Abstract
Small heat shock proteins (sHSPs) are known to exhibit in vitro chaperone activity by suppressing the aggregation of misfolded proteins. The 12‐kDa sHSPs (Hsp12s) subfamily members from Caenorhabditis elegans, including Hsp12.2, Hsp12.3, and Hsp12.6, however, are devoid of such chaperone activity, and their in vivo functions are poorly understood. Here we verified that Hsp12.1, similar to its homologs Hsp12.2, Hsp12.3, and Hsp12.6, hardly exhibited any chaperone activity. Strikingly, we demonstrated that these Hsp12s seem to play crucial physiological roles in C. elegans, for suppressing dauer formation and promoting both longevity and reproduction. A unique sHSP gene from Filarial nematode worm Brugia malayi was identified such that it encodes two products, one as a full‐length Hsp12.6 protein and the other one having an N‐terminal arm of normal length but lacks the C‐terminal extension. This gene may represent an intermediate form in evolution from a common sHSP to a Hsp12. Together, our study offers insights on what biological functions the chaperone‐defective sHSPs may exhibit and also implicates an evolutionary scenario for the unique Hsp12s subfamily.
Keywords: chaperone, dauer, evolution, Hsp12s, longevity, reproduction, small heat shock protein, thermal resistance
1. INTRODUCTION
The protein quality control system is essential for cells to produce functional proteins and remove harmful misfolded or aggregated proteins. Molecular chaperone proteins are able to prevent protein aggregation or misfolding and assist protein folding or assembly, and thus play essential roles in protein quality control.1 Small heat shock proteins (sHSPs), a virtually ubiquitous molecular chaperone family, are characterized in primary structure by possessing a relatively conserved α‐crystallin domain, which is flanked by a highly variable N‐terminal arm and a flexible C‐terminal extension.2 The sHSPs are known to prevent protein aggregation3, 4 and assist protein refolding in cooperation with other molecular chaperones such as Hsp70s and Hsp60s.5, 6, 7 As such, the over‐expression of sHSPs has been found to increase cell tolerance against stress threats.8, 9, 10, 11
The sHSPs are considered as the first line of defense against stress‐induced protein aggregation in cells.12, 13 They have been reported to be involved in a wide range of cellular processes in animals and humans, as exampled by mouse Hsp25 in cell differentiation,14 human Hsp27 in apoptosis10 and fly Hsp22 in longevity.15 The dysfunctions of certain sHSPs have been linked to such diseases as cancer development,9 cardiovascular diseases,16, 17 cataracts,18 myopathy,19 and motor neuron diseases.20, 21 Such physiological and pathologic roles of sHSPs are generally believed to be underlain by their in vitro chaperone activity in suppressing protein aggregation/misfolding.
Nevertheless, Hsp12.3 and Hsp12.6 from the nematode worm Caenorhabditis elegans, though being unable to act as a chaperone in vitro,22, 23 were found to contribute to the hypertonic stress‐resistant phenotype of age‐1 animals.24 This fact raises a possibility that the function of certain sHSPs does not rely on an in vitro chaperone activity. In retrospect, Hsp12.3 and Hsp12.6, together with Hsp12.1 and Hsp12.2, comprise a unique sHSP subfamily (Hsp12s) among the 14 sHSPs of C. elegans.22 Hsp12s, though containing the relatively conserved α‐crystallin domain, are distinguished from the common sHSPs by carrying a much shorter N‐terminal arms and lacking the flexible C‐terminal extension.22, 23 Further, unlike the common sHSPs that usually form large oligomers of 12–40 subunits,25, 26, 27 Hsp12.2 and Hsp12.3 exist as tetramers23 while Hsp12.6 as dimers or monomers.22 Notably, Hsp12.6 was reported to act downstream of DAF‐16, the primary transcription factor required for the profound lifespan extension observed when the insulin‐like receptor daf‐2 is defective in C. elegans.28, 29
Here we attempted to clarify the physiological function of the unique Hsp12s subfamily members that hardly possess in vitro chaperone activity. We found that each of these four Hsp12s acts as a suppressor for dauer formation and as positive effector for promoting both lifespan and reproduction in C. elegans. In addition, we identified a unique sHSP gene representing an intermediate form in evolution from common sHSPs to Hsp12s. Our study offers insights into the functions of sHSPs that hardly exhibit in vitro chaperone activities and also helps to establish the evolutionary scenario for the unique Hsp12s subfamily.
2. RESULTS
2.1. Hsp12.1 exists as small oligomers under in vitro conditions and exhibits little chaperone activity
Given that sHSPs usually exist as large oligomers comprising 12–40 subunits and that Hsp12.2 and Hsp12.3 exist as tetramers23 and Hsp12.6 as dimer or monomer,22 we first examined the oligomeric status of Hsp12.1 under in vitro conditions. For this, we purified the recombinant Hsp12.1 protein expressed in E. coli bacterial cells via affinity‐chromatography and examined its oligomeric state via blue native PAGE analysis, a technique that allows separation of multiprotein complexes in their native conformations. The results displayed in Figure 1a reveals that the purified Hsp12.1 protein (lane 5) mobilizes on the gel at a rate higher than that of the monomeric form of the bovine serum albumin (BSA, 66 kDa; lane 1) standard, similar to that of the monomeric alcohol dehydrogenase (40 kDa, lane 2), or slower than that of either the trypsin inhibitor (6.5 kD; lane 3) or insulin (5.8 kD; lane 4). This indicates that Hsp12.1 exists as small oligomers, being made of 2–4 subunits. Further, formaldehyde‐mediated chemical crosslinking coupled with SDS‐PAGE analysis (Figure 1b) revealed that Hsp12.1 seems to exist as trimers or dimers at low concentration (0.2 mg/ml; lane 2), as tetramers, trimmers or dimers at a higher concentration (1 mg/ml; lane 5). Together, these results indicate that Hsp12.1 exist as small rather than large oligomers under in vitro conditions.
FIGURE 1.
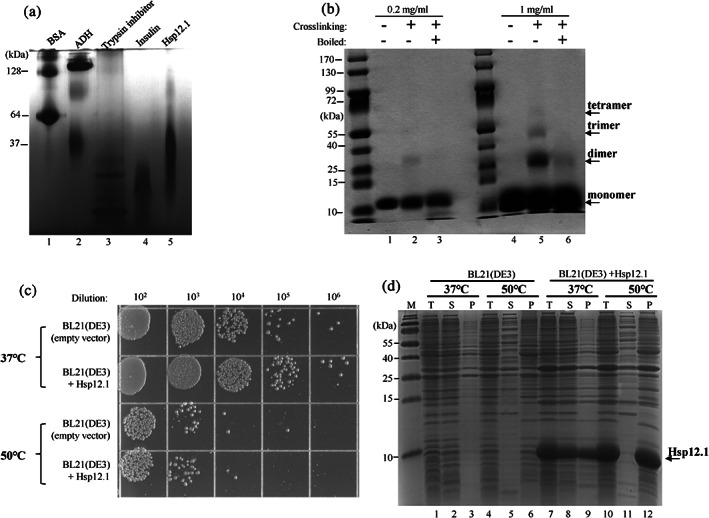
The Hsp12.1 protein exhibits little chaperone activity and exists as small oliogmers. (a) Blue native PAGE analysis of the purified Hsp12.1 protein (lane 5), with other indicated proteins (BSA: bovine serum albumin); ADH: alcohol dehydrogenase) being analyzed in parallel for molecular size comparison. (b) Coomassie blue staining results of SDS‐PAGE analysis of the purified Hsp12.1 protein being crosslinked with formaldehyde. The reaction was performed by incubating Hsp12.1 (at a final concentration of 0.2 mg/ml or 1 mg/ml) with formaldehyde (0.5%) for 5 min and quenched with glycine (1 M). The samples were also boiled in the SDS sample‐loading buffer to disrupt the chemical crosslinkages (lanes 3 and 6). (c) Survival of the E. coli BL21(DE3) cells with or without the heterologous expression of Hsp12.1, after being treated at 37°C or 50°C for 1 hr. (d) Coomassie blue staining results of SDS‐PAGE analysis of proteins present in the supernatant or pellet fractions of the cell lysates of BL21(DE3) cells with or without the heterologous expression of Hsp12.1 after the cells were treated at 37°C or 50°C for 1 hr. T, total cell lysate; S, supernatant; P, pellet
We then demonstrated that the Hsp12.1 protein exhibited little in vitro chaperone activity. To this end, we found that Hsp12.1, unlike common sHSPs as reported by us30 and others,31, 32, 33, 34, 35, 36 did not increase the thermal resistance of E. coli cells against lethal heat shock (50°C, 1 hr) when it was heterologously over‐expressed (Figure 1c). In line with this, we found that the over‐expressed Hsp12.1 protein, unlike C. elegans Hsp17 as we reported earlier,30 hardly prevented the thermal aggregation of cellular proteins when the cells were heated at 50°C (lane 5 vs. 11, lane 6 vs. 12, Figure 1d), although Hsp12.1 was largely detected in the protein aggregates (lane 11 vs. 12). These results thus indicate that Hsp12.1, similarly to Hsp12.2, Hsp12.3, and Hsp12.6 (22; 23; 37), is apparently devoid of any chaperone activity.
2.2. The Hsp12s are constitutively expressed at all developmental stages but absent in the dauer state in the C. elegans worms
We then aimed to investigate the physiological functions of chaperone‐defective Hsp12.1 and its homologs (i.e., Hsp12.2, Hsp12.3, and Hsp12.6). We first examined their expression pattern during different developmental stages, given that the expression of certain sHSPs in animals has been reported to vary during development.37 Immunoblotting analyses, using polyclonal antibodies against Hsp12.1, Hsp12.2, Hsp12.3, and Hsp12.6, respectively, revealed that: (a) none of them was significantly detectable in the embryos (lane 1, Figure 2a); (b) all were clearly detected at the four larvae stages (from L1 to L4; lanes 2–5); (c) Hsp12.1 and Hsp12.6, but hardly Hsp12.2 or Hsp12.3, were detected in the young adult worms (lane 6). These observations suggest that the Hsp12s may play certain roles during the normal development of worms.
FIGURE 2.
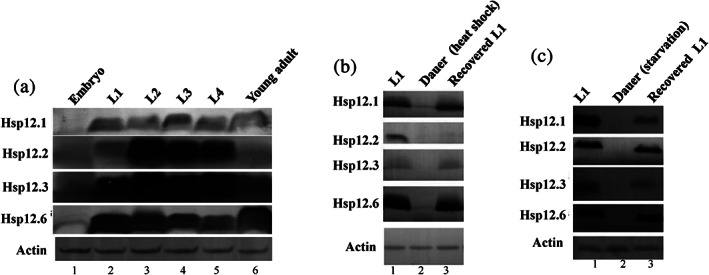
The Hsp12s are constitutively expressed at normal development stages but not in dauer C. elegans worms. (a) Immunoblotting results for analyzing the level of the Hsp12 proteins in different developmental stages of C. elegans worms. (b) Immunoblotting results for analyzing the level of the Hsp12 proteins in the heat shock temperature‐induced dauer (lane 2) and recovered worms (lane 3). (c) Immunoblotting results for analyzing the level of the Hsp12 proteins in the starvation‐induced dauer (lane 2) and recovered worms (lane 3). Actin was analyzed as a loading control
Interestingly, we failed to detect Hsp12s in the C. elegans worms that entered the dauer state as induced either by heat shock (lane 2, Figure 2b), or by starvation (lane 3, Figure 2b). Upon recovery from the dauer state, all of them (except Hsp12.2 for the dauer state induced by heat shock) were re‐appeared (Figure 2c). These observations implicate that the Hsp12s may play a certain role for the C. elegans animals to enter and exit the dauer state. It should be pointed out that some protein/genes are up‐regulated in the dauer stage compared to normal larvae worms as revealed by earlier proteomic and transcriptomic studies.38, 39, 40, 41 Conceivably, those proteins up‐regulated in the dauer state may be involved in maintaining this unique C. elegans developmental stage that endow the nematode an increased longevity, a stress resistance, the nictation behavior, as well as a metabolism different from that of normal worms. In addition, we observed that the time‐dependent re‐expression pattern of Hsp12.1 and Hsp12.6 following recovery from dauer stage was different: the Hsp12.1 keeps increase after 3 hr of recovery while Hsp12.6 becomes significantly decreased again after 3 hr (Figure S1).
2.3. A knockdown of each hsp12 gene alone significantly enhances dauer formation for C. elegans worms
In light of these dramatic changes in the levels of the Hsp 12 proteins during and after the dauer stage, we next investigated the potential functions of Hsp12s in C. elegans dauer formation. First, we tried to decrease the levels of the Hsp12s via RNA interference (RNAi), a technique that has been effectively applied for investigating gene functions in C. elegans.42, 43 Experimentally, the L4 stage of the wild type N2 worms were fed with E. coli cells carrying the hsp12.1, hsp12.2, hsp12.3, or hsp12.6 RNAi plasmid for 3 days. The immunoblotting results presented in Figure 3a indicate that our RNAi manipulations did effectively decrease the protein level of each Hsp12 in the worms.
FIGURE 3.
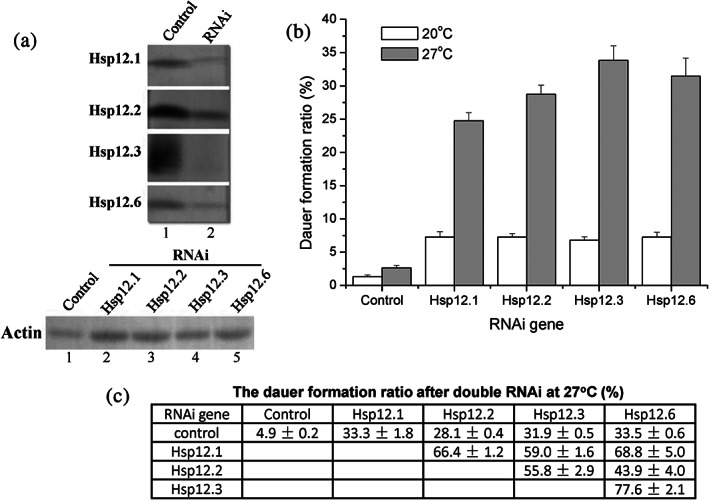
Knockdown of each Hsp12s gene led to an increase of the dauer formation ratio for C. elegans worms. (a) Immunoblotting results for analyzing the levels of the Hsp12 proteins in worms fed on bacteria carrying a Hsp12 RNAi vector (lane 2) or the control vector (lane 1), using antibodies against each Hsp12 (the upper part). Actin was analyzed as an internal control to indicate equal sample loading (the bottom part). (b). Dauer formation ratio of the N2 wild type worms that were fed on RNAi bacteria carrying the control vector or a Hsp12 RNAi vector were grown at 20°C (white bars) or 27°C (gray bars). (c) Dauer formation ratio of the N2 wild type worms fed on E. coli bacterial cells expressing two types of Hsp12 RNAi and grown at 27°C. Results represent mean ± SE of three replicates
In light of our observation that the Hsp12s are not expressed in the dauer form but re‐expressed upon recovery from dauer state (Figure 2b,c), we then analyzed whether their knockdown affected the dauer formation of the C. elegans worms. Remarkably, we observed a significantly enhanced dauer formation ratio for the knockdown worms when cultured at 20°C (white bars, Figure 3b), and such enhancement effect was even stronger when the worms were cultured at a higher temperature of 27°C (gray bars). These results, together with their presence in the larval stages and absence in the dauer stage (Figure 2b), suggest that Hsp12s act as suppressors for dauer formation in worms. Of note, we found that a simultaneous knockdown of any two of the four Hsp12s genes only led to an additive, instead of synergistic, effect (Figure 3c), indicating their function to be somehow redundant.
2.4. A knockdown of each hsp12 gene decreases both the lifespan and reproduction rate of the C. elegans worms
It has been reported that the absence of small heat shock protein Hsp22 in the fruit fly Drosophila led to a 40% decrease in the lifespan of the animals.15 Here, we also observed that a knockdown of each hsp12 gene led to a significant and similar degree of decrease in the average lifespan of the worms from 17 days to 13 days when cultured at the normal temperature of 20°C (Figure 4a, Figure S2A). Of note, the lifespan reduction effect was similarly significant when these hsp12‐knockdown worms were cultured at the heat shock temperature of 25.5°C (Figure 4b), with the average lifespan of the worms being shortened from 10 days to 7 days (Figure S2B).
FIGURE 4.
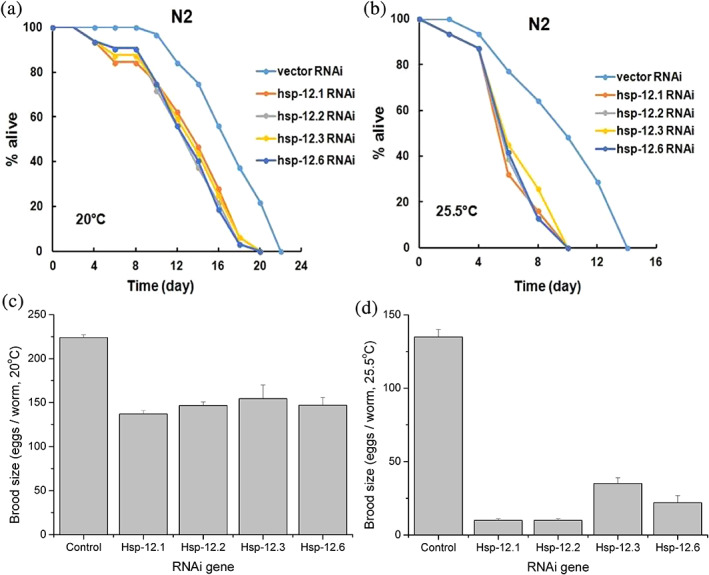
Knockdown of each hsp12 gene decreased the lifespan and reproduction rate of the C. elegans worms. (a,b) Lifespan curves of the N2 wild type C. elegans worms fed on bacteria carrying the indicated Hsp12 RNAi vector or the control vector and grown at either 20°C (Panel a) or 25.5°C (Panel b). (c,d) Brood size of the N2 wild type worms fed on bacteria carrying a Hsp12 RNAi vector or the control vector and grown at 20°C (Panel c) or 25.5°C (Panel d). Results represent mean ± SE of three replicates
It was reported that the Hsp12s are specifically expressed in the reproductive tissues of worms,44 raising the possibility that the Hsp12s are functionally linked to their reproduction. In support of this, here we found that the knockdown of each hsp12 gene decreased the brood size of the worms by about 30% when cultured under normal temperature of 20°C (Figure 4c), and by about 75% when cultured at the heat shock temperature of 25.5°C (Figure 4d). All these results, together with the fact that the Hsp12s are constitutively expressed at all the developmental stages, indicate that the Hsp12s play important roles for the longevity and reproduction of the C. elegans worms.
2.5. Hsp12s and common sHSPs in C. elegans exhibit a monophyletic grouping distinguishable from that of the sHSPs in fruit flies and vertebrates
Although gene duplication and lateral gene transferring are believed to account for the evolutionary production of such a diversified sHSPs molecular chaperone family that is ubiquitously found in all the three domains of life,45, 46, 47 how this unique Hsp12s subfamily has been evolved is largely unknown. To probe this, we first examined whether Hsp12s, like common sHSPs, are ubiquitously present in all three domains of life. To this end, we performed BLAST analysis on the UniProtKB database by using the amino acid sequence of each C. elegans Hsp12 protein as a template. We extracted a total of 250 protein sequences by setting the sequence identity requirement as 30% (with the homologs of Hsp12.6 being listed in Table S1).
Of note, the proteins displayed in Table S1 are exclusively found in metazoans, with none from plants, fungi, bacteria and archaea, consistent with earlier reports showing that the sHSPs of animals overall exhibit a monophyletic grouping.46, 48, 49 It is therefore concluded that Hsp12s are not ubiquitously being present in all life, but present only in such animals as the metazoans. Further phylogenetic analyses reveal that the C. elegans Hsp12s, together with other sHSPs in C. elegans, exhibit a monophyletic grouping, being distinguishable from the sHSPs derived from fruit flies and other vertebrates (Figure 5). This result implicates that the Hsp12 genes in worms should have been evolved from the common sHSPs genes in worms rather than from the common sHSPs genes in metazoans.
FIGURE 5.
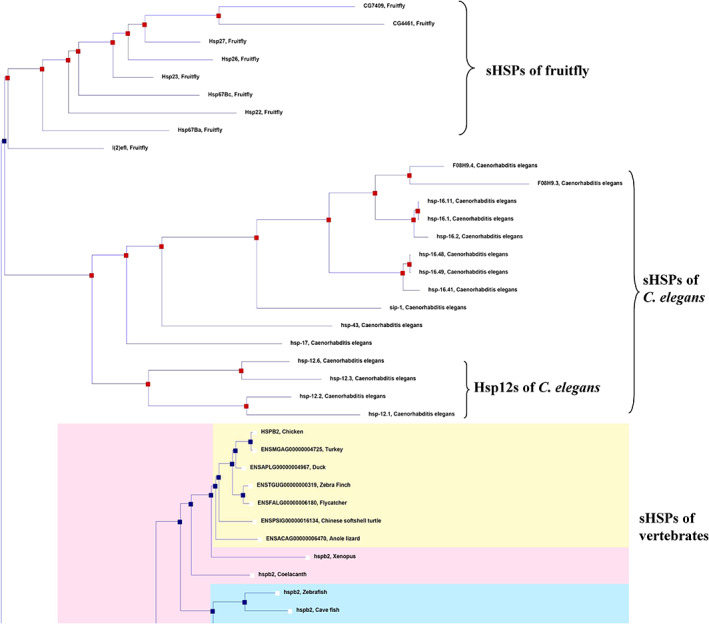
Monophyletic grouping of the Hsp12s subfamily. The phylogenetic tree (shown in part) was generated on the Ensembl database. Apparently, the Hsp12s subfamily in C. elegans is closest to the common sHSPs of C. elegans, being separated from the sHSPs of fruitfly and vertebrates
2.6. Identification of a unique sHSP gene that may represent an evolutionary intermediate form from common sHSPs to Hsp12s
The Hsp12s are unique in their amino acid sequences by having a much shorter N‐terminal arm and lacking the C‐terminal extension, in comparison with the common sHSPs (as illustrated in Figure 6a; also refer to22, 23). It is conceivable that two types of sHSP intermediate gene subgroups might have existed as intermediates during the evolution of the Hsp12 genes from the common sHSP genes of the nematode worms: intermediate subgroup I that possessed a short N‐terminal arm and a full C‐terminal extension and intermediate subgroup II that possessed a normal N‐terminal arm but lacking the C‐terminal extension (as schematically illustrated in Figure 6a).
FIGURE 6.
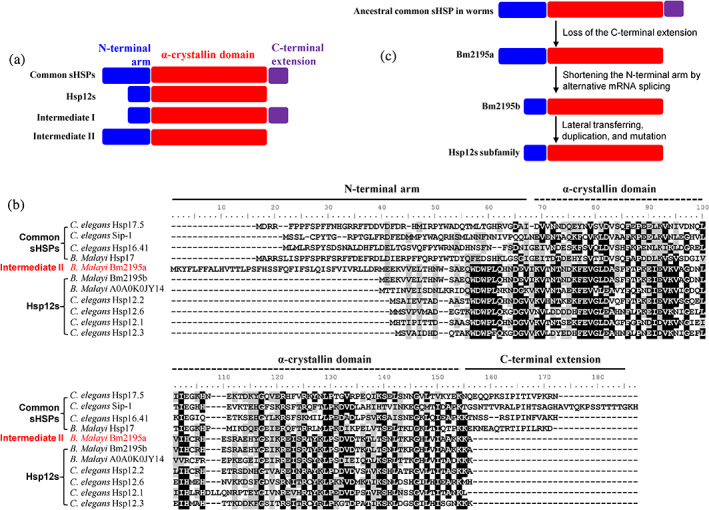
Identification of Bm2195a gene from B. Malayi as an evolutionary intermediate from common sHSPs to the Hsp12s subfamily. Domain organization of the common sHSPs, Hsp12s subfamily, and the proposed evolutionary intermediates I and II. The N‐terminal arm, the α‐crystallin domain and the C‐terminal extension are shown in blue, red, and purple, respectively. (b) Sequence alignment of representative common sHSPs and Hsp12s from worms. The highly conserved residues are indicated by black backgrounds. The potential evolutionary intermediate Bm2195a is highlighted in red. (c) Schematic illustration of the evolution of Hsp12s from the common sHSPs via the intermediate Bm2195a
We attempted to examine whether such intermediate subgroup forms are present in current animal species. To this end, BLAST analyses on the UniProtKB database using the amino acid sequence of Hsp12.1, Hsp12.3, Hsp12.2, or Hsp12.6 of C. elegans revealed one specific gene (ID: Bm2195) from Filarial nematode worm Brugia malayi. This gene encodes a sHSP protein (ID: Bm2195a) that apparently possesses an N‐terminal arm of normal length but lacks the C‐terminal extension (as indicated by data shown in Figure 6b). We propose this gene represents a member of the intermediate subgroup II formed during the evolutionary production of Hsp12s. Notably, there are also a Bm2195a‐like protein in the Filarial nematode worm species Brugia pahangi and Brugia timori (Figure S3), which might be produced due to lateral gene transfer among them.
Interestingly, a splicing mRNA variant lacking the first exon (Figure S4) can be produced from the B. malayi Bm2195 gene for guiding the synthesis of a Hsp12.6 protein (ID: Bm2195b or BmHsp12.6). Apparently, BmHsp12.6 has a much shorter N‐terminal arm and meanwhile lacks the C‐terminal extension (Figure 6b). Of note, BmHsp12.6 was recently identified as a human IL‐10 receptor binding protein50 and may potentially be used as a vaccine for preventing lymphatic filariasis.51 Therefore, the B. malayi Bm2195 gene represents an excellent case in which both intermediate II and mature forms of Hsp12s are manifested via alternative mRNA splicing.
3. DISCUSSION
The physiological functions of sHSPs are generally attributed to their chaperone activity.13, 52, 53, 54 Our work reported here apparently challenges this currently prevailing view, because the C. elegans Hsp12s exhibit little chaperone activity (Figure 1c,d, and also refer to earlier reports22, 23, 55) but apparently play crucial physiological roles in dauer formation, longevity and reproduction (Figures 2, 3, 4). These observations are consistent with a previous report showing that Hsp12 transcriptions occur mainly in the reproductive tissues of worms.44 On the other hand, Giese et al previously reported that a few mutant forms of Hsp16.6 (a sHSP from cyanobacteria Synechocystis), although exhibiting chaperone activity in vitro, were functionally impaired in vivo for protecting the host cells from being killed by heat shock.56 These reported observations suggest that the chaperone activity is not indispensable for explaining the in vivo biological functions of certain sHSPs, even though the chaperone activity accounts for the in vivo functions in most cases.
How the Hsp12s having little in vitro chaperone activity play their physiological functions in vivo remains elusive. It is well established that residues involved in binding substrate proteins are primarily located at the N‐terminal arm and also substantially in the N‐terminus of the α‐crystallin domain for the common sHSPs, as revealed by unnatural amino acid‐mediated protein photo‐crosslinking analyses57, 58 as well as by chemical‐crosslinking analyses.59, 60, 61 Another feature of the common sHSPs is their existence as large oligomers usually comprising 12–24 subunits,25, 26 which may facilitate the formation of soluble sHSP‐substrate complexes that is a prerequisite for them to suppress protein aggregation. In these regards, it is conceivable that Hsp12s are able neither to act as chaperones in suppressing the aggregation of substrate proteins not to protect bacterial cells from being killed by heat shock, likely due to their incapability to form large oligomers (Figure 1c,d; also refer to earlier reports22, 23). More likely, the inability for Hsp12s to exhibit chaperone activity is due to the lack of the major substrate‐binding sites in the N‐terminal arm of Hsp12s, as implicated by our comparative hydropathy plot analysis between the common sHSPs and Hsp12s (Figure S5A vs. Figure S5B).
Nevertheless, Hsp12s may contain substrate‐binding residues in the α‐crystallin domain, as implicated by our hydropathy plot analysis results presented in Figure S5B. As such, they are still capable of interacting with certain specific cellular proteins of worms that are the natural substrate proteins of common sHSPs. It follows that the Hsp12s would not exhibit any chaperone activity toward those artificial substrate proteins commonly analyzed under in vitro conditions. In this regard, the interactions between Hsp12s and the specific cellular proteins in worms would accordingly change the functional and structural status of the Hsp12s‐interacting proteins, which, in turn, affect certain biological processes, including those involved in the dauer formation, longevity and reproduction of worms. Identification of the Hsp12s‐interacting proteins in worms may guide to dissect the molecular mechanism underlying Hsp12s‐mediated changes in the dauer formation, longevity and reproduction of worms. Another approach to uncovering the underlying mechanism is to investigate the functional cross‐talks between Hsp12s and such critical pathways as TGF‐β and insulin‐like signaling pathways that are well known to play crucial roles in stress resistance, dauer formation and longevity.62, 63 It is conceivable that under these circumstances, the Hsp12s do not have to function as chaperones, but to play other roles.
So far, there is no report on the origin and evolution of this unique Hsp12s subfamily since their identification about 20 years ago.22 As a matter of fact, sHSPs are divergent in their primary sequences (especially the highly variable N‐terminal arms), with their evolutionary origin being difficult to be traced.45, 46, 48 Here, our identification of the Bm2195 sHSP gene, which encodes both a Hsp12 protein (i.e., BmHsp12.6) and an intermediate form (Bm2195a) as a result of alternative mRNA splicing (Figure 6b, Figure S4), somehow suggests that this gene represents the ancestral one of the Hsp12s subfamily (as indicated in Figure 6c). The evolution of the Hsp12s subfamily is proposed to take place via three steps (as illustrated in Figure 6c): loss of the C‐terminal extension, shortening of the N‐terminal arm and subsequent lateral transferring, duplication and mutation of the genes. Since the Hsp12s are apparently distinguished from the sHSPs found in fruit flies and vertebrates (Figure 5, Table S1), this subfamily is apparently young in evolutionary terms. Less likely, the Hsp12s subfamily was evolved since worms appear during the evolution history, but they were discarded in such higher organisms as fruit flies and vertebrates because they are no longer needed and thus became lost due to certain deleterious events.
4. MATERIALS AND METHODS
4.1. Nematode strains, maintenance, and culture
The wild type Bristol N2 was obtained from Caenorhabditis Genetics Centre (University of Minnesota) and was used for all analyses. OP50 bacterial cells were inoculated into LB broth and grown overnight at 37°C. The culture was then used to prepare bacterial lawns on standard nematode growth media agar plates. Nematodes were grown on NGM (nematode growth medium) agar plates seeded with Escherichia coli strain OP50 at 20°C unless otherwise specified.
4.2. Heterologous expression of Hsp12.1 in E. coli
The coding sequence of Hsp12.1 was amplified and inserted into the expression vector pET21a after being digested with restriction enzymes NdeI and HindIII. The expression plasmid pET21a‐Hsp12.1 was transformed into E. coli BL21(DE3) (Transgen). Bacterial cells were cultured at 37°C in LB (Luria‐Bertani) broth medium containing antibiotics at final concentrations of 100 μg/ml for ampicillin. Protein expression was induced by 1 mM of IPTG.
4.3. Protein purification
E. coli BL21(DE3) cells expressing Hsp12.1 were harvested and re‐suspended in 20 mM Tris–HCl, pH 8.0 and lysed by high pressure. Cell lysates were centrifuged at 10,000g for 10 min and the His‐tagged Hsp12.1 was purified from the supernatant via Ni‐NTA affinity chromatography and the protein was then dialyzed against 20 mM phosphate sodium buffer (pH 7.4).
4.4. Protein oligomeric structure analyses
The purified Hsp12.1 protein (1 mg/ml) was dissolved in blue native sample‐loading buffer (containing 0.1% Coomassie Blue G‐250) and resolved on the acrylamide gel (8%). The electrophoresis was first performed at 120 V in Anode Buffer (25 mM imidazole, pH 7.0) and Cathode Buffer A (50 mM Tricine, 7.5 mM imidazole, 0.02% Coomassie Blue G‐250, pH 7.0) for 1 hr before switching the Cathode Buffer A to Cathode Buffer B (50 mM Tricine, 7.5 mM imidazole, 0.002% Coomassie Blue G‐250, pH 7.0) for another 2 hr. In addition, the purified Hsp12.1 protein (at a final concentration of 0.2 or 1 mg/ml) was incubated with formaldehyde (at a final concentration of 0.5% v/v) in the phosphate sodium buffer (pH 7.0) at room temperature for 5 min and then mixed with 1 M glycine to stop the crosslinking reaction. The protein samples were mixed with SDS sample‐loading buffer (with or without boiling treatment) and then analyzed by SDS‐PAGE (visualized by coomassie blue staining).
4.5. Thermal resistance analysis
E. coli BL21(DE3) cells expressing Hsp12.1 were heat shocked at 50°C for an hour. Serial dilutions of cell cultures were made with LB medium and 10 μl of each dilution was spotted on dish before incubating overnight at 37°C for colony formation. The control cells (carrying the empty plasmid pET21a) were also treated in a similar manner.
4.6. Protein solubility analysis
E. coli BL21(DE3) cells expressing Hsp12.1, after heat shock treatment at 50°C for an hour, were harvested and re‐suspended in 20 mM phosphate sodium buffer (pH 7.4) and lysed by high pressure. Cell lysates were centrifuged at 800g for 5 min to remove cell debris and unbroken cells, and the supernatant was then subjected to centrifugation at 10,000 g for 30 min, with the resulting supernatant (soluble proteins) and precipitate (aggregated proteins) being analyzed by 10% Tricine SDS‐PAGE and visualized by Coomassie blue staining. In addition, the purified His‐tagged Hsp12.1 protein (at a final concentration of 0.2 mg/ml) was subjected to the heat treatment (at 50°C for an hour), in the absence or presence of insulin (at a final concentration of 0.2 mg/ml) or the extract of worms (1 mg total proteins/ml), and then the protein mixtures were centrifuged at 10,000g for 30 min, with the soluble and pellet fractions before analyzed by SDS‐PAGE.
4.7. RNA‐interference (RNAi) assays
The bacteria carrying RNAi vectors was used to reduce the expression levels of the Hsp12s proteins in C. elegans. HT115 bacteria was transformed with the L440 vector expressing dsRNA of each of the Hsp12 gene or the empty vector, grown at 37°C in LB with 100 μg/ml ampicillin and 10 μg/ml tetracycline, then seeded onto NGM‐ampicillin plates supplemented with 1 mM isopropyl β‐D‐1‐thiogalactopyranoside (IPTG).
4.8. Dauer formation assay
To assay dauer development, strains were synchronized by sodium hypochlorite treatment. The isolated eggs were distributed on 1 mM IPTG‐containing NGM agar plates seeded with hsp12s dsRNA expressing bacterial strains and placed at either 20°C or 27°C. Plates at 27°C were scored for dauers, nondauers, and dead eggs 2–3 days later, while plates at 20°C were scored for dauers, non‐dauers and dead eggs 10–14 days later. Worms that had crawled off the side of the plate were censored. Animals were harvested and suspended in 1% SDS for half an hour, and were then separated from debris by centrifugation through 60% sucrose. The percentage of resulting dauer larvae was scored. Worms that had crawled off the side of the plate were censored. Each experiment was repeated at least two times in triplicates.
4.9. Entry into and exit from dauer larvae
The wild type hermaphrodite from either food exhausted NGM plates at 20°C or plates at 27°C were washed off the plates in M9 buffer. After treatment with SDS (at a final concentration of 1%) at room temperature for 30 min, SDS‐resistant worms were transferred to fresh NGM plates seeded with bacteria and incubated at 20°C to allow for recovery from dauers.
4.10. Lifespan analysis
Strains were synchronized by sodium hypochlorite treatment and eggs were placed on 1 mM IPTG containing NGM agar plates seeded with hsp12s dsRNA expressing bacterial strains. Next, young adult hermaphrodites were transferred onto NGM agar plates containing 20 μg/ml FUdR (Sigma) to inhibit reproduction. Worms were scored every other day until all worms were scored dead by not responding to gentle prodding with a platinum wire. Animals that had crawled off the plate, or became desiccated on the sides of the NGM plate were censored. Lifespan assays were performed in triplicate at 20°C or 25.5°C unless specified, and repeated at least twice.
4.11. Brood size assay
Adult hermaphrodites fed on hsp12s RNAi or control vector RNAi bacteria were picked (6–8 per plate) and allowed to undergo one full generation at 20°C to ensure that the parental strain had not starved or gone through dauer. From these plates, individual L4 or young adult animals were singled to individual plates at 20°C or 25.5°C. The parental animals were transferred every 24 hr to fresh plates.
4.12. Immunoblotting analysis and antibodies
Immunoblotting analysis was performed according to standard protocols. Briefly, equal amounts of protein (100 μg) were loaded into each lane and separated by SDS‐ PAGE (12%) and transferred to polyvinylidene difluoride membranes. Following overnight blocking and washings, the blots were incubated for 1 hr at room temperature with the Rabbit polyclonal antibodies against Hsp12.1, Hsp12.1, Hsp12.3, or Hsp12.6 produced in our own lab at 1:10000 dilution in TBS‐T buffer containing 5% non‐fat dry skim milk. After 3 times washing with TBS‐T, the membranes were incubated for 1 hr at room temperature with secondary antibody goat anti‐rabbit IgG‐alkaline phosphatase conjugate (Transgene) at 1:5000 dilution in the same blocking buffer (TBS‐T buffer containing 5% non‐fat dry skim milk). The protein bands on the PVDF membrane were visualized with NBT (Amresco) and BCIP (Promega). Polyclonal antibodies were made in the Institute of Microbiology of Chinese Academy of Sciences by injecting the purified recombinant Hsp12.1, Hsp 12.2, Hsp12.3, or Hsp12.6 into rabbits.
4.13. Sequence analysis
BLAST search was performed on the UnitProt database (http://www.uniprot.org/). The amino acid sequences of Hsp12s and other common sHSPs proteins were downloaded and subjected to sequence alignment by using the clustalW multiple alignment in BioEdit (version 5.06; Department of Microbiology, North Carolina State University). The α‐crystallin domain sequences were identified according to the sequence alignment result presented by Leroux et al.22 The phylogenetic tree was generated on the Ensembl database (http://asia.ensembl.org/index.html). Hydrophobicity analysis was performed in BioEdit.
AUTHOR CONTRIBUTIONS
Xinmiao Fu: Funding acquisition; supervision; writing ‐ original draft; writing‐review & editing. Anastasia N. Ezemaduka: Investigation; methodology; validation. Xinping Lu: Investigation. Zengyi Chang: Supervision; writing‐review & editing.
Supporting information
Appendix S1. Supporting Information.
ACKNOWLEDGMENTS
We thank Caenorhabditis Genetics Center for the C. elegans N2 wild type strain. This work was supported by the National Natural Science Foundation of China (31770830 and 31770830 to X. F.; 31670775 to Z. C.; 41871095 to A. E.).
Fu X, Ezemaduka AN, Lu X, Chang Z. The Caenorhabditis elegans 12‐kDa small heat shock proteins with little in vitro chaperone activity play crucial roles for its dauer formation, longevity, and reproduction. Protein Science. 2021;30:2170–2182. 10.1002/pro.4160
Xinmiao Fu and Anastasia N. Ezemaduka contributed equally to this work.
Funding information National Natural Science Foundation of China, Grant/Award Numbers: 41871095, 31670775, 31770830
Contributor Information
Xinmiao Fu, Email: xmfu@fjnu.edu.cn.
Zengyi Chang, Email: changzy@pku.edu.cn.
REFERENCES
- 1.Hartl FU, Bracher A, Hayer‐Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. [DOI] [PubMed] [Google Scholar]
- 2.de Jong WW, Leunissen JA, Voorter CE. Evolution of the alpha‐crystallin/small heat‐shock protein family. Mol Biol Evol. 1993;10:103–126. [DOI] [PubMed] [Google Scholar]
- 3.Horwitz J. Alpha‐crystallin can function as a molecular chaperone. Proc Natl Acad Sci U S A. 1992;89:10449–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jakob U, Gaestel M, Engel K, Buchner J. Small heat shock proteins are molecular chaperones. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 5.Ehrnsperger M, Graber S, Gaestel M, Buchner J. Binding of non‐native protein to Hsp25 during heat shock creates a reservoir of folding intermediates for reactivation. EMBO J. 1997;16:221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee GJ, Vierling E. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat‐denatured protein. Plant Physiol. 2000;122:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mogk A, Deuerling E, Vorderwulbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–595. [DOI] [PubMed] [Google Scholar]
- 8.Arata S, Hamaguchi S, Nose K. Effects of the overexpression of the small heat shock protein, HSP27, on the sensitivity of human fibroblast cells exposed to oxidative stress. J Cell Physiol. 1995;163:458–465. [DOI] [PubMed] [Google Scholar]
- 9.Richards EH, Hickey E, Weber L, Master JR. Effect of overexpression of the small heat shock protein HSP27 on the heat and drug sensitivities of human testis tumor cells. Cancer Res. 1996;56:2446–2451. [PubMed] [Google Scholar]
- 10.Salinthone S, Ba M, Hanson L, Martin JL, Halayko AJ, Gerthoffer WT. Overexpression of human Hsp27 inhibits serum‐induced proliferation in airway smooth muscle myocytes and confers resistance to hydrogen peroxide cytotoxicity. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1194–L1207. [DOI] [PubMed] [Google Scholar]
- 11.Balogi Z, Cheregi O, Giese KC, et al. A mutant small heat shock protein with increased thylakoid association provides an elevated resistance against UV‐B damage in synechocystis 6803. J Biol Chem. 2008;283:22983–22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao W, Qian M, Li P, Zhao L, Chang Z. The essential role of the flexible termini in the temperature‐responsiveness of the oligomeric state and chaperone‐like activity for the polydisperse small heat shock protein IbpB from Escherichia coli . J Mol Biol. 2005;347:871–884. [DOI] [PubMed] [Google Scholar]
- 13.Haslbeck M, Vierling E. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J Mol Biol. 2015;427:1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Favet N, Duverger O, Loones MT, Poliard A, Kellermann O, Morange M. Overexpression of murine small heat shock protein HSP25 interferes with chondrocyte differentiation and decreases cell adhesion. Cell Death Differ. 2001;8:603–613. [DOI] [PubMed] [Google Scholar]
- 15.Morrow G, Battistini S, Zhang P, Tanguay RM. Decreased lifespan in the absence of expression of the mitochondrial small heat shock protein Hsp22 in Drosophila. J Biol Chem. 2004;279:43382–43385. [DOI] [PubMed] [Google Scholar]
- 16.Hollander JM, Martin JL, Belke DD, et al. Overexpression of wild‐type heat shock protein 27 and a nonphosphorylatable heat shock protein 27 mutant protects against ischemia/reperfusion injury in a transgenic mouse model. Circulation. 2004;110:3544–3552. [DOI] [PubMed] [Google Scholar]
- 17.Sanbe A, Marunouchi T, Abe T, et al. Phenotype of cardiomyopathy in cardiac‐specific heat shock protein B8 K141N transgenic mouse. J Biol Chem. 2013;288:8910–8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–474. [DOI] [PubMed] [Google Scholar]
- 19.Vicart P, Caron A, Guicheney P, et al. A missense mutation in the alphaB‐crystallin chaperone gene causes a desmin‐related myopathy. Nat Genet. 1998;20:92–95. [DOI] [PubMed] [Google Scholar]
- 20.Evgrafov OV, Mersiyanova I, Irobi J, et al. Mutant small heat‐shock protein 27 causes axonal Charcot‐Marie‐Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. [DOI] [PubMed] [Google Scholar]
- 21.Irobi J, Van Impe K, Seeman P, et al. Hot‐spot residue in small heat‐shock protein 22 causes distal motor neuropathy. Nat Genet. 2004;36:597–601. [DOI] [PubMed] [Google Scholar]
- 22.Leroux MR, Ma BJ, Batelier G, Melki R, Candido EP. Unique structural features of a novel class of small heat shock proteins. J Biol Chem. 1997;272:12847–12853. [DOI] [PubMed] [Google Scholar]
- 23.Kokke BP, Leroux MR, Candido EP, Boelens WC, de Jong WW. Caenorhabditis elegans small heat‐shock proteins Hsp12.2 and Hsp12.3 form tetramers and have no chaperone‐like activity. FEBS Lett. 1998;433:228–232. [DOI] [PubMed] [Google Scholar]
- 24.Lamitina ST, Strange K. Transcriptional targets of DAF‐16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am J Physiol Cell Physiol. 2005;288:C467–C474. [DOI] [PubMed] [Google Scholar]
- 25.Kim KK, Kim R, Kim SH. Crystal structure of a small heat‐shock protein. Nature. 1998;394:595–599. [DOI] [PubMed] [Google Scholar]
- 26.van Montfort RL, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. [DOI] [PubMed] [Google Scholar]
- 27.Aquilina JA, Benesch JL, Bateman OA, Slingsby C, Robinson CV. Polydispersity of a mammalian chaperone: Mass spectrometry reveals the population of oligomers in alphaB‐crystallin. Proc Natl Acad Sci U S A. 2003;100:10611–10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age‐related disease by DAF‐16 and heat‐shock factor. Science. 2003;300:1142–1145. [DOI] [PubMed] [Google Scholar]
- 29.Murphy CT, McCarroll SA, Bargmann CI, et al. Genes that act downstream of DAF‐16 to influence the lifespan of Caenorhabditis elegans . Nature. 2003;424:277–283. [DOI] [PubMed] [Google Scholar]
- 30.Ezemaduka AN, Yu J, Shi X, et al. A small heat shock protein enables Escherichia coli to grow at a lethal temperature of 50°C conceivably by maintaining cell envelope integrity. J Bacteriol. 2014;196:2004–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landry J, Chretien P, Lambert H, Hickey E, Weber LA. Heat shock resistance conferred by expression of the human HSP27 gene in rodent cells. J Cell Biol. 1989;109:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeh CH, Chang PF, Yeh KW, Lin WC, Chen YM, Lin CY. Expression of a gene encoding a 16.9‐kDa heat‐shock protein, Oshsp16.9, in Escherichia coli enhances thermotolerance. Proc Natl Acad Sci U S A. 1997;94:10967–10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo JS, Lee YM, Park HG, Lee JS. The intertidal copepod Tigriopus japonicus small heat shock protein 20 gene (Hsp20) enhances thermotolerance of transformed Escherichia coli . Biochem Biophys Res Commun. 2006;340:901–908. [DOI] [PubMed] [Google Scholar]
- 34.Montero‐Barrientos M, Cardoza RE, Gutierrez S, Monte E, Hermosa R. The heterologous overexpression of hsp23, a small heat‐shock protein gene from Trichoderma virens, confers thermotolerance to T. harzianum. Curr Genet. 2007;52:45–53. [DOI] [PubMed] [Google Scholar]
- 35.Kim KH, Alam I, Kim YG, et al. Overexpression of a chloroplast‐localized small heat shock protein OsHSP26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnol Lett. 2012;34:371–377. [DOI] [PubMed] [Google Scholar]
- 36.Sun L, Liu Y, Kong X, et al. ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep. 2012;31:1473–1484. [DOI] [PubMed] [Google Scholar]
- 37.Morrow G, Tanguay RM. Small heat shock protein expression and functions during development. Int J Biochem Cell Biol. 2012;44:1613–1621. [DOI] [PubMed] [Google Scholar]
- 38.Cherkasova V, Ayyadevara S, Egilmez N, Shmookler Reis R. Diverse Caenorhabditis elegans genes that are upregulated in dauer larvae also show elevated transcript levels in long‐lived, aged, or starved adults. J Mol Biol. 2000;300:433–448. [DOI] [PubMed] [Google Scholar]
- 39.Jones SJ, Riddle DL, Pouzyrev AT, et al. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans . Genome Res. 2001;11:1346–1352. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Kim SK. Global analysis of dauer gene expression in Caenorhabditis elegans . Development. 2003;130:1621–1634. [DOI] [PubMed] [Google Scholar]
- 41.Jones LM, Staffa K, Perally S, LaCourse EJ, Brophy PM, Hamilton JV. Proteomic analyses of Caenorhabditis elegans dauer larvae and long‐lived daf‐2 mutants implicates a shared detoxification system in longevity assurance. J Proteome Res. 2010;9:2871–2881. [DOI] [PubMed] [Google Scholar]
- 42.Sonnichsen B, Koski LB, Walsh A, et al. Full‐genome RNAi profiling of early embryogenesis in Caenorhabditis elegans . Nature. 2005;434:462–469. [DOI] [PubMed] [Google Scholar]
- 43.Schwarz EM, Kato M, Sternberg PW. Functional transcriptomics of a migrating cell in Caenorhabditis elegans . Proc Natl Acad Sci U S A. 2012;109:16246–16251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding L, Candido EP. Association of several small heat‐shock proteins with reproductive tissues in the nematode Caenorhabditis elegans . Biochem J. 2000;351:13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waters ER, Vierling E. Chloroplast small heat shock proteins: Evidence for atypical evolution of an organelle‐localized protein. Proc Natl Acad Sci U S A. 1999;96:14394–14399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu X, Jiao W, Chang Z. Phylogenetic and biochemical studies reveal a potential evolutionary origin of small heat shock proteins of animals from bacterial class a. J Mol Evol. 2006;62:257–266. [DOI] [PubMed] [Google Scholar]
- 47.Waters ER, Nguyen SL, Eskandar R, Behan J, Sanders‐Reed Z. The recent evolution of a pseudogene: Diversity and divergence of a mitochondria‐localized small heat shock protein in Arabidopsis thaliana . Genome. 2008;51:177–186. [DOI] [PubMed] [Google Scholar]
- 48.de Jong WW, Caspers GJ, Leunissen JA. Genealogy of the alpha‐crystallin—small heat‐shock protein superfamily. Int J Biol Macromol. 1998;22:151–162. [DOI] [PubMed] [Google Scholar]
- 49.Munchbach M, Nocker A, Narberhaus F. Multiple small heat shock proteins in rhizobia. J Bacteriol. 1999;181:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gnanasekar M, Anandharaman V, Anand SB, Nutman TB, Ramaswamy K. A novel small heat shock protein 12.6 (HSP12.6) from Brugia malayi functions as a human IL‐10 receptor binding protein. Mol Biochem Parasitol. 2008;159:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dakshinamoorthy G, Samykutty AK, Munirathinam G, et al. Biochemical characterization and evaluation of a Brugia malayi small heat shock protein as a vaccine against lymphatic filariasis. PLoS One. 2012;7:e34077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark JI, Muchowski PJ. Small heat‐shock proteins and their potential role in human disease. Curr Opin Struct Biol. 2000;10:52–59. [DOI] [PubMed] [Google Scholar]
- 53.Zeng L, Tan J, Lu W, Lu T, Hu Z. The potential role of small heat shock proteins in mitochondria. Cell Signal. 2013;25:2312–2319. [DOI] [PubMed] [Google Scholar]
- 54.Fu X. Chaperone function and mechanism of small heat‐shock proteins. Acta Biochim Biophys Sin (Shanghai). 2014;46:347–356. [DOI] [PubMed] [Google Scholar]
- 55.Kokke BP, Boelens WC, de Jong WW. The lack of chaperonelike activity of Caenorhabditis elegans Hsp12.2 cannot be restored by domain swapping with human alphaB‐crystallin. Cell Stress Chaperones. 2001;6:360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giese KC, Basha E, Catague BY, Vierling E. Evidence for an essential function of the N terminus of a small heat shock protein in vivo, independent of in vitro chaperone activity. Proc Natl Acad Sci U S A. 2005;102:18896–18901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc Natl Acad Sci U S A. 2009;106:15604–15609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu X, Shi X, Yin L, et al. Small heat shock protein IbpB acts as a robust chaperone in living cells by hierarchically activating its multi‐type substrate‐binding residues. J Biol Chem. 2013;288:11897–11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma KK, Kumar GS, Murphy AS, Kester K. Identification of 1,1′‐bi(4‐anilino)naphthalene‐5,5′‐disulfonic acid binding sequences in alpha‐crystallin. J Biol Chem. 1998;273:15474–15478. [DOI] [PubMed] [Google Scholar]
- 60.Sharma KK, Kumar RS, Kumar GS, Quinn PT. Synthesis and characterization of a peptide identified as a functional element in alphaA‐crystallin. J Biol Chem. 2000;275:3767–3771. [DOI] [PubMed] [Google Scholar]
- 61.Fu X, Zhang X, Chang Z. 4,4'‐Dianilino‐1,1′‐binaphthyl‐5,5′‐sulfonate, a novel molecule having chaperone‐like activity. Biochem Biophys Res Commun. 2005;329:1087–1093. [DOI] [PubMed] [Google Scholar]
- 62.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf‐2, an insulin receptor‐like gene that regulates longevity and diapause in Caenorhabditis elegans . Science. 1997;277:942–946. [DOI] [PubMed] [Google Scholar]
- 63.Shaw WM, Luo S, Landis J, Ashraf J, Murphy CT. The C. elegans TGF‐beta Dauer pathway regulates longevity via insulin signaling. Curr Biol. 2007;17:1635–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.


