Summary
In systemic lupus erythematosus (SLE), the clearance of apoptotic cells and microparticles (MPs) is reduced. Some MPs contain molecules that can modulate immune responses. This study aimed to evaluate the presence of miR‐126 and miR‐146a in plasma MPs of patients with SLE (SLE MPs) and analyse the ability of MPs to modulate some events in the promonocytic U937 cell line. Circulating MPs were isolated from plasma samples of healthy controls (HCs), patients with SLE and other autoimmune diseases (OAD). MPs were analysed for size and cell origin by flow cytometry and content of miR‐126 and miR‐146a by RT‐qPCR. MPs were then added to U937 cell cultures to evaluate changes in cell phenotype, cytokine expression, content of miR‐126 and miR‐146a, and levels of IRF5. Patients with active SLE (aSLE) showed an increase in concentration of plasma MPs that positively correlated with the SLEDAI (SLE Disease Activity Index) score. CD14+ MPs were significantly more abundant in patients with SLE than HCs. SLE MPs contained decreased levels of miR‐146a, but the miR‐126 content in aSLE MPs was increased. The miR‐126 content in SLE MPs correlated positively with the SLEDAI score. The treatment of U937 cells with MPs from HCs and patients induced reduced expression of HLA‐DR, CD18 and CD119, increased frequency of IL‐6+ and TNF‐α+ cells, accumulation of IL‐8 in culture supernatants, increased miR‐126 levels and decreased miR‐146a content, but no change in the expression of IRF5. These findings suggest that plasma MPs, especially SLE MPs, could modulate some biological events in U937 cells.
Keywords: microparticles, microRNA, systemic lupus erythematosus
SLE‐MPs contained decrease levels of miR‐146a compare with HC. The miR‐126 content in aSLE‐MPs was increase. The treatment of U937 cells with MPs from HCs and patients increased miR‐126 levels and decreased miR‐146a content.
Abbreviations
- miRNA
microribonucleic acid
- MPs
microparticles
- SLE
systemic lupus erythematosus
- OAD
other autoimmune disease
- aSLE
active SLE
- iSLE
inactive
- HCs
healthy controls
- SLEDAI
SLE Disease Activity Index
- DNA
deoxyribonucleic acid
- RNA
ribonucleic acid
- PBMCs
peripheral blood mononuclear cells
- DNMT1
DNA methyltransferase 1
- DPBS
Dulbecco's phosphate‐buffered saline
- CBA
Cytometric bead array
- CMPs
healthy control microparticles
- SLE MPs
microparticles from patients with SLE
- OAD MPs
other autoimmune diseases
- PPP
platelet‐poor plasma
- SS
systemic sclerosis
- RA
rheumatoid arthritis
- MCTD
mixed connective tissue disease
- DM
dermatomyositis
- GPA
granulomatosis with polyangiitis
- TAK
Takayasu's arteritis
- TM
transverse myelitis
- AAV
ANCA‐associated vasculitis
- HSP
Henoch–Schönlein purpura
- PRED
prednisolone
- CQ
chloroquine
- HCQ
hydroxychloroquine
- AZA
azathioprine
- CP
cyclophosphamide
- MTX
methotrexate
INTRODUCTION
Autoimmune diseases such as systemic lupus erythematosus (SLE) is characterized by loss of immune tolerance; this phenomenon is facilitated by increased availability of autoantigens such as DNA, RNA, nuclear proteins and several cytoplasmic components [1, 2]. These autoantigens can circulate freely or be contained in vesicles such as microparticles (MPs) [3, 4, 5]. Microparticles are small extracellular vesicles, heterogeneous in size (0·1 μm−1 µm), cell origin, composition and stability [6]. Microparticles have multiple functions and can mediate intercellular communication, transfer molecules to recipient cells and induce intracellular signalling pathways. These functions imply that MPs contain biologically active mediators derived from their parental cells: immunoreactive molecules, transmembrane receptors, enzymes, transcription factors, cytokines, DNA and RNA [7, 8].
MicroRNAs (miRNAs) such as miR‐126 and miR‐146a that are differentially expressed in peripheral blood mononuclear cells (PBMCs) play essential roles in the pathogenesis of SLE [9]. In patients with SLE, the high miR‐126 content in CD4+ T cells is associated with reduced production of DNA methyltransferase 1 (DNMT1) and DNA hypomethylation at the CD70 and CD11a promoter regions [10]; these events are implicated in the pathogenesis of SLE. Overexpression of CD70 and CD11a in CD4+ T cells relates to their hyperactivity, the overstimulation of B cells and the subsequent production of autoantibodies [11]. miR‐146a expression correlates negatively with plasma levels of IFN type I (IFN‐I) and the expression of IRF5 and STAT1 in PBMCs of patients with SLE [12]; besides, miR‐146 also inhibits the proinflammatory NF‐κB transcriptional pathway, by binding to IRAK1 and TRAF6 mRNAs [13, 14]. In 2015, Perez‐Hernandez et al. found that urinary exosomes of patients with SLE contained higher levels of miRNA than cellular and cell‐free fractions present in urine. In particular, miR‐146a was found selectively in urine exosomes of patients with active lupus nephritis, suggesting its potential utility as a biomarker of renal disease activity [15]. This finding also highlighted the relevance of microvesicles as carriers of miRNAs and their possible involvement in the pathogenesis of inflammatory diseases. Microparticles transport or protect their cargo of miRNAs and other biologically active molecules with potential immunomodulatory capacity [16, 17]; therefore, MPs could modulate the response of recipient cells responsible of their clearance. After interacting with phagocytic cells, MPs could alter the cell miRNA content by two mechanisms: direct transfer of their miRNA cargo or induction/repression of the cellular miRNAs.
In this study, MPs isolated from peripheral blood of control and patients with SLE were shown to contain miR‐146a and miR‐126 and have the ability to modulate the levels of miR‐146a and miR‐126 in recipient U937 cells, but this effect was significant higher with SLE MPs. Based on these findings, we propose a model of the mechanism by which MPs could contribute to the pathogenesis of SLE.
MATERIALS AND METHODS
Blood samples
Microparticles were isolated from blood samples of patients with a diagnosis of SLE (n = 10) or other autoimmune diseases (OAD), under similar treatment (n = 10); also, age‐ and gender‐matched healthy controls (HCs) were enrolled in the study (n = 10). Patients were evaluated by the Grupo de Reumatología de la UDEA (GRUA) at Hospital Universitario San Vicente Fundación. The diagnosis of SLE was established according to the criteria of the American College of Rheumatology [18, 19]; patients were divided into two groups according to the disease activity index (SLEDAI) as follows: active SLE (SLEDAI score ≥4) and inactive SLE (SLEDAI score ≤4) [20]. All individuals agreed to participate in the study voluntarily by signing an informed consent approved by the Ethics Committee of the Institute of Medical Research in the School of Medicine at Universidad de Antioquia.
Reagents
RPMI culture media supplemented with glutamax (RPMI‐1640), fetal calf serum (FCS), and Dulbecco's phosphate‐buffered saline (DPBS) were purchased from Gibco®, Life Technologies (Grand Island, NY, USA); and penicillin and streptomycin from Cambrex/BioWhittaker (Walkersville, MD, USA). Phorbol‐12‐myristate‐13‐acetate (PMA), lipopolysaccharide (LPS from Escherichia coli, serotype O111:B4), 0·04% trypan blue dye, IGEPAL®CA630, protease inhibitor (Complete™ EDTA‐free Protease Inhibitor Cocktail l), 95% ethanol, and Millipore® Immobilon‐P PVDF transfer membranes (0·2 µm pore size) and RNase A were provided by Sigma‐Aldrich (St. Louis, MO, USA). Tris‐HCl, and 3,3′‐dihexyloxacarbocyanineiodide (DIOC6) were obtained from Invitrogen (Carlsbad, CA, USA). Cytometric bead array (CBA) kits, BD FACSFlow™ Sheath Fluid, propidium iodide (PI) and fluorochrome‐conjugated monoclonal antibodies (mAbs) specific for anti‐CD3‐PE‐Cy5 (clone UCHT1), anti‐CD19‐V450 (clone HIB19), anti‐CD18‐FITC (clone 7953), anti‐CD119‐PE (clone GIR‐208), anti‐IL‐6‐FITC (clone MQ2‐6A3) and anti‐TNFα‐PerCP‐Cy5 (clone MAb11) were purchased from BD Biosciences (San Diego, CA, USA). Fluorochrome‐conjugated mAbs specific for anti‐CD41a‐PE, (clone P2) and anti‐CD14‐RD1 (clone 322A‐1(MY4)) were provided by Beckman Coulter (Carlsbad, CA, USA), and anti‐HLA‐DR‐APCCy7 (clone L243) by Biolegend (San Diego, CA, USA). Fluorescent Fluoresbrite® Plain YG latex microspheres (0·1 µm, 0·5 μm and 1·0 µm in diameter) were obtained from Polysciences, Inc. (Warrington PA, USA). Mouse mAbs (all IgG1 isotype) specific for anti‐IRF5 (sc‐56714), anti‐DNMT‐1 (sc‐271729) and anti‐β‐actin (sc‐47778) were provided by Santa Cruz Biotechnology Inc. (Dallas, TX, USA), and IRDye® 800CW‐conjugated goat anti‐mouse IgG secondary polyclonal antibody by LI‐COR (Lincoln, NE, USA). Sodium chloride (NaCl) and sodium orthovanadate (Na3VO4) were acquired from MP Biomedicals, Inc. (Aurora, OH, USA) and tetramethylethylenediamine (Temed) from Amresco, Inc. (Cleveland, OH, USA). Thermo Fisher Scientific (Waltham, MA, USA) provided the following reagents: brefeldin A, sodium dodecyl sulphate (SDS), polysorbate 20 (Tween‐20), 7‐aminoactinomycin D (7‐ADD), sodium fluoride (NaF), nuclease‐free water, primers for 18S rRNA (Hs99999901S1), miR‐126 (hsa‐miR‐126‐3p sequence: UCGUACCGUGAGUAAUAAUGCG; catalogue ID 002228) and miRNA146a (hsa‐miR‐146a‐5p sequence: UGAGAACUGAAUUCCAUGGGUU; catalogue ID 000468), the kits for miRNA isolation (mirVana™ catalogue ID AM1560), PCR (RevertAid H Minus First Strand cDNA Synthesis, catalogue ID K1631), reverse transcription (TaqMan® MicroRNA Reverse Transcription, TaqMan Universal master mix II with UNG, catalogue ID 4440042) and protein quantification (Pierce BCA Protein Assay catalogue ID 23250). Acrylamide/Bis‐acrylamide, ammonium persulphate and molecular weight markers were obtained from Bio‐Rad (Hercules, CA, USA). All reagents for cell culture were endotoxin‐free by the LAL (Limulus amebocyte lysate) assay (Lonza, Visp, SWZ): LPS content was ≤0·1 EU/mL.
U937 cell line
The promonocytic U937 cell line (isolated from a histiocytic lymphoma of a 37‐year‐old male patient; ATCC® CRL1593·2™, Manassas, VA, USA) was cultured in RPMI‐1640 supplemented with 10% FCS, 100 U/mL penicillin and 100 µg/mL streptomycin. U937 cell viability and genome stability were evaluated in terms of mitochondrial membrane potential and cell cycle every 15 days; U937 cells were used up to passage 20 only, according to the published literature [21, 22].
The mitochondrial membrane potential (∆Ψm) of U937 cells was assessed using an adapted version of protocols previously described [23]. DiOC6 (80 nM final concentration) and PI (2 µg/mL final concentration) were pipetted and equally distributed in polystyrene test tubes. Cells were added and incubated for 30 min at 4° and darkness. Then, ∆Ψm was evaluated on a BD LSRFortessa™ (BD Biosciences) flow cytometer using 488‐nm solid‐state laser excitation; fluorescence from DiOC6 and PI was detected at 530/30 nm and 630/30 nm, respectively.
The cell cycle was evaluated by PI staining for determining the DNA relative content in cells, as described by the Darzynkiewicz et al literature [24]. Cells (0·5 × 106) were fixed with 300 μL of 70% ethanol (diluted in PBS pH 7·4) for 30 min at 4℃. Cells were then centrifuged (500 g, 5 min, 4℃), and the resulting pellets were washed twice with 3·0 mL of PBS. After, cells were stained with PI (1 μg/mL in PBS with 0·37% (w/v) EDTA, 0·01% (v/v) Triton‐X‐100 and 200 U/mL RNase A). After 30‐min incubation at darkness and room temperature, cells were acquired on a BD LSRFortessa™ flow cytometer. Cell data were analysed using the FlowJo software version 7·6.2 (Ashland, OR: Becton, Dickinson and Company; 2019).
Isolation of microparticles
Microparticles from patients with SLE (SLE MPs), other autoimmune diseases (OAD MPs) and healthy controls (CMPs) were isolated from 12 mL of citrated venous blood (BD‐Vacutainer® 3·8% sodium citrate tubes; Becton Dickinson). Blood samples were centrifuged at 1,800 x g for 10 minutes at room temperature to obtain platelet‐rich plasma; then, 2 mL of this plasma was centrifuged twice at 3,000 g for 20 minutes at room temperature to obtain platelet‐poor plasma (PPP). Finally, 1 mL of PPP was centrifuged at 16,900 g for 1 hour to allow the sedimentation of MPs. These vesicles were suspended in DPBS and kept at −70℃ until use.
Characterization of microparticles
The isolated vesicles were confirmed to be MPs (0·1 μm ‐ 1 μm in diameter) by flow cytometric analysis using fluorescent reference latex microspheres with diameters of 0·1 μm, 0·5 μm and 1·0 μm. The acquisition threshold for size (FSC‐A) and granularity (SSC‐A) parameters were set with filtered sheath fluid. MP samples (suspended in 200 µL of sheath fluid) were acquired for 1 min on a BD FACSCanto™ II flow cytometer (BD Biosciences) with the FACSDiva™ software version 7·6.1 (BD Biosciences). Data were analysed using the FlowJo software version 7·6.2. Microparticle counts were used for calculating the plasma MP concentration (MPs/mL) for each individual. The protein content of MP samples was quantified by the bicinchoninic acid (BCA) method following the manufacturer's instructions. The absorbance of samples was read at 562 nm using a Universal Microplate Reader BiotekELx800NB spectrophotometer (BioTek Instruments, Winooski, VT). Protein concentrations were calculated using a standard curve made with known concentrations of bovine serum albumin (BSA).
Microparticle phenotyping
Microparticles (100 µL) were incubated with fluorescent mAbs for specific markers of platelets (anti‐CD41a‐PE), monocytes (anti‐CD14 MY4‐RD1), T cells (anti‐CD3‐PECY5) and B cells (anti‐CD19‐V450) for 30 min. Then, MPs were washed with 200 µL of DPBS at 16,900 g for 1 h. After, the supernatant was removed, and the MPs were suspended in 200 µL of DPBS and acquired on a BD FACSCanto™ II flow cytometer; frequency of MPs positive for each marker was recorded.
Treatment of U937 cells with microparticles
U937 cells (1 × 106) were cultured with 30 µg/µL protein of CMPs, SLE MPs or OAD MPs for 2 h. LPS (10 ng/mL) and PMA (0·5 µg/mL) were used as positive controls. For some assays, brefeldin A (1 μg/mL) was added to cell cultures for 2 h. From each experimental condition, cells, cell lysates and culture supernatants were prepared for different analyses as described below.
Cellular activation by MPs
U937 cells (0·5 × 106) previously untreated of treated with MPs were washed with cold FACS buffer at 500 g for 5 min. at 4℃; then, cells were incubated with blocking solution for 15 min at 4℃, centrifuged at 500 g for 5 min. at 4℃, suspended in FACS buffer and stained with fluorescent mAbs: anti‐HLA‐DR, anti‐CD18, anti‐CD119, anti‐CD64, anti‐CCR5, anti‐CD11a, anti‐CD11c and anti‐CD163 at 4℃ for 30 min. Cells were acquired on a BD FACSCanto™ II flow cytometer with the FACSDiva™ software version 7·6.1. Data were analysed using the FlowJo software version 7·6.2; mean fluorescent intensity (MFI) for each molecule and percentage of marker‐positive cells were recorded.
Cytokines
Two approaches were used to evaluate the synthesis of cytokines by U937 cells previously untreated or treated with MPs. In the first one, U937 cells from cultures exposed to brefeldin A were suspended in 250 uL of fixation/ permeabilization solution for 60 min at 4℃ and washed twice with Perm/Wash buffer at 1700 x g, at 4℃, for 5 min. Then, fluorescent mAbs for IL‐6 and TNF‐α were added and cells incubated for 30 min, at 4℃, in darkness. After washing with FACS buffer, cells were acquired on a BD FACSCanto™ II flow cytometer with the FACSDiva™ software version 7·6.1. Data were analysed using the FlowJo software version 7·6.2; the percentage of IL‐6+ and TNF‐α+ U937 cells were recorded.
In the second approach, supernatants were collected from U937 cultures non‐exposed to brefeldin A for measuring IL‐8, IL‐1β, IL‐6, IL‐10, IL‐12p70 and TNF‐α using the BD CBA Human Inflammatory Cytokine Kit. Samples were acquired on a BD FACSCanto™ II flow cytometer with the FACSDiva™ software version 7·6.1. Data were analysed using the FlowJo software version 7·6.2; data were recorded as ng/mL for each cytokine.
miRNA isolation and quantification
miR‐126 and miR‐146a were isolated and quantified in MPs isolated from controls and patients and in U937 cells previously untreated or MP‐treated MPs. The RNA was isolated using the mirVana™ miRNA Isolation Kit following the manufacturer's instructions. However, quantification of the content of miR‐126 and miR‐146a was first necessary to determine the minimal amounts of MPs, U937 and primary monocytes required for optimal RNA isolation and miRNA amplification. To this purpose, different concentrations of MPs (µg/µl protein content) and cells (cells/mL) were screened to obtain at least 10 ng/µL RNA from MPs and 50 ng/µL from cells. The screening showed that 5000 µg/mL MPs and 1x106 U937 cells were necessary for the assay.
RNA isolated from MPs and U937 cells were used to generate cDNAs of interest in a CFX96™ Real‐Time PCR detection System (Bio‐Rad) by reverse transcription–quantitative polymerase chain reaction (RT‐qPCR) using the cDNA RevertAid H Minus First Strand cDNA Synthesis Kit (Applied Biosystems) and the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems; Bedford, MA, USA) with specific primers for hsa‐miR‐126‐3p and hsa‐miR‐146a‐5p. Then, cDNA amplification was performed with the TaqMan® Universal Master Mix II (Applied Biosystems) using specific primers for miR‐126, miRNA146a and 18S (HS99999901S1). Standardization was done according to the relative 18S rRNA expression. The content of miRNAs in MP‐treated U937 cells was expressed in fold change relative to miRNA present in untreated U937 cells using the Livak and Schmittgen method [23].
Protein electrophoresis and Western blotting
U937 cells (1x106) previously untreated or treated with MPs, PMA (0·5 µg/mL) or LPS (10 ng/mL LPS) were suspended in IGEPAL lysis buffer (50mM Tris‐HCl, 120mM NaCl, 0·5% IGEPAL, 50mM NaF, 1mM Na3VO4 and 60mM Complete™ EDTA‐free Protease Inhibitor Cocktail l) at 4℃ for 20 min. Cells were centrifuged at 13,000 x g at 4℃ for 10 min, and the supernatant was collected. The protein content was quantified by the bicinchoninic acid (BCA) method following the manufacturer's instructions. The absorbance of samples was read at 562 nm using a Universal Microplate Reader BiotekELx800NB spectrophotometer (BioTek Instruments, Winooski, VT, USA). Protein concentrations were calculated using a standard curve made with known concentrations of bovine serum albumin (BSA).
Proteins in cell lysates (30 μg/mL) were separated by electrophoresis in 10% polyacrylamide gel with sodium dodecyl sulphate (SDS‐PAGE) at 200 V for 45 min. A wet protein transfer onto polyvinylidene fluoride (PVDF) membranes was performed at 40 V, 4℃, for 1 hour using a Mini Trans‐Blot® Electrophoretic Transfer Cell (Bio‐Rad, Hercules, CA, USA). After transfer, free membrane sites were blocked with 10 ml blocking buffer (5% skim milk, 0·05% Tween‐20 in 1X PBS) for 1 hour at room temperature. The membranes were then incubated for 2 h with mouse anti‐human IRF5 and β‐actin primary mAbs (both diluted 1:200 in blocking buffer). After, the membranes were incubated with IRDye® 800CW‐conjugated goat anti‐mouse IgG1 secondary polyclonal antibody (diluted 1:7000 in blocking buffer) at room temperature for 1 hour. Finally, fluorescent bands were evaluated using the Li‐COR Odyssey® 9120 Infrared Imaging System with the ODYSSEY® application software (LI‐COR Biosciences; Lincoln, NE, USA). The IRF5 expression was estimated by densitometry analysis in relation to β‐actin that was used as a loading control.
Statistical analysis
Parameters of patients and controls are described as absolute frequencies in the case of qualitative variables (sex, autoimmune diseases and frequency of MPs derived from different cell sources); as medians and interquartile ranges for non‐normally distributed quantitative variables (age, SLEDAI score and number of MPs/mL); and as means and standard deviations (SD) for normally distributed quantitative variables. The Anderson–Darling test and the Shapiro–Wilk normality test were used for comparison, and when the data do not have a normal distribution because of the very small sample, all non‐parametric tests were used to compare the data. Frequencies and numbers of MPs/mL positive for each lineage marker were compared with the Kruskal–Wallis test, followed by Dunn's multiple comparison test. Variables in experiments performed with and without MPs were compared with the Wilcoxon test and Dunn's multiple comparison test.
Correlation analyses were performed with Pearson's or Spearman's coefficients. In all cases, a P‐value ≤0·05 was considered statistically significant. Statistical analyses were run in GraphPad Prism version 6 (GraphPad Software; La Jolla, CA, USA).
RESULTS
Demographic characteristics of patients and controls
Each study group (patients with SLE, patients with OAD and healthy controls) included 10 individuals and all individuals aged between 17 and 42 years, and 90% were female. In the SLE group, the median SLEDAI score for patients with active and inactive disease was 9·5 (range = 4‐33) and 0·5 (range = 0‐3), respectively. Treatment of patients with SLE and OAD was similar (Table 1).
TABLE 1.
Characteristics of patients and controls
| Characteristic | Healthy controls | Patients with aSLE | Patients with iSLE | Patients with OAD |
|---|---|---|---|---|
| n | 10 | 10 | 10 | 10 |
| Sex F/M | 9/1 | 9/1 | 9/1 | 9/1 |
| Age (years)a | 28 (18–38) | 30 (17–42) | 27·5 (19–36) | 30 (17–36) |
| Treatment | N/A | PRED, CQ, HCQ, AZA, CP, MTX | PRED, CQ, HCQ | PRED, CQ, AZA, CP, MTX |
| SLEDAIa | N/A | 9·5 (4–33) | 0·5 (0–3) | N/A |
| Diagnosis (n) | N/A | SLE (10) | SLE (10) |
SS (2) RA (1) (MCTD) (1) DM (1) GPA (1) TAK (1) TM (1) AAV (1) HSP (1) |
AAV (ANCA)‐associated vasculitis; aSLE_ active SLE; AZA, azathioprine; CP, cyclophosphamide; CQ, chloroquine; DM, dermatomyositis; GPA, granulomatosis with polyangiitis; HCQ, hydroxychloroquine; HSP, Henoch–Schönlein purpura; iSLE, inactive SLE; MCTD, mixed connective tissue disease; MTX, methotrexate; N/A, not applicable; PRED, prednisolone; RA, rheumatoid arthritis; SLEDAI, systemic lupus erythematosus Disease Activity Index score. SS, systemic sclerosis; TAK, Takayasu's arteritis; TM, transverse myelitis.
Median (interquartile range).
Characterization of plasma microparticles
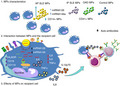
Circulating MPs isolated from patients and healthy controls were analysed by flow cytometry. The MP region was depicted in SSC‐A vs FSC‐A dot plots (Figure 1a), and the size (FSC‐H) of MPs was evaluated in comparison with reference latex beads; MPs ranged in size between 0·1 µm and 1·0 µm being smaller than platelets (Figure 1b).
FIGURE 1.
Identification and characterization of plasma microparticles. Microparticles (MPs) were isolated from plasma of healthy controls (HC) and patients with active SLE (aSLE), inactive SLE (iSLE) and other autoimmune diseases (OAD). (a) The MP region was set in a SSC‐A vs FSC‐A dot plot (0·1 µm to 1·0 µm diameter range). (b) FSC‐H histogram was used to confirm the size of MPs according to fluorescent reference latex spheres. The red line corresponds to the distribution of the reference spheres (0·1 µm to 1·0 µm in diameter) and the blue line to the distribution of MPs. (c) Concentration of plasma MPs in healthy controls and patients. Horizontal bars correspond to median values; the Kruskal–Wallis test and Dunn post‐test, *p < 0·05 (n = 10 per group). (d) Correlation between the SLEDAI score and the concentration of circulating MPs in patients with SLE (r = 0·5530); Spearman's correlation coefficient, p < 0·05 (n = 20). (e) MPs isolated from healthy controls and patients were stained with fluorescent mAbs for specific cell surface lineage markers and analysed by flow cytometer. Frequencies of CD41a+, CD3+ CD19+ and CD14+ MPs are shown for each study group. Columns and bars correspond to mean and SEM values. The Kruskal–Wallis test and Dunn post‐test, * p < 0·05 (n = 10 per group). (f‐h) Microparticles were lysed, miR‐126 and miR‐146a were amplified by RT‐qPCR, and quantified in comparison with 18S rRNA. Relative content of (f) miR‐126 and (g) miR‐146a in MPs isolated from healthy controls and patients. The Kruskal–Wallis test and Dunn post‐test, * p < 0·05 (n = 10 per group). (h) Correlation between the SLEDAI score and the miR‐126 content in circulating MPs from patients with SLE (r = 0·7069); Pearson's correlation coefficient, p < 0·05 (n = 20)
Plasma MP concentration is higher in patients with SLE and correlates with disease activity
The concentration of circulating MPs (MPs/mL) was higher in patients with aSLE in comparison with healthy controls (p ≤ 0·05) (Figure 1c); moreover, the concentration of MPs from patients with SLE correlated positively with the SLEDAI score (Figure 1d). On the other hand, CD41a+ MPs were the most abundant in plasma of patients and controls, and all individuals exhibited similar proportions of circulating CD41a+ and CD3+ MPs; however, patients with aSLE had a higher proportion of CD14+ MPs and patients with OAD a lower proportion of CD19+ MPs (Figure 1e).
Plasma microparticles from aSLE MPs contain higher miR‐126 and lower miR‐146a levels than CMPs
Some miRNAs, such as miR‐126 and miR‐146a, are involved in the pathogenesis of SLE. Therefore, their levels were evaluated in plasma microparticles isolated from healthy controls and patients. The content of miR‐126 was found to be 5·8 times higher and that of miR‐146a 2·8 times lower in aSLE MPs than CMPs (Figure 1f,g). These results evidenced that although miRNAs can be part of the cargo transported by MPs, their levels differ in patients with SLE; it is important to highlight that the content of miR‐126 correlated positively with the SLEDAI score (Figure 1h).
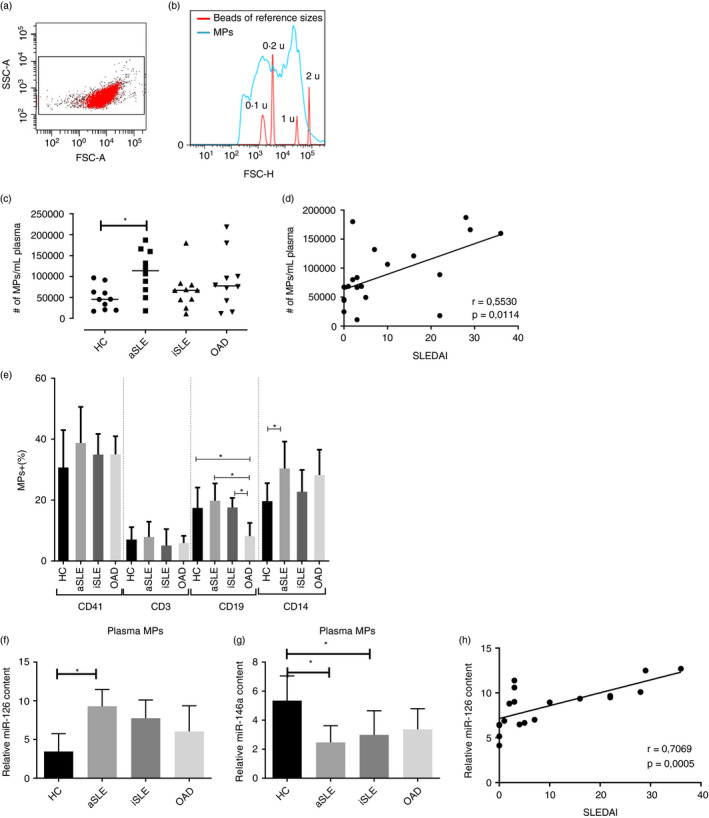
Plasma microparticles induce changes in activation markers of U937 cells
The surface expression of several molecules (HLA‐DR, CD18, CD119, CD64, CCR5, CD11a, CD11c and CD163) was evaluated in U937 cells after treatment with MPs. These proteins are relevant in the functionality of mononuclear phagocytic cells and are modulated during their activation and differentiation processes [25, 26, 27, 28]. To this purpose, U937 cells were exposed to different concentrations of MPs (0·5, 1,5, 10 and 30 µg/µL protein) (Figure S1) isolated from patients and controls for 2 h. The expression (MFI) of HLA‐DR decreased in all cultures of U937 cells independently of the source of MPs (Figure 2a). As decreased HLA‐DR expression could be related to cell death, cell viability was evaluated. Exposure of U937 cells to MPs (30 µg/µl protein) for 2 h did not alter either the ∆Ψm (DIOC6 staining) or the cell membrane permeability (PI uptake) in comparison with untreated cells; similar results were observed even after 72‐h incubation (data not shown). Additionally, MPs, regardless of their source, induced a lower expression of CD18 and CD119 in U937 cells (Figure 2e,f; Figure S2); there were no changes in the MFI for CD64, CCR5, CD11a, CD11c or CD163 (data not shown).
FIGURE 2.
Effect of plasma microparticles on the phenotype of U937 cells. U937 cells were untreated or treated for 2 h with plasma MPs (30 µg/µl protein) isolated from patients with active SLE (aSLE), inactive SLE (iSLE) and other autoimmune diseases (OAD). Cells were then stained with mAbs for molecules involved in cell functionality and analysed by flow cytometry. (a) HLA‐DR expression in U937 cells exposed to MPs from HC and patients; PMA (0·5 µg/µl): positive control. Wilcoxon's test, * p < 0·05 (n = 10 independent experiments). Expression of (b) CD18 and (c) CD119 in U937 cell exposed to MPs from healthy controls (CMPs). Wilcoxon's test, p < 0·05 (n = 3)
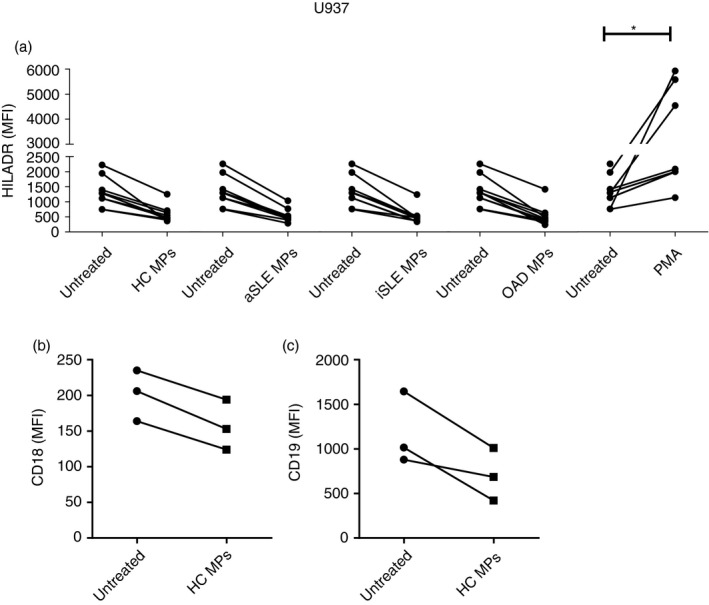
Plasma microparticles alter the production of cytokines by U937 cells
Given the previous results, the next step was to determine the effect of MPs (30 µg/ul protein content) on the production of inflammatory cytokines by U937 cells after 2‐h incubation. To this purpose, CMPs (from different individuals, n = 3) were added to U937 cell cultures in the presence of brefeldin A to evaluate the intracellular accumulation of IL‐6 and TNF‐α. Additionally, CMPs, SLE MPs and OAD MPs (from 3 different individuals per group) were added to U937 cell cultures to analyse the levels of inflammatory cytokines (IL‐8, IL‐1β, IL‐6, IL‐10, IL‐12p70 and TNF‐α) in culture supernatants. Untreated and LPS‐treated cells were used as negative and positive controls, respectively.
In the first approach, CMP‐treated U937 cell cultures showed higher frequencies of IL‐6+ and TNF‐α+ U937 cells than untreated cell cultures (Figure 3a,b; and Figure S3). In the second approach, only IL‐8 increased in response to CMPs, SLE MPs and OAD MPs (Figure 3c); no changes were observed in the levels of IL‐1β, IL‐6, IL‐10, IL‐12p70 and TNF‐α (data not shown).
FIGURE 3.
Effect of plasma microparticles on cytokine production by U937 cells. U937 cells were untreated or treated for 2 h with plasma MPs (30 µg/µl protein) isolated from healthy controls (HCs) and patients. The intracellular accumulation of (a) IL‐6 and (b) TNF‐α was evaluated with fluorescent mAbs and flow cytometry in U937 cells exposed to MPs from HCs (CMPs). Wilcoxon's test, p < 0·05. (n = 3). (c) Concentration of IL‐8, measured by CBA, in supernatants from U937 cultures exposed to MPs from HCs and patients with active SLE (aSLE), inactive SLE (iSLE) or other autoimmune diseases (OAD). LPS (10 ng/mL): positive control. The Kruskal–Wallis test, * p < 0·05. The dotted line represents the detection limit for IL‐8 by CBA (n = 10 independent experiments)
Plasma microparticles alter the expression of miR‐126 and miR‐146 in U937 cells
As previously shown, MPs containing miR‐126 and miR‐146 induced changes in phenotype and synthesis of cytokines by U937 cells. The next step was to evaluate the effect of MPs on the miRNA content of U937 cells. Therefore, the levels of miR‐126 and miR‐146a in U937 cells were evaluated; also, the expression of IRF5 protein, a target of both miRNAs, was analysed. To this purpose, U937 cells were exposed to MPs (30 µg/ul protein) isolated from patients and controls for 2 h. Untreated and LPS‐ or PMA‐treated U937 cells were used as negative and positive controls, respectively. LPS induced a reduction in miR‐126, but PMA and all types of MPs (CMPs, SLE MPs, and OAD MPs) induced an increase in the levels of this miRNA. On the other hand, opposite results were observed for miR‐146a; that is, LPS increased its content in U937 cells, although PMA and all types of MPs induced its reduction; it is important to note that the downregulating effect over miR‐146a was significantly higher for aSLE MPs than for CMPs (Figure 4a,b).
FIGURE 4.
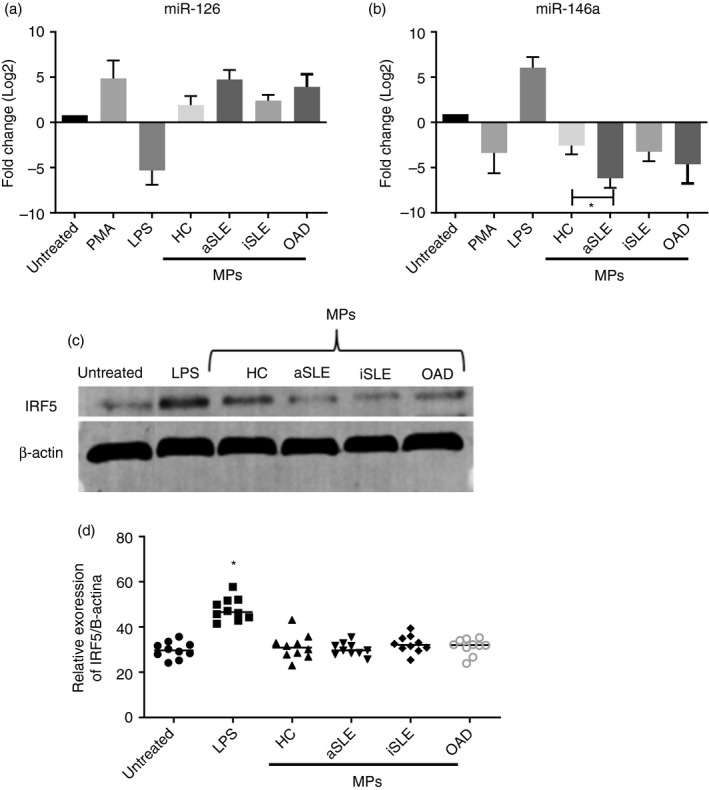
Effect of plasma microparticles on the levels of miR‐126, miR‐146a and IRF5 of U937 cells. U937 cells were untreated or treated for 2 h with plasma MPs (30 µg/µl protein) isolated from healthy controls (HC) and patients. Cell lysates were prepared for miRNA and protein analysis. (a–b) miR‐126 and miR‐146a were amplified by RT‐qPCR, and their quantity was normalised to the relative expression of 18S rRNA. Content of (a) miR‐126 and (b) miR‐146a in cell cultures exposed to MPs isolated from HCs, and patients with active SLE (aSLE), inactive SLE (iSLE) or other autoimmune diseases (OAD). Positive controls: PMA (0·5 µg/µL) and LPS (10 ng/mL). The Kruskal–Wallis test, * p < 0·05. (n = 10 individuals per group). (c) IRF5 expression, evaluated by Western blotting, in cell lysates of U937 cells treated with MPs isolated from HCs and patients with aSLE, iSLE or OAD. Positive control: LPS (10 ng/mL). (d) Densitometry analysis of IRF5 relative to β‐actin. The Kruskal–Wallis test, p < 0·05 (n = 10 individuals per group)
Plasma microparticles did not alter the IRF5 expression in U937 cells
Decreased miR‐146a content in MP‐treated U937 cells could have functional consequences; therefore, the levels of IRF5, a target of miR‐146a, were measured in lysates of U937 cells after treatment with MPs (30 µg/µL protein) for 2 h. Densitometry analysis showed no differences in the IRF5 expression in U937 cells exposed to any type of MPs for 2 h, whereas the IRF5 protein content increased in the presence of LPS (Figure 4c,d).
DISCUSSION
Several studies have reported that both plasma and urine microvesicles can transport miRNAs [29, 30]. Perez‐Hernandez et al. found increased levels of miR‐335, miR‐302d, miR‐200c, and miR‐146a in urine exosomes from patients with SLE; what is more, the high miR‐146a content in urine exosomes of patients with active lupus nephritis differentiates them from patients with SLE but without nephritis [15].
In the present study, and in comparison with CMPs, the aSLE MPs were found to have particular characteristics: higher plasma concentration, higher proportion of monocytic origin, as previously described [31], lower miR‐146a content and higher levels of miR‐126; noticeably, both the plasma concentration and the miR‐126 content correlated positively with the SLEDAI score. As mentioned before, urine exosomes of patients with SLE are enriched in miR‐146a [15], contrary to plasma MPs here observed, suggesting that miRNAs have differential tissue distribution and could be related to lesions in different organs.
The present findings of higher miR‐126/lower miR‐146a levels in circulating MPs from patients with aSLE cannot be attributed to pharmacotherapy; patients with OAD under similar treatment had plasma MPs with levels of miR‐126 and miR‐146a similar to those in CMPs. Therefore, the miRNA composition of aSLE MPs could be inherent to the disease. Interestingly, circulating miRNA profiles were similar in patients with SLE and other conditions who were under similar immunosuppressant treatments [32].
According to previous studies, delivering of a bioactive cargo from extracellular vesicles to recipient cells can alter the content of cell proteins, DNA, RNA and miRNAs [33]. In the present study, when compared to CMPs, aSLE MPs induced a significant decrease in miR‐146a and an increasing trend of miR‐126 in recipient U937 cells; this increment could have resulted from direct transfer from MPs.
The present results suggested that aSLE MPs could regulate the cellular content of miR‐126 and miR‐146a giving rise to values inverse to those of freely circulating miRNAs (lower miR‐126/higher miR‐46a) previously described in patients with SLE. Other studies have shown that extracellular vesicles modulate the expression of miRNAs in recipient cells; for instance, endothelium‐derived vesicles upregulated the expression of miR‐126 and miR‐146b in primary monocytes and THP1 cells, and platelet‐derived MPs induced a higher miR‐126 content in macrophages [34].
The induction of altered miRNA levels in recipient cells could have pathogenic consequences; therefore, the expression of IRF5 was evaluated because in addition to being a target of miR‐126 and miR‐146a, it also participates in the pathogenesis of SLE. IRF5 is a transcription factor involved in IFN‐I induction and cell growth, differentiation, activation and apoptosis [35] and also an important factor in SLE; in fact, the expression of IRF5 is required for MRL/lpr mice to develop SLE. In the absence of IRF5, autoantibodies, activated CD4+ T‐cell counts and production of inflammatory cytokines (TNF‐α, IL‐6, e IL‐10) decreased [36]. Moreover, IRF5 polymorphisms and altered IRF5 expression are associated with Th1 and Th17 responses, macrophage polarization towards the M1 inflammatory phenotype, elevated synthesis of IFN‐α by plasmacytoid dendritic cells and SLE susceptibility [37, 38, 39, 40]. Additionally, IRF5 is increased in macrophages of patients with SLE, and the kinetics of nuclear localization of IRF5 correlates with the production of IFN‐α, TNF‐α and IL‐6 in monocytes stimulated with sera or autoantigens of patients with SLE [41]. Different miRNAs, such as miR3‐02c and miR‐520b, also regulate the expression of IRF5 [12, 42]. However, the IRF5 expression did not change in U937 cells exposed to any type of plasma microparticles here analysed. The transport of miRNA between cells is not the only way that MP could modulate immunological or inflammatory events; previously, evidence has shown that MP also could modulate the activation of NF‐kB or the macrophage differentiation [43, 44].
Additionally, the present study showed that plasma MPs induce changes in recipient U937 cells such as reduced expression of HLA‐DR, CD18 and CD119 and higher synthesis of proinflammatory cytokines such as IL‐6 and TNF‐α. Similar observations are found in the literature; for instance, leucocyte‐derived MPs from healthy controls induced the release of proinflammatory cytokines (IL‐6, IL‐8 and members of the monocyte chemoattractant protein/MCP family) by synovial fibroblasts from patients with arthritis rheumatoid and osteoarthritis [45, 46]. Likewise, it has also been described that circulating SLE MPs can interact with and activate plasmacytoid and myeloid dendritic cells to increase the production of IFN‐α, IFN‐γ, TNF‐α and TNF‐α, IL‐6, respectively [47].
Since a clinical point of view, it is important to note that present data showed that the concentration of plasma MPs and their content of miR‐126a correlated positively with the SLEDAI score.
Finally, based on the present results, we propose a model for the mechanism by which circulating MPs, regardless of their cellular origin, interact with phagocytic cells such as monocytes and induce decreased expression of membrane proteins involved in cell function (HLA‐DR, CD18 and CD119); augment the synthesis of IL‐8, IL‐6 and TNF‐α; reduce the levels of miR‐146a; and increase the content of miR‐126. This interaction could be mediated by surface receptors such as Fc, mannose binding lectin receptor, B‐cell receptor and others or can also occurred by membrane fusion. The cellular uptake or binding may be dependent of the molecules found in the surface of the MP, such as DNA, RNA, or facilitated by the formation of immune complex as our group described previously [44]. These interactions with phagocyte cells, such as monocytes, induce a decrease in the expression of membrane proteins involved in cell function (HLA‐DR, CD18 and CD119). All these events can be relevant in the pathogenesis of SLE because as indicated in the diagram, the concentration, cell origin, miR‐126 and miR‐146a content of plasma microparticles from patients with SLE are different (Figure 5). Moreover, because of their positive correlation with the SLEDAI score, plasma concentration of MPs and their miRNA 126 content could become biomarkers of disease activity in patients with SLE.
FIGURE 5.
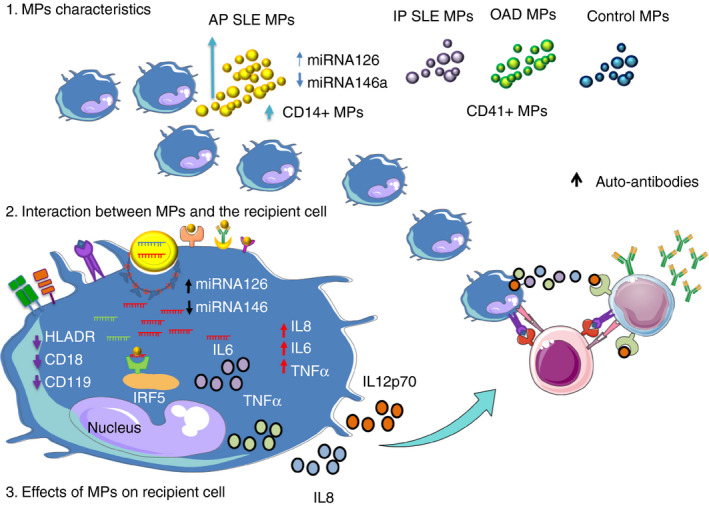
Effect of plasma microparticles of patients with SLE on the expression of miRNAs in phagocytic cells: hypothetical model. 1. Characteristics of microparticles (MPs). Patients with active SLE (aSLE) have a higher concentration of plasma MPs in comparison with patients with inactive SLE (iSLE), other autoimmune diseases (OAD) and healthy controls (HCs). Moreover, aSLE MPs exhibit a higher frequency of monocyte‐derived MPs (CD14+), have higher content of miR‐126 and have lower levels of miR‐146a. The frequency of platelet‐derived MPs (CD41+) is elevated in all study groups. 2. Interaction between MPs and recipient cells. MPs could interact with recipient cells by different mechanisms: linking to cell surface membrane receptors, fusing with cell membranes or undergoing endocytosis/phagocytosis. After, MPs can release their biologically active cargo into the recipient cells. 3. Effect of MPs on recipient cells. Microparticles could mediate intercellular communication, transfer molecular components or induce cellular signalling pathways in the recipient cells. In turn, these events could induce changes in the expression of membrane molecules, cytokine synthesis and/or levels of proteins and miRNAs in the recipient cells. MP‐containing miRNAs induce cellular changes (decreased expression of HLA‐DR, CD18 and CD119, increased synthesis of IL‐8, IL‐6 and TNF‐α, lower levels of miR‐146a, and increasing trend of miR‐126) in recipient U937 cells. However, in comparison with CMPs, aSLE MPs induced significant changes in recipient U937 cells: increased synthesis of IL‐8 and decreased levels of miR‐146a. Therefore, we hypothesized that SLE MPs have particular characteristics that are related to their different effects on recipient cells and could explain their role in the pathogenesis of SLE
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
AUTHOR CONTRIBUTION
L. Carmona‐Pérez performed the experiments. L. Carmona‐Pérez, Rojas M. and Vásquez G. analysed the results shown in this manuscript, contributed to the preparation of the figures and wrote the manuscript. C. Muñoz‐Vahos C and A. Vanegas‐García helped to evaluate patients. All authors approved the final version of the manuscript.
Supporting information
Fig S1
Fig S2
Fig S3
ACKNOWLEDGEMENTS
We want to thank COLCIENCIAS, which, through its Call for Young Researchers and Innovators, promotes the training of professionals with researcher profile. Additionally, we would like to thank COLCIENCIAS for funding the project 645 of 2014, code 1115556933389. We also thank the Flow Cytometry Unit and the Sustainability Program of the Universidad de Antioquia for supporting the development of this project. Finally, a sincere acknowledgement to the Universidad de Antioquia, the GICIG Research Group, the patients and healthy controls, and to Martha Mesa for the edition support.
Carmona‐Pérez L, Rojas M, Muñoz‐Vahos C, Vanegas‐García A, Vásquez G. Plasma microparticles from patients with systemic lupus erythematosus modulate the content of miRNAs in U937 cells. Immunology. 2021;164:253–265. 10.1111/imm.13366
Funding information
This project was financed by COLCIENCIAS (Grant No. 1115‐569‐33389).
REFERENCES
- 1.Colasanti T, Maselli A, Conti F, Sanchez M, Alessandri C, Barbati C, et al. Autoantibodies to estrogen receptor alpha interfere with T lymphocyte homeostasis and are associated with disease activity in systemic lupus erythematosus. Arthritis Rheum. 2012;64:778–87. [DOI] [PubMed] [Google Scholar]
- 2.Arneth B. Systemic lupus erythematosus and DNA degradation and elimination defects. Front Immunol. 2019;10:1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casciola‐Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zirngibl M, Fürnrohr BG, Janko C, Munoz LE, Voll RE, Gregory CD, et al. Loading of nuclear autoantigens prototypically recognized by systemic lupus erythematosus sera into late apoptotic vesicles requires intact microtubules and myosin light chain kinase activity. Clin Exp Immunol. 2015;179:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mobarrez F, Fuzzi E, Gunnarsson I, Larsson A, Eketjäll S, Pisetsky DS, et al. Microparticles in the blood of patients with SLE: Size, content of mitochondria and role in circulating immune complexes. J Autoimmun. 2019;102:142–9. [DOI] [PubMed] [Google Scholar]
- 6.Nieri D, Neri T, Petrini S, Vagaggini B, Paggiaro P, Celi A. Cell‐derived microparticles and the lung. European Respiratory Review. 2016;25:266–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbano C, Rojas M, Vásquez G, Castaño D. microparticles that form immune complexes as modulatory structures in autoimmune responses. Mediators Inflamm. 2015;204:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai J, Han YU, Ren H, Chen C, He D, Zhou L, et al. Extracellular vesicle‐mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J Mol Cell Biol. 2013;5:227–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng LI, Wu J‐L, Liu L‐M, Jiang J‐Q, Wu H‐J, Zhao M, et al. Serum miRNA‐371b‐5p and miRNA‐5100 act as biomarkers for systemic lupus erythematosus. Clinical immunology (Orlando, Fla). 2018;196:103–9. [DOI] [PubMed] [Google Scholar]
- 10.Zhao S, Wang YU, Liang Y, Zhao M, Long H, Ding S, et al. MicroRNA‐126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011;63:1376–86. [DOI] [PubMed] [Google Scholar]
- 11.Oelke K, Lu Q, Richardson D, Wu A, Deng C, Hanash S, et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 2004;50:1850–60. [DOI] [PubMed] [Google Scholar]
- 12.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, et al. MicroRNA‐146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–75. [DOI] [PubMed] [Google Scholar]
- 13.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF‐kappaB‐dependent induction of microRNA miR‐146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou J, Wang P, Lin LI, Liu X, Ma F, An H, et al. MicroRNA‐146a feedback inhibits RIG‐I‐dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183:2150–8. [DOI] [PubMed] [Google Scholar]
- 15.Perez‐Hernandez J, Forner MJ, Pinto C, Chaves FJ, Cortes R, Redon J. Increased urinary exosomal MicroRNAs in patients with systemic lupus erythematosus. PLoS One. 2015;10:e0138618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Distler JH, Huber LC, Gay S, Distler O, Pisetsky DS. Microparticles as mediators of cellular cross‐talk in inflammatory disease. Autoimmunity. 2006;39:683–90. [DOI] [PubMed] [Google Scholar]
- 17.Claßen L, Tykocinski L‐O, Wiedmann F, Birr C, Schiller P, Tucher C, et al. Extracellular vesicles mediate intercellular communication: Transfer of functionally active microRNAs by microvesicles into phagocytes. Eur J Immunol. 2017;47:1535–49. [DOI] [PubMed] [Google Scholar]
- 18.Tan EM, Cohen AS, Fries JF, Masi AT, Mcshane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. [DOI] [PubMed] [Google Scholar]
- 19.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. [DOI] [PubMed] [Google Scholar]
- 20.Mikdashi J, Nived O. Measuring disease activity in adults with systemic lupus erythematosus: the challenges of administrative burden and responsiveness to patient concerns in clinical research. Arthritis Res Ther. 2015;17:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strefford JC, Foot NJ, Chaplin T, Neat MJ, Oliver R, Young BD, et al. The characterisation of the lymphoma cell line U937, using comparative genomic hybridisation and multi‐plex FISH. Cytogenet Cell Genet. 2001;94(1–2):9–14. [DOI] [PubMed] [Google Scholar]
- 22.Olsson I, Gullberg U, Ivhed I, Nilsson K. Induction of differentiation of the human histiocytic lymphoma cell line U‐937 by 1 alpha,25‐dihydroxycholecalciferol. Cancer Res. 1983;43(12 Pt 1):5862–7. [PubMed] [Google Scholar]
- 23.Weinberg JM, Venkatachalam MA, Roeser NF, Saikumar P, Dong Z, Senter RA, et al. Anaerobic and aerobic pathways for salvage of proximal tubules from hypoxia‐induced mitochondrial injury. Am J Physiol Renal Physiol. 2000;279:F927–F943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz MA, Lassota P, et al. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. [DOI] [PubMed] [Google Scholar]
- 25.Ammon C, Meyer SP, Schwarzfischer L, Krause SW, Andreesen R, Kreutz M. Comparative analysis of integrin expression on monocyte‐derived macrophages and monocyte‐derived dendritic cells. Immunology. 2000;100:364–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alpsoy E, Kodelja V, Goerdt S, Orfanos CE, Zouboulis CHC. Serum of patients with Behcet's disease induces classical (pro‐inflammatory) activation of human macrophages in vitro. Dermatology. 2003;206:225–32. [DOI] [PubMed] [Google Scholar]
- 27.Pérez‐García A, Arroyo‐Valerio AG, Zaldivar‐Fujigaki JL, Bustos‐Esquivel MA, Gastelum‐Strozzi A, Padilla‐Castañeda MA, et al. Young adult binge drinkers have immunophenotypic changes in peripheral polymorphonuclear cells and monocytes. Am J Drug Alcohol Abuse. 2017;1–10. [DOI] [PubMed] [Google Scholar]
- 28.Fischer T, Wiegmann K, Bottinger H, Morens K, Burmester G, Pfizenmaier K. Regulation of IFN‐gamma‐receptor expression in human monocytes by granulocyte‐macrophage colony‐stimulating factor. J Immunol. 1990;145:2914–9. [PubMed] [Google Scholar]
- 29.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gracia T, Wang X, Su YA, Norgett EE, Williams TL, Moreno P, et al. Urinary exosomes contain MicroRNAs capable of paracrine modulation of tubular transporters in kidney. Sci Rep. 2017;7:40601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez P, Rodriguez‐Carrio J, Martinez‐Zapico A, Caminal‐Montero L, Suarez A. Circulating microparticle subpopulations in systemic lupus erythematosus are affected by disease activity. Int J Cardiol. 2017;236:138–44. [DOI] [PubMed] [Google Scholar]
- 32.Carlsen AL, Schetter AJ, Nielsen CT, Lood C, Knudsen S, Voss A, et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013;65:1324–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. [DOI] [PubMed] [Google Scholar]
- 34.Laffont B, Corduan A, Rousseau M, Duchez A‐C, Lee CHC, Boilard E, et al. Platelet microparticles reprogram macrophage gene expression and function. Thromb Haemost. 2016;115:311–23. [DOI] [PubMed] [Google Scholar]
- 35.Barnes BJ, Moore PA, Pitha PM. Virus‐specific activation of a novel interferon regulatory factor, IRF‐5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276:23382–90. [DOI] [PubMed] [Google Scholar]
- 36.Tada Y, Kondo S, Aoki S, Koarada S, Inoue H, Suematsu R, et al. Interferon regulatory factor 5 is critical for the development of lupus in MRL/lpr mice. Arthritis Rheum. 2011;63:738–48. [DOI] [PubMed] [Google Scholar]
- 37.Berggren O, Alexsson A, Morris DL, Tandre K, Weber G, Vyse TJ, et al. IFN‐alpha production by plasmacytoid dendritic cell associations with polymorphisms in gene loci related to autoimmune and inflammatory diseases. Hum Mol Genet. 2015;24:3571–81. [DOI] [PubMed] [Google Scholar]
- 38.Budarf ML, Goyette P, Boucher G, Lian J, Graham RR, Claudio JO, et al. A targeted association study in systemic lupus erythematosus identifies multiple susceptibility alleles. Genes Immun. 2011;12:51–8. [DOI] [PubMed] [Google Scholar]
- 39.Wang C, Sandling JK, Hagberg N, Berggren O, Sigurdsson S, Karlberg O, et al. Genome‐wide profiling of target genes for the systemic lupus erythematosus‐associated transcription factors IRF5 and STAT4. Ann Rheum Dis. 2013;72:96–103. [DOI] [PubMed] [Google Scholar]
- 40.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1‐TH17 responses. Nat Immunol. 2011;12:231–8. [DOI] [PubMed] [Google Scholar]
- 41.Stone RC, Feng DI, Deng J, Singh S, Yang L, Fitzgerald‐Bocarsly P, et al. Interferon regulatory factor 5 activation in monocytes of systemic lupus erythematosus patients is triggered by circulating autoantigens independent of type I interferons. Arthritis Rheum. 2012;64:788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahluwalia PK, Pandey RK, Sehajpal PK, Prajapati VK. Perturbed microRNA expression by mycobacterium tuberculosis promotes macrophage polarization leading to pro‐survival foam cell. Front Immunol. 2017;8:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Álvarez K, Villar‐Vesga J, Ortiz‐Reyes B, Vanegas‐García A, Castaño D, Rojas M, et al. Induction of NF‐κB inflammatory pathway in monocytes by microparticles from patients with systemic lupus erythematosus. Heliyon. 2020;6:e05815‐e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burbano C, Villar‐Vesga J, Vásquez G, Muñoz‐Vahos C, Rojas M, Castaño D. Proinflammatory differentiation of macrophages through microparticles that form immune complexes leads to T‐ and B‐Cell activation in systemic autoimmune diseases. Front Immunol. 2019;10:2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Distler JHW, Jungel A, Huber LC, Seemayer CA, Reich CF, Gay RE, et al. The induction of matrix metalloproteinase and cytokine expression in synovial fibroblasts stimulated with immune cell microparticles. Proc Natl Acad Sci U S A. 2005;102:2892–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mesri M, Altieri DC. Endothelial cell activation by leukocyte microparticles. J Immunol. 1998;161:4382–7. [PubMed] [Google Scholar]
- 47.Dieker J, Tel J, Pieterse E, Thielen A, Rother N, Bakker M, et al. Circulating apoptotic microparticles in systemic lupus erythematosus patients drive the activation of dendritic cell subsets and prime neutrophils for NETosis. Arthritis Rheumatol. 2016;68:462–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3


