. In addition, both mouse and human neutrophils can kill helminth larvae in vitro with the cooperation of macrophages158, and neutrophils and macrophages have been found clustered around parasites in vivo160, suggesting that neutrophils play a cooperative role in type II immune responses. On the other hand, recent reports demonstrate that IL-4 signaling suppresses neutrophil function. Woytschak et al. reported that IL-4 treatment of neutrophils inhibited their migration toward CXCL2 and reduced expression of CXCR2161. Furthermore, treatment of mice infected with the bacteria led to reduced neutrophil counts, reduced neutrophil migration, increased bacterial burden, and decreased survival. Neutrophils isolated from IL-4-treated mice had reduced expression of CXCR2 and elevated expression of CXCR4. Similarly, Impellizzieri et al. showed that treatment of human neutrophils with IL-4 inhibited their migration to CXCL2 and reduced the formation of extracellular neutrophil extracellular traps162. Thus, it has been hypothesized that neutrophils assist macrophages early in the course of a nematode infection, but as Th2 cells arrive and produce high levels of IL-4, neutrophil function is downregulated to limit neutrophil-mediated tissue damage163. Future studies will be needed to determine whether these pathways can be targeted in various infectious or inflammatory diseases.
Therapeutic manipulation of the actions of IL-4
Agents that either increase or inhibit IL-4 activity have been generated. Although it is less clear from a therapeutic perspective that augmenting IL-4 would be beneficial, engineered IL-4 “superkines” have been generated. A type I IL-4 receptor–selective IL-4 superkine exhibits marked enhanced binding for γc and exhibits 3- to 10-fold enhanced potency for STAT6 activation, whereas an IL-4 variant with enhanced affinity for IL-13Rα1 more potently drives the differentiation of monocyte-derived DCs164.
One of the most exciting molecules from a therapeutic perspective is a fully humanized IgG4 monoclonal antibody, dupilumab, to IL-4Rα that binds to an epitope on the IL-4Rα chain required for receptor dimer formation and thereby inhibits the actions of both IL-4 and IL-13. Dupilumab was approved by the US Food and Drug Administration in 2017 for the treatment of moderate to severe atopic dermatitis and subsequently for add-on treatment of moderate to severe asthma with eosinophilic phenotype in patients age 12 or older165,166, and there was an 87% decrease in asthma exacerbation in one study167. Another monoclonal antibody, lebrikizumab, which blocks the actions of IL-13 but not IL-4, has been shown to be effective in treating asthma168. Pitrakinra is a variant of human IL-4 with mutations in helix D that are critical for dimerization of IL-4Rα formation with γc or IL-13Rα1 and thus blocks signaling by IL-4 or IL-13. Pitrakinra has shown some efficacy in the treatment of asthma in a subset of patients169. Tralokinumab is another IL-13-based inhibitor. Thus, monoclonal antibodies, engineered cytokines, and potentially small molecules are ways to fine-tune the signal strength of IL-4 or IL-13 (or both) and represent next-generation approaches for treating allergic and inflammatory diseases. It will be interesting to assess the potency and relative utility of agents that selectively target type I versus type II immunity versus the combination of both type I and type II immunity.
Besides anti-IL-4Rα-based therapies, reagents targeting the Fc region of IgE (omalizumab), the α subunit of IL-5 (reslizumab, mepolizumab—phase 2b studies), domain 1 of IL-5Rα (benralizumab), and TSLP (tezepelumab-phase 2a study) are under active investigation170,171.
Finally, because type I IL-4 receptors contain γc as a critical component and IL-4 activates JAK1 and JAK3 via these receptors, one must be cognizant of the inhibition of IL-4 signaling by JAK inhibitors (Jakinibs) that target either of these JAK kinases. Indeed, a number of Jakinibs are being evaluated for atopic dermatitis, with topical delgocitinib having recently been approved in Japan for the treatment of adults with this disease and other agents under ongoing evaluation172.
Conclusions
In a little under 40 years, IL-4 has expanded from being discovered as a factor with biological actions on B cells to a molecularly defined molecule with diverse actions on a range of cell lineages. Accordingly, manipulating the actions of IL-4 has broad ramifications, including related to allergy, cancer, and even the central nervous system. Besides the enormous new basic science information that has accrued, the therapeutic manipulation of the actions of this cytokine has already shown promise in disease states. Better understanding its actions and signaling mechanisms will continue to provide new information and targets to further manipulate its actions.
The peer reviewers who approve this article are:
Graham Le Gros, Malaghan Institute of Medical Research, Wellington, New Zealand
Booki Min, Department of Microbiology and Immunology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA
Ilkka S Junttila, Faculty of Medicine and Health Technology, Tampere University, Tampere, Finland; Fimlab Laboratories, Tampere, Finland
Funding Statement
ADK is supported by Merit Review Award I01 BX001850 from the US Department of Veterans Affairs Biomedical Research and Development Service and National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) grants R01 AI122631and R01 AI143845 (to ADK). WJL is supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute, NIH. JZ is supported by the Division of Intramural Research, NIAID, NIH.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Howard M, Farrar J, Hilfiker M, et al. : Identification of a T cell-derived b cell growth factor distinct from interleukin 2. J Exp Med. 1982; 155(3): 914–23. 10.1084/jem.155.3.914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isakson PC, Puré E, Vitetta ES, et al. : T cell-derived B cell differentiation factor(s). Effect on the isotype switch of murine B cells. J Exp Med. 1982; 155(3): 734–48. 10.1084/jem.155.3.734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul WE, Zhu J: How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol. 2010; 10(4): 225–35. 10.1038/nri2735 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 4.Paul WE: History of interleukin-4. Cytokine. 2015; 75(1): 3–7. 10.1016/j.cyto.2015.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 5.Wirth JJ, Kierszenbaum F, Zlotnik A: Effects of IL-4 on macrophage functions: Increased uptake and killing of a protozoan parasite (Trypanosoma cruzi). Immunology. 1989; 66(2): 296–301. [PMC free article] [PubMed] [Google Scholar]
- 6.Yoshimoto T: The Hunt for the Source of Primary Interleukin-4: How We Discovered That Natural Killer T Cells and Basophils Determine T Helper Type 2 Cell Differentiation In Vivo. Front Immunol. 2018; 9: 716. 10.3389/fimmu.2018.00716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Junttila IS: Tuning the Cytokine Responses: An Update on Interleukin (IL)-4 and IL-13 Receptor Complexes. Front Immunol. 2018; 9: 888. 10.3389/fimmu.2018.00888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu J: T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine. 2015; 75(1): 14–24. 10.1016/j.cyto.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YJ, Holzapfel KL, Zhu J, et al. : Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013; 14(11): 1146–54. 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohara J, Paul WE: Receptors for B-cell stimulatory factor-1 expressed on cells of haematopoietic lineage. Nature. 1987; 325(6104): 537–40. 10.1038/325537a0 [DOI] [PubMed] [Google Scholar]
- 11.Kondo M, Takeshita T, Ishii N, et al. : Sharing of the interleukin-2 (IL-2) receptor gamma chain between receptors for IL-2 and IL-4. Science. 1993; 262(5141): 1874–7. 10.1126/science.8266076 [DOI] [PubMed] [Google Scholar]
- 12.Russell SM, Keegan AD, Harada N, et al. : Interleukin-2 receptor gamma chain: A functional component of the interleukin-4 receptor. Science. 1993; 262(5141): 1880–3. 10.1126/science.8266078 [DOI] [PubMed] [Google Scholar]
- 13.Aman MJ, Tayebi N, Obiri NI, et al. : cDNA cloning and characterization of the human interleukin 13 receptor alpha chain. J Biol Chem. 1996; 271(46): 29265–70. 10.1074/jbc.271.46.29265 [DOI] [PubMed] [Google Scholar]
- 14.LaPorte SL, Juo ZS, Vaclavikova J, et al. : Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008; 132(2): 259–72. 10.1016/j.cell.2007.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Lee HH, Bell JJ, et al. : IL-4 Utilizes an Alternative Receptor to Drive Apoptosis of Th1 Cells and Skews Neonatal Immunity toward Th2. Immunity. 2004; 20(4): 429–40. 10.1016/s1074-7613(04)00072-x [DOI] [PubMed] [Google Scholar]
- 16.Kühn R, Rajewsky K, Müller W: Generation and analysis of interleukin-4 deficient mice. Science. 1991; 254(5032): 707–10. 10.1126/science.1948049 [DOI] [PubMed] [Google Scholar]
- 17.Noben-Trauth N, Shultz LD, Brombacher F, et al. : An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc Natl Acad Sci U S A. 1997; 94(20): 10838–43. 10.1073/pnas.94.20.10838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avery DT, Bryant VL, Ma CS, et al. : IL-21-induced isotype switching to IgG and IgA by human naive B cells is differentially regulated by IL-4. J Immunol. 2008; 181(3): 1767–79. 10.4049/jimmunol.181.3.1767 [DOI] [PubMed] [Google Scholar]
- 19.Pène J, Gauchat JF, Lécart S, et al. : Cutting edge: IL-21 is a switch factor for the production of IgG1 and IgG3 by human B cells. J Immunol. 2004; 172(9): 5154–7. 10.4049/jimmunol.172.9.5154 [DOI] [PubMed] [Google Scholar]
- 20.Ozaki K, Spolski R, Feng CG, et al. : A critical role for IL-21 in regulating immunoglobulin production. Science. 2002; 298(5598): 1630–4. 10.1126/science.1077002 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 21.Suto A, Nakajima H, Hirose K, et al. : Interleukin 21 prevents antigen-induced IgE production by inhibiting germ line C(epsilon) transcription of IL-4-stimulated B cells. Blood. 2002; 100(13): 4565–73. 10.1182/blood-2002-04-1115 [DOI] [PubMed] [Google Scholar]
- 22.Weinstein JS, Herman EI, Lainez B, et al. : TFH cells progressively differentiate to regulate the germinal center response. Nat Immunol. 2016; 17(10): 1197–205. 10.1038/ni.3554 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 23.Heller NM, et al. : Chapter 3: The Type I and Type II receptor complexes for IL-4 and IL-13 differentially regulate allergic lung inflammation. Allergic Diseases. Highlights in the clinic, mechanisms and treatments. Celso Pereira, Ed. Tech, Rijeca, Croatia.: 2012; 43–82. 10.5772/25650 [DOI] [Google Scholar]
- 24.Hou J, Schindler U, Henzel WJ, et al. : An interleukin-4-induced transcription factor: IL-4 Stat. Science. 1994; 265(5179): 1701–6. 10.1126/science.8085155 [DOI] [PubMed] [Google Scholar]
- 25.Nelms K, Keegan AD, Zamorano J, et al. : The IL-4 receptor: Signaling mechanisms and biologic functions. Annu Rev Immunol. 1999; 17: 701–38. 10.1146/annurev.immunol.17.1.701 [DOI] [PubMed] [Google Scholar]
- 26.Keegan AD, Zamorano J, Keselman A, et al. : IL-4 and IL-13 Receptor Signaling From 4PS to Insulin Receptor Substrate 2: There and Back Again, a Historical View. Front Immunol. 2018; 9: 1037. 10.3389/fimmu.2018.01037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelms K, Huang H, Ryan J, et al. : Interleukin-4 receptor signalling mechanisms and their biological significance. Adv Exp Med Biol. 1998; 452: 37–43. 10.1007/978-1-4615-5355-7_5 [DOI] [PubMed] [Google Scholar]
- 28.Chu CC, Paul WE: Fig1, an interleukin 4-induced mouse B cell gene isolated by cDNA representational difference analysis. Proc Natl Acad Sci U S A. 1997; 94(6): 2507–12. 10.1073/pnas.94.6.2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boulland ML, Marquet J, Molinier-Frenkel V, et al. : Human IL4I1 is a secreted L-phenylalanine oxidase expressed by mature dendritic cells that inhibits T-lymphocyte proliferation. Blood. 2007; 110(1): 220–7. 10.1182/blood-2006-07-036210 [DOI] [PubMed] [Google Scholar]
- 30.Yue Y, Huang W, Liang J, et al. : IL4I1 Is a Novel Regulator of M2 Macrophage Polarization That Can Inhibit T Cell Activation via L-Tryptophan and Arginine Depletion and IL-10 Production. PLoS One. 2015; 10(11): e0142979. 10.1371/journal.pone.0142979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadik A, Somarribas Patterson LF, Öztürk S, et al. : IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell. 2020; 182(5): 1252–1270.e34. 10.1016/j.cell.2020.07.038 [DOI] [PubMed] [Google Scholar]
- 32.Ramspott JP, Bekkat F, Bod L, et al. : Emerging Role of IL-4-Induced Gene 1 as a Prognostic Biomarker Affecting the Local T-Cell Response in Human Cutaneous Melanoma. J Invest Dermatol. 2018; 138(12): 2625–34. 10.1016/j.jid.2018.06.178 [DOI] [PubMed] [Google Scholar]
- 33.Psachoulia K, Chamberlain KA, Heo D, et al. : IL4I1 augments CNS remyelination and axonal protection by modulating T cell driven inflammation. Brain. 2016; 139(Pt 12): 3121–36. 10.1093/brain/aww254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu J, Yamane H, Paul WE: Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010; 28: 445–89. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosmann TR, Coffman RL: TH1 and TH2 cells: Different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989; 7: 145–73. 10.1146/annurev.iy.07.040189.001045 [DOI] [PubMed] [Google Scholar]
- 36.Walker JA, McKenzie ANJ: TH2 cell development and function. Nat Rev Immunol. 2018; 18(2): 121–133. 10.1038/nri.2017.118 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 37.Ruterbusch M, Pruner KB, Shehata L, et al. : In Vivo CD4+ T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annu Rev Immunol. 2020; 38: 705–25. 10.1146/annurev-immunol-103019-085803 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 38.Le Gros G, Ben-Sasson SZ, Seder R, et al. : Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J Exp Med. 1990; 172(3): 921–9. 10.1084/jem.172.3.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J, Guo L, Watson CJ, et al. : Stat6 is necessary and sufficient for IL-4's role in Th2 differentiation and cell expansion. J Immunol. 2001; 166(12): 7276–81. 10.4049/jimmunol.166.12.7276 [DOI] [PubMed] [Google Scholar]
- 40.Yamane H, Zhu J, Paul WE: Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med. 2005; 202(6): 793–804. 10.1084/jem.20051304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao W, Lin JX, Wang L, et al. : Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011; 12(6): 551–9. 10.1038/ni.2030 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 42.Zhu J, Huang H, Guo L, et al. : Transient inhibition of interleukin 4 signaling by T cell receptor ligation. J Exp Med. 2000; 192(8): 1125–34. 10.1084/jem.192.8.1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng W, Flavell RA: The Transcription Factor GATA-3 Is Necessary and Sufficient for Th2 Cytokine Gene Expression in CD4 T Cells. Cell. 1997; 89(4): 587–96. 10.1016/s0092-8674(00)80240-8 [DOI] [PubMed] [Google Scholar]
- 44.Zhu J, Min B, Hu-Li J, et al. : Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004; 5(11): 1157–65. 10.1038/ni1128 [DOI] [PubMed] [Google Scholar]
- 45.Siegel MD, Zhang DH, Ray P, et al. : Activation of the interleukin-5 promoter by cAMP in murine EL-4 cells requires the GATA-3 and CLE0 elements. J Biol Chem. 1995; 270(41): 24548–55. 10.1074/jbc.270.41.24548 [DOI] [PubMed] [Google Scholar]
- 46.Wei G, Abraham BJ, Yagi R, et al. : Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011; 35(2): 299–311. 10.1016/j.immuni.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 47.Tanaka S, Motomura Y, Suzuki Y, et al. : The enhancer HS2 critically regulates GATA-3-mediated Il4 transcription in T(H)2 cells. Nat Immunol. 2011; 12(1): 77–85. 10.1038/ni.1966 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 48.Lee HJ, Takemoto N, Kurata H, et al. : GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000; 192(1): 105–15. 10.1084/jem.192.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pai SY, Truitt ML, Ho IC: GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U S A. 2004; 101(7): 1993–8. 10.1073/pnas.0308697100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, Cote-Sierra J, Guo L, et al. : Stat5 Activation Plays a Critical Role in Th2 Differentiation. Immunity. 2003; 19(5): 739–48. 10.1016/s1074-7613(03)00292-9 [DOI] [PubMed] [Google Scholar]
- 51.Liao W, Schones DE, Oh J, et al. : Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol. 2008; 9(11): 1288–96. 10.1038/ni.1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Guo L, Min B, et al. : Growth Factor Independent-1 Induced by IL-4 Regulates Th2 Cell Proliferation. Immunity. 2002; 16(5): 733–44. 10.1016/s1074-7613(02)00317-5 [DOI] [PubMed] [Google Scholar]
- 53.Tortola L, Jacobs A, Pohlmeier L, et al. : High-Dimensional T Helper Cell Profiling Reveals a Broad Diversity of Stably Committed Effector States and Uncovers Interlineage Relationships. Immunity. 2020; 53(3): 597–613.e6. 10.1016/j.immuni.2020.07.001 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 54.Henriksson J, Chen X, Gomes T, et al. : Genome-wide CRISPR Screens in T Helper Cells Reveal Pervasive Crosstalk between Activation and Differentiation. Cell. 2019; 176(4): 882–896.e18. 10.1016/j.cell.2018.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 55.Dewas C, Chen X, Honda T, et al. : TSLP expression: Analysis with a ZsGreen TSLP reporter mouse. J Immunol. 2015; 194(3): 1372–80. 10.4049/jimmunol.1400519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu YJ, Soumelis V, Watanabe N, et al. : TSLP: An epithelial cell cytokine that regulates T cell differentiation by conditioning dendritic cell maturation. Annu Rev Immunol. 2007; 25: 193–219. 10.1146/annurev.immunol.25.022106.141718 [DOI] [PubMed] [Google Scholar]
- 57.Al-Shami A, Spolski R, Kelly J, et al. : A role for TSLP in the development of inflammation in an asthma model. J Exp Med. 2005; 202(6): 829–39. 10.1084/jem.20050199 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 58.He R, Oyoshi MK, Garibyan L, et al. : TSLP acts on infiltrating effector T cells to drive allergic skin inflammation. Proc Natl Acad Sci U S A. 2008; 105(33): 11875–80. 10.1073/pnas.0801532105 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 59.Roan F, Obata-Ninomiya K, Ziegler SF: Epithelial cell-derived cytokines: More than just signaling the alarm. J Clin Invest. 2019; 129(4): 1441–51. 10.1172/JCI124606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.von Moltke J, Ji M, Liang HE, et al. : Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016; 529(7585): 221–5. 10.1038/nature16161 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 61.Kabata H, Moro K, Koyasu S: The group 2 innate lymphoid cell (ILC2) regulatory network and its underlying mechanisms. Immunol Rev. 2018; 286(1): 37–52. 10.1111/imr.12706 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 62.Klose CS, Artis D: Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol. 2016; 17(7): 765–74. 10.1038/ni.3489 [DOI] [PubMed] [Google Scholar]
- 63.Vannella KM, Ramalingam TR, Borthwick LA, et al. : Combinatorial targeting of TSLP, IL-25, and IL-33 in type 2 cytokine-driven inflammation and fibrosis. Sci Transl Med. 2016; 8(337): 337ra65. 10.1126/scitranslmed.aaf1938 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 64.Halim TY, Steer CA, Mathä L, et al. : Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014; 40(3): 425–35. 10.1016/j.immuni.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 65.Oliphant CJ, Hwang YY, Walker JA, et al. : MHCII-mediated dialog between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity. 2014; 41(2): 283–95. 10.1016/j.immuni.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 66.Gurram RK, Zhu J: Orchestration between ILC2s and Th2 cells in shaping type 2 immune responses. Cell Mol Immunol. 2019; 16(3): 225–35. 10.1038/s41423-019-0210-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kubo M: Innate and adaptive type 2 immunity in lung allergic inflammation. Immunol Rev. 2017; 278(1): 162–72. 10.1111/imr.12557 [DOI] [PubMed] [Google Scholar]
- 68.Martinez-Gonzalez I, Steer CA, Takei F: Lung ILC2s link innate and adaptive responses in allergic inflammation. Trends Immunol. 2015; 36(3): 189–95. 10.1016/j.it.2015.01.005 [DOI] [PubMed] [Google Scholar]
- 69.Rochman Y, Kashyap M, Robinson GW, et al. : Thymic stromal lymphopoietin-mediated STAT5 phosphorylation via kinases JAK1 and JAK2 reveals a key difference from IL-7-induced signaling. Proc Natl Acad Sci U S A. 2010; 107(45): 19455–60. 10.1073/pnas.1008271107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leonard WJ, Lin JX, O'Shea JJ: The γ c Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity. 2019; 50(4): 832–50. 10.1016/j.immuni.2019.03.028 [DOI] [PubMed] [Google Scholar]
- 71.Rochman Y, Dienger-Stambaugh K, Richgels PK, et al. : TSLP signaling in CD4 + T cells programs a pathogenic T helper 2 cell state. Sci Signal. 2018; 11(521): eaam8858. 10.1126/scisignal.aam8858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ito T, Wang YH, Duramad O, et al. : TSLP-activated dendritic cells induce an inflammatory T helper type 2 cell response through OX40 ligand. J Exp Med. 2005; 202(9): 1213–23. 10.1084/jem.20051135 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 73.Akiba H, Miyahira Y, Atsuta M, et al. : Critical contribution of OX40 ligand to T helper cell type 2 differentiation in experimental leishmaniasis. J Exp Med. 2000; 191(2): 375–80. 10.1084/jem.191.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amsen D, Blander JM, Lee GR, et al. : Instruction of Distinct CD4 T Helper Cell Fates by Different Notch Ligands on Antigen-Presenting Cells. Cell. 2004; 117(4): 515–26. 10.1016/s0092-8674(04)00451-9 [DOI] [PubMed] [Google Scholar]
- 75.Prout MS, Kyle RL, Ronchese F, et al. : IL-4 Is a Key Requirement for IL-4- and IL-4/IL-13-Expressing CD4 Th2 Subsets in Lung and Skin. Front Immunol. 2018; 9: 1211. 10.3389/fimmu.2018.01211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ochiai S, Jagot F, Kyle RL, et al. : Thymic stromal lymphopoietin drives the development of IL-13 + Th2 cells. Proc Natl Acad Sci U S A. 2018; 115(5): 1033–8. 10.1073/pnas.1714348115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dardalhon V, Awasthi A, Kwon H, et al. : IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat Immunol. 2008; 9(12): 1347–55. 10.1038/ni.1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Veldhoen M, Uyttenhove C, van Snick J, et al. : Transforming growth factor-beta 'reprograms' the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008; 9(12): 1341–6. 10.1038/ni.1659 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 79.Goswami R, Jabeen R, Yagi R, et al. : STAT6-dependent regulation of Th9 development. J Immunol. 2012; 188(3): 968–75. 10.4049/jimmunol.1102840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xue G, Jin G, Fang J, et al. : IL-4 together with IL-1β induces antitumor Th9 cell differentiation in the absence of TGF-β signaling. Nat Commun. 2019; 10(1): 1376. 10.1038/s41467-019-09401-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doherty TA, Khorram N, Lund S, et al. : Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J Allergy Clin Immunol. 2013; 132(1): 205–13. 10.1016/j.jaci.2013.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roediger B, Kyle R, Yip KH, et al. : Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat Immunol. 2013; 14(6): 564–73. 10.1038/ni.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 83.Salimi M, Stöger L, Liu W, et al. : Cysteinyl leukotriene E 4 activates human group 2 innate lymphoid cells and enhances the effect of prostaglandin D 2 and epithelial cytokines. J Allergy Clin Immunol. 2017; 140(4): 1090–1100.e11. 10.1016/j.jaci.2016.12.958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pelly VS, Kannan Y, Coomes SM, et al. : IL-4-producing ILC2s are required for the differentiation of T H 2 cells following Heligmosomoides polygyrus infection. Mucosal Immunol. 2016; 9(6): 1407–17. 10.1038/mi.2016.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moro K, Yamada T, Tanabe M, et al. : Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010; 463(7280): 540–4. 10.1038/nature08636 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 86.Fallon PG, Ballantyne SJ, Mangan NE, et al. : Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J Exp Med. 2006; 203(4): 1105–16. 10.1084/jem.20051615 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 87.Shih HY, Sciumè G, Mikami Y, et al. : Developmental Acquisition of Regulomes Underlies Innate Lymphoid Cell Functionality. Cell. 2016; 165(5): 1120–33. 10.1016/j.cell.2016.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 88.Liang HE, Reinhardt RL, Bando JK, et al. : Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol. 2011; 13(1): 58–66. 10.1038/ni.2182 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 89.van Dyken SJ, Nussbaum JC, Lee J, et al. : A tissue checkpoint regulates type 2 immunity. Nat Immunol. 2016; 17(12): 1381–7. 10.1038/ni.3582 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 90.Ricardo-Gonzalez RR, van Dyken SJ, Schneider C, et al. : Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol. 2018; 19(10): 1093–9. 10.1038/s41590-018-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noval Rivas M, Burton OT, Oettgen HC, et al. : IL-4 production by group 2 innate lymphoid cells promotes food allergy by blocking regulatory T-cell function. J Allergy Clin Immunol. 2016; 138(3): 801–811.e9. 10.1016/j.jaci.2016.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang Y, Guo L, Qiu J, et al. : IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential 'inflammatory' type 2 innate lymphoid cells. Nat Immunol. 2015; 16(2): 161–9. 10.1038/ni.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 93.Huang Y, Paul WE: Inflammatory group 2 innate lymphoid cells. Int Immunol. 2016; 28(1): 23–8. 10.1093/intimm/dxv044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miller MM, Patel PS, Bao K, et al. : BATF acts as an essential regulator of IL-25-responsive migratory ILC2 cell fate and function. Sci Immunol. 2020; 5(43): eaay3994. 10.1126/sciimmunol.aay3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang Y, Mao K, Chen X, et al. : S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science. 2018; 359(6371): 114–9. 10.1126/science.aam5809 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 96.Schraml BU, Hildner K, Ise W, et al. : The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009; 460(7253): 405–9. 10.1038/nature08114 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 97.Li P, Spolski R, Liao W, et al. : BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012; 490(7421): 543–6. 10.1038/nature11530 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 98.Tussiwand R, Lee WL, Murphy TL, et al. : Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012; 490(7421): 502–7. 10.1038/nature11531 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 99.Glasmacher E, Agrawal S, Chang AB, et al. : A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012; 338(6109): 975–80. 10.1126/science.1228309 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 100.Ciofani M, Madar A, Galan C, et al. : A validated regulatory network for Th17 cell specification. Cell. 2012; 151(2): 289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bernink JH, Ohne Y, Teunissen MBM, et al. : c-Kit-positive ILC2s exhibit an ILC3-like signature that may contribute to IL-17-mediated pathologies. Nat Immunol. 2019; 20(8): 992–1003. 10.1038/s41590-019-0423-0 [DOI] [PubMed] [Google Scholar]
- 102.Wang L, Tang J, Yang X, et al. : TGF-β induces ST2 and programs ILC2 development. Nat Commun. 2020; 11(1): 35. 10.1038/s41467-019-13734-w [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 103.Bal SM, Bernink JH, Nagasawa M, et al. : IL-1β, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat Immunol. 2016; 17(6): 636–45. 10.1038/ni.3444 [DOI] [PubMed] [Google Scholar]
- 104.Lim AI, Menegatti S, Bustamante J, et al. : IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J Exp Med. 2016; 213(4): 569–83. 10.1084/jem.20151750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ohne Y, Silver JS, Thompson-Snipes L, et al. : IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat Immunol. 2016; 17(6): 646–55. 10.1038/ni.3447 [DOI] [PubMed] [Google Scholar]
- 106.Symowski C, Voehringer D: Th2 cell-derived IL-4/IL-13 promote ILC2 accumulation in the lung by ILC2-intrinsic STAT6 signaling in mice. Eur J Immunol. 2019; 49(9): 1421–32. 10.1002/eji.201948161 [DOI] [PubMed] [Google Scholar]
- 107.Motomura Y, Morita H, Moro K, et al. : Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity. 2014; 40(5): 758–71. 10.1016/j.immuni.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 108.Stein M, Keshav S, Harris N, et al. : Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J Exp Med. 1992; 176(1): 287–92. 10.1084/jem.176.1.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stout RD, Suttles J: Functional plasticity of macrophages: Reversible adaptation to changing microenvironments. J Leukoc Biol. 2004; 76(3): 509–13. 10.1189/jlb.0504272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mosser DM, Edwards JP: Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008; 8(12): 958–69. 10.1038/nri2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Martinez FO, Helming L, Gordon S: Alternative activation of macrophages: An immunologic functional perspective. Annu Rev Immunol. 2009; 27: 451–83. 10.1146/annurev.immunol.021908.132532 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 112.Martinez FO, Helming L, Milde R, et al. : Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: Similarities and differences. Blood. 2013; 121(9): e57–69. 10.1182/blood-2012-06-436212 [DOI] [PubMed] [Google Scholar]
- 113.Murray PJ, Allen JE, Biswas SK, et al. : Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity. 2014; 41(1): 14–20. 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 114.Ferrante CJ, Pinhal-Enfield G, Elson G, et al. : The adenosine-dependent angiogenic switch of macrophages to an M2-like phenotype is independent of interleukin-4 receptor alpha (IL-4Rα) signaling. Inflammation. 2013; 36(4): 921–31. 10.1007/s10753-013-9621-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ginhoux F, Schultze JL, Murray PJ, et al. : New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016; 17(1): 34–40. 10.1038/ni.3324 [DOI] [PubMed] [Google Scholar]
- 116.Twum DYF, Burkard-Mandel L, Abrams SI: The Dr. Jekyll and Mr. Hyde complexity of the macrophage response in disease. J Leukoc Biol. 2017; 102(2): 307–15. 10.1189/jlb.4MR1116-479R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. : Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018; 233(9): 6425–40. 10.1002/jcp.26429 [DOI] [PubMed] [Google Scholar]
- 118.Kelly-Welch AE, Melo MEF, Smith E, et al. : Complex role of the IL-4 receptor alpha in a murine model of airway inflammation: Expression of the IL-4 receptor alpha on nonlymphoid cells of bone marrow origin contributes to severity of inflammation. J Immunol. 2004; 172(7): 4545–55. 10.4049/jimmunol.172.7.4545 [DOI] [PubMed] [Google Scholar]
- 119.Ford AQ, Dasgupta P, Mikhailenko I, et al. : Adoptive transfer of IL-4Rα+ macrophages is sufficient to enhance eosinophilic inflammation in a mouse model of allergic lung inflammation. BMC Immunol. 2012; 13: 6. 10.1186/1471-2172-13-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dasgupta P, Keegan AD: Contribution of alternatively activated macrophages to allergic lung inflammation: A tale of mice and men. J Innate Immun. 2012; 4(5–6): 478–88. 10.1159/000336025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dasgupta P, Dorsey NJ, Li J, et al. : The adaptor protein insulin receptor substrate 2 inhibits alternative macrophage activation and allergic lung inflammation. Sci Signal. 2016; 9(433): ra63. 10.1126/scisignal.aad6724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Keegan AD, Shirey KA, Bagdure D, et al. : Enhanced allergic responsiveness after early childhood infection with respiratory viruses: Are long-lived alternatively activated macrophages the missing link? Pathog Dis. 2016; 74(5): ftw047. 10.1093/femspd/ftw047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Park GY, Christman JW: Hidden in Plain Sight: The Overlooked Role of Pulmonary Macrophages in the Pathogenesis of Asthma. Am J Respir Cell Mol Biol. 2016; 55(4): 465–6. 10.1165/rcmb.2016-0188ED [DOI] [PubMed] [Google Scholar]
- 124.Girodet PO, Nguyen D, Mancini JD, et al. : Alternative Macrophage Activation Is Increased in Asthma. Am J Respir Cell Mol Biol. 2016; 55(4): 467–75. 10.1165/rcmb.2015-0295OC [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 125.Ganeshan K, Chawla A: Metabolic regulation of immune responses. Annu Rev Immunol. 2014; 32: 609–34. 10.1146/annurev-immunol-032713-120236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Martinez FO, Gordon S: The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014; 6: 13. 10.12703/P6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vannella KM, Barron L, Borthwick LA, et al. : Incomplete deletion of IL-4Rα by LysM(Cre) reveals distinct subsets of M2 macrophages controlling inflammation and fibrosis in chronic schistosomiasis . PLoS Pathog. 2014; 10(9): e1004372. 10.1371/journal.ppat.1004372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Abram CL, Roberge GL, Hu Y, et al. : Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J Immunol Methods. 2014; 408: 89–100. 10.1016/j.jim.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 129.McCubbrey AL, Allison KC, Lee-Sherick AB, et al. : Promoter Specificity and Efficacy in Conditional and Inducible Transgenic Targeting of Lung Macrophages. Front Immunol. 2017; 8: 1618. 10.3389/fimmu.2017.01618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sommerfeld SD, Cherry C, Schwab RM, et al. : Interleukin-36γ-producing macrophages drive IL-17-mediated fibrosis. Sci Immunol. 2019; 4(40): eaax4783. 10.1126/sciimmunol.aax4783 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 131.Sadtler K, Estrellas K, Allen BW, et al. : Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells. Science. 2016; 352(6283): 366–70. 10.1126/science.aad9272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.MacParland SA, Liu JC, Ma XZ, et al. : Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018; 9(1): 4383. 10.1038/s41467-018-06318-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou B, Magana L, Hong Z, et al. : The angiocrine Rspondin3 instructs interstitial macrophage transition via metabolic-epigenetic reprogramming and resolves inflammatory injury. Nat Immunol. 2020; 21(11): 1430–43. 10.1038/s41590-020-0764-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 134.Raimondo TM, Mooney DJ: Functional muscle recovery with nanoparticle-directed M2 macrophage polarization in mice. Proc Natl Acad Sci U S A. 2018; 115(42): 10648–53. 10.1073/pnas.1806908115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Anthony RM, Urban JF, Alem F, et al. : Memory T(H)2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat Med. 2006; 12(8): 955–60. 10.1038/nm1451 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 136.Chen F, Liu Z, Wu W, et al. : An essential role for TH2-type responses in limiting acute tissue damage during experimental helminth infection. Nat Med. 2012; 18(2): 260–6. 10.1038/nm.2628 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 137.Voehringer D, van Rooijen N, Locksley RM: Eosinophils develop in distinct stages and are recruited to peripheral sites by alternatively activated macrophages. J Leukoc Biol. 2007; 81(6): 1434–44. 10.1189/jlb.1106686 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 138.Melgert BN, ten Hacken NH, Rutgers B, et al. : More alternative activation of macrophages in lungs of asthmatic patients. J Allergy Clin Immunol. 2011; 127(3): 831–3. 10.1016/j.jaci.2010.10.045 [DOI] [PubMed] [Google Scholar]
- 139.Borthwick LA, Barron L, Hart KM, et al. : Macrophages are critical to the maintenance of IL-13-dependent lung inflammation and fibrosis. Mucosal Immunol. 2016; 9(1): 38–55. 10.1038/mi.2015.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shirey KA, Pletneva LM, Puche AC, et al. : Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R alpha-, TLR4-, and IFN-beta-dependent. Mucosal Immunol. 2010; 3(3): 291–300. 10.1038/mi.2010.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Krljanac B, Schubart C, Naumann R, et al. : RELMα-expressing macrophages protect against fatal lung damage and reduce parasite burden during helminth infection. Sci Immunol. 2019; 4(35): eaau3814. 10.1126/sciimmunol.aau3814 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 142.Svedberg FR, Brown SL, Krauss MZ, et al. : The lung environment controls alveolar macrophage metabolism and responsiveness in type 2 inflammation. Nat Immunol. 2019; 20(5): 571–80. 10.1038/s41590-019-0352-y [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 143.Schreiber HA, Loschko J, Karssemeijer RA, et al. : Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J Exp Med. 2013; 210(10): 2025–39. 10.1084/jem.20130903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kambara K, Ohashi W, Tomita K, et al. : In vivo depletion of CD206+ M2 macrophages exaggerates lung injury in endotoxemic mice. Am J Pathol. 2015; 185(1): 162–71. 10.1016/j.ajpath.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 145.Takeuch O, Akira S: Epigenetic control of macrophage polarization. Eur J Immunol. 2011; 41(9): 2490–3. 10.1002/eji.201141792 [DOI] [PubMed] [Google Scholar]
- 146.Ivashkiv LB: Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013; 34(5): 216–23. 10.1016/j.it.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 147.Kittan NA, Allen RM, Dhaliwal A, et al. : Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS One. 2013; 8(10): e78045. 10.1371/journal.pone.0078045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ishii M, Wen H, Corsa CAS, et al. : Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009; 114(15): 3244–54. 10.1182/blood-2009-04-217620 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 149.Satoh T, Takeuchi O, Vandenbon A, et al. : The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010; 11(10): 936–44. 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 150.Blacquière MJ, Hylkema MN, Postma DS, et al. : Airway inflammation and remodeling in two mouse models of asthma: Comparison of males and females. Int Arch Allergy Immunol. 2010; 153(2): 173–81. 10.1159/000312635 [DOI] [PubMed] [Google Scholar]
- 151.Keselman A, Fang X, White PB, et al. : Estrogen Signaling Contributes to Sex Differences in Macrophage Polarization during Asthma. J Immunol. 2017; 199(5): 1573–83. 10.4049/jimmunol.1601975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Piccolo V, Curina A, Genua M, et al. : Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat Immunol. 2017; 18(5): 530–40. 10.1038/ni.3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tang MS, Miraldi ER, Girgis NM, et al. : Alternative Activation of Macrophages Is Accompanied by Chromatin Remodeling Associated with Lineage-Dependent DNA Shape Features Flanking PU.1 Motifs. J Immunol. 2020; 205(4): 1070–83. 10.4049/jimmunol.2000258 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 154.Zhou D, Yang K, Chen L, et al. : Promising landscape for regulating macrophage polarization: Epigenetic viewpoint. Oncotarget. 2017; 8(34): 57693–706. 10.18632/oncotarget.17027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Czimmerer Z, Daniel B, Horvath A, et al. : The Transcription Factor STAT6 Mediates Direct Repression of Inflammatory Enhancers and Limits Activation of Alternatively Polarized Macrophages. Immunity. 2018; 48(1): 75–90.e6. 10.1016/j.immuni.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vento-Tormo R, Company C, Rodríguez-Ubreva J, et al. : IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. 2016; 17: 4. 10.1186/s13059-015-0863-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhou X, Yu W, Lyu SC, et al. : A positive feedback loop reinforces the allergic immune response in human peanut allergy. J Exp Med. 2021; 218(7): e20201793. 10.1084/jem.20201793 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 158.Bonne-Année S, Kerepesi LA, Hess JA, et al. : Human and mouse macrophages collaborate with neutrophils to kill larval Strongyloides stercoralis. Infect Immun. 2013; 81(9): 3346–55. 10.1128/IAI.00625-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen F, Wu W, Millman A, et al. : Neutrophils prime a long-lived effector macrophage phenotype that mediates accelerated helminth expulsion. Nat Immunol. 2014; 15(10): 938–46. 10.1038/ni.2984 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 160.Al-Qaoud KM, Pearlman E, Hartung T, et al. : A new mechanism for IL-5-dependent helminth control: Neutrophil accumulation and neutrophil-mediated worm encapsulation in murine filariasis are abolished in the absence of IL-5. Int Immunol. 2000; 12(6): 899–908. 10.1093/intimm/12.6.899 [DOI] [PubMed] [Google Scholar]
- 161.Woytschak J, Keller N, Krieg C, et al. : Type 2 Interleukin-4 Receptor Signaling in Neutrophils Antagonizes Their Expansion and Migration during Infection and Inflammation. Immunity. 2016; 45(1): 172–84. 10.1016/j.immuni.2016.06.025 [DOI] [PubMed] [Google Scholar]
- 162.Impellizzieri D, Ridder F, Raeber ME, et al. : IL-4 receptor engagement in human neutrophils impairs their migration and extracellular trap formation. J Allergy Clin Immunol. 2019; 144(1): 267–279.e4. 10.1016/j.jaci.2019.01.042 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 163.Egholm C, Heeb LEM, Impellizzieri D, et al. : The Regulatory Effects of Interleukin-4 Receptor Signaling on Neutrophils in Type 2 Immune Responses. Front Immunol. 2019; 10: 2507. 10.3389/fimmu.2019.02507 [DOI] [PMC free article] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 164.Junttila IS, Creusot RJ, Moraga I, et al. : Redirecting cell-type specific cytokine responses with engineered interleukin-4 superkines. Nat Chem Biol. 2012; 8(12): 990–8. 10.1038/nchembio.1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chang HY, Nadeau KC: IL-4Rα Inhibitor for Atopic Disease. Cell. 2017; 170(2): 222. 10.1016/j.cell.2017.06.046 [DOI] [PubMed] [Google Scholar]
- 166.Shirley M: Dupilumab: First Global Approval. Drugs. 2017; 77(10): 1115–21. 10.1007/s40265-017-0768-3 [DOI] [PubMed] [Google Scholar]
- 167.Wenzel S, Ford L, Pearlman D, et al. : Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med. 2013; 368(26): 2455–66. 10.1056/NEJMoa1304048 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 168.Corren J, Lemanske RF, Hanania NA, et al. : Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011; 365(12): 1088–98. 10.1056/NEJMoa1106469 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 169.Wenzel S, Wilbraham D, Fuller R, et al. : Effect of an interleukin-4 variant on late phase asthmatic response to allergen challenge in asthmatic patients: Results of two phase 2a studies. Lancet. 2007; 370(9596): 1422–31. 10.1016/S0140-6736(07)61600-6 [DOI] [PubMed] [Google Scholar]; Faculty Opinions Recommendation
- 170.Mavissakalian M, Brady S: The Current State of Biologic Therapies for Treatment of Refractory Asthma. Clin Rev Allergy Immunol. 2020; 59(2): 195–207. 10.1007/s12016-020-08776-8 [DOI] [PubMed] [Google Scholar]
- 171.Chun PIF, Lehman H: Current and Future Monoclonal Antibodies in the Treatment of Atopic Dermatitis. Clin Rev Allergy Immunol. 2020; 59(2): 208–19. 10.1007/s12016-020-08802-9 [DOI] [PubMed] [Google Scholar]
- 172.Li H, Zhang Z, Zhang H, Guo H, Yao Z: Update on the Pathogenesis and Therapy of Atopic Dermatitis. Clin Rev Allergy Immunol. 2021. 10.1007/s12016-021-08880-3 [DOI] [PubMed] [Google Scholar]