Abstract
Covid-19 is a viral disease caused by the virus SARS-CoV-2 that spread worldwide and caused more than 4.3 million deaths. Moreover, SARS-CoV-2 still continues to evolve, and specifically the E484K, N501Y, and South Africa triple (K417N + E484K + N501Y) spike protein mutants remain as the ‘escape’ phenotypes. The aim of this study was to compare the interaction between the receptor binding domain (RBD) of the E484K, N501Y and South Africa triple spike variants and ACE2 with the interaction between wild-type spike RBD-ACE2 and to show whether the obtained binding affinities and conformations corraborate clinical findings. The structures of the RBDs of the E484K, N501Y and South Africa triple variants were generated with DS Studio v16 and energetically minimized using the CHARMM22 force field. Protein-protein dockings were performed in the HADDOCK server and the obtained wild-type and mutant spike-ACE2 complexes were submitted to 200-ns molecular dynamics simulations with subsequent free energy calculations using GROMACS. Based on docking binding affinities and free energy calculations the E484K, N501Y and triple mutant variants were found to interact stronger with the ACE2 than the wild-type spike. Interestingly, molecular dynamics and MM-PBSA results showed that E484K and spike triple mutant complexes were more stable than the N501Y one. Moreover, the E484K and South Africa triple mutants triggered greater conformational changes in the spike glycoprotein than N501Y. The E484K variant alone, or the combination of K417N + E484K + N501Y mutations induce significant conformational transitions in the spike glycoprotein, while increasing the spike-ACE2 binding affinity.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, escape mutants, protein-protein docking, molecular dynamics, MM-PBSA
1. Introduction
Towards the end of December 2019, cases of pneumonia of unknown etiological origin started to emerge in the Hubei region of Wuhan, China (Riou & Althaus, 2020). These cases that occurred in individuals were soon identified as the 2019 novel Coronavirus (2019-nCoV) by Chinese authorities (WHO, 2020). Although these cases, which occurred in China, were not followed very closely by other states, 2019-nCoV spread to the whole world like a storm in a short time and rose from the epidemic category to the pandemic category. Currently, the causative agent of this pandemic is formally named SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2, COVID-19) by the International Committee on Taxonomy of Viruses (Ceraolo & Giorgi, 2020). According to the latest global statistics, there are over 200 million coronavirus cases and more than 4.3 million deaths in the world (Worldometer, 2020).
Shortly after the epidemic in China became a pandemic, many groups began to characterize the structure of the viral proteins and its interaction with the receptor, especially the spike protein, interacting with the human angiotensin-converting enzyme 2, ACE2 (Lan et al., 2020; Luan et al., 2020; Shang et al., 2020; Walls et al., 2020; Wrapp et al., 2020; Yan et al., 2020). In the same time, many computational studies were carried out aiming to investigate the spike-ACE2 interactions (Ali & Vijayan, 2020; Al-Karmalawy et al., 2021; Amin et al., 2020; de Andrade et al., 2020; Ghorbani et al., 2020; Qiao & Olvera de la Cruz, 2020; Sakkiah et al., 2020; Spinello et al., 2020; Verkhivker, 2020; Verkhivker & Di Paola, 2021; Wang, Liu et al., 2020) and feverish bioinformatics-based drug-repurposing (Barros et al., 2020; de Oliveira et al., 2021; Ekins et al., 2020; Idris et al., 2020; Sharma et al., 2020) and, albeit limited, experimental SARS-CoV-2 inhibition studies were initiated all over the world following the statement of National Institutes of Health (NIH) in April 2020 that the basic genomic layout and the biology of MERS, SARS and SARS-CoV-2 viruses were very similar (Harrison, 2020). Repurposed drugs were divided into two groups as ‘acting on the virus’ and ‘acting on the host’, and it was suggested that 20 drugs in the first group and 10 drugs in the second group could be effective in SARS-CoV-2 infections (Singh et al., 2020). At the same time, as a result of WHO's call on accelerating vaccine development studies in April 2020, today a total of 11 vaccines (Moderna mRNA-1273, Pfizer–BioNTech tozinameran, Sputnik Light, Sputnik V, Oxford–AstraZeneca, Novavax, Johnson&Johnson, Covaxin, Convidecia, BBIBP-CorV, CoronaVac) are currently approved for use in the treatment of SARS-CoV-2 (Anonymous, 2021; ClinicalTrials.gov, 2020a, 2020b, 2020c, 2020d, 2020e).
However, as in all other RNA viruses, mutations have been occurring above the expected frequency in SARS-CoV-2 and the problem of resistance against repurposed drugs or vaccines has already emerged (Team, 2021). Today, there are a total of 32 recorded variants of the SARS-CoV-2 and three of these variants (B.1.351 [South Africa], B.1.351 + P384L [South Africa], B.1.351 + E516Q [South Africa]) still maintain their importance in terms of public health, as they are ‘escape mutants’ (Variants of Concern) (European Centre for Disease Prevention and Control (ECDC),), 2021).
Interestingly, most of the mutations that the virus can tolerate occurs on the spike glycoprotein. Although most amino acid substitutions are deleterious for receptor binding domain (RBD) expression and ACE2 binding, a substantial number of mutations in the RBD has been reported to be functional or even enhance ACE2 binding, including ACE2 interface residues (Starr et al., 2020). A very recent study reported that the amino acid substitutions are predominantly found in the spike gene and the RBD, that make up 13% and 2% of the viral genome, respectively (Choi et al., 2020). A recurrent emergence and significant onward transmission of a mutation in the spike gene which result in loss of H69/V70 has recently been reported to co-occur with the receptor binding motif (RBM) amino acid substitutions N501Y, N439K and Y453F (Kemp et al., 2020). It has been reported that the N501Y mutation affects the receptor binding affinity of the spike protein and increases the infectivity of the virus alone or together with the 69/70 deletion in the N-terminal domain of the protein. It was also reported from the same study that SARS-CoV-2 variants with mutations at position 501 showed increased ability to evade monoclonal antibodies (Chand et al., 2020; Rambaut et al., 2020).
Recently, just after N501Y, one of the mutations with the highest frequency nearly in all SARS-CoV-2 variants is the E484K. It has been reported that the experimental induction of E484K mutation alone can provide resistance to many monoclonal antibodies, and is associated with complete elimination of neutralization capacity in convalescent sera tested in some SARS-CoV-2 lineages (Ferrareze et al., 2021; Jangra et al., 2021; Xie et al., 2021). Furthermore, vaccines manufactured by Novavax and Johnson & Johnson which have been tested in recent clinical trials were reported to be less effective against South African variants (B.1.351) harboring the E484K mutation than UK (B.1.1.7) and US (B.1.427 and B.1.429) lineages (Wise, 2021). Mechanistically, the E484K mutation in the flexible loop region of the SARS-CoV-2 receptor binding domain (RBD) causes the formation of novel favorable contacts in the RBD/ACE2 interface (Nelson et al., 2021) and more specifically, it ends up with a stronger ionic interaction between the residue GLU75 of ACE2 and the LYS484 of RBD (Franceschi, Caldana et al., 2021).
In addition to the N501Y and E484K mutations, the substitution that is at least as effective in the escape of SARS-CoV-2 from antibodies and causes greater conformational changes when found in combination with other mutations is the K417N mutation (Nelson et al., 2021). While the K417N mutation alone cannot give the virus the ability to escape from neutralization significantly, it causes a 5- to 12-fold decrease in serum neutralization when found in combination with E484K and N501Y mutations (Chen et al., 2021; Fratev, 2020). Thus, these three mutations (K417N, E484K, N501Y), alone or in combination, most likely play a role as key mutational factors in the virulence and escape potential of the SARS-CoV-2.
The aim of this study was therefore to compare these mutant (N501Y, E484K, K417N) spike glycoprotein/ACE2 interactions with the wild-type RBD/ACE2 interaction and to investigate whether the obtained binding conformations correlate with the clinical data. For this purpose: i) two single mutants (N501Y and E484K) and a multiple-mutant (K417N + N501Y + E484K) were constructed on the initial crystallographic structure of the spike protein, ii) the obtained mutant structures were submitted to energy minimization, iii) protein-protein dockings were performed on the HADDOCK server, iv) the docked spike/ACE2 conformations with the most negative binding energy (including control) were subjected to 200-nanosecond long molecular dynamics simulations in order to observe the stability of these interactions and, v) the binding free energies of all complexes were calculated using the endpoint MM-PBSA method.
2. Material and methods
2.1. Retrieval and mutation of the SARS-CoV-2 spike glycoprotein structure
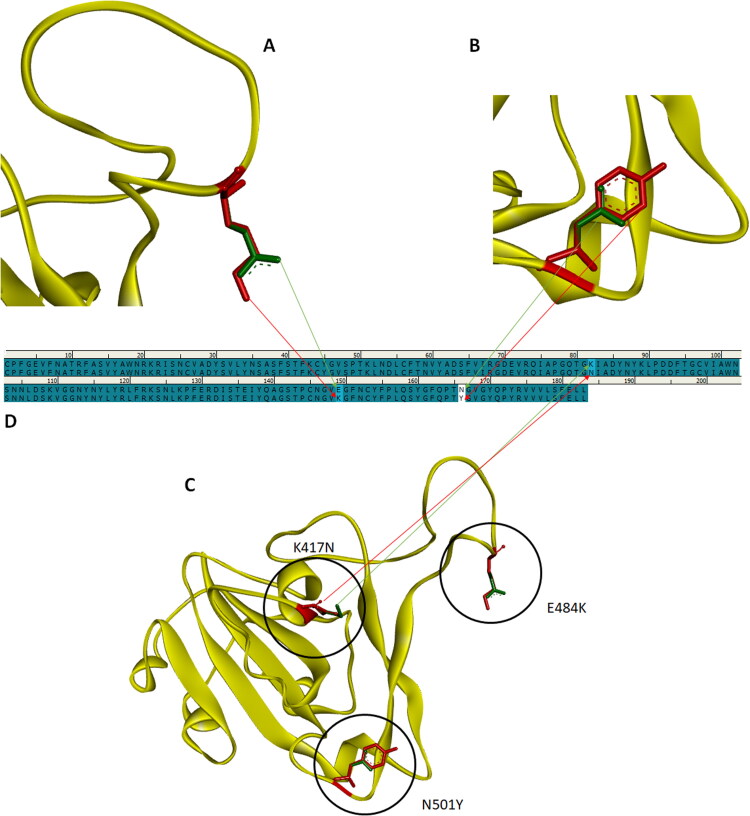
Initially, using the Discovery Studio Visualizer v16, the crystallographic structure of the receptor binding domain (RBD) of spike glycoprotein was gained by removing the ACE2 enzyme from the 2019-nCoV RBD/ACE2-B0AT1 complex (PDB ID: 6M17, Resolution: 2.90 Å) (Yan et al., 2020). Subsequently, using the protein editing tool of Discovery Studio Visualizer v16, the K417, E484 and N501 residues in the receptor binding motif (RBM) of the spike glycoprotein were computationally mutated into residues N417, K484 and Y501 to generate the K417N, E484K and N501Y mutants of spike glycoprotein, respectively (Figure 1). In order to perform energy minimization after mutation, the resulting mutant 3D crystallographic structures were subjected to geometry optimization using CHARMM22 force field implemented in Vega ZZ (Pedretti et al., 2004). Therefore, the protein structural imperfections after the generation of each mutant were refined.
Figure 1.
A. Superposed images of spike glycoproteins SARS-CoV-2 E484K mutant and wild-type SARS-CoV-2. The mutated glutamic acid 484 (GLU484) residue is shown in green, and the substituted lysine 484 (LYS484) residue is shown in red. B. Superposed images of spike glycoproteins SARS-CoV-2 N501Y mutant and wild-type SARS-CoV-2. The mutated asparagine 501 (ASN501) residue is shown in green, and the substituted tyrosine 501 (TYR501) residue is shown in red. C. Superposed images of spike glycoproteins SARS-CoV-2 K417N + E484K + N501Y mutant and wild-type SARS-CoV-2. The mutated lysine 417 (LYS417), glutamic acid 484 (GLU484) and asparagine 501 (ASN501) residues are shown in green, and the substituted asparagine 417 (ASN417), lysine 484 (LYS484) and tyrosine 501 (TYR501) residues are shown in red. D. Sequence representation of the regions of the E484K, N501Y and K417N mutations corresponding to the amino acid sequence of the spike glycoprotein.
2.2. Protein-protein dockings
Following the energy minimization, the control SARS-CoV-2 RBD/ACE2 and mutant (E484K, N501Y, K417N + E484K + N501Y) SARS-CoV-2 RBD/ACE2 protein-protein dockings were performed on the HADDOCK (High Ambiguity Driven protein-protein DOCKing) 2.4 docking server (De Vries et al., 2010). HADDOCK is a docking server that uses biochemical or biophysical interaction data such as chemical shift perturbation obtained from NMR titration experiments or mutagenesis data. Information about the interacting residues is introduced as ambiguous interaction restraints (AIRs) in order to perform docking. According to the HADDOCK docking protocol, the protein-protein interface is set flexible based on an analysis of intermolecular contacts within a 5 Å cut-off. Therefore, each binding pose is allowed to have different flexible regions defined. Residues in this interface region are then allowed to move their side-chains in a second refinement step. Finally, both backbone and side-chains of the flexible interface are granted freedom (De Vries et al., 2010). After the docking calculations are completed, the protein-protein complexes are ranked according to their intermolecular energies (sum of electrostatic, van der Waals and AIR energy terms) (Dominguez et al., 2003). In addition, in order to ensure the interaction of the correct interface residues in protein-protein docking using the HADDOCK server, Lys417, Tyr449, Tyr453, Glu484, Asn487, Tyr489, Gln493, Gly496, Gln498, Thr500, Asn501 and Gly502 were determined as the interacting interface amino acids of the spike protein, whereas the ACE2 interacting residues were restricted to Thr27, Asp30, Lys31, His34, Asp38, Tyr41, Gln42, Glu75, Tyr83, Lys353, Gly354, Asp355 and Arg357. These interacting residues of spike glycoprotein and human ACE2 (both wild-type) were identified by visual inspection of the interface of the 2019-nCoV RBD/ACE2-B0AT1 complex (PDB ID: 6M17) in the DS Visualizer v16 software.
Among the top 10 clusters generated by the HADDOCK server, the cluster with the best HADDOCK score was selected and the number one protein-protein docking pose (the complex with the most favorable binding free energy) in this cluster was saved as the top-ranked (representative) conformation. The protein-protein docking is an important step to investigate if the mutants have the same binding mode as the wild-type spike and to obtain quantitative and qualitative estimates of the interactions.
2.3. Calculation of wild-type SARS-CoV-2 RBD/ACE2 and mutant SARS-CoV-2 RBD/ACE2 binding affinities
The PRODIGY (PROtein binDIng enerGY prediction) web server (https://bianca.science.uu.nl/prodigy/) was used to calculate the binding affinities (kcal/mol) and dissociation constants (Kd) of top-ranked protein-protein complexes (wild-type, E484K, N501Y and K417N + E484K + N501Y) obtained from the HADDOCK server. The predictive model of PRODIGY operates on the assumption that the number of interfacial contacts (ICs) in a protein-protein complex is directly correlated with the free energy of binding (Kastritis et al., 2014).
2.4. Structural (interface) analysis of docking complexes
The structural analysis of the interfaces in the complexes obtained as a result of wild-type and mutant SARS-CoV-2 RBD/ACE2 dockings was performed using the PDBePISA (Proteins, Interfaces, Structures and Assemblies) web server (https://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver) as well as the DS Visualizer v16. Details of the protein-protein interface area (Å2) calculation in the PDBePISA server are described in the study of Shrake and Rupley (Shrake & Rupley, 1973). The other parameters such as the total number of interfacing residues, number of salt bridge-making residues, number of hydrogen bond-making residues and number of hydrogen bonds are calculated automatically by the PDBePISA server.
2.5. Molecular dynamics study
In this study, protein-protein complexes obtained from docking analysis with ACE2 of wild-type spike protein and other variants (E484K, N501Y and K417N + E484K + N501Y) were subjected to 200-ns molecular dynamics simulations to test the stability and to investigate the dynamic evolution of these systems. The lowest binding free energy configurations obtained from the docking of the Spike-hACE2 complexes, either for the wild type or for the mutant type, were used as starting points for the molecular dynamics simulations as follows: The pdb files were used as input geometry for molecular dynamics simulations using standard GROMACS (Abraham et al., 2015) tools, with the AMBER03 force field (Duan et al., 2003). The complexes were solvated using TIP3P water molecules (Jorgensen et al., 1983), sodium and chloride ions corresponding to physiological concentration, employing cubic simulation boxes with periodic boundary conditions. The van der Waals interactions were calculated directly until a 1.2 nm cutoff and the electrostatic interactions were calculated using the Particle Mesh Ewald (PME) method (Darden et al., 1993). The constructed systems were initially energetically minimized using the combination of the conjugate gradients and the steepest descent algorithms. After the minimization they were submitted to 500 ps position restrained simulations allowing the solvent and ions to relax without disturbing the complex geometry. After that followed the thermalization, as a sequence of three unrestricted molecular dynamics simulations with 5 ns each in temperatures of 200 K, 240 K and 280 K. After these steps, 200 ns long production simulation runs, in the NPT ensemble, were carried out, employing a Nosé-Hoover thermostat (Hoover, 1985) and a Parrinello-Rahman barostat (Parrinello & Rahman, 1981). The trajectories were then analyzed visually using VMD (Humphrey et al., 1996) and also using standard GROMACS tools, to quantify the structural and thermodynamic stability of the complexes as well as to investigate the details of the interactions.
2.6. Binding free energy calculations
The binding free energy between the wild-type or mutant spike proteins and the ACE2 protein was estimated using Molecular mechanics Poisson-Boltzmann surface area (MM/PBSA) binding free energy calculations (Baker et al., 2001; Homeyer & Gohlke, 2012). Sets of 200 configurations for each system were obtained as 1ns spaced snapshots, obtained directly from the molecular dynamics trajectories. The calculations were carried out using the g_mmpbsa GROMACS-compatible free energy program (Kumari et al., 2014) with a gridspace of 0.5 A, salt concentration of 0.150 M, solute dielectric constant of 2 and employing the solvent accessible surface area (SASA) as estimate of the nonpolar solvation energy. The same program was also used to estimate the residues’ contributions to the binding free energy.
3. Results and discussion
3.1. Docking and structural analysis
The spike protein is located on the surface of the SARS-CoV-2 virus and its molecular interaction with ACE2 plays a major role in viral entry into host cells (Hattori et al., 2021). Therefore, understanding the conformational effects of receptor binding domain (RBD) amino acid substitutions that occur at this interface, which could increase the infectivity of the virus, has important implications for vaccine- or drug-based treatment strategies. The receptor binding motif (RBM) is the main functional component of RBD and consists of two regions, region 1 and region 2, which form the interface between the spike (S) protein and hACE2 (Yi et al., 2020). Structural analysis of hACE2 recognition by SARS-CoV-2 shows that Leu455, Phe486, Glu493, Ser494 and Asn501 in the RBM of SARS-CoV-2 play critical roles in hACE2 binding. On the other hand, Lys31, Glu35, Asp38, Met82 and Lys353 residues of ACE2 form direct contact with the RBM of SARS-CoV-2 (Wan et al., 2020). However, as the molecular interaction between RBM and hACE2 has a dynamic nature, the type and number of interacting residues can vary considerably (Lan et al., 2020).
In this study, calculated binding energies and dissociation constants (Kd) as a result of the dockings of wild-type spike/ACE2 and mutant spike/ACE2 complexes, obtained using PRODIGY server, are given in Table 1. The binding affinity (kcal/mol) and dissociation constant (Kd, in molar) values obtained from the PRODIGY web server were found as −10.7 and 1.4 × 10−8 for wild-type (control) spike-ACE2, −13.3 and 1.7 × 10 −10 for spike (N501Y)-ACE2, −12.9 and 3.4 × 10 −10 for spike (E484K)-ACE2, and −12.4 and 7.6 × 10−10 for spike South Africa triple mutant (K417N + E484K + N501Y)- ACE2 complex. The dissociation constant (Kd) refers to the concentration of ACE2 at which half of the SARS-CoV-2 spike glycoproteins form complexes with ACE2 in the equilibrium state. The smaller the Kd numerically, the more tightly ACE2 binds to the spike glycoprotein (or vice versa) and the greater the affinity between the ACE2 and the spike glycoprotein. Therefore, considering the Kd, the N501Y mutant binds ACE2 82-fold stronger, the spike E484K mutant binds ACE2 41-fold stronger, and the South African triple mutant (K417N + E484K + N501Y) binds ACE2 18-fold stronger than the wild-type spike protein.
Table 1.
HADDOCK protein-protein docking results of the most common spike variants and the wild-type spike with ACE2.
| Complex | HADDOCK cluster score |
PRODIGY binding affinity (kcal/mol) | PRODIGY dissociation constant Kd (M) |
|---|---|---|---|
| Spike-ACE2_wild_type (Control) | −125.0 ± 5.7 | −10.7 | 1.4 × 10-10 |
| Spike_N501Y-ACE2 | −143.1 ± 7.5 | −13.3 | 1.7 × 10-10 |
| Spike_E484K-ACE2 | −117.7 ± 7.1 | −12.9 | 3.4 × 10-10 |
| Spike_South_Africa_triple (K417N + E484K + N501Y) | −124.9 ± 3.8 | −12.4 | 7.6 × 10-10 |
It has been reported that a 2.1 kcal/mol free energy reduction in SARS-CoV-2 RBD/ACE2 binding causes a 30-fold increase in protein-protein dissociation constant (Wang, Xu et al., 2020). Therefore, our view of how much a reduction in binding energy between 1.7 and 2.6 kcal/mol (or the decrease of Kd between 18 − 82 times) would increase binding affinity, compared to control, is in agreement with the result reported by Wang, Xu et al. (2020).
Thus, the mutations N501Y, E484K and South Africa triple (K417N + E484K + N501Y) greatly increase the strength of spike/ACE2 intermolecular interactions. Based on the interface analysis we performed using EMBL-EBI PDBePISA web server (Krissinel & Henrick, 2007) and DS Visualizer v16, the interface areas of the spike (N501Y)-ACE2, spike (E484K)-ACE2, and spike (K417N + E484K + N501Y)-ACE2 complexes were found to be 1027.3 Å2, 968.8 Å2 and 1026 Å2, respectively, whereas this value was 901.6 Å2 for the wild-type spike-ACE2 complex (Table 2). Another remarkable point is that in our analysis, the N501Y, E484K, and spike triple mutants have been found to have significantly larger numbers of amino acid residues in the spike RBD/ACE2 binding interface: 66, 57 and 62, respectively (Table 2). The residues involved in the interaction, as well as the classification of the type of the interaction are shown in Table S1 in the supplementary file.
Table 2.
Comparison of interface area, total number of interfacing residues, number of salt bridge making residues, number of hydrogen bond-making residues and number of hydrogen bonds at the wild-type spike/ACE2 interface and spike_N501Y/ACE2, spike_E484K/ACE2 and spike_South_Africa_triple (K417N + E484K + N501Y)/ACE2 interfaces obtained from HADDOCK protein-protein docking.
| Interface | Interface area (Å2) |
Total number of interfacing residues | Residues making salt bridges | Residues making H-bonds |
Number of H-bonds |
|---|---|---|---|---|---|
| Wild-type Spike/ACE2 (control) | 901.6 | 54 | 2 | 15 | 10 |
| Spike_N501Y/ACE2 | 1027.3 | 66 | 6 | 21 | 14 |
| Spike_E484K/ACE2 | 968.8 | 57 | 4 | 25 | 14 |
| Spike_South_Africa_triple (K417N + E484K + N501Y)/ACE2 | 1026 | 62 | – | 20 | 11 |
The data in the table were generated using PDBePISA (Proteins, Interfaces, Structures and Assemblies) web server (https://www.ebi.ac.uk/pdbe/pisa/) and DS Studio v16, respectively.
Depending on the increase in the number of amino acids in the interface, the total number of chemical interactions (salt bridges and hydrogen bonds) established at the spike-ACE2 interface in three different mutants increased significantly compared to the control. These were 20, 18 and 11 for the N501Y, E484K and South Africa triple mutant, respectively (Table 2).
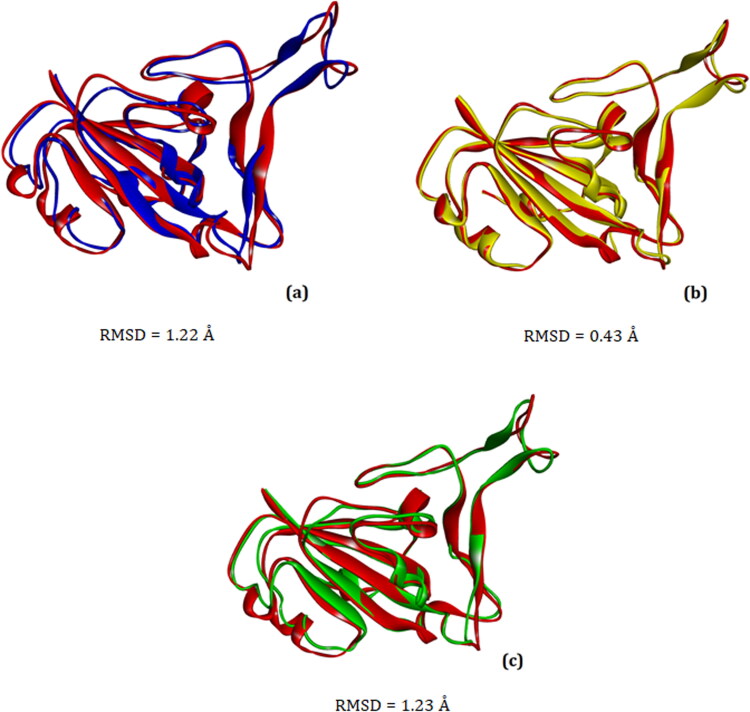
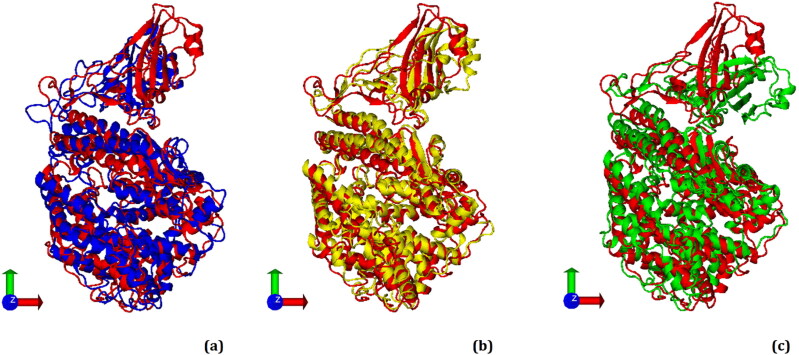
By comparing the structures of the mutated spike proteins after molecular docking with the wild-type spike (Figure 2), we see that the binding mode is essentially the same in all cases, although some structural changes can be observed.
Figure 2.
Superimposed conformations of the spike glycoproteins after molecular docking simulations with ACE2. (a) Native (control) spike (red) and E484K variant (blue), (b) Native spike (red) and N501Y variant (yellow), (c) Native spike (red) and multiple (K417N + E484K + N501Y) spike variant (green). Note that the three spike variants show some conformational change relative to control. Figures were generated with Discovery Studio v16. RMSD: Root-mean-square deviation.
There are significant differences between the amino acids of mutant spike proteins and wild-type spike protein interacting with the ACE2 receptor (supplementary file Tables S1 and S2). The H-bonds and electrostatic interactions between ACE2 and wild-type spike RBD or mutant spike RBD are shown in Table S1 (supplementary file). While Tyr501 in N501Y mutant forms one hydrogen bond with Thr324 and Lys353 of ACE2, Asn501 does not appear to interact with ACE2 in the wild-type spike protein. In addition, two novel electrostatic interactions occurred between Lys458 and Glu484 residues of the N501Y mutant and Glu23 and Lys31 of ACE2, two of which were absent in the wild-type mutant (supplementary file Tables S1 and S2). In the E484K mutant, Lys484 interacted with Glu35 of ACE2 via a salt bridge interaction, whereas in wild-type spike, Glu484 interacted with Lys31 of ACE2 via a carbon-hydrogen bond. Lys417 of E484K also formed a novel electrostatic interaction with Asp30 of the ACE2 receptor (supplementary file Tables S1 and S2). In Spike South Africa triple mutant (K417N + E484K + N501Y), Asn417 and Lys484 did not appear to undergo any bond formation reaction with ACE2, while Tyr501 formed a hydrogen bond contact with Lys353 of ACE2. In contrast, in the wild-type spike, only Glu484 appears to interact with ACE2 and form a carbon-hydrogen bond with Lys31 of ACE2 (supplementary file Table S1 and S2). In particular, novel electrostatic interactions (Glu23-Lys458, and Lys31-Glu484 for N501Y; Glu35-Lys484 for E484K) were observed in docking simulations of spike mutants N501Y and E484K (supplementary file Table S1). When the residues interacting with ACE2 are compared, it was revealed that only 38% of the contacting residues in the N501Y mutant, 45% in the E484K mutant and 30% in the triple mutant resemble the contacting residues of the wild-type spike (supplementary file Table S2). Figure S1 (supplementary file) shows the ‘heatmap’, which counts how many times the contacting residues of wild-type and mutant spike glycoproteins interact with the ACE2 receptor (regardless of bond type), or vice versa. It is clear from the heatmap that mutations N501Y and E484K (except the South Africa triple mutant) resulted in an increase in the frequency of novel contacts between spike protein and ACE2 compared to the wild-type spike. The increased number of chemical contacts (supplementary file Figure S1) shows that the N501Y and E484K mutations cause conformational changes in the spike glycoprotein's receptor binding domain (RBD) which interacts with ACE2, resulting in an increased interface surface area.
The increase on the magnitude of the total interface area (Å2) and in the number of amino acids at the interface and the increase of non-covalent salt bridges and H-bond interactions indicate that the variants of E484K, N501Y and South Africa triple strains induce conformational changes in the spike RBD (Figure 2) and may explain why these ‘variants of concern’ have increased infectious ability. Therefore, these conformational changes were investigated more deeply using molecular dynamics, as discussed in the next subsection.
In line with our views on the induced conformational change in the spike RBD, it has been reported that the E484K mutation reduces or completely abolishes the neutralizing antibody response in convalescent serums, and also weakens the binding of serum polyclonal antibodies to the receptor binding domain (RBD) of the spike glycoprotein (Ferrareze et al., 2021; Jangra et al., 2021; Wang et al., 2021; Wise, 2021; Xie et al., 2021). As can be seen in our study, the spike protein carrying the E484K mutation binds to ACE2 in a slightly different conformation compared to the wild-type (Figure 2a). In our protein-protein docking analysis, the spike N501Y showed a significantly higher binding capacity to the ACE2 receptor compared to the wild-type (Table 1). Consistent with our results, it has been reported that the N501Y mutation increases the hydrophobic interactions between RBD and ACE2, while decreasing the hydrophilic repulsion, thereby increasing the transmissibility of the infection (Li et al., 2021).
In our study, the spike triple variant (K417N + E484K + N501Y) also showed a significantly more favorable binding affinity to the ACE2 than the wild-type (Table 1) and caused some conformational changes in the spike protein (Figure 2c).
3.2. Molecular dynamics and free energy calculations
As already pointed out, the conformational changes and the interaction pattern obtained from the docking motivated a more detailed analysis, using molecular dynamics.
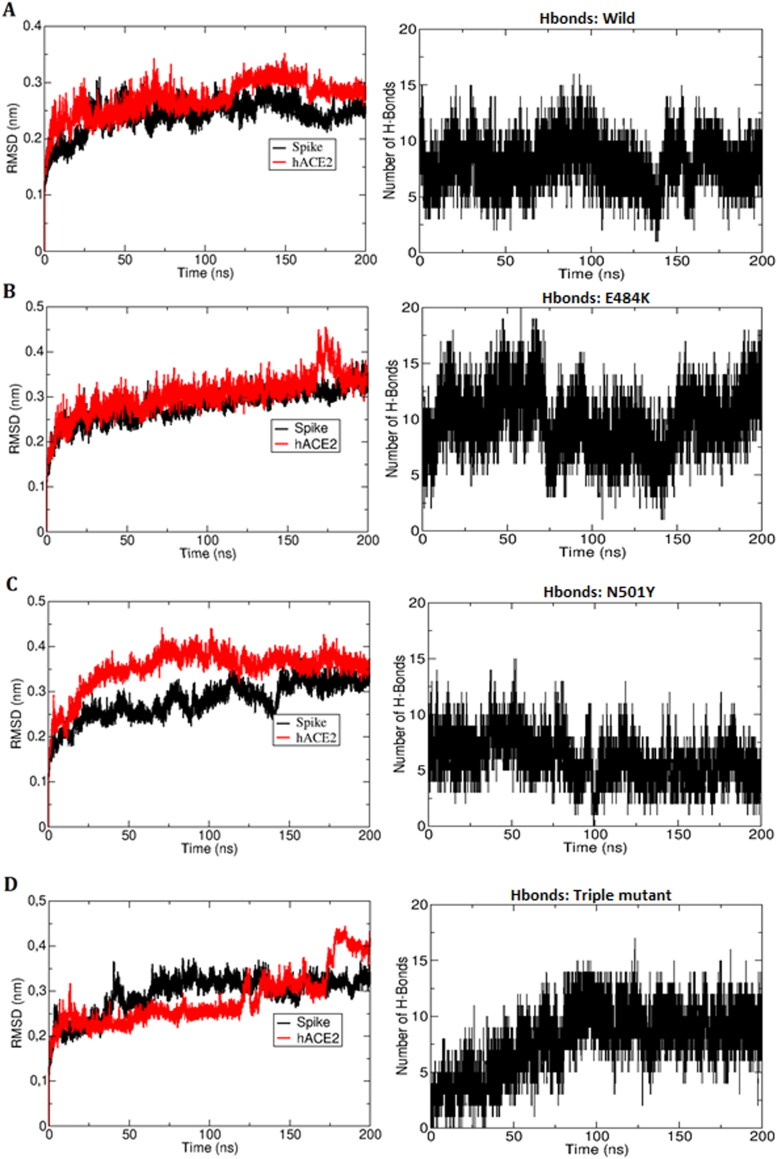
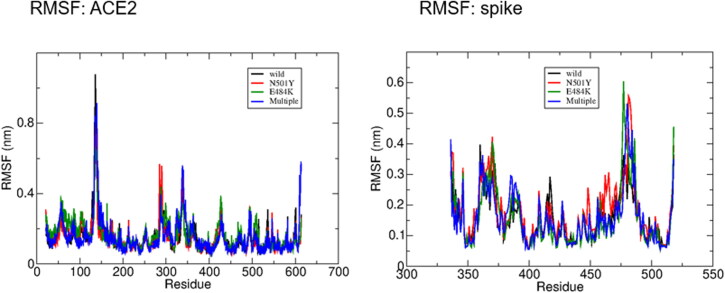
In this study, all mutant spike-ACE2 complexes (including wild-type) obtained from protein-protein docking analyzes showing the most negative binding energy were subjected to 200-ns long molecular dynamics simulations in order to investigate the time-dependent stability and dynamic evolution of the complexes. All systems were found to be stable, showing conservation of the structure, as can be seen by the analysis of the time evolution of the root mean square deviation (RMSDs) and number of hydrogen bonds (Figure 3). The root mean square fluctuation (RMSF), analyzed for each residue, averaged over the whole trajectory, was also computed (Figure 4). According to the RMSF, the mobility pattern is nearly the same for the ACE2 protein in all systems. Besides, a slight enhanced mobility relative to the native spike was found in the spike residues 448, 460-470 and 481 in the case of N501Y mutation, in the residue 477 in the E484K mutation and in the residues 480 and 385-390 in the case of the triple mutation.
Figure 3.
Time evolution of the structure and interactions in the complexes of A- Wild-type Spike/ACE2 (control), B- Spike_E484K/ACE2 C- Spike_N501Y/ACE2, and D- Spike_South_Africa_triple (K417N + E484K + N501Y)/ACE2 in molecular dynamics simulations. In the left the root-mean-square deviation (RMSD) of atomic positions of the ligand (spike glycoprotein, black) and of the receptor (ACE2, red) are shown, whereas in the right the number of hydrogen bonds between spike glycoprotein and hACE2 is shown.
Figure 4.
The RMSF (root mean square fluctuation) showing the mobility pattern for the several residues, averaged along the simulation trajectory. It can be noticed that the RMSF of the ACE2 residues is essentially the same in all systems, whereas there are some (small) differences in the RMSF of the spike residues. The N501Y variant shows enhanced mobility, compared to the wild spike, at the residues 448, 460-470 and 481. The E484K variant shows only a slight enhanced mobility at the residue 477 and the multiple variant also a very slight enhanced mobility in the residues 385, 390 and 480.
The Spike-hACE2 interaction was characterized by the presence of many close contacts and hydrogen bonds which can be monitored along the molecular dynamics trajectory. In the wild-type system there were in average 7.95 ± 2.04 hydrogen bonds. The system with three mutations (K417N + E484K + N501Y) showed roughly the same number of hydrogen bonds: 7.87 ± 2.82, whereas the N501Y mutation system displayed a smaller number of hydrogen bonds (6.05 ± 2.00) and the E484K mutation system displayed the largest number of hydrogen bonds: 9.81 ± 2.64 (Table 3).
Table 3.
Interface area of the complexes (the half of the sum of solvent-accessible areas of spike and ACE2 minus solvent-accessible area of the complex) and number of hydrogen bonds, calculated as average over the MD simulations.
| Complex | Interface area (Å2) | Number of hydrogen bonds |
|---|---|---|
| Wild | 1013.7 ± 65.6 | 7.95 ± 2.04 |
| N501Y | 1008.2 ± 85.1 | 6.05 ± 2.00 |
| E484K | 1068.3 ± 68.1 | 9.81 ± 2.64 |
| Spike_South_Africa_triple | 1215.3 ± 136.7 | 7.87 ± 2.82 |
The interface area was also calculated using the molecular dynamics results, with the same method as for the docking results (Shrake & Rupley, 1973). The results are shown in Table 3. It is clear that the interface area of the multiple mutant is higher than of the other complexes, but the interface area of the E484K mutant is also somewhat higher than the wild-type, whereas the interface area of the N501Y mutant is almost the same as the wild-type complex.
The binding free energy was calculated using the snapshots generated from the molecular dynamics trajectory. The g_mmbpsa program (Kumari et al., 2014) yields not only the binding free energy (total and components – van der Waals, electrostatic, polar solvation and surface energy), see Table 4, but also the binding free energy contributions averaged per residues (see supplementary file Table S3). Despite the apparently weaker hydrogen bond network, the system with the N501Y mutation showed a more negative free energy of binding than the wild-type system. The strongest interaction, however, was found for the system with the mutation E484K, where the binding free energy was remarkably stronger than in the wild-type. This can be easily understood analysing the residues contribution to the binding free energy (supplementary file Table S3). Indeed, the mutation of the negatively charged residue Glu484 to the positively charged residue Lys484 yielded a substantial negative free energy contribution to the stabilization of the complex (the contribution of residue 484 changed from + 250,6 kJ in the wild spike to − 206,6 kJ upon mutation). The mutation in the residue 501 (Asn to Tyr, both polar non-charged residues) also contributes to the stabilization of the complex, but much less than the E484K mutation.
Table 4.
Binding free energies (kcal/mol) and their components, calculated using MM/PBSA, for the interaction between ACE2 and A- Wild-type spike, B- Spike_N501Y, C- Spike_E484K and D- Spike_South_Africa_triple (K417N + E484K + N501Y).
| Complex | Van der Waals Energy (kcal/mol) |
Electrostatic Energy (kcal/mol) |
Polar Solvation Energy (kcal/mol) |
SASA Energy (kcal/mol) |
Binding Energy (kcal/mol) |
|---|---|---|---|---|---|
| Wild-type Spike/ACE2 (control) | −91.56 ± 0.56 | −317.31 ± 1.64 | 132.47 ± 2.48 | −10.75 ± 0.07 | −287.25 ± 2.10 |
| Spike_N501Y/ACE2 | −96.74 ± 0.70 | −324.27 ± 2.00 | 123.98 ± 3.01 | −10.65 ± 0.08 | −307.62 ± 2.41 |
| Spike_E484K/ACE2 | −97.95 ± 0.54 | −560.16 ± 2.37 | 174.02 ± 2.41 | −11.25 ± 0.06 | −495.37 ± 2.64 |
| Spike_South_Africa_triple (K417N + E484K + N501Y)/ACE2 | −120.46 ± 1.06 | −418.06 ± 1.44 | 187.42 ± 3.38 | −12.85 ± 0.11 | −363.98 ± 2.34 |
Although the HADDOCK docking algorithm considered some flexibility of the proteins, the final docking scores and the PRODIGY binding affinities (Table 1) were based on only one (the final) configuration, whereas the binding free energies obtained by the combination of MD and MM/PBSA methods (Table 4) are values based on several configurations, sampled over the entire MD trajectories. Considering the importance of the conformational changes, the MD-MM/PBSA estimates are more reliable. Therefore, the most stable mutant complexes in our study can be ranked as: E484K > South Africa triple (K417N + E484K + N501Y) > N501Y.
A surface plasmon resonance study showed that the N501Y mutation had a faster association rate and slower dissociation constant (Kd) upon binding to ACE2 (Tian et al., 2021). However, the overall interaction pattern of N501Y with ACE2 did not differ greatly from that of the wild-type spike/ACE2 complex (Figure 5b and supplementary file Figure S2a), and it was suggested that this mutation would not pose a significant disadvantage to the vaccine efficacy (Zhang et al., 2021). Indeed, by following the distances between chosen pairs of residues we could show that the pattern of neighboring residues along the simulation of the N501Y mutated system does not differ substantially from the wild spike one (supplementary file Figures S3–S9).
Figure 5.
Superimposed conformations after 200-ns molecular dynamics simulations of the spike glycoprotein/ACE2 complexes. (a) Native spike (red) and E484K (blue) variant, (b) Native spike (red) and N501Y variant (yellow), (c) Native spike and multiple (K417N + E484K + N501Y) spike variant (green). Note that the E484K (a) and multiple (c) spike variants show significant conformational changes relative to control (red).
The conversion of negatively charged Glu484 to positively charged Lys484 in the E484K mutant, which we found to be the most stable complex according to the results of MM-PBSA analysis, has also been shown in another study to have profound effects on a highly flexible loop in the RBD (Franceschi, Caldana et al., 2021; Nelson et al., 2021). The comparison of the conformations of the wild-type spike-ACE2 complex with the E484K mutated spike-ACE2 complex (Figure 5a) shows clearly a conformational transition, where the spike protein is bent toward one of the ACE2 helices (residues 549–560). This conformational change can be also seen in the zoomed figure in the supplementary file Figure S2b). Moreover, the residues 417 (supplementary file Figure S5) and 478 (supplementary file Figure S7) come closer to the ACE2 and there was a clear change of the pattern of interacting residues for the spike residue 487 (supplementary file Figure S8). Specifically, this mutation creates a strong ionic interaction between the lysine (K484) amino acid of the mutant spike protein and the residue E75 of hACE2 (Nelson et al., 2021), and this finding is in excellent agreement with the residual free energy contributions we obtained from MM-PBSA calculations (supplementary file Table S3). Such strong contribution of free energy is not found in the wild-type spike E484.
The spike triple mutant (K417N + E484K + N501Y) we analyzed in our study showed significantly higher binding free energy than wild-type in molecular dynamics simulations and MM-PBSA analysis (Table 4). It has also been reported in other MD simulations that this mutant combination, compared with E484K or N501Y, maximally induces conformational changes when bound to ACE2, resulting in novel contacts between RBD and ACE2, and leading to a viral 'escape' conformation (Franceschi, Ferrareze et al., 2021; Nelson et al., 2021). As can be seen in Figures 2c and 5c and Figure S2c (supplementary file) and supported by MM-PBSA results (Table 4), we also found that the triple mutated spike protein is strongly bent (although in a different way compared to the E484K mutant, as seen in supplementary file Figure S2c) yielding significant conformational changes relative to the wild-type. As can be seen in Figures S3–S9 (supplementary file), for most of the triple mutated spike key residues, the pattern of neighboring ACE2 residues along the simulation is strikingly different from the other systems. Indeed, due to these conformational changes, the residue ASP405 moves closer to ACE2, whereas the residue GLY446 moves away. More details can be found in the supplementary material.
4. Conclusion
From an evolutionary point of view, viral mutations are expected to reduce the mortality of the virus on the host over time and let them survive in a higher harmony. However, the investigated three SARS-CoV-2 variants of concern (E484K, N501Y and triple mutant) in this study have significantly increased the binding affinity of all the mutant spike glycoproteins against ACE2, for which we believe that could pose a potential danger to the efficacy of the currently launched anti-SARS-CoV-2 vaccines. We also showed that among variants, E484K and triple mutant triggered the strongest changes in both the binding energy and the conformation. It is clear that the selection pressure on SARS-CoV-2 further increases the RBD-ACE2 stabilization of variants (Fernández, 2021; Nelson et al., 2021), and the E484K alone or in combination with K417N and N501Y, potentially results in an ‘escape’ phenotype (Franceschi, Ferrareze et al., 2021; Hu et al., 2021). Consistent with our molecular dynamics study, it was reported that especially the K417N + E484K + N501Y (B.1.351) variant of the RBD was more successful in avoiding neutralizing antibodies, and this was due to the high plasticity of the SARS-CoV-2 S and ACE2 interfaces (Hoffmann et al., 2021). In addition, E484K alone or the triple mutant RBD (K417N + E484K + N501Y) abolished binding of the potent class 2 RBD neutralizing antibodies (nAbs) (Wibmer et al., 2021). It has recently been reported that the mechanism underlying the formation of these 'escape phenotypes' is generally the transition from the 'Hook-like' folded RBD tip seen in the native spike to the highly 'Disordered' state, where the RBD tip cycles between a series of conformations (Gobeil et al., 2021). Thus, we are of the opinion that E484K and the triple mutant most likely distort the native conformation of the RBD tip, thereby blocking the binding of RBD-directed SARS-CoV-2 antibodies to this domain, and hence the virus may escape from the vaccine-induced immune response.
Supplementary Material
Acknowledgements
P. A. N. acknowledges financial support from CNPq and CAPES (Financial code 001), as well as the Centro Nacional de Supercomputação (CESUP), Universidade Federal do Rio Grande do Sul (UFRGS) for providing HPC resources.
Funding Statement
Conselho Nacional de Desenvolvimento Científico e Tecnológico.
Author contributions
E.S.I. conceptualized the study, performed protein-protein docking calculations, prepared Figures 1 and 2 and Tables 1 and 2, contributed to the writing and revision of the article; P.A.N. conducted the molecular dynamics simulations and MM/PBSA calculations, prepared Figures 3, 4, and 5 and Tables 3–4 , interpreted the results and contributed to the detailed revision of the manuscript; A.S.T. helped create and interpret Tables 1 and 2; C.S. contributed to the interpretation of protein-protein docking results; B.T. supervised E.S.I. and revised the manuscript. All the authors read and approved the final manuscript.
Disclosure statement
None of the authors declare conflict of interest in this work.
References
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Al-Karmalawy, A. A., Dahab, M. A., Metwaly, A. M., Elhady, S. S., Elkaeed, E. B., Eissa, I. H., & Darwish, K. M. (2021). Molecular docking and dynamics simulation revealed the potential inhibitory activity of ACEIs against SARS-CoV-2 targeting the hACE2 receptor. Frontiers in Chemistry, 9, 661230. 10.3389/fchem.2021.661230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, A., & Vijayan, R. (2020). Dynamics of the ACE2-SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Scientific Reports, 10, 14214. 10.1038/s41598-020-71188-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, M., Sorour, M. K., & Kasry, A. (2020). Comparing the binding interactions in the receptor binding domains of SARS-CoV-2 and SARS-CoV. The Journal of Physical Chemistry Letters, 11, 4897–4900. 10.1021/acs.jpclett.0c01064 [DOI] [PubMed] [Google Scholar]
- Anonymous (2021). COVID-19 vaccine (Harkless D. (Ed.)). Wikipedia. [Google Scholar]
- Baker, N. A., Sept, D., Joseph, S., Holst, M. J., & McCammon, J. A. (2001). Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences United States of America, 98, 10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros, R. O., Junior, F. L., Pereira, W. S., Oliveira, N. M., & Ramos, R. M. (2020). Interaction of drug candidates with various SARS-CoV-2 receptors: An in silico study to combat COVID-19. Journal of Proteome Research, 19, 4567–4575. 10.1021/acs.jproteome.0c00327 [DOI] [PubMed] [Google Scholar]
- Ceraolo, C., & Giorgi, F. M. (2020). Genomic variance of the 2019-nCoV coronavirus. Journal of Medical Virology, 92, 522–528. 10.1002/jmv.25700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand, M., Hopkins, S., Dabrera, G., Achison, C., Barclay, W., Ferguson, N., Volz, E., Loman, N., Rambaut, A., & Barrett, J., 2020. Investigation of novel SARS-COV-2 variant, Variant of Concern 202012/01. Public Health England, United Kingdom, pp. 1–11.
- Chen, R. E., Zhang, X., Case, J. B., Winkler, E. S., Liu, Y., VanBlargan, L. A., Liu, J., Errico, J. M., Xie, X., Suryadevara, N., Gilchuk, P., Zost, S. J., Tahan, S., Droit, L., Turner, J. S., Kim, W., Schmitz, A. J., Thapa, M., Wang, D., … Diamond, M. S. (2021). Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nature Medicine, 27, 717–726. 10.1038/s41591-021-01294-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, B., Choudhary, M. C., Regan, J., Sparks, J. A., Padera, R. F., Qiu, X., Solomon, I. H., Kuo, H. H., Boucau, J., Bowman, K., Adhikari, U. D., Winkler, M. L., Mueller, A. A., Hsu, T. Y., Desjardins, M., Baden, L. R., Chan, B. T., Walker, B. D., Lichterfeld, M., … Li, J. Z. (2020). Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. New England Journal of Medicine, 383, 2291–2293. 10.1056/NEJMc2031364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrials.gov . (2020a). Clinical trial of efficacy, safety, and immunogenicity of Gam-COVID-Vac vaccine against COVID-19.
- ClinicalTrials.gov . (2020b). Clinical trial of recombinant novel coronavirus vaccine (Adenovirus Type 5 Vector) against COVID-19.
- ClinicalTrials.gov . (2020c). Safety and immunogenicity study of inactivated vaccine for prevention of SARS-CoV-2 infection (COVID-19) (Renqiu).
- ClinicalTrials.gov . (2020d). Study to describe the safety, tolerability, immunogenicity, and efficacy of RNA vaccine candidates against COVID-19 in healthy adults.
- ClinicalTrials.gov . (2020e). A study to evaluate efficacy, safety, and immunogenicity of mRNA-1273 vaccine in adults aged 18 years and older to prevent COVID-19.
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. Journal of Chemical Physics, 98, 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- de Andrade, J., Gonçalves, P. F. B., & Netz, P. A. (2020). Why does the novel coronavirus spike protein interact so strongly with the human ACE2? A thermodynamic answer. ChemBioChem, 22, 865–875. 10.1002/cbic.202000455 [DOI] [PubMed] [Google Scholar]
- de Oliveira, O. V., Rocha, G. B., Paluch, A. S., & Costa, L. T. (2021). Repurposing approved drugs as inhibitors of SARS-CoV-2 S-protein from molecular modeling and virtual screening. Journal of Biomolecular Structure and Dynamics, 11, 3924–3933 10.1080/07391102.2020.1772885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries, S. J., Van Dijk, M., & Bonvin, A. M. (2010). The HADDOCK web server for data-driven biomolecular docking. Nature Protocols, 5, 883. 10.1038/nprot.2010.32 [DOI] [PubMed] [Google Scholar]
- Dominguez, C., Boelens, R., & Bonvin, A. M. (2003). HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. Journal of the American Chemical Society, 125, 1731–1737. 10.1021/ja026939x [DOI] [PubMed] [Google Scholar]
- Duan, Y., Wu, C., Chowdhury, S., Lee, M. C., Xiong, G., Zhang, W., Yang, R., Cieplak, P., Luo, R., Lee, T., Caldwell, J., Wang, J., & Kollman, P. (2003). A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. Journal of Computational Chemistry, 24, 1999–2012. 10.1002/jcc.10349 [DOI] [PubMed] [Google Scholar]
- Ekins, S., Mottin, M., Ramos, P. R., Sousa, B. K., Neves, B. J., Foil, D. H., Zorn, K. M., Braga, R. C., Coffee, M., & Southan, C. (2020). Déjà vu: Stimulating open drug discovery for SARS-CoV-2. Drug Discovery Today, 25, 928–941. 10.1016/j.drudis.2020.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (ECDC) . (2021). SARS-CoV-2 variants of concern as of 21 May 2021. file:///C:/Users/ANT%C4%B0OKS%C4%B0DAN/Downloads/2_En_G%C3%BCncel_SARS-CoV-2%20variants%20of%20concern%20as%20of%2021%20May%202021.pdf
- Fernández, A. (2021). COVID-19 evolution in the post-vaccination phase: Endemic or extinct? ACS Pharmacology & Translational Science, 4, 403–405. 10.1021/acsptsci.0c00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrareze, P. A. G., Franceschi, V. B., de Menezes Mayer, A., Caldana, G. D., Zimerman, R. A., & Thompson, C. E. (2021). E484K as an innovative phylogenetic event for viral evolution: Genomic analysis of the E484K spike mutation in SARS-CoV-2 lineages from Brazil. bioRxiv. 10.1101/2021.01.27.426895 [DOI] [PMC free article] [PubMed]
- Franceschi, V. B., Caldana, G. D., de Menezes Mayer, A., Cybis, G. B., Neves, C. A. M., Ferrareze, P. A. G., Demoliner, M., de Almeida, P. R., Gularte, J. S., Hansen, A. W., Weber, M. N., Fleck, J. D., Zimerman, R. A., Kmetzsch, L., Spilki, F. R., & Thompson, C. E. (2021). Genomic epidemiology of SARS-CoV-2 in Esteio, Rio Grande do Sul, Brazil. BMC Genomics, 22, 371. 10.1186/s12864-021-07708-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi, V. B., Ferrareze, P. A. G., Zimerman, R. A., Cybis, G. B., & Thompson, C. E. (2021). Mutation hotspots, geographical and temporal distribution of SARS-CoV-2 lineages in Brazil, February 2020 to February 2021: Insights and limitations from uneven sequencing efforts. MedRxiv. 10.1101/2021.03.08.21253152 [DOI] [PMC free article] [PubMed]
- Fratev, F. (2020). The N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with both hACE2 and human derived antibody: A Free energy of perturbation study. bioRxiv. 10.1101/2020.12.23.424283 [DOI] [PubMed]
- Ghorbani, M., Brooks, B. R., & Klauda, J. B. (2020). Critical sequence hotspots for binding of novel coronavirus to angiotensin converter enzyme as evaluated by molecular simulations. Journal of Physical Chemistry B, 124, 10034–10047. 10.1021/acs.jpcb.0c05994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobeil, S. M. C., Janowska, K., McDowell, S., Mansouri, K., Parks, R., Stalls, V., Kopp, M. F., Manne, K., Saunders, K., Edwards, R. J., Haynes, B. F., Henderson, R. C., & Acharya, P. (2021). Effect of natural mutations of SARS-CoV-2 on spike structure, conformation and antigenicity. bioRxiv: the preprint server for biology. 10.1101/2021.03.11.435037,2021.2003.2011.435037. [DOI] [PMC free article] [PubMed]
- Harrison, C. (2020). Coronavirus puts drug repurposing on the fast track. Nature Biotechnology, 38, 379–381. 10.1038/d41587-020-00003-1 [DOI] [PubMed] [Google Scholar]
- Hattori, T., Koide, A., Noval, M. G., Panchenko, T., Romero, L. A., Teng, K. W., Tada, T., Landau, N. R., Stapleford, K. A., & Koide, S. (2021). The ACE2-binding interface of SARS-CoV-2 spike inherently deflects immune recognition. Journal of Molecular Biology, 433, 166748. 10.1016/j.jmb.2020.166748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, M., Arora, P., Groß, R., Seidel, A., Hörnich, B. F., Hahn, A. S., Krüger, N., Graichen, L., Hofmann-Winkler, H., Kempf, A., Winkler, M. S., Schulz, S., Jäck, H.-M., Jahrsdörfer, B., Schrezenmeier, H., Müller, M., Kleger, A., Münch, J., & Pöhlmann, S. (2021). SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell, 184, 2384–2393.e12. 10.1016/j.cell.2021.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homeyer, N., & Gohlke, H. (2012). Free energy calculations by the molecular mechanics Poisson–Boltzmann surface area method. Molecular Informatics, 31, 114–122. 10.1002/minf.201100135 [DOI] [PubMed] [Google Scholar]
- Hoover, W. G. (1985). Canonical dynamics: Equilibrium phase-space distributions. Physical Review A: Atomic, Molecular, and Optical Physics, 31, 1695–1697. [DOI] [PubMed] [Google Scholar]
- Hu, J., Peng, P., Wang, K., Liu, B.-z., Fang, L., Luo, F.-y., Jin, A.-s., Tang, N., & Huang, A. (2021). Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cellular & Molecular Immunology, 18, 1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: visual molecular dynamics. Journal of Molecular Graphics, 14, 27–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Idris, M. O., Yekeen, A. A., Alakanse, O. S., & Durojaye, O. A. (2020). Computer-aided screening for potential TMPRSS2 inhibitors: a combination of pharmacophore modeling, molecular docking and molecular dynamics simulation approaches. Journal of Biomolecular Structure and Dynamics, 39, 5638–5656. 10.1080/07391102.2020.1792346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra, S., Ye, C., Rathnasinghe, R., Stadlbauer, D., Personalized Virology Initiative study, g., Krammer, F., Simon, V., Martinez-Sobrido, L., Garcia-Sastre, A., & Schotsaert, M. (2021). SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe, 2, e283–e284. 10.1016/S2666-5247(21)00068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics, 79, 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kastritis, P. L., Rodrigues, J. P., Folkers, G. E., Boelens, R., & Bonvin, A. M. (2014). Proteins feel more than they see: Fine-tuning of binding affinity by properties of the non-interacting surface. Journal of Molecular Biology, 426, 2632–2652. 10.1016/j.jmb.2014.04.017 [DOI] [PubMed] [Google Scholar]
- Kemp, S. A., Datir, R. P., Collier, D. A., Ferreira, I. A. T. M., Carabelli, A., Harvey, W., Robertson, D. L., Gupta, & R. K., 2020. Recurrent emergence and transmission of a SARS-CoV-2 spike deletion H69/V70, 15 December 2020 ed. bioRxiv.
- Krissinel, E., & Henrick, K. (2007). Inference of macromolecular assemblies from crystalline state. Journal of Molecular Biology, 372, 774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., Open Source Drug Discovery Consortium , & Lynn, A. (2014). g_mmpbsa – A GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54, 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., & Wang, X. (2020). Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature, 581, 215–220. 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Li, J., Hou, C., Wang, M., Liao, C., Guo, S., Shi, L., Ma, X., Zhang, H., Jiang, S., & Zheng, B. (2021). Hydrophobic interaction determines docking affinity of SARS CoV 2 variants with antibodies. arXiv preprint arXiv:2103.00399.
- Luan, J., Lu, Y., Jin, X., & Zhang, L. (2020). Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochemical and Biophysical Research Communications, 526, 165–169. 10.1016/j.bbrc.2020.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, G., Buzko, O., Spilman, P., Niazi, K., Rabizadeh, S., & Soon-Shiong, P. (2021). Molecular dynamic simulation reveals E484K mutation enhances spike RBD-ACE2 affinity and the combination of E484K, K417N and N501Y mutations (501Y.V2 variant) induces conformational change greater than N501Y mutant alone, potentially resulting in an escape mutant. bioRxiv 2021.2001.2013.426558. 10.1101/2021.01.13.426558 [DOI]
- Parrinello, M., & Rahman, A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics, 52, 7182–7190. 10.1063/1.328693 [DOI] [Google Scholar]
- Pedretti, A., Villa, L., & Vistoli, G. (2004). VEGA – An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. Journal of Computer-Aided Molecular Design, 18, 167–173. 10.1023/B:JCAM.0000035186.90683.f2 [DOI] [PubMed] [Google Scholar]
- Qiao, B., & Olvera de la Cruz, M. (2020). Enhanced binding of SARS-CoV-2 spike protein to receptor by distal polybasic cleavage sites. ACS Nano, 14, 10616–10623. 10.1021/acsnano.0c04798 [DOI] [PubMed] [Google Scholar]
- Rambaut, A., Loman, N., Pybus, O., Barclay, W., Barrett, J., Carabelli, A., Connor, T., Peacock, T., Robertson, D. L., & Volz, E.. 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological.org. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
- Riou, J., & Althaus, C. L. (2020). Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Eurosurveillance, 25, 2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakkiah, S., Guo, W., Pan, B., Ji, Z., Yavas, G., Azevedo, M., Hawes, J., Patterson, T. A., & Hong, H. (2020). Elucidating interactions between SARS-CoV-2 trimeric spike protein and ACE2 using homology modeling and molecular dynamics simulations. Frontiers in Chemistry, 8, 622632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., Geng, Q., Auerbach, A., & Li, F. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature, 581, 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, A., Tiwari, V., & Sowdhamini, R. (2020). Computational search for potential COVID-19 drugs from FDA-approved drugs and small molecules of natural origin identifies several anti-virals and plant products. Journal of Biosciences, 45, 1– 18. 10.1007/s12038-020-00069-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrake, A., & Rupley, J. A. (1973). Environment and exposure to solvent of protein atoms. Lysozyme and insulin. Journal of Molecular Biology, 79, 351–371. 10.1016/0022-2836(73)90011-9 [DOI] [PubMed] [Google Scholar]
- Singh, T. U., Parida, S., Lingaraju, M. C., Kesavan, M., Kumar, D., & Singh, R. K. (2020). Drug repurposing approach to fight COVID-19. Pharmacological Reports, 72, 1479–1508. 10.1007/s43440-020-00155-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinello, A., Saltalamacchia, A., & Magistrato, A. (2020). Is the rigidity of SARS-CoV-2 spike receptor-binding motif the hallmark for its enhanced infectivity? Insights from all-atom simulations. The Journal of Physical Chemistry Letters, 11, 4785–4790. 10.1021/acs.jpclett.0c01148 [DOI] [PubMed] [Google Scholar]
- Starr, T. N., Greaney, A. J., Hilton, S. K., Crawford, K. H. D., Navarro, M. J., Bowen, J. E., Tortorici, M. A., Walls, A. C., Veesler, D., & Bloom, J. D. (2020). Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell, 182, 1295–1310. 10.1016/j.cell.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, E. E. (2021). Updated rapid risk assessment from ECDC on the risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA–First update. Eurosurveillance, 26, 2101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, F., Tong, B., Sun, L., Shi, S., Zheng, B., Wang, Z., Dong, X., & Zheng, P. (2021). Mutation N501Y in RBD of spike protein strengthens the interaction between COVID-19 and its receptor ACE2. bioRxiv. 10.1101/2021.02.14.431117 [DOI] [PMC free article] [PubMed]
- Verkhivker, G. M. (2020). Molecular simulations and network modeling reveal an allosteric signaling in the SARS-CoV-2 spike proteins. Journal of Proteome Research, 19, 4587–4608. 10.1021/acs.jproteome.0c00654 [DOI] [PubMed] [Google Scholar]
- Verkhivker, G. M., & Di Paola, L. (2021). Dynamic network modeling of allosteric interactions and communication pathways in the SARS-CoV-2 spike trimer mutants: Differential modulation of conformational landscapes and signal transmission via cascades of regulatory switches. Journal of Physical Chemistry B, 125, 850–873. 10.1021/acs.jpcb.0c10637 [DOI] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 183, 1735. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y., Shang, J., Graham, R., Baric, R. S., & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. Journal of Virology, 94, e00127-20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Xu, X., Zhou, X., Chen, P., Liang, H., Li, X., Zhong, W., & Hao, P. (2020). Molecular simulation of SARS-CoV-2 spike protein binding to pangolin ACE2 or human ACE2 natural variants reveals altered susceptibility to infection. Journal of General Virology, 101, 921–924. 10.1099/jgv.0.001452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Liu, M., & Gao, J. (2020). Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proceedings of the National Academy of Sciences United States of America, 117, 13967–13974. 10.1073/pnas.2008209117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Schmidt, F., Weisblum, Y., Muecksch, F., Barnes, C. O., Finkin, S., Schaefer-Babajew, D., Cipolla, M., Gaebler, C., Lieberman, J. A., Oliveira, T. Y., Yang, Z., Abernathy, M. E., Huey-Tubman, K. E., Hurley, A., Turroja, M., West, K. A., Gordon, K., Millard, K. G., … & Nussenzweig, M. C. (2021). mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature, 592, 616–622. 10.1038/s41586-021-03324-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . (2020). Coronavirus disease 2019 (COVID-19) situation report , p. 52. https://apps.who.int/iris/handle/10665/331476
- Wibmer, C. K., Ayres, F., Hermanus, T., Madzivhandila, M., Kgagudi, P., Oosthuysen, B., Lambson, B. E., de Oliveira, T., Vermeulen, M., van der Berg, K., Rossouw, T., Boswell, M., Ueckermann, V., Meiring, S., von Gottberg, A., Cohen, C., Morris, L., Bhiman, J. N., & Moore, P. L. (2021). SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nature Medicine, 27, 622–625. 10.1038/s41591-021-01285-x [DOI] [PubMed] [Google Scholar]
- Wise, J. (2021). Covid-19: The E484K mutation and the risks it poses. BMJ, 372, n359. [DOI] [PubMed] [Google Scholar]
- Worldometer . (2020). Covid-19 coronavirus pandemic. Dadax Limited. https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1? [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C. L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367, 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X., Liu, Y., Liu, J., Zhang, X., Zou, J., Fontes-Garfias, C. R., Xia, H., Swanson, K. A., Cutler, M., & Cooper, D. (2021). Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nature Medicine, 27, 620–621. 10.1038/s41591-021-01270-4 [DOI] [PubMed] [Google Scholar]
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 367, 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, C., Sun, X., Ye, J., Ding, L., Liu, M., Yang, Z., Lu, X., Zhang, Y., Ma, L., Gu, W., Qu, A., Xu, J., Shi, Z., Ling, Z., & Sun, B. (2020). Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cellular & Molecular Immunology, 17, 621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., He, X., Man, V. H., Zhai, J., Ji, B., & Wang, J. (2021). Binding profile assessment of N501Y: A more infectious mutation on the receptor binding domain of SARS-CoV-2 spike protein. ChemRxiv. 10.26434/chemrxiv.13710961.v2 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.