Abstract
The CRISPR-based nucleic acid detection systems have shown great potential for point-of-care testing of viral pathogens, especially in the context of COVID-19 pandemic. Here we optimize several key parameters of reaction chemistry and develop a Chemical Enhanced CRISPR Detection system for nucleic acid (termed CECRID). For the Cas12a/Cas13a-based signal detection phase, we determine buffer conditions and substrate range for optimal detection performance, and reveal a crucial role of bovine serum albumin in enhancing trans-cleavage activity of Cas12a/Cas13a effectors. By comparing several chemical additives, we find that addition of L-proline can secure or enhance Cas12a/Cas13a detection capability. For isothermal amplification phase with typical LAMP and RPA methods, inclusion of L-proline can also enhance specific target amplification as determined by CRISPR detection. Using SARS-CoV-2 pseudovirus, we demonstrate CECRID has enhanced detection sensitivity over chemical additive-null method with either fluorescence or lateral flow strip readout. Thus, CECRID provides an improved detection power and system robustness, and helps to develop enhanced reagent formula or test kit towards practical application of CRISPR-based diagnostics.
Keywords: Chemical, CRISPR, Detection, Diagnostics, SARS-CoV-2, COVID-19
1. Introduction
Point-of-care testing (POCT) plays a pivotal role for infectious disease control by ensuring rapid and convenient diagnosis (Chen et al., 2019; Shin et al., 2019). The furious spreading of the novel coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 RNA virus has spurred a huge demand for reliable and field-deployable POCT solutions (Vandenberg et al., 2021). The recent advent of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technology not only revolutionizes genome editing field, but also represents a new paradigm for molecular diagnosis and POCT (Wang et al., 2020b). Several Cas nucleases such as Cas12a, Cas12b, Cas13a and Cas14 possess special collateral cleavage activity to cut nearby single stranded DNA or RNA molecules non-specifically, which is triggered by specific binding of guide RNA (gRNA, a chimeric RNA consisted of target-matching crRNA and trans-acting tracrRNA) along with CRISPR associated (Cas) protein complex to its cognate DNA or RNA target via precise base pairing. Taking advantage of this target-induced trans-cleavage activity as a signal amplifier, people have developed several CRISPR-based DNA or RNA detection methods including SHERLOCK (using Cas13a), DETECTR (Cas12a, Cas14), HOLMES (Cas12a) and CDetection (Cas12b) (Chen et al., 2018; Gootenberg et al. 2017, 2018; Harrington et al., 2018; Kellner et al., 2019; Li et al. 2018b, 2019; Teng et al., 2019). Most of these methods include a target amplification phase to generate ample detection template from a trace amount of start material by varied isothermal amplification approaches such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA). In the following CRISPR detection phase, target-matching gRNA:Cas protein complex undergo conformational change to activate collateral cleavage onto nucleic acid reporter, thereby generating readout signal. The detection results are manifested by either portable fluorescence reader or lateral flow strips for POCT purpose. Inclusion of the target amplification phase can significantly increase the detection sensitivity, specificity and system robustness for CRISPR detection. The two phase reactions can be performed either separately or combined into one-tube reaction, although the latter may have compromised detection power due to constituent complexity and/or promiscuous reaction interference. Using these CRISPR-based methods, people have been trying to develop POCT solutions for monitoring microbial pathogens or genetic variants associated with diseases (van Dongen et al., 2020; Wang et al., 2020a). During the COVID-19 pandemic, several CRISPR-based POCT methods for SARS-CoV-2 RNA detection were proposed (Arizti-Sanz et al., 2020; Broughton et al., 2020; Crone et al., 2020; Ding et al., 2020; Fozouni et al., 2021; Guo et al., 2020; Nguyen et al., 2020; Patchsung et al., 2020; Ramachandran et al., 2020; Wang et al., 2021) and a SHERLOCK-based COVID-19 test has been granted for an Emergency Use Authorization by The U.S. Food and Drug Administration recently (Joung et al., 2020). However, these POCT solutions have not been widely applied yet, compared to the standard reverse transcription-polymerase chain reaction (RT-PCR) approach. Further development or improvement of these CRISPR detection methods is still necessary for broader and more practical applications in POCT settings.
Both CRISPR detection phase and isothermal target amplification phase are in essence composed of multiple enzyme-catalyzing processes. To elevate the detection power, people have been mainly focusing on selecting better effector components (e.g., Cas nuclease, gRNA, reporter, and polymerase), adjusting the stoichiometry of different constituents (e.g., Cas protein:gRNA ratio) (Guo et al., 2020; Joung et al., 2020) or modifying gRNA structure to augment trans-cleavage activity of Cas protein (Nguyen et al., 2020). In addition to manipulating detection components, changing the reaction environment by either optimizing buffer condition (e.g., use Mn2+ instead of Mg2+) or adding chemical additives into reaction systems represent another directions to increase detection capability (Joung et al., 2020; Ma et al., 2020). Chemicals such as polyols, polymers, amino acids and their derivatives have been used as solvent additives to stabilize proteins and/or affect solvent properties, thereby influencing the kinetics of reaction chemistry (Simpson, 2010). Researchers have shown that chemical additives such as DMSO, glycerol and betaine can enhance the efficiency and/or specificity of some PCR or isothermal amplification reactions (Henke et al., 1997; Jensen et al., 2010; Luo et al., 2019; Nagai et al., 1998; Ralser et al., 2006; Varadharajan and Parani, 2021). However, systematic study is still required to examine whether these chemical additives may affect the detection power of CRISPR-based nucleic acid detection, in hope of searching the reaction enhancer for CRISPR diagnostics.
To determine the optimal reaction condition for Cas12a/Cas13a-mediated nucleic acid detection, here we examined several reaction parameters within the systems and evaluated several chemical additives for their effects on both target amplification and CRISPR detection phases. We found that L-proline can consistently enhance the reaction efficiency and specificity for both phases, and could serve as a practical additive for the reaction package of CRISPR-mediated nucleic acid detection. Applying this Chemical Enhanced CRISPR Detection system for nucleic acid (termed CECRID) in detecting SARS-CoV-2 pseudovirus demonstrated an improved detection power over additive-null methods, making CECRID a promising strategy towards further practical use.
2. Materials and methods
2.1. Constructs and reagents
The coding regions of LwaCas13a (Leptotrichia wadeii Cas13a) (Abudayyeh et al., 2017), AsCas12a (Acidaminococcus sp. Cas12a) (Li et al., 2018a) and LbCas12a (Lachnospiraceae bacterium Cas12a) (Li et al., 2018a) were inserted into pET28a expression vector. N gene fragments (two different regions: #1 and #2) from SARS-CoV-2 viral genome were inserted into pHAGE-EF1α-puro vector for pseudovirus detection. Oligonucleotides used in this study (Table S2) were synthesized from HuaGene Biotech (Shanghai, China), Synbio Technologies (Suzhou, China) and GENEWIZ (Suzhou, China). Detailed information about reagents and instruments, including the commercial vendors and item numbers, is provided in Table S3.
2.2. Nucleic acid preparation
For preparation of DNA templates of SARS-CoV-2 ORF1ab, N and S genes, PCR amplification was performed by indicated primers with the forward primer containing an appended T7 promoter sequence using the template prepared through annealing of two synthetic oligonucleotides (Table S2). For preparation of RNA templates and crRNAs, in vitro transcription (IVT) was performed with T7 promoter-inclusive DNA templates. RNA templates and crRNAs were purified by RNA Clean & Concentrator-5 Kit (Zymo research), and quantified by the high sensitivity RNA Qubit fluorometer (Thermo Fisher).
2.3. Cas protein expression and purification
Protein expression plasmids were transformed into Escherichia coli. Rosseta™ 2 (DE3)pLySs competent cells. Isopropyl β-D-thiogalactoside (IPTG) was used to induce protein expression. Cell pellet was resuspended in lysis buffer and sonicated. HisPur™ Cobalt Resin (Thermo Fisher, #89964) was utilized to pull-down the protein. Zeba™ Spin Desalting Columns (Thermo Fisher, #89890) was used to desalt protein. Amicon® Ultra-15 Centrifugal Filter Devices (Millipore, #UFC905008) was used to concentrate the protein and exchange the storage buffer as well. Purified protein was stored at −80 °C as 10 μL aliquots at a concentration of 2 mg/mL.
2.4. CRISPR detection assay
The standard RNA detecting LwaCas13a-based assay was performed at 37 °C with 1x CutSmart buffer, 100 nM LwaCas13a protein, 100 nM crRNA, 250 nM RNA reporter and 1 μL of nucleic acid target in a 20 μL reaction system. For DNA detection system, CRISPR-AsCas12a and CRISPR-LbCas12a systems were applied. The standard reaction systems of Cas12a-based nucleic acid detection consisted of 1x CutSmart buffer, 50 nM AsCas12a protein or LbCas12a protein, 50 nM crRNA, 250 nM DNA reporter, and 1 μL of nucleic acid target or amplification products in a final volume of 20 μL at 37 °C for 1 h. Fluorescent reporter (RNA FQ reporter: 5′ -/6-FAM/UUUUUU/BHQ1/-3′; DNA FQ reporter: 5′ -/6-FAM/TTATT/BHQ1/-3′) was added in CRISPR detection mix. Fluorescence signal was dynamically measured by QuantStudio™ 5 Real-Time PCR System (Thermo Fisher). Visual detection was accomplished by imaging the tubes through E-Gel™ Safe Imager™ Real-Time Transilluminator (Thermo Fisher). For lateral flow strip assays, lateral flow cleavage reporter (5′ -/6-FAM/TTATTATT/Biotin/-3′) was added to the reaction and incubate at 37 °C for 1 h. Lateral flow strip (Milenia Biotec GmbH, # MGHD 1) was used to visualize the results.
2.5. LAMP assay
One-step RT-LAMP mix was assembled with 1 μL 10x isothermal amplification buffer, 1.4 mM of dNTPs, 6.5 mM MgSO4, 3.2 U Bst 2.0 (NEB, #M0538S), 3 U WarmStart RTx Reverse Transcriptase (NEB, #M0380S), 0.2 μM F3/B3 primers, 1.6 μM FIP/BIP primers, 0.8 μM LF/LB primers, and 1 μL of RNA template in a 10 μL volume, and then incubated at 62 °C for 15–30 min. Primers for LAMP are listed in Table S2. The heat inactivation (80 °C for 20 min) of LAMP product was performed to reduce the background signal before proceeding to detection assays.
2.6. RPA assay
RPA assays were set up using commercial RPA kit (TwistDx). One step RT-RPA reaction system was composed of 9 μL of RPA solution (primer-free rehydration buffer), 224 mM of MgOAc, 40 U of ProtoScript II Reverse Transcriptase (NEB, #M0368S), 0.5 μM of forward primer, 0.5 μM of reverse primer and UltraPure water to a total volume of 16 μL. Primers for RPA are listed in Table S2. The mixture was incubated at 40 °C for 30 min, followed by heat inactivation for 20 min at 80 °C.
2.7. Pseudovirus production and detection
HEK293FT cells were employed to pack the SARS-CoV-2 pseudoviral particles. Cells were cultured with DMEM medium supplemented with 10% fetal bovine serum. DNA Transfection was performed in 6-well plates by Lipofectamine™ 2000 Transfection Reagent with a mix of 1.5 μg pHAGE-EF1α-puro plasmid carrying SARS-CoV-2 N gene fragment (#1 and #2), 0.75 μg pCMVR8.74, and 0.5 μg pMD2.G. The viral particle-containing supernatant was harvested at 48 h post transfection. To simulate the actual application, viral particles were firstly transferred into the Viral Transport Media (VTM) of Sample Collection Kit (BEAVER, #43903) as a mimic of nasopharyngeal swab collected sample. Viral RNA was extracted and RT-LAMP was performed followed by AsCas12a-based detection.
2.8. Statistical analysis
Statistic significances were calculated by GraphPad Prism 8.4.0 using two-way ANOVA with Sidak's multiple comparisons test and unpaired two-tailed t-test. All the data were shown as mean ± s.d.
More details of Materials and Methods can be found in Supplementary material.
3. Results
3.1. Optimization of key parameters for CRISPR detection systems
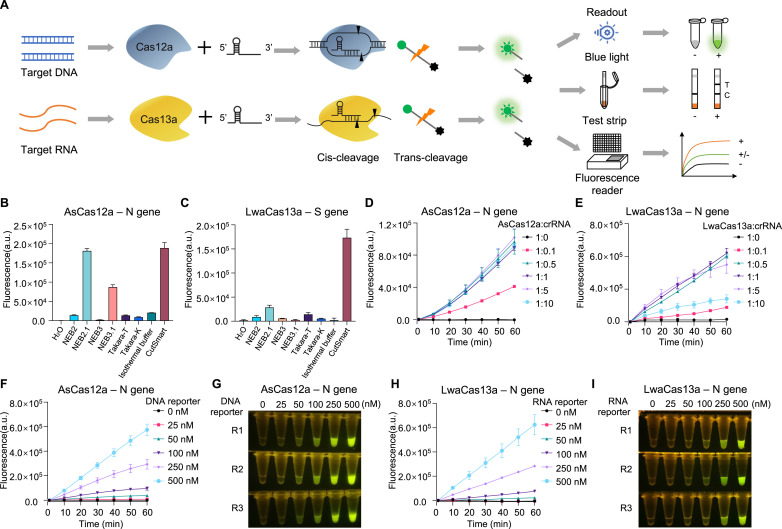
The DNA-targeting Cas12a and RNA-targeting Cas13a systems represent two major branches of the CRISPR machineries for nucleic acid detection (Fig. 1 A). To compare and optimize these CRISPR detection systems, we set up three independent assays with purified AsCas12a, LbCas12a and LwaCas13a proteins (Fig. S1). Fragments of ORF1ab, N and S genes from SARS-CoV-2 RNA genome were chosen as nucleic acid templates for detection. Target amplification regions by either LAMP or RPA method are denoted in Fig. S2A. Different crRNAs for respective Cas effectors were designed and synthesized by in vitro transcription (Fig. S2A). Fluorophore-labeled DNA reporter or RNA reporter were employed as signal readout for Cas12a and Cas13a systems, respectively (Fig. 1A).
Fig. 1.
Optimization of key parameters for CRISPR detection systems.
(A) Schematic description of CRISPR-Cas12a/Cas13a detection systems for nucleic acids.
(B, C) Comparison of the indicated commercial reaction buffers for their effects on AsCas12a- and LwaCas13a-mediated trans-cleavage activity. A synthetic DNA template corresponding to SARS-CoV-2 N gene (B), and a synthetic RNA template corresponding to SARS-CoV-2 S gene (C) are used. (D, E) Evaluation of the indicated molecular ratio of Cas protein:crRNA for the effect on AsCas12a detection (D) and LwaCas13a detection (E). (F–
I) The effect of indicated amount of fluorescence reporter on CRISPR detection assays using either AsCas12a (F and G) or LwaCas13a (H and I). The results are shown by either real-time recording of fluorescence signal (F and H) or endpoint (60
min) visualization (G and I) under blue light illuminator. R1, R2 and R3 indicate three replicates. Error bars represent mean ± s.d. (n = 3). a. u., arbitrary unit. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
With synthetic ORF1ab and S DNA templates, we found that AsCas12a consistently has an enhanced detection sensitivity over LbCas12a in our hands as determined by the fluorescence signal (Fig. S2B and C). We therefore mainly utilized AsCas12a for the following assays. To determine the optimal conditions for CRISPR-based detection, we evaluated several key parameters that affect the assay performance. Firstly, we compared different commercially available buffer systems for their effects on AsCas12a- or LwaCas13a-mediated detection with synthetic DNA or RNA templates. Interestingly, AsCas12a exhibited fair compatibility to several buffers (e.g., NEB2.1, NEB3.1 and CutSmart), whereas LwaCas13a seemed to be quite sensitive to buffer conditions (Fig. 1B and C). The commercial buffer CutSmart outperformed any other buffers tested for both Cas12a and Cas13a systems (Fig. 1B and C, Fig. S2D and E). Secondly, we determined the optimal range of Cas protein:crRNA ratio for the detection capability. AsCas12a displayed a saturated activity when the molecular ratio of Cas protein:crRNA sits between 1:0.5–1:10 (Fig. 1D). More crRNAs did not increase the fluorescence signal as AsCas12a amount is restricted (Fig. 1D). In contrast, LwaCas13a showed a narrower window of Cas protein:crRNA ratio (1:0.5–1:5) for maximal detection activity (Fig. 1E). Too much crRNA over LwaCas13a (at a ratio of 1:10) rather significantly decreased the fluorescence signal (Fig. 1E), suggesting the importance of appropriate stoichiometry of Cas effector and crRNA in Cas13a-mediated assays. Thirdly, we monitored the effect of DNA or RNA reporter amount on the detection signals. As expected, the fluorescence intensity was positively correlated with the amount of reporter within the tested range for both AsCas12a and LwaCas13a (Fig. 1F–I), These results suggest that the detection efficiency and signal strength could be elevated with increased amount of reporter on top of the fixed amount of other components.
3.2. Dynamic detection range of target amount
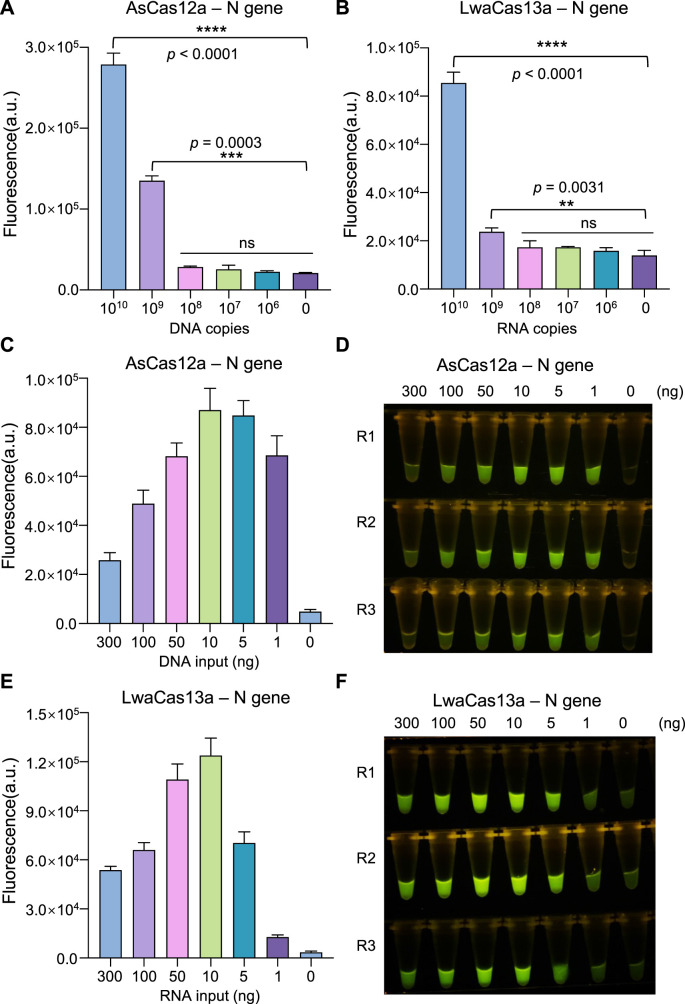
Using the above optimized parameters, we determined the detection sensitivity of AsCas12a and LwaCas13a systems with pure DNA or RNA target templates. Both systems reached a significant signal detection threshold over the background when the target DNA or RNA had 109 copies (~1.66 × 10−9 mol/L in the reaction system) or more under indicated conditions in our hands (Fig. 2 A and B). In practical scenarios of POCT, the start materials are often limited. To improve the detection efficiency and system robustness, people usually incorporate an additional target amplification step by various isothermal amplification methods to generate ample target molecules for CRISPR detection. To determine the relationship between target abundance and CRISPR detection performance, we used differential amount of pure nucleic acid targets as CRISPR substrate and examined the corresponding signal strength. As the target substrate increased gradually, the detection signals from both AsCas12a and LwaCas13a initially correlated well with the target abundance, which is consistent with previous report (Gootenberg et al., 2017). However, the signals then started to drop after reaching the plateau as the targets continued to increase (Fig. 2C–F). It is surprising that too much redundant target substrates rather had an inhibitory effect, if not incremental or stable, on the detection system. We speculate that excess targets may competitively bind to crRNAs before the latter form complex with Cas proteins, thereby interfering the generation of productive target:gRNA:Cas protein complex. This effective target amount window is important for CRISPR detection systems to achieve optimal capacity and should be especially considered in assays conjugated with a target amplification phase.
Fig. 2.
Dynamic detection range of target amount
(A, B) Evaluation of the detection sensitivity of AsCas12a (A) and LwaCas13a (B) systems with indicated amount of synthetic templates. (C, D) AsCas12a-mediated detection power for indicated amount of pure targets using plasmid bearing SARS-CoV-2 N gene fragment. The fluorescence signals are shown by either real-time recording of fluorescence signal (C) or endpoint (60
min) visualization (D) under blue light illuminator. R1, R2 and R3 indicate three replicates. (E, F) LwaCas13a-mediated detection power for indicated amount of pure targets using in vitro transcribed RNAs corresponding to SARS-CoV-2 N gene fragment. Error bars represent mean ± s.d. (n = 3). a. u., arbitrary unit. Unpaired two-tailed t-test, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns means not significant. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Chemical additive enhancement for CRISPR detection phase
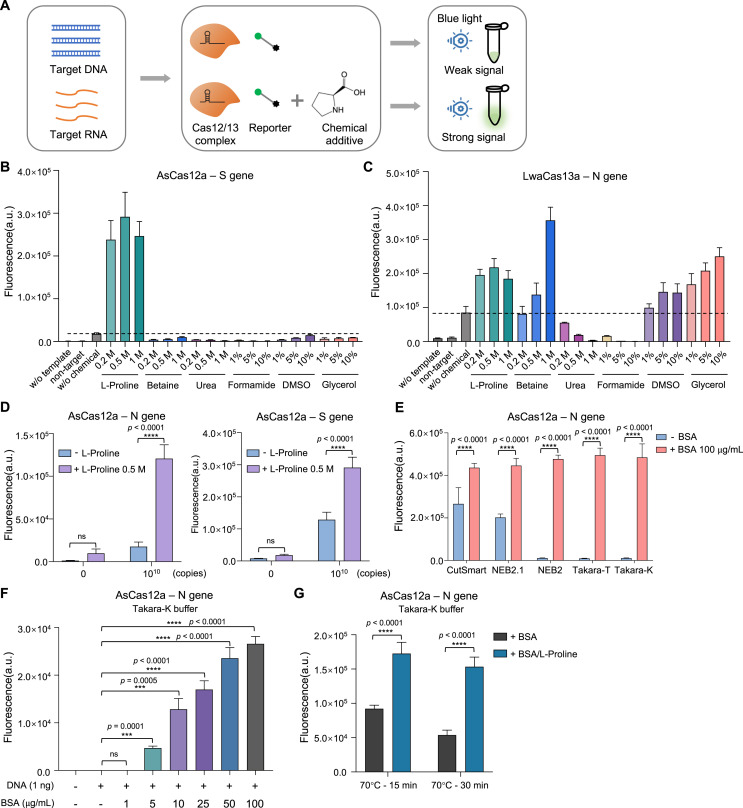
To further improve the efficiency of CRISPR detection systems, we tried to determine whether chemical additives may have some positive effects. Several widely used chemical additives were chosen and added into CRISPR detective reactions (Fig. 3 A). For AsCas12a-mediated S gene DNA detection, addition of L-proline can greatly increase the detection efficiency compared to control samples without chemical additive (Fig. 3B; Fig. S3A). On the other hand, using N gene RNA template as substrate for LwaCas13a system, L-proline, betaine, DMSO and glycerol displayed differential but significant signal enhancement over the control (Fig. 3C).
Fig. 3.
Effects of chemical additives on CRISPR detection phase
(A) Schematic diagram showing the workflow to evaluate chemical additive effect on CRISPR detection system. (B, C) Endpoint (60
min) recording of fluorescence detection signals for either AsCas12a (B) or LwaCas13a (C) system. w
/o template means no input; non-target means non-targeting template (N gene template as S gene non-target, and vice versa); w/o chemical means no chemical addition. (D) Comparison of fluorescent signals resulted from AsCas12a-based detection with or without L-proline addition. Two-way ANOVA test, ****p < 0.0001; ns means not significant. (E) The effect of BSA addition on AsCas12a-based detection system. Two-way ANOVA test, ****p < 0.0001. (F) Comparison of different concentrations of BSA addition for the effects on AsCas12a-mediated detection. Unpaired two-tailed t-test, ***p < 0.001, ****p < 0.0001. (G) Evaluation of L-proline's effect on protecting BSA from heat-induced denature and BSA's capability to enhance AsCas12a detection. BSA along or BSA co-incubated with L-proline are treated at 70 °C for 15 min or 30 min, before serving as additive in AsCas12a assays. Two-way ANOVA test, ****p < 0.0001. Error bars represent mean ± s.d. (n = 3). a. u., arbitrary unit.
Notably, L-proline, among all the tested chemicals, exhibited the most dramatic and consistent effect on both detection systems. We further verified the enhancement effect of 0.5 M L-proline on AsCas12a-based detection assays using both N and S gene DNA templates, suggestive of target- and crRNA-independence of L-proline's enhancement effect (Fig. 3D). We postulate that L-proline may help to maintain or enhance the Cas effector activity as a protein stabilizer and refolding chaperone. To test this hypothesis, we prepared different types of AsCas12a protein with potentially differential basal activities (type 1: fresh protein from frozen stock; type 2: protein left at room temperature for 48 h; and type 3: protein undergone multiple freeze-thaw cycles during 48 h). As expected, the basal activities of AsCas12a protein in CutSmart buffer batch #1 declined gradually from type 1 to type 3 (Fig. S3B). Interestingly, the addition of L-proline can significantly elevate the signal strength for all the three types of proteins in a dose-dependent manner (Fig. S3B). Unexpectedly, when the similar tests were performed in CutSmart buffer batch #2 rather than buffer batch #1, the difference between basal activities of three types of AsCas12a are gone as type 2 and type 3 samples recovered their detection strength to the similar level of type 1 sample (Fig. S3B-C). In addition, L-proline did not show enhancement effect in CutSmart buffer batch #2 (Fig. S3C). These batch deviated results suggest that certain ingredients within CutSmart buffer #2 may retain the activity of AsCas12a even if after mild deterioration treatment, and L-proline's effect on AsCas12a is probably masked by such endogenous signal recovery. In contrast, when AsCas12a protein was harshly denatured under 42 °C heat stress, the decline of detection activities cannot be reversed back to normal range by the CutSmart buffer #2 (Fig. S3D). However, addition of L-proline in detection mix retarded the heat-induced activity decline (Fig. S3D), suggesting that L-proline may protect the activity loss of Cas nuclease against unfavorable stresses or conditions.
3.4. BSA as an essential signal enhancer in CRISPR detection buffer
The significant batch effect of reaction buffers on the detection results and L-proline's effect led us to interrogate the endogenous signal enhancers within the buffer. After scrutinizing and comparing buffer components between active and inactive buffers (Table S1), we found that the presence or absence of bovine serum albumin (BSA) was highly correlated with the detection signal especially for AsCas12a (Fig. 1B and C). To test the potential function of BSA, we examined the detection activities of AsCas12a with N gene DNA template using different buffer systems while adding exogenous BSA. The tested buffers were chosen with similar salt and buffering constituents but mainly differed in the BSA content (BSA-inclusive buffers: CutSmart and NEB2.1; BSA-null buffers: NEB2, Takara-T and Takara-K) (Table S1). As shown in Fig. 3E, the basal detection activity of BSA-inclusive buffers are significantly higher than BSA-null buffers, and addition of BSA into all the three BSA-null buffers drastically elevated their detection signals from the bottom to the saturated level. Moreover, inclusion of BSA alone in custom SHERLOCK buffer can significantly increase the detection signal for both AsCas12a and LwaCas13a systems (Fig. S2D and E). In contrast, adding L-proline alone into BSA-null buffers (Takara-T and Takara-K) cannot enhance the detection signal (Fig. S3E). Furthermore, the enhancement effect of BSA is concentration-dependent, and the signal increase required the presence of target template which exclude the possibility that BSA's addition had direct cleavage activity on the signal reporter (Fig. 3F; Fig. S3F). Thus, BSA is a bona fide signal enhancer for CRISPR detection whereas L-proline may act as a safeguard in the buffer system to protect BSA or Cas protein from unfavorable stresses. Consistently, we did find that L-proline can protect BSA from heat stress and recover the signal enhancement effect of BSA on CRISPR detection (Fig. 3G). Since BSA itself is a protein in essence and therefore requires more stringent manufacturing, transportation and storage conditions, the BSA-inclusive buffers such as CutSmart may display batch effect on CRISPR detection resulted from differential quality of endogenous BSA (Fig. S3G and H). L-proline's effect was more pronounced in buffers with low basal activity and probably deteriorated BSA's function such as batch #3 and #6, and this phenomenon was consistently shown in both AsCas12a- and LwaCas13a-based assays (Fig. S3G and H). The additional inclusion of L-proline in BSA-inclusive buffers may largely maintain the detection potential, antagonize the mild deterioration and represent the optimal reaction buffer system.
3.5. Effects of chemical additives on target amplification phase
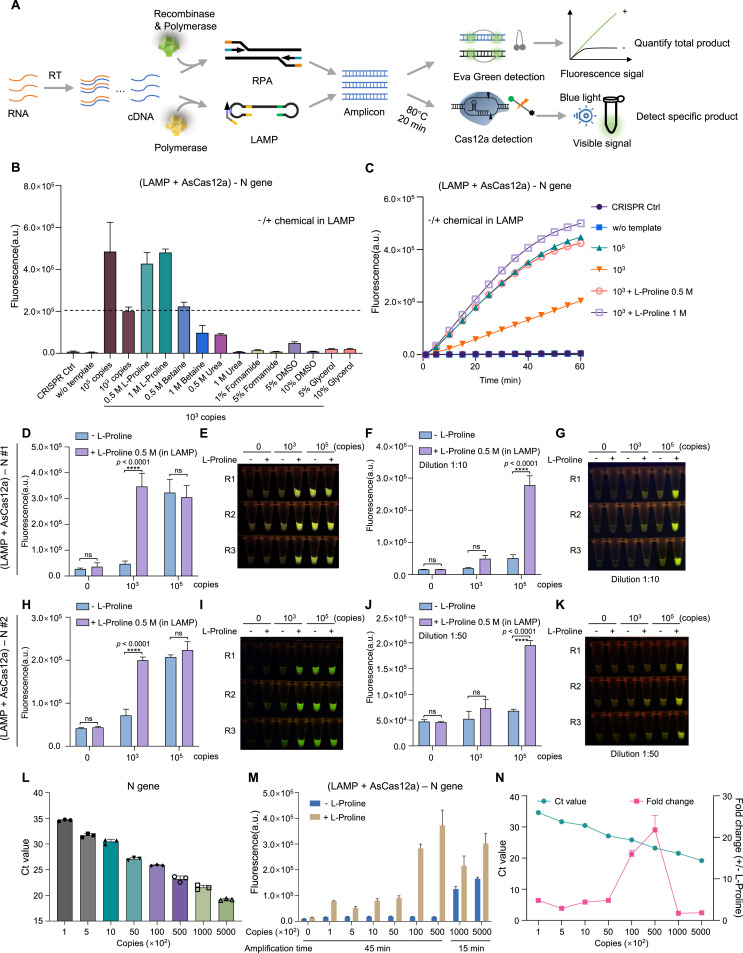
Pre-amplification of target molecules is often a requisite step before CRISPR detection phase to achieve the best detection sensitivity and specificity. LAMP and RPA are the most representative isothermal target amplification methods conjugated with CRISPR detection thus far (Fig. 4 A). Using synthetic RNA template of SARS-CoV-2 N gene fragment, we performed LAMP-based target amplification. Total product yield accumulated quickly as reaction continued and apparent nonspecific product arose in non-template samples as determined by either ethidium bromide staining or fluorescent DNA binding dye quantification (Fig. S4A and B). We then employed AsCas12a system to detect specific target signals, and found that LAMP-based two-step detection can significantly catch the signal from 103 copies of template within 15 min of amplification (Fig. S4C). Next, we tried to determine whether chemical additives could affect the efficiency and specificity of LAMP by inclusion of the tested additives within the reaction mix. As shown in Fig. S4D, none of the tested chemicals can increase the gross yield of LAMP according to the signal kinetics captured by fluorescence reader using sequence-independent DNA-binding dye. Interestingly, when checking out the specific target amplification by CRISPR detection, only addition of L-proline (0.5 M and 1 M), among other tested chemicals, into LAMP reaction mix can significantly enhance the specific signal (Fig. 4B and C). This enhancement effect was further reproduced by two independent LAMP-CRISPR two-step assays targeting SARS-CoV-2 N gene (Fig. 4D–K). Moreover, using precisely quantified standard SARS-CoV-2 reference RNA, we observed a varied but consistent enhancement effect of L-proline on AsCas12a detection over a broad range of input materials (Fig. 4L–N).
Fig. 4.
Effects of chemical additives on target amplification phase
(A) Schematic diagram showing the workflow to evaluate LAMP and RPA detection for target RNAs. (B) Evaluation of different chemical additives for the effect on specific target amplification as determined by AsCas12a detection using purified LAMP products. LAMP is performed for 15
min at 62 °C. Endpoint (60 min) recording of fluorescence detection signals are shown. CRISPR Ctrl indicates no nucleic acid input for AsCas12a detection; w/o template means no input for LAMP reaction. (C) The fluorescence signal dynamics is shown for the selected samples in (B). (D–G) Comparison of L-proline addition in LAMP reaction phase determined by AsCas12a. The template and primer set for this LAMP reaction is quoted from Broughton et al. (N #1). LAMP is performed for 30 min at 62 °C. Purified LAMP products (D and E) or after proper dilution (F and G) are subjected to AsCas12a detection. (H–K) Similar assays are performed using different template and primer set for LAMP reaction according to Joung et al. (N #2). (L) qRT-PCR quantification of varied SARS-CoV-2 reference RNA. (M) Effect of L-proline on AsCas12a detection with varied amount of template. (N) Correlation of Ct values and enhancement fold change of AsCas12a detection when adding L-proline. Two-way ANOVA test, ****p < 0.0001. ns means not significant. Error bars represent mean ± s.d. (n = 3). a. u., arbitrary unit.
We also performed similar analysis for another isothermal amplification method RPA (Fig. 4A). Both RPA and LAMP eventually produced significant background signals in empty control sample without target template as the run lasted for 1 h, possibly due to the trade-off resulted from the easy and super-quick sparking principles of the methods (Fig. S4A and B; S5A). However, these non-specific signals usually came late or weaker than target amplification wells, and target-derived specific signals still showed a time- and concentration-dependent increase by gRNA-mediated CRISPR detection (Fig. S4C; Fig. S5B). Similar to LAMP, we did not find any chemical additive with significant enhancement effect on gross yield of RPA assay to amplify S gene RNA template (Fig. S5C). On the other hand, for specific target amplification determined by two-phase RPA-CRISPR assays, we observed that several chemical additives such as L-proline, betaine, DMSO and glycerol displayed varied but significant boost of specific signals (Fig. S5D-F). Again, L-proline showed the most comprehensive compatibility and effectiveness in reinforcing target amplification by either isothermal method in two-phase CRISPR detection assays. We also examined the effect of BSA addition on LAMP- and RPA-based assays using N gene RNA template. BSA addition into LAMP mix seemed to have little effect on specific target amplification, whereas in RPA assays BSA might have a beneficial role by promoting specific target amplification (Fig. S6).
3.6. Additional factors affecting signal-to-noise ratio of CRISPR detection
Given the high background nature of these fast isothermal amplification methods, it is critical to increase the signal-to-noise ratio for two-step CRISPR detection. In addition to chemical enhancement of the two phases, we also tried nested-RPA by introducing additional pair of RPA primers and performing two rounds of RPA amplification. Using ORF1ab gene RNA fragment as template, we observed significant increase of detection signal from nested-RPA-AsCas12a assay (Fig. S7A). This nested primer approach may be less applicable for LAMP since there are already three pairs of primers with appreciable complexity within LAMP reaction system.
During our practice of two-phase CRISPR detection assay using RNA as template, we occasionally observed apparent background signals in both negative control and tested samples, which is not the case in assays with DNA template. Interestingly, this background signal can be removed if the input material from the pre-amplification step was heat-treated at 80 °C for 20 min before proceeding to AsCas12a-mediated trans-cleavage assay (Fig. S7B). Since DNA-targeting assay also contains the pre-amplification step but without such background signal, we reasoned that the key for this phenomenon might lie in the reverse transcription (RT) step which is a special difference between RNA- and DNA-targeting assays. We inferred that the input materials for CRISPR detection from RT-LAMP or RT-RPA step may contain some leftover of reverse transcriptase and dNTPs, which may cross-react with crRNAs in CRISPR detection mix to initiate promiscuous DNA production, thereby resulting in occasional background signals. To test this hypothesis, we added reverse transcriptase directly into LAMP-AsCas12a assay using N gene DNA template. The fluorescence signal was increased in reverse transcriptase-containing sample compared to control and 80 °C heat-inactivation can effectively blunt this background increase, demonstrating that reverse transcriptase is the primary source of the background interference (Fig. S7B-D). These results suggest that it may be advisable to take precautionary actions such as heat inactivation or DNA purification prior to CRISPR detection for reducing unwanted noise from reverse transcriptase in related assays.
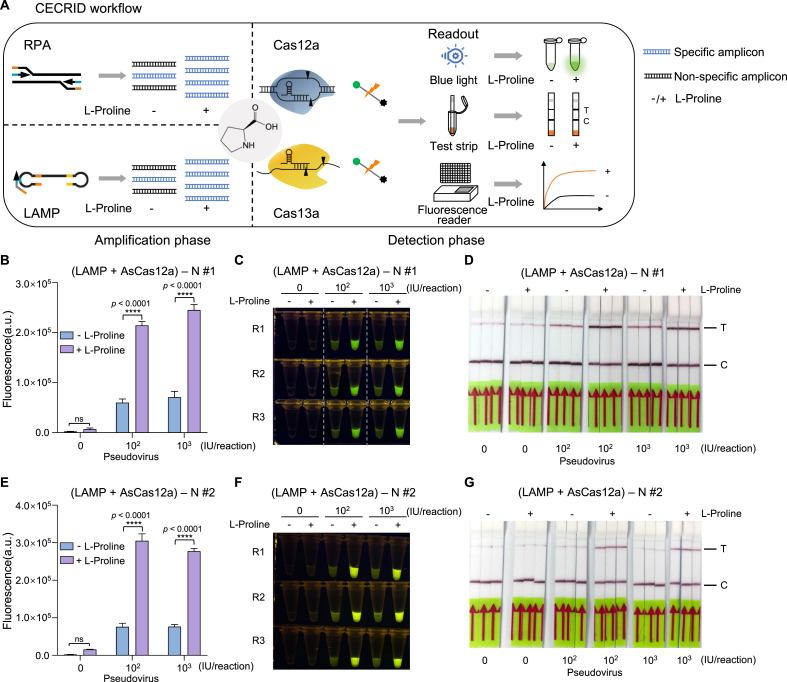
3.7. CECRID deployment for SARS-CoV-2 pseudovirus detection
With above optimized conditions and L-proline embedded in both target amplification phase and CRISPR detection phase, we established a Chemical Enhanced CRISPR Detection system for nucleic acid (termed CECRID) (Fig. 5 A). To further validate the performance of CECRID in more practical settings, we packaged SARS-CoV-2 RNA fragments into VSVG-pseudotyped lentiviral particles to mimic live viruses for detection purpose. Quantified pseudoviral particles were directly added into viral transport media (VTM) to mimic samples collected from nasopharyngeal swab, and the viral RNA extraction was performed with commercial kit according to the standard procedures. The SARS-CoV-2 N gene viral RNA was detected by RT-LAMP-AsCas12a two-phase assays with two independent setup using different pseudoviruses/primers/crRNAs. Compared to standard assay without chemical additive, CECRID can significantly enhance the detection sensitivity of SARS-CoV-2 N gene RNA from pseudoviral particle input by either fluorescence reader or lateral flow strips (Fig. 5B–G). These results highlight the potential of CECRID application in real POCT settings.
Fig. 5.
CECRID deployment for detection of SARS-CoV-2 pseudovirus
(A) Schematic diagram showing the principle and application workflow of CECRID systems.
(B–
D) Comparative analysis of SARS-CoV-2 N gene-bearing pseudovirus detection with CECRID and normal detection platforms. The template and primer set for this LAMP reaction is N#1. Various amount of pseudoviral particles in VTM are collected and viral RNA is purified by commercial RNA purification kit. RT-LAMP is performed with viral RNA at 62 °C for 30 min followed by 20 min of 80 °C inactivation. The purified amplification products (1:50 dilution) are then subjected to AsCas12a-based fluorescence and lateral flow strip detection. For CECRID, L-proline is added in both the LAMP phase and CRISPR detection phase. The endpoint (60 min) fluorescence signal is shown by either bar plot (B), direct visualization (C) or lateral flow strip using biotin-labeled reporter (D). The significant band in test line represents positive result. C: control line. T: test line. (E–G) Similar assays are performed using an independent pseudovirus/template and primer set N #2. Two-way ANOVA test, ****p < 0.0001. ns means not significant. Error bars represent mean ± s.d. (n = 3). a. u., arbitrary unit.
4. Conclusion
This work presents a chemical-enhanced CRISPR detection system CECRID by optimizing the key parameters of reaction chemistry and inclusion of L-proline into the reaction mix. The effect of several chemical additives on isothermal amplification and CRISPR detection systems are evaluated. BSA displays a drastic enhancement for CRISPR detection and should be included as a necessary component in the reaction buffer. L-proline can further reinforce the detection power through both target amplification and CRISPR detection phases. The enhancement effect of our CECRID platform have been reproduced with multiple Cas effectors, different target regions of templates, independent primer/crRNA sets and various vendors/batches of reagents, indicating the well generality and great applicability of the enhancement system.
Despite our restricted access to real clinical samples, we corroborated the improved detection performance of CECRID for VTM-collected swab-like samples using SARS-CoV-2 pseudovirus as well as standard SARS-CoV-2 reference RNA samples. As an improved CRISPR detection system, CECRID can be applied in more broad scenarios of nucleic acid detection. In addition to fluorescence or lateral flow strip readout, CECRID, as an enhanced reaction system, could be combined with other advanced or integrated detection systems such as electrochemical microfluidic biosensor to develop more practical and powerful diagnostic tools (Bruch et al. 2019, 2021). Taken together, our work not only uncovers several key parameters affecting the strength of nucleic acid detection, but also paves the way for developing robust reagent formula or test kit towards practical POCT application of CRISPR-based detection technology.
CRediT author contribution statement
Zihan Li: Conceptualization, Investigation, Roles/Writing - original draft, Writing - review & editing. Wenchang Zhao: Investigation, Roles/Writing - original draft, Writing - review & editing. Shixin Ma: Investigation, Roles/Writing - original draft, Writing - review & editing. Zexu Li: Investigation. Yingjia Yao: Investigation. Teng Fei: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Roles/Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: A patent has been filed through Northeastern University related to this work.
Acknowledgements
We thank Drs. Tengfei Xiao, Yue Feng and Ren Sheng for sharing reagents. This work was supported by the National Natural Science Foundation of China (31871344, 32071441), the Fundamental Research Funds for the Central Universities (N182005005, N2020001), the 111 Project (B16009), and LiaoNing Revitalization Talents Program (XLYC1807212).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113493.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., Lander E.S., Voytas D.F., Ting A.Y., Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.F., Kemball M.E., Uwanibe J.N., Ajogbasile F.V., Eromon P.E., Gross R., Wronka L., Caviness K., Hensley L.E., Bergman N.H., MacInnis B.L., Happi C.T., Lemieux J.E., Sabeti P.C., Myhrvold C. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020;11(1):5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruch R., Baaske J., Chatelle C., Meirich M., Madlener S., Weber W., Dincer C., Urban G.A. CRISPR/Cas13a-Powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 2019;31(51) doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- Bruch R., Johnston M., Kling A., Mattmuller T., Baaske J., Partel S., Madlener S., Weber W., Urban G.A., Dincer C. CRISPR-powered electrochemical microfluidic multiplexed biosensor for target amplification-free miRNA diagnostics. Biosens. Bioelectron. 2021;177:112887. doi: 10.1016/j.bios.2020.112887. [DOI] [PubMed] [Google Scholar]
- Chen H., Liu K., Li Z., Wang P. Point of care testing for infectious diseases. Clinica chimica acta; international journal of clinical chemistry. 2019;493:138–147. doi: 10.1016/j.cca.2019.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone M.A., Priestman M., Ciechonska M., Jensen K., Sharp D.J., Anand A., Randell P., Storch M., Freemont P.S. A role for Biofoundries in rapid development and validation of automated SARS-CoV-2 clinical diagnostics. Nat. Commun. 2020;11(1):4464. doi: 10.1038/s41467-020-18130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Yin K., Li Z., Lalla R.V., Ballesteros E., Sfeir M.M., Liu C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11(1):4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni P., Son S., Diaz de Leon Derby M., Knott G.J., Gray C.N., D'Ambrosio M.V., Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184(2):323–333. doi: 10.1016/j.cell.2020.12.001. e329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R.R., Zhou Q., Li W. SARS-CoV-2 detection with CRISPR diagnostics. Cell discovery. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke W., Herdel K., Jung K., Schnorr D., Loening S.A. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 1997;25(19):3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M.A., Fukushima M., Davis R.W. DMSO and betaine greatly improve amplification of GC-rich constructs in de novo synthesis. PloS One. 2010;5(6) doi: 10.1371/journal.pone.0011024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N.G., Woolley A.E., Segel M., Barretto R.P.J., Ranu A., Macrae R.K., Faure G., Ioannidi E.I., Krajeski R.N., Bruneau R., Huang M.W., Yu X.G., Li J.Z., Walker B.D., Hung D.T., Greninger A.L., Jerome K.R., Gootenberg J.S., Abudayyeh O.O., Zhang F. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. HOLMESv2: a CRISPR-cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Li S.Y., Cheng Q.X., Liu J.K., Nie X.Q., Zhao G.P., Wang J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28(4):491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.Y., Cheng Q.X., Wang J.M., Li X.Y., Zhang Z.L., Gao S., Cao R.B., Zhao G.P., Wang J. CRISPR-Cas12a-assisted nucleic acid detection. Cell discovery. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G.C., Yi T.T., Jiang B., Guo X.L., Zhang G.Y. Betaine-assisted recombinase polymerase assay with enhanced specificity. Anal. Biochem. 2019;575:36–39. doi: 10.1016/j.ab.2019.03.018. [DOI] [PubMed] [Google Scholar]
- Ma P., Meng Q., Sun B., Zhao B., Dang L., Zhong M., Liu S., Xu H., Mei H., Liu J., Chi T., Yang G., Liu M., Huang X., Wang X. MeCas12a, a highly sensitive and specific system for COVID-19 detection. Advanced science. 2020:2001300. doi: 10.1002/advs.202001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M., Yoshida A., Sato N. Additive effects of bovine serum albumin, dithiothreitol, and glycerol on PCR. Biochem. Mol. Biol. Int. 1998;44(1):157–163. doi: 10.1080/15216549800201172. [DOI] [PubMed] [Google Scholar]
- Nguyen L.T., Smith B.M., Jain P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020;11(1):4906. doi: 10.1038/s41467-020-18615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., Eiamthong B., Lakkanasirorat B., Phoodokmai T., Niljianskul N., Pakotiprapha D., Chanarat S., Homchan A., Tinikul R., Kamutira P., Phiwkaow K., Soithongcharoen S., Kantiwiriyawanitch C., Pongsupasa V., Trisrivirat D., Jaroensuk J., Wongnate T., Maenpuen S., Chaiyen P., Kamnerdnakta S., Swangsri J., Chuthapisith S., Sirivatanauksorn Y., Chaimayo C., Sutthent R., Kantakamalakul W., Joung J., Ladha A., Jin X., Gootenberg J.S., Abudayyeh O.O., Zhang F., Horthongkham N., Uttamapinant C. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nature biomedical engineering. 2020;4(12):1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Ralser M., Querfurth R., Warnatz H.J., Lehrach H., Yaspo M.L., Krobitsch S. An efficient and economic enhancer mix for PCR. Biochem. Biophys. Res. Commun. 2006;347(3):747–751. doi: 10.1016/j.bbrc.2006.06.151. [DOI] [PubMed] [Google Scholar]
- Ramachandran A., Huyke D.A., Sharma E., Sahoo M.K., Huang C., Banaei N., Pinsky B.A., Santiago J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117(47):29518–29525. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.J., Andini N., Hsieh K., Yang S., Wang T.H. Emerging analytical techniques for rapid pathogen identification and susceptibility testing. Annu. Rev. Anal. Chem. 2019;12(1):41–67. doi: 10.1146/annurev-anchem-061318-115529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R.J. Stabilization of proteins for storage. Cold Spring Harb. Protoc. 2010;2010(5) doi: 10.1101/pdb.top79. pdb top.79. [DOI] [PubMed] [Google Scholar]
- Teng F., Guo L., Cui T., Wang X.G., Xu K., Gao Q., Zhou Q., Li W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019;20(1):132. doi: 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J.E., Berendsen J.T.W., Steenbergen R.D.M., Wolthuis R.M.F., Eijkel J.C.T., Segerink L.I. Point-of-care CRISPR/Cas nucleic acid detection: recent advances, challenges and opportunities. Biosens. Bioelectron. 2020;166:112445. doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19(3):171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadharajan B., Parani M. DMSO and betaine significantly enhance the PCR amplification of ITS2 DNA barcodes from plants. Genome. 2021;64(3):165–171. doi: 10.1139/gen-2019-0221. [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang R., Li J. CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens. Bioelectron. 2020;165:112430. doi: 10.1016/j.bios.2020.112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., Zhao M., Qian F., Yu H., Liu Z., Ni T., Zheng Y., Wang Y. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shang X., Huang X. Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerg. Microb. Infect. 2020;9(1):1682–1691. doi: 10.1080/22221751.2020.1793689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.