Abstract
Higher-order organisms possess information processing capabilities that are only made possible by their biological complexity. Emerging evidence indicates a critical role for regulatory RNAs in coordinating many aspects of cellular function that are directly involved in experience-dependent neural plasticity. Here, we focus on a structurally distinct class of RNAs known as circular RNAs. These closed loop, single-stranded RNA molecules are highly stable, enriched in the brain, and functionally active in both healthy and disease conditions. Current evidence implicating this ancient class of RNA as a contributor toward higher-order functions such as cognition and memory is discussed.
Keywords: circRNA, translation, RNA transfer, learning, memory, synapse
Introduction
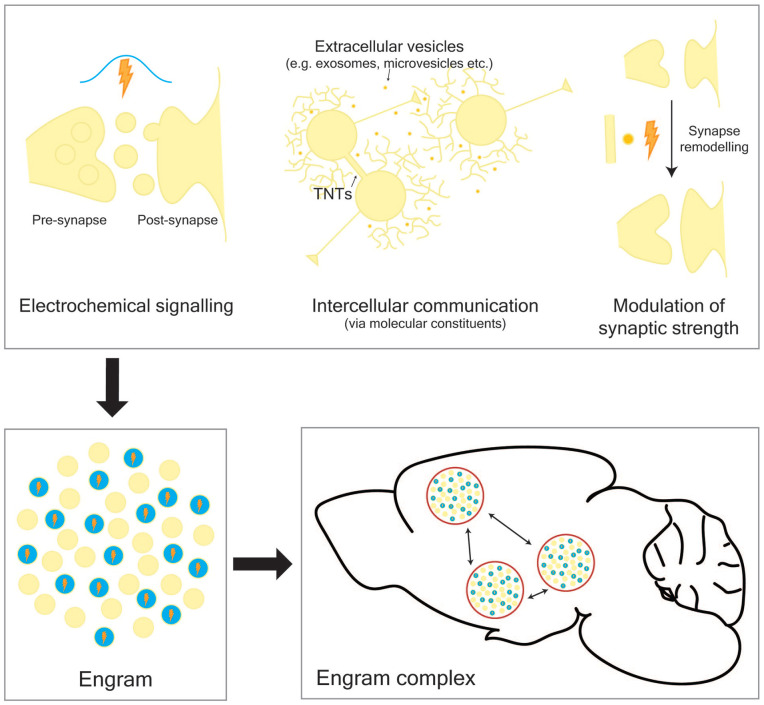
In general terms, learning can be thought of as the ability to adapt in response to both external and internal experience with memory being the long-lasting representation of these learned experiences (Kandel and others 2014; Kukushkin and Carew 2017). During learning, electrochemical, physical, and molecular pathways intersect spatiotemporally in order to form a network of neurons within the brain responsible for storing new memories (Fig. 1). This network of neurons is best described by the conceptual theory of the “engram,” which was first proposed by Richard Semon in 1904 to describe a population of cells that undergo long-lasting chemical and/or physical changes during learning (Schacter and others 1978; Semon 1904). This population of cells can be reactivated, or retrieved, following presentation of only a fraction of the cues present in the original experience. More recently, support has been growing for the idea that memory is supported by multiple cell ensembles that communicate across several brain regions, and together comprise an “engram complex” (Josselyn and Tonegawa 2020). In order to fully understand how memories are formed, the mechanisms of communication within the brain that coordinate and underpin these ensembles must be deciphered.
Figure 1.
Memories are stored in engram complexes that comprise multiple engrams distributed across the brain. During learning, multiple pathways intersect both spatially and temporally to form an engram, a network of connected cells that store new memories. These pathways include electrochemical signaling, intercellular transfer of molecules, and physical alteration to synaptic neural connections. TNTs, tunneling nanotubes.
During a sensory event, both intercellular and intracellular mechanisms of communication contribute toward translating this experience into long-lasting alterations in cellular behavior. Upon stimulation, the following trajectory of events is known to occur: (1) neurotransmitter release at excitatory glutamatergic synapses activates N-methyl-d-aspartate (NMDA) receptors, which leads to influx of Ca++ into the cell, (2) Ca++ influx triggers a variety of downstream signalling and transcription factor pathways, and (3) these Ca++-induced signaling pathways activate a program of gene expression (i.e. activity-dependent gene expression) that is directly required for experience-dependent changes in synaptic plasticity (West and Greenberg 2011).
Rapid, stimulus-induced modification of existing prestimulus transcription factors, such as CREB, mediates the first wave of activity-dependent “immediate early gene” (IEG) expression. IEGs are classically defined as a set of stimulus-induced genes that do not require de novo protein synthesis and are rapidly transcribed following stimulus onset (Sheng and Greenberg 1990). A number of IEGs are also transcription factors and their expression leads to a second delayed wave of transcription, which comprises genes that are regulated in a cell-type- and stimulus-specific manner. The products of these two waves of transcription lead to changes in processes such as postsynaptic receptor expression and dendritic spine formation that alter the strength of synaptic connections and the underlying neural circuitry (Loebrich and Nedivi 2009).
Importantly, activity-dependent gene expression is not limited to the production of messenger RNAs (mRNAs) that act as templates for protein translation. In fact, approximately 98% of the output from the human genome does not code for proteins and is classified as “non-coding” (Mattick 2004). These non-coding RNAs perform regulatory roles that are now known to be critical for driving changes in brain function and behavior (Alberini and Kandel 2015; Mercer and others 2008). Moreover, throughout evolution, the number and percentage of non-coding RNAs within the genome has increased in proportion to organismal complexity whereas the number of protein-coding genes remains about the same (Mattick 2004). The implication of this is that higher-order cognitive abilities may be the direct result of an increase in regulatory architecture rather than effector number.
In a neuron, RNA can act within the nucleus to coordinate gene expression or localize to the synapse to mediate rapid, activity-dependent alterations to local translation and synaptic plasticity (Holt and others 2019; Leighton and others 2018). Importantly, it has recently been shown that RNA does not function exclusively within a single neuron but can also transfer between cells, which enables both locally and distally connected cells to coordinate their behavior over timescales beyond that of the triggering stimulus (Belting and Wittrup 2008; Budnik and others 2016; Dinger and others 2008). However, the activity-dependent processes that occur during learning are short-lived. How then are memories able to persist for longer than a couple of days? One answer to this question may lie in an evolutionarily old class of RNA known as circular RNAs.
Circular RNAs (circRNAs) are closed loop single-stranded RNA molecules that are highly stable and enriched in the brain. As a result of their stability, it has been speculated that circRNAs may serve as “memory molecules” and could potentially transfer information between cells, given that they resemble small virus-like particles called viroids and other circular forms of nucleic acid (e.g., plasmids) that are known to do this (Lasda and Parker 2014). Here, we will review the current evidence implicating circRNAs with the biological processes that underlie neural plasticity, learning, and memory, as well as how perturbations to circRNA regulation and function contribute to neurodegenerative disease and psychiatric disorders.
What Are circRNAs and How Are They Detected?
CircRNAs, which comprise a structurally distinct class of RNA, consist of closed loops of single-stranded RNA molecules that are highly abundant in the brain and enriched within synapses (Rybak-Wolf and others 2015; You and others 2015). As a result of their unique structure, circRNAs are resistant to exonuclease-mediated RNA degradation and are long-lived (Jeck and others 2013). They were first visualised in the cytoplasm of HeLa cells by electron microscopy (Hsu and Coca-Prados 1979). However, until recently, circRNAs were thought to be artefacts of splicing or rare oddities derived from only a few genes (Capel and others 1993; Cocquerelle and others 1993; Zaphiropoulos 1996). With the advent of high-throughput sequencing, thousands of circRNAs have now been identified, many of which are highly conserved and are regulated separately from their linear counterparts (Gokool and others 2020; Ragan and others 2019; Rybak-Wolf and others 2015; Zhou and others 2017). Similar to other classes of RNA, circRNAs exhibit regions of secondary structure (i.e., 16-26 base pairs of double-stranded RNA duplexes) and can be chemically modified (e.g., by N6-methyladenosine, m6A) (Chen and others 2019; Liu and others 2019; Yang and others 2017; Zhou and others 2017).
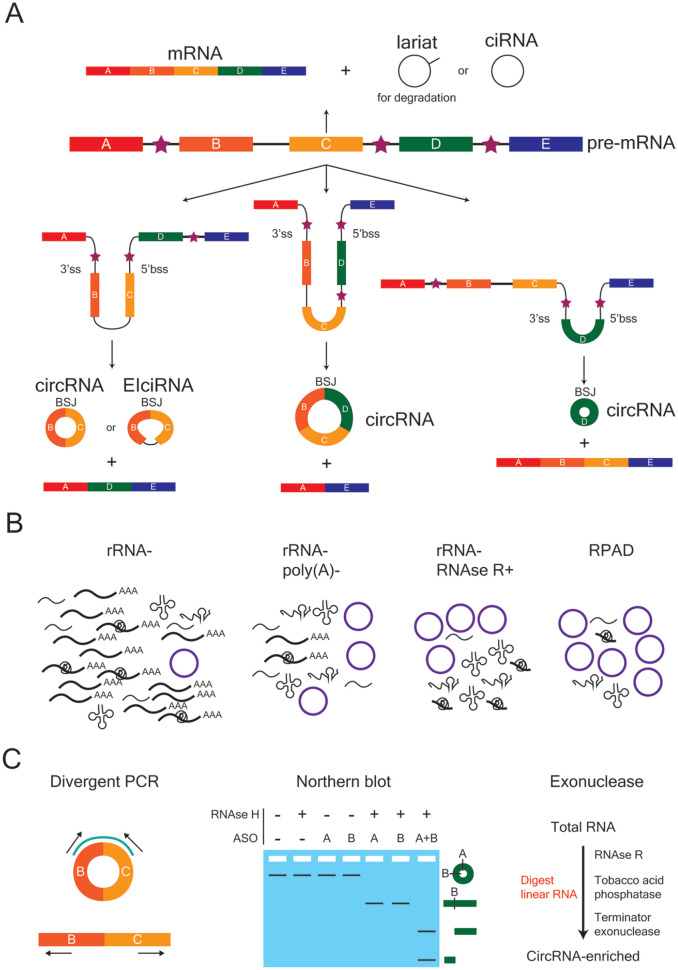
CircRNAs are bioinformatically identified by aligning sequencing reads to the backsplice junction (BSJ), which occurs at the site where a downstream 5′bss (backsplice site) and 3′ss (splice site) are covalently joined (Fig. 2A) (Gao and Zhao 2018; Jeck and Sharpless 2014; Szabo and Salzman 2016). Traditionally, circRNAs were detected by ribosomal RNA (rRNA) depletion followed by digestion with ribonuclease R (RNAse R), an enzyme that digests linear RNAs at their 3′ end (Fig. 2B and C). Newer methods such as RPAD (RNAse R treatment, polyadenylation, and poly(A)+ depletion) (Pandey and others 2019) offer an improvement by polyadenylating all digested RNAs with a 3′ end (i.e., non-circular RNA) before performing a poly(A)+/− selection to enrich for non-polyadenylated RNA (i.e., circular RNA). Multiple validation strategies are then employed to verify the circularity of bioinformatically predicted circRNA targets, including, divergent polymerase chain reaction (PCR), Sanger sequencing, exonuclease treatment (e.g., RNAse R, tobacco acid phosphatase, and terminator exonuclease), and northern blot (Fig. 2C) (Jeck and Sharpless 2014).
Figure 2.
Biogenesis, enrichment, and validation of circular RNAs (circRNAs). (A) CircRNAs can be exon-only (circRNA), intron-only (ciRNA) or a mixture of both (EIciRNA). The backsplice junction (BSJ) is the site where the 5′ back-splice site (5′bss) and 3′ splice site (3′ss) of a linear strand of RNA are covalently joined. Exons are depicted as rectangular boxes, lettered A to E, whereas introns are represented by the black lines between exons. Stars denote the introns that flank a potential BSJ, which can contain either repetitive or non-repetitive sequences that promote circularization. (B) Several enrichment strategies can be employed to increase the fraction of circRNAs within a given RNA population in order to improve their bioinformatic detection following sequencing. From left to right, the enrichment strategies are rRNA depletion (rRNA−), rRNA depletion and removal of poly(A)+ sequences (rRNA−, poly(A)−), rRNA depletion and RNAse R treatment (rRNA−, RNAse R+), and RNAse R treatment followed by polyadenylation and poly(A)+ depletion (RPAD). (C) Strategies used to verify the circularity of a bioinformatically predicted circRNA. Divergent polymerase chain reaction (PCR) uses outward-facing primers to specifically amplify and detect the region around the BSJ of a predicted circRNA target. These primers are not able to generate a PCR product from the linear RNA of the same gene. A northern blot can be used in combination with anti-sense oligonucleotide (ASO) probes (A, B) and RNAse H treatment to verify circularity. Exonuclease treatment with enzymes that digest linear RNA, followed by divergent PCR, is also commonly used.
How Are circRNAs Regulated?
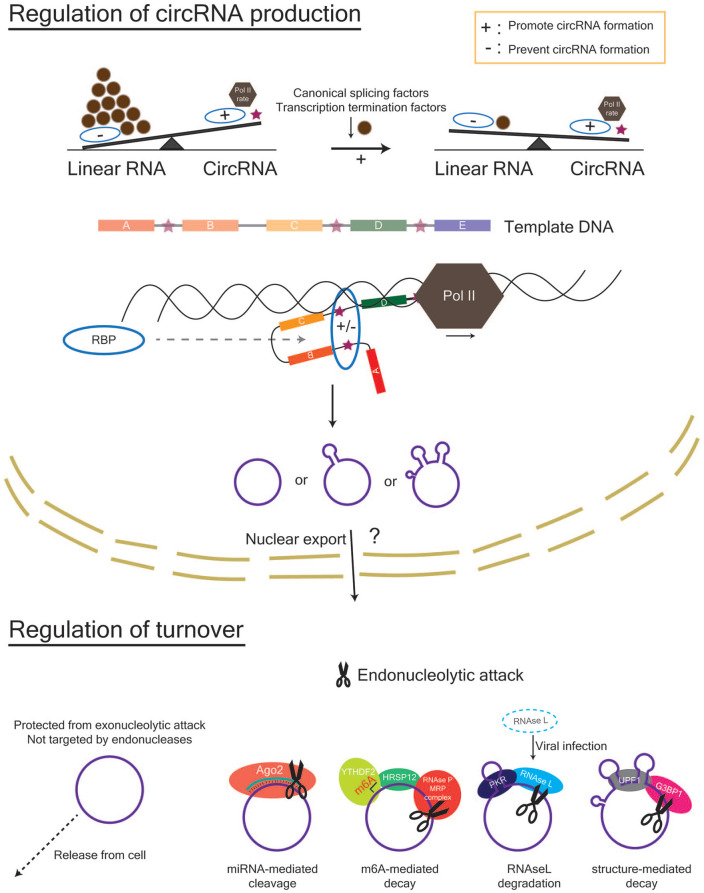
Three different types of eukaryotic circRNAs have been identified (Fig. 2A). “CircRNA” is commonly used to refer to exons that are backspliced from a linear transcript. On occasion, introns are also retained between exons, with this subset of circRNAs referred to as exon-intron circRNAs (EIciRNAs) (Li and others 2015). Finally, circular intronic RNAs (ciRNAs) are formed from intron lariats that have escaped debranching (Zhang and others 2013). CircRNA formation can be facilitated by intronic complementary sequences (ICSs) that flank the putative BSJ and bring it together (Jeck and others 2013; Zhang and others 2014). Both repetitive (e.g., short interspersed nuclear elements such as Alu) and non-repetitive sequences can promote base pairing (Jeck and others 2013). CircRNA production is known to occur both co- and post-transcriptionally (Ashwal-Fluss and others 2014; Zhang and others 2016; Zhang and others 2020), is reliant on canonical splice signals (Starke and others 2015), and can be increased by limiting the pre-mRNA processing machinery (e.g. core spliceosome factors, transcription termination factors) that exists to promote mRNA production (Fig. 3) (Liang and others 2017; Wang and others 2019). Furthermore, the rate of Pol II–mediated transcription can also affect the composition and fraction of circRNAs that are produced during host gene transcription (Ragan and others 2019). In particular, accentuated differences in the Pol II transcription rate between introns and exons (introns > exons) are observed in circRNA host genes, which are hypothesized to promote intronic base pairing between non-sequential introns (backsplicing) rather than within the same intron (linear splicing).
Figure 3.
Regulation of circRNA abundance in the nucleus and cytoplasm. Within the nucleus, the factors that affect the rate and success of circRNA formation include the rate of Pol II transcription, the amount of canonical splicing factors and transcription termination factors, and the presence of RBPs that can bind to sequences within the flanking introns (indicated by stars) of a potential circRNA. Within the cytoplasm, circRNAs can be removed by cellular release via extracellular vesicles or degradation via endonucleolytic attack. Mechanisms for endonucleolytic attack include (1) miRNA-mediated cleavage by Ago2, (2) RNAse L–mediated degradation following viral infection in immune cells, (3) m6A-mediated decay via YTHDF2- HRSP12-RNAseP/MRP interactions, and 4) structure-mediated decay via G3BP1 and UPF1. Ago2, Argonaute 2; G3BP1, G3BP stress granule assembly factor 1; HRSP12, heat-responsive protein 12; m6A, N6-methyladenosine; MRP, multidrug resistance protein; Pol II, polymerase II; RBP, RNA-binding protein; UPF1, UP-Frameshift-1; YTHDF2, YTH N6-methyladenosine RNA binding protein 2.
Trans factors, such as RNA-binding proteins (RBPs), can bind to ICSs (e.g., inverted repeated Alus) and help to facilitate or disrupt base pairing and subsequent covalent bond formation. For instance, circRNA formation is promoted by the immune factors NF90 and/or NF110 (Li and others 2017), whereas ADAR1 (Rybak-Wolf and others 2015) and DHX9 (Aktaş and others 2017) disrupt circRNA formation by RNA editing, which alters base pairing affinity, and unwinding of RNA pairs, respectively. The splicing factors Quaking and Muscleblind also facilitate circRNA formation by recognising and binding their motifs within flanking introns (Ashwal-Fluss and others 2014; Conn and others 2015). Once a circRNA is produced, its high stability leads to its accumulation within cells, particularly those that are non-dividing (e.g., neurons in the brain) (Rybak-Wolf and others 2015; Zhang and others 2016). Current methods to actively remove circRNAs from the cell include endonucleolytic attack and cellular release via extracellular vesicles (Fig. 3). Endonucleolytic attack occurs in the cytoplasm and can be achieved by (1) microRNA (miRNA)-mediated cleavage by Ago2, (2) RNAse L–mediated degradation following viral infection in immune cells, (3) m6A-mediated decay via YTHDF2-HRSP12-RNAseP/MRP interactions, and (4) structure-mediated decay via G3BP1 and UPF1 (Fischer and others 2020; Guo and others 2020; Hansen and others 2011; Liu and others 2019; Park and others 2019). Furthermore, Drosophila GW182 and its human homologues are involved in circRNA degradation, which is mediated via its Mid domain and not its Ago-binding domain or p-body localisation signal (Jia and others 2019).
Another aspect of circRNA regulation to consider is their localisation. Typically, intron-containing circRNAs localise to the nucleus whereas exon-containing circRNAs are found in the cytoplasm and synapse. The nuclear export of circRNAs is known to be length-dependent (Huang and others 2018a); however, the mechanisms for the retention of intron-containing circRNAs in the nucleus vs exclusion of exon-containing circRNAs are still unclear. Thus, a more thorough understanding of the mechanisms that underlie circRNA biogenesis and regulation in different cellular compartments is required in order to manipulate the function of these RNAs.
Functions of circRNAs
miRNA Sponge
Despite only recently being recognized as a functionally relevant class of RNA, it is evident that circRNAs are incredibly diverse and can perform a wide range of functions (Fig. 4). The best understood function of circRNAs is that of a miRNA sponge. miRNAs are a class of small non-coding RNAs (~22 nucleotides) that regulate the transcriptome by post-transcriptional silencing that is mediated by target recognition of complementary base pairs in mRNA. miRNAs are spatiotemporally expressed in the brain and have been implicated in learning and memory (Bredy and others 2011).
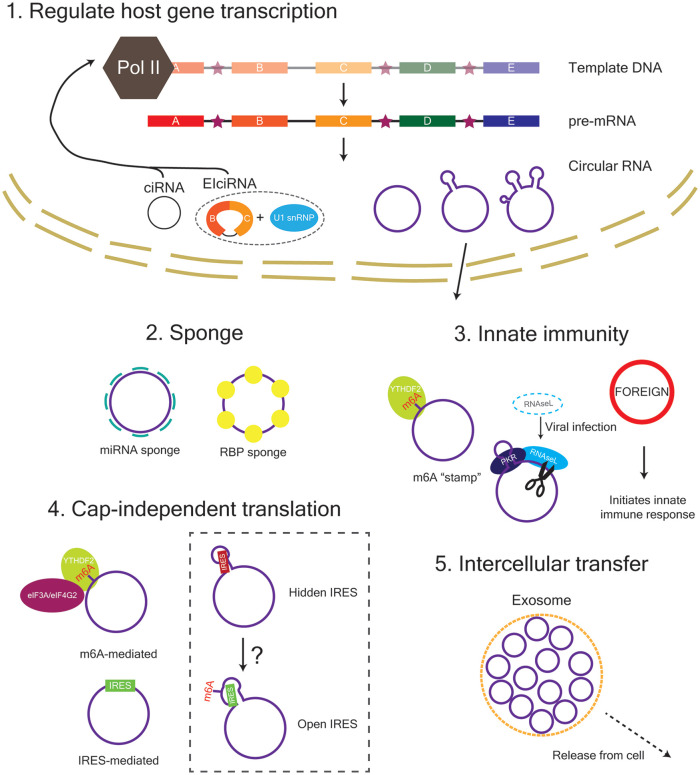
Figure 4.
The function of circRNAs within the cell. ciRNA and EIciRNA are retained in the nucleus and can act to regulate their host gene transcription. Exon-containing circRNAs are exported from the nucleus in a length-dependent manner and can function as a sponge for miRNAs and RNA-binding-proteins (RBPs) as well as a template for translation. CircRNAs are also involved in innate immunity and can be transferred between cells via exosomes. eIF3A, eukaryotic translation initiation factor 3 subunit A; eIF4G2, eukaryotic translation initiation factor 4 gamma 2; IRES, internal ribosome entry site; m6A, N6-methyladenosine; PKR, protein kinase R; YTHDF2, YTH N6-methyladenosine RNA binding protein 2.
The first and most well-studied circRNA implicated in brain function, CDR1as, is a miRNA sponge that forms part of a non-coding RNA regulatory network (long non-coding RNA (lncRNA)-miRNA-circRNA) that acts to regulate neural activity (Kleaveland and others 2018; Piwecka and others 2017). CDR1as contains over 70 partial miR-7 binding sites with one perfectly complementary miR-671 binding site (Hansen and others 2013). These features enable CDR1as to be targeted by miR-671 for degradation as well as acting to “sponge” (i.e., sequester) miR-7 from the cellular milieu. Unchecked miR-7 enhances miR-671-directed cleavage of CDR1as while the lncRNA Cyrano regulates CDR1as accumulation by degrading miR-7 (Kleaveland and others 2018). However, most annotated circRNAs contain few miRNA binding sites (Guo and others 2014), which suggests that sponging miRNAs is not the only function of circRNAs and/or not all circRNAs are regulated by miRNAs. Further dissection of this subset of circRNAs that contain miRNA binding sites is critical to understand their role in regulating neural plasticity, learning, and memory.
Regulation of Transcription
Memory formation requires de novo gene transcription, which is flexibly fine-tuned by transcriptional regulators during the progression of a new memory from a labile to stable state (Alberini and Kandel 2015). Intron-containing circRNAs, such as ciRNAs and EIciRNAs, are predominantly expressed in the nucleus, whereas exon-containing circRNAs are exported to the cytoplasm and synapse (Li and others 2015; Zhang and others 2013). Both ciRNAs and EIciRNAs act in cis to positively regulate transcription of their host genes. In particular, ciRNAs have been found to escape debranching due to the presence of consensus motifs near the 5′ splice site and branchpoint site, and subsequently associate with Pol II at their transcription sites to upregulate host gene transcription (Zhang and others 2013). Similarly, EIciRNAs have been shown to interact with U1 small nuclear ribonucleoproteins (U1A and U1C), which associate with Pol II at the promoters of their host genes to enhance transcription within a positive feedback loop (Li and others 2015). Thus, intron-containing circRNAs are more than just “transcriptional noise” and can act to regulate their own host genes. Little is known, however, about the mechanisms that regulate circRNA alternative splicing, which can result in either the retention of introns for transcriptional regulation or exon-only circRNAs that are exported to the cytoplasm to be potentially translated.
m6A Modification and Translation
m6A is the most prevalent chemical modification on linear RNA and is present on many annotated circRNAs (Yang and others 2017; Zhou and others 2017). Within the brain, m6A is dynamically regulated by experience and acts to enhance long-term memory formation (Widagdo and others 2016). m6A-modified circRNAs share the same methylation machinery as linear RNAs; however, m6A-modified circRNAs are commonly derived from exons that are not methylated in mRNAs (Zhou and others 2017). Within mRNAs, m6A is enriched within the 3′-untranslated region (3′UTR) and around the stop codon whereas circRNAs are usually produced from gene segments closer to the 5′ end or middle (Ragan and others 2019; Zhang and others 2014). It has been reported that mRNAs that are methylated on the same exons as those found in m6A-modified circRNAs tend to be less stable, due to regulation by the m6A reader YTHDF2 (Zhou and others 2017). Thus, promotion of circRNA production over linear RNA transcripts may be one of the reasons that m6A is not observed in high abundance within the 5′ end of mRNA. Further study is required to understand how the m6A machinery identifies targets for methylation and how the context surrounding methylation marks (e.g., sequence and structure context, circular vs linear) affects its regulation of RNA function. Nevertheless, m6A on circRNAs is known to have important functional consequences. For instance, m6A modification enables the cell to recognize circRNAs as belonging to “self” unlike foreign-derived circRNAs that are known to initiate innate immunity (Chen and others 2019). From an evolutionary perspective, immune functions enable a cell to differentiate between “messages” that are derived from itself or friendly neighbours versus foreign invaders trying to hijack the system. Thus, m6A could be acting as a “stamp” for circRNAs that enables their “message” to be safely read without destruction. Furthermore, m6A modification, as well as internal ribosome entry site (IRES) sequences, can promote the translation of circRNAs. IRES sequences can drive direct binding of ribosomes and translation factors whereas m6A can be read by YTHDF3, which then recruits translation initiation factors such as eIF4G2 and eIF3A (Chen and Sarnow 1995; Pamudurti and others 2017; Yang and others 2017). Moreover, m6A is also known to alter the accessibility of structured regions of RNA to enable RNA-protein interactions (Liu and others 2017). Thus, there is a possibility that m6A deposition on structured regions of circRNAs could act as a translation switch to reveal hidden IRES sequences, which then become accessible for translation initiation. Furthermore, as a result of their high stability, circRNAs are ideally positioned to regulate and maintain synaptic status over long periods of time. It is therefore tempting to speculate that reservoirs of translatable circRNAs at the synapse could be switched “ON” to modulate synaptic plasticity on demand. However, under normal conditions, cap-independent translation of both circRNAs and mRNAs remains quite low, although it can serve to enhance the translation of specific mRNAs that contain both IRES sequences and a 5′ cap (Legnini and others 2017; Pamudurti and others 2017). In general, cap-independent mechanisms are promoted/relied on during conditions where cap-dependent mechanisms are reduced/impaired (e.g., stress). As such, it will be critical to understand when periods of increased cap-independent translation arise within the brain during learning and memory formation and under what conditions (e.g., frequency and strength of stimulation). Moreover, the effect of a given small peptide/protein on neural plasticity over a more abundantly expressed protein is not necessarily less pronounced. Further studies into the stoichiometry of small peptides derived from circRNAs and the magnitude of their effects will be required to determine the effect that circRNA translation has on the processes of neural plasticity, learning, and memory formation.
Intercellular Transfer of circRNAs
The ability of viroids and circular genomes to exchange information between cells hints at the possibility that circRNAs may also function as intercellular information carriers. Indeed, circRNAs have been detected within extracellular vesicles and are selectively packaged over their linear counterparts (Fanale and others 2018; Lasda and Parker 2016). The release of circRNAs from the cell via extracellular vesicles potentially contributes to the stoichiometry of circRNAs at the synapse in order to regulate synaptic plasticity and may influence the behavior of neighboring and connected cells to fine-tune engram formation. As yet, there is limited understanding of the composition of extracellular vesicles and how they are targeted and taken up by particular cell-types. For instance, microglia might preferentially bind to extracellular vesicles that contain transcripts that need to be cleared from the cell whereas messages related to memory formation may be preferentially taken up by neurons and potentially other cells involved in neural plasticity (e.g., astrocytes) (Lachenal and others 2011). Thus, circRNA transfer could contribute toward maintaining communication amongst engram cells (i.e., engram maintenance), which would have important consequences for the long-term stability and robustness of a given memory. Further study into the mechanisms underpinning the packaging, uptake mechanisms, and timing of extracellular vesicle release is required in order to fully understand the functional relevance that intercellular circRNA transfer may have for learning and memory formation.
circRNAs Over a Lifetime
Within the brain, circRNAs are spatiotemporally regulated across development, with their expression, independent of their host gene, coinciding with the onset of synaptogenesis (You and others 2015). Indeed, many neural circRNAs are derived from genes related to synaptic function and are differentially expressed during neuronal differentiation and maturation (Rybak-Wolf and others 2015; You and others 2015). For example, a highly conserved circRNA, circSLC45A4, is required to keep neural cells in a progenitor state and its knock-down in the developing mouse cortex results in a depleted basal progenitor pool and an increase in Cajal-Retzius cells over cortical neurons, which leads to improper cortex formation (Suenkel and others 2020). Furthermore, circRNA expression continues throughout adulthood and is dynamically regulated in response to neural activity (You and others 2015). For instance, circHomer1a, a highly conserved circRNA derived from the Homer1 gene, is upregulated during neural activity, and its knock-down within the mouse orbitofrontal cortex (OFC) leads to deficits in OFC-mediated cognitive flexibility during reversal learning (You and others 2015; Zimmerman and others 2020). On the other hand, an unregulated increase in circRNA production by knocking down ADAR1, which edits ICSs and prevents the production of circRNAs, could also impair memory updating (Marshall and others 2020; Rybak-Wolf and others 2015). However, as ADAR1 has many non-circRNA targets, further work on specific circRNA examples is required to explore the relationship between circRNAs and cognitive flexibility. Studies of aged animals, which typically exhibit deficits in cognitive flexibility, could provide additional, albeit indirect, evidence for this relationship; studies across multiple species have observed a general trend of increased expression or accumulation of circRNAs across the genome in an age-dependent manner (Knupp and Miura 2018). However, it remains unclear whether the increased abundance of circRNAs exerts a direct effect on cognitive function or simply represents a marker of aging, especially given that alterations in alternative splicing patterns are known to occur with age (Tollervey and others 2011).
In general, older adults who are cognitively healthy exhibit a reduced rate of acquiring new information whereas their ability to retain previously learned information remains intact (Harada and others 2013). Hence, one possibility is that an increased abundance of circRNAs is a compensatory mechanism that acts to stabilise previously learned information in order to reduce the energetic burden of neural remodeling, which becomes more difficult and risky in advanced age. In simple terms, there is the potential to lose both old and new information by spreading resources too thin within an already distressed cellular environment. Further understanding of the role that circRNAs play over the course of a lifetime will also provide insight into disorders where circRNA expression is dysregulated. In particular, a given circRNA, or a combination of circRNAs, may produce a variety of different behavioural outcomes depending on the surrounding environment (e.g., development vs. aging) and confer either a protective or deleterious effect on neuronal function.
CircRNAs and Cognitive Dysfunction
Neurodegeneration
Alzheimer’s disease (AD) is a neurodegenerative disorder that is characterized by toxic β-amyloid (Aβ) plaques and tau tangles that lead to cell death and progressive decline in cognitive function. Age is the most prominent risk factor for neurodegeneration and, interestingly, age-related changes in alternative splicing patterns found in cognitively healthy adults are also observed in 95% of individuals with frontotemporal lobe dementia or AD patients, irrespective of age (Tollervey and others 2011). Hence, one possibility is that changes in alternative splicing that only occur in AD are contributors toward disease pathogenesis whereas the remainder of changes, which overlap with those in cognitively healthy aged adults, could be the result of a cell’s coping response to suboptimal environmental conditions (e.g., metabolic dysfunction in aging vs. toxic pathology in AD). Changes in alternative splicing patterns may also underlie the accumulation of circRNAs observed during aging, with several studies already demonstrating the dysregulation of circRNAs in AD and their involvement in its pathogenesis (Wang and others 2018). For instance, a known therapeutic agent for AD, Panax notoginseng saponins, alters the expression of several circRNAs that are linked to AD-related pathways and could potentially play a role in AD pathogenesis (Huang and others 2018b). More directly, a recent study showed that one of 17 circRNAs derived from the amyloid precursor protein (APP) gene can be translated into an Aβ-related peptide, which is then further processed into Aβ peptides that aggregate into plaques in vitro (Mo and others 2018). Furthermore, the translatable Aβ-circRNA also promotes the phosphorylation of tau, which is a key feature of tau tangles, by up-regulating glycogen synthase kinase 3β. Hence, Aβ-circRNAs may play a role in initiating or contributing toward AD pathogenesis and should be considered as a target for therapeutic intervention.
The circRNA miR-7 sponge, CDR1as, has a protective effect against AD pathogenesis but is reduced in the brains of AD patients (Lukiw 2013). Ubiquitin conjugating enzyme E2 A (UBE2A) is a target of miR-7 repression and is responsible for ubiquitinating aggregated Aβ42 peptides for proteolysis (Zhao and others 2016). Thus, reduced levels of CDR1as lead to excess miR-7 levels, increased downregulation of UBE2A, and accumulation of Aβ plaques. CDR1as can also prevent the formation of plaques before they accumulate. The cleavage of APP into Aβ peptides by the BACE1 enzyme is a major causative pathway for the pathogenesis of AD. Nuclear factor-κB (NF-κB) represses ubiquitin carboxyl-terminal hydrolase L1 (UCHL1), which is a protein that is able to ubiquitinate and subsequently degrade APP and BACE1. CDR1as expression inhibits the translation of NF-κB and induces its cytoplasmic localisation, thereby lifting the repression of UCHL1 and promoting the degradation of APP and BACE1 (Shi and others 2017). Taken together, circRNAs appear to be key regulatory elements that can influence a variety of cellular pathways, leading to either the progression of or protection against the pathogenesis of neurodegenerative disorders such as AD.
Neuropsychiatric and Neurodevelopmental Disorders
Given that circRNAs influence a wide variety of biological processes involved in cognition, it is not surprising that their dysregulation is also a feature of neuropsychiatric disorders. For instance, despite showing no significant effect on recognition memory, a CDR1as knock-out mouse model presented a strong sensorimotor gating deficit, which is a behavioral phenotype that is associated with schizophrenia and other neuropsychiatric disorders (Piwecka and others 2017). In schizophrenia patients themselves, reduced complexity and abundance of circRNAs is observed within the dorsolateral prefrontal cortex (DLPFC), with many depleted circRNAs predicted to act as sponges for miRNAs that have previously been identified as being increased in schizophrenia (Mahmoudi and others 2019). circHomer1a is a functionally validated example of a circRNA involved in cognitive flexibility and is also known to be dysregulated in neuropsychiatric disease (Zimmerman and others 2020). In particular, circHomer1a is depleted in the OFC of both bipolar disorder and schizophrenia patients as well as the DLPFC of schizophrenia patients. Notably, changes in circHomer1a levels within the OFC and DLPFC of schizophrenia patients is positively correlated with age of clinical onset.
In autism spectrum disorder (ASD), dysregulation of several circRNA-miRNA-mRNA regulatory axes have been found to target known ASD risk genes, as well as inhibitory postsynaptic density proteins, which supports prior observations of increased inhibitory neuron numbers in ASD-derived organoids (Chen and others 2020; Gokool and others 2020). In major depressive disorder (MDD), circDYM and circSTAG1 are reduced in the blood of both MDD patients and a chronic unpredictable stress (CUS) mouse model of depression (Huang and others 2020; Zhang and others 2018). CircDYM is a miR-9 sponge and its overexpression within depressive-like mouse models induced by CUS or lipopolysaccharide leads to an increase in HECTD1 expression, increased HSP90 ubiquitination, and a subsequent decrease in microglial activation that results in attenuated depressive-like behavior (Zhang and others 2018). Similarly, overexpression of circSTAG1 in a CUS mouse model attenuates astrocyte dysfunction and subsequent depressive-like behaviors (Huang and others 2020). However, rather than acting as a miRNA sponge, circSTAG1 functions as an RBP sponge that binds to the demethylase ALKBH5 and prevents its translocation into the nucleus. This leads to increased m6A methylation and subsequent degradation of FAAH mRNA in astrocytes, which reduces astrocyte loss induced by corticosterone in vitro. Taken together, these and other examples of circRNA dysregulation in psychiatric disorders provide considerable evidence for the role of circRNAs in cognitive function.
Concluding Remarks
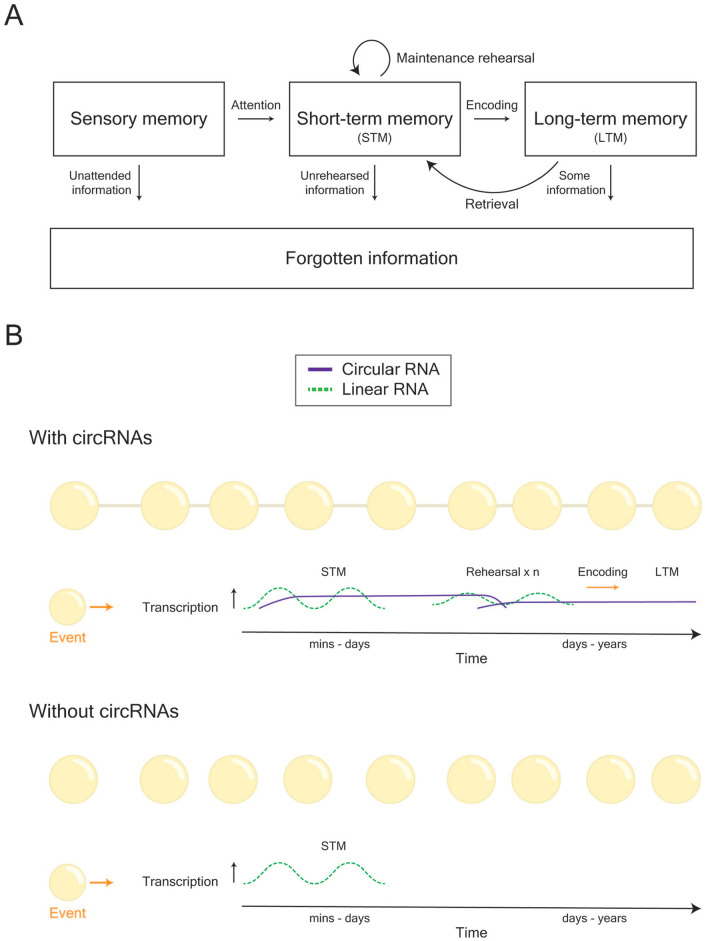
In general, memories can be viewed as a population of cells that communicate across both space and time in order to form connected networks (i.e., the “engram complex”). Previously, the role of RNA as a regulatory architect of cellular behavior was constrained by its short longevity whereas the long-lived DNA blueprint and its protein effectors were typically viewed as the molecular constituents that connect the behavior of a given cell—or network of cells—from one instant to the next. However, with the discovery of circRNAs, our understanding of how molecular networks function and communicate with each other, both intracellularly and intercellularly, may soon be revised (Fig. 5). So far, studies of circRNA function have revealed a great deal of overlap with linear RNA function. Both types of RNA are involved in regulation of transcription, miRNA regulation, can be chemically modified, assume secondary structures, transfer between cells, and some circRNAs can even serve as templates for translation into small peptides. Therefore, the major difference lies in their stability, with circRNAs able to act over broad timescales whereas linear RNAs function within short and specific time windows. CircRNAs may therefore serve to ensure the stability, and hence continuity, of cellular behavior. Linear RNAs could also be preferentially recruited to perform minute adjustments to ongoing cellular processes since circRNAs are liable to continue making adjustments over a long period of time leading to overadjustment. Given their relatively recent discovery, there is still a lot to uncover about the regulation and function of circRNAs and their involvement in cognition. In particular, how is the function of circRNAs regulated across time and how do they act to support cognition both within and between cells as well as across a lifetime in both healthy and disease conditions? Taken together, the diverse roles that circRNAs play in both health and disease, together with their ability to act over broad time scales, highlights the importance of understanding these RNAs in greater detail.
Figure 5.
Conjectured role of circRNAs in learning and memory formation. (A) General model of how memories are formed, stored, and retrieved. (B) During learning, two peaks of gene transcription contribute toward the formation of memory. However, linear RNAs and their actions are relatively short-lived. Given their long life span, there is the possibility that circRNAs may be necessary for the stability, and hence continuity, of cellular behavior that underlies memory. Thus, we propose that circRNAs act as a mechanism to keep track of the history of experiences (i.e., alterations in cellular behavior) that a cell, or network of cells, undergoes.
Acknowledgments
The authors would like thank Ms. Rowan Tweedale for helpful editing of the transcript.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors acknowledge grant support from the ARC (DP180102998-TWB). ELZ is supported by the University of Queensland and is a recipient of a Westpac Future Leaders Scholarship.
ORCID iD: Esmi L. Zajaczkowski  https://orcid.org/0000-0003-3246-6959
https://orcid.org/0000-0003-3246-6959
References
- Aktaş T, Ilık İA, Maticzka D, Bhardwaj V, Rodrigues CP, Mittler G, and others. 2017. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544:115–9. [DOI] [PubMed] [Google Scholar]
- Alberini CM, Kandel ER. 2015. The regulation of transcription in memory consolidation. Cold Spring Harb Perspect Biol 7:a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, and others. 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66. [DOI] [PubMed] [Google Scholar]
- Belting M, Wittrup A. 2008. Nanotubes, exosomes, and nucleic acid–binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: implications in health and disease. J Cell Biol 183:1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Lin Q, Wei W, Baker-Andresen D, Mattick JS. 2011. MicroRNA regulation of neural plasticity and memory. Neurobiol Learn Mem 96:89–94. [DOI] [PubMed] [Google Scholar]
- Budnik V, Ruiz-Cañada C, Wendler F. 2016. Extracellular vesicles round off communication in the nervous system. Nat Rev Neurosci 17:160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, and others. 1993. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73:1019–30. [DOI] [PubMed] [Google Scholar]
- Chen CY, Sarnow P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415–7. [DOI] [PubMed] [Google Scholar]
- Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, and others. 2019. N6-Methyladenosine modification controls circular RNA immunity. Mol Cell 76:96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-J, Chen C-Y, Mai T-L, Chuang C-F, Chen Y-C, Gupta SK, and others. 2020. Genome-wide integrative analysis of circular RNA dysregulation and the corresponding circular RNA-microRNA-mRNA regulatory axes in autism. Genome Res. Epub March 3. doi: 10.1101/gr.255463.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. 1993. Mis-splicing yields circular RNA molecules. FASEB J 7:155–60. [DOI] [PubMed] [Google Scholar]
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, and others. 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–34. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Mercer TR, Mattick JS. 2008. RNAs as extracellular signaling molecules. J Mol Endocrinol 40:151–9. [DOI] [PubMed] [Google Scholar]
- Fanale D, Taverna S, Russo A, Bazan V. 2018. Circular RNA in exosomes. In: Xiao J, editor. Circular RNAs: biogenesis and functions. Advances in experimental medicine and biology. Singapore: Springer Singapore. pp. 109–17. doi: 10.1007/978-981-13-1426-1_9 [DOI] [PubMed] [Google Scholar]
- Fischer JW, Busa VF, Shao Y, Leung AKL. 2020. Structure-mediated RNA decay by UPF1 and G3BP1. Mol Cell 78:70-84.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao F. 2018. Computational strategies for exploring circular RNAs. Trends Genet 34:389–400. [DOI] [PubMed] [Google Scholar]
- Gokool A, Anwar F, Voineagu I. 2020. The landscape of circular RNA expression in the human brain. Biol Psychiatry 87:294–304. [DOI] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP. 2014. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Wei X, Peng Y. 2020. Structure-mediated degradation of circRNAs. Trends Cell Biol 30:501–3. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, and others. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495:384–8. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, and others. 2011. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30:4414–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada CN, Natelson Love MC, Triebel K. 2013. Normal cognitive aging. Clin Geriatr Med 29:737–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Martin KC, Schuman EM. 2019. Local translation in neurons: visualization and function. Nat Struct Mol Biol 26:557. [DOI] [PubMed] [Google Scholar]
- Hsu M-T, Coca-Prados M. 1979. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–40. [DOI] [PubMed] [Google Scholar]
- Huang C, Liang D, Tatomer DC, Wilusz JE. 2018. a. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev 32:639–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J-L, Xu Z-H, Yang S-M, Yu C, Zhang F, Qin M-C, and others. 2018. b. Identification of differentially expressed profiles of Alzheimer’s disease associated circular RNAs in a Panax notoginseng saponins-treated Alzheimer’s disease mouse model. Comput Struct Biotechnol J 16:523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zhang Y, Bai Y, Han B, Ju M, Chen B, and others. 2020. N6-methyladenosine modification of FAAH mRNA in circSTAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol Psychiatry 88:392–404. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sharpless NE. 2014. Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, and others. 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Xiao M-S, Li Z, Shan G, Huang C. 2019. Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov 5:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, Tonegawa S. 2020. Memory engrams: recalling the past and imagining the future. Science 367:eaaw4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Dudai Y, Mayford MR. 2014. The molecular and systems biology of memory. Cell 157:163–86. [DOI] [PubMed] [Google Scholar]
- Kleaveland B, Shi CY, Stefano J, Bartel DP. 2018. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174:350-362.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knupp D, Miura P. 2018. CircRNA accumulation: a new hallmark of aging? Mech Ageing Dev 173:71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukushkin NV, Carew TJ. 2017. Memory takes time. Neuron 95:259–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachenal G, Pernet-Gallay K, Chivet M, Hemming FJ, Belly A, Bodon G, and others. 2011. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci 46:409–18. [DOI] [PubMed] [Google Scholar]
- Lasda E, Parker R. 2014. Circular RNAs: diversity of form and function. RNA 20:1829–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasda E, Parker R. 2016. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One 11:e0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, and others. 2017. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66:22-37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton LJ, Ke K, Zajaczkowski EL, Edmunds J, Spitale RC, Bredy TW. 2018. Experience-dependent neural plasticity, learning, and memory in the era of epitranscriptomics. Genes Brain Behav 17:e12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu C-X, Xue W, Zhang Y, Jiang S, Yin Q-F, and others. 2017. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell 67:214-227.e7. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, and others. 2015. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–64. [DOI] [PubMed] [Google Scholar]
- Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen L-L, and others. 2017. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell 68:940-954.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-X, Li X, Nan F, Jiang S, Gao X, Guo S-K, and others. 2019. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177:865-880.e21. [DOI] [PubMed] [Google Scholar]
- Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. 2017. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 45:6051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loebrich S, Nedivi E. 2009. The function of activity-regulated genes in the nervous system. Physiol Rev 89:1079–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. 2013. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi E, Fitzsimmons C, Geaghan MP, Shannon Weickert C, Atkins JR, Wang X, and others. 2019. Circular RNA biogenesis is decreased in postmortem cortical gray matter in schizophrenia and may alter the bioavailability of associated miRNA. Neuropsychopharmacology 44:1043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PR, Zhao Q, Li X, Wei W, Periyakaruppiah A, Zajaczkowski EL, and others. 2020. Dynamic regulation of Z-DNA in the mouse prefrontal cortex by the RNA-editing enzyme Adar1 is required for fear extinction. Nat. Neurosci 23:718–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. 2004. RNA regulation: a new genetics? Nat Rev Genet 5:316–23. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mariani J, Kosik KS, Mehler MF, Mattick JS. 2008. Noncoding RNAs in long-term memory formation. Neuroscientist 14:434–45. [DOI] [PubMed] [Google Scholar]
- Mo D, Cui D, Li X. 2018. The role of Aβ circRNA in Alzheimer’s disease: alternative mechanism of Aβ biogenesis from Aβ circRNA translation. Neuroscience. BioRxiv preprint. doi: 10.1101/260968 [DOI] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, and others. 2017. Translation of CircRNAs. Mol Cell 66:9-21.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey PR, Rout PK, Das A, Gorospe M, Panda AC. 2019. RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods 155:41–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, and others. 2019. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol Cell 74:494-507.e8. [DOI] [PubMed] [Google Scholar]
- Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, and others. 2017. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526. [DOI] [PubMed] [Google Scholar]
- Ragan C, Goodall GJ, Shirokikh NE, Preiss T. 2019. Insights into the biogenesis and potential functions of exonic circular RNA. Sci Rep 9:2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, and others. 2015. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–85. [DOI] [PubMed] [Google Scholar]
- Schacter DL, Eich JE, Tulving E. 1978. Richard Semon’s theory of memory. J Verbal Learn Verbal Behav 17:721–43. [Google Scholar]
- Semon R. 1904. Die Mneme als erhaltendes Prinzip im Wechsel des organischen Geschehens. Leipzig: Wilhelm Engelmann. [Google Scholar]
- Sheng M, Greenberg ME. 1990. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4:477–85. [DOI] [PubMed] [Google Scholar]
- Shi Z, Chen T, Yao Q, Zheng L, Zhang Z, Wang J, and others. 2017. The circular RNA ciRS-7 promotes APP and BACE1 degradation in an NF-κB-dependent manner. FEBS J 284:1096–109. [DOI] [PubMed] [Google Scholar]
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung L-H, and others. 2015. Exon circularization requires canonical splice signals. Cell Rep 10:103–11. [DOI] [PubMed] [Google Scholar]
- Suenkel C, Cavalli D, Massalini S, Calegari F, Rajewsky N. 2020. A highly conserved circular RNA is required to keep neural cells in a progenitor state in the mammalian brain. Cell Rep 30:2170-2179.e5. [DOI] [PubMed] [Google Scholar]
- Szabo L, Salzman J. 2016. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 17:679–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Wang Z, Hortobágyi T, Witten JT, Zarnack K, Kayikci M, and others. 2011. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res 21:1572–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Hou J, Müller-McNicoll M, Chen W, Schuman EM. 2019. Long and repeat-rich intronic sequences favor circular RNA formation under conditions of reduced spliceosome activity. iScience 20:237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xu P, Chen B, Zhang Z, Zhang C, Zhan Q, and others. 2018. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer’s disease-like rats using microarray analysis. Aging 10:775–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME. 2011. Neuronal activity–regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol 3:a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo J, Zhao Q-Y, Kempen M-J, Tan MC, Ratnu VS, Wei W, and others. 2016. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J Neurosci 36:6771–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, and others. 2017. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27:626–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, and others. 2015. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18:603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG. 1996. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 93:6536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang X-O, Jiang T, Cai L, Huang X, Liu Q, and others. 2020. Comprehensive identification of alternative back-splicing in human tissue transcriptomes. Nucleic Acids Res 48:1779–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. 2014. Complementary sequence-mediated exon circularization. Cell 159:134–47. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Du L, Bai Y, Han B, He C, Gong L, and others. 2018. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol Psychiatry 25:1175–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang J-L, and others. 2016. The biogenesis of nascent circular RNAs. Cell Rep 15:611–24. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, and others. 2013. Circular intronic long noncoding RNAs. Mol Cell 51:792–806. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. 2016. Deficiency in the ubiquitin conjugating enzyme UBE2A in Alzheimer’s disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel) 7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Wittenberghe NV, and others. 2017. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep 20:2262–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AJ, Hafez AK, Amoah SK, Rodriguez BA, Dell’Orco M, Lozano E, and others. 2020. A psychiatric disease-related circular RNA controls synaptic gene expression and cognition. Mol Psychiatry. Epub January 27. doi: 10.1038/s41380-020-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]