Abstract
Current models predict that β-catenin (β-cat) functions in Wnt signaling via activation of Tcf/Lef target genes and that its abundance is regulated by the adenomatous polyposis coli (APC) and glycogen synthase kinase 3β (GSK3β) proteins. In colon and other cancers, mutations in APC or presumptive GSK3β phosphorylation sites of β-cat are associated with constitutive activation of Tcf/Lef transcription. In spite of assumptions about its oncogenic potential, prior efforts to demonstrate that mutated β-cat will induce neoplastic transformation have yielded equivocal results. We report here that mutated, but not wild-type, β-cat proteins induced neoplastic transformation of RK3E, an adenovirus E1A-immortalized epithelial cell line. Analysis of the properties of mutant β-cat proteins and studies with a dominant negative Tcf-4 mutant indicated that the ability of β-cat to bind and activate Tcf/Lef factors is crucial for transformation. c-myc has recently been implicated as a critical Tcf-regulated target gene. However, c-myc was not consistently activated in β-cat-transformed RK3E cells, and a dominant negative c-Myc mutant protein failed to inhibit β-cat transformation. Our findings underscore the role of β-cat mutations and Tcf/Lef activation in cancer and illustrate a useful system for defining critical factors in β-cat transformation.
β-Catenin (β-cat) plays critical roles in axis formation, embryonic patterning, cell fate determination, and tissue homeostasis in the adult (41). The protein was first identified because of its binding to the cytoplasmic domain of E-cadherin (E-cad), a calcium-dependent cell adhesion molecule. β-cat links E-cad to α-cat, a vinculin-like protein which, in turn, links the E-cad/cat adhesive complex to the cytoskeleton (4, 14, 21). In addition to its role in cell adhesion, β-cat functions in Wnt signaling. In brief, the role of β-cat in Wnt signaling is thought to be as follows. Binding of Wnt to the Frizzled receptor activates the disheveled protein, which, in turn, inhibits the function of glycogen synthase kinase 3β (GSK3β) (24, 41). When complexed with the adenomatous polyposis coli (APC) and axin or conductin proteins, GSK3β may phosphorylate specific residues in the amino (N) terminus of β-cat (6, 11, 17, 19, 27). Phosphorylated β-cat, but not the nonphosphorylated form, is rapidly degraded by the ubiquitin-proteasome pathway (2, 42). Following its accumulation, β-cat binds to the transcription factor Tcf (T-cell factor) or Lef (lymphoid enhancer factor) (5, 23, 41). Upon translocation to the nucleus, β-cat serves as a transcriptional coactivator of Tcf/Lef target genes.
Defects in the Wnt/APC/β-cat/Tcf pathway have been implicated in cancer. The first mammalian Wnt gene, initially termed Int-1 and subsequently termed Wnt-1, was identified because of its activation by mouse mammary tumor virus integration events in mouse mammary tumor virus-induced breast cancers (28). More recently, mutational inactivation of the APC tumor suppressor gene has been found in about 70% of colon cancers, and constitutive activation of Tcf-4 transcription results from APC inactivation (20). In some colon cancers lacking APC mutations, mutations in presumptive GSK3β phosphorylation sites in the β-cat N terminus result in constitutive activation of Tcf-4 transcription (25). Missense mutations or in-frame deletions of the β-cat N terminus have been found in a subset of other cancers, such as melanoma (32), medulloblastoma (47), and endometrial (9) and hepatocellular (22) carcinoma. Recent studies indicate that critical consequences of APC or β-cat mutations in colon cancer may be activation of c-MYC and/or cyclin D1 gene expression (12, 37).
The existing data imply that mutant β-cat proteins are oncogenic, because they are resistant to regulation by the APC/axin/GSK3β complex, and, as a result, that mutant β-cat accumulates in the cell. Unfortunately, prior efforts to demonstrate the oncogenic activity of mutant β-cat proteins have yielded, at best, ambiguous results. An initial report suggested that overexpression of an N-terminally truncated form of β-cat or even wild-type β-cat could neoplastically transform NIH 3T3 murine fibroblasts (40). However, results of other studies (46), as well as data presented here, indicate that mutant and wild-type β-cat proteins do not consistently transform rodent fibroblasts. In addition, an N-terminally truncated form of β-cat failed to induce neoplastic changes when expressed at high levels in the intestinal epithelium of transgenic mice (43). One recent study has provided evidence that mutant β-cat alleles may function as oncogenes. Mice carrying a transgene, expressing an N-terminally truncated form of β-cat under the control of a keratin 14 promoter element, developed epithelioid cysts and lesions resembling well-differentiated hair follicle tumors (10).
We initiated studies to assess the oncogenic potential of wild-type and mutated forms of β-cat, and we report here that β-cat proteins with a missense mutation of the type found in human cancer or in-frame deletions of the N terminus efficiently induce neoplastic transformation of RK3E, an adenovirus E1A-immortalized epithelial cell line derived from neonatal rat kidney (33). Although Tcf/Lef factors have been implicated in transducing Wnt and β-cat signals in other systems and deregulation of Tcf/Lef transcription has been associated with APC or β-cat mutations in cancer (41), our findings firmly establish that deregulation of Tcf/Lef transcription is required for neoplastic transformation by mutant β-cat. Furthermore, our data indicate that activation of c-myc gene expression is not required for β-cat-mediated transformation of RK3E.
MATERIALS AND METHODS
Plasmids.
The wild-type and S33Y mutant alleles of β-cat were cloned by PCR with Pfu polymerase (Stratagene, La Jolla, Calif.), using a normal colon cDNA library (Clontech Laboratories Inc., Palo Alto, Calif.) or hexamer-primed cDNA from the colorectal cancer cell line SW48 as templates, respectively. The wild-type and S33Y β-cat cDNAs were then used as templates in further PCR-based approaches to generate the N-terminally truncated forms of β-cat and the deleted forms of the S33Y mutant allele. All PCR-generated cDNAs contained a 12-bp sequence upstream of the initiating ATG codon matching the consensus Kozak initiator sequence, and each of the encoded β-cat proteins had a C-terminal Flag epitope, in addition to the coding sequences outlined in Fig. 1A. All PCR products were initially subcloned into the pcDNA3 vector (Invitrogen, San Diego, Calif.), and their sequences were verified by manual and/or automated DNA sequencing. Further details of the generation of mutated β-cat cDNAs will be provided upon request. The cDNAs with C-terminal Flag epitope tags were then subcloned into the retroviral expression vector pBMN (provided by G. Nolan, Stanford University, Stanford, Calif.). A cDNA encoding a mutant K-ras protein (a valine to cysteine mutation at codon 12) was amplified from the colon cancer cell line SW480 and subcloned into pBMN. The retroviral vector pBMN-Z, carrying the β-galactosidase (lacZ) gene, was provided by G. Nolan. A dominant-negative form of Tcf-4, known as Tcf-4ΔN31 and lacking the N-terminal 31 amino acids (aa) was generated by PCR with the vector pHRhTCF4 (provided by B. Vogelstein, Johns Hopkins Oncology Center, Baltimore, Md.) as a template. Together with a N-terminal Flag epitope tag, the Tcf-4ΔN31cDNA was subcloned into the retroviral vector pPGS-CMV-CITE-neo (provided by G. Nabel, University of Michigan, Ann Arbor, Mich.). The pPGS-CMV-CITE-neo vector allows the expression of chimeric transcripts encoding a gene of interest fused to the neomycin resistance gene. Expression of the neomycin resistance protein results from use of an internal ribosomal entry site upstream of the neomycin open reading frame. In addition to Tcf-4ΔN31, cDNAs for wild-type β-cat and a dominant negative form of c-myc, known as MycΔ106–143 (34), were cloned into the pPGS-CMV-CITE-neo vector. The reporter constructs pTOPFLASH and pFOPFLASH (20), containing either three copies of the optimal Tcf motif CCTTTGATC or three copies of the mutant motif CCTTTGGCC upstream of a minimal c-Fos promoter driving luciferase expression, were kindly provided by B. Vogelstein. The reporter construct pGLDH637Luc contains the myc-responsive elements of the rat LDH-A promoter cloned upstream of luciferase, while the construct pGLDH637-mut/Luc has localized mutations of both c-Myc-responsive elements (E boxes) in the LDH-A promoter (35). Plasmid pCH110 (Pharmacia, Piscataway, N.J.), containing a functional lacZ gene cloned downstream of a cytomegalovirus early-region promoter-enhancer element, was used as control for transfection efficiency in the reporter assays.
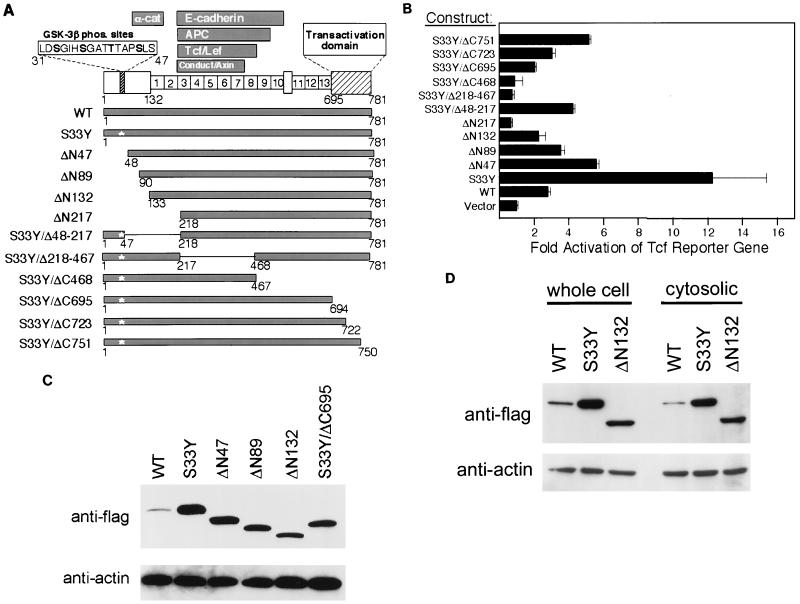
FIG. 1.
Mutated β-cat proteins activate Tcf transcription and accumulate in the cytosol. (A) Schematic outline of β-cat domains and proteins encoded by the retroviral expression constructs. Shown are presumptive GSK3β phosphorylation (phos.) sites in the β-cat N-terminal region; armadillo repeats in the central region; the C-terminal transcriptional activation domain; and the regions required for interaction with α-cat, E-cad, APC, Tcf/Lef factors, and conductin/axin. In addition to wild-type (WT) β-cat, the structures of mutated proteins are indicated. The star indicates the position of the S33Y substitution, the solid boxes represent the β-cat sequences present, and the thin line indicates the in-frame deletions in the constructs. (B) Activation of Tcf transcription by wild-type (WT) and mutated forms of β-cat in RK3E cells following transient transfection of pcDNA3 expression constructs. The ratio of luciferase activities from a Tcf-responsive reporter (pTOPFLASH) and a control luciferase reporter gene construct (pFOPFLASH) was determined 48 h after transfection. The means and standard deviations of three independent experiments are shown. (C) Mutated β-cat proteins accumulate to higher levels than wild-type β-cat (WT). ECL-Western blot analysis with an anti-Flag antibody was carried out on whole-cell lysates prepared 2 days after transient transfection of 293 cells with pcDNA3 constructs encoding wild-type β-cat and the indicated mutant forms. To demonstrate equivalent loading of the lanes, the blot was stripped and ECL-Western blotting with an anti-actin antibody was performed. (D) Mutant forms of β-cat accumulate to higher levels than wild-type β-cat in RK3E cells following infection with retroviruses encoding wild-type (WT) β-cat or mutated forms (S33Y or ΔN132). ECL-Western blot studies with an anti-Flag antibody were carried out on whole-cell or cytosolic lysates. The blot was stripped, and analysis with an anti-actin antibody was performed.
Cell Culture and Retrovirus Infection.
The amphotropic Phoenix packaging cell line was provided by G. Nolan; RK3E cells were provided by J. M. Ruppert (University of Alabama, Birmingham, Ala.); 1811 cells were provided by K. Cho (University of Michigan, Ann Arbor, Mich.); and the IEC-18, NIH 3T3, and 293 cell lines were obtained from the American Type Culture Collection (Rockville, Md.). The Phoenix, 293, and RK3E cells were propagated in Dulbecco’s minimal essential medium (DMEM) supplemented with 10% fetal bovine serum; the IEC-18 cells were grown in DMEM supplemented with 5% fetal bovine serum; the 1811 keratinocytes were propagated in KGM medium (Clonetics); and the NIH 3T3 cells were grown in DMEM containing 10% calf serum. The Phoenix packaging cells were transfected with the particular retroviral expression constructs, using FuGENE6 (Boehringer Mannheim, Indianapolis, Ind.), as described by the manufacturer. Briefly, 2.5 × 106 Phoenix cells were seeded in 60-mm dishes 12 h prior to transfection. They were then transfected with 6 μg of retroviral plasmid DNA and 12 μl of FuGENE6. After 24 h, the growth medium was replaced by 3 ml of fresh medium. After a further 24-h period, the supernatant containing nonreplicating forms of amphotrophic virus was harvested. Target cell lines, incubating in fresh medium, were infected in 100-mm dishes at 70 to 80% confluency with virus supernatant at a ratio of 2:1 in the presence of 4 μg of Polybrene (Sigma, St. Louis, Mo.) per ml. At 24 h later, the medium was changed. The cells were monitored for up to 4 weeks for focus formation. During this period, the medium was changed twice weekly. After 4 weeks, the dishes were fixed and stained with 6 ml of Hanks’ balanced salt solution containing 1.5% glutaraldehyde and 0.06 g of methylene blue per 100 ml. The plates were photographed with a digital camera system (Alpha Innotech Corp., San Leandro, Calif.), and the foci were counted.
Generation and growth of stable RK3E cell lines.
From multiple tissue culture dishes with β-cat-induced foci, more than 25 independent foci were isolated with a micropipette under microscopic visualization and then expanded as clonal lines. Polyclonal, G418-resistant RK3E cell lines expressing wild-type β-cat, the Tcf-4ΔN31 mutant, the c-MycΔ106–143 mutant, or only the neomycin resistance gene were obtained following infection with the particular pPGS-CMV-CITE-neo vector-based retrovirus and subsequent selection of the bulk cell population in G418 at an initial concentration of 1 mg/ml. After 48 h, the G418 concentration was reduced to 0.75 mg/ml. After 1 week, the G418 concentration was further reduced to 250 μg/μl and the expression of the transferred genes was confirmed by Western blot analysis. An H-ras-transformed RK3E line was generated by selection of foci induced following transfection with an H-ras allele with a codon 12 mutation. To measure proliferation in medium with reduced serum, 2 × 104 cells were seeded in 35-mm dishes in the presence of growth medium containing 10% fetal bovine serum. The following day, the medium was exchanged for medium containing 0.5% fetal bovine serum. At specific times following culture in reduced serum, the cells were dissociated by trypsinization and viable cells were counted after trypan blue staining.
Reporter gene assays.
At 12 h prior to transfection, 3 × 105 cells were seeded in 35-mm dishes. Transfections were performed with 2 μl of FuGENE6 per μg of transfected DNA. To assess the ability of wild-type and mutated β-cat proteins to activate Tcf transcription, 293 and RK3E cells were transfected with 1 μg of the respective pcDNA3 expression construct, 0.5 μg of pTOPFLASH or pFOPFLASH reporter, and 0.5 μg of pCH110. To determine Tcf reporter activity in stably transformed RK3E lines, the cells were transfected with 0.5 μg of reporter plasmid pTOPFLASH or pFOPFLASH and 0.5 μg of the control plasmid pCH110. Assays for c-Myc transcription activity were performed in a similar fashion, using the reporter construct pGLDH637Luc, which contains myc-responsive elements of the rat LDH-A promoter cloned upstream of luciferase, and the matched control pGLDH637-mut/Luc. The total mass of transfected DNA per dish was kept constant by adding empty vector plasmid, if necessary. At 2 days after transfection, the cells were collected and resuspended in reporter lysis buffer (Promega, Madison, Wis.) and luciferase activities were measured with luciferase assay reagent (Promega) and a luminometer (model TD-20E; Turner Corp. Mountain View, Calif.). β-Galactosidase activities were determined by standard methods as a control for transfection efficiency.
Colony formation in soft agar.
Assays of colony formation in soft agar were performed essentially as described previously (8). Briefly, 1-ml underlayers of 0.6% agar medium were prepared in 35-mm dishes by combining equal volumes of 1.2% Noble agar and 2× DMEM with 40% fetal bovine serum (Difco, Detroit, Mich.). The cells were trypsinized, centrifuged, and resuspended, and 104 cells were plated in 0.3% agar medium. The surface was kept wet by addition of a small amount of growth medium. After 2 to 3 weeks, dishes were stained with methylene blue and colonies were photographed and counted.
Tumorigenicity in nude mice.
nu/nu-nuBR mice (6 weeks old) were obtained from the Charles River Breeding Laboratories. Into each lower flank, 5 × 106 cells, resuspended in 200 μl of DMEM without serum, were injected subcutaneously. Groups of five mice were injected with each of the following cell lines: β-cat-transformed clones RK3E/S33Y-A and RK3E/S33Y-D and the parental RK3E line. One mouse was injected with RK3E/Kras cells. After 3 weeks, all mice injected with β-cat or K-Ras-transformed cells were sacrificed and tumor sizes were assessed. Mice injected with parental RK3E cells were monitored for 6 weeks for tumor formation, and no tumors were observed.
Western blot assays.
Whole-cell extracts were prepared with RIPA lysis buffer (Tris-buffered saline, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1% Nonidet P-40), and extracts of cytosolic proteins were prepared with Nonidet P-40 lysis buffer (100 mM NaCl, 50 mM Tris-HCl [pH 7.5], 0.5% Nonidet P-40). Both buffers contained the protease inhibitors antipain (final concentration, 50 μg/ml), aprotinin (5 μg/ml), leupeptin (2 μg/ml), and phenylmethylsulfonyl fluoride (10 μg/ml). Lysates were precleared, and the protein concentration was determined by the bicinchoninic acid assay (Pierce Biochemicals, Rockford, Ill.). For electrophoresis, lysates were supplemented with SDS loading buffer and separated on SDS–8% polyacrylamide gels. Proteins were transferred to Immobilon P membranes (Millipore, Bedford, Mass.) by semidry electroblotting. The blots were incubated in Tris-buffered saline containing 0.1% Tween 20 and 10% nonfat dry milk during the blocking step and 0.05% Tween 20 and 5% nonfat dry milk during the antibody incubation steps. Anti-Flag antibody (Sigma) was used at a 1:5,000 dilution, anti-actin (Sigma) antibody was used at 1:1,000, and the horseradish peroxidase-conjugated goat anti-mouse and goat anti-rabbit immunoglobulin (Pierce) were used at a 1:20,000 dilution. Antibody complexes were detected by enhanced chemiluminescence (ECL; Amersham Life Science, Arlington Heights, Ill.) and exposure to X-Omat film (Kodak, Rochester, N.Y.).
Northern blot analysis.
Total RNA was extracted from cells with Trizol (Gibco BRL, Grand Island, N.Y.); 7.5 μg of total RNA was separated on 1.2% formaldehyde–agarose gels and transferred to Zeta-Probe GT membranes (Bio-Rad, Hercules, Calif.) by capillary action. Rat β-cat, c-myc, lactate dehydrogenase A (LDH-A)-ornithine decarboxylase (ODC), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene fragments, ranging from 500 to 800 bp, were generated by PCR and labeled with 32P with the random primer kit (Gibco BRL). Northern blot hybridization to 32P-labeled probes was carried out by standard methods. Signals were detected by exposure to BioMax-MS film (Kodak) at −80°C with intensifying screens.
RESULTS
Mutant β-cat proteins activate Tcf transcription.
The 781 amino acid (aa) β-cat protein functions in E-cad cell adhesion and Wnt signaling. β-cat domains required for binding to E-cad (15, 29) and α-cat (1) have been localized, as have regions that confer binding to APC (15, 31), Tcf/Lef (5, 23), conductin (6), and axin (11, 16) (Fig. 1A). The carboxy (C)-terminal region of β-cat was previously shown to function in transcriptional activation (13, 39), and recent studies indicate N-terminal sequences of β-cat also play an important role in activation of Tcf transcription (13). The β-cat N terminus (aa 31 to 47) contains four presumptive GSK3β phosphorylation sites (45). We generated retroviral expression constructs encoding wild-type and 11 different mutated human β-cat proteins (Fig. 1A). A β-cat allele, containing a missense mutation of tyrosine for serine at codon 33 (S33Y), was isolated from the human colon cancer cell line SW48 (25). To define domains required for the ability of mutated β-cat proteins to potently activate Tcf transcription, the S33Y mutant allele was further altered in vitro, generating a series of deletion mutants lacking specific domains (e.g., S33Y/Δ148-217 [Fig. 1A]). Because mutant β-cat proteins with N-terminal deletions are found in some cancers, a series of N-terminal deletion mutants was also generated to explore effects on Tcf transcription, β-cat protein stability, and neoplastic transformation.
As expected, the S33Y mutant β-cat allele strongly activated Tcf-dependent transcription in RK3E cells (Fig. 1B). Consistent with prior studies (20, 25), the S33Y mutant activated Tcf transcription about 12-fold over basal levels and wild-type β-cat activated Tcf transcription only about 3-fold. The effect of the S33Y mutation on Tcf transcription was largely abrogated by deletion of certain β-cat domains, such as the C-terminal 85 aa (S33Y/ΔC695) or armadillo repeats 3 to 8 (S33Y/Δ218–467) (Fig. 1B). A mutant β-cat protein carrying a deletion of aa 48 to 217 in the context of the S33Y mutation (S33Y/Δ48–217) activated Tcf transcription more strongly than did wild-type β-cat but much less potently than did the intact S33Y protein (Fig. 1B). Nearly identical results for wild-type β-cat and the various β-cat mutants were also obtained in transient-transfection assays of Tcf-dependent transcription in 293 cells, an immortal human kidney line (data not shown).
Following transient transfection, expression of wild-type and mutated forms of β-cat was monitored by Western blotting with a Flag epitope tag present at the C terminus of each protein. The S33Y mutant and N-terminally truncated forms of β-cat accumulated to much higher levels than did wild-type β-cat, consistent with prior data indicating that the N-terminal GSK3β sites are critical in the rapid degradation of β-cat (41). The S33Y mutant protein was expressed at somewhat higher levels than were N-terminally truncated forms of β-cat (Fig. 1C), perhaps reflecting effects of the N-terminal deletions on β-cat protein folding and stability. Similar findings for the relative levels of protein expression were also obtained following infection of cells with retroviruses encoding wild-type and mutants β-cat (Fig. 1D and data not shown). Northern blot studies did not reveal any differences in the expression of transcripts for wild-type or mutated constructs (data not shown), suggesting that the differential protein expression was attributable to posttranscriptional events. In toto, the data indicate that β-cat proteins with mutation or deletion of the presumptive GSK3β phosphorylation sites are resistant to normal regulation, leading to their accumulation in the cells and their ability to activate Tcf transcription.
Our data are consistent with a model in which β-cat binds to Tcf through armadillo repeats 3 to 8 and activates transcription via its C-terminal 85 aa-domain and apparently also via sequences in the N terminus. As indicated above, N-terminally truncated β-cat proteins have been found in several cancers, and transgenic mice expressing a β-cat protein with an N-terminal truncation (ΔN87) under control of the keratin K14 promoter developed benign skin tumors (10). In our studies, several N-terminally truncated forms of β-cat, including ΔN47 and ΔN89, displayed more potent Tcf activation than did wild-type β-cat (Fig. 1B). However, none of the N-terminally truncated β-cat proteins was as active in the transient-transfection assay of Tcf transcription as the S33Y mutant was, even though their expression in transient-transfection assays and following infection of cells with retroviral constructs was much higher than that of wild-type β-cat and nearly equivalent to the levels of the S33Y mutant. Therefore, we suggest that the reduced activity of some N-terminally deleted forms of β-cat in the transient-transfection Tcf assay may be due, at least in part, to the function of β-cat N-terminal sequences in transcriptional activation (13), a role which our data with the S33Y/Δ48–217 mutant also support (Fig. 1B).
Mutant β-cat proteins induce morphologically transformed foci.
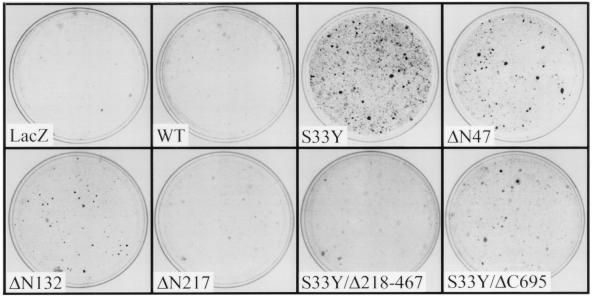
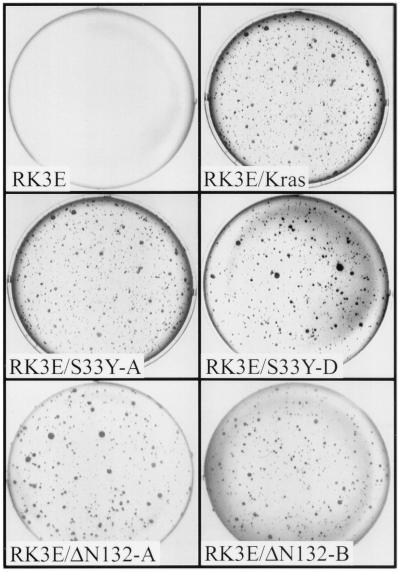
We assessed the ability of replication-defective retroviruses expressing wild-type or mutated forms of β-cat to generate macroscopic foci of morphologically transformed cells (i.e., focus formation) following infection of four different cell lines: NIH 3T3 mouse fibroblasts, IEC-18 rat intestinal epithelial cells (30), 1811 human squamous epithelial cells (18), and RK3E cells (33). A prior study reported that NIH 3T3 cells were neoplastically transformed following overexpression of wild-type β-cat or an N-terminally truncated β-cat mutant (40). However, we failed to induce focus formation following infection of NIH 3T3, IEC-18, or 1811 cells with retroviruses encoding wild-type β-cat, the S33Y mutant, or various N-terminal truncation mutants of β-cat. In contrast, in the RK3E line, dense foci of transformed cells were readily detected within 3 weeks of infection with retroviruses encoding different mutated β-cat proteins (Fig. 2). No foci were induced by retroviruses encoding wild-type β-cat or a control β-galactosidase (lacZ) gene. The S33Y β-cat mutant was more potent in focus formation than were the N-terminally truncated forms of β-cat, and β-cat proteins with more substantial truncations, such as the ΔN217 mutant, lacked focus-forming activity (Fig. 2 and Table 1). Deletion of armadillo repeats 3 to 8 or the C-terminal 85 aa inhibited the focus-forming activity conferred by the S33Y mutation (Fig. 2 and Table 1).
FIG. 2.
Induction of macroscopic foci in RK3E cells following infection with retroviruses encoding mutated forms of β-cat but not wild-type (WT) β-cat. The specific proteins encoded by the retroviruses used for infection, including a control LacZ virus, are indicated in each panel. Schematic representations of the β-cat proteins are shown in Fig. 1A. Four weeks after infection with the retroviruses, the plates were fixed, stained, and photographed, as described in the text.
TABLE 1.
Comparison of focus formation in RK3E cells by replication-defective retroviruses expressing wild-type or mutated forms of β-cat with the ability of the proteins to activate Tcf transcription
| Proteina | Fold Tcf activationb | No. of foci formedc in:
|
|||
|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Expt 4 | ||
| WT | 2.8 ± 0.15 | 1 | 4 | 6 | 5 |
| S33Y | 12.3 ± 3.1 | 408 | 296 | 440 | 276 |
| ΔN47 | 5.6 ± 0.12 | 192 | 168 | 160 | 104 |
| ΔN89 | 3.55 ± 0.17 | 180 | 144 | 240 | 160 |
| ΔN132 | 2.3 ± 0.35 | 96 | 68 | 48 | 60 |
| ΔN217 | 0.7 ± 0.05 | 7 | 3 | 6 | 3 |
| S33Y/Δ48-217 | 4.32 ± 0.10 | 102 | 74 | 88 | NDd |
| S33Y/Δ218-467 | 0.75 ± 0.07 | 9 | 6 | 1 | 4 |
| S33Y/ΔC468 | 0.92 ± 0.4 | 11 | 8 | 5 | 3 |
| S33Y/ΔC695 | 2.05 ± 0.1 | 24 | 27 | 21 | 32 |
| S33Y/ΔC723 | 3.07 ± 0.15 | 76 | 86 | 88 | 72 |
| S33Y/ΔC751 | 5.2 ± 0.1 | 164 | 128 | 124 | 132 |
β-cat proteins encoded by the retroviral constructs are illustrated in Fig. 1A.
Mean Tcf transcriptional activation ± standard deviation was measured in triplicate transient-transfection assays with pcDNA3 expression vectors and RK3E cells. The data are displayed graphically in Fig. 1B. The empty-vector control was assigned a value of 1.0.
Number of macroscopic foci at 4 weeks after infection of RK3E cells in 100-mm dishes at 70 to 80% confluency with retroviral supernatants. Results of four independent experiments are shown.
ND, not determined.
A correlation was observed between the focus-forming ability of the mutants and their strength of Tcf activation in the transient-transfection assay. Nonetheless, while wild-type β-cat had greater activity in the transient-transfection assay of Tcf transcription than did some β-cat mutants with focus-forming activity (Fig. 1B and Table 1), wild-type β-cat failed to generate transformed foci in RK3E. Based on our studies and those of others, β-cat mutants with missense mutations or deletions of the GSK3β phosphorylation sites can no longer be regulated appropriately by the APC/GSK3β complex. As a result, β-cat proteins with N-terminal missense mutations or deletions accumulate in the cytosol whereas wild-type β-cat does not (Figs. 1C and 1D). These observations on the stability of various β-cat proteins provide insights into why we found that overexpression of β-cat proteins with N-terminal mutations transformed RK3E cells but overexpression of wild-type β-cat did not.
Lines with stable expression of mutant β-cat manifest malignant growth properties.
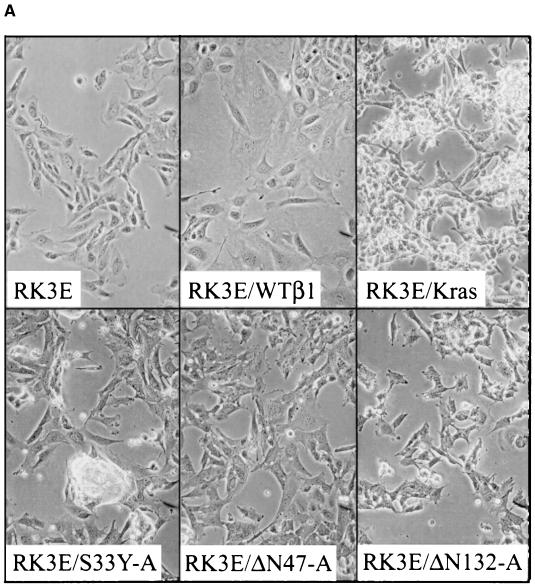
More than 25 stable cell lines were generated by selection and expansion of independent foci of transformed RK3E cells, including cells transformed by the S33Y mutant and N-terminally truncated β-cat proteins. Parental RK3E cells and a polyclonal RK3E line with stable overexpression of wild type β-cat (e.g., RK3E/WTβ1) displayed a flat, polygonal appearance (Fig. 3A). In contrast, β-cat-transformed RK3E cells had a different morphology, akin to that seen in a polyclonal RK3E line transformed by a K-Ras protein with a codon 12 mutation (RK3E/Kras) (Fig. 3A). Common features in β-cat-transformed lines included a more refractile appearance, an increased number of membrane extensions, and a tendency to form multicellular aggregates. Expression of the Flag epitope-tagged wild-type and mutated forms of β-cat was confirmed in the stable cell lines, and lines expressing mutated β-cat proteins had much higher expression of β-cat protein than did the RK3E/WTβ1 line (Fig. 3B and data not shown). All lines transformed by mutated β-cat had markedly elevated Tcf transcriptional activity relative to parental RK3E, RK3E/Kras, or RK3E/WTβ1 cells (Fig. 3C and data not shown). Like Ras-transformed RK3E cells, β-cat-transformed RK3E lines proliferated robustly under low-serum conditions (Fig. 3D). In contrast to mutant K-Ras, which conferred immediate growth in soft agar in transduced RK3E cells, mutated β-cat proteins failed to induce growth in soft agar directly upon their introduction into RK3E cells. Nevertheless, after initiation from transformed foci, all β-cat-transformed RK3E lines readily formed colonies in soft agar (Fig. 4 and data not shown). Several β-cat-transformed RK3E lines were tested and found to form rapidly progressive tumors in nude mice, with growth rates similar to those of K-Ras-transformed RK3E cells. In these experiments, all mice injected with 5 × 106 RK3E/S33Y-A (10 mice), RK3E/S33Y-D (10 mice), and RK3E/Kras (2 mice) cells had tumors ranging from 0.8 to 3.0 cm in diameter at 3 weeks, whereas no tumors were detected in mice injected with an identical number of RK3E parental cells (10 mice) during the 6-week monitoring period.
FIG. 3.
Altered phenotypic properties of stable RK3E cells transformed by mutated β-cat proteins. (A) Morphology of parental RK3E cells and a polyclonal RK3E line with overexpression of wild-type β-cat are shown in the top left and top center panels, respectively. A polyclonal RK3E line transformed by K-Ras (RK3E/Kras) is shown at the top right. Three different RK3E lines transformed by mutant β-cat are shown at the bottom. Magnification for all panels, ×200. (B) ECL-Western blot analysis of Flag-epitope tagged β-cat proteins in stable RK3E cell lines and the negative control RK3E parental line. The blot was stripped, and ECL-Western blot analysis with anti-actin antibody was carried out to control for loading of the lanes. (C) RK3E lines stably transformed by mutated β-cat proteins have markedly elevated Tcf transcription activity compared to control lines (parental RK3E, RK3E/Kras, or RK3E/WTβ1). The ratio of luciferase activities from a Tcf-responsive reporter (pTOPFLASH) and a control luciferase reporter gene construct (pFOPFLASH) was measured for each cell line 48 h after transfection. Mean values and standard deviations from three independent experiments are shown. (D) RK3E lines transformed by mutated β-cat proliferate in 0.5% serum. A total of 2 × 104 cells were seeded in 35-mm dishes in the presence of growth medium containing 10% fetal bovine serum. The following day, the medium was exchanged for medium containing 0.5% fetal bovine serum. Cell numbers were determined at specific time points after the switch to 0.5% serum. Values shown represent the means and standard deviations of triplicate experiments.
FIG. 4.
RK3E lines transformed by mutated β-cat form colonies in agar. Colony formation in soft agar was assessed for parental RK3E cells, a polyclonal RK3E line transformed by mutant K-Ras, and four representative β-cat-transformed lines. A total of 104 cells of each line were plated in 0.3% agar medium over agar underlayers. After 3 weeks, the dishes were stained with methylene blue and the colonies were photographed.
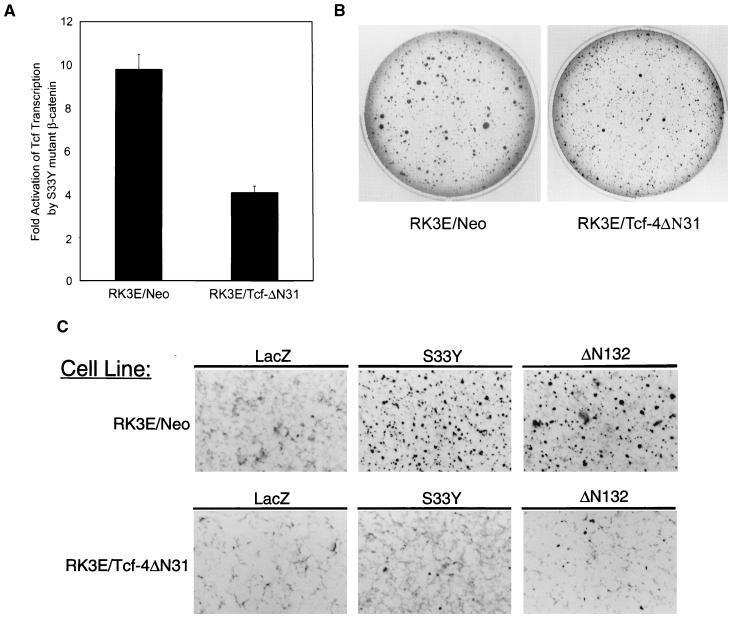
Tcf/Lef deregulation is required for β-cat transformation.
As noted above, all β-cat-transformed cell lines had markedly elevated Tcf transcriptional activity. Previous studies have shown that the Tcf N terminus is required for binding to β-cat and that Tcf mutant proteins lacking N-terminal sequences retain DNA binding activity but function in a dominant negative fashion (20). Hence, we sought to test the effects of such a dominant negative Tcf-4 mutant protein on β-cat-induced transformation of RK3E cells. We prepared an expression construct encoding a mutant Tcf-4 protein in which the N-terminal 31 aa were deleted, termed Tcf-4ΔN31. We then generated a polyclonal RK3E line with constitutive expression of the Tcf-4ΔN31 mutant protein, termed RK3E/Tcf-4ΔN31. The ability of the S33Y β-cat mutant protein to activate Tcf transcription was strongly inhibited in RK3E/Tcf-4ΔN31 cells compared to a polyclonal control RK3E line (RK3E/Neo) (Fig. 5A). The RK3E/Tcf-4ΔN31 cells were readily transformed by mutant K-Ras, although modest effects on colony size in agar were seen in some experiments (Fig. 5B). In contrast, the ability of the S33Y and ΔN132 mutant β-cat proteins to induce focus formation in the RK3E/Tcf-4ΔN31 line was nearly completely inhibited (Fig. 5C). These data establish that Tcf/Lef deregulation is required for RK3E transformation by mutant β-cat but not for transformation by mutant K-Ras.
FIG. 5.
Expression of a dominant negative Tcf-4 mutant protein, lacking the N-terminal 31 aa of Tcf-4 (i.e., Tcf-4ΔN31), inhibits transformation of RK3E by mutated β-cat but not mutated K-Ras. (A) Activation of Tcf transcription by the S33Y mutated form of β-cat is substantially inhibited in a G418-resistant polyclonal RK3E line expressing Tcf-4ΔN31 (RK3E/Tcf-4DN31) compared to a control G418-resistant RK3E line (RK3E/Neo). (B) K-Ras-mediated colony formation in soft agar is not inhibited by expression of the dominant negative Tcf-4 mutant. Following infection with the K-Ras retrovirus, colony formation assays in RK3E/Neo control and RK3E/Tcf-4DN31 cells were carried out as described in the legend to Fig. 4 and the text. (C) β-cat-induced focus formation in RK3E is inhibited by expression of a dominant negative Tcf-4 mutant. Focus formation assays in the control RK3E/Neo and the RK3E/Tcf-4DN31 lines was carried out with retroviruses expressing the S33Y and ΔN132 β-cat mutants.
Role of c-myc in β-cat transformation.
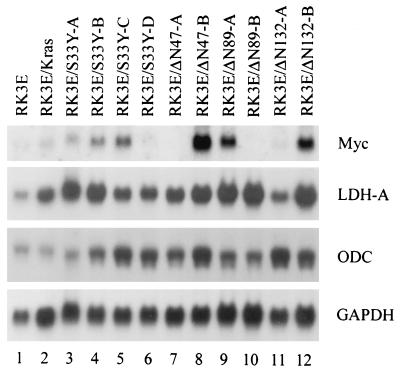
The c-MYC gene has recently been suggested to be a critical downstream target of the APC/β-cat/Tcf pathway in human colorectal cancer (12). We found that c-myc expression was transiently induced by about twofold in RK3E cells 4 to 6 days after infection of the cells with retroviruses encoding mutated β-cat, although the time course of c-myc induction was delayed relative to β-cat accumulation and Tcf transcriptional activation (data not shown). More significantly, we found that c-myc expression was not consistently increased in β-cat-transformed RK3E lines compared to the RK3E parental line or RK3E/Kras cells. Only 9 of 17 β-cat-transformed cell lines studied had elevated c-myc expression (Fig. 6 and data not shown). Furthermore, a direct comparison of β-cat-transformed RK3E lines with elevated c-myc expression to lines lacking increased c-myc expression revealed no differences in critical aspects of the transformed phenotype, such as growth in reduced serum (Fig. 3D), growth in soft agar (Fig. 4), and tumorigencity in mice (see above). Expression of presumptive c-Myc-regulated genes, such as those encoding lactate dehydrogenase A and ornithine decarboxylase (7), was elevated in many of the β-cat-transformed cell lines but was not consistently associated with c-myc levels (Fig. 6). The absence of elevated c-myc expression in roughly half of the β-cat-transformed lines argues that c-myc may not be a specific target gene in the APC/β-cat/Tcf pathway, particularly because all β-cat-transformed lines had grossly deregulated Tcf transcriptional activities, ranging from 30- to 700-fold higher than the levels seen in control lines (e.g., parental RK3E or RK3E/WTβ1 [Fig. 3C]).
FIG. 6.
c-myc expression is not uniformly activated in stable RK3E lines transformed by mutated β-cat. Northern blot studies of c-myc, candidate c-Myc-regulated genes (lactate dehydrogenase A [LDH-A] and ornithine decarboxylase [ODC]), and a loading control (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) were carried out on total RNA from parental RK3E cells, a polyclonal RK3E line transformed by activated K-Ras (RK3E/Kras), and 10 different RK3E lines transformed by mutated β-cat. All β-cat-transformed lines form colonies in soft agar, and several of the lines were tested and found to grow in nude mice (see the text for details).
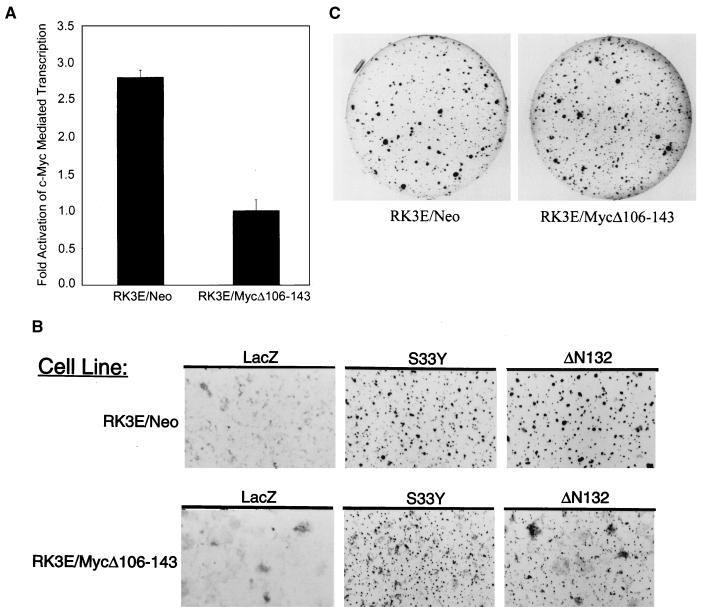
The lack of elevated c-myc expression in multiple independent β-cat-transformed RK3E lines provides clear, albeit correlative, data arguing against a critical role for c-myc activation in β-cat transformation. To further assess the role of c-myc in β-cat-mediated transformation of epithelial cells, we examined the ability of a dominant negative c-Myc mutant protein to inhibit transformation by β-cat. The dominant negative c-Myc mutant chosen for our studies, termed MycΔ106–143, lacks critical sequences in the amino-terminal c-Myc activation domain. This mutant protein has previously been used to define the role of c-Myc in transformation by the Abl protein (34). The MycΔ106–143 mutant retains the ability to oligomerize with Max and mediate DNA binding. However, because it fails to regulate transcription appropriately, expression of c-Myc target genes is altered. The MycΔ106–143 mutant is also inactive in several assays for functions of c-Myc unrelated to its ability to transcriptionally activate genes (44). A polyclonal RK3E line, termed RK3E/MycΔ106–143, with stable expression of the MycΔ106–143 mutant was generated and c-Myc-dependent transcription of a reporter gene construct containing c-Myc-responsive promoter elements (i.e., E-box elements) was largely abrogated (Fig. 7A). In support of the hypothesis that β-cat transformation of RK3E is independent of c-Myc, mutant β-cat proteins generated roughly equivalent numbers of foci in the RK3E/MycΔ106–143 line and the control RK3E/Neo line. In addition, we found that the stable introduction of the c-Myc dominant negative mutant (Δ106–143) construct into several β-cat-transformed RK3E lines, including lines with elevated c-myc expression as well as others lacking c-myc activation, failed to affect the ability of the cells to form colonies in agar (data not shown). We did note that the foci induced by β-cat were smaller in size in the RK3E/MycΔ106–143 line than in the control RK3E/Neo line (Fig. 7B). The specificity of the effect of the c-Myc mutant on β-cat-induced foci was demonstrated by the fact that mutant K-ras had essentially identical transforming activity in both the RK3E/MycΔ106–143 and RK3E/Neo lines (Fig. 7C). Therefore, while our findings suggest that β-cat-induced neoplastic transformation of RK3E is independent of c-Myc, increased c-myc expression may contribute to the phenotype of some β-cat-transformed lines.
FIG. 7.
c-myc activation is not required for transformation by mutated β-cat, but increased c-myc expression can contribute to a more aggressive phenotype. (A) A polyclonal RK3E line with stable expression of a dominant negative c-Myc mutant protein (i.e., MycΔ106–143) was generated and termed RK3E/MycΔ106–143. Activation of a c-Myc-responsive reporter (pGLDH637Luc) is substantially inhibited in the RK3E/MycΔ106–143 line. (B) Mutated β-cat proteins induce focus formation in RK3E/MycΔ106–143, but the size of the foci is reduced compared to those induced by mutated β-cat in the control RK3E/Neo line. (C) Colony formation in soft agar following infection with a K-Ras retrovirus is not inhibited in RK3E/MycΔ106–143 cells.
DISCUSSION
Our decision to pursue detailed studies of the means by which mutated β-cat proteins contribute to neoplastic transformation was motivated by recent evidence that defects in β-cat regulation are found in nearly all colon cancers and a subset of other cancers (9, 20, 22, 25, 26, 32, 36, 47). In normal epithelial cells, the bulk of β-cat is complexed at the plasma membrane with the E-cad cell adhesion molecule (4, 21, 41). The abundance of β-cat in the cytosol and nucleus is regulated by a multiprotein complex containing the GSK3β and APC proteins (24, 41). Activation of the Wnt pathway inhibits GSK3β, thus inhibiting β-cat degradation. In colon cancer, regulation of the cytosolic and nuclear pools of β-cat is most often disrupted as a result of APC inactivation (25, 36). In some colon and other cancers, missense mutations or deletions of presumptive GSK3β phosphorylation sites in the β-cat N terminus render the protein resistant to regulation by the GSK3β/APC/axin complex (9, 22, 25, 32, 36). Regardless of the underlying cause, the consequences appear to be increased levels of β-cat in the cytosol and nucleus, constitutive interaction of β-cat with Tcf/Lef transcription factors, and activation of Tcf/Lef-regulated genes (25, 36, 41).
Here, we have shown that mutated β-cat proteins, harboring either a human cancer-derived missense mutation in a presumptive GSK3β phosphorylation site (S33Y) or deletions of up to the first 132 N-terminal amino acids will induce neoplastic transformation of RK3E, an E1A-immortalized epithelial cell line derived from neonatal rat kidney. The reason why mutated β-cat proteins transformed RK3E cells but not the NIH 3T3, IEC-18, or 1811 lines is not known. In contrast to mutated β-cat, activated Ras proteins readily transform NIH 3T3 and IEC-18. Further work is required to determine whether the transforming activity of mutated β-cat in RK3E is attributable to the presence of the adenovirus E1A protein, the specific constellation of other gene defects in the line, or another aspect of the phenotype of RK3E, such as its particular cell of origin. NIH 3T3 cells may not be transformed by mutated β-cat, because they lack critical targets of β-cat action, such as Lef-1 or certain Tcfs (13, 37).
Mutated β-cat proteins capable of transforming RK3E cells were able to activate a Tcf reporter gene, although some N-terminally truncated β-cat mutants (e.g., ΔN132) had only a modest ability to activate Tcf transcription in our transient-transfection assay. The reduced transcriptional activity of N-terminally truncated forms, particularly the ΔN132 form, may be attributable, at least in part, to the fact that the N-terminal region of β-cat plays a role in activation of Tcf transcription (13). It is also possible that the transient-transfection assay of Tcf transcriptional activity does not entirely reflect the biological activity of the β-cat mutants. The apparent common theme we observed among β-cat proteins with transforming activity was that the mutated proteins accumulated to higher levels in the cytosol than did wild-type β-cat. As suggested previously, this characteristic is most probably due to the inability of APC and GSK3β to regulate N-terminally mutated forms of β-cat. Wild-type β-cat fails to transform RK3E even when overexpressed, because cells with wild-type APC and GSK3β function can appropriately regulate the abundance of the wild-type β-cat protein. Our observation that the S33Y mutant form of β-cat accumulated to higher levels in the cytosol in transient-transfection assays and was more active in the focus formation assay than were the N-terminally truncated forms is consistent with the fact that β-cat proteins with single-amino-acid substitutions and single-amino-acid deletions in the GSK3β consensus are more frequently found in colon cancer than are proteins with larger N-terminal truncations (25, 36). Nonetheless, after expansion of the β-cat-transformed foci into stable cell lines, regardless of whether transformation of the line was initiated by the S33Y mutant β-cat protein or an N-terminal truncation (e.g., ΔN47 or ΔN132), the stably transformed RK3E lines expressed high levels of the mutant β-cat protein and displayed essentially uniform growth and tumorigenicity properties. These findings are consistent with the notion that other secondary genetic and epigenetic changes collaborate with the mutated β-cat proteins to induce the full neoplastic phenotype in RK3E. The reduced transcriptional and focus-forming activities of the N-terminally truncated forms of β-cat compared to the S33Y mutant may be of consequence only in initiation of RK3E neoplastic transformation, not in its maintenance.
Studies of the β-cat domains required for the transforming activity of the S33Y mutant protein revealed that armadillo repeats 3 to 8 (aa 218 to 467) and the C-terminal 85 aa were particularly critical, although deletion of N-terminal aa 48 to 217 also had a clear effect on the activity of the S33Y mutant protein. The requirement of armadillo repeats 3 to 8 implies that interaction of mutated β-cat with Tcf/Lef transcription factors is required for transformation. The C-terminal 85 aa of β-cat have previously been implicated in transcriptional activation (13, 39), as have sequences at the N terminus (13), indicating that β-cat transformation is probably dependent on the transcriptional activation of Tcf/Lef-regulated genes. Other data support the view that activation of Tcf/Lef transcription is critical in β-cat-induced transformation. First, all cell lines stably transformed by mutated β-cat proteins displayed markedly elevated Tcf transcription activity, ranging from 30- to 700-fold higher than that of parental RK3E or K-Ras-transformed RK3E. Second, the polyclonal RK3E/Tcf-4ΔN31 line with stable expression of a dominant negative Tcf-4 mutant protein was resistant to transformation by mutated β-cat but could be transformed by activated K-Ras. Finally, recent studies indicated that a chimeric protein in which Lef-1 sequences are fused to potent transcriptional activation domains will transform chicken embryo fibroblasts (3).
The identity of Tcf/Lef-regulated genes in mammalian cells, particularly of genes responsible for neoplastic transformation, is poorly understood. Recently, He et al. (12) found that c-MYC expression was suppressed in a colorectal cancer cell line with an endogenous APC gene defect, following induction of an exogenous APC gene. The critical elements in the c-MYC promoter responsible for APC-mediated suppression included Tcf-4 binding sites. Wild-type APC suppressed the activity of heterologous reporter gene constructs containing the Tcf-4 regulatory elements from the c-MYC promoter, and mutated β-cat strongly activated gene expression from constructs containing the regulatory elements. Thus, the data of He et al. (13) imply that c-MYC is a critical downstream target of the APC/β-cat/Tcf pathway in cancer cells.
Our studies failed to provide evidence that c-myc was a critical target in β-cat-mediated neoplastic transformation of RK3E. In transient-transfection assays in RK3E, we found that mutated β-cat proteins could modestly increase c-myc gene expression, although the time course of c-myc activation was delayed relative to accumulation of mutant β-cat proteins and Tcf activation. More significantly, roughly half of the RK3E lines stably transformed by mutant β-cat had no detectable increase in c-myc gene expression relative to control RK3E lines, even though all β-cat-transformed lines had markedly elevated, constitutive Tcf transcription activity. Expression of c-myc was increased, relative to that in parental RK3E or RK3E/Kras cells, in several transformed lines with the highest Tcf transcriptional activity. Nevertheless, a clear correlation between Tcf transcriptional activity and c-myc expression was not observed in the β-cat-transformed lines. As such, it is possible that increased c-myc expression in some RK3E lines simply reflects the transformed phenotype. Additional data on the role of c-myc in β-cat transformation was obtained in studies in which a polyclonal RK3E line with stable expression of a dominant negative c-Myc mutant protein (i.e., RK3E/MycΔ106–143) could still be transformed by mutated β-cat proteins. The presence of the E1A protein in the RK3E cells may have substituted for c-Myc in neoplastic transformation by β-cat. However, while E1A might substitute for the functional requirement for c-Myc in β-cat transformation of RK3E, if the c-myc gene were truly a direct transcriptional target of the Tcf/β-cat complex, then we should have observed uniformly elevated c-myc gene expression levels in all of the β-cat-transformed RK3E lines. Because we failed to obtain such results, our findings imply that c-myc is not a common downstream target of the APC/β-cat/Tcf pathway in all neoplastic cells. The studies described here illustrate the value and utility of the RK3E system for evaluation of effector proteins and candidate target genes in β-cat transformation of epithelial cells. Future research with the RK3E system should offer additional insights into the means through which defects in β-cat regulation contribute to cancer.
ACKNOWLEDGMENTS
This work was supported by Deutsche Forschungsgemeinschaft grant KO1826/1 (F.T.K.) and NIH grants CA70097 (E.R.F.) and CA57341 (C.V.D.).
We thank J. M. Ruppert for providing RK3E cells, B. Vogelstein and K. W. Kinzler for providing the Tcf-4 cDNA and the pTOPFLASH and pFOPFLASH reporter constructs, G. P. Nolan for providing the Phoenix retroviral system, G. Nabel for providing the pPGS-CMV-CITE-neo vector, K. R. Cho for providing the 1811 cells, and K. R. Cho and S. Weiss for critical comments on the manuscript.
REFERENCES
- 1.Aberle H, Butz S, Stappert J, Weissig H, Kemler R, Hoschuetzky H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J Cell Sci. 1994;107:3655–3663. doi: 10.1242/jcs.107.12.3655. [DOI] [PubMed] [Google Scholar]
- 2.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki M, Hecht A, Kruse U, Kemler R, Vogt P K. Nuclear endpoint of Wnt signaling: neoplastic transformation induced by transactivating lymphoid-enhancing factor 1. Proc Natl Acad Sci USA. 1999;96:139–144. doi: 10.1073/pnas.96.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth A I, Nathke I S, Nelson W J. Cadherins, catenins and APC protein: interplay between cytoskeletal complexes and signaling pathways. Curr Opin Cell Biol. 1997;9:683–690. doi: 10.1016/s0955-0674(97)80122-6. [DOI] [PubMed] [Google Scholar]
- 5.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of β-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 6.Behrens J, Jerchow B A, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 7.Dang C V. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freshney R I. Culture of animal cells: a manual of basic technique. 3rd ed. New York, N.Y: Wiley-Liss, Inc.; 1994. pp. 166–169. [Google Scholar]
- 9.Fukuchi T, Sakamoto M, Tsuda H, Maruyama K, Nozawa S, Hirohashi S. β-Catenin mutation in carcinoma of the uterine endometrium. Cancer Res. 1998;58:3526–3528. [PubMed] [Google Scholar]
- 10.Gat U, DasGupta R, Degenstein L, Fuchs E. De novo hair follicle morphogenesis and hair tumors in mice expressing a truncated β-catenin in skin. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 11.Hart M J, de los Santos R, Albert I N, Rubinfeld B, Polakis P. Downregulation of β-catenin by human Axin and its association with the APC tumor suppressor, β-catenin, and GSK3β. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 12.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 13.Hsu S-C, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with β-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber O, Bierkamp C, Kemler R. Cadherins and catenins in development. Curr Opin Cell Biol. 1996;8:685–691. doi: 10.1016/s0955-0674(96)80110-4. [DOI] [PubMed] [Google Scholar]
- 15.Hülsken J, Birchmeier W, Behrens J. E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J Cell Biol. 1994;127:2061–2069. doi: 10.1083/jcb.127.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh K, Krupnik V E, Sokol S Y. Axis determination in Xenopus involves biochemical interactions of axin, glycogen synthase kinase 3 and β-catenin. Curr Biol. 1998;8:591–594. doi: 10.1016/s0960-9822(98)70229-5. [DOI] [PubMed] [Google Scholar]
- 18.Kaur P, McDougall J K. HPV-18 immortalization of human keratinocytes. Virology. 1989;173:302–310. doi: 10.1016/0042-6822(89)90247-x. [DOI] [PubMed] [Google Scholar]
- 19.Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, Kikuchi A. Axin, a negative regulator of the wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of β-catenin. J Biol Chem. 1998;273:10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 20.Korinek V, Barker M, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 21.Mareel M, Boterberg T, Noe V, Van Hoorde L, Vermeulen S, Bryneel E, Bracke M. E-cadherin/catenin/cytoskeleton complex: a regulator of cancer invasion. J Cell Physiol. 1997;173:271–274. doi: 10.1002/(SICI)1097-4652(199711)173:2<271::AID-JCP34>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 22.Miyoshi Y, Iwao K, Nagasawa Y, Aihara T, Sasaki Y, Imaoka S, Murata M, Shimano T, Nakamura Y. Activation of the β-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res. 1998;58:2524–2527. [PubMed] [Google Scholar]
- 23.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 24.Moon R T, Miller J R. The APC tumor suppressor protein in development and cancer. Trends Genet. 1997;13:256–258. doi: 10.1016/s0168-9525(97)01196-7. [DOI] [PubMed] [Google Scholar]
- 25.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 26.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular β-catenin levels by the adenomatous polyposis (APC) tumor suppressor protein. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura T, Hamada F, Ishidate T, Anai K, Kawahara K, Toyoshima K, Akiyama T. Axin, an inhibitor of the Wnt signaling pathway, interacts with β-catenin, GSK-3β and APC and reduces the β-catenin level. Genes Cells. 1998;3:395–403. doi: 10.1046/j.1365-2443.1998.00198.x. [DOI] [PubMed] [Google Scholar]
- 28.Nusse R, Varmus H E. Wnt genes. Cell. 1992;69:1073–1087. doi: 10.1016/0092-8674(92)90630-u. [DOI] [PubMed] [Google Scholar]
- 29.Orsulic S, Peifer M. An in vivo structure-function study of armadillo, the β-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quaroni A, Wands J, Trelstad R L, Isselbacher K J. Epitheloid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubinfeld B, Souza B, Albert I, Munemitsu S, Polakis P. The APC protein and E-cadherin form similar but independent complexes with α-catenin, β-catenin, and plakoglobin. J Biol Chem. 1995;270:5549–5555. doi: 10.1074/jbc.270.10.5549. [DOI] [PubMed] [Google Scholar]
- 32.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of β-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 33.Ruppert J M, Vogelstein B, Kinzler K W. The zinc finger protein GLI transforms primary cells in cooperation with adenovirus E1A. Mol Cell Biol. 1991;11:1724–1728. doi: 10.1128/mcb.11.3.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawyers C L, Callahan W, Witte O N. Dominant negative MYC blocks transformation by ABL oncogenes. Cell. 1992;70:901–910. doi: 10.1016/0092-8674(92)90241-4. [DOI] [PubMed] [Google Scholar]
- 35.Shim H, Dolde C, Lewis B C, Wu C S, Dang G, Jungmann R A, Dalla-Favera R, Dang C V. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sparks A B, Morin P J, Vogelstein B, Kinzler K W. Mutational analysis of the APC/β-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- 37.Tetsu O, McCormick F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 38.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor-α enhancer function. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 39.van de Wetering M, Davallo R, Booijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 40.Whitehead I, Kirk H, Kay R. Expression cloning of oncogenes by retroviral transfer of cDNA libraries. Mol Cell Biol. 1995;15:704–710. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willert K, Nusse R. β-catenin: a key regulator of Wnt signaling. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 42.Winston J T, Strack P, Beer-Romano P, Chu C Y, Elledge S J, Harper J W. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong M H, Rubinfeld B, Gordon J I. Effects of forced expression of an NH2-terminal truncated β-catenin on mouse intestinal epithelial homeostasis. J Cell Biol. 1998;141:765–777. doi: 10.1083/jcb.141.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao Q, Claassen G, Shi J, Adachi S, Sedivy J, Hann S R. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 1998;12:3803–3808. doi: 10.1101/gad.12.24.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yost C, Torres M, Miller J R, Huang E, Kimelman D, Moon R T. The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 46.Young C S, Kitamura M, Hardy S, Kitajewski J. Wnt-1 induces growth, cytosolic β-catenin, and Tcf/Lef transcriptional activation in Rat-1 fibroblasts. Mol Cell Biol. 1998;18:2474–2485. doi: 10.1128/mcb.18.5.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zurawel R H, Chiappa S A, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic β-catenin mutations. Cancer Res. 1998;58:1344–1347. [PubMed] [Google Scholar]