A CRISPR-engineered bacteriophage T4-COVID vaccine delivers multiple SARS-CoV-2 antigens for potential broader protection.
Abstract
A “universal” platform that can rapidly generate multiplex vaccine candidates is critically needed to control pandemics. Using the severe acute respiratory syndrome coronavirus 2 as a model, we have developed such a platform by CRISPR engineering of bacteriophage T4. A pipeline of vaccine candidates was engineered by incorporating various viral components into appropriate compartments of phage nanoparticle structure. These include expressible spike genes in genome, spike and envelope epitopes as surface decorations, and nucleocapsid proteins in packaged core. Phage decorated with spike trimers was found to be the most potent vaccine candidate in animal models. Without any adjuvant, this vaccine stimulated robust immune responses, both T helper cell 1 (TH1) and TH2 immunoglobulin G subclasses, blocked virus-receptor interactions, neutralized viral infection, and conferred complete protection against viral challenge. This new nanovaccine design framework might allow the rapid deployment of effective adjuvant-free phage-based vaccines against any emerging pathogen in the future.
INTRODUCTION
Rapid discovery of safe and effective vaccines against emerging and pandemic pathogens such as the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1, 2) requires a “universal” vaccine design platform that can be adapted to any infectious agent (3). It should allow incorporation of diverse targets, DNAs, and proteins (multicomponent)—full-length proteins as well as peptides and domains—in various combinations (multivalent). Such a multiplex platform would not only compress the timeline for vaccine discovery but also offers critical choices for selecting the most effective vaccine candidate(s) without going through iterative design cycles (3, 4).
Numerous SARS-CoV-2 vaccine candidates have been developed at record-breaking pace to quell this devastating pandemic, and many others are in clinical trials (4–11). Two of these, both mRNA-based and developed by Moderna and Pfizer-BioNTech, were found to have an extraordinary, up to 95%, efficacy in preventing symptomatic coronavirus disease 2019 (COVID-19) (12) and have been approved by Food and Drug Administration (FDA) for emergency use. However, other innovative platforms are still desperately needed because future pathogens might be more complex and their vaccine targets may not be as well defined (3). In addition, the current platforms focus on a single vaccine target, the viral spike in the case of SARS-CoV-2, hence at risk of losing some or most of their efficacy when variants emerge during a pandemic, as has been reported with the alpha (B.1.1.7), beta (B.1.351), gamma (P.1), and delta (B.1.617.2) variants (13). Furthermore, vaccine escape variants could also emerge even after mass vaccination, potentially requiring the design of a series of variant-specific vaccines. One way to overcome these concerns is to include multiple and, if possible, conserved antigen targets in a vaccine formulation. In addition, most of the platforms require the inclusion of chemicals and adjuvants to formulate and/or boost immune responses that often lead to a degree of reactogenicity following vaccination (7, 9, 14). Here, we present a new universal adjuvant-free nanovaccine platform by CRISPR (clustered regularly interspaced short palindromic repeats) engineering of bacteriophage (phage) T4 in which multiple targets can be incorporated into a single-phage scaffold to rapidly generate multiplex vaccine candidates against any emerging pathogen during epidemic or pandemic situations. Further, it may also be possible to deliver the vaccine by the mucosal route (e.g., intranasal) rather than the parenteral (intramuscular) route that would be highly beneficial from the public health prospective.
Tailed bacteriophages such as T4 are the most abundant and widely distributed organisms on Earth. T4 belongs to the Myoviridae family, infects Escherichia coli, and has served as an extraordinary model organism in molecular biology and biotechnology (15). It consists of a 1200-Å-long and 860-Å-wide prolate head (or capsid) that encapsidates ~170-kb linear DNA genome and a ~1400-Å-long contractile tail with six long tail fibers emanating from a baseplate present at the tip of the tail (Fig. 1) (15, 16). The head, the principal component for vaccine design, is assembled with 155 hexameric capsomers of the major capsid protein gp23* (“*” represents cleaved mature form), 11 pentamers of gp24* at 11 of 12 vertices, and 1 dodecameric portal protein gp20 at the unique 12th vertex (Fig. 1I) (16, 17).
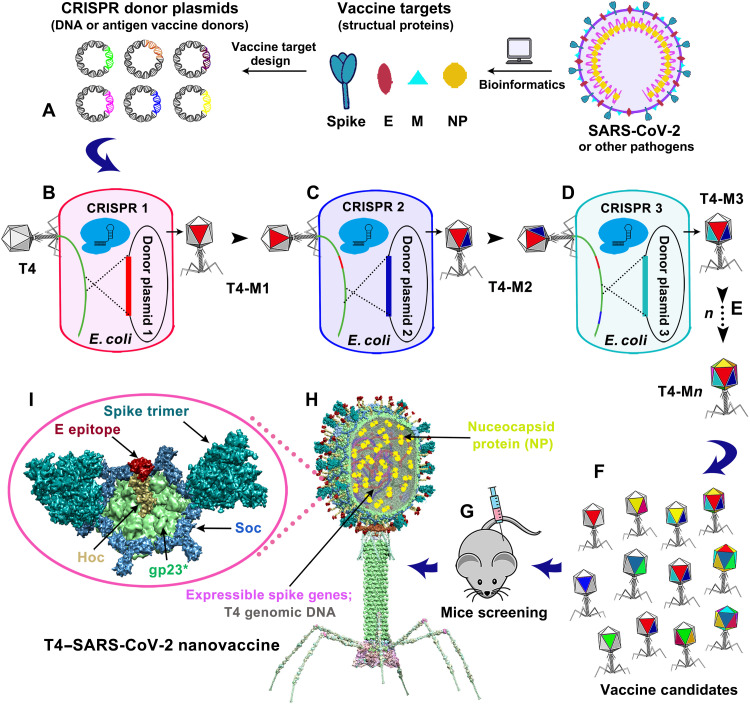
Fig. 1. Design of T4–SARS-CoV-2 nanovaccine by CRISPR engineering.
Engineered DNAs corresponding to various components of SARS-CoV-2 virion are incorporated into bacteriophage T4 genome. Each DNA was introduced into E. coli as a donor plasmid (A), recombined into injected phage genome through CRISPR-targeted genome editing (B). Different combinations of CoV-2 inserts were then generated by simple phage infections and identifying the recombinant phages in the progeny (C to E). For example, recombinant phage containing CoV-2 insert #1 (red; T4-mutant 1 or T4-M1) can be used to infect CRISPR E. coli containing CoV-2 insert containing donor plasmid 2 (blue). The progeny plaques obtained will contain recombinant phage mutant 2 (T4-M2) with both inserts #1 and #2 (red plus blue) in the same genome. This process was repeated to rapidly construct a pipeline of multiplex T4–SARS-CoV-2 vaccine phages (F). Selected vaccine candidates were then screened in a mouse model (G) to identify the most potent vaccine (H). Structural model of T4–SARS-CoV-2 nanovaccine showing an enlarged view of a single hexameric capsomer (I). The capsomer shows six subunits of major capsid protein gp23* (green), trimers of Soc (blue), and a Hoc fiber (yellow) at the center of capsomer. The expressible spike genes are inserted into phage genome, the 12–amino acid E external peptide (red) is displayed at the tip of Hoc fiber, S-trimers (cyan) are attached to Soc subunits, and NP proteins (yellow) are packaged in genome core. See Results, Materials and Methods, and movie S1 for additional details.
The T4 capsid is coated with two nonessential proteins: Soc (small outer capsid protein) (9.1 kDa; 870 copies per capsid) and Hoc (highly antigenic outer capsid protein) (40.4 kDa; 155 copies per capsid) (Fig. 1I) (17). Soc is a trimer, bound to quasi-threefold axes, and acts as a “molecular clamp” by clasping adjacent capsomers (18). Hoc is a 170-Å-long fiber containing a string of four immunoglobulin (Ig)–like domains with its N-terminal domain exposed at the tip of the fiber (19). Soc reinforces an already stable T4 capsid, while Hoc helps phage to adhere to host surfaces (20).
The above constitutes an ideal architecture to develop a universal vaccine design template. Further, our four decades of genetic, biochemical, and structural studies on phage T4 including our recently developed CRISPR phage engineering (21, 22) provide an extraordinary resource. The atomic structures of all the capsid proteins including Soc, Hoc, and the entire capsid have been determined (16, 18, 19, 23). We have also shown that Soc and Hoc can be used as efficient adapters to tether foreign proteins to T4 capsid (24, 25). Both have nanomolar affinities and exquisite specificity, allowing up to ~1025 molecules of full-length proteins, domains, and peptides to be arrayed on capsid (26, 27). T4 capsids decorated with pathogen epitopes mimic PAMPs (pathogen-associated molecular patterns) of natural viruses and stimulate strong innate and adaptive immune responses (28). Furthermore, phage nanoparticles displayed in vitro with purified 83-kDa anthrax protective antigen (PA) or 56-kDa plague F1-V antigen fused to Soc and/or Hoc provided complete protection against lethal anthrax and plague challenges by their respective pathogens Bacillus anthracis and Yersinia pestis in mouse, rat, rabbit, and macaque animal models (29–31).
Although our previous studies demonstrated phage T4 as an effective vaccine delivery platform, it lacked the engineering flexibility to rapidly generate multiplex vaccine candidates against an emerging or pandemic pathogen from which the most effective candidate can be selected using an animal model. The successful vaccine candidate can then be transitioned to human clinical trials, manufactured, and distributed in a relatively short period of time. Such a strategy would provide a powerful multiplex vaccine alternative for controlling a pandemic. However, traditional genetic approaches to inserting foreign genetic elements into phages and viruses are tedious, and it is further difficult to combine more than one insertion into the same genome (32, 33). Moreover, since T4 genome is circularly permuted and terminally redundant (15), insertion of more than ~3 kb will result in loss of terminal redundancy and infectivity. Therefore, we have developed a CRISPR genome engineering concept by which numerous combinations of recombinant phages can be rapidly constructed as vaccine candidates and tested in an animal model (Fig. 1).
To develop a universal vaccine design template, we took advantage of a large amount of nonessential genetic space available in different parts of T4 phage genome. Using SARS-CoV-2 as a model pathogen, we inserted a number of viral components into the phage, including spike (S) (34, 35), envelope (E) (36), and nucleocapsid (NP) proteins (2, 37) by CRISPR engineering as DNA and/or as protein (Fig. 1, A to F, and movie S1). These were then combined by simple phage infections (Fig. 1, B to E), using CRISPR genome cleavage as a strong selection, to create a collection of recombinant phages harboring different combinations of vaccine targets (Fig. 1F). In addition, mammalian cell–expressed and –glycosylated spike trimer molecules were efficiently displayed on T4 capsid via the well-established SpyTag-SpyCatcher bridging system (6, 7, 14, 38). In a few weeks, a pipeline of vaccine candidates in dozens of combinations was generated, demonstrating the unprecedented engineering power and flexibility of this approach.
When tested in mouse and rabbit models, one of the candidates—T4 phage decorated with S-trimers (Fig. 1, G to I, and movie S1), without the use of an adjuvant—elicited robust immunogenicity and ACE2 (angiotensin-converting enzyme-2) receptor (39) blocking and virus-neutralizing antibodies that conferred complete protection against virus challenge in a mouse model. In addition, this vaccine also elicited strong NP-specific antibodies against the NP packaged in the core of phage and E-specific antibodies against the putative externalized N-terminal 12–amino acid epitope of the E protein. Thus, the T4-COVID vaccine generated immune responses against multiple viral targets that might potentially lead to broader protection and minimize the emergence of escape and/or lethal and highly transmission variants, such as the delta. While these need to be further investigated, the current studies established a “blueprint” for a new type of nanovaccine framework for rapid and multiplex design of effective vaccine candidates that can be potentially applied to any emerging pathogen in the future.
RESULTS
Construction of T4–SARS-CoV-2 recombinant phages by CRISPR engineering
A series of CRISPR E. coli strains were constructed by inserting SARS-CoV-2 gene segments into T4 phage genome. Each strain harbored two plasmids (Fig. 2A): a “spacer” plasmid expressing the genome-editing nuclease, either type II Cas9 or type V Cas12a, and CRISPR RNAs (crRNAs or spacer RNAs) corresponding to target protospacer sequence(s) in phage genome and a second “donor” plasmid containing the SARS-CoV-2 sequence. The latter also has ~500–base pair (bp) homologous flanking arms of phage genome corresponding to the point of insertion. Four nonessential regions in the genome were identified for insertion of various SARS-CoV-2 genes (Fig. 2B, I to IV). When these E. coli strains were infected by T4, a double-stranded break would occur in the protospacer sequence by Cas9 or Cas12a that inactivates the phage genome, and no phage should be produced. However, the T4 phage allows efficient recombination between the cleaved DNA and the donor plasmid through the flanking homologous arms, transferring the CoV-2 gene into phage genome and propagating it as part of phage infection (Fig. 2A). The same strategy was used to introduce many other genetic modifications, including deletions by simply creating that modification in the donor plasmid, and various modifications were combined as desired by simple phage infections of appropriate CRISPR E. coli strains (Fig. 1).
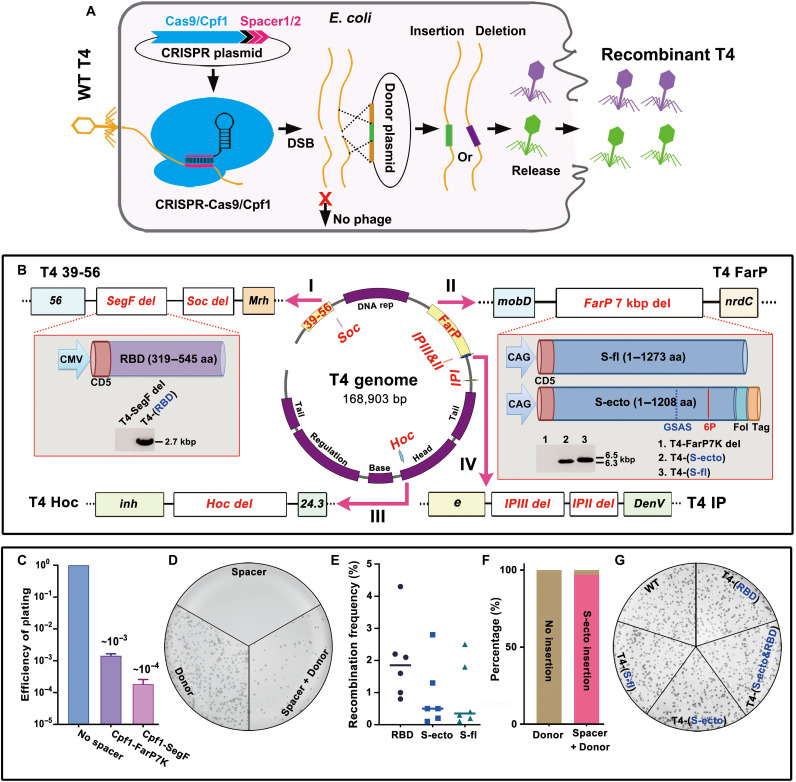
Fig. 2. Construction of T4–SARS-CoV-2 recombinant phages by CRISPR engineering.
(A) Schematic of T4 CRISPR engineering. DSB, double-strand break. (B) Four nonessential regions of T4 genome are chosen for deletion and insertion of various SARS-CoV-2 genes [shown in red; SegF/Soc, FarP, internal protein (IP), and Hoc]. 6P, six proline substitutions in S-ecto (F817P, A892P, A899P, A942P, K986P, and V987P); Fol, T4 fibritin motif Foldon for efficient trimerization; Tag, octa-histidine and twin-strep tags. Furin cleavage site RRAR was mutated to GSAS to stabilize trimers in a prefusion state (35). aa, amino acid. (C) Efficiency of plating (EOP) of representative Cas12a-FarP7K and Cas12a-SegF spacers. (D) Plate showing plaques from phage infection of bacteria containing Cas12a-FarP7K spacer only, S-ecto donor only, or Cas12a-FarP7K spacer plus S-ecto donor. (E) Recombination frequency of three spike gene (RBD, S-ecto, and S-fl) insertions. (F) DNA sequencing of 30 independent plaques showed that >95% of the plaques generated in S-ecto recombination contained the correct S-ecto insert. (G) Plate showing that the wild-type (WT), T4-(RBD), T4-(S-fl), T4-(S-ecto), and T4-(S-ecto&RBD) recombinant phages had similar plaque size.
Initially, we constructed “acceptor” phages by deleting certain known nonessential segments of the phage genome (40)—~18 kb of FarP, ~11 kb of 39-56, or both (~29 kb)—that created space for CoV-2 insertions (fig. S1, A and B). However, the yields of these phages were low, ~1 to 2 orders of magnitude lower than the wild-type (WT) phage (fig. S1, A and B). Since yield is critical for vaccine manufacture, we then constructed shorter deletions in which ~700 bp of SegF within 39-56 and ~7 kb of segment within FarP were deleted (Fig. 2B, I and II). The yields of these phages (7del.SegFdel.T4) were similar to the WT phage, suitable for SARS-CoV-2 vaccine design.
Next, three spike gene variants (35) corresponding to (i) 1273–amino acid WT full-length (S-fl), (ii) 1208–amino acid ectodomain (S-ecto; amino acids 1 to 1208), and (iii) 227–amino acid receptor binding domain (RBD; amino acids 319 to 545) were engineered as expressible cassettes and inserted into 7del.SegFdel.T4 (Fig. 2B and fig. S1C). Previous studies showed that these gene cassettes carrying reporter or antigen genes [green fluorescent protein (GFP), luciferase, anthrax PA, and plague F1-V] could be packaged in vitro into empty T4 capsids using a packaging motor and could be delivered and expressed in human embryonic kidney (HEK) 293 cells or in vivo in mice by intramuscular administration. In the latter case, antigen-specific antibody titers and T cell responses were also elicited (26). The spike gene segments were codon-optimized and kept under the control of a strong mammalian expression promoter, either cytomegalovirus (CMV) or CAG (CMV early enhancer/chicken β-Actin/rabbit beta-Globin splice acceptor site), and a human CD5 signal peptide fused to the N terminus for efficient secretion (Fig. 2B, I and II, and fig. S1D). The S-ecto recombinant contained additional mutations including six proline substitutions that imparted greater stability and ~10-fold greater expression, as was described by Hsieh et al. (35) (Fig. 2B and fig. S1D).
The control CRISPR E. coli strain containing Cas9/Cas12a spacer plasmid but lacking the spike gene donor plasmids yielded very few or no plaques when infected with 7del.SegFdel phage (plating efficiency, <10−4 to 10−3) (Fig. 2, C and D, and fig. S1E), whereas those containing both the plasmids produced plaques at a recombination frequency of up to ~4.5% (Fig. 2, D and E). DNA sequencing confirmed that >95% of these plaques were true recombinants with correct insert (Fig. 2F) and showed similar plaque-forming ability as the WT phage (Fig. 2G). A similar CRISPR strategy was used for creating deletions and/or insertions at the other sites [internal protein (IP) II, IPIII, Hoc, and Soc] (Fig. 2B and fig. S1F).
Encapsidation of SARS-CoV-2 NP
SARS-CoV-2–infected patients have been reported to generate robust NP-specific immune responses including cytotoxic T cells that might be important for protection and virus clearance (37, 41). We incorporated NP into the T4 nanoparticle by designing a CRISPR strategy that packaged NP molecules inside phage capsid along with the genome. As NP is a nucleic acid–binding protein, the packaged phage genome might provide an appropriate environment to localize this protein (42).
During T4 phage morphogenesis, the major capsid protein gp23 assembles around a scaffolding core formed by a cluster of proteins including three nonessential, highly basic, internal proteins IPI, IPII, and IPIII. Following assembly, while most of the scaffold proteins are degraded and expelled from the capsid (Fig. 3A), the IPs are cleaved only once, next to a ~10–amino acid N-terminal capsid targeting sequence (CTS; MKTYQEFIAE). The cleaved CTS leaves the capsid, but ~1000 molecules of IPs, or any foreign protein fused to CTS, remain in the “expanded” capsid (Fig. 3A) (43). To replace IPs with NP, we first created an IPIII deletion phage using appropriate spacer and donor (Fig. 3B and fig. S2A). Then, a CTS-NP fusion sequence was transferred into this phage under the control of the native IPIII promoter (fig. S2A). Next, IPII was deleted to reduce protein packaging competition and increase the copy number of NP (Fig. 3, B and C, and fig. S2A). We have also introduced an amber mutation into the CTS sequence [TTT (Phe) at amino acid 7] because, for unknown reasons, the donor plasmid containing the WT CTS sequence was found to be toxic to E. coli. This CTSam-NP phage when grown on a tRNA amber suppressor–containing E. coli (Sup1) expressed NP and encapsidated it as demonstrated by Western blotting (WB) with NP-specific monoclonal antibodies (mAbs) (Fig. 3C and fig. S2, B and C). The copy number is about 70 NP molecules per phage capsid (fig. S2D).
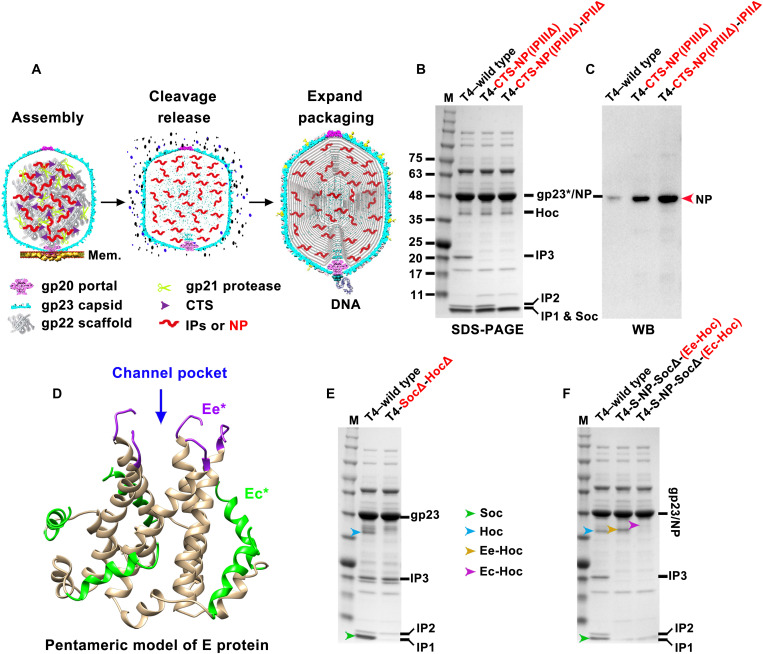
Fig. 3. Encapsidation of SARS-CoV-2 NP and display of SARS-CoV-2 E epitopes on T4 phage.
(A) Schematic showing steps in T4 phage head morphogenesis. Mem., E. coli membrane. The major capsid protein gp23 assembles around a scaffolding core formed by a cluster of proteins including gp22 scaffold and three IPs. Following assembly, while most of the gp22 scaffold proteins are degraded by gp21 protease and expelled from capsid, the IPs are cleaved only once, next to a ~10–amino acid N-terminal CTS. The cleaved CTS leaves the capsid, but ~1000 molecules of IPs, or any foreign protein (such as NP here) fused to CTS, remain in the expanded and packaged head. (B and C) SDS–polyacrylamide gel electrophoresis (PAGE) and WB analysis of phage particles with IPII and IPIII deletions (IPIIΔIPIIIΔ) and NP encapsidation. Since NP has a very similar molecular size to T4 major capsid protein gp23*, an NP-specific antibody was used to detect NP. (D) Structural model of viroporin-like tetrameric assembly of CoV-2 E protein (36). The N-terminal 7 residues and C-terminal 10 residues are not shown due to the lack of a corresponding segment in the structural template used for homology modeling. Ee* indicates amino acids 8 to 12, and Ec* indicates amino acids 53 to 65. (E) SDS-PAGE of Hoc deletion and Soc deletion phage (HocΔSocΔ). (F) SDS-PAGE of recombinant phages displaying Ee-Hoc or Ec-Hoc fusion proteins.
Display of SARS-CoV-2 epitopes on T4 phage
Next, we incorporated SARS-CoV-2 antigens onto the nanoparticle surface. We first deleted Hoc and Soc genes from the above recombinant phages and then inserted Hoc- and Soc-fused CoV-2 genes under the control of their respective native promoters. Upon infection, these phages would express and assemble the epitopes on T4 capsid surface (Fig. 3, D and E, and fig. S2E). We fused the gene segments corresponding to the N-terminal 12–amino acid “external domain” peptide (Ee) or the 18–amino acid peptide from the C-terminal domain (Ec) of E protein to the N terminus of Hoc (Fig. 3D). These peptides are predicted to be exposed on the SARS-CoV-2 virion and shown to elicit T cell immune responses in humans (36). By virtue of fusion to the N terminus of Hoc, these epitopes would be exposed at the tip of ~170-Å-long Hoc fiber (19). The Ee and Ec recombinant phages showed an upward shift of the Hoc band upon SDS–polyacrylamide gel electrophoresis (PAGE) (Fig. 3F, yellow and magenta arrowheads). However, only the 12–amino acid Ee peptide was displayed at the maximum copy number, up to ~155 copies per capsid, while the 18–amino acid Ec peptide showed lower epitope copies and also negatively affected the phage yield (Fig. 3F).
Display of SARS-CoV-2 RBD on T4 phage
We used a similar strategy to display CoV-2 RBD (39) on the capsid surface as a Soc fusion (fig. S3, A and B). However, the copy number of the displayed RBD was very low (fig. S3C). RBD contains ~82.5% nonhydrophilic amino acid residues and formed insoluble inclusion bodies in E. coli (fig. S3, D and E). Numerous N- and C-terminal truncations of RBD were constructed (fig. S4A); however, none [including the shortest 67–amino acid receptor binding motif (39)] had shown notable improvement in solubility and copy number (fig. S4B). We therefore resorted to alternative strategies to display RBD on phage capsid.
First, we constructed E. coli–expressing Soc-RBD from a plasmid under the control of the phage T7 promoter to determine whether preexpression of Soc-RBD for a short period of time (~10 min) would keep it sufficiently soluble, which could then assemble on capsids produced during phage infection. Phage isolated from these infections showed improved display, ~100 copies of RBD (referred to as soluble RBD or sRBD) per phage particle (Fig. 4, A and B).
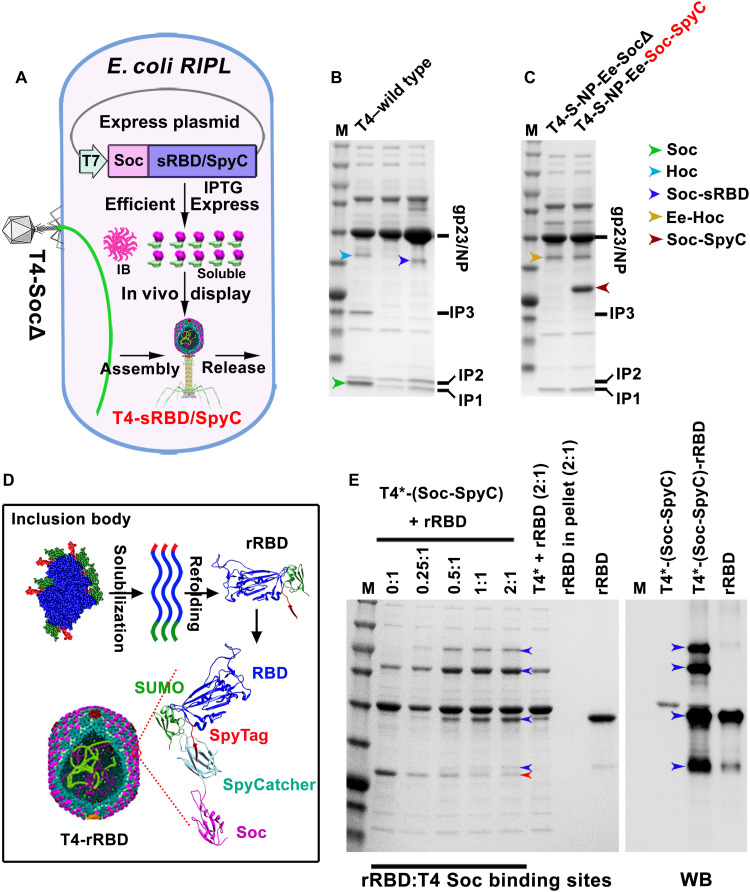
Fig. 4. Display of SARS-CoV-2 RBD on T4 phage.
(A) Schematic showing Soc-sRBD or Soc–SpyCatcher (SpyC) in vivo display on T4-SocΔ capsid. Soc-sRBD or Soc-SpyCatcher expression under the control of phage T7 promoter was induced by isopropyl-β-d-thiogalactopyranoside (IPTG). Most of the expressed Soc-RBD was in the inclusion body (IB). Soluble Soc-sRBD (minor amount) or Soc-SpyCatcher can be efficiently displayed on capsid. (B) SDS-PAGE showing ~100 copies of Soc-sRBD displayed on T4 capsid. (C) SDS-PAGE showing ~500 copies of Soc-SpyCatcher displayed on T4 capsid. (D) Schematic diagram showing the solubilization and refolding of SUMO (small ubiquitin-like modifiers)–RBD–SpyTag inclusion body. Refolded SUMO-RBD-SpyTag (rRBD) protein was efficiently displayed on T4-SpyCatcher phage via SpyTag-SpyCatcher bridging. (E) Display of rRBD on the T4-SpyCacher surface at increasing ratios of rRBD molecules to capsid Soc binding sites (0:1 to 2:1). RBD-specific antibody was used to verify the displayed rRBD and rRBD-SpyCatcher-Soc complexes. T4* indicates T4–S-ecto–NP-Ec-SocΔ recombinant phage. Blue and red arrowheads indicate rRBD/complexes and Soc-SpyCatcher, respectively.
Second, we deployed the well-established SpyTag-SpyCatcher technology (38) to display RBD on T4 phage. The optimized SpyCatcher and SpyTag from Streptococcus pyogenes interact with more than picomole affinity (approaching “infinite” affinity; second-order rate constant, 5.5 × 105 M−1 s−1) and exquisite specificity that then leads to covalent linkage (38). To display RBD, phage decorated with the 12.6-kDa soluble SpyCatcher was produced by growing the T4-Spike-Ee-NP-SocΔ phage on E. coli–expressing Soc-SpyCatcher fusion protein from the T7 expression plasmid (Fig. 4A and fig. S3F). Phage prepared from these infections contained up to ~600 copies of Soc-SpyCatcher per capsid (Fig. 4C).
Third, RBD was expressed as SUMO (small ubiquitin-like modifier)–RBD–SpyTag fusion protein in E. coli. The SUMO domain is expected to enhance the solubility of RBD (44) but resulted in only a small improvement (fig. S3G). Therefore, the SUMO-RBD-SpyTag protein was purified from insoluble inclusion bodies by urea denaturation and refolding (referred to as refolded RBD or rRBD) and was displayed in vitro on the SpyCatcher phage (Fig. 4D). SDS-PAGE and WB of the phage particles showed that the SUMO-RBD-SpyTag was efficiently captured by the SpyCatcher phage as shown by the disappearance of the SpyCatcher band and appearance of higher molecular weight band(s) (Fig. 4E). The copy number was ~300 rRBD molecules per capsid (Fig. 4E).
The sRBD and rRBD phages produced as above (T4-Spike-Ee-CTSam-NP-sRBD and T4-Spike-Ee-CTSam-NP-rRBD) were bound to human ACE2 receptor protein (fig. S5A) (39) and to some of the RBD-specific mAbs and polyclonal antibodies, but not to all (fig. S5, B to D). However, these RBDs exhibited considerably lower binding when compared to mammalian-expressed RBD, suggesting that the E. coli–produced RBDs were not properly folded (fig. S5, E and F). This was also consistent with the copurification of a 65-kDa E. coli GroEL chaperone with RBD indicating the presence of partially folded and/or misfolded RBD (fig. S3E).
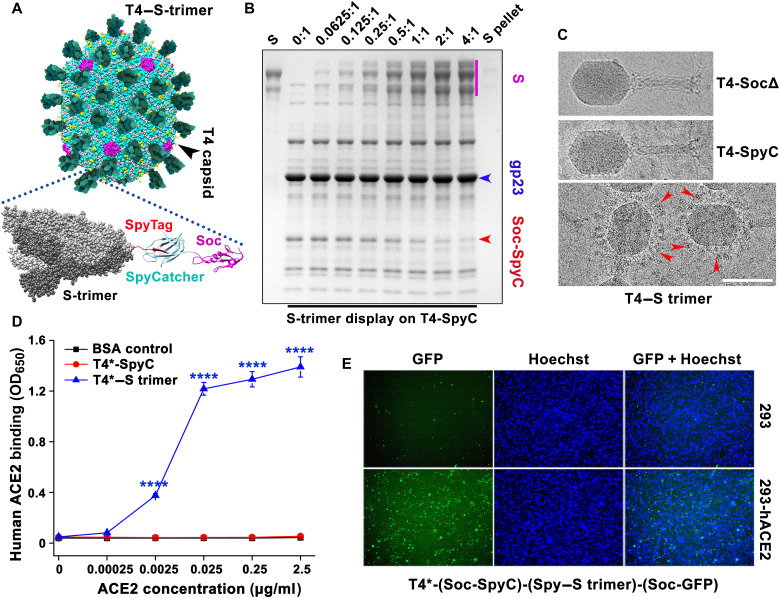
Decoration of phage T4 nanoparticles with S-ecto trimers
Next, we displayed S-ecto trimers (S-trimer) (433.5 kDa) (35) on T4-Spike-Ee-NP-SocΔ phage. The prefusion stabilized hexa-Pro S-ecto construct described as above (35) was fused to a 16–amino acid SpyTag at the C terminus and expressed in ExpiCHO cells. The ectodomain trimers secreted into the culture medium were purified by HisTrap affinity chromatography and size exclusion chromatography (fig. S6, A and B). These trimers appeared authentic and native-like because (i) they migrated predominantly as a single species and showed no nonspecific aggregation, which usually appears as a peak (or shoulder) near void volume and as a smeary high–molecular weight species on native gel (fig. S6, A and B), and (ii) they bound efficiently to human ACE2 receptor (fig. S6C) and to conformation-specific RBD mAbs (fig. S5F).
Several criteria were used to display these trimers in an appropriate spatial orientation that mimic the spikes on SARS-CoV-2 virion: (i) Since our atomic structures (16) showed that the C termini of trimeric Soc assembled at the quasi-threefold axes of the icosahedral capsid are solvent-exposed and projected out, these were used to fuse the SpyCatcher domain. The trimeric SpyTagged S-ecto is expected to land on the capsid with the neutralization epitopes of RBD and N-terminal domain well exposed and facing away from the capsid wall. (ii) The 16–amino acid SpyTag was fused to the C terminus of S-ecto such that the trimer would be anchored to the T4 capsid “membrane” through its “stem,” similar to the SARS-CoV-2 spike anchored to the viral E membrane; and (iii) flexible linkers were included between the S-ecto C terminus and the SpyTag to introduce sufficient flexibility for efficient conjugation to the capsid-bound SpyCatcher domains.
The S-trimers so designed were efficiently captured by the SpyCatcher phage produced as above. Binding was so strong that efficient assembly occurred by simple mixing of trimers and phage even at an equimolar ratio of S-trimers to T4-SpyCatcher molecules (Fig. 5, A and B). The copy number was ~100 S-trimers per phage capsid (Fig. 5B). Cryo–electron microscopy (cryo-EM) showed decoration of T4 phage capsids with S-trimers (Fig. 5C), mimicking the orientation of spikes on SARS-CoV-2 virion (45). The trimer-decorated T4 phage efficiently bound to human ACE2 receptor (Fig. 5D and fig. S7A), and when codisplayed with GFP, it decorated the ACE2-expressing HEK293 cells (Fig. 5E and fig. S7, B and C).
Fig. 5. Decoration of phage T4 nanoparticles with S-ecto trimers.
(A) Schematic showing the decoration of phage T4 nanoparticles with S-ecto trimers via SpyTag-SpyCatcher bridges. (B) In vitro assembly of S-trimers on T4-SpyCatcher phage at increasing ratios of S-trimer molecules to Soc binding sites (0:1 to 4:1). Phage and S-trimer were incubated at 4°C for 1 hour, followed by centrifugation to remove the unbound material. After two washes, the pellet was resuspended in buffer, and SDS-PAGE was performed. (C) Representative cryo-EM images showing T4-SocΔ, T4-(Soc-SpyCatcher), and T4–(Soc-SpyCatcher)–S-trimer phages. The red arrowheads indicate the representative S-trimer displayed on phage. Scale bar, 100 nm. (D) Enzyme-linked immunosorbent assay (ELISA) analysis of T4–S-trimer phage binding to ACE2 at various ACE2 concentrations. OD650, optical density at 650 nm. ****P < 0.0001. (E) Binding of T4–S-trimer–GFP phage to HEK293 cells expressing human ACE2. The nucleus was stained with Hoechst. T4* indicates T4-(S-ecto)-RBD-NP-Ee-SocΔ phage used for display (S-ecto and RBD refer to insertions of gene expression cassettes).
Immunogenicity and protective efficacy of T4–SARS-CoV-2 vaccine candidates
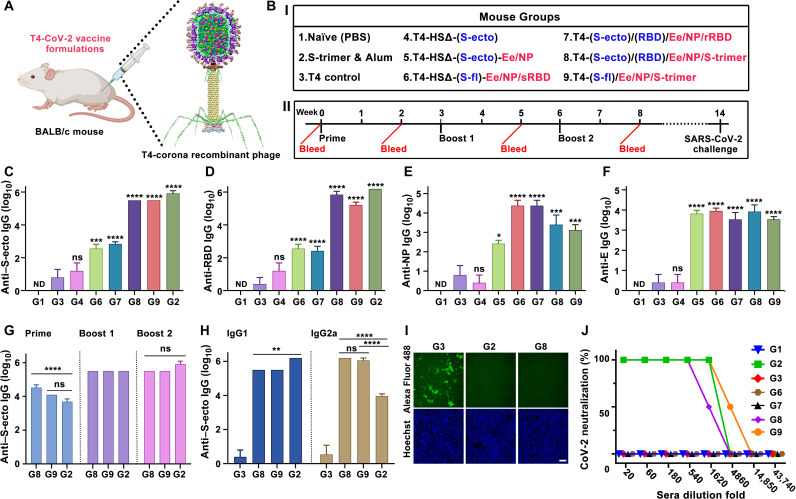
The T4–SARS-CoV-2 vaccine candidates generated as above by sequential engineering (Fig. 1, B to G) were screened for their immunogenicity and protective efficacy in a mouse model. BALB/c mice were immunized at weeks 0, 3, and 6 with CsCl-purified phage particles (Fig. 6, A and B), and sera were analyzed by a series of immunological assays.
Fig. 6. Immunogenicity of T4–SARS-CoV-2 vaccine candidates in mice.
(A and B) Schematic showing BALB/c mice immunized with various T4–SARS-CoV-2 (T4-CoV-2) vaccine formulations. Photo credit: The mouse image was created with BioRender.com. (I) HSΔ indicates Hoc and Soc deletion. Blue color indicates insertion of expression cassette into T4 genome as DNA vaccine. Red color indicates capsid-displayed antigens. (II) Prime-boost immunization and challenge scheme. (C to F) Boost 2 sera (week 8 bleeding) were assessed by ELISA for antigen-specific IgG antibody titers (end point) against S-ecto (C), RBD (D), NP (E), and E (F). *P < 0.05, ***P < 0.001, and ****P < 0.0001, compared with phage control group G3. ns, not significant, P > 0.05; ND, not detected. (G and H) Comparison of anti–S-ecto IgG antibody titers (G) (****P < 0.0001) and of IgG1 and IgG2a subtype antibody titers in boost 2 sera (H) (**P < 0.01 and ****P < 0.0001). (I) Blocking of RBD binding to HEK293-ACE2 by 500× diluted sera. Scale bar, 20 μm. (J) Neutralization antibody measurements. See Materials and Methods for additional details.
SARS-CoV-2–specific antibody titers were determined by enzyme-linked immunosorbent assay (ELISA) using purified proteins (S-ecto, RBD, NP, or E) as coating antigens. Recombinant phages that delivered only the CoV-2 spike gene (no protein antigens; equivalent to a DNA vaccine), either the S-ecto gene and/or the RBD gene, did not induce significant spike protein–specific antibody titers (Fig. 6, C and D, and fig. S9, A to D) or provide any protection (see below). This is probably because the transgenes were not sufficiently expressed to stimulate an immune response. In vitro transduction of isolated recombinant phage DNA into HEK293 cells also did not show notable expression of the spike genes as analyzed by binding to ACE2 receptor or spike-specific mAbs. This might be because of inefficient delivery and/or intracellular trafficking and translocation of the large ~170-kb phage DNA into the nucleus, since only a single copy of transgene is present in the genome. This was in contrast to the in vitro–packaged heads used in previous studies in which on an average 8 to 10 copies of the transgene were efficiently delivered and expressed both in vitro and in vivo (26, 27). However, when these mice were boosted once with phage nanoparticles displayed with protein antigen, the S-trimers, significant antibody titers were elicited (fig. S10A).
Without any adjuvant, the T4 nanoparticles stimulated strong antibody titers to phage-delivered proteins and peptides, either displayed on the surface or packaged inside. These included RBD- and spike-specific antibodies, E-specific antibodies, and NP-specific antibodies (Fig. 6, C to F). However, the highest titers, up to an endpoint titer of ~1.5 × 106, were obtained with phage nanoparticles decorated with S-trimers (Fig. 6, C to F). No significant difference was observed between the endpoint titers obtained by using either RBD or S-trimers as the coating antigens (Fig. 6, C and D). This result is consistent with recent reports that the immunodominant RBD comprises multiple and distinct antigenic sites and is the target of most neutralizing activity in COVID-19 convalescent sera (46, 47).
Furthermore, the antibodies elicited were conformation specific. For instance, the antibodies elicited against sRBD or rRBD displayed on phage reacted poorly with the mammalian-expressed S-trimer or RBD (Fig. 6, C and D), consistent with the antigenicity data described above that these also reacted poorly with ACE2 and conformation-specific mAbs (fig. S5, E and F). Similarly, the antibodies elicited against T4-dispalyed S-trimers reacted poorly with the E. coli–produced RBD (endpoint titer of ~102 using E. coli RBD as the coating antigen versus ~105 using mammalian RBD) (fig. S10D).
It is recognized that the spike trimer alone is poorly immunogenic and used as a vaccine candidate by formulating with strong chemical adjuvants such as AS03 or CpG/Alum (48). The spike-specific titers elicited by T4 phage–decorated trimers without any adjuvant were as high as those generated with Alhydrogel adjuvant (Fig. 6, C, D, and G). Furthermore, notably, IgG subclass data indicated that phage nanoparticles stimulated both T helper cell 2 (TH2)–biased and TH1-biased immune responses. The adjuvant-free T4 nanoparticles mounted high levels of both IgG1 and IgG2a classes against all three SARS-CoV-2 antigens—spike/RBD, E, and NP—whereas the Alhydrogel-adjuvanted trimers predominantly elicited TH2-derived IgG1 class antibodies (Fig. 6H and fig. S9, A to J). The TH1-derived IgG2a antibodies were higher than the TH2-derived IgG1 antibodies in T4 groups, whereas the Alhydrogel-adjuvanted mice induced ≥2 orders of magnitude lower than IgG2a antibodies (Fig. 6H and fig. S9, I and J). While TH2-biased responses may lead to lung injury via eosinophilic infiltrates, the TH1-type responses are proposed to alleviate potential lung immunopathology and reduce the potential for disease enhancement (49). The T4 nanoparticle vaccine with balanced TH1 and TH2 responses might therefore be better for safety and virus clearance. However, this point requires further investigation, which is currently underway.
The T4-stimulated spike-specific antibodies blocked the binding of RBD to human ACE2 expressed on HEK293 cells in a dose-dependent manner (Fig. 6I and fig. S9K). These antibodies also exhibited strong virus-neutralizing activity as determined by Vero E6 cell cytopathic assay using the live biosafety level 3 (BSL-3) SARS-CoV-2 US-WA-1/2020 strain, the first patient isolate obtained through the U.S. Centers for Disease Control and Prevention (Fig. 6J) (50). The neutralization titers correlated well with protective efficacy when mice were challenged with the mouse-adapted (MA) SARS-CoV-2 MA10 virus (Fig. 7) (51).
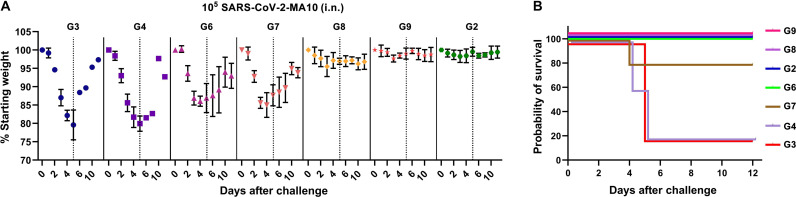
Fig. 7. Protective efficacy of T4–SARS-CoV-2 vaccine candidates in mice.
See Fig. 6 for the immunization scheme and mouse groups (G2 to G9) immunized with various T4–SARS-CoV-2 vaccine candidates. (A) Percentage starting body weight of immunized mice at days after challenge with 105 plaque-forming units (PFU) of SARS-CoV-2 MA10 (intranasal). Dotted lines represent the percentage starting weight at day 5 after infection when mice showed maximum weight loss in control groups. In groups G3 and G4, only 20% of mice survived after day 5. Thus, data presented after day 5 are biased toward minor survivors. The data were presented as means ± SD. (B) Survival rate of mice against SARS-CoV-2 MA10 challenge. See Materials and Methods for additional details.
Upon challenge, the naïve and T4 control mice showed a rapid weight decline, up to 25% of their starting weight in 5 days due to acute viral infection, and were moribund (humanely euthanized) or succumbed to infection. After day 5, the surviving mice began to regain weight and recover from infection during the next several days (Fig. 7A). This rapid weight loss resulted in ~80% mortality rate in the control animals (Fig. 7, A and B). None of the groups receiving the spike DNA vaccine alone and/or CoV-2 antigens other than spike trimers showed significant protection, closely correlating to the lack of, or extremely weak, neutralization antibody responses. However, E. coli RBD groups [group 6 (G6) and G7] showed less weight loss and higher survival rate than the susceptible control groups (G3 and G4) (Fig. 7, A and B). This is an interesting finding considering that these groups elicited high titers of sRBD- or rRBD-specific antibodies but did not exhibit significant virus neutralization activity. Since non-neutralizing antibodies are generally not associated with protection or severity of infection in humans (52), it might be that this partial protection was due to spike-specific T cell responses. However, this finding requires further investigation and forms, in part, our future studies.
On the other hand, mice vaccinated with T4-decorated trimers exhibited profound neutralization antibody responses and were fully protected from the acute onset of morbidity (weight loss and other signs of illness) and mortality. The weight loss, if any, was insignificant for both these groups, as well as the positive control group vaccinated with Alhydrogel-adjuvanted trimers (Fig. 7). In addition, those mice boosted with one dose of T4 trimers also showed partial protection. The weight loss was in between unprotected and protected groups, with a milder weight loss than the unimmunized and challenged controls, clearly correlating protection with spike-specific antibodies (fig. S10, B and C).
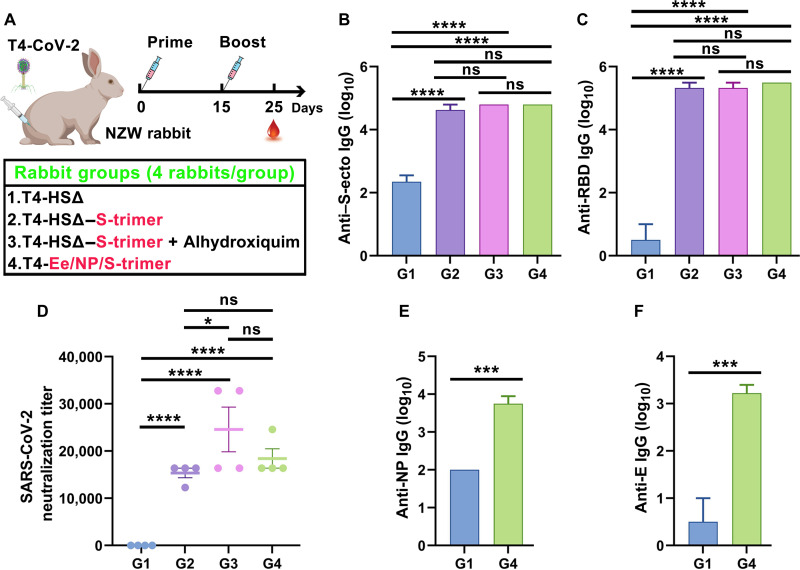
Next, we evaluated the most effective T4 phage–decorated S-trimers vaccine in a second animal model, the New Zealand White rabbit (Fig. 8A). This vaccine, again without any adjuvant, by only two immunizations at days 0 and 15, elicited robust spike- and RBD-specific antibodies and virus neutralization titers in rabbits that were ~5 to 6 times greater than those obtained in mice (Fig. 8, B to D, and fig. S11). Likewise, inclusion of displayed Ee peptide and packaged NP into the nanoparticle broadened the immune responses (Fig. 8, E and F), while addition of a TH1-biased adjuvant Alhydroxiquim-II only slightly enhanced the antibody titers (Fig. 8, B to D). Together, these datasets allowed the selection of a highly effective T4 phage vaccine candidate that generated robust virus neutralization titers in two different animal models, mouse and rabbit, and conferred complete protection against acute viral infection in mice.
Fig. 8. Immunogenicity and virus neutralization responses of the T4–S-trimer vaccine in New Zealand White rabbit model.
(A) Schematic diagram showing formulations, groups, and prime–single boost immunization scheme for intramuscular vaccinations of New Zealand White (NZW) rabbits. Photo credit: The rabbit image was created with BioRender.com. HSΔ indicates Hoc deletion and Soc deletion recombinant phage. Red color indicates the capsid-displayed Ee, S-trimers, or the capsid-encapsidated NP. Rabbits immunized with HSΔ phage served as negative control. (B and C) Boost sera (10 days after boost) were assessed by ELISA for antigen-specific IgG antibody titers against S-ecto (B) and RBD (C). ****P < 0.0001. ns, not significant, P > 0.05. (D) Serial dilutions of serum from immunized rabbits were assessed for neutralization of live BSL-3 strain SARS-CoV-2 US-WA-1/2020. Neutralization titers were calculated as the reciprocal dilution where infection [cytopathic effect (CPE)] was reduced by more than 90% relative to infection in the absence of serum. *P < 0.05, ****P < 0.0001. (E and F) Comparison of G1 (control phage) and G4 (Ee-NP–S-trimer–displayed phage) for anti-NP (E) and anti-E (F) IgG antibody titers. ***P < 0.001.
DISCUSSION
We have developed a universal vaccine platform centered on bacteriophage T4 nanoparticle and CRISPR engineering (Fig. 1 and movie S1). A number of features demonstrated here establish this as a new and alternative vaccine design platform to rapidly generate vaccine candidates against any emerging and pandemic pathogen in the future.
First, a series of recombinant phages containing SARS-CoV-2 gene insertions were created in days, by CRISPR genome engineering using a combination of type II Cas9 and type V Cas12a nucleases. This combination provided built-in choices for spacers and for efficient cleavage of T4 genome that is extensively modified by cytosine hydroxymethylation and glycosylation to attain near 100% success (21, 22). Such a CRISPR-based engineering is central to T4 vaccine design because, otherwise, it would be nearly impossible to generate multiplex vaccine candidates through classical genetics in the time frame necessary to deal with an emerging pandemic.
Second, a large amount of genetic and structural space available in phage T4 (40) was exploited to incorporate payloads containing CoV-2 DNAs, peptides, proteins, and/or complexes into the same nanoparticle. For instance, we have inserted ~6.5-kb full-length spike gene expression cassette, 2.7-kb RBD gene expression cassette, and 1.3-kb NP gene into the same genome by replacing the nonessential genetic material of T4. It was further expanded by inserting Hoc and Soc fusions and/or replacing additional nonessential segments that span across the phage genome. In addition, up to 155 copies of a 12–amino acid Ee peptide and ~100 copies of 433-kDa S-trimers were displayed on the same nanoparticle, while ~70 molecules of 50-kDa NP were copackaged with genome core. These represent unprecedented engineering capability and payload complexity by a vaccine delivery vehicle.
Third, different compartments of phage nanostructure were used for placing different vaccine cargos. The tips of Hoc fibers that are at ~170 Å away from the capsid were used to display short 12–amino acid Ee peptide epitopes, as this would allow efficient capture by antigen-presenting cells and B and T cells (28). These elicited strong antibody titers. At the same time, NP was copackaged with genome, as this mimics NP’s natural nucleic acid environment (2, 42).
Fourth, the T4 platform was readily adapted to mammalian-expressed proteins through highly efficient SpyCatcher-SpyTag bridging (38). This might be essential for proper folding and glycosylation of some pathogen antigens, as in the case of the spike trimers (34). As the cryo-EM images showed, these spikes anchored to capsid mimic the spikes on SARS-CoV-2 virion (45), providing a native-like context for stimulating effective immune responses.
Fifth, alternative strategies such as preexpression from a plasmid and display through subsequent phage infection provided additional advantages to enhance copy number and better control over folding. As demonstrated, this approach yielded a three- to sixfold higher copy number of Soc-SpyCatcher on phage capsid. However, the hydrophobic RBD remained poorly folded.
Sixth, our sequential engineering generated a pipeline of SARS-CoV-2 vaccine candidates in mere weeks and allowed down-selection of the best vaccine candidate, the phage-decorated trimers, in a single-mouse experiment. This is particularly critical for complex pathogens and when the vaccine target(s) is not adequately defined. However, the spike genes engineered into phage failed to elicit detectable immune responses probably because delivery of a single copy of transgene present as part of large phage genome was inefficient.
Seventh, all the phage-delivered CoV-2 peptides and proteins, whether displayed on the surface or packaged inside, generated robust immune responses. The strong virus neutralization titers and ACE2 blocking titers elicited by T4-delivered trimers in two different animal models, mouse and rabbit, are particularly noteworthy. These responses correlated with protective efficacy where the vaccinated mice were completely protected from acute viral infection.
Eighth, T4 nanoparticle vaccines generated balanced TH1- and TH2-derived antibody responses against all three CoV-2 antigens tested. T4 seems to have a TH1 bias (27, 29, 30). These are desirable properties because the TH1-type immunity is essential for mediating more sustainable immune protection against SARS-CoV-2 infection and disease (37, 41).
Ninth, the T4 nanoparticle vaccine does not require an adjuvant to stimulate robust anti–CoV-2 immune responses, as demonstrated in two animal models. In addition to reducing cost and manufacturing complexity, the adjuvant-free T4 vaccine formulations might provide a safe alternative, especially in pandemic situations where most of the global populations get vaccinated. Various degrees of reactogenicity are often associated with chemicals and adjuvants used in many vaccine formulations. In extensive preclinical animal studies including mice (26, 27, 29, 30, 32), rats (29, 30), rabbits (29, 30), and rhesus macaques (31), no significant adverse reactions were noted when diverse T4 phage–associated antigens were administered by intramuscular, intranasal, intravenous, or oral routes. In addition, no side effects were reported in a human trial where T4 phage was given orally (53, 54).
Tenth, in addition to spike-specific antibodies, our best T4-COVID vaccine elicited broad antibody responses against two additional SARS-CoV-2 virion components. These include NP-specific antibodies, which are abundantly present on SARS-CoV-2–infected host cells and hence an important target for T cell immunity (36, 37), and E-specific antibodies against the capsid-displayed E peptide that might also be a potential target (55). Thus, the T4-CRISPR platform allows incorporation of combinations of antigens into a single-phage backbone leading to broader immune responses against multiple viral targets. This is particularly relevant to the current pandemic because broad immune responses against multiple conserved regions of S-ecto, Ee, and NP (1, 2) would minimize the emergence of vaccine escape mutants as it would be difficult for the virus to overcome diverse immune pressures mounted by the host. Furthermore, it might also confer broader protection against the emerging lethal and highly transmissible variants of the pandemic. This question is currently under investigation.
Last, the SpyCatcher phage could serve as a “plug-and-play” backbone to capture ectodomain trimers from other coronaviruses such as SARS-CoV-1 and Middle East respiratory syndrome (pan-coronavirus vaccine) (2, 7) or any SpyTagged antigen(s) from other infectious agents. The versatile SpyCatcher system has been extensively used by other nanoparticle scaffolds, both synthetic and natural, for conjugation of spike RBD or trimers, which also elicited strong virus-neutralizing antibody titers and protection (6, 7, 9, 56–58). Some of these have also been used for generating pan-coronavirus vaccine candidates (7). However, these nanoscaffolds are considerably smaller than the 120 nm by 86 nm size of T4 capsid lattice that can accommodate the 433-kDa S-trimers at higher density. Furthermore, the nanoscaffolds still need adjuvants to boost their immunogenicity, whereas the T4 vaccine is adjuvant free, and the spatial disposition of the displayed epitopes can be flexibly manipulated by structural and genetic engineering. It is thus conceivable that different T4 vaccine formulations can be “created” at the site of administration by mixing the SpyCatcher T4 phage backbone with the desired antigen combinations. Cocarrying multiple and distinct antigens in the same formulation also increases the breadth of immune responses (9).
The above features make phage T4 a versatile new and alternative nanovaccine design platform using which multiplex vaccine candidates can be rapidly generated against a pandemic pathogen, evaluated in animal models, and transitioned to human clinical trials. Some of the advantages of this platform might stem from the symmetric, repetitive, and high-density arrays of antigens exposed on a large phage capsid that mimic the PAMPs of human viral pathogens. The resulting T4-COVID vaccine so generated is now being good manufacturing practice–manufactured and undergoing additional preclinical animal studies such as dosage optimization, route of administration (intranasal versus intramuscular), and breadth of immune responses (T cell, B cell, and mucosal) to transition this vaccine to phase 1 human clinical trials. Our current estimates are that several hundred thousand to a million T4–SARS-CoV-2 vaccine doses can be produced in a 100-liter fermenter, allowing large-scale manufacturing possible. It is anticipated that this T4–SARS-CoV-2 vaccine will be a strong candidate for a broadly protective next-generation coronavirus vaccine and a booster vaccine to individuals already vaccinated by the first-generation vaccines. As T4 is a highly stable nanoparticle, has good safety profile, and can be manufactured at a relatively low cost, it provides a new and robust alternative platform to rapidly generate effective vaccines against any epidemic or pandemic pathogen in the future, particularly when multivalent vaccines are essential to control future pandemics and protect global communities.
MATERIALS AND METHODS
DNA, bacteria, and bacteriophages
The expression vector pET28b (Novagen, MA) was used for donor plasmid construction and protein expression plasmid construction. The spacer plasmids LbCas12a and SpCas9 were constructed as described previously (21, 22). The DNA fragments containing NP and RBD were codon-optimized for E. coli expression and synthesized by GeneArt (Thermo Fisher Scientific). The plasmids containing WT SARS-CoV-2 spike (S) gene and S-ecto-6P gene were provided by B. Graham and K. S. Corbett [National Institutes of Health (NIH)] and J. S. McLellan (University of Texas, Austin), respectively. The RBD gene for mammalian expression was amplified from the WT spike (S) gene. SpyCatcher/SpyTag and SUMO containing plasmids were purchased from Addgene (#133449 and #111560).
E. coli DH5α [hsdR17 (rK–mK+) sup2] (New England Biolabs) was used for all the clone constructions. The E. coli BL21-CodonPlus (DE3)-RIPL (Novagen) was used for the expression of recombinant proteins. E. coli P301 (sup0) and E. coli B834 (hsdRB hsdMB met thi sup0) were used for the propagation and recombination of phages without amber mutations. The E. coli BL21-CodonPlus (DE3)-RIPL transformed with amber suppressor-1 plasmid, and E. coli B40 (sup1) was used for propagation and recombination of phages with amber mutations (CTS-amber-NP). WT T4 phage was propagated on E. coli P301 or B834 and used as a starting phage for CRISPR engineering.
Plasmid construction
CRISPR-LbCas12a/SpCas9 plasmid was constructed on the basis of the streptomycin-resistant plasmid DS-SPCas (Addgene no. 48645) as described previously (21, 22). The spacer-containing fragments were prepared by annealing and extension of two amplified DNA fragments containing 26-bp complementary nucleotides [overlap extension polymerase chain reaction (PCR)]. The spacer fragment digested with restriction enzymes Xho I and Eag I was cloned into linearized LbCas12a/SpCas9 plasmid. Sequences of the spacers are shown in table S1.
The donor plasmids for deletion/insertion—including Hoc-del, Soc-del, FarP7K-del, FarP18K-del, 39-56 11K-del, SegF-del, IPIII-del, IPII-del, E insertion (Hoc site), Soc-SpyCatcher/RBD insertion (Soc site), CTS-amber-NP insertion (IPIII site), CAG-S-fl/S-ecto insertion (FarP7K site), and CMV-RBD insertion (SegF site)—were constructed using overlap extension PCR. Briefly, the DNAs including ~500-bp homology arms (left and right) with a desired deletion in the middle were amplified from T4 genome DNA, stitched, and cloned into pET28b linearized with Bgl II and Xho I to generate the deletion donor plasmid. For the insertion donor plasmid, the insertion fragment, left homology arm, and right homology arm, which contains 25-bp complementary nucleotides were stitched by annealing and overlap extension. The Bgl II– and Xho I–digested donor fragment was cloned into pET28b plasmid.
The Soc fusion expression plasmids—including Soc-SpyCatcher, Soc-RBD, RBD-Soc, SUMO-Soc-Spy, and Soc-truncated RBDs (RBD67, RBD106, RBD135, RBD162, RBD181, and RBD197)—were constructed by two rounds of cloning. First, the multiple cloning site (MCS) (Nco I, Nde I, Nhe I, and Bmt I)–linker (4GGS)-Soc-linker (2GGGGS)–MCS (Hind III, Eag I, Not I, and Xho I) was amplified using Soc plasmid template and cloned into the pET28b DNA linearized with Nco I and Xho I restriction enzymes to generate pET28b-MCS-L-Soc-L-MCS. Second, DNAs corresponding to SpyCatcher, SUMO, Spy, and various RBDs were amplified and inserted to 3′ or 5′ MCS of pET28b-MCS-L-Soc-L-MCS as needed. CTS-NP and Ee-Hoc expression fragments were amplified using synthesized NP and T4 genomic DNA, respectively, and cloned into Nco I– and Xho I–linearized pET28b.
The plasmids for expression in mammalian cells—including pCMV-CD5-RBD, pCAG-CD5–S-fl, pCAG-CD5–S-ecto–6P, and pCAG-CD5–S-ecto–6P-SpyTag—were constructed using standard protocols. The RBD fragment was amplified using the WT spike gene, and CD5 secretion–leading peptide (MPMGSLQPLATLYLLGMLVASVLA) was added to the N terminus of RBD by PCR. The CD5-RBD was directionally cloned into pAAV vector (Cell Biolabs) using the Hind III and Xho I restriction enzyme sites (pCMV-CD5-RBD). For the construction of pCAG-CD5–S-fl, pCAG-CD5–S-ecto–6P, and pCAG-CD5–S-ecto–6P-SpyTag, plasmids pCAG–S-fl and pCAG–S-ecto–6P were used as template and backbone. The CD5 fragment was cloned into the N terminus of S-fl or S-ecto-6P using Kpn I and Eco RI restriction sites. Similarly, SpyTag (RGVPHIVMVDAYKRYK) was cloned into the C terminus of S-ecto using Bam HI and Xho I restriction sites. All the constructed plasmids were sequenced to confirm 100% accuracy of the recombinant fragment (Retrogen, CA).
Plaque assay
Plaque assays were performed to determine the efficiency of the individual spacers to restrict T4 phage infection. The CRISPR-Cas12a/Cas9 spacer plasmid was transformed into E. coli strains B834 or B40. Serially diluted T4 phages, in the range of 101 to 107 plaque-forming units (PFU) in 100 μl of Pi-Mg buffer [26 mM Na2HPO4, 68 mM NaCl, 22 mM KH2PO4, and 1 mM MgSO4 (pH 7.5)] were mixed with 350 μl of spacer-containing E. coli (~108 cells\). E. coli cells without spacer were used as a control. After incubation at 37°C for 7 min, 3.5 ml of 0.75% top agar with spectinomycin (50 μg/ml) was added to each tube, mixed, and poured onto LB plate. The plates were incubated at 37°C overnight to allow the formation of plaques. The PFU were counted on each plate, and the efficiency of plating (EOP) was determined by dividing the PFU produced from infection of E. coli containing a spacer with the input PFU.
CRISPR-mediated phage T4 genome editing and recombination
The CRISPR-Cas12a/Cas9 spacer plasmid and the corresponding donor plasmid were cotransformed into E. coli strains, either B834/P301 without amber suppressor or B40/RIPL with amber suppressor. E. coli cells transformed with single plasmids, either the donor plasmid or the CRISPR spacer plasmid, were used as controls. An appropriate amount of T4 phages, as determined by the EOP as described above, were added to E. coli and incubated for 7 min at 37°C. After adding 3.5 ml of 0.75% top agar containing spectinomycin (50 μg/ml) and kanamycin (50 μg/ml), the infection mixture was poured onto the LB plate and incubated overnight. Single plaques, namely, G1, were picked using a sterile Pasteur glass pipet and transferred into a 1.5-ml Eppendorf tube containing 200 μl of Pi-Mg buffer. After 20 min of incubation at room temperature (RT) with gentle vortexing every 5 min, serially diluted G1 phages were used to infect spacer-containing E. coli cells (spectinomycin, 50 μg/ml). This eliminated any parental phage background under CRISPR pressure. The resultant single G2 plaques were picked and used to infect E. coli cells (without spacer or donor) to produce purified G3 phages. Single G3 plaques were then picked into 200 μl of Pi-Mg buffer. PCR analysis was performed to confirm DNA deletion or foreign gene insertion. One microliter of G3 phages were denatured at 94°C for 8 min and used as a template for PCR using the Phusion High-Fidelity PCR Master Mix (Thermo Fisher Scientific). The amplified DNA fragment was purified using the QIAquick Gel Extraction Kit (QIAGEN) and sequenced (Retrogene). The G3 phages in which the recombinant DNA sequence was confirmed with 100% accuracy were selected, and a few drops of chloroform were added. These plaque-purified “zero stocks” were stored at 4°C. More rounds of CRISPR gene editing were similarly introduced into the same phage as needed as described above.
Phage production and purification
E. coli strains B40 or B834 were used for the production of amber phage or non–amber phage, respectively. Fresh overnight E. coli cells were inoculated in 1 liter of Moore’s medium (20 g of tryptone, 15 g of yeast extract powder, 2 g of dextrose, 8 g of NaCl, 2 g of Na2HPO4, and 1 g of KH2PO4, dissolved in 1 liter of Milli-Q water) at 1/50 dilution and then grown at 37°C for 2 to 2.5 hours in a shaker incubator at 200 rpm. When the cells reached a density of ~4 × 108/ml, phages were added at a multiplicity of infection (MOI) of 0.5. The infection mixture was cultured at 37°C, 200 rpm, for another 2.5 to 3 hours and periodically checked under the microscope. As the cells get infected with phage, the shape of cells changes from long bacilli to short dumbbell shape. When the cell number started dropping, the culture was transferred to a Sorvall GSA bottle and centrifuged at 27,504g for 1 hour at 4°C. The supernatant was discarded, and the pellet was resuspended in 50 ml of Pi-Mg buffer containing deoxyribonuclease I (10 μg/ml) (or Benzonase) and one tablet of protease inhibitor cocktail. The resuspended pellet was added with 5 ml of chloroform and incubated at 37°C for 1 hour to lyse the bacteria and release the phage. The debris was removed by low-speed centrifugation at 4302g for 10 min. The phage-containing supernatant was transferred to a sterile Falcon tube, and the pellet was discarded. The titer of this phage stock was determined using E. coli B40 or B834. The working stock of phages can be stored at 4°C for long periods of time with a few drops of chloroform added to prevent microbial contamination, used as seed phage to make SpyCatcher- or Soc-RBD–displayed phage, or purified as a vaccine candidate for animal studies.
For purification of phage, ~50 ml of phage stock was distributed into five Corex glass tubes (10 ml each) and sealed with parafilm. The phage stock was centrifuged at 34,540g for 1 hour using a Sorvall SS34 rotor. The supernatant was discarded, and the phage-containing pellet was resuspended with 1 to 3 ml of Pi-Mg buffer plus 5 μl of Benzonase overnight at 4°C. Next, the resuspended phages were loaded onto a CsCl gradient and centrifuged at 152,000g for 1 hour in a Beckman ultracentrifuge using a SW 55 Ti swinging bucket rotor. The purified phage band localized between CsCl densities of 1.46 and 1.55 was collected by inserting a syringe right below the phage band and aspirating the band. The phages were dialyzed in high-salt tris-Mg buffer [10 mM tris-HCl (pH 7.5), 200 mM NaCl, and 5 mM MgCl2] for 4 hours, followed by low-salt tris-Mg buffer [10 mM tris-HCl (pH 7.5), 50 mM NaCl, and 5 mM MgCl2] overnight. The second-round CsCl centrifugation and dialysis of purified phages were applied to obtain purer phages. Two-round CsCl-purified phages were further purified by passing through a 0.22-μm filter to remove any minor bacterial contaminants. The phage concentration and copy numbers of displayed antigens were quantified by 4 to 20% SDS-PAGE.
Production of Soc-SpyCatcher– and Soc-RBD–displayed phages
E. coli BL21 (DE3)-RIPL transformed with T7-Soc-SpyCatcher (or Soc-RBD) plasmid were used for Soc-SpyCatcher (or Soc-RBD) in vivo–displayed phage production. E. coli BL21 (DE3)-RIPL cotransformed with T7-Soc-SpyCatcher (or Soc-RBD) plasmid and amber suppressor plasmid was used for the production of Soc-SpyCatcher (or Soc-RBD)–displayed and NP-packaged phage. Briefly, the RIPL cells were inoculated into 1 liter of Moore’s medium at 1/50 dilution with appropriate antibiotics [RIPL-Soc-SpyCatcher, kanamycin (50 μg/ml) + chloramphenicol (37 μg/ml); RIPL-Soc-SpyCatcher-Amber suppressor, kanamycin (50 μg/ml) + chloramphenicol (37 μg/ml) + ampicillin (100 μg/ml)]. The culture was incubated at 37°C, 200 rpm, for 2.5 to 3 hours. When the cells reached a density of ~4 × 108/ml, 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added for induction of recombinant protein expression. At 10 min after IPTG addition, the corresponding phages (Hoc-del/Soc-del/IPII-del/IPIII-del) were added to infect cells at an MOI of 0.5. The culture was further incubated at 37°C, 200 rpm, for 3 hours. The following phage production and purification procedures are the same as described above.
Lipopolysaccharide endotoxin measurement
E. coli lipopolysaccharide (LPS), an agonist of Toll-like receptor 4, can associate with phage and be present in the phage preparations. To minimize this contamination, the recombinant phages were extensively purified by differential centrifugations, two rounds of CsCl density gradient centrifugations, and 0.22 μm of membrane filtration. Furthermore, as part of our optimized in vitro display protocol, the phage particles were extensively washed with excess phosphate-buffered saline (PBS) (see below). All these procedures greatly reduced the LPS level to ~10 to 30 endotoxin units per dose and also freed any bacterial contamination as determined by their enumeration on tryptic soy agar (TSA) plates with sheep blood. This LPS level is below the FDA-recommended level. Furthermore, no weight reduction or reactogenicity was observed upon administration of T4 vaccines to mice or rabbits.
The endotoxin quantitation kit (Thermo Fisher Scientific) was used to measure the amount of LPS endotoxin in phage samples using the Limulus Amebocyte Lysate (LAL) assay according to the manufacturer’s instructions. Phage samples were diluted in a twofold dilution series beginning with 1010 particles in 50 μl of endotoxin-free water. LAL reagent was added, and the mixture was incubated at 37°C, followed by the addition of chromogenic substrate solution. After 6 min of incubation, the stop reagent was added, and the absorbance was measured at 405 nm. The endotoxin concentration of the phage sample was determined using the formulated standard curve.
S-trimer expression and purification
Plasmid pCAG-CD5–S-ecto–6P-SpyTag was transiently transfected into ExpiCHO cells using ExpiFectamine CHO transfection kit (Thermo Fisher Scientific). After 18 to 22 hours of transfection, cells were supplemented with the ExpiCHO Feed and Enhancer and grown at 32°C according to the manufacturer’s high-titer protocol. Cultures were harvested 8 to 10 days after transfection by centrifuging the cells at 3000g for 20 min at 4°C. The supernatant was clarified through a 0.22-μm filter and then loaded on a HisTrap HP column (Cytiva) previously equilibrated with wash buffer [50 mM tris-HCl (pH 8.0), containing 300 mM NaCl and 20 mM imidazole], at a flow rate of 1 ml/min, using ÄKTAprime plus liquid chromatography system (GE Healthcare). Protein-bound column was washed with wash buffer until the ultraviolet absorbance reached the baseline to remove nonspecifically bound proteins. The trimers were eluted using a 20 to 300 mM linear gradient of imidazole. HisTrap-eluted peak fractions were pooled and applied to a HiLoad 16/600 Superdex 200 (preparation grade) size exclusion column (GE Healthcare) equilibrated with the gel filtration buffer [50 mM tris-HCl (pH 8) and 150 mM NaCl] to obtain purified trimers, using the ÄKTA FPLC system. Eluted fractions were collected, filtered through 0.22-μm filter unit, flash-frozen, and stored at −80°C until use.
Quantification of S-trimer and rRBD display on T4-SpyCatcher phage
In vitro display of S-trimer/rRBD on the T4-SpyCatcher phage was assessed by cosedimentation as described previously with some modifications (26, 27). Briefly, two-round CsCl-purified and 0.22-μm filtered phage particles were sedimented for 45 min at 34,000g in Protein-LoBind Eppendorf tubes, washed twice with sterilized PBS buffer (pH 7.4), and resuspended in PBS buffer (pH 7.4). S-trimer/rRBD was sedimented for 25 min at 34,000g to remove any possible aggregates present in the sample. T4-SpyCatcher phages were incubated with S-trimer/rRBD proteins at 4°C for 1 hour. The mixtures were sedimented by centrifugation at 34,000g for 45 min, and unbound protein in the supernatants was removed. After washing twice with excess PBS to further remove the unbound protein and any other minor contaminants, the phage pellets containing the displayed proteins were incubated at 4°C overnight and then resuspended in PBS. For rabbit animal studies, 50 μl of phage trimer particles were added to blood agar (TSA with sheep blood) to examine any contamination of a wide variety of fastidious microorganisms. The resuspended pellets were analyzed using Novex 4 to 20% SDS-PAGE mini gel (Thermo Fisher Scientific) to quantify the S-trimer/rRBD copies. After Coomassie Blue R-250 (Bio-Rad, CA) staining and destaining, the protein bands on SDS-PAGE gels were scanned and quantified by ChemiDoc MP imaging system (Bio-Rad) and ImageJ. The copy numbers of SpyCatcher and displayed S-trimer/rRBD molecules per capsid were calculated using gp23 (major capsid protein; 930 copies) or gp18 (major tail sheath protein; 138 copies) as internal controls and S-trimer protein standard.
SUMO-RBD-Spy protein expression and purification
The E. coli expression, denaturation, refolding, and purification of SUMO-RBD-Spy (rRBD) were performed using a similar procedure described previously (59). Briefly, the BL21-CodonPlus (DE3)-RIPL cells containing PET28b-SUMO-RBD-Spy expression plasmid were induced with 0.5 mM IPTG for 3 hours at 28°C. Cells were harvested and resuspended in buffer A [20 mM tris-HCl, 500 mM NaCl, 5 mM imidazole, and 5 mM β-mercaptoethanol (pH 7.9)] containing protease inhibitor cocktail (Roche, Indianapolis, IN, USA) and Benzonase. After the cells were lysed using a French press (Aminco, Urbana, IL) and centrifuged, the pellet containing the inclusion body proteins was resuspended and washed with buffer B (buffer A + 0.5% Triton X-100). Then, the inclusion bodies were solubilized in buffer C (buffer A + 8 M urea) by incubating/stirring at 4°C overnight, followed by centrifugation and clarification. The supernatant containing the denaturing protein was loaded on HisTrap column (ÄKTAprime, GE Healthcare), followed by washing with buffer C. The rRBD was refolded on HisTrap column using a linear gradient of 8 to 0 M urea containing buffer D [20 mM tris-HCl, 500 mM NaCl, 5 mM imidazole, 1 mM glutathione, 0.1 mM oxidized glutathione, and 20% glycerol (pH 7.9)]. Last, the column was washed with buffer E [20 mM tris-HCl, 500 mM NaCl, 100 mM imidazole, 20% glycerol, and 5% glucose (pH 7.9)], and the refolded rRBD was eluted with buffer F [20 mM tris-HCl, 500 mM NaCl, 800 mM imidazole, 20% glycerol, and 5% glucose (pH 7.9)] and dialyzed to remove imidazole. The proteins were then quantified, aliquoted, and stored at −80°C until use.
WB analysis
After treatment with multiple freeze-thaw cycles and Benzonase, phage particles were boiled in SDS loading buffer for 10 min, separated by 4 to 20% SDS-PAGE, and transferred to nitrocellulose membrane polyvinylidene difluoride (PVDF) (Bio-Rad). The PVDF was then blocked with 5% bovine serum albumin (BSA)–PBS (pH 7.4) buffer at RT for 1 hour with gentle shaking. Anti-NP or anti-RBD primary antibodies were added to the blots and incubated overnight at 4°C in PBS–5% BSA, followed by five times rinsing in PBST buffer [1× PBS (pH 7.4) and 0.05% Tween 20]. Goat anti-mouse or goat anti-rabbit horseradish peroxidase (HRP)–conjugated antibody (Thermo Fisher Scientific) was applied at a 1:5000 dilution in 5% BSA-PBST for 1 hour at RT with gentle shaking. After rinsing five times in PBST, binding was visualized with an enhanced chemiluminescence substrate (Bio-Rad) using the Bio-Rad Gel Doc XR+ System and Image Lab software according to the manufacturer’s instructions (Bio-Rad).
Cell culture and transfection
HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco–Thermo Fisher Scientific) supplemented with 1% antibiotics (Thermo Fisher Scientific), 1× Hepes (Thermo Fisher Scientific), and 10% fetal bovine serum (Thermo Fisher Scientific). Cells were passaged with 0.25% (w/v) trypsin/0.53 mM EDTA at a subcultivation ratio of 1:5 at 80 to 90% confluence. Cultures were incubated in a humidified atmosphere at 37°C and 5% CO2. The recombinant plasmid containing human ACE2 gene (Addgene, #1786) was transfected into HEK293T cells using the Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. Two days after ACE2 plasmid transfection, the cells were used for RBD or phage binding assay.
Inhibition by mice sera of RBD binding to cell surface ACE2
Human ACE2-transfected HEK293T cells were washed with PBS twice and then fixed with 4% formaldehyde for 15 min at RT. After rinsing twice in PBS for 5 min each, cells were blocked in blocking buffer (5% BSA-PBS) for 1 hour at RT. Recombinant SARS-CoV-2 RBD protein (Sino Biological) was added to the cells to a final concentration of 0.2 μg/ml in the presence or absence of the sera with a series of dilutions. The unbound RBD was removed by washing cells five times with PBST (PBS and 0.1% Tween 20) for 5 min each. The 1/1000 diluted human anti-RBD mAb (Thermo Fisher Scientific) was added to cells and incubated in a humidified chamber for 1 hour at RT or overnight at 4°C. After rinsing five times in PBST for 5 min each, Alexa Fluor 488– or rhodamine-conjugated goat anti-human secondary antibody was added (1/500 dilution) (Thermo Fisher Scientific) and incubated for 2 to 3 hours at RT in the dark. The cells were then rinsed five times in PBST for 5 min each and counterstained with Hoechst 33342 (1 μg/ml) (Thermo Fisher Scientific) for 5 min. The fluorescent signals were recorded by fluorescence microscopy (Carl Zeiss).
Binding of T4–S-trimer–GFP phages to cell surface ACE2
Soc-GFP protein was produced as described previously (26). In vitro display of Soc-GFP on the T4-SpyCatcher or T4–S-trimer phage was assessed by the cosedimentation and SDS-PAGE, similar to the procedures of S-trimers displayed on T4 phage. The T4-SpyCatcher-GFP or T4–S-trimer–GFP phages were resuspended in Opti-MEM medium (Thermo Fisher Scientific) and then added to ACE2-transfected HEK293T cells. After 6 hours of incubation, the unbound phages were removed by rinsing three times in PBS for 5 min each. The GFP fluorescence was recorded by fluorescence microscopy (Carl Zeiss).
Mouse immunizations
We followed the recommendations of the NIH about mouse study (the Guide for the Care and Use of Laboratory Animals). All mouse experiments were approved by the Institutional Animal Care and Use Committee of the Catholic University of America (Washington, DC) (Office of Laboratory Animal Welfare assurance number A4431-01) and the University of Texas Medical Branch (Galveston, TX) (Office of Laboratory Animal Welfare assurance number A3314-01). The SARS-CoV-2 virus challenge study was conducted in the animal BSL-3 suite.
Six- to 8-week-old female BALB/c mice (the Jackson laboratory) were randomly grouped (five mice per group) and allowed to acclimate for 14 days. The phage vaccine candidates were administered by intramuscular injections into their hind legs. A total of 6 × 1011 phages carrying approximately 20 μg of SARS-CoV-2 antigen(s) were injected on days 0 (prime), 21 (boost 1), and 42 (boost 2). Negative control mice received the same volume of PBS buffer (naïve) or the same amount of T4 control phage. A group of mice immunized with purified S-trimer (20 μg) adjuvanted with Alhydrogel was included as the positive control. Blood was drawn from each animal on days 0 (prebleed), 14, 35, and 56, and the isolated sera were stored at −80°C.
Rabbit immunizations
All experiments were performed at Envigo/Cocalico Biologicals (Reamstown, PA) in accordance with institutional guidelines. Adult New Zealand White rabbits were immunized intramuscularly in the flank region with 3 × 1011 PFU of T4 phages per dose in 0.2 ml of saline (n = 4 for each group). Preimmune test bleeds were first obtained via venipuncture of the marginal vein of the ear on day 1. Animals were immunized on days 1 and 15 (prime + one-boost regimen). Immune sera were obtained on day 25.
ELISA determination of IgG and IgG subtype antibodies
ELISA plates (Evergreen Scientific) were coated with 100 μl per well of SARS-CoV-2 S-ecto protein (1 μg/ml; Sino Biological), SARS-CoV-2 RBD-untagged protein (Sino Biological), SARS-CoV-2 NP (Sino Biological), or SARS-CoV-2 E protein (1 to 75 amino acids) (Thermo Fisher Scientific) in coating buffer [0.05 M sodium carbonate–sodium bicarbonate (pH 9.6)]. After overnight incubation at 4°C, the plates were washed twice with PBS buffer and blocked for 2 hours at 37°C with 200 μl per well of PBS–5% BSA buffer. Serum samples were diluted with a 5-fold dilution series beginning with an initial 100-fold dilution in PBS–1% BSA. One hundred microliters of diluted serum samples was added to each well, and the plates were incubated at 37°C for 1 hour. After washing five times with PBST (PBS + 0.05% Tween 20), the secondary antibody was added at 1:10,000 dilution in PBS–1% BSA buffer (100 μl per well) using either goat anti-mouse IgG-HRP, goat anti-mouse IgG1-HRP, goat anti-mouse IgG2a-HRP (Thermo Fisher Scientific), or goat anti-rabbit IgG-HRP (Abcam). After incubation for 1 hour at 37°C and five washes with PBST buffer, plates were developed using the TMB (3,3′,5,5′-tetramethylbenzidine) Microwell Peroxidase Substrate System (KPL). After 5 to 10 min, the enzymatic reaction was stopped by adding TMB BlueSTOP (KPL) solution. The absorbance was read within 30 min at 650 nm on a VersaMax spectrophotometer. The endpoint titer was defined as the highest reciprocal dilution of serum that gives an absorbance more than twofold of the mean background of the assay.
Binding of T4 displayed RBD or S-trimer to human ACE2 protein
An ELISA to analyze the binding of RBD, S-ecto-6P-SpyTagged trimer, and T4 that displayed RBD/S-trimer to human ACE2 protein was performed as described above. Briefly, 100 ng of protein or 1 × 1010 phages were coated on plates overnight at 4°C. After blocking with PBS–5% BSA buffer, recombinant human ACE2-mouse Fc protein (Sino Biological) with a series of dilution was added and incubated for 1 hour at 37°C. Plates were then incubated with the secondary goat anti-mouse IgG-HRP antibody and developed with TMB substrate. Reactions were stopped, and the absorbance was measured at 650 nm on a VersaMax spectrophotometer.
Virus neutralization assay using BSL-3 live SARS-CoV-2
Neutralizing antibody titers in mouse immune sera were quantified by Vero E6 cell–based microneutralization assay using SARS-CoV-2 US-WA-1/2020 strain as previously described (50). Briefly, serially 1:3 downward diluted mouse sera that were decomplemented at 56°C for 60 min in a 60-μl volume were incubated for 1 hour at RT in duplicate wells of 96-well microtiter plates that contained 120 infectious SARS-CoV-2 viruses in 60 μl in each well. After incubation under BSL-3 conditions, 100 μl of the mixtures in individual wells was transferred to Vero E6 cell monolayer grown in 96-well microtiter plates containing 100 μl of MEM/2% fetal bovine serum medium in each well and was cultured for 72 hours at 37°C before assessing the presence or absence of cytopathic effect (CPE). Neutralizing antibody titers of the tested specimens were calculated as the reciprocal of the highest dilution of sera that completely inhibited virus-induced CPE in at least 50% of the wells and expressed as 50% neutralizing titer.
Neutralizing antibody titers in rabbit immune sera were quantified using an automated, liquid handler–assisted, high-throughput, microfocus neutralization/high-content imaging methods developed at ViroVax. Briefly, rabbit sera (paired preimmune and immune) were first decomplemented at 56°C for 60 min and were then serially diluted in 384-well plates, in duplicate, using a BioTek Precision 2000 liquid handler, along with two reference sera. Twenty-microliter aliquots of SARS-CoV-2 US-WA-1/2020 were added to all test wells and positive control wells to yield a final MOI of 10 under BSL-3 conditions. Vero (American Type Culture Collection, CCL-81) cells were maintained in a high-glucose DMEM supplemented with 10% fetal bovine serum (HyClone Laboratories, South Logan, UT) and 1% penicillin/streptomycin at 37°C with 5% CO2. After preincubating the plates for 1 hour, 20 μl of Vero cells (106/ml), containing propidium iodide (PI) (50 μg/ml), was added to all wells using the liquid handler. Plates were then loaded in an IncuCyte S3 high-content imaging system (Essen BioScience/Sartorius, Ann Arbor, MI). Longitudinal image acquisition and processing for virus-induced CPE and cell death (PI uptake) were performed every 6 hours, until cell death profiles had crested and stabilized (3.5 days). Neutralizing antibody titers [expressed as median inhibitory concentration (IC50) or IC90] were obtained from four-parameter logistic curve fits of cell death profiles using OriginPro 9 (OriginLab Corp., Northampton, MA).
Challenge of the mice with MA live BSL-3 SARS-CoV-2 virus
Immunized mice were challenged with the MA SARS-CoV-2 MA10 strain (51), a gift from R. Baric at the University of North Carolina, by the intranasal route as previously described (60). Briefly, mice were inoculated with 60 μl of SARS-CoV-2 MA10 at a dose of ~105 median tissue culture infectious dose. The animals were weighed every day over the indicated period of time for monitoring the onset of morbidity (weight loss and other signs of illness) and mortality, as the end points for evaluating the vaccine efficacy.
Statistics
All the data were presented as means ± SEM except where indicated. Statistical analyses were performed by GraphPad Prism 9.0 software using one-way or two-way analysis of variance (ANOVA) according to the data. Tukey’s multiple comparisons posttest was used to compare individual groups. Significant differences between two groups were indicated by *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. ns indicates not significant. P < 0.05 was considered significant.
Acknowledgments
We thank V. Padilla-Sanchez and Frontera Supercomputer in Texas for assistance with the preparation of the video and Fig. 1; Z. Wang and T. Close at Purdue University for the cryo-EM images; B. Graham, K. S. Corbett (NIH), and J. S. McLellan (University of Texas, Austin) for providing recombinant plasmids containing SARS-CoV-2 spike (S) genes; and R. Kempaiah (UTMB) for providing the inoculum of the MA SARS-CoV-2 virus. Funding: This research was supported by NIAID/NIH supplement grant 3R01AI095366-07S1 (subaward: 1100992-100) and, in part, by NIAID/NIH grants AI111538 and AI081726 and NSF grant MCB-0923873 to V.B.R. ViroVax LLC acknowledges the NIAID/NIH supplement contract HHSN272201800049C to S.A.D. Special funding provided by the IHII-COVID19 pilot grant to A.K.C. is acknowledged. Author contributions: V.B.R. and A.K.C. designed and directed the project. J.Z. and V.B.R. designed the research. J.Z., N.A., S.J., H.B., and W.-C.T. conducted most of the experiments. D.A.L., M.L.R., and S.A.D. performed the rabbit immunization and neutralization antibody titer assay. A.D. performed neutralization assay using the live virus with immune sera derived from mice in consultation with C.-T.K.T. A.K.C., P.B.K., and J.S. conducted mice challenge experiments. J.Z., V.B.R., C.-T.K.T., and A.K.C. analyzed and interpreted the data. V.B.R. and J.Z. wrote the manuscript, and A.K.C. and C.-T.K.T. edited the manuscript. Competing interests: J.Z. and V.B.R. are inventors on a provisional patent application related to this work filed by The Catholic University of America (no. 63/126047, filed 16 December 2020). The authors declare that they have no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The recombinant plasmid containing SARS-CoV-2 WT spike gene can be provided by Vaccine Research Center, NIH pending scientific review and a completed material transfer agreement. Requests for the plasmid should be sent to B. Graham, Deputy Director, Vaccine Research Center.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Table S1
Legend for movie S1
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Wu F., Zhao S., Yu B., Chen Y. M., Wang W., Song Z. G., Hu Y., Tao Z. W., Tian J. H., Pei Y. Y., Yuan M. L., Zhang Y. L., Dai F. H., Liu Y., Wang Q. M., Zheng J. J., Xu L., Holmes E. C., Zhang Y. Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.V’Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V., Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin M. D., Shukla S., Chung Y. H., Beiss V., Chan S. K., Ortega-Rivera O. A., Wirth D. M., Chen A., Sack M., Pokorski J. K., Steinmetz N. F., COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 15, 646–655 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Krammer F., SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Corbett K. S., Edwards D. K., Leist S. R., Abiona O. M., Boyoglu-Barnum S., Gillespie R. A., Himansu S., Schafer A., Ziwawo C. T., DiPiazza A. T., Dinnon K. H., Elbashir S. M., Shaw C. A., Woods A., Fritch E. J., Martinez D. R., Bock K. W., Minai M., Nagata B. M., Hutchinson G. B., Wu K., Henry C., Bahl K., Garcia-Dominguez D., Ma L., Renzi I., Kong W. P., Schmidt S. D., Wang L., Zhang Y., Phung E., Chang L. A., Loomis R. J., Altaras N. E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M. K., Shi W., Leung K., Yang E. S., West A., Gully K. L., Stevens L. J., Wang N., Wrapp D., Doria-Rose N. A., Stewart-Jones G., Bennett H., Alvarado G. S., Nason M. C., Ruckwardt T. J., McLellan J. S., Denison M. R., Chappell J. D., Moore I. N., Morabito K. M., Mascola J. R., Baric R. S., Carfi A., Graham B. S., SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan T. K., Rijal P., Rahikainen R., Keeble A. H., Schimanski L., Hussain S., Harvey R., Hayes J. W. P., Edwards J. C., McLean R. K., Martini V., Pedrera M., Thakur N., Conceicao C., Dietrich I., Shelton H., Ludi A., Wilsden G., Browning C., Zagrajek A. K., Bialy D., Bhat S., Stevenson-Leggett P., Hollinghurst P., Tully M., Moffat K., Chiu C., Waters R., Gray A., Azhar M., Mioulet V., Newman J., Asfor A. S., Burman A., Crossley S., Hammond J. A., Tchilian E., Charleston B., Bailey D., Tuthill T. J., Graham S. P., Duyvesteyn H. M. E., Malinauskas T., Huo J., Tree J. A., Buttigieg K. R., Owens R. J., Carroll M. W., Daniels R. S., McCauley J. W., Stuart D. I., Huang K. A., Howarth M., Townsend A. R., A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nat. Commun. 12, 542 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen A. A., Gnanapragasam P. N. P., Lee Y. E., Hoffman P. R., Ou S., Kakutani L. M., Keeffe J. R., Wu H. J., Howarth M., West A. P., Barnes C. O., Nussenzweig M. C., Bjorkman P. J., Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinbuck M. P., Seenappa L. M., Jakubowski A., McNeil L. K., Haqq C. M., DeMuth P. C., A lymph node–targeted Amphiphile vaccine induces potent cellular and humoral immunity to SARS-CoV-2. Sci. Adv. 7, eabe5819 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A. C., Fiala B., Schäfer A., Wrenn S., Pham M. N., Murphy M., Tse L. V., Shehata L., O’Connor M. A., Chen C., Navarro M. J., Miranda M. C., Pettie D., Ravichandran R., Kraft J. C., Ogohara C., Palser A., Chalk S., Lee E. C., Guerriero K., Kepl E., Chow C. M., Sydeman C., Hodge E. A., Brown B., Fuller J. T., Dinnon K. H. III, Gralinski L. E., Leist S. R., Gully K. L., Lewis T. B., Guttman M., Chu H. Y., Lee K. K., Fuller D. H., Baric R. S., Kellam P., Carter L., Pepper M., Sheahan T. P., Veesler D., King N. P., Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell 183, 1367–1382.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung Y. H., Beiss V., Fiering S. N., Steinmetz N. F., COVID-19 vaccine frontrunners and their nanotechnology design. ACS Nano 14, 12522–12537 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Klasse P. J., Nixon D. F., Moore J. P., Immunogenicity of clinically relevant SARS-CoV-2 vaccines in nonhuman primates and humans. Sci. Adv. 7, eabe8065 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson L. A., Anderson E. J., Rouphael N. G., Roberts P. C., Makhene M., Coler R. N., McCullough M. P., Chappell J. D., Denison M. R., Stevens L. J., Pruijssers A. J., McDermott A., Flach B., Doria-Rose N. A., Corbett K. S., Morabito K. M., O’Dell S., Schmidt S. D., Swanson P. A. II, Padilla M., Mascola J. R., Neuzil K. M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J. E., Graham B. S., Beigel J. H.; mRNA-1273 Study Group , An mRNA vaccine against SARS-CoV-2—Preliminary report. N. Engl. J. Med. 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakraborty C., Bhattacharya M., Sharma A. R., Present variants of concern and variants of interest of severe acute respiratory syndrome coronavirus 2: Their significant mutations in S-glycoprotein, infectivity, re-infectivity, immune escape and vaccines activity. Rev. Med. Virol. 2021, e2270 (2021). [Google Scholar]
- 14.Zhang B., Chao C. W., Tsybovsky Y., Abiona O. M., Hutchinson G. B., Moliva J. I., Olia A. S., Pegu A., Phung E., Stewart-Jones G. B. E., Verardi R., Wang L., Wang S., Werner A., Yang E. S., Yap C., Zhou T., Mascola J. R., Sullivan N. J., Graham B. S., Corbett K. S., Kwong P. D., A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 10, 18149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller E. S., Kutter E., Mosig G., Arisaka F., Kunisawa T., Ruger W., Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67, 86–156 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Sun L., Zhang Z., Fokine A., Padilla-Sanchez V., Hanein D., Jiang W., Rossmann M. G., Rao V. B., Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-Å resolution and its relevance to the assembly of icosahedral viruses. Proc. Natl. Acad. Sci. U.S.A. 114, E8184–E8193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fokine A., Chipman P. R., Leiman P. G., Mesyanzhinov V. V., Rao V. B., Rossmann M. G., Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 101, 6003–6008 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin L., Fokine A., O’Donnell E., Rao V. B., Rossmann M. G., Structure of the small outer capsid protein, Soc: A clamp for stabilizing capsids of T4-like phages. J. Mol. Biol. 395, 728–741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fokine A., Islam M. Z., Zhang Z., Bowman V. D., Rao V. B., Rossmann M. G., Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J. Virol. 85, 8141–8148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barr J. J., Auro R., Furlan M., Whiteson K. L., Erb M. L., Pogliano J., Stotland A., Wolkowicz R., Cutting A. S., Doran K. S., Salamon P., Youle M., Rohwer F., Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 10771–10776 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao P., Wu X., Tang W. C., Zhu J., Rao V., Engineering of bacteriophage T4 genome using CRISPR-Cas9. ACS Synth. Biol. 6, 1952–1961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Dai L., Dong J., Chen C., Zhu J., Rao V. B., Tao P., Covalent modifications of the bacteriophage genome confer a degree of resistance to bacterial CRISPR systems. J. Virol. 94, e01630-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang Q., Tang W. C., Tao P., Mahalingam M., Fokine A., Rossmann M. G., Rao V. B., Structural morphing in a symmetry-mismatched viral vertex. Nat. Commun. 11, 1713 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q., Shivachandra S. B., Zhang Z., Rao V. B., Assembly of the small outer capsid protein, Soc, on bacteriophage T4: A novel system for high density display of multiple large anthrax toxins and foreign proteins on phage capsid. J. Mol. Biol. 370, 1006–1019 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shivachandra S. B., Rao M., Janosi L., Sathaliyawala T., Matyas G. R., Alving C. R., Leppla S. H., Rao V. B., In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc-capsid interactions: A strategy for efficient display of large full-length proteins. Virology 345, 190–198 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Tao P., Mahalingam M., Marasa B. S., Zhang Z., Chopra A. K., Rao V. B., In vitro and in vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proc. Natl. Acad. Sci. U.S.A. 110, 5846–5851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J., Tao P., Mahalingam M., Sha J., Kilgore P., Chopra A. K., Rao V., A prokaryotic-eukaryotic hybrid viral vector for delivery of large cargos of genes and proteins into human cells. Sci. Adv. 5, eaax0064 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao P., Zhu J., Mahalingam M., Batra H., Rao V. B., Bacteriophage T4 nanoparticles for vaccine delivery against infectious diseases. Adv. Drug Deliv. Rev. 145, 57–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao P., Mahalingam M., Zhu J., Moayeri M., Sha J., Lawrence W. S., Leppla S. H., Chopra A. K., Rao V. B., A bacteriophage T4 nanoparticle-based dual vaccine against anthrax and plague. MBio 9, e01926-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao P., Mahalingam M., Kirtley M. L., van Lier C. J., Sha J., Yeager L. A., Chopra A. K., Rao V. B., Mutated and bacteriophage T4 nanoparticle arrayed F1-V immunogens from Yersinia pestis as next generation plague vaccines. PLOS Pathog. 9, e1003495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao M., Peachman K. K., Li Q., Matyas G. R., Shivachandra S. B., Borschel R., Morthole V. I., Fernandez-Prada C., Alving C. R., Rao V. B., Highly effective generic adjuvant systems for orphan or poverty-related vaccines. Vaccine 29, 873–877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren Z. J., Lewis G. K., Wingfield P. T., Locke E. G., Steven A. C., Black L. W., Phage display of intact domains at high copy number: A system based on SOC, the small outer capsid protein of bacteriophage T4. Protein Sci. 5, 1833–1843 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang J., Abu-Shilbayeh L., Rao V. B., Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun. 65, 4770–4777 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandra Y.-J. P., Walls C., Alejandra Tortorici M., Wall A., McGuire A. T., Veesler D., Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsieh C. L., Goldsmith J. A., Schaub J. M., DiVenere A. M., Kuo H. C., Javanmardi K., Le K. C., Wrapp D., Lee A. G., Liu Y., Chou C. W., Byrne P. O., Hjorth C. K., Johnson N. V., Ludes-Meyers J., Nguyen A. W., Park J., Wang N., Amengor D., Lavinder J. J., Ippolito G. C., Maynard J. A., Finkelstein I. J., McLellan J. S., Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar M., Saha S., Structural insight into the role of novel SARS-CoV-2 E protein: A potential target for vaccine development and other therapeutic strategies. PLOS ONE 15, e0237300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Bert N., Tan A. T., Kunasegaran K., Tham C. Y. L., Hafezi M., Chia A., Chng M. H. Y., Lin M., Tan N., Linster M., Chia W. N., Chen M. I., Wang L. F., Ooi E. E., Kalimuddin S., Tambyah P. A., Low J.-G., Tan Y.-J., Bertoletti A., SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature 584, 457–462 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Keeble A. H., Turkki P., Stokes S., Khairil Anuar I. N. A., Rahikainen R., Hytonen V. P., Howarth M., Approaching infinite affinity through engineering of peptide-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 26523–26533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215–220 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Kutter E., Bryan D., Ray G., Brewster E., Blasdel B., Guttman B., From host to phage metabolism: Hot tales of phage T4’s takeover of E. coli. Viruses 10, 387 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., Marrama D., de Silva A. M., Frazier A., Carlin A. F., Greenbaum J. A., Peters B., Krammer F., Smith D. M., Crotty S., Sette A., Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., Mohammed A., Zhao C., Yang Y., Xie J., Ding C., Ma X., Weng J., Gao Y., He H., Jin T., Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 527, 618–623 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullaney J. M., Black L. W., Capsid targeting sequence targets foreign proteins into bacteriophage T4 and permits proteolytic processing. J. Mol. Biol. 261, 372–385 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Zuo X., Mattern M. R., Tan R., Li S., Hall J., Sterner D. E., Shoo J., Tran H., Lim P., Sarafianos S. G., Kazi L., Navas-Martin S., Weiss S. R., Butt T. R., Expression and purification of SARS coronavirus proteins using SUMO-fusions. Protein Expr. Purif. 42, 100–110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C. J., Cerikan B., Lu J. M., Peukes J., Xiong X., Krausslich H. G., Scheres S. H. W., Bartenschlager R., Briggs J. A. G., Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 588, 498–502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piccoli L., Park Y. J., Tortorici M. A., Czudnochowski N., Walls A. C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L. E., Bowen J. E., Acton O. J., Jaconi S., Guarino B., Minola A., Zatta F., Sprugasci N., Bassi J., Peter A., De Marco A., Nix J. C., Mele F., Jovic S., Rodriguez B. F., Gupta S. V., Jin F., Piumatti G., Presti G. L., Pellanda A. F., Biggiogero M., Tarkowski M., Pizzuto M. S., Cameroni E., Havenar-Daughton C., Smithey M., Hong D., Lepori V., Albanese E., Ceschi A., Bernasconi E., Elzi L., Ferrari P., Garzoni C., Riva A., Snell G., Sallusto F., Fink K., Virgin H. W., Lanzavecchia A., Corti D., Veesler D., Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell 183, 1024–1042.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robbiani D. F., Gaebler C., Muecksch F., Lorenzi J. C. C., Wang Z., Cho A., Agudelo M., Barnes C. O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T. Y., Viant C., Hurley A., Hoffmann H. H., Millard K. G., Kost R. G., Cipolla M., Gordon K., Bianchini F., Chen S. T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A. W., Waltari E., Pak J. E., Huey-Tubman K. E., Koranda N., Hoffman P. R., West A. P. Jr., Rice C. M., Hatziioannou T., Bjorkman P. J., Bieniasz P. D., Caskey M., Nussenzweig M. C., Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richmond P., Hatchuel L., Dong M., Ma B., Hu B., Smolenov I., Li P., Liang P., Han H. H., Liang J., Clemens R., Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: A phase 1, randomised, double-blind, placebo-controlled trial. Lancet 397, 682–694 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jordan M. B., Mills D. M., Kappler J., Marrack P., Cambier J. C., Promotion of B cell immune responses via an alum-induced myeloid cell population. Science 304, 1808–1810 (2004). [DOI] [PubMed] [Google Scholar]
- 50.J. Harcourt, A. Tamin, X. Lu, S. Kamili, S. K. Sakthivel, J. Murray, K. Queen, Y. Tao, C. R. Paden, J. Zhang, Y. Li, A. Uehara, H. Wang, C. Goldsmith, H. A. Bullock, L. Wang, B. Whitaker, B. Lynch, R. Gautam, C. Schindewolf, K. G. Lokugamage, D. Scharton, J. A. Plante, D. Mirchandani, S. G. Widen, K. Narayanan, S. Makino, T. G. Ksiazek, K. S. Plante, S. C. Weaver, S. Lindstrom, S. Tong, V. D. Menachery, N. J. Thornburg, Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. bioRxiv 2020.03.02.972935 [Preprint]. 3 March 2020. 10.1101/2020.03.02.972935. [DOI]
- 51.Leist S. R., Dinnon K. H. III, Schäfer A., Tse L. V., Okuda K., Hou Y. J., West A., Edwards C. E., Sanders W., Fritch E. J., Gully K. L., Scobey T., Brown A. J., Sheahan T. P., Moorman N. J., Boucher R. C., Gralinski L. E., Montgomery S. A., Baric R. S., A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183, 1070–1085.e12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson E. M., Goodwin E. C., Verma A., Arevalo C. P., Bolton M. J., Weirick M. E., Gouma S., McAllister C. M., Christensen S. R., Weaver J., Hicks P., Manzoni T. B., Oniyide O., Ramage H., Mathew D., Baxter A. E., Oldridge D. A., Greenplate A. R., Wu J. E., Alanio C., D’Andrea K., Kuthuru O., Dougherty J., Pattekar A., Kim J., Han N., Apostolidis S. A., Huang A. C., Vella L. A., Kuri-Cervantes L., Pampena M. B.; The UPenn COVID Processing Unit, Betts M. R., Wherry E. J., Meyer N. J., Cherry S., Bates P., Rader D. J., Hensley S. E., Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell 184, 1858–1864.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruttin A., Brussow H., Human volunteers receiving Escherichia coli phage T4 orally: A safety test of phage therapy. Antimicrob. Agents Chemother. 49, 2874–2878 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarker S. A., Berger B., Deng Y., Kieser S., Foata F., Moine D., Descombes P., Sultana S., Huq S., Bardhan P. K., Vuillet V., Praplan F., Brussow H., Oral application of Escherichia coli bacteriophage: Safety tests in healthy and diarrheal children from Bangladesh. Environ. Microbiol. 19, 237–250 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Mandala V. S., McKay M. J., Shcherbakov A. A., Dregni A. J., Kolocouris A., Hong M., Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat. Struct. Mol. Biol. 27, 1202–1208 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fougeroux C., Goksøyr L., Idorn M., Soroka V., Myeni S. K., Dagil R., Janitzek C. M., Søgaard M., Aves K. L., Horsted E. W., Erdogan S. M., Gustavsson T., Dorosz J., Clemmensen S., Fredsgaard L., Thrane S., Vidal-Calvo E. E., Khalifé P., Hulen T. M., Choudhary S., Theisen M., Singh S. K., Garcia-Senosiain A., Van Oosten L., Pijlman G., Hierzberger B., Domeyer T., Nalewajek B. W., Strøbæk A., Skrzypczak M., Andersson L. F., Buus S., Buus A. S., Christensen J. P., Dalebout T. J., Iversen K., Harritshøj L. H., Mordmüller B., Ullum H., Reinert L. S., de Jongh W. A., Kikkert M., Paludan S. R., Theander T. G., Nielsen M. A., Salanti A., Sander A. F., Capsid-like particles decorated with the SARS-CoV-2 receptor-binding domain elicit strong virus neutralization activity. Nat. Commun. 12, 324 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saunders K. O., Lee E., Parks R., Martinez D. R., Li D., Chen H., Edwards R. J., Gobeil S., Barr M., Mansouri K., Alam S. M., Sutherland L. L., Cai F., Sanzone A. M., Berry M., Manne K., Bock K. W., Minai M., Nagata B. M., Kapingidza A. B., Azoitei M., Tse L. V., Scobey T. D., Spreng R. L., Rountree R. W., DeMarco C. T., Denny T. N., Woods C. W., Petzold E. W., Tang J., Oguin T. H. III, Sempowski G. D., Gagne M., Douek D. C., Tomai M. A., Fox C. B., Seder R., Wiehe K., Weissman D., Pardi N., Golding H., Khurana S., Acharya P., Andersen H., Lewis M. G., Moore I. N., Montefiori D. C., Baric R. S., Haynes B. F., Neutralizing antibody vaccine for pandemic and pre-emergent coronaviruses. Nature 594, 553–559 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arunachalam P. S., Walls A. C., Golden N., Atyeo C., Fischinger S., Li C., Aye P., Navarro M. J., Lai L., Edara V. V., Roltgen K., Rogers K., Shirreff L., Ferrell D. E., Wrenn S., Pettie D., Kraft J. C., Miranda M. C., Kepl E., Sydeman C., Brunette N., Murphy M., Fiala B., Carter L., White A. G., Trisal M., Hsieh C. L., Russell-Lodrigue K., Monjure C., Dufour J., Spencer S., Doyle-Meyer L., Bohm R. P., Maness N. J., Roy C., Plante J. A., Plante K. S., Zhu A., Gorman M. J., Shin S., Shen X., Fontenot J., Gupta S., O’Hagan D. T., Van Der Most R., Rappuoli R., Coffman R. L., Novack D., McLellan J. S., Subramaniam S., Montefiori D., Boyd S. D., Flynn J. L., Alter G., Villinger F., Kleanthous H., Rappaport J., Suthar M. S., King N. P., Veesler D., Pulendran B., Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 594, 253–258 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Zhao J. C., Zhao Z. D., Wang W., Gao X. M., Prokaryotic expression, refolding, and purification of fragment 450-650 of the spike protein of SARS-coronavirus. Protein Expr. Purif. 39, 169–174 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y., Tai W., Yang J., Zhao G., Sun S., Tseng C. K., Jiang S., Zhou Y., Du L., Gao J., Receptor-binding domain of MERS-CoV with optimal immunogen dosage and immunization interval protects human transgenic mice from MERS-CoV infection. Hum. Vaccin. Immunother. 13, 1615–1624 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Table S1
Legend for movie S1
Movie S1