Abstract
Background
Accurate microbiologic diagnosis is important for appropriate management of infectious diseases. Sequencing-based molecular diagnostics are increasingly used for precision diagnosis of infections. However, their clinical utility is unclear.
Methods
We conducted a retrospective analysis of specimens that underwent 16S ribosomal RNA (rRNA) gene polymerase chain reaction (PCR) followed by Sanger sequencing at our institution from April 2017 through March 2019.
Results
A total of 566 specimens obtained from 460 patients were studied. Patients were considered clinically infected or noninfected based on final diagnosis and management. In 17% of patients, 16S rRNA PCR/sequencing was positive and in 5% of patients, this test led to an impact on clinical care. In comparison, bacterial cultures were positive in 21% of patients. Specimens with a positive Gram stain had 12 times greater odds of having a positive molecular result than those with a negative Gram stain (95% confidence interval for odds ratio, 5.2–31.4). Overall, PCR positivity was higher in cardiovascular specimens (37%) obtained from clinically infected patients, with bacterial cultures being more likely to be positive for musculoskeletal specimens (P < .001). 16S rRNA PCR/sequencing identified a probable pathogen in 10% culture-negative specimens.
Conclusion
16S rRNA PCR/sequencing can play a role in the diagnostic evaluation of patients with culture-negative infections, especially those with cardiovascular infections.
Keywords: molecular diagnostics, 16S rRNA gene PCR, Sanger sequencing, broad range bacterial PCR, bacterial infections
Our study suggests the 16S ribosomal RNA gene polymerase chain reaction and Sanger sequencing is a clinically useful test and can play an important role in diagnostic evaluation of patients with culturenegative infections, especially with cardiovascular infections.
Accurate identification of microbial pathogens informs targeted therapy, and ultimately increases the likelihood of favorable clinical outcomes [1]. Standard identification of bacteria in clinical specimens involves Gram stain, followed by growth of organisms using appropriate culture media. Despite using standard microbiologic practices and testing, in patients with high underlying clinical suspicion of infection, cultures often remain negative. This may be related to prior antimicrobial therapy or the inability of fastidious organisms to grow on standard culture media [2–5]. Lack of microbiologic diagnosis in patients with a high index of suspicion for infection often leads to use of broad-spectrum antimicrobial therapy, which results in increased risk of selection for antimicrobial resistance and other antibiotic-associated side effects and does not invariably “cover” the pathogen present [6–8].
In the last decade, sequencing-based molecular diagnostics have been increasingly used in complex cases to overcome limitations of culture-based diagnostics. One such tool, based on amplification and sequencing of the 16S ribosomal RNA (rRNA) gene, is becoming more widely available, with improvements in turnaround time, over the last few years. The 16S rRNA gene, present in all bacteria, has variable and conserved regions that provide for identification of most bacteria to the genus or species level, depending on the design of the assay. Species-level identification has been reported to be achievable in 65%–91% of cases in the literature, depending on the region used for sequencing [9–11]. 16S rRNA gene polymerase chain reaction (PCR) followed by Sanger sequencing (16S rRNA PCR/sequencing) was initially used in clinical laboratories to identify isolates not easily identifiable by phenotypic means [12]. More recently, 16S rRNA PCR/sequencing is being used directly on clinical specimens, especially to identify difficult-to-cultivate bacteria or those rendered noncultivatable by antimicrobial therapy [13–15].
Studies comparing 16S rRNA PCR/sequencing with culture-based diagnostics have reported low sensitivity and specificity [16–18]. Despite known limitations of 16S rRNA PCR/sequencing, there is growing interest in evaluating its clinical use in culture-negative infections [19–22]. Few studies have examined the clinical utility and impact of 16S rRNA PCR/sequencing on patient management. Given the cost and technical complexity involved, attempts have been made to target use of 16S rRNA PCR/sequencing to patients who are most likely to benefit from it to maximize diagnostic yield and cost effectiveness. The aim of the current study was to assess real-world performance of 16S rRNA PCR/sequencing, especially in culture-negative infected patients and to identify specimen and patient factors associated with the highest yield, as well as to assess the impact of 16S rRNA PCR/sequencing on clinical management.
METHODS
We performed a retrospective chart review of patients whose specimens underwent 16S rRNA PCR/sequencing testing at our institution from April 2017 to March 2019. These were identified by searching a microbiologic database. All patients whose specimens underwent 16S rRNA PCR/sequencing regardless of final diagnosis were included, including those with fungal or viral infection. Demographic, clinical, microbiologic and histopathologic data were extracted by review of the electronic medical record and collected in a REDCap database. The study was approved by the Mayo Clinic Institutional Review Board. If multiple specimens were collected from the same patient during a single procedure, specimens with the same source and the same 16S rRNA PCR/sequencing result were counted as a single specimen for the purposes of analysis. There was no restriction on ordering 16S rRNA PCR/sequencing in place at the time of this study, aside from an option to send a specimen to the laboratory to hold for 16S rRNA PCR/sequencing to be performed in the 2 weeks after specimen receipt if that testing were to be ordered in that time frame.
Definitions
Synovial fluid, pleural fluid, pericardial fluid, and abscess aspirates were considered “fluid” specimens, with other specimens (eg, valve and other surgical specimens) considered “tissue” specimens. Patients were classified as having clinical infection if a final diagnosis by the treatment team was deemed infectious and there was resolution of symptoms with antimicrobial therapy. Patients were classified as having “no infection” if the final diagnosis was not infection and they did not receive antimicrobial therapy (beyond initial empiric therapy). Prior antibacterial therapy was defined as any antimicrobial therapy administered in the 2 weeks preceding 16S rRNA PCR/sequencing. 16S rRNA PCR/sequencing was deemed to have an “effect on clinical management” if the result led to escalation or de-escalation, or initiation or discontinuation, of antimicrobial therapy.
Sample Processing Methods
Sample processing and cultures were conducted in the Initial Processing and Clinical Bacteriology Laboratories of the Division of Clinical Microbiology, Mayo Clinic, Minnesota. Isolated bacteria were identified using conventional biochemical identification and/or matrix-assisted laser desorption ionization–time-of-flight mass spectrometry. 16S rRNA PCR/sequencing and sequencing was performed in the Mayo Clinic Bacteriology Laboratory, as described elsewhere [23]. Briefly, specimens were processed by incubation with proteinase K followed by lysis through rapid shaking with silica/zirconium beads at 100oC. Nucleic acid extraction was performed manually using Zymo Genomic DNA Clean & Concentrator-10. PCR was performed on a LightCycler 480II instrument (Roche Diagnostics) and used to amplify approximately 530 base pairs of the bacterial 16S rRNA gene (V3–V4) with SYBR Green detection of amplified product.
The following primers were used: forward, 5′-CGGCCCAGACTCCTACGGGAGGCAGCA-3′; reverse, 5′-GCGTGGACTACCAGGGTATCTAATCC-3′. Five microliters of extracted DNA was added to the reaction mixture that contained 0.08 µL of each primer, 10 µL of LightCycler 480 SYBR green I Master, 0.5 µL of double-stranded DNase, 0.5 µL of dithiothreitol, and 3.84 µL of sterile water. Amplification inhibition was detected by means of a second PCR reaction performed using the extracted specimen spiked with a low concentration of positive control DNA. Samples with cycle threshold values ≤32 cycles underwent bidirectional Sanger sequencing using an Applied Biosystems 3500xL instrument. Consensus sequences of ≥400 base pairs were used for identification.
Statistical Analysis
Student t and Mann-Whitney tests were used to compare means and medians for continuous variables with χ 2 tests used for independent proportions. McNemar’s test was used to calculate sensitivity and specificity, where applicable, and of confidence intervals were calculated using the binomial exact method. Differences were considered statistically significant at P < .05. Statistical analyses were performed using JMP 15.2.1 software.
RESULTS
A total of 566 specimens that underwent 16S rRNA PCR/sequencing testing were identified from 460 patients during the study period (April 2017 to March 2019). The median patient age was 63 years (interquartile range, 2–93 years); 46% of patients were female. Comorbid conditions included diabetes mellitus (18%), solid cancer (10%), hematologic cancer (8%), hematopoietic stem cell transplantation (3%), solid organ transplantation (5%), human immunodeficiency virus infection (1%), and other causes of immunosuppression (19%). The most common specimen source was the musculoskeletal (MSK) system (69%), followed by the central nervous system (8%), and cardiovascular system (7%), and skin and soft tissues (4%). Of the specimens, 56% were tissues and 44% fluids.
The overall 16S rRNA PCR/sequencing positivity rate was 17.1% (97 of 566), with 90% (88 of 97) of positive results being from patients with clinical infections (Table 1). DNA amplification was not achieved in 2% of specimens (10 of 566) owing to the presence of inhibitors in the sample. In 22% of specimens with positive results (n = 21), the bacterial 16S rRNA gene was detected by amplification; however an organism could not be identified by Sanger sequencing, for the most part owing to mixed sequences.
Table 1.
Characteristics of Specimens and Comparison Between Infected and Noninfected Groups
| Specimens, No. (%)a | ||||
|---|---|---|---|---|
| Characteristics | All specimens (N = 566) | Infection (n = 303) | No Infection (n = 263) | P Value |
| ESR, mean, mm/h | 36 | 42 | 29 | <.001 |
| CRP, mean, mg/L | 44 | 53.9 | 31.8 | <.001 |
| Specimen type | ||||
| Fluid | 252 (44) | 95 (31) | 157 (60) | … |
| Tissue | 314 (56) | 208 (69) | 106 (40) | |
| Type of specimen | ||||
| Fresh | 524 (93) | 279 (92) | 245 (93) | … |
| FFPE | 35 (6) | 20 (7) | 15 (6) | |
| Unknown | 5 (1) | 3 (1) | 2 (1) | |
| Gram stain (n = 536) | ||||
| Positive | 24 (4.5) | 22/282 (7.8) | 2/254 (0.8) | <.001 |
| Negative | 512 (95.5) | 260/282 (92) | 252/254 (99) | |
| Bacterial culture | ||||
| Positive | 114 (20) | 99 (33) | 15 (6) | … |
| Negative | 437 (77) | 194 (64) | 243 (92) | |
| Not performed | 15 (2.6) | 10 (3) | 5 (2) | |
| Culture concordance with PCRb | 429/556 (77) | 198/298 (66.4) | 231/258 (89.5) | <.001 |
| PCR positive | ||||
| Positive with specific ID | 76 (13) | 72 (23.7) | 4 (1.5) | <.001 |
| DNA detected, no ID | 21 (4) | 16 (5.3) | 5 (1.9) | |
| Total | 97 (17.5) | 88 (29) | 9 (3.4) | |
| PCR negative | 459 (81) | 210 (69.3) | 249 (94.6) | <.001 |
| PCR inhibitor present | 10 (1.7) | 5 (1.6) | 5 (1.9) | |
| Pathology performed | ||||
| Yes | 249 (44) | 157 (51.8) | 92 (35) | … |
| No | 312 (55) | 141 (46.5) | 171 (65) | |
| Specimen source | ||||
| MSK | 391(69) | 206 (68) | 185 (70.3) | … |
| Cardiovascular | 41 (7) | 27 (9) | 14 (5) | |
| CNS | 44 (7.2) | 19 (6) | 25 (9.5) | |
| Skin or soft tissue | 23 (4) | 15 (5) | 8 (3) | |
| Other | 67 (12) | 36 (12) | 31 (12) | |
| Prior antibiotics | 182 (32) | 146 (48) | 36 (13.7) | <.001 |
| Impact on clinical care | 31 (5) | 25 (8) | 6 (2) | |
| Changes in therapy | (n = 31) | (n = 25) | (n = 6) | .002 |
| Escalation | 6 (1) | 6 (2) | 0 | |
| De-escalation | 14 (2.5) | 13 (4.2) | 1 (0.3) | |
| Initiation | 6 (1) | 6 (2) | 0 | |
| Discontinuation | 5 (0.8) | 0 | 5 (2) | |
Abbreviations: CNS, central nervous system; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; FFPE, formalin-fixed paraffin-embedded; ID, identification; MSK, musculoskeletal; PCR, polymerase chain reaction.
aData represent no. (%) of specimens unless otherwise specified.
bPositive and negative concordance.
Bacterial cultures of 20% of specimens (114 of 566) were positive; 87% of culture-positive specimens (99 of 114) were from patients with clinical infection. The overall concordance between PCR and culture was 77% (429 of 556) (Table 1). The sensitivity and specificity of 16S rRNA PCR/sequencing, considering clinical diagnosis as the reference standard, were 30% and 97%, respectively. In comparison, the sensitivity and specificity of bacterial cultures were 34% and 94%, respectively.
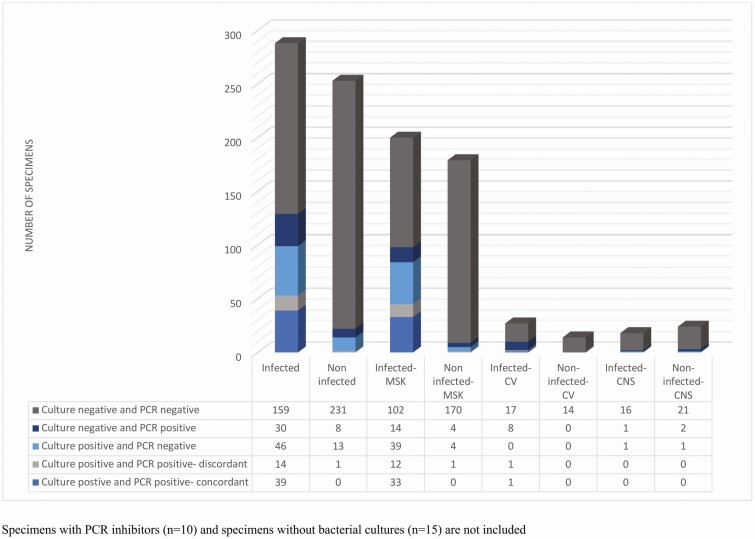
Among the 54% of specimens (303 of 566) from patients with a final diagnosis of infection, bacterial cultures were positive in 33% (99 of 303). In 18% of patients (53 of 303), both bacterial cultures and PCR were positive (Figure 1). There was concordance between culture and PCR in 74% (39 of 53), whereas in 26% (14 of 53), PCR was either discordant (n = 4) or detected bacterial DNA only without identification of a particular organism owing to mixed sequences (n = 10) (Supplementary Table 1).
Figure 1.
Culture and polymerase chain reaction (PCR) results by clinical diagnosis. Specimens with PCR inhibitors (n = 10) and specimens without bacterial cultures (n = 15) are not included. Abbreviations: CNS, central nervous system; CV, cardiovascular; MSK, musculoskeletal.
In 15% of clinically infected patients (46 of 303), 16S rRNA PCR was negative despite there being positive cultures. In almost half of these patients, cultures were positive for coagulase-negative Staphylococcus, Micrococcus, or Corynebacterium species or there was polymicrobial growth (Supplementary Table 2).
In 10% of patients (30 of 303) with clinical infection, 16S rRNA PCR/sequencing was positive, whereas bacterial cultures were negative. In 5 of these patients, only bacterial DNA was detected but a particular organism could not be identified owing to mixed sequences (Supplementary Table 3). In 10 patients with clinical infection, cultures were not obtained; 16S rRNA PCR/sequencing was positive in 5 (Supplementary Table 3). In 53% of infected patients (159 of 303), both bacterial cultures and 16S rRNA PCR were negative; in 12 of these, a fungal organism was identified by culture.
Results of 16S rRNA PCR/sequencing were positive in 4 noninfected patients. Bacteria identified in these noninfected patients included Lactobacillus species, and Streptococcus species in 1 patient each, and Staphylococcus species in 2 patients. Bacterial cultures were positive in 15 patients without clinical infection, with cultures yielding Cutibacterium acnes in 6, Micrococcus species in 1, coagulase-negative Staphylococcus species in 5, and polymicrobial growth in 3.
Inflammatory markers, including erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP) were higher in infected versus noninfected patients overall (both P < .001). Similarly, patients with positive 16S rRNA PCR/sequencing results had higher mean CRP and ESR values than those with negative 16S rRNA PCR results (both P < .001). In the clinically infected subgroup, there was a difference in CRP levels between patients with positive versus negative 16S rRNA PCR/sequencing results (P = .005); however, ESR values did not differ (P = .12). Specimens with positive Gram stains obtained in the clinical microbiology laboratory had a 12 times greater odds of positive 16S rRNA PCR/sequencing results than those with negative Gram stains (95% confidence interval for odds ratio, 5.2–31.4).
Overall, although more tissue than fluid specimens were collected from clinically infected versus noninfected patients (P < .001) (Table 1), there was no difference in PCR positivity between fluid and tissue specimens (P = .8) (Table 2). Subgroup analysis of the clinically infected group showed that 16S rRNA PCR/sequencing positivity was higher for fluid than for tissue specimens (40 vs 25%; P = .009). Six percent of samples were formalin-fixed paraffin-embedded (FFPE) tissues; although there was no difference in PCR positivity rates between FFPE and fresh tissue specimens within the clinically infected group, fresh specimens were more likely to be PCR positive than specimens overall (18% vs 12%; P = .04).
Table 2.
Characteristics of Polymerase Chain Reaction (PCR)–Positive Versus PCR-Negative Specimens
| Specimens, No. (%)a | |||
|---|---|---|---|
| Characteristic | PCR Positive (n = 97) | PCR Negative (n = 459) | P Value |
| ESR, mean, mm/h | 46 | 34 | <.001 |
| CRP, mean, mg/L | 74 | 37 | <.001 |
| Specimen source | |||
| MSK | 65 (17) | 319 (83) | .21 |
| Cardiovascular | 10 (24) | 31 (76) | |
| CNS | 4 (9) | 40 (91) | |
| Skin or soft tissue | 6 (27) | 16 (73) | |
| Other | 12 (18) | 53 (82) | |
| Specimen type | |||
| Fluid | 42 (43) | 206 (45) | .8 |
| Tissue | 55 (57 | 253 (55) | |
| Specimen | |||
| Fresh | 91 (94) | 424 (93) | .6 |
| FFPE | 4 (4.17) | 30 (7) | |
| Unknown | 2 (2) | 5 (1) | |
| Gram stain (n = 536) | |||
| Positive | 16 (18) | 8 (2) | <.001 |
| Negative | 70 (81) | 432 (98) | |
| Bacterial cultures | |||
| Positive | 54 (59) | 59 (13) | <.001 |
| Negative | 38 (41) | 390 (87) | |
| Final microbiologic diagnosis | |||
| Monomicrobial | 67 (69) | 55 (12) | … |
| Polymicrobial | 16 (17) | 10 (2) | |
| Mycobacterial | 1 (1) | 16 (4) | |
| Fungal | 0 | 12 (3) | |
| Microbiology undefined | 4 (4) | 117 (25) | |
| Noninfectious | 9 (9) | 249 (54) | |
| Infectious syndrome | 88 (91) | 210 (46) | <.001 |
| Prior antimicrobial therapy | 39 (40) | 139 (30) | .07 |
| Impact on clinical care | 23 (24) | 8 (1.7) | <.001 |
| Changes in therapy | |||
| Escalation | 6 (6) | 0 | … |
| De-escalation | 11 (11) | 3 (0.6) | |
| Initiation | 6 (6) | 0 | |
| Discontinuation | 0 | 5 (1) | |
Abbreviations: CNS, central nervous system; CRP, C-reactive protein; CSF, cerebrospinal fluid; ESR, erythrocyte sedimentation rate; FFPE, formalin-fixed paraffin-embedded; MSK, musculoskeletal; PCR, polymerase chain reaction.
aData represent no. (%) of patients unless otherwise specified.
Overall, 16S rRNA PCR/sequencing was more likely to be positive with cardiovascular specimens (10 of 27 [37%]) than with other specimen sources (50 of 175 [28%]) in clinically infected patients (Table 3). In contrast, rates of Gram stain positivity (4%) and bacterial culture positivity (7%) were much lower with cardiovascular specimens than with other specimen types. Among clinically infected patients, cardiovascular specimens were less likely to be culture positive than other specimen sources (P = .002). In contrast, bacterial cultures were more likely to be positive from MSK compared with non-MSK specimens (P < .001). Most central nervous system specimens were cerebrospinal fluid (CSF) (77%); 16S rRNA PCR/sequencing positivity was higher for brain biopsy specimens than for CSF specimens (3 vs 1 specimen; P = .01). Two of 3 brain biopsy results led to a change in clinical management.
Table 3.
Characteristics and Sources of Musculoskeletal, Cardiovascular, and Central Nervous System Specimens
| Characteristic | Musculoskeletal (n = 391) | Cardiovascular (n = 41) | CNS (n = 44) |
|---|---|---|---|
| Specimen source | Synovial fluid, 50%; periprosthetic joint tissue, 18%; native joint tissue, 8%; bone 4%; spine, 10%; hardware sonicate fluid, 2%; other, 9% | Heart valve, 66%; aortic graft, 15%; other, 20% | CSF, 77%; brain biopsy, 23% |
| Specimen type, (%) | |||
| Fluid | 50 | 5a | 23a |
| Tissue | 50 | 95 | 77 |
| Gram stain positivity rate, no. positive/total (%) | |||
| Infected | 17/195 (9) | 1/27 (4) | 1/17 (6) |
| Uninfected | 1/182 (0.5) | 0/13 (0) | 0/23 (0) |
| Total | 18/377 (5) | 1/40 (2.5) | 2.5 (3) |
| Clinically diagnosed infectious syndrome, (%) | 52 | 66 | 43 |
| Culture positivity rate, no. positive/total (%) | |||
| Infected | 84/204 (41) | 2/27 (7) | 1/18 (5.5) |
| Uninfected | 5/182 (3) | 0/14 (0) | 1/24 (4) |
| Total | 89/386 (23)a | 2/41 (5) | 2/42 (5) |
| PCR positivity rate, no. positive/total (%) | |||
| Infected | 60/202 (30) | 10/27 (37) | 2/19 (10.5) |
| Uninfected | 5/182 (3) | 0/14 (0) | 2/25 (8) |
| Total | 65/384 (17) | 10/41 (24) | 4/44 (9) |
| Prior antibiotics, no./total (%) | |||
| Infected | 88/206 (43) | 19/27 (70) | 15/19 (80) |
| Uninfected | 21/185 (11) | 5/14 (36) | 3/25 (12) |
| Total | 109/391(28) | 24/41 (59) | 18/44 (41) |
| Impact on clinical care, no./total (%) | |||
| Infected | 12/205 (6) | 7/27 (26) | 2/19 (10.5) |
| Uninfected | 4/185 (2) | 2/14 (14) | 0/25 (0) |
| Total | 16/390 (4) | 9/41 (22)a | 2/44 (5) |
Abbreviations: CNS, central nervous system; CSF, cerebrospinal fluid; PCR, polymerase chain reaction.
aP < .05
Overall, 32% of patients had received antimicrobial therapy in the 2 weeks preceding testing by 16S rRNA PCR/sequencing. More infected compared with uninfected patients received antecedent antimicrobial therapy (48 vs 14%; P < .001). There was no difference in receipt of antecedent antimicrobial therapy with regard to PCR or culture positivity overall. 16S rRNA PCR/sequencing results led to a change in clinical management in 31 patients (5% overall), including escalation in therapy in 6, de-escalation in 14, initiation in 6, and discontinuation in 5. Positive 16S rRNA PCR/sequencing results were more likely than negative results to lead to clinical management changes (P < .001).
DISCUSSION
In this study, 16S rRNA PCR/sequencing identified a potentially pathogenic organism in 14% of clinically infected patients who either had negative cultures or did not have cultures performed. Among infected patients, the 16S rRNA PCR/sequencing positivity rate was higher for cardiovascular specimens than for other specimen types. Bacterial culture positivity rates were highest in MSK specimens.
In several cases, 16S rRNA PCR/sequencing identified a difficult to culture or uncultivable organism, including Ureaplasma species, Mycoplasma hominis, Lawsonella clevelandensis, Treponema species, and Mycobacterium lepromatosis (single cases of each). In the M. lepromatosis case, which has been described elsewhere, acid-fast bacilli were seen in skin biopsy and the clinical picture matched the 16S rRNA PCR/sequencing result [23]. The patient had clinical improvement after receiving treatment directed against M. lepromatosis. In a case of M. hominis endocarditis, pathogen-directed therapy led to resolution of infection without recurrence. Similarly, in the Ureaplasma native joint septic arthritis case, Ureaplasma urealyticum–specific PCR results were positive and there was clinical improvement after treatment with azithromycin and doxycycline. Likewise, in the syphilitic panuveitis case, the rapid plasma reagin titer was 1:1024. L. clevelandensis, which was identified in a case of endovascular graft infection, is increasingly recognized as a cause of purulent infections. It is a slow-growing, anaerobic gram-variable branching rod that requires incubation beyond the usual standard number of days of incubation and hence may be missed by standard cultures [24].
Previous studies and our unpublished experience have demonstrated identification by 16S rRNA PCR/sequencing of other organisms that are challenging to cultivate, including Bartonella species, Tropheryma whipplei, and Coxiella burnetti [25, 26]. In clinically infected cases with positive cultures and negative 16S rRNA PCR results, approximately 50% had coagulase-negative Staphylococcus species or Micrococcus or Corynebacterium species identified, which may represent contaminants rather than pathogens.
Although more clinically infected compared with noninfected patients had received antibacterial therapy before 16S rRNA PCR/sequencing, treatment did not obviously affect rates of 16S rRNA PCR/sequencing or culture positivity. The decision to administer empiric therapy is usually dictated by the clinical picture; in patients with a final diagnosis of an infectious syndrome, a high suspicion of infection at presentation may have prompted initiation of empiric therapy.
The diagnostic yield of 16S rRNA PCR/sequencing has been variable in the literature and may depend on patient and specimen as well as assay characteristics. Previous studies have suggested that 16S rRNA PCR/sequencing on tissue-based specimens may be more likely to yield a clinically significant result than fluid samples [27]; however, in this cohort, fluid type specimens were more likely to be 16S rRNA PCR/sequencing positive than tissue specimens in clinically infected patients. In addition, mean CRP and ESR levels were higher in those with positive than in those with negative 16S rRNA PCR/sequencing results (P < .001). This is in contrast to previous findings in which histopathologic findings of inflammation rather than systemic inflammation were related to the high rates of 16S rRNA PCR/sequencing positivity [28].
When blood and tissue cultures fail to identify a pathogen in culture-negative infective endocarditis, 16S rRNA PCR/sequencing may be a useful diagnostic tool [18]. High yield has been reported with 16S rRNA PCR/sequencing compared with bacterial cultures in blood culture-negative infective endocarditis, with detection of fastidious organisms leading to a change in management in up to 15% of the cases [29]. In the current study, rates of 16S rRNA PCR/sequencing positivity were highest for cardiovascular specimens, whereas bacterial cultures in this group were less likely than other specimen sources to be positive. In 2 cases, difficult-to-culture organisms (M. hominis and L. clevelandensis) were detected which may explain these findings. Another reason may be the high frequency of prior antimicrobial therapy, given in 70% of the cases with cardiovascular infection (Table 3). The high yield of 16S rRNA PCR/sequencing from cardiac valves from patients with infective endocarditis may be related to persistence of bacterial DNA in valves for months to years even with successful treatment [30, 31]; 16S rRNA PCR/sequencing of valve tissue has been incorporated in the diagnostic algorithm of infective endocarditis given the high sensitivity and specificity with this specimen type [32].
Past studies have shown that molecular diagnostics may lead to changes in clinical management in infectious syndromes in up to 4%–15% of cases [27, 28]. Overall, in the current study, 16S rRNA PCR/sequencing led to a change in management in 5% of cases. This effect was proportionally higher in cases involving testing of cardiovascular specimens (22%), compared with other specimen types. Among MSK specimens, the overall PCR positivity rate was 30% in the clinically infected group. However, it led to change in management in only 6% of the cases. The previously reported yield of 16S rRNA PCR/sequencing in the diagnosis of prosthetic joint infection was 23%–32%, with an impact on clinical care in 15%–72% of cases [18, 20, 33]. Overall, positive 16S rRNA PCR/sequencing results affected clinical care more than negative results as patients with a high suspicion for underlying infection may receive treatment regardless of culture or 16S rRNA PCR/sequencing results. However, even in 16S rRNA PCR negative cases, antimicrobial therapy was either de-escalated or discontinued in 8 patients, highlighting the potential utility of such testing as an antimicrobial stewardship tool [34].
The overall sensitivity of bacterial cultures and 16S rRNA PCR/sequencing for diagnosis of infection was low. Previous studies have compared the sensitivity and specificity of 16S rRNA PCR/sequencing with culture as a reference standard; however, it is known that culture results are affected by prior antimicrobial therapy and may not detect fastidious organisms [35–38]. Studies comparing 16S rRNA PCR/sequencing to cultures have reported varying sensitivities and specificities, ranging from 43% to 96% and from 72% to 95%, respectively, depending on the tissue type and the patient population [3, 16, 39–41]. In the current study, we calculated sensitivity based on final clinical diagnosis, and with that criterion, the sensitivity of 16S rRNA PCR/sequencing and cultures were 30% and 34%, respectively, highlighting the limitation of culture-based tests as diagnostic tools and a concern with their use as a reference standard.
There are several limitations to 16S rRNA PCR/sequencing. Neither cultures (as typically performed clinically) nor 16S rRNA PCR/sequencing produce truly quantitative results (although 16S rRNA PCR/sequencing could be developed as a quantitative PCR-type assay by leveraging cycle thresholds and incorporating quantitative standards). Another limitation is that results do not provide information about antimicrobial susceptibility; 16S rRNA may detect nonviable organisms, which may or may not be clinically relevant. Moreover, as with microbiologic cultures, there is a risk of contamination. Depending on the sequencing technique used, 16S rRNA PCR/sequencing may (next-generation sequencing) or may not (Sanger sequencing, as used here) be able to detect multiple organisms in a sample. In addition, this technique is limited to bacterial pathogens and even in these cases, may not discriminate well between certain species (eg, Mycobacterium species) owing to high sequence similarities. The specific region of the 16S rRNA gene sequence could be changed to increase differentiation between related species.
Our study has several limitations. There may be selection bias influencing interpretation of results and generalizability; Mayo Clinic is a referral center, and the patient population was complex, with prolonged hospitalizations, multiple comorbid conditions, and antimicrobial exposure. In 22% of 16S rRNA PCR/sequencing positive cases, a specific organism could not be identified by sequencing. This is a limitation of Sanger sequencing that allows detection of only a single organism or 16S rRNA gene copy variant. This can be overcome by next-generation sequencing of the amplified 16S rRNA gene, but that approach can yield a complicated interpretive scenario and is more costly and generally slower than Sanger sequencing. Finally only samples with cycle threshold of <32 cycles underwent sequencing. Those with higher cycle threshold values (considered negative here) might yield clinically valuable results, especially with next generation sequencing.
This study suggests that 16S rRNA PCR/sequencing is clinically useful with an important role in diagnostic evaluation of patients with culture-negative infections, especially cardiovascular infections. Optimizing specimen selection based on clinical suspicion of infection, infectious syndrome and systemic inflammation may increase the likelihood of a positive result. These advantages and limitations should be considered on an individualized basis before performing 16S rRNA PCR/sequencing in clinical practice.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Disclaimer. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant R01AR056647).
Potential conflicts of interest. R. P. reports grants from ContraFect, TenNor Therapeutics, Hylomorph, and Shionogi and is a consultant to Curetis, Specific Technologies, Next Gen Diagnostics, PathoQuest, Selux Diagnostics, 1928 Diagnostics, PhAST, and Qvella; monies are paid to Mayo Clinic. R. P. is also a consultant to Netflix; has a patent on Bordetella pertussis/parapertussis polymerase chain reaction, a patent on a device/method for sonication with royalties paid by Samsung to Mayo Clinic, and a patent on an antibiofilm substance; and receives an editor’s stipend from the Infectious Diseases Society of America and honoraria from the National Board of Medical Examiners, Up-to-Date, and the Infectious Diseases Board Review Course. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Harbarth S, Nobre V, Pittet D. Does antibiotic selection impact patient outcome? Clin Infect Dis 2007; 44:87–93. [DOI] [PubMed] [Google Scholar]
- 2.Fournier PE, Gouriet F, Casalta JP, et al. Blood culture-negative endocarditis: Improving the diagnostic yield using new diagnostic tools. Medicine (Baltimore) 2017; 96:e8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fournier PE, Thuny F, Richet H, et al. Comprehensive diagnostic strategy for blood culture-negative endocarditis: a prospective study of 819 new cases. Clin Infect Dis 2010; 51:131–40. [DOI] [PubMed] [Google Scholar]
- 4.Tan TL, Kheir MM, Shohat N, et al. Culture-negative periprosthetic joint infection. Jbjs Open Access 2018; 3:e0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon HK, Cho SH, Lee DY, et al. A review of the literature on culture-negative periprosthetic joint infection: epidemiology, diagnosis and treatment. Knee Surg Relat Res 2017; 29:155–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teshome BF, Vouri SM, Hampton N, Kollef MH, Micek ST. Duration of exposure to antipseudomonal β-lactam antibiotics in the critically ill and development of new resistance. Pharmacother J Hum Pharmacol Drug Ther 2019; 39:261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kollef MH, Burnham JP. Antibiotic thresholds for sepsis and septic shock. Clin Infect Dis 2019; 69:938–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S, Sakhuja A, Kumar G, McGrath E, Nanchal RS, Kashani KB. Culture-negative severe sepsis: nationwide trends and outcomes. Chest 2016; 150:1251–9. [DOI] [PubMed] [Google Scholar]
- 9.Fontana C, Favaro M, Pelliccioni M, Pistoia ES, Favalli C. Use of the MicroSeq 500 16S rRNA gene-based sequencing for identification of bacterial isolates that commercial automated systems failed to identify correctly. J Clin Microbiol 2005; 43:615–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mignard S, Flandrois JP. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods 2006; 67:574–81. [DOI] [PubMed] [Google Scholar]
- 11.Patel JB. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol Diagn 2001; 6:313–21. [DOI] [PubMed] [Google Scholar]
- 12.Clarridge JE 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 2004; 17:840–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bador J, Nicolas B, Chapuis A, Varin V, et al. 16S rRNA PCR on clinical specimens: impact on diagnosis and therapeutic management. Med Mal Infect 2020; 50:63–73. [DOI] [PubMed] [Google Scholar]
- 14.Larsen LH, Khalid V, Xu Y, et al. Differential contributions of specimen types, culturing, and 16S rRNA sequencing in diagnosis of prosthetic joint infections. J Clin Microbiol 2018; 56:e01351-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra D, Satpathy G, Wig N, Fazal F, Ahmed NH, Panda SK. Evaluation of 16S rRNA broad range PCR assay for microbial detection in serum specimens in sepsis patients. J Infect Public Health 2020; 13:998–1002. [DOI] [PubMed] [Google Scholar]
- 16.Bémer P, Plouzeau C, Tande D, et al. ; Centre de Référence des Infections Ostéo-articulaires du Grand Ouest (CRIOGO) Study Team . Evaluation of 16S rRNA gene PCR sensitivity and specificity for diagnosis of prosthetic joint infection: a prospective multicenter cross-sectional study. J Clin Microbiol 2014; 52:3583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panousis K, Grigoris P, Butcher I, Rana B, Reilly JH, Hamblen DL. Poor predictive value of broad-range PCR for the detection of arthroplasty infection in 92 cases. Acta Orthop 2005; 76:341–6. [PubMed] [Google Scholar]
- 18.Akram A, Maley M, Gosbell I, Nguyen T, Chavada R. Utility of 16S rRNA PCR performed on clinical specimens in patient management. Int J Infect Dis 2017; 57:144–9. [DOI] [PubMed] [Google Scholar]
- 19.Borde JP, Häcker GA, Guschl S, et al. Diagnosis of prosthetic joint infections using UMD-Universal Kit and the automated multiplex-PCR Unyvero i60 ITI® cartridge system: a pilot study. Infection 2015; 43:551–60. [DOI] [PubMed] [Google Scholar]
- 20.Grif K, Heller I, Prodinger WM, Lechleitner K, Lass-Flörl C, Orth D. Improvement of detection of bacterial pathogens in normally sterile body sites with a focus on orthopedic samples by use of a commercial 16S rRNA broad-range PCR and sequence analysis. J Clin Microbiol 2012; 50:2250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothman R, Ramachandran P, Yang S, et al. Use of quantitative broad-based polymerase chain reaction for detection and identification of common bacterial pathogens in cerebrospinal fluid. Acad Emerg Med 2010; 17:741–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marín M, Garcia-Lechuz JM, Alonso P, et al. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J Clin Microbiol 2012; 50:583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Virk A, Pritt B, Patel R, et al. Mycobacterium lepromatosis lepromatous leprosy in US citizen who traveled to disease-endemic areas. Emerg Infect Dis 2017; 23:1864–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldenberger D, Naegele M, Steffens D, Eichenberger R, Egli A, Seth-Smith HMB. Emerging anaerobic and partially acid-fast Lawsonella clevelandensis: extended characterization by antimicrobial susceptibility testing and whole genome sequencing. Clin Microbiol Infect 2019; 25:1447–8. [DOI] [PubMed] [Google Scholar]
- 25.Voldstedlund M, Nørum Pedersen L, Baandrup U, Klaaborg KE, Fuursted K. Broad-range PCR and sequencing in routine diagnosis of infective endocarditis. APMIS 2008; 116:190–8. [DOI] [PubMed] [Google Scholar]
- 26.Podglajen I, Bellery F, Poyart C, et al. Comparative molecular and microbiologic diagnosis of bacterial endocarditis. Emerg Infect Dis 2003; 9:1543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kerkhoff AD, Rutishauser RL, Miller S, Babik JM. Clinical utility of universal broad-range polymerase chain reaction amplicon sequencing for pathogen identification: a retrospective cohort study. Clin Infect Dis 2020; 71:1554–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basein T, Gardiner BJ, Andujar Vazquez GM, et al. Microbial identification using DNA target amplification and sequencing: clinical utility and impact on patient management. Open Forum Infect Dis 2018; 5:ofy257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsch G, Orszag P, Mashaqi B, Kuehn C, Haverich A. Antibiotic therapy following polymerase chain reaction diagnosis of infective endocarditis: a single centre experience. Interact Cardiovasc Thorac Surg 2015; 20:589–93. [DOI] [PubMed] [Google Scholar]
- 30.Lang S, Watkin RW, Lambert PA, Littler WA, Elliott TS. Detection of bacterial DNA in cardiac vegetations by PCR after the completion of antimicrobial treatment for endocarditis. Clin Microbiol Infect 2004; 10:579–81. [DOI] [PubMed] [Google Scholar]
- 31.Branger S, Casalta JP, Habib G, Collard F, Raoult D. Streptococcus pneumoniae endocarditis: persistence of DNA on heart valve material 7 years after infectious episode. J Clin Microbiol 2003; 41:4435–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liesman RM, Pritt BS, Maleszewski JJ, Patel R. Laboratory diagnosis of infective endocarditis. J Clin Microbiol 2017; 55:2599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alraddadi B, Al-Azri S, Forward K. Influence of 16S ribosomal RNA gene polymerase chain reaction and sequencing on antibiotic management of bone and joint infections. Can J Infect Dis Med Microbiol 2013; 24:85–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Donnell S, Gaughan L, Skally M, et al. The potential contribution of 16S ribosomal RNA polymerase chain reaction to antimicrobial stewardship in culture-negative infection. J Hosp Infect 2018; 99:148–52. [DOI] [PubMed] [Google Scholar]
- 35.Bereza P, Ekiel A, Auguściak-Duma A, et al. Comparison of cultures and 16S rRNA sequencing for identification of bacteria in two-stage revision arthroplasties: preliminary report. BMC Musculoskelet Disord 2016; 17:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoo IY, Kang OK, Lee MK, et al. Comparison of 16S ribosomal RNA targeted sequencing and culture for bacterial identification in normally sterile body fluid samples: report of a 10-year clinical laboratory review. Ann Lab Med 2020; 40:63–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhoads DD, Cox SB, Rees EJ, Sun Y, Wolcott RD. Clinical identification of bacteria in human chronic wound infections: culturing vs. 16S ribosomal DNA sequencing. BMC Infect Dis 2012; 12:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esparcia O, Montemayor M, Ginovart G, et al. Diagnostic accuracy of a 16S ribosomal DNA gene-based molecular technique (RT-PCR, microarray, and sequencing) for bacterial meningitis, early-onset neonatal sepsis, and spontaneous bacterial peritonitis. Diagn Microbiol Infect Dis 2011; 69:153–60. [DOI] [PubMed] [Google Scholar]
- 39.Bosshard PP, Kronenberg A, Zbinden R, Ruef C, Böttger EC, Altwegg M. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin Infect Dis 2003; 37:167–72. [DOI] [PubMed] [Google Scholar]
- 40.Marín M, Muñoz P, Sánchez M, et al. ; Group for the Management of Infective Endocarditis of the Gregorio Marañón Hospital (GAME) . Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine (Baltimore) 2007; 86:195–202. [DOI] [PubMed] [Google Scholar]
- 41.Faraji R, Behjati-Ardakani M, Moshtaghioun SM, et al. The diagnosis of microorganism involved in infective endocarditis (IE) by polymerase chain reaction (PCR) and real-time PCR: a systematic review. Kaohsiung J Med Sci 2018; 34:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.