Abstract
During murine spermatogenesis, beginning in late pachytene spermatocytes, the β4-galactosyltransferase-I (β4GalT-I) gene is transcribed from a male germ cell-specific start site. We had shown previously that a 796-bp genomic fragment that flanks the germ cell start site and contains two putative CRE (cyclic AMP-responsive element)-like motifs directs correct male germ cell expression of the β-galactosidase reporter gene in late pachytene spermatocytes and round spermatids of transgenic mice (N. L. Shaper, A. Harduin-Lepers, and J. H. Shaper, J. Biol. Chem. 269:25165–25171, 1994). We now report that in vivo expression of β4GalT-I in developing male germ cells requires an essential and previously undescribed 14-bp regulatory element (5′-GCCGGTTTCCTAGA-3′) that is distinct from the two CRE-like sequences. This cis element is located 16 bp upstream of the germ cell-specific start site and binds a male germ cell protein that we have termed TASS-1 (transcriptional activator in late pachytene spermatocytes and round spermatids 1). The presence of the Ets signature binding motif 5′-GGAA-3′ on the bottom strand of the TASS-1 sequence (underlined sequence) suggests that TASS-1 is a novel member of the Ets family of transcription factors. Additional transgenic analyses established that an 87-bp genomic fragment containing the TASS-1 regulatory element was sufficient for correct germ cell-specific expression of the β-galactosidase reporter gene. Furthermore, when the TASS-1 motif was mutated by transversion, within the context of the original 796-bp fragment, transgene expression was reduced 12- to 35-fold in vivo.
β1,4-Galactosyltransferase-I (β4GalT-I; EC 2.4.1.38) is a constitutively expressed, trans-Golgi resident, type II membrane-bound glycoprotein that is widely distributed in vertebrates. This enzyme catalyzes the transfer of galactose (Gal) to N-acetylglucosamine (GlcNAc) residues, forming the β4-N-acetyllactosamine (Galβ4-GlcNAc) or poly-β4-N-acetyllactosamine structures found in glycoconjugates (1). In mammals, β4GalT-I has been recruited for a second biosynthetic function, the tissue-specific production of lactose (Galβ4-Glc), which takes place exclusively in the mammary gland during lactation (reviewed in reference 37; see also references 25 and 40). The synthesis of lactose is carried out by a heterodimer assembled from β4GalT-I and the abundant milk protein α-lactalbumin, which is expressed de novo only in the mammary gland during lactation (3, 4, 16).
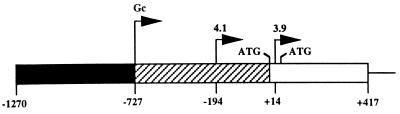
The organization of the 5′ end of the murine β4GalT-I gene is unusual in that three transcriptional start sites are contained within an ∼725-bp contiguous piece of DNA (Fig. 1). In somatic cells and tissues, the β4GalT-I gene specifies two transcripts of 4.1 and 3.9 kb. These two transcripts arise as a consequence of initiation at two different start sites separated by ∼200 bp (35, 39). Since the two start sites are positioned either upstream of the first two in-frame ATGs (4.1 kb) or between these two in-frame ATGs (3.9 kb), translation of each mRNA results in the synthesis of two catalytically identical, structurally related protein isoforms that differ only in the lengths of their respective short, NH2-terminal cytoplasmic domains. In somatic tissues, the 4.1-kb start site is used preferentially. The only exception to this pattern is found in the mammary gland during lactation, where there is a switch to the preferential use of the 3.9-kb start site. Expression from this start site is controlled by a stronger promoter (relative to the constitutive promoter governing transcription from the 4.1-kb start site) that is, in part, regulated by lactating mammary gland-restricted transcription factors (31). This results in an estimated 10-fold increase in steady-state β4GalT-I mRNA levels (14). This increase in mRNA levels, combined with the demonstration that the 3.9-kb transcript is translated in vivo three- to fivefold more efficiently than the 4.1-kb mRNA, results in the ∼50-fold increase in β4GalT-I enzyme levels needed for lactose biosynthesis (7).
FIG. 1.
Schematic representation of the 5′ end of the murine β4GalT-I gene. Shown is the upstream genomic DNA (solid box), the corresponding 5′-untranslated region (727 nt) of the male germ cell-specific transcript (hatched box), the coding sequence of exon 1 (open box), and a portion of the first intron (thin line). The bent arrows denote the three β4GalT-I transcriptional start sites. The male germ cell-specific start site (Gc) is used exclusively in late pachytene spermatocytes and round spermatids. The 4.1-kb start site (4.1) is used exclusively in spermatogonia and predominantly in all somatic cells. The 3.9-kb start site (3.9) is used predominately in the mammary gland during lactation. The locations of the first two in-frame ATGs are shown. Numbering is relative to the first in-frame ATG, which is designated +1. Exon 1 is not drawn to scale. Translation of the 4.1- and 3.9-kb β4GalT-I mRNAs results in two catalytically identical, trans-Golgi resident protein isoforms with NH2-terminal cytoplasmic domains of 24 and 11 amino acids, respectively.
The third, most distal transcriptional start site (Gc in Fig. 1) is used exclusively during the later stages of spermatogenesis (15). Spermatogenesis refers to the process in which spermatogonia, through a series of mitotic and meiotic divisions, give rise to spermatozoa. In spermatogonia, only the 4.1-kb transcript can be detected and it is identical in structure to the 4.1-kb mRNA found in somatic cells. As spermatogonia develop into early pachytene spermatocytes, transcription of the β4GalT-I gene is reduced to barely detectable levels. Continued differentiation to late pachytene spermatocytes and haploid round spermatids is coincident with renewed β4GalT-I expression to levels comparable to that observed in spermatogonia. However, the 4.1-kb β4GalT-I transcript is replaced with two truncated germ cell-specific transcripts of 2.9 and 3.1 kb (15, 41). These two transcripts encode the same open reading frame as the 4.1-kb transcript; however, their respective 3′-untranslated regions are only ∼0.8 or ∼1.0 kb in length, due to the utilization of either one of two alternative polyadenylation signals positioned ∼200 nucleotides apart within the long 3′-untranslated region (2.7 kb) of the somatic transcript. The 2.9- and 3.1-kb germ cell-specific transcripts are also distinguished from the 4.1-kb transcript by the presence of an additional ∼560 nucleotides of 5′-untranslated sequence.
We previously reported that a 796-bp TATA-less genomic fragment that flanks the β4GalT-I male germ cell start site mediates expression of the β-galactosidase reporter gene in late pachytene spermatocytes and round spermatids of transgenic mice in a manner comparable to that of the endogenous β4GalT-I gene (38). In this study, by using additional transgenic constructs in combination with DNase I footprinting and electrophoretic mobility shift assays (EMSAs), we demonstrated that a 14-bp regulatory element (5′-GCCGGTTTCCTAGA-3′), that we have termed the TASS-1 (transcriptional activator in late pachytene spermatocytes and round spermatids 1) motif, is essential for in vivo expression of β4GalT-I in developing male germ cells. Observations that suggest that TASS-1 is a member of the Ets family are discussed.
MATERIALS AND METHODS
DNA constructs.
All constructs were derived from the promoterless pLacI vector, which contains the Escherichia coli lacZ gene plus an intron and a polyadenylation sequence from the mouse metallothionein II gene. The β4GalT[−1270/−474]LacZ construct was described in reference 38, where it was referred to as β4GT-LacZ. It contains a 796-bp PvuII-NruI fragment that includes 543 bp of the genomic sequence upstream of the β4GalT-I germ cell start site and 253 bp of the flanking downstream sequence (spanning −1270 to −474) fused to the bacterial LacZ coding sequence. The male germ cell start site is located at −727 relative to the first in-frame ATG, designated +1 (Fig. 1). The β4GalT[−1085/−474]LacZ construct was produced by subcloning a 611-bp Eco47III-NruI fragment, spanning −1085 to −474, into the blunt-ended SalI site of pLacI. A 651-bp PvuII-HphI fragment, spanning −1270 to −619, was isolated, blunt ended with Klenow DNA polymerase, and subcloned into the blunt-ended SalI site of pLacI to produce the β4GalT[−1270/−619]LacZ construct. To produce the β4GalT[−1085/−628]LacZ construct, two primers 5′-ATTGTCGACGCTGTGTACAACGGGCTGTTC-3′ and 5′-TTTGTCGACACCACTTCCTGACGTAC-3′, containing a SalI site at each end (underlined sequences) were used to amplify a 457-bp fragment spanning −1085 to −628. After digestion with SalI, the amplified fragment was subcloned into the SalI site of pLacI. To produce the β4GalT[−793/−707]LacZ construct, two primers, 5′-ATAGGTACCAGAGAATCCGTCCGCT-3′ and 5′-AGCGTCGACTTGGGATAGTGAGAAA-3′, containing KpnI and SalI sites, respectively (underlined sequences), were used to amplify an 87-bp fragment spanning −793 to −707. After digestion with KpnI and SalI, the amplified fragment was subcloned into pLacI digested with KpnI and SalI. To assemble the β4GalT[mut−756/−743]LacZ construct, the forward primer 5′-GCCGGTACCTGTGAGATTTTACAGGCCATTCATTCT-3′ and the reverse primer 5′-ATTAGGCCTATTATTCGACGTCCCGCGAGGCCAGCG-3′ were used to amplify a 531-bp fragment spanning −1270 to −757. The PCR product was then digested with KpnI and StuI (underlined sequences). Next, the forward primer 5′-CCGTTTAAAGCTCCGACCTTTCTCCACTCCATTTTC-3′ and the reverse primer 5′-ATAGTCGACCGACACTTGGGGACTAAACGTGGCGCT-3′ were used to amplify a 287-bp fragment spanning −742 to −474. The PCR fragment was then digested with DraI and SalI (underlined sequences). A three-way ligation of the KpnI-StuI fragment, the DraI-SalI fragment, and pLacI digested with KpnI and SalI yielded the final construct, in which the −756/−743 sequence 5′-GCCGGTTTCCTAGA-3′ was mutated to 5′-TAATAGGAAAGCTC-3′. The sequences of all constructs were confirmed by DNA sequencing.
Production and identification of transgenic mice.
DNA inserts containing the relevant β4GalT-I genomic fragment inserted into pLacI were excised by KpnI/PstI digestion of plasmid DNA, isolated on agarose gels, purified by using an Elutip-d minicolumn (Schleicher & Schuell), and resuspended in injection buffer (10 mM Tris-HCl [pH 7.4] and 0.1 mM EDTA filtered through a 0.22-μm-pore-size membrane). Each transgene was injected into single-cell C57B6/BALBC3 embryos (at the Johns Hopkins University Transgenic Core Laboratory). Identification of transgenic founders and copy number determination were carried out essentially as previously described (38). Founders were bred to obtain heterozygous male transgenic offspring. At least three independent transgenic lines were generated for each construct. Transmission of the transgene proceeded according to Mendelian laws in the transgenic lines studied.
β-Galactosidase assays and histology.
β-Galactosidase assays and histology were performed as previously described (38). Fresh tissues were homogenized for 20 s in 100 mM potassium phosphate (pH 7.8)–0.2% Triton X-100–1 mM dithiothreitol–5 μg of leupeptin per ml (the last component was added just before use). The homogenate was centrifuged at 12,500 × g for 10 min, and the supernatant was heated at 48°C for 60 min to inactivate the endogenous eukaryotic β-galactosidase. Following centrifugation at 12,500 × g for 10 min, 10 μl of the supernatant was assayed for β-galactosidase activity by using the Galacto-Light chemiluminescence assay kit (Tropix, Inc., Bedford, Mass.). Protein concentrations were estimated by using the Bio-Rad Protein Assay kit. For histology, tissues were removed from transgenic and nontransgenic mice, fixed and incubated in buffer containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and paraffin embedded. Sections (3 to 5 μm) were cut, mounted on glass slides, deparaffinized, counterstained for 30 s with nuclear fast red, and photographed by using a Zeiss Axiophot microscope.
Cells and culture conditions.
Mouse L cells were obtained from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% horse serum, 100 U of penicillin per ml, and 50 μg of streptomycin per ml at 37°C in 5% CO2.
Preparation of nuclear extracts.
Pooled male germ cells were prepared by digesting adult mouse testes with collagenase and hyaluronidase (41). The final preparation contained >95% male germ cells. Within this pooled male germ cell population, ∼50% of the cells are round spermatids. Nuclear extracts from mouse L cells and from pooled male germ cells were prepared by the method of Dignam et al. (10). Nuclear extracts were made from frozen, decapsulated testes of adult mice by a combination of the methods of Roy et al. (34) and Dignam et al. (10) as previously described (31). Protein concentrations were estimated by using the Bio-Rad Protein Assay kit.
DNase I footprinting analysis.
DNase I footprinting was performed as previously described by using two DNA probes (31). To amplify the 396-bp region spanning −870 to −474, the forward primer 5′-TGCAGAATTCTCAATAAGCAGCTCCT-3′ and the reverse primer 5′-TTTGTCGACTGTAAAACGACGGG-3′ were used. To amplify the 380-bp region spanning −1150 to −770, the forward primer 5′-TGCAGAATTCGATTACAGGTGTTCA-3′ and the reverse primer 5′-TTTGTCGACAGCGGACGGATTCTC-3′ were used. Primers were designed so that PCR fragments contained an EcoRI site at the 5′ end and a SalI site at the 3′ end (underlined sequences). After digestion with EcoRI and SalI, the fragments were subcloned into the EcoRI/SalI site of pSL301 (Invitrogen). The sequences of the two inserts were confirmed by DNA sequencing. DNA fragments were excised by using EcoRI and SalI and purified as described above. To analyze the interactions of nuclear proteins with the coding (top) strand, probes were end labeled at the SalI site with [α-32P]dCTP and the remaining deoxynucleoside triphosphates by using the Klenow DNA polymerase (the EcoRI site is not labeled, since it lacks cytosine). To investigate the interactions of nuclear proteins with the noncoding (bottom) strand, probes were end labeled at the EcoRI site with [α-32P]dATP by using the Klenow DNA polymerase in the absence of additional nucleotides (thus, the SalI site is not labeled). DNase I (5-mg/ml stock solution) was diluted in 10 mM HEPES-NaOH (pH 7.6)–25 mM CaCl2. The dilutions used were 1:4,000 for bovine serum albumin (BSA), and 1:50 for mouse L-cell and testis nuclear extracts.
EMSAs.
Single-stranded oligomers were synthesized by the Johns Hopkins DNA Synthesis Facility, and complementary strands were annealed before use. Each double-stranded oligomer contained a recessed 3′ end which was filled in with either [α-32P]dATP or [α-32P]dCTP and the remaining deoxynucleoside triphosphates by using the Klenow DNA polymerase. The 32P-labeled probes were separated from the unincorporated nucleotides by chromatography on Sephadex G-50. EMSAs were performed essentially as previously described, by using 10 μg of nuclear extract (31). For competition experiments, a 100-fold molar excess of an unlabeled, double-stranded probe was incubated with 10 μg of nuclear extract for 20 min prior to the addition of the labeled probe. For the sequences of the double-stranded oligomers, see Table 2.
TABLE 2.
Oligomers used in EMSAs
| Probe | Sequencea |
|---|---|
| CRE consensus | 5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′ |
| CRE-like −645/−638 | 5′-AGTCACTGGGTACGTCAGGAAGTGGTGAGCA-3′ |
| CRE-like −766/−759 | 5′-GCCTCGCGGGACGTCGAAGCCGGTTTC-3′ |
| −756/−734 | 5′-GCCGGTTTCCTAGACGACCTTTCTCCA-3′ |
The sense sequences of the double-stranded oligomers are shown. Putative protein binding motifs are underlined.
RESULTS
Experimental approach.
One major limitation in studying promoters regulating the expression of genes during murine spermatogenesis is the lack of established male germ cell lines. To circumvent this deficiency, investigators have relied on in vitro transcription assays (6, 46) or transgenic mice (22, 30, 33, 52). We have chosen the latter approach, since the functional significance of putative promoter elements can be established in vivo.
Initially, a computer analysis was carried out to identify potential transcription factor binding sites within the 796-bp fragment containing the β4GalT-I male germ cell promoter (38). We noted that the ∼170 bp at either end of this fragment contained relatively few potential binding sites. In contrast, the ∼450-bp core region contained 13 potential cis motifs, including two cyclic AMP-responsive element (CRE)-like motifs (38). These motifs were of interest, since it had been shown that consensus CRE sites or variants (CRE-like sites) bind the transcriptional activator CREMτ, a spermatogenic cell-specific product of the CRE modulator (CREM) gene (reviewed in reference 36).
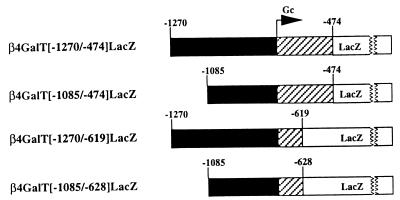
To determine if the β4GalT-I male germ cell promoter was confined to this ∼450-bp region, we initially designed three deletion constructs (Fig. 2). The first construct (β4GalT[−1085/−474]LacZ) lacks the 5′ sequence from −1270 to −1086, whereas the second (β4GalT[−1270/−619]LacZ) lacks the 3′ sequence from −618 to −474. If both constructs were found to exhibit β-galactosidase activity in the late pachytene spermatocytes and round spermatids of transgenic mice, then a third construct (β4GalT[−1085/−628]LacZ) containing both 5′ and 3′ deletions would be generated and tested. A positive result with this construct would allow us to rule out the possibility that transcriptional activation is mediated by synergistic interactions between cis elements located on either deleted fragment and the ∼450-bp core region.
FIG. 2.
Schematic representation of the LacZ-based constructs used to generate the first three transgenic lines. The original β4GalT[−1270/−474]LacZ construct contains a 796-bp fragment that includes 543 bp of the genomic sequence upstream of the male germ cell start site (Gc) and 253 bp of the flanking downstream sequence (38). In the β4GalT[−1085/−474]LacZ construct, the sequence spanning −1270 to −1086 was deleted, while in the β4GalT[−1270/−619]LacZ construct, the sequence spanning −618 to −474 was deleted. The β4GalT[−1085/−628]LacZ construct combines both 5′ and 3′ deletions. The solid, hatched, and open boxes represent the upstream genomic DNA, the 5′-untranslated region of the male germ cell transcript, and the LacZ coding sequence, respectively.
Promoter activity is retained in vivo after removal of 185 bp from the 5′ end, or 145 bp from the 3′ end, of the 796-bp fragment.
DNA fragments derived from the β4GalT[−1085/−474]LacZ and β4GalT[−1270/−619]LacZ constructs were used to generate transgenic mice. A total of four founders were identified for β4GalT[−1085/−474]LacZ, while a total of three founders were identified for β4GalT[−1270/−619]LacZ (Table 1, groups B and C, respectively). Subsequent breeding with wild-type mice produced heterozygous F1 and F2 progeny for analysis. In order to assess if expression of the reporter gene was observed exclusively in the testes of the transgenic mice, β-galactosidase activity was measured in extracts from various tissues (brain, heart, intestine, kidney, liver, lung, skeletal muscle, spleen, and testis) by using a chemiluminescence assay (Tropix Inc.). Levels of β-galactosidase enzymatic activity in the tissues tested were found to be comparable to background levels in nontransgenic control mice (data not shown), except for the testis tissue, which exhibited high levels of enzyme activity. As seen in Table 1, the enzyme levels in the testes of mice from groups B and C were similar to that of the previously characterized transgenic line in group A, which contained the entire 796-bp fragment. It can also be seen that levels of activity were independent of the transgene copy number. The testes of mice from transgenic lines 310, 915, and 930 exhibited the same level of β-galactosidase activity as found in nontransgenic controls (Table 1, group E), presumably due to the insertion of the transgene at a chromosomal location that suppresses expression.
TABLE 1.
β-Galactosidase activity in testes of transgenic mice
| Group and transgenic linea | Copy no. | β-Galactosidase activity (U min−1 mg of protein−1)b |
|---|---|---|
| A | ||
| β4GalT[−1270/−474]LacZ-140c | 49,000 (3) | |
| B | ||
| β4GalT[−1085/−474]LacZ-310 | 53 | 650 (1) |
| β4GalT[−1085/−474]LacZ-328 | 3 | 45,000 (2) |
| β4GalT[−1085/−474]LacZ-330 | 6 | 50,000 (2) |
| β4GalT[−1085/−474]LacZ-346 | 3 | 50,000 (3) |
| C | ||
| β4GalT[−1270/−619]LacZ-915 | 7 | 650 (1) |
| β4GalT[−1270/−619]LacZ-923 | 3 | 47,000 (2) |
| β4GalT[−1270/−619]LacZ-930 | 53 | 240 (1) |
| D | ||
| β4GalT[−1085/−628]LacZ-42 | 12 | 43,000 (3) |
| β4GalT[−1085/−628]LacZ-58 | 2 | 53,000 (3) |
| β4GalT[−1085/−628]LacZ-641 | 3 | 400 (2) |
| β4GalT[−1085/−628]LacZ-642 | 8 | 700 (2) |
| β4GalT[−1085/−628]LacZ-648 | 5 | 800 (1) |
| E | ||
| Nontransgenic | 0 | 600 (3) |
The terminal number denotes the individual founder.
Except for that in testis tissue, the β-galactosidase enzymatic activity in all of the tissues analyzed was comparable to the background level in nontransgenic mice. The number in parentheses is the number of F1 and/or F2 mice analyzed.
The transgene β4GalT[−1270/−474]LacZ was previously referred to as β4GT-LacZ (38).
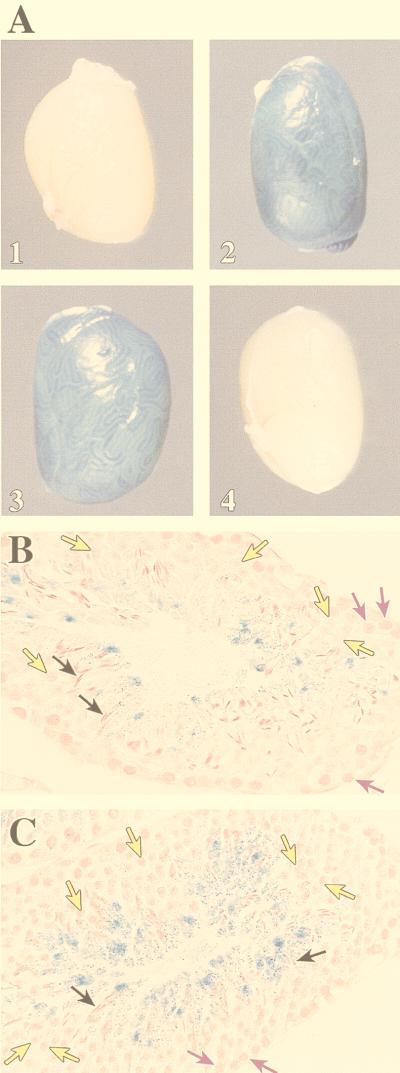
Following immersion in buffer containing the substrate X-Gal, the testes from a β4GalT[−1085/−474]LacZ-346 male mouse stained blue, indicating expression of the β-galactosidase reporter gene (Fig. 3A, part 2). Moreover, expression of the reporter gene was confined predominately to the seminiferous tubules. An identical result was also obtained with testes of mice from lines 328, 330, and 923 (data not shown), whereas the testes from a nontransgenic control mouse failed to stain blue (Fig. 3A, part 1). To establish which germ cell populations were expressing the reporter gene, the testes of mice from the β4GalT[−1085/−474]LacZ-346 line were embedded in paraffin, sectioned, and counterstained with nuclear fast red. Examination of these sections by light microscopy revealed that expression of the reporter gene was not detectable until the late pachytene spermatocyte stage (Fig. 3B). Expression was clearly evident in round spermatids. In the tissue section shown, the elongated spermatids, although transcriptionally inactive, are also stained due to accumulation of the β-galactosidase reaction product. A similar pattern of specific cell type staining was also observed in sections of testes of mice from lines 328, 330, and 923 (data not shown). Therefore, removal of either 185 bp from the 5′ end or 145 bp from the 3′ end of the 796-bp fragment resulted in the same germ cell type-specific expression of the reporter gene as observed with the original 796-bp fragment (β4GalT[−1270/−474]LacZ transgene) and with the endogenous β4GalT-I gene (38, 41).
FIG. 3.
Expression of the β-galactosidase reporter gene in the testes of transgenic mice. Testes from a nontransgenic control and the indicated transgenic lines were paraformaldehyde fixed, incubated in X-Gal for 48 h, and visualized directly (parts 1 to 4). To establish which male germ cell populations expressed the β-galactosidase reporter gene, the fixed and stained testes were paraffin embedded, sectioned, counterstained with nuclear fast red, and inspected under a microscope. (A) Parts: 1, a testis from a nontransgenic mouse; 2, a testis from a β4GalT[−1085/−474]LacZ-346 mouse; 3, a testis from a β4GalT[−793/−707]LacZ-320 mouse; 4, a testis from a β4GalT[mut−756/−743]LacZ-374 mouse. Note that the blue staining, indicative of the expression of the β-galactosidase reporter gene (LacZ) is concentrated in the seminiferous tubules. (B) Section of a seminiferous tubule obtained from the testis of a β4GalT[−1085/−474]LacZ-346 mouse. The purple arrows indicate the pachytene spermatocytes, the yellow arrows indicate the round spermatids, and the black arrows indicate the elongated spermatids (final magnification, ×625). (C) Section of a seminiferous tubule obtained from the testis of a β4GalT[−793/−707]LacZ-320 mouse (final magnification, ×625). Note that this 87-bp genomic fragment which contains the TASS-1 regulatory element is sufficient to drive male germ cell-specific expression of the reporter gene in a pattern that is comparable to that of the endogenous β4GalT-I gene.
A 457-bp core fragment spanning the β4GalT-I germ cell start site is sufficient to confer correct germ cell-specific expression in transgenic mice.
Since male germ cell promoter activity was maintained in transgenic mice harboring either a 5′ or a 3′ deletion of the 796-bp promoter region, the third construct (β4GalT[−1085/−628]LacZ), combining both 5′ and 3′ deletions, was generated and analyzed. As seen in Table 1, group D, testes of mice from two of the five lines (lines 42 and 58) showed high levels of β-galactosidase enzymatic activity similar to that seen in the animals in groups A to C. Levels of β-galactosidase in all of the somatic tissues analyzed were comparable to the background levels in nontransgenic control mice (data not shown). Background levels of β-galactosidase activity were found in the testes of mice from the other three lines (641, 642, and 648), presumably due to the insertion of the transgene at a chromosomal location that suppresses expression.
As anticipated from the results of the enzymatic assays, only the testes of transgenic males from lines 42 and 58 stained blue when incubated in X-Gal. Again, the staining was confined primarily to the seminiferous tubules and examination of testis sections by light microscopy revealed that the cell-specific pattern of expression of the reporter gene was comparable to that seen in the testes of mice from the β4GalT[−1270/−474]LacZ line (data not shown).
DNase I footprinting analysis of the 457-bp β4GalT-I male germ cell promoter region.
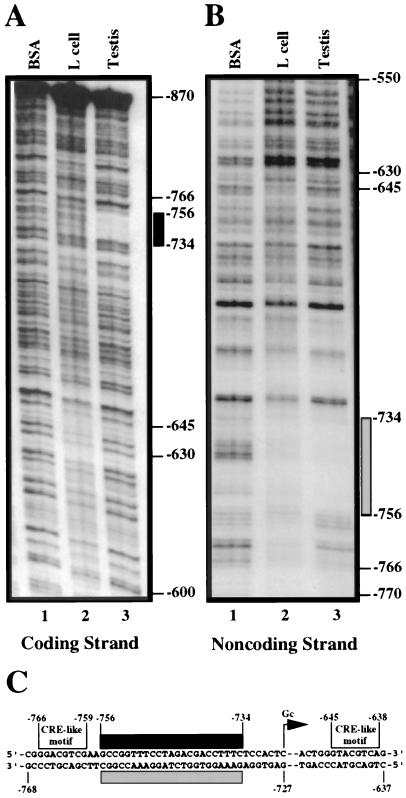
The functional analysis using transgenic mice demonstrated that a 457-bp genomic fragment spanning −1085 to −628 contains the cis elements necessary for correct male germ cell-specific expression of the transgene. Of the several motifs within this region identified by computer analysis, two putative CRE-like elements, spanning −766 to −759 and −645 to −630, had been noted (38). To directly assess whether the two CRE-like motifs and/or other sequences within the 457-bp fragment bind testis-specific nuclear proteins, DNase I footprinting assays were carried out. A restriction fragment spanning either the −1150 to −770 or the −870 to −474 sequence was end labeled on either the coding or the noncoding strand. Each probe was then incubated with 25 μg of a testis nuclear extract or an equivalent amount of BSA. Partial digestion with DNase I revealed only a single protected region spanning −756 to −734 on both the coding and noncoding strands (Fig. 4A and B, lanes 3). The sequence of the protected region is shown in Fig. 4C. To determine whether this region was also protected by nuclear proteins derived from somatic cells, probes were incubated with 25 μg of mouse L-cell nuclear extract. A footprint extending across the same sequence was observed, but only on the noncoding strand (Fig. 4B, lane 2). This observation suggests that the binding activity in the mouse L-cell nuclear extract is due to a different protein or protein complex.
FIG. 4.
Identification of a single 23-bp protected region within the 457-bp promoter region by DNase I footprinting analysis. Protected regions on the coding strand (A) and noncoding strand (B) are indicated by the black and gray bars, respectively. A DNA fragment corresponding to the region between −870 to −474 was end labeled with 32P and digested with DNase I in the presence of BSA (lanes 1) or nuclear proteins from mouse L cells (lanes 2) or testes (lanes 3). (C) Sequence of the region spanning −768 to −637. The black rectangle indicates the region protected by the testis nuclear extract, while the gray rectangle indicates the region protected by both testis and L-cell nuclear extracts. The locations of the two CRE-like motifs are shown. The male germ cell transcription start site (Gc) is located at position −727.
Neither CRE-like sequence within the 457-bp promoter region binds nuclear protein.
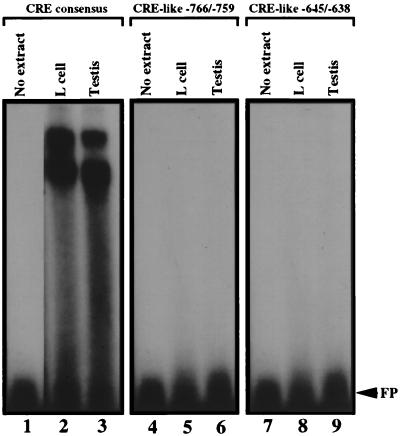
The DNase I footprinting analysis showed that the two CRE-like motifs did not bind protein, as the region containing these motifs was not protected after incubation with either the testis (Fig. 4A and B, lanes 3) or mouse L-cell (Fig. 4A and B, lanes 2) nuclear extract. To confirm this result, two double-stranded oligomers containing either the CRE-like motif spanning −766 to −759 or −645 to −638 were synthesized (Table 2) and tested by EMSA. Both failed to bind nuclear proteins from the testis (Fig. 5, lanes 6 and 9) or mouse L cells (Fig. 5, lanes 5 and 8). As a positive control, incubation of an oligomer containing the CRE consensus sequence with either the mouse L-cell or the testis nuclear extract produced two retarded bands (Fig. 5, lanes 2 and 3). Thus, even though CRE binding factors are present in both nuclear extracts, the inability to demonstrate complex formation with either of the CRE-like motifs present in the 457-bp promoter region suggests that members of the CRE family do not play a role in the regulation of β4GalT-I expression in male germ cells.
FIG. 5.
The two CRE-like sequences within the 457-bp promoter fail to bind a nuclear protein(s) when analyzed by EMSA. Double-stranded oligomers containing the CRE consensus binding site (left panel) or the CRE-like motif spanning −766 to −759 (middle panel) or −645 to −638 (right panel) were end labeled with 32P and incubated with 10 μg of testis (lanes 3, 6, and 9) or mouse L-cell (lanes 2, 5, and 8) nuclear extract. FP designates the migration position of the free probe.
Analysis of the DNase I-protected region by EMSA.
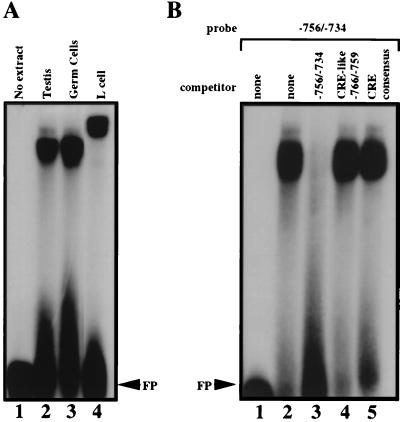
By using the information from the DNase I protection assays, we synthesized a double-stranded oligomer that corresponded to the sequence of the single footprint identified within the 457-bp core promoter region. Incubation of the mouse testis nuclear extract with this −756/−734 oligomer (Table 2) resulted in the formation of one very intense retarded band (Fig. 6A, lane 2). To confirm the specificity of protein binding, we also performed EMSAs by using specific and nonspecific DNA competitors. As seen in Fig. 6B, complex formation did not occur in the presence of a 100-fold molar excess of the unlabeled −756/−734 oligomer (lane 3) whereas two nonspecific oligomers had no effect on complex formation (lanes 4 and 5).
FIG. 6.
Nuclear factor(s) binding to the DNA sequence spanning −756 to −734. (A) The −756/−734 oligomer was end labeled with 32P using Klenow DNA polymerase and incubated with nuclear extracts from testes (lane 2), pooled male germ cells (lane 3), or mouse L cells (lane 4). Upon a lighter exposure, the band present in lane 4 can be resolved into two bands. (B) Specificity of the complex obtained with the testis nuclear extract (lane 2) was demonstrated by addition of a 100-fold molar excess of the unlabeled −756/−734 oligomer (lane 3) prior to addition of the labeled probe. Incubation with a 100-fold molar excess of a nonspecific oligomer containing the CRE-like motif spanning −766 to −759 (lane 4) or the CRE consensus motif (lane 5) did not prevent complex formation. FP indicates the migration position of the free probe.
In order to verify that a nuclear extract derived from somatic cells did not give rise to a similar migration pattern, the −756/−734 oligomer was next incubated with a mouse L-cell nuclear extract. As seen in Fig. 6A, lane 4, although a DNA-protein complex was seen, its migration pattern differed from that produced by the testis nuclear extract. Additional EMSAs, using specific and nonspecific DNA competitors, established that the binding activity in the mouse L-cell nuclear extract was specific (data not shown). Therefore, even though a DNA binding protein(s) present in somatic cells interacts with the noncoding strand of the −756/−734 oligomer, it differs from the nuclear protein(s) present in the testis.
The −756/−734 oligomer binds a protein factor(s) present in a male germ cell nuclear extract.
Since the testis is composed of both germ and somatic cell types, an unequivocal confirmation that the gel shift pattern seen with the testis nuclear extract is due to proteins in male germ cells can be established by preparing nuclear extracts from isolated, pooled male germ cells, in which round spermatids comprise ∼50% of the cell population. Incubation of the male germ cell nuclear extract with the −756/−734 oligomer gave rise to the identical band observed when the testis nuclear extract was used (Fig. 6A, compare lane 3 with lane 2). This result clearly demonstrates that the testis nuclear protein(s) that recognizes the binding site in the −756/−734 oligomer is present in male germ cells.
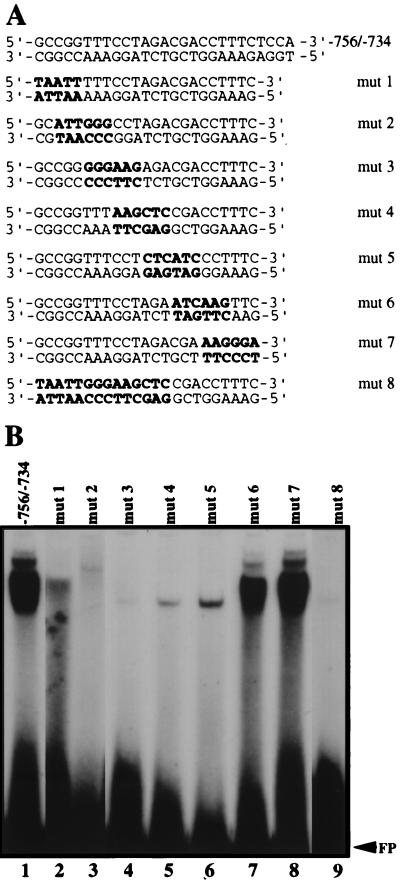
Mutation analysis defines a 14-bp DNA binding site.
The DNA binding site within the −756 to −734 sequence was determined by using a series of oligomers containing overlapping groups of 5- or 6-bp mutations (Fig. 7A). Complex formation equivalent to that seen with the −756/−734 oligomer (Fig. 7B, lane 1) was only observed when oligomers mut 6 and mut 7 were used (Fig. 7B, lanes 7 and 8). This shows that the 13 bp at the 3′ end of the sequence are not critical for protein binding. The other oligomers tested either abolished (mut 2 and mut 3) or greatly reduced (mut 1, mut 4, and mut 5) complex formation (Fig. 7B, lanes 2 to 6), showing that the 14-bp sequence (5′-GCCGGTTTCCTAGA-3′) at the 5′ end of the sequence is involved in protein binding. As anticipated, an oligomer (mut 8) in which the 14 bp were mutated by transversion also abrogated complex formation (Fig. 7B, lane 9).
FIG. 7.
Effect of mutations within the −756 to −743 region on complex formation. (A) Sequences of the wild-type and mutated (mut 1 to mut 8) double-stranded oligomers used for EMSAs. The mutated bases are in boldface. (B) Eight mutated double-stranded oligomers (lanes 2 to 9) labeled with 32P at their respective 5′ ends and incubated with 10 μg of testis nuclear extract. Complex formation observed with each mutated oligomer was compared with that observed with the −756/−734 oligomer (lane 1). FP designates the migration position of the free probe.
Because no significant match was obtained when the 14-bp sequence was used as the input query sequence for the TRANSFAC or TFD database (44a), we have assigned the acronym TASS-1 to the factor binding this regulatory motif.
An 87-bp fragment containing the TASS-1 binding motif is sufficient to direct correct germ cell-specific β4GalT-I expression in transgenic mice.
The in vitro DNA-protein binding assays established that only one DNA binding site, located 16 bp upstream of the transcriptional start site, was recognized by a protein(s) present in the male germ cell nuclear extract. If the TASS-1 binding motif is the only regulatory element required for male germ cell-specific expression of β4GalT-I, the following predictions can be made. (i) Mice harboring a transgenic construct containing this motif within a short genomic fragment should express β-galactosidase in late pachytene spermatocytes and round spermatids. (ii) Mice harboring a transgenic construct in which the TASS-1 binding motif has been mutated, within the context of the original 796-bp fragment, should not express β-galactosidase in these cells.
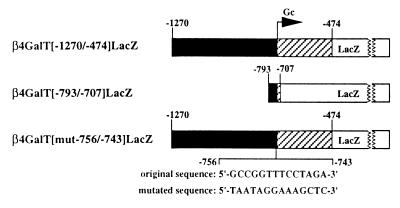
To test the first prediction, an 87-bp fragment containing 67 bp of the genomic sequence upstream of the male germ cell start site and 20 bp of the flanking downstream sequence was fused to the bacterial LacZ coding sequence, generating the β4GalT[−793/−707]LacZ construct (Fig. 8). A total of five transgenic founders were identified (Table 3, group F). Two lines (320 and 337) showed levels of β-galactosidase enzymatic activity similar to those seen in the mice in groups A to D, while a third line (339) showed levels of β-galactosidase activity about threefold higher. Again, there appeared to be no correlation between expression levels and transgene copy number. Reporter gene activity in all of the other tissues analyzed from these lines was similar to the background value measured in nontransgenic controls (data not shown). Testes of mice from lines 330 and 349 showed reduced levels of β-galactosidase activity (∼10-fold over the background; Table 3, group F), presumably due to the insertion of the transgene at a chromosomal location that suppresses expression.
FIG. 8.
Schematic representation of the β4GalT[−793/−707]LacZ and β4GalT[mut−756/−743]LacZ constructs. The original β4GalT[−1270/−474]LacZ construct is shown for comparison. In the β4GalT[−793/−707]LacZ construct, an 87-bp fragment containing 67 bp of the genomic sequence upstream from the β4GalT-I male germ cell start site and 20 bp of the flanking downstream sequence was fused to the bacterial LacZ coding sequence. In the β4GalT[mut−756/−743]LacZ construct, the nucleotides within the 14-bp TASS-1 motif (spanning −756 to −743) were mutated by transversion. The positions and sequences of the original and mutated sites are shown. The solid, hatched, and open boxes represent the upstream genomic DNA, the 5′-untranslated region of the male germ cell transcript, and the LacZ coding sequence, respectively.
TABLE 3.
β-Galactosidase activity in testes of transgenic mice
| Group and transgenic linea | Copy no. | β-Galactosidase activity (U min−1 mg of protein−1)b |
|---|---|---|
| F | ||
| β4GalT[−793/−707]LacZ-320 | 25 | 56,000 (3) |
| β4GalT[−793/−707]LacZ-330 | 4 | 5,500 (3) |
| β4GalT[−793/−707]LacZ-337 | 55 | 68,000 (3) |
| β4GalT[−793/−707]LacZ-339 | 64 | 161,000 (2) |
| β4GalT[−793/−707]LacZ-349 | 21 | 6,700 (2) |
| G | ||
| β4GalT[mut−756/−743]LacZ-369 | 25 | 4,200 (2) |
| β4GalT[mut−756/−743]LacZ-374 | 8 | 4,000 (3) |
| β4GalT[mut−756/−743]LacZ-377 | 20 | 400 (2) |
| β4GalT[mut−756/−743]LacZ-378 | 8 | 2,400 (1) |
| β4GalT[mut−756/−743]LacZ-380 | 3 | 3,300 (3) |
| β4GalT[mut−756/−743]LacZ-2211 | 72 | 2,600 (2) |
| β4GalT[mut−756/−743]LacZ-2226 | 5 | 1,400 (3) |
The terminal number denotes the individual founder.
The number in parentheses is the number of F1 and/or F2 mice analyzed. Except for that in testis tissue, the β-galactosidase enzymatic activity in all of the tissues analyzed was comparable to the background level in nontransgenic mice.
As seen in Fig. 3A, part 3, the staining seen when the testis of a mouse from line 320 was incubated in X-Gal was comparable to that observed with the four previous transgenic constructs. Examination of sections by light microscopy revealed that the X-Gal staining pattern in male germ cells was comparable to that of the four previous transgenic constructs (Fig. 3C). Identical results were also obtained when testes of mice from lines 337 and 339 were used. These results demonstrate that a short genomic fragment of only 87 bp which contains the TASS-1 binding motif is sufficient to direct expression of the β-galactosidase reporter gene in late pachytene spermatocytes and round spermatids of transgenic mice.
The TASS-1 binding motif is essential for transgene expression in late pachytene spermatocytes and round spermatids in vivo.
The second prediction was tested by generating the β4GalT[mut−756/−743]LacZ construct (Fig. 8), in which the TASS-1 binding site was mutated by transversion within the context of the original 796-bp sequence. Seven transgenic founders were identified (Table 3, group G), and in all of the lines analyzed, β-galactosidase activity was found to be 12- to 35-fold lower than that measured in the testes of mice from group A.
To determine if β-galactosidase was expressed in the late pachytene spermatocytes and round spermatids of these transgenic mice, testes were collected and incubated in X-Gal. As seen in Fig. 3A, part 4, the testis of a mouse from line 374 (or from any of the remaining six lines) did not stain blue nor was any germ cell staining seen after examination of testis sections by light microscopy (data not shown). Collectively, the results presented demonstrate that the TASS-1 regulatory element located 16 bp upstream of the male germ cell start site is essential for β4GalT-I expression in late pachytene spermatocytes and round spermatids in vivo.
DISCUSSION
One of the most intriguing results from this study is the finding that a short 87-bp genomic fragment that flanks the β4GalT-I male germ cell-specific start site and contains a single 14-bp regulatory element (termed the TASS-1 motif) drives the in vivo expression of a reporter gene in the later stages of spermatogenesis. It is instructive to compare the relative sizes of the respective promoter regions that govern the expression of β4GalT-I in male germ cells or somatic cells. As summarized in the Introduction, DNase I footprinting and EMSAs have been used to elucidate the DNA binding motifs in an ∼1.3-kb genomic fragment positioned upstream of the two somatic transcriptional start sites (Fig. 1; see references 14 and 31). When this genomic fragment was evaluated for the ability to drive somatic cell expression of a reporter gene in transgenic mice, we failed to detect expression in six different lines (unpublished data). Thus, in contrast to the 87-bp male germ cell promoter, the β4GalT-I somatic cell promoter region extends beyond the 1.3-kb genomic fragment analyzed.
While we find the compactness of the β4GalT-I male germ cell promoter remarkable, this appears to be an emerging general rule and not the exception, based on the limited subset of male germ cell promoters that have been analyzed in vivo. In addition to β4GalT-I, other examples of short, compact promoter regions that regulate male germ cell-specific gene expression include the 91-bp angiotensin-converting enzyme promoter (17), the 330-bp calmegin promoter (50), the 100-bp lactate dehydrogenase c promoter (23), the 187-bp promoter of the E1α subunit of the pyruvate dehydrogenase complex (18), the 328-bp phosphoglycerate kinase 2 (Pgk-2) promoter (33), the 116-bp proenkephalin promoter (24), and the 113-bp protamine 1 promoter (53).
Most of these male germ cell-specific promoters, including β4GalT-I, do not have a TATA binding site immediately upstream of their respective start sites. This suggests that binding of RNA polymerase II may occur via an initiator (Inr) element (YCANTYY) (19). Inspection of the sequence surrounding the β4GalT-I male germ cell start site (5′-CACTC+1CATTTT-3′) does reveal a potential Inr consensus motif (underlined sequence), with the requisite A nucleotide positioned near the start site (C+1). Thus, the Inr and TASS-1 binding sites may be all that is required for β4GalT-I expression in late pachytene spermatocytes and round spermatids.
Does TASS-1 associate with a cofactor(s) to activate transcription of the β4GalT-I gene in male germ cells?
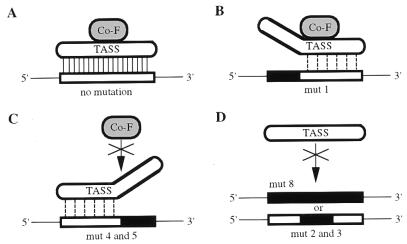
While it is proposed that only the TASS-1 and Inr binding motifs are required for expression of β4GalT-I in male germ cells, it is possible that an additional protein(s) is needed to mediate communication between TASS-1 and the basal transcription machinery (positioned at the Inr site). In fact, the presence of a cofactor interacting directly with TASS-1 could explain the differing electrophoretic mobilities of the DNA-protein complexes formed by using the various mutated oligomers (Fig. 7). In the schematic in Fig. 9A, we propose that binding of TASS-1 to the 14-bp motif leads to the recruitment of a cofactor (Co-F), which gives rise to the intense band seen in Fig. 7B, lane 1. While mutation at the 5′ end of the binding motif (Fig. 9B, mut 1) results in reduced TASS-1 binding, Co-F can still bind the complex; thus, the migration position of the complex seen with mut 1 is identical to that seen with the 14-bp sequence (Fig. 7, compare lanes 1 and 2). Mutation at the 3′ end of the binding motif (Fig. 9C, mut 4 and mut 5) also results in reduced TASS-1 binding, but in this case, Co-F cannot bind the DNA–TASS-1 complex. Therefore, the migration pattern seen in Fig. 7, lanes 5 and 6, reflects the binding of TASS-1 alone. Finally, mutation of the core sequence (Fig. 9D, mut 2 and mut 3) or of the 14-bp motif (Fig. 9D, mut 8) abrogates the binding of TASS-1 and Co-F (Fig. 7, lanes 3, 4, and 9).
FIG. 9.
Model depicting the binding of TASS-1 and a putative cofactor (Co-F) to the 14-bp regulatory element. (A) High-affinity binding of TASS-1 to the 14-bp motif (open rectangle) is shown by the solid vertical lines. This binding leads to the recruitment of a putative Co-F which interacts with TASS-1 via protein-protein interactions. (B) TASS-1 binds weakly, as shown by the dashed vertical lines, to oligomer mut 1, in which the 5′ end of the binding motif has been mutated (solid box). Co-F binding to DNA-bound TASS-1 is unaffected, but the complex can easily dissociate; thus, the steady-state level of the complex is greatly reduced. (C) TASS-1 binds weakly to oligomers mut 4 and mut 5, in which the 3′ end of the binding motif has been mutated (solid box); however, Co-F cannot bind to the DNA–TASS-1 complex. (D) TASS-1 is unable to bind an oligomer in which the core sequence is mutated (mut 2 or mut 3) or the entire 14-bp motif is mutated (mut 8).
Transcriptional activation of the β4GalT-I male germ cell promoter is CREMτ independent.
Recent studies have begun to provide insight into the factors responsible for the transcriptional activation of genes expressed in a stage-specific manner during spermatogenesis. The genes studied can be divided into two categories, i.e., those that are activated by CREMτ and those that are not. The genes regulated by CREMτ include those for the angiotensin-converting enzyme (17, 20, 55), calspermin (43), RT7 (9), transition protein 1 (21), and protamines 1 and 2 (13, 44, 53). As anticipated, none of these genes are expressed in the testes of CREM knockout mice (2, 28).
While we had speculated that the β4GalT-I male germ cell promoter was also regulated by CREMτ, due to the presence of two putative CRE-like motifs in the 796-bp genomic fragment initially analyzed (38), the DNase I footprinting and EMSA results clearly demonstrate that neither site binds testis nuclear proteins (Fig. 4 and 5). In addition, when steady-state levels of the β4GalT-I male germ cell-specific transcripts in the testes of CREM knockout mice were examined by Northern blot analysis, they were comparable to those of wild-type mice (10a). Collectively, these results show that the regulation of the β4GalT-I gene during spermatogenesis is CREMτ independent.
Other examples of genes activated in developing male germ cells by a CREMτ-independent mechanism include those for histone H1 (8, 47, 48, 51), lactate dehydrogenase c (23, 54), proenkephalin (24), and Pgk-2 (11, 12). Transgenic constructs, in combination with DNase I footprinting and EMSAs, have been used to delineate functional regulatory elements in the male germ cell promoter of these genes. With the exception of that for Pgk-2 (discussed below), the transcriptional activation of each of these genes apparently requires unique regulatory elements that are unrelated to the TASS-1 motif.
A comparison of the murine Pgk-2 male germ cell promoter with the β4GalT-I promoter proved interesting. Male germ cell-specific expression of both genes is seen in pachytene spermatocytes and postmeiotic haploid round spermatids. Cell-free transcription assays, DNase I footprinting, and EMSAs were used to identify a testis protein designated TAP-1 that binds the sequence 5′-AATTTGAAAGGAAATCCAG-3′ in the Pgk-2 promoter (12). Furthermore, it was shown by mutation analysis that Tap-1 recognizes the core 5′-GGAA-3′ sequence, which is the signature binding motif for the Ets family of transcription factors (reviewed in reference 49; see also references 29 and 45). From an inspection of the bottom strand of the TASS-1 binding site, we noted the presence of this same 4-bp Ets motif (sequence in Fig. 4C); in addition, three of the surrounding eight nucleotides in the TASS-1 binding site were identical to those in the Tap-1 binding site.
Is TASS-1 an Ets protein?
The presence of the Ets core motif in the TASS-1 binding site suggested that TASS-1 is an Ets family member. To date, ∼30 different Ets proteins have been identified; each interacts specifically with its respective cis element via a conserved DNA binding domain ∼85 amino acids in length (reviewed in references 42 and 49). The 5-bp flanking sequence on either side of the 5′-GGAA-3′ core appears to determine the affinity of a specific Ets protein for its target sequence, although it has been shown that different Ets proteins can bind with comparable affinity to the same target sequence in vitro (5).
To determine if the Tap-1 and TASS-1 motifs bind the same Ets transcriptional activator, an oligomer shown to bind Tap-1 in vitro (12) was used as a competitor in an EMSA using a testis nuclear extract and the −756/−734 oligomer (containing the TASS-1 binding site). Complex formation was not affected, even at a 400-fold molar excess of the unlabeled Tap-1 oligomer (data not shown). This result strongly suggests that the protein binding to the TASS-1 regulatory element is not Tap-1.
We have also examined a subset of Ets family members reported to be expressed in murine testes. This group includes Elk-1 (32), ER71 and ER81 (5), ERM (27), ERP (26), GABPα (5), and PEA3 (27). We subsequently established that these seven Ets family members are expressed in pachytene spermatocytes and round spermatids by using a reverse transcription-PCR-based assay (data not shown). ER71 was of particular interest, since Northern analysis of a panel of 10 murine tissues revealed that ER71 mRNA was only found in testes and the 10.5-day-old embryo (5). EMSAs were carried out by using the appropriate Ets oligomers as competitors with the −756/−734 oligomer. The inability of each of the oligomers to affect complex formation, even at a 400-fold molar excess (data not shown), suggests that TASS-1 is not one of these proteins.
Finally, Ets-1, Ets-2, Ets-3, Erg-1, Erg-2, and Fli-1 were ruled out as candidates, since an antibody (Santa Cruz Biotechnology Inc.) that recognizes a region of the conserved DNA binding domain of these proteins failed to abrogate or supershift the complex observed after incubation of the −756/−734 oligomer with the testis nuclear extract (data not shown). Further experiments are necessary to establish if TASS-1 is (i) a novel Ets protein, (ii) a member of the Ets family, or (iii) a novel transcriptional activator that is unrelated to the Ets family but also recognizes a 5′-GGAA-3′ motif. If TASS-1 proved to be an Ets family member, this would be the first demonstration of gene regulation in male germ cells in vivo by an Ets protein.
ACKNOWLEDGMENTS
We thank P. A. Coulombe, A. D. Friedman, and W. W. Wright for many insightful discussions and Jane Scocca and Neng-Wen Lo for critical review of the manuscript. We are grateful to W. W. Wright for his help in preparing the pooled male germ cells; S. Brust, A. Chen, C. Hawkins, D. Blesh, and M. Cowan (Johns Hopkins University Transgenic Mouse Core Laboratory) for the production of transgenic mice; and H. Chapman and the animal facility staff of the Johns Hopkins Oncology Center for animal care.
This work was supported in part by National Institutes of Health grant CA45799 (to J.H.S.). Postdoctoral support of M.C. was provided by Human Frontiers Science Program grant RG-414/94 M (to J.H.S.).
REFERENCES
- 1.Beyer T A, Hill R L. Glycosylation pathways in the biosynthesis of nonreducing terminal sequences in oligosaccharides of glycoproteins. III. New York, N.Y: Academic Press, Inc.; 1982. pp. 25–45. [Google Scholar]
- 2.Blendy J A, Kaestner K H, Weinbauer G F, Nieschlag E, Schutz G. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature. 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 3.Brew K, Vanaman T C, Hill R L. The role of α-lactalbumin and the A protein in lactose synthetase: a unique mechanism for the control of a biological reaction. Proc Natl Acad Sci USA. 1968;59:491–497. doi: 10.1073/pnas.59.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodbeck U, Denton W L, Tanahashi N, Ebner K E. The isolation and identification of the B protein of lactose synthetase as α-lactalbumin. J Biol Chem. 1967;242:1391–1397. [PubMed] [Google Scholar]
- 5.Brown T A, McKnight S L. Specificities of protein-protein and protein-DNA interaction of GABPα and two newly defined ets-related proteins. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 6.Bunick D, Johnson P A, Johnson T R, Hecht N B. Transcription of the testis-specific mouse protamine 2 gene in a homologous in vitro transcription system. Proc Natl Acad Sci USA. 1990;87:891–895. doi: 10.1073/pnas.87.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charron M, Shaper J H, Shaper N L. The increased level of β1,4-galactosyltransferase required for lactose biosynthesis is achieved in part by translational control. Proc Natl Acad Sci USA. 1998;95:14805–14810. doi: 10.1073/pnas.95.25.14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clare S E, Hatfield W R, Fantz D A, Kistler W S, Kistler M K. Characterization of the promoter region of the rat testis-specific histone H1t gene. Biol Reprod. 1997;56:73–82. doi: 10.1095/biolreprod56.1.73. [DOI] [PubMed] [Google Scholar]
- 9.Delmas V, van der Hoorn F, Mellstrom B, Jegou B, Sassone-Corsi P. Induction of CREM activator proteins in spermatids: down-stream targets and implications for haploid germ cell differentiation. Mol Endocrinol. 1993;7:1502–1514. doi: 10.1210/mend.7.11.8114765. [DOI] [PubMed] [Google Scholar]
- 10.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Fimia, G. M., F. Nantel, and P. Sassone-Corsi. Personal communication.
- 11.Gebara M M, McCarrey J R. Protein-DNA interactions associated with the onset of testis-specific expression of the mammalian Pgk-2 gene. Mol Cell Biol. 1992;12:1422–1431. doi: 10.1128/mcb.12.4.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goto M, Masamune Y, Nakanishi Y. A factor stimulating transcription of the testis-specific Pgk-2 gene recognizes a sequence similar to the binding site for a transcription inhibitor of the somatic-type Pgk-1 gene. Nucleic Acids Res. 1993;21:209–214. doi: 10.1093/nar/21.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ha H, van Wijnen A J, Hecht N B. Tissue-specific protein-DNA interactions of the mouse protamine 2 gene promoter. J Cell Biochem. 1997;64:94–105. [PubMed] [Google Scholar]
- 14.Harduin-Lepers A, Shaper J H, Shaper N L. Characterization of two cis-regulatory regions in the murine β1,4-galactosyltransferase gene. Evidence for a negative regulatory element that controls initiation at the proximal site. J Biol Chem. 1993;268:14348–14359. [PubMed] [Google Scholar]
- 15.Harduin-Lepers A, Shaper N L, Mahoney J A, Shaper J H. Murine β1,4-galactosyltransferase: round spermatid transcripts are characterized by an extended 5′-untranslated region. Glycobiology. 1992;2:361–368. doi: 10.1093/glycob/2.4.361. [DOI] [PubMed] [Google Scholar]
- 16.Hill R L, Brew K, Vanaman T C, Trayer I P, Mattock P. The structure, function, and evolution of α-lactalbumin. Brookhaven Symp Biol. 1968;21:139–154. [PubMed] [Google Scholar]
- 17.Howard T, Balogh R, Overbeek P, Bernstein K E. Sperm-specific expression of angiotensin-converting enzyme (ACE) is mediated by a 91-base-pair promoter containing a CRE-like element. Mol Cell Biol. 1993;13:18–27. doi: 10.1128/mcb.13.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iannello R C, Young J, Sumarsono S, Tymms M J, Dahl H H, Gould J, Hedger M, Kola I. Regulation of Pdha-2 expression is mediated by proximal promoter sequences and CpG methylation. Mol Cell Biol. 1997;17:612–619. doi: 10.1128/mcb.17.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessler S P, Rowe T M, Blendy J A, Erickson R P, Sen G C. A cyclic AMP response element in the angiotensin-converting enzyme gene and the transcription factor CREM are required for transcription of the mRNA for the testicular isozyme. J Biol Chem. 1998;273:9971–9975. doi: 10.1074/jbc.273.16.9971. [DOI] [PubMed] [Google Scholar]
- 21.Kistler M K, Sassone-Corsi P, Kistler W S. Identification of a functional cyclic adenosine 3′,5′-monophosphate response element in the 5′-flanking region of the gene for transition protein 1 (TP1), a basic chromosomal protein of mammalian spermatids. Biol Reprod. 1994;51:1322–1329. doi: 10.1095/biolreprod51.6.1322. [DOI] [PubMed] [Google Scholar]
- 22.Langford K G, Shai S Y, Howard T E, Kovac M J, Overbeek P A, Bernstein K E. Transgenic mice demonstrate a testis-specific promoter for angiotensin-converting enzyme. J Biol Chem. 1991;266:15559–15562. [PubMed] [Google Scholar]
- 23.Li S, Zhou W, Doglio L, Goldberg E. Transgenic mice demonstrate a testis-specific promoter for lactate dehydrogenase, LDHC. J Biol Chem. 1998;273:31191–31194. doi: 10.1074/jbc.273.47.31191. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Tokeson J, Persengiev S P, Ebert K, Kilpatrick D L. Novel repeat elements direct rat proenkephalin transcription during spermatogenesis. J Biol Chem. 1997;272:5056–5062. doi: 10.1074/jbc.272.8.5056. [DOI] [PubMed] [Google Scholar]
- 25.Lo N W, Shaper J H, Pevsner J, Shaper N L. The expanding β4-galactosyltransferase gene family: messages from the databanks. Glycobiology. 1998;8:517–526. doi: 10.1093/glycob/8.5.517. [DOI] [PubMed] [Google Scholar]
- 26.Lopez M, Oettgen P, Akbarali Y, Dendorfer U, Libermann T A. ERP, a new member of the ets transcription factor/oncoprotein family: cloning, characterization, and differential expression during B-lymphocyte development. Mol Cell Biol. 1994;14:3292–3309. doi: 10.1128/mcb.14.5.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monte D, Baert J L, Defossez P A, de Launoit Y, Stehelin D. Molecular cloning and characterization of human ERM, a new member of the Ets family closely related to mouse PEA3 and ER81 transcription factors. Oncogene. 1994;9:1397–1406. [PubMed] [Google Scholar]
- 28.Nantel F, Monaco L, Foulkes N S, Masquilier D, LeMeur M, Henriksen K, Dierich A, Parvinen M, Sassone-Corsi P. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature. 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 29.Nye J A, Petersen J M, Gunther C V, Jonsen M D, Graves B J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 30.Peschon J J, Behringer R R, Brinster R L, Palmiter R D. Spermatid-specific expression of protamine 1 in transgenic mice. Proc Natl Acad Sci USA. 1987;84:5316–5319. doi: 10.1073/pnas.84.15.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajput B, Shaper N L, Shaper J H. Transcriptional regulation of murine β1,4-galactosyltransferase in somatic cells. Analysis of a gene that serves both a housekeeping and a mammary gland-specific function. J Biol Chem. 1996;271:5131–5142. doi: 10.1074/jbc.271.9.5131. [DOI] [PubMed] [Google Scholar]
- 32.Rao V N, Huebner K, Isobe M, ar-Rushdi A, Croce C M, Reddy E S. Elk, tissue-specific ets-related genes on chromosomes X and 14 near translocation breakpoints. Science. 1989;244:66–70. doi: 10.1126/science.2539641. [DOI] [PubMed] [Google Scholar]
- 33.Robinson M O, McCarrey J R, Simon M I. Transcriptional regulatory regions of testis-specific PGK2 defined in transgenic mice. Proc Natl Acad Sci USA. 1989;86:8437–8441. doi: 10.1073/pnas.86.21.8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy R J, Gosselin P, Guerin S L. A short protocol for micropurification of nuclear proteins from whole animal tissue. BioTechniques. 1991;11:770–777. [PubMed] [Google Scholar]
- 35.Russo R N, Shaper N L, Shaper J H. Bovine β1,4-galactosyltransferase: two sets of mRNA transcripts encode two forms of the protein with different amino-terminal domains. In vitro translation experiments demonstrate that both the short and the long forms of the enzyme are type II membrane-bound glycoproteins. J Biol Chem. 1990;265:3324–3331. [PubMed] [Google Scholar]
- 36.Sassone-Corsi P. CREM: a master-switch governing male germ cell differentiation and apoptosis. Semin Cell Dev Biol. 1998;9:475–482. doi: 10.1006/scdb.1998.0200. [DOI] [PubMed] [Google Scholar]
- 37.Shaper N L, Charron M, Lo N-W, Shaper J H. β1,4-Galactosyltransferase and lactose biosynthesis: recruitment of a housekeeping gene from the nonmammalian vertebrate gene pool for a mammary gland specific function. J Mammary Gland Biol Neoplasia. 1998;3:315–324. doi: 10.1023/a:1018719612087. [DOI] [PubMed] [Google Scholar]
- 38.Shaper N L, Harduin-Lepers A, Shaper J H. Male germ cell expression of murine β4-galactosyltransferase. A 796-base pair genomic region, containing two cAMP-responsive element (CRE)-like elements, mediates male germ cell-specific expression in transgenic mice. J Biol Chem. 1994;269:25165–25171. [PubMed] [Google Scholar]
- 39.Shaper N L, Hollis G F, Douglas J G, Kirsch I R, Shaper J H. Characterization of the full length cDNA for murine β-1,4-galactosyltransferase. Novel features at the 5′-end predict two translational start sites at two in-frame AUGs. J Biol Chem. 1988;263:10420–10428. [PubMed] [Google Scholar]
- 40.Shaper N L, Meurer J A, Joziasse D H, Chou T D, Smith E J, Schnaar R L, Shaper J H. The chicken genome contains two functional nonallelic β1,4-galactosyltransferase genes. Chromosomal assignment to syntenic regions tracks fate of the two gene lineages in the human genome. J Biol Chem. 1997;272:31389–31399. doi: 10.1074/jbc.272.50.31389. [DOI] [PubMed] [Google Scholar]
- 41.Shaper N L, Wright W W, Shaper J H. Murine β1,4-galactosyltransferase: both the amounts and structure of the mRNA are regulated during spermatogenesis. Proc Natl Acad Sci USA. 1990;87:791–795. doi: 10.1073/pnas.87.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharrocks A D, Brown A L, Ling Y, Yates P R. The ETS-domain transcription factor family. Int J Biochem Cell Biol. 1997;29:1371–1387. doi: 10.1016/s1357-2725(97)00086-1. [DOI] [PubMed] [Google Scholar]
- 42a.Signal Scan. 1991, copyright date. [Online.] National Institutes of Health. http://bimas.dcrt.nih.gov/molbio/signal. [16 June 1999, last date accessed.]
- 43.Sun Z, Sassone-Corsi P, Means A R. Calspermin gene transcription is regulated by two cyclic AMP response elements contained in an alternative promoter in the calmodulin kinase IV gene. Mol Cell Biol. 1995;15:561–571. doi: 10.1128/mcb.15.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura T, Makino Y, Mikoshiba K, Muramatsu M. Demonstration of a testis-specific trans-acting factor Tet-1 in vitro that binds to the promoter of the mouse protamine 1 gene. J Biol Chem. 1992;267:4327–4332. [PubMed] [Google Scholar]
- 45.Thompson C C, Brown T A, McKnight S L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- 46.van der Hoorn F A, Tarnasky H A. Factors involved in regulation of the RT7 promoter in a male germ cell-derived in vitro transcription system. Proc Natl Acad Sci USA. 1992;89:703–707. doi: 10.1073/pnas.89.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.vanWert J M, Panek H R, Wolfe S A, Grimes S R. The TE promoter element of the histone H1t gene is essential for transcription in transgenic mouse primary spermatocytes. Biol Reprod. 1998;59:704–710. doi: 10.1095/biolreprod59.3.704. [DOI] [PubMed] [Google Scholar]
- 48.vanWert J M, Wolfe S A, Grimes S R. Binding of nuclear proteins to a conserved histone H1t promoter element suggests an important role in testis-specific transcription. J Cell Biochem. 1996;60:348–362. doi: 10.1002/(SICI)1097-4644(19960301)60:3%3C348::AID-JCB7%3E3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 49.Wasylyk B, Hahn S L, Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe D, Okabe M, Hamajima N, Morita T, Nishina Y, Nishimune Y. Characterization of the testis-specific gene ’calmegin’ promoter sequence and its activity defined by transgenic mouse experiments. FEBS Lett. 1995;368:509–512. doi: 10.1016/0014-5793(95)00729-s. [DOI] [PubMed] [Google Scholar]
- 51.Wolfe S A, van Wert J M, Grimes S R. Expression of the testis-specific histone H1t gene: evidence for involvement of multiple cis-acting promoter elements. Biochemistry. 1995;34:12461–12469. doi: 10.1021/bi00038a045. [DOI] [PubMed] [Google Scholar]
- 52.Zambrowicz B P, Harendza C J, Zimmermann J W, Brinster R L, Palmiter R D. Analysis of the mouse protamine 1 promoter in transgenic mice. Proc Natl Acad Sci USA. 1993;90:5071–5075. doi: 10.1073/pnas.90.11.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zambrowicz B P, Palmiter R D. Testis-specific and ubiquitous proteins bind to functionally important regions of the mouse protamine-1 promoter. Biol Reprod. 1994;50:65–72. doi: 10.1095/biolreprod50.1.65. [DOI] [PubMed] [Google Scholar]
- 54.Zhou W, Goldberg E. A dual-function palindromic sequence regulates testis-specific transcription of the mouse lactate dehydrogenase c gene in vitro. Biol Reprod. 1996;54:84–90. doi: 10.1095/biolreprod54.1.84. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Y, Sun Z, Means A R, Sassone-Corsi P, Bernstein K E. cAMP-response element modulator tau is a positive regulator of testis angiotensin converting enzyme transcription. Proc Natl Acad Sci USA. 1996;93:12262–12266. doi: 10.1073/pnas.93.22.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]