ABSTRACT
Gene-editing tools such as CRISPR-Cas9 have created unprecedented opportunities for genetic studies in plants and animals. We designed a course-based undergraduate research experience (CURE) to train introductory biology students in the concepts and implementation of gene-editing technology as well as develop their soft skills in data management and scientific communication. We present two versions of the course that can be implemented with twice-weekly meetings over a 5-week period. In the remote-learning version, students performed homology searches, designed guide RNAs (gRNAs) and primers, and learned the principles of molecular cloning. This version is appropriate when access to laboratory equipment or in-person instruction is limited, such as during closures that have occurred in response to the COVID-19 pandemic. In person, students designed gRNAs, cloned CRISPR-Cas9 constructs, and performed genetic transformation of Arabidopsis thaliana. Students learned how to design effective gRNA pairs targeting their assigned gene with an 86% success rate. Final exams tested students’ ability to apply knowledge of an unfamiliar genome database to characterize gene structure and to properly design gRNAs. Average final exam scores of ∼73% and ∼84% for in-person and remote-learning CUREs, respectively, indicated that students met learning outcomes. The highly parallel nature of the CURE makes it possible to target dozens to hundreds of genes, depending on the number of sections. Applying this approach in a sensitized mutant background enables focused reverse genetic screens for genetic suppressors or enhancers. The course can be adapted readily to other organisms or projects that employ gene editing.
KEYWORDS: course-based undergraduate research experience, CURE, remote learning, plant biology, CRISPR-Cas9
INTRODUCTION
Course-based undergraduate research experiences (CUREs) are credit-based classes in which students investigate an unresolved research question rather than carry out predetermined experiments with a well-defined outcome (1). CUREs can have powerful positive impacts on students by providing a more accurate representation of the process of science and introducing them to problem solving required to answer open-ended questions (2, 3). Students that experience CUREs are more likely to remain in science, technology, engineering, and mathematics (STEM) during college, enter STEM graduate programs, and identify as scientists (4, 5). CUREs can increase diversity in STEM by promoting higher levels of retention of traditionally underrepresented students (6). Because CUREs require few or no prerequisites, they provide unique opportunities to students from diverse backgrounds to experience research, serving more students than do traditional undergraduate research experiences (7). Despite the benefits, CUREs are not yet widely implemented for several reasons. CUREs require flexibility and coordination between lab instructors and the researcher(s) providing the research project. Further, CUREs lack the predictability of traditional lab courses because the outcomes of experiments are unknown (8, 9).
Several strategies are recommended to promote a successful CURE. The direct participation of professors and graduate students who are knowledgeable about the research project encourages students to learn the scientific process (2, 10). Using simple laboratory techniques and building redundancy within or between class sections ensures that research goals are met (11). Frequently monitoring student understanding with quizzes, lab notebook entries, and writing assignments highlights when concepts need clarification (12).
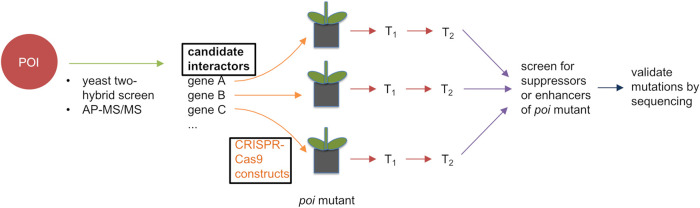
We envisioned a CURE enabling high-throughput mutagenesis of genes using simple gene-editing techniques. Our goals were to discover genes that function in division-plane orientation or in karrikin signaling (13–15). Here, we describe a reverse genetic approach to characterize a set of candidate genes associated with a gene of interest by biochemical screens for protein interactors. Vast collections of defined Arabidopsis thaliana mutants have been generated through insertional mutagenesis (16, 17). Mutant collections are enviable resources for reverse genetic studies but are not well-suited to high-throughput tests for genetic modifiers, which would require extensive crossing and isolation of double or higher-order combinations of mutants. In contrast, clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR-Cas9) can introduce mutations into two or more related genes at once. Biallelic or homozygous mutations are common in the first transformed generation (18). Much effort in CRISPR-Cas9 approaches goes toward identification of useful alleles and deriving homozygous mutant lines. We reasoned that this effort could be reduced by applying a forward genetics strategy where the pooled progeny of CRISPR-Cas9 transgenic plants are screened for phenotypes and then focusing further studies on individuals with phenotypes and their targeted candidate gene(s) (Fig. 1).
FIG 1.
Strategy to identify genetic modifiers through CRISPR-Cas9. Proteins that potentially interact with a protein of interest (POI) are identified through yeast two-hybrid library screens or affinity-purification mass spectrometry. Each candidate interactor gene is assigned to two students. Students identify homologs that may be functionally redundant with a candidate interactor and select two guide RNA (gRNA) sequences to target the candidate and its close homologs, if any. Students use PCR and Golden Gate cloning to insert both gRNAs into a CRISPR-Cas9 construct. Correct constructs are identified after E. coli transformation with colony PCR, plasmid preparation, and sequencing. Constructs are then transformed into the Arabidopsis thaliana poi mutant background. Transformed seed (T1) are selected and selfed to produce T2 seed. Pooled T2 seed from different T1 lines carrying a single construct are phenotyped for either suppression or synthetic enhancement of the poi mutant phenotype. CRISPR-induced mutations are then validated by sequencing of the target gene(s). Black boxes around “candidate interactors” and “CRISPR-Cas9 constructs” indicate CURE contributions specific to this research.
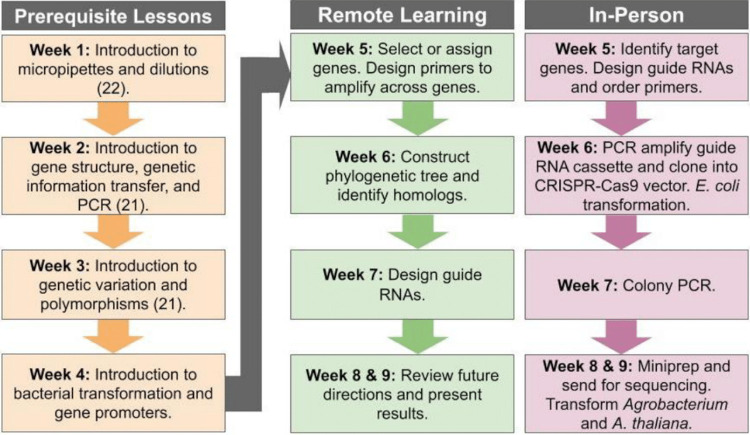
We adopted this approach to evaluate a list of proteins identified as candidate interactors with TANGLED1 (TAN1) or KARRIKIN UPREGULATED F-BOX1 (KUF1) through yeast two-hybrid library screens, affinity-purification mass spectrometry, or predicted function (19, 20). In the in-person CURE, students were asked to use CRISPR-Cas9 to target 64 genes encoding candidate protein interactors of KUF1. Students designed guide RNAs (gRNAs), generated CRISPR-Cas9 constructs, and transformed A. thaliana kuf1 mutants. In the remote-learning CURE, we asked students to design gRNAs to mutate 46 target genes encoding potential TAN1 interactors. Later, these gRNAs will be cloned into vectors and transformed into plants to generate mutants as part of the independent research project. Here, we provide detailed protocols to implement either version of the CURE depending upon the availability of laboratory facilities (Fig. 2). This innovative CURE uses a reverse genetics approach to generate higher-order mutants in a sensitized mutant background using CRISPR-Cas9 gene editing.
FIG 2.
Timeline of the CURE. Steps shown in the left column are recommended prerequisite lessons before beginning the research project. Middle column steps outline the remote-learning CURE while right column steps outline the in-person CURE and require laboratory facilities.
Intended audience
This course is intended for first-year biology students or other STEM majors. It was implemented in-person in Spring 2019 for 6 sections and online in Spring 2020 for 8 sections with 18 to 24 students enrolled in each section. Student demographics were ∼50% Asian and ∼25% Hispanic for both courses (see Appendix S1 in the supplemental material). The CURE is a long-running course offered to first-year biology students as an alternative to the traditional first-year lab course (21). During the first 4 to 5 weeks of the 10-week quarter, class instructors introduce students to common laboratory techniques and basic biological concepts. University of California, Riverside faculty then offer 5- to 6-week-long CUREs in the second half of the class while course administration, grading, and most technical support are handled by instructors, teaching assistants, and staff. This organization allows faculty to deliver a focused research project experience. The remote-learning option was a direct result of the cancelation of in-person instruction due to the COVID-19 pandemic. However, instructions presented here can be applied to online courses or used in settings where laboratory space for students is unavailable.
Prerequisite student knowledge
Students are first-year undergraduates. Students are introduced to basic experimental and computational genomics techniques during the first 4 to 5 weeks. Structured lab activities include pipetting and dilutions (22), PCR, bacterial transformation, and introduction to gene structure and polymorphisms (21). Videos demonstrating techniques were provided for remote instruction. Because the class is for first-year students, techniques and concepts taught before the start of the CURE are necessary for students to perform the experiments and understand the key concepts of genetic information transfer and genome organization.
Learning time
The course is 10 weeks, with a 5-week CURE consisting of class meetings twice a week on either Mondays and Wednesdays or Tuesdays and Thursdays. Online classes’ meetings are 1 to 1.5 h each. For in-person classes, lab periods are 3 h each. Because students cannot access the laboratory outside class, all protocols are designed to fit in 3-h sessions.
Learning outcomes
Three of six core competencies (the ability to apply the process of science, the ability to tap into the interdisciplinary nature of science, and the ability to use modeling and simulation) and three of five core concepts for biological literacy (information flow, exchange, and storage, evolution, and systems) outlined by the Vision and Change framework are addressed (Table 1; Appendix S22).
TABLE 1.
Assessment of key learning outcomesa
| Learning outcomes assessed | Exam question | Rubric |
Outcomes | |
|---|---|---|---|---|
| Points | Example | |||
| 9. Design gRNAs to specifically target different regions of a gene to create a null mutation. | Design gRNAs that will remove the whole gene. Draw a diagram with gRNA locations indicated and show the size of the deletion. Justify why you chose those guides. | 14 | gRNAs shown with location, target whole gene, PCR primers shown, gRNAs are inside the PCR priming sites, deletion shown and size is correct. Diagram is drawn correctly. | Points possible, 14; mean, 13.6 (97%); SD, 1.26 |
| 10. Evaluate and select gRNAs based on a given set of criteria to selectively target a single gene. | 7 | gRNAs are wrong, are outside PCR primers, and/or do not target whole gene. | ||
| 1 | Much is missing, weak attempt. | |||
| 2. Design PCR primers with the aid of online tools and databases to amplify their assigned gene. | Design a PCR experiment that would show that the CRISPR gRNAs were effective in making the deletion. | 6 | Prediction that the genomic DNA size will be smaller in the deletion plant, mRNA from wild type and mutant, mRNA copied to cDNA by reverse transcriptase (RT), PCR with primers, run gel. | Points possible, 6; mean, 5.1 (85%); SD, 0.32 |
| 11. Design PCR experiment to detect successful gRNA guided deletions. | 3 | Prediction is missing or RT is missing. | ||
| 1 | Incomplete description of setting up the reactions is given. | |||
Assessment of learning outcomes from 10 random exams taken in Spring 2020. A full list of learning outcomes can be found in Appendix S2, and mapping of learning outcomes to other exam questions can be found in Appendix S42.
By the end of the CURE, students should be able to
-
1.
Use online resources and databases to research genes from A. thaliana or another organism.
-
2.
Design PCR primers with the aid of online tools and databases to amplify their assigned A. thaliana gene.
-
3.
Identify homologs of their gene and design gRNAs targeting their gene and any close homologs.
-
4.
Define homology and identify gene homologs through DNA database searches.
-
5.
Discuss the evolutionary relationships between paralogs and orthologs.
-
6.
Create a phylogenetic tree for a gene family using online tools and identify paralogs and orthologs for their assigned gene.
-
7.
Discuss the mechanism of CRISPR-Cas9 mutagenesis.
-
8.
Form a hypothesis about the potential consequences of mutating a gene and what that suggests about the gene’s function.
-
9.
Design gRNAs to specifically target different regions of a gene to create a null mutation.
-
10.
Evaluate and select gRNAs based on a given set of criteria to selectively target a single gene.
-
11.
Design PCR experiment to detect successful gRNA guided deletions in A. thaliana.
PROCEDURE
Student and faculty instructions
To promote peer learning and replicability, students work in pairs on a single gene. This increases the likelihood of meeting research goals by producing multiple unique gRNAs to target each gene and CRISPR-Cas9 constructs in case one student is not successful in their cloning. All other lab work and assignments are completed individually by students. We present an overview of the remote-learning and in-person workflow (see Appendix S3, remote learning, and Appendix S4, in person, in the supplemental material). The in-person timeline differs from the online version because gRNA selection is done during the first week; however, students write a report, record their results in an online lab notebook, complete quizzes, and present their results as described in the remote-learning version.
Remote-learning CURE
Week 1. A video provides background information and the scientific rationale for the project (Appendix S5). Specific guidelines are provided for the written project report due at the end of the course (Appendix S6). Because one common error in student-generated reports is lack of familiarity with formatting and terminology, examples of published papers relating to the project are provided for students and discussed in class to help students conceptualize how to format and write a scientific report. Students start working on a draft of their report introduction, which is due at the end of the week. Teaching assistants grade the draft and offer guidance on formatting in their feedback, providing opportunities for students to improve their scientific writing.
Primer design concepts are introduced, emphasizing using primers to detect insertions and deletions (indels). A step-by-step guide for primer design is provided using the maize ACTIN-1 gene (Appendix S7). Students are assigned their genes and asked to create primers to amplify the coding sequence using online resources (Appendix S8). Students record their primer design results in an online lab notebook, which is graded for completeness by teaching assistants who also monitor the quality of the results obtained (rubric in Appendix S9 and example notebook entry in Appendix S10). We used WordPress to keep all lab notebook entries in a single, accessible place (23). Student understanding of the project goals and how to use BLAST to examine gene structure is assessed with Quiz 1 (Appendix S11, answer key Appendix S12).
Week 2. The concepts of gene evolution and homologs are explored with a prerecorded lecture (Appendix S5). Students create a phylogenetic tree for their assigned gene using Plaza 4.0 (https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v4_dicots/) (Appendix S13). Students use their phylogenetic tree to identify homologs for their assigned gene and record their findings in an electronic lab notebook entry (example in Appendix S14).
Week 3. CRISPR-Cas9 methodologies are explained and students design dual gRNAs for their assigned genes using the E-CRISP (http://www.e-crisp.org/E-CRISP/) tool (Appendix S15). The instructor explains features of acceptable gRNAs, including location of the gRNAs within the gene, the presence of a protospacer-adjacent motif (PAM) sequence, and lack of off-target sequences. To gain experience designing gRNAs, students create two pairs targeting a small region of their gene and two pairs targeting the entire gene for deletion. In their notebooks, students identify which gRNA pairs likely render their assigned gene nonfunctional (example in Appendix S16). Students then add their best gRNA pairs into a class spreadsheet (example in Appendix S17). Students begin working on a draft for the methods and data sections of their reports to be turned in for feedback at the start of the following week (Appendix S6). Teaching assistants address common pitfalls such as writing bullet points instead of using complete sentences. Student understanding of how to create a phylogenetic tree and design gRNAs is assessed with Quiz 2 (Appendix S18, answer key Appendix S19).
Weeks 4 and 5. Students practice communicating scientific ideas by preparing a 5-min presentation about the project rationale, their assigned gene, and their results that is presented to the class in week 5 (presentation guidelines in Appendix S20, presentation rubric in Appendix S21, example score sheet in Appendix S22, and example presentation slides in Appendix S23). Presentations are scored by the instructor and teaching assistants. Future project plans are discussed in class (Appendix S24). Students are then expected to complete their project report by writing conclusions and future directions (example in Appendix S25 and rubric in Appendix S26). The report helps solidify their understanding of the experiments and form their own hypothesis about the next steps and potential project outcomes. Additionally, scientific writing improves students’ overall academic performance and critical thinking skills (24). During their final exam, students are asked to locate the gene sequence and design gRNAs of an assigned rice gene (Appendix S27, answer key Appendix S28, and exam rubric Appendix S29).
In-person CURE
Week 1. Students are introduced to the project with a slide presentation. This is followed by an introduction to Arabidopsis thaliana as a genetic model and use of forward and reverse genetic strategies to understand gene function. The metaphor “how a biologist would fix a radio” makes these concepts accessible to introductory biology students (25). We introduce CRISPR-Cas9 as a gene-editing tool, and each student is assigned a target gene. Students use BLAST and phylogenetic comparisons to identify close homologs of their gene that may be functionally redundant (Appendix S30). Students use CRISPR-P 2.0 to select two gRNA sequences that target their assigned gene and its homolog(s) (Appendix S31) (26). Students submit their selected 23-nucleotide guide plus protospacer-adjacent motif (PAM) sequence with their gRNA selection rationale to an online form. Instructors order four oligonucleotide primers for each student that will be used to incorporate the two guide sequences into a CRISPR-Cas9 construct in the following week (Appendix S32). A materials and equipment list is provided (Appendix S33).
Week 2. A brief lecture introduces students to CRISPR-Cas9 components and the PCR product they will generate (27, 28). PCRs are performed using four primers to incorporate both the RNA-encoding sequences into a cassette containing a U6-26 terminator and a U6-29 promoter (Appendix S34). After students check them for successful amplification, the PCR products are purified and quantified. This is followed by a Golden Gate cloning reaction to incorporate the gRNA cassette into a CRISPR-Cas9 vector backbone such as pHEE401E or pYUU (28–30). If time permits, students transform Escherichia coli with the Golden Gate reaction (Appendix S35).
Week 3. If not completed during week 2, students transform E. coli with the Golden Gate reaction. A short lecture is given on selectable markers and transformation approaches (e.g., electroporation and infection with Agrobacterium tumefaciens) for bacteria and plants. The next lab day, students use colony PCR to confirm insertion of the gRNA cassette into the plant transformation vector (Appendix S36).
Week 4. Teaching assistants inoculate two PCR-positive colonies for overnight cultures the evening before the first lab day. During class, students prepare glycerol stocks and plasmid miniprep isolations from the cultures (Appendix S37). After the plasmid DNA concentration is measured, the constructs are sent for sequencing. The next class period, A. tumefaciens (e.g., strain GV3101) (31) is transformed, as it requires 2 days to grow colonies (Appendix S38). Rather than wait for the results of sequencing, it may be expedient to transform all constructs, discarding those with errors later after sequencing results are available. Sequences are visualized using chromatograms and compared to a template sequence using free tools provided by Benchling (Appendix S39) to ensure that both guide sequences are incorporated without errors. We typically observe a high rate of success from PCR-positive colonies. If some students are unable to generate clones, they use other students’ constructs to participate in Arabidopsis transformation the following week.
Week 5.A. tumefaciens cultures for each construct are initiated by teaching assistants the evening before the first lab day. We used the floral dip transformation method (32) because it is simple, effective, and tolerant of experimental variation with modifications for small culture volumes (Appendix S40). Students transform one pot of five healthy, flowering A. thaliana plants each. We typically obtained about 16 transformants. Because the students’ transformations will not be ready for several weeks, a demonstration sample can be prepared in advance to show students what will happen next to their samples.
Safety concerns
No additional safety training is required when the course is conducted remotely.
For in-person activities, the course instructors review lab safety protocols on the first day. Students are instructed how to appropriately dispose of hazardous and biohazardous waste, and the locations of biohazard waste bins and fire extinguishers are reviewed. In the lab, students wear proper personal protective equipment at all times, including lab coat, eye protection, gloves, closed-toe shoes, and long pants or skirts. Because the mutagen ethidium bromide is incorporated into agarose gels, all ethidium bromide-contaminated materials (gels, gloves, pipette tips) are collected and disposed of by environmental health and safety personnel. Materials containing transgenic E. coli and A. tumefaciens are classified as BSL-1 and are placed in biohazard bins and later autoclaved. Bleach is added to bacterially contaminated solutions. Finally, after each laboratory session, students disinfect bench surfaces with antibacterial wipes.
DISCUSSION
Evidence of student learning
Assessment of this CURE and dissemination of the data were performed in accordance with UC Riverside Institutional Review Board (IRB) approval (HS-14-085). Student learning outcomes were assessed by open-answer exam questions. We randomly selected 10 final exams each from one in-person CURE (Spring 2019) and two remote-learning CURES (Spring 2020 and Fall 2020), representing ∼6% of total exams, to measure mastery of learning outcomes. During the Spring 2020 exam, students demonstrated their understanding of the principles of guide RNA design by applying what they learned in the CURE to a new organism (rice) and using an unfamiliar, yet similar, genome database to acquire gene information and sequences. Students scored ≥85% on related questions, demonstrating mastery of key learning outcomes 2, 9, 10, and 11 (Table 1). For all exams analyzed, average scores were between 75 and 100% for each question (see Appendix S42 in the supplemental material), and total exam scores were ∼73% and ∼84% for in-person and remote-learning CUREs, respectively (Appendix S41), demonstrating that students learned the course material.
Likert-style surveys designed by course instructors focused on student perceptions of learning are completed at the beginning and end of the course (Appendix S43). For those completing either the in-person or the remote-learning CURE, results indicate increased interest in participating in research on campus, with values rising from ∼24% presurvey to an impressive ∼90% postsurvey. When asked to rate perceptions of their understanding of certain lab skills (on a scale of 1 to 7, where 1 is strongly disagree and 7 is strongly agree), overall scores increased 1 point from the presurveys to the postsurveys for both CUREs. Additionally, student perceptions of the online project were favorable (ratings of 5 to 7), with 84% reporting that the project was enjoyable and the research meaningful. Taken together, these results indicate that participation in primary research (whether in-person or remote) increased student understanding of modern lab techniques and their likely future participation in research.
During the Spring 2020 quarter, 8 sections containing 142 students total participated in the remote-learning CURE by designing gRNAs for 46 A. thaliana genes. Students generated a total of 285 gRNA pairs. A total of 245 pairs correctly and specifically targeted the assigned genes, representing an 86% success rate for gRNA design. All assigned genes had at least one correctly designed pair of gRNAs. The most common error made by students was selecting overlapping gRNA pairs (9/285 guide RNA pairs overlapped). In the future, instructors will mention a minimum expected deletion size so students will not select guides that overlap. In two instances, gRNAs were predicted to target both the gene of interest and a different gene. Altogether, successful gRNA design demonstrates that first-year undergraduates learned how to use free online software to identify homologs and design gRNAs.
Possible modifications
Specific goals and organisms used in the project can be modified, making this type of CURE adaptable and generalizable. This CURE is best suited to organisms with sequenced and annotated genomes that are amenable to gene editing and transformation. To identify candidate genes and potential interactors, genome databases are available for other organisms such as zebrafish (http://zfin.org) or Drosophila (http://flybase.org). Other online tools such as STRING (http://string-db.org) may be used to identify interacting proteins.
There are many free gRNA design programs available online that do not require downloads. The program chosen for the in-person course was CRISPR-P 2.0 (http://crispr.hzau.edu.cn/CRISPR2/). The CRISPR-P 2.0 output shows all potential gRNAs, which is useful if the goal of the project includes mutating more than one gene per gRNA. The program chosen for the remote-learning course was E-CRISP (http://www.e-crisp.org/E-CRISP/index.html). The interface, including simple bar graphs that show the specificity, annotation score, and efficiency of each gRNA, provides intuitively simple parameters for judging gRNAs.
Challenges specific to remote-learning classes
Remote learning has several unique challenges. Students may occasionally experience technical issues, including slow Internet connection or lack of computers altogether. To promote inclusion, we identified online tools supported across multiple platforms, including tablets and Chromebooks. We provided detailed protocols for students to follow and demonstrated the protocols with example genes. Online polls are useful to determine whether students need more time to complete steps. Online quiz tools such as Kahoot can be used to assess conceptual understanding. We offered additional one-on-one help from teaching assistants or instructors during office hours to clarify concepts and help students with activities missed due to absence or technical issues.
ACKNOWLEDGMENTS
We thank BIOL20 students, teaching assistants, undergraduate lab assistants, and laboratory preparation staff at the University of California, Riverside (UCR) for their hard work, dedication, and feedback. We thank undergraduate students Dorothy Nguyen and Kruti Seethammagari for allowing us to use their exceptional work as examples. We thank Lindy Allsman (UCR) and Aimee Uyehara (UCR) for helpful manuscript comments. We thank Tarek Azzam (UC Santa Barbara) for survey information. We gratefully acknowledge funding from the National Science Foundation (CAREER MCB-1942734 to C.G.R., IOS-1856741 to D.C.N., CAREER IOS-1751385 to J.M.V.N., IOS-1027542 to S.R.W.), HHMI Institutional Grant (52008110) to S.R.W., and Neil and Rochelle Campbell Presidential Chair for Innovation in Science Education to S.R.W.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
David C. Nelson, Email: david.nelson@ucr.edu.
Carolyn G. Rasmussen, Email: carolyn.rasmussen@ucr.edu..
REFERENCES
- 1.Auchincloss LC, Laursen SL, Branchaw JL, Eagan K, Graham M, Hanauer DI, Lawrie G, McLinn CM, Pelaez N, Rowland S, Towns M, Trautmann NM, Varma-Nelson P, Weston TJ, Dolan EL. 2014. Assessment of course-based undergraduate research experiences: a meeting report. CBE Life Sci Educ 13:29–40. doi: 10.1187/cbe.14-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brewer CA, Smith D. 2011. Vision and change in undergraduate biology education: a call to action. 2009 National Conference by AAAS, Washington, DC, USA. American Association for the Advancement of Science, Washington, DC. [Google Scholar]
- 3.Linn MC, Palmer E, Baranger A, Gerard E, Stone E. 2015. Education. Undergraduate research experiences: impacts and opportunities. Science 347:1261757. doi: 10.1126/science.1261757. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez PR, Woodcock A, Estrada M, Schultz PW. 2018. Undergraduate research experiences broaden diversity in the scientific workforce. Bioscience 68:204–211. doi: 10.1093/biosci/bix163. [DOI] [Google Scholar]
- 5.Seymour E, Hunter A-B, Laursen SL, DeAntoni T. 2004. Establishing the benefits of research experiences for undergraduates in the sciences: first findings from a three-year study. Sci Ed 88:493–534. doi: 10.1002/sce.10131. [DOI] [Google Scholar]
- 6.Bangera G, Brownell SE. 2014. Course-based undergraduate research experiences can make scientific research more inclusive. CBE Life Sci Educ 13:602–606. doi: 10.1187/cbe.14-06-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowland SL, Lawrie GA, James BY, Gillam EMJ. 2012. Is the undergraduate research experience (URE) always best?: The power of choice in a bifurcated practical stream for a large introductory biochemistry class. Biochem Mol Biol Educ 40:46–62. doi: 10.1002/bmb.20576. [DOI] [Google Scholar]
- 8.Weaver GC, Russell CB, Wink DJ. 2008. Inquiry-based and research-based laboratory pedagogies in undergraduate science. Nat Chem Biol 4:577–580. doi: 10.1038/nchembio1008-577. [DOI] [PubMed] [Google Scholar]
- 9.Gormally C, Brickman P, Hallar B, Armstrong N. 2009. Effects of inquiry-based learning on students’ science literacy skills and confidence. IJ-SoTL 3:16. doi: 10.20429/ijsotl.2009.030216. [DOI] [Google Scholar]
- 10.Hensel NH. 2018. Course-based undergraduate research: educational equity and high-impact practice. Stylus Publishing, LLC. [Google Scholar]
- 11.Kloser MJ, Brownell SE, Chiariello NR, Fukami T. 2011. Integrating teaching and research in undergraduate biology laboratory education. PLoS Biol 9:e1001174. doi: 10.1371/journal.pbio.1001174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shortlidge EE, Brownell SE. 2016. How to assess your CURE: a practical guide for instructors of course-based undergraduate research experiences. J Microbiol Biol Educ 17:399–408. doi: 10.1128/jmbe.v17i3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson DC, Flematti GR, Ghisalberti EL, Dixon KW, Smith SM. 2012. Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu Rev Plant Biol 63:107–130. doi: 10.1146/annurev-arplant-042811-105545. [DOI] [PubMed] [Google Scholar]
- 14.Waters MT, Gutjahr C, Bennett T, Nelson DC. 2017. Strigolactone signaling and evolution. Annu Rev Plant Biol 68:291–322. doi: 10.1146/annurev-arplant-042916-040925. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen CG, Bellinger M. 2018. An overview of plant division-plane orientation. New Phytol 219:505–512. doi: 10.1111/nph.15183. [DOI] [PubMed] [Google Scholar]
- 16.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 17.O’Malley RC, Ecker JR. 2010. Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J 61:928–940. doi: 10.1111/j.1365-313X.2010.04119.x. [DOI] [PubMed] [Google Scholar]
- 18.Minkenberg B, Xie K, Yang Y. 2017. Discovery of rice essential genes by characterizing a CRISPR-edited mutation of closely related rice MAP kinase genes. Plant J 89:636–648. doi: 10.1111/tpj.13399. [DOI] [PubMed] [Google Scholar]
- 19.Nelson DC, Flematti GR-A, Riseborough J, Ghisalberti EL, Dixon KW, Smith SM. 2010. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci U S A 107:7095–7100. doi: 10.1073/pnas.0911635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mir R, Morris VH, Buschmann H, Rasmussen CG. 2018. Division plane orientation defects revealed by a synthetic double mutant phenotype. Plant Physiol 176:418–431. doi: 10.1104/pp.17.01075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.BurnetteJM, 3rd, Wessler SR. 2013. Transposing from the laboratory to the classroom to generate authentic research experiences for undergraduates. Genetics 193:367–375. doi: 10.1534/genetics.112.147355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burnette J, Kanizay L, Chester N, Wessler SR. 2016. Dilution and pipetting lesson using food dyes. Cs 3. doi: 10.24918/cs.2016.5. [DOI] [Google Scholar]
- 23.Robb S, BurnetteJM, III, Chapovskya A, Palmer K, Wessler SR. 2015. An open source, collaborative electronic notebook for undergraduate laboratory classes. CourceSource 2. [Google Scholar]
- 24.Gupta T, Burke KA, Mehta A, Greenbowe TJ. 2015. Impact of guided-inquiry-based instruction with a writing and reflection emphasis on chemistry students’ critical thinking abilities. J Chem Educ 92:32–38. doi: 10.1021/ed500059r. [DOI] [Google Scholar]
- 25.Lazebnik Y. 2002. Can a biologist fix a radio?—Or, what I learned while studying apoptosis. Cancer Cell 2:179–182. doi: 10.1016/S1535-6108(02)00133-2. [DOI] [PubMed] [Google Scholar]
- 26.Liu H, Ding Y, Zhou Y, Jin W, Xie K, Chen L-L. 2017. CRISPR-P 2.0: an improved CRISPR-Cas9 tool for genome editing in plants. Mol Plant 10:530–532. doi: 10.1016/j.molp.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Xing H-L, Dong L, Wang Z-P, Zhang H-Y, Han C-Y, Liu B, Wang X-C, Chen Q-J. 2014. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z-P, Xing H-L, Dong L, Zhang H-Y, Han C-Y, Wang X-C, Chen Q-J. 2015. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. 2011. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res 39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angulo J, Astin CP, Bauer O, Blash KJ, Bowen NM, Chukwudinma NJ, Dinofrio AS, Faletti DO, Ghulam AM, Gusinde-Duffy CM, Horace KJ, Ingram AM, Isaack KE, Jeong G, Kiser RI, Kobylanski JS, Long MR, Manning GA, Morales JM, Nguyen KH, Pham RT, Phillips MH, Reel TW, Seo JE, Vo HD, Wukuson AM, Yeary KA, Zheng GY, Lukowitz W. 2020. Targeted mutagenesis of the Arabidopsis GROWTH-REGULATING FACTOR (GRF) gene family suggests competition of multiplexed sgRNAs for Cas9 apoprotein. bioRxiv. doi: 10.1101/2020.08.16.253203. [DOI] [PMC free article] [PubMed]
- 31.Hellens R, Mullineaux P, Klee H. 2000. Technical focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5:446–451. doi: 10.1016/s1360-1385(00)01740-4. [DOI] [PubMed] [Google Scholar]
- 32.Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download JMBE00155-21_Supp_1_seq1.pdf, PDF file, 8.4 MB (8.6MB, pdf)