Abstract
The incidence of colon cancer continues to increase annually, and it is the leading cause of cancer-associated mortality worldwide. Altering cell metabolism and inducing autophagic cell death have recently emerged as novel strategies in preventing tumor growth. Autophagy plays an essential role in energy production by degrading damaged cellular components and is also associated with tumor proliferation suppression. Itraconazole is an FDA-approved drug used as an antifungal medication and has been reported to induce autophagic cell death in breast cancer. However, the effects of itraconazole on cell metabolism and induction of apoptosis in colon cancer remain unclear. The present study analyzed extensive data from patients diagnosed with colon cancer using itraconazole between January 2011 and December 2015, from the Taiwanese National Health Insurance Research Database. The underlying molecular mechanisms of itraconazole in autophagy-induced cell death were also investigated. The results demonstrated that the 5-year survival rate was significantly higher in patients with colon cancer who received itraconazole treatment. In addition, itraconazole decreased the viability and cell colony formation, and induced cleaved caspase-3 expression and G1 cell cycle arrest of COLO 205 and HCT 116 cells. Notably, itraconazole induced autophagy by enhancing LC3B and p62 expression. Following LC3 knockdown, the viability of itraconazole-treated COLO 205 and HCT 116 cells notably improved. Taken together, the results of the present study suggest that itraconazole may have a beneficial effect on patients with colon cancer, and its underlying molecular mechanisms may be associated with the induction of autophagic cell death.
Keywords: colon cancer, autophagy, apoptosis, itraconazole, transketolase
Introduction
Advancing techniques for detection and targeted therapy of colon cancer have increased patient 5-year survival rates to 65% and estimated newly diagnosed cases per year was 134,490 in the United states in 2016 (1). Despite medical advancements, colon cancer remains the fifth most common type of cancer and is the fourth-leading cause of cancer-associated mortality worldwide (2). The direct cost per patient with colon cancer during the primary treatment period was estimated to be >24,000 US dollars (3). Current treatment strategies for colon cancer include surgery, radiation therapy, chemotherapy and immunotherapy (4). However, a proportion of patients with colon cancer require alternative treatments due to the adverse effects of conventional therapy. Thus, it is important to identify and develop novel therapeutic strategies and pathways to improve treatment efficacy and reduce side effects.
Autophagy is a highly conserved metabolic process that can occur in all cell types and is induced by increasing oxidative stress, drug stimulation and nutrient depletion (5). Recently, autophagy has become a novel target for cancer treatment and adjuvant therapy. The molecular mechanisms underlying autophagy involve regulating the expression of related proteins, including LC3B and p62 (6). LC3B expression is associated with autophagosome formation, and p62 expression is involved in autophagy substrate selection and serves as an adaptor for intracellular targets (7). LC3B and p62 expression are used to evaluate autophagy activity during tumor progression (8). However, autophagy plays a dual role in cancer. During the initial progression phase, it serves as a tumor suppressor (9,10). Conversely, in hypoxic environments, autophagy serves as a developer (11). Thus, it is important to identify the role of drug-induced autophagy in cancer.
Direct inhibition of the key enzymes in glycolysis to decrease ATP production is also a novel cancer treatment strategy. For example, transketolase (TKT) is an enzyme of the pentose phosphate pathway (PPP), which connects PPP to glycolysis (12). In cancer cells, ATP is produced by glycolysis rather than oxidative phosphorylation (13). Furthermore, overexpression of TKT is associated with tumorigenesis, such as the development of hepatocellular carcinoma (14) and peritoneal metastases of ovarian cancer (15). Recent studies have reported that anti-TKT therapy inhibits cell migration and invasion in esophageal cancer (16) and inhibits the expression of the TKT-like-1 gene, which significantly decreases TKT expression in human colon cancer cells and substantially inhibits cell proliferation (17). Furthermore, cryptotanshinone, a traditional oriental medicinal agent, has been demonstrated to decrease non-small cell lung cancer proliferation by suppressing TKT expression (18).
Itraconazole has been used as an antifungal drug in the past few decades (19). The primary mechanism by which itraconazole inhibits microorganism growth is interference with ergosterol synthesis, which damages the cell membrane integrity of fungal cells (20). Recently, induction of apoptosis or autophagic apoptosis by itraconazole has been widely used as an anticancer therapy (21,22). Preclinical data have demonstrated that itraconazole can suppress tumor growth, and many clinical trials have been performed to evaluate the effects of itraconazole on different cancer treatments (23,24). Itraconazole can induce autophagic cell death by activating the hedgehog pathway to inhibit breast cancer cell proliferation (25). However, the potential effect of itraconazole on colon cancer and the role of autophagy in colon cancer cell proliferation remain unclear.
The present study aimed to investigate the effects of itraconazole on the antitumor development of colon cancer, determine its underlying molecular mechanisms, and used a nationwide database analysis to determine the associations between basic experiments and clinical manifestations. The results presented here provide evidence that itraconazole may act as a promising drug for colon cancer therapy.
Materials and methods
Data collection
This observational cohort study used the cancer registry dataset of the ‘2010 Longitudinal Generation Tracking Database (LGTD 2010),’ established by the Health and Welfare Data Science Center (HWDC), Ministry of Health and Welfare (MOHW) in Taiwan. The Taiwan National Health Insurance (NHI) program has been implemented since 1995 and offers comprehensive medical care coverage to >99% of the country's 23 million inhabitants (26).
The present study collected data from the LGTD 2010, which were randomly selected from a cohort of 2 million individuals covered by the NHI Program, between January 2011 and December 2011. The LGTD 2010 contains all information on diagnostic codes, detailed prescriptions, dates of clinical visits, dates of admission and discharge, and expenditures for the enrollees of 2 million beneficiaries (26). No statistical differences was observed in the age and sex distributions between the cohorts in the LGTD and Taiwan NHI enrollees. All patients were regularly monitored after diagnosis until death or at the last follow-up date. Follow-up was completed on December 31, 2015 (27). From the Taiwanese National Health Insurance Research Database (https://www.mohw.gov.tw/mp-2.html), data from patients newly diagnosed with colon cancer were identified using the International Classification of Diseases for Oncology codes (C180-C189), treated with or without itraconazole, according to the Anatomical Therapeutic Chemical code: J02AC02, between January 2011 and December 2015. A total of 5,221 patients newly diagnosed with colon cancer were included in the present study, and confounding factors, such as age, sex, clinical stage, surgery, radiation therapy and chemotherapy were adjusted accordingly. Patients were pathologically diagnosed with primary colon cancer based on Taiwan Cancer Registry, a population-based cancer registry enrolled. The exclusion criteria for this study were patients' age at diagnosis was unknown; incomplete data of tumor stage, pathology and treatment history; lacking clear records on survival status; previous history of cancer; incomplete data on the American Joint Committee on Cancer (AJCC) stage and cause of death. All staging was performed according to the AJCC Staging System (7th edition) (28). To protect the privacy of individuals included in the database, the NHI only released data with encoded identification numbers so that personnel without authorization would not be able to associate any direct information to the enrollees. The Bureau of NHI approved the application after reviewing all the required medical documents. The present study was approved by the Institutional Review Board of Kaohsiung Veterans General Hospital (IRB no. KSVGH18-CT10-07) and performed in accordance with the tenets of the Declaration of Helsinki (1975) and its later amendments (2013). The requirement for written informed consent was waived.
Cell culture
The human colon adenocarcinoma cell line, COLO 205, was purchased from the Food Industry Research and Development Institute, and the colorectal cell line, HCT 116, was kindly gifted by Dr Tzu-Ming Jao from Kaohsiung Veterans General Hospital (Kaohsiung, Taiwan). Both cell lines were maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Sigma-Aldrich; Merck KGaA), 2 mM L-glutamine (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin and streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.), at 37°C with 5% CO2.
To determine the role of autophagy on itraconazole induced apoptosis in COLO 205 cells, 5 mM 3-MA (3-methyladenine, Sigma-Aldrich; Merck KGaA) was co-cultured with 50 µM itraconazole (Sigma-Aldrich; Merck KGaA) at 37°C for 24 h.
MTT assay
The MTT assay (Sigma-Aldrich; Merck KGaA) was performed to assess the cytotoxic effect of itraconazole on COLO-205 and HCT 116 cells. A total of 1×104 cells were seeded into 96-well plates and cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Sigma-Aldrich; Merck KGaA), 2 mM L-glutamine (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin and streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.), at 37°C with 5% CO2 for overnight. Following incubation, 1, 2, 5, 10, 25, 30, 50, 60, 100, 125, 250 and 500 µM itraconazole were added into each well and cells were incubated for an additional 24 h at 37°C. The culture medium was subsequently replaced with complete medium (DMEM, Gibco; Thermo Fisher Scientific, Inc.) containing 5 mg/ml MTT and incubated for 2 h at 37°C. The purple formazan crystals were dissolved using DMSO and data were subsequently measured at a wavelength of 570 nm, using an ELISA reader (BioTek Instruments, Inc.).
Small interfering (si)RNA transfection
The commercial siRNA gene silencer (LC3B) and scrambled siRNA oligo were purchased from Cell Signaling Technology, Inc. (cat. nos. 6212S and 6568S). Briefly, 1×106 COLO 205 and HCT 116 cells were seeded into 6-well plates, respectively, and cultured in complete medium overnight at 37°C. siRNA (100 nM) and scrambled siRNA (100 nM) were transfected into cells using Lipofectamine® RNAiMAX (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following incubation at 37°C for 24 h, the supernatant was removed, washed with PBS and incubated with different concentrations of itraconazole in complete medium for an additional 24 h at 37°C. Representative images were obtained from three independent experiments.
Cell Counting Kit-8 (CCK-8) assay
Following transfection with LC3B or scrambled siRNA, COLO 205 and HCT 116 cells were treated with different concentrations of itraconazole in triplicate wells at 37°C for 24 h. Cell viability was assessed via the CCK-8 assay. Briefly, 100 µl of cell suspension was seeded into a 96-well plate, and 10 µl of CCK-8 reagent (Sigma-Aldrich; Merck KGaA) was added to each well and incubated for 1 h. Optical density was measured at a wavelength of 450 nm, using an enzyme microplate reader (Synergy HTX; BioTek Instruments, Inc.).
Trypan blue exclusion assay
Following transfection with LC3B or scrambled siRNA, COLO 205 and HCT 116 cells were treated with different concentrations of itraconazole in triplicate wells for 24 h. Cells were collected and resuspended in the culture medium. Subsequently, cells were mixed with trypan blue at a ratio of 1:1 (Gibco; Thermo Fisher Scientific, Inc.) and counted using a hemocytometer (Yung Yuen Scientific Instrument Co., Ltd.).
Annexin V/PI staining
Itraconazole-induced cell apoptosis was detected via Annexin V/PI staining. A total of 1×106 cells were seeded into 6-well plates and incubated in complete medium overnight at 37°C. Following attachment to the plate, cells were treated with different concentrations of itraconazole for 24 h. Annexin V/PI staining was performed according to the manufacturer's instructions (AAT Bioquest, Inc.). The percentage of apoptotic cells was detected using FACSCalibor (BD Biosciences). Representative images are shown for three independent experiments.
Cell cycle analysis
A total of 1×106 COLO 205 and HCT 116 cells were seeded into 6-well plates and treated with different concentrations of itraconazole for 24 h. Following treatment, cells were fixed with 100% ethanol overnight at −20°C. DNA in the nuclei was stained with PI (50 µg/ml) and RNase A (0.5 µg/ml) for 30 min at 4°C. Cell cycle distribution was analyzed using an Attune NxT flow cytometer (Thermo Fisher Scientific, Inc.). Representative images were obtained from three independent experiments.
Reverse transcription-quantitative (RT-q)PCR
Following treatment with itraconazole for 24 h, mRNA was extracted from COLO 205 and HCT 116 cells using the GENEzol™ TriRNA Pure kit (Geneaid, http://www.geneaid.com/Tri-RNA/GZXD), according to the manufacturer's instructions. A total of 1 µg RNA was reverse transcribed into cDNA using the cDNA reverse transcription kit (Morrebio, http://www.light-biotech.com/pcr), according to the manufacturer's instructions. The temperature protocol for RT was as follows: 50°C for 15 min followed by 85°C for 5 sec, and the cDNA products were used for the gene expression assay. The PCR primer and the TaqMan probe were purchased from Thermo Fisher Scientific, Inc. The primer and probe sequences used in the present study were as follows: LC3B (Gene ID: Hs00797944_s1), p62 (Gene ID: Hs02621445_s1) and GAPDH (Gene ID: Hs02786624_g1). Gene expression was detected via qPCR (StepOnePlus™; Thermo Fisher Scientific, Inc.) and the protocol was as follows: 10 µl 2× PCR master mix, 2 µl cDNA, 7 µl dd H2O and 1 µl probe were mixed in each reaction. The following thermocycling conditions were used for qPCR: 50°C for 2 min, 95°C for 2 min followed by 40 cycles of 95°C for 1 sec and 60°C for 20 sec. LC3B and p62 expression levels were calculated using the 2−ΔΔCq method and normalized to β-actin (29).
Western blotting
A total of 1×106 COLO 205 and HCT 116 cells, were seeded into 6-well plates and treated with different concentrations of itraconazole for 24 h. Following treatment, total protein was extracted using RIPA buffer (Biomed, http://www.bio-protech.com.tw/products_detailed.php?id=347) and quantified using the BCA kit (Invitrogen; Thermo Fisher Scientific, Inc.). Equal amounts (25 µg/lane) of protein extracts were subjected in 12% polyacrylamide gel and separated by electrophoresis then transferred onto PVDF membranes and blocked with 5% skim milk for 1 h at room temperature. Membranes were washed three times with PBST (PBS with 0.05% Tween-20) and incubated with primary antibodies against TKT (1:1,000; cat. no. SC-390179; Santa Cruz Biotechnology, Inc.), Cleaved caspase-3 (1:1,000; cat. no. 9664; Cell Signaling Technology, Inc.), Caspase-3 (1:1,000; cat. no. E-AB-30756; Elabscience, http://www.elabscience.com/PDF/Cate81/E-AB-30756-Elabscience.pdf), Bax (1:1,000; cat. no. 2772; Cell Signaling Technology, Inc.), LC3B (1:1,000; cat. no. 83506; Cell Signaling Technology, Inc.), p62 (1:1,000; cat. no. 5114; Cell Signaling Technology, Inc.), Beclin-1 (1:1,000; cat. no. 3738; Cell Signaling Technology, Inc.) and β-actin (1:5,000; cat. no. E-AB-20034; Elabscience) overnight at 4°C. The membranes were washed three times with PBST and subsequently incubated with anti-mouse IgG, HRP-linked antibody (1:5,000; cat. no. 7076P2; Cell Signaling Technology, Inc.) or goat anti-rabbit IgG antibody (1:5,000; cat. no. A0545; Sigma-Aldrich; Merck KGaA) for 1 h at room temperature. Protein bands were visualized using the ECL western blot detection kit (Thermo Fisher Scientific, Inc.) and analyzed using Alliance Q9 software (V18.11; UVITEC; http://www.uvitec.co.uk/alliance-q9-advanced). Representative images and band quantification were obtained from three independent experiments.
Confocal microscopy
To detect autophagosome formation, 1×105 COLO 205 and HCT 116 cells were seeded onto cover glasses in a 6-well plate. Following treatment with itraconazole for 24 h, cells were fixed with 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) for 15 min at room temperature and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich; Merck KGaA) in PBS for 15 min at room temperature. Cells were subsequently blocked with 1% BSA for 1 h at room temperature. Cells were incubated with LC3B primary antibody (Cell Signaling Technology, Inc.) overnight at 4°C, followed by incubation with FITC-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature. DAPI was used for nuclear staining. Cells were observed under a Zeiss LSM 5 Pascal confocal microscope (Carl Zeiss). Number of LC3 puncta per cell was further quantified using ImageJ software (Version 1.50i; National Institutes of Health) (30). Representative images were obtained from three independent experiments.
Statistical analysis
Statistical analysis was performed using SAS software (version 9.3; SAS Institute, Inc.) and SPSS software (version 20; IBM Corp.). Descriptive statistics were used to analyze baseline demographic data and the distribution of each variable among the study population. Continuous variables are presented as the mean ± SD. Survival analysis was performed using the Kaplan-Meier method and log-rank test. Univariate Cox regression analysis was performed to assess the association between survival and the impact of itraconazole. Hazard ratios (HRs) and their 95% confidence intervals (CIs) from the Cox regression analyses were used to estimate the relative risk. Then stratified survival analyses were also performed for different groups based on multivariate Cox regression analysis. The results of the control and treatment with different concentrations of itraconazole were analyzed via one-way ANOVA followed by Tukey's post hoc test, and performed using GraphPad Prism software (version 6.0; GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Baseline demographic characteristics
In the present study, a total of 5,221 patients with colon cancer were identified using ICD-O3 codes between 2011 and 2015, and data were analyzed. The demographic characteristics of all patients, including age, sex, primary tumor location, AJCC clinical stage, treatment, itraconazole use and mortality are presented in Table I. The average age of all patients was 65.6±13.7 years (age range, 21–85 years). The proportions of stage I+II and stage III+IV were 42 and 58%, respectively. Most patients underwent surgery, and the proportions of patients who received radiation therapy and chemotherapy were 2 and 38%, respectively. Notably, only 1% of all patients received itraconazole treatment. The mortality rate of all patients with colon cancer was 34%.
Table I.
Baseline characteristics of patients with colon cancer (n=5,221).
| Characteristic | Patients, n (%) |
|---|---|
| Age, years (mean ± SD) | 65.6±13.7 |
| Sex | |
| Male | 2,921 (56) |
| Female | 2,300 (44) |
| AJCC clinical stage | |
| I+II | 2,206 (42) |
| III+IV | 3,015 (58) |
| Surgery | |
| Yes | 4,095 (78) |
| No | 1,126 (22) |
| Radiation therapy | |
| Yes | 88 (2) |
| No | 5,133 (98) |
| Chemotherapy | |
| Yes | 1,977 (38) |
| No | 3,244 (62) |
| Drug | |
| Yes | 62 (1) |
| No | 5,159 (99) |
| Mortality | |
| Yes | 1,780 (34) |
| No | 3,441 (66) |
AJCC, American Joint Committee on Cancer.
Itraconazole increases the 5-year survival rate in patients with late-stage colon cancer who receive chemotherapy
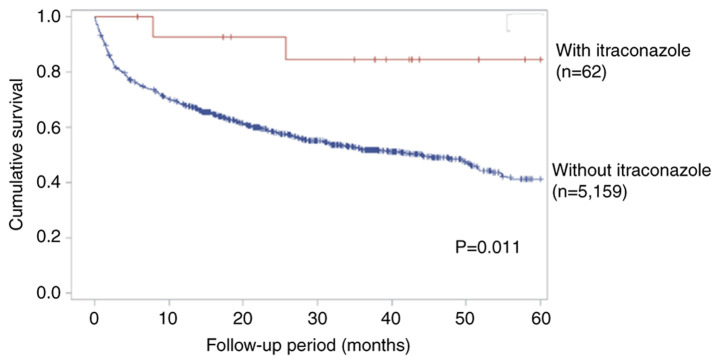
The univariate Cox proportional hazard model was used to assess the HR for patients with colon cancer, in the presence and absence of itraconazole treatment. Statistically significant HRs for the 5-year survival rate were observed for age (HR, 1.03; 95% CI, 1.01–1.04; P<0.001), AJCC clinical stage (HR, 3.08; 95% CI, 2.75–3.44; P<0.001), surgery (HR, 4.15; 95% CI, 3.77–4.57; P<0.001) and radiation therapy (HR, 0.43; 95% CI, 0.33–0.58; P<0.001) (Table II). Subgroup analysis was performed to assess the effect of itraconazole on patients with late-stage colon cancer who received chemotherapy. Following adjustment for age, sex, surgery and radiation therapy, the benefit of itraconazole treatment in reducing the risk of mortality was observed (HR, 0.27; 95% CI, 0.07–1.09; P=0.067; Table III). Although there was not statistically significant, however the trend in reducing the risk of mortality could still be noted. The 5-year survival rate also increased in patients treated with itraconazole group, according to Kaplan-Meier curve by log-rank test with significant statistical difference. (P<0.05; Fig. 1). Taken together, these results suggest that itraconazole can increase the 5-year survival rate in patients with late-stage colon cancer.
Table II.
Univariate Cox regression analysis in patients with colon cancer.
| Variable | HR (95% CI) | P-value |
|---|---|---|
| Age, years | 1.03 (1.01–1.04) | <0.001a |
| Sex | 0.541 | |
| Male | Reference | |
| Female | 0.97 (0.88–1.07) | |
| AJCC clinical stage | <0.001a | |
| I+II | Reference | |
| III+IV | 3.08 (2.75–3.44) | |
| Surgery | <0.001a | |
| Yes | Reference | |
| No | 4.15 (3.77–4.57) | |
| Radiation therapy | <0.001a | |
| Yes | Reference | |
| No | 0.43 (0.33–0.58) | |
| Chemotherapy | 0.493 | |
| Yes | Reference | |
| No | 0.97 (0.88–1.06) | |
| Drug | 0.434 | |
| Yes | Reference | |
| No | 0.83 (0.52–1.32) |
P<0.001. HR, hazard ratio; CI, confidence interval; AJCC, American Joint Committee on Cancer.
Table III.
Stratification multivariate Cox regression analysis in patients with colon cancer, with advanced AJCC clinical stage (III+IV).
| Variable | Adjusted HR (95% CI) | P-value |
|---|---|---|
| Chemotherapy | ||
| Drug | ||
| Yes | 0.27 (0.07–1.09) | 0.067 |
| No | Reference | |
| No chemotherapy | ||
| Drug | ||
| Yes | 1.06 (0.47–2.38) | 0.883 |
| No | Reference |
AJCC, American Joint Committee on Cancer; HR, hazard ratio; CI, confidence interval.
Figure 1.
Kaplan-Meier analysis for the 5-year survival rate of patients with colon cancer.
Itraconazole inhibits the proliferation of colon cancer cells and induces apoptosis and cell cycle arrest
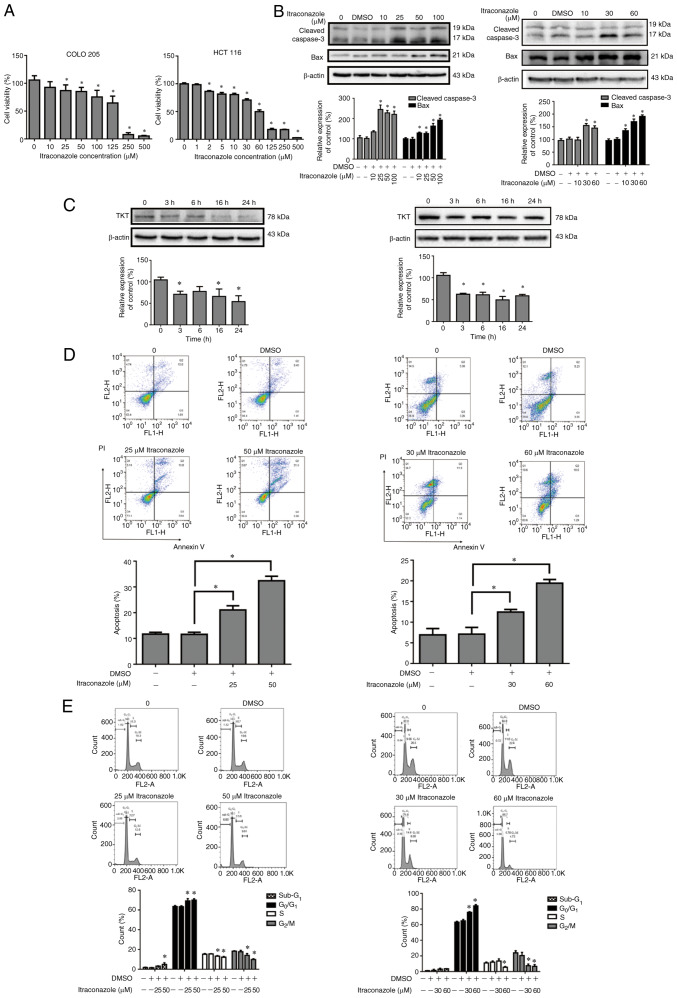
To further investigate the effects and underlying molecular mechanisms of itraconazole on colon cancer, the effect of itraconazole on the proliferation of COLO 205 and HCT 116 cells was assessed. Cells were treated with different concentrations of itraconazole for 24 h, and cell viability was assessed via the MTT assay. The results demonstrated that cell viability significantly decreased following treatment with itraconazole, in a dose-dependent manner (P<0.05; Fig. 2A). Apoptosis-related proteins and TKT expression were detected in COLO 205 and HCT 116 cells treated with itraconazole. The results demonstrated that the expression levels of cleaved caspase-3 and Bax increased after itraconazole treatment, in a dose-response manner for 24 h (P<0.05; Fig. 2B). TKT expression was decreased following treatment with itraconazole in a time-dependent manner (P<0.05; Fig. 2C). Annexin V-PI staining demonstrated that COLO 205 and HCT 116 cells treated with itraconazole significantly induced cell apoptosis (P<0.05; Fig. 2D). Furthermore, treatment with itraconazole significantly induced subG1 phase, G1 arrest and decreased the number of cells in the G2/M phase (P<0.05; Fig. 2E). Collectively, these results suggest that itraconazole decreases colon cancer proliferation and TKT expression and induces apoptosis.
Figure 2.
Treatment with itraconazole inhibits the proliferation of colon cancer cells and induces apoptosis, autophagy and cell cycle arrest. (A) COLO 205 and HCT 116 cells were treated with different concentrations of itraconazole for 24 h, and cell viability was assessed via the MTT assay. (B) COLO 205 and HCT 116 cells were treated with itraconazole in a dose-dependent manner for 24 h, and the expression levels of Cleaved caspase-3, Bax and β-actin were detected via western blotting. Protein expression was quantified using ImageJ software (Version 1.50i; National Institutes of Health). (C) COLO 205 and HCT 116 cells were respectively treated with 50 and 60 µM itraconazole in a time-dependent manner for 24 h, and the expression levels of TKT and β-actin were detected via western blotting. Protein expression was quantified using ImageJ software (Version 1.50i; National Institutes of Health). COLO 205 and HCT 116 cells were treated with 50 and 60 µM itraconazole for 24 h, and (D) Annexin V/PI staining and (E) cell cycle distribution were detected via flow cytometry. *P<0.05 vs. control or vehicle control group. TKT, transketolase.
Itraconazole induces autophagy in colon cancer
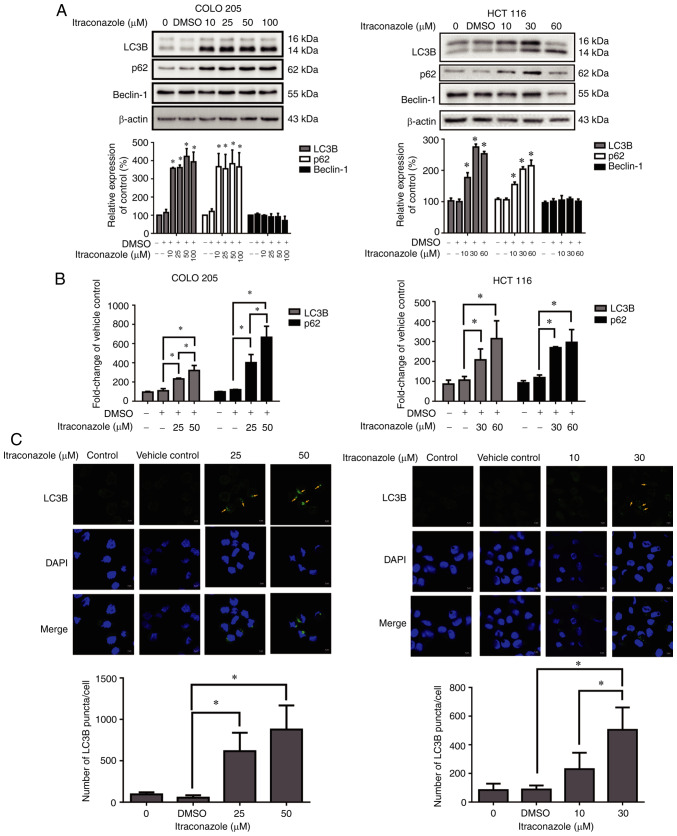
Autophagic cell death plays an important role in the treatment of cancer (31); therefore, the present study investigated the effect of itraconazole on inducing autophagy in colon cancer cells. The results demonstrated that the expression levels of the autophagy-related proteins, LC3B and p62, significantly increased in COLO 205 and HCT 116 cells following treatment with itraconazole for 24 h (P<0.05; Fig. 3A), and the mRNA expression levels of LC3B and p62 significantly increased following treatment with itraconazole, in a concentration-dependent manner (P<0.05; Fig. 3B). In addition, the number of LC3B puncta/cell increased following treatment with itraconazole (P<0.05; Fig. 3C). Taken together, these results suggest that itraconazole can induce autophagy in colon cancer cells.
Figure 3.
Itraconazole induces autophagic proteins and LC3B puncta expression. COLO 205 and HCT 116 cells were treated with different concentrations of itraconazole, and the (A) protein expression levels of LC3B, p62, Beclin-1 and β-actin were detected via western blot analysis, while (B) the mRNA expression levels were detected via reverse transcription-quantitative PCR analysis. (C) COLO 205 and HCT 116 cells were treated with itraconazole for 24 h and LC3B puncta was detected via confocal microscopy (software: ZEN, version: 2.6) (yellow arrows). The number of LC3B puncta/cell were quantified using ImageJ software (Version 1.50i; National Institutes of Health). *P<0.05 vs. vehicle control group.
Itraconazole-induced autophagy increases cell death in colon cancer cells
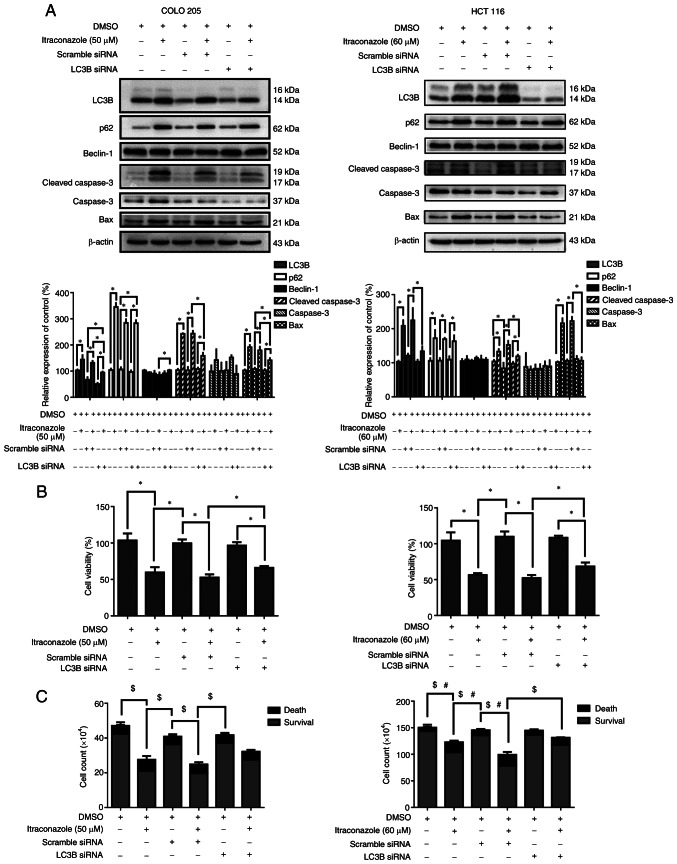
A previous study has reported that itraconazole can induce both apoptosis and autophagy in colon cancer cells (32). The role of autophagy in itraconazole-induced cell death remains controversial; thus, the present study used autophagy siRNA targeting LC3B to determine the role of autophagy in itraconazole-induced cell death. The results demonstrated that the expression levels of the autophagy-related proteins, LC3B and p62, decreased following transfection of COLO 205 and HCT 116 cells with LC3B siRNA (P<0.05; Fig. 4A). Furthermore, the expression levels of the apoptosis-related proteins, cleaved caspase-3 and Bax, also decreased (P<0.05; Fig. 4A). The viability of cells transfected with LC3B siRNA was assessed via the CCK-8 and trypan blue exclusion assays. The results demonstrated that cell viability decreased following treatment with itraconazole and transfection with the scrambled siRNA. Notably, these effects were reversed following transfection with LC3B siRNA (P<0.05; Fig. 4B and C). Collectively, these results suggest that autophagy plays an important role in itraconazole-induced cell death.
Figure 4.
Itraconazole-induced autophagy promotes cell death by activating LC3B. (A) COLO 205 and HCT 116 cells were transfected with scrambled siRNA or LC3B siRNA and subsequently treated with 50 or 60 µM itraconazole for 24 h. Western blot analysis was performed to detect the protein expression levels of LC3B, p62, Beclin-1, Cleaved caspase-3, caspase-3, Bax and β-actin. Protein expression was quantified using ImageJ software (Version 1.50i; National Institutes of Health). Cell viability was assessed via the (B) Cell Counting Kit-8 and (C) trypan blue exclusion assays. *P<0.05. $P<0.05 vs. survival cell counts. #P<0.05 vs. death cell counts. si, small interfering.
Discussion
The results of the present study demonstrated that itraconazole increased the 5-year survival rate of patients with late-stage colon cancer. Furthermore, treatment with itraconazole decreased TKT expression and induced apoptosis in vitro. Taken together, these results suggest that autophagy plays an important role in itraconazole-induced cell death. Itraconazole is a well-known medication for treating fungal infection and giving new indications to the FDA proofed drug is safe and cost-effective (33,34). Notably, itraconazole is associated with clinical outcomes and survival rates in patients with advanced gastric cancer (35). The results of the present study suggest that itraconazole may be a promising adjuvant therapeutic agent for colon cancer, based on the analyses of medical records in Taiwan.
As an FDA-approved antifungal drug, itraconazole has been demonstrated to inhibit the proliferation of gastric and liver cancer cells (36,37). Itraconazole targets AMP-activated protein kinase activation in the Hedgehog pathway, and inhibition of this signaling pathway can significantly decrease cell proliferation (32). Itraconazole induces cell cycle arrest at G1 and G2 phases in non-small cell lung cancer and also induces apoptosis (38). Alterations in the energy metabolism of cancer cells may be due to impaired mitochondrial function (39). Given that the glycolytic pathway serves as an intermediate and signaling complex, key enzymes in the signaling network, such as TKT, may be used as promising therapeutic candidates (40). The results of the present study demonstrated that itraconazole decreased cell viability and induced cell apoptosis, and in the cell cycle, induced subG1 phase and G1 arrest. Furthermore, cleaved caspase-3 expression increased, while TKT expression decreased following treatment with itraconazole. Collectively, these results suggest that itraconazole inhibits the proliferation and induces the apoptosis of colon cancer cells.
Autophagy plays a distinct role in tumor progression. In cancer cell biology, disruption of autophagy-related gene expression can enhance spontaneous tumor progression and increase the expansion of hepatitis B virus-induced premalignant lesions (41). Furthermore, autophagy-related genes, such as mTOR, class I PI3K and AKT, activate oncogenes to inhibit autophagy and enhance tumor formation (42). The results of the present study demonstrated that itraconazole increased the expression levels of the autophagy-related proteins, LC3B and p62, and LC3B puncta formation. Notably, p62 expression increased following treatment with itraconazole, and autophagy was also enhanced. A recent study demonstrated that p62 expression can serve as a predictor for drug-induced autophagic cell death (43). Furthermore, p62 depletion suppresses the recruitment of LC3B to autophagosomes, which increases the basal level of LC3B in cells overexpressing p62 (44). Taken together, these results suggest that p62 expression is associated with autophagic activity.
It is well-known that autophagy is an evolutionarily conserved intracellular recycling system; thus, with increased intracellular oxidative stress and organ damage, autophagy begins to self-degrade the injured organelle, producing more ATP to maintain cell function and survival (31). Given that 3-MA can induce undesired cytotoxicity on COLO 205 cells, it is difficult to identify the exact role of autophagy on itraconazole-induced apoptosis (Fig. S1); thus, the present study used siRNA to silence LC3B expression and identify the role of autophagy in itraconazole-treated colon cancer cells. The results demonstrated that decreased autophagy significantly increased cell viability compared with the itraconazole-only group. Taken together, these results suggest that itraconazole can induce both apoptosis and autophagy to inhibit the proliferation of colon cancer cells.
In the present study, the effect of itraconazole on the inhibition of colon cancer cell proliferation was reflected by an increased 5-year survival rate. However, the present study is not without limitations. First, itraconazole is not the standard treatment for colon cancer (45); thus, the number of patients with colon cancer treated with itraconazole was extremely small. Increasing the sample size of the database will increase the reliability of the data to confirm the clinical outcome of itraconazole treatment. Secondly, the present study only performed in vitro experiments to determine the effects of itraconazole on colon cancer cells. Thus, prospective studies will perform in vivo experiments to determine whether itraconazole directly inhibits colon cancer growth in mice. Currently, clinical trials assessing the effects of itraconazole on patients with colon cancer are ongoing. Thirdly, HCT 116 cell proliferation was inefficient in the matrix gel, thus the colony formation assay was unable to be performed to determine whether itraconazole induces apoptosis in both COLO 205 and HCT 116 cells (data not shown). Alternatively, the present study performed Annexin V/PI staining analysis to determine the effect of itraconazole on the apoptosis of colon cancer cells. Lastly, the associations between TKT, autophagy and apoptosis are highly complicated, involving gene-gene interactions and a specific sequence of gene expression. Thus, further studies are required to determine the potential role of itraconazole-induced autophagy in protecting cell survival or promoting cell death.
To the best of our knowledge, the present study was the first to assess the in vitro effects of itraconazole on inhibiting the proliferation of colon cancer cells. The results presented here offer a potential mechanism of action of itraconazole in inducing apoptosis and autophagy; thus, itraconazole may be used as a therapeutic target for the treatment of colon cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was partly supported by the Kaohsiung Veterans General Hospital (grant nos. VGHKS109-D04-1 and VGHKS109-180).
Funding
The present study was partly supported by the Kaohsiung Veterans General Hospital (grant nos. VGHKS109-D04-1 and VGHKS109-180).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
PWS, YMC, and SJY designed the experiments. CLL, ECL and HSH performed the experiments and molecular biology experiment results statistics. CHY and CLC acquired and analyzed the data. PWS, YMC, CLL and SJY drafted the initial manuscript. HSH and SJY confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Review Board of Kaohsiung Veterans General Hospital (IRB no. KSVGH18-CT10-07) and performed in accordance with the tenets of the Declaration of Helsinki (1975) and its later amendments (2013). The requirement for written informed consent was waived.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Färkkilä N, Torvinen S, Sintonen H, Saarto T, Järvinen H, Hänninen J, Taari K, Roine RP. Costs of colorectal cancer in different states of the disease. Acta Oncol. 2015;54:454–462. doi: 10.3109/0284186X.2014.985797. [DOI] [PubMed] [Google Scholar]
- 4.Aiello P, Sharghi M, Mansourkhani SM, Ardekan AP, Jouybari L, Daraei N, Peiro K, Mohamadian S, Rezaei M, Heidari M, et al. Medicinal plants in the prevention and treatment of colon cancer. Oxid Med Cell Longev. 2019;2019:2075614. doi: 10.1155/2019/2075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Yp, Hu Lf, Zheng Hf, Mao Cj, Hu Wd, Xiong Kp, Wang F, Liu Cf. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sin. 2013;34:625–635. doi: 10.1038/aps.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307–3318. doi: 10.1002/cncr.31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam MA, Sooro MA, Zhang P. Autophagic regulation of p62 is critical for cancer therapy. Int J Mol Sci. 2018;19:1405. doi: 10.3390/ijms19051405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauren P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 9.Jin S, Wei J, You L, Liu H, Qian W. Autophagy regulation and its dual role in blood cancers: A novel target for therapeutic development (Review) Oncol Rep. 2018;39:2473–2481. doi: 10.3892/or.2018.6370. [DOI] [PubMed] [Google Scholar]
- 10.Huang F, Wang BR, Wang YG. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24:4643–4651. doi: 10.3748/wjg.v24.i41.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang Y, Zhao J, Zhao M, Wang K. Allicin activates autophagic cell death to alleviate the malignant development of thyroid cancer. Exp Ther Med. 2018;15:3537–3543. doi: 10.3892/etm.2018.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stepanova NG, Demcheva MV. Formation of a pentose phosphate cycle metabolite, erythrose-4-phosphate, from initial compounds of glycolysis by transketolase from the rat liver. Biokhimiia. 1987;52:1907–1913. (In Russian) [PubMed] [Google Scholar]
- 13.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 14.Qin Z, Xiang C, Zhong F, Liu Y, Dong Q, Li K, Shi W, Ding C, Qin L, He F. Transketolase (TKT) activity and nuclear localization promote hepatocellular carcinoma in a metabolic and a non-metabolic manner. J Exp Clin Cancer Res. 2019;38:154. doi: 10.1186/s13046-019-1131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ricciardelli C, Lokman NA, Cheruvu S, Tan IA, Ween MP, Pyragius CE, Ruszkiewicz A, Hoffmann P, Oehler MK. Transketolase is upregulated in metastatic peritoneal implants and promotes ovarian cancer cell proliferation. Clin Exp Metastasis. 2015;32:441–455. doi: 10.1007/s10585-015-9718-1. [DOI] [PubMed] [Google Scholar]
- 16.Chao YK, Peng TL, Chuang WY, Yeh CJ, Li YL, Lu YC, Cheng AJ. Transketolase serves a poor prognosticator in esophageal cancer by promoting cell invasion via epithelial-mesenchymal transition. J Cancer. 2016;7:1804–1811. doi: 10.7150/jca.15467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu LH, Yang JH, Zhang DT, Zhang S, Wang L, Cai PC, Zheng JF, Huang JS. The TKTL1 gene influences total transketolase activity and cell proliferation in human colon cancer LoVo cells. Anticancer Drugs. 2007;18:427–433. doi: 10.1097/CAD.0b013e328013d99e. [DOI] [PubMed] [Google Scholar]
- 18.Cao L, Hong W, Cai P, Xu C, Bai X, Zhao Z, Huang M, Jin J. Cryptotanshinone strengthens the effect of gefitinib against non-small cell lung cancer through inhibiting transketolase. Eur J Pharmacol. 2021;890:173647. doi: 10.1016/j.ejphar.2020.173647. [DOI] [PubMed] [Google Scholar]
- 19.Bhatia A, Kanish B, Badyal DK, Kate P, Choudhary S. Efficacy of oral terbinafine versus itraconazole in treatment of dermatophytic infection of skin-A prospective, randomized comparative study. Indian J Pharmacol. 2019;51:116–119. doi: 10.4103/ijp.IJP_578_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pantziarka P, Sukhatme V, Bouche G, Meheus L, Sukhatme VP. Repurposing drugs in oncology (ReDO)-itraconazole as an anti-cancer agent. Ecancermedicalscience. 2015;9:521. doi: 10.3332/ecancer.2015.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi CH, Ryu JY, Cho YJ, Jeon HK, Choi JJ, Ylaya K, Lee YY, Kim TJ, Chung JY, Hewitt SM, et al. The anti-cancer effects of itraconazole in epithelial ovarian cancer. Sci Rep. 2017;7:6552. doi: 10.1038/s41598-017-06510-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Li J, Zhang T, Zou L, Chen Y, Wang K, Lei Y, Yuan K, Li Y, Lan J, et al. Itraconazole suppresses the growth of glioblastoma through induction of autophagy: Involvement of abnormal cholesterol trafficking. Autophagy. 2014;10:1241–1255. doi: 10.4161/auto.28912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudin CM, Brahmer JR, Juergens RA, Hann CL, Ettinger DS, Sebree R, Smith R, Aftab BT, Huang P, Liu JO. Phase 2 study of pemetrexed and itraconazole as second-line therapy for metastatic nonsquamous non-small-cell lung cancer. J Thorac Oncol. 2013;8:619–623. doi: 10.1097/JTO.0b013e31828c3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonarakis ES, Heath EI, Smith DC, Rathkopf D, Blackford AL, Danila DC, King S, Frost A, Ajiboye AS, Zhao M, et al. Repurposing itraconazole as a treatment for advanced prostate cancer: A noncomparative randomized phase II trial in men with metastatic castration-resistant prostate cancer. Oncologist. 2013;18:163–173. doi: 10.1634/theoncologist.2012-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Wei S, Zhao Y, Shi C, Liu P, Zhang C, Lei Y, Zhang B, Bai B, Huang Y, Zhang H. Anti-proliferation of breast cancer cells with itraconazole: Hedgehog pathway inhibition induces apoptosis and autophagic cell death. Cancer Lett. 2017;385:128–136. doi: 10.1016/j.canlet.2016.10.034. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh CY, Su CC, Shao SC, Sung SF, Lin SJ, Yang YH, Lai EC. Taiwan's national health insurance research database: Past and future. Clin Epidemiol. 2019;11:349–358. doi: 10.2147/CLEP.S196293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin CC, Lai MS, Syu CY, Chang SC, Tseng FY. Accuracy of diabetes diagnosis in health insurance claims data in Taiwan. J Formos Med Assoc. 2005;104:157–163. [PubMed] [Google Scholar]
- 28.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 29.Zonoobi E, Saeedfar K, Pourdowlat G, Masjedi MR, Behmanesh M. The study of IL-10 and IL-17A genes expression in patients with different stages of asthma: A case-control study. Tanaffos. 2018;17:146–154. [PMC free article] [PubMed] [Google Scholar]
- 30.Runwal G, Stamatakou E, Siddiqi FH, Puri C, Zhu Y, Rubinsztein DC. LC3-positive structures are prominent in autophagy-deficient cells. Sci Rep. 2019;9:10147. doi: 10.1038/s41598-019-46657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun CW, Lee SH. The roles of autophagy in cancer. Int J Mol Sci. 2018;19:3466. doi: 10.3390/ijms19113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng H, Huang L, Liao Z, Liu M, Li Q, Xu R. Itraconazole inhibits the Hedgehog signaling pathway thereby inducing autophagy-mediated apoptosis of colon cancer cells. Cell Death Dis. 2020;11:539. doi: 10.1038/s41419-020-02742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitburn J, Edwards CM, Sooriakumaran P. Metformin and prostate cancer: A new role for an old drug. Curr Urol Rep. 2017;18:46. doi: 10.1007/s11934-017-0693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellmann R, Smuszkiewicz P. Pharmacokinetics of antifungal drugs: Practical implications for optimized treatment of patients. Infection. 2017;45:737–779. doi: 10.1007/s15010-017-1042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan K, Yan R, Zhu K, Li W, Xu Z, Dang C, Li K. Itraconazole inhibits the proliferation of gastric cancer cells in vitro and improves patient survival. Oncol Lett. 2018;16:3651–3657. doi: 10.3892/ol.2018.9072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Dong X, Liu Y, Ni B, Sai N, You L, Sun M, Yao Y, Qu C, Yin X, Ni J. Itraconazole exerts anti-liver cancer potential through the Wnt, PI3K/AKT/mTOR, and ROS pathways. Biomed Pharmacother. 2020;131:110661. doi: 10.1016/j.biopha.2020.110661. [DOI] [PubMed] [Google Scholar]
- 37.Hu Q, Hou YC, Huang J, Fang JY, Xiong H. Itraconazole induces apoptosis and cell cycle arrest via inhibiting Hedgehog signaling in gastric cancer cells. J Exp Clin Cancer Res. 2017;36:50. doi: 10.1186/s13046-017-0526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alhakamy NA, Md S. Repurposing itraconazole loaded PLGA nanoparticles for improved antitumor efficacy in non-small cell lung cancers. Pharmaceutics. 2019;11:685. doi: 10.3390/pharmaceutics11120685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review) Oncol Lett. 2012;4:1151–1157. doi: 10.3892/ol.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gill KS, Fernandes P, O'Donovan TR, McKenna SL, Doddakula KK, Power DG, Soden DM, Forde PF. Glycolysis inhibition as a cancer treatment and its role in an anti-tumour immune response. Biochim Biophys Acta. 2016;1866:87–105. doi: 10.1016/j.bbcan.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi AMK, Ryter SW, Levine B. Autophagy in human health and disease. New Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 43.Chu CW, Ko HJ, Chou CH, Cheng TS, Cheng HW, Liang YH, Lai YL, Lin CY, Wang C, Loh JK, et al. Thioridazine enhances P62-mediated autophagy and apoptosis through wnt/beta-catenin signaling pathway in glioma cells. Int J Mol Sci. 2019;20:473. doi: 10.3390/ijms20030473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe T, Muro K, Ajioka Y, Hashiguchi Y, Ito Y, Saito Y, Hamaguchi T, Ishida H, Ishiguro M, Ishihara S, et al. Japanese society for cancer of the colon and rectum (JSCCR) guidelines 2016 for the treatment of colorectal cancer. Int J Clin Oncol. 2018;23:1–34. doi: 10.1007/s10147-017-1101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.