Abstract
COVID-19 disease caused by the SARS-CoV-2 virus has shaken our health and wealth foundations. Although COVID-19 vaccines will become available allowing for attenuation of disease progression rates, distribution of vaccines can create other challenges and delays. Hence repurposed drugs against SARS-CoV-2 can be an attractive parallel strategy that can be integrated into routine clinical practice even in poorly-resourced countries. The present study was designed using knowledge of viral pathogenesis and pharmacodynamics of broad-spectrum antiviral agents (BSAAs). We carried out the virtual screening of BSAAs against the SARS-CoV-2 spike glycoprotein, RNA dependent RNA polymerase (RdRp), the main protease (Mpro) and the helicase enzyme of SARS-CoV-2. Imatinib (a tyrosine kinase inhibitor), Suramin (an anti-parasitic), Glycyrrhizin (an anti-inflammatory) and Bromocriptine (a dopamine agonist) showed higher binding affinity to multiple targets. Further through molecular dynamics simulation, critical conformational changes in the target protein molecules were revealed upon drug binding which illustrates the favorable binding conformations of antiviral drugs against SARS-CoV-2 target proteins. The resulting drugs from the present study in combination and in cocktails from the arsenal of existing drugs could reduce the translational distance and could offer substantial clinical benefit to decrease the burden of COVID-19 illness. This also creates a roadmap for subsequent viral diseases that emerge.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, drug repurposing, virtual screening, molecular dynamics simulation, SARS-CoV-2
Introduction
The coronavirus pandemic, also known as coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is creating a severe global health crisis which is one of the worst the world has faced since world war II. An unexplained virus, SARS-CoV-2, derived out of Wuhan, China in late 2019 (Yang et al., 2020; Zhou et al., 2020) and has since spread to every continent of the world with over 72,692,481 confirmed cases and over 1,619,801 confirmed deaths worldwide as of December 14, 2020 (www.WHO.org). Due to the lack of any demonstrable cure against this devastating virus, researchers around the globe are trying to develop or repurpose antiviral drugs through experimental and computational means to alleviate the pain of this pandemic.
SARS-CoV-2 the major pathogen of the COVID-19 illness is closely related (89.1%) to SARS-CoV and it’s a positive single-stranded RNA virus (+ssRNA) (StatPearls, 2021). The interaction of SARS-CoV-2 spike protein with the host receptor angiotensin-converting enzyme 2 (ACE2) drives the infection cycle of the virus by releasing the viral genome. The SARS-CoV-2 genome is comprised of variable number of 6-11 ORFs among which encodes the 16 non-structural proteins and other ORFs encode structural proteins including spike (S), envelope (E), membrane (M), and nucleocapsid (N) (Shereen et al., 2020). ORF1a and ORF1b contain a frameshift in between which produces two polypeptides: pp1a and pp1ab. These polypeptides are processed by virally encoded chymotrypsin-like protease (3CLpro) or main protease (Mpro) and one or two papain-like proteases into 16 NSPs (Mousavizadeh & Ghasemi, 2021). All NSPs perform their specific functions like NSP1 and NSP2 suppress the expression of host gene, formation of a multidomain complex by NSP3, NSP5 the main protease (Mpro) with critical role in replication form the multidomain complex (Stobart et al., 2013). Furthermore, the transmembrane proteins NSP4 and NSP6 (Wang et al., 2016) as well as NSP7 and NSP8 are primases (Te Velthuis et al., 2012). The dimeric form of NSP9 (an RNA-binding protein) is crucial for viral infection (Egloff et al., 2004). NSP10 acts as a cofactor for the activation of the replicative enzyme (Bouvet et al., 2014), NSP12 known as RNA-dependent RNA polymerase (Jiang et al., 2021) and NSP13 as helicase (Jang et al., 2020). While NSP14 and NSP15 shows exoribonuclease and endoribonuclease activities, respectively, and NSP16 with methyltransferase activity (Wang et al., 2016). NSP10-NSP16 complex is crucial to evade from the host immune system (Rosas-Lemus et al., 2020). Conclusively, all structural and nonstructural proteins critical for the virus entry, replication and transcription process may serve an important role from drug design perspectives.
Currently, there are no specific and appropriate antiviral therapy for the treatment of SARS-CoV-2 induced illness. Every country needs an immediate response and recovery from this crisis in this critical time, drug repurposing is an attractive approach with reduced time and cost to treat COVID-19. Broad spectrum antiviral agents (BSAAs) are small-molecules which may inhibit different types of human viruses that exploit similar pathways and host factors to replicate inside the cells. Instead of individual drug, combination of drugs could serve as front-line therapeutics for newly emerging viruses (SARS-CoV-2) and remerging viruses (Andersen et al., 2020).
The current study is focused on a drug repurposing strategy to find potential drug candidates suitable for targeting possible SARS-CoV-2 infection pathways. Targeting virus at entry and replication stage is attractive and credible strategy to treat covid-19. Therefore, spike (entry protein) and RdRp, Mpro and helicase (replication proteins) are selected for the present study as drug target. Through structure based virtual screening and molecular dynamics simulations. We screened all the existing BSAAs (those reported in multiclinical trials against COVID-19 and other approved, investigational and experimental) against SARS-CoV-2 druggable targets to iteratively explore the SARS-CoV-2-BSAAs interactions. The most probable binding mode for selected target-drug candidate from docking experiments were subjected to molecular dynamics (MD) simulations to rationalize the flexibility of their binding sites, mechanism of action of BSAAs and target molecules stability. With the aim to impact the COVID-19 outbreak, drug repurposing through virtual screening, molecular modeling and molecular dynamics (MD) simulation can facilitate and accelerate the search for appropriate antiviral drugs for SARS-CoV-2.
Materials and methods
Target dataset collection and optimization
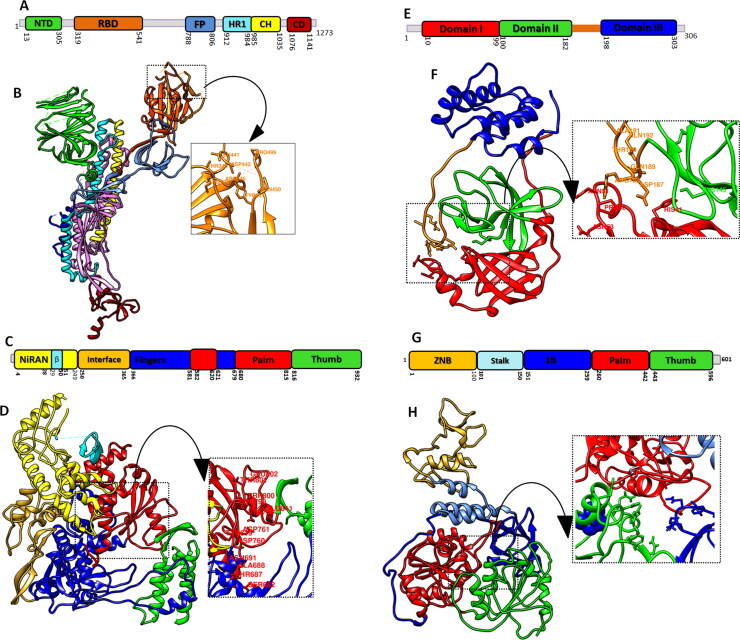
The recently resolved three-dimensional crystal structure of SARS-CoV-2 spike protein (PDB ID: 7BWJ) (Ju et al., 2020), RNA-Directed RNA polymerase (RdRp; PDB ID: 6M71) (Gao et al., 2020), main protease (Mpro) (PDB ID: 6LU7) (Jin et al., 2020), were retrieved from Protein Databank (PDB) (www.rcsb.org) and helicase protein (QHD43415_12) was retrieved from zhang lab (zhanglab.ccmb.med.umich.edu/COVID-19/). The structures of four target proteins were subjected to energy minimization (steepest descent steps of 100, with step size of 0.02, and with 10 conjugate gradient steps with step size of 0.02 (Å) followed by detachment of any ligand attached and nonstandard residues through UCSF Chimera (Pettersen et al., 2004) and Discovery Studio Viewer (Pazel, 1989). Structural features of selected target molecules have been elucidated along with binding site identification for inhibitor binding (Figure 1).
Figure 1.
Structural representation of SARS-CoV-2 selected target proteins: (A) Domain architecture of Spike protein homo-trimers (B) domain representation on 3 D structure whereas, N-terminal domain (NTD), receptor binding domain (RBD), fusion peptide (FP), heptad repeat (HR), central helix (CH) and connector domain (CD) and binding site residues are labeled (C) linear schematic description of domain architecture of RdRp domains and features followed by (D) three-dimensional structure of palm, thumb and fingers domains (E) Mpro of SARS-CoV2 schematics domain representation and (F) ribbon representation of Mpro composed of N-terminal domain I (red) domain II (green) and C-terminal domain III (blue) (G) and (H) domain architecture and ribbon representation of helicase protein of SARS-CoV2. The binding pockets of all the studied proteins are labelled and zoomed.
Electrostatic potential and pKa calculation
Electrostatic potential and pKa values for spike, RdRp, Mpro and helicase protein were calculated using Bluues Method (Walsh et al., 2012), that compute the electrostatic properties through generalized Born radii comparable to Poisson-Boltzmann equation. Electrostatic potential of the target proteins was estimated in molecular context. As a gauge of protein stability and function, pKa values for four target molecules were also calculated. Following parameters were used for electrostatic potential and pKa calculation: Solvent probe radius (Å):1.4 Å, Salt radius (Å): 2.0 Å, Inner dielectric constant: 4, outer dielectric constant: 78.5, and ionic strength:0.150 mol/L.
Virtual screening and molecular docking
BSAAs are small-molecules which may inhibit different types of human viruses that exploit similar pathways and host factors to replicate inside cells. In the present study a total of 872 BSAAs were retrieved from the DrugVirus.info database (https://drugvirus.info/). 3 D coordinates of all the BSAAs were obtained using Open Babel (https://sourceforge.net/projects/openbabel/) followed by energy minimization. The dataset of BSAAs were subjected to structure based virtual screening against selected set of receptors, spike protein, RdRp, Mpro and helicase using Autodock vina wizard of PyRx0.8 (Dallakyan & Olson, 2015). Default parameters in PyRx was used during docking except grid size and spacing. The grid size and spacing were set according to each target’s binding cavity (Table S1). The virtually screened best compounds were then docked with the target receptors again to ensure the conformation poses and binding affinities. The interactions were visualized using UCSF Chimera (Pettersen et al., 2004) and Discovery Studio (Pazel, 1989).
Table 1.
Electrostatic energies and potential of spike, RdRp, Mproand helicase.
| Born self-energy | Coulomb energy | Electrostatic solvation energy | Total energy | |||
|---|---|---|---|---|---|---|
| Spike | −38488.987079 (kJ) | −249351.024900 (kJ) | −4764.570203 (kJ/mol) | −248313.684306 (kJ/mol) | ||
| RdRp | −31424.949160 (kJ) | −215404.830067 (kJ) | −4012.361552 (kJ/mol) | −215602.297744 (kJ/mol) | ||
| Helicase | −23640.005076 (kJ) | −158501.639705 (kJ) | −701.425606 (kJ/mol) | −155943.188269 (kJ/mol) | ||
| Mpro | −11722.878621 (kJ) | −77686.237198 (kJ) | −1572.369138 (kJ/mol) | −77600.619632 (kJ/mol) | ||
| Generalized Born Radii | Surface potential | pKa shifts | ||||
| Residue | Score | Residue | Score | Residue | Score | |
| Spike | Thr874 | 19.424 | Leu977 | 14.32 | Arg457 | 13.534 |
| RdRp | Leu663 | 24.678 | Asp304 | 16.32 | Arg457 | 13.738 |
| Mpro | Met162 | 17.821 | Lys336 | 13.37 | Tyr161 | 12.632 |
| Helicase | Val372 | 18.976 | Lys347 | 24.96 | Arg579 | 13.582 |
Statistical analysis
Statistical analysis of the screening results was performed in Origin 2018. Box plot for four different groups with normal distribution was constructed. Histogram was constructed and linear curve fitting was performed to define the best fit model. Normal probability plot was constructed to assess the approximate normal distribution of four datasets. Cystoscope 3.8.2 (https://cytoscape.org/) was used to construct the drug-receptor network of top ten selected drugs for each target molecule. Network analyzer was used with following statistical parameters: clustering coefficient of 0.072, network diameter of 4 and radius of 1, network centrality of 0.286 and characteristic path length of 2.862.
Molecular dynamics simulations
Molecular dynamics (MD) simulations were performed for best ranked protein-ligand complexes for all studied targets(drug-protein) using GROMACS package version 5.0.5 (Van Der Spoel et al., 2005). PRODRG server (Schüttelkopf & van Aalten, 2004) was used to pick up the topologies of top binding affinity BSAAs. The parameters of all the target and drugs molecules were optimized by using the GROMOS96 54a7 force field (Schmid et al., 2011) having better propensities and free energy of hydration. Periodic cubic box equipped with solvent molecules was used for all the systems. For non-bonded interactions periodic boundary condition was used and similarly for strong electrostatic interactions particle mesh Ewald method was used (Essmann et al., 1995). Systems were neutralized with Na + Cl counter ions. The prepared systems in the periodic box containing protein, ligand, ions and solvent were minimized using a steepest-descent algorithm for 50,000 steps. NVT (number of atoms, volume, temperature) ensemble was used to add constraint to protein-ligand complex for 100ps during heating. NPT (constant pressure, constant temperature) ensemble was used at constant pressure (1 bar) and temperature (300 K) to simulate the systems for 100 ns. To the end of simulations all the systems were analyzed for stability and fluctuations using VMD (Humphrey et al., 1996), PyMol (http://www.pymol.org) and GROMACS tools.
MM-PBSA calculations
In biomolecular simulations binding free energy calculation is a state function based on predicted affinities of the receptor-ligand complex. Using Molecular mechanics Poisson-Boltzmann Surface Area (MM-PBSA) (Iqbal & Iqbal, 2014; Wrapp et al., 2020), binding free energy calculations were performed to measure the affinities of shortlisted drugs with respective target molecules. Overall, 100 frames from the trajectories were processed to estimate the net energy of the systems through the following equation,
All above mentioned terms involves the computation of several energy components: electrostatic energy, van der Waals energy and internal energy summed from molecular mechanics and polar contribution towards solvation energy. The analysis also takes into account the contribution from non-polar term towards solvation energy and inhibitor entropy. In the calculations of the MM-PBSA, the external dielectric constant was set to 80.0, the internal dielectric constant was 1.0 and the reciprocal grid spacing of 1.0 Å was used.
Results
Electrostatic potential and pKa energies of selected target proteins
Electrostatic potential helps to understand the contribution of electrostatic potential on structural aspects of the protein and protein-ligands interactions through analysis of positive and negative potential. pKa values of the protein determine its pH characteristics which is measure of protein flexibility and structural feature to accept or release the ligand.
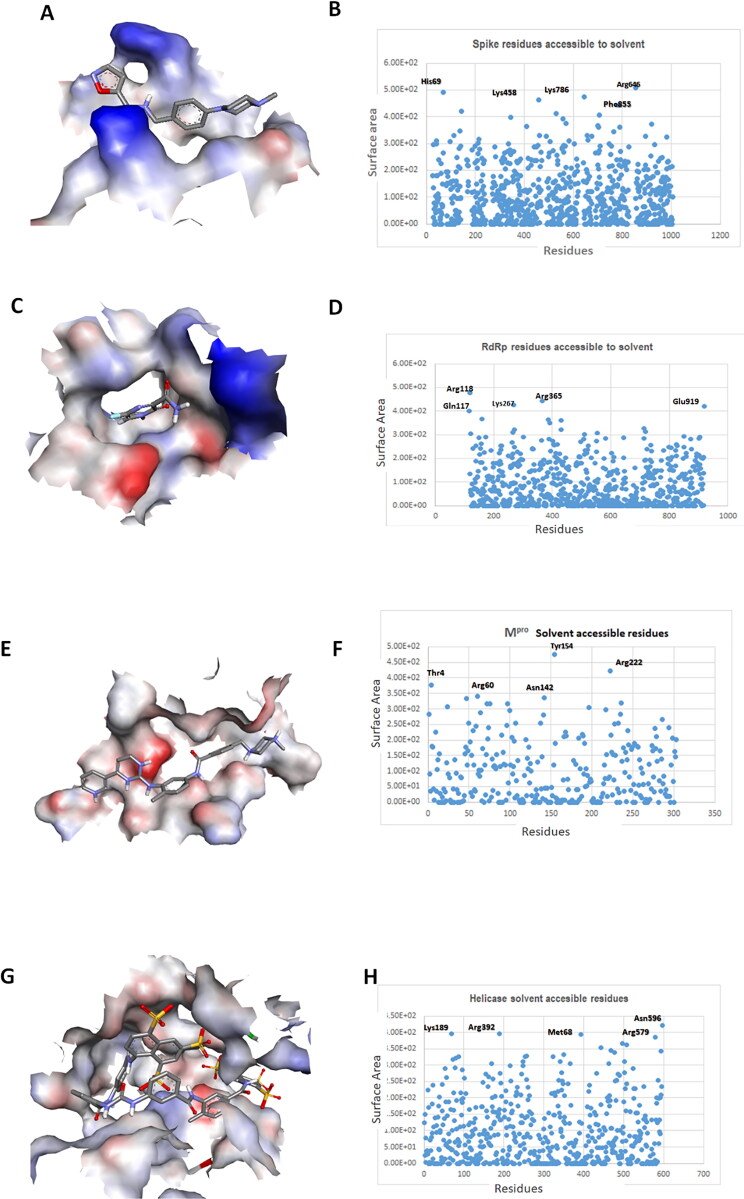
The spike protein of SARS-CoV-2 biding site showed more positive potential. His69, Lys458, Arg664, Lys786 and Phe855 are the highly solvent accessible residues of spike protein (Figure 2A and B). Thr874 is the most buried residue, H (hydrogen atom) is the most positive potential atom of protein belonging to Leu977 and the most positive pKa atom is CZ (carbon atom) belong to Arg457 (Table1). The electrostatic potential of binding site of RdRp enzyme of SARS-CoV-2, showed positive and negative potential. Gln117, Arg118, Lys267, Arg365 and Glu919 are the highly solvent accessible residues of RdRp (Figure 2C and D). HD12 is the most buried atom belong to Leu663, HB3 (hydrogen atom) is the most positive potential atom of RdRp belonging to Asp304 and the most positive pKa atom is CZ (carbon atom) belong to Arg457 (Table1). The electrostatic potential of Mpro binding site showed more negative potential. Tyr154, Arg222, Thr4, Arg60 and Asn142 are the most solvent accessible residues of the Mpro (Figure 2E and F) HE1 (hydrogen atom) is the most buried atom of Met162, HZ3 is the most positive potential atom of Lys336 and OH is the most positive pKa atom of Tyr161 (Table 1). Helicase enzyme binding cavity showed both positive and negative potential. Whereas, Asn596, Met68, Lys189, Arg392 and Arg579 are the highly solvent accessible residues (Figure 1G and H). CE2 is the most buried atom belong to residue Val372. NZ is the most positive potential atom belong to Lys347. CZ is the most positive pKa atom belong to Arg579 (Table 1).
Figure 2.
Electrostatic potential and pKa properties of spike, RdRp, Mpro and helicase: (A) Electrostatic energies distribution of spike protein. Blue color represents positive and red color negative energies charge potential of receptor binding site of spike protein, (B) surface point representing the residues and atom accessible to solvent, (C) RdRp, charge potential of catalytic binding pocket (D) most solvent accessible residues of RdRp (E) electrostatic potential and charge potential of Mpro binding cavity (F solvent accessible residues (G) helicase potential charge of binding pocket (H) solvent accessible residues.
Structure-based virtual screening of broad-spectrum antiviral agents
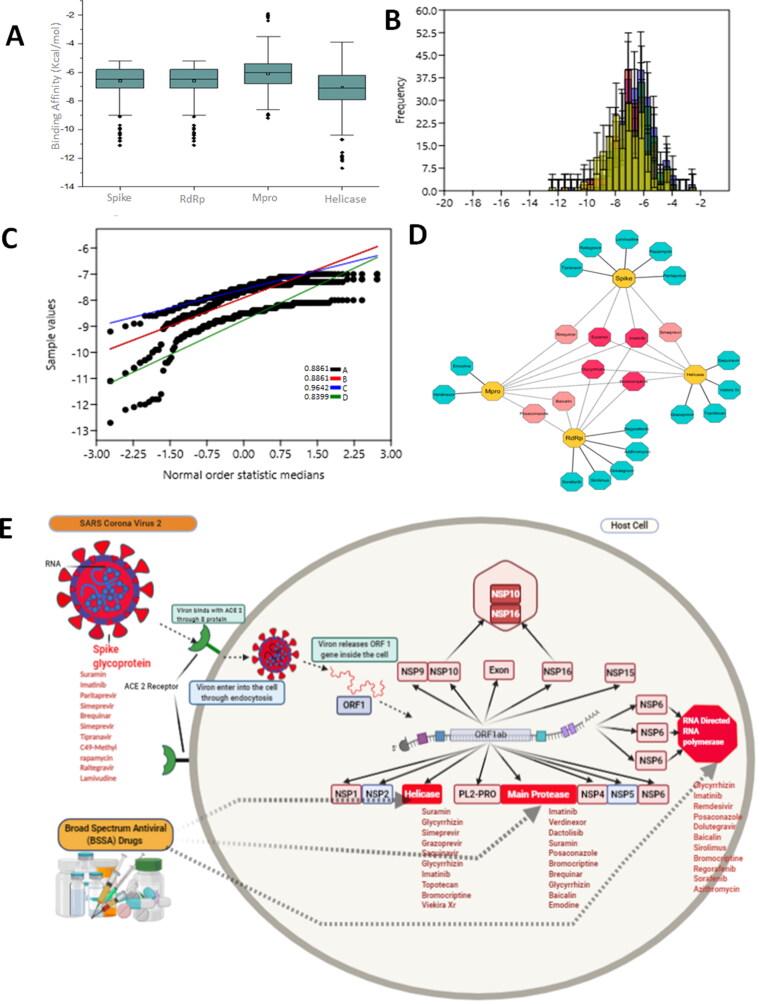
Structure based drug design strategy was used to screen the dataset of BSAAs (Table S1) selected against the promising druggable targets (spike, RdRp, Mpro and helicase) of SARS-CoV-2. High throughput screening using molecular docking approach resulted in broad range of BSAAs showing broad range of binding affinities towards four target molecules (Figure 3A–C), ranging from −1 to −13.0 kcal/mol. Among all BSAAs Imatinib showed strong binding affinity towards multiple targets molecules. Imatinib a cancer growth blocker showed high binding affinity towards RdRp, Mpro and spike protein of SARS-CoV-2. Suramin, a 100-year-old multifunctional drug with broad array of action against parasites to virus and cancer, showed the high binding affinity towards spike and helicase. Glycyrrhizin an anti-inflammatory drug and Bromocriptine showed affinity towards RdRp, Mpro and helicase (Figure 3D). Drugs targeting various stages of SARS-CoV-2 life cycle are shown in Figure 3E and Table 2. These finding from the present study can reduce the translational distance to repurpose the BSAAs against SARS-CoV-2 induced disease.
Figure 3.
Virtual screening of potential BSAAs against spike, RdRp, Mpro and helicase. (A) Box plot representation of binding affinities of BSAAs for four different groups with 95% confidence interval, the outliers are shown in black sphere (B) Histogram for four different groups with fit curve (C) Normal probability plot for spike, RdRp, Mpro and Helicase with correlation coefficient values. (D) Drug target interaction of top ten shortlisted drugs. Deep pink nodes represent drugs with more than 3 targets and light pink with two target molecules. While blue nodes represent those BSAAs that showed affinity towards a single target molecule.
Table 2.
Binding residues and types of interactions of BSAAs with SARS-CoV-2 proteins.
| S.NO | Proteins | Binding energies | Drug name | Interacting residues |
|---|---|---|---|---|
| 1 | Spike | −9.1 kcal/mol | Imatinib | TYR: 451, ASN: 440, ASN:450, GLU: 484, GLY:485, ASP: 442, ILE: 468, SER: 469, THR: 470, GLU: 471, ILE: 472 |
| −9.7 kcal/mol | Suramin | ASN: 440, LEU: 441, ARG: 346, TYR: 451, THR: 345, PHE: 347, ASN: 439, ASP: 442, SER: 443, LYS: 444, ASN: 448, ASN: 450, PRO: 499, ARG: 509 | ||
| 2 | RdRp | −12 kcal/mol | Glycyrrhizin | TYR: 619, ASN: 691, ASP: 760, ASP: 761, LYS: 798, GLU: 811, SER: 814, ILE: 589, ALA: 688, TRP: 617, CYS: 622, SER: 682, THR: 687, TYR: 689, SER:759, TRP: 800, PHE: 812, CYS: 813 |
| −8.8 kcal/mol | Imatinib | ALA: 797, CYS: 799, GLU:802, GLU: 811, HIS: 810, ASP: 706, ASP:761, GLU: 811, ASP: 618, LEU: 758, GLU: 796, LYS: 798, TRP: 800, THR: 801, CYS: 813, SER: 814 | ||
| 3 | Mpro | −9.0 kcal/mol | Verdinexor | LEU:27, HIS: 41, MET: 49, CYS: 145, MET: 165, THR: 26, GLN: 189, SER: 46, THR: 25, LEU: 27, VAL: 42, TYR: 54, ASN: 142, HIS: 164, GLU: 166, ASP: 187, ARG: 188 |
| −8.5 kcal/mol | Imatinib | ARG: 40, VAL: 186, CYS: 145, GLY:183, GLN: 192, HIS: 41, ASN: 84, CYS: 85, GLY: 179, ASN: 180, PHE: 181, PRO: 184, PHE: 185, ARG: 188, ALA:193 | ||
| 4 | Helicase | −11.6 kcal/mol | Suramin | ASN: 179, ASN: 516, ASP: 539, ARG: 560, TYR: 515, ARG: 560, PRO: 175, ARG: 178, TYR: 180, MET: 378, PRO: 406, PRO: 408, ARG: 409, THR: 410, LEU: 412, THR: 416 |
| −10.7 kcal/mol | Glycyrrhizin | ASN: 179 ASN: 516 ARG: 560, ASN: 177, ARG: 178, PRO: 406, PRO: 408, THR: 416, LEU: 417, ASN: 519, THR: 532, ASP: 534, SER: 535, ASN: 557 |
Molecular docking analysis of screened drugs
Potential drugs against spike protein
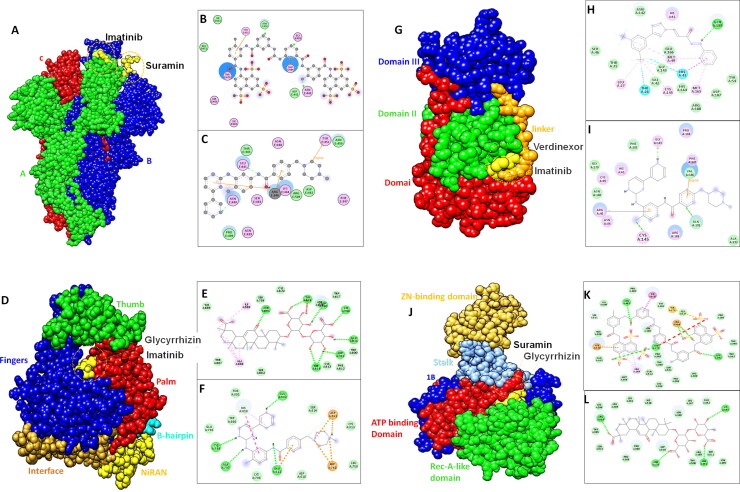
The forefront of SARS-CoV-2 infection is spike protein with important receptor binding domain (RBD) vital for host attachment (Assaad & Assaad-Khalil, 2020). As an ideal drug target for COVID-19 to stop the viral entry into host cell, we identified some BSAAs as potential drugs against spike-RBD to interfere the binding of virus with host receptor. Screening of BSAAs library resulted in broad range of inhibitors with binding energy ranging from −2.7 to −9.7 kcal/mol. Among all the BSAAs, Suramin showed binding affinity with binding free energy of −9.7 kcal/mol. Asn440, Asn450, Glu484 and Gly485 showed conventional hydrogen bond with the Suramin and Tyr451make pi-alkyl interaction along with number of hydrophobic interactions by Asp442, Ile468, Thr470, Glu471 and Ile472 (Figure 4A and B). Imatinib also showed good binding affinity with binding free energy of −9.1 kcal/mol. Imatinib is an anticancer drug (Chen et al., 1997) used for the treatment of covid-19 (Assaad & Assaad-Khalil, 2020). Binding interaction analysis revealed that Asn440 and Leu441made conventional hydrogen binding while two pi-alkyl interactions by Arg436 and one pi-sigma interaction by Tyr451 was observed (Figure 4C and Table 2). The best ten BSAAs for spike protein on the basis of binding energy and RMSD score are given in the table S2A along with mode of action and drug status.
Figure 4.
Binding modes of shortlisted BSAAs against spike, RdRp, Mpro and helicase. (A) surface representation of structural domains of spike protein with Suramin and Imatinib shown in yellow (B and C) represent 2 D interactions of Suramin and Imatinib with spike, drug molecule is shown in grey ball and stick with interacting residues shown in disc green dotted lines represents hydrogen bonding(D) Surface representation of RdRp structural domains represented in their respective colors (E and F) 2 D representation of molecular interactions of Glycyrrhizin and Imatinib with RdRp catalytic cavity. Green dotted lines represent hydrogen bond while yellow and pink dotted lines represent pi-anion and pi-Sulphur interactions. (G) Surface representation of Mpro domains and features with bound drugs (Verdinexor and Imatinib) in yellow color (H and I) representation of molecular interaction in 2 D form. (J) Surface representation of helicase with domain features in respective colors (K and L) 2 D representation of molecular interaction of Suramin and Glycyrrhizin with ATP binding domain of helicase enzyme.
Potential drugs against RdRP
RNA-dependent RNA polymerase (RdRP) is crucial for viral life cycle, and as catalytic site of the RdRp is the most conserved and accessible, so could be used as an attractive drug candidate to inhibit viral replication. Virtual screening of BSAAs against RdRp of SARS-CoV-2 showed high binding affinity of Glycyrrhizin with binding free energy of −12 kcal/mol. Imatinib also showed good binding affinity towards catalytic cavity of RdRp enzyme. Glycyrrhizin drug interaction with RdRp revealed binding within the palm domain conserved motif (Figure 4D). Asp618, Tyr618, Asn691, Asp760, Asp761, Lys798, Glu811 and Ser814 formed conventional hydrogen bond with the oxygen group of different functional moieties of Glycyrrhizin. Ile589 and Ala688 made alkyl bond. Whereas, Imatinib showed conventional carbon-hydrogen bond with Ala797, Cys799, Glu802 and Glu811 along with alkyl bond formed by Asp760 and Asp761 with N atom of drug (Figure 4E and F and Table2). The best screened antiviral drug along with status and therapeutic potential is given in supplementary Table S2B.
Potential drugs against Mpro
Main protease (Mpro) of SARS-CoV-2 is a promising drug target as it plays critical role in viral replication and transcription. Mpro showed high binding with Imatinib with binding free energy of −9.0 kcal/mol. Verdinexor, which is an investigational drug and has shown inhibition of influenza virus replication was bound with Mpro of SARS-CoV-2 with binding free energy of −8.5 kcal/mol. The detailed interaction analysis of high affinity drugs showed that Imatinib formed three conventional hydrogen bonds with the Cys145, Gly183 and Glu192, and pi-alkyl and pi-sigma interactions with the Arg40 and Val186 while number of hydrophobic interactions were observed with following residues Asn180, Phe181, Pro184, Pro185 and Ala193. Whereas, Verdinexor showed one conventional hydrogen bonding with the Gln189, His41 and Thr26 made halogen interactions, Met49, Cys145, Glu166 showed alkyl interactions. Apart from this number of hydrophobic interactions were observed with Asn142, Asp187 and Arg188 (Figure 4G–I and Table 2). The final screened best ten BSAAs with virtual screening high binding affinity score for Mpro have been provided in data file S2C.
Potential drugs against helicase
Nsp13 (helicase), is a multi-functional protein with metal binding N-terminal domain and C-terminal helicase domain. Due to essential role helicase in replication, transcription, and translation of SARS-COV-2 it is promising druggable target. In the virtual screening analysis, Suramin showed high binding affinity towards helicase with the binding free energy of 11.6 kcal/mol. Asn179, Asn516, Asp534 and Arg560 made conventional hydrogen bond with the Suramin. Apart from this Tyr515 and Arg560 formed pi-interactions along with number of hydrophobic interactions by Asn177, Arg178, Thr410, Leu417, Thr532, Gln537, His554 and Asn557. Apart from Suramin, Glycyrrhizin also returned good binding affinity with binding energy of −10.7 kcal/mol. Asn179, Asn516, Arg560 made conventional hydrogen bond along with number of hydrophobic interactions by Asn177, Arg178, Pro406, Pro408, Thr532, Asp534 and Asn557 (Figure 4J–L and Table 2). The final screened best ten BSAAs with virtual screening high binding affinity score for helicase have been provided in data file S2D Therapeutics implications of top screening drugs are also shown in the supplementary table.
Molecular dynamics simulations
To validate the conformational behavior of the best screened BSAAs bound with respective target protein molecule, GROMACS package (Pazel, 1989) was used for all atom MD simulations for 100 ns. We have taken following protein-drug conjugates; spike-Suramin, RdRp-glycyrrhizin, Mpro-Imatinib and helicase-Suramin along with their apo state to measure the conformational changes through comparative analysis.
Structural stability
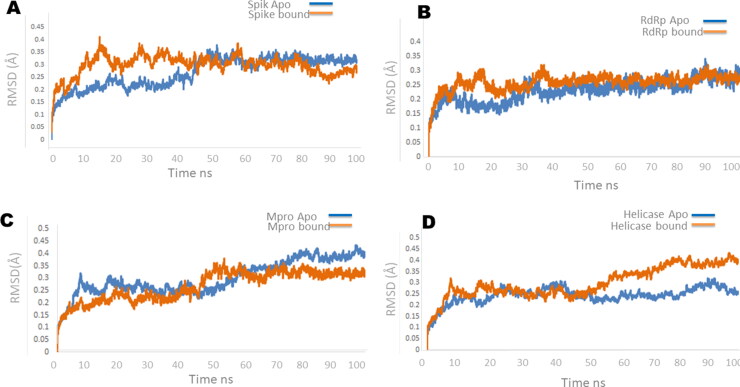
The stability of protein is the description of all the net forces to determine whether the protein will remain in folded state or assume non-native congregating structures. Therefore, the stability of protein is important to study the function of protein as alteration in protein stability would lead to misfolding or degradation of protein. Root Mean Square Deviation (RMSD) is the measure of conformational difference of the protein backbone from start to end state or comparison between the two poses of same molecule.
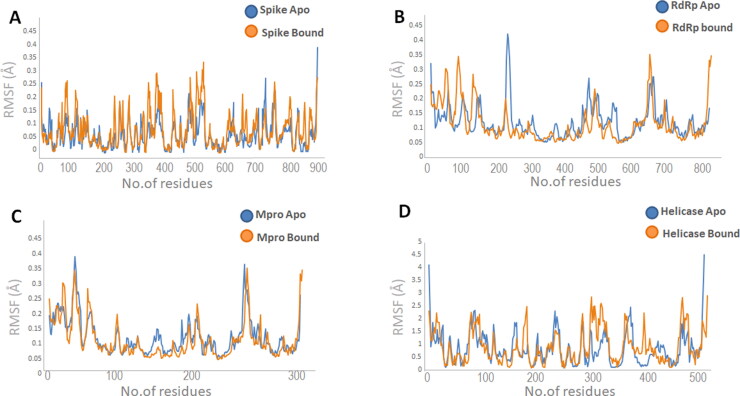
In spike protein apo protein RMSD staring from ∼0nm stabilize between ∼0.2-∼0.4 nm till 40 ns with little increase in stability to ∼0.35 and showed stability between ∼0.35 to∼0.4 for the rest of simulation experiment. In spike-Suramin complex an increased RMSD was observed from ∼0nm to ∼0.4 in the first 40 ns of the simulations and stabilize between ∼0.35 to ∼0.4 for the remaining simulations experiments (Figure 5A). In RdRp apo and bound state both the systems showed RMSD variation in the first 20 ns of simulations and get stabilize between ∼0.2 to ∼0.3 throughout the simulations (Figure 5B). RMSD measures of Mpro apo and bound state (Mpro-Imatinib) showed stability in the RMSD value from start of simulations till 40 ns. The system showed increase in RMSD value from ∼0.2-∼0.35 nm between 45 ns-55ns and for rest of simulations system get stabilized (Figure 5C). RMSD measure of helicase apo and bound state showed increased RMSD value in the start of simulation and different behavior throughout experiment. Helicase apo starting from ∼0nm stabilize between ∼0.2-∼0.25 nm till 20 ns and showed increased RMSD of ∼0.25 nm between 30-40ns and get stabilize rest of the time. In helicase bound state (helicase-Suramin complex) the value of RMSD increased in the beginning form ∼0nm to ∼0.3 nm and get stabilize between ∼0.2-∼0.25 nm for 50 ns of simulations and increased stability was monitored between 0.25 to 0.4 nm for rest of the simulations (Figure 5D). Binding pocket RMSD of each target molecule Spike, RdRp, Mpro and helicase is calculated as shown in Table S3. Root mean square fluctuation (RMSF) plot of spike, RdRp, Mpro and helicase showed overall stability in the region encompassing the binding cavity (Figure 6A–D).
Figure 5.
Root mean square deviation plot of the backbone atoms calculated by MD simulations for (A)spike, (B) RdRp, (C) Mpro (D) Helicase. Blue curves represent apo and orange curves represents bound state of four target proteins.
Table 3.
The calculated MM-PBSA binding free energies of Spike, RdRp. Mpro and helicase complexes.
| Protein ligand complex | MM-PBSA (kcal/mol) | Electrostatic energy (kcal/mol) | ΔG Bind vdWc (kcal/mol) | ΔG Solv GBd (kcal/mol) |
|---|---|---|---|---|
| Spike -Suramine | −12.36 | −10.452 | −20.962 | 8.087 |
| RdRp-Glycyrrhizin | −13.54 | −12.083 | −24.326 | 9.442 |
| Mpro-Imatinib | −11.25 | −9.301 | −20.322 | 7.330 |
| Helicase-Suramin | −9.28 | −8.567 | −18.231 | 6.550 |
Figure 6.
Root mean square fluctuation plot of (A) spike, (B) RdRp, (C) Mpro (D) Helicase calculated through MD simulations. Blue curves represent apo and orange curves represents bound state of four target proteins.
Conformational adjustments of spike, RdRp, mpro and helicase upon drug binding
SARS-CoV-2 spike
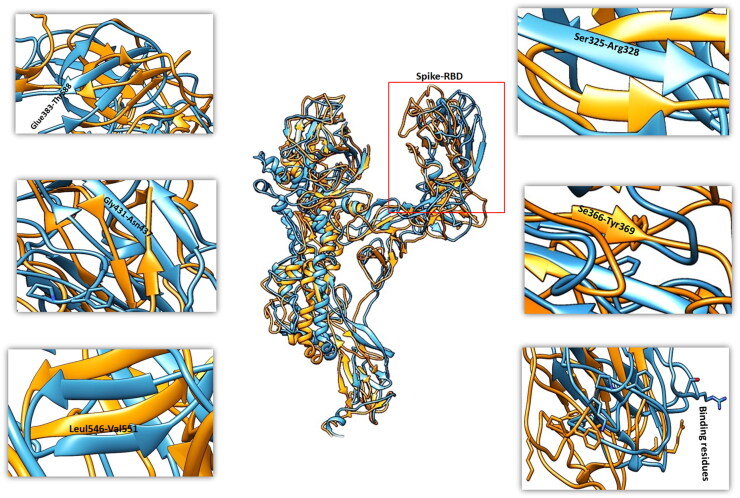
Structural changes were captured for the spike protein at different time intervals upon binding of Suramin compound. A loop region in apo state (residue range: Ser366-Tyr369) was converted to β-sheet in bound state of spike protein RBD region. Gly431-Asn437 a β-sheet in apo form was converted to loop in bound sate. Tyr508 and Arg509 the part of β-sheet in apo form was changed to loop in bound sate of spike protein. Leu546-Val551 was extended β-sheet observed in bound sate and Ser325-Arg328, β-sheets shortening was monitored in bound sate in comparison to apo state of spike protein. The Suramin bound spike protein showed significant fluctuation to aid in binding (Figure 7). Significant conformational adjustments were observed in and around the RBD of spike protein at secondary structure and residue level that are main determinant of interactions. The analysis of spike protein conformational adjustments upon binding of small molecule inhibitor suggests that it might reduce chances of binding with host receptor and will not trigger the signaling cascade for viral infection.
Figure 7.
Fluctuation of MD trajectories for spike-apo (blue ribbon), and spike-bound (orange ribbon).
RdRp SARS-CoV-2
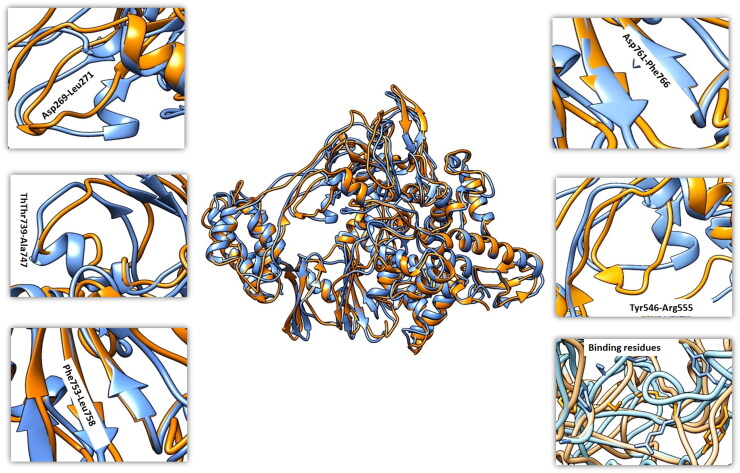
The dynamic trajectories of apo and drug bound state of RdRp enzyme revealed significant conformational changes. In unbound state there are 31 alpha helices of RdRp and loss of two helices were observed at Asp269-Leu271 and Thr739-Ala747 in bound form of RdRp. Apart from these alterations in helix length was also observed throughout the protein. Overall, the topology of β-sheet showed minor fluctuation with increase in sheet length at few places (residue range: Phe753-Leu758, Asp761, Phe766) (Figure 8). Most of the conformational changes were observed in the palm, thumb, and finger motifs that are main contributor of RdRp active site for drug binding. So, these predictions conclude that binding of BSAAs in the active site will not allow the binding of RNA and will stop RNA synthesis. These results are consistent with Weisberg et al. (2020) and Al-Kamel and Grundmann (2021). Moreover, the docking analysis of shortlisted BSAAs and RNA with RdRp revealed strong binding of drugs with binding free energy of -12kcal/mol and −8.8 kcal/mol as compared to −7.8 kcal/mol binding free energy of RMA molecule (Figure S1).
Figure 8.
Fluctuation of MD trajectories for RdRp-apo (blue ribbon), and RdRp-bound (orange ribbon).
Mpro SARS-CoV-2
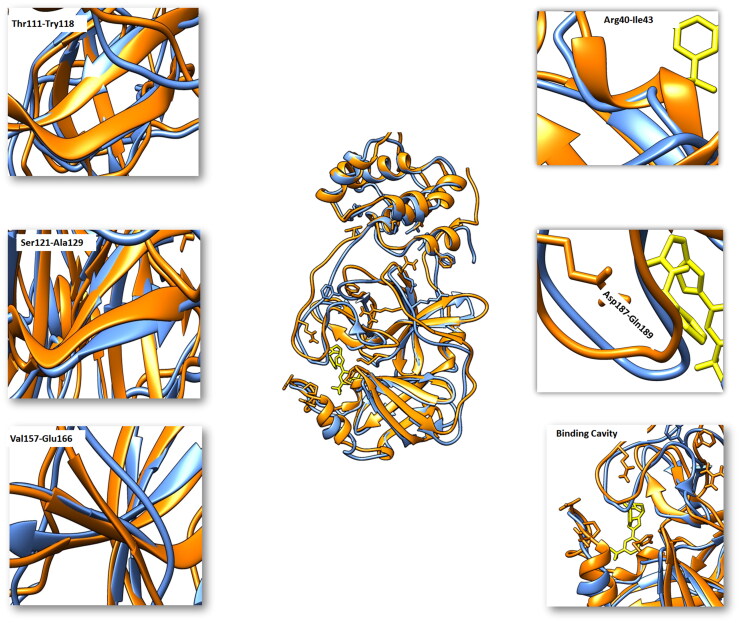
Comparative analysis of the trajectories generated at different ns (Mpro-Imatinib) with apo state of Mpro showed important structural twists. A short β-sheet in apo state was extended four residues long sheet from Thr111-Tyr118. Another short β-sheet in apo state is extended by five residues long sheet from Ser121 to Ala129 in bound state. A loop region in the apo state is converted to β-sheet comprising of residues ranging from Val157-Glu166. A loop region in unbound state encompassing Arg40-Ile43 was converted to an extended helix in the bound sate of Mpro. Asp187-Gln189 a loop region at the cavity mouth was moved upward to widen the cavity opening. His41, Tyr54, Asn142 and Cys145 a loop region was tilted towards the binding cavity to make covalent interactions with the Imatinib (Figure 9).
Figure 9.
Fluctuation of MD trajectories for Mpro-apo (blue ribbon), and Mpro-bound (orange ribbon).
Helicase SARS-CoV-2
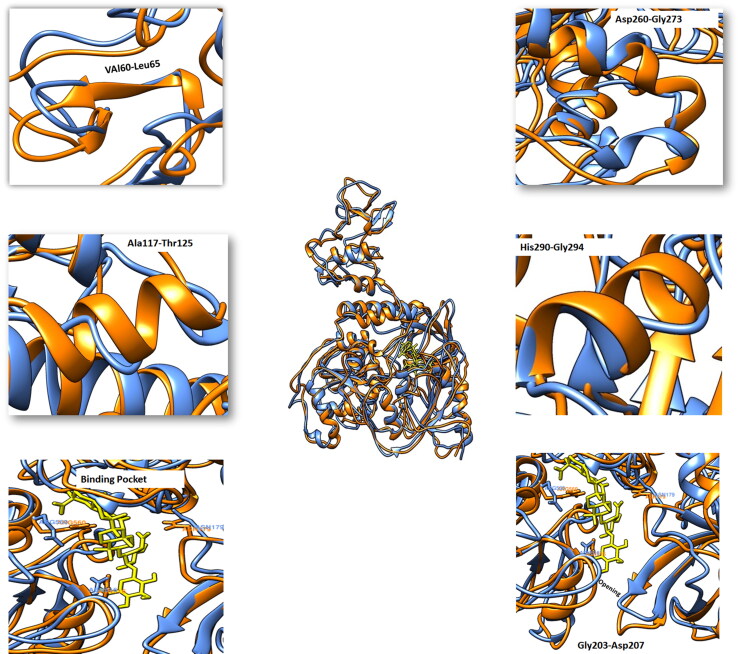
Comparative analysis of the trajectories generated at different ns with helicase apo and helicase bound (helicase-Suramin) state showed significant structural turns. Some secondary structure elements were built and while the few loss of regular secondary structure was observed in bound state in comparison to unbound form. From Val60-Leu65 a loop region a loop region in apo state was converted to short β-sheet and up to nine residues long helix was created form Ala117-Thr125 as compared to loop region in the apo state. Two short helices with inter loop region of seven residues was converted to one long helix from Asp260-Gly273 in bound state. Helix6 was extended form His290 to Gly294 in bound state as compared to apo state. A loop at the cavity opening encompassing Asp204-Asp207 was extended to open the mouth of cavity. Asn179, Asn516 and Arg560 showed interesting conformational switches towards the cavity to aid in hydrogen bonding with drug molecule (Figure 10).
Figure 10.
Fluctuation of MD trajectories for helicase-apo (blue ribbon), and helicase-bound (orange ribbon).
MM-PBSA calculations
After molecular dynamics simulations trajectories for all the protein ligand complexes from last 10 ns were extracted to calculate the binding free energy MM-PBSA. While the Suramin bound with spike protein depicted the binding energy of −12.63 kcal/mol, similarly the MM-PBSA energy calculation of Glycyrrhizin in complex with RdRp enzyme revealed relatively higher binding free energy of −13.54 kcal/mol. Imatinib in complex with Mpro showed binding energy of −11.25 kcal/mol, while Suramin bound to helicase showed binding free energy of −9.28 kcal/mol (Table 3).
Discussion
The COVID-19 disease caused by the SARS-CoV-2 virus has disrupted modern global infrastructure. The development of rapid and effective antiviral agents is remarkably challenging due to evident cost and time-consuming nature. In this era of COVID-19 mostly public discussion is centered on vaccine, but vaccine use may not defeat the virus completely especially as mutant forms emerge which are not responsive to the vaccines (https://www.ft.com/content/c2aa5ea4-66b9-4f64-9e74-7c89c12f9461), as well as vaccine distribution challenges. Drug repurposing is an alternate approach to expedite the identification of potential drugs for rapid management of emerging and remerging infections and offer an immediate integration into routine clinical practice.
The present study is designed for expeditious screening and identification of BSAAs against spike, RdRp, Mpro and helicase of the SARS-CoV-2 to combat the diseases at various stages. Initial screening showed that Imatinib, Suramine, Glycyrrhizin and Bromocriptine have affinity towards multiple targets. Remdesivir and Azithromycin already approved by FDA and NIH for COVID-19 treatment also came up among the top ten high binding affinity drugs for RdRp that further strengthen our strategy. Suramin is approved antiprotozoal drug with broad spectrum antiviral activity against Zika virus, Ebola virus, Hepatitis C virus and Human influenza virus. Suramin also inhibits the binding of dengue virus to host cells through a direct effect on the viral envelope protein (Chen et al., 1997). Inhibition of host cell attachment was also found for herpes simplex (Aguilar et al., 1999) and hepatitis C (Garson et al., 1999) viruses, which explained the previously reported protective effects of Suramin against in vitro herpes simplex virus infections (Alarcón et al., 1984) and in vivo infections of ducks with duck hepatitis B virus (Offensperger et al., 1993). Imatinib is an approved anticancer drug also known as ABL1-Tyrosine kinase inhibitor of Abelson murine leukemia virus oncogene 1 (ABL1) pathway which is critical for many viral replications. In vitro studies confirmed the role of Imatinib activity against MERS-CoV and SARS-CoV by inhibiting the kinase signaling pathway which is important for replication (Dyall et al., 2014). Imatinib have the potential to become broad-spectrum antivirals for the treatment of SARS-CoV-2 as a number of clinical trials demonstrated that Imatinib will interfere with release or replication of SARS-CoV-2 and would have significant impact on COVID-19 patients in intensive care units (https://clinicaltrials.gov/ct2/show/NCT04394416). Imatinib showed affinity towards all targets (Spike, RdRp, Mpro and Helicase), and most preferable residues were Asn, Asp, Arg and Thr. Glycyrrhizin was reported as the most active drug inhibiting replication of the SARS-CoV (Numazaki, 2003) and can be repurposed to stop SARS-CoV-2 replication. Bromocriptine is dopamine receptor agonist and can be used for the treatment of diabetes (DeFronzo, 2011). Fumihiro Kato in 2016 and Chan JF et al in 2017 reported the antiviral activity of Bromocriptine against dengue virus and zika virus, respectively (Chan et al., 2017; Kato et al., 2016). Instead of individual drug, combination of drugs against viruses could serve as front line therapeutics for newly emerging viruses (SARS-CoV-2) and remerging viruses (Bibi et al., 2020; Shyr et al., 2020). Therefore, the cocktail of antiviral drugs from the present study with explicit antiviral effect can be used for various stages of SARS-CoV-2 infection. Due to the lack of experimental support, there are limitations in validating these in silico proposed work. However, the data provide support for experimental validation. Combination of drugs that can attack the virus differently through different mechanism can be an optimum approach.
Conclusion
In conclusion BSAAs revealed to have potential against SARAS-CoV-2 as antiviral agents. Particularly, Imatinib, Glycyrrhizin Bromocriptine, Suramin revealed good affinity towards multiple targets. MD simulations of shortlisted high biding affinity drugs against Spike, RdRp, Helicase and Mpro revealed a formation of stable complex throughout the simulations. RMSD and RMSF analysis revealed that the interaction of shortlisted BSAAs with respective target were very stabilizing. Therefore, BSAAs could offer significant clinical benefit to decrease the burden of COVID-19 illness. These molecules can either be used as main drug or combination as multitargeted therapy against SARS-CoV-2.
Future prospects
Due to the speedy outburst of the SARS-CoV-2 and high mortality rate, there is burning needs to control this highly transmissible disease either by novel therapeutic development or repurposing the existing antivirals to inhibit this virus are essential. Throughout the world several clinical trials are running by targeting structural and/or nonstructural proteins of SARS-CoV-2 to find efficient drug. The future goal of this study is to complete clinical trial of shortlisted BSAAs and find the most effective antiviral drugs for SARS-CoV-2 infection.
Supplementary Material
Acknowledgements
The schematic figure was created in BioRender.com.
Authors contribution
NB: conceived and designed the experiments, performed experiments and manuscript writing, AF and SG: made a significant contribution by performing virtual screening experiments. FA, JA, UK and TH: made a substantial contribution in revising the manuscript for intellectual content.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data and material availability
The data supporting the findings of the article are available in the Protein Data Bank (PDB) at http://www.rcsb.org. The data needed to evaluate the article is present in the article and supplementary material.
References
- Aguilar, J. S., Rice, M., & Wagner, E. K. (1999). The polysulfonated compound Suramin blocks adsorption and lateral difusion of herpes simplex virus type-1 in vero cells. Virology, 258(1), 141–151. 10.1006/viro.1999.9723 [DOI] [PubMed] [Google Scholar]
- Alarcón, B., Lacal, J. C., Fernández-Sousa, J. M., & Carrasco, L. (1984). Screening for new compounds with antiherpes activity. Antiviral Research, 4(5), 231–244. 10.1016/0166-3542(84)90029-9 [DOI] [PubMed] [Google Scholar]
- Al-Kamel, H., & Grundmann, O. (2021). Glycyrrhizin as a potential treatment for the novel coronavirus (COVID-19). Mini Reviews in Medicinal Chemistry, 21(16), 2204-2208. 10.2174/1389557521666210210160237 [DOI] [PubMed] [Google Scholar]
- Andersen, P. I., Ianevski, A., Lysvand, H., Vitkauskiene, A., Oksenych, V., Bjørås, M., Telling, K., Lutsar, I., Dumpis, U., Irie, Y., Tenson, T., Kantele, A., & Kainov, D. E. (2020). Discovery and development of safe-in-man broad-spectrum antiviral agents. International Journal of Infectious Diseases, 93, 268–276. 10.1016/j.ijid.2020.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad, H. S., & Assaad-Khalil, S. (2020). Imatinib a Tyrosine Kinase Inhibitor: A potential treatment for SARS- COV-2 induced pneumonia. Alexandria Journal of Medicine, 56(1), 68–72. 10.1080/20905068.2020.1778417 [DOI] [Google Scholar]
- Bibi, N., Gul, S., Ali, J., & Kamal, M. A. (2020). Viroinformatics approach to explore the inhibitory mechanism of existing drugs repurposed to fight against COVID-19. European Journal of Pharmacology, 885, 173496. 10.1016/j.ejphar.2020.173496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet, M., Lugari, A., Posthuma, C. C., Zevenhoven, J. C., Bernard, S., Betzi, S., Imbert, I., Canard, B., Guillemot, J. C., Lécine, P., Pfefferle, S., Drosten, C., Snijder, E. J., Decroly, E., & Morelli, X. (2014). Coronavirus Nsp10, a critical co-factor for activation of multiple replicative enzymes. The Journal of Biological Chemistry, 289(37), 25783–25796. 12 10.1074/jbc.M114.577353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F., Chik, K. K., Yuan, S., Yip, C. C., Zhu, Z., Tee, K. M., Tsang, J. O., Chan, C. C., Poon, V. K., Lu, G., Zhang, A. J., Lai, K. K., Chan, K. H., Kao, R. Y., & Yuen, K. Y. (2017). Novel antiviral activity and mechanism of Bromocriptine as a Zika virus NS2B-NS3 protease inhibitor. Antiviral Research, 141, 29–37. 10.1016/j.antiviral.2017.02.002 (2017). [DOI] [PubMed] [Google Scholar]
- Chen, Y., Maguire, T., Hileman, R. E., Fromm, J. R., Esko, J. D., Linhardt, R. J., & Marks, R. M. (1997). Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nature Medicine, 3(8), 866–871. 10.1038/nm0897-866 [DOI] [PubMed] [Google Scholar]
- Dallakyan, S., & Olson, A. (2015). Small-molecule library screening by docking with PyRx. Methods in Molecular Biology (Clifton, N.J.), 1263, 243–250. [DOI] [PubMed] [Google Scholar]
- DeFronzo, R. A. (2011). Bromocriptine: A sympatholytic, D2-dopamine agonist for the treatment of type 2 diabetes. Diabetes Care, 34(4), 789–794. 10.2337/dc11-0064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall, J., Coleman, C. M., Hart, B. J., Venkataraman, T., Holbrook, M. R., Kindrachuk, J., Johnson, R. F., Olinger, G. G., Jr, Jahrling, P. B., Laidlaw, M., Johansen, L. M., Lear-Rooney, C. M., Glass, P. J., Hensley, L. E., & Frieman, M. B. (2014). Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrobial Agents and Chemotherapy, 58(8), 4885–4893. 10.1128/AAC.03036-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff, M. P., Ferron, F., Campanacci, V., Longhi, S., Rancurel, C., Dutartre, H., Snijder, E. J., Gorbalenya, A. E., Cambillau, C., & Canard, B. (2004). The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proceedings of the National Academy of Sciences of the United States of America, 101(11), 3792–3796. 10.1073/pnas.0307877101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., Yang, X., … Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, N.Y.), 368(6492), 779–782. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garson, J. A., Lubach, D., Passas, J., Whitby, K., & Grant, P. R. (1999). Suramin blocks hepatitis C binding to human hepatoma cells in vitro. Journal of Medical Virology, 57(3), 238–242. 10.1002/(SICI)1096-9071(199903)57: [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Iqbal, N., & Iqbal, N. (2014). Imatinib: A breakthrough of targeted therapy in cancer. Chemotherapy Research and Practice, 2014, 357027. 10.1155/2014/357027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, K. J., Jeong, S., Kang, D. Y., Sp, N., Yang, Y. M., & Kim, D. E. (2020). A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Scientific Reports, 10(1), 4481. 10.1038/s41598-020-61432-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., Yin, W., & Xu, H. E. (2021). RNA-dependent RNA polymerase: Structure, mechanism, and drug discovery for COVID-19. Biochemical and Biophysical Research Communications, 538, 47–53. S0006-291X(20)31721-31726) 10.1016/j.bbrc.2020.08.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. June 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Ju, B., Zhang, Q., Ge, J., Wang, R., Sun, J., Ge, X., Yu, J., Shan, S., Zhou, B., Song, S., Tang, X., Yu, J., Lan, J., Yuan, J., Wang, H., Zhao, J., Zhang, S., Wang, Y., Shi, X., … Zhang, L. (2020). Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature, 584(7819), 115–119. 10.1038/s41586-020-2380-z [DOI] [PubMed] [Google Scholar]
- Kato, F., Ishida, Y., Oishi, S., Fujii, N., Watanabe, S., Vasudevan, S. G., Tajima, S., Takasaki, T., Suzuki, Y., Ichiyama, K., Yamamoto, N., Yoshii, K., Takashima, I., Kobayashi, T., Miura, T., Igarashi, T., & Hishiki, T. (2016). Novel antiviral activity of Bromocriptine against dengue virus replication. Antiviral Research, 131, 141–147. 10.1016/j.antiviral.2016.04.014 [DOI] [PubMed] [Google Scholar]
- Mousavizadeh, L., & Ghasemi, S. (2021). Genotype and phenotype of COVID-19: Their roles in pathogenesis. Journal of Microbiology, Immunology, and Infection = Wei Mian yu Gan Ran za Zhi, 54(2), 159–163. 10.1016/j.jmii.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numazaki, K. (2003). Glycyrrhizin therapy for viral infections. African Journal of Biotechnology, 2(10), 392–393. [Google Scholar]
- Offensperger, W. B., Offensperger, S., Walter, E., Blum, H. E., & Gerok, W. (1993). Suramin prevents duck hepatitis B virus infection in vivo. Antimicrobial Agents and Chemotherapy, 37(7), 1539–1542. 10.1128/aac.37.7.1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazel, D. P. (1989). DS-Viewer- interactive graphical data structure presentation facility. IBM Systems Journal, 28(2), 307–323. in 10.1147/sj.282.0307 [DOI] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Rosas-Lemus, M., Minasov, G., Shuvalova, L., Inniss, N. L., Kiryukhina, O., Wiersum, G., Kim, Y., Jedrzejczak, R., Maltseva, N. I., Endres, M., Jaroszewski, L., Godzik, A., Joachimiak, A., & Satchell, K. (2020). The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine. bioRxiv: the preprint server for biology. 10.1101/2020.04.17.047498 [DOI] [Google Scholar]
- Schmid, N., Eichenberger, A., Choutko, A., Riniker, S., Winger, M., Mark, A., & van Gunsteren, W. (2011). Definition and testing of the GROMOS force-field versions 54A7 and 54B7. European Biophysics Journal : EBJ, 40(7), 843–856. 10.1007/s00249-011-0700-9 [DOI] [PubMed] [Google Scholar]
- Schüttelkopf, A. W., & van Aalten, D. M. F. (2004). PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallographica. Section D, Biological Crystallography, 60(Pt 8), 1355–1363. 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- Shereen, M. A., Khan, S., Kazmi, A., Bashir, N., & Siddique, R. (2020). COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research, 24, 91–98. 10.1016/j.jare.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyr, Z. A., Gorshkov, K., Chen, C. Z., & Zheng, W. (2020). Drug discovery strategies for SARS-CoV-2. The Journal of Pharmacology and Experimental Therapeutics, 375(1), 127–138. 10.1124/jpet.120.000123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- StatPearls . (2021). Features, Evaluation and Treatment Coronavirus (COVID19). https://www.ncbi.nlm.nih.gov/books/NBK554776/. [PubMed]
- Stobart, C. C., Sexton, N. R., Munjal, H., Lu, X., Molland, K. L., Tomar, S., Mesecar, A. D., & Denison, M. R. (2013). Chimeric exchange of coronavirus nsp5 proteases (3CLpro) identifies common and divergent regulatory determinants of protease activity. Journal of Virology, 87(23), 12611–12618. 10.1128/JVI.02050-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis, A. J., van den Worm, S. H., & Snijder, E. J. (2012). The SARS-coronavirus nsp7 + nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Research, 40(4), 1737–1747. 10.1093/nar/gkr893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. C. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Walsh, I., Minervini, G., Corazza, A., Esposito, G., Tosatto, S. C. E., & Fogolari, F. (2012). Bluues server: Electrostatic properties of wild-type and mutated protein structures. Bioinformatics (Oxford, England), 28(16), 2189–2190. Pages 10.1093/bioinformatics/bts343 [DOI] [PubMed] [Google Scholar]
- Wang, H., Xue, S., Yang, H., & Chen, C. (2016). Recent progress in the discovery of inhibitors targeting coronavirus proteases. Virologica Sinica, 31(1), 24–30. 10.1007/s12250-015-3711-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg, E., Parent, A., Yang, P. L., Sattler, M., Liu, Q., Liu, Q., Wang, J., Meng, C., Buhrlage, S. J., Gray, N., & Griffin, J. D. (2020). Repurposing of kinase inhibitors for treatment of COVID-19. Pharmaceutical Research, 37(9), 167 10.1007/s11095-020-02851-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Peng, F., Wang, R., Yange, M., Guan, K., Jiang, T., Xu, G., Sun, J., & Chang, C. (2020). The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. Journal of Autoimmunity, 109, 102434–102434. 10.1016/j.jaut.2020.102434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., & Huang, C.-L. (2020). Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv, 2020.01.22.914952. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of the article are available in the Protein Data Bank (PDB) at http://www.rcsb.org. The data needed to evaluate the article is present in the article and supplementary material.