Abstract
Five keratin-associated protein 6 genes (KRTAP6) have been identified in sheep and variation in some KRTAP6 has been associated with wool fiber diameter-related traits, but none of these homologues have been identified in goats. In this study, we reported the identification of the sheep KRTAP6-5 homologue on goat chromosome 1 and polymerase chain reaction (PCR)-single strand conformation polymorphism analysis in 300 Longdong cashmere goats revealed the existence of 12 variant sequences. Both coding region and 3′UTR of the putative caprine KRTAP6-5 displayed a biggest sequence similarity to ovine KRTAP6-5 gene. This suggested that the gene represents caprine KRTAP6-5 sequences, and these sequences composed 23 genotypes, which was the most polymorphism gene in KRTAPs that have been studied. Among these sequences, 15 nucleotide substitutions and a 24-bp insertion/detection were identified. Variation in goat KRTAP6-5 was associated with variation in mean-fiber diameter, suggesting that KRTAP6-5 is worthy of further study in the context of variation in cashmere traits.
Keywords: goat, keratin-associated protein KAP6-5 gene, mean-fiber diameter, PCR-SSCP, variation
Introduction
Cashmere fiber is a main product of cashmere goats that is produced by secondary hair follicles. The fiber of cashmere is soft, warm, and strong and is probably the finest and lightest among all animal fibers. Like wool, the cashmere fiber is composed of hair-keratins and keratin-associated proteins (KAPs) (Rogers, 2006). KAPs serve as a matrix that cross-links hair-keratins (Rogers et al., 2002) and are hence believed to play an important role in defining the physical and mechanical properties of the fiber (Powell and Rogers, 1997; Gong et al., 2016).
KAPs are a diverse group of proteins encoded by small intron less genes called KRTAPs (Gong et al., 2012). To date, over 100 KRTAPs have been identified across a range of mammalian species (Gong et al., 2010 and 2016; Khan et al., 2014), and human has reported 88 functional KRTAPs (Rogers and Schweizer, 2005; Rogers et al., 2007, and 2008). In sheep, while many of the human KRTAP orthologues await discovery, 30 KRTAPs have been identified. In goats, only 17 KRTAPs have been characterized to date, and these are KRTAP1-1 (Andrews et al., 2017), KRTAP1-2 (Zhao et al., 2021), KRTAP1-4 (Shah et al., 2013), KRTAP3-1 (Parris and Swart, 1975), KRTAP7-1 (Jin et al., 2011), KRTAP8-1(Zhao et al., 2009), KRTAP8-2 (Jin et al., 2011), KRTAP9-2 (Wang et al., 2012),KRTAP11-1 (Jin et al., 2017), KRTAP13-1 (Fang et al., 2010), KRTAP13-3 (Andrews et al., 2017), KRTAP15-1 (Zhao et al., 2019), KRTAP20-1 (Wang et al., 2018), KRTAP20-2 (Wang et al., 2017), KRTAP24-1 (Wang et al., 2019), KRTAP27-1 (Zhao et al., 2020), and KRTAP28-1 (Wang et al., 2020). Despite there being KRTAP sequences reported as caprine KRTAP6-2 (AY316158 and EU145019; Yin et al., 2004; Zhao et al., 2008), these sequences are very different to KRTAP6-n sequences from sheep and human and are unlikely to represent caprine KRTAP6-n, suggesting that not any gene from KAP6 family has been actually identified in goats.
KAP6 is a multigene subfamily with five family members having been identified in sheep (Zhou et al., 2016). Variation in KRTAP6-1 and KRTAP6-3 has been reported to affect fiber diameter-associated traits in sheep (Zhou et al., 2015; Li et al., 2017b; Li et al., 2019). Given the potential effect of KRTAP6 on wool traits (Plowman et al., 2019), investigation is needed to identify KRTAP6-n in goats and to determine whether variation in KRTAP6s affects cashmere traits. In this study, we attempted to identify the caprine KRTAP6-5, to search for potential variation in the gene, and to investigate its effect on cashmere fleece traits.
Materials and Methods
The collection of blood from of goats was approved by the Animal Care Committee of Gansu Agricultural University and was conducted under the guidelines for the care and use of experimental animals established by the Ministry of Science and Technology of China (Approval number 2006-398).
Goats investigated and data collection
Three hundred Longdong cashmere goats from 11 unrelated sires-lines farmed at Yusheng Cashmere Goat Breeding Company in Huan County of China were investigated. Cashmere fibers were collected at 1-yr-old goats by combing, and their greasy fleece weight (GFW) was measured. Cashmere samples were collected from the mid-side region of each goat and then the mean crimped fiber length (MCFL) and mean fiber diameter (MFD) were analyzed using an optical-based fiber length and diameter analyzer OFDA4000 (EPCO, Shanghai, CHN), at Inner Mongolia Agricultural University, China.
The blood samples from all Longdong cashmere goats were collected onto FTA cards (Whatman BioScience, Middlesex, UK), and genomic DNA from dried blood spots was purified using the two-step method described in Zhou et al. (2006).
Search for the Caprine KRTAP6-5
Using the ovine KRTAP6-5 sequence (GenBank KT725841.1), a BLASTN search in GenBank of the Caprine Genome Assembly ASM170441 V1 (NC030808.1) was undertaken. Of all the sequences found by the BLASTN search, the sequence with the greatest similarity to the ovine KRTAP6-5 sequence was assumed to be caprine KRTAP6-5.
Polymerase chain reaction (PCR) amplification of caprine KRTAP6-5
Using the goat sequence identified above, two PCR primers were designed to amplify the putative caprine KRTAP6-5. The sequences of the primers were 5′-AATGGCATGAAGGTGTGGTG-3′and 5′-TGCTGTGGGATAAGCATCAC-3′.
The PCR amplifications were performed in a 20-µL reaction including the purified genomic DNA from one 1.2-mm punch of dried blood spot, 0.25 μM of each primer, 1 × PCR buffer, 150 μM of each deoxyribonucleoside triphosphate (dNTP) (Bioline, London, UK), 2.5 mM Mg2+, and 0.5 U of Taq DNA polymerase (Takara). Reaction was carried out in an Applied Biosystems 2720 thermal cycler (Bio-Rad, Hercules, CA, USA). The thermal profile consisted of 2 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C, with a final extension of 5 min at 72 °C.
Screening for sequence variation
The PCR amplicons were screened for DNA sequence variation using SSCP analysis. For each amplicon, a 0.7-µL aliquot was add to 7 µL of loading dye (0.025% bromophenol blue, 98% formamide, 10 mM EDTA, and 0.025% xylene-cyanol) and after denaturation at 95 °C for 5 min, the mixtures were then cooled on ice and loaded on 16 × 18 cm, 10% acrylamide: bisacrylamide (37.5:1) (Bio-Rad) gels. Electrophoresis was performed in 0.5× TBE using ProteanII xi cells (Bio-Rad) and the electrophoretic conditions were 290 V for 16 h under the temperature of 20 °C. Then the SSCP gels were silver-stained using a method described by Byun et al. (2009).
Sequencing of amplicons and sequence analysis
If the amplicons produced PCR-SSCP banding patterns that suggested the goat was homozygous in the amplified region, then a representative sample for each pattern was sequenced in both directions at Shanghai Shenggong biological Co., LTD (China). While for heterozygous, PCR amplicons were sequenced using an approach described by Gong et al. (2011). Briefly, a band corresponding to the allele was excised as a gel slice from the polyacrylamide gel, macerated, and then used as a template for re-amplification with the original primers. This second amplicon was then sequenced. Sequence assembly, alignment, and translation were carried out using DNAMAN version 8 (Lynnon BioSoft, Vaudreuil, Canada).
Statistical analyses
Statistical analyses were performed using IBM SPSS Statistics version 24.0. General linear mixed-effects models (GLMMs) were used to compare the various cashmere traits among KRTAP6-5 genotypes and with a Bonferroni correction being applied to reduce the chances of obtaining false positive results during the multiple comparisons. Gender and sire were found to affect (P < 0.05) all the fiber traits and were included in the models as fixed and random factors, respectively. All P values were considered statistically significant when P < 0.05.
Results
Identification of KRTAP6-5 in Longdong cashmere goats
Using coding sequences of ovine KRTAP6-5 homologous regions, a BLAST search of the caprine Genome Assembly ASM170441 V1 (NC030808.1) revealed a 252-bp open reading frame (ORF) with 99% sequence identity on caprine chromosome 1 (nt3527973_nt3528224). Twelve previously identified caprine KAP genes were also found near this ORF and these from centromere to telomere were KRTAP11-1, KRTAP7-1, KRTAP8-1, KRTAP8-2, KRTAP20-2, KRTAP20-1, KRTAP15-1, KRTAP13-1, KRTAP13-3, KRTAP27-1, KRTAP28-1, and KRTAP24-1.
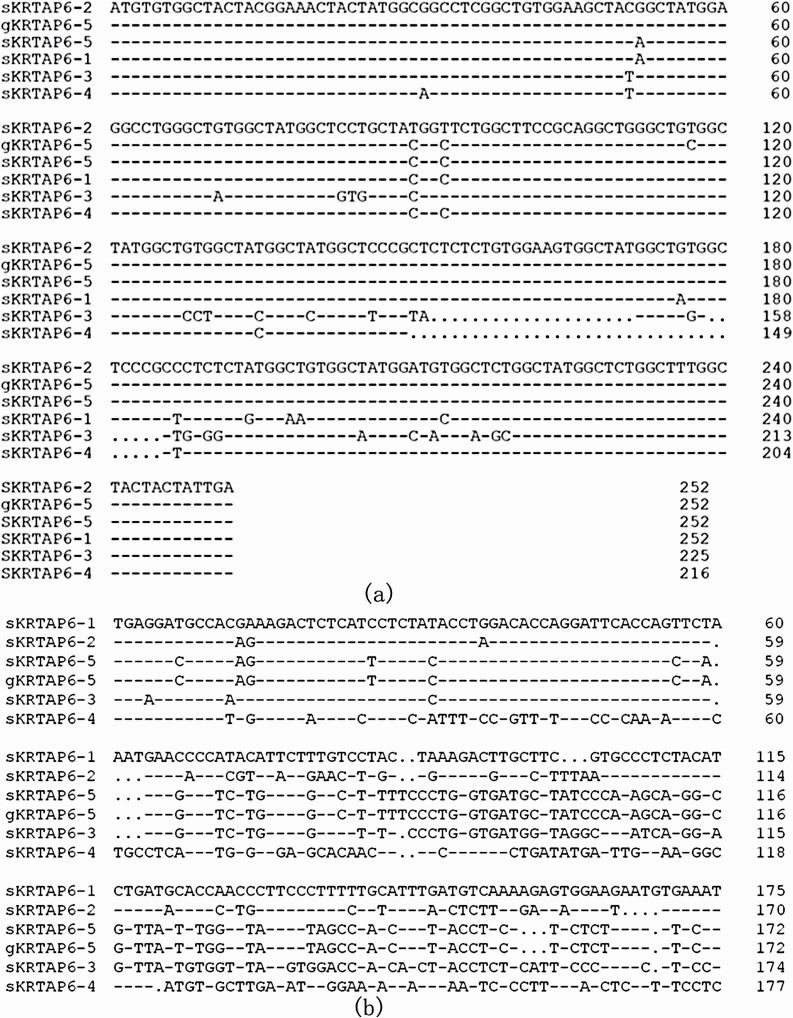
Neucleotide sequence homology analysis revealed that the ORF sequences were 96.8%, 98.6%, 89.4%, 97.7%, and 99.1% identity with ovine KRTAP6-1, KRTAP6-2, KRTAP6-3, KRTAP6-4, and KRTAP6-5, respectively (Figure 1a). The comparison of the 3′ flanking sequences of ovine KRTAP6-s with that of putative caprine KRTAP6-5 showed that the sequence of goat was exactly the same as that of ovine KRTAP6-5 (Figure 1b). This suggests that this ORF represents the caprine KRTAP6-5.
Figure 1.
Comparison of the ORF (a) and flanking sequences (b) of the new identified gene with other ovine KRTAP6-s. Dashes mean sequences identical to the first sequence and dots are used as spacers to improve the alignments. The GenBank accession numbers for ovine KRTAP6-s are NM_001193399 (KRTAP6-1), KT725832 (KRTAP6-2), KT725837 (KRTAP6-3), KT725840 (KRTAP6-4), and KT725845 (KRTAP6-5).
Amino acid composition of caprine KAP6-5
The caprine KAP6-5 sequences would encode a polypeptide of 83 amino acid residues. This polypeptide would have a high content of glycine (38.5 mol%) and tyrosine (21.7 mol%), and moderate levels of cysteine (12.1 mol%) and serine (10.8 mol%). The putative caprine KAP6-5 protein would therefore appear to be a basic protein, with a predicted isoeletric point (pI) value of approximately 8.2.
Sequence variation in caprine KRTAP6-5
PCR-SSCP analysis of the 300 goats revealed 12 unique SSCP patterns, with either one or a combination of two banding patterns observed for each goat. DNA sequencing revealed that these 12 PCR-SSCP patterns represented 12 nucleotide acid sequences (named A-L) and were deposited into GenBank with accession numbers MW762477–MW762488. Twenty-three SSCP bands represent 23 genotypes (AA, BB, CC, DD, GG, AB, AC, AD, AE, AF, AG, AI, AJ, AK, BD, BG, CG, CE, CK, CH, CI, CL, and DG; Supplementary Figure 1).
Fifteen nucleotide substitutions (c.-36A>C, c.-25A>G, c.-3A>C, c.27T>C, c.45A>G, c.51C>T, c.59G>C, c.83C>G, c.84C>T, c.85T>G, c.103C>T, c.117C>T, c.135T>C, c.238G>A, and C.*77C>A) and a 24-bp insertion/deletion were detected when comparing the 12 sequences (Supplementary Figure 2). Of these SNPs, five of them were non-synonymous (c.59G>C, c.83C>T, c.85T>G, c.103C>T, and c.238G>A) and would nominally result in amino acid changes (p.Gly20Aly, p.Ser28Cys, p.Cys29Gly, p.Arg35Cys, and p.Gly80Ser). The 24-bp insertion/deletion was located in a region encoding a repeated amino acid sequence “SRPLYGCG.”
Variant frequencies and genetic polymorphism of KRTAP6-5 in Longdong cashmere goat
The variant frequencies of caprine KRTAP6-5 in Longdong cashmere goat were A: 61.83%; B: 4.67%; C: 19.16%; D: 6.33%, E: 0.50%; F: 0.17%; G: 5.33%; H: 0.17%; I: 0.67%; J: 0.17%; K: 0.83%; and L: 0.17%. Of the 23 genotypes, with AA being the most common genotype (at a frequency of 38.00%), and to ensure adequate sample size (>10), only AA, AB, AC, AD, AG, and CC were used in the genotype association analyses.
Associations between KRTAP6-5 variation and cashmere traits
Goats with genotype AA, AB, AC, and AD produced cashmere fibers with higher MFD than CC (P < 0.05); meanwhile, MFD of goats with genotype AA was also higher than that of AG (P < 0.05; Table 1).
Table 1.
The association of KRTAP6-5 genotype and various cashmere traits
| Traits1 | Mean ± SE2 | P-value | |||||
|---|---|---|---|---|---|---|---|
|
AA (n = 116) |
AB (n = 17) |
AC (n = 80) |
AD (n = 22) |
AG (n = 12) |
CC (n =13) |
||
| GFW (g) | 418.13 ± 4.35 | 417.47 ± 10.38 | 415.25 ± 5.23 | 411.30 ± 8.86 | 413.33 ± 12.31 | 395.94 ± 11.79 | 0.590 |
| MFD (μm) | 13.62 ± 0.04a | 13.61 ± 0.09ab | 13.63 ± 0.05ab | 13.59 ± 0.08ab | 13.57 ± 0.11bc | 13.25 ± 0.11c | 0.030 |
| MCFL (cm) | 4.23 ± 0.05 | 4.26 ± 0.12 | 4.23 ± 0.06 | 4.22 ± 0.10 | 4.13 ± 0.14 | 4.28 ± 0.14 | 0.979 |
1GFW, greasy fleece weight; MFD, mean fiber diameter; MCFL, mean crimped fiber length.
2Estimated marginal means and standard errors (SE) of those means derived from general linear mixed-effects models. “Gender” and “sire” were included in the models as a fixed factor and a random factor, respectively.
Discussion
This study reports the identification of a new caprine high glycine-tyrosine KAP (HGT-KAP). This gene was located between KRTAP 20-2 and KRTAP 20-1, and this is consistent with the location of ovine KRTAP6-s. Both coding region and 3′UTR of the putative caprine KRTAP6-5 displayed a biggest sequence similarity to ovine KRTAP6-5 gene. This suggests that the gene represents caprine KRTAP6-5 sequence. The identification of caprine KRTAP6-5 brings the total number of HGT-KAPs identified in goats, from five to six.
If transcribed and translated, the amino acid sequence that would be produced from the caprine KAP6-5 sequence would contain 83 amino acids and more than half of these amino acids would be glycine and tyrosine and contain moderate levels of cysteine and serine. This is consistent with the defining characteristic of the HGT-KAPs.
Fifteen nucleotide substitutions and a 24-bp insertion/deletion were detected in caprine KRTAP6-5 gene and produced 12 unique variant sequences. Till now, caprine KRTAP6-5 is the most polymorphism gene among KRTAPs that have been studied, and six alleles were ovine KRTAP6-5 in 96 Merino sheep and Romney sheep. Deletions/insertions that maintain the reading frame have been reported for other KRTAPs, such as ovine KRTAP1-1 (Rogers et al., 1994), KRTAP5-4 (Gong et al., 2010), KRTAP 6-n (Zhou et al., 2016; Li et al., 2017b), and human KRTAP1-n (Shimomura et al., 2002) and KRTAP4-n (Kariya et al., 2005). The identification of this new deletion in KRTAP6-5 supports the notion that length variation is a structural hallmark of the KRTAPs (Zhou et al., 2015).
The mutation c.59G>C (p. Gly20Ala), c.85T>G (p.Cys29Gly), c.238G>A (p.Gly80Ser), and insertion/deletion mutation would result in the change of glycine. The presence of glycine may affect the secondary structure of proteins, which mainly destroy the helical structure in the polypeptide chain. Glycine is also known as the “helix-breaker,” which leads to a more flexible structure of HGT-KAPs and form a compact protein structure (Monné et al., 1999). The SNPs c.84C>T (p.Ser28Cys), c.85T>G (p.Cys29Gly), c.103C>T (p.Arg35Cys), and insertion/deletion mutation would result in the gain or loss of cysteine. Cysteine is considered to be a rate-limiting amino acid for sheep wool growth (Wickham, 1970), and it is a significant form of disulphide bonds between wool and cashmere fiber growth. Eventually, cysteine is very important in fiber growth and the ability of cashmere fiber protein to cross-link. On behalf of these, it can be predicted that the mutations of KRTAP6-5 may have influence on fiber traits in Longdong cashmere goat, and this is supported by subsequent analysis.
In the six genotypes analyzed, the CC goats showed a lower MFD than four genotype (AA, AB, AC, and AD) goats. Among AA, AC, and CC goats, as there was no difference in the marginal means for these traits between AA and AC goats, this suggests that C has a recessive effect, while A has a dominant effect on these traits. The presence of A was significantly associated with an increase in MFD. The fiber of lower MFD would be more desirable in the market, which means that reducing the frequency of A on goat group would fit the market demand. But as the number of CC goats was a little small, more goats samples will be needed to conform whether the association identified could have utility as a gene-marker for cashmere traits. The effect of other variants (E, F, H, I, J, K, and L) on cashmere traits still needs to explore in the future, especially the J and L, which were produced by length variation.
Given that the only difference between variants A and C is a single synonymous SNP, the difference in wool traits between genotypes AA and CC may be due to this synonymous SNP. Although synonymous mutations lead to no change in amino acid sequence, they may affect the gene function in other ways. There is some evidence showing that synonymous mutation may have a direct impact on gene function because they affect mRNA stability, folding, transport or translation; protein folding; and miRNA-based regulation of expression (Goymer, 2007; Gotea et al., 2015). Research on sheep KRTAP22-1 and KRTAP6-3 also found that the synonymous SNP had effect on wool traits which confirmed this viewpoint again (Li et al., 2017a; 2017b).
The effect of KRTAP6-5 on cashmere traits detected in this study is consistent with the previous finding of a quantitative trait locus (QTL) on chromosome 1 for MFD in medium wool Merinos (Beh et al., 2001), and is similar to the findings reported for ovine KRTAP6-1 and KRTAP6-3 in which variation in these genes was also found to be associated with variation in MFD (Zhou et al., 2015; Li et al., 2019; 2017b). This is probably not surprising given that both genes are clustered next to each other on chromosome 1, and that they belong to the same subfamily gene with a very similar amino acid sequence. The possibility exists that the effects observed for caprine KRTAP6-5 may be due to its linkage to other KRTAPs on the same chromosome. Together, this suggests that KRTAP6-5 is worthy of further study in the context of variation in cashmere traits.
Supplementary Material
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (32060140 and 31860688), Fuxi Young Talents Fund of Gansu Agricultural University (Gaufx-03Y04), Key R & D projects in Gansu Province (18YF1WA082), and Basic Research Creative Groups of Gansu Province (18JR3RA190).
Conflict of interest statement. The authors declare no real or perceived conflicts of interest.
Glossary
Abbreviations
- HGT
high glycine–tyrosine
- KAP
keratin-associated protein
- KRTAP
keratin-associated protein gene
- ORF
open reading frame
- PCR-SSCP
PCR-single strand conformation polymorphism
Literature Cited
- Andrews, M., Visser C., and Van Marle-Köster E.. . 2017. Identification of novel variants for KAP 1.1, KAP 8.1 and KAP 13.3 in South African goats. Small. Rumin. Res. 149:176–180. doi: 10.1016/j.smallrumres.2017.02.014 [DOI] [Google Scholar]
- Beh, K. J., Callaghan M. J., Leish Z., Hulme D. J., Lenane I., and Maddox J. F.. . 2001. A genome scan for QTL affecting fleece and wool traits in Merino sheep. Wool Technol. Sheep Breeding. 49:88–97 [Google Scholar]
- Byun, S. O., Fang Q., Zhou H., and Hickford J. G.. . 2009. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 385:174–175. doi: 10.1016/j.ab.2008.10.024 [DOI] [PubMed] [Google Scholar]
- Fang, Y., Liu W. J., Zhang F. Q., Shao Y. G., and Yu S. G.. . 2010. The polymorphism of a novel mutation of KAP13.1 gene and its associations with cashmere traits on Xinjiang local goat breed in China. Asian. J. Anim. Vet. Adv. 5(1):34–42. doi: 10.3923/ajava.2010.34.42 [DOI] [Google Scholar]
- Gong, H., Zhou H., Dyer J. M., and Hickford J. G.. . 2011. Identification of the ovine KAP11-1 gene (KRTAP11-1) and genetic variation in its coding sequence. Mol. Biol. Rep. 38:5429–5433. doi: 10.1007/s11033-011-0697-2 [DOI] [PubMed] [Google Scholar]
- Gong, H., Zhou H., McKenzie G. W., Hickford J. G., Yu Z., Clerens S., Dyer J. M., and Plowman J. E.. . 2010. Emerging issues with the current keratin-associated protein nomenclature. Int. J. Trichology 2:104–105. doi: 10.4103/0974-7753.77519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, H., Zhou H., McKenzie G. W., Yu Z., Clerens S., Dyer J. M., Plowman J. E., Wright M. W., Arora R., Bawden C. S., Chen Y., Li J., and Hickford J. G.. . 2012. An updated nomenclature for keratin-associated proteins (KAPs). Int J Biol Sci 8(2):258–264. doi: 10.7150/ijbs.3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, H., Zhou H., Forrest R. H. J., Li S., Wang J., Dyer J. M., Luo Y., and Hickford J. G. H.. . 2016. Wool keratin-associated protein genes in sheep—A review. Genes. 7(6):24. doi: 10.3390/genes7060024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goymer, P. 2007. Synonymous mutations break their silence. Nat Rev Genet. 8, 92. doi: 10.1038/nrg2056 [DOI] [Google Scholar]
- Gotea, V., Gartner J. J., Qutob N., Elnitski L., and Samuels Y.. . 2015. The functional relevance of somatic synonymous mutations in melanoma and other cancers. Pigment Cell Melanoma Res. 28:673–684. doi: 10.1111/pcmr.12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M., Cao Q., Wang R., Piao J., Zhao F., and Piao J.. . 2017. Molecular characterization and expression pattern of a novel Keratin-associated protein 11.1 gene in the Liaoning cashmere goat (Capra hircus). Asian-Australas. J. Anim. Sci. 30:328–337. doi: 10.5713/ajas.16.0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, M., Wang L., Li S., Xing M. X., and Zhang X.. . 2011. Characterization and expression analysis of KAP7.1, KAP8.2 gene in Liaoning new-breeding cashmere goat hair follicle. Mol. Biol. Rep. 38:3023–3028. doi: 10.1007/s11033-010-9968-6 [DOI] [PubMed] [Google Scholar]
- Kariya, N., Shimomura Y., and Ito M.. . 2005. Size polymorphisms in the human ultrahigh sulfur hair keratin-associated protein 4, KAP4, gene family. J. Invest. Dermatol. 124:1111–1118. doi: 10.1111/j.0022-202X.2005.23662.x [DOI] [PubMed] [Google Scholar]
- Khan, I., Maldonado E., Vasconcelos V., O’Brien S. J., Johnson W. E., and Antunes A.. . 2014. Mammalian keratin associated proteins (KRTAPs) subgenomes: disentangling hair diversity and adaptation to terrestrial and aquatic environments. BMC Genomics 15:779. doi: 10.1186/1471-2164-15-779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Gong H., Zhou H., Wang J., Li S., Liu X., Luo Y., and Hickford J. G. H.. . 2019. Variation in KRTAP6-1 affects wool fibre diameter in New Zealand Romney ewes. Arch. Anim. Breed. 62:509–515. doi: 10.5194/aab-62-509-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Zhou H., Gong H., Zhao F., Wang J., Liu X., Luo Y., and Hickford J. G. H.. . 2017a. Identification of the ovine keratin-associated protein 22-1 (KAP22-1) gene and its effect on wool traits, Genes. 8(1):27. doi: 10.3390/genes8010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Zhou H., Gong H., Zhao F., Wang J., Luo Y., and Hickford J. G. H.. . 2017b. Variation in the ovine KAP6-3 gene (KRTAP6-3) is associated with variation in mean fibre diameter-associated wool traits. Genes (Basel). 8(8):204. doi: 10.3390/genes8080204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monné, M., Nilsson I., Elofsson A., and Von Heijne G.. . 1999. Turns in transmembrane helices: determination of the minimal length of a “helical hairpin” and derivation of a fine-grained turn propensity scale. J. Mol. Biol. 293:807–814. doi: 10.1006/jmbi.1999.3183 [DOI] [PubMed] [Google Scholar]
- Parris, D., and Swart L. S.. . 1975. Studies on the high-sulphur proteins of reduced mohair. The isolation and amino acid sequence of protein scmkb-m1.2. Biochem. J. 145:459–467. doi: 10.1042/bj1450459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman, J., Thomas A., Perloiro T., Clerens S., and de Almeida A. M.. . 2019. Characterisation of white and black merino wools: a proteomics study. Animal 13:659–665. doi: 10.1017/S1751731118001647 [DOI] [PubMed] [Google Scholar]
- Powell, B. C., and Rogers G. E.. . 1997. The role of keratin proteins and their genes in the growth, structure and properties of hair. Exs 78:59–148. doi: 10.1007/978-3-0348-9223-0_3 [DOI] [PubMed] [Google Scholar]
- Rogers, G. E. 2006. Biology of the wool follicle: an excursion into a unique tissue interaction system waiting to be re-discovered. Exp. Dermatol. 15:931–949. doi: 10.1111/j.1600-0625.2006.00512.x [DOI] [PubMed] [Google Scholar]
- Rogers, M. A., and Schweizer J.. . 2005. Human KAP genes, only the half of it? Extensive size polymorphisms in hair keratin-associated protein genes. J. Invest. Dermatol. 124:vii–vix. doi: 10.1111/j.0022-202X.2005.23728.x [DOI] [PubMed] [Google Scholar]
- Rogers, G. R., J. G. Hickford, and R. Bickerstaffe. 1994. Polymorphism in two genes for B2 high sulfur proteins of wool. Anim. Genet. 25(6):407–415. doi: 10.1111/j.1365-2052.1994.tb00531.x [DOI] [PubMed] [Google Scholar]
- Rogers, M. A., Langbein L., Winter H., Ehmann C., Praetzel S., and Schweizer J.. . 2002. Characterization of a first domain of human high glycine-tyrosine and high sulfur keratin-associated protein (KAP) genes on chromosome 21q22.1. J. Biol. Chem. 277:48993–49002. doi: 10.1074/jbc.M206422200 [DOI] [PubMed] [Google Scholar]
- Rogers, M. A., Langbein L., Praetzel-Wunder S., and Giehl K.. . 2008. Characterization and expression analysis of the hair keratin associated protein KAP26.1. Br. J. Dermatol. 159: 725–729. doi: 10.1111/j.1365-2133.2008.08743.x [DOI] [PubMed] [Google Scholar]
- Rogers, M. A., Winter H., Langbein L., Wollschläger A., Praetzel-Wunder S., Jave-Suarez L. F., and Schweizer J.. . 2007. Characterization of human KAP24.1, a cuticular hair keratin-associated protein with unusual amino-acid composition and repeat structure. J. Invest. Dermatol. 127:1197–1204. doi: 10.1038/sj.jid.5700702 [DOI] [PubMed] [Google Scholar]
- Shah, R. M., Ganai T. A., Sheikh F. D., Shanaz S., Shabir M., and Khan H. M.. . 2013. Characterization and polymorphism of keratin associated protein 1.4 gene in goats. Gene 518: 431–442. doi: 10.1016/j.gene.2012.12.021 [DOI] [PubMed] [Google Scholar]
- Shimomura, Y., Aoki N., Schweizer J., Langbein L., Rogers M. A., Winter H., and Ito M.. . 2002. Polymorphisms in the human high sulfur hair keratin-associated protein 1, KAP1, gene family. J. Biol. Chem. 277:45493–45501. doi: 10.1074/jbc.M206398200 [DOI] [PubMed] [Google Scholar]
- Wang, J., Che L., Hickford J. G. H., Zhou H., Hao Z., Luo Y., Hu J., Liu X., and Li S.. . 2017. Identification of the caprine keratin-associated protein 20-2 (KAP20-2) gene and its effect on cashmere traits. Genes (Basel). 8(11):328. doi: 10.3390/genes8110328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Hao Z., Zhou H., Luo Y., Hu J., Liu X., Li S., and Hickford J. G. H.. . 2018. A keratin-associated protein (KAP) gene that is associated with variation in cashmere goat fleece weight. Small. Rumin. Res. 167:104–109. doi: 10.1016/j.smallrumres.2018.08.014 [DOI] [Google Scholar]
- Wang, X., Zhao Z. D., Xu H. R., Qu L., Zhao H. B., Li T., and Zhang Z. Y.. . 2012. Variation and expression of KAP9.2 gene affecting cashmere trait in goats. Mol. Biol. Rep. 39:10525–10529. doi: 10.1007/s11033-012-1937-9 [DOI] [PubMed] [Google Scholar]
- Wang, J., Zhou H., Luo Y., Zhao M., Gong H., Hao Z., Hu J., and Hickford J. G. H.. . 2019. Variation in the Caprine KAP24-1 Gene Affects Cashmere Fibre Diameter. Animals (Basel). 9(1):15. doi: 10.3390/ani9010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Zhou H., Hickford J. G. H., Zhao M., Gong H., Hao Z., Shen J., Hu J., Liu X., Li S., and Luo Y.. . 2020. Identification of caprine KRTAP28-1 and its effect on cashmere fiber diameter. Genes (Basel). 11(2):121. doi: 10.3390/genes11020121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, G. A. 1970. Wool growth in relation to sulphur-containing amino acid administration to sheep. Proceedings of the New Zealand Society of Animal Production 30: 209–215. New Zealand Society of Animal Production [Google Scholar]
- Yin, J., Hu T., Li J., Zhang C., Guo Z., Zhou H.. . 2004. Construction of a skin cDNA library of cashmere goat and cloning of KAP6-2 full-length cDNA. Zool. Res. 25(2):166–171 [Google Scholar]
- Zhao, M., Chen H., Wang X., Yu H., Wang M., Wang J., Lan X. Y., Zhang C. F., Zhang L. Z., Guo Y. K., Zhang B., and Hu S. R.. . 2009. aPCR-SSCP and DNA sequencing detecting two silent SNPs at KAP8.1 gene in the cashmere goat. Mol Biol Rep. 36(6):1387–1391. doi: 10.1007/s11033-008-9325-1 [DOI] [PubMed] [Google Scholar]
- Zhao, M., H. Zhou, J. G. H. Hickford, H. Gong, J. Wang, J. Hu, X. Liu, S. Li, Z. Hao, and Y. Luo. 2019. Variation in the caprine keratin-associated protein 15-1 (KAP15-1) gene affects cashmere fibre diameter. Arch. Anim. Breed. 62(1):125–133. doi: 10.5194/aab-62-125-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M., Zhou H., Luo Y., Wang J., Hu J., Liu X., Li S., Hao Z., Jin X., Song Y., Wu X., Hu L., and Hickford J. G. H.. . 2020. Variation in the caprine keratin-associated protein 27-1 gene is associated with cashmere fiber diameter. Genes (Basel). 11(8):934. doi: 10.3390/genes11080934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M., Zhou H., Luo Y., Wang J., Hu J., Liu X., Li S., Zhang K., Zhen H., and Hickford J. G. H.. . 2021. Variation in a newly identified caprine KRTAP gene is associated with raw cashmere fiber weight in Longdong cashmere goats. Genes. 12(5):625. doi: 10.3390/genes12050625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Gong H., Li S., Luo Y., and Hickford J. G.. . 2015. A 57-bp deletion in the ovine KAP6-1 gene affects wool fibre diameter. J. Anim. Breed. Genet. 132:301–307. doi: 10.1111/jbg.12138 [DOI] [PubMed] [Google Scholar]
- Zhou, H., Gong H., Wang J., Dyer J. M., Luo Y., and Hickford J. G.. . 2016. Identification of four new gene members of the KAP6 gene family in sheep. Sci. Rep. 6:24074. doi: 10.1038/srep24074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Hickford J. G., and Fang Q.. . 2006. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chain reaction amplification. Anal. Biochem. 354:159–161. doi: 10.1016/j.ab.2006.03.042 [DOI] [PubMed] [Google Scholar]
- Zhao, M., Wang X., Chen H., Lan X. Y., Guo Y. K., Li J. Y., Wei T. B., Jing Y. J., Liu S. Q., Zhang M. H., and Gao Q. W.. . 2008. The PCR-SSCP and DNA sequencing methods detecting a large deletion mutation at KAP6.2 locus in the cashmere goat. Small Rumin. Res. 75(2):243–246. doi: 10.1016/j.smallrumres.2007.10.007 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.