A new CRISPR screening strategy allows characterizing enhancers for multiple genes associated with converging phenotypes.
Abstract
Current pooled CRISPR screens for cis-regulatory elements (CREs), based on transcriptional output changes, are typically limited to characterizing CREs of only one gene. Here, we describe CRISPRpath, a scalable screening strategy for parallelly characterizing CREs of genes linked to the same biological pathway and converging phenotypes. We demonstrate the ability of CRISPRpath for simultaneously identifying functional enhancers of six genes in the 6-thioguanine–induced DNA mismatch repair pathway using both CRISPR interference (CRISPRi) and CRISPR nuclease (CRISPRn) approaches. Sixty percent of the identified enhancers are known promoters with distinct epigenomic features compared to other active promoters, including increased chromatin accessibility and interactivity. Furthermore, by imposing different levels of selection pressure, CRISPRpath can distinguish enhancers exerting strong impact on gene expression from those exerting weak impact. Our results offer a nuanced view of cis-regulation and demonstrate that CRISPRpath can be leveraged for understanding the complex gene regulatory program beyond transcriptional output at scale.
INTRODUCTION
Cis-regulatory elements (CREs) are key regulators for spatial-temporal control of gene expression. Mutations in CREs can contribute to complex diseases by modulating gene expression over long genomic distances (1–3). Thus, functionally characterizing CREs can provide important insight into gene regulation mechanisms and enable us to better interpret noncoding genetic variants associated with diseases. Despite the fact that tremendous numbers of candidate CREs have been mapped by biochemical signature (4), our knowledge of whether, how, and how much these putative CREs are functional on gene expression remain scarce in the human genome. Pooled CRISPR screens have been developed for testing CREs in their native chromatin context by monitoring the transcriptional levels for the gene of interest (5–11). Although results from these studies have made notable contributions to the annotation of functional DNA elements, challenges remain in pooled CRISPR screens of CREs. First, CRISPR screens for enhancers based on gene expression levels largely depend on generating reporter knock-in cell lines (7) or using FlowFISH signals (8). These procedures, involving generation of reporter lines and selection of cells with positive hits by flow cytometry, are time consuming and difficult to scale up to multiple genes in the same experiment. Second, the approaches of using gene expression as the screening phenotype (9, 10) fail to connect the functions of DNA elements from transcriptional regulation at the molecular level to interpretable cellular and physiological functions. Third, in cases of CRE screens using phenotypes such as cell proliferation and survival (11, 12), they fail to quantify the effect sizes of enhancers on transcriptional output.
To address these limitations, we developed CRISPRpath, a pooled CRISPR screening approach to simultaneously characterize CREs for multiple target genes involved in the same biological pathway. CRISPRpath allows us to screen functional DNA elements based on phenotypes associated with well-defined biological pathways. We demonstrate the capacity of CRISPRpath by performing CRISPR interference (CRISPRi) and nuclease (CRISPRn) screens for six genes in human induced pluripotent stem cells (iPSCs) and reveal different strengths of enhancer functions by imposing varying levels of selection pressure on the cells.
RESULTS
Leveraging CRISPRpath for parallel characterization of CREs for multiple genes in iPSCs
To characterize candidate CREs for multiple genes within the same pooled CRISPR screening, we designed and applied CRISPRpath to six genomic loci containing six genes (HPRT1, MSH2, MSH6, MLH1, PMS2, and PCNA) involved in the 6-thioguanine (6TG)–induced mismatch repair (MMR) (Fig. 1A). The MMR pathway is highly conserved and essential for the maintenance of genome stability (13). The MMR pathway recognizes DNA mismatches caused by 6TG treatment and induces cell apoptosis (14, 15). On the other hand, cells with a malfunctioning MMR pathway, due to aberrant expression levels of 6TG metabolism genes or MMR genes, may survive during 6TG treatment. Using the properties of the MMR pathway, we used cell survival for selecting cells with the reduced expression of MMR genes due to defects in enhancer activities (Fig. 1B). To design the screening library, we first identified open chromatin regions by performing assay for transposase-accessible chromatin using sequencing (ATAC-seq) in WTC11 iPSCs. We included all open chromatin regions defined by ATAC-seq peaks located 1 Mb upstream and 1 Mb downstream of each of the six genes (spanning a total of 10.6-Mb genomic regions) as candidate CREs for functional characterization (fig. S1, A and B, and table S1). We then designed a single-guide RNA (sgRNA) library with 32,383 distal sgRNAs targeting 294 distal ATAC-seq peaks, 2755 proximal sgRNAs targeting 81 ATAC-seq peaks overlapped with transcription start site (TSS) and coding regions of the six genes, and 625 nontargeting sgRNAs with genomic sequences in the same genomic loci but are not followed by PAM (protospacer adjacent motif) sequences (fig. S1C and table S2). In total, we included 35,763 sgRNAs in the library with an average of 110 sgRNAs per ATAC-seq peak (figs. S1D and S2, A and B). We generated a lentiviral library expressing these sgRNAs and transduced this library into two engineered WTC11 iPSC lines, one expressing doxycycline-inducible dCas9-KRAB (CRISPRi) and the other doxycycline-inducible Cas9 (CRISPRn) (16), both at a multiplicity of infection (MOI) of 0.5 (Fig. 1B).
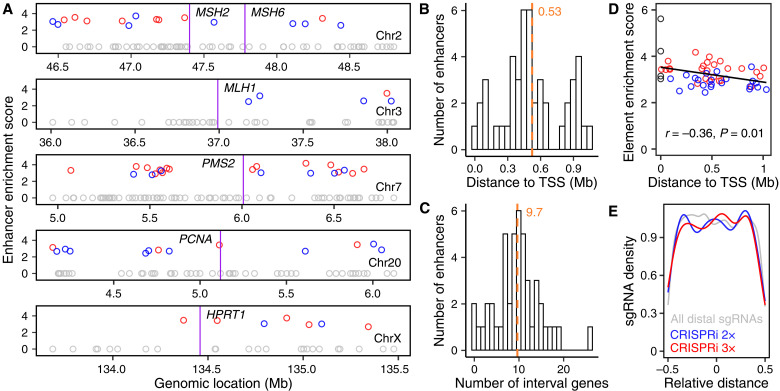
Fig. 1. CRISPRpath for identifying enhancers of multiple genes.
(A) Six genes (HPRT1, MSH2, MSH6, MLH1, PMS2, and PCNA) in the 6TG-induced mismatch repair (MMR) process were used for CRISPRpath screen in this study. (B) Schematic of the CRISPRpath screening strategy with 6TG treatment in iPSCs. Cell survival was used as readout for the screen. (C) Spearman correlation analysis of sgRNA ranking based on fold changes for CRISPRpath screens with different 6TG concentrations (1×, 2×, and 3×). (D) Venn diagram shows the overlapping enriched sgRNAs identified from the screens with 2× and 3× 6TG treatments. (E) Box plots show the fold changes of the enriched distal and proximal sgRNAs from 2× and 3× CRISPRi and CRISPRn screens. Asterisk indicates that no enriched distal sgRNA was identified from 3× CRISPRn screen. Box plots indicate the median, interquartile range (IQR), Q1 − 1.5 × IQR, and Q3 + 1.5 × IQR. (F) Bar plot shows the number of enriched distal and proximal sgRNAs from 2× and 3× CRISPRi and CRISPRn screens. Asterisk indicates that no enriched distal sgRNA was identified from 3× CRISPRn screen. (G) Venn diagram shows the identified enhancers from each CRISPRpath screen.
To carry out the screening, we predetermined the minimal lethal concentration of 6TG at 80 ng/ml for CRISPRi and CRISPRn iPSC lines (see Materials and Methods for details) and applied three different 6TG concentrations (1×: 80 ng/ml, 2×: 160 ng/ml, and 3×: 240 ng/ml) in both CRISPRi and CRISPRn screens. We extracted and sequenced DNA samples from the survival cells 7 days after 6TG treatment to determine enriched sgRNAs by comparing the results to that of the control cells taken after sgRNA library infection before the 6TG treatment (Fig. 1B). To avoid confounding signals generated by off-target effects of low-quality sgRNAs (17), we only used sgRNAs with high specificity [defined as specificity score >0.2 (18) and without any off-target sites with sequence similarity of ≤2 mismatches] for data analysis. This led to the use of a total of 12,702 high-quality sgRNAs with an average of 38 sgRNAs per ATAC-seq peak for analysis (figs. S1, E and F, and S2, C and D). We performed each screen in two biological replicates, with each pair of replicates exhibiting high reproducibility (fig. S2E). We compared the abundance of each sgRNA between the 6TG-treated population and the control population using a negative binomial model and computed the fold change and P value to quantify the effect size and the significance of enrichment of each sgRNA. We used the 5% percentile of the P values from nontargeting control sgRNAs as the empirical significance threshold to achieve a false discovery rate (FDR) of 5%. sgRNAs with P values less than the empirical significance threshold and with fold changes >2 were defined as enriched (fig. S3). As expected, sgRNAs targeting TSS and coding region were identified as positive hits from both CRISPRi and CRISPRn screens, exhibiting greater fold changes in CRISPRn screens compared to the CRISPRi screens (fig. S4A). We also observed enrichment of sgRNA bias toward coding regions over TSS regions for CRISPRn screen (fig. S4B). These results are consistent with CRISPRi functioning best near TSS by inhibiting transcription, and CRISPRn can disrupt gene function by generating indels downstream of TSS (19, 20).
Further sgRNA fold change ranking analysis revealed strong positive correlation between the screens with 2× and 3× 6TG treatment for both CRISPRi and CRISPRn screen (Spearman correlation, CRISPRi = 0.97, CRISPRn = 0.84) (Fig. 1C), with the correlations for proximal sgRNAs being higher than for distal sgRNAs (fig. S4C). On the contrary, results from the 1× screen correlated poorly with either 2× or 3× screens (Fig. 1C), suggesting that more substantial selection pressure (2× and 3×) can reduce background noise in CRISPRpath screens. Thus, we used sgRNAs enriched from 2× and 3× screens data for identifying active enhancers in the following section (Fig. 1, D and E).
CRISPRi is more efficient than CRISPRn in pooled CRISPR screens of CREs
Performing CRISPRpath with CRISPRi and CRISPRn in the same genetic background with an identical sgRNA library offers a unique opportunity for comparing the efficacies of CRISPRi and CRISPRn in pooled CRISPR screens of CREs. We noticed that CRISPRn screens recovered fewer enriched distal sgRNAs than CRISPRi screens (Fig. 1F). This is possibly due to the fact that CRISPRi-mediated heterochromatin formation can more effectively perturb CREs compared to CRISPRn-mediated genetic perturbations. We then called a candidate element as an enhancer if there are at least three enriched sgRNAs in that CRE. On the basis of this criterion, we identified 62 and 33 enhancers from the 2× and 3× CRISPRi screen, respectively, and 19 enhancers from the 2× CRISPRn screen (Fig. 1G and table S3). However, no enhancer was identified from the 3× CRISPRn screen, indicating that either the CRISPRn-induced mutations did not lead to any strong effect on gene expression to make the cells survive the 3× 6TG treatment or there are insufficient numbers of sgRNAs exhibiting deleterious effects on the tested DNA elements to satisfy our criterion of calling functional enhancers. In total, 66 unique enhancers were identified for the six target genes with CRISPRpath under different 6TG treatments (Fig. 1G). Together, we demonstrate that CRISPRpath can simultaneously identify enhancers for multiple target genes, with CRISPRi outperforming CRISPRn. For the following analysis, we focused on the 63 enhancers identified from the 2× and 3× CRISPRi screens (Fig. 1G).
Genomic feature of CRISPRpath identified enhancers
To determine the genomic feature of the enhancers, we plotted all the tested elements by their genomic locations and enrichment scores [average of log2(fold change) of enriched sgRNAs of each element] (Fig. 2A). Not surprisingly, our data suggest that each gene can be regulated by multiple enhancers, with the identified functional enhancers having no position bias relative to the TSS. The average distance between an enhancer and its paired TSS is about 530 kb (Fig. 2B), with an average of 10 interval genes between an identified enhancer and its target gene pairs (Fig. 2C). We observed a weak negative correlation between the enhancer enrichment score and the distance between an enhancer and its paired TSS (Pearson correlation, ρ = −0.36, P = 0.01; Fig. 2D), suggesting that enhancers near to TSS tend to have higher regulatory activity compared to enhancers further away from their target genes. Note that the relative positions for the enriched sgRNAs exhibited no preference relative to ATAC-seq peaks (Fig. 2E and fig. S5A) and no preference for the strand on which the sgRNAs were designed (fig. S5B), consistent with our knowledge that CRISPRi-mediated heterochromatin spreads over hundreds of base pairs in distance (21).
Fig. 2. Genomic features of identified enhancers from CRISPRpath using CRISPRi.
(A) Genomic locations of identified enhancers relative to TSS. Circles indicate enhancers identified from the CRISPRi 3× screen (red), enhancers uniquely identified from the CRISPRi 2× screen (blue), and tested CREs that are not identified as enhancers (gray). Purple lines label the location of each target gene. (B) Histogram shows the distance distribution between identified enhancers and their paired TSS. (C) Histogram shows the number of interval genes between enhancers and their target gene TSS. Mean is indicated with an orange dashed line in (B) and (C). (D) A weak negative correlation is observed between enrichment score and genomic distance between enhancers and their target genes (Pearson correlation, r = −0.36, P = 0.01). Black circles indicate TSS regions. The red and blue circles are enhancers shown in (A). Only the enhancers for HPRT1, MLH1, PMS2, and PCNA are included in (B) to (D). (E) Density plot shows no significant difference (two-tailed two-sample Kolmogorov-Smirnov test) for the distribution of all distal sgRNAs (gray) and enriched distal sgRNAs from 2× (blue) and 3× (red) CRISPRi screens.
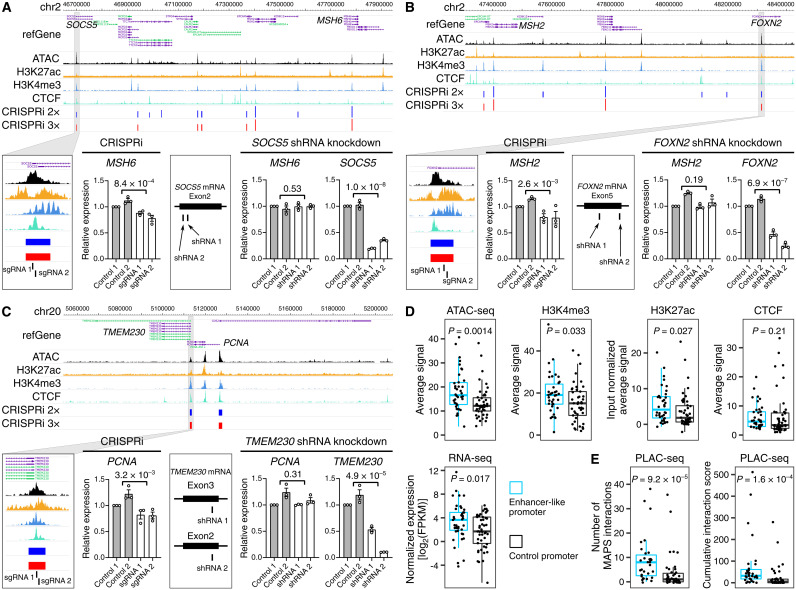
Previous studies have revealed that promoters can function as enhancers (7, 22). Sixty percent (38 of 63) of the functional enhancers identified in CRISPRi screens overlapped with annotated promoters, providing an excellent opportunity to further explore the genomic features of these enhancer-like promoters. To validate whether these promoters function as bona fide enhancers, we targeted three enhancer-like promoters with CRISPRi. We confirmed significant down-regulation of their target genes including MSH6, MSH2, and PCNA (Fig. 3, A to C, and fig. S6A). In contrast, short hairpin RNAs (shRNAs) against the transcripts from these promoters (SOCS5, FOXN2, and TMEM230) only led to a significant downregulation of its own transcripts and did not affect their target gene expression (Fig. 3, A to C). These results suggest that these promoter sequences identified by CRISPRpath can function as enhancers. It is possible that other mechanisms play a role at a subset of these promoters, such as promoter pairs in the same transcriptional factories because we observed subtle but significant decreases in SOCS5 and TMEM230 expression upon perturbation at MSH6 and MSH2 promoters (fig. S6B) (23).
Fig. 3. Enhancer-like promoters act as functional enhancers.
(A to C) Three examples of promoters that function as enhancers. CRISPRi silencing of the promoter region of SOCS5, FOXN2, and TMEM230 results in significant downregulation of MSH6, MSH2, and PCNA, respectively. shRNA knockdown of SOCS5, FOXN2, and TMEM230 can only down-regulate SOCS5, FOXN2, and TMEM230 expression. Three independent replicates per condition and two independent sgRNAs or shRNAs per replicate were used for each experiment. P values are from two-tailed two-sample t test. (D) Average signal enrichment of ATAC-seq, gene transcription, H3K4me3, H3K27ac, and CTCF binding for enhancer-like promoters (n = 38) and control promoters (n = 47). P values are from Wilcoxon test. Box plots indicate the median, IQR, Q1 − 1.5 × IQR, and Q3 + 1.5 × IQR. (E) Number of H3K4me3-mediated chromatin interactions and cumulative interaction score for enhancer-like promoters (n = 31) and control promoters (n = 43). Box plots indicate median, IQR, Q1 − 1.5 × IQR, and Q3 + 1.5 × IQR. P values are calculated from Wilcoxon test.
Although it has been shown that enhancer-like promoters are enriched with active chromatin marks and physically close to target genes (7), it is not clear whether enhancer-like promoters have unique genome features that can differentiate them from other regular active promoters. To this end, we compared chromatin accessibility; occupancy of histone 3 lysine 4 trimethylation (H3K4me3), histone 3 lysine 27 acetylation (H3K27ac), and CTCF (CCCTC-binding factor); transcription; and chromatin interactivity levels between enhancer-like promoters and all other active promoters that did not show enhancer activity in our CRISPRi screens. We show that enhancer-like promoters exhibit higher chromatin accessibility, higher level of transcription, and stronger H3K4me3 and H3K27ac signals than those at other active promoters (Fig. 3D). On the other hand, we did not observe a significant difference for CTCF binding signals between enhancer-like promoters and control promoters (Fig. 3D). Furthermore, by evaluating chromatin interaction data using H3K4me3 proximity ligation-assisted chromatin immunoprecipitation sequencing (ChIP-seq) (PLAC-seq), we show that enhancer-like promoters have significantly more and stronger interactions compared to control promoters (Fig. 3E).
CRISPRpath is capable of distinguishing enhancers with distinct effect sizes
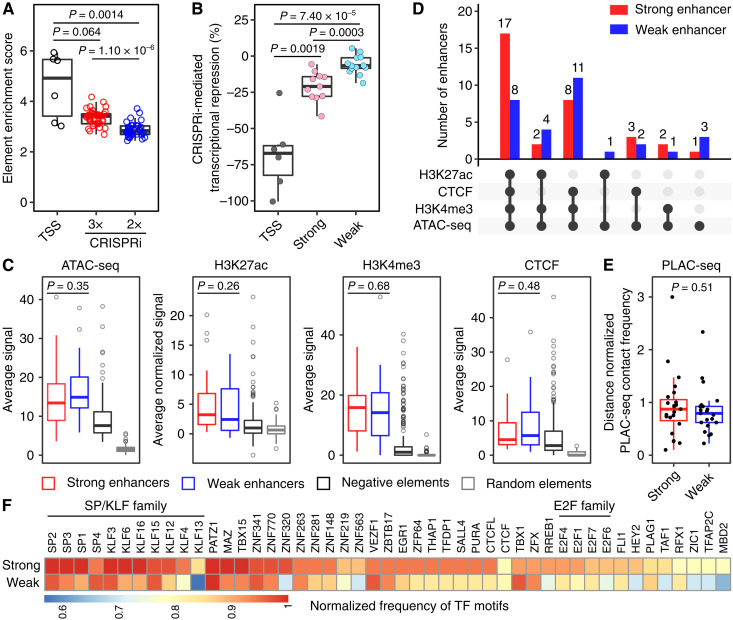
Gene expression is often a result of combinatorial regulatory effects from multiple CREs (11, 24). Understanding how individual enhancers contribute to gene expression in a quantitative manner is an important first step in dissecting how enhancers orchestrate precise transcriptional control. We seek a new strategy to differentiate enhancers based on their effect sizes on gene expression using CRISPRpath. We hypothesized that cells with marked down-regulation of MMR genes have a fitness advantage under higher 6TG concentration than cells with modest down-regulation of MMR genes. Consistent with this hypothesis, proximal sgRNAs exhibit larger fold changes than distal sgRNAs (Fig. 1E) because perturbing proximal regions has more profound effects on gene down-regulation than perturbing distal regulatory regions. On the basis of these observations, we hypothesize that enhancers identified under different selection pressure represent distinct regulatory strengths on transcriptional activation. We noticed that the enriched sgRNAs for enhancers identified under strong selection pressure (3×) have bigger fold changes compared to those for enhancers uniquely identified under weak selection pressure (2×) (fig. S7A). Similarly, enrichment scores for these two groups of enhancers are significantly different, with the TSS regions manifesting the highest enrichment scores (Fig. 4A). sgRNA fold changes (fig. S7B) and element enrichment scores (fig. S7C) both have no significant differences between distal enhancers and enhancer-like promoters (fig. S7, B and C). Therefore, enhancers identified in the 3× screen are strong enhancers (n = 33), while enhancers uniquely identified in the 2× screen are weak enhancers (n = 30) (Fig. 1G).
Fig. 4. CRISPRpath can distinguish weak and strong enhancers by imposing different selection pressures.
(A) Box plots show the enrichment score of the tested elements. TSS regions (black circles) show highest enrichment scores. Enhancers uniquely identified from the lower selection pressure (CRISPRi 2×, blue circles) exhibit lower enrichment scores compared to the enhancers identified from the higher selection pressure (CRISPRi 3×, red circles). Each circle represents an individual element. P values are from Wilcoxon test. (B) Box plots show that the CRISPRi perturbation at enhancers induced various degrees of transcriptional repression of target genes measured with RT-qPCR. Each dot represents the average value from three biological replicates. CRISPRi targeting TSS regions (dark gray) achieved the highest transcriptional repression. CRISPRi targeting strong enhancers (pink) leads to a more substantial transcription silencing of the target gene compared to CRISPRi targeting weak enhancers (cyan). P values are from Wilcoxon test. (C) Enrichment analysis of ATAC-seq, H3K27ac, H3K4me3, and CTCF binding signals for strong (n = 33) and weak (n = 30) enhancers. P values for the difference between strong and weak enhancers are from Wilcoxon test; see table S7 for P values of all pairwise comparisons. Box plots (A to C) indicate the median, IQR, Q1 − 1.5 × IQR, and Q3 + 1.5 × IQR. (D) Intersection of genomic features for weak enhancers (blue bar) and strong enhancers (red bar). (E) Distance normalized H3K4me3 PLAC-seq contact frequency for strong (n = 23) and weak (n = 21) enhancers. Only the enhancers for HPRT1, MLH1, PMS2, and PCNA are included (see Materials and Methods for details). Box plots indicate the median, IQR, Q1 − 1.5 × IQR, and Q3 + 1.5 × IQR. P value is from Wilcoxon test. (F) Heatmap shows the normalized frequency of transcription factor (TF) motifs found in strong and weak enhancers.
To confirm the quantitative effect of enhancers on target gene expression, we tested 11 strong and 10 weak enhancers using CRISPRi followed by quantitative reverse transcription polymerase chain reaction (RT-qPCR) measurement of the corresponding target gene expression (Fig. 4B and fig. S8A). We show that perturbations of strong enhancers led to significantly more down-regulation of target gene expression (mean down-regulation of target gene by 21%) than perturbations of weak enhancers (mean down-regulation of target gene by 6%), with the perturbations of TSS regions achieving the strongest down-regulation of target genes, by an average of 68% reduction in gene expression (Fig. 4B and fig. S8, A and B). These quantitative effects on target gene expression are consistent with the enrichment scores from our CRISPRpath screens (fig. S8C) and demonstrate the capacity of distinguishing enhancers with different effect sizes by imposing different levels of selection pressures. These CRISPRi-validated enhancers also demonstrated enhancer activities in luciferase reporter assays with only one exception (fig. S9A). Luciferase activities did not show the same level of strong anti-correlation with the levels of CRISPRi-mediated transcriptional repression compared to element enrichment scores (figs. S8C and S9, B and C) and cannot distinguish weak and strong enhancers (fig. S9D), possibly because it tests enhancers out of their native genomic context with a heterogeneous promoter.
We further explored chromatin features of strong and weak enhancers by analyzing chromatin accessibility, H3K4me3, H3K27ac, and CTCF binding signals in these regions. At individual chromatin mark level, while CRISPRpath-identified enhancers were more accessible and enriched with active chromatin marks, such as H3K4me3 and H3K27ac, and CTCF binding compared to negative elements or random elements (Fig. 4C), we did not observe significant differences between strong and weak enhancers in the chromatin features that we individually examined. However, strong enhancers tend to have more active chromatin signatures than weak enhancers (Fig. 4D), suggesting that combined signatures of active chromatin can be a better indicator of enhancer strength. Strong enhancers tend to have higher distance normalized PLAC-seq contact frequencies with their target promoters than weak enhancers, although not statistically significant, possibly due to the small sample size in this study (Fig. 4E). We obtained similar results by expanding this analysis for characterized enhancers in K562 cells and mouse embryonic stem cells (mESCs) (fig. S10) (6, 8), which reinforces the idea that enhancers with larger effects on gene expression tend to have higher chromatin interactions with their cognate promoters. To explore the possible mechanisms that drive enhancer activities in a quantitative manner, we evaluated potential transcription factor (TF) binding motifs in strong and weak enhancer sequences. Both strong and weak enhancers are enriched with CTCF binding motif (Fig. 4F). Most of strong and weak enhancers are bound by CTCF (Fig. 4D), consistent with the notion that CTCF-mediated chromatin loops are essential for gene activation (25). Furthermore, strong enhancers and weak enhancers have differential enrichment with TF binding motifs. For example, the binding motifs for SP/KLF family (26) and E2F family (27, 28) appear more frequently in strong enhancers compared to weak enhancers, suggesting that these strong enhancers could be major docking sites for master regulators in iPSCs (Fig. 4F).
DISCUSSION
CRISPR-mediated high-throughput screening using bulk cells allows the functional characterization of regulatory elements in their native genomic context. However, current approaches are limited to validating a small number of regulatory elements for a single gene (5, 7, 9, 12, 29, 30). To overcome this bottleneck, we developed CRISPRpath, a strategy for functional characterization of enhancers for multiple genes simultaneously by leveraging the genes involved in the same biological pathway so that the effects can be measured via a define phenotype. For example, α-toxin resistance phenotype can be used to identify CREs for 17 genes in glycosylphosphatidylinositol (GPI) anchor synthesis pathway (31). CRISPRpath can also be leveraged to identify CREs for protein folding regulators that contribute to the endoplasmic reticulum stress response pathway (32) using UPRE (unfolded protein response element) reporter in mammalian cells. Because CRISPR screen technology is widely used, the CRISPRpath strategy is readily applicable to simultaneously identifying enhancers for genes converging in the defined biological processes and pathways across different cell types. Compared to the existing pooled CRISPR screens of CREs (5, 7, 8, 10–12, 29, 30, 33), CRISPRpath is scalable with additional benefits of connecting DNA elements to cellular function, beyond the most standard molecular phenotype of gene expression.
Multiple factors can contribute to the observed low effects on gene expression upon enhancer perturbation in this study. First, enhancers tend to have big impacts on genes with cell type–specific expression patterns. For example, perturbing HBG1/2 enhancers at the locus control region of the human β-globin locus leads to the marked reduction of HBG1/2 expression in K562 cells (34). On the other hand, enhancer disruptions for broadly expressed genes such as GATA1, HDAC6, and MYC genes only resulted in 9 to 62% expression reduction in K562 cells (11), comparable to our results on perturbing MMR gene enhancers. Second, on the technical level, it has been demonstrated that CRISPRi cannot completely silence enhancers compared to the CRISPR deletion approach (6).
Promoters can function as enhancers more widespread than expected, with more than half of the enhancers identified for MMR genes in our study being previously annotated promoters. This is consistent with previous reports that enhancer-like promoters are more prevalent for ubiquitously expressed genes (35). Enhancer-like promoters are more accessible compared to other promoters, possibly because these regions are required to be more open to accommodate additional transcriptional machinery such as TF for activating target gene expression besides their own transcription (36). Enhancer-like promoters also exhibit significantly higher levels of chromatin interactions with distal regions compared to other active promoters. This observation can be explained by the fact that enhancer-like promoters will form chromatin loops not only with their distal target genes but also with CREs for controlling the expression of their own genes.
Genomic studies of chromatin marks have revealed hundreds of thousands of candidate CREs in the human genome but with very little quantitative information regarding how CREs contribute to gene regulation (37, 38). Using CRISPRpath, we can systematically classify enhancers based on their effect sizes on transcription. Identifying and characterizing the effect size for each individual enhancer is the critical first step to future studies of their combinatory effects on target gene expression. Strong and weak enhancers cannot be distinguished by individual epigenetic marks that we examined. One possible explanation for this observation is that chromatin features only mark enhancer’s identity but do not quantify enhancer activity. On the other hand, the strong and weak enhancers that we identified may regulate other genes differently from regulating the MMR gene. Strong enhancers tend to harbor more than one active chromatin signature, which indicates that enhancer activities are regulated by multiple epigenetic factors, for example, TF-mediated transcriptional regulation. Differential TF binding motifs observed within strong and weak enhancers suggest that enhancer strength is modulated by TF binding. Future studies that further integrate TF binding datasets with functional data of enhancers will shed light on the molecular mechanisms that drive enhancers’ effect sizes on gene regulation.
MATERIALS AND METHODS
Cell culture
Doxycycline-inducible CRISPRi and CRISPRn WTC11 iPSC lines were purchased from Gladstone Stem Cell Core. Both CRISPRi and CRISPRn WTC11 iPSCs were cultured on Matrigel-coated (Corning, 354277) plates with Essential 8 Medium (Life Technologies, A1517001). iPSCs were passaged using Accutase (STEMCELL Technologies, 07922) and 10 μM ROCK inhibitor Y-27632 (STEMCELL Technologies, 72302). Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, 11995065) with 10% fetal bovine serum (CPS Serum, FBS-500). HEK293T cells were passaged with trypsin-EDTA (Gibco, 25200072). All the cells were grown with 5% CO2 at 37°C and verified mycoplasma free using the MycoAlert Mycoplasma Detection Kit (Lonza, LT07-218).
sgRNA library design
CRISPRpath sgRNA library was designed to screen CREs for HPRT1, MSH2, MSH6, MLH1, PMS2, and PCNA. ATAC-seq peaks within the region of 1 Mb upstream and 1 Mb downstream of each target gene including TSS and coding regions were selected as targeting regions for the sgRNA library design (table S1). We generated a genome-wide sgRNA database containing all the available unique sgRNAs, each followed by an “NGG” PAM sequence. All the designed unique sgRNAs in the target regions were added in the sgRNA library, excluding sgRNAs containing AATAAA, AAAAA, TTTTT, or TTTTTT sequences. Unique 20–base pair (bp) sequences in the target regions that were not followed by the NGG or “NAG” PAM sequences were taken as nontargeting control sgRNAs, excluding nontargeting sgRNAs containing TTT, TTNTT, AATAAA, AAAAA, TTTTT, or TTTTTT sequences. Then, a guanine nucleotide was added to all the sgRNAs if the sequence did not start with G to increase efficiency of transcription from U6 promoter. Final sgRNA oligos adhered to the following template: 5′-ATATCTTGTGGAAAGGACGAAACACC-[20- or 21-bp sgRNA sequence]-GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC-3′. In total, 35,763 sgRNAs were included in the library (fig. S1 and table S2). We retrieved specificity score and off-target site for each sgRNA from GuideScan (18) and assigned the specificity score of sgRNAs not existing in the GuideScan database to 0. The high-quality sgRNAs were filtered with specificity score >0.2 and without perfectly matched or one to two mismatched off-target sites.
Oligo synthesis and library cloning
sgRNA library oligos were synthesized by Twist Bioscience and amplified with the forward primer 5′-TCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACAC-3′ and the reverse primer 5′-AACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′. We replaced the Cas9 sequence in lentiCRISPR v2 plasmid (Addgene, 52961) with blasticidin S deaminase sequence to construct the lentiCRISPR-v2-Blast-Puro plasmid (Addgene, 167186). The PCR products were purified via gel excision and column purification (Promega, A9282) and then inserted into the Bsm BI–digested lentiCRISPR-v2-Blast-Puro vector by Gibson assembly [New England Biolabs (NEB), E2621L]. The assembled products were transformed into NEB 5-α electrocompetent Escherichia coli cells (NEB, C2989K) by electroporation. About 40 million independent bacterial colonies were cultured, and sgRNA library plasmids were extracted with the Qiagen EndoFree Plasmid Mega Kit (Qiagen, 12381). The recovery rate and distribution of the sgRNA library were checked with next-generation sequencing (fig. S2, A to D).
Lentivirus production and titration
To make the lentiviral library, 5 μg of sgRNA plasmid library was cotransfected with 3 μg of psPAX (Addgene, 12260) and 1 μg of pMD2.G (Addgene, 12259) lentivirus packaging plasmids into 8 million HEK293T cells in a 10-cm dish with PolyJet (SignaGen Laboratories, SL100688). For each individual sgRNA, 3.75 μg of sgRNA plasmid was cotransfected with 2.25 μg of psPAX (Addgene, 12260) and 0.75 μg of pMD2.G (Addgene, 12259) plasmids into 4 million HEK293T cells in a T25 flask with PolyJet (SignaGen Laboratories, SL100688). The medium was replaced 12 hours after transfection and harvested every 24 hours for a total of three harvests. Harvested media containing the desired virus were filtered through Millex-HV 0.45-μm polyvinylidene difluoride filters (Millipore, SLHV033RS) and further concentrated with 100,000 NMWL (nominal molecular weight limit) Ultra-15 centrifugal filter units (Amicon, UFC910008).
The titer of lentivirus was determined by transducing 500,000 cells with varying amounts (0, 0.5, 1.0, 2.0, 4.0, and 8.0 μl) of concentrated virus and polybrene (8 μg/ml; Millipore, TR-1003-G). Viral transduction was performed by centrifuging the lentivirus and cell combination at 1000 relative centrifugal force (RCF) for 90 min at 37°C. Three to 4 hours later, virus-containing medium was replaced with fresh medium. Twenty-four hours after the transduction, transduced cells were dissociated with Accutase and seeded as duplicates. One replicate was treated with blasticidin (4 μg/ml; Gibco, A1113903), and the other replicate was not treated with blasticidin. Four days later, the blasticidin-resistant cells and control cells were counted to calculate the ratio of infected cells and the viral titer.
Determining 6TG concentration via killing curve titration
Both CRISPRi and CRISPRn WTC11 iPSCs were used to determine the minimal lethal concentration of 6TG. Cells were seeded in 24-well plates. When the cells reached around 50% confluence (day 0), they were treated with 6TG concentrations of 0 (control), 20, 40, 60, 80, 100, 120, 140, and 160 ng/ml. Two wells were allocated for each condition. The cells were examined daily and cultured for 7 days. The medium was replaced daily with the specified 6TG concentration. After 3 days, wells with 6TG concentration greater than or equal to 100 ng/ml had no surviving cells. On day 4 of treatment, the wells with 6TG treatment of 80 ng/ml had no surviving cells. On the last day of treatment, the wells with treatments of 40 and 60 ng/ml had very few surviving cells, while the treatment of 20 ng/ml had many surviving cells. On the basis of these results, we set 80 ng/ml as the minimal lethal concentration for 6TG.
CRISPRpath screening and sequencing library preparation
CRISPRpath screens were carried out with 72 million doxycycline-inducible CRISPRi or CRISPRn iPSCs in biological replicate. The cells for lentiviral transduction were seeded into six-well plates with 1 million cells per well, and the lentiviral library (MOI = 0.5) was transduced into the iPSCs with polybrene (8 μg/ml) (Millipore, TR-1003-G) and spun at 1000 RCF at 37°C for 90 min. The transduced cells were treated with doxycycline (2 μM; Sigma-Aldrich, D9891) and blasticidin (4 μg/ml; Gibco, A1113903) for 4 days. After this doxycycline and blasticidin treatment, 10 million cells were reserved as a control population, and 100 million cells were used for CRISPRpath screen with doxycycline and 6TG (Sigma-Aldrich, A4660) treatment for 7 days. Last, survival cells were collected from the 6TG-treated population.
The genomic DNA was extracted from each sample via cell lysis and digestion [100 mM tris-HCl (pH 8.5), 5 mM EDTA, 200 mM NaCl, 0.2% SDS, and proteinase K (100 μg/ml)], phenol:chloroform (Thermo Fisher Scientific, 17908) extraction, and isopropanol (Fisher Scientific, BP2618500) precipitation. To amplify the sgRNA sequences from each sample, thirty-two 50-μl PCRs were performed using 500 ng of genomic DNA for each reaction and NEBNext High-Fidelity 2× PCR Master Mix (NEB, M0541S). The purified libraries were sequenced on NovaSeq 6000 with 150-bp paired-end sequencing. The detailed protocol is available at the ENCODE portal (https://www.encodeproject.org/documents/2e6451a9-3b98-4d95-922e-a3d8d2100ddf/).
CRISPRpath data analysis
The sequence files were down-sampled to the same amount of total reads and then mapped to the sgRNA library with the requirement of exact match of designed sgRNA sequences in the following pattern: 5′-CCG-[N19 or N20]-GTT-3′. Only the highly specific sgRNAs (specificity score >0.2, without perfectly matched or one to two mismatched off-target sites) were used for downstream data analysis. The sgRNA enrichment for each screen was calculated by comparing 6TG-treated samples with the associated control samples with edgeR and TMM (trimmed mean of M values) normalization. We first used edgeR (39) to calculate the P value based on negative binomial model for both targeting sgRNAs and nontargeting control sgRNAs. To achieve empirical FDR less than 5%, we then selected a P value cutoff corresponding to the 5% percentile of P values from nontargeting control sgRNAs. Last, we defined enriched sgRNAs with a P value less than the selected P value cutoff and a fold change >2. The ATAC-seq peaks were identified as functional enhancers for the six MMR genes by having at least three significantly enriched sgRNAs. Analysis scripts are available at https://github.com/MichaelMW/crispy.
Analysis of genomic feature and chromatin signature of identified enhancers
Genomic distances between enhancer and TSS pairs were calculated on the basis of the distance from the center of enhancers to the TSSs of the target genes. The number of interval genes is the number of all the RefSeq annotated genes between each enhancer and paired target gene. The signal of chromatin signatures, including ATAC-seq, H3K27ac, H3K4me3, CTCF binding, and RNA-seq, was calculated by deeptools (v3.4.3) (40). The enhancer-like promoters are the enhancers that overlap with the region 500 bp upstream and downstream of a RefSeq annotated TSS.
Validation of identified enhancers using CRISPRi
We cloned lentiCRISPR-v2-HygR-EGFP (Addgene, 167188) and lentiCRISPR-v2-HygR-mCherry (Addgene, 167189) vectors by replacing the Cas9 and puromycin N-acetyltransferase sequences in lentiCRISPR v2 plasmid (Addgene, 52961) with hygromycin B phosphotransferase and enhanced green fluorescent protein (EGFP) or mCherry sequences. To validate the identified enhancers, individual sgRNAs targeting identified enhancers were cloned into the lentiCRISPR-v2-HygR-GFP or lentiCRISPR-v2-HygR-mCherry vector. The doxycycline-inducible CRISPRi WTC11 iPSCs were infected with the lentivirus expressing sgRNAs for three replicates per sgRNA. The sgRNA-infected cells were grown with hygromycin-containing (150 μg/ml; Gibco, 10687010) and doxycycline-containing (2 μM; Sigma-Aldrich, D9891) medium. Seven days later, the cells were collected and total RNA was extracted from the cells using the Qiagen RNeasy Plus Kit (Qiagen, 74134). One microgram of RNA was then used to synthesize complementary DNA (cDNA) using the iScript cDNA Synthesis Kit (Bio-Rad, 1708840). RT-qPCRs for targeted genes were performed with the Luminaris HiGreen qPCR Master Mix (Thermo Fisher Scientific, K0993) on the Roche LightCycler 96 System. The RT-qPCR primers are listed in table S4, and the sgRNA sequences are listed in table S6. For each tested element in Fig. 3 (A to C) and fig. S8B, we performed CRISPRi experiments with two independent sgRNAs and used the results from the sgRNA with stronger transcriptional repression in Fig. 4B.
shRNA-mediated RNA interference
shRNAs were designed by using DSIR tool (http://biodev.extra.cea.fr/DSIR/DSIR.html) targeting SOCS5, FOXN2, and TMEM230. The sequences of shRNAs are listed in table S5. The shRNAs were cloned into lentiCRISPR-v2-HygR-mCherry vector under the control of human U6 promoter and packaged into lentivirus for cell transduction. The WTC11 iPSCs transduced with shRNA lentivirus were treated with hygromycin (150 μg/ml; Gibco, 10687010) for 7 days and then collected for RNA extraction and RT-qPCR.
ATAC sequencing
ATAC-seq was carried out using the Nextera DNA Library Prep Kit (Illumina, FC-121-1030) as previously described (41). The detailed protocol is available on the ENCODE portal (www.encodeproject.org/documents/0317894c-5a42-4f03-b865-c2a2d08708ef/). Briefly, each library started with 100,000 fresh iPSCs, and the cells were incubated with ice-cold nuclei extraction buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 0.1% Igepal CA630, and 1× protease inhibitor] for 5 min on ice and then centrifuged at 500 RCF for 5 min. A total of 50,000 resulting nuclei were treated with tagmentation buffer (25 μl of buffer TD with 50,000 nuclei, 22.5 μl of water, and 2.5 μl of TDE1) for 30 min at 37°C. The transposed DNA was purified using the MinElute PCR Purification Kit (Qiagen, 28006), amplified using Nextera primers, and then size-selected for fragments between 150 and 1000 bp using SPRISelect beads (Beckman Coulter, B23319). Libraries were sent for single-end sequencing on HiSeq 4000 (50-bp single-end reads). Reads were mapped to GRCh38/hg38 and processed using the ENCODE pipeline (https://github.com/kundajelab/atac_dnase_pipelines, V1.8.0), which ran on the default settings. The ATAC-seq peaks were filtered with an FDR cutoff of 0.1%, and adjacent peaks were merged if they are less than 1 kb apart.
RNA sequencing
RNA was extracted from fresh cells using the RNeasy Plus Mini Kit (Qiagen, 74134). Approximately 1000 ng of extracted RNA was used to prepare libraries for sequencing using the TruSeq Stranded mRNA Library Prep Kit (Illumina, 20020594). Libraries were sent for paired-end sequencing on NovaSeq 6000 (100-bp paired-end reads). Reads were aligned to GRCh38/hg38 using STAR 2.7.0f (42) with the standard ENCODE settings, and transcript quantification was performed in a strand-specific manner using RSEM 1.3.1 (43) with the annotation from GENCODE v32. Only the first read was used, and all reads were trimmed to 51 bp using Trim Galore 0.4.5 running the following options: -q 20 --length 20 -- stringency 3 --trim-n. The edgeR package in R (3.20.9) (39) was used to calculate TMM-normalized FPKM (fragments per kilobase of transcript per million mapped reads) values for each gene based on the expected counts and gene lengths for each library. The mean gene expression across all replicates was used for analysis.
ChIP sequencing
ChIP-seq libraries were constructed from 2 million WTC11 iPSCs. Cells were cross-linked in 1% formaldehyde at room temperature for 20 min and then quenched with 2.5 M glycine at room temperature for 5 min. Fixed cells were lysed and chromatin was sonicated using Covaris S220 focused-ultrasonicator with the following parameters: duty factor, 2%; peak incident power, 105 W; cycles per burst, 200, for 30 min. Input chromatin was removed and stored at −20°C for later processing. Magnetic beads (Invitrogen, Dynabeads Protein A, 10001D) were preincubated with H3K27ac antibody (Active Motif, 39133, lot 22618011) for 2 hours at 4°C before being added to sheared chromatin. Samples were incubated overnight at 4°C. Beads were washed three times, and chromatin was then eluted. Samples were incubated at 65°C overnight to reverse the crosslinking. DNA was treated with ribonuclease A for 1 hour at 37°C and proteinase K (NEB, 8107) for 1 hour at 55°C. DNA was purified by phenol-chloroform extraction and ethanol precipitation. Libraries were prepared using TruSeq adapters and size-selected using SPRIselect beads before amplification and paired-end sequencing. Libraries were sent for paired-end sequencing on NovaSeq 6000 (150-bp paired-end reads). Sequencing reads were trimmed to 50 bp and mapped to GRCh38/hg38 using bowtie2 with the following options: --local --very-sensitive-local --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700. Picard Tools was used to remove blacklisted regions and duplicate reads, and MACS2 was used to call peaks on merged replicates at an FDR cutoff of 1%.
CUT&Tag
CUT&Tag libraries were constructed from 150,000 WTC11 iPSCs according to previously described methods (44). Cells were lysed in nuclei extraction buffer [20 mM Hepes-KOH (pH 7.9), 10 mM MgCl2, 0.1% Triton X-100, 20% glycerol, and 1× protease inhibitor] on ice for 10 min. The samples were spun and resuspended in 100 μl of nuclei extraction buffer. Meanwhile, 10 μl of BioMag Plus Concanavalin A (Bangs Laboratories, BP531) was equilibrated in binding buffer (1× phosphate-buffered saline, 1 mM CaCl2, 1 mM MgCl2, and 1 mM MnCl2). The equilibrated beads were added to the samples and incubated with rotation for 15 min at 4°C. Nuclei-bound beads were washed with buffer 1 [20 mM Hepes-KOH (pH 7.9), 150 mM NaCl, 2 mM EDTA, 0.5 mM spermidine, 0.1% bovine serum albumin (BSA), and 1× protease inhibitor] and buffer 2 [20 mM Hepes-KOH (pH 7.9), 150 mM NaCl, 0.5 mM spermidine, 0.1% BSA, and 1× protease inhibitor]. After washing, nuclei-bound beads were resuspended in 50 μl of buffer 2 with 0.5 μl of antibody (H3K4me3 from Millipore, 04-745, lot 3543820 and CTCF from Millipore, 07-729, lot 3059608) and incubated with rotation overnight at 4°C. Samples were washed twice with buffer 2 and resuspended in 50 μl of buffer 2 with antibody (antibodies-online Inc., guinea pig anti-rabbit IgG, ABIN101961, lot 42323) and incubated for 1 hour at room temperature with rotation. Samples were washed again with buffer 2 and resuspended in 100 μl of buffer 3 [20 mM Hepes-KOH (pH 7.9), 300 mM NaCl, 0.5 mM spermidine, 0.1% BSA, and 1× proteinase inhibitor] containing 0.04 μM pA-Tn5. Samples were incubated for 1 hour at room temperature, washed three times with buffer 3, and resuspended in tagmentation buffer [20 mM Hepes-KOH (pH 7.9), 300 mM NaCl, 0.5 mM spermidine, 10 mM MgCl2, 0.1% BSA, and 1× proteinase inhibitor]. Samples were incubated for 1 hour at 37°C. Samples were treated with proteinase K (NEB, 8107) for 1 hour at 50°C. DNA was purified by phenol-chloroform extraction and ethanol precipitation. Libraries were prepared using TruSeq adapters and size-selected using SPRIselect beads before amplification and paired-end sequencing. Libraries were sent for paired-end sequencing on MiniSeq (37-bp paired-end reads, H3K4me3 libraries) or NovaSeq 6000 (150-bp paired-end reads, CTCF libraries). Sequencing reads (CTCF libraries were trimmed to 50 bp) were mapped to GRCh38/hg38 using bowtie2 with the following options: --local --very-sensitive-local --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700. Picard Tools was used to remove blacklisted regions and duplicate reads, and SEACR (45) was used to call peaks on merged replicates.
H3K4me3 PLAC-seq
H3K4me3 PLAC-seq data in WTC11 cells were generated as previously described (46) in biological replicates (clones 6 and 28) (https://data.4dnucleome.org/experiment-set-replicates/4DNESDRL4ZKM/ and https://data.4dnucleome.org/experiment-set-replicates/4DNESIZ5TTHO/). We combined the two biological replicates and applied the MAPS (model-based analysis of PLAC-seq and HiChIP) pipeline (47) to identify significant long-range chromatin interactions at 5-kb bin resolution for the genomic distance 10 kb to 1 Mb. The reference genome is GRCh38/hg38. In addition, for each 5-kb bin pair anchored at H3K4me3 peaks, the MAPS pipeline outputs the normalized contact frequency, which adjusts for the biases from effective fragment length, GC content, sequence mappability, H3K4me3 enrichment level, and one-dimensional genomic distance effect.
Comparison between strong enhancers and weak enhancers using H3K4me3 PLAC-seq data
For four genes HPRT1, MLH1, PMS2, and PCNA, there are 23 enhancer-promoter pairs between strong enhancers and their target genes and 21 enhancer-promoter pairs between weak enhancers and their target genes. We mapped each enhancer and promoter of target gene into 5-kb bins and obtained the distance normalized H3K4me3 PLAC-seq contact frequency for 5-kb bin pairs containing the enhancer-promoter pairs. Because MSH2 and MSH6 are located within 407-kb linear genomic distance with each other and we cannot assign enhancers to either gene reliably, enhancers identified near MSH2 and MSH6 were excluded from this analysis.
Comparison between enhancer-like promoters and control promoters using H3K4me3 PLAC-seq data
For this analysis, control promoters are active promoter regions with annotated ATAC-seq peaks and tested negative as enhancers for the MMR genes. We mapped each promoter into a 5-kb bin that was used in the PLAC-seq analysis. We only choose the bins with one annotated active promoter, which gave us 31 enhancer-like promoters and 43 control promoters in this analysis. We counted the number of significant H3K4me3 PLAC-seq interactions anchored at the 5-kb bins with these promoter sequences. In addition, as described in our previous study (24), for promoters with at least one significant interaction, we calculated the summation of −log10 FDR of significant interactions, which is a measure of the overall interaction strength.
Chromatin contact frequency comparison between strong enhancers and weak enhancers in K562 cells and mESCs
For the chromatin contact frequency comparison of enhancers in K562 cells and mESCs, we downloaded the identified enhancers from each publication (6, 8) and defined strong enhancer with cutoff of 50% ≤ transcriptional contribution ≤ 100% and weak enhancer with cutoff of 0% < transcriptional contribution ≤ 20%. H3K27ac HiChIP data in K562 cells (48) and H3K4me3 PLAC-seq data in mESCs (47) were used for comparison. The comparisons were performed in 10-kb resolution.
Motif scan and TF identification
The FASTA files were first generated in the GRCh38/hg38 genome for the identified strong enhancers and weak enhancers separately. For each strong enhancer and weak enhancer, the FIMO software (version 5.1.0) (49) with human motif database HOCOMOCO (v11 FULL) (50) was used to scan the motifs. All the FIMO motif scans were in default settings. We then filtered the TFs in each strong and weak enhancer loci by FDR cutoff of 0.05, P value cutoff of 0.0001, and gene expression cutoff of FPKM >1. By taking the TFs with TF motif appearing in more than 80% enhancers, 47 TFs were considered as commonly appearing in the strong enhancers, and 35 TFs were in the weak enhancers.
Dual-luciferase reporter assay
Dual-luciferase reporter assay system (Progema, E1910) was used to test the enhancer activity of weak and strong enhancers. The weak enhancers, strong enhancers, and negative control elements (table S8) were PCR-amplified from WTC11 iPSC genomic DNA with NEBNext High-Fidelity 2× PCR Master Mix (NEB, M0541S). The amplified DNA elements and synthesized minimal promoter were cloned into Xho I– and Nco I–digested pGL4.13 vector (Progema, E6681) by Gibson assembly (NEB, E2621L). After validation of the sequence by Sanger sequencing, the vectors were cotransfected with pRL-CMV-Renilla luciferase vector (Promega, E2261) in WTC11 iPSCs with FuGENE HD (Promega, E2311) at a 3:1 reagent to DNA ratio. pGL4.13-SV40-Firefly luciferase vector (Progema, E6681) was used as a positive control. The luciferase activity was measured 48 hours after transfection with a BioTek Synergy 2 multi-mode microplate reader. The relative firefly luciferase activity of each sample was normalized to the average of the activities of minimal promoter.
Acknowledgments
Funding: This work was supported by NIH grants UM1HG009402 (to Y.S., B.R., and W.W.) and U54DK107977 (to B.R. and M.H.). Author contributions: X.R. and Y.S. designed the study. X.R. and Bin Li designed the sgRNA library under the supervision of Y.S. and B.R. X.R., K.J., I.R.J., M.A.T., J.L., L.M., and T.W.T. performed the experiments. M.W. designed the CRISPY under the supervision of W.W. X.R., Bingkun Li, L.Z., G.L., and Y.L. performed data analysis. M.Y. and R.H. constructed the H3K4me3 PLAC-seq datasets. L.L., A.A., and M.H. analyzed PLAC-seq and HiChIP data. X.R. and Y.S. prepared the manuscript with input from all other authors. Competing interests: B.R. is cofounder and shareholder of Arima Genomics and Epigenome Technologies. The other authors declare that they have no competing interests. Data and materials availability: The CRISPRpath screen datasets used in this study are available at the ENCODE portal [www.encodeproject.org, accession numbers: ENCSR617AZY (sgRNA plasmid library), ENCSR427OPP (CRISPRi control), ENCSR900AXT (CRISPRi 1×), ENCSR254SJU (CRISPRi 2×), ENCSR793DSE (CRISPRi 3×), ENCSR250ZWC (CRISPRn control), ENCSR117YGQ (CRISPRn 1×), ENCSR071ZGB (CRISPRn 2×), and ENCSR482PHH (CRISPRn 3×)]. WTC11 iPSC H3K4me3 PLAC-seq datasets are available at the 4DN data portal (data.4dnucleome.org, accession numbers: 4DNESIZ5TTHO and 4DNESDRL4ZKM). ATAC-seq, ChIP-seq, CUT&Tag, and RNA-seq datasets in WTC11 iPSCs are available at the Gene Expression Omnibus under the accession number GSE166839. Data can be visualized on the WashU Epigenome Browser using the following session: https://epigenomegateway.wustl.edu/browser/?genome=hg38&sessionFile=https://shen-xren.s3-us-west-1.amazonaws.com/CRISPRpath/eg-session-QRXJ0218-4d710b60-6ea7-11eb-8d8d-03c7189570c0.json. Tracks include ATAC-seq, H3K27ac, H3K4me3, and CTCF signals and the identified enhancers from CRISPRi 2× and 3× screens. The plasmids generated in this study are available from Addgene (#167186, #167188, and #167189). The computer code used for analyzing CRISPRpath datasets is available at https://zenodo.org/record/5138151 and https://github.com/MichaelMW/crispy. The WTC11 CRISPRi and CRISPRn iPSC lines can be provided by the Gladstone Institutes pending scientific review and a completed material transfer agreement. Requests for the WTC11 iPSC lines should be submitted to B. Conklin at bconklin@gladstone.ucsf.edu.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Legends for tables S1 to S8
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S8
REFERENCES AND NOTES
- 1.Soldner F., Stelzer Y., Shivalila C. S., Abraham B. J., Latourelle J. C., Barrasa M. I., Goldmann J., Myers R. H., Young R. A., Jaenisch R., Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visel A., Rubin E. M., Pennacchio L. A., Genomic views of distant-acting enhancers. Nature 461, 199–205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long H. K., Osterwalder M., Welsh I. C., Hansen K., Davies J. O. J., Liu Y. E., Koska M., Adams A. T., Aho R., Arora N., Ikeda K., Williams R. M., Sauka-Spengler T., Porteus M. H., Mohun T., Dickel D. E., Swigut T., Hughes J. R., Higgs D. R., Visel A., Selleri L., Wysocka J., Loss of extreme long-range enhancers in human neural crest drives a craniofacial disorder. Cell Stem Cell 27, 765–783 e714 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.E. P. Consortium, Moore J. E., Purcaro M. J., Pratt H. E., Epstein C. B., Shoresh N., Adrian J., Kawli T., Davis C. A., Dobin A., Kaul R., Halow J., Van Nostrand E. L., Freese P., Gorkin D. U., Shen Y., He Y., Mackiewicz M., Pauli-Behn F., Williams B. A., Mortazavi A., Keller C. A., Zhang X. O., Elhajjajy S. I., Huey J., Dickel D. E., Snetkova V., Wei X., Wang X., Rivera-Mulia J. C., Rozowsky J., Zhang J., Chhetri S. B., Zhang J., Victorsen A., White K. P., Visel A., Yeo G. W., Burge C. B., Lecuyer E., Gilbert D. M., Dekker J., Rinn J., Mendenhall E. M., Ecker J. R., Kellis M., Klein R. J., Noble W. S., Kundaje A., Guigo R., Farnham P. J., Cherry J. M., Myers R. M., Ren B., Graveley B. R., Gerstein M. B., Pennacchio L. A., Snyder M. P., Bernstein B. E., Wold B., Hardison R. C., Gingeras T. R., Stamatoyannopoulos J. A., Weng Z., Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanjana N. E., Wright J., Zheng K., Shalem O., Fontanillas P., Joung J., Cheng C., Regev A., Zhang F., High-resolution interrogation of functional elements in the noncoding genome. Science 353, 1545–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gasperini M., Hill A. J., McFaline-Figueroa J. L., Martin B., Kim S., Zhang M. D., Jackson D., Leith A., Schreiber J., Noble W. S., Trapnell C., Ahituv N., Shendure J., A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176, 377–390.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diao Y., Fang R., Li B., Meng Z., Yu J., Qiu Y., Lin K. C., Huang H., Liu T., Marina R. J., Jung I., Shen Y., Guan K. L., Ren B., A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods 14, 629–635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulco C. P., Nasser J., Jones T. R., Munson G., Bergman D. T., Subramanian V., Grossman S. R., Anyoha R., Doughty B. R., Patwardhan T. A., Nguyen T. H., Kane M., Perez E. M., Durand N. C., Lareau C. A., Stamenova E. K., Aiden E. L., Lander E. S., Engreitz J. M., Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet. 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simeonov D. R., Gowen B. G., Boontanrart M., Roth T. L., Gagnon J. D., Mumbach M. R., Satpathy A. T., Lee Y., Bray N. L., Chan A. Y., Lituiev D. S., Nguyen M. L., Gate R. E., Subramaniam M., Li Z., Woo J. M., Mitros T., Ray G. J., Curie G. L., Naddaf N., Chu J. S., Ma H., Boyer E., Van Gool F., Huang H., Liu R., Tobin V. R., Schumann K., Daly M. J., Farh K. K., Ansel K. M., Ye C. J., Greenleaf W. J., Anderson M. S., Bluestone J. A., Chang H. Y., Corn J. E., Marson A., Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549, 111–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klann T. S., Black J. B., Chellappan M., Safi A., Song L., Hilton I. B., Crawford G. E., Reddy T. E., Gersbach C. A., CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 35, 561–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulco C. P., Munschauer M., Anyoha R., Munson G., Grossman S. R., Perez E. M., Kane M., Cleary B., Lander E. S., Engreitz J. M., Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasperini M., Findlay G. M., McKenna A., Milbank J. H., Lee C., Zhang M. D., Cusanovich D. A., Shendure J., CRISPR/Cas9-mediated scanning for regulatory elements required for HPRT1 expression via thousands of large, programmed genomic deletions. Am. J. Hum. Genet. 101, 192–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pecina-Slaus N., Kafka A., Salamon I., Bukovac A., Mismatch repair pathway, genome stability and cancer. Front. Mol. Biosci. 7, 122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan T., Berry S. E., Desai A. B., Kinsella T. J., DNA mismatch repair (MMR) mediates 6-thioguanine genotoxicity by introducing single-strand breaks to signal a G2-M arrest in MMR-proficient RKO cells. Clin. Cancer Res. 9, 2327–2334 (2003). [PubMed] [Google Scholar]
- 15.Li G. M., Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Mandegar M. A., Huebsch N., Frolov E. B., Shin E., Truong A., Olvera M. P., Chan A. H., Miyaoka Y., Holmes K., Spencer C. I., Judge L. M., Gordon D. E., Eskildsen T. V., Villalta J. E., Horlbeck M. A., Gilbert L. A., Krogan N. J., Sheikh S. P., Weissman J. S., Qi L. S., So P. L., Conklin B. R., CRISPR interference efficiently induces specific and reversible gene silencing in human iPSCs. Cell Stem Cell 18, 541–553 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tycko J., Wainberg M., Marinov G. K., Ursu O., Hess G. T., Ego B. K., Aradhana A. L., Truong A., Trevino A. E., Spees K., Yao D., Kaplow I. M., Greenside P. G., Morgens D. W., Phanstiel D. H., Snyder M. P., Bintu L., Greenleaf W. J., Kundaje A., Bassik M. C., Mitigation of off-target toxicity in CRISPR-Cas9 screens for essential non-coding elements. Nat. Commun. 10, 4063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez A. R., Pritykin Y., Vidigal J. A., Chhangawala S., Zamparo L., Leslie C. S., Ventura A., GuideScan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 35, 347–349 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radzisheuskaya A., Shlyueva D., Muller I., Helin K., Optimizing sgRNA position markedly improves the efficiency of CRISPR/dCas9-mediated transcriptional repression. Nucleic Acids Res. 44, e141 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenbluh J., Xu H., Harrington W., Gill S., Wang X., Vazquez F., Root D. E., Tsherniak A., Hahn W. C., Complementary information derived from CRISPR Cas9 mediated gene deletion and suppression. Nat. Commun. 8, 15403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li K., Liu Y., Cao H., Zhang Y., Gu Z., Liu X., Yu A., Kaphle P., Dickerson K. E., Ni M., Xu J., Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat. Commun. 11, 485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engreitz J. M., Haines J. E., Perez E. M., Munson G., Chen J., Kane M., McDonel P. E., Guttman M., Lander E. S., Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sutherland H., Bickmore W. A., Transcription factories: Gene expression in unions? Nat. Rev. Genet. 10, 457–466 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Song M., Pebworth M. P., Yang X., Abnousi A., Fan C., Wen J., Rosen J. D., Choudhary M. N. K., Cui X., Jones I. R., Bergenholtz S., Eze U. C., Juric I., Li B., Maliskova L., Lee J., Liu W., Pollen A. A., Li Y., Wang T., Hu M., Kriegstein A. R., Shen Y., Cell-type-specific 3D epigenomes in the developing human cortex. Nature 587, 644–649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubo N., Ishii H., Xiong X., Bianco S., Meitinger F., Hu R., Hocker J. D., Conte M., Gorkin D., Yu M., Li B., Dixon J. R., Hu M., Nicodemi M., Zhao H., Ren B., Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 28, 152–161 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-Zapico M. E., Lomberk G. A., Tsuji S., DeMars C. J., Bardsley M. R., Lin Y. H., Almada L. L., Han J. J., Mukhopadhyay D., Ordog T., Buttar N. S., Urrutia R., A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth. Biochem. J. 435, 529–537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R. A., Dynlacht B. D., E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16, 245–256 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polager S., Kalma Y., Berkovich E., Ginsberg D., E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21, 437–446 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Canver M. C., Smith E. C., Sher F., Pinello L., Sanjana N. E., Shalem O., Chen D. D., Schupp P. G., Vinjamur D. S., Garcia S. P., Luc S., Kurita R., Nakamura Y., Fujiwara Y., Maeda T., Yuan G. C., Zhang F., Orkin S. H., Bauer D. E., BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajagopal N., Srinivasan S., Kooshesh K., Guo Y., Edwards M. D., Banerjee B., Syed T., Emons B. J., Gifford D. K., Sherwood R. I., High-throughput mapping of regulatory DNA. Nat. Biotechnol. 34, 167–174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koike-Yusa H., Li Y., Tan E. P., Velasco-Herrera Mdel C., Yusa K., Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 32, 267–273 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Adamson B., Norman T. M., Jost M., Cho M. Y., Nunez J. K., Chen Y., Villalta J. E., Gilbert L. A., Horlbeck M. A., Hein M. Y., Pak R. A., Gray A. N., Gross C. A., Dixit A., Parnas O., Regev A., Weissman J. S., A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diao Y., Li B., Meng Z., Jung I., Lee A. Y., Dixon J., Maliskova L., Guan K. L., Shen Y., Ren B., A new class of temporarily phenotypic enhancers identified by CRISPR/Cas9-mediated genetic screening. Genome Res. 26, 397–405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie S., Duan J., Li B., Zhou P., Hon G. C., Multiplexed engineering and analysis of combinatorial enhancer activity in single cells. Mol. Cell 66, 285–299.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Zabidi M. A., Arnold C. D., Schernhuber K., Pagani M., Rath M., Frank O., Stark A., Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dao L. T. M., Spicuglia S., Transcriptional regulation by promoters with enhancer function. Transcription 9, 307–314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikhaylichenko O., Bondarenko V., Harnett D., Schor I. E., Males M., Viales R. R., Furlong E. E. M., The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32, 42–57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas H. F., Kotova E., Jayaram S., Pilz A., Romeike M., Lackner A., Penz T., Bock C., Leeb M., Halbritter F., Wysocka J., Buecker C., Temporal dissection of an enhancer cluster reveals distinct temporal and functional contributions of individual elements. Mol. Cell 81, 969–982.e13 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramirez F., Ryan D. P., Gruning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dundar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaya-Okur H. S., Wu S. J., Codomo C. A., Pledger E. S., Bryson T. D., Henikoff J. G., Ahmad K., Henikoff S., CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meers M. P., Tenenbaum D., Henikoff S., Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang R., Yu M., Li G., Chee S., Liu T., Schmitt A. D., Ren B., Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 26, 1345–1348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Juric I., Yu M., Abnousi A., Raviram R., Fang R., Zhao Y., Zhang Y., Qiu Y., Yang Y., Li Y., Ren B., Hu M., MAPS: Model-based analysis of long-range chromatin interactions from PLAC-seq and HiChIP experiments. PLOS Comput. Biol. 15, e1006982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mumbach M. R., Satpathy A. T., Boyle E. A., Dai C., Gowen B. G., Cho S. W., Nguyen M. L., Rubin A. J., Granja J. M., Kazane K. R., Wei Y., Nguyen T., Greenside P. G., Corces M. R., Tycko J., Simeonov D. R., Suliman N., Li R., Xu J., Flynn R. A., Kundaje A., Khavari P. A., Marson A., Corn J. E., Quertermous T., Greenleaf W. J., Chang H. Y., Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulakovskiy I. V., Vorontsov I. E., Yevshin I. S., Sharipov R. N., Fedorova A. D., Rumynskiy E. I., Medvedeva Y. A., Magana-Mora A., Bajic V. B., Papatsenko D. A., Kolpakov F. A., Makeev V. J., HOCOMOCO: Towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Legends for tables S1 to S8
Tables S1 to S8