No evidence of vaccine-enhanced disease was observed following inactivated SARS-CoV-2 vaccine in ferrets or rhesus macaques.
Abstract
There is an urgent requirement for safe and effective vaccines to prevent COVID-19. A concern for the development of new viral vaccines is the potential to induce vaccine-enhanced disease (VED). This was reported in several preclinical studies with both SARS-CoV-1 and MERS vaccines but has not been reported with SARS-CoV-2 vaccines. We have used ferrets and rhesus macaques challenged with SARS-CoV-2 to assess the potential for VED in animals vaccinated with formaldehyde-inactivated SARS-CoV-2 (FIV) formulated with Alhydrogel, compared to a negative control vaccine. We showed no evidence of enhanced disease in ferrets or rhesus macaques given FIV except for mild transient enhanced disease seen 7 days after infection in ferrets. This increased lung pathology was observed at day 7 but was resolved by day 15. We also demonstrate that formaldehyde treatment of SARS-CoV-2 reduces exposure of the spike receptor binding domain providing a mechanistic explanation for suboptimal immunity.
INTRODUCTION
Novel coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a global pandemic with a cumulative total of more than 143 million cases and 3.0 million deaths reported as of 21 April 2021 (1). Consequently, there is an urgent requirement to develop safe and effective vaccines to prevent COVID-19 (2). Currently, 91 vaccine candidates are in clinical evaluation with 184 listed as in preclinical evaluation (World Health Organization draft landscape of COVID-19 vaccines–22 April 2021). Nine vaccines have been approved for emergency use in various countries including two nonreplicating viral vector vaccines, three inactivated virus vaccines, and two vaccines based on mRNA technology (3). Efficacy l results from phase 3 studies are available for mRNA-based vaccines (4, 5) and viral vector vaccines (6, 7), which expresses a codon-optimized full-length spike protein (S). Whole virus vaccines grown in Vero cells and inactivated with β-propiolactone are also in use (8–10). These vaccines have also been evaluated for protection in nonhuman primates following challenge with SARS-CoV-2. The virus induces only mild to moderate disease in macaques, but these vaccines reduce viral loads and pathology in the upper and lower respiratory tracts to varying degrees (8, 9, 11, 12).
A concern for the development of new viral vaccines is the potential to induce vaccine-enhanced disease (VED) (13), which has been associated with prior preclinical studies of both SARS and Middle East respiratory syndrome (MERS) vaccines. The most studied mechanism of VED is antibody-dependent enhancement (ADE) of disease, reviewed recently by Arvin et al. (14) and Lee et al. (15). It has been suggested that ADE could be a consequence of low-affinity antibodies that bind to viral proteins but have limited neutralizing activity (16). The vaccine enhancement of disease by ADE mechanisms was described in children given formaldehyde (FA)–inactivated respiratory syncytial virus (RSV) vaccines in the 1960s (17), measles vaccines (18), and in dengue hemorrhagic fever due to secondary infection with a heterologous dengue serotype (19).
There is limited evidence of ADE with SARS-CoV-1 vaccines in animal models and, while it has not been reported in the majority of vaccine studies, a study that used formalin or ultraviolet-inactivated SARS-CoV-1 observed that older mice developed pulmonary pathology with an eosinophil infiltrate (20). A further study demonstrated protection in mice following immunization with formalin or ultraviolet light–inactivated SARS-CoV-1, but animals developed eosinophilic pulmonary infiltrates (21). A modified vaccinia virus Ankara expressing S protein (MVA-S) was not protective in ferrets challenged with SARS-CoV-1, but liver inflammation was noted (22). Formalin-inactivated SARS-CoV-1 vaccines were protective in rhesus macaques (23) but also promoted lymphocytic infiltrates and alveolar oedema with fibrin deposition after challenge (24). Likewise, MVA-S showed protection in one study (25) but greater occurrence of diffuse alveolar damage than seen in control animals following challenge. Fortuitously, VED has not been reported in the numerous SARS-CoV-2 efficacy animal challenge vaccine studies published to date. However, these studies were primarily designed to assess efficacy and not VED.
We have developed a ferret intranasal SARS-CoV-2 infection model where viral shedding and mild lung pathology is observed and rechallenged animals are fully protected (26). We have also evaluated both rhesus and cynomolgus macaques for their susceptibility to SARS-CoV-2 challenge and showed the development of pulmonary lesions in both species which are equivalent to those seen in mild clinical cases in humans (27). To interrogate the potential for VED in ferrets and rhesus macaques to support future safety studies on novel COVID-19 vaccines, we prepared a FA-inactivated SARS-CoV-2 vaccine (FIV), formulated in Alhydrogel. In this study design, we aim to induce a suboptimal immune response that may promote VED, immunized animals are challenged with SARS-CoV-2 14 days after vaccination. Clinical signs, viral shedding, and pathology are monitored following challenge, and immune responses are characterized before and after infection. No enhanced pathology is observed in either species except for transient enhanced pathology at 7 days after infection in ferrets, and we present a possible mechanism for suboptimal immunity induced by FA-inactivated SARS-CoV-2 spike.
RESULTS
Characterization of FA-inactivated vaccine
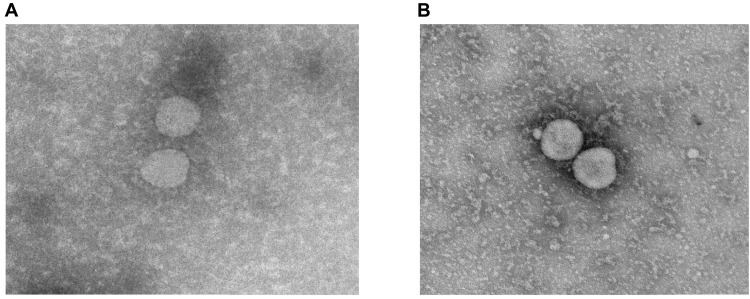
Transmission electron microscopy (Fig. 1) showed that the washing and FA inactivation procedure resulted in virus particles that appeared similar to typical coronavirus morphology with a complete ring of peplomers/spikes on each particle (Fig. 1, A and B). SDS–polyacrylamide gel electrophoresis (PAGE) analysis (fig. S1A) shows that most of the protein bands seen in the medium only (lane 2) are also seen in the live wild-type virus (lane 3) and the FIV (lane 4), indicating that these proteins are likely to be components of the culture medium, including fetal bovine serum (FBS) and host cell proteins. The medium-only protein species are visibly reduced in intensity following washing using the centrifugal concentrator in the FIV although this was sixfold concentrated. Notably, protein species in the 62- to 100-kDa range were almost absent in FIV. The Western blot analysis (fig. S1, B and C) confirms that both wild-type live virus and FIV react with antibodies to both SARS-CoV-2 spike receptor binding domain (RBD) and nucleocapsid. Western blot with the NIBSC (National Institute of Biological Standards and Control, UK), SARS-CoV-2 neutralizing, MERS convalescent serum (S3) standard has limited reactivity with proteins found in the medium (fig. S1B). However, a virus-specific band corresponding to the spike-RBD band on the specific antisera blot is detected at approximately 160 kDa in unwashed virus preparation and at 180 kDa in FIV with the increased molecular weight likely to be a consequence of FA fixation cross-linking this protein.
Fig. 1. Electron microscopy of SARS-CoV-2 virus.
Representative transmission electron microscopy images of (A) the initial SARS-CoV-2 virus preparation and (B) following FA inactivation and washing to remove medium constituents.
SARS-CoV-2 infection in control and FIV-vaccinated ferrets
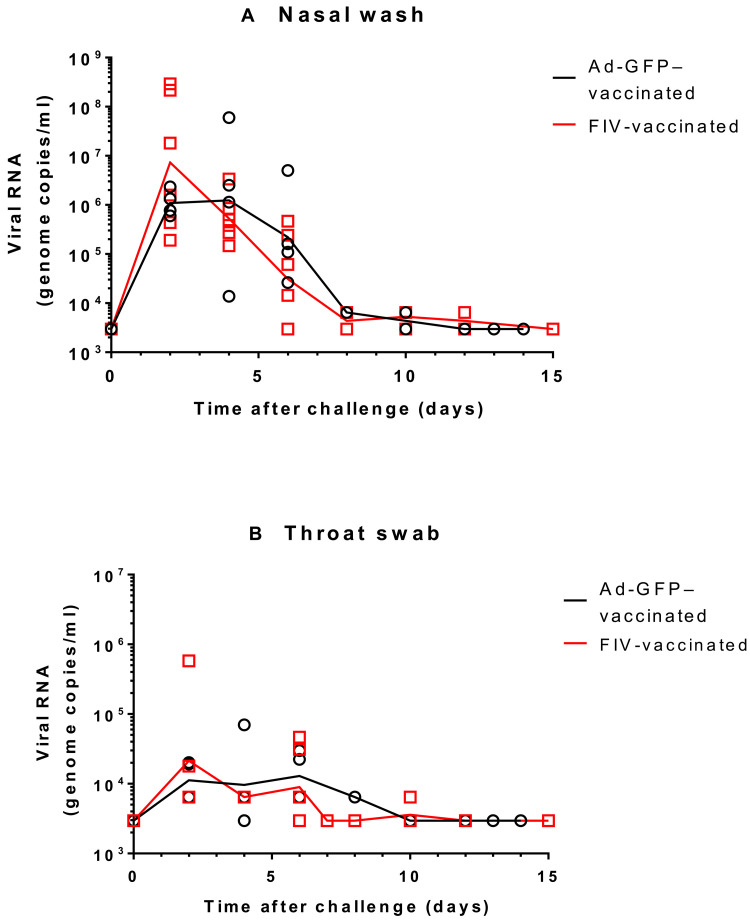
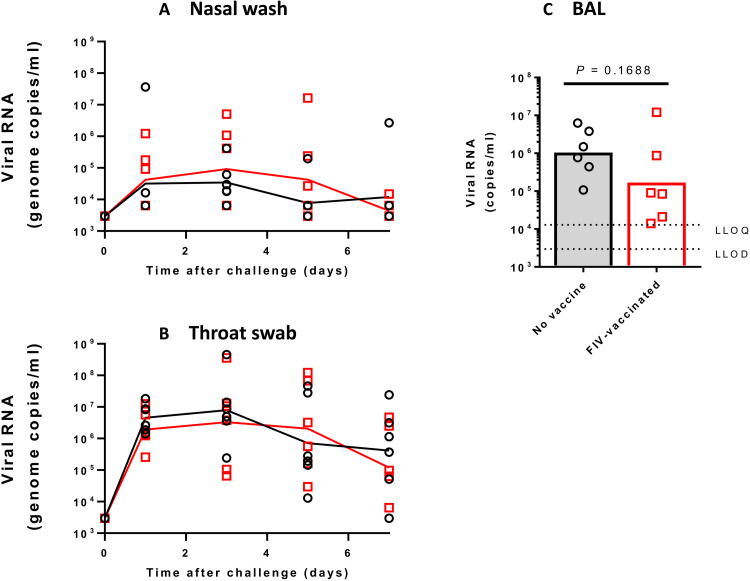
Ferrets vaccinated with either a recombinant adenovirus expressing green fluorescent protein (Ad-GFP) or FIV were challenged intranasally with 1 ml of Victoria/1/2020 SARS-CoV-2 at 5 × 106 plaque-forming units (PFU) (28). The sampling schedule is shown in Table 1. The high-titer stock of challenge virus was prepared (passage 3), and quality control sequencing showed that it was identical to the original stock received from the Doherty Institute and did not contain a commonly reported 8–amino acid deletion in the furin cleavage site (29). Both groups displayed similar viral genome copies in nasal wash samples, which continued to be detected until the end of the experiment at day 15 after challenge (Fig. 2A). Similar to a previous study (26), the peak in viral RNA shedding was seen between days 2 and 4 after challenge for all ferrets in both groups. The majority of virus detected in nasal wash occurred between challenge and day 8 after challenge. Viral RNA detected in nasal wash and throat swab samples was also shown to be approximately threefold higher in the FIV group at day 2 after challenge. A similar trend was seen in both groups during the first week after challenge although the RNA genome copies measured were substantially lower in the throat swab than in the nasal wash samples (Fig. 2B). Overall, there were no substantial differences between groups in virus shedding from either the nose or throat. Neither group of animals showed weight loss due to the infection (fig. S2). The apparent difference in weight gain in the FIV-vaccinated group was due to the necropsy sampling of the two lightest animals from this group on day 7. No fever was seen at any time in either group of ferrets after infection (fig. S2).
Table 1. Experimental animal groups.
| Group | Number of animals |
Vaccination—days before
challenge |
Sampling |
Euthanasia days after
challenge |
| Ferret – Ad-GFP | 4 females | 28 | Blood for serology before vaccination, on day of challenge, and at necropsy. |
6 days; n = 2 |
| 13 days; n = 1 | ||||
| 14 days; n = 1 | ||||
| Ferret – FIV | 6 females | 14 | Nasal washes and throat swabs on days 0, 2, 4, 6, 8, 10, and 12 after challenge and necropsy |
7 days; n = 2 |
| 14 days; n = 4 | ||||
| Rhesus macaque – no vaccine | 3 males | 14 | Blood serology before vaccination, on day of challenge and day 7 after challenge. |
7 days; n = 6 |
| 3 females | ||||
| Rhesus macaque – FIV | 3 males | – | Nasal wash and throat swab on days 1, 3, 5, and 7 after challenge. BAL at necropsy. |
7 days; n = 6 |
| 3 females |
Fig. 2. Detection of SARS-CoV-2 RNA in ferret respiratory samples.
Viral RNA in ChAdOx-1-GFP– and FIV-vaccinated ferrets was quantified by reverse transcription polymerase chain reaction (RT-PCR) in (A) nasal washes and (B) throat swabs. Lines plotted are the geometric mean genome copies/ml.
Pathology following SARS-CoV-2 infection in ferrets
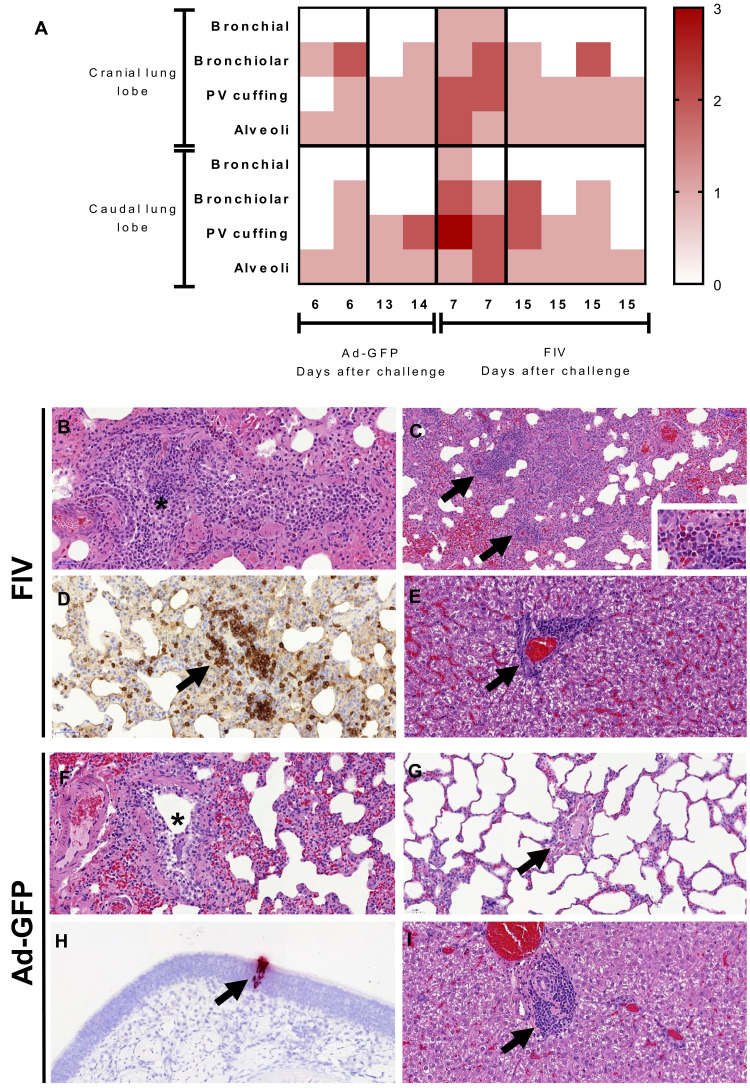
We performed sequential culls on days 6 to 7 and 13 to 14 to study the potential for VED during and after resolution of SARS-CoV-2 infection. The lung histopathology scores for individual Ad-GFP– and FIV-vaccinated ferrets are shown in the heatmap in Fig. 3A. Samples were obtained from two animals from each group early in the infection (Ad-GFP at day 6 after challenge and FIV at day 7 pc). The remaining animals were euthanized at days 13 to 14 pc (Ad-GFP) and day 15 pc (FIV). All assessments, including bronchiolar, bronchial, and interstitial infiltrates together with perivascular cuffing, were scored as minimal or mild in the Ad-GFP–vaccinated animals with a greater number of mild or moderate scores in the FIV-vaccinated ferrets at the early time point (6 to 7 days after challenge). One animal from the Ad-GFP group showed mild lesions compatible with acute bronchiolitis and perivascular/peribronchiolar cuffing (Fig. 3, F and G). The other animal from this group showed only occasional minimal bronchiolar infiltrates. Both animals from the FIV group at 7 days pc showed more remarkable changes, with mild to moderate bronchiolitis (infiltrates within the bronchioles and occasionally bronchi) and inflammatory foci within the parenchyma (Fig. 3B). Moreover, perivascular cuffing was observed frequently (Fig. 3C), with the infiltrates being mostly mononuclear cells, including CD3+ T lymphocytes identified by immunohistochemistry staining (Fig. 3D). Occasionally, neutrophils and eosinophils were also present (Fig. 3C, inset). The cuffing also affected numerous airways (Fig. 3C). Because of the small numbers of animals, the differences in scores observed between FIV- and Ad-GFP–vaccinated groups did not reach significance. In contrast, at 13 to 15 days after challenge, the lesions observed were minimal to mild with no obvious differences between groups (Fig. 3A).
Fig. 3.
(A) Heatmap showing the individual lung histopathology scores for each ferret and parameter following FIV or Ad-GFP vaccination and challenged with SARS-CoV-2 and culled at 6/7 days and 13/15 days after challenge. Histopathology of FIV-vaccinated (B to E) and Ad-GFP–vaccinated (F to I) ferrets. (B) Inflammatory infiltrates within a bronchiole (*), with abundant mononuclear cells but also some neutrophils and eosinophils. Hematoxylin and eosin (H&E), 200×. (C) Multiple inflammatory infiltrates surrounding blood vessels (perivascular cuffing, arrows). H&E, 100×. The infiltrates are composed mostly of macrophages and lymphocytes, but abundant eosinophils can also be observed in some areas within the infiltrates (inset; H&E, 400×). (D) Perivascular cuff (arrow) with abundant mononuclear cells, many of them identified as CD3+ T lymphocytes. Immunohistochemistry, 200×. (E) Periportal mononuclear inflammatory infiltrate in the liver (mild multifocal hepatitis). H&E, 400×. (F) Mild inflammatory infiltrate within a bronchiole (*). H&E, 200×. (G) Blood vessel (arrow) within the lung parenchyma not showing any perivascular cuffing. H&E, 200×. (H) In situ hybridization (ISH) detection of SARS-CoV-2 RNA in a small focus of epithelial and sustentacular cells within the nasal cavity. ISH, 200×. (I) Periportal mononuclear inflammatory infiltrate in the liver (mild multifocal hepatitis). H&E, 400×.
RNAScope in situ hybridization (ISH) technique was used to detect viral RNA in lung and nasal cavity tissue sections. Only very few occasional scattered cells were found positive to viral RNA in the lung at day 6/7, which were within the alveolar walls and not related to the presence of lesions. No differences were observed between groups. Viral RNA was also found only as small foci of positive cells (epithelial and or sustentacular) within the olfactory and respiratory mucosa in only one animal from the Ad-GFP group at day 6 pc (Fig. 3H). No obvious lesions were observed in any other organ except for the liver, which showed a variable degree of multifocal hepatitis, mild to moderate in all animals (Fig. 3, E and I).
Immune responses to FIV in ferrets
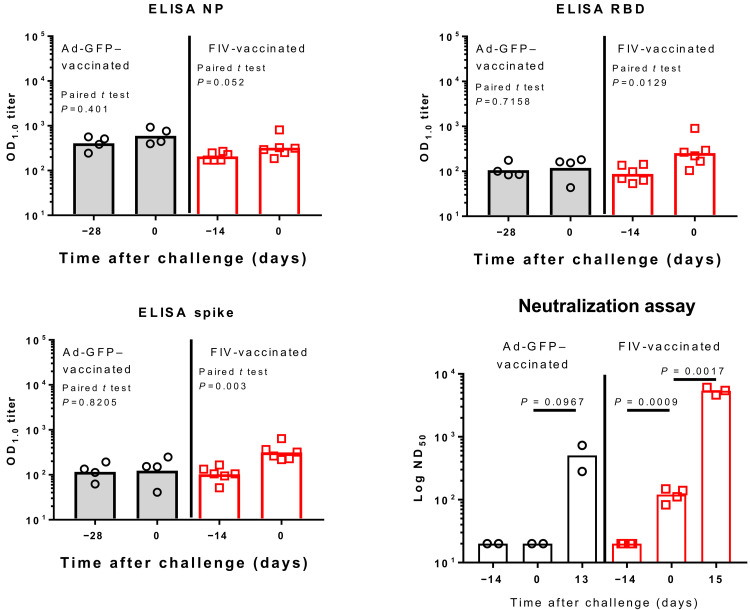
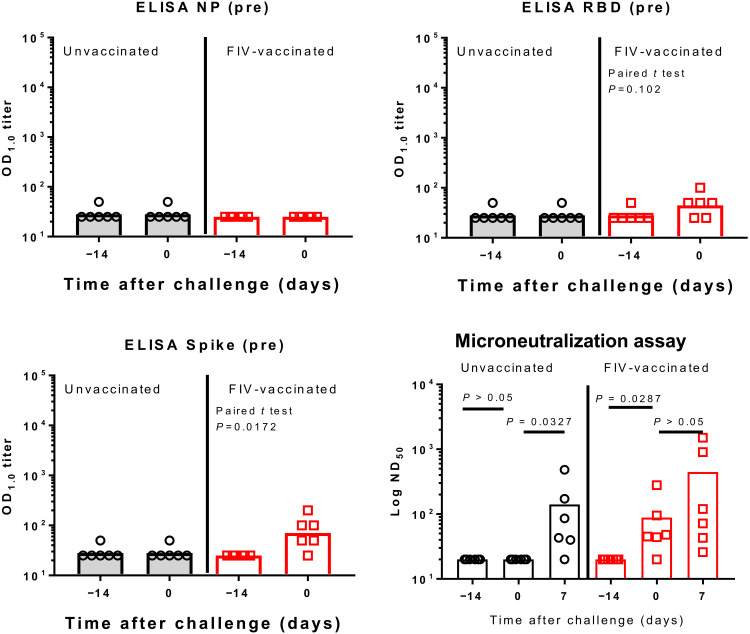
Ad-GFP–vaccinated animals showed no immune responses before SARS-CoV-2 challenge (Fig. 4). FIV-vaccinated animals produced significant increases in immunoglobulin G (IgG) after vaccination against SARS-CoV-2 spike and spike RBD. The response to nucleoprotein (N) was not significant. Modest rises [geometric mean titer (GMT) = 89, P = 0.002] in neutralizing antibody titers were seen in sera from the FIV-vaccinated animals with a rapid rise in neutralizing antibody titers after challenge indicating immune priming by the FIV. At termination, the GMTs for FIV-vaccinated animals were 5356 and 453 for the two remaining Ad-GFP–vaccinated animals. Enzyme-linked immunospot (ELISpot) assays applied to splenocytes isolated on day 6 pc (Ad-GFP–vaccinated) or day 7 pc (FIV-vaccinated) show that animals vaccinated with FIV made more pronounced responses to whole live virus, membrane, and nucleocapsid peptide pools with both groups showing similar low responses to virus spike peptides (fig. S3A). This early interferon-γ (IFNγ) response after infection is consistent with an anamnestic response in the FIV-vaccinated animals following priming with viral antigens. The lack of cellular immune response to spike is interesting and might indicate that FIV promotes a skewed immune response. Responses measured in peripheral blood mononuclear cells (PBMCs) were much lower than those seen in splenocytes.
Fig. 4. Serological response to Ad-GFP and FIV in ferrets.
IgG was quantified by enzyme-linked immunosorbent assay (ELISA) to recombinant nucleocapsid protein (NP), receptor binding domain (RBD), and full-length trimeric and stabilised spike protein (Spike). Bars are geometric mean titers. The significance of any difference from pre- to postvaccination is shown, determined by a paired t test. The plaque reduction neutralization 50% titer (PRNT50) is also shown with samples obtained pre- and postvaccination and following SARS-CoV-2 challenge. Bars are geometric mean PRNT50 titers. ND50, 50% neutralizing dilution; OD1.0, optical density at 1.0 nm.
SARS-CoV-2 infection in control and FIV-vaccinated rhesus macaques
Following infection with 5 × 106 PFU Victoria/1/2020 SARS-CoV-2 given in 2 ml by the intratracheal route and 1 ml intranasally, viral RNA was quantified in nasal wash and throat swab samples. Viral RNA was detected in both samples during the experiment with peak values detected by day three and a decline thereafter (Fig. 5, A and B). There was no difference between viral copies detected in macaques that received FIV or no vaccine. Bronchoalveolar lavage was obtained on necropsy at day 7, and lower geometric mean viral RNA copies/ml (P = 0.168) were measured in macaques that received FIV than no-vaccine controls (Fig. 4C). No significant changes in body temperature (fig. S2C) were observed. Slight weight loss was observed in both groups (fig. S2D), but no adverse clinical signs were recorded for any macaque despite frequent monitoring during the study period.
Fig. 5. Detection of SARS-CoV-2 RNA in macaque respiratory samples.
Viral RNA in unvaccinated and FIV-vaccinated macaques was quantified by RT-PCR in (A) nasal washes, (B) throat swabs, and (C) bronchoalveolar lavage (BAL). Lines plotted are the geometric mean genome copies/ml. LLOD, lower limit of detection; LLOQ, lower limit of quantification.
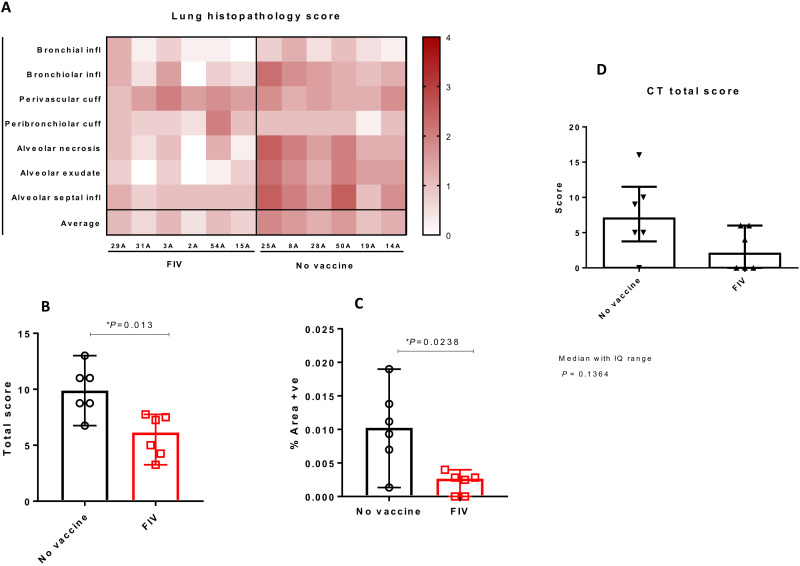
Images from computed tomography (CT) scans collected 5 days after challenge were examined by an expert thoracic radiologist with experience of nonhuman primate prior CT interpretation and human COVID-19 CT features, blinded to the clinical status. Pulmonary abnormalities that involved less than 25% of the lung and reflected those characteristics of SARS-CoV-2 infection in humans were identified in three of the FIV group and five of the unvaccinated group. Where reported, disease was predominantly bilateral (two of three FIV and five of six unvaccinated) with a similar peripheral distribution through the lung lobes reported in the FIV vaccinated and unvaccinated macaques. Ground glass opacity (GGO) was observed in all the macaques showing abnormal lung structure, with the exception of one FIV-vaccinated animal in which consolidation was identified. Other featured characteristics of human COVID (reverse halo, perilobular, nodules, and pulmonary embolus) were not observed in any of the macaques in either group. Evaluation of pulmonary disease burden using a scoring system designed to discriminate differences between individual macaques with low disease volume revealed a nonsignificant trend (P = 0.1364) for a reduction in the total CT score in the FIV group compared to the scores attributed to macaques in the unvaccinated group (Fig. 6D). Similarly, the FIV vaccine reduced both the amount of abnormalities induced (pattern score) and distribution of disease (zone score) (fig. S4, H and I).
Fig. 6. Histopathology and CT scan analysis in rhesus macaques.
(A) Heatmap showing the individual lung histopathology scores for each animal and parameter and the average severity for each animal. (B) Total histopathology scores in nonvaccinated and FIV animals showing a significant reduction in severity in the FIV group [P = 0.013, Mann-Whitney U tests; boxes and whiskers show median ± 95% confidence interval (CI)]. (C) Percentage of area positively stained with ISH RNAScope (viral RNA) in nonvaccinated and FIV animals showing a significant reduction in the FIV group (P = 0.0238, Mann-Whitney U tests; boxes and whiskers show median ± 95% CI). (D) Plot shows the total CT scores in nonvaccinated and FIV-vaccinated animals showing a nonsignificant trend for reduction in severity in the FIV group (P = 0.1364, Mann-Whitney U test); box plots show the experimental group median with ±interquartile (IQ) range indicated by box whiskers, and symbols show scores measured in individual animals.
Pathology following SARS-CoV-2 infection in rhesus macaques
Pathological changes were found in the lungs of all SARS-CoV-2–infected macaques and consisted of multiple areas of mild to moderate bronchiolo-alveolar necrosis, inflammatory cell infiltration, and type II pneumocyte proliferation. Mild perivascular and peribronchiolar cuffing was also observed. The lung pathology scores for individual macaques are shown in Fig. 6 and with milder pathological changes observed in the FIV-vaccinated macaques. The total pathology score for the no-vaccine group was greater than the FIV group (P = 0.013; Fig. 6B). RNAScope analysis of the percentage of area positively stained for SARS-CoV-2 RNA showed a greater lung area infected in the no-vaccine group than FIV-vaccinated macaques (P = 0.0238; Fig. 6C).
Immune responses to FIV in rhesus macaques
Serum from control macaques obtained on the day of challenge did not show any N-, RBD-, or S-specific IgG, but rises in RBD- and S-specific IgG were detected in serum from the FIV-vaccinated macaques (Fig. 7). FIV-vaccinated animals also showed a rise in neutralizing antibody titer on the day of challenge (P = 0.0287). Both groups showed a rise in neutralizing antibody titer 7 days following challenge (Fig. 7). Similarly, on the day of challenge, a higher frequency of spike-specific IFNγ-secreting cells was measured by ELISpot assay in the FIV group compared to that determined in the unvaccinated group suggesting the induction of a modest but significant (P = 0.0433) SARS-CoV-2–specific cellular response (fig. S3B). The trend reversed 6 to 8 days after challenge when frequencies were assessed at the end of the study, with higher frequencies of spike-specific IFNγ-secreting cells measured in the unvaccinated group compared to the FIV group in both PBMCs and spleen cells.
Fig. 7. Serological response in unvaccinated and FIV-vaccinated macaques.
IgG was quantified by ELISA to recombinant NP, RBD, and full-length trimeric and stabilized spike protein (Spike). Bars are geometric mean titers. The significance of any difference from pre- to postvaccination is shown, determined by a paired t test. The microneutralization 50% titer (ND50) is also shown with samples obtained pre- and postvaccination and following SARS-CoV-2 challenge. Bars are geometric mean ND50 titers.
Immunophenotyping flow cytometry assays were applied to whole blood samples collected immediately before and 14 days after FIV vaccination, as well as at days 3 and 7 after SARS-CoV-2 challenge to explore potential vaccine-induced changes in the cellular immune compartment that might influence the course of the immune response following infection (fig. S3). CD4 and CD8 T cells expressing the immune checkpoint signaling receptor programmed cell death protein 1 (PD-1) increased significantly following FIV vaccination (P = 0.03) with further significant increases observed in PD-1 expressing CD4 T cell populations following SARS-CoV2 infection in both FIV-vaccinated and unvaccinated groups (both P = 0.03) (fig. S3D). Similarly, FIV vaccination led to a significant increase in CD4 regulatory T cells expressing CD25 and CD127 (fig. S3E), indicating that, alongside the proinflammatory cellular response evident in the antigen-specific IFNγ ELISpot profiles, FIV vaccination also induced T cell populations with a more tolerogenic phenotype.
Effects of FA on SARS-CoV-2 spike
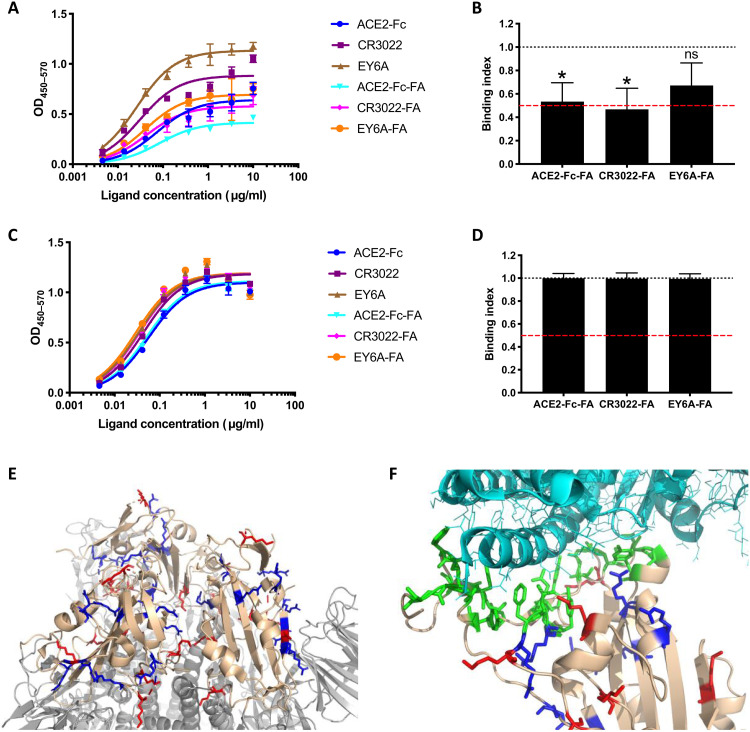
FA treatment of SARS-CoV-2 virus will cross-link viral proteins, of which the S glycoprotein trimer is the target of most neutralizing antibodies. Cross-linking of S may modify its antigenicity, potentially altering elicitation of neutralizing antibodies. To analyze the effects of FA on the S trimer and the isolated RBD, soluble antigens were captured onto enzyme-linked immunosorbent assay (ELISA) plates using either anti-Myc tag (S-Myc) or anti-Fc (RBD-Fc), respectively, to maintain their native conformation and treated or not with FA using the same protocol as for inactivation of whole virus: 0.02% for 72 hours at room temperature. Samples were then tested for binding of RBD ligands, either soluble (s)ACE2-Fc or the RBD binding monoclonal antibodies (mAbs) CR3022 and EY6A that interact with RBD surfaces nonoverlapping the angiotensin-converting enzyme 2 (ACE2) binding site. Binding curves revealed that ligands binding to FA-treated S protein gave substantially lower maximum binding than that to the untreated S counterpart (Fig. 8A). Area under the curve (AUC) analysis revealed that binding was significantly reduced for sACE2-Fc and CR3022 and had a trend to reduction for EY6A (Fig. 8B). The reduction for sACE2 and CR3022 was almost precisely twofold, suggesting either that the FA treatment had reduced the binding activity of a subset of RBD domains or that half of the FA-treated S trimers were in a non-RBD available conformation. To differentiate between these two possibilities, we tested binding directly to the isolated untreated or FA-treated RBD (Fig. 8C). Notably, FA treatment had no effect on RBD ligand binding, with AUC analysis showing near identical values for FA-treated and untreated RBD (Fig. 8D). It therefore seems most likely that FA treatment is stabilizing a population of S trimers in the “RBD down” conformation which would be unable to engage ACE2. Recent cryo-EM structures imply that the S trimer is 50% one RBD up and 50% all RBD down at equilibrium (30). Therefore, cross-linking of a population of trimers would most likely fix this 1:1 equilibrium, allowing only half of the trimers to expose the one up RBD. By contrast, untreated S trimer would be free to sample both conformations, allowing progressive ACE2 occupancy to maximum of the one up RBD trimer conformation over time. This would explain the twofold decrease in occupancy of FA-treated S trimers shown above. To further interrogate this, we modeled the location of lysine and arginine residues, the side chains of which are targets for FA attack, in the S trimer and RBD structures. Figure 6E focuses on the location of lysines and arginines in the RBD-down S trimer, revealing a large number proximal to the RBD-trimer interface that might be cross-linked to prevent RBD movement. Figure 6F shows that, while there are lysine and arginine residues proximal to the RBD-ACE2 binding interface (green), there are none within the interface, implying that FA treatment would not directly affect RBD-ACE2 binding. This modeling is therefore consistent with the idea that FA cross-linking will lock 50% of trimers into the all RBD-down state, reducing access of the RBD to ligand binding and B cell recognition. Such modified antigenicity would probably translate into reduced RBD immunogenicity, reducing antibody production against this neutralizing antibody-eliciting surface.
Fig. 8. FA treatment reduces S trimer binding to ACE2.
(A) Representative ELISA with S trimer captured onto the ELISA plate using anti-Myc mAb 9E10, untreated or treated with FA for 72 hours at room temperature before addition of ligands at the concentrations shown and ligand detection using anti-human Fc-HRP. (B) Area under the curve (AUC) analysis was used to determine the binding index, where 1 is equivalent to binding between untreated and FA-treated S trimer, and 0 is equivalent to zero binding to FA-treated trimer. The red dashed line represents 50% binding. n = means of pooled data from three independent ELISAs, each performed in triplicate. *P < 0.05 compared to unmodified condition; Student’s t test after normality testing; ns, not significant. (C) Representative ELISA with RBD-Fc captured onto ELISA plate with anti-human-Fc, untreated or treated with FA as for the S trimer. (D) AUC analysis was used to determine the binding index as for S trimer above, n = means of pooled data from three independent ELISAs, each performed in triplicate. Student’s t test after normality testing revealed no significant differences. (E) Model prepared from PBD6LZG with 3 RBD “down.” RBD rendered in beige, S trimer in gray, lysines in red, arginines in blue. Close-up of RBD-ACE2 interface with RBD in beige, ACE2 in light cyan, binding site in green, lysines in red, arginines in blue.
DISCUSSION
Rapid development of vaccines to prevent COVID-19 disease is in progress (2). Safety is of primary importance for vaccines that are administered to healthy people. Thus, vaccines not only must be thoroughly assessed for reactogenicity and longer-term safety (31) but also to ensure that they do not cause any enhancement of disease (32). VED, which can be mediated by ADE, have been described for respiratory virus vaccines. The potential risks from COVID-19 vaccines have been described by Graham (33). Enhanced disease can potentially be mediated by antibodies that bind virus without neutralizing activity, which cause disease through increased viral replication or via formation of immune complexes that deposit in tissues and activate complement pathways associated with inflammation (13). T helper 2 cell (TH2)–biased immune responses have also been associated with ineffective vaccines that can lead to enhanced disease following infection (34) (35). To understand the risks of VED caused by SARS-CoV-2 vaccines, it will be of great benefit to produce a positive control vaccine that enhances disease in an animal model following challenge so this endpoint can be defined and the mechanisms can be understood and avoided in vaccines prepared for human use.
As examples of VED have been observed following the use of an FIV, e.g., RSV (36) and measles (18), we have prepared a killed SARS-CoV-2 vaccine by FA fixation of the virus. FA-fixed SARS-CoV-1 vaccines have been shown to induce enhanced disease (20, 21, 23, 24), although the mechanism for this is not understood. It has been suggested that nonviral components of FA-inactivated preparations, such as cellular components and debris or medium constituents, may also play a role in enhanced disease. In a cotton rat model of RSV VED, cell culture contaminants were a major driver of lung pathology, which was exacerbated by the FIV and RSV challenge (37). The SARS-CoV-2 FIV prepared in this study also contained cell and medium components, although the content was significantly reduced by washing using a centrifugal concentrator. Another factor in the FIV vaccine design was to use Alhydrogel as an adjuvant that is known to induce TH2-biased immune responses (38). In addition, as a suboptimal immune response has been suggested to be associated with VED (16), we chose to challenge the ferrets and rhesus macaques 14 days after intramuscular delivery of a single dose of FIV.
A study investigating the potential of the hamster model of SARS-CoV-2 for VED has recently been published (39). These workers also used a formalin-inactivated SARS-CoV-2 virus vaccine given either 3 days before challenge or 2 days following challenge. The hamster provides a more severe model of disease, but the opportunities for detailed pathology and immunology investigations are more limited than for ferrets and primates. VED was not observed in the hamsters following challenge with this suboptimal vaccine regimen. The pneumonia was not more severe, and no excess eosinophilic infiltration was observed in the vaccinated animals.
The SARS-CoV-2 infection in the Ad-GFP– or FIV-vaccinated ferrets followed a similar course to that observed in our previous study (26) with peak viral RNA shedding between 2 and 4 days after challenge. Note that higher viral loads were detected in the upper respiratory tract of FIV-vaccinated animals at day 2 after challenge, but after this sampling time, very similar genome copy values were obtained. There were no differences in temperature, weight (fig. S2), or any other clinical signs between the two groups.
Consistent with the higher viral load at day 2, the lung histopathology from the two FIV-vaccinated ferrets necropsied at day 7 was more severe than the two Ad-GFP–vaccinated animals necropsied at day 6. The semiquantitative scoring system was used to discriminate the severity of lesions between animals and groups. Although the number of animals was small, and although lung pathology was not severe in any case, we observed some unique differences in the FIV-vaccinated ferrets. At 6 to 7 days after challenge, a higher severity was observed in animals from the FIV group (combined total score, 24) compared to the Ad-GFP control (combined total score, 10). This included eosinophilic infiltrate and perivascular cuffing that were not observed in the control-vaccinated ferrets. The lung pathology in the ferret model (26) was quite transient, and thus, at 13 to 15 days after challenge, while there was some individual variability present, both groups showed mild pathology. Eosinophilic infiltrates have previously been observed for SARS in mice and rabbits (40, 41).
Multifocal mild to moderate hepatitis has also been described as a potential adverse effect of SARS-CoV-1 vaccines (42). However, these lesions are found as a background finding for this species in many experimental studies, although viral infections, systemic or in the gastrointestinal tract, have also been related to the presence of these periportal inflammatory infiltrates (43). Because of the variability in severity and the fact that naïve ferrets frequently show some degree of hepatitis, the interpretation of this lesion must be taken cautiously.
Following this observation of mild transient enhanced disease in the two FIV-vaccinated ferrets culled at day 7, we have tested the same FIV in six macaques along with six unvaccinated controls. We have previously compared the course of SARS-CoV-2 infection in both rhesus and cynomolgus macaques and showed virus replication in the upper and lower respiratory tract with pulmonary lesions resembling mild COVID-19 in humans (27). Macaques allow for a more detailed examination of lung pathology using precise scoring system devised in our recent study. We also have considerable experience of in-life CT scanning of macaques (27, 44, 45), which allows further aspects of lung pathology to be characterized at time points before necropsy. This detailed analysis has revealed no evidence of enhanced disease in macaques at any time point but rather that the FIV provided some protection against the mild-to-moderate lung pathology observed in the unvaccinated control macaques.
The presence of inflammatory infiltrates, and particularly perivascular cuffing, has been described as a feature potentially related to VED in SARS-CoV-1 preclinical vaccine trials (21, 23, 24). In our study, these infiltrates were always of mild to moderate severity.
Characterization of the immune response to the FIV vaccine before challenge in both species confirmed the expectation of modest immunity to SARS-CoV-2 spike. A significant rise in anti-S IgG, but not anti-N IgG was detected by ELISA. The geometric mean neutralizing titer of 89 seen in ferrets and 61 in macaques was low compared to that observed in primate studies with candidate vaccines (8, 9, 11, 12) and in clinical trials (8, 10, 46–48). However, the larger rise in neutralizing antibody titer in FIV-vaccinated ferrets and macaques following challenge compared to control animals indicated that priming had been mediated by the FIV.
The importance of antinucleocapsid immune responses for severe SARS-CoV-1 pneumonia in mice has been highlighted (49). Vaccinia containing the S, N, and other structural SARS-CoV-1 genes was used to immunize mice before SARS-CoV-1 challenge. Mice given vaccines that contained nucleocapsid exhibited severe pneumonia, and vaccinia-N–vaccinated mice showed up-regulation of both TH1 [IFN-β and interleukin-2 (IL-2)] and TH2 (IL-4 and IL-5) cytokines and down-regulation of anti-inflammatory cytokines (IL-10 and transforming growth factor–β), which resulted in infiltration of neutrophils, eosinophils, and lymphocytes in to the lung. Thus, suggesting an excessive host response to the SARS-CoV-1 nucleocapsid was involved in the severe pneumonia caused by SARS-CoV-1 infection. This parallels the observation by Atyeo et al. (50) who noted that functional antibody responses to nucleocapsid were elevated in deceased individuals, while elevated anti-S responses were measured in survivors. Thus, it may be that vaccines that produce an excessive nucleocapsid immune response may be useful for a model to explore the potential for enhanced disease.
The SARS-CoV-2–specific IFNγ response measured in splenocytes using an ELISpot assay 6 or 7 days after challenge showed a greater response in the FIV-vaccinated animals to live virus, membrane, and nucleocapsid peptide pools but very little response to the S protein pools, indicating a poor cellular response to the S antigen with FIV. Conversely, spike peptide–specific IFNγ spot-forming units (SFU) measured in macaques increased significantly following FIV vaccination and evaluation of the spike-specific IFNγ response in splenocytes and PBMC collected from macaques at a similar time after SARS-CoV2 challenge revealed the reverse pattern to that observed in ferrets, with greater responses measured in the unvaccinated group relative to the FIV-vaccinated animals. This may reflect that improved priming of the T cell–mediated response contributed to the protection afforded by the FIV vaccine in this species if response magnitude is driven by antigenic load following infection.
The expression of checkpoint inhibitory receptors is often considered a marker of T cell exhaustion, although more recently, PD-1 signaling has also been linked to improved effector T cell priming and enhanced clearance of acute viral infections (51). Similarly, the induction of regulatory CD4 T cells, and the inhibitory influence they are likely to exert, may be considered counterproductive to vaccine induced immunity. However, the increased frequency of T regulatory T cells observed following FIV vaccination is likely to reflect the immune response typically induced by coronaviruses in this species, as similar increases were also seen in the unvaccinated animals early after SARS-CoV-2 infection and thus may help to explain the relatively mild disease that develops in this species and the improved outcome observed in FIV vaccine primed macaques (52) (53).
There are limitations to the current study, the principal of which is the small number of ferrets in the early culled group. The transient nature of the pathology meant that these differences resolved by the second necropsy time point.
Some insight into the weak anti–S-neutralizing response induced by the FIV was gained using a capture ELISA, which preserved the conformation of the S trimer on the solid phase, unlike direct coating onto the ELISA plate that can modify antigenicity, as has been observed for soluble forms of the HIV-1 envelope glycoprotein trimer (54). FA treatment of S trimer in this format was the same as that used to inactivate the vaccine, allowing extrapolation of ligand binding to the ELISA-captured FA-treated S trimer to that on the virus. FA cross-linking resulted in a twofold reduction in binding of ACE2-Fc and two RBD-specific mAbs (CR3022 and EY6A) to FA-treated compared to untreated S trimer. By contrast, FA treatment of recombinant RBD did not affect binding of ACE2-Fc or these mAbs, implying that FA treatment cross-linked a proportion of S trimer into a nonligand binding conformation. These results are consistent with the location of lysine and arginine residues and suggest that RBD exposure may be limited by cross-linking, reducing exposure of neutralizing antibody epitopes on the RBD. This result may be of more general interest, since other viral envelope glycoproteins, such as those of HIV-1, are metastable and sample different conformational states, some of which are more relevant to neutralization than others (55). Cross-linking may trap these different conformational states, modifying exposure of neutralizing antibody epitopes to B cell recognition (56).
FA inactivation has been widely used to prepare inactivated viral vaccines (57) and as a toxoiding agent for bacterial toxin vaccines. Some of the considerations for inactivated SARS-CoV-2 vaccines are discussed in a recent commentary (58). The study reported here has confirmed that caution should be used if FA is the inactivation reagent for COVID-19 vaccines. Several vaccines are in development that use β-propiolactone as inactivation agent. One such vaccine has been shown to be protective in rhesus macaques following SARS-CoV-2 challenge without induction of VED (8). A preliminary report of phase 1 and 2 studies with another β-propiolactone vaccine indicated that it is tolerated, safe, and produced neutralizing antibodies in phase 1 and 2 studies (10). In addition, the authors mention in the discussion that enhancement of disease was not observed in primates following SARS-CoV-2 challenge but no pathology results are presented.
In conclusion, we have prepared an experimental SARS-CoV-2 vaccine based on previous inactivated virus studies that induced VED. We showed no evidence of enhanced disease at later time points in ferrets or at any time in a more in-depth analysis in rhesus macaques which included CT imaging. However, we did observe increased pathology scores early in the infection of FIV-vaccinated ferrets that resolved by the later necropsy time point. It is reassuring that, even with a vaccine deliberately designed to induce enhanced disease, no enhancement was seen apart from at 7 days after infection in ferrets. Future studies to investigate the potential of SARS-CoV-2 vaccines to cause enhanced disease should examine lung pathology at multiple time points including soon after challenge. Formalin-inactivated virus can be used as a suboptimal comparator to determine the potential of SARS-CoV-2 vaccine candidates to induce VED so that unsuitable vaccines are identified at an early stage of development before significant clinical studies commence.
MATERIALS AND METHODS
Study design
The goal of the study was to determine the potential for a suboptimal SARS-CoV-2 vaccine to induce VED in ferrets and rhesus macaques. An FIV that contained cell culture impurities was prepared. Animals were immunized with FIV 14 days before challenge to induce a weak immune response. Ferrets were euthanized at 6, 13, or 14 days after challenge. Following the indication of transient enhanced disease at 6 days in ferrets, all rhesus macaques were euthanized at day 7 after infection so the potential for enhanced disease at early time points could be determined. The humoral and cellular immune responses to FIV were characterized and clinical signs and viral replication were measured. A detailed examination of postmortem pathology was performed by three veterinary pathologists who were blinded to the treatment and group details. In addition, in-life CT of rhesus macaques was performed with images analyzed by a radiography blinded to the treatment groups.
Viruses and cells
SARS-CoV-2 Victoria/01/2020 (28) was provided by The Doherty Institute, Melbourne, Australia at P1 and passaged twice in Vero/hSLAM cells (ECACC 04091501). Briefly, confluent monolayers of hSLAM cells were infected at a multiplicity of infection of approximately 0.0005 for 60 min in medium (see below) containing no FBS at 37°C. The flasks were then filled with media supplemented with 4% heat-inactivated FBS. Virus was harvested at 72 hours after infection by removal of any remaining attached cells with sterile 5-mm borosilicate glass beads, clarification of the cell/media supernatant by centrifugation at 1000g for 10 min, followed by dispensing and storage at ≥−65°C. Whole-genome sequencing was performed, on the challenge isolate, using both nanopore and Illumina as described previously (26). Virus titer was determined by plaque assay on Vero/E6 cells (ECACC 85020206). Cell cultures were maintained at 37°C in minimum essential medium (Life Technologies, California, USA) supplemented with 10% FBS (Sigma-Aldrich, Dorset, UK) and 25 mM Hepes (Life Technologies). All Vero/hSLAM cell cultures were also supplemented with geneticin (0.4 μg/ml) (Gibco).
Preparation of formalin-inactivated virus vaccine
Centrifugal concentrators (VivaSpin20; 300-kDa cut off) were sterilized with 20 ml of 70% ethanol for 10 min followed by a wash with 20 ml of Dulbecco’s PBS (DPBS; Gibco). To reduce the concentration of calf serum components in the material, concentrators were loaded with 120 ml of SARS-CoV-2 at a titer of 8.45 × 106 PFU/ml and centrifuged at 3000g for 60 to 80 min (until the retained volume was ≤2 ml); the concentrators were refilled with 20 ml of DPBS and centrifugation was repeated for a total of three washes. After the final wash, the material was pooled and made up to 30 ml in sterile DPBS. Methanol-free FA solution at 36% (w/v) was added to a final FA concentration of 0.02% at room temperature for 72 hours. The inactivated virus was subjected to further three 20-ml DPBS washes to remove the residual FA, made up to 20 ml in DPBS, and aliquoted and stored below −15°C. To confirm inactivation, virus was seeded onto Vero/hSLAM cells in three flasks (100 μl per flask), which were serially passaged for a total of 27 days. Microscopic examination for signs of cytopathic effect, and reverse transcription quantitative polymerase chain reaction (RT-qPCR) was used to confirm that no viable virus remained.
Assessment of inactivated SARS-CoV-2
SDS–polyacrylamide gel electrophoresis
Samples were added to Laemmli buffer (Sigma-Aldrich, S3401) and heated at 90°C for 5 min and loaded onto a 10-well NuPAGE 4 to 12% bis-tris gel, 1.0 mm (Thermo Fisher Scientific). Five microliters of SeeBlue Plus2 (Thermo Fisher Scientific) ladder was loaded as a marker, and gels were stained with SimplyBlue SafeStain (Thermo Fisher Scientific).
Western blot
Samples were processed as described for SDS-PAGE and transferred to polyvinylidene difluoride membrane with iBlot2 (Thermo Fisher Scientific). After transfer, membranes were washed with tris-buffered saline 0.1% Tween 20 (TBST) for 5 min at room temperature, followed by 1 hour in blocking buffer (TBST, 5% skimmed milk powder). Membranes were washed three times for 5 min with TBST. MERS convalescent neutralizing serum (NIBSC S3) was diluted 1:1000 in blocking buffer and incubated for 1 hour at room temperature and then at 4°C overnight. Membranes were washed three times for 5 min with TBST and then incubated with either anti-human IgG-alkaline phosphatase (AP) or anti-rabbit IgG-AP (1:5000 in blocking buffer) for 1 hour with agitation. Membranes were washed three times as above and then developed with bromochloroindolyl phosphate–nitro blue tetrazolium liquid substrate system (Sigma-Aldrich). The protein concentration of the FIV was determined using a bicinchoninic acid assay (Pierce, #23227) (859 μg/ml). Densitometry analysis (ImageQuant TL; GE Healthcare) of the Western blot revealed a band of approximately 180 kDa that was only present in wild-type virus and FIV preparations. The relative density of this band (20.7%) permitted estimation of the proportion of the FIV total protein that was coronavirus-specific (178 μg/ml).
Transmission electron microscopy
Live virus was inactivated and fixed with final concentrations of 4% (w/v) FA and 2.5% (w/v) glutaraldehyde at ambient temperature for >16 hours before processing. Inactivated virus was processed without any additional fixation steps. Samples (approximately 10 μl) were placed directly on to electron microscopy grids (400 mesh copper grid, covered with a carbon reinforced plastic film). After 5-min adsorption, the sample was removed, and the grids were negatively stained using 2% methylamine tungstate. The grids were examined using a CM100 transmission electron microscope (Philips/FEI/Thermo Fisher Scientific) operated at 80 kV.
Animals
Ferrets
Ten healthy, female ferrets (Mustela putorius furo) aged 5 to 7 months were obtained from a U.K. Home Office accredited supplier (Highgate Farm, UK). The mean weight at the time of challenge was 1002 g per ferret (range, 871 to 1150 g). Animals were housed as described previously (26).
Rhesus macaques
Twelve rhesus macaques of Indian origin (Macaca mulatta) were used in the study. Study groups comprised three males and three females, and all were adults aged 2 to 4 years and weighing between 3.73 and 5.52 kg at the time of challenge. Animals were housed as described previously (27). All experimental work was conducted under the authority of a U.K. Home Office–approved project license that had been subject to local ethical review at PHE Porton Down by the Animal Welfare and Ethical Review Body.
Vaccinations
Animals were randomly assigned to control (Ad-GFP for ferrets, no vaccine for rhesus macaques) and FIV-vaccinated groups. The weight distribution of the ferrets was tested to ensure that there was no difference between groups (t test, P > 0.05). An identifier chip (Bio-Thermo Identichip, Animalcare Ltd., UK) was inserted subcutaneously into the dorsal cervical region of each animal. Macaques were stratified for sex and into socially compatible cohorts and then randomly assigned into treatment groups. FIV was diluted in PBS to coronavirus-specific protein (133 μg/ml) and mixed 1:1 in 2% Alhydrogel (Invivogen vac-alu-250) to give a final concentration of 66.7 μg/ml in 1% Alhydrogel. Ferrets were immunized with a single intramuscular dose of 10 μg of coronavirus-specific protein in 150 μl divided over two sites, and macaques were immunized with 25 μg in 300 μl administered into the quadriceps femoris muscle of the right leg. Vaccination was 14 days before challenge. Control ferrets were immunized with a single intramuscular dose of 2.5 × 1010 virus particles of Ad-GFP (59), a replication-deficient simian adenovirus vector containing an insert unrelated to coronavirus (GFP) 28 days before challenge. Control macaques received no vaccine.
SARS-CoV-2 challenge
Before challenge, ferrets were sedated by intramuscular injection of ketamine/xylazine (17.9 and 3.6 mg/kg bodyweight) and macaques with ketamine hydrochloride (Ketaset, 100 mg/ml, Fort Dodge Animal Health Ltd., UK; 10 mg/kg). SARS-CoV-2 Victoria/01/2020 (28) was prepared as described previously (26). It was delivered to ferrets by intranasal instillation (1.0 ml in total, 0.5 ml per nostril) diluted in PBS. A single dose of virus (5 × 106 PFU per ferret) was delivered to Ad-GFP–vaccinated (n = 4) and FIV-vaccinated (n = 6) ferrets. Macaques were challenged with 5 × 106 delivered by the intratracheal route (2 ml) and intranasal instillation (1 ml in total, 0.5 ml per nostril). The schedule of euthanization and sampling is shown in Table 1.
Nasal washes were obtained by flushing the nasal cavity with 2 ml of PBS. Throat swabs were collected using a standard swab (Sigma Virocult) gently stroked across the back of the pharynx in the tonsillar area. Throat swabs were processed, and aliquots were stored in viral transport media and AVL (Qiagen) at ≤−60°C until assay. Clinical signs of disease were monitored as described previously (26, 27). The necropsy procedures were also as described previously (26, 27).
SARS-CoV-2 virology
RNA was isolated from nasal wash and throat swabs. Samples were inactivated in AVL and ethanol. Downstream extraction was then performed using the BioSprint96 One-For-All vet kit (Indical) and KingFisher Flex platform as per manufacturer’s instructions.
RT-qPCR targeting a region of the SARS-CoV-2 nucleocapsid (N) gene was used to determine viral loads and was performed using TaqPath 1-Step RT-qPCR Master Mix, CG (Applied Biosystems), 2019-nCoV CDC RUO Kit (Integrated DNA Technologies), and QuantStudio 7 Flex Real-Time PCR System. Sequences of the N1 primers and probe were as follows: 2019-nCoV_N1-forward, 5′-GACCCCAAAATCAGCGAAAT-3′; 2019-nCoV_N1-reverse, 5′-TCTGGTTACTGCCAGTTGAATCTG-3′; 2019-nCoV_N1-probe, 5′-FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1-3′. The cycling conditions were 25°C for 2 min, 50°C for 15 min, 95°C for 2 min, followed by 45 cycles of 95°C for 3 s, and 55°C for 30 s. The quantification standard was in vitro transcribed RNA of the SARS-CoV-2 N ORF (accession no. NC_045512.2) with quantification between 1 × 101 and 1 × 106 copies/μl. Positive samples detected below the limit of quantification were assigned the value of 5 copies/μl, while undetected samples were assigned the value of 2.3 copies/μl, equivalent to the assay’s lower limit of detection.
ELISA to quantify anti-S, RBD, and N IgG
A full-length trimeric and stabilized version of the SARS-CoV-2 spike protein (amino acids 1 to 1280, GenBank: MN MN908947) was developed by F. Krammer’s lab as described (60). Recombinant SARS-CoV-2 RBD (319-541) Myc-His was provided by MassBiologics. Recombinant SARS-CoV-2 nucleocapsid phosphoprotein (GenBank: MN908947, isolate Wuhan-Hu-1) was expressed and purified from Escherichia coli as full-length nucleoprotein (amino acids 1 to 419) with a C-terminal 6xHis-Tag (Native Antigen Company). High-binding 96-well plates (Nunc Maxisorp, 442404) were coated with 50 μl per well of Spike trimer (2 μg/ml), RBD, or N in 1× PBS (Gibco) and incubated overnight at 4°C. The ELISA plates were washed five times with wash buffer (PBS 0.05% Tween 20) and blocked with 5% FBS (100 μl per well) (Sigma-Aldrich, F9665) in PBS 0.1% Tween 20 for 1 hour at room temperature. After washing, serum samples were serially diluted in 10% FBS in PBS 0.1% Tween 20, and each dilution (50 μl per well) was added to the antigen-coated plate and incubated for 2 hours at room temperature. Following washing, anti-ferret IgG–horseradish peroxidase (HRP) (Novus Biologics, NB7224) diluted (1:1000) or anti-monkey IgG-HRP (diluted 1:10,000; Invitrogen, PA-84631) in 10% FBS in 1× PBS/0.1% Tween 20 and 100 μl per well was added to each plate and then incubated for 1 hour at room temperature. After washing, O-phenylenediamine dihydrochloride solution (1 mg/ml) (Sigma-Aldrich, P9187) was prepared and 100 μl per well was added. The development was stopped with 1 M hydrochloric acid (50 μl per well) (Fisher Chemical, J/4320/15), and the absorbance at 490 nm was measured. The serum dilution where an OD of 1.0 was reached was used to calculate the titers using Softmax Pro 7.0.
SARS-CoV-2 neutralization assays
The plaque reduction neutralization test was performed as described previously (61). The microneutralization assay was performed with macaque serum as described for human sera (61).
Isolation of immune cells
Similar to as described previously (26), heparinised blood and spleens were removed for the isolation of immune cells: PBMCs and splenocytes. The spleens were dissected into small pieces. Dissected spleen was dissociated using a gentleMACS. The tissue solution was passed through two cell sieves (100 and 70 μm) and then layered with Ficoll-Paque Premium (GE Healthcare, Hatfield, UK). Density gradient centrifugation was carried out at 400g for 30 min on dissociated tissue and on whole blood. Buffy coats containing lymphocytes were collected and washed with medium by pelleting cells via centrifugation at 400g for 10 min. The cells were counted using a vial-1 cassette and a Nucleocounter-200 before cryopreservation in 95% fetal calf serum/5% (v/v) dimethyl sulfoxide. Cryopreserved cells were then frozen at −80°C in controlled rate freezer containers overnight, before transfer to liquid nitrogen (vapor phase).
IFNγ ELISpot assay
An IFNγ ELISpot assay was performed as described previously for ferrets (26) and macaques (27).
Immunophenotyping
Whole blood immunophenotyping assays were performed using 50 μl of heparinised blood incubated for 30 min at room temperature with optimal dilutions of the following antibodies: anti–CD3-AF700, anti–CD4-allophycocyanin (APC)–H7, anti–CD8-PerCP-Cy5.5, anti–CD95-Pe-Cy7, anti–CD14-PE, anti–HLA-DR-BUV395, anti–CD25-FITC (all from BD Biosciences, Oxford, UK); anti–CD127-APC (eBioscience); anti–γδ-TCR-BV421, anti–CD16-BV786, anti–PD-1-BV711, anti–CD20-PE-Dazzle (all from BioLegend); and amine reactive fixable viability stain red (Life Technologies); all prepared in brilliant stain buffer (BD Biosciences). Red blood cell contamination was removed using a Cal-lyse reagent kit as per the manufacturer’s instructions (Thermo Fisher Scientific). BD CompBeads (BD Biosciences) were labeled with the above fluorochromes for use as compensation controls. Following antibody labeling, cells and beads were fixed in a final concentration of 4% paraformaldehyde solution (Sigma-Aldrich, Gillingham, UK) before flow cytometric acquisition.
Cells were analyzed using a five laser LSRII Fortessa instrument (BD Biosciences), and data were analyzed using FlowJo (version 10, Treestar, Ashland, US). Immediately before flow cytometric acquisition, 50 μl of Truecount bead solution (Beckman Coulter) was added to each sample. Leukocyte populations were identified using a forward scatter-height versus side scatter-area dot plot to identify the lymphocyte, monocyte, and granulocyte populations to which appropriate gating strategies were applied to exclude doublet events and nonviable cells. Lymphocyte subpopulations including T cells, natural killer (NK) cells, NKT cells, and B cells were delineated by the expression pattern of CD3, CD20, CD95, CD4, CD8, CD127, CD25, and CD16 and the activation and inhibitory markers HLA-DR and PD-1. GraphPad Prism (version 8.0.1) was used to generate graphical representations of flow cytometry data.
Histopathology
The following samples from each ferret were fixed in 10% neutral-buffered formalin, processed to paraffin wax and 4-μm thick sections cut, and stained with hematoxylin and eosin; respiratory tract (left cranial and caudal lung lobes; three sections from each lung lobe: proximal, medial, and distal to the primary lobar bronchus), trachea (upper and lower), larynx, tonsil, liver, kidney, spleen, mediastinal lymph node, and small (duodenum) and large intestines (colon). Nasal cavity samples were also taken and decalcified in an EDTA solution for 3 weeks before embedding. These tissues above were examined by light microscopy and evaluated subjectively. Three qualified veterinary pathologists examined the tissues independently and were blinded to treatment and group details, and the slides were randomized before examination to prevent bias. A semiquantitative scoring system was developed to compare the severity of the lung lesions for each individual animal and among groups. This scoring system was applied independently to the cranial and caudal lung lobe tissue sections using the following parameters: (i) bronchial inflammation with the presence of exudates and/or inflammatory cell infiltration; (ii) bronchiolar inflammation with the presence of exudates and/or inflammatory cell infiltration; (iii) perivascular inflammatory infiltrates (cuffing); and (iv) infiltration of alveolar walls and spaces by inflammatory cells, mainly mononuclear. The severity of the histopathological lesions was scored as follows: 0 = none (within normal limits), 1 = minimal, 2 = mild, 3 = moderate, and 4 = severe.
Tissue sections of both lung lobes, nasal cavity, and gastrointestinal tract from animals culled at the early time point (day 6/7 after challenge) were stained using the RNAscope ISH technique to identify SARS-CoV-2 RNA. Briefly, tissues were pretreated with hydrogen peroxide for 10 min (room temperature), target retrieval for 15 min (98° to 101°C), and protease plus for 30 min (40°C) (Advanced Cell Diagnostics). A V-nCoV2019-S probe (catalog no. 848561, Advanced Cell Diagnostics) was incubated on the tissues for 2 hours at 40°C. Amplification of the signal was carried out following the RNAscope protocol using the RNAscope 2.5 HD Detection kit–Red (Advanced Cell Diagnostics).
In addition, immunohistochemistry was used to identify T cells (CD3+) in lung tissue sections. Samples were cut at 4 μm onto adhesive slides and stained using the Leica Bond RxM (Leica Biosystems, Germany). Briefly, slides were dewaxed and treated with peroxide block for 5 min. Epitope retrieval was performed using Epitope Retrieval solution 2 (Leica Biosystems, Germany) for 20 min. A polyclonal rabbit anti-human CD3 antibody (1:200; Agilent Technologies Inc., CA) was applied for 15 min and used with a Leica Polymer Refine Detection kit to complete the staining. The following samples from each rhesus macaque were fixed, processed, cut, and stained as described above: left cranial and caudal lung lobes, trachea, larynx, mediastinal lymph node, tonsil, spleen, liver, kidney, duodenum, and colon.
For the lung, three sections from each left lung lobe were sampled from different locations: proximal, medial, and distal to the primary lobar bronchus. A scoring system was used to evaluate objectively the histopathological lesions observed in the lung tissue sections (27). The scores for each histopathological parameter were calculated as the average of the scores observed in the six lung tissue sections evaluated per animal.
Sections from the lung lobes, duodenum, and colon were stained with RNAScope ISH as described above. For the lung sections, digital image analysis was carried out with Nikon NIS-Ar software to calculate the total area of the lung section positive for viral RNA.
In-life imaging of macaques by CT
CT scans were collected 4 weeks before vaccination and 5 days after challenge. CT imaging was performed on sedated animals using a 16-slice Lightspeed CT scanner (General Electric Healthcare, Milwaukee, WI, USA) in both the prone and supine positions to assist the differentiation of pulmonary changes at the lung bases caused by gravity-dependent atelectasis from GGO caused by SARS-CoV-2. All axial scans were performed at 120 kilovolt peak with auto-mA (ranging between 10 and 120) and were acquired using a small scan field of view. Rotation speed was 0.8 s. Images were displayed as an 11-cm field of view. To facilitate full examination of the cardiac and pulmonary vasculature, lymph nodes, and extrapulmonary tissues, Niopam 300 (Bracco, Milan, Italy), a nonionic, iodinated contrast medium, was administered intravenously at 2 ml/kg body weight, and scans were collected immediately after injection and 90 s from the midpoint of injection.
Scans were evaluated by a medical radiologist expert in respiratory diseases, including in nonhuman primates (45), blinded to the animal’s clinical status for the presence of disease features characteristic of COVID-19 in humans (GGO, consolidation, crazy paving, nodules, and peri-lobular consolidation; distribution: upper, middle, lower, central 2/3, bronchocentric); pulmonary embolus and the extent of any abnormalities estimated (<25%, 25 to 50%, 51 to 75%, and 76 to 100%).
CT score system
To provide the power to discriminate differences between individual NHP’s with low disease volume (i.e., <25% lung involvement), a score system was applied in which scores were attributed for possession of abnormal features characteristic of COVID in human patients (COVID pattern score) and for the distribution of features through the lung (Zone score). The COVID pattern score was calculated as sum of scores assigned for the number of nodules identified and the possession and extent of GGO and consolidation according to the following system: nodule(s): score 1 for 1, 2 for 2 or 3, 3 for 4, or more; GGO: each affected area was attributed with a score according to the following: score 1 if area measured <1 cm, 2 if 1 to 2 cm, 3 if 2 to 3 cm, and 4 if >3 cm, and scores for each area of GGO were summed to provide a total GGO score; consolidation: each affected area was attributed with a score according to the following: 1 if area measured <1 cm, 2 if 1 to 2 cm, 3 if 2 to 3 cm, and 4 if >3 cm. Scores for each area of consolidation are summed to provide a total consolidation score. To account for estimated additional disease impact on the host of consolidation compared to GGO, the score system was weighted by doubling the score assigned for consolidation. To determine the zone score, the lung was divided into 12 zones, and each side of the lung was divided (from top to bottom) into three zones: the upper zone (above the carina), the middle zone (from the carina to the inferior pulmonary vein), and the lower zone (below the inferior pulmonary vein). Each zone was further divided into two areas: the anterior area (the area before the vertical line of the midpoint of the diaphragm in the sagittal position) and the posterior area (the area after the vertical line of the midpoint of the diaphragm in the sagittal position). This results in 12 zones in total where a score of one is attributed to each zone containing structural changes. The COVID pattern score and the zone are summed to provide the total CT score.
ELISA to characterize ligand binding to FA-treated and untreated S trimer and RBD
Antigens and ligands
The S trimer expression plasmid was obtained from R. Shattock and P. McCay (Imperial College London, UK) and expressed the Wuhan-Hu-1 sequence (NCBI Reference NC_045512.2) in the context of a functional S1S2 cleavage site and with the addition of a C-terminal trimerization domain and 6x his and Myc tags. The RBD-Fc expression plasmid was also based on the Wuhan-Hu-1 sequence and was obtained from the Krammer lab (Mount Sinai, NY, USA). Proteins were expressed in 293F cells and purified by nickel column (Thermo Fisher Scientific) for S trimer and protein A column (Thermo Fisher Scientific) for RBD-Fc. Soluble ACE2-Fc was based on a published sequence (62) and was obtained from H. Waldmann and manufactured by Absolute Antibody Inc., and RBD binding mAbs CR3022 (63) and EY6A3 (64) were obtained from T. Tan and K.-Y. Huang, respectively, and were biotinylated using NHS-LC-biotin following manufacturer’s instructions (Thermo Fisher Scientific).
For S trimer capture ELISA, high protein binding ELISA plates (PerkinElmer) were coated overnight at 4°C with anti-Myc antibody 9E10 at 4 μg/ml. After washing and blocking in PBS/2% BSA/0.05% Tween 20, S trimer at 1 μg/ml was added in 50 μl per well in PBS/1% BSA/0.05% Tween 20 (ELISA buffer, EB) for 2 hours at room temperature. For RBD capture ELISA, high-protein binding ELISA plates (PerkinElmer) were coated overnight at 4°C with rabbit anti-human IgG (the Jackson laboratory) at 5 μg/ml in PBS. After washing and blocking, RBD-Fc at 1 μg/ml was added in 50 μl per well in EB for 2 hours at room temperature. After washing, wells requiring FA treatment were incubated with 50 μl of 0.02% methanol-free FA (Thermo Fisher Scientific) in PBS for 72 hours and untreated wells with PBS for the same amount of time. After washing, S-captured plates were incubated with soluble ACE2-Fc and mAbs CR3022 (63) and EY6A (64) in EB, and binding was detected using donkey anti-human HRP (Jackson Immunoresearch) at 1:5000 in 50 μl of EB. RBD-captured plates were incubated with biotinylated ligands soluble ACE2-Fc, CR3022, and EY6A, and binding was detected using streptavidin-HRP (GE Healthcare) diluted 1:4000 in 50 μl of EB. Plates were incubated for 1 hour at room temperature, washed, and developed in 50 μl of 3,3’,5,5’-tetramethylbenzidine (TMB) ELISA substrate (Thermo Fisher Scientific), and the reaction was stopped with 50 μl of 0.5 M H2SO4. Absorbance at 450 and 570 nm was read on a SpectraMax M5 plate reader (Molecular Devices), and data were analyzed in GraphPad Prism v7.
Molecular modeling
The structure shown is PDB 6lzg, and the views shown were generated in Pymol 2.3.5 (Schrodinger LLC).
Acknowledgments
We acknowledge the contributions of all staff within the PHE Biological Investigations Group for assistance with the delivery of the in vivo study and B. Cavell, J. Gouriet, V. Lucas, D. Ngabo, S. Thomas, and R. Watson for assistance with processing of in vivo samples. We thank S. Findlay-Wilson, T. Hender, N. McLeod, and C. Turner for assistance with RNA extraction and PCR; E. Penn for assistance with neutralization assays; and T. Tipton for preparation of ELISpot peptides. A. Morrison, A. Mabbut, and D. Gkolfinos assisted with ELISPot assays. We thank M. Elmore and M. Matheson for assistance with data analysis. We also acknowledge the donation of the S trimer expression construct by R. Shattock and P. McCay, the ACE2-Fc construct by H. Waldmann, and the RBD-specific monoclonal antibodies CR3022 and EY6A by T. Tan and K.-Y. Huang, respectively. We thank T. Schiffner for assistance with the molecular modeling, S. Zhang for assistance with S trimer production, and the Jenner Institute Viral Vector Core Facility for the production of Ad-GFP. Q.J.S., S.G., and T.L. are Jenner Institute Investigators. Q.J.S. is a James Martin senior fellow. Funding: This work was supported by UKRI Grant MC_PC_19080 and MRC UKRI Grant MC_PC_19055. Viral stock preparation was funded by the Coalition for Epidemic Preparedness Innovations. Author contributions: K.R.B.: Development of virus growth, plaque, and neutralization assay methods. K.R.B.: Design of virus inactivation strategy, vaccine preparation, drafting manuscript, and data analysis. K.G.: Study plan and management, data analysis, and manuscript review. K.M.T: Study plan and management, vaccine preparation, data collation and analysis, PBMC preparation, and manuscript review. S.Lo.: ELISA design, assay, and analysis; N.Wi.: In vivo study supervisor. L.H.: Histopathology slide preparation and staining. K.C.: SDS-PAGE and Western blots. P.B.: ELISpot assay and analysis and processing of in vivo samples. R.A.R.: Performed experimental procedures, analysis, and preparation of figures. C.H.: Preparation of in vivo samples and sera. G.S.: PCR data analysis. H.E.H.: PRNT assays and analysis. L.A.: In vivo study management. L.A.: Performed PRNT assays. M.A.: Performed PCR assays. N.B.: Virus inactivation and confirmation. E.B.: Performed PRNT assays. R.C.: Processing of in vivo samples. S.Fo.: In vivo study management. D.H.: In vivo study management. C.K.: Histology analysis including RNAscope. S.Le.: Performed PRNT assay. K.R.: Processed in vivo samples. H.T.: Performed electron microscopy. N.Wa.: RNA extraction and PCR assays. A.W., L.S., and C.S.: Designed and performed the macaques ELISpot assays. X.X.: Prepared and characterized proteins and manuscript drafting. Y.H.: Preparation and characterization of inactivated virus. T.L.: In vivo study design and provision of control vaccine. S.C.: In vivo study design and preparation of inactivated vaccine. S.Fu.: study design and manuscript preparation. S.G.: In vivo study design and provision of control vaccine. Q.J.S.: Designed and performed experiments to characterize the effect of FA on SARS-CoV-2 spike protein and drafted manuscript. F.J.S.: Lead the histopathology analysis and reporting and drafted manuscript. G.P.: Histopathology analysis. E.R.: Histopathology analysis. S.S.: CT scanning and analysis and manuscript preparation. F.G.: Analysis of CT scans. A.G.: Scientific direction and lead preparation of the manuscript. M.C.: Established the project, in vivo study design, scientific direction, and manuscript preparation. Competing interests: S.G. and T.L. are named on a patent application covering a vaccine ChAdOx1 nCoV-19. S.G. has been a board member and paid consultant to Vaccitech. The other authors declare that they have no competing interests. The funders played no role in the conceptualisation, design, data collection, analysis, decision to publish, or preparation of the manuscript. Data and materials availability statement: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Materials from the study can be provided by Public Health England pending scientific review and a completed material transfer agreement. Requests should be submitted to the corresponding authors.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
REFERENCES AND NOTES
- 1.Coronovirus Disease (2019) Weekly Epidemiological Update; www.who.int/docs/default-source/coronaviruse/situation-reports/20200824-weekly-epi-update.pdf?sfvrsn=806986d1_4.
- 2.Lurie N., Saville M., Hatchett R., Halton J., Developing covid-19 vaccines at pandemic speed. N. Engl. J. Med. 382, 1969–1973 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Kumar A., Dowling W. E., Román R. G., Chaudhari A., Gurry C., Le T. T., Tollefson S., Clark C. E., Bernasconi V., Kristiansen P. A., Status report on COVID-19 vaccines development. Curr. Infect. Dis. Rep. 23, 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baden L. R., El Sahly H. M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S. A., Rouphael N., Creech C. B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B. S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C.; C4591001 Clinical Trial Group , Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voysey M., Clemens S. A. C., Madhi S. A., Weckx L. Y., Folegatti P. M., Aley P. K., Angus B., Baillie V. L., Barnabas S. L., Bhorat Q. E., Bibi S., Briner C., Cicconi P., Collins A. M., Colin-Jones R., Cutland C. L., Darton T. C., Dheda K., Duncan C. J. A., Emary K. R. W., Ewer K. J., Fairlie L., Faust S. N., Feng S., Ferreira D. M., Finn A., Goodman A. L., Green C. M., Green C. A., Heath P. T., Hill C., Hill H., Hirsch I., Hodgson S. H. C., Izu A., Jackson S., Jenkin D., Joe C. C. D., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A. M., Lelliott A., Libri V., Lillie P. J., Mallory R., Mendes A. V. A., Milan E. P., Minassian A. M., McGregor A., Morrison H., Mujadidi Y. F., Nana A., O’Reilly P. J., Padayachee S. D., Pittella A., Plested E., Pollock K. M., Ramasamy M. N., Rhead S., Schwarzbold A. V., Singh N., Smith A., Song R., Snape M. D., Sprinz E., Sutherland R. K., Tarrant R., Thomson E. C., Török M. E., Toshner M., Turner D. P. J., Vekemans J., Villafana T. L., Watson M. E. E., Williams C. J., Douglas A. D., Hill A. V. S., Lambe T., Gilbert S. C., Pollard A. J.; Oxford COVID Vaccine Trial Group , Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397, 99–111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Logunov D. Y., Dolzhikova I. V., Zubkova O. V., Tukhvatullin A. I., Shcheblyakov D. V., Dzharullaeva A. S., Grousova D. M., Erokhova A. S., Kovyrshina A. V., Botikov A. G., Izhaeva F. M., Popova O., Ozharovskaya T. A., Esmagambetov I. B., Favorskaya I. A., Zrelkin D. I., Voronina D. V., Shcherbinin D. N., Semikhin A. S., Simakova Y. V., Tokarskaya E. A., Lubenets N. L., Egorova D. A., Shmarov M. M., Nikitenko N. A., Morozova L. F., Smolyarchuk E. A., Kryukov E. V., Babira V. F., Borisevich S. V., Naroditsky B. S., Gintsburg A. L., Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: Two open, non-randomised phase 1/2 studies from Russia. Lancet 396, 887–897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., Liang H., Bao L., Xu Y., Ding L., Zhou W., Gao H., Liu J., Niu P., Zhao L., Zhen W., Fu H., Yu S., Zhang Z., Xu G., Li C., Lou Z., Xu M., Qin C., Wu G., Gao G. F., Tan W., Yang X., Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 182, 713–721.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C., Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369, 77–81 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia S., Duan K., Zhang Y., Zhao D., Zhang H., Xie Z., Li X., Peng C., Zhang Y., Zhang W., Yang Y., Chen W., Gao X., You W., Wang X., Wang Z., Shi Z., Wang Y., Yang X., Zhang L., Huang L., Wang Q., Lu J., Yang Y., Guo J., Zhou W., Wan X., Wu C., Wang W., Huang S., Du J., Meng Z., Pan A., Yuan Z., Shen S., Guo W., Yang X., Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: Interim analysis of 2 randomized clinical trials. JAMA 324, 951–960 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J. N., Port J. R., Avanzato V. A., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E. R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Pérez-Pérez L., Edwards N. J., Wright D., Bissett C., Gilbride C., Williamson B. N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P. W., Scott D., Saturday G., de Wit E., Gilbert S. C., Munster V. J., ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586, 578–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett K. S., Flynn B., Foulds K. E., Francica J. R., Boyoglu-Barnum S., Werner A. P., Flach B., O’Connell S., Bock K. W., Minai M., Nagata B. M., Andersen H., Martinez D. R., Noe A. T., Douek N., Donaldson M. M., Nji N. N., Alvarado G. S., Edwards D. K., Flebbe D. R., Lamb E., Doria-Rose N. A., Lin B. C., Louder M. K., O’Dell S., Schmidt S. D., Phung E., Chang L. A., Yap C., Todd J.-P. M., Pessaint L., Van Ry A., Browne S., Greenhouse J., Putman-Taylor T., Strasbaugh A., Campbell T.-A., Cook A., Dodson A., Steingrebe K., Shi W., Zhang Y., Abiona O. M., Wang L., Pegu A., Yang E. S., Leung K., Zhou T., Teng I.-T., Widge A., Gordon I., Novik L., Gillespie R. A., Loomis R. J., Moliva J. I., Stewart-Jones G., Himansu S., Kong W.-P., Nason M. C., Morabito K. M., Ruckwardt T. J., Ledgerwood J. E., Gaudinski M. R., Kwong P. D., Mascola J. R., Carfi A., Lewis M. G., Baric R. S., McDermott A., Moore I. N., Sullivan N. J., Roederer M., Seder R. A., Graham B. S., Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 383, 1544–1555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smatti M. K., Al Thani A. A., Yassine H. M., Viral-induced enhanced disease illness. Front. Microbiol. 9, 2991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arvin A. M., Fink K., Schmid M. A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H. W., A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 584, 353–363 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Lee W. S., Wheatley A. K., Kent S. J., DeKosky B. J., Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 5, 1185–1191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki A., Yang Y., The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 20, 339–341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapikian A. Z., Mitchell R. H., Chanock R. M., Shvedoff R. A., Stewart C. E., An epidemiologic study of altered clinical reactivity to Respiratory Syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am. J. Epidemiol. 89, 405–421 (1969). [DOI] [PubMed] [Google Scholar]
- 18.Polack F. P., Hoffman S. J., Crujeiras G., Griffin D. E., A role for nonprotective complement-fixing antibodies with low avidity for measles virus in atypical measles. Nat. Med. 9, 1209–1213 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Katzelnick L. C., Gresh L., Halloran M. E., Mercado J. C., Kuan G., Gordon A., Balmaseda A., Harris E., Antibody-dependent enhancement of severe dengue disease in humans. Science 358, 929–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R. S., A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 85, 12201–12215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Te Tseng C., Sbrana E., Iwata-Yoshikawa N., Newman P. C., Garron T., Atmar R. L., Peters C. J., Couch R. B., Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLOS ONE 7, e35421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. Czub, H. Weingartl, S. Czub, R. He, J. Cao, in Vaccine (Elsevier BV, 2005), vol. 23, pp. 2273–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J., Wang W., Zhong Q., Hou W., Yang Z., Xiao S. Y., Zhu R., Tang Z., Wang Y., Xian Q., Tang H., Wen L., Immunogenicity, safety, and protective efficacy of an inactivated SARS-associated coronavirus vaccine in rhesus monkeys. Vaccine 23, 3202–3209 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q., Zhang L., Kuwahara K., Li L., Liu Z., Li T., Zhu H., Liu J., Xu Y., Xie J., Morioka H., Sakaguchi N., Qin C., Liu G., Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates. ACS Infect. Dis 2, 361–376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z., Zhang L., Qin C., Ba L., Yi C. E., Zhang F., Wei Q., He T., Yu W., Yu J., Gao H., Tu X., Gettie A., Farzan M., Yuen K., Ho D. D., Recombinant modified vaccinia virus Ankara expressing the spike glycoprotein of severe acute respiratory syndrome coronavirus induces protective neutralizing antibodies primarily targeting the receptor binding region. J. Virol. 79, 2678–2688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan K. A., Bewley K. R., Fotheringham S. A., Slack G. S., Brown P., Hall Y., Wand N. I., Marriott A. C., Cavell B. E., Tree J. A., Allen L., Aram M. J., Bean T. J., Brunt E., Buttigieg K. R., Carter D. P., Cobb R., Coombes N. S., Findlay-Wilson S. J., Godwin K. J., Gooch K. E., Gouriet J., Halkerston R., Harris D. J., Hender T. H., Humphries H. E., Hunter L., Ho C. M. K., Kennard C. L., Leung S., Longet S., Ngabo D., Osman K. L., Paterson J., Penn E. J., Pullan S. T., Rayner E., Skinner O., Steeds K., Taylor I., Tipton T., Thomas S., Turner C., Watson R. J., Wiblin N. R., Charlton S., Hallis B., Hiscox J. A., Funnell S., Dennis M. J., Whittaker C. J., Catton M. G., Druce J., Salguero F. J., Carroll M. W., Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat. Commun. 12, 81 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salguero F. J., White A. D., Slack G. S., Fotheringham S. A., Bewley K. R., Gooch K. E., Longet S., Humphries H. E., Watson R. J., Hunter L., Ryan K. A., Hall Y., Sibley L., Sarfas C., Allen L., Aram M., Brunt E., Brown P., Buttigieg K. R., Cavell B. E., Cobb R., Coombes N. S., Darby A., Daykin-Pont O., Elmore M. J., Garcia-Dorival I., Gkolfinos K., Godwin K. J., Gouriet J., Halkerston R., Harris D. J., Hender T., Ho C. M. K., Kennard C. L., Knott D., Leung S., Lucas V., Mabbutt A., Morrison A. L., Nelson C., Ngabo D., Paterson J., Penn E. J., Pullan S., Taylor I., Tipton T., Thomas S., Tree J. A., Turner C., Vamos E., Wand N., Wiblin N. R., Charlton S., Dong X., Hallis B., Pearson G., Rayner E. L., Nicholson A. G., Funnell S. G., Hiscox J. A., Dennis M. J., Gleeson F. V., Sharpe S., Carroll M. W., Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19. Nat. Commun. 12, 1260 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caly L., Druce J., Roberts J., Bond K., Tran T., Kostecki R., Yoga Y., Naughton W., Taiaroa G., Seemann T., Schultz M. B., Howden B. P., Korman T. M., Lewin S. R., Williamson D. A., Catton M. G., Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 212, 459–462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson A. D., Williamson M. K., Lewis S., Shoemark D., Carroll M. W., Heesom K. J., Zambon M., Ellis J., Lewis P. A., Hiscox J. A., Matthews D. A., Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 12, 68 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walls A. C., Park Y. J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonhoeffer J., Bentsi-Enchill A., Chen R. T., Fisher M. C., Gold M. S., Hartman K., Heininger U., Hoet B., Jefferson T., Khuri-Bulos N., Kohl K. S., Marcy S. M., Nalin D., Pless R., Sanabria-Rojas H., Sleeman K., Wise R.; Brighton Collaboration Methods Working Group , Guidelines for collection, analysis and presentation of vaccine safety data in pre- and post-licensure clinical studies. Vaccine 27, 2282–2288 (2009). [DOI] [PubMed] [Google Scholar]
- 32.P. H. Lambert, D. M. Ambrosino, S. R. Andersen, R. S. Baric, S. B. Black, R. T. Chen, C. L. Dekker, A. M. Didierlaurent, B. S. Graham, S. D. Martin, D. C. Molrine, S. Perlman, P. A. Picard-Fraser, A. J. Pollard, C. Qin, K. Subbarao, J. P. Cramer, in Vaccine (Elsevier Ltd, 2020), vol. 38, pp. 4783–4791. [Google Scholar]
- 33.Graham B. S., Rapid COVID-19 vaccine development. Science 368, 945–946 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Connors M., Giese N. A., Kulkarni A. B., Firestone C. Y., Morse H. C. III, Murphy B. R., Enhanced pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of interleukin-4 (IL-4) and IL-10. J. Virol. 68, 5321–5325 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moghaddam A., Olszewska W., Wang B., Tregoning J. S., Helson R., Sattentau Q. J., Openshaw P. J. M., A potential molecular mechanism for hypersensitivity caused by formalin-inactivated vaccines. Nat. Med. 12, 905–907 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Acosta P. L., Caballero M. T., Polack F. P., Brief history and characterization of enhanced respiratory syncytial virus disease. Clin. Vaccine Immunol. 23, 189–195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw C. A., Galarneau J. R., Bowenkamp K. E., Swanson K. A., Palmer G. A., Palladino G., Markovits J. E., Valiante N. M., Dormitzer P. R., Otten G. R., The role of non-viral antigens in the cotton rat model of respiratory syncytial virus vaccine-enhanced disease. Vaccine 31, 306–312 (2013). [DOI] [PubMed] [Google Scholar]
- 38.He P., Zou Y., Hu Z., Advances in aluminum hydroxide-based adjuvant research and its mechanism. Hum. Vaccin. Immunother. 11, 477–488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li C., Chen Y.-X., Liu F.-F., Lee A. C.-Y., Zhao Y., Ye Z.-H., Cai J.-P., Chu H., Zhang R.-Q., Chan K.-H., Chiu K. H.-Y., Lung D. C., Sridhar S., Hung I. F.-N., To K. K.-W., Zhang A. J.-X., Chan J. F.-W., Yuen K.-Y., Absence of vaccine-enhanced disease with unexpected positive protection against SARS-CoV-2 by inactivated vaccine given within three days of virus challenge in Syrian hamster model. Clin. Infect. Dis. 73, e719–e734 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwata-Yoshikawa N., Uda A., Suzuki T., Tsunetsugu-Yokota Y., Sato Y., Morikawa S., Tashiro M., Sata T., Hasegawa H., Nagata N., Effects of toll-like receptor stimulation on eosinophilic infiltration in lungs of BALB/c mice immunized with UV-inactivated severe acute respiratory syndrome-related coronavirus vaccine. J. Virol. 88, 8597–8614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houser K. V., Broadbent A. J., Gretebeck L., Vogel L., Lamirande E. W., Sutton T., Bock K. W., Minai M., Orandle M., Moore I. N., Subbarao K., Enhanced inflammation in New Zealand white rabbits when MERS-CoV reinfection occurs in the absence of neutralizing antibody. PLOS Pathog. 13, e1006565 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weingartl H., Czub M., Czub S., Neufeld J., Marszal P., Gren J., Smith G., Jones S., Proulx R., Deschambault Y., Grudeski E., Andonov A., He R., Li Y., Copps J., Grolla A., Dick D., Berry J., Ganske S., Manning L., Cao J., Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J. Virol. 78, 12672–12676 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li T. C., Yang T., Yoshizaki S., Ami Y., Suzaki Y., Ishii K., Kishida N., Shirakura M., Asanuma H., Takeda N., Wakita T., Ferret hepatitis E virus infection induces acute hepatitis and persistent infection in ferrets. Vet. Microbiol. 183, 30–36 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Dennis M. J., Parks S., Bell G., Taylor I., Lakeman J., Sharpe S. A., A flexible approach to imaging in ABSL-3 laboratories. Appl. Biosaf. 20, 89–99 (2015). [Google Scholar]
- 45.Sharpe S. A., Smyth D., McIntyre A., Gleeson F., Dennis M. J., Refinement and reduction through application of a quantitative score system for estimation of TB-induced disease burden using computed tomography. Lab. Anim 52, 599–610 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Folegatti P. M., Ewer K. J., Aley P. K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E. A., Dold C., Faust S. N., Finn A., Flaxman A. L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A. M., Pollock K. M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A. D., Hill A. V. S., Lambe T., Gilbert S. C., Pollard A. J.; Oxford COVID Vaccine Trial Group , Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: A preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet 396, 467–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jackson L. A., Anderson E. J., Rouphael N. G., Roberts P. C., Makhene M., Coler R. N., McCullough M. P., Chappell J. D., Denison M. R., Stevens L. J., Pruijssers A. J., McDermott A., Flach B., Doria-Rose N. A., Corbett K. S., Morabito K. M., O’Dell S., Schmidt S. D., Swanson P. A., Padilla M., Mascola J. R., Neuzil K. M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J. E., Graham B. S., Beigel J. H.; mRNA-1273 Study Group , An mRNA vaccine against SARS-CoV-2 — Preliminary report. N. Engl. J. Med. 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mulligan M. J., Lyke K. E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K. A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K. R., Walsh E. E., Frenck R., Falsey A. R., Dormitzer P. R., Gruber W. C., Şahin U., Jansen K. U., Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586, 589–593 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., Kase R., Sekiguchi S., Morita K., Hishima T., Suzuki H., Karamatsu K., Yasutomi Y., Shida H., Kidokoro M., Mizuno K., Matsushima K., Kohara M., Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J. Immunol. 181, 6337–6348 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Atyeo C., Fischinger S., Zohar T., Slein M. D., Burke J., Loos C., McCulloch D. J., Newman K. L., Wolf C., Yu J., Shuey K., Feldman J., Hauser B. M., Caradonna T., Schmidt A. G., Suscovich T. J., Linde C., Cai Y., Barouch D., Ryan E. T., Charles R. C., Lauffenburger D., Chu H., Alter G., Distinct early serological signatures track with SARS-CoV-2 survival. Immunity 53, 524–532.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ahn E., Araki K., Hashimoto M., Li W., Riley J. L., Cheung J., Sharpe A. H., Freeman G. J., Irving B. A., Ahmed R., Role of PD-1 during effector CD8 T cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 115, 4749–4754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cecere T. E., Todd S. M., LeRoith T., Regulatory T cells in arterivirus and coronavirus infections: Do they protect against disease or enhance it? Viruses 4, 833–846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meckiff B. J., Ramírez-Suástegui C., Fajardo V., Chee S. J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., Sette A., Ay F., Seumois G., Ottensmeier C. H., Vijayanand P., Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell 183, 1340–1353.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiffner T., de Val N., Russell R. A., de Taeye S. W., de la Peña A. T., Ozorowski G., Kim H. J., Nieusma T., Brod F., Cupo A., Sanders R. W., Moore J. P., Ward A. B., Sattentau Q. J., Chemical cross-linking stabilizes native-like HIV-1 envelope glycoprotein trimer antigens. J. Virol. 90, 813–828 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guttman M., Cupo A., Julien J. P., Sanders R. W., Wilson I. A., Moore J. P., Lee K. K., Antibody potency relates to the ability to recognize the closed, pre-fusion form of HIV Env. Nat. Commun. 6, 6144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schiffner T., Pallesen J., Russell R. A., Dodd J., de Val N., LaBranche C. C., Montefiori D., Tomaras G. D., Shen X., Harris S. L., Moghaddam A. E., Kalyuzhniy O., Sanders R. W., McCoy L. E., Moore J. P., Ward A. B., Sattentau Q. J., Structural and immunologic correlates of chemically stabilized HIV-1 envelope glycoproteins. PLOS Pathog. 14, e1006986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delrue I., Verzele D., Madder A., Nauwynck H. J., Inactivated virus vaccines from chemistry to prophylaxis: Merits, risks and challenges. Expert Rev. Vaccines 11, 695–719 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Mulligan M. J., An inactivated virus candidate vaccine to prevent COVID-19. JAMA 324, 943–945 (2020). [DOI] [PubMed] [Google Scholar]
- 59.van Doremalen N., Haddock E., Feldmann F., Meade-White K., Bushmaker T., Fischer R. J., Okumura A., Hanley P. W., Saturday G., Edwards N. J., Clark M. H. A., Lambe T., Gilbert S. C., Munster V. J., A single dose of ChAdOx1 MERS provides protective immunity in rhesus macaques. Sci. Adv. 6, eaba8399 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amanat F., White K. M., Miorin L., Strohmeier S., McMahon M., Meade P., Liu W. C., Albrecht R. A., Simon V., Martinez-Sobrido L., Moran T., García-Sastre A., Krammer F., An in vitro microneutralization assay for SARS-CoV-2 serology and drug screening. Curr. Protoc. Microbiol. 58, e108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bewley K. R., Coombes N. S., Gagnon L., McInroy L., Baker N., Shaik I., St-Jean J. R., St-Amant N., Buttigieg K. R., Humphries H. E., Godwin K. J., Brunt E., Allen L., Leung S., Brown P. J., Penn E. J., Thomas K., Kulnis G., Hallis B., Carroll M., Funnell S., Charlton S., Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat. Protoc. 16, 3114–3140 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Kruse R. L., Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res. 9, 72 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuan M., Wu N. C., Zhu X., Lee C.-C. D., So R. T. Y., Lv H., Mok C. K. P., Wilson I. A., A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou D., Duyvesteyn H. M. E., Chen C.-P., Huang C.-G., Chen T.-H., Shih S.-R., Lin Y.-C., Cheng C.-Y., Cheng S.-H., Huang Y.-C., Lin T.-Y., Ma C., Huo J., Carrique L., Malinauskas T., Ruza R. R., Shah P. N. M., Tan T. K., Rijal P., Donat R. F., Godwin K., Buttigieg K. R., Tree J. A., Radecke J., Paterson N. G., Supasa P., Mongkolsapaya J., Screaton G. R., Carroll M. W., Gilbert-Jaramillo J., Knight M. L., James W., Owens R. J., Naismith J. H., Townsend A. R., Fry E. E., Zhao Y., Ren J., Stuart D. I., Huang K.-Y. A., Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 27, 950–958 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4