Abstract
Background
Raw materials composed of easily assimilated monosaccharides have been employed as carbon source for production of microbial lipids. Nevertheless, agro-industrial wastes rich in galactose-based carbohydrates have not been introduced as feedstocks for oleaginous yeasts.
Results
In this study, Aureobasidium namibiae A12 was found to efficiently accumulate lipid from soy molasses and whey powder containing galactose-based carbohydrates, with lipid productions of 5.30 g/L and 5.23 g/L, respectively. Over 80% of the fatty acids was C16:0, C18:0, C18:1, and C18:2. All kinds of single sugar components in the two byproducts were readily converted into lipids, with yields ranging between 0.116 g/g and 0.138 g/g. Three α-galactosidases and five β-galactosidases in the strain were cloned and analyzed. Changes of transcriptional levels indicated GalB and GalC were key α-galactosidases, and GalG was key β-galactosidase. In 10 L fermentor, lipid production from SM and WP achieved 6.45 g/L and 6.13 g/L, respectively. β-galactosidase was responsible for lactose hydrolysis; sucrase and α-galactosidase both contributed to the efficient hydrolysis of raffinose and stachyose in a cooperation manner.
Conclusions
This is a new way to produce lipids from raw materials containing galactose-based carbohydrates. This finding revealed the significance of sucrase in the direct hydrolysis of galactose-based carbohydrates in raw materials for the first time and facilitated the understanding of the efficient utilization of galactose-based carbohydrates to manufacture lipid or other chemicals in bioprocess.
Graphic abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13068-021-02031-8.
Keywords: Lipid, Soy molasses, Whey powder, α-Galactosidase, β-Galactosidase
Background
Microbial lipid can be produced with high efficiency and has no competition from edible oil. Thus, microbial lipid has become important sources for producing biodiesel and other chemicals now [15, 39]. Among the lipid-producing microorganisms, oleaginous yeasts are attractive lipid producers for their rapid growth, large biomass, and high lipid yield [19, 33]. The reported oleaginous yeasts mainly belong to several genera, such as Aureobasidium, Candida, Cryptococcus, Lipomyces, Rhodotorula, Rhodosporidium, Trichosporon, and Yarrowia [8]. Among them, Aureobasidium pullulans, Lipomyces starkeyi, Yarrowia lipolytica, and Rhodosporidium toruloides can accumulate high level of neutral lipids, over 60% of their dry cell weight, from different feedstocks [8, 28, 31].
At present, the high production cost, especially the cost of carbon source for fermentation, impedes the industrial applications of microbial lipids [8]. In view of this, accumulating lipid from low-cost substrates is a feasible strategy to lower the production cost. A variety of raw materials or biowastes, including corncob, corn stover, wheat straw, cassava starch, waste oils, inulin, sugarcane bagasse, and sugarcane molasses, have been employed as carbon source for the lipid production by oleaginous yeasts [8]. Generally, raw materials can be converted into available monosaccharides using pretreatment methods, including acid hydrolysis, enzymatic hydrolysis, microbiological pretreatment, and detoxification process [8]. Corncob, corn stover, wheat straw, cassava starch, and sugarcane bagasse were converted into hydrolysate containing glucose as main component [8]. Inulin can be hydrolyzed into fructose [34].
Overall, the carbohydrates in these raw materials above mainly rely on glucose or fructose as monosaccharide unit, which can be easily assimilated. However, some agro-industrial by-products contain plenty of galactose-based carbohydrates, such as lactose and raffinose family oligosaccharides (RFOs) [35]. The galactoside bonds in the sugars are difficult to break up, thereby leading to limited applications and lowered commercial value. Due to lacking galactosidase synthesis and intracellular galactose assimilation pathway, most oleaginous yeasts cannot naturally consume galactose and RFO [35]. In addition, construction of galactose metabolic pathways in most yeast was not easy to achieve [18]. The abundant agro-industrial wastes containing galactose-based carbohydrates, such as whey powder (WP) and soy molasses (SM), have not been introduced as carbon source for lipid production [18, 35].
SM is a main by-product of the soy protein extraction from soybean. As soybean is one kind of staple crops providing plant proteins and lipid, SM is produced in large amount. Generally, SM has a high carbohydrate content of above 30% (w/v), mainly containing stachyose, raffinose, sucrose, monosaccharides, and other micronutrients [10, 22, 26]. Owing to the existence of α-(1,6) glycosidic bonds in RFO, SM cannot even be efficiently utilized by non-ruminant animals [35]. WP is a by-product in cheese production, and most of the production was concentrated in European and North America, with an annual worldwide production of about 165 million tons [16]. Despite whey containing many fermentable nutrients, such as lactose, proteins, and lipids, it has not efficiently utilized now [3]. In Europe, it was estimated that only 75% of the whey was recovered and developed as resources. While in the rest of the world, only than 50% of the whey was recovered [16].
Aureobasidium spp. has recently been evaluated as an excellent lipid producer and reported to efficiently assimilate various monosaccharides [31]. The genus was found to secrete many kinds of hydrolytic enzymes involved in biomass degradation [31]. In our previous study, several galactosidases were detected among extracellular substances of Aureobasidium sp., indicating the ability of assimilating galactose-based carbohydrates [41]. In this study, Aureobasidium namibiae strain A12 derived from mangrove was screened to accumulate high content of lipid directly from WP and SM.
Results
Evaluation of lipid producers of the yeast strains from SM
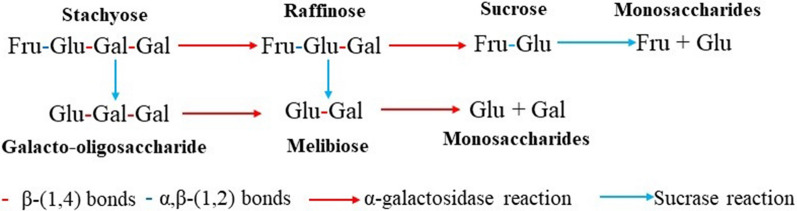
Apart from raffinose and stachyose, SM contained high level of conventional sugars, such as glucose, fructose, and sucrose, accounting for more than 40% of total carbohydrates [35]. These conventional sugars provided enough carbon sources for lipid accumulation of L. starkeyi and R. toruloides, with the lipid production of 1.80 g/L and 1.93 g/L, respectively (Fig. 1a). The highest lipid content (47.3%, w/w) was observed in the strain A12 system, with the lipid production of 5.30 g/L and the biomass of 11.20 g/L (Fig. 1a). Obviously, only the conventional sugars in SM cannot support the large-scale lipid synthesis of strain A12. In the final culture of typical oleaginous yeasts, the galactose-based carbohydrates, including stachyose, raffinose, and galactose, in the culture of typical oleaginous yeasts remained the same with the original concentrations in the medium (Fig. 1b). Stachyose and raffinose was totally utilized by strain A12 (Fig. 1b). This indicated that the RFO should be converted into lipid in strain A12. The low content of residual sugar also provided another evidence for this.
Fig. 1.
a Lipid productions of typical oleaginous yeasts and strain A12 from SM. b Contents of the residual galactose-based carbohydrates in the SM culture of typical oleaginous yeasts and strain A12. Data are given as means ± standard deviation, n = 3
Evaluation of lipid producers of the yeast strains from WP
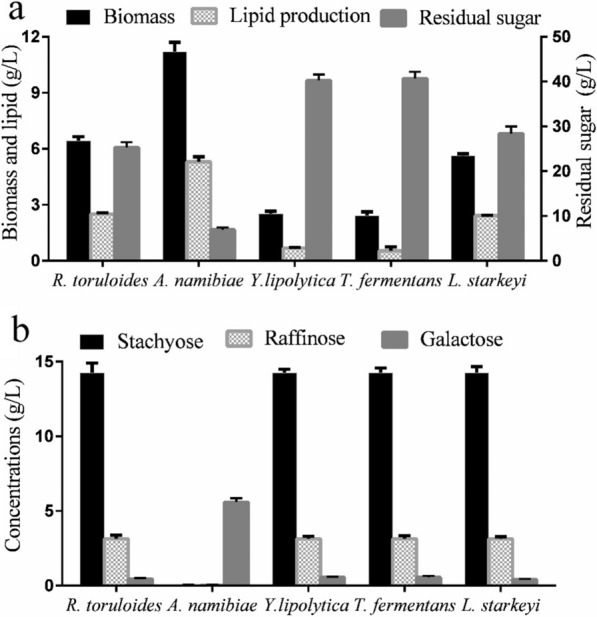
Many researchers have made efforts to convert WP into citric acid, lactic acid, and lipases [2, 25, 36]. WP also has received a wide attention to serve as the raw material for the production of biofuels recently [18]. A. namibiae strain A12 and other four typical oleaginous yeasts were evaluated from the perspective of the capability to produce lipid from WP, whose main component is lactose. As shown in Fig. 2a, strain A12 possessed the highest lipid content (48.6%, w/w) and was able to accumulate 5.23 g/L lipid and 10.72 g/L biomass from WP. On the contrary, L. starkeyi, Y. lipolytica, and R. toruloides can only accumulate a small amount of biomass and lipid compared with strain A12. As shown in Fig. 2b, four typical oleaginous yeasts can just utilize little amount of the lactose. This indicates that strain A12 can utilize lactose as the main component in WP, while the other three oleaginous strains can only assimilate the monosaccharides with low concentrations as carbon source.
Fig. 2.
a Lipid productions of typical oleaginous yeasts and strain A12 from WP. b Contents of the residual galactose-based carbohydrates in the WP culture of typical oleaginous yeasts and strain A12. Data are given as means ± standard deviation, n = 3
Composition of the fatty acids
Triacylglycerols (TAGs), the reported main components of lipid in oleaginous yeasts, were stored in intracellular lipid droplets: One to three droplets in each cell of strain A12 (data not shown). After being stained by Nile Red, the lipid droplets presented yellow fluorescence which can be observed under a fluorescent microscope with blue excitation light. GC analysis of the fatty acids showed that the content of linoleic acid (C18:2), oleic acid (C18:1), stearic acid (C18:0), palmitoleic acid (C16:1), and palmitic acid (C16:0) in lipid converted from SM were 9.83%, 57.42%, 10.51%, 2.1%, and 20.14%, respectively (Table 1). Oleic acid (C18:1) and palmitic acid (C16:0) in lipid of A. namibiae converted from SM account for high proportion (78.94%). The composition of the fatty acids in lipid converted from glucose and WP was rather similar to that converted from SM.
Table 1.
Composition of the fatty acids cultivated in different carbon sources
| Fatty acid composition (%) | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 |
|---|---|---|---|---|---|
| Glucose | 20.56 ± | 3.10 ± 0.35 | 10.43 ± 0.16 | 58.38 ± 0.43 | 7.53 ± 0.27 |
| SM | 20.14 ± 0.43 | 2.10 ± 0.12 | 10.51 ± 0.61 | 57.42 ± 0.89 | 9.83 ± 0.42 |
| WP | 20.24 ± 0.11 | 2.62 ± 0.24 | 10.56 ± 0.26 | 57.89 ± 0.31 | 8.69 ± 0.23 |
Analysis on the utilization of different sugars
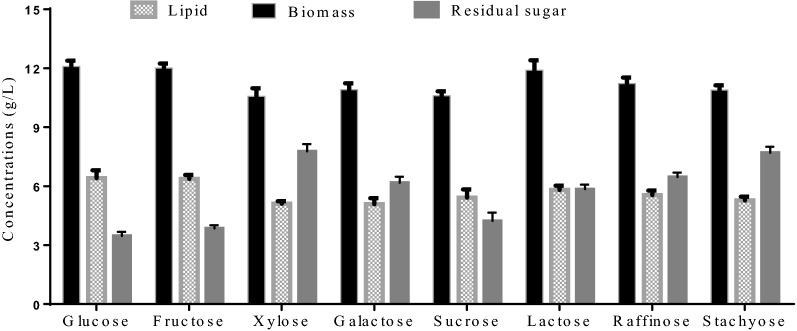
To further demonstrate the ability of strain A12 to utilize the waste sugars with galactoside bonds, it was cultured in a lipid-producing medium in which a single component in SM and WP was employed as the sole carbon source, including glucose, galactose, fructose, lactose, sucrose, xylose, stachyose, and raffinose. As shown in Fig. 3, the lipid production, biomass, and residual sugars in these fermentations have been compared. The test monosaccharides, which are present in SM and WP or exist as monosaccharide units of other sugars, were all readily converted to lipid. The lipid yield from them was not much different, ranging from 0.116 g/g to 0.138 g/g. This suggests that an efficient galactose and metabolism pathway indeed exists in the strain A12. It should be noted that this strain can even convert xylose into lipid, which is quite challenging for many other yeasts [8]. Besides, it can utilize the sucrose, commonly used as a conventional carbon source, and galactose-based sugars in an efficient manner, revealing the existence of a developed galactosidase system [6, 30].
Fig. 3.
Lipid productions of strain A12 from single sugar components in SM and WP. Data are given as means ± standard deviation, n = 3
Galactosidase system in strain A12
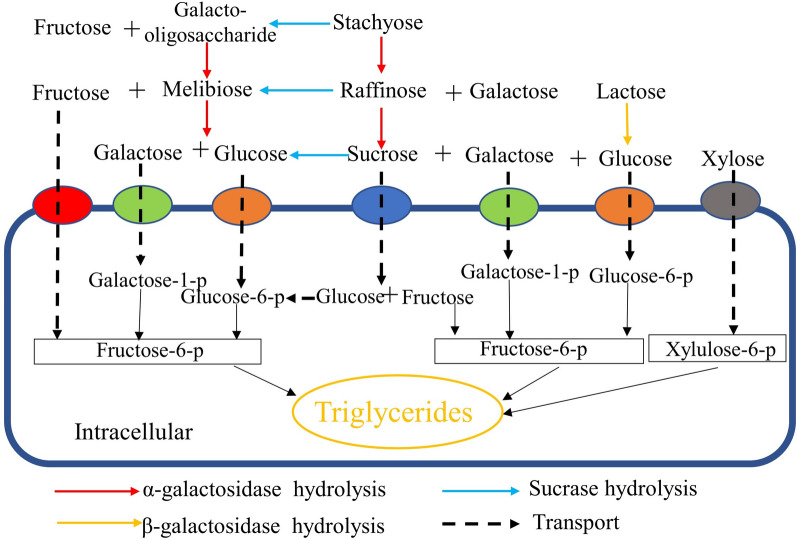
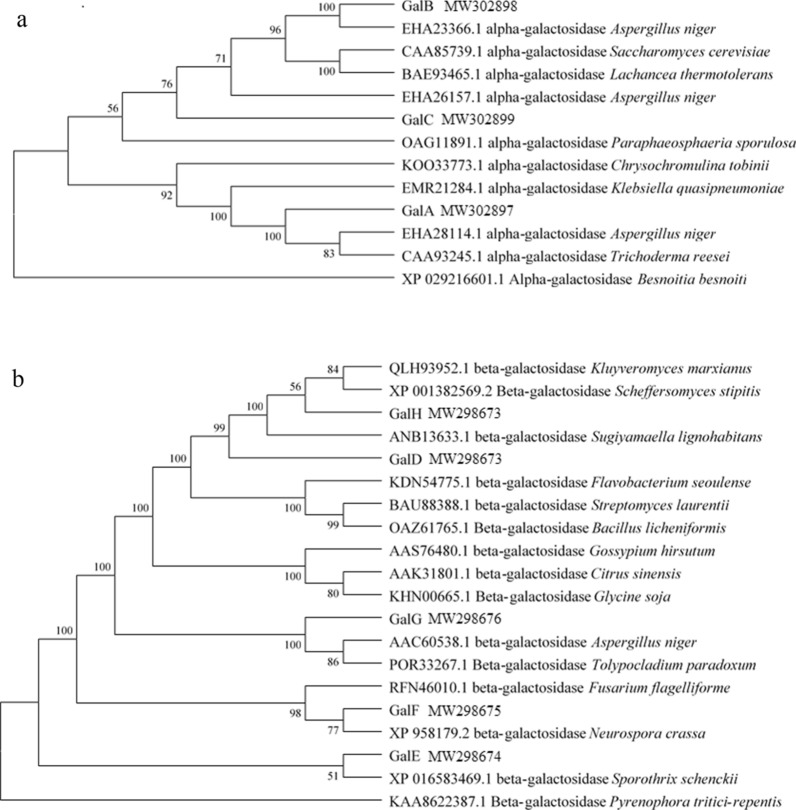
α-Galactosidase is effective in catalyzing the generation of α-linked galactose residues from various substrates, thus being crucial for the conversion of RFO into conventional carbon source sucrose, while β-galactosidase is able to cut the β-linked galactose residue of lactose to yield glucose [18, 35]. Based on the genome sequence of the type strain for this species, A. namibiae CBS 147.97, the potential galactosidase coding gene in strain A12 was cloned and analyzed. As listed in Table 2, diverse α-galactosidases and β-galactosidases were constituted by different numbers of amino acids. Through analysis, three potential α-galactosidases (namely GalA, GalB, and GalC) and five potential β-galactosidases (i.e., GalD, GalE, GalF, GalG, and GalH) were found. α-Galactosidases in strain A12 have been classified into two glycoside hydrolase families GH27 and GH36. GH27 and GH36 are perceived to have a common ancestral gene and hence they share a common catalytic mechanism. The phylogenic tree showed that GalB and GalC from GH27 are clustered into the same branch, while GalA belongs to another branch (Fig. 4). Each α-galactosidase of A. namibiae A12 is found very close to that of Aspergillus niger. This is in accordance with the recognition that the genus Aureobasidium is close to A. niger in terms of evolutionary relationship [21]. Nevertheless, β-galactosidases of A. namibiae A12 from two glycoside hydrolase families are much more diverse. Although the five β-galactosidases in A. namibiae A12 are all separated from β-galactosidases from plants and bacteria, they diverge in four branches close to specific basidiomycetes or ascomycetes, forming subclasses of specialized enzymes.
Table 2.
Potential α-galactosidases, β-galactosidases in strain A12 and the basic characteristics
| Proteins | Accession | Function | Super family | Amino acids | Signal peptide |
|---|---|---|---|---|---|
| GalA | MW302897 | α-galactosidase | GH36 | 750 | Yes |
| GalB | MW302898 | α-galactosidase | GH27 | 452 | Yes |
| GalC | MW302899 | α-galactosidase | GH27 | 522 | No |
| GalD | MW298673 | β-galactosidase | GH2 | 858 | No |
| GalE | MW298674 | β-galactosidase | GH35 | 1001 | Yes |
| GalF | MW298675 | β-galactosidase | GH35 | 1002 | Yes |
| GalG | MW298676 | β-galactosidase | GH35 | 1009 | Yes |
| GalH | MW298673 | β-galactosidase | GH7 | 1071 | No |
Fig. 4.
The phylogenetic tree generated with the neighbor-joining method based on α-galactosidase (a) and β-galactosidase sequences (b). Branch-related numbers are bootstrap values (confidence limits)
When A. namibiae A12 was cultivated in the LP medium containing SM, the transcriptional levels of GALB and GALC increased to 326.3% and 276.1% of those cultivated in the LP medium containing glucose (Fig. 5). The transcriptional levels of the genes coding β-galactosidases were not obviously changed (Fig. 5). GALA cannot be detected by qRT-PCR. When A. namibiae A12 was cultivated in the LP medium containing WP, the transcriptional levels of GALG increased to 456.3%. The results above indicated that GalB and GalC were key α-galactosidases for utilizing galactose-based carbohydrates in SM; GalG was key β-galactosidase for utilizing galactose-based carbohydrates in WP. To explore the characteristics of key galactosidases, GalB, GalC, and GalG were heterologously expressed. The molecular weights of GalB, GalC, and GalG were determined as 50.0 kDa, 55.3 kDa, and 107.2 kDa, respectively (Additional file 1: Fig. S1–S3). The molecular weights were consistent with those deduced from the amino acid sequences. Recombinant GalB and GalC can efficiently hydrolyze stachyose and raffinose (Additional file 1: Fig. S1, Fig. S2). Recombinant GalB peaked at 40 °C and was maintained higher than 60% at 30 °C–60 °C; GalB showed high activity and stability in a broad pH range (4.0–7.0) (Fig.S1). The characteristics of GalC were similar with those of GalB (Additional file 1: Fig. S1, S2). GalG showed higher in lower pH; the optimal pH for activity and stability was pH 4.0 (Additional file 1: Fig. S3). The three galactosidases can catalyze efficiently in the condition of lipid fermentation.
Fig. 5.
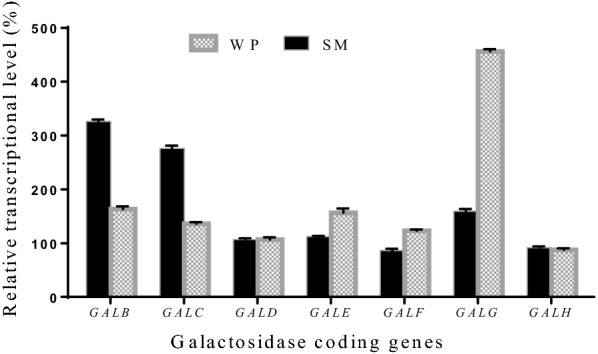
Changes of transcriptional levels of galactosidase coding genes
Coordination of glycoside hydrolases during the lipid production
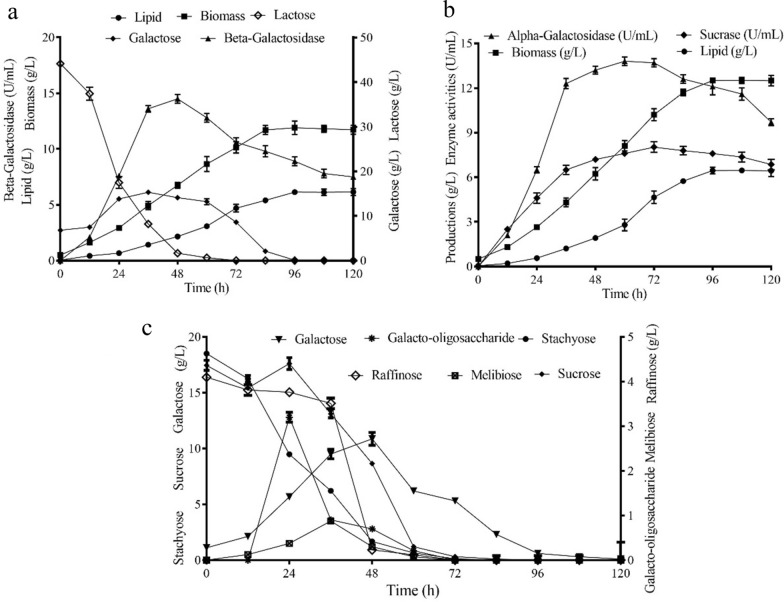
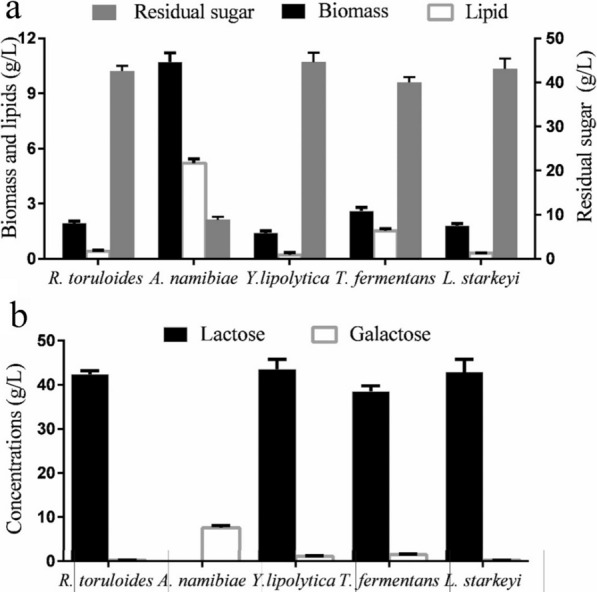
The hydrolysis of carbohydrates in the medium can be realized by inducing microorganisms to generate corresponding glycoside hydrolases. To elucidate the role of glycoside hydrolases in the medium containing SM or WP as the carbon source, their activities and the concentrations of different sugars were monitored at a 12-h interval in a 10 L fermentor. The different activities of these glycoside hydrolases can be attributed to the different concentrations of the corresponding enzyme inducers, the substrates of the enzymes in the medium. As shown in Fig. 6a, β-galactosidase activity was 7.6 U/mL at 24 h in WP medium, while the activities of α-galactosidase and sucrase were at low levels of 0.6 U/mL and 1.3 U/mL, respectively. Galactose and glucose derived from lactose hydrolysis were then directed to lipid and biomass synthesis. After 72 h, no lactose was detected and the β-galactosidase started to decrease. At 96 h, galactose was completely consumed, with a maximal lipid production of 6.13 g/L. Adequate β-galactosidase enables the utilization of lactose in WP medium.
Fig. 6.
a Time course of lipid, biomass, galactosidases, and sugar contents in the 10L bioreactor during fermentation by strain A12 from WP. b Time course of lipid, biomass, sucrase and α-galactosidase in the 10 L bioreactor during fermentation by strain A12 from SM. c Time course of sugar contents during fermentation by strain A12 from SM. Data are given as means ± standard deviation, n = 3
α-Galactosidase is responsible for converting raffinose family oligosaccharides into conventional sucrose and galactose. As shown in Fig. 6b and c, α-galactosidases and sucrase were induced much more than those in WP, with 22.6 g/L RFO detected. At 24 h, the activity of α-galactosidase reached 6.52 U/mL, with reduced content of stachyose and increased galactose; the activity of sucrase reached 4.60 U/mL. However, the content of raffinose just decreased slightly to 3.51 g/L at 36 h, as a result of the supplement through the release of raffinose from stachyose cut by α-galactosidase (Fig. 6b, c). Besides, the content of sucrose even increased to the level before 24 h. This indicated that the hydrolysis of sucrose by sucrase was slower than its generation from stachyose and raffinose catalyzed by α-galactosidase. At 48 h, the activity of α-galactosidase and sucrase reached 13.2 U/mL and 7.2 U/mL, respectively. Under the reaction of α-galactosidase and sucrase, sucrose and RFO were both not detected after 96 h, and the lipid achieved a maximal yield of 6.45 g/L (Fig. 6b, c).
Discussion
As far as we know, this is the first time for the introduction of SM as low-cost carbon source in lipid production [8, 10, 22, 26]. Benefiting from the low cost and easy availability, SM has become a promising feedstock in China [35]. Moreover, the high carbohydrate content makes SM attractive among researchers and biorefinery industrialists [8, 10, 22, 26]. However, the current SM-derived fermentations suffer from limited efficiency, as most of them still focused on the utilization of the conventional constituents [7, 10, 27, 37]. Up to now, only the synthesis of poly-hydroxyalkanoate, propionic acid, and polymalic acid from RFO in SM was achieved [7, 12, 37].
The tested typical oleaginous yeasts in this study all have been proved capable of accumulating high level of lipid from various carbohydrates with glucose or fructose as the monosaccharide unit. R. toruloides accumulates lipid from cassava starch [20]. Inulin, sucrose, and tuber meal of Jerusalem artichoke have been used for lipid production in Aureobasidium pullulans [31]. The lipid content and lipid production of Y. lipolytica Po1g grown in sugarcane bagasse hydrolysate were 58.5% and 6.68 g/L, respectively [32]. Moreover, R. toruloides, L. starkeyi, and Y. lipolytica can produce lipid with the hydrolysate derived from the pretreatment of wheat straw [38]. As for galactose-based carbohydrates, only A. namibiae among the oleaginous yeasts can convert them into lipid. The composition of the fatty acids in lipid produced by A. namibiae was similar to that of plant oils [8]. C18:1 was also the predominant fatty acid in lipid of R. toruloides, L. starkeyi, and Y. lipolytica, with a content of more than 40% [8, 28, 31]. As revealed in previous studies, the long-chain fatty acids constituted by 16 and 18 carbon atoms were ideal candidates for biodiesel production via methylation.
Generally, very few yeasts possess galactosidases and galactose metabolism pathway, thus resulting in the impossibility to utilize lactose by most yeasts [9]. Actually, A. namibiae was the first oleaginous microorganism reported to produce lipid using SM as the feedstock. In previous studies, only two oleaginous yeasts have been reported able to accumulate lipid from WP with a content of more than 40% [6, 30]. Cystobasidium oligophagum has been characterized to efficiently convert WP into lipid. The biomass and the lipid content were 43.1% and 6.38 g/L, respectively [30]. In another study, the biomass and lipid production of Cryptococcus curvatus from WP achieved 10.77 g/L and 63.4%, respectively [6]. Moreover, a secreted β-galactosidase was introduced in Y. lipolytica, along with the enhanced cellular galactose metabolism. The engineered strain could achieve the rapid conversion of acid whey, producing 6.61 g/L of fatty acids [18]. In 10 L fermentor, lipid production by A. namibiae in the medium containing SM or WP as the carbon source achieved 6.45g/L and 6.13 g/L, respectively, with lipid content of 53.3% and 51.5%, respectively (Fig.6a, b). As shown in Fig. 6a and b, lipid production increased slowly than biomass accumulation in the first 48 h, but lipid production increased significantly in the 48–72 h. This indicated the lipid yield enhanced after 48-h culture. Due to the slow growth of the yeast cell, more carbon source was partitioned into lipid synthesis. The similar trend was also be confirmed in the lipid production of R. toruloides [20, 34].
The lipid productivity of A. namibiae A12 was slightly lower than that of C. curvatus strain, and much higher than that of C. oligophagum strain, using WP as the carbon source (Table 3). However, the lipid productivity of engineered Y. lipolytica strain achieved 0.092 g/L/h, remarkably higher than those of other strains. Lipid synthesis flux was significantly enhanced by overexpressing acetyl-CoA carboxylase and diacylglycerol acyltransferase in the engineered strains, thus caused a shorter fermentation period and higher lipid production [3]. The engineering strategy in Y. lipolytica can be adopted in A. namibiae A12 to optimize lipid production. And fed-batch fermentation will be another method to enhance the lipid production.
Table 3.
Comparison of lipid productions by different strains from galactose-based carbohydrates
Generally, similar compositions were found in the composition of fatty acids in the majority of oleaginous yeasts, including Y. lipolytica, L. starkeyi, and strain A12 [8, 28]. These results confirmed that SM and WP would be the ideal substrates for lipid production. Although the fermentations in SM and WP were probably slowed down due to the hydrolysis of raffinose and stachyose, lower osmotic pressures were allowed and the biomass was increased. Thus, as largely untapped resources currently, SM and WP deserve arousing attention from A. namibiae system to be a self-sufficient nutrient for numerous potential bioprocesses [8]. This galactosidases system provides molecular basis for strain A12 to utilize sugars with galactoside bonds. Based on the excellent the characteristics of GalB, GalC, and GalG (Additional file 1: Fig. S1; Fig. S2; Fig. S3), the three key galactosidases in Aureobasidium can be a valuable untapped industrial enzyme resource [21]. Besides, the sucrose-utilizing capacity has been proved among Aureobasidium strains to accumulate biopolymer and other products [1, 13, 24, 37]. In the genome of Aureobasidium namibiae CBS 147.97, ten potential sucrase coding genes have been identified, giving support for the capacity of A. namibiae A12 to utilize sucrose in SM.
Unexpectedly, two kinds of sugars not present in SM medium have been detected during the fermentation, which were identified as melibiose and galacto-oligosaccharide. Their existence revealed the neglected fact that the sucrase also functions in the hydrolysis of stachyose and raffinose apart from α-galactosidase [29]. Sucrase has been proved to catalyze the release of fructose residues from different substrates, including stachyose and raffinose. Thus, sucrase and α-galactosidase both contribute to efficient hydrolysis of RFO in a cooperation manner, rather than in a sequential way [8, 29]. The sequence of cutting the two kinds of glycosidic bonds influences the generation of intermediate sugars, as shown in Fig. 7. In the case of sole α-galactosidase, RFO was converted into sucrose with the release of galactose; when only sucrose exists, melibiose and galacto-oligosaccharide were generated with the release of fructose (Fig. 7). This finding revealed the significance of sucrase in the direct hydrolysis of RFO in raw materials for the first time and facilitated the understanding of the efficient utilization of RFO to manufacture lipid or other chemicals in bioprocess [8, 35].
Fig. 7.
The contribution of sucrase and α-galactosidase to the hydrolysis of RFO. Glu: Glucose; Fuc: Fructose; Gal: Galactose
Conclusions
A. namibiae A12 efficiently accumulate lipid from galactose-based SM and WP. β-Galactosidase was responsible for lactose hydrolysis in WP; sucrase and α-galactosidase both contributed to the efficient hydrolysis of RFO in a cooperation manner. These results demonstrate the feasibility of using SM and WP as a previously untapped resource for biotechnology. This platform strain can be further improved to produce high-value lipid-derived chemicals from waste sugars with galactoside bonds.
Methods
Strains and media
Four typical oleaginous yeast strains, i.e., R. toruloides R2, Trichosporon fermentans T1, Y. lipolytica URA, and L. starkeyi AM, were preserved at − 80 ℃ in our laboratory. Strain A12 was isolated from a plant in Zhanjiang Mangrove National Nature Reserve, and identified as A. namibiae. Yeast strains were cultivated in YPD medium (containing 20.0 g/L glucose, 20.0 g/L peptone, and 10.0 g/L yeast extract) [17]. The composition of lipid production (LP) medium included 50 g/L glucose, 3.0 g/L K2HPO4, 2.0 g/L KH2PO4, 0.1 g/L MgSO4·7H2O, and 2 g/L corn steep liquor (CSL), with pH 6.0. When other pure carbohydrates were selected as the carbon source, the concentrations of the carbohydrates remained 50 g/L. GPPB medium was used for SIase production and contained 30.0 g/L glucose, 1.0 g/L (NH4)2SO4, 6.0 g/L yeast extract, 2.0 g/L KH2PO4, 3.0 g/L K2HPO4, and 0.1 g/L MgSO4·7H2O with a pH of 6.0 [42].
Pretreatment of SM
SM was adjusted to pH 10.0 with Ca(OH)2 powder which was held for 1 h. Subsequently, the liquid was filtered to remove excess Ca(OH)2 and adjusted to pH 6.0 with H2SO4, which was kept for 1 h. After 10 min of boiling, the solution was centrifuged at 7000 × g, followed by being stored at 4 °C [41]. The pretreated SM contained 3.32% (w/w) glucose, 2.81% (w/w) fructose, 13.42% (w/w) sucrose, 1.73% (w/w) xylose, 3.15% (w/w) raffinose, 14.26% (w/w) stachyose, and 0.78% (w/w) galactose.
Lipid production at flask level
Strain A12 was cultivated in 5 mL YPD medium at 30 ℃ for 20 h which was then switched to 50 mL LP medium at the same temperature. The fermentation proceeded for a total of 120 h, after which the broth was centrifuged at 5000 × g. The yeast cells were harvested by centrifugation at 5000 × g, washed with sterile water, and dried at 80 ℃ overnight for the biomass detection. Nile Red (0.5 mg/L in DMSO, GenMed Scientifics Inc., USA) was adopted to stain the washed cells (10 μL) which were then observed under an Olympus U-LH100HG fluorescent microscope with blue excitation light [31]. In the experiments checking the lipid production from the wastes, glucose in LP medium was absent; SM and low protein WP concentrations were set at 130 g/L and 60 g/L, respectively. So that the sugar content in the medium can be maintained at about 50 g/L. The initial lactose and galactose in LP medium containing WP were maintained as 44.1 g/L and 6.8 g/L, respectively. The initial LP medium containing SM contained 4.3 g/L glucose, 3.7 g/L fructose, 17.5 g/L sucrose, 2.2 g/L xylose, 4.1 g/L raffinose, 18.5 g/L stachyose, and 1.0 g/L galactose, respectively.
Molecular manipulations
The extraction of genomic DNA from the A. namibiae strain was conducted with the TIANamp Yeast DNA Kit (TIANGEN BIOTECH, China). The DNA polymerase was utilized according to the recommendation by the manufacturer (New England BioLabs, USA). The transformation of E. coli was realized by the heat shock method of Sambrook et al. [23]. The primers were designed based on the potential gene encoding galactosidase in A. namibiae CBS 147.97, and this potential gene in strain A12 was subjected to PCR amplification. The amplified DNA fragments were then transformed into E. coli DH5α (TaKaRa Biotechnology, China). The E. coli transformants were screened on LB agar which contained 100.0 μg/mL ampicillin and play a role in the plasmid amplification and DNA sequencing.
Bioinformatics analysis of galactosidases
The gene protein sequence was compared using HMMER3 based on the CAZy (Carbohydrate-Active enZYmes) database. In this way, the annotation information of carbohydrate-active enzymes became available [4]. The genes coding for galactosidases and sucrases were amplified using primers listed in Additional file 1: Table S1. E-value < 1e-5 was adopted as the filter condition. SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP-4.1/) was applied to the signal peptide analysis. The phylogenetic tree was constructed on the basis of reported α-galactosidase and β-galactosidase sequences by virtue of the neighbor-joining method in MEGA version 7.0.
Gene expression level analyses with qRT-PCR assays
Strain A12 was inoculated for 36 h at 30 °C in the LP medium containing glucose, SM, and WP, respectively. Total RNA was isolated using TRIzol reagents described by the manufacturer. RNA concentration was analyzed with NanoDrop2000c spectrophotometer (Thermo Fisher, Germany) and reversed transcribed into cDNA. Real-time PCR was performed in triplicate using a SYBR green assay kit (Toyobo, Japan), with the SYBR real-time PCR master mix (Applied Biosystems). The primers for transcription-quantitative PCR (qRT-PCR) were designed according to the sequences of the genes coding galactosidases; 18S rDNA gene was employed as an internal reference (Additional file 1: Table S2). Relative gene expression level changes in LP medium containing SM and WP were analyzed by using the comparative CT method with a 10 μL reaction system. Samples from LP medium containing glucose was used as control. All of the amplifications were performed in triplicate from biological triplicates.
Secretory expression of galactosidases
After codons were optimized, the genes coding galactosidases containing the XPR2 signal peptide gene was synthesized in Synbio Technologies, China. The DNA fragment synthesized in this study was transformed into the URA− strain [42]. After the cultivation in GPPB liquid medium at 30 °C for 48 h, the positive transformants were detected for their galactosidase activities. The supernatant of positive transformant was adjusted to pH 7.5 and then loaded on gel filtration chromatography column as well as anion exchange chromatography column (GE Healthcare, USA). The Mw and purity of recombinant galactosidases were verified based on SDS-PAGE on 12% (w/v) gel.
Effects of temperature and pH on activity and stability of recombinant galactosidases
The enzymatic reaction mediated by recombinant galactosidases proceeded in 10 mM glycine–NaOH buffer (pH 6.0) at temperatures ranging from 10 °C to 70 °C, the results of which showed the optimal reaction temperature. To investigate the thermal stability of recombinant galactosidases, the enzyme after purification was first incubated at temperatures ranging from 10 °C to 70 °C for 12 h and then the remaining activity at 40 °C was detected. Tannic acid solutions were prepared with 10 mM buffer at different pH levels (Na2HPO4–citric acid, pH 2.0–8.0; glycine–NaOH, pH 8.5–0.0) to act as the substrate for the determination of the optimal reaction pH. pH stability was estimated according to the remaining activity after the incubation at 4 °C for 12 h in buffer at different pH levels. All reactions were allowed to proceed in triplicate.
Determination of fatty acid composition of the extracted oil
The cellular lipids were extracted and weighed with the method of Folch et al. [11]. For the sake of determining fatty acid composition, the oil obtained by Soxhlet extraction was directly subjected to transmethylation and fatty acid esters were then analyzed using gas chromatography (GC) according to the literature procedure.
Enzymatic activity assay and sugar concentration determination
The α-galactosidase activity was evaluated with pNPG as substrate [5]. The reaction was terminated by Na2CO3 and the solution was filtered through a 0.22 μm membrane, followed by high-performance liquid chromatography (HPLC) analysis. The carbohydrate concentration was calculated depending on the retention time and peak area. One unit of the α-galactosidase activity (U) was defined as the amount of the enzyme that generated 1 μM p-nitrophenol per minute at 40 °C in buffer at pH 4.5.
The detection of β-galactosidase activity was carried out by the similar method [14]. The mixture of the culture (0.5 mL) and o-nitrophenyl-β-D-galactopyranoside (ONPG, 20.0 mM, 0.5 mL) in 100 mM citrate buffer (pH 4.0) underwent the incubation at 40 °C for 10 min. The β-galactosidase in the mixture was inactivated by 2.0 mL of 0.5 M Na2CO3 solution. One unit of the β-galactosidase activity (U) was defined as the amount of the enzyme producing 1.0 μM ONP per minute in the conditions of this study.
The sucrase activity was detected as reported [40]. The carbohydrate content was determined on an Agilent 1200 system (Agilent Technologies, USA) with amino (NH2) column (Thermo Scientific, USA).
Lipid production in 10 L fermentor
Large-scale fermentation was performed in Biostat B 10 L fermentors (B. Braun, Germany) for the lipid production from WP and SM, respectively. Strain A12 was first inoculated in YPD medium (600 mL) as a seed culture and then transferred to the fermentor which contained 6 L of lipid production medium. In the next step, the fermentation began and lasted for 120 h under the following conditions: agitation speed: 300 rpm, aeration rate: 50 L/min, temperature: 30 ℃, and pH 6.0. The samples were also taken every 12 h to determine the biomass, lipid yield, and activities of α-galactosidase, β-galactosidase, and sucrase. Besides, the sugar content was monitored as well.
Supplementary Information
Additional file 1: Figure S1. (a) SDS-PAGE analysis of recombinant GalB. Lane M, standard Mw markers; lane 1, purified GalB; lane 2, Bovine albumin. (b) Substrate specificity of GalB (c) The pH activity and stability of GalB. (d) Thetemperature activity and stability of GalB. Figure S2. (a) SDS-PAGE analysis of recombinant GalC. Lane M, standard Mw markers; lane 1, purified GalC; lane 2, Bovine albumin. (b) Substrate specificity of GalB (c) The pH activity and stability of GalC. (d) Thetemperature activity and stability of GalC. Figure S3. (a) SDS-PAGE analysis of recombinant GalG. Lane M, standard Mw markers; lane 1, purified GalG; lane 2, Bovine albumin. (b) The pH activity and stability of GalG. (c) Thetemperature activity and stability of GalG. Table S1. Primers for amplifying genes coding galactosidases. Table S2. Primers used to perform qRT-PCR assays.
Authors’ contributions
ZW conceived the research, performed the fermentation experiments, and wrote the original draft. XZ performed the molecular experiments. YM analyzed the data. JY performed gene sequence analysis. JJ performed purification of the recombinant enzymes. HW supervised the study. WC reviewed and edited the article. All authors read and approved the final manuscript.
Funding
This research was supported by the Natural Science Foundation of Shandong Province (ZR2020MC003), National Natural Science Foundation of China (41806163), National Key R&D Program of China (2019YFD0901902), “First Class Fishery Discipline” Programme in Shandong Province, a special talent programme, “One Thing One Decision (YishiYiyi)” Programme in Shandong Province, China, and Breeding Plan of Shandong Provincial Qingchuang Research Team (2019).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All the authors consent for publications.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhi-Peng Wang, Email: wangzpmbio@163.com.
Wei Chen, Email: chenwei@qau.edu.cn.
References
- 1.Al-Araimi SH, Elshafie A, Al-Bahry SN, Al-Wahaibi YM, Al-Bemani AS. Biopolymer production by Aureobasidium mangrovei SARA-138H and its potential for oil recovery enhancement. Appl Microbiol Biotechnol. 2021;105(1):105–117. doi: 10.1007/s00253-020-11015-x. [DOI] [PubMed] [Google Scholar]
- 2.Arslan NP, Aydogan MN, Taskin M. Citric acid production from partly deproteinized whey under non-sterile culture conditions using immobilized cells of lactose-positive and cold-adapted Yarrowia lipolytica B9. J Biotechnol. 2016;231:32–39. doi: 10.1016/j.jbiotec.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 3.Blazeck J, Hill A, Liu L, Knight R, Miller J, Pan A, Otoupal P, Alper HS. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat Commun. 2014;5:3131. doi: 10.1038/ncomms4131. [DOI] [PubMed] [Google Scholar]
- 4.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao YN, Yuan TZ, Shi PJ, Luo HY, Ning L, Meng K, Bai YG, Yang PL, Zhou ZG, Zhang ZF. Properties of a novel α-galactosidase from Streptomyces sp. S27 and its potential for soybean processing. Enzyme Microb Technol. 2010;47(7):305–312. doi: 10.1016/j.enzmictec.2010.09.007. [DOI] [Google Scholar]
- 6.Carota E, Crognale S, D'Annibale A, Gallo AM, Stazi SR, Petruccioli M. A sustainable use of Ricotta Cheese Whey for microbial biodiesel production. Sci Total Environ. 2017;584–585:554–560. doi: 10.1016/j.scitotenv.2017.01.068. [DOI] [PubMed] [Google Scholar]
- 7.Cheng C, Zhou Y, Lin M, Wei P, Yang ST. Polymalic acid fermentation by Aureobasidium pullulans for malic acid production from soybean hull and soy molasses: Fermentation kinetics and economic analysis. Biores Technol. 2016;223:166–174. doi: 10.1016/j.biortech.2016.10.042. [DOI] [PubMed] [Google Scholar]
- 8.Cho HU, Park JM. Biodiesel production by various oleaginous microorganisms from organic wastes. Bioresour Technol. 2018;256:502–508. doi: 10.1016/j.biortech.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Domingues L, Guimarães PMR, Oliveira C. Metabolic engineering of Saccharomyces cerevisiae for lactose/whey fermentation. Bioeng Bugs. 2010;1:164–171. doi: 10.4161/bbug.1.3.10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong J, Du Y, Zhou Y, Yang ST. Butanol production from soybean hull and soy molasses by acetone-butanol-ethanol fermentation. In: Brentin R, editor. SoyBased Chemicals and Materials, ACS Symposium Series 1178, Chapter 2. Washington: American Chemical Society; 2014. pp. 25–41. [Google Scholar]
- 11.Folch J, Lees M, Slane-Stanley J. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. doi: 10.1016/S0021-9258(18)64849-5. [DOI] [PubMed] [Google Scholar]
- 12.Full TD, Jung DO, Madigan MT. Production of poly-beta-hydroxyalkanoates from soy molasses oligosaccharides by new, rapidly growing Bacillus species. Lett Appl Microbiol. 2006;43(4):377–384. doi: 10.1111/j.1472-765X.2006.01981.x. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H, Xue SJ, Li YF, Liu GL, Chi ZM, Hu Z, Chi Z. Efficient transformation of sucrose into high pullulan concentrations by Aureobasidium melanogenum TN1-2 isolated from a natural honey. Food Chem. 2018;15(257):29–35. doi: 10.1016/j.foodchem.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Juers DH, Matthews BW, Huber RE. LacZ β-galactosidase: structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012;221(12):1792–1807. doi: 10.1002/pro.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma Y, Gao Z, Wang Q, Liu Y. Biodiesels from microbial oils: opportunity and challenges. Bioresour Technol. 2018;263:631–641. doi: 10.1016/j.biortech.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 16.Macwan SR, Dabhi BK, Parmer SC, Aparnathi KD. Whey and its utilization. Int J Curr Microbiol Appl Sci. 2016;5:134–155. doi: 10.20546/ijcmas.2016.508.016. [DOI] [Google Scholar]
- 17.Madzak C. Yarrowia lipolytica: recent achievements in heterologous protein expression and pathway engineering. Appl Microbiol Biotechnol. 2015;99(11):4559–4577. doi: 10.1007/s00253-015-6624-z. [DOI] [PubMed] [Google Scholar]
- 18.Mano J, Liu N, Hammond JH, Currie DH, Stephanopoulos G. Engineering Yarrowia lipolytica for the utilization of acid whey. Metab Eng. 2020;57:43–50. doi: 10.1016/j.ymben.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Munch G, Sestric R, Sparling R, Levin DB, Cicek N. Lipid production in the under-characterized oleaginous yeasts, Rhodosporidium babjevae and Rhodosporidium diobovatum, from biodiesel-derived waste glycerol. Bioresour Technol. 2015;185:49–55. doi: 10.1016/j.biortech.2015.02.051. [DOI] [PubMed] [Google Scholar]
- 20.Park YK, Nicaud JM, Ledesma-Amaro R. The Engineering Potential of Rhodosporidium toruloides as a Workhorse for Biotechnological Applications. Trends Biotechnol. 2018;36(3):304–317. doi: 10.1016/j.tibtech.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Prasongsuk S, Lotrakul P, Ali I, Bankeeree W, Punnapayak H. The current status of Aureobasidium pullulans in biotechnology. Folia Microbiol (Praha) 2018;63(2):129–140. doi: 10.1007/s12223-017-0561-4. [DOI] [PubMed] [Google Scholar]
- 22.Romão BB, da Silva FB, de Resende MM, Cardoso VL. Ethanol production from hydrolyzed soybean molasses. Energy Fuels. 2012;26:2310–2316. doi: 10.1021/ef201908j. [DOI] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. Preparation and analysis of eukaryotic genomic DNA. In: Michael RG, editor. Molecular cloning: A laboratory manual. 2. Beijing: Cold Spring Harbor Laboratory Press; 1989. pp. 367–370. [Google Scholar]
- 24.Sheng L, Tong Q, Ma M. Why sucrose is the most suitable substrate for pullulan fermentation by Aureobasidium pullulans CGMCC1234? Enzyme Microb Technol. 2016;92:49–55. doi: 10.1016/j.enzmictec.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Silva MDA, Silva TAL, Campos-Takaki GM, Amorim Salgueiro A, Tambourgi EB. Stability of lipases produced by Yarrowia lipolytica in the presence of cheese whey. Chem Eng Trans. 2014;37:703–708. [Google Scholar]
- 26.Siqueira PF, Karp SG, Carvalho JC, Sturm W, Rodriguez-Leon JA, Tholozan JL, Singhania RR, Pandey A, Soccol CR. Production of bio-ethanol from soybean molasses by Saccharomyces cerevisiae at laboratory, pilot and industrial scales. Bioresour Technol. 2008;99:8156–8163. doi: 10.1016/j.biortech.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 27.Solaiman DK, Ashby RD, Hotchkiss AT, Jr, Foglia TA. Biosynthesis of medium-chain-length poly(hydroxyalkanoates) from soy molasses. Biotechnol Lett. 2006;28(3):157–162. doi: 10.1007/s10529-005-5329-2. [DOI] [PubMed] [Google Scholar]
- 28.Takaku H, Matsuzawa T, Yaoi K, Yamazaki H. Lipid metabolism of the oleaginous yeast Lipomyces starkeyi. Appl Microbiol Biotechnol. 2020;104(14):6141–6148. doi: 10.1007/s00253-020-10695-9. [DOI] [PubMed] [Google Scholar]
- 29.Trindade MI, Abratt VR, Reid SJ. Induction of sucrose utilization genes from Bifidobacterium lactis by sucrose and raffinose. Appl Environ Microbiol. 2003;69(1):24–32. doi: 10.1128/AEM.69.1.24-32.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vyas S, Chhabra M. Assessing oil accumulation in the oleaginous yeast Cystobasidium oligophagum JRC1 using dairy waste cheese whey as a substrate. 3Biotech. 2019;9(5):173. doi: 10.1007/s13205-019-1701-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G, Bai T, Miao Z, Ning W, Liang W. Simultaneous production of single cell oil and fumaric acid by a newly isolated yeast Aureobasidium pullulans var. aubasidani DH177. Bioprocess Biosyst Eng. 2018;41(11):1707–1716. doi: 10.1007/s00449-018-1994-0. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Ledesma-Amaro R, Wei Y, Ji B, Ji XJ. Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica-A Review. Bioresour Technol. 2020;313:123707. doi: 10.1016/j.biortech.2020.123707. [DOI] [PubMed] [Google Scholar]
- 33.Wang ZP, Xu HM, Wang GY, Chi Z, Chi ZM. Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA-DC 50109. Biochim Biophys Acta. 2013;1831:675–682. doi: 10.1016/j.bbalip.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Wang ZP, Fu WJ, Xu HM, Chi ZM. Direct conversion of inulin into cell lipid by an inulinase-producing yeast Rhodosporidium toruloides 2F5. Bioresour Technol. 2014;161:131–136. doi: 10.1016/j.biortech.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 35.Wang ZP, Zhang LL, Liu S, Liu XY, Yu XJ. Whole Conversion of Soybean Molasses into Isomaltulose and Ethanol by Combining Enzymatic Hydrolysis and Successive Selective Fermentations. Biomolecules. 2019;9(8):353. doi: 10.3390/biom9080353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yalcin SK, Bozdemir MT, Ozbas ZY. Utilization of whey and grape must forcitric acid production by two Yarrowia lipolytica strains. Food Biotechnol. 2009;23:266–283. doi: 10.1080/08905430903106860. [DOI] [Google Scholar]
- 37.Yang H, Wang Z, Lin M, Yang ST. Propionic acid production from soy molasses by Propionibacterium acidipropionici: Fermentation kinetics and economic analysis. Bioresour Technol. 2018;250:1–9. doi: 10.1016/j.biortech.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Yu X, Zheng Y, Dorgan KM, Chen S. Oil production by oleaginous yeasts using the hydrolysate from pretreatment of wheat straw with dilute sulfuric acid. Bioresour Technol. 2011;102(10):6134–6140. doi: 10.1016/j.biortech.2011.02.081. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y, Wang HP, Han B, Yu X. Coupling of abiotic stresses and phytohormones for the production of lipids and high-value by-products by microalgae: A review. Bioresour Technol. 2019;274:549–556. doi: 10.1016/j.biortech.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 40.Zhang CY, Lin X, Feng B, Liu XE, Bai XW, Xu J, Pi L, Xiao DG. Enhanced leavening properties of baker's yeast by reducing sucrase activity in sweet dough. Appl Microbiol Biotechnol. 2016;100(14):6375–6383. doi: 10.1007/s00253-016-7449-0. [DOI] [PubMed] [Google Scholar]
- 41.Zhang LL, Li J, Wang YL, Liu S, Wang ZP, Yu XJ. Integrated Approaches to Reveal Genes Crucial for Tannin Degradation in Aureobasidium melanogenum T9. Biomolecules. 2019;9(9):439. doi: 10.3390/biom9090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Wang ZP, Sheng J, Zheng Y, Ji XF, Zhou HX, Liu XY, Chi ZM. High and efficient isomaltulose production using an engineered Yarrowia lipolytica strain. Bioresour Technol. 2018;265:577–580. doi: 10.1016/j.biortech.2018.06.081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. (a) SDS-PAGE analysis of recombinant GalB. Lane M, standard Mw markers; lane 1, purified GalB; lane 2, Bovine albumin. (b) Substrate specificity of GalB (c) The pH activity and stability of GalB. (d) Thetemperature activity and stability of GalB. Figure S2. (a) SDS-PAGE analysis of recombinant GalC. Lane M, standard Mw markers; lane 1, purified GalC; lane 2, Bovine albumin. (b) Substrate specificity of GalB (c) The pH activity and stability of GalC. (d) Thetemperature activity and stability of GalC. Figure S3. (a) SDS-PAGE analysis of recombinant GalG. Lane M, standard Mw markers; lane 1, purified GalG; lane 2, Bovine albumin. (b) The pH activity and stability of GalG. (c) Thetemperature activity and stability of GalG. Table S1. Primers for amplifying genes coding galactosidases. Table S2. Primers used to perform qRT-PCR assays.