An antifungal drug reveals a dependency of glioblastoma on intracellular biotin distribution.
Abstract
Glioblastoma (GBM) is a uniformly lethal disease driven by glioma stem cells (GSCs). Here, we use a chemical biology approach to unveil previously unknown GBM dependencies. By studying sulconazole (SN) with anti-GSC properties, we find that SN disrupts biotin distribution to the carboxylases and histones. Transcriptomic and metabolomic analyses of SN-treated GSCs reveal metabolic alterations that are characteristic of biotin-deficient cells, including intracellular cholesterol depletion, impairment of oxidative phosphorylation, and energetic crisis. Furthermore, SN treatment reduces histone biotinylation, histone acetylation, and expression of superenhancer-associated GSC critical genes, which are also observed when biotin distribution is genetically disrupted by holocarboxylase synthetase (HLCS) depletion. HLCS silencing impaired GSC tumorigenicity in an orthotopic xenograft brain tumor model. In GBM, high HLCS expression robustly indicates a poor prognosis. Thus, the dependency of GBM on biotin distribution suggests that the rational cotargeting of biotin-dependent metabolism and epigenetic pathways may be explored for GSC eradication.
INTRODUCTION
Glioblastoma (GBM) is the most common and malignant adult brain tumor with an abysmal patient prognosis. The current standard of care for GBM remains to be aggressive surgery followed by radiotherapy, in combination with adjuvant temozolomide treatment. Tumor recurrence is almost inevitable due to a subpopulation of tumor cells, commonly known as glioma stem cells (GSCs), which exhibit stem cell–like traits, robust proliferation, invasiveness, and therapy resistance (1–4). The intimate link between GBM pathogenesis and GSCs has popularized the GSCs as a clinically invaluable experimental model to identify GBM vulnerabilities. GBM emerges within a hypoxic and harsh environment and relies on a plethora of metabolic pathways, including glycolysis, oxidative phosphorylation (OXPHOS), amino acid (e.g., glutamine), and lipid metabolism (e.g., cholesterol) for survival and growth (5). Notably, many of these metabolic pathways directly or indirectly depend on biotin, suggesting that GBM may exploit aberrant biotin regulation for its proliferation and tumorigenicity (6–8).
In noncancerous cells, a major function of biotin is to serve as a coenzyme for biotin-dependent carboxylases, including acetyl–coenzyme A (CoA) carboxylase 1 (ACC1), ACC2, methylcrotonoyl-CoA carboxylase 1 (MCCC1), propionyl-CoA carboxylase (PCC), and pyruvate carboxylase (PC), which play crucial roles in fatty acid metabolism, gluconeogenesis, and amino acid catabolism (6). These carboxylases are synthesized in an inactive form (apocarboxylases) and require activation by the holocarboxylase synthetase (HLCS) enzyme via biotinylation (9). Subsequently, biotin must be released from the holocarboxylases, which is catalyzed by the enzyme biotinidase (10). Although less well understood, histone proteins can also be biotinylated by HLCS, and the levels of biotinylated histones may affect cell proliferation, epigenetic, and gene expression alterations (11, 12). The importance of HLCS and biotinidase is underscored by biotin deficiency disorders that arise from loss-of-function mutations in the HLCS or biotinidase genes (6). In patients with such disorders, the activity of multiple carboxylases is severely compromised, leading to various symptoms, including seizures and neurological disorders. That fibroblasts from patients with HLCS deficiency exhibited a profound reduction in carboxylase and histone biotinylation strengthens the view that histone biotinylation is a physiologically relevant phenomenon (13). As expected, cancer cells appear to have hijacked biotin metabolism to sustain their proliferation. Biotin levels were higher in tumor versus nontumor tissues, which correlated with the overexpression of biotin receptors (14). These observations have also motivated efforts to develop biotin-based drug delivery systems for cancer treatment (15). Given that regulated biotin metabolism is ubiquitous and essential across nature, targeting biotin regulation within deregulated pathways may represent a potential approach to disable GBM.
In an attempt to uncover new GBM dependency using a chemical biology approach, we generated an anti-GBM query signature, which comprises the low expression of genes in the epidermal growth factor receptor (EGFR; related to cell cycle) and mesenchymal subtype of GBM (Mes; related to invasiveness) gene sets, and high expression of genes in the generation of neurons gene sets (GN; reflecting increased tumor differentiation), for the interrogation of anti-GBM compounds through Connectivity Map Analysis (CMA) (16–18). Briefly, CMA is a catalog of drug-induced gene expression profiles that allows one to explore compounds that elicit highly similar (activators) or dissimilar (inhibitors) gene expression profiles to the query signature (19). The mode of action of the hits can subsequently be revealed by a repertoire of experimental approaches, including chemical pull-down and enzymatic assays, in silico docking studies, and transcriptomic and metabolomic analyses. Here, we hypothesize that GBM is dependent on biotin distribution and that targeting of the biotin-dependent metabolism and epigenetic pathways may represent a potential node of intervention for GBM.
RESULTS
An anti-GBM signature identifies SN that impairs GSC proliferation and invasiveness
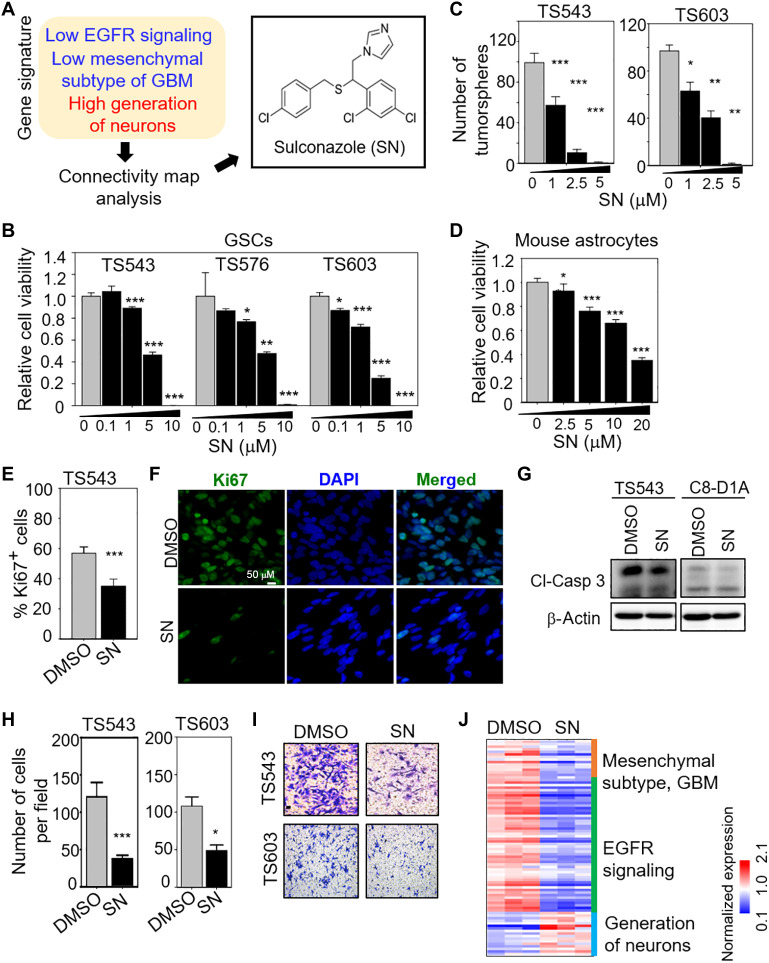
Given that GBM is characterized by high proliferation and invasiveness and is intimately linked to GSCs, we reasoned that an EGFRlowMeslowGNhigh query signature should guide us to find anti-GBM compounds through CMA (table S1). Unsupervised clustering of all patients with glioma in The Cancer Genome Atlas (TCGA) dataset based on our query signature confirmed that the EGFRlowMeslowGNhigh signature correlated with low-grade gliomas [mostly IDH (isocitrate dehydrogenase) mutants] and favorable patient outcome, while the EGFRhighMeshighGNlow signature correlated with GBM (mostly IDH wild type) and inferior patient survival (fig. S1, A and B). When we applied the EGFRlowMeslowGNhigh signature to CMA, sulconazole (SN) emerged as one of the top hits (fig. S1C and Fig. 1A). SN is an antifungal medication that is used for the treatment of skin infections, including athlete’s foot and ringworm (20). As SN has not been studied in GBM, and its mode of action in mammalian cells is poorly understood, we selected it for further investigation. SN treatment significantly reduced cell viability and tumorsphere formation of multiple patient-derived GSC lines in a dose-dependent manner (Fig. 1, B and C). In contrast, noncancerous mouse astrocytes were less sensitive to SN treatment than the GSCs (20 μM SN for astrocytes versus 5 μM SN for GSCs to achieve similar cell viability reduction) (Fig. 1D). To determine whether the loss of cell viability in SN-treated GSCs was due to an antiproliferative response, we performed Ki67 immunostaining of GSCs that were treated with SN or vehicle control. There was a significant reduction in the Ki67+ GSCs upon SN treatment (Fig. 1, E and F). To assess whether the loss of cell viability in SN-treated GSCs was due to a cell death response, we performed Western blot analysis of cleaved-caspase 3 in SN- versus DMSO-treated GSCs. There was no change in cleaved-caspase 3 levels in SN-treated GSC (and mouse astrocytes), indicating no increase in apoptosis (Fig. 1G). SN treatment also significantly reduced GSC invasiveness in the Transwell migration and invasion assay (Fig. 1, H and I), and up-regulated genes in the GN pathway, suggestive of increased GSC differentiation (Fig. 1J). Thus, we conclude that the loss of cell viability in SN-treated GSCs was mainly attributed to an antiproliferative response that may be linked to increased GSC differentiation.
Fig. 1. Identification of SN as a previously unknown anti-GSC compound.
(A) CMA using the EGFRlowMeslowGNhigh signature identifies SN. (B and D) Cell viability assay of GSCs and mouse astrocytes with 3 days of SN treatment (n = 8) (means ± SD). (C) Tumorsphere formation of GSCs following SN treatment for 5 days (n = 6) (means ± SD). (E) Ki67 immunostaining of GSC with or without 5 μM SN treatment for 3 days (n = 5) (mean ± SD). (F) Representative images of (E). (G) Western blot analysis of cleaved-caspase 3 from DMSO or 5 μM SN-treated cells (3 days). β-Actin serves as the loading control. (H) Transwell migration and invasion assay of GSCs with 5 μM SN pretreatment (3 days) (n = 3) (means ± SD). (I) Representative images of (H). (J) Heatmap for selected genes in the EGFR, Mes, and GN gene sets after treatment of GSC TS543 with 5 μM SN versus DMSO control (3 days). Gene expressions [TPM (transcripts per million) values] were mean-normalized. Two-tailed unpaired Student’s t test. ***P < 1 × 10−4; **P < 0.001; *P < 0.05.
SN disrupts biotin distribution to the carboxylases
SN belongs to the family of azoles that inhibit lanosterol 14α-demethylase (Cyp51A1), which is involved in ergosterol (the equivalent of cholesterol in mammalian cells) biosynthesis in fungi (fig. S2A) (21). Thus, we explored this possible mode of action of SN by measuring GSC viability upon treatment with two well-established Cyp51A1 inhibitors, namely, fluconazole and tebuconazole (fig. S2B). Fluconazole did not affect GSC viability even up to a concentration of 50 μM, while the same concentration of tebuconazole significantly reduced GSC viability by ~30% (compared to 60% reduction in GSC viability with 5 μM SN) (fig. S2C). Furthermore, the small interfering RNA–mediated knockdown (KD) of Cyp51A1 did not significantly alter GSC viability (fig. S2, D and E). It is unlikely that SN exerts its effect on GSC via Cyp51A1 inhibition.
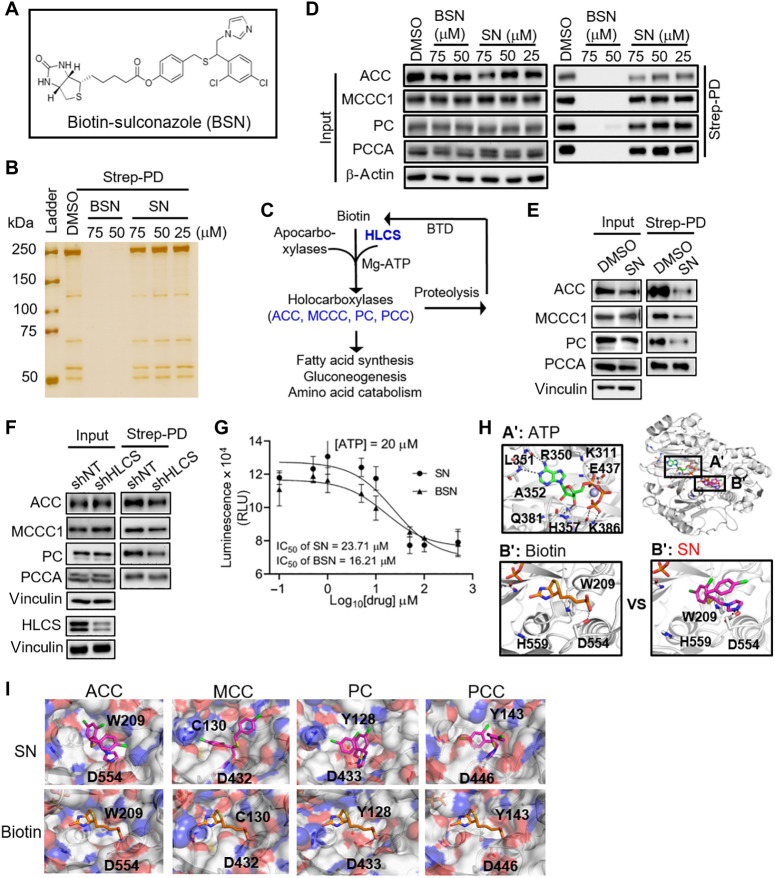
To capture the primary protein targets of SN using streptavidin pull-down assay, we synthesized biotin-conjugated SN (BSN) (Fig. 2A, fig. S3A, and data S1 to S4). We confirmed that BSN retains the anti-GBM activity of SN in reducing GSC viability at 5 μM (fig. S3B). Consistent with our cell viability data, SN is slightly more potent in reducing GSC tumorsphere formation than BSN (fig. S3C). Unexpectedly, a 6-hour BSN or SN, but not biotin, treatment compromised the association between biotin-bound proteins and the streptavidin beads when compared to the dimethyl sulfoxide (DMSO) control, although this effect was much less with SN than with BSN treatment (Fig. 2B and fig. S3D). This unexpected result resembled that reported in biotin deficiency models (7), suggesting that both compounds may compete with biotin for binding to the biotin carboxylation (BC) component of the carboxylases (Fig. 2C) (9). Western blot analysis of the streptavidin pull-down lysates validated that BSN markedly reduced the association between the carboxylases (ACC/MCC/PCC/PC) and streptavidin relative to the DMSO control without altering their total protein levels, indicating reduced biotinylated carboxylases (Fig. 2D). Given the modest decrease in biotinylated carboxylases with SN treatment, we performed a similar experiment with 293T cells that were cultured in biotin-free medium. There was a more pronounced reduction in biotinylated carboxylases in SN-treated cells under biotin-deficient conditions, especially for ACC and PC (Fig. 2E). Since HLCS is involved in carboxylase biotinylation, we repeated the streptavidin pull-down experiment using lysates from HLCS-depleted cells grown under biotin-free culture conditions (3 days). As expected, there was less biotinylated carboxylases (particularly for ACC and PC) upon HLCS KD, although the reduction was less marked than that with SN treatment (Fig. 2, E and F). If SN indeed reduces biotinylation of the carboxylases, one would predict the decreased enzymatic activity of these enzymes. As a case in point, we showed that both SN and BSN inhibited the activity of recombinant ACC1 in a dose-dependent manner (Fig. 2G). Notably, this inhibition was independent of the adenosine triphosphate (ATP) levels, strengthening the view that SN inhibits ACC1 by competing with biotin (fig. S3E). Given that efficient carboxylase biotinylation requires the action of the HLCS enzyme, the higher concentration of SN required to inhibit the ACC1 enzyme in vitro (IC50 ~ 20 μM) when compared to that needed to reduce GSC viability by ~60% (i.e., 5 μM SN) would suggest that the inhibitory effect of SN on the carboxylases may involve the HLCS enzyme (Figs. 1B and 2G).
Fig. 2. SN inhibits biotinylation of carboxylases by competing with biotin for the BC components of these enzymes.
(A) Chemical structure of BSN. (B) Silver-stained SDS–polyacrylamide gel electrophoresis gel of streptavidin pull-down lysates (Strep-PD) from 293T cells after 6 hours of DMSO, BSN, or SN treatment. (C) Schematic view of the biotin cycle involving the HLCS, carboxylases, and biotinidase, and their roles in cellular metabolism. (D) Western blot analysis of the carboxylases in streptavidin pull-down lysates from BSN-, SN-, or DMSO-treated 293T cells (6-hour treatment). β-Actin serves as the loading control. (E and F) Western blot analysis of the carboxylases in streptavidin pull-down lysates from 75 μM SN–treated (6 hours) (E) or HLCS KD (F) 293T cells (3 days) that were cultured in biotin-free medium. Vinculin serves as the loading control. (G) Enzymatic activity of recombinant ACC1 protein in vitro in the presence of SN or BSN (n = 5) (means ± SD). (H and I) In silico docking of ATP, biotin, and SN into the BC component of ACC1 (H), MCC1, PC, and PCCA (I) using the Glide program.
Docking studies support a model wherein SN competes with biotin for binding to the carboxylases
Next, we carried out in silico docking studies using a widely used docking program Glide to identify potential SN binding site on the carboxylases. SN was docked into the biotin-binding pocket of the models of these enzymes that we constructed (Fig. 2, H and I). The biotin-binding pocket was highly conserved in these enzymes, with one side of the pocket hydrophobic and the other charged. The imidazole group of SN occupied the charged region in the pocket of the BC component of ACC1, with the free nitrogen forming hydrogen bond interactions with Asp (D554) and with the side chain of Trp (W209) through stacking interactions (Fig. 2H). The hydrophobic groups (4-chlorophenyl and 2,4-dichlorophenyl) occupied the hydrophobic region in the pocket, with the 2,4-dichlorophenyl group buried into the pocket formed by residues Ile128, Trp209, Val444, and Phe557, while the 4-chlorophenyl group faced toward the solvent front side of the pocket (Fig. 2H). A similar interaction pattern was observed for the imidazole group of SN with the side chains of respective amino acids in the BC component of MCCC1 (Asp432 and Cys130), PCC-alpha (Asp446 and Tyr143), and PC (Asp433 and Tyr128) (Fig. 2I). We also confirmed that SN and the BC components of the carboxylases remained stably bound relative to the docked starting conformations (root mean square deviation ~2 Å and ~ 3–4 Å, respectively) by refining the protein-ligand models using molecular dynamics (MD) simulations (fig. S3F).
To strengthen our proposal that SN disrupts biotin distribution to the carboxylases, we performed cell viability assay of HLCS-depleted GSC, with or without SN treatment. HLCS KD resulted in an average of ~0.3- to 0.5-fold reduction in cell viability [based on two independent short hairpin–mediated RNAs (shRNAs) against HLCS] relative to the shNT (non-targeting shRNA) control, and the addition of 5 μM SN further decreased cell viability by ~0.1- to 0.3-fold, resulting in a reduction of cell viability that is quite similar to that observed with 5 μM SN treatment alone (fig. S3G). This is consistent with our view that SN functions in part by impairing biotin distribution, similar to HLCS inhibition (hence, no additive effect). Collectively, our chemical pull-down, enzymatic, and molecular docking data support a model wherein both BSN and SN can inhibit the carboxylases by competing with biotin for binding to the BC components of these enzymes, and that BSN may alter the quaternary structures of these enzymes (which typically function as dimers or oligomers), hence blocking streptavidin access (fig. S3H) (9).
SN treatment depletes intracellular cholesterol in GSC
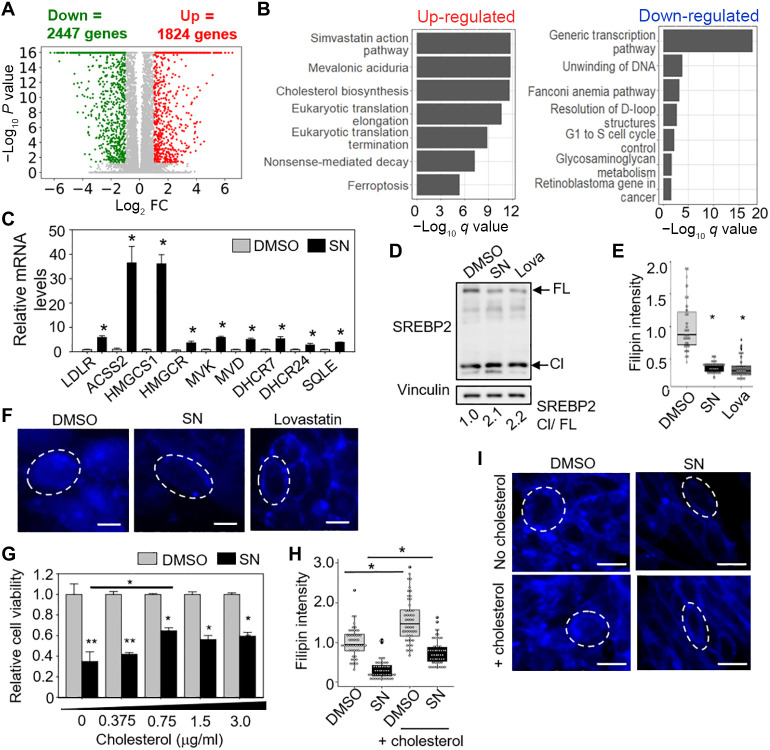
To gain further insights into the mechanisms underlying SN’s effect on GSC, we performed RNA sequencing (RNA-seq) analysis of SN- versus DMSO-treated GSC. ConsensusPathDB analysis of the differentially expressed genes revealed the enrichment of “simvastatin action pathway” as one of the top pathways for the up-regulated genes (suggesting cholesterol depletion) and the “generic transcription pathway” as one of the top pathways for the down-regulated genes (Fig. 3, A and B, and table S2). In response to low levels of intracellular cholesterol, sterol regulatory element–binding protein 2 (SREBP2) translocates from endoplasmic reticulum membranes to the Golgi apparatus. Upon cleavage by S1P (site-1 protease) and S2P (site-2 protease) proteases, the mature form of SREBP2 enters the nucleus and activates gene transcription in cholesterol uptake and de novo synthesis (mevalonate pathway) (22). We validated most of the up-regulated genes in the cholesterol homeostasis pathways by quantitative reverse transcription polymerase chain reaction
Fig. 3. RNA-seq analysis reveals cholesterol depletion in SN-treated GSC.
(A) Volcano plot showing the number of genes that are differentially expressed in 5 μM SN- versus DMSO-treated GSC (3 days). (B) ConsensusPathDB analysis of up- and down-regulated genes upon SN treatment. (C) Validation of the up-regulated genes in the cholesterol homeostasis pathway by qRT-PCR analysis (n = 4) (means ± SD). HSP70 and TBP serve as the housekeeping genes. *P < 0.01. (D) Western blot analysis of full-length (FL) and cleaved (Cl) SREBP2 protein after treatment of GSC TS543 with DMSO, 5 μM SN, or 5 μM lovastatin (3 days). Vinculin serves as the loading control. SREBP2 Cl/FL ratios are indicated. (E) Filipin staining of GSC TS543 after treatment with DMSO, 5 μM SN, or 5 μM lovastatin (3 days) (n = 10) (means ± SD). *P < 1 × 10−10. (F) Representative images of (E). Scale bar, 20 μm. (G) Cell viability of GSC TS543 treated with DMSO or 5 μM SN, in the presence exogenous cholesterol (n = 8) (means ± SD). **P < 1 × 10−8; *P < 1 × 10−4. (H) Filipin staining of 5 μM SN-treated GSC TS543 with or without cholesterol (0.75 μg/ml) (3 days) (n = 10) (means ± SD). *P < 1 × 10−7. (I) Representative images of (H). Two-tailed unpaired Student’s t test. Scale bar, 20 μm.
(qRT-PCR) analysis (Fig. 3C) and the increase in cleaved SREBP2 upon SN treatment (similar to lovastatin) (Fig. 3D). Furthermore, detection of cholesterol using filipin, a cholesterol-specific fluorophore, revealed a significant decrease in filipin intensity in SN-treated GSCs, relative to the vehicle control (Fig. 3, E and F). Since cholesterol depletion can impair GSC proliferation (23), we performed a rescue experiment by adding exogenous cyclodextrin-complexed cholesterol in the presence of SN. There was a modest but significant increase in cell viability when cholesterol (0.75 μg/ml) was added to SN-treated GSC, which was accompanied by a partial increase in filipin staining (Fig. 3, G to I). Thus, SN exerts its effect on GSCs in part by causing intracellular cholesterol depletion.
Previous studies showed that the carboxylase activity of fibroblasts from patients with mutant HLCS proteins and metabolic defects of biotin deficiency models can only be revealed under biotin deprivation conditions (8, 24, 25). Hence, we next evaluated the effect of HLCS KD on intracellular cholesterol levels in 293T cells under biotin-free culturing conditions as there is no biotin-free GSC medium to perform similar analysis in the GSC. Unlike SN treatment, HLCS KD did not result in cholesterol depletion as there was no up-regulation of genes in the cholesterol homeostasis pathway and increase in cleaved SREBP2 protein level (under 3 days of biotin-free condition) (fig. S4, A and B), consistent with a modest decrease in carboxylase biotinylation upon HLCS depletion (Fig. 2F). These results suggest that cholesterol depletion will manifest only when the carboxylases are severely deprived of biotin.
Metabolic reprogramming of SN-treated GSC toward glycolysis
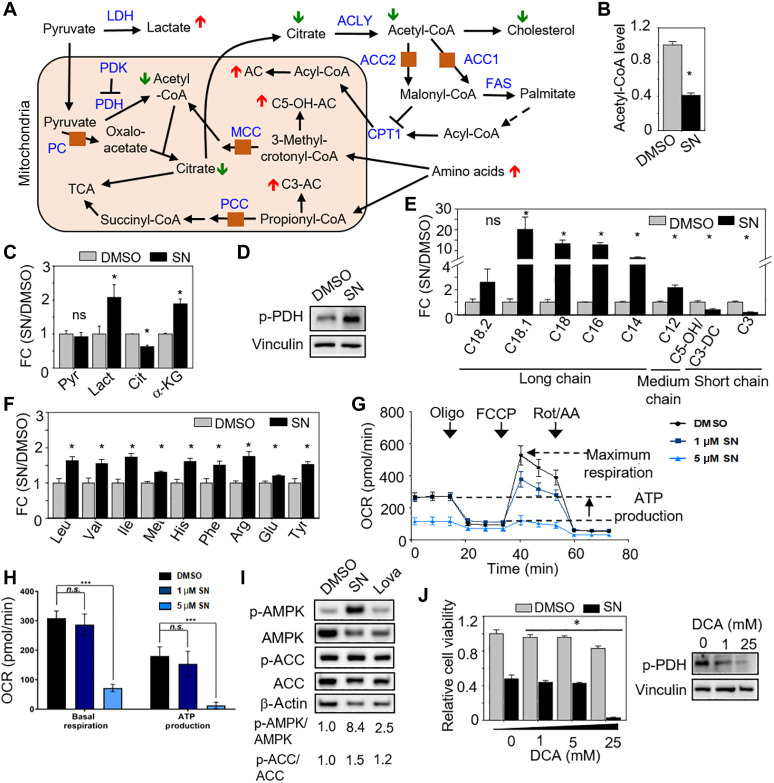
To begin tracing how cholesterol levels may be depleted in SN-treated GSC, we quantified acetyl-CoA, an important precursor of cholesterol and a by-product of the carboxylases, and found a significant reduction of acetyl-CoA levels upon SN treatment (Fig. 4, A and B). Next, we performed metabolomic analyses to quantify the levels of organic acids, acylcarnitines, and amino acids in SN- versus DMSO-treated GSCs. There was no significant change in pyruvate level, but there was a significant decrease in citrate (downstream product of oxaloacetate) in SN-treated GSC, along with an increase in lactate, supporting PC inhibition (Fig. 4, A to C, and table S3) (26). Accordingly, there was an increase in phosphorylated pyruvate dehydrogenase (p-PDH) levels with SN treatment, indicating a block in PDH activity (Fig. 4D). The decrease in citrate would contribute to reduced acetyl-CoA levels and, hence, cholesterol biosynthesis. Combined inhibition of ACC1 and ACC2 decreases malonyl-CoA, an allosteric inhibitor of carnitine palmitoyltransferase (CPT-1), leading to reduced de novo lipogenesis and enhanced fatty acid oxidation (27). SN treatment significantly increased the levels of long- and medium-chain acylcarnitines, but not short-chain acylcarnitines, indicating an increase in CPT-1 activity with no enhanced fatty acid oxidation, as more acylcarnitines are generated from acyl-CoAs, consistent with the inhibition of ACC1 and ACC2 (Fig. 4, A and E, and table S3) (26).
Fig. 4. Metabolic reprogramming of SN-treated GSC toward glycolysis.
(A) Schematic showing the predicted metabolomic changes if multiple carboxylases were inhibited. LDH, lactate dehydrogenase; PDK, pyruvate dehydrogenase kinase; PDH, pyruvate dehydrogenase; ACLY, ATP-citrate synthase; FAS, fatty acid synthase. (B, C, E, and F) Relative intracellular levels of acetyl-CoA (B), organic acids (C), acylcarnitines (E), and amino acids (F) in GSC TS543 with or without 5 μM SN treatment (3 days) (n = 3) (means ± SD). *P < 0.05. (D) Western blot analysis of p-PDH levels with or without 5 μM SN treatment (3 days). Vinculin serves as the loading control. (G and H) Seahorse analysis of GSC TS543 with or without 5 μM SN treatment (20 hours) (n = 15) (means ± SD). ***P < 1.0 × 10−3. (I) Western blot analysis of p-AMPK, AMPK, p-ACC, and ACC levels in GSC TS543 with DMSO, 5 μM SN, or 5 μM lovastatin (3 days). β-Actin serves as the loading control. (J) Relative cell viability of GSC TS543 upon 5 μM SN or DMSO treatment, combined with increasing concentration of DCA (3 days) (n = 6) (mean ± SD). *P < 0.05. Western blot analysis of p-PDH levels in GSC TS543 with DCA treatment shown on the right. Vinculin serves as the loading control. Two-tailed unpaired Student’s t test. ns, not significant.
The inhibition of MCCC and PCC is predicted to result in an increase in amino acids, C3- and C5-OH acylcarnitines (26). SN treatment significantly increased the levels of leucine, valine, methionine, isoleucine, and threonine, but not C3- and C5-OH acylcarnitines, suggesting that SN does not inhibit MCCC and PCC in GSC at 5 μM (Fig. 4, A, E, and F, and table S3). The metabolic changes seen in SN-treated GSC were specific as these were not recapitulated in SN-treated astrocytes (fig. S5, A to F, and table S3), consistent with the modest reduction of astrocyte viability (Fig. 1D). To explore the possibility that SN exerts its anti-GSC effect by inhibiting a single carboxylase, we depleted each carboxylase individually and measured the resulting cell viability. The efficient KD of each carboxylase significantly decreased GSC viability by ~31 to 45% on average, which is lower than that achieved with 5 μM SN (~60%). Collectively, all these data indicate that SN likely inhibits multiple carboxylases (at least PC, ACC1, and ACC2) in GSC (Fig. 2E and fig. S6, A and B).
A reduction in carbon flux into the tricarboxylic acid (TCA) cycle is predicted to impair OXPHOS. Thus, we measured the oxygen consumption rate in SN- versus DMSO-treated GSC, revealing a significant decrease in basal respiration and ATP production upon SN treatment (Fig. 4, G and H). In response to OXPHOS inhibition, adenosine monophosphate–activated protein kinase (AMPK) (the energy sensor) is activated in GBM cells (28). Accordingly, we found the robust phosphorylation of AMPK (p-AMPK) but a slight increase in the phosphorylation of ACC1 (p-ACC-79), a downstream substrate of AMPK (Fig. 4I) (29). Last, we asked whether the SN-treated GSCs are more dependent on glycolysis than the control cells since OXPHOS is inhibited. To this end, we exposed the SN- or DMSO-treated GSCs to dichloroacetic acid (DCA), which would activate PDH [by inhibiting pyruvate dehydrogenase kinase (PDK)] and direct pyruvate away from glycolysis. As expected, the SN-treated GSCs were exquisitely sensitive to DCA treatment (Fig. 4J). In contrast to SN treatment, HLCS KD of 293T cells (under 3 days of biotin-free culturing conditions) did not alter acetyl-CoA and p-AMPK levels, as well as cellular respiration (fig. S4, C to E). Again, these results corroborate with a modest disruption of carboxylase biotinylation upon HLCS KD (Fig. 2F). Collectively, our metabolic studies support the view that SN inhibits multiple carboxylases in GSC, resulting in the metabolic reprogramming of GSC toward glycolysis and that this only occurs when the carboxylases are severely deprived of biotin.
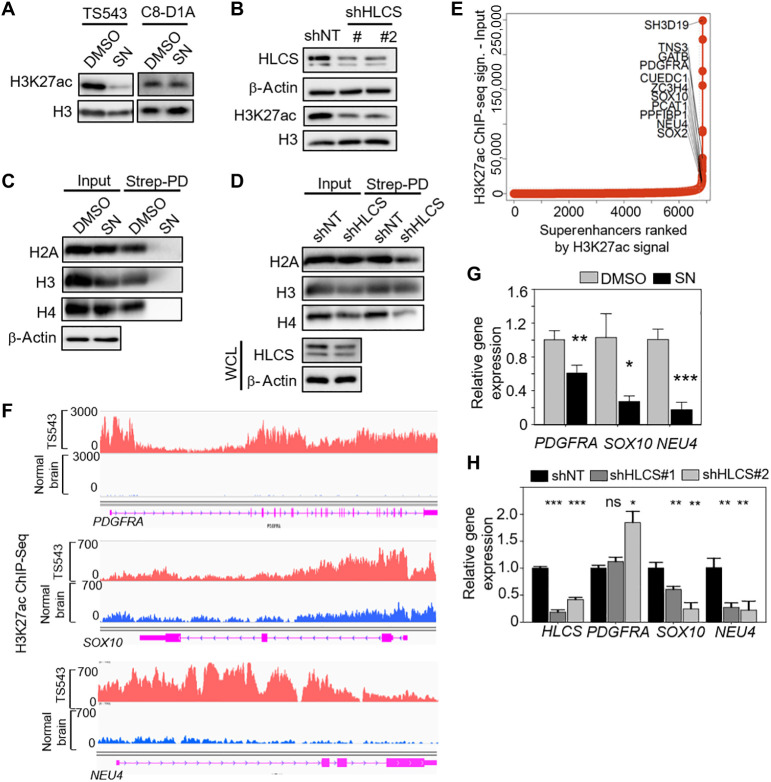
Reduced histone biotinylation correlates with decreased H3K27ac levels
Our RNA-Seq analysis revealed an enrichment of the “generic transcription pathway” among the down-regulated genes in SN-treated GSCs (Fig. 2B). Thus, we explored the possibility that histone 3 lysine 27 acetylation (H3K27ac) levels (an active enhancer mark; also associated with chromatin accessibility) may be reduced in SN-treated GSCs. SN treatment led to a selective reduction of H3K27ac levels in GSCs, but not mouse astrocytes (Fig. 5A). HLCS depletion also decreased H3K27ac levels in GSCs, indicating that this epigenetic change is related to the disruption of biotin distribution (Fig. 5B). Notably, we found that both SN treatment and HLCS KD reduced the levels of biotinylated histones, particularly H2A and H4 (Fig. 5, C and D), consistent with a previous report that HLCS can be found in the nucleus where it regulates histone biotinylation (13). Recently, cancer-associated superenhancers have emerged as key oncogenic drivers of tumor cells (30). These superenhancers are characterized by large clusters of transcriptional enhancers with high H3K27ac occupancy that promotes active transcription. Given the dramatic reduction of H3K27ac levels with SN treatment, we asked whether this epigenetic change may affect the transcription of superenhancer-associated genes that play a crucial role in GSC proliferation. To this end, we performed H3K27ac ChIP-seq (chromatin immunoprecipitation followed by next-generation sequencing) of GSC TS543 followed by the identification of the superenhancer-associated genes (Fig. 5E). SN treatment significantly reduced the mRNA levels of PDGFRA, SOX10, and NEU4, which play key roles in cancer stemness and tumorigenicity and are identified as superenhancer-associated genes in GSC TS543 (Fig. 5, F and G). Similarly, qPCR analysis of HLCS KD GSC also showed a significant decrease in SOX10 and NEU4, but not PDGFRA, mRNA levels (Fig. 5H). Thus, biotin distribution to histones can influence the epigenetic regulation of GSC critical genes.
Fig. 5. Reduced histone biotinylation correlates with decreased H3K27ac levels.
(A and B) Western blot analysis of H3K27ac levels in 5 μM SN–treated GSC TS543 and mouse astrocytes (3-day treatment) (A), as well as that in HLCS-depleted GSC (B). Total H3 and β-actin serve as the loading control, while HLCS serve as the positive control. (C and D) Western blot analysis of histone proteins in streptavidin pull-down histone extracts from 25 μM SN (24-hour treatment) or HLCS KD 293T cells, under biotin-free culturing conditions. β-Actin serves as the loading control, while HLCS serves as the positive control. (E) Hockey-stick plot display ranked superenhancers with their nearest gene neighbors detected from TS543 GSC. Superenhancers were defined by the ROSE software and ranked on the basis of the difference between the H3K27ac signal from ChIP-seq data and the respective matched ChIP input file. (F) H3K27ac signal at the superenhancer loci of representative superenhancer-associated genes. ChIP-seq signal from normal brain is used as a negative control. (G and H) qPCR analysis of PDGFRA, SOX10, and NEU4 mRNA levels in 5 μM SN- versus DMSO-treated GSC (G), as well as in shNT- versus shHLCS-transduced GSC TS543 (H). HSP70 and TBP serve as the housekeeping genes, while HLCS serves as the positive control (H) (n = 3) (means ± SD). ***P < 0.001; **P < 0.01; *P < 0.05. Two-tailed unpaired Student’s t test.
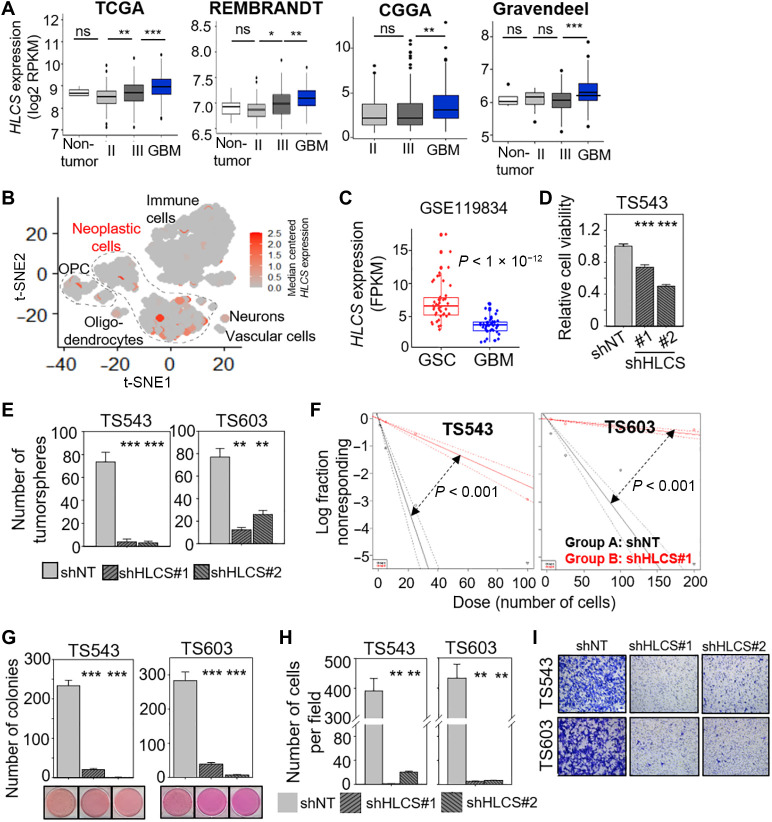
Genetic disruption of biotin distribution by HLCS depletion impairs GSC proliferation and invasiveness
Extending from our knowledge of SN’s mode of action and the heightened sensitivity of the GSCs to SN relative to the noncancerous mouse astrocytes, we then asked whether HLCS may be deregulated in GBM. HLCS is overexpressed in GBM in multiple patient cohorts and single-cell RNA-seq analysis when compared to other tumor cellular components (including immune cells, neurons, and oligodendrocytes) (Fig. 6, A and B) (31). Furthermore, HLCS expression was also higher in GSCs compared to bulk GBM, reinforcing the idea that the GSCs may be dependent on HLCS for their survival (Fig. 6C). Next, we evaluated whether HLCS is critical for GSC viability and found an average of 0.3- to 0.5-fold reduction of cell viability upon HLCS KD, which was quite similar to that achieved with SN treatment (Fig. 6D). HLCS KD also significantly reduced the number of tumorspheres and tumor-initiating cell frequency as determined using the extreme limiting dilution assay (Fig. 6, E and F). Moreover, HLCS-depleted GSC generated significantly less colonies than shNT-transduced GSCs in the soft agar assay, indicating a loss of transforming potential (Fig. 6G). Similar to SN treatment, HLCS KD also compromised GSC invasiveness when compared to the shNT control (Fig. 6, H and I). Thus, both pharmacologic and genetic disruption of biotin distribution impaired GSC proliferation and invasiveness, although HLCS KD appears to exert its effect mainly by affecting histone biotinylation and, hence, gene expression.
Fig. 6. Genetic disruption of biotin distribution by HLCS depletion impairs GSC proliferation and invasiveness.
(A) Correlative analysis of HLCS mRNA levels with glioma grades in four different glioma patient cohorts. Wilcoxon-Mann-Whitney exact test. *P < 0.05; **P < 0.01; ***P < 1 × 10−4. (B) scRNA-seq data from GBM specimens derived from GSE84465 (31) presented as a t-distributed stochastic neighbor embedding (t-SNE) plot demonstrating expression of HLCS across GBM cells, neurons, oligodendrocyte precursor cells (OPC), oligodendrocytes, and immune cells. Data are presented as median-centered mRNA expression in counts per million (CPM). (C) Correlative analysis of HLCS expression in GSC versus bulk GBM tumors from GSE119834 (45). (D) Cell viability assay of GSC with or without HLCS KD (n = 6) (means ± SD). (E) Tumorsphere formation of GSCs following HLCS KD (n = 6) (means ± SD). (F) Extreme limiting dilution assay of GSCs following HLCS KD (means ± SD). (G) Colony formation of GSC following HLCS KD (n = 4) (means ± SD). Representative images shown below. (H) Transwell migration and invasion assay of GSCs with HLCS KD (n = 3) (means ± SD). (I) Representative images of (H). (D), (E), (G), and (H): **P < 0.01; ***P < 0.001. Two-tailed unpaired Student’s t test.
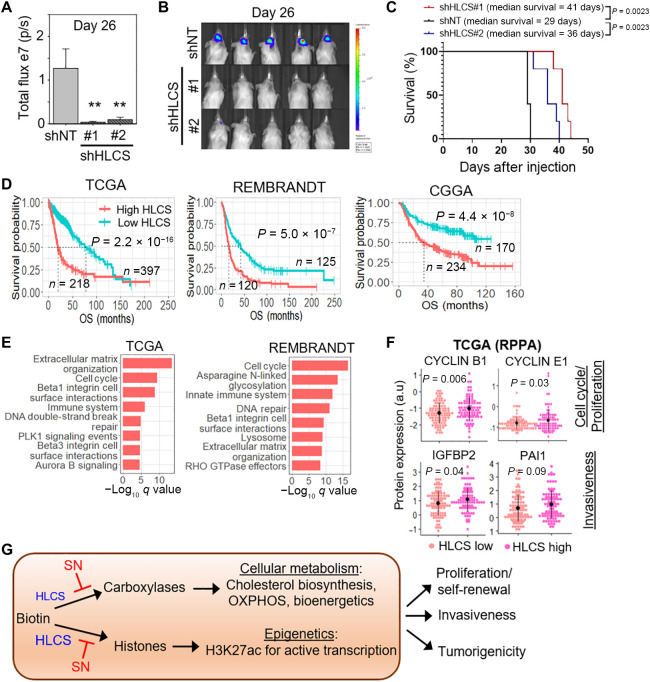
HLCS levels affect GSC tumorigenicity and aggressiveness of GBM
Next, we assessed the importance of optimal biotin distribution in GSC tumorigenicity by transplanting GSC, with or without HLCS KD, into immunocompromised mice. HLCS depletion resulted in a significant reduction of tumor volume and extended median mouse survival by ~7 to 12 days relative to that with shNT control (Fig. 7, A to C). Last, we interrogated the relevance of HLCS levels in influencing human GBM tumorigenicity. We first showed that high HLCS expression correlated with inferior glioma patient outcome in multiple patient cohorts (Fig. 7D). HLCS expression was also the highest in the Mes that was associated with poorest patient survival and high STAT3 (signal transducer and activator of transcription 3) signaling (fig. S7, A and B). Next, we compared HLCS expression in IDH1 wild type versus mutant GBM and found HLCS expression to be significantly higher in IDH1 wild type (worse outcome) than mutant GBM (fig. S7C). In contrast, there was no significant difference in HLCS expression between GBM with methylated or nonmethylated MGMT (O-6-methylguanine-DNA methyltransferase) promoter, suggesting that HLCS expression is unlikely to influence tumor response to temozolomide (fig. S7D). We did not observe any correlation between HLCS expression with PTEN gene status, TP53 gene status, and EGFR copy number, which are common genetic alterations in GBM (fig. S7, E to G).
Fig. 7. HLCS levels affect GSC tumorigenicity and aggressiveness of GBM.
(A and B) In vivo bioluminescence-based imaging of postorthotropic injection of GSC TS543 transduced with NT/control or HLCS shRNAs. Quantification of tumor volume based on bioluminescence (A) and representative images of the tumor-bearing mice (B) (n = 5) (means ± SD). (C) Survival curves of mice implanted with TS543 with or without HLCS KD. (D) Correlative analysis of HLCS levels with glioma patient survival in multiple glioma patient cohorts. OS: overall survival. Wald test. (E) Pathways that are enriched in HLCS correlated genes using ConsesusPathDB analysis. (F) Correlative analysis of HLCS mRNA levels with protein levels of cell cycle (CCNB1 and CCNE2) and invasiveness (IGFBP2 and PAI1) genes in GBM patients from TCGA. (G) Proposed model of how HLCS and SN may regulate biotin distribution to the carboxylases and histones, resulting in metabolic and epigenetic alterations in GSC.
In both TCGA and REMBRANDT datasets, we observed that genes that positively correlated with HLCS were significantly enriched in the “extracellular matrix organization,” “cell cycle,” and “beta1-integrin cell surface interaction” pathways (Fig. 7E), in agreement with our data that HLCS promotes GSC proliferation, invasiveness, and tumor-initiating cell frequency (Fig. 6). HLCS-high GBM was also associated with significantly higher levels of proteins that are involved in the cell cycle (cyclin B1 and cyclin E1) and invasiveness [IGFBP2 (insulin-like growth factor-binding protein 2) and PAI1 (plasminogen activator inhibitor 1] when compared to HLCS-low GBM in the TCGA reverse-phase protein array dataset (Fig. 7F). Thus, we conclude that GBM is dependent on optimal biotin distribution for its proliferation, invasiveness, and tumorigenicity.
DISCUSSION
In this study, we first identified an anti-GSC compound using CMA and thereafter exploited this small molecule to find previously unknown GBM dependencies, taking advantage of its poorly understood mode of action. On the basis of multiple lines of evidence—chemical pull-down, enzymatic, molecular docking, transcriptomic, and metabolomic data—we propose that SN impairs GSC proliferation and invasiveness by disrupting intracellular biotin distribution to biotin-dependent proteins, including the carboxylases and histones. By severely depriving the carboxylases of biotin, SN prevents the activation of multiple carboxylases resulting in metabolic alterations, including depletion of intracellular cholesterol, reduced TCA cycle flow, impaired OXPHOS, severe ATP deficit, and activation of AMPK (Fig. 7G). The metabolic changes in SN-treated GSC mirror that reported in experimental models of biotin-deficiency disorders, supporting the idea that SN treatment induces a biotin-deficient–like cellular state (7, 8, 25, 32). Biotin (vitamin H) is an essential nutrient that is obtained through the diet of higher organisms. Although numerous vitamins have been implicated in carcinogenesis, the role of biotin in cancer remains unclear. The importance of biotin in GBM metabolism is subtlely implied by previous findings showing that GBM relies on PC and PDH for glucose-dependent anaplerosis (33); inhibition of ACC1 and ACC2 reduced proliferation and de novo lipogenesis of GBM cells (34); and blocking fatty acid synthesis via fatty acid synthase (that functions downstream of ACC1) suppressed GBM growth (35). The aforementioned metabolic enzymes share a commonality in their reliance on biotin for activity, either directly or indirectly.
Despite the expectation that SN would create metabolic chaos in GSC, cholesterol depletion and OXPHOS impairment are prominently featured in SN-treated GSC, reinforcing the importance of these metabolic pathways in regulating GBM proliferation (23, 28). The selectivity of SN for GBM cell viability over that of mouse astrocytes may be explained by the greater dependency of GBM cells on cholesterol and OXPHOS. Cancer cells have an increased demand for cholesterol, and glioma cells take up significantly more cholesterol than normal human astrocytes (36). Accordingly, GBM is uniquely sensitive to liver X receptor (LXR) agonists, which deplete intracellular cholesterol by effluxing cholesterol through up-regulated ABCA1 (ATP binding cassette subfamily A member 1) transporter and reducing cholesterol uptake by degrading the low-density lipoprotein receptor (23). In the case of OXPHOS, glioma cells have up-regulated OXPHOS, and OXPHOS inhibitors have been shown to inhibit GBM cell proliferation in vitro and in vivo (5, 33).
Although it is widely accepted that HLCS plays a major role in carboxylase activation, we found that HLCS depletion (at least under 3 days of biotin-free culturing conditions) modestly deprives the carboxylases of biotin and did not recapitulate the metabolic defects of SN-treated cells. This suggests that HLCS may play a minor role in regulating cellular metabolism at least under short-term in vitro conditions or that HLCS may be preferentially localized to the nucleus instead of cytoplasm as previously shown for HeLa cells (Fig. 7G) (13). Given that GBM metabolism may differ in vitro versus in vivo (where conditions are harsher), it would be interesting to evaluate whether HLCS deficiency may influence tumor metabolism in vivo. Our results are consistent with patient data whereby the metabolic defects only manifest when mutant HLCS proteins have extremely low affinity for biotin (e.g., 70-fold reduction that leads to a severe disruption in carboxylase activation), and that the biochemical and clinical manifestations of the disease can be reversed with pharmacological doses of biotin (10 to 100 mg/day) (6).
The cross-talk between metabolism and epigenetics can influence cancer cell state and fate, but this is poorly understood in GBM. In addition to its central role in metabolism, biotin bioavailability can affect gene expression, although the underlying mechanism remains obscure (37). We established a link between reduced histone biotinylation and decreased histone acetylation in SN-treated and HLCS KD GSC, indicating that a major role of HLCS lies in its distribution of biotin to histones in GBM (Fig. 7G). Numerous studies have used the analysis of superenhancers, characterized by dense H3K27ac occupancy, to identify genes and pathways that indicate the cell of origin and maintain the malignant state of gliomas (38, 39). Notably, both SN treatment and HLCS KD reduced the expression of superenhancer-associated genes, including SOX10 and NEU4, which are crucial for tumor stemness and malignancy (40, 41). Therefore, the dependence of GBM on HLCS-regulated histone biotinylation serves to sustain specific epigenetic demands of GBM. In multiple patient cohorts, high HLCS expression reliably portended a negative prognosis in GBM. HLCS is also overexpressed in breast cancer and associated with metastasis (42). Thus, our studies with SN illuminate a previously unrecognized role of HLCS in GBM pathogenesis and suggest that HLCS deregulation may be broadly relevant to other cancer types.
HLCS loss-of-function mutations lead to biotin deficiency disorders, indicating that HLCS inhibitors (no such compound to date) are predicted to be unsuitable for GBM treatment. That SN only exhibits anti-GSC property at micromolar concentrations would also exclude its potential use in the clinic. Extending from the present work, we envision that the rational inhibition of biotin-dependent metabolism and epigenetic pathways may represent new approaches in the design of anti-GBM treatment options. For instance, one may consider cotargeting multiple metabolism pathways, including cholesterol biosynthesis [e.g., via statins and LXR-623 (23)] and OXPHOS (e.g., via IACS-010759) or cholesterol biosynthesis and ACC1 (e.g., TOFA), to deliver a synergistic anti-GBM effect. As proof of concept, we found that lovastatin and TOFA synergistically reduced cell viability of GSCs, but not noncancerous astrocytes (fig. S8, A to C). Alternatively, we may consider cotargeting specific metabolism (e.g., cholesterol biosynthesis and OXPHOS) and epigenetic [e.g., inhibiting bromodomain and extraterminal (BET) proteins that regulate superenhancers via JQ1 or dBET6] pathways, which would partially mimic the effect of SN treatment. Of immediate clinical translation, our findings raise an important consideration of regulating biotin consumption in patients with GBM.
MATERIALS AND METHODS
For details of standard lab techniques, unsupervised clustering of patients with glioma based on query gene signature, chemical synthesis of BSN, model generation for MD simulation, metabolomic analysis, RNA-seq analysis, ChIP-seq analysis, and superenhancer analysis, see the Supplementary Materials.
Connectivity Map Analysis
CMA (Build2; Broad Institute, http://broad.mit.edu/cmap) was used for the in silico drug screen (19) with our query signature (table S1). Gene symbols for each of three CMA gene lists were converted into the U133A&B Affymetrix probe set lists (NetAffx Query, https://affymetrix.com/analysis/index.affx) required as an input for CMA database. Last, “EGFR signaling,” “Mesenchymal subtype of GBM,” and “Generation of neuron” gene sets were represented by 132, 176, and 155 Affymetrix probe sets, respectively. They were used to query up to 7000 unique gene expression profiles (and representing 1309 compounds) stored in the CMA database (19). We considered the top 10 hits as potential drug candidates (cutoff permutation P < 0.05 and enrichment score > 0.8).
Cell lines and compounds
Patient-derived GSCs were provided by C. Brennan (Memorial Sloan Kettering Cancer Center) and R. A. DePinho at MD Anderson Cancer Center. The GSCs were cultured in human neural stem cell maintenance media (Millipore), 1% penicillin-streptomycin (PS), and supplemented with EGF and β fibroblast growth factor (20 ng ml−1 each). Noncancerous mouse astrocytes (C8-D1A) were provided by T. V. Arumugam (La Trobe University) and cultured with Dulbecco’s modified Eagle’s medium/nutrient mixture F12 (DMEM/F12) with 10% fetal bovine serum (FBS) and 1% PS. Human embryonic kidney–293 (HEK-293) T cells were cultured with DMEM with 10% FBS and 1% PS. The following inhibitors were used in this study: sulconazole nitrate (Selleckchem), fluconazole and tebuconazole (Sigma-Aldrich), lovastatin hydroxy acid (Cayman), and DCA (Sigma-Aldrich). Cholesterol–methyl-β-cyclodextrin (Sigma-Aldrich) was used in rescue experiments.
Filipin staining
Filipin staining was done using a Cholesterol Assay Kit (Cell-Based, ab133116, Abcam). Briefly, cells were seeded and treated on the following day with DMSO, SN, or lovastatin. After 3 days of incubation, the cells were fixed, stained with the diluted Filipin III, washed, and imaged at least 20 fields at ×10 magnification with an excitation of 340 to 380 nm and emission of 385 to 470 nm. Image J was used for determination of stain intensity. Data presented are from triplicate experiments.
Streptavidin pull-down assay
HEK-293T cells were treated with the indicated drugs for 6 hours and lysed in IP lysis buffer [50 mM tris-Cl (pH 7.5), 150 mM NaCl, and 1% Triton X-100] with protease inhibitor and phosphatase inhibitor. Equal amount of proteins was incubated with streptavidin beads (20347, Thermo Fisher Scientific) at 4°C for 16 hours. The beads were washed with IP lysis buffer five times, and the streptavidin-bound proteins were eluted with 2× SDS sample loading dye. The same volume of eluted proteins was resolved using SDS electrophoresis detected using either silver staining or Western blot analysis. The isolation of histone proteins was performed according to an Abcam online protocol. Briefly, cells were incubated with Triton extraction buffer [PBS containing 0.5% Triton X-100 (v/v) with protease inhibitor] for 10 min and the nuclei were collected by centrifugation. After overnight incubation of the pellet in 0.2 N HCl to extract histone proteins, the samples were centrifuged, neutralized with NaOH, and used for streptavidin pull-down assay.
ACC1 enzyme activity assay
The measurement of the recombinant ACC1 enzyme was performed using the ACC1 Assay Kit (79315, BPS Bioscience) following the manufacturer’s instructions.
Measurement of acetyl-CoA levels
After 3 days of drug treatment, the cells were lysed with immunoprecipitation assay buffer, and the resulting lysates were deproteinized using the Deproteinizing Sample Preparation Kit (B808-200, BioVision). Next, the measurement of acetyl-CoA levels was performed using the PicoProbe Acetyl-CoA Fluorometric Assay Kit (K317-100, BioVision) following the manufacturer’s instructions.
Mitochondrial oxygen consumption rates measurement (Seahorse assay)
Mitochondrial oxygen consumption rates (OCRs) were measured using XFe96 Seahorse Biosciences Extracellular Flux Analyzer (Agilent Technologies). After 20 hours of the indicated drug treatment, the cells were replated onto the Seahorse 96-well plate at 125,000 cells per well to ensure that only the OCR of viable cells were recorded. On the next day, the culture medium was replaced with 175 μl of fresh Seahorse DMEM basal medium 45 min before the assay. Seahorse analyzer injection ports were filled with 1 μM oligomycin, 1 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone, or 0.5 μM each of rotenone and antimycin A for OCR, following the manufacturer’s instructions. Levels of OCR were recorded, normalized, and quantified according to the manufacturer’s instructions.
Intracranial tumor formation in vivo
GSCs (1 × 105 viable cells) were grafted intracranially into NSG (NOD scid gamma mouse) mice (InVivos) aged 6 to 8 weeks. Tumor incidence was determined at indicated time points by luciferase imaging of mice using Xenogen IVIS (PerkinElmer) according to the manufacturer’s instructions. Animals were maintained until neurological signs were apparent, at which point they were euthanized. All mouse manipulations were performed with the approval of National University of Singapore Institutional Animal Care and Use Committee.
Statistical analyses
Student’s t test and Mann-Whitney test were performed using either R3.4.1 or Cytel Studio (version 9.0.0). Significance was defined as P < 0.05. For multiple testing correction, Benjamini-Hochberg statistic was applied to estimate the false discovery rate. One-dimensional data-driven grouping method (43, 44) was used to estimate the significance of association of HLCS gene with cancer patient’s survival. Details are shown in the Supplementary Materials. SigmaPlot (version 11.0) or a set of R packages (ggplot2, ggpubr, and survminer) were implemented for plot generation. For all experiments with error bars, the standard error of the mean (SEM) was calculated to indicate the variation within each experiment and data, and values represent means ± SEM or means ± SD, as indicated in the figure legends.
Acknowledgments
We thank C. Brennan, R. A. DePinho, and T. V. Arumugam for the cell lines; the Duke-NUS metabolomic facility (H. N. Wee, K. V. Chua, and L. S. Lee) for the metabolomic analyses; and C. Tang (NNI, Singapore) for training on the intracranial injection procedure. Funding: This work was supported by the National Research Foundation Fellowship NRF-NRFF2017-01 (D.S.T.O.), National University of Singapore (NUS) start-up grant (D.S.T.O. and M.J.K.), NUS President’s Assistant Professorship (D.S.T.O.), and NUS Research Scholarships (M.J.Y.A., B.W.L.L., Y.H.C., and R.T.M.). S.K. and C.S.V. thank A*STAR and NSCC for support. Author contributions: D.S.T.O. conceived the project and provided project leadership. J.Y. performed experiments involving GSCs and the streptavidin pull-down assay. O.V.G., R.T.M., and T.B. provided bioinformatics support. S.K. and C.S.V. assisted in the molecular docking and MD simulation experiments. C.L., H.Z., and M.J.K. carried out the synthesis of BSN. J.H.H. and S.-Y.N. assisted in the Seahorse analysis. J.J.L., T.B.T., and E.K.-H.C. assisted in the drug combination experiments. M.J.Y.A., Z.L., E.X.Y.T., K.Z.L., B.W.L.L., Y.H.C., K.C., and X.L. assisted with various GSC assays. J.C. and J.-P.K. assisted in the metabolomic analyses. Competing interests: S.K. and C.S.V. are founders of Sinopsee Therapeutics and Aplomex. The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Raw data from RNA-seq of DMSO- and SN-treated GSC TS543 and H3K27ac ChIP-seq of GSC TS543 are available on the Gene Expression Omnibus database (GSE149366 and GSE152862).
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S8
Tables S1 to S4
Data S1 to S4
References
REFERENCES AND NOTES
- 1.Hu B. L., Wang Q., Wang Y. A., Hua S., Sauvé C.-E. G., Ong D., Lan Z. D., Chang Q., Ho Y. W., Monasterio M. M., Lu X., Zhong Y., Zhang J., Deng P., Tan Z., Wang G., Liao W.-T., Corley L. J., Yan H., Zhang J., You Y., Liu N., Cai L., Finocchiaro G., Phillips J. J., Berger M. S., Spring D. J., Hu J., Sulman E. P., Fuller G. N., Chin L., Verhaak R. G. W., De Pinho R. A., Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell 167, 1281–1295.e18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ong D. S. T., Hu B., Ho Y. W., Sauvé C. E. G., Bristow C. A., Wang Q., Multani A. S., Chen P., Nezi L., Jiang S., Gorman C. E., Monasterio M. M., Koul D., Marchesini M., Colla S., Jin E. J., Sulman E. P., Spring D. J., Yung W. K. A., Verhaak R. G. W., Chin L., Wang Y. A., DePinho R. A., PAF promotes stemness and radioresistance of glioma stem cells. Proc. Natl. Acad. Sci. U.S.A. 114, E9086–E9095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J., Li Y., Yu T.-S., McKay R. M., Burns D. K., Kernie S. G., Parada L. F., A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488, 522–526 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bao S. D., Wu Q., Lendon R. E. M., Hao Y., Shi Q., Hjelmeland A. B., Dewhirst M. W., Bigner D. D., Rich J. N., Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Bi J., Chowdhry S., Wu S., Zhang W., Masui K., Mischel P. S., Altered cellular metabolism in gliomas—An emerging landscape of actionable co-dependency targets. Nat. Rev. Cancer 20, 57–70 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Leon-Del-Rio A., Biotin in metabolism, gene expression, and human disease. J. Inherit. Metab. Dis. 42, 647–654 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Hernández-Vázquez A., Wolf B., Pindolia K., Ortega-Cuellar D., Hernández-González R., Heredia-Antúnez A., Ibarra-González I., Velázquez-Arellano A., Biotinidase knockout mice show cellular energy deficit and altered carbon metabolism gene expression similar to that of nutritional biotin deprivation: Clues for the pathogenesis in the human inherited disorder. Mol. Genet. Metab. 110, 248–254 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Ochoa-Ruiz E., Díaz-Ruiz R., de J Hernández-Vázquez A., Ibarra-González I., Ortiz-Plata A., Rembao D., Ortega-Cuéllar D., Viollet B., Uribe-Carvajal S., Corella J. A., Velázquez-Arellano A., Biotin deprivation impairs mitochondrial structure and function and has implications for inherited metabolic disorders. Mol. Genet. Metab. 116, 204–214 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Tong L., Structure and function of biotin-dependent carboxylases. Cell. Mol. Life Sci. 70, 863–891 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hymes J., Wolf B., Biotinidase and its roles in biotin metabolism. Clin. Chim. Acta 255, 1–11 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Kothapalli N., Zempleni J., Biotinylation of histones depends on the cell cycle in NCI-H69 small cell lung cancer cells. Faseb J. 19, A55 (2005). [Google Scholar]
- 12.Hassan Y. I., Zempleni J., Epigenetic regulation of chromatin structure and gene function by biotin. J. Nutr. 136, 1763–1765 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narang M. A., Dumas R., Ayer L. M., Gravel R. A., Reduced histone biotinylation in multiple carboxylase deficiency patients: A nuclear role for holocarboxylase synthetase. Hum. Mol. Genet. 13, 15–23 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Russell-Jones G., McTavish K., McEwan J., Rice J., Nowotnik D., Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumours. J. Inorg. Biochem. 98, 1625–1633 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Chen S., Zhao X., Chen J., Chen J., Kuznetsova L., Wong S. S., Ojima I., Mechanism-based tumor-targeting drug delivery system. Validation of efficient vitamin receptor-mediated endocytosis and drug release. Bioconjug. Chem. 21, 979–987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan C. W., Verhaak R. G. W., McKenna A., Campos B., Noushmehr H., Salama S. R., Zheng S., Chakravarty D., Sanborn J. Z., Berman S. H., Beroukhim R., Bernard B., Wu C. J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L., Vegesna R., Shukla S. A., Ciriello G., Yung W. K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D. D., van Meir E. G., Prados M., Sloan A., Black K. L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D. W., Guha A., Iacocca M., O’Neill B. P., Foltz G., Myers J., Weisenberger D. J., Penny R., Kucherlapati R., Perou C. M., Hayes D. N., Gibbs R., Marra M., Mills G. B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P. W., Haussler D., Getz G., Chin L., Benz C., Barnholtz-Sloan J., Barrett W., Ostrom Q., Wolinsky Y., Black K. L., Bose B., Boulos P. T., Boulos M., Brown J., Czerinski C., Eppley M., Iacocca M., Kempista T., Kitko T., Koyfman Y., Rabeno B., Rastogi P., Sugarman M., Swanson P., Yalamanchii K., Otey I. P., Liu Y. S., Xiao Y., Auman J. T., Chen P. C., Hadjipanayis A., Lee E., Lee S., Park P. J., Seidman J., Yang L., Kucherlapati R., Kalkanis S., Mikkelsen T., Poisson L. M., Raghunathan A., Scarpace L., Bernard B., Bressler R., Eakin A., Iype L., Kreisberg R. B., Leinonen K., Reynolds S., Rovira H., Thorsson V., Shmulevich I., Annala M. J., Penny R., Paulauskis J., Curley E., Hatfield M., Mallery D., Morris S., Shelton T., Shelton C., Sherman M., Yena P., Cuppini L., DiMeco F., Eoli M., Finocchiaro G., Maderna E., Pollo B., Saini M., Balu S., Hoadley K. A., Li L., Miller C. R., Shi Y., Topal M. D., Wu J., Dunn G., Giannini C., O’Neill B. P., Aksoy B. A., Antipin Y., Borsu L., Berman S. H., Brennan C. W., Cerami E., Chakravarty D., Ciriello G., Gao J., Gross B., Jacobsen A., Ladanyi M., Lash A., Liang Y., Reva B., Sander C., Schultz N., Shen R., Socci N. D., Viale A., Ferguson M. L., Chen Q. R., Demchok J. A., Dillon L. A. L., Shaw K. R. M., Sheth M., Tarnuzzer R., Wang Z., Yang L., Davidsen T., Guyer M. S., Ozenberger B. A., Sofia H. J., Bergsten J., Eckman J., Harr J., Myers J., Smith C., Tucker K., Winemiller C., Zach L. A., Ljubimova J. Y., Eley G., Ayala B., Jensen M. A., Kahn A., Pihl T. D., Pot D. A., Wan Y., Eschbacher J., Foltz G., Hansen N., Hothi P., Lin B., Shah N., Yoon J. G., Lau C., Berens M., Ardlie K., Beroukhim R., Carter S. L., Cherniack A. D., Noble M., Cho J., Cibulskis K., DiCara D., Frazer S., Gabriel S. B., Gehlenborg N., Gentry J., Heiman D., Kim J., Jing R., Lander E. S., Lawrence M., Lin P., Mallard W., Meyerson M., Onofrio R. C., Saksena G., Schumacher S., Sougnez C., Stojanov P., Tabak B., Voet D., Zhang H., Zou L., Getz G., Dees N. N., Ding L., Fulton L. L., Fulton R. S., Kanchi K. L., Mardis E. R., Wilson R. K., Baylin S. B., Andrews D. W., Harshyne L., Cohen M. L., Devine K., Sloan A. E., VandenBerg S. R., Berger M. S., Prados M., Carlin D., Craft B., Ellrott K., Goldman M., Goldstein T., Grifford M., Haussler D., Ma S., Ng S., Salama S. R., Sanborn J. Z., Stuart J., Swatloski T., Waltman P., Zhu J., Foss R., Frentzen B., Friedman W., McTiernan R., Yachnis A., Hayes D. N., Perou C. M., Zheng S., Vegesna R., Mao Y., Akbani R., Aldape K., Bogler O., Fuller G. N., Liu W., Liu Y., Lu Y., Mills G., Protopopov A., Ren X., Sun Y., Wu C. J., Yung W. K. A., Zhang W., Zhang J., Chen K., Weinstein J. N., Chin L., Verhaak R. G. W., Noushmehr H., Weisenberger D. J., Bootwalla M. S., Lai P. H., Triche T. J. Jr., van den Berg D. J., Laird P. W., Gutmann D. H., Lehman N. L., VanMeir E. G., Brat D., Olson J. J., Mastrogianakis G. M., Devi N. S., Zhang Z., Bigner D., Lipp E., McLendon R., The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verhaak R. G. W., Hoadley K. A., Purdom E., Wang V., Qi Y., Wilkerson M. D., Miller C. R., Ding L., Golub T., Mesirov J. P., Alexe G., Lawrence M., O’Kelly M., Tamayo P., Weir B. A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H. S., Hodgson J. G., James C. D., Sarkaria J. N., Brennan C., Kahn A., Spellman P. T., Wilson R. K., Speed T. P., Gray J. W., Meyerson M., Getz G., Perou C. M., Hayes D. N., Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzoleni S., Politi L. S., Pala M., Cominelli M., Franzin A., Sergi Sergi L., Falini A., de Palma M., Bulfone A., Poliani P. L., Galli R., Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 70, 7500–7513 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., Golub T. R., The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Fromtling R. A., Overview of medically important antifungal azole derivatives. Clin. Microbiol. Rev. 1, 187–217 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lepesheva G. I., Friggeri L., Waterman M. R., CYP51 as drug targets for fungi and protozoan parasites: Past, present and future. Parasitology 145, 1820–1836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad F., Sun Q., Patel D., Stommel J. M., Cholesterol metabolism: A potential therapeutic target in glioblastoma. Cancer 11, 146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Villa G. R., Hulce J. J., Zanca C., Bi J., Ikegami S., Cahill G. L., Gu Y., Lum K. M., Masui K., Yang H., Rong X., Hong C., Turner K. M., Liu F., Hon G. C., Jenkins D., Martini M., Armando A. M., Quehenberger O., Cloughesy T. F., Furnari F. B., Cavenee W. K., Tontonoz P., Gahman T. C., Shiau A. K., Cravatt B. F., Mischel P. S., An LXR-cholesterol axis creates a metabolic co-dependency for brain cancers. Cancer Cell 30, 683–693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burri B. J., Sweetman L., Nyhan W. L., Heterogeneity of holocarboxylase synthetase in patients with biotin-responsive multiple carboxylase deficiency. Am. J. Hum. Genet. 37, 326–337 (1985). [PMC free article] [PubMed] [Google Scholar]
- 25.Hernández-Vázquez A., Ochoa-Ruiz E., Ibarra-González I., Ortega-Cuellar D., Salvador-Adriano A., Velázquez-Arellano A., Temporal development of genetic and metabolic effects of biotin deprivation. A search for the optimum time to study a vitamin deficiency. Mol. Genet. Metab. 107, 345–351 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Mock D. M., Biotin: From nutrition to therapeutics. J. Nutr. 147, 1487–1492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim C.-W., Addy C., Kusunoki J., Anderson N. N., Deja S., Fu X., Burgess S. C., Li C., Ruddy M., Chakravarthy M., Previs S., Milstein S., Fitzgerald K., Kelley D. E., Horton J. D., Acetyl CoA carboxylase inhibition reduces hepatic steatosis but elevates plasma triglycerides in mice and humans: A bedside to bench investigation. Cell Metab. 26, 394–406.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y., Lim S. K., Liang Q., Iyer S. V., Wang H.-Y., Wang Z., Xie X., Sun D., Chen Y.-J., Tabar V., Gutin P., Williams N., De Brabander J. K., Parada L. F., Gboxin is an oxidative phosphorylation inhibitor that targets glioblastoma. Nature 567, 341–346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta B., Chhipa R. R., Evolving lessons on the complex role of AMPK in normal physiology and cancer. Trends Pharmacol. Sci. 37, 192–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang F., Yang Z., Tan Y., Li Y., Super-enhancer function and its application in cancer targeted therapy. npj Precis. Oncol. 4, 2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darmanis S., Sloan S. A., Croote D., Mignardi M., Chernikova S., Samghababi P., Zhang Y., Neff N., Kowarsky M., Caneda C., Li G., Chang S. D., Connolly I. D., Li Y., Barres B. A., Gephart M. H., Quake S. R., Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 21, 1399–1410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suchy S. F., Wolf B., Effect of biotin deficiency and supplementation on lipid metabolsim in rats: Cholesterol and lipoproteins. Am. J. Clin. Nutr. 43, 831–838 (1986). [DOI] [PubMed] [Google Scholar]
- 33.Marin-Valencia I., Yang C., Mashimo T., Cho S., Baek H., Yang X.-L., Rajagopalan K. N., Maddie M., Vemireddy V., Zhao Z., Cai L., Good L., Tu B. P., Hatanpaa K. J., Mickey B. E., Matés J. M., Pascual J. M., Maher E. A., Malloy C. R., DeBerardinis R. J., Bachoo R. M., Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 15, 827–837 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones J. E. C., Esler W. P., Patel R., Lanba A., Vera N. B., Pfefferkorn J. A., Vernochet C., Inhibition of acetyl-CoA carboxylase 1 (ACC1) and 2 (ACC2) reduces proliferation and de novo lipogenesis of EGFRvIII human glioblastoma cells. PLOS ONE 12, e0169566 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Röhrig F., Schulze A., The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Murakami M., Ushio Y., Mihara Y., Kuratsu J.-I., Horiuchi S., Morino Y., Cholesterol uptake by human glioma cells via receptor-mediated endocytosis of low-density lipoprotein. J. Neurosurg. 73, 760–767 (1990). [DOI] [PubMed] [Google Scholar]
- 37.Smith E. M., Hoi J. T., Eissenberg J. C., Shoemaker J. D., Neckameyer W. S., Ilvarsonn A. M., Harshman L. G., Schlegel V. L., Zempleni J., Feeding Drosophila a biotin-deficient diet for multiple generations increases stress resistance and lifespan and alters gene expression and histone Biotinylation patterns. J. Nutr. 137, 2006–2012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagaraja S., Vitanza N. A., Woo P. J., Taylor K. R., Liu F., Zhang L., Li M., Meng W., Ponnuswami A., Sun W., Ma J., Hulleman E., Swigut T., Wysocka J., Tang Y., Monje M., Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell 31, 635–652.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gimple R. C., Kidwell R. L., Kim L. J. Y., Sun T., Gromovsky A. D., Wu Q., Wolf M., Lv D., Bhargava S., Jiang L., Prager B. C., Wang X., Ye Q., Zhu Z., Zhang G., Dong Z., Zhao L., Lee D., Bi J., Sloan A. E., Mischel P. S., Brown J. M., Cang H., Huan T., Mack S. C., Xie Q., Rich J. N., Glioma stem cell-specific superenhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov. 9, 1248–1267 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dravis C., Spike B. T., Harrell J. C., Johns C., Trejo C. L., Southard-Smith E. M., Perou C. M., Wahl G. M., Sox10 regulates stem/progenitor and mesenchymal cell states in mammary epithelial cells. Cell Rep. 12, 2035–2048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silvestri I., Testa F., Zappasodi R., Cairo C. W., Zhang Y., Lupo B., Galli R., Di Nicola M., Venerando B., Tringali C., Sialidase NEU4 is involved in glioblastoma stem cell survival. Cell Death Dis. 5, e1381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sukjoi W., Siritutsoontorn S., Chansongkrow P., Waiwitlikhit S., Polyak S. W., Warnnissorn M., Charoensawan V., Thuwajit C., Jitrapakdee S., Overexpression of holocarboxylase synthetase predicts lymph node metastasis and unfavorable prognosis in breast cancer. Anticancer Res. 40, 4557–4565 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Chen L., Jenjaroenpun P., Pillai A. M. C., Ivshina A. V., Ow G. S., Efthimios M., Zhiqun T., Tan T. Z., Lee S.-C., Rogers K., Ward J. M., Mori S., Adams D. J., Jenkins N. A., Copeland N. G., Ban K. H.-K., Kuznetsov V. A., Thiery J. P., Transposon insertional mutagenesis in mice identifies human breast cancer susceptibility genes and signatures for stratification. Proc. Natl. Acad. Sci. U.S.A. 114, E2215–E2224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grinchuk O. V., Yenamandra S. P., Iyer R., Singh M., Lee H. K., Lim K. H., Chow P. K.-H., Kuznetsov V. A., Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol. Oncol. 12, 89–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mack S. C., Singh I., Wang X., Hirsch R., Wu Q., Villagomez R., Bernatchez J. A., Zhu Z., Gimple R. C., Kim L. J. Y., Morton A., Lai S., Qiu Z., Prager B. C., Bertrand K. C., Mah C., Zhou W., Lee C., Barnett G. H., Vogelbaum M. A., Sloan A. E., Chavez L., Bao S., Scacheri P. C., Siqueira-Neto J. L., Lin C. Y., Rich J. N., Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J. Exp. Med. 216, 1071–1090 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowman R. L., Wang Q., Carro A., Verhaak R. G. W., Squatrito M., GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 19, 139–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Šali A., Blundell T. L., Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993). [DOI] [PubMed] [Google Scholar]
- 49.Kannan S., Pradhan M. R., Tiwari G., Tan W.-C., Chowbay B., Tan E. H., Tan D. S.-W., Verma C., Hydration effects on the efficacy of the epidermal growth factor receptor kinase inhibitor afatinib. Sci. Rep. 7, 1540 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lescar J., Meyer I., Akshita K., Srinivasaraghavan K., Verma C., Palous M., Mazier D., Datry A., Fekkar A., Aspergillus fumigatus harbouring the sole Y121F mutation shows decreased susceptibility to voriconazole but maintained susceptibility to itraconazole and posaconazole. J. Antimicrob. Chemother. 69, 3244–3247 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Darden T., York D., Pedersen L., Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 52.Miyamoto S., Kollman P. A., Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992). [Google Scholar]
- 53.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muoio D. M., Noland R. C., Kovalik J.-P., Seiler S. E., Davies M. N., DeBalsi K. L., Ilkayeva O. R., Stevens R. D., Kheterpal I., Zhang J., Covington J. D., Bajpeyi S., Ravussin E., Kraus W., Koves T. R., Mynatt R. L., Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 15, 764–777 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newgard C. B., An J., Bain J. R., Muehlbauer M. J., Stevens R. D., Lien L. F., Haqq A. M., Shah S. H., Arlotto M., Slentz C. A., Rochon J., Gallup D., Ilkayeva O., Wenner B. R., Yancy W. S. Jr., Eisenson H., Musante G., Surwit R. S., Millington D. S., Butler M. D., Svetkey L. P., A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowman S. K., Simon M. D., Deaton A. M., Tolstorukov M., Borowsky M. L., Kingston R. E., Multiplexed Illumina sequencing libraries from picogram quantities of DNA. BMC Genomics 14, 466 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madhavan S., Zenklusen J.-C., Kotliarov Y., Sahni H., Fine H. A., Buetow K., Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 7, 157–167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gravendeel L. A. M., Kouwenhoven M. C. M., Gevaert O., de Rooi J. J., Stubbs A. P., Duijm J. E., Daemen A., Bleeker F. E., Bralten L. B. C., Kloosterhof N. K., De Moor B., Eilers P. H. C., van der Spek P. J., Kros J. M., Sillevis Smitt P. A. E., van den Bent M. J., French P. J., Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 69, 9065–9072 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Wang Y., Qian T., You G., Peng X., Chen C., You Y., Yao K., Wu C., Ma J., Sha Z., Wang S., Jiang T., Localizing seizure-susceptible brain regions associated with low-grade gliomas using voxel-based lesion-symptom mapping. Neuro Oncol. 17, 282–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roadmap Epigenomics Consortium , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan M. S. Y., Sandanaraj E., Chong Y. K., Lim S. W., Koh L. W. H., Ng W. H., Tan N. S., Tan P., Ang B. T., Tang C., A STAT3-based gene signature stratifies glioma patients for targeted therapy. Nat. Commun. 10, 3601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S8
Tables S1 to S4
Data S1 to S4
References