Abstract
Many recent studies have reported the onset of a robust antibody response to SARS-CoV-2 infection and highlighted produced antibodies’ specific qualitative and quantitative aspects, relevant for developing antibody-based diagnostic and therapeutic options. In this review, firstly we will report main information acquired so far regarding the humoral response to COVID-19; we will concentrate, in particular, upon the observed levels and the kinetics, the specificity spectrum and the neutralizing potential of antibodies produced in infected patients. We will then discuss the implication of humoral response’s characteristics in the development and correct use of serologic tests, as well as the efficacy and safety of convalescent plasma therapy and of neutralizing monoclonal antibodies for treating infected patients and preventing new infections. An update of the list of newly isolated specific neutralizing antibodies and suggestions for vaccine evaluation and development will be also provided.
Keywords: Convalescent plasma, humoral response to COVID-19, neutralizing monoclonal antibodies, SARS-CoV-2, serologic tests, vaccine design
1. Introduction
In March 2020, WHO declared Severe Acute Respiratory Syndrome CoronaVirus 2 (SARS-CoV-2) infection to be a worldwide pandemic, originating from Wuhan in China in late 2019. After about one year, precisely in late April 2021, WHO reported 148,329,348 confirmed cases worldwide, including 3,128,962 deaths. This pulmonary disease, named Coronavirus disease 2019 (COVID-19), is proving to be a major global health burden [1].
COVID-19 shows a complex profile with many different clinical manifestations. Patients may be asymptomatic, or may experience a variety of mild to severe symptoms, sometimes along with pneumonia. A substantial proportion of cases progress to severe disease, where approximately 5% of patients require intensive care and over 20% of such cases prove fatal [2].
Attempting to limit the rapid spread of the infection, many countries have implemented various forms of lockdown, with unavoidable economic and social consequences. In the meantime, the scientific community as a whole is exerting a great deal of effort to combat the infection by employing the maximum resources in order to study the infection spread modalities, virus characteristics, immunopathogenesis and patients’ immune responses.
A full understanding of all these aspects is a prerequisite, first for early diagnosis, second for effective treatment particularly of those patients suffering from the severe form of the disease and third for adequate prevention of new transmissions.
As in many other viral infections, specific antibodies play a central role in the infection; in addition to being an important protective tool for the immune system, they do represent the most easily identifiable elements for diagnostic purposes. In this review, we summarize the studies that show the contribution of antibodies in the main fight areas against the COVID-19 pandemic:
early diagnosis: knowledge of the humoral response in different types of patients is essential for the implementation of serological tests. Even if to date antigenic rapid tests are preferred for rapid diagnosis, serologic tests have been developed as first most rapid tool to monitor the evolution of the pandemic at its start. Now, serologic tests are also an essential tool for monitoring the response to vaccines.
effective therapeutic treatment: convalescent plasma therapy and specific neutralizing monoclonal antibodies (mAbs), both synthetic and derived from patient B cell clones, can be developed as therapeutics for patients with severe disease. Some neutralizing mAbs are already approved by regulatory agencies;
evaluation of the response to new vaccines and the design of more broadly effective ones. The measure of neutralizing antibodies titers can be used as correlate of protection against original SARS-CoV-2 and its variants, while the analysis of specific neutralizing antibodies epitopes could be useful for the identification of key SARS-CoV-2 antigenic regions useful for vaccine design.
2. Antibody response to SARS-CoV-2 infection
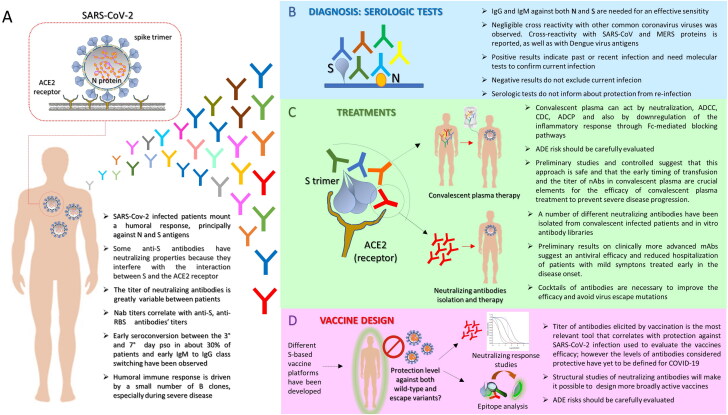
In last few months, many studies have analyzed the antibody response in various type of patients. Here we summarize the most common observations (Figure 1A). Almost all COVID-19 patients mounted IgA, IgM and IgG responses to SARS-CoV-2, which are especially directed against nucleocapsid (N) and spike (S) proteins. N is the virus nucleocapsid protein, is highly immunogenic and abundantly expressed in vivo after the virus infects human being. S is responsible for binding the virus to the human angiotensin converting enzyme 2 (ACE2) and its subsequent cellular uptake [3–5].
Figure 1.
Humoral response against SARS-CoV-2 and its implication: key messages. A. SARS-CoV-2 is a single-stranded positive-sense RNA virus which infects human cells expressing ACE2 receptor through its trimeric surface spike (S). Glycoprotein Nucleocapsid (N) protein is a highly immunogenic structural protein which participate in RNA package and virus particle release. The most common observations regarding the humoral response mounted by SARS-CoV-2 infected patients are summarized. B. Different serologic tests have been promptly implemented for the diagnosis of SARS-CoV-2 infection diagnosis. At present, serum antibodies against S and N proteins are the ones mainly detected. Potentialities and drawbacks of this kind of assay are summarized. C. Antibody response against SARS-CoV-2 has been used for the implementation of convalescent plasma therapy (up) and the isolation and characterization of neutralizing mAbs for therapy (down). Salient points concerning these two treatment options are reported. D. Both analysis of neutralizing response against SARS-CoV-2 and epitope characterization of isolated monoclonal Nabs are useful for vaccine design. Focal points of the design of a vaccine aimed to induce an effective antibody response are highlighted.
2.1. The antibody response and its kinetics
Stronger total antibodies, as well as single Ig classes IgA, IgM and IgG responses are all reported in both severe patients and non-severe patients [6–8]. Importantly, these clinical observations are independent of the SARS-CoV-2 antigens used for the immunoassay, that include Region-binding Domain (RBD), S1 subunits, full-length S or N proteins [4–7, 9]. However, some studies that went into the details of the quality of humoral response, revealed that some features could be associated to a more severe disease or death.
Firstly, it seems that an antibody response directed mostly versus N antigen in respect to S antigen may predict the disease progression toward a more severe illness. A study [10] which analyzed 38 patients divided in Intensive Care Unit (ICU) (=11) and non-ICU patients (=27), reported lower anti-S IgG titers in the 3rd week after symptom onset and a higher anti-N IgG/anti-S IgG ratio during all the weeks analyzed (1, 2 and 3 weeks after symptom onset) in ICU patients than in non-ICU patients. In addition, anti-N IgG and anti-N IgM were higher in ICU patients in respect to non-ICU patients at all-time points; anti-S-IgG positively correlated with the decrease of C-reactive protein (CRP) in non-ICU patients, while anti-N IgG did not. Another more recent study [11] which analyzed 22 hospitalized patients, all recruited within the first 20 days following symptom onset, showed a N-focused humoral response in patients who ultimately deceased and an S-focused response, in particular implying an S-specific antibody dependent complement deposition and phagocytosis, in patients who survived. They validated this observation in a larger cohort of acutely infected patients and found that the higher ratio of S/N antibody response was significantly associated to a protection against severe disease.
Secondly, regarding the Ig subclasses induced by the infection, de Campos Mata L et al. [5], interestingly showed how patients with a more pronounced IgA2 response, in particular RBD-specific more than N-specific, respect to RBD-IgG1 and IgG3 response, experienced a more favorable clinical course, with shorter hospitalization time and lower inflammation markers. These patients also experienced more gut-associated symptoms, suggesting the possible involvement of the more tolerogenic gut immune system.
Regarding the kinetics of antibody response, studies have generally observed an increase of both IgG and IgM during the first 3 weeks post-symptoms onset (pso); at 3-4 weeks pso almost all patients presented seroconversion while both antibodies types reach the peak [10]. After the third week, IgM begin to decrease, while IgG remain stable for longer time and sustained levels were detected until 6-7 months so far, even if a great variability was observed among patients [9, 12–16]. However, interestingly, some studies that focused on the analysis of patients’ serum starting from first days pso, have reported that most SARS-CoV-2 infected patients mount an early antibody response. Both IgG and IgM seroconversion was detected during the first week pso. In another recent study on 38 samples from 13 COVID-19 patients with known date of symptom onset, 33% of patients seroconverted for IgG between 3rd and 7th day pso [17]. Although, in this regard, some studies didn’t report any significant difference among different type of patients (asymptomatic, pre-symptomatic, and symptomatic) [10, 12, 18], another two analytic studies underlined a major proportion of early responders among hospitalized [6, 19] and severe patients [20]. In particular Iyer et al. [19] reported that the median time to seroconversion to SARS-CoV-2 RBD was, on average,4 days earlier for ospitalized patients compared with non-hospitalized patients, suggesting an association between antibody kinetics and disease severity.
Another interesting aspect of the anti-SARS-CoV-2 Ab response is the common early IgM to IgG class switching, as suggested by the rapid appearance of IgG, often synchronous to the IgM seroconversion or, sometimes, even earlier [13]. Interestingly, Sun et al. [10] showed that non-ICU patients had a faster and higher IgM to IgG class switch than ICU patients. Moreover, in ICU patients, anti-S IgM maintained a stable high levels, while anti-S IgG appeared to increase slowly compared to anti- N IgG levels. Observations on asymptomatic carriers have shown that these patients had constantly low levels of IgM, but high levels of IgG, together with an earlier viral clearance. Very early class switching was also detected in the pediatric population, characterized by milder symptoms and better recovery rate than adults. These observations suggest that high and persistent IgM played a negative role in SARS-CoV-2 infection, while efficient and rapid IgM to IgG class switch, especially for S-specific antibodies is important for viral clearance.
Significantly, there are cases of patients that experience no seroconversion, as for 4 out 5 cases of asymptomatic patients reported by Zhang Yongchen et al. [20], with no humoral response up to 4 weeks of observation. It is not known whether they become seropositive later. Such seronegative asymptomatic carriers may have resulted from low level of viral load. Likewise, Quan-Xin Long et al. [13] reported the case of a mother and a daughter of 11 years who maintained IgG and IgM seronegative status during hospitalization and other cases where IgM or IgG response are lacking [3, 6, 21]. Again, Ger Rijkers et al. [6] reported two cases of patients with severe disease who failed to show an IgG response at day 21 after disease onset (although the total antibody assay was positive). These patients, 69 and 87 years of age, had persistently positive PCR test results, respectively, on day 28 and day 37 after disease onset. The authors hypothesize that patients with an inadequate IgG antibody response may exhibit prolonged viral shedding, and thus longer periods of infectivity
2.2. Neutralizing humoral response profiling
Many studies analyzed in particular anti-S-RBD antibodies for their putative pivotal involvement in neutralizing activity of serum [3–5].
Recently, with the attempt to isolate and characterize B cell clones expressing neutralizing antibodies (NAbs) from recovered infected patients, many researchers have identified interesting data regarding the sequences of SARS-CoV-2 specific antibodies. With some exceptions, the data agree firstly that humoral immune response is driven by a small number of B clones [22–25], and secondly that, heavy (VH) and light (VL) variable gene classes utilized are often shared among different patients [23, 26]. These clones have been referred as public clonotypes and are associated to different Abs classes in reference to the portion of S-protein that is recognized [27]. Variability was observed in somatic hypermutations and complementarity determining region 3 (CDR3) lengths, even though there are indications that longer CDR3 sequences were represented in SARS-CoV-2 infections [22, 24]. In particular, Kuri-Cervantes L et al. [22], analyzed these characteristics in relation to the severity of the disease. They found a significant expansion of plasmablasts during severe COVID-19; this is directly correlated with an oligoclonal expansion of antibody clones within the overall B cell repertoire, suggesting that many of these large clonal expansions reside within the plasmablast pool. By contrast, mild patients, with one exception, showed a reduced clone expansion with more diverse repertoires. Moreover, the analysis of antibody sequences of the largest B cell clones in severe COVID-19 individuals were surprisingly variable in terms of somatic hypermutation levels, although they consistently had long CDR3 regions compared to healthy donors and those with moderate COVID-19.
It is already known that there is a great variability in neutralization potential of antibody response [28]. A study on 175 COVID-19 recovered patients with mild symptoms reported about 17%, 39%, and 14% of medium-low (ID50: 500-999), medium-high (ID50: 1000-2500), and high (ID50: > 2500) NAb titers, respectively [29]. The remaining 30% of analyzed convalescent patients generated a very low level of NAb titers, this suggesting that other immune responses, including T cells or cytokines, may contribute to overcoming the infection. How these patients recover without the help of NAbs and whether they are at risk of re-infection of SARS-CoV-2 should be explored further. The same observations were reported also by Ling Ni et al. who found a variability in NAb titers of serum from 14 analyzed patients [3], with one patient remaining completely negative, and by Robbiani et al. [23], who measured a half-maximal NAb titer (NT50) less than 50 in 33% of cases and below 1,000 in 79% with only 2 individuals reaching NT50s above 5,000.
Regarding the link between SARS-CoV-2-specific NAbs titers and levels of S-binding antibodies targeting RBD, S1, and S2 subunities, different studies reported various degrees of correlation that range from strong [21] (r = 0,9) to moderate (r = 0,4/0,5) [3, 14, 18], but not with levels of anti-N IgG [3].
Interestingly, a study analyzing the rapid generation of NAb in a cohort of 44 acutely infected patients, focused the attention on anti-RBD antibody levels; they found that the majority of patients develop Nabs as well as anti-RBD antibodies around 8 days pso (many patients also between 2-6 days); class-switching, that is dominated by RBD-specific IgG1 and IgG3 responses, occurs early during infection. In the observation period of the study, both RBD-specific IgM and IgA responses were detected at relatively lower levels as compared to IgG and the magnitude of RBD-specific IgG titers correlated positively with neutralization titers (r2 = 0,9) [18].
In parallel with overall humoral responses, neutralization titer is surprisingly higher in severe patients [30]; the actual effect on disease progression of NAbs and binding antibodies, even the latter potentially involved in viral clearance through antibody-dependent cellular phagocytosis (ADCP), complement-dependent cytotoxicity (CDC) or antibody-dependent cellular cytotoxicity (ADCC), should be subject to deeper comprehensive in further studies evaluating the different aspects of the disease and of the overall immune response in critical patients. Finally, Ling Ni et al. [3] described one case of a patient who exhibited no significant serum virus-neutralizing (VN) activity. This patient, though with anti-N and S-RBD IgM, did not have significant IgG or IgG1 production. Interestingly, the same patient had detectable virus-specific T cell function.
3. Serologic antibody tests for SARS-CoV-2 infection detection
Since almost all SARS-CoV-2 infection patients develop an antibody response, serologic tests (in particular CLIA and ELISA methods) for detecting SARS-CoV-2 have been promptly implemented for the diagnosis of the infection.
Based on the knowledge regarding the kinetics and the dynamic of the antibody response against SARS-CoV-2 infection, we would like to highlight some aspects of the relevant tests, in order to better understand the potentialities and the drawbacks of this kind of assay (Figure 1B).
3.1. Serologic tests for early diagnosis?
Even if IgM and IgG ELISA have been found to be positive even as early as the fourth day after symptom onset, the use of RT-PCR remains more suitable for diagnosing an early acute SARS-CoV-2 infection. Serologic tests, in fact, represent an indirect measure of the host response to the infection that can be variable among individuals, in terms of kinetics, as well as of antibody levels at peaks and targeted antigens. Generally, SARS-CoV-2 antibodies (both IgM and IgG) reach the peak between the 2nd and the 3rd week pso. Differently from other infections, where IgM/IgG detection is used for establish a recent or past infection, the dynamic of appearance of these two classes of antibodies is quite similar in SARS-CoV-2 infection. Indeed, IgG production occurs concurrently to or sometimes before IgM antibodies, the latter decreasing after the 4th week pso. Therefore, serologic assays can be useful to support and confirm molecular diagnosis of infection, especially after 5.5 days pso [31]. They are unable, however, to establish the actual infectious status of the patient: negative result does not exclude a recent infection, while a positive result does not indicate if the patient has stopped viral shedding or not (RT-PCR is a necessary complement assay in this case).
3.2. Sensibility and specificity of serologic tests
Like all other diagnostic tests, also serologic antibody tests should be sensitive and specific. Various studies report that the detection of anti-N antibodies is more sensitive respect to anti-S and anti-RBD antibodies, due to the higher abundancy of N protein during the infection [32, 33]. For example, Bubelo PB et al. [32], analyzed samples from PCR + confirmed COVID-19 cases and showed that seropositive N- antibodies were detected in a major number of samples collected both at >14 days after onset of symptoms (35/35 for anti-N antibodies, 100% sensitivity, and 32/35 for anti-S antibodies, 91% sensitivity) and at <14 days after onset of symptoms (33/65 for anti-N antibodies, 51% sensitivity, and 28/65 for anti-S antibodies, 43% sensitivity). This suggests that anti-N antibodies appear earlier than anti-S antibodies and are present at more elevated levels. Interestingly, the authors also analyzed five immunocompromised patients, who showed a general delayed production of antibodies and seropositivity for anti-N antibodies even when anti-S antibodies were undetected.
Moreover, it is clear that a sensitive test needs to detect both IgG and IgM classes of antibodies, against at least two specific antigens, especially if samples are taken during the first two weeks of infection. In their analytic study of ICU and non –ICU patients, Sun et al. [10] showed that the combined detection of N and specific IgM and IgG could identify up to 75% of SARS-CoV-2 infected patients in the first week and 94,7% in the first two weeks, while single and double antibody classes or specificities produced minor seropositive rates.
Regarding the serologic tests’ specificity, to avoid false positive results it is important to take into account the eventual cross- reactivity of antibodies specific for other endemic human CoV infections including alpha-CoV (229E and NL63) and other beta-CoV (OC42 and HKU1), against SARS-CoV-2 antigens. Multiple groups have shown limited or no cross-reactivity of antibodies to NL63, 229E, OC42 and HKU1 CoV against recombinant forms of SARS-CoV-2 N, S and RBD proteins by Western blot or ELISA analysis [34]. In this regard, Phipps WS et al. [17] in their recent study analyzed 23 cytomegalovirus IgG positive samples, several cases associated with prior influence A+ (n = 8) and influence B+ (n = 7) viruses, respiratory syncytial virus + (n = 6), and all 4 types of human CoV (n = 47); none of these patients were COVID-19 IgG positive. They also tested samples from patients with multiple autoantibodies, obtaining the same results. Generally, almost all recent studies confirmed negligible cross-reactivity [21, 35] of antibodies specific for common human CoVs, whereas cross-reactivity it is observed with SARS-CoV and, at lesser extent, MERS-CoV infection because of the higher similarity and the phylogenetic affinity between these two other CoVs and SARS-CoV-2. In particular, considering the 90% similarity of SARS-CoV and SARS-CoV-2 N proteins, cross-reactivity for this antigen is very high, while cross-reactivity with MERS-CoV infection was not observed (49% of homology between N proteins) in different studies [21, 35]. By contrast, when anti-S or S1, S2, RBD antibodies were individually detected, Nisreen M.A. Okba et al. [33], observed cross-reactivity of SARS-CoV-2 antibodies against SARS-CoV S and S1 proteins, and to a lesser extent with MERS-CoV S protein, but not with the MERS-CoV S1 protein. More importantly, the authors analyzed sera from healthy blood donors, PCR-confirmed acute respiratory non-CoV infections, acute to convalescent PCR-confirmed alpha- and beta-HCoV infections, and PCR-confirmed MERS-CoV and SARS-CoV infections. None of these sera were reactive against SARS-CoV-2 S1 protein, RBD antigen and N protein except for SARS-CoV sera. This cross-reactivity pattern is not unexpected if we consider the higher homology levels of S and S1 proteins between SARS-CoV-2 and SARS-CoV that reach 77% for the entire S, 66% for the S1 portion and 73% for the restricted RBD region (for MERS-CoV homology were respectively 33%, 25% and 16%) [18]. S2 region is more conserved with 90% similarity with SARS-CoV and 43% with MERS-CoV. It is therefore likely that this portion of the protein is responsible for the cross-reactivity observed with MERS-CoV patients’ sera. On the other hand, recently Ng K et al. [35] have reported cases (5 of 34 SARS-CoV-2-uninfected individuals with recent HCoV infection, as well as in 1 out of 31 individuals without recent HCoV infection) of weak preexisting humoral immunity in uninfected and unexposed humans to the new CoV. This phenomenon was particularly prevalent in children and adolescents, predominantly for the IgG class and targeted the S2 subunit and N protein, but not with the S1 subunit or the RBD of S. Notably, these sera also exhibited specific neutralizing activity against SARS-CoV-2 and SARS-CoV-2 S pseudotypes, according to levels of SARS-CoV-2 S-binding IgG and with efficiencies comparable to those of COVID-19 patient sera. In the same way, Morgenlander et al. [36], showed that a small minority of pre-COVID plasma contain antibodies reactive to CoV2 FP peptide, which represents a pan-CoV conserved antibody epitope.
All these data suggest that using an S1 subunit-based immunoassay may be more specific than the entire S antigen for diagnosing SARS-CoV-2 infections. That said, considering that SARS-CoV, which is the major source of cross-reactive antigens, has not circulated in the human population since 2003, and studies reported an undetectable level of anti-SARS-CoV antibodies in 91% of SARS infected patients after 6 years after infection, false-positive results of SARS-CoV-2 serologic tests are unlikely.
Differently, a remarkable issue arises from the confirmed cross-reactivity between SARS-CoV-2 and Dengue virus (DV) antigens and vice versa. Even if the two viruses belong to different families, two cases in Singapore with confirmed SARS-CoV-2 infection and false positive in DV serological tests [37] have been reported. At the same time, also the reverse scenario was reported with 5 out of 30 DV positive sample found to produce false-positive in SARS-CoV-2 rapid test serologic test. This cross-reactivity seems to be linked to S protein: a very interesting study by Nath et al. [38] showed how DV Envelope antibodies could recognize SARS-CoV-2 S protein, and in particular the RBD amino acids residues.
The last point to be noted regarding specificity of antibody tests is that, even if IgA are produced at higher levels and their detection is more sensitive, they are also less specific [34, 39].
Clearly, all these considerations on sensibility and specificity and test performances are subordinate to the correct interpretation of results (especially weak signals); therefore, along with development of robust serologic tests, guidelines for appropriate utilization and interpretation of clinical and epidemiological data, are essential.
3.3. Serologic tests can predict the level of protection to re-infection?
Another important focus of the serologic tests is their possible use for establishing if patient is protected from re-infection. It had even been thought that a plan for large scale serological testing could provide results upon which governments could formulate both social policies and the return to work. However, this information needs to be backed up by further research as we will discuss later and also by NAbs measurement. Currently, the only assay able to establish if a patient serum contains NAb (and in which quantity), and as such usable as a correlate of protective immunity, is the plaque reduction neutralization test (PRNT); unfortunately, the test demands biosafety level 3 (BSL3) containment facilities and therefore is impracticable by clinical laboratories. On the other hand, as already previously discussed in this review, the presence of NAb is high variable among patients, even if almost all of them develop an antibody response against SARS-CoV-2. In this context different studies focused on the possible correlations between PRNT results and levels of SARS-CoV-2 specific antibodies. Since the protein S, in particular the S1 region, RBD, was implicated in the binding to the cellular angiotensin converting enzyme 2 receptor for viral entry, some antibodies specific for these antigens are supposed to be neutralizing. Accordingly, various studies found correlations between the neutralizing power of sera and the levels of anti-S antibodies. Ni L et al. [3] analyzed 14 patients’ sera: 13 presented various degrees of neutralizing capabilities with 5 patients displaying high titers of NAb, with a significant correlation between NT50 and area under the curve (AUC) (calculated performing serum diluitions/OD ELISA curves) of anti-S-RBD IgG, but not of anti-N IgG. Wu F et al. [29], through plasma analysis from 175 discharged patients, interestingly observed that NAbs titers moderately correlated with S- binding antibodies targeting RBD (r = 0.51, p < 0.0001), S1 (r = 0.42, p < 0.0001), and also S2 (r = 0.435, p < 0.0001). This suggests that the S2 region could also be targeted by NAb, although this region is not directly involved in virus-cellular receptor interaction. Finally, Suthar MS et al. [18] analyzed the range of anti-RBD-specific and Nab responses across their cohort of 44 acutely infected COVID-19 patients and found a positive correlation between the magnitude of this two measures (r2 = 0.7; p < 0.0001). Subsequently, robust validation of this RBD-specific IgG ELISA by high-throughput testing at the Emory Medical Laboratories, on 231 PCR-confirmed COVID-19 patient serum samples demonstrated that this test also resulted highly sensitive and specific (97.5% and 98%, respectively), and perfectly discriminatory, above all for group of patients on day 7 or later after PCR confirmation (AUC of 1.00 (n = 83)). Another very interesting study [40] conducted on 68 COVID-19 patients, confirmed a strong positive correlation between both plasma anti-S ectodomain (ECD) IgG titers and, better, anti-RBD and in vitro VN titer. The same study also establishes that the probability of a VN titer ≥160 (the titer firstly recommended by FDA for convalescent plasma therapy) was 80% or greater when anti-RBD or anti-ECD titers are ≥1:1350. This is further evidence that anti-RBD IgG and anti-ECD IgG titers measured by ELISA serve as a very reliable surrogate of VN.
As a whole, all the above data suggest that future studies could make it possible to develop serologic tests allowing the measure of neutralizing power of sera without performing more complicated neutralization tests with clinically isolated virus or pseudovirus. This will pave the way for a series of important objectives: e.g. to evaluate the general protection to re-infection on large population basis, to promptly evaluate the usability of convalescent plasma for quickly available therapy and to evaluate efficacy of the different newly developed vaccines.
Clearly, to achieve information on the possibility of re-infection, a validated serologic test, but also the actual PRNT, should derive form a deeper knowledge of Nabs’ real capacity (also in relation to their titers) to protect the recovered patient and, if so, for how long. Regarding Nabs’ ability to protect patients from re-infection, currently we lack the data to answer this question, even if preliminary results from studies in Rhesus Macaques show a lack of re-infection after SARS-CoV-2 re-challenge in animals recovered from first infection [41]. Whether persons can be reinfected with SARS-CoV and MERS-CoV is unknown; SARS has not reemerged since 2004 and MERS cases remain sporadic. However, at present, NAbs titer necessary to confer protective immunity is unknown; thus future studies are required to determine which titer has therapeutic benefit. Finally, as yet, stable persistence of Nab, mainly IgG, has been described up to a variable time from four to seven months [14–16] and SARS-CoV-2 re-infection is estimated as a rare event [14]. However, due to the high similarity between the two diseases, lessons can be reasonable drawn from SARS-CoV and MERS infections, for which concentrations of IgG remained high for approximately 4 to 5 months before declining slowly over the next 2 to 3 years [42, 43].
4. Implications for SARS-CoV-2 THERAPY
4.1. Convalescent plasma and hyperimmune globulins
All data regarding the neutralizing power of sera from convalescent and recovered patients in vitro, have paved the way for the use of convalescent sera and hyper immune globulins for the treatment of severe COVID-19 cases (Figure 1C). Certainly, passive administration of antibodies is not new. The first example dates back to 1890 when antimicrobial therapies were unknown. Its utility was successively demonstrated during other outbreaks such as poliomyelitis, measles, mumps, and influenza (clearly with variable outcomes depending on the virus and the study), even if the treatment was not always associated with the measurement of NAb titers and the knowledge of viral sierotypes [31]. More recently, convalescent serum was used during Ebola (2013) and H5N1(2007) outbreaks and also in the cases of the two CoV infections, SARS-CoV and MERS-CoV, demonstrating its real efficacy especially if administered during the disease’s early stages [44–46].
In August, the FDA has allowed the treatment with convalescent plasma with an “expanded access-program” [47] for patients at high risk of developing a severe form of the disease and various clinical trials are being carried out on COVID-19 patients worldwide. Primary endpoints of all these studies are mostly homogenous; this will facilitate comparison and meta-analysis of data, while diversity in secondary endpoints will allow the individual studies to answer more specific questions about the use of convalescent plasma in COVID-19 treatment [48].
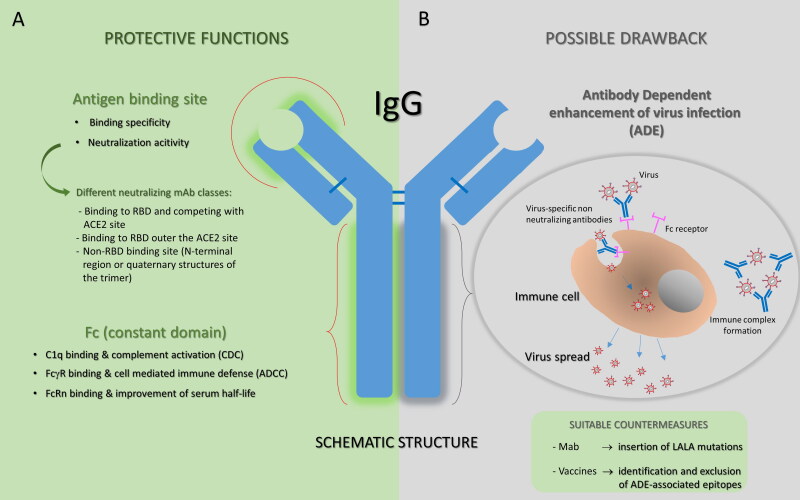
Critical factors to be considered for analysis of results should include the severity of patients’ illness, timing of administration (taking into account also the seroconversion timing and the titers of endogenous Nabs developed by the patient himself), severity illness of the donor, timing for the donation and last but not least, the NAbs titers of the donor. Obviously, the aspect of Ab titers needs to be clarified by future efficacy studies, taking into account that serum antibodies can fight infection not only by direct inhibition of the virus attachment to the host cells receptor (presumably the mode of action of anti-RBD antibodies), but also by ADCC and activation of the complement system (Figure 2).
Figure 2.
Antibody structure and function. A. Protective functions of antibodies are summarized. Antibodies are able to deploy a plethora of effector functions over the course of an infection: the antigen binding site of a neutralizing mAb is specifically directed against the viral surface antigen and directly interferes with virus-cell receptor interaction (on the top); human IgGs, particularly IgG1 and IgG3, bound to to the viral antigen exposed on the target cell, can subsequently interact through their Fc region with FcγRs expressed by effector cells or with complement component 1q, potentially supporting the destruction of target cells through ADCC or CDC, respectively. In addition, the Fc region of IgG can bind the salvage receptor FcRn after fluid-phase uptake by vascular endothelial cells and other cells, an interaction that contributes to the long (∼21 day) half-life of human IgG. B. The ADE phenomenon is illustrated as a possible adverse event that could occur with some antibodies. ADE has been documented to occur through two distinct mechanisms in viral infections: by enhanced antibody-mediated virus uptake into FcγR-expressing phagocytic cells leading to increased viral infection and replication, or by excessive antibody Fc-mediated effector functions or immune complex formation causing enhanced inflammation and immunopathology. Both ADE pathways can occur when non-neutralizing antibodies or antibodies at sub-neutralizing levels bind to viral antigens without blocking or clearing the infection. Identification and exclusion of ADE-associated epitopes in vaccine design or modification of the amino acid sequence of IgG reducing the interaction with one or more binding partners (for example by inserting LALA mutations) could be promising strategies to improve the clinical potential of antibodies.
Results from different interesting studies about the clinical use of plasma from convalescent donors are summarized in Table 1.
Table 1.
Clinical studies on convalescent plasma therapy: characteristics and results.
| Reference and trial characteristic | Participants characteristics and treatment groups | Administration time | Doses and CP nAbs titers | Primary outcome (PO) data | Secondary outcome data |
|---|---|---|---|---|---|
| Ling et al. [49] Randomized trial |
103 hospitalized adults > 18y with severe disease. Randomization: 52 patients with CP + SOC treatment + and 51 patients with SOC treatment |
Not specified | CP ≥ 1:640 of anti S-RBD IgG titers. Two doses of 200 mL, 24 hrs apart |
Clinical improvement within 28 days observed in 51.9% CP group versus 43.1% in the ctr group, no statistical significance If stratified for disease severity: PO reached in 91.3% in CP group and 68.2% in ctr group p > 0,03 excluding critically ill patients |
No differences in mortality rate at 28 days and time of discharge Rate of negative PCR in CP group 87.2% vs 37.5% in ctr group, P < .001 at 72 hours |
| Agarwal et al. [50] PLACID trial, randomized | 464 hospitalized adults > 18y with moderate ill patients Randomization: 235 patients with CP + SOC treatment and 229 patients with SOC treatment |
Not specified | nAbs titer not known at the time of transfusion; Two doses of 200 mL, 24 hrs apart |
Disease progression and mortality at 28 days, no statistical difference between group. Same results after nAbs titers stratification |
Higher proportion of patients in CP group showed resolution of shortness of breath and fatigue at day 7; No differences in resolution of fever and cough Negative conversion of SARS-CoV-2 RNA at day 7 was significantly higher in CP arm compared with ctr arm |
| Joyner M et al. [51]. EAP three months results, open label study | 35,322 hospitalized adults > 18y with severe acute infection with high risk of severe disease progression all receiving CP treatment | Variable. Administration time was used for stratified analysis. | nAbs titer not known at the time of transfusion 200 ml, at least one dose. |
7 days mortality rate was 8.7% in patients transfused within 3 days of COVID-19 diagnosis but 11.9% in patients transfused 4 or more days after diagnosis (p < 0.001). Same observations for 30 days mortality with 21.6% vs. 26.7%, p < 0.0001 7-days mortality was 8.9% recipients of high IgG plasma, 11,6% for recipient of medium IgG plasma, 13,7% for recipients of low IgG plasma(p = 0.048). |
|
| Libster R. et al. [51]. Randomized, double-blind trial |
160 older patients > 75 y Or >65 and < 74y with at least one defined coexisting condition |
72 hours after the onset of mild Covid-19 symptoms |
Anti-S IgG titer ≥ 1:1000. One 250 ml dose |
Severe respiratory disease developed in 16% cases in CP group versus 31% in ctr group, p = 0,03. In CP group, 73% patients treated with CP with SARS-CoV-2 S IgG titer ≥ 1:3200 showed a reduced risk of COVID worsening versus 31,4% of patients treated with CP with SARS-CoV-2 S IgG titer < 1:3200 |
Combined secondary end-point event (life-threatening respiratory disease, critical systemic illness, and death, or any of these outcomes) occurred in 9% of patients in CP group and in 15% of patients in ctr group. |
| Shenoy A et al. [52]. Retrospective matched control study | 576 hospitalized adults > 18y with severe acute infection. 263 treated with CP + SOC and 263 controls receiving SOC matched by gender, age and clinical characteristics |
Not specified | nAbs titer not known at the time of transfusion One or two doses of 200 ml of CP according the FDA guidelines |
No statistical difference in 28-day mortality was seen in CP group (25,5%) compared to controls (27%, P = 0,06). 7-day mortality was statistically better for CP group (9,1%) than controls (19,8%, P < 0,001) and continued at 14 days (14,8% vs. 23,6%, P = 0,01). |
After 72 h since transfusion transition from nasal cannula to room air was superior in CP group (median 4 days vs. 1 day, P = 0,02). The length of stay was longer in CP group than controls (14,3 days vs. 11,4 days, P < 0,001). |
Beyond the limitations of the single study, it seems clear that the timing of transfusion (more precisely within 72 hr since the symptoms onset) and the titer of nAbs in convalescent plasma used for transfusions (that should be at least ≥1:640) are crucial elements for the efficacy of convalescent plasma treatment to prevent severe disease progression in hospitalized patients. On the contrary, scarse efficacy in terms of patients’ survival and disease progression was found when convalescent plasma is administered later in the course of the infection. This is true even if the antiviral activity is present, probably because the infection has already led to the exacerbation of immune response and to eventual multiorgan damage.
Importantly, all these results do serve to stimulates ideas for implementation of the future clinical trials with a specific target population (severe patients), selection of convalescent plasma with specific NAb titers and earlier administration times.
Alongside efficacy, the safety is another priority. In general, possible risks due to administration of convalescent plasma are those known to be associated to the transfusion of blood derived substances; they notoriously include potential infections or host’s immunological reactions, but also theorical risks such as the antibody dependent enhancement (ADE), that means inflammatory and dangerous mechanisms mediated by pathogen specific antibodies, mostly of low quality, non-neutralizing or with low concentrations through Fcγ-receptors on immune cells or via complement activation [1,53,54] (Figure 2). Since the phenomenon of ADE is described for other CoV, including SARS-CoV and MERS-CoV, the possibility that this mechanism could also intensify SARS-CoV-2 infection cannot be excluded. However, provided that donor sera selection is carefully evaluated for the presence of high titer of NAbs, this risk could be considered negligible.
Regarding convalescent plasma safety, a large study on 5000 COVID-19 patients was conducted in USA, it reporting a final incidence of serious adverse events <1% in the first four hours after transfusion and the seven-day incidence of mortality was 14.9%. Considering the large population of critically-ill patients included in these analyses (for which mortality rate is 15-20% for hospitalized cases and 57% for ICU admitted cases), the mortality rate does not appear excessive. The authors conclude that transfusion of convalescent plasma is safe in hospitalized patients with COVID-19 [55].
In the two clinical trials previously cited [49, 56], a minimum number of patients experienced minor adverse events due to convalescent plasma transfusion. In three patients, mortality was possibly linked to transfusion [47].
Next to convalescent plasma, there is also the use of hyperimmune globulins concentrated from a pool of convalescent plasma and purified for IVIG use; this would probably prove more safe and more effective because of specific and accurately defined NAb titers. Moreover, IVIG are not subjected to ABO match blood groups, and the infusion volume would be lower. However, IVIG takes more months to prepare and to distribute among clinics and hospitals [44, 57, 58].
In conclusion, given the current absence of selective drugs against SARS-CoV-2 infection as well as the uncertainty about the long-term protection of newly approved vaccines and their effect on emerging viral variants, convalescent plasma could represent a treatment option in combination with standard of care protocols, whose use made feasible by the high number of convalescent and infected patients. However, it is necessary to test and, if possible, optimize a series of clinical and laboratory aspects.
4.2. Neutralizing monoclonal antibodies
4.2.1. Structure of the spike protein
Considering convalescent sera’s ability to inhibit SARS-CoV-2 infections, and the well-established clinical use of mAbs for therapeutic and prophylactic purposes, various research groups have isolated and characterized mAbs neutralizing SARS-CoV-2 infection (Figure 1C). These studies have also clarified structural aspects of SARS-CoV and SARS-CoV-2 S-proteins.
Both SARS-CoV and SARS-CoV2 S-proteins are glycoproteins of the viral surface that mediate the viral entry into the host cell and are active in trimeric form [59, 60]. S protein is composed of two subunities, named S1 and S2, the first of which binds the cellular receptor ACE2, thus promoting the cleavage of the S2 subunit into S2’ and S2” by cell-surface proteases; this in turn enables fusion and internalization of the virus. When the trimer is in the closed conformation, the three RBDs sites located at S1 level are inaccessible and, the S1 opening is clearly necessary for receptor binding.
Cryo-EM structures of the trimer have revealed the presence of multiple conformational states of SARS-CoV-2 S corresponding to distinct organization of the S1 RBD domains with a prevalence of the state harboring one of the three S1 RBD domains in receptor-accessible conformation. Stochastic RBD movements were observed also in MERS-CoV and SARS-CoV, for which, by contrast, two of three S1 RBD domains seem to be prevalently in an open conformation.
Receptor binding to the exposed RBD leads to an unstable three-RBD up conformation of S1 and initiate subsequent conformational changes that precede S1/S2 and then S2’/S2” cleavages by host proteases; these, in turn, have been proposed as activating the protein for membrane fusion via extensive irreversible conformational changes in the S2 subunits.
Clearly, all these conformational movements transiently hide or expose epitopes that can be targeted by neutralizing mAbs, both in S1 and in S2 domains.
4.2.2. Anti-S neutralizing monoclonal antibodies
SARS-CoV-2 S and SARS-CoV full-length S share 75% sequence homology (73% for the single RBD site and 88% for the S2 subunit), and are structurally similar. On the basis of these considerations, in order to speed the isolation of mAbs for therapeutic and prophylactic use against COVID-19, scientists promptly screened a number of SARS-CoV specific antibodies and sera from SARS-CoV patients for the ability to recognize SARS-CoV-2 S protein and neutralize SARS-CoV-2 infection.
However, despite the elevated homology between the two spike proteins, most of mAbs targeting SARS-CoV RBD did not exhibit evident binding to SARS-CoV-2 RBD [61] and, even when they did, they did not show protective activity. Noteworthy in this regard is the case of CR3022 mAb, that despite being able to strongly bind to SARS-CoV-2 RBD, it did not neutralize SARS-CoV-2 at the highest concentration tested [62]. Pursuing this interesting observation, structural modeling demonstrated that the binding epitope can only be accessed by CR3022 when at least two RBDs on the same trimeric S protein are in the “up” conformation and slightly rotated. These results provide molecular explanation about CR3022 behavior against SARS-CoV-2.
From these studies, interesting data has emerged on the different exposition of neutralizing epitopes between the S proteins of the two viruses and the effective utility of SARS-CoV specific mAbs against SARS-CoV-2 infection.
Table 2 summarizes the characteristics of the two mAbs isolated from SARS-CoV infected patients that are also active against SARS-CoV-2 infection.
Table 2.
SARS-CoV cross-reactive monoclonal antibodies.
| Reference | Name | Isolation | Epitope information | Functionality | Clinical study level |
|---|---|---|---|---|---|
| Pinto et al. [63] | S309 (VIR-7831 or GSK4182136 was obtained engineering S309 to improve its pharmacokinetics properties) |
SARS-CoV infected patient | RBD-not competing with ACE2 receptor-highly conserved, glycan containing epitope accessible in both open and closed S states | Neutralization ADCC ADCP |
Phase 2/3 |
| Wang et al. [64] | 47D11 | SARS-CoV –S immunized transgenic mouse expressing human VH and VL domains and rat constant regions | Conserved epitope on S1 subunity, external to RBD | Neutralization | Phase I (fully human version of the antibody) |
Color of lines distinguishes mAbs for the region of S-protein recognized.
Successively, much effort was and is being focused on isolating SARS-CoV-2 specific mAbs from both convalescent patients and synthetic antibody libraries.
Many neutralizing mAbs have been isolated that differ also for the region of S protein that is recognized.
Table 3 reports some examples of neutralizing mAbs isolated against SARS-CoV-2, divided according to the recognized region.
Table 3.
SARS-CoV-2 specific monoclonal antibodies.
| Reference of isolation | Name | Isolation | Epitope information | Functionality | Clinical study level |
|---|---|---|---|---|---|
| Jones et al. [65] | Ly-CoV555 | SARS-CoV-2 convalescent patients | RBD- competing with ACE2 receptor | Neutralization | Phase 3 |
| Shi et al. [66] | CB-6 CB-6-LALA version is also known as JS016 or Ly-CoV016 or Etesevimab |
SARS-CoV-2 convalescent patients | RBD-epitope partially overlapping with ACE2 receptor | Neutralization | Phase 2/3 (Etesevimab) in combination therapy |
| Hansen et al. [67] | REGN10933 | Joined approach using both immunized humanized mice and B cells from convalescent patients | RBD- competing with ACE2 receptor | Neutralization ADCC ADCP |
Phase 3 |
| Rogers et al. [25] | CC12.1, CC6.29, CC6.30 | SARS-CoV-2 convalescent patients | RBD- competing with ACE2 receptor | Neutralization | Preclinical for therapeutic efficacy (for CC12.1) |
| Zost et al. [26] | COV2196, COV-2130 | SARS-CoV-2 convalescent patients | RBD- competing with ACE2 receptor-two different epitopes | Neutralization, synergistic activity | Preclinical for therapeutic and prophylactic efficacy |
| Cao et al. [68] |
BD-368-2 | SARS-CoV-2 convalescent patients | RBD- competing with ACE2 receptor | Neutralization |
Preclinical for therapeutic and prophylactic efficacy |
| Miersh et al. [69] | IgG 15033 | Human Fab phage library | RBD- competing with ACE2 receptor | Neutralization | NA |
| Gai et al. [70] | Nb11-59 | Phage library of camelid nanobodies | RBD- competing with ACE2 receptor | Neutralization, stable after nebulization | NA |
| Barnes et al. [71] | C121 and others | SARS-CoV-2 convalescent patients | RBD- competing with ACE2 receptor | Neutralization | NA |
| Hansen et al. [67] | REGN10987 | Joined approach using both immunized humanized mice and B cells from convalescent patients | RBD- not competing with ACE2 receptor | Neutralization ADCC ADCP |
Phase 3 |
| Wu et al. [72] | n3088, n3031 and others | naive library of human 3 66*01 human VH genes | RBD- not competing with ACE2 receptor | Neutralization | NA |
| Barnes et al. [71] | C135 and others | SARS-CoV-2 convalescent patients | RBD- not competing with ACE2 receptor | Neutralization | NA |
| Chi et al. [73] | 4A8 | SARS-CoV-2 convalescent patients | S1 NTD | Neutralization | NA |
| Liu et al. [74] | 5-7, 5-24, 2-17 and others | SARS-CoV-2 convalescent patients | S1 NTD | Neutralization | NA |
| Liu et al. [74] | 2-43 | SARS-CoV-2 convalescent patients | Quaternary epitope of S1 trimer | Neutralization | NA |
Color of lines distinguishes mAbs for the region of S-protein that is recognized.
The ability of isolate new antibodies against different regions of S protein is extremely important in view of the emerging viral variants that can affect the therapeutic efficacy. Cocktails of antibodies against different regions of the S-proteins will be crucial to avoid the escape mutations of the virus (Figure 2).
The list of anti SARS-CoV-2 mAbs is not exhaustive, but it gives a good idea of the effort that researchers are putting into this field. Several antibodies have already been isolated and tested very quickly and many other are in early stage of development.
Currently, data are already available from most advanced clinical trials on LY-CoV555 antibody isolated by AbCellera (Vancouver, British Columbia, Canada) and the NIAID Vaccine Research Center, and developed by Eli Lilly and Company and the REGN10933 + REGN10987 antibody cocktail, sponsored jointly by NIAID and Regeneron Pharmaceuticals.
LY-CoV555 (Bamlanivimab) was evaluated in phase 3 study BLAZE-1 [75] at three different concentrations (700, 2800 and 7000 mg) and in combination with Ly-Cov016 (Etesevimab) at 2800 mg, administered in patients with mild and moderate symptoms within a median of 4 days of symptoms onset. Chen et al., reported decreased viral loads at day 11 in treated patients versus placebo, interestingly showing that the antiviral activity was highest in the middle dose. However, the most promising data is the 70% reduction of COVID-19 related hospitalization in Ly-CoV555 treated patients, in particular at 700 mg dose. Both antiviral activity and reduced hospitalizations were significantly higher in combination group. Ely Lilly and company also announced preliminary results of BLAZE-4 with lower doses of antibodies in combination which seem to have the same results than the combination doses in BLAZE-1 study.
First results with REGN10933 (Casirivimab)+REGN10987 (Imdevimab) antibody cocktail (REGN-CoV-2) are referred to 275 symptomatic not hospitalized patients divided into those who received 2,4 g or 8 g of REGN-CoV-2 and those who received placebo [76]. Patients received treatments within 72 hours since positive molecular test and no more than 7 days since the symptoms onset. The main result is a significant reduced viral load in REGN-CoV-2 groups in patients who were seronegative at baseline. Moreover, the patients who had higher viral loads obtained better results from the treatment. Same observations were obtained for the secondary endpoint which was at least one COVID-19 related medical visit at day 29.
Actually, FDA has approved REGN-Cov-2 for emergency use in USA in patients who are at high risk for progressing to severe COVID-19 and/or hospitalization, while Lilly has requested revocation of emergency use authorization for Ly-Cov555 alone to complete transition to bamlanivimab and etesevimab together for treatment of COVID-19 in the U.S.
As mentioned earlier, an extremely important issue is to understand how emerging viral variant can affect the neutralization efficacy of the mAbs isolated so far. Different studies are being conducted and other have already provided first indications [27, 77]. The interesting study of Wang et al. [77], has shown, for example, how the efficacy of Ly-CoV555 and REGN10933 are remarkably affected by the SAΔ9 variant, while 2-7 [74], REGN10987 and S309 (all of them binding the external side of RBD without competition with ACE2) are still active against both UKΔ8 and SAΔ variants. The authors also evaluated the impact of single mutations found in the two variants and mapped the most relevant ones. For example, for the UkΔ8 variant deletion at the position 144 affects the efficacy of anti-NTD mAbs such as 5-24, 4-8, 4A8, but not 5-7 [74] (Table 3); for the SAΔ9 variant, mutations in RBD portion K417N and E484K affect the efficacy of Ly-CoV555 and REGN10933, while mutations in NTD portion Del242-244 and R246I affect anti-NTD mAbs such as 5-24, 4-8, 4A8, but not 5-7 [74] (Table 3) as the UK variant.
The identification of antibodies that are active against all variants is extremely important, and the usage of one or other antibody cocktail shall be carefully evaluated also in relation to the territorial diffusion of known strains. In this perspective an interesting study [78] describes the development of a bispecific antibody in IgG1-like format containing two neutralizing antibodies directed against non –overlapping regions of RBD and isolated from convalescent patients (Table 3); C121 directly competes with ACE2 receptor binding, while C135 recognize the outer side of the RBD and is accessible in up and down S trimer conformation. The bispecific construct (Cov-X2) is able to protects mice from disease and prevent viral escape and neutralizes also all SARS-CoV-2 variants. The construct also contains LALA mutations in Fc portion in order to avoid possible ADE phenomenon. In this case, the construct retains all the advantages of an antibody cocktails in one molecule to be developed and administered.
5. Role of antibodies in vaccine evaluation and design
The joint effort of pharmaceutical companies and regulatory bodies has made it possible to develop and authorize several COVID-19 vaccines in an extraordinarily short period of time (about a year). The objective of this review goes beyond a detailed description of the different vaccines approved and in development, with the relative clinical results, for which the EMA online website [79] and other excellent recently published works are recommended [80–82]. Instead, starting from what is currently known about the protective response against SARS-CoV2, we will trace a brief picture of the current scenario concerning the anti-Covid19 vaccines, focusing on the main features of the first approved vaccines and on key aspects could be critical for the development of future ones.
The need to give a rapid response to the emergency triggered by the pandemic combined with the evidence about key role of neutralizing antibodies in protecting against infection, have pushed many pharmaceutical companies to develop vaccines whose common denominator is the “whole” S-protein as the main target for an effective immune response, giving up for now to more targeted strategies based on more restricted portions of viral proteins. Indeed, all vaccines to date approved by regulatory agencies are designed for an expression in the host cells of the entire S protein, even if the delivery platforms are different.
The vaccine Pfizer/BioNTech, as well as the Moderna one, is based on a molecule of "messenger RNA" protected by lipid nanoparticles, containing the genetic instructions for the expression of the spike protein inside human cells. The AstraZeneca/Oxford vaccine, as well as the one developed by Janssen (J&J) and the Russian Sputnik V vaccine, is instead based on a harmless adenovirus that contains a DNA sequence useful for making the patient’s body produce the spike protein. Again, the American vaccine NVX-CoV2373 contains a full-length, prefusion spike protein (produced in insect cells) made using Novavax’ recombinant nanoparticle technology and the company’s proprietary saponin-based Matrix-M™ adjuvant [80].
Several other vaccines against covid-19 with alternative platforms are in clinical and preclinical development. For example, the Beijing Institute for Biological Products has created an inactivated vaccine against the coronavirus (BBIBP-CorV) that has been claimed to be approximately 80% effective, prompting the Chinese government to grant approval [83].
All the current vaccine candidates are being evaluated for their capacity to induce humoral immunity. A special focus is put on the quality of antibody response intended as neutralizing antibodies level, especially RBD-specific ones, which is considered the leading correlate of protection to be used to verify the response to the vaccine and its durability over-time neutralizing capacity; in addition, it also represents a key element to evaluate the possible impact of the emerging viral variants on the vaccination efficacy.
Actually, the most effective developed vaccines, stimulate strong cellular and humoral responses, the latter notably found superior to that measured in convalescent severe patients, in terms of neutralization titers levels.
An interesting study by Wang Z et al. [84] has evidenced how mRNA vaccines induce a robust S-RBD-specific B cell memory response. They examined the nature of the produced antibodies and found that they are similar to that produced during natural infection in terms of IGVH and IGVL genes usage and of IGVH/IGVL combinations, CDR3 length and hydrophobicity. In few words also in vaccinated individuals, an expansion of a few number of recurrent clones was observed and some of these antibodies are the same found in natural infected patients. This information let us suppose that S-trimer produced in human cells by mRNA inoculation adopt a range of conformations similar to that observed during viral infection.
The debate is still open about the efficacy of vaccines currently available against the new variants - in particular against the English and South African ones. Results from more recent studies on both mRNA vaccines generally confirm that neutralizing antibodies elicited by vaccination are able to neutralize also these two variants, even if with variable reduction titers against the South African one [85–87], suggesting an antibody response directed against different epitopes of the S-protein. Moreover, an interesting study by Stamatatos et al. [88], have highlighted the importance of vaccinating also previously infected persons since they showed how a single immunization with mRNA vaccines was able to boost cross-neutralizing antibodies titers against all variants (even if, again, with a reduced potency against the South African variant) and that antibodies involved in the cross-neutralization target the RBD. Interestingly, this effect was up-to 1000 fold higher in previously infected patients after a single vaccine dose than in uninfected persons after two vaccine doses. However, to date the neutralizing titer levels that can be considered protective against infection or disease severity by original and variant SARS-CoV-2 haven’t yet been determined and only real-world experience can provide this answers. In this perspective, the challenge that the above mentioned pharmaceutical pharma are facing is to adapt and modify the current vaccines in order to be maximally efficacious also against the most resistant variants. Moderna and Pfizer/Biontech are developing, for example, multivalent mRNA combining the codes of all S-proteins and are planning, together with Johnson and Johnson, a third boost of their original vaccines.
Given the current scenario, understanding the impact of the mutations present in the emerging variants on the efficacy of the available antibody-based therapeutics – namely neutralizing mAbs, convalescent plasma and sera from vaccinated individuals [77]- could give precious information for an effective re-modeling of the current vaccine platforms, even with the ultimate goal to design more broadly efficacious vaccines.
Moreover, detailed studies focused on the epitopes recognized by Nabs isolated from B cells of convalescent patients as well as on the immunodominant portions of the S-protein capable to elicit a strong neutralizing humoral response are certainly key elements for finely guiding also new vaccine design. In this regard, the isolation of natural infection-elicited potent neutralizing antibodies against the S1 N-terminal domain or quaternary epitopes of the S1 trimer [74], can drive the choice of the antigen for vaccine development.
It is also important to note that, at the moment, there are no reliable data on the duration of coverage offered by approved vaccines. Precisely in this regard, the study and monitoring of antibody responses may still prove to be key elements to adequately face the evolution of the pandemic.
Finally, studies regarding the Ig classes and subclasses which identified a possible correlation between the IgA2 response, rather than IgA1 or IgG responses, and a better prognosis of the COVID-19 infection [5], and the evidence of the anti-inflammatory role of mucosal response [89], pose important questions for the future development of vaccines also in terms of the best administration site to obtain a strong barrier to the infection.
All the mentioned aspects once again point out how humoral response has a great importance in fighting SARS-CoV-2 infection, as well as how the capability to measure neutralizing potency of plasma is extremely useful to spin the development of vaccines and to evaluate their effectiveness (Figure 1D).
In this context, further evaluation of the presence of dangerous antibodies that can induce ADE or the important contribution of cellular responses to protection is also recommended.
6. Discussion
To combat the SARS-CoV-2 infection it is important to define virus’s pathogenetic mechanisms and to fully understand how our immune system responds. This review summarizes the most important results from numerous relevant studies regarding the humoral response against SARS-CoV2, also focusing their role in supporting the development of diagnostic tests, treatment options and preventive vaccines.
The first important issue is surely to better understand how a stronger and earlier antibody response could be correlated with a more severe disease. In other words, it is necessary to establish the extent to which a robust antibody response to SARS-CoV-2 results in virus neutralization or contrariwise contributes to the pathology in severe COVID-19 disease.
Some scientists have interpreted early antibody response and rapid class switching as the immune response’s recall of previous recent infection by one of the four common CoVs (i.e hCoV-OC43, hCoV-NL63,142 hCoV-HKU1 and hCoV-229E). And, even if first studies have detected no change in antibody levels against hCoV-OC43 and hCoV-HKUI in early responder patients compared to other patients and during the course of COVID-19 [6], other authors have showed that SARS-CoV-2 infection can boost a preexisting anti-CoV immunity which can confer some degree protection [28, 35, 36].
Notably, except for one study that found no correlation between IgG and IgM concentration and disease severity [17], all the others agree that increased COVID-19 disease severity is associated with a more robust humoral immune response and more interestingly, with neutralizing titers, which, on the contrary, usually correlate with host protection.
Different hypothesis have been formulated to explain how higher humoral responses can promote disease progression; one of these takes into account the possibility that ADE phenomenon can occur [1]. ADE was shown to promote virus uptake into macrophages resulting in impaired antiviral response, elevated production of inflammatory cytokines and acute lung injury, and it was observed with dengue virus, Zika virus, Ebola virus and, importantly in the context of COVID-19 and CoVs, including SARS-CoV and MERS-CoV. Higher levels of SARS-CoV-2 antibodies in severe cases prompt the hypothesis that a similar mechanism could occur in some cases for COVID-19, but a more recent study tends to exclude it [30] showing that ADE is not involved in severe forms experienced by the patients analyzed. However, further studies will be necessary to establish ADE’s real contribution to SARS-CoV-2 immunopathology.
Meanwhile, some researchers speculate that the high viral load in early infection of severe patients can drive strong and immediate extrafollicular B cell responses leading to rapid antibody responses which do not follow the sequence of IgM to IgG development stages. Such high quantity of antibodies produced from extrafollicular B cells may contribute greatly to the inflammatory responses by promoting monocyte and macrophage accumulation and the massive cytokine storm (including IL-8 and MCP-1), possibly responsible for fatal acute lung injury, as indicated during SARS-CoV infection. On the other hand, a gradual development of viral antigen specific B cells undergoes somatic hypermutation and affinity maturation at the traditional germinal center; this ultimately leads to high affinity protective antibody responses as observed in non-severe COVID-19 patients [20]. In another study, Cervantes LK et al. set out to elucidate their observation that severe COVID-19 patients displayed longer CDR3: given that B cells harboring antibodies with long CDR3 sequences are often multi-reactive and counter-selected during B cell development [22], they propose a contribution of longer CDR3 sequences as part of severe COVID-19 immunopathology.
As already discussed elsewhere in this review, other studies identify in an N-focused rather than S-centric humoral response a distinct feature for disease progression [11], while a higher mucosal response as RBD-specific-IgA2 mediated by gut immune system respect to pro-inflammatory RBD-IgG1 and IgG3 seems to be associated with a more favorable outcome [5].
However, the efficacy and safety studies carried out to date with convalescent plasma, obtained from recovered patients regardless of the disease’s severity, but only in relation to the neutralization potential, show that natural humoral response promotes viral clearance and prevents disease progression if administered early in the disease onset, and it is not dangerous.
This observation leads us to suppose that the association between the strength of humoral response and severity of disease is due not to the putative contribution of produced antibodies to the disease progression: rather it derives from the fact that the higher production of antibodies is a consequence of the disease’s severity. This could be caused by an elevated immune activation, and to higher exposure to the virus and its antigens. Moreover, higher exposure to S protein could induce, in a probabilistic way, more NAb titers. In most cases, these antibodies are effective, but other independent factors could contribute to the severity of the disease: excessive T responses, cytokine storms or other physiological elements, such as imbalance in the ACE2-angiotensin I-angiotensin II axis [90]. All the hypothesis above need verification; in depth studies would clarify the relationship between the degree of humoral response and a more severe disease.
A final point of interest deserves detailed study in IgG seroconversion durability along with Nabs ability to protect convalescent patients (or vaccinated people) from possible re-infection or simply mitigate the consequences of the infection. Both factors could benefit the immunity of the population as a whole, while also preventing infection among fragile communities and the categories most exposed, with social and financial implications. A detailed evaluation of all the multiple factors involved is evidently necessary, from the pathogenetic to the immune ones, in order to fill the knowledge gaps still remaining, also taking into account cellular response’s already established role in fighting SARS-CoV-2 infection.
Understanding the quality and the potency of the humoral response to SARS-CoV-2 natural infection is essential in order to design specific diagnostic and therapeutic strategies.
We have described the strength and limitations of antibody-based testing for SARS-CoV-2. However, concerns remain when testing patient populations differ from those used to validate the assay are tested, e.g. asymptomatic patients and immunocompromised patients, whose humoral responses to SARS-CoV-2 infection remain to be delineated. Moreover, as described in this review, some RNA-positive patients display no antibody responses; alternatively, they could produce antibodies that currently developed tests were unable to detect. On the other hand, actually, setting-up of serologic, in particular RBD-based, and neutralizing tests has extreme importance in the evaluation of the efficacy of emerging vaccines against SARS-CoV-2 and the response to vaccines at whole population level. An important issue is the reported cross-reactivity between SARS-CoV-2 antibodies with DV antigens (probably Envelope protein) which can cause misdiagnosis in region where the DV infection is endemic, considering also that the two virus can cause diseases with overlapping symptoms. However, an interesting hypothesis arose from the observed cross-reactivity and the reported less severity of COVID-19 in regions with relevant frequency of DV infections: it is supposed that preexisting DV antibodies could mask S-RBD and reduce the binding to ACE2 receptor, thus protecting from the infection [38]. This issue certainly need to be further evaluated and verified.
Concerning the therapeutic aspect, the ability of most patients’ plasma to neutralize SARS-CoV-2 infection, has not only encouraged researchers to evaluate convalescent plasma therapy, it has also helped isolate and characterize an increasing number of neutralizing mAbs. Regarding convalescent plasma infusions, significant evidence shows that this treatment is able to accelerate viral clearance and that diminishes the respiratory symptoms. The ability of this treatment to impact on mortality and disease progression seems strictly dependent on the early time of administration and nAbs titer of the donor; interestingly and, probably as awainted, these results match also with the efficacy results of monoclonal nAbs, thus suggesting a fundamental role of nAbs in the acute infection in order to prevent severe disease progression, more than the induction of severe forms of the diseases.
Actually, an important point of discussion will be the exact evaluation of the impact of emerging viral variants on convalescent plasma and neutralizing mAbs therapy efficacy. It seems that South Africa and UK variants affect at various degrees the neutralizing capacity of both convalescent sera and isolated monoclonal nAbs [27, 77].
Since it seems that the neutralizing response against SARS-CoV-2 natural infection is led by a small number of public clonotypes, shared across most infected patients, it is presumable that this selective pressure has accelerated the appearance of these resistant variants. Unfortunately, monoclonal nAbs were isolated exactly from these clonotypes. Clearly, we need to take into consideration the spread of these and other new viral variants and we need to take advantage from our scientific knowledge to face this aspect.
For example, sequencing of the B-cells repertoire of convalescent patients whose sera neutralize also the new variants should give us useful information.
Moreover, the development of different neutralizing mAbs makes it possible both to select the best SARS-CoV-2 infection’s treatment option and prophylaxis, and to identify the effective mAbs cocktails. The cocktail could boost the effect of single mAbs and, importantly, limit the number of mutations that confer resistance to neutralization, as known in the history of antibody-based therapies. This information will be also useful for vaccines development.
Finally, another debated issue is the possibility that specific antibodies could induce ADE, both in the case of the natural infection, and of neutralizing mAbs or vaccine-induced responces. It is debatable whether strong NAbs induce ADE in respect to weak NAbs; further research is also needed to determine conditions (affinity, dosage, mechanisms of interaction with the S protein) [54, 91] augmenting the risk for the occurrence of a such pathogenic process increases. This said, various biotechnological strategies are already in place to avoid the onset of this mechanism for mAbs, e.g. the introduction of LALA mutations in Fc region (Figure 2), as performed with CB6 mAb [66]. Obviously, since LALA mutations reduce mAb Fc’s interaction with the Fc receptors on immune cells, one should also take into consideration that ADCC and ADCP will also be reduced, thereby limiting important mAb-mediated mechanisms that may contribute to viral clearance. Therefore, the risks of ADE should be determined accurately by specific testing [91] case by case, before deciding to adopt these protein engineering strategies.
Accordingly, analysis of patients’ sera by high-throughput peptide-based scanning, sequencing of patients’ antibodies repertoires and computational approaches could help identify ADE-associated epitopes; indeed, studies on mouse antibodies produced against immunogenic SARS-CoV epitopes suggest that regions of the S protein vary in their propensity to cause ADE [92]. These studies will also support the development of safer vaccines, which, as in the case of convalescent plasma and mAbs, could induce dangerous suboptimal antibody responses [53] (Figure 2).
Certainly issues regarding the SARS-CoV-2 specific antibody response that are relevant for implementing fully effective antibody-based diagnostic and therapeutic strategies are still pending. Nevertheless, the scientific effort has been exceptional and results are promising.
Funding Statement
This work was supported by intramural fund of Istituto Superiore di Sanità.
Authors’ contribution
AM wrote the manuscript; AA contributed to the revision of the data and decided the structure of the figure; FM contributed to the design of the review and data discussion.
Declaration of Interest
The authors declare no conflicts of interest.
References
- 1.de Alwis R, Chen S, Gan ES, Ooi EE.. Impact of immune enhancement on Covid-19 polyclonal hyperimmune globulin therapy and vaccine development. EBioMedicine. 2020;55:102768. 10.1016/j.ebiom.2020.102768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75(7):1564–1581. 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ni L, Ye F, Cheng ML, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977.e3. 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padoan A, Sciacovelli L, Basso D, et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: A longitudinal study. Clin Chim Acta. 2020;507:164–166. 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Campos Mata L, Piñero J, Vaquero ST, et al. SARS-CoV-2-specific antibody profiles distinguish patients with moderate from severe COVID-19. medRxiv 2020.12.18.20248461. 10.1101/2020.12.18.20248461. [DOI] [Google Scholar]
- 6.Rijkers G, Murk JL, Wintermans B, et al. Differences in antibody kinetics and functionality between severe and mild SARS-CoV-2 infections. J Infect Dis. 2020;222(8):1265–1269. 10.1093/infdis/jiaa463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynch KL, Whitman JD, Lacanienta NP, et al. Magnitude and kinetics of anti-SARS-CoV-2 antibody responses and their relationship to disease severity. Clin Infect Dis. 2021;72(2):301–308. 10.1093/cid/ciaa979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carnicelli AFiori B, Ricci R, et al. Characteristic of IgA and IgG antibody response to SARS-CoV-2 infection in an Italian referral Covid-19 hospital. medRxiv 2020.11.16.20232470. 10.1101/2020.11.16.20232470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan W, Lu Y, Zhanget J, et al. Viral kinetics and antibody responses in patients with COVID-19 medRxiv 2020. 10.1101/2020.03.24.20042382. [DOI]
- 10.Sun B, Feng Y, Mo X, et al. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microb Infect. 2020;9(1):940–948. 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atyeo C, Fischinger S, Zohar T, et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;1553(3):524–532.e4. 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao T, Wang Y, Yuan J, et al. Early viral clearance and antibody kinetics of COVID-19 among asymptomatic carriers. Front Med (Lausanne). 2021;8:595773. 10.3389/fmed.2021.595773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long Q, Deng H, Chenet J, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020; 10.1101/2020.03.18.20038018. [DOI] [Google Scholar]
- 14.Abu-Raddad LJ, Chemaitelly H, Coyle P, et al. ARS-CoV-2 reinfection in a cohort of 43,000 antibody-positive individuals followed for up to 35 weeks. medRxiv 2021.01.15.21249731. 10.1101/2021.01.15.21249731. [DOI] [Google Scholar]
- 15.Tan Y, Liu F, Xu X, et al. Durability of neutralizing antibodies and T-cell response post SARS-CoV-2 infection. Front Med. 2020;14(6):746–751. 10.1007/s11684-020-0822-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford KHD, Dingens AS, Eguia R, at al. Dynamics of neutralizing antibody titers in the months after SARS-CoV-2 infection. J Infect Dis. 2021;223(2):197–205. 10.1093/infdis/jiaa618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phipps WS, SoRelle JA, Li QZ, et al. SARS-CoV-2 antibody responses do not predict COVID-19 disease severity. Am J Clin Pathol. 2020;154(4):459–465. 10.1093/ajcp/aqaa123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suthar MS, Zimmerman MG, Kauffman RC, et al. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med. 2020;1(3):100040. 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iyer AS, Jones FK, Nodoushani A, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. 2020;5(52):eabe0367. 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yongchen Z, Shen H, Wang X, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg Microb Infect. 2020;9(1):833–836. 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu F, Wang A, Liuet M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv 2020. 10.1101/2020.03.30.20047365 [DOI] [Google Scholar]
- 22.Kuri-Cervantes L, Pampena MB, Meng W, et al. Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection. Preprint. bioRxiv. 2020;2020.05.18.101717. Published 2020 May 18. 10.1101/2020.05.18.101717. [DOI] [Google Scholar]
- 23.Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brouwer PJM, Caniels TG, van der Straten K, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369(6504):643–650. 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zost SJ, Gilchuk P, Case JB, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584(7821):443–449. 10.1038/s41586-020-2548-6[32668443] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv [Preprint]. 2021; Update in: Nat Med. 2021 Mar 2; PMID: 33501446; PMCID: PMC7836116. 10.1101/2021.01.18.427166 [DOI] [PubMed] [Google Scholar]
- 28.Ruetalo N, Businger R, Althauset K, al. Neutralizing antibody response in non-hospitalized SARS-CoV-2 patients. medRxiv 2020.08.07.20169961. 10.1101/2020.08.07.20169961. [DOI] [Google Scholar]
- 29.Okba NMA, Müller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478–1488. 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legros V, Denolly S, Vogrig M, et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol. 2021;18(2):318–327. 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71(15):778–785. 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burbelo PD, Riedo FX, Morishima C, et al. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis. 2020;222(2):206–213. 10.1093/infdis/jiaa273[32427334]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okba N, Müller MA, Li W, et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. Emerg Infect Dis. 2020;26(7):1478–1488. 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Theel ES, Slev P, Wheeler S, Couturier MR, Wong SJ, Kadkhoda K.. The role of antibody testing for SARS-CoV-2: is there one? J Clin Microbiol. 2020;58(8):e00797–20. Published 2020 Jul 23. 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng KW, Faulkner N, Cornish GH, et al. Pre-existing and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370(6522):1339–1343. 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgenlander W, Henson S, Monaco D, et al. Antibody responses to endemic coronaviruses modulate COVID-19 convalescent plasma functionality. medRxiv 2020.12.16.20248294. 10.1101/2020.12.16.20248294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan G, Lee CK, Lam LTM, et al. Covert COVID-19 and false-positive dengue serology in Singapore. Lancet Infect Dis. 2020;20(5):536. 10.1016/S1473-3099(20)30158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nath H, Mallick A, Roy S, et al. Computational modelling supports that dengue virus envelope antibodies can bind to SARS-CoV-2 receptor binding sites: Is pre-exposure to dengue virus protective against COVID-19 severity? Comput Struct Biotechnol J. 2021;19:459–466. 10.1016/j.csbj.2020.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CY, Lin RTP, Renia L, Ng LFP.. Serological approaches for COVID-19: epidemiologic perspective on surveillance and control. Front Immunol. 2020;11:879. Published. 2020;11:879. 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salazar E, Kuchipudi SV, Christensen PA, et al. Relationship between anti-spike protein antibody titers and SARS-CoV-2 in vitro virus neutralization in convalescent plasma. Preprint. bioRxiv. 2020;2020.06.08.138990. Published 2020 Jun 9. 10.1101/2020.06.08.138990. [DOI] [Google Scholar]
- 41.Bao L, Deng W, Gao H, et al. Lack of reinfection in rhesus macaques infected with SARS-CoV-2. bioRxiv 2020. 10.1101/2020.03.13.990226. [DOI] [Google Scholar]
- 42.Wu LP, Wang NC, Chang YH, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. 2007;13(10):1562–1564. 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Payne DC, Iblan I, Rha B, et al. Persistence of antibodies against middle east respiratory syndrome coronavirus. Emerg Infect Dis. 2016;22(10):1824–1826. 10.3201/eid2210.160706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casadevall A, Pirofski LA.. The convalescent sera option for containing COVID-19. J Clin Invest. 2020;130(4):1545–1548. 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kazatchkine MD, Goldman M, Vincent JL.. Antibody-based therapies for COVID-19: can Europe move faster? PLoS Med. 2020;17(5):e1003127. Published 2020 May 5. 10.1371/journal.pmed.1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar S, Sharma V, Priya K.. Battle against COVID-19: efficacy of convalescent plasma as an emergency therapy. Am J Emerg Med. 2021;41:244–246. 10.1016/j.ajem.2020.05.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.US Food and Drug administration (FDA) . Expanded Access Program, Version 10.0. https://www.uscovidplasma.org/pdf/COVID-19%20Plasma%20EAP.pdf. Published August 3, 2020.
- 48.Barone P, DeSimone RA.. Convalescent plasma to treat coronavirus disease 2019 (COVID-19): considerations for clinical trial design. Transfusion. 2020;60(6):1123–1127. 10.1111/trf.15843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarwal A, Mukherjee A, Kumar G.et al. Convalescent plasma in the management of moderate COVID-19 in India: An open-label parallel-arm phase II multicentre randomized controlled trial (PLACID Trial). Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial). BMJ. 2020;371:m3939. 10.1136/bmj.m3939. Erratum in: BMJ. 2020 Nov 3;371:m4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joyner MJ, Senefeld JW, Klassen SA, et al. Effect of convalescent plasma on mortality among hospitalized patients with COVID-19: initial three-month experience. medRxiv 2020.08.12.20169359. 10.1101/2020.08.12.20169359. [DOI] [Google Scholar]
- 51.Libster R, Pérez Marc G, Wappner D, et al. Early high-titer plasma therapy to prevent severe Covid-19 in older adults. N Engl J Med. 2021;384(7):610–618. 10.1056/NEJMoa2033700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shenoy AG, Hettinger AZ, Fernandez SJ, et al. Early mortality benefit with COVID-19 convalescent plasma: a matched control study. Br J Haematol. 2021;192(4):706–713. Epub 2021 Jan 22. 10.1111/bjh.17272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eroshenko N, Gill T, Keaveney MK, et al. Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat Biotechnol. 2020;38(7):789–791. 10.1038/s41587-020-0577-1.PMID:32504046. [DOI] [PubMed] [Google Scholar]
- 54.Iwasaki A, Yang Y.. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20(6):339–341. 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joyner M, Scott Wright R, Fairweather D, et al. Early safety indicators of COVID-19 convalescent plasma in 5,000 patients. Preprint. medRxiv. 2020;2020.05.12.20099879. Published 2020 May 14. 10.1101/2020.05.12.20099879. [DOI] [Google Scholar]
- 56.Li L, Zhang W, Hu Y, et al. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. JAMA. 2020;324(5):460–470. 10.1001/jama.2020.10044. Erratum in: JAMA. 20204;324(5):519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roback JD, Guarner J.. Convalescent plasma to treat COVID-19: possibilities and challenges. JAMA. 2020;323(16):1561–1562. 10.1001/jama.2020.4940.PMID:32219429. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Z, Xie YW, Hong J, et al. Purification of severe acute respiratory syndrome hyperimmune globulins for intravenous injection from convalescent plasma. Transfusion. 2005;45(7):1160–1164. 10.1111/j.1537-2995.2005.00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microb Infect. 2020;9(1):382–385. Published 2020 Feb 17. 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan M, Wu NC, Zhu X, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pinto D, Park YJ, Beltramello M, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 64.Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS-CoV-2 infection [published correction appears in Nat Commun. 2020 May 14;11(1):2511]. Nat Commun. 2020;11(1):2251. Published 2020 May 4. 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jones BE, Brown-Augsburger PL, Corbett KS, et al. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv 2020.09.30.318972. 10.1101/2020.09.30.318972. [DOI] [Google Scholar]
- 66.Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 67.Hansen J, Baum A, Pascal KE, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369(6506):1010–1014. 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182(1):73–84.e16. 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miersch S, Ustav M, Liet Z, et al. Synthetic antibodies neutralize SARS-CoV-2 infection of mammalian cells. bioRxiv. 10.1101/2020.06.05.137349. [DOI] [Google Scholar]
- 70.Gai J, Ma L, Li G, et al. A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential. MedComm (Beijing). 2021;2(1):101–113. 10.1002/mco2.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barnes CO, Jette CA, Abernathy ME, et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. 10.1038/s41586-020-2852-1.Epub2020Oct12.PMID:33045718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu Y, Li C, Xia S, et al. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27(6):891–898.e5. 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. Epub 2020 Jun 22. 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu L, Wang P, Nair MS, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 75.Gottlieb RL, Nirula A, Chen P, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. 2021;325(7):632–644. 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weinreich DM, Sivapalasingam S, Norton T, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2021;384(3):238–251. 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. 10.1038/s41586-021-03398-2.Epub2021Mar8.PMID:33684923. [DOI] [PubMed] [Google Scholar]
- 78.De Gasparo R, Pedotti M, Simonelli L, et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature. 2021;593(7859):424–428. 10.1038/s41586-021-03461-y.Epub2021Mar25.PMID:33767445. [DOI] [PubMed] [Google Scholar]
- 79.European Medicines agencies website: https://www.ema.europa.eu/en/human-regulatory/overview/public-healththreats/coronavirus-disease-covid-19/treatments-vaccines/covid-19-vaccines.
- 80.Tumban E.Lead SARS-CoV-2 candidate vaccines: expectations from phase III trials and recommendations post-vaccine approval. Viruses. 2021;13(1):54. Published online 2020 Dec 31. 10.3390/v13010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ura T, Yamashita A, Mizuki N, et al. New vaccine production platforms used in developing SARS-CoV-2 vaccine candidates. Vaccine. 2021;39(2):197–201. 10.1016/j.vaccine.2020.11.054.Epub2020Nov24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wouters OJ, Shadlen KC, Salcher-Konrad M, et al. Challenges in ensuring global access to COVID-19 vaccines: production, affordability, allocation, and deployment. Lancet. 2021;397(10278):1023–1034. Published online 2021 Feb 12. 10.1016/S0140-6736(21)00306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia S, Zhang Y, Wang Y, et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis. 2021;21(1):39–51. Epub 2020 Oct 15. 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv 2021.01.15.426911. 10.1101/2021.01.15.426911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu K, Werner AP, Moliva JI, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv 2021.01.25.427948. 10.1101/2021.01.25.427948. [DOI] [Google Scholar]
- 86.Edara VV, Norwood C, Floyd K, et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microb. 2021;29(4):516–521.e3. 10.1016/j.chom.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Liu J, Xia H, et al. Neutralizing activity of BNT162b2-elicited serum. N Engl J Med. 2021;384(15):1466–1468. 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stamatatos L, Czartoski J, Wan YH, et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 25:eabg9175. 10.1126/science.abg9175.Epubaheadofprint.PMID:33766944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Russell MW, Moldoveanu Z, Ogra PL, Mestecky J.. Mucosal immunity in COVID-19: a neglected but critical aspect of SARS-CoV-2 infection. Front Immunol. 2020;11:611337. 10.3389/fimmu.2020.611337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Verdecchia P, Cavallini C, Spanevello A, Angeli F.. COVID-19: ACE2centric Infective Disease? Hypertension. 2020. Aug;76(2):294–299. 10.1161/HYPERTENSIONAHA.120.15353.Epub2020Jun1.PMID:32476472. [DOI] [PubMed] [Google Scholar]
- 91.Wan Y, Shang J, Sun S, et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol. 2020;94(5):e02015–19. Published 2020 Feb 14. 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Q, Zhang L, Kuwahara K, et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates [published correction appears in ACS Infect Dis. 2020 May 8;6(5):1284-1285]. ACS Infect Dis. 2016;2(5):361–376. 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]