Abstract
α‐Synuclein (αS) plays a key role in Parkinson's disease (PD). The αS nuclear role, its binding affinity and specificity to histones and dsDNA remains unknown. Here, we have measured the binding affinity () between αS wild‐type (wt) and PD‐specific αS S129‐phosphorylation mimicking (S129E) mutant with full‐length and flexible tail truncated individual core histones (H2a, H2b, H3, and H4), linker histone (H1), and carried out αS‐dsDNA interaction studies. This study revealed that αS(wt) interacts specifically with N‐terminal flexible tails of histone H3, H4, and flexible tails of H1. The αS(S129E) mutant recognizes histones similar to αS(wt) but binds with higher affinity. Intriguingly, αS(S129E) showed a binding affinity for control proteins (bovine serum albumin and lysozyme), while no interaction was seen for αS(wt). Based on our above observation, we contemplate that the physio‐chemical properties of αS with S129‐phosphorylation has changed compared to αS(wt), resulting in interaction for other proteins, which is the basis for Lewy body formation. Besides, this study showed αS binding to dsDNA is weak and nonspecific. Overall, αS specificity for histone binding suggests that its nuclear role is possibly driven through histone interaction.
Keywords: dsDNA, histone, Parkinson's disease, posttranslational modification, α‐Synuclein
1. INTRODUCTION
Parkinson's disease (PD) is the second most common neurodegenerative disorder in humans associated with the loss of dopaminergic neurons.1 The key pathological hallmark of PD being intracellular abnormal filamentous α‐Synuclein (αS) protein aggregates in the form of Lewy bodies (LBs).2, 3 αS is a small acidic protein of 140 amino acids belongs to the Intrinsically Disordered Protein (IDP) family comprises of three regions: (1) N‐terminal amphipathic region (1–60 residues). (2) The central nonamyloid‐β component of Alzheimer's disease (NAC) region (61–95 residues) forms the hydrophobic core. (3) C‐terminal acidic tail (104–140 residues).4
αS is an abundant neuronal protein with presynaptic and nuclear localization.5 Its cellular function is not entirely understood and is known to interact with several proteins.6, 7, 8, 9, 10, 11, 12 The nuclear role of αS is largely overlooked for several years, but the recent growing evidence associates αS toxicity to its nuclear function.13, 14, 15, 16, 17 For over a decade, αS is known to interact with histones and double‐standard DNA (dsDNA), the eukaryotic chromatin assembly intermediate.13, 18 Nevertheless, the specific nuclear role of αS, its binding affinity and specificity to histones and dsDNA are not well understood. The αS S129‐phosphorylation is shown to enhance the shuttling of αS between nucleus and cytoplasm.19 In addition, 90% of αS filamentous protein aggregates in LBs are phosphorylated at Ser129 position.20, 21 In contrast to αS, the histones are highly basic proteins belongs to the IDP family and are grouped into core histones (H2a, H2b, H3, and H4) and linker histone (H1).22 The individual core histones comprise of N‐terminal flexible tails consisting of many posttranslational modifications (PTMs) sites,23 and the C‐terminal histone‐fold region has a high‐degree of hydrophobic amino acids and is buried when individual core histones assemble into heterodimer with complementary histones.24 In comparison, linker histone has a central globular domain (~80 residues) with N‐terminal (~40 residues) and C‐terminal (~100 residues) flexible tails.22
In the present study, we have measured the binding affinity () between αS wild‐type (wt) and PD‐specific αS‐S129‐phosphorylation mimicking (S129E) mutant with individual core histones and linker histones of human origin. Additionally, we have carried out αS interaction studies with dsDNA using Electrophoretic Mobility Shift Assay (EMSA). This study gave detailed insights into αS interactions with histones and DNA.
2. RESULTS
Here, we have measured αS‐histones interaction in a solution using both MST and ITC. MST requires labeling of one binding partner using labels specific to either Lys/Cys‐residues or (His)6‐tag and it follows the diffusion of molecules along a microscopic temperature gradient.25 There is a limitation to Lys‐labeling; histones are basic proteins consists of several Lys‐residues; likewise, the αS N‐terminal region (1–103) has an isoelectric point (pI) of 9.5 consists of 15 Lys‐residues. Moreover, we have noticed that too high a Lys‐labeled dye density in αS might have steric interference in histone interaction. The drawback is that both proteins have no Cys‐residue except for histone H3. To overcome the above challenge, we have introduced one Cys‐residue to the C‐terminal end of αS constructs for labeling purposes and named them accordingly: αS(wt)‐141C and αS(S129E)‐141C. Such an approach was previously reported where αS(N122C) mutant was generated for labeling purposes.26 Furthermore, the above study showed that Cys‐labeled αS mutant protein does not interfere in the aggregation process than the unlabeled protein. For this study, we have individually expressed and purified the αS, core histones, and linker histone constructs using a bacterial expression system for biophysical studies (Figure S1a–c). Additionally, we carried out Dynamic Light Scattering (DLS) and Circular Dichroism (CD) spectra studies on the purified αS(wt) and αS(S129E) mutant protein (Figure S2c). Above studies suggest that purified protein is homogeneous and monomeric in solution, and has characteristic minima in the vicinity of 198 nm suggesting that both proteins exist typical in random coil conformations.
2.1. Standardization of αS‐histone interaction using MST and ITC
To validate that Cys‐labeled αS do not interfere in MST binding studies, we performed Cys‐labeled αS(wt)‐141C and αS(S129E)‐141C interaction with full‐length (FL) histone H3 (H3 FL) and the value are 4.2 and 24.4 μM, respectively. The control protein bovine serum albumin (BSA) showed no binding with αS(wt)‐141C and has a value of 5.4 μM for αS(S129E)‐141C (Figure S3a). Next, we have cross‐validated by reverse Cys‐labeling of histone H3 FL and BSA and studied their interaction with αS(wt) and αS(S129E) proteins having no cysteine residues at the C‐terminal end. As expected, both the experiments yield similar values suggesting that the addition of Cys‐residue to αS C‐terminal end does not interfere in binding measurement (Figure S3b). The raw MST curves for the monomeric αS‐histone complex shows no aggregation, and it correlates with previously reported monomeric αS‐nanobody binding data.27 Besides, the addition of histones to fluorescently labeled αS showed changes in thermophoresis indicating binding between both molecules (Figure S4). To extend the conclusion from MST studies, we explored αS‐histone interaction using ITC experiments between αS(wt) with histones H2a FL and H3 FL. Binding between αS (wt) protein with H2a FL and H3 FL has a value of 259 and 11 μM. The raw data indicate an endothermic interaction, based on the positive values observed for the peaks. The calculated Binding enthalpy ΔH, the , the stoichiometry, n, Gibb's free energy, ΔG and entropy, ΔS reported from the ITC experiment (Figure S5). Due to very weak binding between αS (wt) and H2a FL, the resultant isotherm is linear, making difficult in the reliable determination of both the binding affinity and enthalpy. However, the estimated values from ITC were comparable to that of the MST values (Figure 1). Since MST offered a reasonable throughput using less sample, we have carried out detailed αS‐histones interaction using this technique.
FIGURE 1.
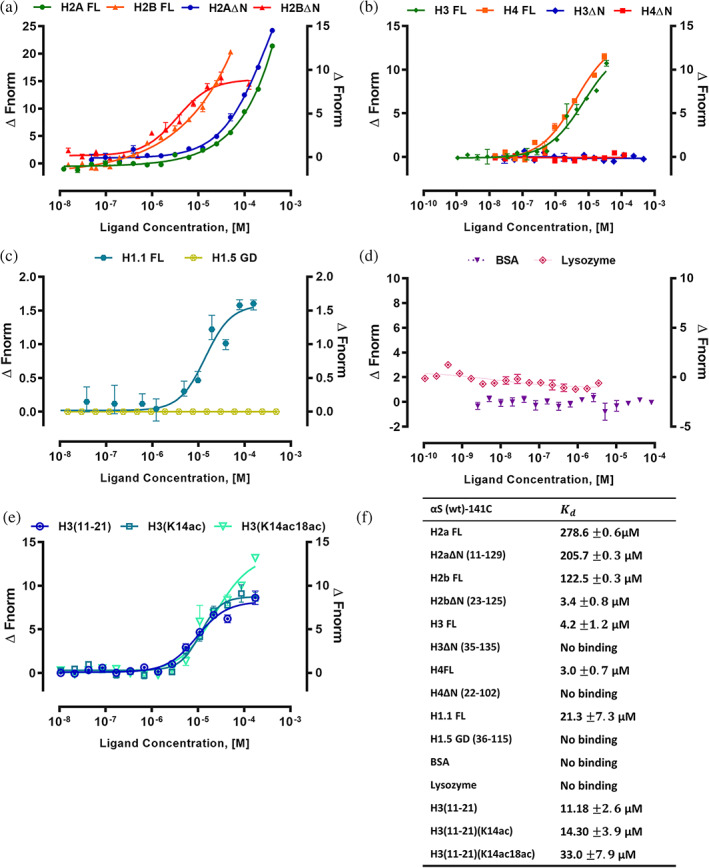
MST analysis of αS(wt)‐141C interactions with histones. Binding curves for the interactions of fluorescently labeled αS(wt)‐141C with (a) H2a FL, H2aΔN (12–129), H2b FL and H2bΔN (24–125). (b) H3 FL, H3ΔN (36–135), H4FL, and H4ΔN (23–102). (c) H1.1 FL and H1.5‐GD (37–116), (d) BSA and lysozyme as controls, and (e) The histone H3(11–21) peptide (unmodified), H3(11–21) with K14ac, and H3(11–21) with K14acK18ac were shown as the change in normalized fluorescence (ΔFnorm). (f) The values are summarized in the table. Error bars represent SEM (n = 3)
2.2. αS(wt) interacts specifically to flexible tails of histone H3, H4, and H1
The histone H2a FL and H2b FL have a value of 278.6 and 122.5 μM for αS(wt)‐141C. Intriguingly, histone H3 FL and H4 FL showed high‐affinity binding with values of 4.2 and 3.0 μM, which is 40 to 66‐fold higher compared to H2a FL and H2b FL. Likewise, the linker histone H1.1 FL interaction with αS(wt)‐141C has a value of 21.3 μM, which is five‐fold lower than histone H3 FL and H4 FL. To further explore the specific region of interaction between αS with core histones and linker histones, we have used N‐terminal tail truncated core histones, and linker histone H1.5‐globular domain (GD) with truncated N‐ and C‐terminal tails. The H2aΔN and H2bΔN have a value of 205 and 3.4 μM, respectively (Figure 1a). Among core histones, only H2a has a shorter N‐terminal tail length, and its truncation did not change its affinity towards αS(wt)‐141C compared to H2a FL. Intriguingly, H3ΔN and H4ΔN showed no binding for αS(wt)‐141C (Figure 1b). This study suggests that αS(wt) has a selective and possibly sequence‐specific binding for N‐terminal flexible tails of histone H3 and H4. Likewise, the H1.5‐GD showed no binding indicating that in the case of linker histone its N‐ and C‐terminal flexible tails are involved in an interaction with αS (Figure 1c). For this study, we have used both BSA (pI 5.6) and chicken egg‐white lysozyme (pI 9.3) as control proteins, and they both showed no binding to αS(wt)‐141C (Figure 1d). During MST studies, we noticed that thermophoresis values of the αS‐histone complex in the fully bound state do not reach a stable plateau. The above effect was previously reported for αS monomer‐nanobody interaction and is possibly caused by electrostatic influence of excess of free nanobodies on the thermophoresis.27 Overall, this study provided an insight into the binding specificity and critical regions within individual histones involved in αS(wt) interaction.
Earlier studies showed αS reduces histone H3 acetylation, and PD brain has change in H3K14 and H3K18 acetylation status in the primary motor cortex.14, 17, 28 Based on the above reports, we have tested H3 peptide corresponding to 11–21 residues for interaction with αS(wt)‐141C. This study showed a value of 11.1 μM, suggesting that the above tail region is a likely interaction site for αS (Figure 1e). Next, we have tested whether αS binds with varying affinity to acetylated compared to an unacetylated peptide. For this study, we have employed covalently modified H3(11–21) peptides with single acetyl‐lysine K14ac and with two acetyl‐lysine K14acK18ac for interaction studies with αS(wt)‐141C, and their values are 14.3 and 33.0 μM, respectively. This study showed two‐fold reduced binding affinities for H3(11–21) K14acK18ac acetylated peptides compared to an unmodified peptide (Figure 1e). Since the histone H3(11–21) tail segment undergoes various PTM modifications, the possible role of other PTM marks and neighboring H3K9 residue in αS interaction may not be ruled out.
2.3. Mapping of the α‐synuclein residues that interacts with histones
The 1H‐15N heteronuclear single quantum correlation (HSQC) spectrum was measured for the uniformly 15N‐labelled sample of αS(wt) in the presence of histones H2A, H2B, H3, and H4. An overlay of the spectrum of free αS(wt) (Figure S6) and in complex with histones are shown in Figure 2. On addition of H2A, the peak positions of the C‐terminal residues, that is, E110 to A140 of αS(wt) is significantly shifted, indicate relatively weak interaction between the αS(wt) and H2A where the binding process is in the fast exchange regime. On addition of H2B, H3, and H4, peak intensity of the C‐terminal region residues of αS(wt) is significantly reduced compared with the peak intensity of the unbound αS(wt), indicate a strong interaction and the process in intermediate exchange regime. These findings are in general agreement with the binding affinity obtained from other biophysical methods, and further confirm that the positively charged histones interact with the dynamically unstructured and highly negatively charged C‐terminal region of the α‐Synuclein.
FIGURE 2.
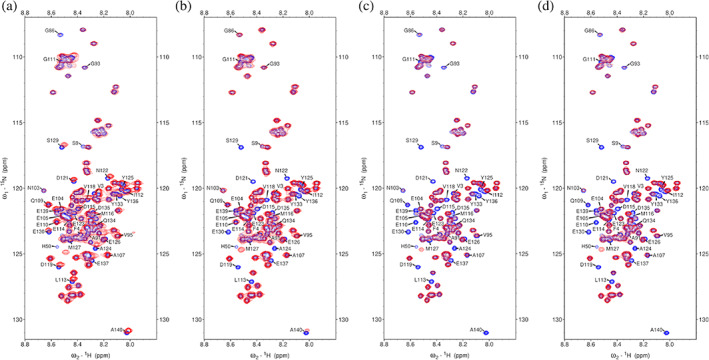
1H‐15N HSQC spectra of αS(wt) in the absence (blue) and presence of (red); (a) H2A (1:0.9), (b) H2B (1:0.9), (c) H3 (1:0.9), and (d) H4 (1:0.5). The labeled residues are those in αS(wt) that undergo significant chemical shift perturbations on addition of various histones
2.4. αS(S129E) has an increased binding affinity for core histones and linker histones compared to αS(wt)
To gain insights into how αS‐Ser129 phosphorylation influences its interaction with histones compared to αS(wt), we have studied the αS(S129E)‐141C binding affinity with histones. The MST trace showed ligand‐dependent fluorescence changes with increasing concentration of full‐length core histone proteins. The SDS‐denaturation test (SD‐test) identifies fluorescence loss as specific and ligand‐induced quenching, so initial fluorescence change was used for determination (Figure S4b).
Histone H2a FL and H2b FL has value of 22.7 and 19.3 μM for αS(S129E)‐141C. Whereas H2aΔN and H2bΔN have a value of 11.8 μM and 1.7 μM. Intriguingly, αS(S129E)‐141C showed 2 to 17‐fold higher affinity for full‐length and tailless core histones H2a and H2b compared to αS(wt). The full‐length histone H3 and H4 has a value of 24.4 and 2.5 μM, while H3ΔN and H4ΔN showed no binding for αS(S129E)‐141C. Likewise, the linker histone H 1.1 FL interaction with αS(S129E)‐141C has a value of 7.6 μM, whereas H1.5‐GD showed no/weak binding (Figure 3a–c). Overall, this study revealed that αS(S129E) mutant recognizes and binds to both core‐ and linker histones in parallel to αS(wt) protein but with higher affinity. We have also studied the αS(S129E)‐141C interaction with control protein BSA and lysozyme. Intriguingly, they both showed interaction compared to no binding for αS(wt)‐141C and their value is 5.4 and 249.4 μM, respectively (Figure 3d).
FIGURE 3.
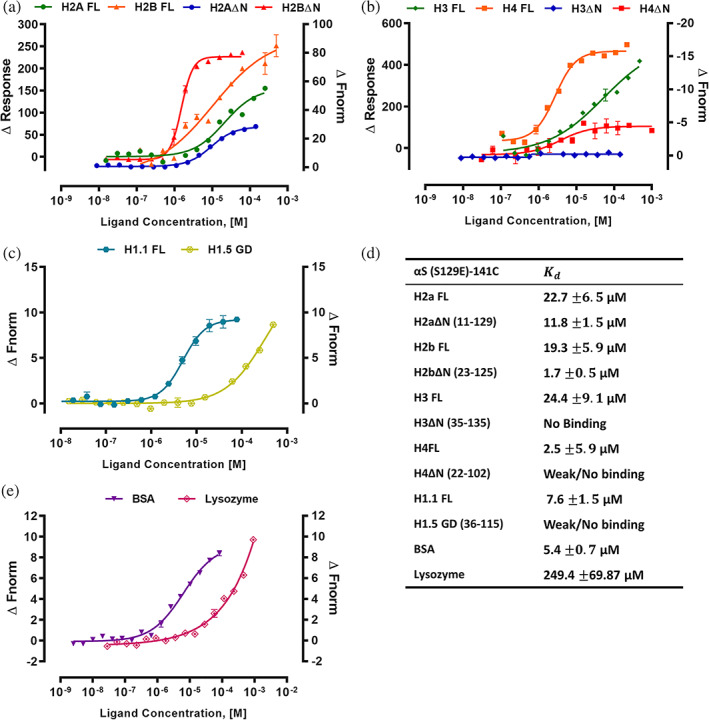
MST analysis of αS(S129E)‐141C interactions with histones. (a) H2a FL, H2aΔN (12–129), H2b FL, and H2bΔN (24–125). (b) H3 FL, H3ΔN (36–135), H4 FL and H4ΔN (23–102). (c) H1.1 FL and H1.5‐GD (37–116). (d) BSA and Lysozyme as controls. Binding curves were shown as a change in initial fluorescence (ΔResponse) for all four full‐length core histones, and the change in normalized fluorescence (ΔFnorm) for all N‐terminal truncated core histones constructs and control proteins. (e) The values are summarized in the table. Error bars represent SEM (n = 3)
2.5. αS interaction with dsDNA is weak and nonspecific
The proposed αS nuclear mechanism involves direct or indirect interaction with DNA or modulating histone PTM state.17, 29, 30 The fundamental questions remain on how αS is recruited to the site of DNA damage and gene expression either through interaction with dsDNA/histone. Second, though αS is known to interact with both dsDNA and histones, its interaction preference towards the aforementioned molecules is not well understood. We have studied αS(wt), αS(S129E) and αSΔC interaction with dsDNA using EMSA with increasing concentration to address the above questions. The PCR‐amplified dsDNA of αS (436 bp, 50% GC content) and mouse Nucleoplasmin‐2 (563 bp, 50% GC content) used as DNA substrates. For αS(wt) and αS(S129E), we have noticed the shift of dsDNA band begins at 18.5 μM and the complete shift was seen at 300 μM protein concentration (Figure 4a–d). In the past, most studies suggested a potential DNA binding property for αS is based on its N‐terminus (1–103) primary sequence, which has several positively charged residues.18, 31 Surprisingly, the EMSA studies using αSΔC, which lack the acidic tail showed no binding to dsDNA substrates even at the highest concentration tested (1200 μM) (Figure 4e,f). Additionally, we have also observed that the DNA length is important to see mobility shift with increasing concentration of αS(wt) and noticed no major shift for 145 bp dsDNA (Figure S4). Overall, this study suggests that αS binding to dsDNA is nonspecific in comparison to histones.
FIGURE 4.
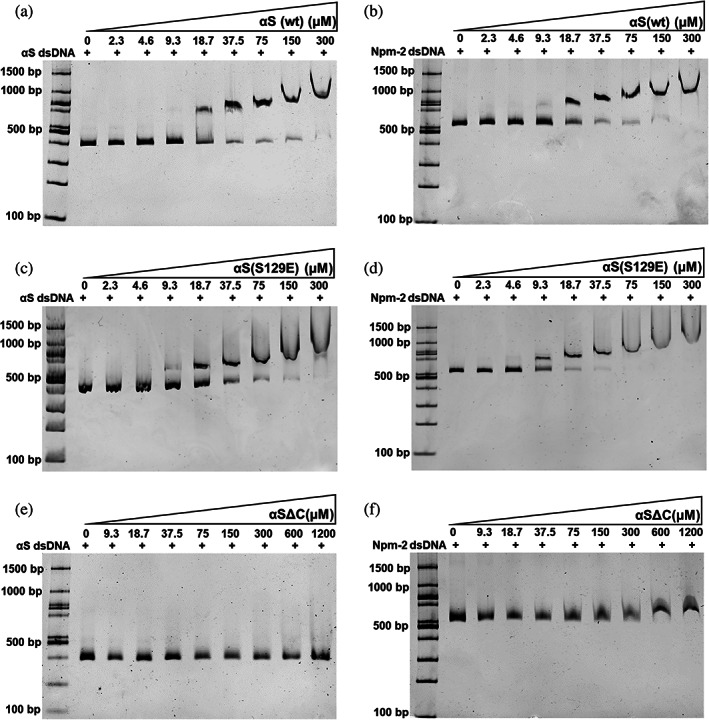
αS(wt), αS(S129E), and αSΔC interaction studies with dsDNA using EMSA. (a–b) αS(S129E) with αS dsDNA (left) and mNpm‐2 (right). (c–d) αS(S129E) with αS dsDNA (left) and mNpm‐2 (right). (e–f) αSΔC with αS dsDNA (left) and mNpm‐2 (right)
3. DISCUSSION
Here, we report the first extensive binding study on αS‐histone interaction. This study showed αS specificity for interaction with N‐terminal flexible tail of H3 and H4, which is of great importance since its epigenetic modification is associated with learning and memory, and neurological disorder.23, 32 The binding of a regulatory protein to histones can inhibit or enhance the histones' access by PTM‐modifying enzyme/modulators.33, 34 Although the role of epigenetics in PD has not been extensively studied,32, 35 many previous studies have reported that αS(wt) and PD‐specific familial mutant (A30P and A53T) co‐localize with histone H3 and reduce its acetylation level, thereby induce neurotoxicity.14, 17 Moreover, the nuclear αS was proposed to function as an inhibitor of acetyltransferase (INHAT) by directly interacting with histone H3.14 The INHAT domain was first reported in Set/TAF‐Iβ protein, which binds specifically to histone H3 and H4, thereby masking them from being acetyltransferases substrate.33 The INHAT domain is rich in acidic Asp/Glu‐residues, and sequence analysis of peptide 2 of Set/TAF‐Iβ used in this study showed 38% sequence identity with αS (117–137). The above analysis suggests that αS C‐terminal tail may function as INHAT by selectively interacting with both N‐terminal tails of histone H3 and H4 and it correlates with our biophysical studies.
This study showed phosphorylation mimicking αS(S129E) mutant recognizes and binds histones with higher affinity than αS(wt). However, the intriguing part of this study is the increased binding affinity of phosphomimicking αS(S129E) mutant's interaction with BSA and Lysozyme. Earlier NMR studies on α‐Synuclein with phospho‐modification have shown conformational flexibility and alteration in the position of few residues compared to wild‐type.36 Based on the above study, we speculate that changes in the positions of few residues compared to wild‐type possibly made them adhesive and is a likely reason for S129E mutants' good binding with BSA and lysozyme compared to wild‐type. Since LBs are composed of several different proteins in addition to αS fibrils, lipids, and membrane organelles,37 the adhesive property of α‐synuclein with S129‐phosphorylation towards other proteins might explain its abundance in the LBs.
Our study showed that αS interaction with dsDNA is weak and nonspecific. The above result agrees with single‐molecule techniques study on αS‐DNA interaction, which suggested that the binding of αS to DNA is weak.38 Interestingly no binding was seen for αSΔC with DNA. αS monomer as transient intramolecular interactions between basic N‐terminal and acidic C‐terminal tail, which holds αS in particular structural conformation. The partially folded αS conformation might be important for binding of αS(wt) to DNA. The deletion of the acidic tail might have affected the αS conformation resulting in loss of DNA binding for αSΔC. In conclusion, we propose that histones are the preferred substrate over dsDNA for αS regulatory role in the nucleus. However, any sequence‐specific dsDNA interaction for αS in the human genome may not be ruled out.
In conclusion, most studies in the literature have extensively explored the aggregation properties of PD‐associated mutations or posttranslational modified α‐Synuclein and compared them with αS(wt). However, the above studies have not produced a unifying explanation to PD‐specific α‐Synuclein modifications role in disease mechanism. So, it will be interesting to explore how the PD‐associated αS modification differ in their interaction with other proteins than the wild‐type. Above study might provide an insights on whether the PD‐specific modification made α‐Synuclein gluey, which could be a basis for LB formation.
4. MATERIALS AND METHODS
4.1. Cloning, expression, and purification of human core histone and linker histone proteins
Human full‐length core histones H2a, H2b, H3, and H4 constructs were generously provided by Prof. Curt A. Davey, NTU, Singapore. Above core histone plasmids were used as a template to PCR amplify the N‐terminal tailless core histones H2aΔN (11–126), H2bΔN (23–125), H3ΔN (35–135), and H4ΔN (22–102) constructs, and sub‐cloned them into pET28a vector at NdeI and BamHI restriction sites. Likewise, human linker histone H1.1 (Addgene ID #32894) and H1.5 (Addgene ID #32898) plasmids were purchased and used as a template to PCR amplify the H1.1 (1–214) full‐length and H1.5‐GD (36–115) constructs and sub‐cloned into pET28a vector at NdeI and BamHI restriction sites.
The full‐length and N‐terminal tail truncated core histones with (His)6‐tag at N‐terminus were expressed in E.coli expression strains BL21(DE3) (H2a FL, H2b FL, H3 FL, H2aΔN, H2bΔN, and H3ΔN) or JM109(DE3) (H4 FL and H4ΔN) as inclusion body and purified under denaturing condition.39 The only exception was for H3ΔN and H4ΔN constructs, which required the addition of 0.5% Triton X‐100 to buffers in all the purification steps. The purified core histones were lyophilized and stored in −80°C until further use. The linker histone variants H1.1‐FL and H1.5‐GD proteins were expressed and purified as previously reported.40 The linker histones were treated with thrombin (Sigma) to cleave the (His)6‐tag and the final purified proteins stored −80°C in small aliquots.
4.2. αS cloning, expression and purification
The human αS wild‐type (αS(wt); Addgene ID #36046), αS(wt) with cysteine residue at C‐terminus (αS(wt)‐141C; Addgene ID #108866), and αS(S129E) mutant (Addgene ID #36050) constructs were purchased. The αS(S129E) mutant construct was used as a template to PCR amplify to introduce a cysteine residue at its C‐terminus (αS(S129E)‐141C) and sub‐cloned into pET28a vector (Novagen) at NdeI and BamHI restriction sites. Likewise, αS(wt) was used as a template to PCR amplify αS(1–103) without cysteine residue (αSΔC), and sub‐cloned them into pET28a vector at NdeI and BamHI restriction sites.
The αS(wt), αS(wt)‐141C, αS(S129E), and αS(S129E)‐141C proteins were expressed and purified from periplasmic space as described previously.41 The αSΔC construct is expressed as cytosolic protein during bacterial expression. Except for bacterial cell lysis using sonication all other purification steps were similar to above αS constructs. In case of αS(S129E)‐141C and αSΔC constructs, the (His)6‐tag is cleaved with thrombin. During αS purification, we have noticed a high 260/280 ratio suggesting that the proteins are bound to a molecule having high absorbance at 260 nm. Analysis of the sample using native‐PAGE suggested no nucleic acid contamination. We were able to get two distinct peaks during size‐exclusion chromatography, indicating the separation of the bound molecule from αS. The final purified αS proteins has a 260/280 ratio of 0.6 and were lyophilized and stored at −80°C until further use.
4.3. Dynamic light scattering
Lyophilized αS(wt) and αS(S129E) mutant proteins were freshly dissolved in assay buffer (20 mM Tris–HCl pH 8.0 and 150 mM NaCl) and concentration was estimated by measuring absorbance at 280 nm using a molar extinction co‐efficient of 5960 M−1 cm−1. DLS was performed using Delsa Max PRO (Beckman Coulter, Brea, California) using 70 μM sample concentration at room temperature. The Polydispersity percent (Pd%) and hydrodynamic radius in nanometer (Rh) were analyzed with DynaMax v1.0 analysis software (Beckman Coulter, Brea, California) using the manufacturer's standard operating procedure size measurements. Data collected in triplicates with 10 acquisitions per each sample and results expressed in terms of the mass distribution.
4.4. Circular dichroism spectroscopy
CD spectra for αS(wt) and αS(S129E) mutant were recorded on a JASCO (J‐1500) CD Spectrometer at 296 K. Spectra for 10 μM samples were measured in 1 mm path‐length quartz cuvette from 250 to 190 nm wavelength with a step size of 0.2 nm and a bandwidth of 1 nm. Each data point represents the average of three experiments. 10 μM αS(wt) and αS(S129E) mutant samples for the experiments were prepared by diluting from 165 μM protein (20 mM Tris–HCl, pH = 8.0, 150 mM NaCl) with water.
4.5. MicroScale thermophoresis
The MST experiments were performed according to the NanoTemper technologies protocol and affinities calculated using Monolith NT.115 (Red/blue) instrument (NanoTemper Technologies GmbH, Munich, Germany). For MST experiments, the αS(wt)‐141C, αS(S129E)‐141C, H3 FL and BSA were labeled using cysteine reactive Monolith NT™ Protein Labeling Kit RED‐MALEIMIDE (NanoTemper Technologies).
The final concentrations of NT‐647 labeled αS(wt)‐141C and αS(S129E)‐141C were 366 and 220 nM, respectively, and H3FL and BSA were 125 nM each. Above labeled concentrations were chosen based on fluorescence intensities from the pretest assay setup. For this study, the samples were freshly prepared in the assay buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl supplemented with 0.05% Tween‐20) before MST studies. The experiments were performed by incubating varying concentrations of target proteins with a constant concentration of respective labeled αS constructs at room temperature for 30 min before recording the measurement using NT.115 standard treated capillaries (NanoTemper Technologies). The data were acquired at 25°C using LED power in the range of 20%–90% with MO. Control 1.5.3 (NanoTemper Technologies GmbH, Munich, Germany). The MST setup for each binding affinity check was analyzed and no sign of aggregation was observed. The reported measured values have two effects: MST dependent responses of the fluorophore to the temperature jump and Ligand‐dependent change in fluorescence. Further analyzed the recorded data with MO. Affinity Analysis 2.2.7 NanoTemper Technologies GmbH) and determined the values. The manuscript figures were prepared using Graphpad Prism 7.0 (GraphPad, San Diego, California).
4.6. Isothermal titration calorimetry
αS(wt), H2A FL and H3 FL protein were freshly prepared in the assay buffer containing 20 mM Tris–HCl pH 8.0, 150 mM NaCl. ITC experiments were performed using MicroCal ITC 200 instrument. All experiments were carried out at 25°C on high feedback mode with stirring speed 800 rpm and filter period time 5 s. 200 μl of 90–100 μM αS(wt) was titrated with 20 injections of 1–1.5 mM H2A FL and H3 FL with each 2 μl injections with 150 s intervals between each injection. Each measurement was paired with a control experiment to account for the heat of dilution and subtracted from the raw titration data. The data was analyzed by origin 7 software and the results were fit by using one set of sites model.
4.7. Nuclear magnetic resonance spectroscopy
Uniformly 15N‐isotopically labeled αS(wt) was produced by expression in minimal media containing 15NH4Cl and purified by using the protocol described for the full‐length protein, later buffer exchanged with 10 mM PBS (pH = 7.4), concentrated to 0.60 mM, snap frozen in LN2 and stored at 195 K.
NMR experiments with 0.25 mM of αS(wt) in PBS buffer and 10% (v/v) D2O were performed on a 600 MHz Bruker Avance III HD spectrometer equipped with a cryoprobe head, processed with Bruker TopSpin software, and analyzed with NMRFAM‐SPARKY.42 Typical 1H‐15N HSQC spectra were obtained at 290 K with 2048 points and 256 t1 increments, four scans per t1 point, and a 1.5 s recycle delay with a with sweep widths of 7,212 Hz (1H) and 1702 Hz (15N). Backbone amides resonance assignment was performed based on the reported structure (BMRB 19337).43 For titration experiments, required volume of lyophilized samples of histones dissolved in PBS buffer were added to 0.60 mM of αS(wt) sample, working concentration of αS(wt) was 0.25 mM.
4.8. Electrophoretic mobility shift assay
The dsDNA template αS(wt) (1–140; 436 bp) and mouse nucleoplasmin‐2 (1–179; 563 bp) were used for interaction studies with αS(wt), αS(S129E), and (αSΔC) proteins using EMSA. For this study, we have PCR amplified the above‐mentioned dsDNA, agarose gel extracted using QIAquick gel extraction kit (Qiagen) and concentration was determined using Nanodrop OneC (Thermo Scientific). αS(wt) dsDNA (60 ng) and nucleoplasmin‐2 dsDNA fragment (60 ng) were mixed with increasing concentration of αS(wt), αS(S129E), and (αSΔC) proteins separately in a 10 μL reaction in the assay buffer (50 mM Tris pH 8.0, 25 mM NaCl) and incubated on ice for 20 min. After the addition of 2 μl of loading buffer, the total 12 μl reaction was loaded in 6% PAGE ×0.5 TBE gel and ran at 100 V for 110 min in the cold cabinet. The gels were stained with ethidium bromide for 30 min in ×0.5 TBE before visualization using the Chemidoc MP system (Bio‐Rad).
AUTHOR CONTRIBUTIONS
Sneha Jos: Data curation; formal analysis; investigation; validation. Hemanga Gogoi: Data curation; formal analysis; investigation; validation. Thazhe Kootteri Prasad: Investigation. Manjunath A. Hurakadli: Investigation. Neelagandan Kamariah: Investigation; validation. Balasundaram Padmanabhan: Formal analysis; validation. Sivaraman Padavattan: Conceptualization; funding acquisition; investigation; supervision; validation; visualization; writing ‐ original draft.
Supporting information
APPENDIX S1: aS‐histone/dsDNA‐supplementary figures.docx
ACKNOWLEDGMENTS
This study is supported by the Science and Engineering Research Board, Government of India (Grant No: ECR/2018/002219) awarded to Sivaraman Padavattan. Hemanga Gogoi is supported by the fellowship program DBT‐JRF (DBT/2019/NIMHANS/1275) from the Department of Biotechnology, Ministry of Science and Technology, Government of India. We thank the Department of Science & Technology (DST), Government of India (DST‐FIST: SR/FST/LS‐I/2017(C)). We also thank NIMHANS Intramural Research Grant (NIMH/PROJ/PS/575) awarded to Sivaraman Padavattan. We are grateful to Saranya Ganesan, Swathi. V and Elakkiya. R for their assistance in the project. We thank Saji. M, Nanotemper, India. The Nuclear Magnetic Resonance (NMR) data were acquired at the National Center for Biological Science‐Tata Institute of Fundamental Research NMR Facility.
Jos S, Gogoi H, Prasad TK, Hurakadli MA, Kamariah N, Padmanabhan B, et al. Molecular insights into α‐synuclein interaction with individual human core histones, linker histone, and dsDNA . Protein Science. 2021;30:2121–2131. 10.1002/pro.4167
Sneha Jos and Hemanga Gogoi have contributed equally to this study.
Funding information NIMHANS Intramural Research Grant, Grant/Award Number: NIMH/PROJ/PS/575; Department of Science & Technology (DST), Government of India, Grant/Award Number: DST‐FIST: SR/FST/LS‐I/2017(C); Department of Biotechnology, Ministry of Science and Technology, Government of India, Grant/Award Number: DBT/2019/NIMHANS/1275; Science and Engineering Research Board, Government of India, Grant/Award Number: ECR/2018/002219
REFERENCES
- 1.Goedert M. Alpha‐synuclein and neurodegenerative diseases. Nat Rev Neurosci. 2001;2:492–501. [DOI] [PubMed] [Google Scholar]
- 2.Bendor JT, Logan TP, Edwards RH. The function of α‐synuclein. Neuron. 2013;79:1044–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha‐synuclein in Lewy bodies. Nature. 1997;388:839–840. [DOI] [PubMed] [Google Scholar]
- 4.Stephens AD, Zacharopoulou M, Kaminski Schierle GS. The cellular environment affects monomeric α‐synuclein structure. Trends Biochem Sci. 2019;44:453–466. [DOI] [PubMed] [Google Scholar]
- 5.Maroteaux L, Campanelli JT, Scheller RH. Synuclein: A neuron‐specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8:2804–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hernandez SM, Tikhonova EB, Karamyshev AL. Protein–protein interactions in alpha‐synuclein biogenesis: New potential targets in Parkinson's disease. Front Aging Neurosci. 2020;12:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payton JE, Perrin RJ, Clayton DF, George JM. Protein–protein interactions of alpha‐synuclein in brain homogenates and transfected cells. Brain Res Mol Brain Res. 2001;95:138–145. [DOI] [PubMed] [Google Scholar]
- 8.Emamzadeh FN. Alpha‐synuclein structure, functions, and interactions. J Res Med Sci. 2016;21:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savolainen MH, Yan X, Myöhänen TT, Huttunen HJ. Prolyl oligopeptidase enhances α‐synuclein dimerization via direct protein–protein interaction. J Biol Chem. 2015;290:5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar R, Kumar S, Hanpude P, et al. Partially oxidized DJ‐1 inhibits α‐synuclein nucleation and remodels mature α‐synuclein fibrils in vitro. Commun Biol. 2019;2:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumari M, Hanpude P, Maiti TK. α‐Synuclein exhibits differential membrane perturbation, nucleation, and TLR2 binding through its secondary structure. ACS Chem Neurosci. 2020;11:4203–4214. [DOI] [PubMed] [Google Scholar]
- 12.Shi X, Sun Y, Wang P, et al. The interaction between calcineurin and α‐synuclein is regulated by calcium and calmodulin. Biochem Biophys Res Commun. 2018;496:1109–1114. [DOI] [PubMed] [Google Scholar]
- 13.Goers J, Manning‐Bog AB, McCormack AL, et al. Nuclear localization of alpha‐synuclein and its interaction with histones. Biochemistry. 2003;42:8465–8471. [DOI] [PubMed] [Google Scholar]
- 14.Kontopoulos E, Parvin JD, Feany MB. α‐Synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet. 2006;15:3012–3023. [DOI] [PubMed] [Google Scholar]
- 15.Fares M‐B, Ait‐Bouziad N, Dikiy I, et al. The novel Parkinson's disease linked mutation G51D attenuates in vitro aggregation and membrane binding of α‐synuclein, and enhances its secretion and nuclear localization in cells. Hum Mol Genet. 2014;23:4491–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinho R, Paiva I, Jercic KG, et al. Nuclear localization and phosphorylation modulate pathological effects of alpha‐synuclein. Hum Mol Genet. 2019;28:31–50. [DOI] [PubMed] [Google Scholar]
- 17.Paiva I, Pinho R, Pavlou MA, et al. Sodium butyrate rescues dopaminergic cells from alpha‐synuclein‐induced transcriptional deregulation and DNA damage. Hum Mol Genet. 2017;26:2231–2246. [DOI] [PubMed] [Google Scholar]
- 18.Vasquez V, Mitra J, Wang H, Hegde PM, Rao KS, Hegde ML. A multi‐faceted genotoxic network of alpha‐synuclein in the nucleus and mitochondria of dopaminergic neurons in Parkinson's disease: Emerging concepts and challenges. Prog Neurobiol. 2020;185:101729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonçalves S, Outeiro TF. Assessing the subcellular dynamics of alpha‐synuclein using photoactivation microscopy. Mol Neurobiol. 2013;47:1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujiwara H, Hasegawa M, Dohmae N, et al. Alpha‐synuclein is phosphorylated in synucleinopathy lesions. Nat Cell Biol. 2002;4:160–164. [DOI] [PubMed] [Google Scholar]
- 21.Oueslati A. Implication of alpha‐synuclein phosphorylation at S129 in synucleinopathies: What have we learned in the last decade? J Parkinsons Dis. 2016;6:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen JC, Lu X, Ross ED, Woody RW. Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem. 2006;281:1853–1856. [DOI] [PubMed] [Google Scholar]
- 23.Gräff J, Tsai L‐H. Histone acetylation: Molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14:97–111. [DOI] [PubMed] [Google Scholar]
- 24.McGinty RK, Tan S. Nucleosome structure and function. Chem Rev. 2015;115:2255–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seidel SAI, Dijkman PM, Lea WA, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinotsi D, Buell AK, Galvagnion C, Dobson CM, Kaminski Schierle GS, Kaminski CF. Direct observation of heterogeneous amyloid fibril growth kinetics via two‐color super‐resolution microscopy. Nano Lett. 2014;14:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolff M, Mittag JJ, Herling TW, et al. Quantitative thermophoretic study of disease‐related protein aggregates. Sci Rep. 2016;6:22829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gebremedhin KG, Rademacher DJ. Histone H3 acetylation in the postmortem Parkinson's disease primary motor cortex. Neurosci Lett. 2016;627:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaser AJ, Osterberg VR, Dent SE, et al. Alpha‐synuclein is a DNA binding protein that modulates DNA repair with implications for Lewy body disorders. Sci Rep. 2019;9:10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidi D, Schechter M, Elhadi SA, Matatov A, Nathanson L, Sharon R. α‐Synuclein translocates to the nucleus to activate retinoic‐acid‐dependent gene transcription. iScience. 2020;23:100910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegde ML, Vasudevaraju P, Rao KJ. DNA induced folding/fibrillation of alpha‐synuclein: new insights in Parkinson's disease. Front Biosci (Landmark Ed). 2010;15:418–436. [DOI] [PubMed] [Google Scholar]
- 32.Berson A, Nativio R, Berger SL, Bonini NM. Epigenetic regulation in neurodegenerative diseases. Trends Neurosci. 2018;41:587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. [DOI] [PubMed] [Google Scholar]
- 34.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core‐histone‐binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. [DOI] [PubMed] [Google Scholar]
- 35.Pavlou MAS, Pinho R, Paiva I, Outeiro TF. The yin and yang of α‐synuclein‐associated epigenetics in Parkinson's disease. Brain. 2017;140:878–886. [DOI] [PubMed] [Google Scholar]
- 36.Paleologou KE, Schmid AW, Rospigliosi CC, et al. Phosphorylation at Ser‐129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha‐synuclein. J Biol Chem. 2008;283:16895–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia Q, Liao L, Cheng D, et al. Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci. 2008;13:3850–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang K, Rocha S, Westling A, et al. Alpha‐synuclein modulates the physical properties of DNA. Chemistry. 2018;24:15685–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka Y, Tawaramoto‐Sasanuma M, Kawaguchi S, et al. Expression and purification of recombinant human histones. Methods. 2004;33:3–11. [DOI] [PubMed] [Google Scholar]
- 40.Adhireksan Z, Sharma D, Lee PL, Davey CA. Near‐atomic resolution structures of interdigitated nucleosome fibres. Nat Commun. 2020;11:4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang C, Ren G, Zhou H, Wang C. A new method for purification of recombinant human alpha‐synuclein in E. coli . Protein Expr Purif. 2005;42:173–177. [DOI] [PubMed] [Google Scholar]
- 42.Lee W, Tonelli M, Markley JL. NMRFAM‐SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1,327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang L, Janowska MK, Moriarty GM, Baum J. Mechanistic insight into the relationship between N‐terminal acetylation of α‐synuclein and fibril formation rates by NMR and fluorescence. PLoS One. 2013;8:e75018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1: aS‐histone/dsDNA‐supplementary figures.docx


