Interferons have been shown to exert antiviral, cell growth regulatory, and immunomodulatory effects on target cells (16, 37). Both type I (α and β) and type II (γ) interferons regulate cellular activities by specifically inducing the expression or activation of endogenous proteins that perform distinct biological functions. The HIN-200 (hematopoietic interferon-inducible nuclear proteins with a 200-amino-acid repeat) gene family found on human and mouse chromosome 1 is positively regulated by type I and II interferons (15, 27, 30). The HIN-200 family of proteins consists of a number of highly homologous human and murine proteins with similar primary amino acid sequences and biological characteristics. The human HIN-200 family members include IFI 16 (39), the myeloid nuclear differentiation antigen (MNDA) (2, 18), and AIM-2 (absent in melanoma) (17), while the mouse HIN-200 family members include p202 (3), p203 (20), p204 (3), and D3 (38). There are a number of reviews that elegantly outline the biochemical characteristics of these proteins and their patterns of expression (15, 27, 30). Recently, there have been a number of reports examining possible cellular functions of HIN-200 proteins. It is now clear that these proteins play a role in modulating cell growth and perhaps in vivo differentiation, and the newest member of the HIN-200 family, AIM-2, has been identified as a possible tumor suppressor protein. Some family members are transcriptional regulators, acting either directly by binding DNA at a promoter or indirectly by modulating the function of other cellular transcription factors. Clearly, there is much still to learn about the biological functions of these proteins and how significant a role they play during an interferon response. While it is clear that the human and mouse family members share many biochemical and structural characteristics, it is unclear which, if any, of the currently identified murine HIN-200 proteins are functional homologs of the different human HIN-200 family members. This review will briefly cover the structural and biochemical properties of HIN-200 proteins but will concentrate on the molecular and biological functions of these molecules.
STRUCTURE AND EXPRESSION OF THE HIN-200 PROTEINS
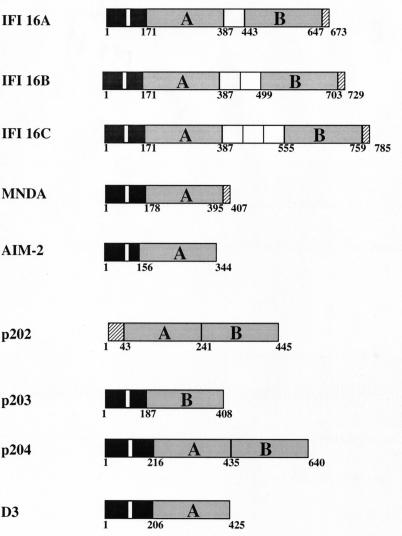
A structural motif found in all members of the HIN-200 family is a 200-amino-acid domain present singly or in duplicate (Fig. 1). There are two contiguous 200-amino-acid domains (A and B) in p202 and p204, while the two repeats are separated by a serine-threonine-proline (S/T/P)-rich spacer region in IFI 16. The size of the spacer region in IFI 16 is regulated by mRNA splicing and can contain one, two, or three copies of a highly conserved 56-amino-acid S/T/P domain encoded by distinct exons (23). In contrast, MNDA, AIM-2, p203, and D3 contain only one 200-amino-acid domain (3, 17, 18, 20). The amino acid composition of the A domains expressed in different HIN-200 proteins is highly conserved, as are the B domains expressed in different family members. For example, there is approximately 55 and 50% sequence identity between the A and B domains, respectively, found in p202 and p204 (30). In contrast, sequence comparison reveals only 27% identity between the A and B domains of p202 and 34% identity between the p204 A and B segments. These findings are consistent with the duplication of a primordial gene encoding the ancestral 200-amino-acid segment giving rise to the second domain. These 200-amino-acid regions are unique to the HIN-200 proteins and contain no known functional motifs that could provide some clue to their physiological relevance. However, there are stretches of amino acids, such as the sequence MFHATVAT, that exhibit almost complete identity across the A and B domains of all family members. The conservation of these domains throughout evolution points to a functional or structural requirement for these regions. Recent studies have investigated the functional significance of the 200-amino-acid domains, and the findings from these studies will be discussed later in this review.
FIG. 1.
Schematic structural representation of HIN-200 proteins. Black boxes indicate amino-terminal regions with amino acid sequence homologies, and putative nuclear localization sequences are indicated by a white line within this region. The type A and B 200-amino-acid domains are shaded grey. For IFI 16, the S/T/P-rich spacer domains between the A and B 200-amino-acid repeats are shown as white boxes. The IFI 16A, -B, and -C isoforms arise due to alternative RNA splicing in exons encoding the S/T/P domains. Amino acid numbers are shown below each structure.
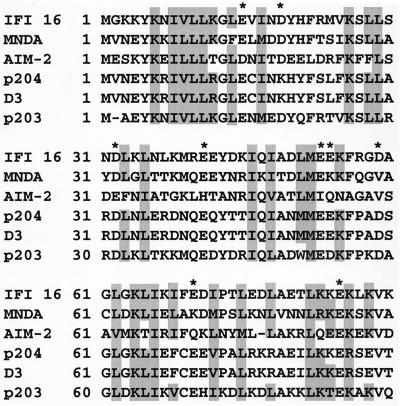
In addition to the 200-amino-acid repeat they have in common, all members of the HIN-200 family, except p202, show a high degree of amino acid homology in the amino-terminal region. Of particular interest are the highly conserved leucine and basic residues (Fig. 2). The leucine residues are thought to form an imperfect leucine zipper motif. Both IFI 16 and MNDA have been shown to homodimerize via their amino-terminal leucine zipper and basic regions (23, 42), and the conservation of these residues throughout the HIN-200 proteins indicates the possibility of homo- and heterodimerization of all family members except, possibly, p202. Interestingly, p202 was recently shown to be able to self-associate and a highly conserved motif (MFHATVAT) within the two 200-amino-acid domains of p202 was necessary and sufficient for the homodimerization (26). This report is in contrast to two others showing that the 200-amino-acid repeats of IFI 16 and MNDA are dispensable for self-interaction (23, 42). Consensus nuclear localization signal sequences are found within the amino-terminal regions of IFI 16, MNDA, p204, and D3, consistent with the constitutive nuclear expression of these molecules (4, 15). In contrast, p202 does not contain such a motif and the expression of p202 differs from that of other family members in that it is expressed within the cytoplasm following interferon induction and then translocates to the nucleus over time (5). The subcellular expression of p203 and AIM-2 has yet to be determined.
FIG. 2.
Conservation of leucine, isoleucine, and basic amino acids within the N-terminal region of the HIN-200 family. The amino-terminal 90 amino acids of all known HIN-200 proteins except p202 are aligned. Amino acid residues capable of forming a functional leucine zipper (including leucine, isoleucine, valine, and methionine residues) and basic amino acids are shaded. Conservation of acidic residues is denoted by an asterisk.
All HIN-200 family members are expressed in hematopoietic cells, with some molecules showing a tightly regulated expression pattern in certain cell types. For example, IFI 16 is expressed in precursor CD34+ stem cells and remains strongly expressed throughout lymphoid development and within monocyte precursors and peripheral blood monocytes. However, IFI 16 is not expressed in mature macrophages, nor is it found in cells of the erythroid and polymorphonuclear lineages. This is in contrast to MNDA, which is found in mature granulocytes and monocytes but not in lymphoid cells or undifferentiated myeloid cell lines (11, 12). This expression pattern is similar to that of p204, which is constitutively expressed in myelomonocytic cells of the mouse (19).
As their name suggests, all HIN-200 genes can be positively regulated by type I and/or type II interferons (Table 1). The responses of individual HIN-200 genes to the two classes of interferon can differ greatly, and the molecular basis for these differences has yet to be clearly defined. In addition, genes such as IFI 16 can be strongly upregulated in promyelocytic cell lines treated with differentiation agents such as dimethyl sulfoxide or retinoic acid (13) and proliferative agents such as lipopolysaccharide (LPS) and phorbol myristate acetate have been used in vitro to upregulate the expression of D3 (38) and MNDA (1), respectively. These data further suggest a possible role for HIN-200 proteins in the regulation of cell growth and differentiation in vivo.
TABLE 1.
Physical and biochemical properties of HIN-200 moleculesa
| Protein | Chromosomal localization | No. of 200-amino-acid repeat copies | Molecular mass (kDa) | Gene inductionb | DNA binding | Subcellular localization | Cellular expression |
|---|---|---|---|---|---|---|---|
| IFI 16 | 1q22 | 2 | 85–95 | IFN-α, -γ, RA, DMSO, vitamin D3 | Yes | Nucleus | Lymphocytes, CD34+ stem cells, monocytes |
| MNDA | 1q21-22 | 1 | 55 | IFN-α, PMA, PHA | Yes | Nucleus | Granulocytes, monocytes |
| AIM-2 | 1q22 | 1 | 40c | IFN-α, -γ | Unknown | Unknown | Spleen, peripheral blood leukocytes, small intestine |
| p202 | 1q21-23 | 2 | 52 | IFN-α, -β, -γ, LPS, poly(rI-rC) | Yes | Cytoplasm/nucleus | Spleen, heart, brain, skeletal muscle, liver, kidney |
| p203 | 1q21-23 | 1 | 48 | IFN-α | Unknown | Nucleus | Unknown |
| p204 | 1q21-23 | 2 | 72 | IFN-α, -β, -γ, LPS, poly(rI-rC) | Unknown | Nucleus | Monocytes, megakaryocytes |
| D3 | 1q21-23 | 1 | 47 | IFN-α, -β, -γ, LPS, poly(rI-rC) | Unknown | Nucleus | Monocytes, granulocytes |
The biochemical properties shown are described in published reports and may change or expand. The cellular expression does not include that seen in tumor cell lines and is based on protein expression data for all molecules but p202 and AIM-2, whose expression was analyzed by Northern blotting.
RA, retinoic acid; DMSO, dimethyl sulfoxide; PHA, phytohemagglutinin; PMA, phorbol myristate acetate.
Estimated mass.
TRANSCRIPTION REGULATION BY HIN-200 PROTEINS
Clues to some of the possible cellular functions of HIN-200 proteins can be drawn from the physical and biochemical characteristics of the molecules. All HIN-200 proteins translocate to the nucleus following induction of expression by interferons, although there do appear to be some differences in the kinetics of nuclear translocation, as p202 can initially be detected in the cytosol after interferon treatment. While the molecular mechanisms involved in nuclear translocation of the HIN-200 family members have not been extensively studied, recent evidence suggests that IFI 16 has a bipartite nuclear localization sequence whose activity may be regulated by phosphorylation of a key serine residue located within the nuclear localization signal by protein kinase CK2 (1a). In addition, IFI 16 (14), p202 (30), and MNDA (18) can all bind double-stranded DNA through their amino-terminal leucine or charged regions. However, specific DNA elements capable of binding these molecules have yet to be defined and it is possible that these molecules can bind DNA in a nonspecific manner or are bound to a specific DNA element via their interaction with a cellular transcription factor(s). Thus, it seems reasonable to postulate that HIN-200 molecules might regulate gene expression and it appears that some HIN-200 proteins do, albeit possibly by different mechanisms.
Full-length IFI 16 fused to the heterologous GAL4 DNA binding domain can act as a potent transcriptional repressor when positioned in proximity to a promoter containing consensus GAL4 DNA elements (22). Each of the two 200-amino-acid repeat regions was independently capable of directing this repression activity; however, the amino-terminal 160 amino acids were dispensable for the repression function (22). Thus, IFI 16, like many cellular and viral transcription factors, has a modular structure. There is a distinct DNA binding domain (amino terminus) which also contains a functional nuclear localization signal sequence and two “active” transcription regulatory domains (A and B 200-amino-acid domains) that are fully functional when fused to a heterologous DNA binding domain. The molecular mechanisms underpinning transcriptional repression by IFI 16, such as chromatin remodelling, are unknown but under investigation.
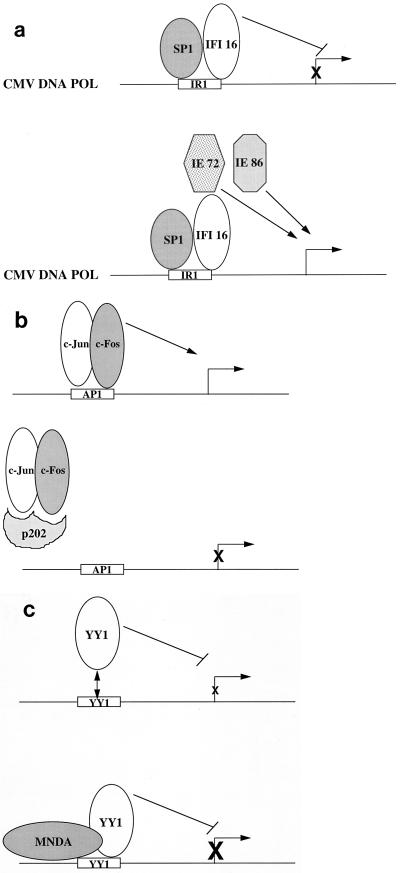
Significantly, wild-type IFI 16 could also function as a transcriptional repressor. It had been shown that IFI 16 forms a stable complex with the cellular transcriptional activator SP1 at a key DNA element (inverted repeat 1 [IR1]) located within the cytomegalovirus (CMV) DNA polymerase gene promoter (32). A functional IR1 is necessary for transcription of the CMV DNA polymerase gene and efficient viral replication (24). IFI 16 was able to repress transcription of a reporter gene containing wild-type IR1 within the upstream promoter, while mutation of IR1, resulting in loss of binding of the SP1/IFI 16 complex, caused a loss of IFI 16-mediated transcriptional repression (22). These studies showed for the first time that a member of the HIN-200 family of proteins is capable of actively modulating gene expression. From these studies, one might think that IFI 16 could negatively regulate CMV DNA polymerase gene expression and therefore inhibit viral replication. However, also necessary for CMV DNA polymerase gene activation are the immediate-early CMV gene products IE72 and IE86, which function synergistically as transcriptional activators of the CMV DNA polymerase gene yet do not bind IR1, and SP1, which can bind IR1 but does not interact with IE72 or IE86 (32). Thus, a current working model is that in the absence of IE72 and IE86, IFI 16 can repress transcription of CMV DNA polymerase when complexed with SP1 at IR1 (Fig. 3A). However, expression of IE72 and/or IE86, possibly resulting in direct binding of these molecules to IFI 16, could result in activation of the CMV DNA polymerase gene (Fig. 3A). In this model, viral proteins (IE72 and/or IE86) modulate the function of a cellular protein (IFI 16) to the advantage of the pathogen. There are precedents for reversal by viral proteins of transcriptional repression mediated by cellular factors. For example, the cellular protein YY1 can repress transcription when bound to its cognate DNA binding sequence at a promoter (37). However, coexpression of the adenovirus oncoprotein E1A converts YY1 from a transcriptional repressor to a transcriptional activator (37). Studies are currently under way to determine if IE86 and/or IE72 can modulate IFI 16-mediated transcriptional repression of the CMV DNA polymerase gene and, if so, how this occurs.
FIG. 3.
(a) Transcriptional repression by IFI 16 and potential modulation by viral transcription factors. IFI 16 and SP1 can bind the IR1 element within the CMV DNA polymerase (POL) promoter. Binding of IFI 16 at this site results in transcriptional repression. The expression of the CMV immediate-early proteins IE72 and/or IE86 is necessary for activation of the CMV DNA polymerase gene, possibly due to interaction of IE72 and/or IE86 with IFI 16, resulting in modulation of the repression activity of IFI 16. (b) Transcriptional regulation by p202. p202 can interact either directly or indirectly with a range of different transcription factors, including E2F-1/DP-1, E2F-4/DP-1, MyoD, NF-κB, p53, and c-Jun/c-Fos. In most cases, this results in a block in the binding of the transcription factor to its cognate DNA element. Shown is an example of p202 binding to the Jun/Fos heterodimer and blocking the binding of this complex to an AP1 site, thereby inhibiting transcriptional activation of the target gene. (c) MNDA stimulates YY1 DNA binding. YY1 can bind DNA in a sequence-specific manner to direct transcriptional repression or, in some cases, transcriptional activation. Binding of MNDA to YY1 increases the affinity of YY1 for its cognate DNA element by increasing the rate of YY1-DNA association and decreasing the rate of dissociation.
These experiments indicate that IFI 16 can function as a transcriptional repressor at a specific promoter. The sequence of IR1 (AGGCTCCG) is a nonconsensus GC box, the prototype of which is capable of binding SP1 with high affinity. It is unclear whether functional IR1 elements or related sequences are present within the promoter regions of cellular genes, but if they are, this raises the possibility of the regulation of those cellular genes by SP1/IFI 16.
In contrast to IFI 16, the GAL4-p202 and GAL4-p204 fusion proteins had no effect on the expression of a reporter gene containing GAL4 DNA elements within the promoter (30). However, p202 can modulate transcriptional activation mediated by the transcription factors E2F-1/DP-1 (7), E2F-4/DP-1 (8), MyoD (10), NF-κB (33), and p53 via interaction with p53-binding protein (9) and the AP-1 complex consisting of c-Jun/c-Fos heterodimers (33). The effect of p202 on transcription activation by E2F-1/DP-1, E2F-4/DP-1, MyoD, p53, and AP-1 appears to reside in its ability to inhibit the binding of these transcription factors to their cognate DNA elements (7–10, 33). Thus, p202 can regulate the ability of distinct transcription factors to bind to specific DNA sequences and can therefore regulate the expression of particular cellular genes (Fig. 3B). However, binding of p202 to NF-κB also inhibits the ability of NF-κB to transactivate but does not prevent NF-κB from binding double-stranded DNA (33). It is unclear how p202 inhibits the transcriptional activity of NF-κB, but it can be postulated that p202 may modulate transcription by interfering with the assembly of the basal transcription machinery at the promoter site. Likewise, it is possible that under certain circumstances and in particular promoter conformations, p202, like IFI 16, may function as a transcriptional repressor and can therefore affect transcriptional activation by NF-κB.
Unlike IFI 16 and p202, MNDA appears to play a positive role in transcription regulation by modulating the activity of a known cellular transcription factor. MNDA can physically associate with the transcriptional activator-repressor YY1 and stimulates the binding of YY1 to its cognate DNA element (43). Binding of MNDA to YY1 increased the rate of DNA-protein association and decreased the rate of YY1 dissociation from DNA (Fig. 3C). It is unclear whether MNDA increases the affinity of YY1 for its cognate DNA element by also forming a protein-DNA interaction or whether binding of MNDA to YY1 alters the conformation of YY1, thereby enabling a higher-affinity interaction between YY1 and DNA. Interestingly, YY1 has been shown to physically associate with the nuclear proteins nucleolin and nucleophosmin (NPM/B23), which can regulate the transcriptional activities of YY1. Nucleolin can function as a transcriptional repressor (45), while NPM can relieve YY1-mediated transcriptional repression (21). Independently, it was shown that MNDA can also bind nucleolin and NPM (41) and thus it is possible that these four molecules may form large multimeric complexes containing various combinations of these proteins to positively or negatively regulate the expression of cellular genes. YY1 has been shown to regulate the expression of key growth regulatory genes, including those for c-myc, p53, c-fos, and gamma interferon (IFN-γ) (36). The necessity for MNDA to induce the YY1-mediated regulation of these genes encoding proteins that regulate cell growth and differentiation is not known but warrants further investigation.
REGULATION OF CELL PROLIFERATION AND THE CELL CYCLE BY HIN-200 PROTEINS
Treatment of human and mouse cell lines with interferons causes a decrease in cell cycle progression with a reduced rate of transition of cells from G1 into S phase (38). Although the growth regulatory effects of type I and II interferons have been well documented in a variety of different human and mouse cell lines, the molecular mechanisms and proteins responsible for this regulatory function remain largely unknown. As stated above, p202 can interact with many key transcription factors that are known to regulate the cell cycle. In addition, p202 has been shown to interact with hypophosphorylated pRb (6), p107 (8), and p130 (8), all of which are members of the retinoblastoma tumor suppressor family which have previously been shown to play a fundamental role in cellular growth and transcription regulation (34). These findings, together with those of many previous studies demonstrating cell growth regulation by interferons, led to experiments designed to determine the possible growth regulatory function of p202. It has been shown that overexpression of p202 in transfected cell lines results in a decrease in the growth rate of these cells (5, 28, 44) and loss of the ability of transfected cells to grow in soft agar (44). Further studies revealed that constitutive expression of p202 inhibited progression of the cell cycle from G0/G1 to S phase. It is important to note that members of the E2F/pRb family of proteins are key players in the regulation of the progression of cells through the G1/S checkpoint and thus the functional consequence of p202 interacting with these cell cycle regulatory proteins and with the E2F/DP-1 transcription factors may be to enforce cell cycle arrest at this point.
p202 can regulate transcriptional activation mediated by p53, and overexpression of p202 resulted in an increase in p53 protein levels (9). Activation of p53 usually leads to the induction of one of two cellular events: (i) cell cycle inhibition with a block in the transition from G1 to S phase or (ii) programmed cell death (31). As overexpression of p202 can also inhibit cell cycle progression, in particular, movement of cells from G1 to S phase, it was possible that in addition to the role of p202 in regulating the function of E2F and pRb family members to affect the cell cycle, it might also modulate cell proliferation via p53. The molecular mechanisms underlying cell cycle regulation by p53 have been well defined. p53 transcriptionally upregulates the p21/waf1/cip1 gene, the protein product of which inhibits the kinase activity of cdk4/cyclin D (31). Activation of cdk4/cyclin D results in phosphorylation of pRb, release of pRb from the pRb/E2F/DP-1 complex, and movement from the cell from G1 to S phase via the activation of genes regulated by E2F/DP-1 (34). It therefore follows that an increase in the levels or activity of p53 by p202 could increase transcription of p21, resulting in a cell cycle block at G1/S. This, however, was shown not to be the case, as various p202-overexpressing cell lines demonstrated unaltered p21 expression even though there was a significant decrease in the growth rate of p202-expressing cells (9). These experiments indicate that p21 is not a mediator of retarded cell proliferation by p202, although it is possible that other genes regulated by p53/p202 may be involved. Surprisingly, inhibition of endogenous p202 production in murine cell lines using tetracycline-induced expression of antisense RNA to the p202 gene did not result in an increase in cell proliferation (25). In fact, decreased endogenous levels of p202 in asynchronously growing cells inhibited cell proliferation and increased the susceptibility of cells to programmed cell death (25). Thus, tightly regulated levels of p202 are necessary to maintain cellular homeostasis and altered expression of the protein can result in cell cycle arrest or apoptosis.
In contrast to p202, p204 does not appear to bind key cellular transcription factors such as E2F proteins, c-Jun/c-Fos, MyoD, or NF-κB. However, constitutive or induced overexpression of p204 in mouse cell lines led to growth retardation with a block in the G1/S phase transition (29). Overexpression of p204 correlated with an increase in the relative levels of hypophosphorylated pRb, a form of Rb that binds E2F and inhibits cell cycle progression. That p204 does not bind pRb, p107, or p130 indicates that p204 probably has an indirect effect on this family of proteins that induces cell cycle arrest. Interestingly, constitutive overexpression of p204 in transgenic mice leads to a block in embryonic development after the four-cell stage (27). This highlights the potential physiological importance of the tightly regulated lineage and stage-specific expression of HIN-200 proteins, indicating that coordinated expression of these proteins at certain times of development is necessary for accurate programming of cell growth and differentiation.
We have shown that like p202, IFI 16 can bind p53 and pRb both in vitro and in vivo (23a). When overexpressed in human cell lines that do not normally express the protein, IFI 16 retards cell growth by delaying progression from G1 to S phase (23a). The effect of IFI 16 on p53- or E2F-mediated transcriptional activation is still not clear; however, IFI 16 now joins the growing list of HIN-200 proteins that can regulate the cell cycle. In addition, the most recent addition to the HIN-200 family, AIM-2, was cloned by using a genetic screen for potential tumor suppressor proteins. A human melanoma cell line with a deletion of part of chromosome 6 was transfected with a wild-type copy of chromosome 6, resulting in a reduction of cell growth in vitro and a loss of in vivo tumorigenicity (40). Cloning of the genes positively or negatively regulated in response to introduction of chromosome 6 into the melanoma line resulted in the identification of AIM-2 as a putative tumor suppressor protein (17, 35). It remains to be determined whether AIM-2 is indeed necessary and/or sufficient for modulation of the transformed phenotype and whether AIM-2 can also regulate cell cycle progression. That AIM-2 might be expressed in melanocytes and the possibility of downregulation of expression in melanoma are currently under investigation in our laboratory.
CONCLUSIONS
The cellular effects induced by type I and II interferons are brought about by the functional activities of the proteins which respond to stimulation by interferon. The HIN-200 gene family appears to be one of probably many which operate as functional proteins in response to the binding of interferon to its cellular receptor. Clearly, both human and mouse HIN-200 proteins can regulate gene transcription and cell growth. It is unclear whether cell cycle regulation by HIN-200 proteins occurs as a direct consequence of transcription regulation (i.e., modulation of transcriptional activities of, say, p53 or E2F/DP-1) or whether this is induced indirectly by binding of HIN-200 proteins to cell regulatory proteins such as pRb. While some of the biochemical properties and molecular interactions of many HIN-200 proteins have been delineated, the biological roles of these proteins have not been determined. The future production of gene knockout mice in which one or more of the murine HIN-200 genes have been functionally deleted should provide great insight into the roles of HIN-200 proteins in the physiological control of cell growth and differentiation.
ACKNOWLEDGMENTS
R.W.J. is an R. D. Wright Fellow of the National Health and Medical Research Council of Australia. J.A.T. is a Principal Research Fellow of the NHMRC.
We thank M. J. Smyth and S. M. Russell for helpful discussions.
REFERENCES
- 1.Briggs R C, Kao W Y, Dworkin L L, Dessypris E N, Clark J. Regulation and specificity of MNDA expression in monocytes, macrophages and leukemia/B lymphoma cell lines. J Cell Biochem. 1994;56:559–567. doi: 10.1002/jcb.240560417. [DOI] [PubMed] [Google Scholar]
- 1a.Briggs, L. J., R. W. Johnstone, R. M. Elliot, C.-H. Xiao, M. J. Dawson, J. A. Trapani, and D. A. Jans. Unpublished data.
- 2.Burrus G R, Briggs J A, Briggs R C. Characterisation of the human myeloid cell nuclear differentiation antigen: relationship to interferon-induced proteins. J Cell Biochem. 1992;48:190–202. doi: 10.1002/jcb.240480210. [DOI] [PubMed] [Google Scholar]
- 3.Choubey D, Snoddy J, Chaturvedi V, Toniato E, Opdenakker G, Thakur A, Samanta H, Engel D A, Lengyel P. Interferons as gene activators: indications for repeated gene duplication during the evolution of a cluster of interferon-activatable genes on murine chromosome 1. J Biol Chem. 1989;264:17182–17189. [PubMed] [Google Scholar]
- 4.Choubey D, Lengyel P. Interferon action: nucleolar and nucleoplasmic localization of the interferon-inducible 72kD protein that is encoded by the Ifi204 gene from the gene 200 cluster. J Cell Biol. 1992;116:1333–1341. doi: 10.1083/jcb.116.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choubey D, Lengyel P. Interferon action: cytoplasmic and nuclear localization of the interferon-inducible 52kD protein that is encoded by the Ifi202 gene from the gene 200 cluster. J Interferon Res. 1993;13:43–52. doi: 10.1089/jir.1993.13.43. [DOI] [PubMed] [Google Scholar]
- 6.Choubey D, Lengyel P. Binding of the interferon-inducible protein (p202) to the retinoblastoma protein. J Biol Chem. 1995;270:6134–6140. doi: 10.1074/jbc.270.11.6134. [DOI] [PubMed] [Google Scholar]
- 7.Choubey D, Li S-J, Datta B, Gutterman J U, Lengyel P. Inhibition of E2F-mediated transcription by p202. EMBO J. 1996;15:5668–5678. [PMC free article] [PubMed] [Google Scholar]
- 8.Choubey D, Gutterman J U. Inhibition of E2F-4/DP-1-stimulated transcription by p202. Oncogene. 1997;15:291–301. doi: 10.1038/sj.onc.1201184. [DOI] [PubMed] [Google Scholar]
- 9.Datta B, Li B, Choubey D, Nallur G, Lengyel P. p202, an interferon-inducible modulator of transcription, inhibits transcriptional activation by the p53 tumor suppressor protein, and a segment from the p53-binding protein 1 that binds to p202 overcomes this inhibition. J Biol Chem. 1996;271:27544–27555. doi: 10.1074/jbc.271.44.27544. [DOI] [PubMed] [Google Scholar]
- 10.Datta B, Min W, Burma S, Lengyel P. Increase in p202 expression during skeletal muscle differentiation: inhibition of MyoD protein expression and activity by p202. Mol Cell Biol. 1998;18:1074–1083. doi: 10.1128/mcb.18.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson M D, Elwood N J, Johnstone R W, Trapani J A. Lineage-specific expression of the nucleoprotein IFI 16 in monocyte but not polymorphonuclear leukocyte precursors in human bone marrow. J Leukoc Biol. 1998;64:546–554. doi: 10.1002/jlb.64.4.546. [DOI] [PubMed] [Google Scholar]
- 12.Dawson M J, Trapani J A, Briggs R C, Nicholl J K, Sutherland G R, Baker E. The closely linked genes encoding the myeloid nuclear differentiation antigen (MNDA) and Ifi 16 exhibit contrasting hemopoietic expression. Immunogenetics. 1994;41:40–43. doi: 10.1007/BF00188431. [DOI] [PubMed] [Google Scholar]
- 13.Dawson M J, Trapani J A. Ifi 16 gene encodes a nuclear protein whose expression is induced by interferons in human myeloid leukemia ell lines. J Cell Biochem. 1995;57:39–51. doi: 10.1002/jcb.240570106. [DOI] [PubMed] [Google Scholar]
- 14.Dawson M J, Trapani J A. The interferon-inducible nuclear antigen Ifi 16: localization to the nucleolus and identification of a DNA-binding domain. Biochem Biophys Res Commun. 1995;214:152–162. doi: 10.1006/bbrc.1995.2269. [DOI] [PubMed] [Google Scholar]
- 15.Dawson M J, Trapani J A. HIN-200: a novel family of interferon-inducible nuclear proteins expressed in leukocytes. J Leukoc Biol. 1996;60:1–7. doi: 10.1002/jlb.60.3.310. [DOI] [PubMed] [Google Scholar]
- 16.DeMaeyer E M, DeMaeyer-Guignard J. Interferons and other regulatory cytokines. New York, N.Y: Wiley-Interscience; 1988. [Google Scholar]
- 17.DeYoung K L, Ray M E, Johnstone R W, Su Y A, Anzick S L, Meltzer P S, Trapani J A, Trent J M. Cloning of a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15:453–457. doi: 10.1038/sj.onc.1201206. [DOI] [PubMed] [Google Scholar]
- 18.Duhl D M, Gaczynski M, Olinski R, Briggs R C. Intranuclear distribution of the human myeloid cell nuclear differentiation antigen in HL-60 cells. J Cell Physiol. 1989;141:148–153. doi: 10.1002/jcp.1041410122. [DOI] [PubMed] [Google Scholar]
- 19.Gariglio M, De Andrea M, Lembo M, Ravotto M, Zappador C, Valente G, Landolfo S. The murine homolog of the HIN 200 family, Ifi 204, is constitutively expressed in myeloid cells and selectively induced in the monocyte/macrophage lineage. J Leukoc Biol. 1998;64:608–614. doi: 10.1002/jlb.64.5.608. [DOI] [PubMed] [Google Scholar]
- 20.Gribaudo G, Ravaglia S, Guandalini L, Riera L, Gariglio M, Landolfo S. Molecular cloning and expression of a novel interferon-inducible protein encoded by a 203 gene from the gene 200 cluster. Eur J Biochem. 1997;249:258–264. doi: 10.1111/j.1432-1033.1997.t01-1-00258.x. [DOI] [PubMed] [Google Scholar]
- 21.Inouye C J, Seto E. Relief of YY1-induced transcriptional repression by protein-protein interaction with the nuclear phosphoprotein B23. J Biol Chem. 1994;269:6506–6511. [PubMed] [Google Scholar]
- 22.Johnstone R W, Kerry J A, Trapani J A. IFI 16 can function as a potent transcriptional repressor. J Biol Chem. 1998;273:17172–17177. doi: 10.1074/jbc.273.27.17172. [DOI] [PubMed] [Google Scholar]
- 23.Johnstone R W, Kershaw M H, Trapani J A. Isotypic variants of the interferon-inducible transcriptional repressor IFI 16 arise through differential mRNA splicing. Biochemistry. 1998;37:11924–11931. doi: 10.1021/bi981069a. [DOI] [PubMed] [Google Scholar]
- 23a.Johnstone, R. W., and J. A. Trapani. Unpublished data.
- 24.Kerry J A, Priddy M A, Stenberg R M. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J Virol. 1994;68:4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koul D, Obeyesekere N U, Gutterman J U, Mills G B, Choubey D. p202 self-associates through a sequence conserved among the members of the 200-family proteins. FEBS Lett. 1998;438:21–24. doi: 10.1016/s0014-5793(98)01263-0. [DOI] [PubMed] [Google Scholar]
- 26.Koul D, Lapushin R, Xu H J, Mills G B, Gutterman J U, Choubey D. p202 prevents apoptosis in murine AKR-2B fibroblasts. Biochem Biophys Res Commun. 1998;247:379–382. doi: 10.1006/bbrc.1998.8804. [DOI] [PubMed] [Google Scholar]
- 27.Landolfo S, Gariglio M, Gribaudo G, Lembo D. The Ifi 200 genes: an emerging family of interferon-inducible genes. Biochimie. 1998;80:721–728. doi: 10.1016/s0300-9084(99)80025-x. [DOI] [PubMed] [Google Scholar]
- 28.Lembo D, Angeretti A, Benefazio S, Hertel L, Gariglio M, Novelli F, Landolfo S. Constitutive expression of the interferon-inducible protein p202 in NIH 3T3 cells affects cell cycle progression. J Biol Regul Homeost Agents. 1995;9:42–46. [PubMed] [Google Scholar]
- 29.Lembo D, Sacchi C, Zappador C, Bellomo G, Gaboli M, Pandolfi P P, Gariglio M, Landolfo S. Inhibition of cell proliferation by the interferon-inducible 204 gene, a member of the Ifi 200 cluster. Oncogene. 1998;16:1543–1551. doi: 10.1038/sj.onc.1201677. [DOI] [PubMed] [Google Scholar]
- 30.Lengyel P, Choubey D, Li S-J, Datta B. The interferon-activatable gene 200 cluster: from structure toward function. Sem Virol. 1995;6:203–213. [Google Scholar]
- 31.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 32.Luu P, Flores O. Binding of SP1 to the immediate-early protein-responsive element of the human cytomegalovirus DNA polymerase promoter. J Virol. 1997;71:6683–6691. doi: 10.1128/jvi.71.9.6683-6691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Min W, Ghosh S, Lengyel P. The interferon-inducible p202 protein as a modulator of transcription: inhibition of NF-κB, c-Fos, and c-Jun activities. Mol Cell Biol. 1996;16:359–368. doi: 10.1128/mcb.16.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 35.Ray M E, Su Y A, Meltzer P S, Trent J M. Isolation and characterization of genes associated with chromosome 6-mediated tumor suppression in human malignant melanoma. Oncogene. 1996;12:2527–2533. [PubMed] [Google Scholar]
- 36.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GL1-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 37.Stark G R, Kerr I M, Williams B R G, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 38.Tannenbaum C S, Major J, Ohmori Y, Hamilton T A. A lipopolysaccharide-inducible macrophage gene (D3) is a new member of an interferon-inducible gene cluster and is selectively expressed in mononuclear phagocytes. J Leukoc Biol. 1993;53:563–568. doi: 10.1002/jlb.53.5.563. [DOI] [PubMed] [Google Scholar]
- 39.Trapani J A, Browne K A, Dawson M J, Ramsay R G, Eddy R L, Shows T B, White P C, Dupont B. A novel gene constitutively expressed in human lymphoid cells is induced with interferon-γ in myeloid cells. Immunogenetics. 1992;36:369–376. doi: 10.1007/BF00218044. [DOI] [PubMed] [Google Scholar]
- 40.Trent J M, Stanbridge E J, McBride H L, Meese E U, Casey G, Araujo D E, Witkowski C M, Nagle R B. Tumorigenicity in human melanoma cell lines controlled by introduction of human chromosome 6. Science. 1990;247:568–571. doi: 10.1126/science.2300817. [DOI] [PubMed] [Google Scholar]
- 41.Xie J, Briggs J A, Morris S W, Olson M O J, Kinney M C, Briggs R C. MNDA binds NPM/B23 and NPM-MLF1 chimera generated by the t(3;5) associated with myelodysplastic syndrome and acute myeloid leukemia. Exp Hematol. 1997;25:1111–1117. [PubMed] [Google Scholar]
- 42.Xie J, Briggs J A, Briggs R C. MNDA dimerizes through a complex motif involving an N-terminal basic region. FEBS Lett. 1997;408:151–155. doi: 10.1016/s0014-5793(97)00404-3. [DOI] [PubMed] [Google Scholar]
- 43.Xie J, Briggs J A, Briggs R C. Human hematopoietic cell specific nuclear protein MNDA interacts with the multifunctional transcription factor YY1 and stimulates YY1 DNA binding. J Cell Biochem. 1998;70:489–506. [PubMed] [Google Scholar]
- 44.Yan D H, Wen Y, Spohn B, Choubey D, Gutterman J U, Hung M C. Reduced growth rate and transformation phenotype of the prostate cancer cells by an interferon-inducible protein, p202. Oncogene. 1999;18:807–811. doi: 10.1038/sj.onc.1202369. [DOI] [PubMed] [Google Scholar]
- 45.Yang T H, Tsai W H, Lee Y M, Lei H Y, Lai M Y, Chen D S, Yeh N H, Lee S C. Purification and characterization of nucleolin and its identification as a transcription repressor. Mol Cell Biol. 1994;14:6068–6075. doi: 10.1128/mcb.14.9.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]