Abstract
The development of open computational pipelines to accelerate the discovery of treatments for emerging diseases allows finding novel solutions in shorter periods of time. Consensus molecular docking is one of these approaches, and its main purpose is to increase the detection of real actives within virtual screening campaigns. Here we present dockECR, an open consensus docking and ranking protocol that implements the exponential consensus ranking method to prioritize molecular candidates. The protocol uses four open source molecular docking programs: AutoDock Vina, Smina, LeDock and rDock, to rank the molecules. In addition, we introduce a scoring strategy based on the average RMSD obtained from comparing the best poses from each single program to complement the consensus ranking with information about the predicted poses. The protocol was benchmarked using 15 relevant protein targets with known actives and decoys, and applied using the main protease of the SARS-CoV-2 virus. For the application, different crystal structures of the protease, and frames obtained from molecular dynamics simulations were used to dock a library of 79 molecules derived from previously co-crystallized fragments. The ranking obtained with dockECR was used to prioritize eight candidates, which were evaluated in terms of the interactions generated with key residues from the protease. The protocol can be implemented in any virtual screening campaign involving proteins as molecular targets. The dockECR code is publicly available at: https://github.com/rochoa85/dockECR.
Keywords: Molecular docking, Exponential consensus ranking, Open source, Virtual screening, SARS-CoV-2 main protease
Graphical abstract
1. Background
Open initiatives for drug discovery purposes have become a priority to tackle neglected and emerging diseases affecting vulnerable populations [1,2]. From a computational perspective, various initiatives are available to analyze public information and predict outcomes useful from a biological and chemical viewpoint [[3], [4], [5]]. Fields such as cheminformatics and chemogenomics, allow the assessment of molecular candidates based on their physico-chemical properties and potential mechanism of action towards a target of interest [6,7]. Many of these methods rely on curated data and open source software to plan, perform and share the results with the community. In critical situations, the massive sharing of scientific findings around novel treatments, or repositioning of known alternatives is crucial to advance in the fight against the causative agents [[8], [9], [10]].
In this scenario, alternatives like molecular docking are useful to screen and rank chemical libraries in a fast and massive way [11,12]. With molecular docking it is possible to find the most favorable position, orientation and conformation (pose) for the binding of a molecule to, for example, a protein target, assigning a score that is the estimate of the likelihood of binding of each molecule and pose [13]. However, the ability of docking software to accurately predict the docking pose can be affected by system-bias effects provided by parameter training or over-fitting [14]. To overcome this limitation, the exponential consensus ranking (ECR) methodology was proposed, which can also include the flexibility of the biological target to increase the success rate of virtual screening in systems where little information is known [15].
Other protocols for consensus docking and scoring have been reported in the literature. One example is the DockBox, a package that facilitates the implementation of multiple docking programs and scoring functions for virtual screening purposes [16]. The protocol proposes the score-based consensus docking as an alternative to classic consensus docking, with reported higher success rates on predicting poses based on enrichment factors of known active and decoy molecules. Similarly, other methodologies have implemented multiple docking approaches to filter a major range of false positives during virtual screening campaigns [17,18], as well as combining multiple scoring functions with trajectories obtained from molecular dynamics (MD) simulations for similar purposes [19]. However, the ranking methodologies can discard molecular actives that are not necessarily detected by all the programs included in the consensus. Additionally, when ranking the molecules using traditional scoring functions, only the score and not the pose predicted by the docking program is taken into account. Therefore, we are implementing the already validated ECR method [15] to provide a different metric to combine the results of widely used docking programs, and a metric based on the RMSD of the best ranked poses in a protocol publicly available for the community.
Here we present dockECR, an open source consensus docking and ranking protocol for virtual screening campaigns. The code allows the parallelisation of the docking runs for multiple ligands, and applies the ECR method to find the most promising candidates. A set of active/decoys benchmarks of the protocol are included using 15 protein targets from the DUD-E dataset [20]. As an application, we implemented the protocol with the main protease from SARS-CoV-2. A total of eight molecules were prioritized as an effort to share the computational findings with other researchers working in the field.
2. Methods
2.1. Molecular docking and consensus ranking protocol
The consensus molecular docking used by dockECR was configured with four open source docking programs: LeDock [21], rDock [22], Smina [23], and AutoDock Vina [24]. Each program has a different scoring function and search algorithm, and the combination of their results by means of a consensus ranking can avoid the bias given by the training set of each docking program [15]. Other studies, inspired by the consensus docking, have also included the docked poses as a criterion to rank the molecules [16,17,25]. For that reason, and in addition to the docking scores, we created the RMSD-based scoring (RBS), a metric to rank the molecular candidates based on the docking poses. This involves calculating per molecule the RMSD by pairs between the best poses obtained by the different molecular docking software. The average RMSD is calculated to rank the molecules according to the most conserved pose (i.e. the lower the average RMSD value the better). The RMSD is calculated using the RDKit package in python (https://www.rdkit.org/) between each pair of molecules.
dockECR uses the ECR method to combine the results of the different docking programs/scoring functions, and considers the effect of the pose prediction by including the results of the RBS in the consensus. The result of the virtual screening given by each docking program/scoring function (j) consists of a ranking in which the molecules are sorted according to their docking score. For each scoring function, the molecules predicted to have a higher affinity for the target will have a position at the top of the rank. The ECR takes the position in the rank of each molecule (i) to assign a score , using the rank of the molecule () given by each individual docking program, . The parameter can be different for each scoring function j and can be interpreted as the weight assigned to each scoring function in the consensus. This parameter is set to the desired x% percentage of filtered molecules at the end of the screening, for instance x = 5% of the dataset (i.e., for a dataset of 10000 molecules where we want to prioritize -filter- only 5% of the data as a result of our consensus docking and scoring, we set ). With this approach, the final score of each molecule i is defined as the sum of all of the scores :
| (1) |
We remark that the notation for σ in eq. (1) is slightly different from Ref. [15], to emphasize the possibility of giving different weights to the scoring functions, a possibility that had not been explored before. The score obtained with the ECR is used to make the final ranking and to prioritize the best candidates for further steps. A summary of the protocol implemented by dockECR is shown in Supplementary Fig. S1.
2.2. Code organization and benchmark systems
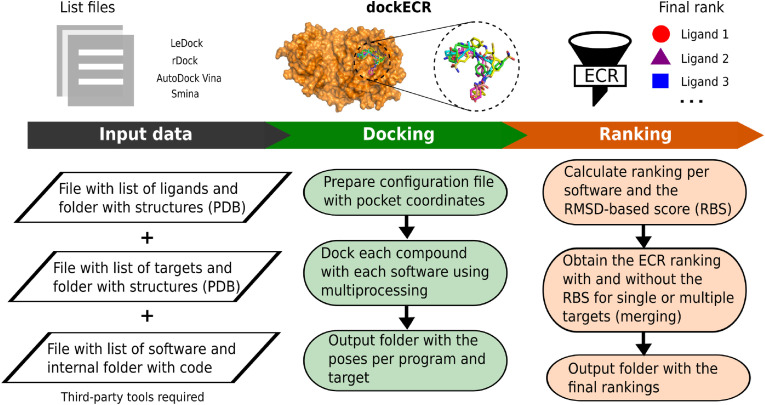
The code is available as a python script with calls to the system through Shell commands. This means the protocol has been configured to be run under a Unix terminal. Each docking run is calculated using a single core, with all the runs controlled by the multiprocessing module available in python [26]. In this sense, the code uses the parallel CPU architecture available in the computer or server where it is located. The results are stored in different folders, including the poses obtained from the docking programs and the calculated ranks of the ligands. A flowchart of dockECR is shown in Fig. 1 .
Fig. 1.
Flowchart of the dockECR protocol, describing the required input data, the molecular docking steps, and the ranking using the ECR approach.
The script was configured to run the virtual screening using one single target or multiple targets based on a merging and shrinking methodology [27,28] with multiple ligands, all of them in PDB format [29]. Third-party tools are provided in an auxiliary folder available at the GitHub repository (https://github.com/rochoa85/dockECR). However, it is recommended to install the docking programs according to the source instructions to guarantee the correct localization of the required libraries. Finally, to submit a run, a configuration file should be provided with all the necessary parameters for the docking search box defined based on the target binding site of interest. An example of the configuration file options and the dockECR folder architecture is provided in the README file of the code repository.
The consensus protocol used by dockECR was validated in a previous publication with multiple protein targets having available bioactivity data [15]. Here, we extend the applicability of the method by implementing only free docking software into the dockECR pipeline. Moreover, we added the possibility to combine the rankings obtained through the scoring functions with the information related to the best pose predicted by the docking software, including the RBS metric into the ECR.
To evaluate the impact of the new protocol code based on known active and decoys of studied molecular targets, we selected 15 protein systems: angiotensin-converting enzyme (ace), beta-lactamase (ampc), caspase-3 cysteine protease (casp3), coagulation factor VII (fa7), fatty acid binding protein adipocyte (fabp4), human immunodeficiency virus type 1 protease (hiv), heat shock protein HSP 90-alpha (hsp90a), hexokinase type IV (hxk4), leukocyte adhesion glycoprotein LFA-1 alpha (ital), human thymidine kinase (kith), tyrosine-protein kinase (lck), neuraminidase (nram), phospholipase A2 group IIA (pa2ga), poly [ADP-ribose] polymerase-1 (parp1) and trypsin I (try). For all the systems, a number of active and decoy molecules from the DUD-E decoy database [20] were docked using the consensus protocol. The numbers of molecules per system are provided in the Supplementary Table S1. All the files are publicly available in the DUD-E database: http://dude.docking.org/targets.
The docking parameters for each program were set as follows: The docking box size for all the programs was set to 30 × 30 × 30 Å, and the box centroids were defined based on the coordinates of a co-crystallized ligand of reference (see Supplementary Table S2). 50 poses per molecule were requested for each of the docking programs. After the docking we collected a ranking from each program/scoring function sorting the molecules according to the docking score of the best scored pose. Then, we combined all the rankings using the ECR method to obtain a single final rank. The only external parameter needed to use the ECR is the σ value in eq. (1). Taking into account that can represent the weight assigned to each scoring function j, we tested the effect of the inclusion of the RBS metric under 3 different scenarios, using values for with x = 0, 2.5 and 5% of the total number of molecules in the dataset. In all cases, the σ value for the other scoring functions x was kept as 5% of the database. Enrichment factors (EF) of the screening were calculated using 2% and 5% of the database to assess how dockECR can benefit the overall results. The calculated EF are the ratio of actives/decoys found in the top-2% and top-5% of the virtual screening ranked results in comparison with the original dataset.
2.3. Application of dockECR with the SARS-CoV-2 main protease Mpro
2.3.1. Selection of ligands and Mpro representative structures
A library of 60 fragments obtained from the XChem screening experiment at Diamond, combined with a mass spectrometry screen of covalent fragments in the London Lab at the Weizmann Institute (Israel), was the motivation to select a list of non-covalent fragments to combine and optimize novel molecular entities against the SARS-CoV-2 main protease. Specifically, a new library was constructed based on the visual inspection and thorough interaction analysis of a list of 21 non-covalent fragments within the active site of the enzyme. We designed a molecular scaffold based on key features and the interaction patterns observed for the fragments 161, 426 and 434 reported on the website (https://covid.postera.ai/covid). 79 derivatives were then constructed based on observations reported in the literature and different strategies to achieve better drug-like physicochemical and pharmacokinetic properties. More details about the library construction are provided in Results section 3.3.1. Regarding the Mpro structures, two strategies were followed up to obtain a structural representation of the flexibility in the protease binding site. The first involved the selection of four distinct Mpro crystals, including one apo form (PDB id 6y2e), and three others in complex with potential inhibitors (PDB ids 5re4, 6fv2, 6lu7). The multiple conformations were used to capture fluctuations of key amino acids within the region of interest. As a second alternative, a MD simulation was run to obtain representative snapshots of the protein during the trajectory. Specifically, the apo crystal structure with PDB id 6y2e was used as initial configuration for the protein.
2.3.2. Preparation of protease and MD setup
The system was prepared as follows. The tautomeric states of the histidines were estimated employing the Gromacs software [30], which performs a hydrogen bond net analysis to determine the most probable tautomer. Special attention was given to His41 and His163, based on their roles in the catalytic mechanism and protein-ligand interactions, respectively. The system was solvated with the TIP3P water model [31] in a truncated octahedric box, extending 10 Å from the protein. A physiological salt concentration of 0.15 M was used, employing Na + an Cl − ions. Finally, hydrogens were added using the Tleap module of the Amber simulation suite [32].
The simulation was performed using the PMEMD cuda module of the Amber simulation suite, and consisted of the following steps: an initial 1 picosecond (ps) run with a 0.01 femtosecond (fs) timestep to eliminate bad contacts, followed by an energy minimization; heating from 0 to 10K over 10 ps with a 0.1 fs time step with strong restraints (50 kcal/mol/Å2) on the protein residues, and then from 10 to 300K over 90 ps with a 0.5 fs timestep and weaker restraints (10 kcal/mol/Å2). The system was then equilibrated for 400 ps at constant temperature and pressure with weak restraints on the CA atoms of the protein (1 kcal/mol/Å2). The Langevin thermostat was used with a collision frequency of 2.0 ps−1 [33], the SHAKE algorithm was used to constrain bonds, allowing a 2 fs timestep, and an 8.0 Å cutoff was used for non-bonded interactions. Finally, a 100 nanoseconds (ns) production simulation was performed under the NPT conditions described above, and ten equidistant snapshots from the second half of the simulation were saved as representative protein structures for the virtual screening process.
2.3.3. Consensus docking and ranking
We used dockECR on the mentioned groups of Mpro structures. For Smina, the Vinardo scoring function was used [34]. The sampling space for docking was defined after aligning the target structures, placing the center of the box in the catalytic site region between domains I and II, where ligands have been previously co-crystallized. The box size remained the same for all programs: 30 × 30 × 30 Å, except for rDock, which does not allow the definition of a search box, but takes as the docking volume the free space that a ligand can occupy in the binding pocket. The rDock docking site was built automatically using the ligand-based method [22], where we randomly selected the best ranked pose for one of the molecules studied (molecule 5) to build the docking space. All other sampling parameters were taken as default.
After screening the database over each target structure, we used the merging and shrinking strategy to obtain a unique rank per scoring function for each molecule. We combined the rankings obtained for each crystal structure and selected the best rank per scoring function and the RBS. We repeated the same process for the MD frames. The best 20 molecules from the two final rankings were selected. The molecules present in both final lists were prioritized for further analysis.
3. Results and discussion
3.1. Open source software and reproducibility
Thanks to the large amount of open source software and the sharing of protocols and best practices, it is possible to configure code projects that can be useful in the search of novel therapeutic alternatives to tackle emerging diseases as quickly and rationally as possible. In this work, we focused on a structure-based open source protocol, dockECR, which implements a consensus docking and ranking approach to propose novel molecular entities. The protocol can be easily reproduced by other researchers based on the provided code. A list of the open software used and the versions implemented for this project is available in Table 1 .
Table 1.
List of open source software implemented by the dockECR protocol and the reported application.
| Name | Version/Year |
|---|---|
| AutoDock Vina | 1.1.2 |
| rDock | 2013.1 |
| LeDock | 2015 |
| Smina | 1.0 |
| Gromacs | 5.1.4 |
| RDKit | 2020.03.1 |
| BioPython | 1.77 |
| OpenBabel | 2.3.2 |
The ECR method for consensus ranking implemented in dockECR is a novel approach that uses similar docking software than other consensus docking and ranking alternatives [35,36]. However, as discussed in Ref. [15], the simple mathematical formulation of the ECR avoids bias due to the training-set dependencies and requires less computational resources than other consensus strategies that have recently been proposed based on machine learning [35, 36]. Additionally, besides the similarities with other consensus strategies regarding the docking programs implemented, we aim to provide a fully efficient code, including the ECR as a method able to prioritize candidates with an overall better performance than individual docking programs for large compound libraries. To reproduce the results of our benchmark and application, we provide the code to run the consensus docking analysis with the subsequent rankings by the ECR method. The code is available in the GitHub repository: https://github.com/rochoa85/dockECR.
Regarding the computational time, one advantage of dockECR is the possibility to parallelize the runs in one single core jobs, which expands depending on the available infrastructure. In addition, the four docking/scoring software selected are very fast, and using the consensus instead of only one program scales the required resources linearly. For example, running a consensus docking campaign of 6000 molecules can take around 6 h in a 24 core server. In the following section, we describe the results of the dockECR benchmarking study using different systems with a list of active/decoy molecules available.
3.2. Benchmark analysis
3.2.1. Comparison with single docking programs
We tested the implementation of the consensus method used by dockECR with and without the inclusion of the RBS metric, and compared the results by means of the enrichment factors using the top-2% of the virtual screening ranking. The higher the enrichment factors, the more hits are prioritized using the selected protocol. A list of the EF for the top-2% of the 15 diverse target systems for the RBS, and for the ECR using different weights (σ) in the consensus to include the RBS metric, are shown in Table 2 . The EF values for the single scoring functions are also included, and a similar table with all the calculated EF for the top-5% is available in Supplementary Table S3.
Table 2.
EF2% for the RBS, the single docking programs and the consensus ranking. The ECR strategies were calculated for a σ threshold equal to the 5% of the database for all the scoring functions, except the RBS where we used and , i.e., thresholds of 5% and 2.5%, respectively.
| Targets | ECR-noRBS | ECR-RBS | ECR-weighted | Vina | Smina | LeDock | rDock | RBS |
|---|---|---|---|---|---|---|---|---|
| ace | 14.61 | 14.23 | 13.1 | 11.52 | 15.14 | 8.36 | 1.13 | 0.98 |
| ampc | 0 | 0.81 | 1.61 | 0.81 | 0 | 0 | 4.03 | 1.61 |
| casp3 | 3.01 | 2.29 | 1.86 | 2.15 | 4.73 | 2.15 | 0.72 | 0.29 |
| fa7 | 12.16 | 13.24 | 10.81 | 6.76 | 7.57 | 9.19 | 1.62 | 1.89 |
| fabp4 | 19.3 | 20.18 | 18.42 | 20.17 | 21.05 | 5.26 | 2.63 | 2.63 |
| hiv | 3.15 | 3.19 | 2.44 | 3.44 | 4.62 | 0.47 | 0.89 | 1.25 |
| hsp90a | 2.4 | 2.8 | 2 | 0 | 0 | 2.40 | 7.20 | 2.8 |
| hxk4 | 3.15 | 1.97 | 1.18 | 0.79 | 3.94 | 7.87 | 0 | 0 |
| ital | 3.65 | 3.22 | 2.79 | 0.64 | 4.08 | 2.15 | 0.21 | 1.29 |
| kith | 4.17 | 4.17 | 3.77 | 2.98 | 4.56 | 3.97 | 0.19 | 3.18 |
| lck | 12.37 | 12.88 | 12.23 | 5.19 | 9.81 | 9.59 | 3.37 | 5.86 |
| nram | 0 | 0.45 | 2.48 | 0 | 0 | 0 | 2.70 | 4.05 |
| pa2ga | 6.69 | 11.02 | 14.17 | 1.18 | 7.87 | 2.76 | 3.54 | 11.42 |
| parp1 | 12.6 | 14.08 | 15.43 | 11.46 | 8.56 | 4.38 | 4.04 | 7.14 |
| try | 7.39 | 6.14 | 4.95 | 4.22 | 5.08 | 8.11 | 0.39 | 0.79 |
The diversity of the targets allows observation of system-bias effects and overfitting that can be present in the individual docking programs, as some programs show outstanding results for some systems, but poor results for others. For instance, rDock shows an enrichment of the dataset for ampc and nram targets, where all the other docking programs show very poor performance, while all the other programs outperform rDock for ace, casp3, kith and try systems. Given this dependency of the outcome of the docking programs on the target, the ECR method allows the combination of the results of different scoring functions, and retrieves a new score based on the rank of the molecules, which has the advantage of being independent of the variable units, scales and offsets of the different scoring functions.
We show that using ECR avoids the problems of individual docking programs with respect to the dependency of the performance on the target. We noted that, from the individual docking programs, Smina has the best results. However, if we compare the EF2% of the consensus including the four scoring functions and the RBS metric, we found that our method is better for 8 of the 15 targets with respect to Smina. Now, if we compare the EF values with the consensus using the top-5% (the ones from Table S3), the consensus is better for 9 of the 15 targets included. In such cases where the EF is better for the consensus, the values are much greater than those from Smina. If we use only the Smina scoring function we will have 3 targets with EF2% = 0 (ampc, hsp90a, nram), while using the ECR (with and without including the RBS) only 2 targets present no enrichment, and when giving more weight to the RBS (ECR-) all the targets present enrichment.
Consensus docking and scoring strategies aim to provide an enrichment of the dataset independently of the target, docking programs, or molecular libraries used in the virtual screening. Even if we get good performances for this set of targets with Smina, the use of a single docking program/scoring function limits the outcome. By using the ECR, we increase the probability of getting enrichment for most of the targets, even if in some cases the EF slightly decreases.
3.2.2. Impact of the RBS metric in the consensus
Considering the conservation of the best ranked pose among the docking programs by means of the RBS metric, a correlation is observed between the scores given by all the scoring functions and the predicted pose. Table 2 shows the EF2% for the RBS in the last column. Interestingly, the RBS scoring displays an enrichment of the top-2% of the dataset for nram, ampc and hsp90a, where at least 2 of the other scoring functions have EF2% < 1. This justifies its inclusion in the ECR procedure to improve the results, especially for these difficult targets.
To test the impact of adding the RBS results in the consensus ranking, we started by using the ECR including only the scoring function of the docking programs (ECR-noRBS in Table 2). As expected, the ECR metric presents an enrichment for most of the benchmark systems. The same tendency can be seen in the EF5% (see Supplementary Table S3), and in the enrichment plots of Fig. S2, where it is possible to analyze the results for other percentages of the dataset. The only two cases where the ECR presents no enrichment are those where most of the programs fail in discriminating between active molecules and decoys (EF2% < 1), which lead to poor results even after the consensus.
Taking into account that the RBS present an enrichment for those systems where most of the programs present poor performance, we compared the results of the ECR without and with inclusion of the RBS scoring function (ECR-noRBS and ECR- in Table 2, respectively). In general, we found an improvement in the EF2% by including the RBS metric in the ECR. Additionally, given the different nature of the RBS result in contrast to the docking score given by a docking program, we can exploit a feature of the ECR not tested before: the use of different weights for a given scoring function. This weight can be set by changing the value for its ranking (j) in eq. (1). We assigned a higher weight to the RBS metric inside the ECR, using and keeping for all the other scoring functions (j = LeDock, rDock, Smina, Vina). By giving more weight to the RBS function, we find a significant improvement in the enrichment for ampc and nram systems. However, for most of the other systems, the enrichment factor is lower for this case.
3.2.3. General advantages of the dockECR consensus method
Overall, we find that the setup that best performs for most systems is given when we use the ECR and include the RBS metric with the same weight as the other docking programs. This finding shows that even if all the docking programs find a similar pose for a molecule, the score assigned, and therefore the rank of the molecule, can be uncorrelated with the docking pose, and in general, its inclusion is relevant to incorporate the impact of the poses in the formation of the interactions and the calculated scores. Despite having similar results in the benchmark with one of the scoring functions (Smina), the overall advantage of using consensus docking for a given system is the statistical assurance that a better, no-biased outcome, is achieved using consensus when applied to less known systems. We also highlight that the docking programs implemented so far in dockECR are diverse, free and easy to use. However, we noted that for DUD-E targets, better enrichment factors are possible to obtain using other docking programs [20]. This is why we provide the option to add different docking software in the dockECR pipeline, depending on the application in hand.
This makes dockECR a suitable and easy-to-use alternative to obtain a good enrichment during docking campaigns for three main reasons: i) the combination of the results of several docking programs through the ECR improves the outcome and avoid system-bias dependencies in comparison with single programs, ii) the RBS metric allows the inclusion of docking pose effects in the consensus ranking, which can be important for systems where several individual docking programs present no enrichment and, iii) dockECR uses only open software which make it more accessible with an implementation that saves computational time by using parallel computations. We also compared the ECR results with a Z-score [37] obtained from the calculated scores, and a basic average ranking using the four docking programs plus the RBS. We found that the ECR method was superior against the average for 13 of the 15 targets based on the EF2%. Regarding the Z-score, the performances are very similar but with a slight improvement for the ECR method (see Supplementary Table S4).
In addition, the results of dockECR with the hiv protease and the cysteine protease casp3 (see Table 2), motivates the application of the protocol with the main cysteine protease from SARS-CoV-2 (Mpro), given that casp3 is from the same protease family of Mpro [38], and the hiv protease is also expressed as a viral protein [39]. Based on this, we can rank a set of molecules with the potential to modulate the enzymatic activity through reported interactions obtained from experimentally-resolved structures.
3.3. Application: SARS-CoV-2 main protease Mpro
The coronavirus disease 2019 (COVID-19), caused by the Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2), is a pandemic disease affecting millions of people around the world [40,41]. The availability of structural data regarding virus-related proteins and human receptors has motivated the implementation of in silico alternatives to accelerate the search for new inhibitor scaffolds and hits [[42], [43], [44]]. This is the case of the 3CL protease or main protease (Mpro) characterized for SARS-CoV-2, which since its description in literature has motivated the publication of various studies aiming to understand its mechanism of action and screening for potential inhibitors [45,46].
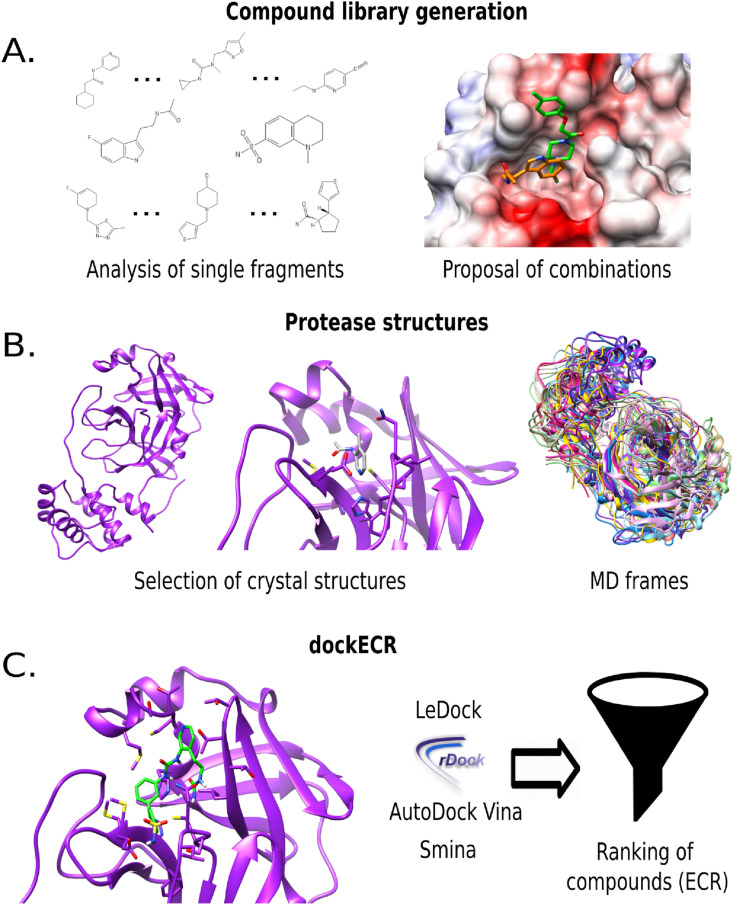
The SARS-CoV-2 main protease is structurally organized in three domains; with the substrate binding site located between domains I and II surrounded by six stranded anti-parallel B-barrels. Domain III is a cluster of five helices involved in the enzyme dimerization that is crucial for enzyme activity [45]. Public consortia are providing valuable insights into the search of molecules that would interact with the binding site, including an international project that published a set of crystal structures of Mpro bound to covalent and non-covalent fragments (https://covid.postera.ai/covid). In order to design powerful inhibitors, one challenge is to implement in silico approaches to combine these fragments and propose better candidates. A summary of the dockECR strategy for Mpro is shown in Fig. 2 . The main results per step are explained in the following sections.
Fig. 2.
General methodology of the protocol. (A) Analysis of single fragments and proposal of new molecular entities based on the combination of some fragments. (B) Selection of Mpro crystal structures and frames from MD simulations of the apo form. (C) dockECR approach using four open source docking programs, with a subsequent ECR ranking of the ligand library.
3.3.1. Generation of libraries
After a visual inspection of the 21 Mpro-fragments crystals, the fragments were classified into three groups according to their position inside the active site (Fig. 3 ), in order to combine fragments from different groups to maximize the interaction with the protein.
Fig. 3.
Fragments 161, 426 and 434 within the active site of Mpro. Interactions with the residues inside the cavity are shown as 2D plots. The colored areas correspond to the zone-classification made for the fragments, according to their position within the active site: group 1 (blue area), group 2 (red area) and group 3 (yellow area). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
From the interaction analysis performed, fragments 161, 426 and 434 were chosen as the basis for the design of the library. The fragments within the active site of Mpro and their main interactions are shown in Fig. 3. The position of the pyridine ring in fragments 426 and 434 was conserved, possibly due to a hydrogen bond interaction with His163, whereas the benzene ring in fragments 434 and 161 were next to each other. Based on these observations, we propose molecule 1 (see Supplementary Fig. S3) as an inhibitor candidate. The fluorobenzene ring in fragment 426 was replaced by a hydroxycyclohexyl group to avoid molecular staking and aggregation, and the two benzene rings were linked by a methylene group to gain flexibility. Several derivatives were then constructed from molecule 1 based on observations reported in the literature and different strategies to achieve better drug-like physicochemical and pharmacokinetic properties.
Despite the library being small and less diverse than those from the benchmark systems, we looked to design a specific library that can satisfy the interactions found in the available crystal structures. With this hypothesis, we can increase the chances of finding an active compound derived from the most promising fragments.
3.3.2. Preparation of targets and MD simulations
The availability of multiple Mpro structures in apo form, or bound to different ligands, allows us to include flexible changes within the active site in our study, which are crucial to prioritize compounds maintaining key interactions. In this case, we observed subtle changes in amino acid orientations in the binding site for the four crystals included.
Simulations were run using a single monomer of the protease. However, the interactions and inferences can be contextualized in the form of the active dimer previously reported [45]. After 100 ns MD production, the protein remained stable with RMSD values below 2.5 Å (Supplementary Fig. S4B), so an equidistant set of frames was chosen for the consensus docking analysis. Most Mpro fluctuations were associated to domain III, which is responsible for mediating the dimerization (Supplementary Fig. S4A).
3.3.3. Consensus docking results
Two ECR rankings using (i) the pool of crystal structures and (ii) the pool of MD frames were constructed, and the molecules present in the top20 of both rankings were prioritized for further analysis (Fig. 4 ). The SMILES representations of the selected structures are reported in the Supplementary Table S5 along with the resulting rankings after applying the consensus strategy with the ECR in Supplementary Tables S6 and S7.
Fig. 4.
Chemical structures of the eight molecules resulting from the combination of the two strategies implemented with the consensus docking analysis.
The docking pose of compounds 2, 20 and 40 along with their predicted interactions are shown in Fig. 5 . These compounds reported suitable docking scores and interactions with several residues involved in the Mpro activity: the catalytic dyad His41 and Cys145 [47], and residues Gly143, His164, Met165, Glu166, Leu167, Arg188, Gln189, Thr190 and Gln192, which are reported as playing significant roles in substrate binding [48], and Phe140, identified as important for Mpro dimerization [48]. Specifically, interactions with Glu166, a key residue that is known to have an effect on shaping the S1 pocket and keeping the enzyme in the active conformation [45] were conserved in all the top candidates.
Fig. 5.
Molecules 2 (A), 20 (B) and 40 (C) in their predicted poses within the active site of Mpro. Their main interactions with the residues inside the cavity are shown as 2D plots.
4. Conclusions
Open source alternatives to accelerate the discovery of novel drugs and vaccines is crucial to tackle multiple diseases, including those caused by the emerging viruses. Among the multiple tools available to identify novel hits, the use of consensus open approaches for docking and ranking ligands in virtual screening campaigns, helps to prioritize molecular candidates that can be openly shared with the scientific community. In this work we describe the protocol called dockECR, which is able to perform consensus docking with a previously published exponential consensus ranking to reduce the number of false positive hits. Moreover, dockECR permits inclusion of the RBS metric in the consensus, which helps to consider the docking pose in the ranking of the molecules and improves the overall performance of the method over the benchmark systems. The computational method is reproducible based on the scripts provided to run similar analysis with any protein system of interest.
For our application with SARS-CoV-2, we found eight molecules that based on the literature and the chemical foundations of ligand interactions with the Mpro enzyme, can become interesting starting points for further optimization steps.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank Dr. Pilar Cossio and Dr. Claudio Cavasotto for the development of the ECR method that inspired this pipeline. We thank Dr. Piraveen Gopalasingam for proofreading of the manuscript. We also thank the Centro de Calculo de Alto Desempeño (Universidad Nacional de Cordoba) and the high performance cluster of INQUIMAE for granting the use of their computational resources. The computations were also performed in a local server of the Max Planck tandem group with an NVIDIA Titan X GPU. This work has been funded and supported by Minciencias, University of Antioquia and Ruta N, Colombia, and the Max Planck Society, Germany.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmgm.2021.108023.
Data availability
The dockECR code is publicly available at: https://github.com/rochoa85/dockECR.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Årdal C., Røttingen J.-A. Open source drug discovery in practice: a case study. PLoS Neglected Trop. Dis. 2012;6(9) doi: 10.1371/journal.pntd.0001827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Voorhis W.C., Adams J.H., Adelfio R., Ahyong V., Akabas M.H., Alano P., Alday A., Resto Y.A., Alsibaee A., Alzualde A., et al. Open source drug discovery with the malaria box compound collection for neglected diseases and beyond. PLoS Pathog. 2016;12(7) doi: 10.1371/journal.ppat.1005763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geldenhuys W.J., Gaasch K.E., Watson M., Allen D.D., Van der Schyf C.J. Optimizing the use of open-source software applications in drug discovery. Drug Discov. Today. 2006;11(3–4):127–132. doi: 10.1016/S1359-6446(05)03692-5. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj A., Scaria V., Raghava G.P.S., Lynn A.M., Chandra N., Banerjee S., Raghunandanan M.V., Pandey V., Taneja B., Yadav J., et al. Open source drug discovery–a new paradigm of collaborative research in tuberculosis drug development. Tuberculosis. 2011;91(5):479–486. doi: 10.1016/j.tube.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Sud M. Mayachemtools: an open source package for computational drug discovery. J. Chem. Inf. Model. 2016;56(12):2292–2297. doi: 10.1021/acs.jcim.6b00505. [DOI] [PubMed] [Google Scholar]
- 6.Riniker S., Landrum G.A. Open-source platform to benchmark fingerprints for ligand-based virtual screening. J. Cheminf. 2013;5(1):26. doi: 10.1186/1758-2946-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Boyle N.M., Guha R., Willighagen E.L., Adams S.E., Alvarsson J., Bradley J.-C., Filippov I.V., Hanson R.M., Hanwell M.D., Hutchison G.R., et al. Open data, open source and open standards in chemistry: the blue obelisk five years on. J. Cheminf. 2011;3(1):37. doi: 10.1186/1758-2946-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slusher B.S., Conn P.J., Frye S., Glicksman M., Arkin M. Bringing together the academic drug discovery community. Nat. Rev. Drug Discov. 2013;12(11):811–812. doi: 10.1038/nrd4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim M.D. Consortium sandbox: building and sharing resources. Sci. Transl. Med. 2014;6(242) doi: 10.1126/scitranslmed.3009024. 242cm6–242cm6. [DOI] [PubMed] [Google Scholar]
- 10.Simpson P.B., Wilkinson G.F. 2020. What Makes a Drug Discovery Consortium Successful? [DOI] [PubMed] [Google Scholar]
- 11.Schneider G. Automating drug discovery. Nat. Rev. Drug Discov. 2018;17(2):97. doi: 10.1038/nrd.2017.232. [DOI] [PubMed] [Google Scholar]
- 12.Chang M.W., Ayeni C., Breuer S., Torbett B.E. Virtual screening for hiv protease inhibitors: a comparison of autodock 4 and vina. PloS One. 2010;5(8) doi: 10.1371/journal.pone.0011955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavasotto C.N. CRC Press; 2015. In Silico Drug Discovery and Design: Theory, Methods, Challenges, and Applications. [Google Scholar]
- 14.Chen Y.-C. Beware of docking! Trends Pharmacol. Sci. 2015;36(2):78–95. doi: 10.1016/j.tips.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Palacio-Rodríguez K., Lans I., Cavasotto C.N., Cossio P. Exponential consensus ranking improves the outcome in docking and receptor ensemble docking. Sci. Rep. dec 2019;9:5142. doi: 10.1038/s41598-019-41594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preto J., Gentile F. Assessing and improving the performance of consensus docking strategies using the dockbox package. J. Comput. Aided Mol. Des. 2019;33(9):817–829. doi: 10.1007/s10822-019-00227-7. [DOI] [PubMed] [Google Scholar]
- 17.Tuccinardi T., Poli G., Romboli V., Giordano A., Martinelli A. Extensive consensus docking evaluation for ligand pose prediction and virtual screening studies. J. Chem. Inf. Model. 2014;54(10):2980–2986. doi: 10.1021/ci500424n. [DOI] [PubMed] [Google Scholar]
- 18.Plewczynski D., Łażniewski M., Grotthuss M.V., Rychlewski L., Ginalski K. “Votedock: consensus docking method for prediction of protein–ligand interactions. J. Comput. Chem. 2011;32(4):568–581. doi: 10.1002/jcc.21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochoa R., Laio A., Cossio P. Predicting the affinity of peptides to major histocompatibility complex class ii by scoring molecular dynamics simulations. J. Chem. Inf. Model. 2019;59(8):3464–3473. doi: 10.1021/acs.jcim.9b00403. [DOI] [PubMed] [Google Scholar]
- 20.Mysinger M.M., Carchia M., Irwin J.J., Shoichet B.K. Directory of useful decoys, enhanced (dud-e): better ligands and decoys for better benchmarking. J. Med. Chem. 2012;55(14):6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N., Zhao H. Enriching screening libraries with bioactive fragment space. Bioorg. Med. Chem. Lett. 2016;26(15):3594–3597. doi: 10.1016/j.bmcl.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Ruiz-Carmona S., Alvarez-Garcia D., Foloppe N., Garmendia-Doval A.B., Juhos S., Schmidtke P., Barril X., Hubbard R.E., Morley S.D. rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS Comput. Biol. 2014;10(4):1–8. doi: 10.1371/journal.pcbi.1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koes D.R., Baumgartner M.P., Camacho C.J. Lessons learned in empirical scoring with smina from the CSAR 2011 benchmarking exercise. J. Chem. Inf. Model. 2013;53(8):1893–1904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scardino V., Bollini M., Cavasotto C. ChemRxiv; 2021. Combination of Pose and Rank Consensus in Docking-Based Virtual Screening: the Best of Both Worlds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marowka A. On parallel software engineering education using python. Educ. Inf. Technol. 2018;23(1):357–372. [Google Scholar]
- 27.Cavasotto C.N., Abagyan R.A. Protein flexibility in ligand docking and virtual screening to protein kinases. J. Mol. Biol. 2004;337(1):209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 28.Cavasotto C., Orry A., Abagyan R. The challenge of considering receptor flexibility in ligand docking and virtual screening. Curr. Comput. Aided Drug Des. 2005;1(4):423–440. [Google Scholar]
- 29.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 31.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79(2):926–935. [Google Scholar]
- 32.Case D.A., Betz R., Cerutti D., Cheatham T., Darden T., Duke R., Giese T., Gohlke H., Goetz A., Homeyer N. University of California; San Francisco: 2016. Amber 2016 Reference Manual; pp. 1–923. [Google Scholar]
- 33.Davidchack R.L., Handel R., Tretyakov M. Langevin thermostat for rigid body dynamics. J. Chem. Phys. 2009;130(23):234101. doi: 10.1063/1.3149788. [DOI] [PubMed] [Google Scholar]
- 34.Quiroga R., Villarreal M.A. Vinardo: a scoring function based on autodock vina improves scoring, docking, and virtual screening. PloS One. 2016;11(5):1–18. doi: 10.1371/journal.pone.0155183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren X., Shi Y.-S., Zhang Y., Liu B., Zhang L.-H., Peng Y.-B., Zeng R. Novel consensus docking strategy to improve ligand pose prediction. J. Chem. Inf. Model. 2018;58(8):1662–1668. doi: 10.1021/acs.jcim.8b00329. [DOI] [PubMed] [Google Scholar]
- 36.Ericksen S.S., Wu H., Zhang H., Michael L.A., Newton M.A., Hoffmann F.M., Wildman S.A. Machine learning consensus scoring improves performance across targets in structure-based virtual screening. J. Chem. Inf. Model. 2017;57(7):1579–1590. doi: 10.1021/acs.jcim.7b00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S., Fu R., Zhou L.-H., Chen S.-P. Application of consensus scoring and principal component analysis for virtual screening against β-secretase (bace-1) PloS One. 2012;7(6) doi: 10.1371/journal.pone.0038086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers C., Erkes D.A., Nardone A., Aplin A.E., Fernandes-Alnemri T., Alnemri E.S. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat. Commun. 2019;10(1):1–17. doi: 10.1038/s41467-019-09397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miao Y., Huang Y.-m. M., Walker R.C., McCammon J.A., Chang C.-e. A. Ligand binding pathways and conformational transitions of the hiv protease. Biochemistry. 2018;57(9):1533–1541. doi: 10.1021/acs.biochem.7b01248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (covid-19) outbreak. J. Autoimmun. 2020:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong E., Du H., Gardner L. An interactive web-based dashboard to track covid-19 in real time. Lancet Infect. Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mirza M.U., Froeyen M. Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J. Pharmaceut. Anal. 2020;10(4):320–328. doi: 10.1016/j.jpha.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1758788. just-accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of sars-cov-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macchiagodena M., Pagliai M., Procacci P. Identification of potential binders of the main protease 3clpro of the covid-19 via structure-based ligand design and molecular modeling. Chem. Phys. Lett. 2020:137489. doi: 10.1016/j.cplett.2020.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rani R., Singh A., Pareek A., Tomar S. 2020. In Silico Guided Drug Repurposing to Combat Sars-Cov-2 by Targeting Mpro, the Key Virus Specific Protease. [Google Scholar]
- 48.Goyal B., Goyal D. Targeting the dimerization of main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb. Sci. 2020;22(6):297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dockECR code is publicly available at: https://github.com/rochoa85/dockECR.