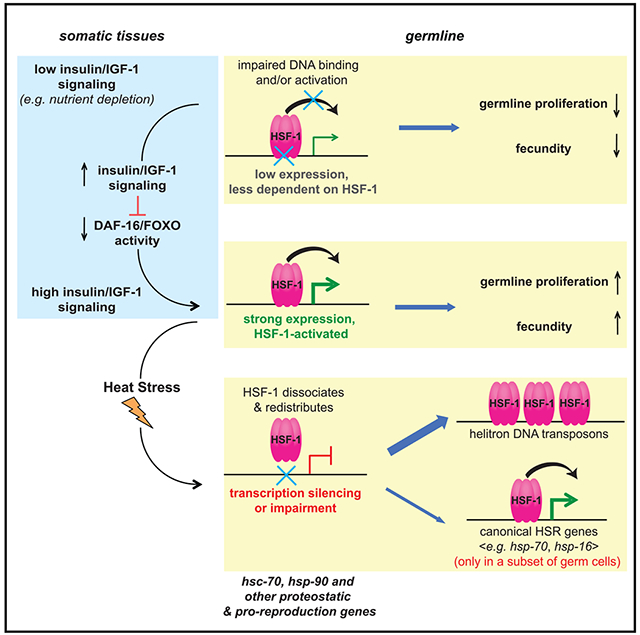
SUMMARY
Germline development is sensitive to nutrient availability and environmental perturbation. Heat shock transcription factor 1 (HSF1), a key transcription factor driving the cellular heat shock response (HSR), is also involved in gametogenesis. The precise function of HSF1 (HSF-1 in C. elegans) and its regulation in germline development are poorly understood. Using the auxin-inducible degron system in C. elegans, we uncovered a role of HSF-1 in progenitor cell proliferation and early meiosis and identified a compact but important transcriptional program of HSF-1 in germline development. Interestingly, heat stress only induces the canonical HSR in a subset of germ cells but impairs HSF-1 binding at its developmental targets. Conversely, insulin/insulin growth factor 1 (IGF-1) signaling dictates the requirement for HSF-1 in germline development and functions through repressing FOXO/DAF-16 in the soma to activate HSF-1 in germ cells. We propose that this non-cell-autonomous mechanism couples nutrient-sensing insulin/IGF-1 signaling to HSF-1 activation to support homeostasis in rapid germline growth.
Graphical abstract
In brief
Using auxin-inducible degradation in Caenorhabditis elegans, Edwards et al. uncover germline-specific activities of HSF-1 that only induce the canonical heat shock response in a subset of germ cells upon stress but are coupled to nutrient-sensing insulin/IGF-1 signaling during development to enhance expression of key proteostatic genes in support of rapid germline proliferation.
INTRODUCTION
Heat shock transcription factor 1 (HSF1) is best known for its roles in the heat shock response (HSR). Upon proteotoxic stress such as heat shock, HSF1 rapidly induces expression of genes encoding molecular chaperones, detoxification enzymes, and protein clearance machineries to restore proteostasis (Morimoto, 2008; Vihervaara et al., 2018). HSF1 is also required for gametogenesis in invertebrates and vertebrates (Abane and Mezger, 2010; Akerfelt et al., 2010; Christians et al., 2000; Jedlicka et al., 1997), but the precise roles of HSF1 and its regulation in germline development are not well understood.
At the cellular level, knockout of HSF1 from mouse oocytes results in difficulty resuming meiosis when reaching reproductive maturity (Metchat et al., 2009). However, the potential contributions of HSF1 to earlier steps of germline development, including proliferation of progenitor cells, have not been explored. At the molecular level, the requirement for HSF1 in germ cells could be linked to its function in stress response because HSF1 knockout leads to oxidative stress and reduces expression of certain chaperone genes in mouse oocytes (Bierkamp et al., 2010; Metchat et al., 2009). It is unclear whether HSF1’s activities in germline development are triggered by proteotoxic stress, as in the HSR. It has been shown during mouse gametogenesis that HSF1 binds to promoters and regulates transcription of genes that are not involved in the canonical HSR. These include meiotic genes in mouse oocytes (Le Masson et al., 2011) and sex chromosomal multicopy genes that are expressed in postmeiotic cells from mouse testis (Akerfelt et al., 2010). Therefore, it remains to be answered how HSF1 is activated in germline development and how HSF1 responds to stress in germ cells.
Germline development and maintenance are energy demanding, and thus sensitive to environmental and metabolic cues (Hubbard et al., 2013). As an essential coordinator of nutrient availability, energy metabolism, and growth, the insulin/insulin growth factor 1 (IGF-1) signaling pathway has evolutionarily conserved roles in male and female gamete development and quality control (Ipsa et al., 2019; Neirijnck et al., 2019; Templeman and Murphy, 2018). In addition, insulin/IGF-1 signaling (IIS) is known to regulate multiple stress response pathways. Among those, it has been shown in C. elegans that IIS suppresses HSF-1 activity in the HSR (Chiang et al., 2012). Whether IIS regulates HSF1 in germline development remains unknown.
Here we used C. elegans as a model organism to interrogate the molecular and cellular functions of HSF-1 in germline development and its regulation by heat stress and IIS. We found that HSF-1 is required for germline progenitor cell proliferation and early meiosis. Using an inducible degron system, we identified the germline-specific binding profiles of HSF-1 and its effects on transcriptomes in germline development and upon heat shock. We found distinct activities of HSF-1 in these two processes, with the canonical HSR being induced in only a small subset of germ cells and heat stress impairing HSF-1 binding at its developmental targets in the germline. Multiple lines of evidence emerged from our experiments, suggesting that HSF-1’s functions in germline development are dictated by IIS. First, HSF-1 becomes partially dispensable for germline development when IIS is reduced. Second, the reproduction-promoting effect of IIS is dependent on HSF-1 in the germline. Finally, IIS represses the transcription factor FOXO/DAF-16 in somatic tissues, which, in turn, non-cell-autonomously activates HSF-1 in germline development. Our findings suggest a mechanism by which HSF-1 activities are coupled to the nutrient-sensing IIS to regulate homeostasis and development of the germline.
RESULTS
HSF-1 has tissue-specific roles in C. elegans larval development and reproduction
Hermaphroditic C. elegans is a good model for studying the process of germline development from progenitor cell proliferation to meiosis and gamete maturation (sperm and oocytes). HSF-1 is essential for larval development (Li et al., 2016; Morton and Lamitina, 2013), so to investigate the functions of HSF-1 in C. elegans germline development, we employed the auxin-inducible degron (AID) system (Zhang et al., 2015) to control HSF-1 protein levels in a spatiotemporal manner. We inserted AID into the C terminus of the endogenous HSF-1 gene using CRISPR, enabling depletion of HSF-1 upon auxin treatment in tissues with transgenic expression of the plant E3 ligase TIR1 (Figure 1A). In our transgenic animals, HSF-1 was depleted to an undetectable level within 2 h in adults (Figures 1B and S1A) and more quickly in larvae (data not shown). Continuous depletion of HSF-1 from the soma after egg lay resulted in larval arrest, as seen in the hsf-1 null mutant (Figure 1C). Depletion of HSF-1 from the germline, however, did not stop or significantly delay larval development (Figures S1B and S1C) but led to sterility (Figure 1D). Neither insertion of AID into HSF-1 nor auxin treatment without AID tagging significantly changed larval development or brood size (Figures 1C and 1D), indicating specificity of the AID system. To confirm the requirement for HSF-1 in the germline in reproduction, we made a transgene that drives pan-somatic expression of HSF-1 in the hsf-1 null mutant. This transgene successfully rescued larval arrest (Figure S1D) but not sterility (Figure S1E). Furthermore, depletion of HSF-1 from the germline but not the soma of young adults significantly reduced brood size (Figure S1F). Our data indicate a function of germline HSF-1 in C. elegans reproduction.
Figure 1. The auxin-inducible degron (AID) system reveals tissue-specific roles of HSF-1 in C. elegans larval development and reproduction.
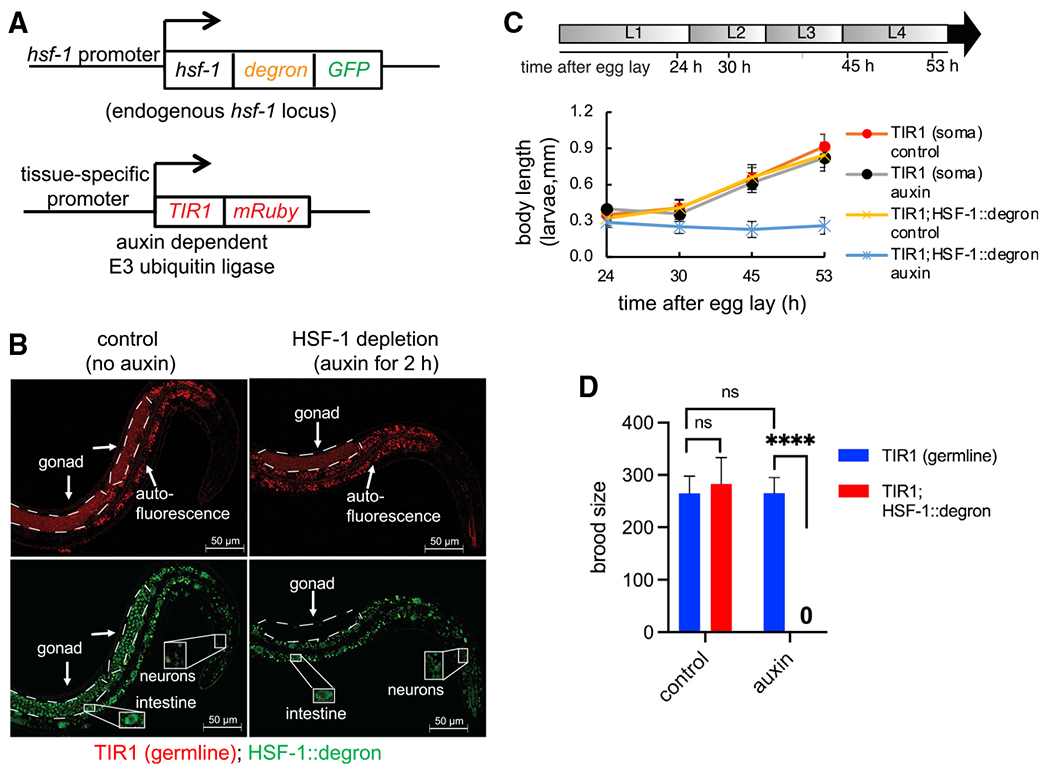
(A) Schematic of the AID system: endogenous HSF-1 tagged with AID and GFP and a transgene expressing the plant E3 ligase TIR1 that recognizes AID in the presence of auxin under control of a tissue-specific promoter.
(B) Live-animal images of young adults with germline-specific HSF-1 depletion by AID for 2 h. Dashed lines outline gonads. Scale bars, 50 μM.
(C) Size tracking of developing larvae with continuous HSF-1 depletion in the soma initiated at egg lay. Data are represented as mean ± standard deviation (n ≥ 15). The timeline represents larval stages (L1–L4) in control animals without HSF-1 depletion. The size of animals with HSF-1 depletion from the soma did not increase after 24 h (Dunnett’s test, p ≥ 0.05) and was significantly smaller than the control without auxin treatment at 30 h (unpaired, two-tailed Student’s t test, P: 4.3E–15) and after, suggesting larval arrest at L1/L2.
(D) Brood size of animals with continuous HSF-1 depletion in the germline initiated at egg lay. Data are represented as mean ± standard deviation (n ≥ 10). Statistical significance was calculated by unpaired, two-tailed Student’s t test. ****p < 0.0001; ns, not significant (p > = 0.05).
HSF-1 is required for mitotic proliferation of germline progenitor cells and early meiosis
To test the contribution of HSF-1 to different steps of germline development, we took advantage of the well-defined temporal progression of this process (Hubbard and Greenstein, 2005) and initiated HSF-1 depletion from germ cells at different times in development. Although HSF-1 depletion starting at any time in embryogenesis and larval development reduced brood size, HSF-1 depletion before mid-L3 led to sterility (Figure S2A). This time point correlates with the beginning of meiotic prophase in the germline, suggesting that HSF-1 may be required for the progenitor cell proliferation that precedes meiosis and/or early stages of meiosis. We then examined the germ cell composition in dissected gonads from young adult animals with HSF-1 depleted from the germline throughout the entire period of external development. Compared with the mock-treated control, HSF-1 depletion resulted in a dramatic decrease in total germline nuclei and meiotic arrest at the pachytene/diplotene stages (Figures 2A–2C and S2B). The decrease in germline nuclei number could be due to inhibition of progenitor cell proliferation or meiotic arrest stopping the flux of germline development. Therefore, we directly examined progenitor cell proliferation at mid-L3 before the start of meiosis. We found significant decreases in total germline nuclei, S phase nuclei (5-ethynyl-2’-deoxyuridine [EdU] positive), and the S index (the portion of cells in S phase) with HSF-1 depletion (Figures S2C and S2D), indicating that HSF-1 is important for proliferation of germline progenitor cells.
Figure 2. HSF-1 is required for mitotic proliferation of germline progenitor cells and early meiosis.
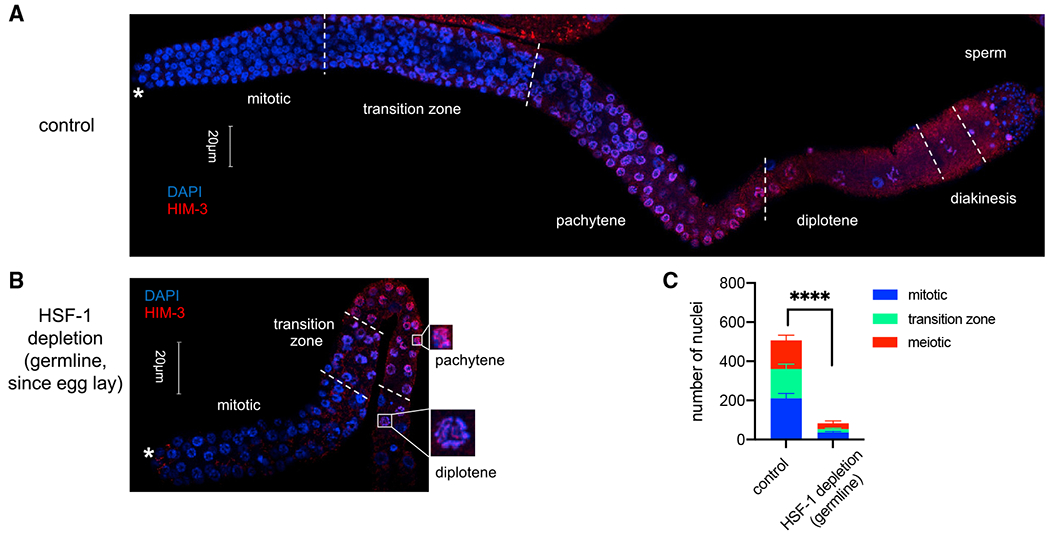
(A and B) Representative images showing immunofluorescence (IF) of the meiotic marker HIM-3 in dissected gonads with DAPI staining of DNA. Young adult animals without auxin treatment (control, A) or with HSF-1 continuously depleted in the germline since egg lay (B) were used. Different stages of germline development are labeled. Germline progenitors are located in the “mitotic” zone. The white asterisks indicate the distal end of gonads where progenitor cell proliferation occurs. Scale bars, 20 μM.
(C) Quantification of nuclei (per gonad arm) in mitosis (progenitor cells), meiosis (from pachytene to diakinesis), or in the transition zone based on IF of Him-3 and the crescent-shaped transition zoon nuclei in (A) and (B). Mean and standard deviation are shown (control, n = 10; HSF-1 depletion, n = 8). Statistical significance on the total number of nuclei was calculated by unpaired, two-tailed Student’s t test. ****p < 0.0001.
HSF-1 exhibits tissue-specific genomic occupancy in young adult animals
To understand the molecular basis of HSF-1’s functions in germline development, we analyzed the germline-specific transcriptional program driven by HSF-1. Because HSF-1 activities are highly sensitive to stress, we developed a strategy to identify tissue-specific HSF-1 binding and transcriptional activities without tissue isolation that may introduce cellular stress. To do so, we coupled tissue-specific HSF-1 depletion by AID with genome-wide transcriptional analyses in whole animals. First, we performed chromatin immunoprecipitation sequencing (ChIP-seq) analyses following acute depletion of HSF-1 from the soma or the germline. 2 h of HSF-1 depletion only changed expression of a handful of genes (Figure S3F), which minimized the potential effects on endocrine/paracrine that may influence HSF-1 binding in non-targeted tissues. The germline-specific binding profiles of HSF-1 could then be inferred by the remaining HSF-1 occupancy upon depletion from the soma or the decrease of HSF-1 occupancy upon depletion from the germline (Figure 3A). Young adult animals were chosen for these analyses because they contain cells at different stages of germline development (Figure 2A) and undergo robust germline proliferation and gametogenesis. HSF-1 binding profiles by ChIP-seq were almost identical in the two AID models before auxin-induced HSF-1 depletion (Figure S3A). However, after 2 h of auxin treatment, the remaining HSF-1 occupancy became quite different (Figure S3B), indicating differential HSF-1 binding in post-mitotic somatic cells and proliferating/differentiating germ cells. We identified three groups of HSF-1 binding sites at the promoters of protein coding genes: those enriched for HSF-1 binding in the soma or the germline and those that display substantial HSF-1 occupancy in both (Figure 3B; Table S1). Genes with “germline-enriched” and “shared” HSF-1 peaks at their promoters are considered HSF-1-bound genes in germ cells.
Figure 3. HSF-1 directs a compact transcriptional program important for homeostasis and development of the germline.
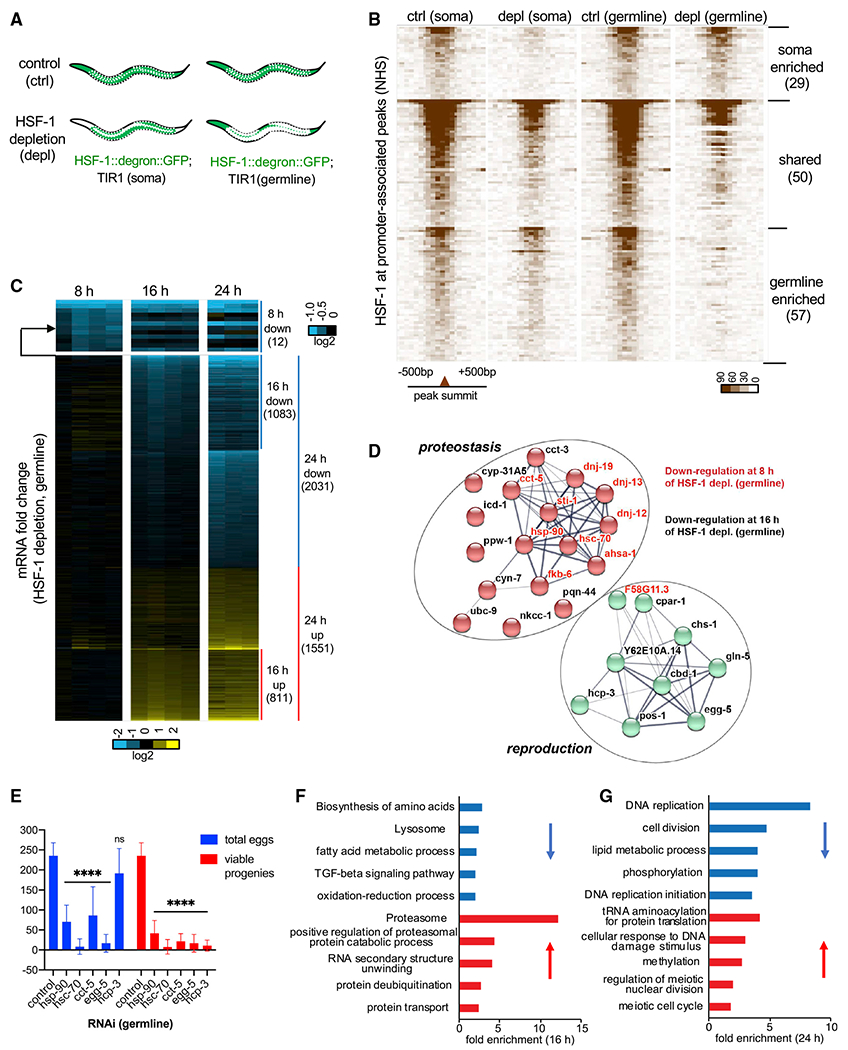
(A) Schematic showing tissue distribution of HSF-1 upon acute depletion in the two AID models. White dashed lines outline gonads.
(B) Heatmaps of HSF-1 occupancy at ChIP-seq peaks (normalized HSF-1 reads mapped to 50-bp bins, ±500 bp from peak summits) that are at promoters (within 1,000 bp from the transcription start sites) of protein-coding genes (PCGs). The two AID models were grown to young adults at 20°C (non-heat-shock [NHS] condition)and treated with auxin for 2 h (depletion [depl]) or mock-treated (control [ctrl]). HSF-1 peaks were grouped into “soma enriched” or “germline enriched” when HSF-1 occupancy at the peaks decreases significantly upon depl from the corresponding tissue but not the other or grouped into “shared” when HSF-1 occupancy decreases significantly upon depl in both (P: 10e–10). The peaks in each group were ranked by the average HSF-1 occupancy under the two “ctrl” conditions. The number of peaks in each group is shown in parentheses.
(C) Heatmap of mRNA fold change upon depl of HSF-1 from the germline of young adults for 8, 16, and 24 h. Whole-animal RNA-seq analyses were done in four biological replicates for the 8-h and 16-h time points and in triplicates for the 24-h time point. TIR1; HSF-1::degron::GFP (auxin) was used as the HSF-1 depl condition, and TIR1; HSF-1::degron::GFP(mock treated), TIR1 (auxin), and TIR1 (mock treated) were pooled together as the ctrl. The mRNA fold change (HSF-1 depl/ctrl) is shown. Differentially expressed (DE) genes (false discovery rate [FDR], 0.05; fold change ≥ 1.25) at any time point were included and ranked by fold change at 8 h, then 16 h, and last, 24 h. The number of PCGs in each group is shown in parentheses.
(D) The gene network directly activated by HSF-1 in germline development. Genes included are those with HSF-1 binding peaks at the promoters and significantly decreased expression upon HSF-1 depl from the germline for 8 h (gene names in red) or 16 h (gene names in black). The protein-protein interaction network was retrieved from the STRING database and grouped by k-means clustering (n = 2). The node color represents the cluster to which the gene belongs (red, proteostasis; green, reproduction). The color saturation of edges represents the confidence score of a functional interaction.
(E) Brood size analyses with germline-specific RNAi. The numbers of eggs and viable progeny were scored. EGG-5 RNAi likely knocked down its paralog EGG-4 as well, based on sequence similarity. Data are represented as mean ± standard deviation (n ≥ 12). Statistical significance was calculated by unpaired, two-tailed Student’s t test relative to the ctrl RNAi (empty vector of L4440). ****p < 0.0001.
(F and G) Gene Ontology (GO) analyses of DE genes by HSF-1 depl from the germline for 16 h (F) and 24 h (G). The top5 GO terms based on enrichment score are shown for downregulated genes (blue bars) and upregulated genes (red bars).
HSF-1 directs a compact transcriptional program important for homeostasis and development of the germline
We then examined the transcriptional effect of HSF-1 depletion by AID via RNA sequencing (RNA-seq). To determine HSF-1-dependent changes in gene expression, we examined the effects of auxin treatment and AID insertion at HSF-1 under all tested conditions (Figure S3C). Either of them only caused a small number of differentially expressed (DE) genes (Figures S3D and S3E). Global transcriptome changes occurred when HSF-1 was depleted from the germline for 24 h (Figures 3C and S3F). These results contrasted with depletion of HSF-1 from the soma, which resulted in much fewer DE genes (Figure S3F).
We next examined the transcriptional changes at 8 h and 16 h to help distinguish primary from secondary effects of HSF-1 depletion from germ cells (Figure 3C; Table S2). At 8 h, all of the 12 DE genes were downregulated. Remarkably, HSF-1 bound to 10 of their promoters in germ cells (Figure 3D, gene names in red), strongly arguing that they are direct targets of HSF-1 in the germline. These results are also consistent with the established role of HSF-1 as a transcriptional activator. Of the 10 “first responders” to HSF-1 depletion, 9 genes encode constitutively expressed chaperones (HSP-90, HSC70/HSP-1 [referred as HSC-70 in this study] and the chaperonin subunit CCT-5) or co-chaperones, which have important roles in protein folding and protein complex assembly. Sixteen additional HSF-1-bound genes in germ cells were downregulated significantly at 16 h (Figure 3D, gene names in black). One of them, egg-5, showed a faster decrease in mRNA upon HSF-1 depletion than its paralog, egg-4, which functions redundantly to egg-5 but has no HSF-1 binding at the promoter (Figure S3G). This suggests that downregulation of these HSF-1-bound genes at 16 h could not exclusively result from a secondary “passenger effect” of germline defects but was likely caused by loss of HSF-1 from the promoters.
We therefore propose that the 26 HSF-1-bound genes that were downregulated at 8 h and/or 16 h are the primary targets of HSF-1 in germ cells. They form a compact transcriptional program consisting of two functional clusters (Figure 3D). One is enriched for genes encoding players in proteostasis (red nodes). In addition to chaperones and co-chaperones, this cluster includes the peptidyl-prolyl isomerase CYN-7, which catalyzes cis-trans isomerization of proline imidic peptide bonds and assists protein folding, and the ubiquitin-conjugating enzyme UBC-9, which functions in protein degradation. The second group is enriched for genes whose encoded proteins function in different processes of reproduction (green nodes). Among these, the centromere protein A (CENP-A) orthologs HCP-3 and CPAR-1 are important for mitotic sister chromatid segregation and nuclear division (Buchwitz et al., 1999; Green et al., 2011; Kalis et al., 2010). EGG-5, an ortholog of human protein tyrosine phosphatase receptor type J (PTPRJ), is required for oocyte-to-zygote transition (Cheng et al., 2009). CBD-1 and CHS-1 are known to make the vitelline and chitin layers of the eggshell (Stein and Golden, 2018). We selected three chaperone genes (hsp-90, hsc-70, and cct-5) and two non-chaperone genes (hcp-3 and egg-5) from the primary targets of germline HSF-1 for a functional test. Knockdown of each of these genes by germline-specific RNAi dramatically decreased the number of eggs and/or viable progenies (Figure 3E), supporting their requirement for fecundity.
To understand the roles of this HSF-1 transcriptional program in germline development, we examined the cascade of transcriptome changes upon germline HSF-1 depletion. Gene Ontology (GO) analyses revealed alterations in cellular redox balance and proteostasis at 16 h of HSF-1 depletion (Figure 3F). These alterations are evident by downregulation of genes functioning in the oxidation-reduction process and upregulation of genes involved in protein degradation by the ubiquitin-proteasome. The latter change is often seen in proteotoxic stress. The genes downregulated at 16 h were also enriched for those involved in amino acid biosynthesis and fatty acid metabolism, two metabolic processes sensitive to proteostasis and cellular redox state (Arnsburg and Kirstein-Miles, 2014; Lismont et al., 2015). Importantly, differential expression of these homeostatic and metabolic genes preceded the massive transcriptome changes in mitotic and meiotic genes after 24 h of HSF-1 deletion (Figure 3G). At 24 h, genes functioning in DNA replication and cell division were downregulated, whereas genes involved in meiosis were upregulated, correlating well with inhibition of progenitor cell proliferation and cell arrest at meiotic prophase caused by HSF-1 depletion. Our data suggest that HSF-1 regulates a compact transcriptional program to maintain cellular homeostasis and support germline development.
Heat stress does not induce the canonical HSR in most germ cells but suppresses HSF-1 activities in germline development
Because chaperone and co-chaperone genes make up a big portion of the germline transcriptional program of HSF-1 (11 of 26 genes), we next wanted to find out whether HSF-1 is activated in germline development by proteotoxic stress as in the HSR. We therefore examined the HSR by RNA-seq analyses in heat-shocked young adult animals following acute depletion of HSF-1 from the soma or the germline by AID. Interestingly, we found that depletion of HSF-1 from the soma abolished almost all heat-shock-induced transcription at HSF-1-bound genes, whereas depletion of HSF-1 from the germline had a minimal effect on the whole-animal HSR (Figure 4A, columns of “%mRNA induction by HS”; Table S3). Because there are similar numbers of cells in the soma and the germline of young adults, our results suggest that the canonical HSR was induced very weakly in the germline or only occurred in a small subset of germ cells. We then chose inducible hsp-70 (at F44E5.4/.5 loci) to probe the spatial pattern of the HSR using RNA fluorescence in situ hybridization (FISH). Transcription of hsp-70 was induced robustly in somatic tissues by 15 min of heat shock (HS) at 34° C and reduced to the non-HS (NHS) level upon HSF-1 depletion from the soma (Figures 4B and S4A). HS induced hsp-70 in only a subset of meiotic prophase nuclei from the germline (Figures 4B and 4C), which is consistent with a previous report (Das et al., 2020). This induction was still dependent on HSF-1 because depletion of HSF-1 from the germline abolished hsp-70 transcription in these selected germ cells without affecting the HSR in the soma (Figures 4C and S4B).
Figure 4. HSF-1 exhibits germline-specific response to heat stress.
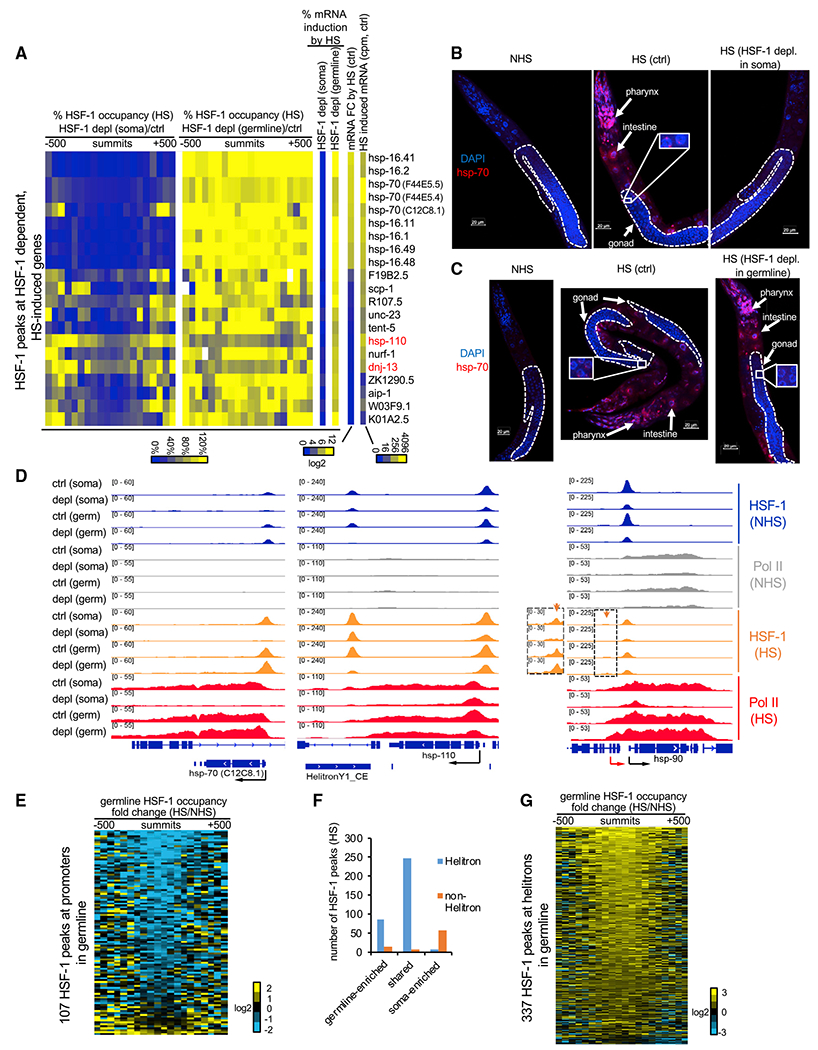
(A) Heatmaps of HSF-1 occupancy and mRNA changes in heat-shocked young adults with acute HSF-1 depl for 2 h. HSF-1 dependent, HS-induced genes are shown and defined by HSF-1 binding at promoters, significantly increasing mRNA levels (FDR, 0.05) upon HS at 34°C for 30 min in both AID models without auxin treatment (ctrl) and significantly decreasing mRNA levels upon HSF-1 depl in at least one AID model. Percent change of HSF-1 occupancy was calculated as the ratio of normalized HSF-1 ChIP-seq reads from HSF-1 depleted to those from the ctrl in 50-bp bins, ± 500bp from peak summits. Percent mRNA induction by HS is the ratio of the increased mRNA after HS from the HSF-1-depleted condition to that from the ctrl condition. The mRNA fold change (FC) by HS and HS-induced mRNA (read counts per million [cpm]) were calculated as the ratio or the difference of mRNA between HS and NHS, respectively, and by pooling the data of ctrl conditions (no auxin) from two AID models.
(B and C) Representative images of RNA fluorescence in situ hybridization (FISH) of hsp-70 (F44E5.4/.5) in young adult AID animals without heat shock or subjected to a heat shock (HS) at 34°C for 15 min following 2 h of mock treatment as the ctrl or auxin treatment to deplete HSF-1 (depl) from the soma (B) or the germline (C). Scale bars, 20 μM.
(D) Gbrowser views of HSF-1 and RNA Pol II occupancy at thehsp-70 (C12C8.1), hsp-110 (and the downstream helitron), and hsp-90 gene loci under NHS and HS conditions with ctrl and HSF-1 depl from the soma or germline (germ). At the hsp-90 locus, the red and black arrows show the distal, inducible and proximal, basal promoters, respectively. The orange arrows and the magnified image show the small, HS-inducible HSF-1 peaks at the distal promoter.
(E) Heatmap showing HS-induced FCs of germline HSF-1 occupancy at the NHS HSF-1 peaks located at promoters of PCGs. Germline HSF-1 occupancy was determined by HSF-1 ChIP-seq reads obtained in animals with acute depl of HSF-1 from the soma. The FC was calculated as the ratio of normalized HSF-1 reads from the HS condition to those from the NHS condition in 50-bp bins, ± 500 bp from peak summits.
(F) Histograms showing the number of HSF-1 binding peaks during HS in the context of their tissue specificity and locations in helitron repeats.
(G) Heatmap showing HS-induced FCs of germline HSF-1 occupancy at the HSF-1 peaks located in helitron repeats. Calculations were done as in (E).
To gain mechanistic insights into the tissue-specific HSR, we performed ChIP-seq analyses to compare HSF-1 binding in the soma and the germline upon HS. For most of the HSF-1-dependent HSR genes, HSF-1 binding at the promoters occurred largely in the somatic tissues but not in the germline during HS. This tissue specificity was evident because HSF-1 occupancy obtained from whole animals was decreased dramatically upon depletion of HSF-1 from the soma but changed modestly by depletion of HSF-1 from the germline (Figure 4A, columns of “ %HSF-1 occupancy”). This observation was illustrated by the classical HSR genes hsp-70 (at the C12C8.1 locus; Figure 4D) and hsp-16s (Figure S4C) as well as the terminal nucleotidyltransferase gene tent-5 (Figure S4C), where HSF-1 binding was induced upon HS. Two HSR genes, hsp-110 and dnj-13 (Figure 4A, names labeled in red), are exceptions that retained substantial levels of HSF-1 in germ cells during HS. We found that their promoters were bound by HSF-1 in the soma and the germline before HS and that HS did not further induce HSF-1 binding (Figures 4D and S4C). Our results indicate that HS-induced binding of HSF-1 at the HSR genes is limited in germ cells. Importantly, HSF-1 was required in the soma for RNA polymerase II (RNA Pol II) transcription during HS regardless of whether HSF-1 binding was induced further. Depletion of HSF-1 from the soma abolished recruitment of RNA Pol II, causing a decrease in RNA Pol II across the genes (hsp-70, hsp-16s, and tent-5) or impaired transcription elongation, resulting in a more prominent decrease in RNA Pol II in the body of genes than in the promoters (hsp-110 and dnj-13).
The effects of HS on HSF-1-mediated transcription extended to HSF-1 targets under NHS conditions. As an example, the hsp-90 gene has two separate promoters for basal (NHS) and HS-inducible transcription (Figure 4D, black and red arrows, respectively), as shown by our previous work (Li et al., 2016). HSF-1 binding at the distal, HS-inducible promoter (orange arrow) was exclusive in the soma, and increased RNA Pol II transcription in HS as it did at hsp-70 and hsp-16s but to a lesser extent. However, binding of HSF-1 at the proximal, basal promoter occurred in the soma and the germline and decreased upon HS (Figure 4D). Importantly, when HSF-1 was depleted from the soma, the remaining HSF-1 in germ cells was not sufficient to induce transcription upon HS. Instead, the decrease in HSF-1 binding to the proximal promoter in HS was accompanied by repression of RNA Pol II elongation. The HS-induced loss of HSF-1 at its basal binding sites was not unique for the hsp-90 proximal promoter. Germline-specific HSF-1 occupancy obtained by ChIP-seq in animals with acute depletion of HSF-1 from the soma revealed that, upon HS, HSF-1 binding decreased from most of its targeted promoters of protein-coding genes in germline development (Figure 4E). This result indicates repression of the germline transcriptional program of HSF-1 by HS.
Notably, the decrease in HSF-1 occupancy at its germline developmental target genes by HS was not due to HSF-1 losing DNA binding activity. Our ChIP-seq analyses revealed hundreds of HSF-1 binding peaks enriched in germ cells or shared between the germline and the soma upon HS. Strikingly, these germline HSF-1 binding sites were almost exclusively located in helitron repeats (Figure 4F), the rolling circle DNA transposons in C. elegans (Kapitonov and Jurka, 2007). As an example, binding of HSF-1 at HelitronY1 near the hsp-110 gene was induced by HS and enriched in the germline rather than the soma (Figure 4D). HSF-1 binding sites used uniquely by somatic cells in HS tend not to be associated with helitrons (Figure 4F). Our ChIP-seq data showed that HSF-1 occupancy in germ cells increased at almost all of the 337 binding sites in helitrons upon HS (Figure 4G), suggesting that preferential binding of HSF-1 to helitrons over promoters of protein coding genes underlies the unique response of HSF-1 to heat stress in the C. elegans germline.
Our results indicate that HSF-1 activity in germline development is not triggered by proteotoxic stress as in the canonical HSR but repressed by heat stress.
HSF-1 is required for IIS-promoted reproduction
To gain more insight into the germline-specific roles of HSF-1 and its regulation, we wanted to find out how HSF-1 may interact with cellular pathways that have established roles in germline development. We performed Ingenuity Pathway Analysis (IPA) on the transcriptomes after 16 h of HSF-1 depletion in the germline. This time point provides abundant expression changes for IPA but precedes the global transcriptome alterations at 24 h. IPA revealed gene expression signatures that are consistent with inhibition of the linked IIS and mTORC2 signaling pathways (Figures 5A and 5B). HSF-1 depletion led to expression changes in genes downstream of the IIS and mTORC2 pathways, as seen upon inhibition of AKT (AKT-1 and AKT-2 in C. elegans) and RICTOR (RICT-1 in C. elegans) and upon activation of NFE2L1 and NFE2L2 (orthologs of the C. elegans transcription factor SKN-1). Overall, these transcriptome features imply that HSF-1 may support the IIS and mTORC2 pathways in germline development.
Figure 5. HSF-1 is required for IIS-promoted reproduction.
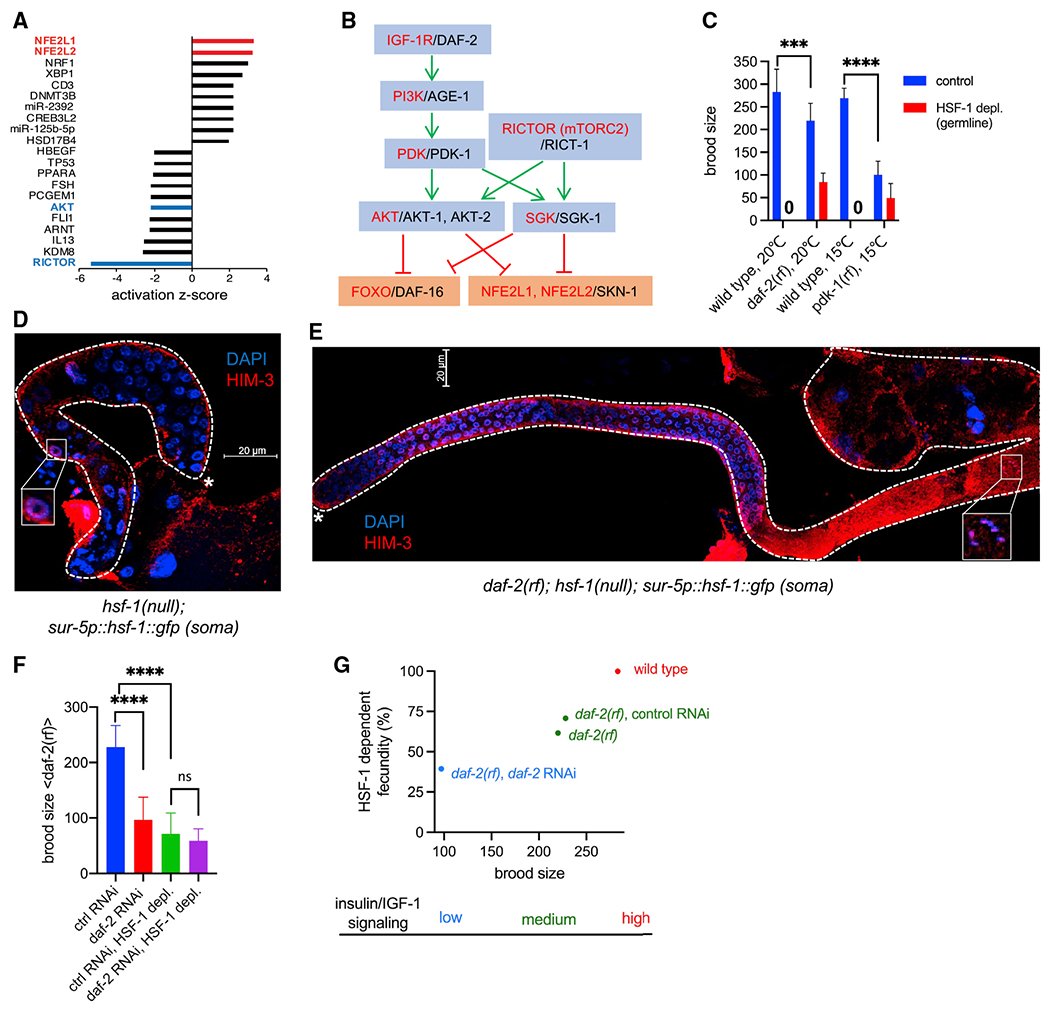
(A) Bar graph of activation Z scores from IPA of transcriptome changes at 16 h of HSF-1 depl from the germline. The activation Z score is a statistical measurement of the activation status of upstream regulators of DE genes (positive Z score, activation; negative Z score, inhibition).
(B) Schematic of the linked IIS and mTORC2 signaling transduction pathway (Blackwell et al., 2019; Hertweck et al., 2004; Murphy and Hu, 2013; Sarbassov et al., 2005). Human protein names are shown in red, and C. elegans ortholog names are shown in black. Green pointed arrows denote activation, and red blunt-ended arrows denote repression.
(C) Brood size of the wild-type, daf-2(rf), and pdk-1(rf) animals upon depl of HSF-1 from the germline since egg lay. The pdk-1(rf) mutants were grown at 15°C to avoid developmental defects at higher temperature. Data are represented as mean ± standard deviation (n ≥ 15). Statistical significance was calculated by unpaired, two-tailed Student’s t test. ***p < 0.001, ****p < 0.0001.
(D and E) Representative images showing IF of the meiotic marker HIM-3 in dissected gonads with DAPI staining of DNA. hsf-1(null) animals that express the hsf-1::gfp transgene from the pan-soma sur-5 promoter (D) showed meiotic arrest at pachytene (boxed image), whereas animals carrying an additional daf-2(rf) allele (E) successfully entered oogenesis (boxed image showing a nucleus at diakinesis). Dashed lines outline gonads, and the white asterisks indicate the distal end of gonads. Scale bars, 20 μM.
(F) Brood size of daf-2(rf) animals treated with daf-2 RNAi or the empty vector of L4440 as ctrl (ctrl RNAi) upon depl of HSF-1 from the germline. Depl of HSF-1 and RNAi started since egg lay. Data are represented as mean ± standard deviation (n ≥ 20). Statistical significance was calculated by unpaired, two-tailed Student’s t test. ****p < 0.0001; ns, p > = 0.05.
(G) Correlation of total brood size and HSF-1-dependent fecundity in animals with different IIS activities. HSF-1-dependent fecundity is calculated as the percent of brood size decrease upon depl of HSF-1 from the germline (as in C and F).
In C. elegans, activities of the single insulin/IGF-1 receptor DAF-2 are important for germline progenitor cell proliferation (Michaelson et al., 2010) and meiotic I progression (Lopez et al., 2013), the two processes of germline development to which HSF-1 also contributes (Figure 2). RICT-1 in mTORC2 is required for C. elegans fecundity predominantly through its downstream kinase SGK-1 (Jones et al., 2009; Soukas et al., 2009), but their roles in germline development are not well characterized. To understand how HSF-1 may interact with the IIS and mTORC2 pathways in germline development, we tested the role of HSF-1 in reproduction upon changes in IIS and mTORC2 signaling. Interestingly, the reduction-of-function (rf) mutants daf-2 and pdk-1 suppressed the sterility caused by chronic depletion of HSF-1 from the germline (Figure 5C) despite their brood sizes being significantly smaller than those of wild-type animals when HSF-1 was present. These results indicate that IIS dictates the requirement for HSF-1 in reproduction via phosphatidylinositol 3-kinase (PI3K)-PDK1. Knockout of SGK-1, the major target of mTORC2 for fecundity, partially suppressed sterility by germline HSF-1 depletion (in 9 of 63 animals). Given the common processes IIS and HSF-1 mediate in germline development and their strong genetic interaction, we decided to focus on understanding this new functional interaction between IIS and HSF-1.
We confirmed that suppression of HSF-1 depletion-induced sterility by daf-2(rf) was not an artifact from incomplete depletion by AID. First, the GFP signal from HSF-1::degron::GFP disappeared from the germline of the daf-2(rf) mutant within 2 h of auxin treatment (Figure S5A). Second, loss of the GFP signal was not due to cleavage of degron::GFP from the C terminus of HSF-1. By inserting a 3xFLAG tag into the N terminus of HSF-1::degron::GFP (Figure S5B) and measuring HSF-1 levels via immunofluorescence (IF) against FLAG, we showed that HSF-1 was depleted to the background level throughout different regions of the germline in wild-type and daf-2(rf) animals (Figures S5C–S5E). Finally, we confirmed the observation in the daf-2(rf) mutant using an AID-independent model. Introducing the daf-2(rf) allele into hsf-1(null) animals with transgenic expression of HSF-1 only in the soma (Figures S1D and S1E) enabled those sterile animals to produce viable progenies (Figure S5F). Our data clearly indicate that lowering IIS can partially bypass the requirement for HSF-1 in germline development. Consistent with this notion, at the cellular level, reducing IIS alleviated the germline progenitor proliferation defects and meiotic arrest caused by the lack of germline HSF-1 (Figures 5D and 5E; compare Figure S5G with Figure 2B).
The daf-2(rf) allele we used (e1370) altered one residue in the tyrosine kinase domain, leading to partial loss of activity (Patel et al., 2008) and slightly reduced brood size (Figure 5C). Further reduction of IIS in daf-2(rf) animals using RNAi against DAF-2 resulted in another ~60% decrease of brood size (Figure 5F, blue bar versus red bar). Importantly, the fecundity-promoting effect of IIS was HSF-1 dependent because daf-2 RNAi did not make a significant difference when HSF-1 was depleted from the germline (Figure 5F, green bar versus purple bar). Our data revealed a correlation between animal brood size and HSF-1’s contribution to fecundity, which are dictated by IIS activity (Figure 5G), and suggest that HSF-1 is required specifically by IIS for promoting reproduction.
IIS activates HSF-1-dependent expression of hsp-90 and hsc-70 in germline development
Given the important roles of HSF-1 target genes in germline homeostasis and development, how could animals with reduced IIS reproduce successfully without HSF-1 in the germline? Because IIS represses multiple stress-responsive transcription factors through the AKT and SGK-1 kinases, we tested whether the daf-2(rf) mutant suppresses the germline defects associated with HSF-1 depletion by enhancing stress responses in germ cells. Single knockout mutants of the AKT and SGK-1 kinases did not rescue (AKT-1 and AKT-2) or very modestly rescued (SGK-1) reproduction upon HSF-1 depletion in the germline (Figure S6A). This is not surprising, given that these kinases have been reported to function in parallel downstream of IIS in various contexts (Hertweck et al., 2004). We therefore tested SKN-1 and DAF-16 (the C. elegans ortholog of mammalian FOXO), two major regulators of cellular responses to proteotoxic and oxidative stress (An and Blackwell, 2003; Li et al., 2011; Murphy and Hu, 2013; Vilchez et al., 2012) that are repressed by AKT and SGK-1 (Brunet et al., 1999; Chen et al., 2013; Hertweck et al., 2004; Lin et al., 2001; Mizunuma et al., 2014; Ruf et al., 2013;Tullet et al., 2008; Figure 5B). Knockdown of DAF-16 in the daf-2(rf) mutant by RNAi resulted in sterility when HSF-1 was depleted from the germline but did not decrease the brood size when HSF-1 was present (Figures 6A and S6B). Similar results were observed in the daf-2(rf); daf-16(null) double mutant (Figure S6C). Knockdown of SKN-1 did not alter the egg numbers regardless of HSF-1 status but caused embryonic lethality (Figures 6A and S6B), consistent with its role in embryogenesis (Bowerman et al., 1992). These results indicate that DAF-16, but not SKN-1, is required for fertility of the daf-2(rf) mutant in the absence of germline HSF-1. Surprisingly, this activity of DAF-16 is required from the somatic tissues because knockdown of DAF-16 in the soma but not in the germline led to sterility of the daf-2(rf) mutant upon HSF-1 depletion (Figure 6B). These results contrast the results from tissue-specific RNAi against EGG-5, which was required from the germline (Figure S6D). Our results argue against the hypothesis that reduced IIS suppresses infertility associated with HSF-1 depletion by activating the DAF-16- and SKN-1-mediated stress response in germ cells. Instead, they suggest that DAF-16 functions downstream of IIS in somatic tissues non-cell-autonomously dictating the requirement for HSF-1 in germline development.
Figure 6. IIS activates HSF-1-dependent expression of hsp-90 and hsc-70 in germline development.
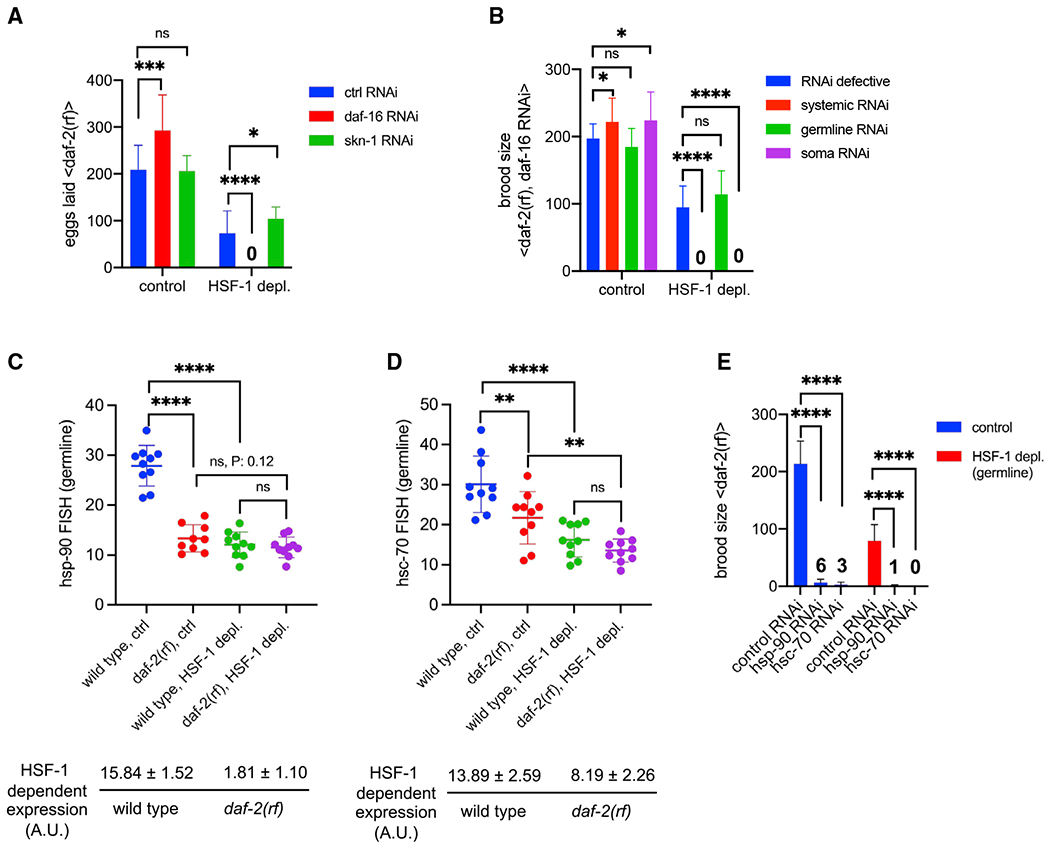
(A) Histograms showing the number of eggs laid by daf-2(rf) animals with systemic RNAi treatment against DAF-16 or SKN-1. The empty vector of L4440 was used as the Ctrl (ctrl RNAi). Depl of HSF-1 from the germline and RNAi started since egg lay. Data are represented as mean ± standard deviation (n ≥ 15). Statistical significance was calculated by unpaired, two-tailed Student’s t test. *p < 0.05, ***p < 0.001, ****p < 0.0001; ns, p > = 0.05.
(B) Histograms showing brood size of the daf-2(rf) animals with RNAi treatment against DAF-16. RNAi was done in genetic models that are RNAi defective (negative Ctrl) or enable RNAi to occur systemically only in the germline or in the soma. Depl of HSF-1 from the germline and RNAi started since egg lay. Data are represented as mean ± standard deviation (n > 15). Statistical significance was calculated by unpaired, two-tailed Student’s t test. *p < 0.05, ***p < 0.001, ****p < 0.0001; ns, p > = 0.05.
(C and D) Scatter dot plot showing the fluorescence intensity of hsp-90 (C) and hsc-70 (D) RNA-FISH in the germline of wild-type and daf-2(rf) animals that were mock treated as the Ctrl or treated with auxin to deplete HSF-1 from the germline since egg lay (HSF-1 depl). Mean and standard deviation are plotted. Statistical significance was calculated by unpaired, two-tailed Student’s t test. **p < 0.01, ****p < 0.0001; ns, p > = 0.05. HSF-1-dependent expression in the germline was calculated as fluorescence intensity in the Ctrl minus HSF-1-depleted animals, a.u., arbitrary units of fluorescence.
(E) Brood size analyses with germline-specific RNAi in the daf-2(rf) mutant. Depl of HSF-1 from the germline and RNAi started since egg lay. Data are represented as mean ± standard deviation (n > 15). Statistical significance was calculated by unpaired, two-tailed Student’s t test. ****p < 0.0001. The average brood sizes of animals with hsp-90 and hsc-70 RNAi are labeled.
Consistent with a previous report (Michaelson et al., 2010), we found that reduced IIS significantly impaired larval germline proliferation, resulting in decreased numbers of mitotic nuclei in day 1 adults (Figure S6E). We wondered whether the slower germline flux would lower the demands for the germline transcriptional program of HSF-1 (Figure 3D), making HSF-1 partially dispensable for reproduction. To test this idea, we took hsp-90 and hsc-70 as examples, which are essential for C. elegans germline development (Green et al., 2011) and support cell proliferation by promoting protein synthesis and the cell cycle (Mayer and Bukau, 2005; Schopf et al., 2017; Truman et al., 2012). Compared with the wild type, daf-2(rf) animals had lower levels of hsp-90 and hsc-70 transcripts in the germline when HSF-1 was present (Figures 6C and 6D, blue versus red dots, S6F, and S6G), supporting our hypothesis that, upon IIS reduction, less hsp-90 and hsc-70 expression is needed to support fecundity. Upregulation of hsp-90 and hsc-70 by IIS is HSF-1 dependent because IIS activity did not significantly affect their expression when HSF-1 was depleted from germ cells (Figures 6C and 6D, green versus purple dots). Importantly, germline-specific RNAi against HSP-90 and HSC-70 reduced the brood size of daf-2(rf) animals regardless of the status of HSF-1 (Figure 6E), suggesting that hsp-90 and hsc-70 were expressed in germ cells in the absence of HSF-1. This low-level, HSF-1-independent expression (Figures 6C and 6D, purple dots) was sufficient and necessary for the daf-2(rf) animals to reproduce without HSF-1 in the germline (Figure 6E). However, in wild-type animals, HSF-1 is indispensable for fertility (Figures 2 and 5C), likely because HSF-1 is required for upregulating hsp-90 and hsc-70 expression (Figures 6C and 6D) to support more rapid germline growth. Consistently, although germline proliferation in wild-type animals is faster, it is more sensitive to HSF-1 depletion compared with daf-2(rf) animals (Figure S6H). Because HSF-1 directly activates transcription of hsp-90 and hsc-70, our data suggest that IIS enhances HSF-1 activity in the germline.
IIS and FOXO/DAF-16 non-cell-autonomously regulate HSF-1 activity in the germline
IIS determines HSF-1’s contributions to fecundity (Figure 5G) and germline expression of hsp-90 and hsc-70 (Figures 6C and 6D). Because DAF-16 is repressed by IIS and its activity in the soma dictates the requirement for HSF-1 in reproduction (Figure 6B), we wanted to find out whether somatic DAF-16 is also involved in regulating HSF-1-dependent gene expression in germ cells. Although germline hsp-90 transcripts decreased in the daf-2(rf) mutant, they were restored to wild-type levels in the daf-2(rf); daf-16(null) double mutant (Figure S7A), suggesting that IIS activates hsp-90 expression in germ cells by repressing DAF-16. Furthermore, knocking down DAF-16 specifically in somatic tissues significantly increased hsp-90 and hsc-70 expression from the germline of the daf-2(rf) mutant in an HSF-1-dependent manner (Figures 7A and 7B). These results suggest that repression of DAF-16 in the soma by IIS non-cell-autonomously activates HSF-1 in the germline. In addition, we found that knockdown of DAF-16 in the soma is sufficient to increase germline proliferation in the daf-2(rf) mutant (Figure S7B). This result is consistent with the idea that HSF-1 is activated to support IIS-promoted germline growth and suggests that DAF-16 non-cell-autonomously coordinates HSF-1 activation and germline growth in response to IIS.
Figure 7. IIS and FOXO/DAF-16 non-cell-autonomously regulate HSF-1 activities in the germline.
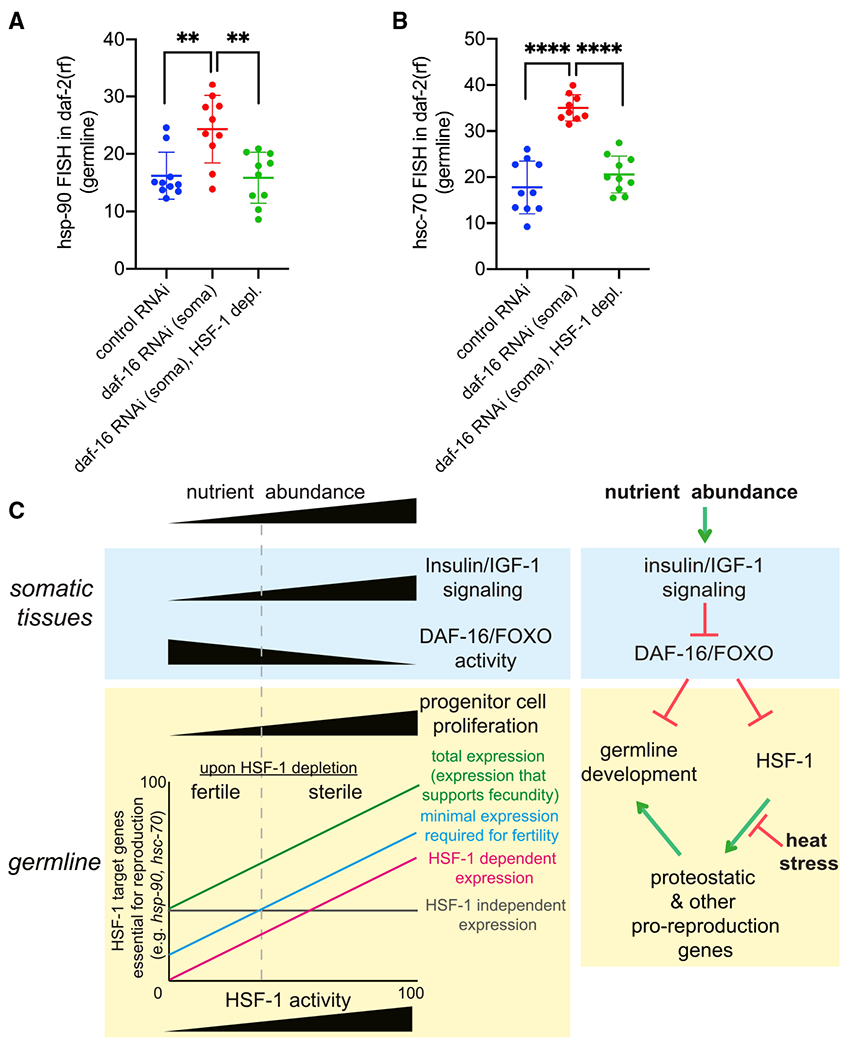
(A and B) Scatter dot plot showing the fluorescence intensity of hsp-90 (A) and hsc-70 (B) RNA-FISH in the germline of the daf-2(rf) animals that are only sensitive to RNAi in somatic tissues. Depl of HSF-1 from the germline by AID and RNAi (daf-16 or empty vector of L4440 as the ctrl) started since egg lay. Mean and standard deviation are plotted. Statistical significance was calculated by unpaired, two-tailed Student’s t test. **p < 0.01, ****p<0.0001.
(C) A proposed model for HSF-1 to support germline homeostasis and development in response to nutrient-sensing insulin/IGF-1 signaling (IIS) and heat stress. Left: correlation of IIS and DAF-16 activities in the soma with progenitor cell proliferation and HSF-1 activities in the germline. The demands for expression of HSF-1 target genes (e.g., hsp-90 and hsc-70) in germ cells of animals with different IIS activities dictate HSF-1’s requirement for fertility and fecundity. Right: model for regulation of germline HSF-1 by somatic IIS and heat stress. Green pointed arrows denote activation, and red blunt-ended arrows denote repression.
DISCUSSION
Germline development is energetically costly, making this process sensitive to environmental perturbation and nutrient availability. HSF1 has evolutionarily conserved roles in proteotoxic stress response and gametogenesis and has emerged as a regulator of energy metabolism (Gomez-Pastor et al., 2018; Li et al., 2017). Here we applied the AID system in C. elegans to understand the precise roles of HSF-1 in germline development and how HSF1 responds to stress and nutrient cues in germ cells. Our results revealed an important role of HSF-1 in germline progenitor cell proliferation in addition to its previously reported functions in meiosis (Le Masson et al., 2011; Metchat et al., 2009). Furthermore, our data demonstrated that, during germline development, HSF-1 is not activated in the same way as during the HSR but is rather repressed by heat stress. Our findings support a model (Figure 7C) where the activities of HSF-1 in germ cells are coupled to the nutrient sensing IIS at the organismal level: HSF-1 supports IIS-promoted germline development, and IIS represses FOXO/DAF-16 in the soma to non-cell-autonomously activate HSF-1 in the germline.
Using the AID system, our study overcame two big challenges of studying HSF-1 in germline development: (1) to robustly deplete HSF-1 protein specifically in germ cells and (2) to probe the transcriptional activities of HSF-1 genome-wide in the germline without tissue isolation. We successfully paired tissue-specific depletion of HSF-1 with whole-animal ChIP-seq and RNA-seq analyses to infer tissue-specific HSF-1 binding profiles and HSF-1-dependent gene expression. Importantly, auxin treatment in animals expressing TIR1 without degron insertion had minimal effects on the transcriptome (Figure S3), indicating target specificity of AID. We established AID as a useful system for studying transcription regulation with spatiotemporal specificity.
Despite the sensitivity of germ cells to heat stress (Abane and Mezger, 2010), how the HSR is regulated and how heat stress affects the physiological functions of HSF1 in germ cells were largely unknown. Our genomic data and RNA-FISH results for hsp-70 show that the canonical HSR is not induced robustly in the germline except for a subset of cells at meiotic prophase. This is consistent with a recent study in C. elegans (Das et al., 2020) and several studies in vertebrates reporting that HS induction of candidate chaperone genes in the germline is developmental stage specific (Curci et al., 1991; Hayashida et al., 2006; Le Goff and Michel, 1999). This germline HSR, although occurring only in a subset of cells, is proposed to have a protective role in C. elegans because hsf-1 RNAi in the germline increased embryonic lethality upon HS (Das et al., 2020). In mouse spermatogenesis, stress-induced HSF1 activity can protect germ cells or promote apoptosis depending on the cell type (Hayashida et al., 2006). It has yet to be determined in C. elegans which portion of germ cells develops into gametes and what portion may undergo apoptosis after HS and the potential roles of HSF-1 in decision making.
We found that preferential binding of HSF-1 to helitron repeats over promoters of protein-coding genes underlies the germline-specific HSR in C. elegans (Figures 4E–4G). Helitron repeats contain clusters of HS elements (HSEs) (Garrigues et al., 2019) that are recognized by the HSF-1 trimer. Given the high number of HSEs at each helitron repeat and the cooperative binding between multiple HSF-1 trimers in heat stress (Xiao et al., 1991), helitrons are expected to outcompete the classical HSR promoters for HSF-1 binding if both are accessible. We found that, in HS, HSF-1 was able to bind at the classical HSR promoters in the soma while almost exclusively bound to helitrons in germ cells (Figure 4F). It is possible that, in proliferating germline progenitor cells, which are enriched in S phase in fed animals (Seidel and Kimble, 2015), DNA replication-coupled nucleosome disassembly may expose more helitrons to compete for HSF-1 binding than in post-mitotic somatic cells. Because of the preferential binding of HSF-1 to helitrons, HS did not induce the canonical HSR robustly in most germ cells but led to loss of HSF-1 from its developmental targets (Figure 4E). Our findings provide a potential explanation for hypersensitivity of germ cells to proteotoxic stress.
Our studies revealed that the functions of HSF-1 in germline development are coupled to IIS. In germ cells, HSF-1 is not serving as an on/off switch of its target genes that are essential for reproduction (e.g., hsp-90 and hsc-70; Figures 6C–6E). The low, HSF-1-independent expression is sufficient and necessary to support fertility when IIS activity is reduced (e.g., in the daf-2(rf) mutant; Figures 5C and 6E). Instead, HSF-1 is activated in germ cells by IIS as HSF-1-dependent expression of hsp-90 and hsc-70 is upregulated with increased IIS (Figures 6C and 6D). IIS promotes germline proliferation (Michaelson et al., 2010) and organismal protein synthesis in young adults (Stout et al., 2013). Activation of HSF-1 by IIS provides a plausible mechanism that couples protein folding capacity to rapid flux of germline development through enhanced expression of key proteostatic genes (Figure 3D). Conversely, in animals of high IIS (e.g., the wild type), insufficient expression of these genes upon depletion of germline HSF-1 led to proteotoxic stress, as implicated by induction of proteasomal genes (Figure 3F) and defects of germline development (Figure 2). In addition, HSP-90 and HSC-70 have evolutionarily conserved roles in cell proliferation and meiosis through regulating activities of cyclins, AKT, and ERK (Basso et al., 2002; Green et al., 2011; Metchat et al., 2009; Schopf et al., 2017; Truman et al., 2012). AKT and ERK kinases function in the C. elegans germline downstream of IIS to promote proliferation and meiosis (Lopez et al., 2013; Michaelson et al., 2010). Therefore, HSF-1 could also support the signaling pathways downstream of IIS in germline development by upregulating hsp-90 and hsc-70. Consistently, we found that HSF-1 is required by IIS for promoting fecundity (Figures 5F, 5G, and 7C).
Our finding that IIS enhances HSF-1 activities in germline development contrasts with the reported repression of HSF-1 by IIS in the canonical HSR (Chiang et al., 2012). This discovery joins the observation that HS impairs HSF-1 binding at its developmental targets in germ cells (Figure 4E), indicating that regulation of HSF-1 in germline development is distinct from that in the canonical HSR. In C. elegans, IIS is one major nutrient-sensing pathway that regulates diapause entry, energy metabolism, and reproduction based on nutrient availability (Templeman and Murphy, 2018). Our discoveries suggest that, through the IIS pathway, HSF-1 could integrate stress and nutritional cues to regulate homeostasis and development of the germline (Figure 7C).
HSF-1 and FOXO/DAF-16 are known to function jointly to enhance animal stress responses and extend the lifespan (Hsu et al., 2003). Our studies revealed a different functional interaction of the two in which repression of DAF-16 in the soma by IIS is important for activation of germline HSF-1 (Figures 7A and 7B). In addition to its cell-autonomous functions in germ cells (Michaelson et al., 2010; Pinkston-Gosse and Kenyon, 2007), DAF-16 acts non-autonomously in the soma, influencing the germline, including progenitor cell proliferation (Michaelson et al., 2010; Qi et al., 2012, 2017) and reproductive longevity (Luo et al., 2010; Qin and Hubbard, 2015; Wang et al., 2014). Our findings add another non-cell-autonomous regulation by DAF-16 in reproduction. Future studies will determine where and how DAF-16 in the soma sends signals to germ cells and identify the germline autonomous pathway that regulates HSF-1 activities. In certain somatic tissues (e.g., the intestine), DAF-16 regulates insulin expression and thus affects IIS in other tissues (Murphy et al., 2007). It is possible that IIS and its downstream PI3K-AKT/SGK-1 and/or RAS-MEK-ERK pathways in the germline (Lopez et al., 2013) are involved in regulating HSF-1. Neither HSF-1 protein levels nor its nuclear localization in the germline were changed significantly in daf-2(rf) animals (Figures S5C–S5E), implying that somatic IIS and DAF-16 may affect germline HSF-1 via its DNA binding and/or transcriptional activities. AKT and MEK have been reported to promote HSF1 transcriptional activities through post-translational modifications to support rapid proliferation of cancer cells (Frezzato et al., 2019; Tang et al., 2015). It will be interesting in future work to determine whether similar mechanisms are in place to coordinate HSF-1 activation and germline development.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jian Li (jian-li@omrf.org).
Materials availability
Worm strains generated in this study as listed in the Key resources table will been deposited to the Caenorhabditis elegans genetics center at University of Minnesota upon publication.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-HIM-3 | Novus | Novus Cat# 53470002; RRID:AB_11013811 |
| Rabbit polyclonal anti-REC-8 | Novus | Novus Cat# 49230002; RRID:AB_10717927 |
| Mouse monoclonal anti-FLAG (M2) | Sigma | Sigma Cat# F3165; RRID:AB_259529 |
| Mouse monoclonal anti-RNA Polymerase II (8WG16) | BioLegend | BioLegend Cat# 664906; RRID:AB_2565554 |
| Rabbit polyclonal anti-GFP | Clontech | Takara Bio Cat# 632592; RRID:AB_2336883 |
| Goat anti-Mouse IgG (H+L) Alexa Fluor Plus 647 | Thermofisher Scientific | Invitrogen Cat# A32728; RRID:AB_2633277 |
| Goat anti-Rabbit IgG (H+L) Alexa Fluor Plus 488 | Thermofisher Scientific | Invitrogen Cat# A32731; RRID:AB_2633280 |
| Bacterial and virus strains | ||
| E. coli OP50 | CGC | OP50 |
| E. coli HT115 | CGC | HT115 |
| E. coli MG1693 | CGSC | MG1693 |
| Chemicals, peptides, and recombinant proteins | ||
| Indole-3-acetic acid (auxin) | Sigma | Cat# I2886-5G; Cas# 87-51-4 |
| IPTG | Thermofisher Scientific | Cat# R0393; Cas# 367-93-1 |
| Trizol reagent | Thermofisher Scientific | Cat #15596026 |
| Protein-G Dynabeads | Thermofisher Scientific | Cat # 10004D |
| EdU (5-ethynyl-2′-deoxyuridine) | Thermofisher Scientific | Cat # A10044 |
| Paraformaldehyde | Fisher Scientific | Cat# AA433689M; Cas# 30525-89-4 |
| iTaq Universal SYBR Green Supermix | BioRad | Cat# 1725124 |
| NEBuilderr HiFi DNA Assembly Master Mix | New England BioLabs | Cat# E2621 |
| Alt-R HiFi S.p. Cas9 NLS | IDT | Cat# 1078727 |
| Critical commercial assays | ||
| iScript cDNA synthesis kit | BioRad | Cat# 1708891 |
| Click-iT EdU Cell Proliferation Kit for Imaging, Alexa Fluor 594 dye | Thermofisher Scientific | Cat# C10339 |
| Zymo Research Direct-zol RNA MiniPrep kit | Zymo Research | Cat # R2051 |
| ZymoPURE Plasmid Miniprep Kit | Zymo Research | Cat # R4208T |
| QIAGEN MinElute PCR Purification Kit | QIAGEN | Cat# 28004 |
| Deposited data | ||
| Tissue-specific HSF-1 occupancy at 20°C or upon heat shock (HS) at 34°C for 30 min (ChIP-seq) | This study | GEO: GSE162063 |
| Transcriptomic analyses of the heat shock response upon HSF-1 depletion from the somatic or germline tissue (RNA-seq) | This study | GEO: GSE162064 |
| Transcriptomic analyses of HSF-1 in germline development (RNA-seq) | This study | GEO: GSE162066 |
| Experimental models: Organisms/strains (C. elegans) | ||
| Wild type, Bristol | CGC | N2 |
| hsf-1(ok600) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III) | CGC | OG576 |
| unc-119(ed3)III;ieSi38[sun-1p::TIR1::mRuby::sun-1 3’UTR+Cbr-unc-119(+)]IV | CGC | CA1199 |
| ieSi57[eft-3p::TIR1::mRuby::unc-54 3’UTR+Cbr-unc-119(+)]II;unc-119(ed3)III | CGC | CA1200 |
| mkcSi13 II; rde-1(mkc36) V | CGC | DCL569 |
| daf-2(e1370)III | CGC | CB1370 |
| pdk-1(sa680)X | CGC | JT9609 |
| daf-16(mgDf50) I; daf-2(e1370) III | CGC | HT1890 |
| akt-1(ok525)V | CGC | RB759 |
| akt-2(ok393)X | CGC | VC204 |
| sqk-1(ok538)X | CGC | VC345 |
| hsf-1(ok600) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III); ljtEx3[sur-5p::HSF-1 (cDNA)::gfp+ttx-3p::rfp] | This study | JTL804 |
| hsf-1(ljt3[hsf-1::degron::gfp])I; unc-119(ed3)III; ieSi38[sun-1p::TIR1 ::mRuby::sun-1 3′UTR+Cbr-unc-119(+)]IV | This study | JTL621 |
| hsf-1(ljt3[hsf-1::degron::gfp])I; ieSi57[eft-3p::TIR1:: mRuby::unc-54 3′ UTR+Cbr-unc-119(+)]II;unc-119(ed3)III | This study | JTL611 |
| daf-2(e1370) III; hsf-1(ljt4[3xFLAG::hsf-1::degron:: gfp])I; ieSi38[sun-1p::TIR1::mRuby::sun-1 3′UTR+Cbr-unc-119(+)]IV | This study | JTL708 |
| hsf-1(ok600) I; daf-2(e1370)III; ljtEx3[sur-5p::HSF-1 (cDNA)::gfp] | This study | JTL806 |
| hsf-1(ljt3[hsf-1::degron::gfp])I; daf-2(e1370)III; ieSi38[sun-1p::TIR1 ::mRuby::sun-1 3′UTR+Cbr-unc-119(+)]IV;rde-1(mkc36)V; ljtEx5[sur-5p::rde-1+ttx-3p::gfp+ myo-2p::rfp] | This study | JTL809 |
| Oligonucleotides | ||
| Please see Table S4 for a list of guide RNAs used in this study. | N/A | N/A |
| Please see Table S5 for a complete list of primers used in this study. | N/A | N/A |
| RNAi clones (multiple) | Ahringer Library; Bioscience | Cat# 3318_Cel_ RNAi_complete |
| Stellaris FISH Probes, Custom Assay with Quasar 670 Dye (multiple, please see Table S6 for sequences) | Biosearch Technologies | Cat# SMF-1065-5 |
| Recombinant DNA | ||
| pLZ29: pCFJ151_Peft-3_degron_EmGFP_unc-54 3′UTR | Zhang et al., 2015 | Addgene Plasmid #71719 |
| pSLGCV: a plasmid that contains sur-5p::luc+::gfp in pPD95.79 | Lagido et al., 2008 | Addgene Plasmid #49862 |
| pNP160: a plasmid that contains ttTi5605_SEC_ mtl-2p_RDE-1_3′rde-1 | a gift from Jonathan Ewbank, Aix-Marseille University | Addgene Plasmid #106362 |
| pPD97_75: empty backbone | a gift from Andrew Fire, Stanford University | Addgene Plasmid #1494 |
| sur-5p::HSF-1 (cDNA)::gfp in pPD95.79 | This study | N/A |
| sur-5p::rde-1 in pPD97_75 | This study | N/A |
| Software and algorithms | ||
| Imaris Image Analysis software | Bitplane | https://imaris.oxinst.com |
| Prism 8 | GraphPad Software | https://www.graphpad.com:443/ |
| Bowtie 2.3.5.1 | Langmead and Salzberg, 2012 | www.cs.jhu.edu/~langmea |
| Bedtools | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| MACS 2.2.7.1 | Zhang et al., 2008 | https://github.com/macs3-project/MACS |
| RNA STAR 2.6 | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Rsubread | Liao et al., 2019 | https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| edgR | Robinson et al., 2010 | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| IGV 2.5.0 | Robinson et al., 2011 | https://software.broadinstitute.org/software/igv/download |
| Ingenuity Pathway Analysis (IPA) | QIAGEN | https://digitalinsights.qiagen.com/products/ingenuity-pathway-analysis |
Data and code availability
The RNA-seq and ChIP-seq datasets from this study have been deposited at Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Unless stated, C. elegans strains were maintained at 20°C on NGM plates seeded with OP50 bacteria and were handled using standard techniques (Brenner, 1974).
The HSF-1 AID models were made by CRISPR knock-in of degron::gfp to the C terminus of endogenous hsf-1 gene through microinjection of chemically modified synthetic sgRNA (Synthego; Table S4) along with Cas9 Nuclease (Integrated DNA Technologies, IDT) following the previously published protocol (Prior et al., 2017). The repair template for C-terminal insertion of degron::gfp was made by NEB assembly of synthetic gene fragments from IDT that contain HSF-1 sequences flanking the insertion site and the PCR fragment of degron::gfp from pLZ29 (Zhang et al., 2015). The repair template for N-terminal insertion of 3XFLAG tag was a single strand oligo (IDT) containing the FLAG tag sequence flanked by 39 bp upstream and 37 bp downstream sequence from the insertion site. The HSF-1 AID models were outcrossed 6 times before use.
Transgenic worms expressing HSF-1 only in the soma were generated by microinjection of sur-5p::HSF-1 (cDNA)::gfp with ttx-3p::rfp co-injection marker into OG576 (hsf-1(ok600) I/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III)). The sur-5p::HSF-1 (cDNA)::gfp construct was made by replacing the luciferase gene in pSLGCV (sur-5p::luc+::gfp) with HSF-1 cDNA through the SmaI and KpnI restriction sites. Transgenic worms that are sensitive to RNAi only in the somatic tissues were made by microinjection of sur-5p::rde-1 with two co-injection markers (ttx-3p::gfp and myo-2p::rfp) into a strain carrying the null allele of rde-1 (mkc36). The sur-5p::rde-1 construct was made by NEB assembly of the pPD97_75 vector digested by EcoRI and BamHI, the sur-5 promoter PCR amplified from the sur-5p::HSF-1 (cDNA)::gfp plasmid, and the rde-1 genomic DNA PCR amplified from pNP160. A list of strains used in this study can be found in the Key resources table.
METHOD DETAILS
Auxin Treatment, RNAi and Heat Shock
Auxin treatment was performed by transferring worms to bacteria-seeded NGM plates containing 1mM auxin (indole-3-acetic acid, Sigma). The preparation of auxin stock solution and auxin containing NGM plates was done as previously described (Zhang et al., 2015). Briefly, auxin was dissolved to 400 mM in ethanol as the stock solution, which can be stored for up to 1 month at 4°C. Auxin was diluted into the NGM agar, cooled to about 50°C, to a final concentration of 1 mM before pouring plates. Plates were seeded with fresh OP50 culture and left at room temperature for 1-2 days to dry. In all experiments, worms were also transferred to NGM plates containing 0.25% of ethanol (EtOH) to serve as the mock treated control.
RNAi was performed by feeding, and all RNAi clones from the Ahringer Library were sequence verified before use. Overnight cultures of RNAi bacteria in LB media containing 100 μg/ml ampicillin were diluted and allowed to grow for another 4-5 hours at 37°C to reach OD600 of 1.0-1.2. Following this, 5 mM IPTG was added and the cultures were incubated for another 3 hours to induce expression of double-stranded RNAs. Cultures were then seeded onto NGM plates containing 100 μg/ml ampicillin and 1 mM IPTG, and allowed to dry at room temperature for 2 days. All RNAi experiments were done by doing egg lay directly on freshly prepared RNAi plates (within one week of seeding). RNAi bacteria in HT115 strain as obtained from the Ahringer Library were used in Figure 3E. Upon HSF-1 depletion, both the reduction-of-function (rf) mutants daf-2 (e1370) and pdk-1(sa680) showed noticeable difference of brood size when fed by OP50 and HT115. To allow direct comparison of brood size in these rf mutants upon HSF-1 depletion and upon RNAi treatment, we therefore conducted all the other RNAi experiments that involved these rf mutants in a RNAi compatible OP50 strain (Xiao et al., 2015). Tissue-specific RNAi were done in transgenic worms that express the Argonaute protein gene rde-1 specifically in either the germline (Zou et al., 2019) or the soma (as described above) in the null mutant rde-1 (mkc36). To control the separation of co-injection markers with rde-1 transgenes in our soma RNAi model, we picked animals that carried both of the co-injection markers in experiments.
Heat shock was done by immersing parafilm wrapped plates in a pre-heated water bath at 34°C. For RNA-seq and ChIP-seq experiments, worms were heat shocked on 10 cm plates for 30 min, and for RNA FISH experiments, worms were heat shocked on 6 cm plates for 15 min.
Measurements of Body Length and Brood Size
The HSF-1 AID animals (JTL611 and JTL621) and the corresponding control animals that only express TIR1 (CA1200 and CA1199) were age-synchronized by egg lay for 1 hour on EtOH or Auxin plates. Larvae were grown to the indicated stages (Figures 1C and S1B) and crawling animals were recorded using a Leica M205 FA microscope. Videos were imported into ImageJ and analyzed for the size of larvae (body length, mm) using the wrMTrck plugin.
For brood size analyses (except Figure S2A), animals were synchronized by egg lay on plates containing auxin and/or RNAi bacteria as specified in figure legends, and singled at L4/young adult stage onto the same type of plates to lay eggs for 24 hours. Worms were then transferred to new plates every day and eggs were allowed to hatch and grow to L3 stage, at which point the number of progeny was counted. In a subset of experiments, significant eggs did not hatch, and we counted and reported the number of dead eggs as specified in figure legends. For Figure S2A, age synchronization of JTL621 was done by egg lay on EtOH plates, and larvae were transferred onto auxin plates at indicated time points. Brood size of each singled animal was counted on auxin plates.
RNA Extraction, cDNA Synthesis and qPCR
The HSF-1 AID animals (JTL611 and JTL621) and the corresponding control animals that only express TIR1 (CA1200 and CA1199) were synchronized by treatment of alkaline hypochlorite solution (bleach). Synchronized L1 larvae were grown on 10 cm normal NGM plates (~500 worms per plate) to develop into young adults (appearance of vulva). Extra few hours were given to CA1199 and JTL621 since they develop slightly slower than CA1200 and JTL611. Approximately 120 young adult worms were picked onto 10 cm NGM plates containing either EtOH or auxin, and kept for indicated time before collection. For each condition, RNA was extracted using 300 μL Trizol reagent. Worms were vortexed continuously for 20 minutes at 4°C and then went through one cycle of freeze-thaw to help release RNA. Following this, RNA was purified using Direct-zol RNA MiniPrep kit (Zymo Research) as per manufacturer’s instructions using on column DNase I digestion to remove genomic DNA. RNA was used in library preparation for sequencing or to synthesize cDNA for qPCR analysis.
cDNA was synthesized using BioRad iScript cDNA synthesis kit as per manufacturer’s instructions. Relative mRNA levels were then determined by real-time quantitative PCR using iTaq Universal SYBR Green Supermix (BioRad) and a Roche Lightcycler 96 thermocycler. Relative mRNA levels were calculated by standard curve method and gene expression was normalized to the mean of the housekeeping genes cdc-42 and rpb-2. All primers used can be found in Table S5.
Chromatin Immunoprecipitation (ChIP)
For each condition, approximately 20,000 – 25,000 bleach synchronized L1 larvae (JTL611 or JTL621) were grown on 10 cm normal NGM plates to develop into young adults. The animals were transferred onto 10 cm plates (~500 worms per plate) containing either EtOH or auxin. After two hours, we either directly collected the worms for crosslinking or subjected them for heat shock at 34°C for 30 min. ChIP experiments were performed as previously described (Li et al., 2016). Briefly, animals were collected from NGM plates, washed with M9 and crosslinked with 2% formaldehyde in PBS at room temperature for 15 min. Crosslinking was quenched by incubation with 0.1M Tris (pH 7.5) for 5 min. Worms were then washed three times in M9 and once in cold FA buffer (50 mM HEPES/KOH pH7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxyholate, 150 mM NaCl with Roche Complete protease inhibitors). Worms were then resuspend in 600 μL of FA buffer and lysed by douncing in a Kontes 2 mL glass dounce, and sonicated in Bioruptor to yield 200 bp–600 bp size DNA fragments.
Immunoprecipitation (IP) was set up using 0.5 μg of chromatin, and either 5 μL of anti-full length GFP antibody (Clontech, living colors), or 3 μg of anti-RNA Pol II antibody (8WG16, BioLegend) in FA buffer (1 mL total volume), and incubated at 4°C overnight on a rotating wheel. The next day, 30 μL of protein-G Dynabeads were added to each IP. Following 2 hours of incubation, protein-G Dynabeads were thoroughly washed and DNA was eluted and purified as previously described (Boehm et al., 2003). To have enough DNA for ChIP-seq, we set up 5 IPs of GFP and 3 IPs of Pol II for each condition, and pool the DNA to do library preparation for sequencing.
ChIP-seq Analysis
ChIP-seq libraries were prepared using Accel-NGS 2S kit as per manufacturer’s instructions. Indexed libraries were treated by Illumina Free Adaptor Blocking Reagent to minimize index hopping and the pooled libraries were sequenced at a NovaSeq 6000 sequencer with 150bp paired-end sequencing at OMRF clinical genomics core.
Sequencing reads were mapped to a non-repeat-masked version of the C. elegans WS235 genome using bowtie2 (Langmead and Salzberg, 2012) with the command bowtie2–no-unal–very-sensitive –dovetail. Mapped reads in coordinated pairs, and with fragment size no bigger than 600bp were kept for downstream analyses. Duplicate reads were filtered for each replicate using MACS2 (Zhang et al., 2008) command macs2 filterdup -g ce–keep-dup auto. HSF-1 peaks were called for each ChIP replicate of the control (no HSF-1 depletion) paired with the corresponding input sample using the command macs2 callpeak -g ce -f BEDPE–call-summits -q 1e-6. Common HSF-1 peaks in the biological duplicates were kept for subsequent analyses. To generate a list of HSF-1 peak summit positions in non-heat-shock (NHS) and heat-shock (HS) conditions, common HSF-1 peaks called in both the JTL611 and JTL621 strains were kept if the peak summits are within 100bp in the two strains, and the midpoints were used as the consensus peak summits. To assign HSF-1 ChIP-seq peaks to promoters, transcription start sites (TSSs) determined by GRO-cap (Kruesi et al., 2013) were used where available. To compare genomic occupancy, filtered reads from biological replicates were combined to generate the bedgragh files using the command macs2 pileup -B. Given average fragment size of ChIP DNA is almost identical in all conditions for either HSF-1 or Pol II, pair-end reads were shifted half of the fragment length toward the center to generate the bedgraph files. To determine the genomic regions with significant HSF-1 occupancy change by HSF-1 depletion, the bedgraph files in the control condition (EtOH) and the depleted condition (auxin) were compared using the command macs2 bdgdiff -l 180 -C 10. To visualize and compare the ChIP-seq data in genome browser views, the bedgraph files were normalized to reads per million using MACS2 callpeak -B –SPMR, and visualized using Integrative Genomics Viewer (IGV) (Robinson et al., 2011) with WS235 genome. Quantification of genomic occupancy were done by mapping the center of ChIP fragments to a reference point (e.g., HSF-1 peak summits) using windowBed in bedtools (Quinlan and Hall, 2010) and Matrix in R. For quantitative comparison of HSF-1 occupancy between conditions, all HSF-1 ChIP-seq data were normalized to 6 millions reads, corresponding to the lowest coverage after duplicate filtering among all conditions. Heatmaps were generated with the Java TreeView package (Saldanha, 2004).
RNA-seq Analysis
Total RNAs were polyA enriched, and directional RNA-seq libraries were prepared using NEBNext Ultra II RNA library prep Kit. Paired-end sequencing was done at a NovaSeq 6000 sequencer at OMRF clinical genomics core. The majority of samples were sequenced by 50 bp. A subset of samples that were sequenced with longer reads were trimmed to 50 bp to make all downstream mapping and analyses consistent.
RNA-seq reads were mapped to Ensembl WBcel235 genome using RNA STAR (Dobin et al., 2013) with–alignIntronMax 120000 to set the intron size, and–outFilterMultimapNmax 200 to allow multi-mapped reads. The mapped reads were then subject to FeatureCounts in Rsubread (Liao et al., 2019) for quantification with the setting -p -B -P -C -M -O–fraction –largestOverlap. The settings in STAR and FeatureCounts enabled proper quantification of those heat shock genes (e.g., hsp-70 and hsp-16s) that are duplicated in C. elegans genome. Differential expression (DE) analyses were then done using edgeR (Robinson et al., 2010) with default settings except of using Likelyhood Ratio Test and filtering out those lowly expressed genes with CPM (counts per million) value less than 1 in more than 75% samples. For the analyses in Figure 3C, to control for effects by auxin and by insertion of AID:GFP to HSF-1, at each time point, we pooled mock treated TIR1 and mock treated HSF-1::degron;TIR1 with auxin treated TIR1 to calculate the expression levels in control condition, and then compare them to auxin treated HSF-1::degron;TIR1 (see Figure S3C for sample description). This analysis reports only DE genes caused by HSF-1 depletion.
Ingenuity Pathway Analysis (IPA) was done by inputting the DE genes and their fold changes at 16 hour of HSF-1 depletion from the germline to QIAGEN IPA. Worm genes were converted into their human orthologs, and compared to published datasets in the database to predict potential upstream regulators and their activation status that interpret the observed gene expression changes. Gene ontology analysis (GO) was done using the program DAVID (https://david.abcc.ncifcrf.gov/) with functional annotation clustering to collapse redundant GO terms. The enrichment score for each cluster was shown with the corresponding GO_BP (Biological Processes) term representing the cluster.
RNA Fluorescence in situ hybridization (FISH)
FISH probes were designed against the worm hsp-70 (F44E5.4/.5), hsp-90 and hsc-70 (hsp-1) genes by using the Stellaris FISH Probe Designer (Biosearch Technologies Inc) at http://www.biosearchtech/com/stellarisdesigner. To increase the sensitivity and specificity, a set of 47-48 Quasar 670 Dye modified probes (Biosearch Technologies Inc) were used for each gene, and the sequences can be found in Table S6. The FISH experiments were done by following the published procedure (Das et al., 2020) with slight modifications. For FISH against hsp-90 and hsc-70, age-synchronized Day 1 adult worms that were at the young adult (YA) and gravid adult (GA) transition (adult vulva and a few if any embryos in the uterus) grown at 20°C were harvested directly by picking off the plates that contained either ethanol or auxin and were seeded with OP50 bacteria or indicated RNAi bacteria. For FISH against hsp-70 (F44E5.4/.5), worms were either kept at 20°C or exposed acutely to 34°C heat shock for 15 min before collection. For each condition, about 20 animals were picked into 1X RNase-free PBS buffer, fixed in freshly prepared 4% paraformaldehyde, and subsequently permeabilized in 70% ethanol at 4°C for about 24 hours. Samples were washed using Stellaris Wash Buffer A (catalog# SMF-WA1-60, Biosearch Technologies Inc) before 100 μL of the hybridization solution (catalog# SMF-HB1-10, Biosearch Technologies Inc) containing 1 μL of 10 mM probe was added to each sample. After 16 hours of hybridization at 37°C, the samples were washed three times with Wash Buffer A, and then incubated for 30 min in Wash Buffer A with 0.5 μg/ml of DAPI. Worms were then washed with Wash Buffer B (catalog no. SMF-WB1-20, Biosearch Technologies Inc) and mounted on slides in about 16 μl of Vectashield mounting medium (catalog no. H-1000, Vector Laboratories).
Immunofluorescence (IF) and Germline Nuclei Quantification
Except for Figures S5C–E, IF was performed with dissected gonads as previously described (Kocsisova et al., 2018) with slight modifications. Animals were synchronized by egg lay and grown to young adult stage (adult vulva start appearing). Approximately 30 animals were dissected in M9 buffer containing 0.1% Tween-20 (PBST) and 0.25 mM levamisole. The samples were then fixed in PBST buffer containing 3% paraformaldehyde for 10 min at room temperature, and subsequently incubated with cold methanol for 15 min at −20°C. After washes with PBST, the samples were blocked for 30 min at room temperature in PBST with 1% bovine serum albumin before rabbit anti-HIM-3 (Novus, Cat# 53470002) was added at 1:200 dilution. After overnight incubation and four washes with the blocking solution, Goat anti-Rabbit IgG (H+L) Alexa Fluor Plus 488 (Invitrogen Cat# A32731) was added to the samples at 1:1000 dilution and incubated for 2 hours in the dark at room temperature. DNA staining was done in 100 ng/ml of DAPI for 20 min, and the samples were mounted on agarose pad for confocal imaging. Mitotic, transition and meiotic zones were defined based on staining of the meiotic marker HIM-3 and the crescent-shaped DAPI staining in the transition zone. Nuclei counting was done using IMARIS 9.3.1 software following the published procedure (Gopal and Pocock, 2018).
For Figures S5C–E, we prepared freeze-cracked worms, fixed in 4% paraformaldehyde, as described (Charlie et al., 2006) with the following changes. We prepared JTL708 (daf-2(e1370) III; 3xFLAG::hsf-1::degron::GFP I; ieSi38[sun-1p::TIR1::mRuby::sun-1_3’UTR+Cbr-unc-119(+)] IV) worms by bleach synchronization, and L1 larvae were grown on NGM plates containing ethanol at 20°C for approximately 75 hours. Young adult worms were rinsed off in M9 buffer and plated on either ethanol or auxin plates for 2 hours before rinsing for freeze-cracking. A methanol incubation for 15 min at 4°C after fixation was added to eliminate the TIR1:RFP signal in the worms. The samples were co-stained with anti-FLAG M2 (mouse, Sigma Cat# F3165) at 1:250 and anti-REC-8 (rabbit, Novus Cat# 49230002) at 1:100 at 15°C overnight. We then labeled the anti-FLAG with anti-mouse-Alexa 647 (Invitrogen Cat# A32728) and anti-REC-8 with goat anti-rabbit-Alexa 488 (Invitrogen Cat# A32731) by incubating for 2 hours at room temperature.
EdU labeling
EdU labeling was performed by feeding animals with EdU-labeled MG1693 bacteria. EdU-labeled bacteria and M9 agar plates were prepared as described (Kocsisova et al., 2018). For the experiments in larvae (as in Figures S2C and S2D), JTL621 animals were synchronized by egg lay on ethanol or auxin plates, and grown for 38 hours at 20°C to L3 stage. The Larvae were then transferred onto M9 plates with EdU bacteria, and fed with EdU for 3 hours. Fixation and rehydration of whole animals were done as described (Michaelson et al., 2010). Briefly, animals were fixed for 10 minutes in 3.7% paraformaldehyde (Fisher Scientific, Cat#AA433689M) in PBST, washed in PBST, incubated for 5 minutes in −20°C methanol, and washed three times in PBST. For experiments in Figure S6E, age-synchronized Day 1 adult worms that were at the young adult (YA) and gravid adult (GA) transition (adult vulva and a few if any embryos in the uterus) were used. The experiments were done by following the published protocol (Kocsisova et al., 2018). Briefly, the animals were fed with EdU bacteria for 30 min, and dissected gonads were fixed in 3% paraformaldehyde, washed in PBST and incubated overnight in −20°C methanol before washing three times in PBST to rehydrate the germlines. EdU click reaction using Click-iT EdU Cell Proliferation Kit with Alexa Fluor 594 dye (Thermofisher, Cat# C10339) and DAPI staining were performed as described (Kocsisova et al., 2018). Worms were then mounted on 2% agarose pad for confocal imaging. Nuclei with any EdU labeling (individual chromosomes or all chromosomes) were scored as positive.
Fluorescence Imaging and Quantification of Fluorescence Intensity
Imaging of live animals as in Figures 1B, S1A, and S5A was done by immobilizing age-synchronized young adult animals in a drop of M9 buffer containing 6mM levamisole on a 2% agarose pad. Images were acquired immediately using a Zeiss LSM710 Confocal Microscope with a 20X objective. Fluorescent imaging for RNA FISH and IF was performed using a Zeiss LSM710 Confocal Microscope through a 63X oil objective. Zen software was used to obtain z- stacks, stitch image tiles, and perform subsequent analyses. Fluorescence intensity was quantified in individual worms after maximal intensity projection. Regions of interest were outlined within individual worms and the arithmetic mean of fluorescence intensity per area was determined. For quantification of IF and FISH signals in the germline, germ cells in mitotic region, transition zone and meiotic prophase were included.
QUANTIFICATION AND STATISTICAL ANALYSIS
Two-tailed, unpaired Student’s t test were used for candidate gene expression analyses (RT-qPCR and RNA FISH), nuclei quantification and brood size analyses. Two-way ANOVA test was used for developmental size analyses in Figure S1C. P values from these statistical analysis were calculated using GraphPad Prism, and are declared in figure legends. Error bars represent SEM or standard deviation as specified in figure legends. Benjamini and Hochberg FDR was used to calculate the adjusted P value for differential expression analyses by RNA-seq.
Supplementary Material
Highlights.
HSF-1 activates proteostatic and pro-reproduction genes in germline development
Heat stress relocates HSF-1 from its developmental targets to helitrons in germ cells
Insulin/IGF-1 signaling in the soma non-autonomously activates HSF-1 in the germline
HSF-1 is required for insulin/IGF-1-promoted germline proliferation and fecundity
ACKNOWLEDGMENTS
We are grateful to Drs. Linda Thompson, Benjamin Miller, Roberto Pezza, and Tali Gidalevitz as well as Li Lab members for helpful comments and discussions regarding our manuscript. We thank the imaging core facility at OMRF for assistance of confocal imaging. We thank the OMRF clinical genomics core for library preparation and sequencing of RNA-seq and ChIP-seq samples. We also thank Dr. Veena Prahlad for helpful discussions regarding the manuscript and the RNA-FISH experiment. This work is supported by NIH grants 2P20GM103636 and 1R35GM138364 (to J. L.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109623.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abane R, and Mezger V (2010). Roles of heat shock factors in gametogenesis and development. FEBS J. 277, 4150–4172. [DOI] [PubMed] [Google Scholar]
- Akerfelt M, Vihervaara A, Laiho A, Conter A, Christians ES, Sistonen L, and Henriksson E (2010). Heat shock transcription factor 1 localizes to sex chromatin during meiotic repression. J. Biol. Chem. 285, 34469–34476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, and Blackwell TK (2003). SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17, 1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnsburg K, and Kirstein-Miles J (2014). Interrelation between protein synthesis, proteostasis and life span. Curr. Genomics 15, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, and Rosen N (2002). Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J. Biol. Chem. 277, 39858–39866. [DOI] [PubMed] [Google Scholar]
- Bierkamp C, Luxey M, Metchat A, Audouard C, Dumollard R, and Christians E (2010). Lack of maternal Heat Shock Factor 1 results in multiple cellular and developmental defects, including mitochondrial damage and altered redox homeostasis, and leads to reduced survival of mammalian oocytes and embryos. Dev. Biol. 339, 338–353. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Sewell AK, Wu Z, and Han M (2019). TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging. Genetics 213,329–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, and Lis JT (2003). Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 23, 7628–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, and Priess JR (1992). skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68, 1061–1075. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, and Greenberg ME (1999). Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868. [DOI] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, and Henikoff S (1999). A histone-H3-like protein in C. elegans. Nature 401, 547–548. [DOI] [PubMed] [Google Scholar]
- Charlie NK, Schade MA, Thomure AM, and Miller KG (2006). Presynaptic UNC-31 (CAPS) is required to activate the G alpha(s) pathway of the Caenorhabditis elegans synaptic signaling network. Genetics 172, 943–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AT, Guo C, Dumas KJ, Ashrafi K, and Hu PJ (2013). Effects of Caenorhabditis elegans sgk-1 mutations on lifespan, stress resistance, and DAF-16/FoxO regulation. Aging Cell 12, 932–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KC, Klancer R, Singson A, and Seydoux G (2009). Regulation of MBK-2/DYRK by CDK-1 and the pseudophosphatases EGG-4 and EGG-5 during the oocyte-to-embryo transition. Cell 139, 560–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang WC, Ching TT, Lee HC, Mousigian C, and Hsu AL (2012). HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell 148, 322–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, and Benjamin IJ (2000). Maternal effect of Hsf1 on reproductive success. Nature 407, 693–694. [DOI] [PubMed] [Google Scholar]
- Curci A, Bevilacqua A, Fiorenza MT, and Mangia F (1991). Developmental regulation of heat-shock response in mouse oogenesis: identification of differentially responsive oocyte classes during Graafian follicle development. Dev. Biol. 144, 362–368. [DOI] [PubMed] [Google Scholar]
- Das S, Ooi FK, Cruz Corchado J, Fuller LC, Weiner JA, and Prahlad V (2020). Serotonin signaling by maternal neurons upon stress ensures progeny survival. eLife 9, e55246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezzato F, Raggi F, Martini V, Severin F, Trimarco V, Visentin A, Scomazzon E, Accordi B, Bresolin S, Piazza F, et al. (2019). HSP70/HSF1 axis, regulated via a PI3K/AKT pathway, is a druggable target in chronic lymphocytic leukemia. Int. J. Cancer 145, 3089–3100. [DOI] [PubMed] [Google Scholar]
- Garrigues JM, Tsu BV, Daugherty MD, and Pasquinelli AE (2019). Diversification of the Caenorhabditis heat shock response by Helitron transposable elements. eLife 8, e51139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, and Thiele DJ (2018). Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal S, and Pocock R (2018). Computational Analysis of the Caenorhabditis elegans Germline to Study the Distribution of Nuclei, Proteins, and the Cytoskeleton. J. Vis. Exp. (134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Kao HL, Audhya A, Arur S, Mayers JR, Fridolfsson HN, Schulman M, Schloissnig S, Niessen S, Laband K, et al. (2011). A high-resolution C. elegans essential gene network based on phenotypic profiling of a complex tissue. Cell 145, 470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida N, Inouye S, Fujimoto M, Tanaka Y, Izu H, Takaki E, Ichikawa H, Rho J, and Nakai A (2006). A novel HSF1-mediated death pathway that is suppressed by heat shock proteins. EMBO J. 25, 4773–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertweck M, Göbel C, and Baumeister R (2004). C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell 6, 577–588. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, and Kenyon C (2003). Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, and Greenstein D (2005). Introduction to the germ line. WormBook, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard EJ, Korta DZ, and Dalfó D (2013). Physiological control of germline development. Adv. Exp. Med. Biol. 757, 101–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsa E, Cruzat VF, Kagize JN, Yovich JL, and Keane KN (2019). Growth Hormone and Insulin-Like Growth Factor Action in Reproductive Tissues. Front. Endocrinol. (Lausanne) 10, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, and Wu C (1997). Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 16, 2452–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KT, Greer ER, Pearce D, and Ashrafi K (2009). Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 7, e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalis AK, Kroetz MB, Larson KM, and Zarkower D (2010). Functional genomic identification of genes required for male gonadal differentiation in Caenorhabditis elegans. Genetics 185, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, and Jurka J (2007). Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 23, 521–529. [DOI] [PubMed] [Google Scholar]
- Kocsisova Z, Mohammad A, Kornfeld K, and Schedl T (2018). Cell Cycle Analysis in the C. elegans Germline with the Thymidine Analog EdU. J. Vis. Exp. (140), 58339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, and Meyer BJ (2013). Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2, e00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagido C, Pettitt J, Flett A, and Glover LA (2008). Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol. 8, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff P, and Michel D (1999). HSF1 activation occurs at different temperatures in somatic and male germ cells in the poikilotherm rainbow trout. Biochem. Biophys. Res. Commun 259, 15–20. [DOI] [PubMed] [Google Scholar]
- Le Masson F, Razak Z, Kaigo M, Audouard C, Charry C, Cooke H, Westwood JT, and Christians ES (2011). Identification of heat shock factor 1 molecular and cellular targets during embryonic and adult female meiosis. Mol. Cell. Biol 31, 3410–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, and Blackwell TK (2011). Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 7, e1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chauve L, Phelps G, Brielmann RM, and Morimoto RI (2016). E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 30, 2062–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Labbadia J, and Morimoto RI (2017). Rethinking HSF1 in Stress, Development, and Organismal Health. Trends Cell Biol. 27, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, and Kenyon C (2001). Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet 28, 139–145. [DOI] [PubMed] [Google Scholar]
- Lismont C, Nordgren M, Van Veldhoven PP, and Fransen M (2015). Redox interplay between mitochondria and peroxisomes. Front. Cell Dev. Biol 3, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AL 3rd, Chen J, Joo HJ, Drake M, Shidate M, Kseib C, and Arur S (2013). DAF-2 and ERK couple nutrient availability to meiotic progression during Caenorhabditis elegans oogenesis. Dev. Cell 27, 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Kleemann GA, Ashraf JM, Shaw WM, and Murphy CT (2010). TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell 143, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, and Bukau B (2005). Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metchat A, Akerfelt M, Bierkamp C, Delsinne V, Sistonen L, Alexandre H, and Christians ES (2009). Mammalian heat shock factor 1 is essential for oocyte meiosis and directly regulates Hsp90alpha expression. J. Biol. Chem 284, 9521–9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Korta DZ, Capua Y, and Hubbard EJ (2010). Insulin signaling promotes germline proliferation in C. elegans. Development 137, 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma M, Neumann-Haefelin E, Moroz N, Li Y, and Blackwell TK (2014). mTORC2-SGK-1 acts in two environmentally responsive pathways with opposing effects on longevity. Aging Cell 13, 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI (2008). Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton EA, and Lamitina T (2013). Caenorhabditis elegans HSF-1 is an essential nuclear protein that forms stress granule-like structures following heat shock. Aging Cell 12, 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, and Hu PJ (2013). Insulin/insulin-like growth factor signaling in C. elegans. WormBook, 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, Lee SJ, and Kenyon C (2007). Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104, 19046–19050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neirijnck Y, Papaioannou MD, and Nef S (2019). The Insulin/IGF System in Mammalian Sexual Development and Reproduction. Int. J. Mol. Sci 20, 4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel DS, Garza-Garcia A, Nanji M, McElwee JJ, Ackerman D, Driscoll PC, and Gems D (2008). Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics 178, 931–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkston-Gosse J, and Kenyon C (2007). DAF-16/FOXO targets genes that regulate tumor growth in Caenorhabditis elegans. Nat. Genet 39,1403–1409. [DOI] [PubMed] [Google Scholar]
- Prior H, Jawad AK, MacConnachie L, and Beg AA (2017). Highly Efficient, Rapid and Co-CRISPR-Independent Genome Editing in Caenorhabditis elegans. G3 (Bethesda) 7, 3693–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Huang X, Neumann-Haefelin E, Schulze E, and Baumeister R (2012). Cell-nonautonomous signaling of FOXO/DAF-16 to the stem cells of Caenorhabditis elegans. PLoS Genet. 8, e1002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Yan Y, Pfeifer D, Donner V Gromoff E, Wang Y, Maier W, and Baumeister R (2017). C. elegans DAF-16/FOXO interacts with TGF-ß/BMP signaling to induce germline tumor formation via mTORC1 activation. PLoS Genet. 13, e1006801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, and Hubbard EJ (2015). Non-autonomous DAF-16/FOXO activity antagonizes age-related loss of C. elegans germline stem/progenitor cells. Nat. Commun 6, 7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf V, Holzem C, Peyman T, Walz G, Blackwell TK, and Neumann-Haefelin E (2013). TORC2 signaling antagonizes SKN-1 to induce C. elegans mesendodermal embryonic development. Dev. Biol 384, 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ (2004). Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, and Sabatini DM (2005). Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101. [DOI] [PubMed] [Google Scholar]
- Schopf FH, Biebl MM, and Buchner J (2017). The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol 18, 345–360. [DOI] [PubMed] [Google Scholar]
- Seidel HS, and Kimble J (2015). Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. eLife 4, e10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, and Ruvkun G (2009). Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 23, 496–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KK, and Golden A (2018). The C. elegans eggshell. WormBook 2018, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout GJ, Stigter EC, Essers PB, Mulder KW, Kolkman A, Snijders DS, van den Broek NJ, Betist MC, Korswagen HC, Macinnes AW, and Brenkman AB (2013). Insulin/IGF-1-mediated longevity is marked by reduced protein metabolism. Mol. Syst. Biol 9, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Dai S, He Y, Doty RA, Shultz LD, Sampson SB, and Dai C (2015). MEK guards proteome stability and inhibits tumor-suppressive amyloidogenesis via HSF1. Cell 160, 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeman NM, and Murphy CT (2018). Regulation of reproduction and longevity by nutrient-sensing pathways. J. Cell Biol 217, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman AW, Kristjansdottir K, Wolfgeher D, Hasin N, Polier S, Zhang H, Perrett S, Prodromou C, Jones GW, and Kron SJ (2012). CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell 151, 1308–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, and Blackwell TK (2008). Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Duarte FM, and Lis JT (2018). Molecular mechanisms driving transcriptional stress responses. Nat. Rev. Genet 19, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, and Dillin A (2012). RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489, 263–268. [DOI] [PubMed] [Google Scholar]
- Wang MC, Oakley HD, Carr CE, Sowa JN, and Ruvkun G (2014). Gene pathways that delay Caenorhabditis elegans reproductive senescence. PLoS Genet. 10, e1004752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Perisic O, and Lis JT (1991). Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell 64, 585–593. [DOI] [PubMed] [Google Scholar]
- Xiao R, Chun L, Ronan EA, Friedman DI, Liu J, and Xu XZ (2015). RNAi Interrogation of Dietary Modulation of Development, Metabolism, Behavior, and Aging in C. elegans. Cell Rep. 11, 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, and Dernburg AF (2015). The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Wu D, Zang X, Wang Z, Wu Z, and Chen D (2019). Construction of a germline-specific RNAi tool in C. elegans. Sci. Rep 9, 2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and ChIP-seq datasets from this study have been deposited at Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the Key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


