Protection in vaccinated hosts occurs quickly upon pathogen-specific memory CD8+ T cell–mediated activation of myeloid cells.
Abstract
While cognate antigen drives clonal expansion of memory CD8+ T (CD8+ TM) cells to achieve sterilizing immunity in immunized hosts, not much is known on how cognate antigen contributes to early protection before clonal expansion occurs. Here, using distinct models of immunization, we establish that cognate antigen recognition by CD8+ TM cells on dendritic cells initiates their rapid and coordinated production of a burst of CCL3, CCL4, and XCL1 chemokines under the transcriptional control of interferon (IFN) regulatory factor 4. Using intravital microscopy imaging, we reveal that CD8+ TM cells undergo antigen-dependent arrest in splenic red pulp clusters of CCR2+Ly6C+ monocytes to which they deliver IFNγ and chemokines. IFNγ enables chemokine-induced microbicidal activities in monocytes for protection. Thus, rapid and effective CD8+ TM cell responses require spatially and temporally coordinated events that quickly restrict microbial pathogen growth through the local delivery of activating chemokines to CCR2+Ly6C+ monocytes.
INTRODUCTION
CD8+ T cells have the unique ability to sense and recognize antigens (Ags) derived from intracellular pathogens and tumors (1–3). Live attenuated vaccines using viral backbones [e.g., vaccinia and vesicular stomatitis virus (VSV)] or intracellular bacteria such as Listeria monocytogenes (Lm) and mycobacteria (Bacille Calmette-Guérin) are known to promote robust CD8+ T cell responses and establish a pool of systemic and tissue-resident long-lived memory CD8+ T (CD8+ TM) cells. These CD8+ TM cells can rapidly react against immunizing Ags expressed in live vectors and provide immunity against life-threatening diseases (4–6). However, much investigation remains to be conducted to achieve a detailed understanding of (i) the role of cognate Ag and (ii) the sequences of events that need to take place for host protection.
It is well established that cognate Ag needs to be presented on dendritic cells (DCs) for optimal clonal reexpansion of both systemic and tissue-resident CD8+ TM cells (7–10). Rapid clonal expansion ensures that sufficient numbers of pathogen-specific effector memory cells are generated to effectively sterilize an infection (1, 3). We and others have shown in models of systemic bacterial and viral infections that both DCs and CCR2+Ly6C+ inflammatory monocytes also provide inflammatory signals that contribute to the early reactivation of CD8+ TM cells in situ (11, 12). Through the production of multiple inflammatory cytokines—i.e., interleukin-12 (IL-12), IL-18, IL-15, and type I interferon (IFN)—these cells can orchestrate rapid Ag-independent activation of CD8+ TM cells (also known as “bystander” activation), including their differentiation into IFNγ-secreting natural killer group 2D (NKG2D+) effector cells (11, 13–16). While this early cytokine-driven activation of CD8+ TM cells contributes to innate protection, cognate Ag recognition is nevertheless required to achieve high levels of microbial pathogen-specific immunity, before clonal expansion occurs. Several mechanisms are likely to account for the rapid Ag-dependent CD8+ TM cell–mediated protection, which include not only direct cytolysis of infected cells but also secretion of cytokines [IFNγ and tumor necrosis factor–α (TNFα)] and chemokines (CCL3) (1, 17, 18). However, how exactly cognate Ag versus inflammation programs CD8+ TM cells during reactivation is unknown, and no studies to date have provided a comprehensive picture of these processes. The early transcriptional gene expression and effector program that is specifically triggered in CD8+ TM cells upon early cognate Ag recognition is not known. This information is essential to further understand how cognate Ag enables CD8+ TM cells to achieve immunized host protection early on and mediate the rapid control of pathogen growth and spreading in situ.
IFNγ is known to be an essential effector cytokine produced by activated effector CD8+ (and CD4+) T cells, which has complex and pleiotropic effects on immune cells (19). These include, for example, favoring T helper 1 cell and M1-type macrophage differentiation, promoting Ag presentation, and the production of microbicidal molecules. In this context, we have shown that IFNγ signaling to CCR2+Ly6C+ monocytes and, to some extent, neutrophils is key to induce them to produce effector molecules such as TNFα and CXCL9 (20). TNFα is absolutely required for immunized host protection during a recall Lm infection (17, 21–23) through the potent induction of reactive oxygen species (ROS) by both CCR2+Ly6C+ monocytes and neutrophils (17). However, whether IFNγ signals are sufficient, or other signals are needed in conjunction, for effective protection to take place is also not known. In several models of infection including Lm, IFNγ is produced independently from cognate Ag (11, 12, 14), which further underscores the need to understand how cognate Ag may potentiate or contribute to IFNγ-mediated protection.
One notion illustrated across multiple studies and that accounts for how quick CD8+ TM cells can protect immunized hosts is that these cells enable the rapid containment and effective elimination of microbial pathogens at portals of entry (24, 25). CD8+ TM cells can rapidly traffic to the sites of infection via chemotaxis (e.g., CXCR3 and CCR5) and adhesion [lymphocyte function-associated antigen 1 (LFA-1) and loss of L-selectin]. Proof-of-concept studies have used models of systemic viral (vaccinia, VSV, and lymphocytic choriomeningitis virus) and bacterial (Lm) infections in which microbial pathogens are rapidly captured in subcapsular draining lymph nodes (dLNs) or splenic marginal zone CD169+ macrophages and drive subsequent homing of CD8+ TM cells in response to chemotactic cues (e.g., CXCL9 and CXCL10) produced by innate immune and structural cells (24–27). The massive Ag-independent recruitment of memory cells also leads to inflammation-driven activation of Ag-irrelevant CD8+ TM cells (26). While comparable chemotactic mechanisms are also documented in the case of CD8+ and CD4+ tissue-resident T (TRM) cells in models of skin and vaginal viral infections, initiation of the rapid mucosal immune response by TRM cells is largely dependent on initial cognate Ag recognition, leading to the establishment of a rapid antiviral state that restricts pathogen spreading (28–31). It is likely that initial chemotactic cues from tissue-resident cells involved in microbial pathogen capture (DCs and macrophages) enable CD8+ TM cell recruitment to infectious foci and that the memory cells then help to quickly amplify and guide the recruitment of more immune effector cells through the secretion of both Ag-dependent and Ag-independent cytokines and chemokines. However, the exact sequences of events and whether memory cells represent the major orchestrators of the rapid amplification of the immune response and associated host protection are not known.
In the current work, we provided a comprehensive analysis of the cellular and molecular mechanisms by which cognate Ag programs and orchestrates early CD8+ TM cell–mediated pathogen-specific protection in vaccinated hosts undergoing a recall infection. We reveal the cognate Ag–driven transcriptome of reactivated CD8+ TM cells and precisely dissect the link between cognate Ag stimulation and CD8+ TM cell–derived IFNγ production, a major protective cytokine produced independently from cognate Ag. Our results show that cognate Ag on DCs mediates CD8+ TM cell arrest in infection foci where blood-derived CCR2+Ly6C+ monocytes have accumulated via CD8+ TM cell–independent chemotactic cues. Here, CD8+ TM cells deliver localized IFNγ and a set of cognate Ag–triggered chemokines, CCL3, CCL4, and XCL1. We also reveal that IFNγ signals, while necessary to drive full CCR2+Ly6C+ monocyte activation (TNFα and CXCL9), are not sufficient. We found that IFNγ signals are required for CCR2+Ly6C+ monocytes to become responsive to chemokine signals that drive their activation and license them with highly effective microbicidal functions for rapid pathogen containment and killing.
RESULTS
Cognate Ag versus inflammation triggers a broad range of functional pathways in CD8+ TM cells
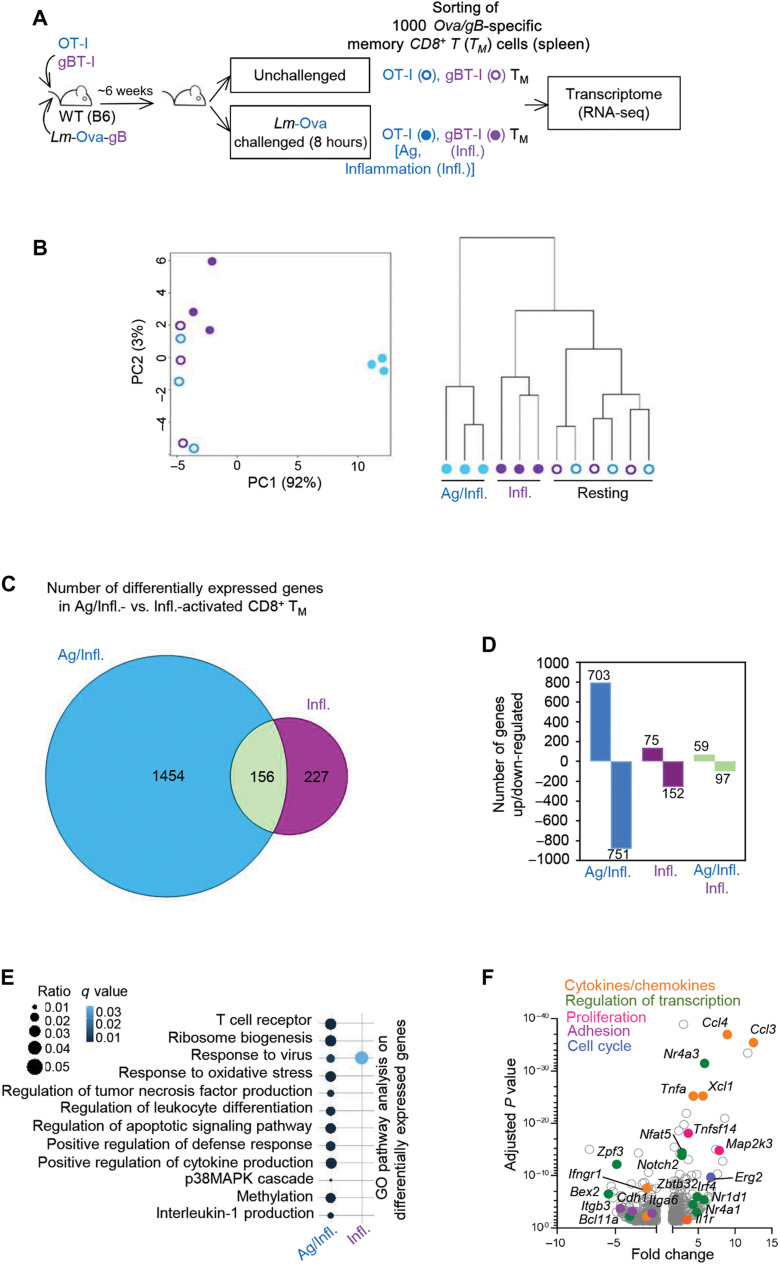
To understand how cognate Ag orchestrates CD8+ TM cell early reactivation and programming in situ, we conducted a genome-wide transcriptional analysis of pathogen-specific memory cells undergoing reactivation in the presence or absence of their cognate Ag (Fig. 1). Naïve Ova257–264/Kb-specific OT-I and gB498–505/Kb-specific gBT-I T cell receptor (TCR) transgenic T cells were adoptively transferred to wild-type (WT) C57BL/6 (B6) mice that were immunized the next day with Lm expressing both T cell epitopes (Lm-Ova-gB). Six weeks later, immunized mice were challenged with Lm expressing Ova only (Lm-Ova), and we monitored OT-I and gBT-I TM cell activation (Fig. 1A). This experimental setup enabled us to track memory cells that either “see” (OT-I, Ag/inflammation (Infl.)-activated) or do not see (gBT-I, Infl.-activated) their cognate Ag. The memory cells were flow-sorted from 8-hour-challenged or control unchallenged mice and subjected to transcriptomic analysis (Fig. 1B). Two-dimensional principal components analysis (PCA) (Fig. 1B, left) and hierarchical clustering (Fig. 1B, right) revealed that OT-I TM cells (Ag/Infl.-activated) clustered separately from gBT-I (Infl.-activated) and resting TM (unchallenged) cells that grouped close together. Thus, cognate Ag triggering drives a significantly distinct transcriptional profile in the CD8+ TM cells. A total of 1837 genes were differentially expressed (P < 0.05; fold change, >1.5) in activated (Ag/Infl. + Infl.) versus resting TM cells, with the vast majority (1454, i.e., ~79%) driven by Ag stimulation only and a smaller proportion triggered by inflammatory signals only (227, i.e., ~12%) (Fig. 1C and table S1). Only 156 genes (i.e., ~9%) among the differentially expressed genes were common between Ag-activated and inflammation-activated CD8+ TM cells. While Ag stimulation induced similar numbers of up- and down-regulated genes, respectively 703 and 751, inflammation favored the expression of a higher proportion of down-regulated genes (152 versus 75 genes) including genes involved in cell adhesion and migration (Cd44, Cd27, Itgax, and S1pr5; Fig. 1D and table S1). Common genes were more similarly distributed between up-regulation and down-regulation. Further analysis of the genes differentially expressed in Ag-stimulated versus inflammation-stimulated CD8+ TM cells using biological process gene ontology (BP-GO) pathway analysis revealed that cognate Ag, but not inflammation, promoted a wide range of biological functions related to TCR signaling, leukocyte differentiation, apoptosis, and cytokine expression (Fig. 1E and table S2). To achieve deeper understanding into the molecular mechanisms by which Ag stimulation modulates the early programming of CD8+ TM cells, we plotted the fold change over respective adjusted P values of all differentially expressed genes (Fig. 1F). The most highly expressed genes in Ag-activated CD8+ TM cells encoded for chemokines and cytokines (Ccl4, Ccl3, Xcl1, and Tnfa), important transcriptional regulators (Nr4a3, Nr4a1, Nfat5, Zbtb32, and Irf4), and proteins involved in proliferation/survival (Tnfsf14 and Map2k3) and cell cycle (Erg2). Notably, expression of genes encoding adhesion molecules was largely down-regulated (Itgb6, Itgb3, and Cdh1) (Fig. 1F). In summary, cognate Ag stimulation endows CD8+ TM cells with a robust early multifunctional gene expression program, among which the most significantly up-regulated genes encode for chemokines and a specific set of transcription factors.
Fig. 1. Transcriptomic profiling of Ag/inflammation-activated versus inflammation-activated CD8+ TM cells.
(A) Schematic of experimental design. Age-matched WT B6 female mice (CD45.2+/+) grafted with tomato-expressing (Td+) OT-I and CD45.1+/+ gBT-I cells were immunized with 104 Lm-Ova-gB and ~6 weeks later challenged or not with 106 Lm-Ova. After 8 hours, 1000 OT-I TM (Ag/Infl., blue) and gBT-I TM (Infl., purple) were flow-sorted from harvested mouse spleens based on CD8, CD3, Tomato (Td+), and CD45.1 expression, and samples were prepared for RNA sequencing (RNA-seq) analysis. (B) PCA plot (left) and clustering tree (right) of Ag/Infl. (OT-I)–stimulated versus Infl. (gBT-I)–stimulated TM cells at steady state and after challenge. Each dot represents an individual mouse, and the number in parentheses indicates the percent of variance. Each set of samples (OT-I and gBT-I) was processed in three biologically independent replicate experiment. (C) Venn diagrams comparing the numbers of differentially expressed genes Ag/Infl. (OT-I)–stimulated versus Infl. (gBT-I)–stimulated TM cells from secondary challenged mice (fold change, ±1.5; adjusted P < 0.05). The number of overlapping genes is specified in the green circle. (D) Bar graphs representing the number of genes up- and down-regulated from the Venn diagram analysis. (E) Representation of the top GO pathway analysis between Ag/Infl.-activated versus Infl-activated TM cells. The size and color of dots are proportional to the number of genes under a specific term and the adjusted P value, respectively. MAPK, mitogen-activated protein kinase. (F) Volcano plot representing the significantly up- and down-regulated genes in Ag/Infl-activated (OT-I) TM cells 8 hours after the recall challenge infection.
Memory CD8+ T cells produce an early and coordinated burst of chemokines upon cognate Ag recognition
To validate chemokine-encoding gene up-regulation in cognate Ag–stimulated CD8+ TM cells (Fig. 1F), we monitored CCL3, CCL4, and XCL1 chemokine accumulation in Ag (OT-I)– versus inflammation (gBT-I)–triggered TM cells from mice primary immunized with Lm-Ova-gB and challenged 6 weeks later with Lm-Ova for 8, 16, 32, and 72 hours (Fig. 2). As early as ~4 hours after challenge infection, OT-I, but not gBT-I, TM cells accumulated detectable levels of the three chemokines, peaking between 12 and 16 hours after infection with 30 to 40% chemokine+ OT-I TM cells (Fig. 2A). As expected (11, 12, 14, 15), both TM cells expressed IFNγ independent of cognate Ag stimulation. Substantial levels of chemokines (CCL3) and IFNγ were measured in short-term culture supernatants of splenocytes (without Golgi Plug/Stop) isolated from 8-hour Lm-Ova–challenged versus unchallenged mice, indicative of their active secretion (fig. S1A). By 32 hours, chemokine secretion was terminated, and OT-I TM cells underwent robust clonal expansion (Fig. 2B). To further define which subset of CD8+ TM cells (32) among central [CX3CR1lowCD27hi (TCM)], peripheral [CX3CR1intCD27hi (TPM)], or effector [CX3CR1hiCD27low (TEM)] CD8+ TM cells produced chemokines and IFNγ, we flow-sorted these populations and incubated them with their cognate Ag in vitro (Fig. 2C and fig. S1B). While both OT-I TCM and TPM accumulated significantly more chemokines and IFNγ than TEM counterparts, they could nevertheless all produce them. To validate results in endogenous non-TCR transgenic CD8+ TM cells and for naturally presented epitopes, we immunized WT B6 mice (H2b) that also express the Kd molecule (B6-Kd) with Lm-gB, allowing for the tracking of both Lm-derived LLO91–99/Kd and p60217–225/Kd as well as herpes simplex virus 2 (HSV-2)–derived gB497–505/Kb–specific CD8+ TM cells, using the corresponding tetramers (Tet) (Fig. 2D). Six weeks after vaccination, mice were challenged with either Lm-gB or Lm-LLOS92 that lacks the LLO91–99 epitope, and we monitored endogenous Tet-specific CD8+ TM cell production of chemokines in the presence or absence of their respective cognate Ags. After Lm-gB challenge, e.g., when all TM cell cognate Ags were present, gB498–505/Kb, p60217–225/Kd, and LLO91–99/Kd Tet+ CD8+ TM cells expressed CCL3. However, when mice were challenged with Lm-LLOS92, inflammation-only–stimulated LLO91–99/Kd-specific and gB498–505/Kb-specific CD8+ TM cells expressed IFNγ but no chemokines, while Ag-triggered p60217–225/Kd-specific CD8+ TM cells accumulated both CCL3 and IFNγ. We next extended findings to CD8+ TM cells induced with a different vaccination model, by immunizing mice grafted with OT-I cells with Ova-expressing VSV (VSV-Ova), challenged them 6 weeks later with either Lm-Ova or Lm, and quantified chemokine and IFNγ production (Fig. 2E). Likewise, upon immunization with Lm, CD8+ TM cells induced after VSV vaccination also induced a rapid and coordinated burst of Ag-dependent chemokines and Ag-independent IFNγ accumulation, peaking at ~16 hours after challenge infection, with 40 to 60% of chemokine/IFNγ+ OT-I TM cells. Thus, together, these data establish that across distinct mouse models of immunization (bacteria and virus) and multiple CD8+ T cell epitopes, cognate Ag recognition triggers a rapid and early coordinated burst of chemokine production by CD8+ TM cells.
Fig. 2. Cognate Ag recognition triggers the rapid and coordinated production of chemokines by CD8+ TM cells.
(A to C) WT mice transferred with OT-I Td+ and CD45.1+/+ gBT-I cells were immunized with 104 Lm-Ova-gB and challenged ~6 weeks later with 106 Lm-Ova for 2, 4, 6, 8, 16, 32, and 72 hours. At these times, spleen cells were incubated with GolgiPlug/Stop for 4 hours and stained for cell surface CD8, CD3, CD45.1, and intracellular CCL3, CCL4, XCL1, and IFNγ. (A) Kinetics of chemokines and IFNγ accumulation in OT-I (blue) and gBT-I (purple) TM cells and representative overlaid dot plots of the staining. (B) Number of OT-I and gBT-I TM cells at indicated times after recall infection. (C) OT-I TM cell subsets flow-sorted from the spleens of OT-I–transferred Lm-Ova–immunized mice, based on CX3CR1 and CD27 expression (TEM, CX3CR1hiCD27low; TPM, CX3CR1intCD27hi; TCM, CX3CR1lowCD27hi). OT-I TM cell subsets were next stimulated for 4 hours with SIINFEKL peptide (10−8 M) before staining. Fluorescence-activated cell sorting (FACS) histograms and graphs show expression of indicated chemokine+ and IFNγ+ OT-I TM cell subsets (n = 2 mice). (D) B6-Kd mice immunized with Lm-gB were challenged ~6 weeks later with Lm-gB or Lm-LLOS92 for 16 hours. Polyclonal CD8+ TM cells were monitored using indicated Tet. The frequency of Tet+ cells among CD8+ TM cells and their expression of CCL3 and IFNγ after challenge with Lm-gB or Lm-LLOS92 are shown. (E) Mice grafted with OT-I cells were immunized with VSV-Ova and ~6 weeks later challenged with Lm or Lm-Ova. Frequencies of chemokine+ and IFNγ+ cells among OT-I TM cells after challenge are shown. Panels pool data from either three (A and C) or two (B, D, and E) independent replicate experiments with n = 4 to 8 mice with P values (*P < 0.05, **P < 0.005, ***P < 0.0005, and ****P < 0.0001). MFI, mean fluorescence intensity.
IFN regulatory factor 4 exerts transcriptional control over chemokine production by CD8+ TM cells
Cognate Ag stimulation induces up-regulation of CCL3, CCL4, and XCL1 chemokine-encoding genes in CD8+ TM cells and their subsequent secretion, suggesting a common transcriptional mechanism of regulation. Our transcriptomic analysis revealed several genes involved in the regulation of transcription, such as the transcription factor IRF4 (IFN regulatory factor 4), that are up-regulated upon cognate Ag recognition. Since IRF4 expression in T cells is directly proportional to the strength of TCR signals (33, 34), we expected that, if IRF4 controlled chemokine expression, lowering TCR signaling should lead to a proportional and concomitant loss of IRF4 and chemokine expression by CD8+ TM cells. To test this possibility, we used Lm expressing three different Ova257–264 (SIINFEKL) altered peptide ligands (APLs) in which the original asparagine amino acid in position 4 of the peptide (N4) is replaced by either a glutamine (Q4), a threonine (T4), or a valine (V4), decreasing OT-I TCR signaling by factors of ~20, 70, and 700 times, respectively (35). Mice grafted with OT-I and gBT-I cells were immunized with Lm-Ova-gB and, 6 weeks later, either left unchallenged or challenged with Lm expressing each Ova APL or control Lm-Ova (N4). We next monitored the secretion of chemokines and IFNγ 16 hours later (Fig. 3A and fig. S1C). Decreasing OT-I TCR signaling led to a proportional loss of chemokine-producing T cells (CCL3, CCL4, and XCL1), which also directly correlated with the loss of IRF4 expression (Fig. 3B). As expected, inflammation-stimulated gBT-I TM cells neither produced chemokines nor up-regulated IRF4, while IFNγ production remained comparable across all challenge conditions, in both cognate Ag (OT-I)– and inflammation (gBT-I)–triggered CD8+ TM cells. To ensure that IRF4 up-regulation during endogenous pathogen-specific polyclonal CD8+ TM cell response was comparable to that of OT-I TCR transgenic T cells, we next used the same immunization/challenge approach as in Fig. 2C. Here, gB498–505/Kb, p60217–225/Kd, and LLO91–99/Kd Tet+ CD8+ TM cells underwent the most robust up-regulation of IRF4 expression during challenge infection in the presence of their respective cognate Ag (Fig. 3C). These results collectively indicate a direct correlation between the strength of TCR signaling and the proportion of chemokine-producing CD8+ TM cells. Furthermore, “in vitro challenge” of OT-I TM cells isolated from Lm-Ova–immunized mice with the SIINFKEL epitope in the presence or absence of broad inhibitors of either translation (cycloheximide) or transcription (actinomycin D) suggested that most of the CCL3 in Ag-stimulated TM cells was being rapidly transcribed (>60%) (Fig. 3D), while only a smaller proportion was stored as mRNA (~30%) but none as protein, a result also consistent with recent reports (36, 37). Together, these data support the hypothesis that IRF4 acts as a transcriptional regulator of chemokine expression downstream of TCR signaling.
Fig. 3. The transcription factor IRF4 orchestrates chemokine production by CD8+ TM cells downstream TCR signaling.
(A and B) Mice grafted with OT-I Td+ and CD45.1+/+ gBT-I cells were immunized with Lm-Ova-gB, challenged or not ~6 weeks later for 16 hours with Lm-Ova N4, Lm-Ova Q4, Lm-Ova T4, or Lm-Ova V4. Spleen cells were next incubated with GolgiPlug/Stop and stained for cell surface CD8, CD3, CD45.1, and CCL3 and IFNγ (A) or IRF4 (B). Proportions of OT-I or gBT-I TM cells expressing indicated intracellular markers are shown, with representative overlaid dot plots of IRF4 intracellular staining in OT-I and gBT-I TM cells (B). (C) B6-Kd mice were immunized with Lm-gB and challenged ~6 weeks later with Lm-gB or Lm-LLOS92. Polyclonal CD8+ TM cells were quantified using indicated Tet. Bar graphs indicate the proportion of IRF4+ cells among tetramer+ (Tet+) cells. (D and E) Splenocytes from 6-week-immunized mice as depicted in (A) were incubated with SIINFEKL with or without either cycloheximide or actinomycin D (D) or with the IRF4 inhibitor SCG-CBP30 (E) and further stained. Graphs show the proportions and/or expression level of indicated chemokine+ and IRF4+ OT-I TM cells. (F) Rosa26CreERT2Irf4flox/floxCd45.2+/+ and Irf4wtCd45.1+/− OT-I cells were cotransferred to Cd45.1+/+ WT mice and immunized with Lm-Ova. Six weeks later, mice received Tx or vehicle before Lm-Ova challenge infection. CCL3 expression was determined 16 hours later. (G) Rosa26CreERT2Irf4flox/flox Cd45.2+/+ and Irf4wtCd45.1+/− OT-I cells were cotransferred to Cd45.1+/+ WT mice, immunized with VSV-Ova, and boosted with Lm-Ova 2 weeks later. After 4 weeks, mice received Tx or vehicle before sorting spleen OT-I TM cells. A total of 2 × 105 OT-I TM cells of either genotype were transferred to recipient mice, challenged with Lm-Ova, and spleens/livers were plated to enumerate Lm titers. Control groups included age-matched WT mice either primary [phosphate-buffered saline (PBS)/Lm)] or secondary (Lm/Lm) challenged with Lm. Panels pool the result of two independent replicate experiments with n = 6 (A and B), 5 (C to E), and 7 (F) mice. In (G), experiment was performed once with n = 3 to 4 mice per group (one symbol, one mouse). P values are indicated. ns, not significant. *P < 0.1, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To establish whether IRF4 controls CCL3, CCL4, and XCL1 chemokine expression in CD8+ TM cells, we blocked IRF4 in OT-I TM cells and quantified their production of chemokines. We first challenged OT-I TM cells isolated from Lm-Ova–immunized mice with the SIINFKEL peptide in vitro challenge in the presence of the chemical inhibitor SCG-CBP30, which selectively inhibits bromodomain-containing transcription factors such as IRF4 (Fig. 3E). We found that IRF4 expression in OT-I TM cells was prevented and the proportion of chemokine+ cells was significantly decreased (by ~80%) compared to incubation with peptide only. To next confirm and validate findings in vivo, we generated OT-I+ Rosa26CreERT2Irf4flox/flox mice in which Irf4 could be inducibly deleted in OT-I TM cells (Fig. 3F). Naïve Rosa26CreERT2Irf4flox/flox or WT Cd45.1/2 OT-I cells were cotransferred to WT Cd45.1+/+ recipient mice and then immunized with Lm-Ova, and 6 weeks later, mice received either tamoxifen (Tx) or vehicle for 5 days before Lm-Ova recall infection. In Tx-treated groups, the proportion of Rosa26CreERT2Irf4flox/flox OT-I TM cells secreting chemokines (CCL3) was significantly decreased compared to that of WT counterparts (by ~45%), yet both of these genotypes secreted comparable amounts of CCL3 in mock-treated mice. Expression levels of IRF4 were also diminished in Tx-treated but not mock-treated Rosa26CreERT2 Irf4flox/flox OT-I TM cells, further validating this result (fig. S1D). Last, we determined the contribution of CD8+ TM cell–derived chemokines to immunized host protection. For this, we tested whether deleting IRF4 in OT-I TM cells, which prevents rapid chemokine secretion, altered their ability to confer protection to challenged mice compared to nondeleted counterpart (Fig. 3G). WT recipient mice were cotransferred with Rosa26CreERT2Irf4fl/fl or Irf4fl/fl OT-I cells that were immunized with VSV-Ova the next day. After 2 weeks, mice were boosted with Lm-Ova and, 4 weeks later, treated with Tx daily for 5 days. Then, both genotypes of OT-I TM cells were flow-sorted and transferred to new recipient mice further challenged with Lm-Ova. Bacterial titers were enumerated 24 hours later and showed that memory cells lacking IRF4 conferred ~50% (in spleen and liver) less protection than that of WT counterparts. Thus, cognate Ag signaling via IRF4 is required to achieve full protection. In conclusion, our data establish that IRF4 contributes to the transcriptional regulation of the coordinated and simultaneous burst of CCL3, CCL4, and XCL1 chemokines produced by Ag-activated CD8+ TM cells in vitro and in vivo and subsequent immunized host protection.
Monocyte clustering occurs independently from cognate Ag or IFNγ signaling
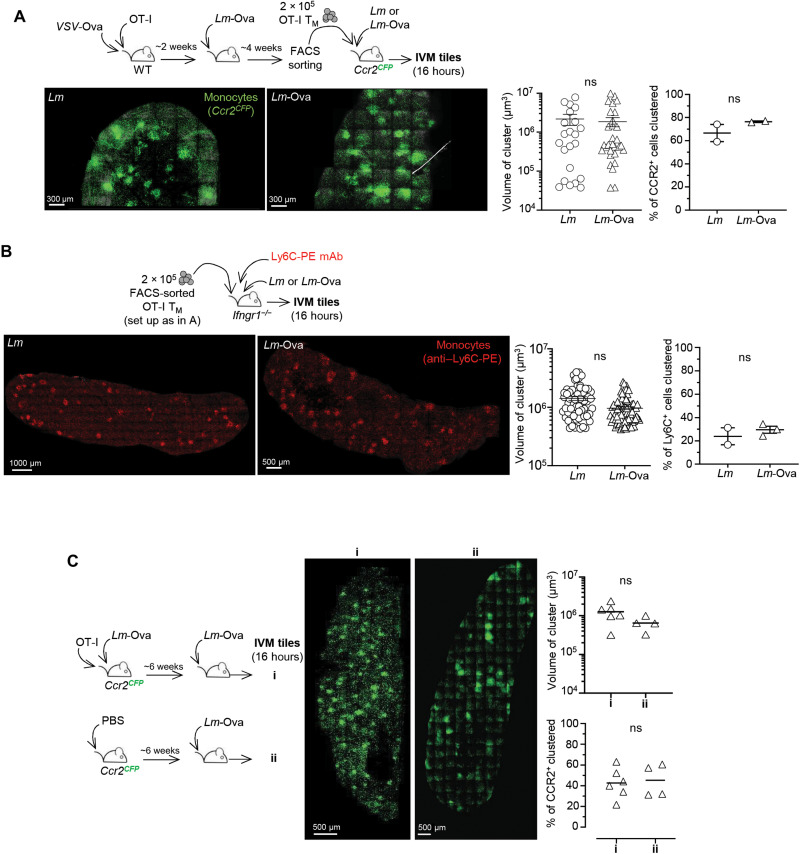
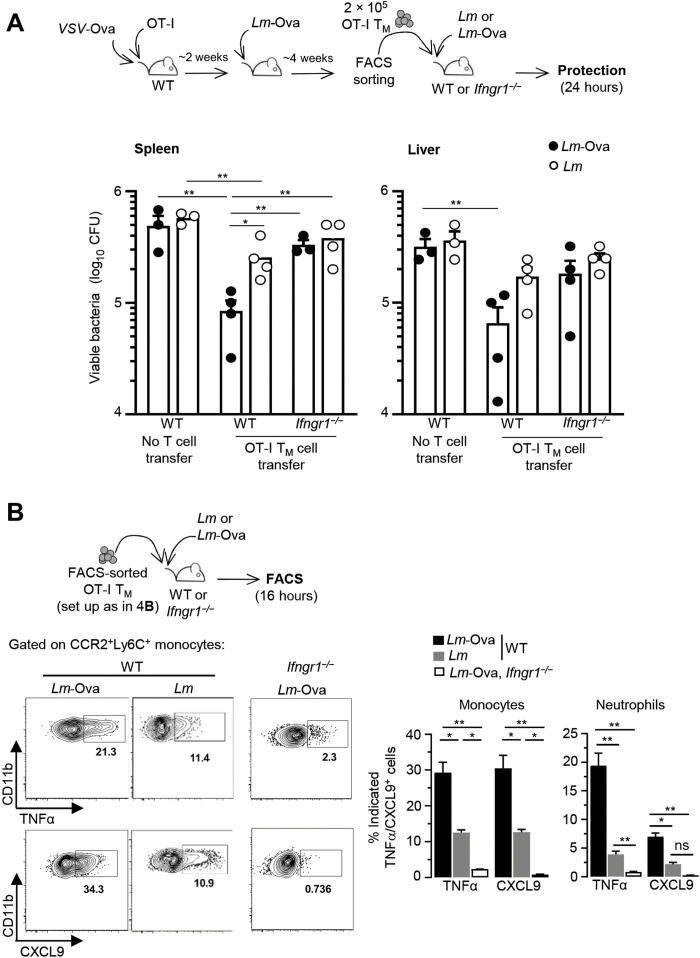
We previously showed that CD8+ TM cell–mediated control of Lm growth during recall infection occurs within only a few hours (~6 to 8 hours) and correlates with their rapid localization with clustered CCR2+Ly6C+ monocytes and neutrophils in the splenic red pulp (RP) of infected mice, at portal of bacterial entry (20, 27, 38). Thus, we hypothesized that CD8+ TM cell–derived chemokines produced in response to cognate Ag recognition orchestrate monocyte homing and clustering to rapidly prevent pathogen spreading and help deliver local IFNγ. To gain deeper understanding of this process, we first monitored the kinetics of Ly6C+ monocyte clustering in the RP of Lm-vaccinated mice undergoing a recall infection (fig. S2A). Ccr2CFP mice, in which all CCR2+Ly6C+ monocytes express the cyan fluorescent protein (CFP) reporter protein, were grafted with OT-I cells and immunized with Lm-Ova. Six weeks later, mice were left unchallenged or challenged with Lm-Ova, and spleens were harvested 3, 6, 16, and 40 hours later for whole-organ tile reconstruction using multiphoton laser scanning microscopy that only enables to visualize splenic RP (fig. S2A). Already by 3 hours after challenge infection, few clusters of CCR2+Ly6C+ monocytes were detected, with proportions increasing from 6 hours to peaking at 16 hours and with some clusters still present by 40 hours. Notably, peak clustering of monocytes at 16 hours correlated with that of chemokines produced by the TM cells (Fig. 2A and fig. S2A). To better investigate the role of CD8+ TM cells and cognate Ag in CCR2+Ly6C+ monocyte cluster formation, we next adoptively transferred OT-I TM cells in Ccr2CFP WT mice subsequently challenged with Lm or Lm-Ova and monitored monocyte clustering in spleen RP (Fig. 4A). For this, we took advantage of a heterologous prime/boost immunization strategy of mice grafted with OT-I cells, primed with VSV-Ova, and challenged with Lm-Ova to generate sufficiently high numbers of OT-I TM cells for purification and transfer. Whether cognate Ag was present, the proportion and volume of monocyte clusters at the peak (16 hours) remained comparable, a result that we also confirmed in WT Ccr2CFP mice grafted with OT-I cells, immunized with VSV-Ova, and challenged with either Lm or Lm-Ova 6 weeks later (fig. S3A). Next, since IFNγ signaling is an essential contributor to vaccinated host protective responses (20), we tested whether it may direct CCR2+Ly6C+ monocyte clustering. For this, we adoptively transferred OT-I TM cell in Ifngr1−/− mice that we next challenged with Lm or Lm-Ova (Fig. 4B). Since CCR2+Ly6C+ monocytes in Ifngr1−/− mice did not express CFP, we tracked them using intravenous injection of anti–Ly6C–PE (phycoerythrin) monoclonal antibody (mAb) 16 hours before imaging, which colabels all detectable clustered CFP+ monocytes (fig. S3B). As before, whether cognate Ag (Ova) and IFNγ signaling were present, the proportion and volume of CCR2+Ly6C+ monocyte clusters at 16 hours were also comparable. Last, we tested whether CD8+ TM cells were required for CCR2+Ly6C+ monocyte clustering to occur (Fig. 4C). Unimmunized mice challenged with Lm-Ova, which do not control the infection compared to immunized counterpart (fig. S2B), developed comparable numbers and volume of CCR2+Ly6C+ monocyte clusters 16 hours after infection. This result indicated that the presence of immunization-induced memory cells is not essential for monocyte homing and clustering to occur, although they may still alter CCR2+Ly6C+ monocyte functions. Hence, together, these data establish that CCR2+Ly6C+ monocyte homing and clustering mostly occur independently of the presence of Ag-specific CD8+ TM cells and IFNγ signaling.
Fig. 4. CCR2+Ly6C+ monocyte clusters form in the splenic RP independent from cognate Ag, IFNγ, and CD8+ TM cells.
(A and B) A total of 2 × 105 OT-I TM flow-sorted cells generated upon immunization with 2 × 105 plaque-forming units (PFUs) of VSV-Ova and challenge with 106 Lm-Ova were transferred to naïve Ccr2CFP (A) or Ifngr1−/− (B) recipient mice subsequently challenged with 106 Lm or Lm-Ova for 16 hours. Representative intravital imaging microscopy (IVM) tiles of reconstructed mouse spleens with CCR2+ (A) or Ly6C+ (B) monocytes in spleen’s RP. In (B), Ifngr1−/− mice were also coinjected with the anti–Ly6C-PE Ab (10 μg). Graphs in (A) and (B) show the volume of individual clusters and the average proportion of clustered CCR2+Ly6C+ monocytes in each mouse spleen analyzed across two independent replicate experiments (n = 2 to 3). (C) Mice were transferred with OT-I cells and primary and secondary challenged with Lm-Ova (i) or only primary immunized with Lm-Ova. Ccr2CFP monocytes are in green, and scales are indicated. Representatives IVM tiles of reconstructed Ccr2CFP mouse spleens. Graphs show the average volume of clusters and proportion of clustered monocytes in each mouse spleen analyzed.
Cognate Ag on DCs, but not monocytes, controls CD8+ TM cell production of chemokines and arrest in CCR2+Ly6C+ monocyte clusters
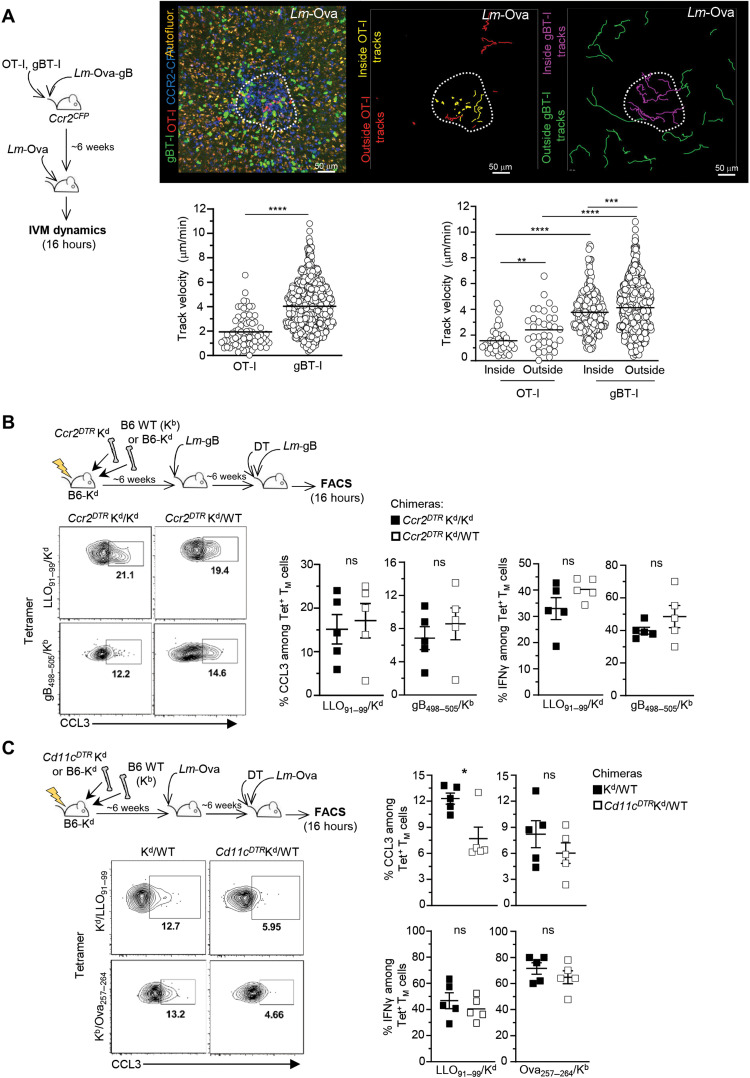
While CCR2+Ly6C+ monocyte homing and clustering still occurred in unimmunized mice, these clusters could nevertheless be necessary for a protective recall response in immunized mice. Thus, we pursued the hypothesis that CCR2+Ly6C+ monocyte clustering is functionally important and that clusters may act as local “hubs” in which CD8+ TM cells arrest and deliver IFNγ and other effector molecules to them and to other innate immune cells recruited to these clusters, i.e., neutrophils and natural killer (NK) cells (20, 27). We used intravital imaging microscopy (IVM) of spleen RP in Ccr2CFP living mice undergoing a recall infection (Fig. 5A and movies S1 to S3). Mice transferred with OT-I (Td+) and gBT-I [green fluorescent protein (GFP)+] cells were immunized with Lm-Ova-gB and challenged 6 weeks later with either Lm-Ova, Lm, or Lm-Ova-gB. In Lm-Ova–challenged mice, in which only OT-I TM cells recognize their cognate Ag, most OT-I TM cells localized in the cluster of monocytes (CFP+) and arrested or only exhibited very limited motility (track velocity, 1.93 μm/min) (Fig. 5A, movie S1, and fig. S4A). In contrast, gBT-I TM cells were more motile (track velocity, 4.01 μm/min), but they were enriched in the monocyte clusters similarly to OT-I cell counterparts (fig. S4B). Both TM cells’ speed also decreased inside compared to outside monocyte clusters, collectively suggesting that non–cognate Ag signals affect their homing to and motility in the clusters (Fig. 5A). As expected, in Lm-Ova-gB–challenged mice, where both TM cells recognize their cognate Ag, OT-I and gBT-I TM cells arrested in the clusters while simultaneously exhibiting higher motility outside of clusters (movie S2 and fig. S4B). Moreover, in Lm-challenged mice in which no cognate Ag was present, both TM cells exhibited the same pattern of enriched localization inside versus outside the clusters and comparable speeds (movie S3 and fig. S4, A and B). Thus, cognate Ag signals induce Ag-specific TM cell arrest in CCR2+Ly6C+ monocyte clusters where IFNγ is detected in TM cells (20, 27), indicating that a functional interaction between TM cells and CCR2+Ly6C+ monocytes may occur in these clusters. In addition, the fact that even non–cognate Ag–specific TM cell speed is reduced inside compared to outside of clusters suggests that the clusters are conductive of a qualitatively distinct, possibly hypoxic, local microenvironment (39).
Fig. 5. Memory CD8+ T cells arrest upon cognate Ag recognition presented by DCs but not CCR2+Ly6C+ monocytes.
(A) Ccr2CFP mice cotransferred with naïve OT-I Td+ and gBT-I GFP+ cells and immunized with 104 Lm-Ova-gB were challenged ~6 weeks later with 106 Lm-Ova, and spleens from live mice were exposed and imaged 16 hours later using IVM imaging. A representative image (left) of OT-I (red) and gBT-I (green) TM cell localization in a cluster (white dotted line) of CCR2+Ly6C+ monocytes (blue) is shown. Autofluorescence appears in yellow. Also shown (center and right images) are OT-I TM cell tracks (outside, red and inside, yellow) and gBT-I TM cell tracks (outside, green and inside, purple) inside/outside the same cluster of CCR2CFP monocytes. Graphs represent the speed of individual OT-I and gBT-I TM cells in the monocyte cluster area (left) and inside/outside the cluster. (B and C) Lethally irradiated (1200 rads) B6-Kd recipient mice were reconstituted with (B) B6-Kd or WT B6 (Kb) and Ccr2DTR Kd BM or (C) Cd11cDTRKd or Kd and WT B6 BM. Six weeks after reconstitution, mice were immunized with 104 Lm-gB (B) or Lm-Ova (C) and, 6 weeks later, challenged with 106 Lm-gB 12 hours after DT treatment. Endogenous CD8+ TM cells were monitored using LLO91–99/Kd, gB498–505/Kb, or Ova257–264/Kb Tet. Graphs show the expression of CCL3 and IFNγ among Tet+ cells after challenge. Each symbol corresponds to one individual mouse in one of two replicate experiments, and P values are shown. *P < 0.1, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Since TM cells arrest in CCR2+Ly6C+ monocyte clusters in the presence of cognate Ag and T cells arrest in response to Ag recognition (40–42), we postulated that monocytes may present Ag to them. To test this possibility, we generated mixed bone marrow (BM) chimera mice in which selective elimination of Kd-dependent cognate Ag presentation by CCR2+Ly6C+ monocytes can be achieved. Here, lethally irradiated B6-Kd mice were reconstituted with Ccr2DTR Kd BM and either (i) B6-Kd (Kd) or (ii) B6 (WT) BM (1:1 ratio), producing Ccr2DTR Kd/WT mice and Ccr2DTR Kd/Kd chimeras. In these mice, diphtheria toxin (DT) injection eliminates CCR2+ Kd monocytes, while DT receptor (DTR)− (Kd or WT) CCR2+Ly6C+ monocytes remain, respectively (fig. S4C). Chimeras were immunized with Lm-gB and treated with DT before Lm-gB challenge infection, and we monitored both LLO91–99/Kd and gB498–505/Kb Tet+ CD8+ TM cells for Ag-dependent chemokine (CCL3) and Ag-independent IFNγ production (Fig. 5B). The proportion of Ag-stimulated (CCL3+) LLO91–99/Kd Tet+ CD8+ TM cells was the same whether CCR2+Ly6C+ monocytes could present the LLO91–99/Kd Ag (in DT-treated Ccr2DTR Kd/Kd chimeras) or not (in DT-treated Ccr2DTR Kd/WT chimeras). However, the frequency of IFNγ+ cells was equivalent, confirming that LLO91–99/Kd Tet+ CD8+ TM cells underwent comparable Ag-independent activation in all groups. No differences in the proportion of CCL3+ and of IFNγ+ gB498–505/Kb Tet+ CD8+ TM cells were measured between the various experimental conditions, ruling out a possible impact of DT-induced deletion on TM cell activation. Thus, Ag presentation by splenic CCR2+Ly6C+ monocytes is not required for Ag-dependent CD8+ TM cell activation during recall infection.
DCs quickly uptake Lm (43, 44) and contribute to CD8+ TM cell reactivation (7). Using Kd/WT and Cd11cDTR Kd/WT chimera mice, in which DT injection eliminates CD11c+Kd DCs while DTR− (WT or Kd) CD11c+ DCs remain (fig. S4D), we tested whether CD11c+ DC presented cognate Ag to TM cells after immunization/challenge with Lm-Ova (Fig. 5C). A significant decrease (~40%) in CCL3+ CD8+ TM cells was only measured for LLO91–99/Kd but not Ova257–264/Kb Tet+ CD8+ TM cells, while the proportion of IFNγ+ cells remained equivalent between the different groups of chimeras. Thus, together, these data indicate that splenic CD11c+ DCs, but not CCR2+Ly6C+ monocytes, selectively present cognate Ag to CD8+ TM cells and that CCR2+Ly6C+ monocytes cannot substitute for DCs in this task.
Cognate Ag stimulation of CD8+ TM cells potentiates CCR2+Ly6C+ monocyte effector functions in the clusters
Cognate Ag enables CD8+ TM cell arrest in CCR2+Ly6C+ monocyte clusters and their concomitant production of a chemokine burst. If, as hypothesized, CD8+ TM cell arrest in these clusters is functionally important for local delivery of chemokines and IFNγ, then we predicted that in the presence of cognate Ag, these cells should produce more effector cytokines (Fig. 6A). To test this model, we immunized mice transferred with OT-I TM cells with VSV-Ova. Six weeks later, mice were challenged with either Lm-Ova or Lm, and we monitored TNFα and CXCL9 production in CCR2+Ly6C+ monocytes and neutrophils. With this experimental setup, the only Lm-induced memory cells are the OT-I cells, allowing to specifically assess how the presence of cognate Ag (Lm-Ova challenge) affects myeloid cell activation. In the presence of cognate Ag recognition, the proportion of CCR2+Ly6C+ monocytes and neutrophils producing TNFα and CXCL9 was significantly increased (factor of ~3) compared to mice challenged without cognate Ag (Lm challenge), consistent with our proposed model.
Fig. 6. Cognate Ag stimulation enhances CCR2+Ly6C+ monocyte effector functions in the clusters.
(A) WT mice transferred with OT-I cells were immunized with 2 × 105 VSV-Ova and challenged or not 6 weeks later with 106 Lm or Lm-Ova. Spleens from 16-hour-challenged or unchallenged mice were harvested, and cells were incubated for 4 to 6 hours with Golgi Plug/Stop before staining for expression of CD11b, Ly6C, and Ly6G cell surface markers and indicated intracellular effector and chemotactic markers. (B) Lm-Ova (104)–immunized mice were challenged with 106 Lm-Ova or not, and 1 hour before euthanasia, 5 μg of Ly6C-PE mAb was intravenously injected to the hosts. Spleens were harvested, and cells were stained for cell surface CD11b, Ly6C-PerCpCy5.5, ICAM-1, CD86, and intracellular TNFα and CXCL9. After gating on Ly6C-PerCpCy5.5+ monocytes, Ly6C-PEhi and Ly6C-PElow monocytes were identified and further analyzed for indicated marker expression. Representative FACS dot plots are shown, and bar graphs pool two independent replicate experiments with n = 6 (A) and 5 (B) mice. P values are indicated. *P < 0.1, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To provide further support that CCR2+Ly6C+ monocyte activation in vivo was spatially restricted to their clusters, we sought to measure the activation of CCR2+Ly6C+ monocytes inside versus outside the clusters. Similar Lm-induced clusters of myeloid cells have been reported to exclude dextrans, suggesting that they were not diffusive (45). Therefore, we stained monocytes in vivo using anti–Ly6C-PE mAb injected 1 hour before spleen harvest (1-hour labeling), which, we found, labels all Ly6C+ splenocytes that are not within established clusters, in contrast to injecting anti–Ly6C-PE mAb at the time of challenge infection (16-hour labeling), before cluster formation (figs. S5A and S3B). With the 1-hour labeling approach, >90% of CCR2+Ly6C+ monocytes exhibited equivalent Ly6C-PE staining in unchallenged mice (no clusters), while ~40% of them had lower Ly6C-PE staining in challenged mice, a proportion consistent with that of clustered CCR2+Ly6C+ monocytes in our microscopy quantifications (Fig. 4A and figs. S5B and S2A). With this approach, we could determine whether CCR2+Ly6C+ monocyte activation was dependent on localization within clusters during recall infection (Fig. 6B and fig. S5C). A significantly higher proportion of Ly6C-PElow (clustered) compared to Ly6C-PEhi (nonclustered) CCR2+Ly6C+ monocytes expressed higher levels of intercellular adhesion molecule–1 (ICAM-1), CD86, and intracellular CXCL9 and TNFα, demonstrating that CCR2+Ly6C+ monocytes undergo robust activation within the clusters, consistent with a spatially restricted delivery of activating cues by arrested CD8+ TM cells.
Blocking GαI-dependent chemotaxis in CX3CR1+ cells prevents CCR2+Ly6C+ spleen monocyte clustering and optimal host protection
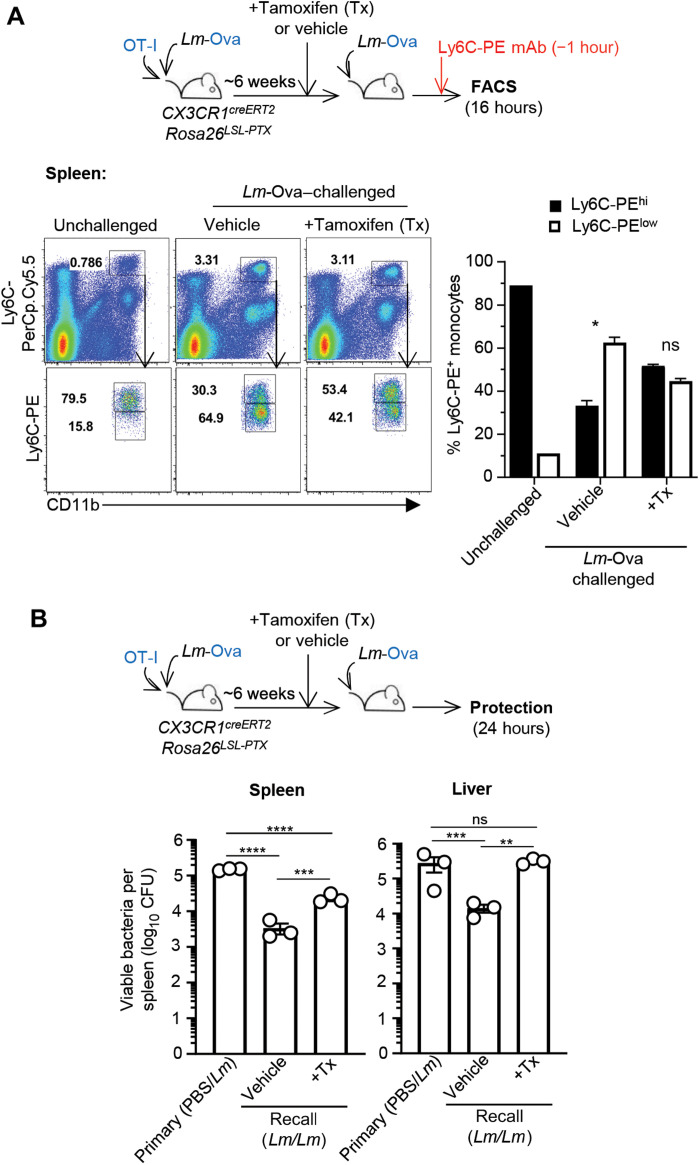
To further explore whether CCR2+Ly6C+ monocyte clustering is important for immunized host protection, we took advantage of a mouse model (46) in which expression of the pertussis toxin (PTX), which blocks GαI protein–coupled receptor signal transduction and related chemotaxis, can be induced upon Tx injection in monocytes (Fig. 7A). We used CX3CR1ERT2Cre mice in which ~50% of CCR2+Ly6C+ monocytes express the Tx-inducible estrogen receptor T2 (ERT2)-Cre recombinase, crossed to mice that carry a Cre-inducible PTX-encoding gene in the Rosa26 locus (Rosa26LoxP-STOP-LoxP(LSL)-PTX). Mice were next transferred with OT-I cells, immunized with Lm-Ova, and treated with Tx for 5 days before Lm-Ova secondary challenge. One hour before euthanizing mice, we injected the anti–Ly6C-PE Ab to label clustered CCR2+Ly6C+ monocytes in the challenged or unchallenged control mice. As expected, CCR2+Ly6C+ monocytes were not clustered in unchallenged mice (>80% Ly6C-PEhi), while they clustered in vehicle-treated challenged groups (~60% Ly6C-PElow). In Tx-treated mice, in which PTX is induced in CX3CR1+ cells, clustering was reduced by ~70% compared to vehicle-treated mice (~44% Ly6Clow), showing that CCR2+Ly6C+ monocytes required GαI-dependent chemotaxis to cluster. While most of the spleen CCR2+Ly6C+ monocytes failed to cluster, twice as many CCR2+Ly6C+ monocytes also could not egress from the BM (47) in Tx-treated compared to vehicle-treated mice (fig. S6). We next used this experimental system to evaluate the impact on host protection (Fig. 7B). CX3CR1ERT2CreRosa26LSL)-PTX mice transferred with OT-I and either immunized with Lm-Ova or injected with phosphate-buffered saline (PBS) were treated with Tx or vehicle 6 weeks later before challenge with Lm-Ova. After 24 hours, we harvested and plated spleens and livers and enumerated Lm titers. Protected mice exhibited ~50 (spleen) and ~20 (liver) times less bacteria compared to primary challenged mice. Tx-treated mice had ~7 (spleen) and ~19 (liver) times higher bacterial loads than vehicle-treated protected mice. These results show that blocking GαI-dependent chemotaxis in CX3CR1+ cells leads to a significant loss of protection in immunized mice undergoing a recall infection, suggesting that splenic CCR2+Ly6C+ monocyte clustering contributes to protection.
Fig. 7. Blocking GαI-dependent chemotaxis in CX3CR1+ cells inhibits CCR2+Ly6C+ splenic monocyte clustering and impairs protection.
CX3CR1CreERT2Rosa26LSL-PTX mice transferred with OT-I cells were immunized with 104 Lm-Ova. Six weeks later, mice received Tx or vehicle intraperitoneally daily for 5 days before 106 Lm-Ova recall infection. (A) One hour before harvesting spleens and 16 hours after challenge, 5 μg of Ly6C-PE mAb was injected intravenously to the hosts. Cells were then stained for cell surface CD11b and Ly6C-PerCpCy5.5. After gating on Ly6C-PerCpCy5.5+ monocytes, the proportion of Ly6C-PEhi and Ly6C-PElow monocytes was quantified. Representative FACS dot plots are shown, and bar graphs pool two independent replicate experiments with n = 6. (B) Twenty-four hours after Lm infection (106), spleens and livers were harvested and plated to enumerate Lm titers. A nonimmunized group of age-matched mice (“primary”) were also infected as control. Bar graphs show one of two independent experiments with n = 3 mice. P values are indicated. *P < 0.1, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Cognate Ag stimulation and IFNγ signaling are both required for CD8+ TM cell–dependent protection of immunized mice
Since both cognate Ag stimulation and IFNγ signaling are required for CD8+ TM cell–dependent protection of immunized hosts against challenge infection, we next assessed the relative contribution of both mechanisms. We adoptively transferred OT-I TM cells to naïve WT or Ifngr1−/− mice that were further challenged with a lethal dose of Lm (no cognate Ag) or Lm-Ova (with cognate Ag) (Fig. 8A). Control groups did not receive any OT-I TM cells. Bacterial titers in spleens and livers were quantified 24 hours later. While as expected, transfer of OT-I TM cells conferred significant levels of protection to WT recipient mice against Lm-Ova challenge (considered 100%), and protection was reduced to ~40% in both organs when challenged with Lm (no cognate Ag). We also recapitulated these findings in WT mice primary immunized with VSV-Ova and challenged 6 weeks later with either Lm or Lm-Ova (fig. S7A). However, OT-I TM cell transfer in Ifngr1−/− mice only conferred modest protection against challenge with Lm or Lm-Ova, with more than 60% protection loss compared to WT mice. However, in Lm-Ova (but not Lm)–challenged Ifngr1−/− mice, OT-I TM cells still efficiently recognized their cognate Ag and produced chemokines (fig. S7B). Consistent with these results, CCR2+Ly6C+ monocyte and neutrophil production of TNFα and CXCL9 effector cytokine/chemokine in WT or Ifngr1−/− mice that received OT-I TM cells was significantly reduced when cognate Ag was absent (Lm challenge) or IFNγ signaling (Ifngr1−/−) was disrupted (Fig. 8B). Together, these results indicate that cognate Ag stimulation and IFNγ signaling are both required to achieve optimal protection and that neither of these signals is individually sufficient.
Fig. 8. Memory CD8+ T cell–mediated protection of vaccinated hosts requires both cognate Ag and IFNγ signaling.
A total of 2 × 105 OT-I TM cells induced using the depicted experimental setup (as in Fig. 4B) were transferred in age- and sex-matched WT B6 or Ifngr1−/− mice, and mice were next challenged with 106 Lm or Lm-Ova. (A) Control groups did not receive OT-I TM cells. Spleens and livers from challenged mice were harvested 24 hours later, and Lm colony-forming units (CFUs) were determined after plating. Bar graphs show one of two representative experiments with each symbol corresponding to one individual mouse. (B) Spleens from WT or Ifngr1−/− mice transferred with OT-I TM cells and challenged with 106 of indicated Lm were harvested, and cells were stained for expression of cell surface CD11b, Ly6C, and Ly6G and intracellular TNFα and CXCL9. Representative FACS dot plots are shown, and bar graphs pool two representative experiments (n = 7 mice) with P values indicated. *P < 0.1 and **P < 0.01.
Memory CD8+ T cell–derived chemokines are necessary but not sufficient to activate monocyte effector functions
We established in Fig. 3G that, despite Ag-independent production of IFNγ, cognate Ag signaling via IRF4 is required to achieve full protection of immunized hosts. A possible interpretation of this result is that the failure of IRF4-deficient OT-I TM cells to protect is not due to their inability to secrete chemokines but rather to stop in CCR2+Ly6C+ monocyte clusters to deliver activating IFNγ. Thus, to directly test the role of chemokines in CCR2+Ly6C+ monocyte activation, we developed an in vitro assay where purified OT-I TM cells and CCR2+Ly6C+ monocytes were incubated together, with or without chemokine-neutralizing polyclonal antisera (Fig. 9A). Here, OT-I TM cells flow-sorted from primed/boosted mice were incubated overnight (OVN) with their cognate peptide, and the next day, CCR2+Ly6C+ monocytes, also flow-sorted from naïve WT mice, were added to the culture in the presence of Golgi PLug/Stop, with or without polyclonal neutralizing antisera against each of the three chemokines. A substantially higher proportion of CCR2+Ly6C+ monocytes produced CXCL9 and TNFα when coincubated with cognate Ag–activated OT-I TM cells than without (factor of 4 and 2, respectively). CCR2+Ly6C+ monocyte activation was abrogated when cells were incubated in the presence of neutralizing antisera against CCL3, CCL4, and XCL1, demonstrating that memory T cell–derived chemokines are required to trigger monocyte production of TNFα and CXCL9. We next asked whether addition of the CCL3, CCL4, and XCL1 recombinant chemokines, which respectively bind CCR5/CCR1 and XCR1, to CCR2+Ly6C+ monocytes is sufficient to drive their activation, but they failed to do so (Fig. 9A). However, when CCR2+Ly6C+ monocytes were isolated from Lm-challenged rather than naïve mice, addition of exogenous chemokines enhanced their production of TNFα ex vivo (Fig. 9B and fig. S8, A and B). After incubation with rCCL3, rCCL4, and rXCL1, CCR2+Ly6C+ monocytes from challenged mice accumulated intracellular TNFα in 15, 20, and 40% of total CCR2+Ly6C+ monocytes, respectively, and in a dose-dependent manner. Blocking CCR5 and CCR1 with chemical inhibitors during coincubation with the corresponding recombinant chemokines prevented TNFα production by CCR2+Ly6C+ monocytes, ruling out any CCR1/CCR5-independent activation mechanisms. Incubation with heat-killed Listeria monocytogenes (HKLm) induced 40 to 50% of them to express TNFα, a proportion similar to that measured in CCR2+Ly6C+ monocytes incubated with rXCL1 or the combination of chemokines. In contrast to CCR2+Ly6C+ monocytes, neutrophils largely failed to respond to chemokine restimulation ex vivo, rather implicating a chemokine-independent mechanism for their activation (fig. S8C). In summary, these data establish that CD8+ TM cell–derived chemokines are necessary but not sufficient to promote CCR2+Ly6C+ monocyte production of effector molecules. Other signals derived from Ag-stimulated CD8+ TM cells, most likely IFNγ, are needed in conjunction.
Fig. 9. Chemokines secreted by CD8+ TM cells enable CCR2+Ly6C+ monocyte activation in an IFNγ-dependent manner.
(A) WT mice transferred with OT-I cells were immunized with VSV-Ova and boosted with Lm-Ova 2 weeks later. After 4 weeks, FACS-sorted OT-I TM cells were stimulated with SIINFEKL peptide (10−6 M) OVN in complete media (37°C, CO2). The next day, CCR2+Ly6C+ monocytes, FACS-sorted from a naïve spleen, were added with GolgiPlug/Stop to the T cells with or without CCL3, CCL4, and XCL1 chemokine-neutralizing antisera. Controls included monocytes only with or without the recombinant chemokines. Intracellular accumulation of CXCL9 and TNFα in the monocytes was quantified by FACS 4 hours later. (B) CCR2+Ly6C+ monocytes were FACS-sorted from the spleen of OT-I–transferred immunized mice challenged 6 weeks later with Lm-Ova. Recombinant chemokines and HKLm were added or not to purified monocytes with GolgiPlug/Stop. Four hours later, TNFα accumulation was quantified by FACS. (C) WT mice transferred with OT-I cells were immunized with Lm-Ova and challenged or not 6 weeks later with Lm-Ova. Spleens from challenged or unchallenged mice were incubated for 4 to 6 hours with GolgiPlug/Stop before staining for expression of CD11b, Ly6C, and Ly6G cell surface markers and expression of CCR1, CCR5, and XCR1 chemotactic receptors. (D) BM from WT (CD45.1+/−) and Ifngr1−/− (CD45.2+/+) mice was cotransferred to CD45.1+/+ mice immunized with Lm-Ova 6 weeks before and challenged with Lm. Sixteen hours later, splenocytes were stained for expression of CCR5 and XCR1 on monocytes. Representative FACS dot plots are shown, and bar graphs pool one in two (A) (n = 2 mice) or two (B to D) (n = 5 to 6 mice) independent replicate experiments. P values are indicated. *P < 0.1 and **P < 0.01.
IFNγ signaling controls up-regulation of CCR5 and XCR1 chemokine receptors, enabling CCR2+Ly6C+ monocyte responsiveness to chemokines and full activation
Our results show that both IFNγ and antigenic signals, specifically chemokines, are required to achieve optimal host protection (Figs. 3G and 8). We also found that CD8+ TM cell–derived signals are essential to make CCR2+Ly6C+ monocytes responsive to chemokine signals and that CCR2+Ly6C+ monocytes are only responsive to chemokines after infection (Fig. 9, A and B). Upon challenge infection, we noted that CCR5 and XCR1 expression on CCR2+Ly6C+ monocytes was significantly increased (Fig. 9C). Thus, we further hypothesized that IFNγ signaling triggers chemokine receptor up-regulation on CCR2+Ly6C+ monocytes, making them responsive to the chemokines released upon cognate Ag stimulation. We tested this idea by cotransferring WT and Ifngr1−/− BM into WT recipient mice immediately challenged with Lm and monitored CCR5 and XCR1 chemokine receptor expression 16 hours later (Fig. 9D). While we could detect CCR5- and XCR1-expressing WT CCR2+Ly6C+ monocytes, Ifngr1−/− monocytes failed to up-regulate these receptors, indicating that IFNγ signaling controls cell surface up-regulation of CCR5 and XCR1 on CCR2+Ly6C+ monocytes, making them responsive to CD8+ TM cell–derived chemokines. Hence, these results collectively support a model where IFNγ signaling in CCR2+Ly6C+ monocytes enables up-regulation of CCR5 and XCR1 chemokine receptors, making them responsive to CCL3, CCL4, and XCL1 chemokines that orchestrate their production of TNFα, a cytokine absolutely required for host protective memory responses against secondary Lm infection (17, 21).
DISCUSSION
This study provides an in-depth cellular and molecular analysis of how cognate Ag orchestrates and programs the activation of CD8+ TM cells for rapid protection against a recall infection in vaccinated hosts in vivo. We show that cognate Ag recognition by CD8+ TM cells leads to a broad gene expression program targeting multiple pathways within only a few hours after stimulation. We also highlight that IRF4, downstream and proportional to TCR signaling strength, transcriptionally controls the most significantly up-regulated cluster of genes in cognate Ag–stimulated CD8+ TM cells that encode for the chemotactic molecules CCL3, CCL4, and XCL1. Production of these chemokines requires Ag presentation by CD11chi DCs and cannot be substituted by CCR2+Ly6C+ monocytes. We reveal that cognate Ag recognition enables CD8+ TM cell arrest in splenic RP clusters of CCR2+Ly6C+ monocytes for the local delivery of IFNγ and chemokines, efficiently restraining microbial pathogen spreading and growth. Our results support a model in which CD8+ TM cell–derived IFNγ enables immunized host protection by controlling CCR2+Ly6C+ monocyte responsiveness to chemokines. Chemokines, in turn, signal to induce TNFα and CXCL9 production by the monocytes. These results suggest a refined model for IFNγ-dependent protection in which CD8+ TM (i) IFNγ enables protection by enhancing chemokine signaling and (ii) chemokines act as key effector molecules activating CCR2+Ly6C+ monocytes, a role that is distinct from their usual role in chemotaxis (Fig. 10).
Fig. 10. Mechanism of early CD8+ TM cell–mediated protection.
CD8+ TM cells arrest upon cognate Ag recognition presented by DCs in CCR2+Ly6C+ monocyte splenic RP clusters. Upon local delivery of IFNγ, monocytes up-regulate chemokine receptors (CCR5 and XCR1) and become responsive to cognate Ag–secreted chemokines (CCL3, CCL4, and XCL1). Chemokines then promote CCR2+Ly6C+ monocyte activation and TNFα production promoting microbicidal effector functions and host protection.
The current results highlight the importance of rapid microbial pathogen containment, a notion that has been elegantly illustrated in prior reports (24, 25). First-line cellular responders such as splenic marginal zone and LN subcapsular macrophages were reported to rapidly uptake and/or sense microbial pathogens (bacteria and viruses) to subsequently provide chemotactic cues that attract prepositioned memory—but not naïve—CD8+ T cells rapidly to the sites of infection. Consistent with these results, intravenously inoculated Lm bacteria are rapidly cleared from the blood by marginal zone CD169+ macrophages and DCs localized in the splenic RP (27, 43, 44, 48, 49). Following rapid pathogen capture by tissue-resident sentinel cells, a body of evidence suggests that CD8+ TM cells home to infectious foci via CXCR3 and/or CCR5 and associated CXCL10, CXCL9, and CCL5 chemokines produced in response to local inflammatory cues such as IFNs (50–52). A large majority of CD8+ TM cells express CXCR3 and CCR5 and thus can be readily mobilized for rapid migration, independent from cognate Ag encounter (26). These cells can produce IFNγ in response to cytokines (11, 12, 14), further increasing local chemokine levels in a feedforward positive loop leading to the rapid amplification of the CD8+ TM cell response. We report in the current study that cognate Ag recognition promotes a broad activation program in CD8+ TM cells, which includes the early expression of a potent set of chemokines. This finding led us to propose that cognate Ag–triggered CD8+ TM cells would amplify the initial chemotactic cues and act as powerful recruiting orchestrators of both adaptive and innate immune cells, setting the stage for more effective microbial pathogen clearance. Unexpectedly, however, our results did not support such a model. Rather, we revealed that CCR2+Ly6C+ monocytes form clusters in the splenic RP independently from cognate Ag and CD8+ TM cells, most likely in response to other infection-driven chemotactic cues. In addition to chemotaxis, adhesion molecules such as ICAM-1, CD11b, and CD44 could be mediating CCR2+Ly6C+ monocyte trafficking to sites of infection as it was shown in the liver of primary Lm-infected mice (53). Consistent with this idea, we also noted a strong up-regulation of ICAM-1 on clustered monocytes in spleen RP. Using IVM imaging, we further revealed that, as expected (40–42), CD8+ TM cells arrest upon cognate Ag recognition, which occurs in CCR2+Ly6C+ monocyte clusters, where they promote their activation through the localized delivery of chemokines and IFNγ. Rather than acting as chemoattractants, the CD8+ TM cell–derived chemokines directly enable CCR2+Ly6C+ monocyte effector functions. While chemokines are necessary, they are not sufficient. IFNγ enables CCR2+Ly6C+ monocytes responsiveness to the chemokines. These data collectively support a model where CD8+ TM cells, through localized delivery of IFNγ and chemokines, function to license and boost monocyte microbicidal function in a targeted manner within monocyte clusters, rather than as initial orchestrators of the early steps of the recall response. Ag presentation also selectively occurs on DC and cannot be substituted by CCR2+Ly6C+ monocytes.
In a previous study using IVM imaging to explore Lm infection foci that form in subcapsular DCs (scDCs) of the splenic RP following primary infection (day 5 after infection), Lm-specific effector CD8+ T cells were shown to migrate to sites of infection where a mixture of myelomonocytic cells (MMCs), which include CCR2+Ly6C+ monocytes and neutrophils, accumulates (39). These MMCs markedly reduced blood flow access to the sites of infection and restricted Lm growth. Lm-specific CD8+ T cells were also shown by IVM imaging to undergo both Ag-dependent arrest and Ag-independent reduced motility in the scDC/MMC Lm-containing clusters, similarly to our observations. However, this study did not address the role that arrested effector CD8+ T cells may play in these clusters. Disappearance of Lm was associated with effector CD8+ T cells regaining motility, but evidence for direct Lm-infected killing could not be documented. Together with the large dependence on MMC for Lm clearance, these data suggested that, like in the setting of the recall response, noncytolytic T cell–dependent effector mechanisms were essential. In addition to promoting the local expression of microbicial activities in clustered CCR2+Ly6C+ monocytes, it seems therefore conceivable that the delivery of effector molecules by Ag-arrested CD8+ TM cells may also restrict permeability and blood flow in these clusters to ultimately enhance rapid and effective Lm containment and killing.
Lm killing and vaccinated host protection during recall infection require TNFα, which CCR2+Ly6C+ monocytes are a major source (17, 21, 54). TNFα directly triggers microbicidal ROS both from CCR2+Ly6C+ monocytes and neutrophils, and ROS promotes antimicrobial autophagy (38). While we previously showed that IFNγ signaling to CCR2+Ly6C+ monocytes induces TNFα production by these cells (20), we now refine this model by reporting that IFNγ signaling, while necessary, is not sufficient. IFNγ-primed CCR2+Ly6C+ monocytes become highly responsive to chemokine signals by up-regulating their chemokine receptors, enabling chemokine signals to trigger enhanced microbicidal functions for rapid Lm clearance. The fact that both IFNγ and chemokines need to be spatially and temporally targeted on CCR2+Ly6C+ monocytes at sites of infection to promote their effector responses may represent an evolutionarily conserved mechanism to prevent systemic tissue damages to the host. While we did not monitor neutrophil dynamics here, neutrophils are well known to be massively recruited and undergo activation in infected spleens, and we and others have previously shown that they cluster with CCR2+Ly6C+ monocytes and CD8+ TM cells at infection foci (16, 20, 27). However, and in contrast to CCR2+Ly6C+ monocytes, neutrophils neither express nor up-regulate high levels of CCR1, CCR5, or XCR1, suggesting that CD8+ TM cell–derived chemokines are unlikely to account for neutrophil activation in this setting. Fine-tuning of CCR2+Ly6C+ monocyte activation in response to local chemokine levels and chemokine receptor up-regulation may regulate their ability to secrete TNFα, which directly promotes ROS production and pathogen killing.
Another important finding in our study relates to the rapid, transcriptionally controlled, and coordinated production of CCL3, CCL4, and XCL1 chemokines by CD8+ TM cells induced upon vaccination with both Lm and VSV in response to cognate Ag recognition. These results are consistent with two recent reports that used multiple models of acute and chronic infections, as well as ex vivo stimulation assays, outlining that the robust chemokine signature is a key and important feature of both Ag-stimulated effector and CD8+ TM cells (36, 37). CD8+ TM cells undergoing repetitive in vivo stimulations were also reported to significantly up-regulate genes encoding for these chemokines (55). Our study further reveals that CCL3, CCL4, and XCL1 chemokines produced by cognate Ag–stimulated CD8+ TM cells are under the control of the IRF4 transcriptional regulator, a known amplifying rheostat downstream of TCR signaling that has been shown to control the size of T cell clonal expansion (33). On the basis of our data, IRF4 may also enable the graded production of these chemokines by CD8+ TM cells, proportionally to the strength of TCR signaling. Further investigations would be needed to determine whether this involves direct IRF4 transcriptional regulation where IRF4 binds to the promoter or enhancer regions of the chemokine-encoding genes. This is likely to represent a mechanism to limit tissue-associated damages, when weak epitopes are presented to Ag-specific CD8+ TM cells. In the context of strong epitope stimulation, however, our results establish that chemokines are secreted concomitantly to CD8+ TM cell arrest in CCR2+Ly6C+ monocyte clusters, promoting their increased production of TNFα and CXCL9 both in vitro and in vivo. These findings also highlight that chemokines can trigger innate immune cell effector functions, delineating a role distinct from usual chemotaxis.
While our study focuses on systemic and secondary lymphoid organ (SLO)-derived CD8+ TM cell responses, multiple evidence suggest that the current mechanisms are also relevant in the context of tissue-resident CD8+ TM cell responses. In several models of viral infection (skin, vagina, and lung), TRM cells—both CD8+ and CD4+—quickly initiate and orchestrate a rapid mucosal response upon cognate Ag encounter, through local production of IFNγ and subsequent CXCL9 (28–30, 51, 56, 57). As discussed earlier, CXCL9 enables migration of more circulating TM cells to sites of infection, enhancing the activation of local DCs and NK cells and the establishment of an IFNγ-driven antiviral state providing broad protective immunity against unrelated microbial pathogens. In these studies, reactivation of CD8+ TM cells and the production of activating IFNγ required cognate Ag recognition, yet many reports monitoring systemic CD8+ TM cells have also established that CD8+ TM cells in SLO undergo cytokine-mediated activation (11, 13–15, 26). This seemingly discrepant result may be a reflection of tissue-specific mechanisms. It was recently shown that LN CD8+ TM cells strictly require cognate Ag to be presented by XCR1+ DCs, while lung TRM cells can be reactivated by both hematopoietic and nonhematopoietic cells (8). Here, cognate Ag presentation by hematopoietic versus nonhematopoietic-derived cells to CD8+ TRM cells was also proposed to dictate distinct functional outcomes with hematopoietic-derived Ag-presenting cells restraining an excessive inflammatory program in CD8+ TRM cells, presumably as a safeguard mechanism against collateral tissue damages. Nonhematopoietic Ag presentation was associated with a proliferative program and largely prevented cytokine-mediated activation of CD8+ TRM cells. Note that this study used the Nur77GFP reporter system, a readout of TCR-dependent cognate Ag stimulation, thereby only focusing on early Ag-dependent CD8+ TM cell expression programs. Other reports using complex biological readouts (e.g., proliferation and protection) have supported a more prominent role for recruited or tissue-resident DCs in the reactivation of CD8+ TRM cells, raising the possibility that different memory cell–intrinsic mechanisms of regulation may be specifically programmed upon DC-mediated versus nonhematopoietic cell–mediated activation (9, 10).
In conclusion, this work provides a comprehensive analysis of cognate Ag–induced early transcriptional and functional changes in reactivated CD8+ TM cells and how these changes enable the rapid control of microbial pathogen in vaccinated hosts in situ. Perhaps contrasting with the widely accepted view, our results favor a model in which CD8+ TM cells mediate host protection during a recall infection through a targeted, “surgical” intervention. The initiating response is largely regulated by tissue-specific cues and innate immune cells before CD8+ TM cells intervene to make the innate cellular effector response highly effective. Another unexpected finding of this work is related to IFNγ, which, at least in this context, is not sufficient to achieve full activation of CCR2+Ly6C+ monocytes and requires chemokine signaling. This result suggests that many levels of fine-tuning are involved to make a memory response most effective while preventing excessive damages to vaccinated host.
MATERIALS AND METHODS
Ethics statement
This study was carried out in strict accordance with the recommendations by the animal use committee at the Albert Einstein College of Medicine. All efforts were made to minimize suffering and provide humane treatment to the animals included in the study.
Mice
All mice were bred in our specific pathogen–free animal facility at the Albert Einstein College of Medicine. We used 6- to 8-week-old WT B6 male or female mice, congenic CD45.1+/+ (JAX#002014), B6-Kd (58), OT-I+ (JAX#003831) crossed to Rosa26Actin-tomato-loxP-STOP-loxP (LSL)–GFP (Td+) (JAX#007576), gBT-I+ [gift from F. Carbone (59)] crossed to UBCGFP/GFP (JAX#004353) or to CD45.1+/+ mice, Ccr2DTR-CFP/WT [gift from E. Pamer (60)], Itgax/Cd11cDTR/WT (JAX#004509), Rosa26CreERT2 (JAX#008463), Irf4loxP/loxP (or fl/fl) (JAX#009380), Ifngr1−/− (JAX#003288), and CX3CR1ERT2Cre (JAX#020940) crossed to Rosa26LSL-PTX [gift from S. Coughlin (46)] purchased from the Jackson laboratories unless otherwise indicated. All mice are on the B6 genetic background unless otherwise specified.
Microbial pathogens and mouse infections
Listeria monocytogenes
Mice were inoculated with Lm, Lm expressing the ovalbumin (Lm-Ova; gift from H. Shen, University of Pennsylvania) or Ova, and HSV-2 glycoprotein B 498-505 epitope (Lm-Ova-gB; gift from D. Zehn, Technical University of Munich), all expressed under the LLO/Hly promoter. All Lm were prepared after passaging into WT B6 mice, by growing to log phase [optical density at 600 nm (OD600), ~0.3 to 0.4], and kept as frozen aliquots for single use at −80°C. For infections, bacteria were grown to a logarithmic phase (OD600, ~0.05 to 0.15) in broth heart infusion medium, diluted in PBS to infecting concentration, and intravenously injected. We used 0.1 × median lethal dose, i.e., 104 Lm colony-forming units (CFUs) for primary immunizations and 106 Lm CFUs for secondary/recall challenge infections (~6 weeks later). All Lm are on the 10403s genetic background.
Vesicular stomatitis virus
Single-use frozen aliquots of VSV encoding Ova (VSV-Ova; gift from K. Khanna, New York University) kept at −80°C were thawed and diluted in cold PBS right before mouse primary intravenous infections with 2 × 105 plaque-forming units (PFUs). For secondary challenge infections of VSV-immunized mice (~6 weeks later), we used 106 Lm CFUs.
Preparation of cell suspensions for flow cytometry and adoptive transfers
Spleens were dissociated on a nylon mesh and lysed in red blood cell lysis buffer (0.83% NH4Cl, v/v), before incubation in Hanks’ balanced salt solution medium with collagenase I (4000 U/ml) and deoxyribonuclease I (0.1 mg/ml). BM cells were obtained by flushing femur with complete medium [RPMI 1640, 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 55 μM β-mercaptoethanol, 1 mM sodium pyruvate, 1× GlutaMAX, and 1× nonessential amino acids] containing 10% FBS.
Cell staining for fluorescence-activated cell sorting analysis and cell sorting
Cell suspensions were incubated with 2.4G2 Fc block and stained in PBS, 1% FBS, 2 mM EDTA, and 0.02% sodium azide with fluorescently tagged Abs purchased from eBioscience, BD Biosciences, R&D Systems, or BioLegend (see details in table S3) or major histocompatibility complex class I (MHCI) (Kd or Kb) Tet. For Tet, biotinylated monomers (1 mg/ml) obtained from the National Institutes of Health (NIH) Tetramer Core Facility were conjugated with PE-labeled streptavidin (1 mg/ml) as follows: 6.4 μl of PE-streptavidin was added to 10 μl of monomers every 15 min, four times on ice. Newly generated Tet were then used to stain spleen cells for 1 hour at 4°C (1:400 to 1:500 dilution). To stain for expression of the IRF4 transcription factor, cells were fixed in eBioscience Fixation/Permeabilization buffer before anti-IRF4 mAb staining in eBioscience Permeabilization buffer for 30 min. For intracellular cytokines, cells were first incubated for 4 hours at 37°C and 5% CO2 in complete medium, 10% FBS, with GolgiPlug/GolgiStop (brefeldin A/monensin A). Next, cells were stained for cell surface marker expression and fixed in intracellular fixation buffer (eBioscience) before permeabilization for ~1 hour in the presence of Abs/sera against intracellular cytokines (IFNγ and TNFα) and chemokines (CXCL9, CCL3, CCL4, and XCL1). For CCL3, CCL4, and XCL1, a donkey anti-goat (2 μg/ml) secondary Ab was used. Data acquisition was performed on a FACSAria III or LSRII flow cytometer. All flow cytometry data were analyzed using FlowJo v9 software (TreeStar).
Cell sorting was performed using a four-laser (405, 488, 561, and 640 nanometer) FACSAria III cell sorter from BD equipped with FACSDiva version 6.1.3. The instrument was set up with a 100-μm nozzle at 20 psi, and the samples were introduced to the system at the lowest flow rate to minimize shear stress. The sorted populations were gated to exclude double and dead cells. The sort was performed with a purity precision mode.
Cell sorting for adoptive T cell transfers
For naïve OT-I and gBT-I cells, WT mice were adoptively transferred with ~1000 OT-I Td+ and 50,000 gBT-I cells isolated from the spleen of OT-I Td+ and gBT-I CD45.1+/+ mice. The next day, mice were immunized with Lm-Ova-gB. Immunized mice were next used ~6 weeks later to investigate OT-I and gBT-I TM cell reactivation by fluorescence-activated cell sorting (FACS) and IVM. For adoptive transfers of OT-I memory cells, WT mice were first adoptively transferred with ~1000 naïve OT-I Td+ cells as above, immunized the next day with VSV-Ova, and challenged 2 weeks later with 106 Lm-Ova. After ~4 weeks, spleens were harvested, and CD8+ T cells were negatively selected using anti-CD4, anti-CD11b, anti-MHCII, anti-TER119, anti-B220, and anti-CD19 mAbs (table S3), which were all added and incubated at 5 μg/ml for 30 min at 4°C. Cells were then washed and incubated with anti-rat Ab magnetic beads at one bead per target cell for 40 min at 4°C [Dynabeads sheep anti-rat immunoglobulin G (IgG), Invitrogen]. CD8+ T cell purity was ~70%. Cells were next sorted into 3 ml of complete media (RPMI 1640, 10% FBS, 1% penicillin/streptomycin, 55 μM β-mercaptoethanol, 1 mM sodium pyruvate, 1× GlutaMAX, and 1× nonessential amino acids) using our four-laser BD FACSAria III cell sorter. A total of 2 × 105 OT-I TM cells (purity, >98.5%) were transferred to either WT, Ccr2CFP, or Ifngr1−/− recipient mice further challenged with 106 Lm or Lm-Ova for analysis of memory functions, protection, or IVM.
BM transfers for CCR2+Ly6C+ monocyte chemokine receptor expression analysis
A total of 5 × 106 BM cells from WT CD45.1+/− and Ifngr1−/− CD45.2+/+ donors were cotransferred to mice immunized with Lm-Ova 6 weeks before and further challenged or not with Lm. Spleen cells were next stained for chemokine receptor expression on monocytes from BM donor-derived cells.
Generation of BM chimera mice
Lethally irradiated 12 Gy B6-Kd mice were immediately reconstituted with a total of 2 × 106 BM cells isolated from (i) Ccr2DTRKd and Kd, (ii) Ccr2DTRKd and WT, (iii) Cd11cDTRKd and WT, and (iv) Kd and WT mice, at a 7:3 ratio, respectively. Donor BM cells were depleted of CD8 and CD4 T cells from WT BM cells using anti-CD8β (clone H35) and anti-CD4 (clone GK1.5) mAbs before reconstitution. Chimerism of reconstituted mice was checked ~6 weeks later in the blood, before immunizations.
In vivo treatments
Monocyte and DC depletion
CD11c+ or CCR2+ cells were respectively depleted in mice expressing the DTR under the CD11c/Integrin Subunit Alpha X (Itgax) or the CCR2 promoter upon intraperitoneal injection of 10 ng/g of mouse body weight of DT (Calbiochem) 12 hours before Lm challenge infection.
Tx treatments to induce Irf4 depletion or to express PTX in CX3CR1+ cells
4-Hydroxytamoxifen (#T5648, Sigma-Aldrich) was dissolved in sunflower oil to a concentration of 10 mg/ml for intraperitoneal injection. For Irf4 depletion, 3000 OT-I-Rosa26-CreERT2Irf4fl/fl and 1000 OT-I Irf4WT were cotransferred to WT B6 mice that were next immunized with Lm-Ova the day after. Six weeks later and before challenge, mice were treated with Tx (1 mg per injection in 100 μl) or vehicle (100 μl of sunflower oil) for 5 days, and 24 hours after the last Tx injection, mice were challenged with Lm-Ova. For induction of PTX expression in CX3CR1+ cells, 1000 OT-I were transferred to CX3CR1creERT2Rosa26LSL-PTX mice that were immunized with Lm-Ova the next day. Six weeks later and before challenge, mice were intraperitoneally treated with Tx or vehicle as above and, the next day, challenged with Lm-Ova.
In vivo Ly6C-PE Ab labeling for CCR2+Ly6C+ monocyte staining
For IVM analysis, 10 μg of Ly6C-PE (clone HK1.4, rat IgG2a, BioLegend) mAb was inoculated to mice intravenously at the time of Lm challenge or 1 hour before euthanizing mice. For FACS analysis, 5 μg of Ly6C-PE mAb was injected to challenged mice 1 hour before the euthanasia.
In vitro activation assays
Quantification of CCL3 and IFNγ secretion
A total of 106 splenocytes from mice immunized with Lm-Ova and challenged or not 6 weeks later with Lm-Ova for 16 hours were incubated in 96-well flat-bottom plate with complete medium only or in the presence of Golgi Plug/Stop for 4 hours at 37°C. CCL3 (Thermo Fisher Scientific) and IFNγ (BioLegend) production in culture supernatants was then quantified by enzyme-linked immunosorbent assay.
Measure of chemokine expression by ex vivo restimulated OT-I TM cells
Splenocytes from mice immunized with Lm-Ova 6 weeks prior were coincubated with SIINFEKL peptide (10−8 M) with Golgi Plug/Stop and (i) with or without cycloheximide (translation inhibitor; 10 μg/ml; Sigma-Aldrich) or actinomycin D (transcription inhibitor; 8 μM; Sigma-Aldrich) for 1, 2, 3, and 4 hours in complete medium at 37°C and (ii) with the SCG-CBP30 IRF4 inhibitor (20 μM; Selleckchem). Cells were next stained as described above including for intracellular expression of CCL3 and/or CCL4 and XCL1.
Measure of TNFα expression in ex vivo stimulated CCR2+Ly6C+ monocytes and neutrophils
A total of 104 monocytes or neutrophils were FACS-sorted (as above; purity, >98%) from mice primary immunized and challenged with Lm-Ova 6 weeks later for 16 hours. Cells were next coincubated in 96-well round-bottom plate and complete medium, with HKLm, rCCL3, rCCL4, rXCL1, or the combination of the three recombinant chemokines, in the presence of GolgiPlug/Stop for 4 hours at 37°C before staining for cell surface and intracellular markers for FACS analysis. In CCR5 and CCR1 blocking experiments, cells were incubated with both CCR5 (1 μM; Maraviroc, Cayman Chemical Company) and CCR1 (1 μM; J113863, Santa Cruz Biotechnology) chemical inhibitors or control dimethyl sulfoxide 30 min before adding recombinant chemokines or HKLm.
Measure of TNFα and CXCL9 expression in in vitro cocultured CCR2+Ly6C+ monocytes and OT-I TM cells
A total of 8 × 104 OT-I TM cells were FACS-sorted (as above; purity, >98%) and stimulated with SIINFEKL peptide (10−6 M) OVN in complete media (37°C, CO2). The next day, we sorted 2 × 104 CCR2+Ly6C+ monocytes from a naïve spleen based on the cell surface expression of CD11b and Ly6C and CD8 exclusion. Sorted monocytes (purity, >98.5%) were next added in the presence of Golgi Plug/Stop to the T cells with or without anti-CCL3, CCL4, and XCL1 chemokine-neutralizing antisera (table S3). As control, naïve sorted monocytes were also incubated with the three recombinant chemokines rCCL3, rCCL4, and rXCL1 (table S3) before staining for cell surface and intracellular markers for FACS analysis.
RNA sequencing
Samples and library preparation
A total of 1000 OT-I Td+ and 1000 gBT-I CD45.1+/+ TM cells were flow-purified (FACSAria III) following the same procedure as for adoptive TM cell transfers described before, from 6-week Lm-Ova-gB–immunized mice either left unchallenged or challenged with Lm-Ova. However, here, TM cells were directly sorted into 1× lysis buffer, and complementary DNA (cDNA) was synthesized and amplified directly from intact cells using the SMART-Seq v4 Ultra Low Input RNA Kit for sequencing (Takara Bio, USA) according to the manufacturer’s protocol. cDNA was isolated using the Agencourt AMPure XP Kit (Beckman Coulter, Brea, CA) and quantified using the Qubit dsDNA High Sensitivity Assay Kit (Life Invitrogen) on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The library preparation was performed using the Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA). Samples were sequenced to depths of up to 16.7 million single-end 75-nucleotide length reads per sample using the Illumina NextSeq 500/550 High Output v2 kit (75 cycles) on an Illumina NextSeq 500 Sequencing System. Image analysis, base calling, and generation of sequence reads were produced using the NextSeq Control software v2.0 and Real-Time Analysis software v2. Data were converted to FASTQ files using the bcl2fastq2 v2.20 software (Illumina Inc.). Sequencing data were initially quality-checked using FastQC, before alignment and initial analysis. Reads were aligned to the mouse reference mm10 using STAR aligner (v2.4.2a) (61). Quantification of genes annotated in Gencode vM5 was performed using featureCounts (v1.4.3), and quantification of transcripts was performed using Kalisto (62). Quality check was collected with Picard (v1.83) and RSeQC (63) (http://broadinstitute.github.io/picard/). Normalization of feature counts was performed using the DESeq2 package version 1.10.1. Before analysis, nonrelevant batch effect (such as library preparation or sequencing batch) was identified using unsupervised PCA, and analysis was corrected for batch effects through our model. Differentially expressed genes were identified using negative binomial distribution as implemented in DESeq 2 [R package (64)]. Significantly up-regulated and down-regulated genes (differentially expressed gene) were defined with a false discovery rate step-up P ≤ 0.05 and a fold change of ≥±1.5. The raw data from the National Center for Biotechnology Information database (GEO GSE160280) were subsequently analyzed for enrichment of GO terms and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, implemented in the clusterProfiler (R package, function enrichGO or enrichKEGG); a pathway is considered significantly enriched if the enrichment score is ≥1.5 (equivalent to P ≤ 0.05).
Intravital and explant imaging
For intravital imaging, mice were anesthetized with isoflurane, and the spleen was surgically exposed and elevated above the body of the mouse. A glass coverslip was carefully applied to the top of the spleen to create an imaging window. Mice were kept at 37°C using a custom heating platform. Imaging was performed on an Olympus FVE-1200 upright microscope using a 25× 1.04–numerical aperture objective and a Deepsee MaiTai Ti-Sapphire pulsed laser (Spectra Physics) tuned to 870 nm. To maintain temperature and limit infiltrating light, the microscope was fitted with a custom-built incubator chamber heated to 37°C. Z-stack images (512 by 512) were acquired every 60 s with 5-μm steps. For explant imaging, mice were euthanized with CO2, and spleens were immediately harvested. Spleens were affixed to coverglass using Vetbond (3M) on the medulla. Tiled images were acquired using 320 × 320 Z-stack images with 15-μm steps. Tiled images were stitched using Olympus Fluoview software. Cell tracking, drift correction, and monocyte volume analysis were carried out using Imaris 9.2 (Bitplane).
Measure of protective immunity
Protection was measured by enumerating Lm titers in spleens and livers of challenged mice. For this, spleens and livers were harvested 24 hours after infection and dissociated on metal screens in 10 ml of water/0.1% Triton X-100 (Sigma-Aldrich). Serial dilutions were performed in the same buffer, and 100 μl was plated onto brain heart infusion (BHI) medium plates. Lm CFU numbers were counted 24 hours later and reported to the whole spleen and liver. This procedure was the same whether mice were (i) primary challenged and transferred or not with OT-I memory cells, (ii) primary/boosted or not, and secondary challenged treated or not with Tx (CX3CR1creERT2Rosa26LSL-PTX, OT-I Rosa26-CreERT2 Irf4fl/fl).
Statistics
Statistical significance was calculated using an unpaired Student’s t test with GraphPad Prism software, and two-tailed P values are given as follows: *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001, and not significant (ns) for P > 0.1. All P values of 0.05 or less were considered significant.
Acknowledgments
We thank the Einstein FACS and genomic core facilities. Funding: This work was funded by the NIH AI103338, Hirschl Caulier Award to G.L., and NIH HL141491 to D.F. M.B. received fellowships from FRM and ZB NIH F32 fellowship HL149155. E.G. received support from NIH PREP R25GM104547 and MSTP T32GM007288. Core resources for FACS were supported by the Einstein Cancer Center (NCI cancer center support grant 2P30CA013330). Author contributions: M.B. designed, performed, and interpreted all experiments, assembled all figures, and wrote the manuscript with G.L. Z.B. and D.F. designed, conducted, analyzed, and interpreted IVM experiments and contributed to scientific discussions and manuscript editing. F.D. with M.B. and G.L. analyzed and interpreted all transcriptomic data. E.G. and C.K. contributed to multiple mouse experiments with M.B. S.M.S. generated the Lm-LLOS92-Ova strain. G.L. designed and interpreted experiments with M.B. and all other authors, contributed to figure design and editing, and wrote the paper with M.B. Competing interests: The authors declare that they have no competing interests. Data materials and availability: The accession number for the microarrays and RNA sequencing data reported in this paper is GEO GSE160280. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Legends for tables S1 to S3
Legends for movies S1 to S3
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S3
Movies S1 to S3
REFERENCES AND NOTES
- 1.Harty J. T., Tvinnereim A. R., White D. W., CD8+ T cell effector mechanisms in resistance to infection. Annu. Rev. Immunol. 18, 275–308 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Vesely M. D., Kershaw M. H., Schreiber R. D., Smyth M. J., Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 29, 235–271 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Wong P., Pamer E. G., CD8 T cell responses to infectious pathogens. Annu. Rev. Immunol. 21, 29–70 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Sallusto F., Lanzavecchia A., Araki K., Ahmed R., From vaccines to memory and back. Immunity 33, 451–463 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schenkel J. M., Masopust D., Tissue-resident memory T cells. Immunity 41, 886–897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin M. D., Badovinac V. P., Defining memory CD8 T Cell. Front. Immunol. 9, 2692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zammit D. J., Cauley L. S., Pham Q. M., Lefrancois L., Dendritic cells maximize the memory CD8 T cell response to infection. Immunity 22, 561–570 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low J. S., Farsakoglu Y., Amezcua Vesely M. C., Sefik E., Kelly J. B., Harman C. C. D., Jackson R., Shyer J. A., Jiang X., Cauley L. S., Flavell R. A., Kaech S. M., Tissue-resident memory T cell reactivation by diverse antigen-presenting cells imparts distinct functional responses. J. Exp. Med. 217, e20192291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakim L. M., Waithman J., van Rooijen N., Heath W. R., Carbone F. R., Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science 319, 198–202 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Shin H., Kumamoto Y., Gopinath S., Iwasaki A., CD301b+ dendritic cells stimulate tissue-resident memory CD8+ T cells to protect against genital HSV-2. Nat. Commun. 7, 13346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soudja S. M., Ruiz A. L., Marie J. C., Lauvau G., Inflammatory monocytes activate memory CD8+ T and innate NK lymphocytes independent of cognate antigen during microbial pathogen invasion. Immunity 37, 549–562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kupz A., Guarda G., Gebhardt T., Sander L. E., Short K. R., Diavatopoulos D. A., Wijburg O. L., Cao H., Waithman J. C., Chen W., Fernandez-Ruiz D., Whitney P. G., Heath W. R., Curtiss R. III, Tschopp J., Strugnell R. A., Bedoui S., NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8+ T cells. Nat. Immunol. 13, 162–169 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Bedoui S., Whitney P. G., Waithman J., Eidsmo L., Wakim L., Caminschi I., Allan R. S., Wojtasiak M., Shortman K., Carbone F. R., Brooks A. G., Heath W. R., Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat. Immunol. 10, 488–495 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Raue H. P., Beadling C., Haun J., Slifka M. K., Cytokine-mediated programmed proliferation of virus-specific CD8+ memory T cells. Immunity 38, 131–139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berg R. E., Crossley E., Murray S., Forman J., Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J. Exp. Med. 198, 1583–1593 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandre Y. O., Ghilas S., Sanchez C., Le Bon A., Crozat K., Dalod M., XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J. Exp. Med. 213, 75–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narni-Mancinelli E., Campisi L., Bassand D., Cazareth J., Gounon P., Glaichenhaus N., Lauvau G., Memory CD8+ T cells mediate antibacterial immunity via CCL3 activation of TNF/ROI+ phagocytes. J. Exp. Med. 204, 2075–2087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook D. N., Smithies O., Strieter R. M., Frelinger J. A., Serody J. S., CD8+ T cells are a biologically relevant source of macrophage inflammatory protein-1α in vivo. J. Immunol. 162, 5423–5428 (1999). [PubMed] [Google Scholar]
- 19.Hu X., Ivashkiv L. B., Cross-regulation of signaling pathways by interferon-γ: Implications for immune responses and autoimmune diseases. Immunity 31, 539–550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soudja S. M., Chandrabos C., Yakob E., Veenstra M., Palliser D., Lauvau G., Memory-T-cell-derived interferon-gamma instructs potent innate cell activation for protective immunity. Immunity 40, 974–988 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neighbors M., Xu X., Barrat F. J., Ruuls S. R., Churakova T., Debets R., Bazan J. F., Kastelein R. A., Abrams J. S., O'Garra A., A critical role for interleukin 18 in primary and memory effector responses to Listeria monocytogenes that extends beyond its effects on interferon γ production. J. Exp. Med. 194, 343–354 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White D. W., Harty J. T., Perforin-deficient CD8+ T cells provide immunity to Listeria monocytogenes by a mechanism that is independent of CD95 and IFN-γ but requires TNF-α. J. Immunol. 160, 898–905 (1998). [PubMed] [Google Scholar]
- 23.White D. W., Badovinac V. P., Kollias G., Harty J. T., Cutting edge: Antilisterial activity of CD8+ T cells derived from TNF-deficient and TNF/perforin double-deficient mice. J. Immunol. 165, 5–9 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Kastenmuller W., Brandes M., Wang Z., Herz J., Egen J. G., Germain R. N., Peripheral prepositioning and local CXCL9 chemokine-mediated guidance orchestrate rapid memory CD8+ T cell responses in the lymph node. Immunity 38, 502–513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung J. H., Zhang H., Moseman E. A., Alvarez D., Iannacone M., Henrickson S. E., de la Torre J. C., Groom J. R., Luster A. D., von Andrian U. H., Chemokine guidance of central memory T cells is critical for antiviral recall responses in lymph nodes. Cell 150, 1249–1263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurice N. J., McElrath M. J., Andersen-Nissen E., Frahm N., Prlic M., CXCR3 enables recruitment and site-specific bystander activation of memory CD8+ T cells. Nat. Commun. 10, 4987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bajenoff M., Narni-Mancinelli E., Brau F., Lauvau G., Visualizing early splenic memory CD8+ T cells reactivation against intracellular bacteria in the mouse. PLOS ONE 5, e11524 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ariotti S., Hogenbirk M. A., Dijkgraaf F. E., Visser L. L., Hoekstra M. E., Song J. Y., Jacobs H., Haanen J. B., Schumacher T. N., Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Iijima N., Iwasaki A., T cell A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science 346, 93–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schenkel J. M., Fraser K. A., Beura L. K., Pauken K. E., Vezys V., Masopust D., T cell memory resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 346, 98–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenkel J. M., Fraser K. A., Vezys V., Masopust D., Sensing and alarm function of resident memory CD8+ T cells. Nat. Immunol. 14, 509–513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerlach C., Moseman E. A., Loughhead S. M., Alvarez D., Zwijnenburg A. J., Waanders L., Garg R., de la Torre J. C., von Andrian U. H., The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45, 1270–1284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Man K., Miasari M., Shi W., Xin A., Henstridge D. C., Preston S., Pellegrini M., Belz G. T., Smyth G. K., Febbraio M. A., Nutt S. L., Kallies A., The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 14, 1155–1165 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Yao S., Buzo B. F., Pham D., Jiang L., Taparowsky E. J., Kaplan M. H., Sun J., Interferon regulatory factor 4 sustains CD8+ T cell expansion and effector differentiation. Immunity 39, 833–845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zehn D., Lee S. Y., Bevan M. J., Complete but curtailed T-cell response to very low-affinity antigen. Nature 458, 211–214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eberlein J., Davenport B., Nguyen T. T., Victorino F., Jhun K., van der Heide V., Kuleshov M., Ma'ayan A., Kedl R., Homann D., Chemokine signatures of pathogen-specific T cells I: Effector T cells. J. Immunol. 205, 2169–2187 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davenport B., Eberlein J., Nguyen T. T., Victorino F., van der Heide V., Kuleshov M., Ma'ayan A., Kedl R., Homann D., Chemokine signatures of pathogen-specific T cells II: Memory T cells in acute and chronic infection. J. Immunol. 205, 2188–2206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narni-Mancinelli E., Soudja S. M., Crozat K., Dalod M., Gounon P., Geissmann F., Lauvau G., Inflammatory monocytes and neutrophils are licensed to kill during memory responses in vivo. PLOS Pathog. 29, e1002457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waite J. C., Leiner I., Lauer P., Rae C. S., Barbet G., Zheng H., Portnoy D. A., Pamer E. G., Dustin M. L., Dynamic imaging of the effector immune response to listeria infection in vivo. PLOS Pathog. 7, e1001326 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mempel T. R., Henrickson S. E., Von Andrian U. H., T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427, 154–159 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Bousso P., Robey E., Dynamics of CD8+ T cell priming by dendritic cells in intact lymph nodes. Nat. Immunol. 4, 579–585 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Stoll S., Delon J., Brotz T. M., Germain R. N., Dynamic imaging of T cell-dendritic cell interactions in lymph nodes. Science 296, 1873–1876 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Neuenhahn M., Kerksiek K. M., Nauerth M., Suhre M. H., Schiemann M., Gebhardt F. E., Stemberger C., Panthel K., Schroder S., Chakraborty T., Jung S., Hochrein H., Russmann H., Brocker T., Busch D. H., CD8α+ dendritic cells are required for efficient entry of Listeria monocytogenes into the spleen. Immunity 25, 619–630 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Edelson B. T., Bradstreet T. R., Hildner K., Carrero J. A., Frederick K. E., Kc W., Belizaire R., Aoshi T., Schreiber R. D., Miller M. J., Murphy T. L., Unanue E. R., Murphy K. M., CD8α+ dendritic cells are an obligate cellular entry point for productive infection by Listeria monocytogenes. Immunity 35, 236–248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waite J. C., Vardhana S., Shaw P. J., Jang J. E., McCarl C. A., Cameron T. O., Feske S., Dustin M. L., Interference with Ca2+ release activated Ca2+ (CRAC) channel function delays T-cell arrest in vivo. Eur. J. Immunol. 43, 3343–3354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Regard J. B., Kataoka H., Cano D. A., Camerer E., Yin L., Zheng Y. W., Scanlan T. S., Hebrok M., Coughlin S. R., Probing cell type-specific functions of Gi in vivo identifies GPCR regulators of insulin secretion. J. Clin. Invest. 117, 4034–4043 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serbina N. V., Pamer E. G., Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 7, 311–317 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Muraille E., Giannino R., Guirnalda P., Leiner I., Jung S., Pamer E. G., Lauvau G., Distinct in vivo dendritic cell activation by live versus killed Listeria monocytogenes. Eur. J. Immunol. 35, 1463–1471 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Perez O. A., Yeung S. T., Vera-Licona P., Romagnoli P. A., Samji T., Ural B. B., Maher L., Tanaka M., Khanna K. M., CD169+ macrophages orchestrate innate immune responses by regulating bacterial localization in the spleen. Sci. Immunol. 2, eaah5520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kohlmeier J. E., Reiley W. W., Perona-Wright G., Freeman M. L., Yager E. J., Connor L. M., Brincks E. L., Cookenham T., Roberts A. D., Burkum C. E., Sell S., Winslow G. M., Blackman M. A., Mohrs M., Woodland D. L., Inflammatory chemokine receptors regulate CD8+ T cell contraction and memory generation following infection. J. Exp. Med. 208, 1621–1634 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohlmeier J. E., Cookenham T., Roberts A. D., Miller S. C., Woodland D. L., Type I interferons regulate cytolytic activity of memory CD8+ T cells in the lung airways during respiratory virus challenge. Immunity 33, 96–105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slutter B., Pewe L. L., Kaech S. M., Harty J. T., Lung airway-surveilling CXCR3hi memory CD8+ T cells are critical for protection against influenza A virus. Immunity 39, 939–948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi C., Velazquez P., Hohl T. M., Leiner I., Dustin M. L., Pamer E. G., Monocyte trafficking to hepatic sites of bacterial infection is chemokine independent and directed by focal intercellular adhesion molecule-1 expression. J. Immunol. 184, 6266–6274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakane A., Minagawa T., Kohanawa M., Chen Y., Sato H., Moriyama M., Tsuruoka N., Interactions between endogenous gamma interferon and tumor necrosis factor in host resistance against primary and secondary Listeria monocytogenes infections. Infect. Immun. 57, 3331–3337 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wirth T. C., Xue H. H., Rai D., Sabel J. T., Bair T., Harty J. T., Badovinac V. P., Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8+ T cell differentiation. Immunity 33, 128–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beura L. K., Mitchell J. S., Thompson E. A., Schenkel J. M., Mohammed J., Wijeyesinghe S., Fonseca R., Burbach B. J., Hickman H. D., Vezys V., Fife B. T., Masopust D., Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19, 173–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kohlmeier J. E., Cookenham T., Miller S. C., Roberts A. D., Christensen J. P., Thomsen A. R., Woodland D. L., CXCR3 directs antigen-specific effector CD4+ T cell migration to the lung during parainfluenza virus infection. J. Immunol. 183, 4378–4384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lauvau G., Vijh S., Kong P., Horng T., Kerksiek K., Serbina N., Tuma R. A., Pamer E. G., Priming of memory but not effector CD8 T cells by a killed bacterial vaccine. Science 294, 1735–1739 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Mueller S. N., Heath W., McLain J. D., Carbone F. R., Jones C. M., Characterization of two TCR transgenic mouse lines specific for herpes simplex virus. Immunol. Cell Biol. 80, 156–163 (2002). [DOI] [PubMed] [Google Scholar]
- 60.Hohl T. M., Rivera A., Lipuma L., Gallegos A., Shi C., Mack M., Pamer E. G., Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe 6, 470–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Wang L., Wang S., Li W., RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Legends for tables S1 to S3
Legends for movies S1 to S3
Tables S1 to S3
Movies S1 to S3