Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineages that are more transmissible and resistant to currently approved antibody therapies poses a considerable challenge to the clinical treatment of coronavirus disease (COVID-19). Therefore, the need for ongoing discovery efforts to identify broadly reactive monoclonal antibodies to SARS-CoV-2 is of utmost importance. Here, we report a panel of SARS-CoV-2 antibodies isolated using the linking B cell receptor to antigen specificity through sequencing (LIBRA-seq) technology from an individual who recovered from COVID-19. Of these antibodies, 54042-4 shows potent neutralization against authentic SARS-CoV-2 viruses, including variants of concern (VOCs). A cryoelectron microscopy (cryo-EM) structure of 54042-4 in complex with the SARS-CoV-2 spike reveals an epitope composed of residues that are highly conserved in currently circulating SARS-CoV-2 lineages. Further, 54042-4 possesses uncommon genetic and structural characteristics that distinguish it from other potently neutralizing SARS-CoV-2 antibodies. Together, these findings provide motivation for the development of 54042-4 as a lead candidate to counteract current and future SARS-CoV-2 VOCs.
Keywords: COVID-19, SARS-CoV-2, antibody discovery, LIBRA-seq, Delta variant, cryo-EM, monoclonal antibodies biology
Graphical abstract
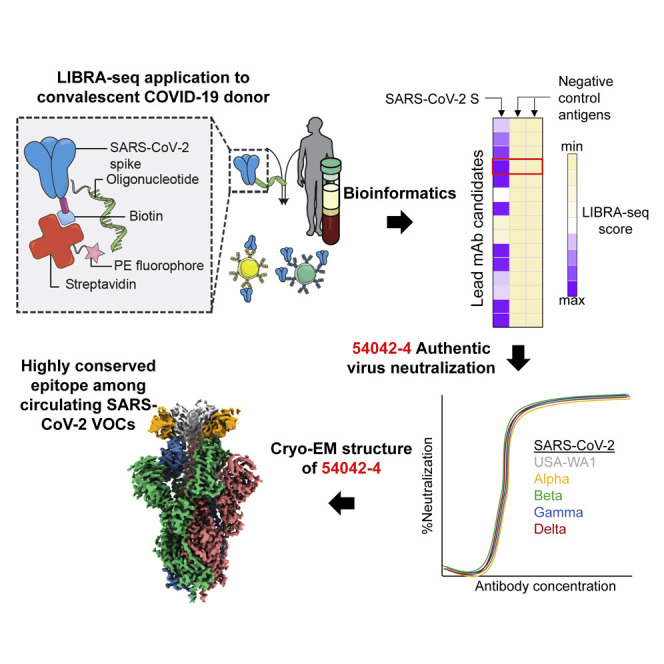
Kramer et al. demonstrate that antibody 54042-4 recognizes residues highly conserved across global SARS-CoV-2 isolates. Antibody 54042-4 potently neutralizes all known circulating variants of concern (VOCs) and could be developed as a clinical candidate to treat COVID-19 infection.
Introduction
The coronavirus disease (COVID-19) pandemic caused by a novel coronavirus from the Sarbecovirus genus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), spawned an unprecedented global research effort dedicated to therapeutic countermeasure development, resulting in rapid United States Food and Drug Administration (US FDA) emergency use authorization (EUA) for vaccines and monoclonal antibodies (Jones et al., 2021; Weinreich et al., 2021). The primary target for vaccine and antibody therapeutic development is the SARS-CoV-2 spike (S) protein, which facilitates host-cell attachment and entry (Wrapp et al., 2020). The emergence of distinct viral lineages that accumulate substitutions in S pose a significant threat to the countermeasures currently approved for clinical use (Chen et al., 2021; Mlcochova et al., 2021; Wang et al., 2021). Continued genomic surveillance and persistent efforts to identify antibodies with distinct binding modes and mechanisms of action are crucial to maintain availability of therapeutics in the event of further neutralization-escape by SARS-CoV-2 variants of concerns (VOCs).
SARS-CoV-2 spike is a class I viral fusion protein that is a trimer of heterodimers composed of S1 and S2 subunits (Wrapp et al., 2020). S1, which includes both the receptor-binding domain (RBD) and the N-terminal domain (NTD), initiates attachment to the receptor angiotensin-converting enzyme 2 (ACE2), whereas S2 drives membrane fusion by refolding from a prefusion to postfusion conformation (Li, 2016; Tortorici and Veesler, 2019). The primary contact of ACE2 and spike is in the RBD of the S1 subunit, which is composed of a receptor binding motif (RBM) and RBD core. The three RBDs within each spike can adopt an ACE2-accessible “up” conformation and an ACE2-inaccessible “down” conformation via a hinge-like motion (Shang et al., 2020). As a result, the RBD serves as the dominant target of neutralizing antibodies via antagonism of ACE2 binding (Piccoli et al., 2020), although other neutralizing epitopes have been identified (Brouwer et al., 2020; Chi et al., 2020; Suryadevara et al., 2021; Tortorici et al., 2021; Zost et al., 2020).
Neutralizing antibodies targeting the RBD have been characterized extensively and partition into different classes based on binding mode, ACE2 interface overlap, and cross-reactivity with other Sarbecoviruses. For example, neutralizing antibodies predominantly encoded by IGHV3-53 and IGHV3-66 have epitopes directly covering the ACE2 interaction footprint in the RBM (Yuan et al., 2020a). Examples of this class of antibodies are clinical EUA candidates REGN10933 and COV2-2196 (Hansen et al., 2020; Zost et al., 2020). Antibodies that bind the RBM but are more distal to the ACE2 interface form another distinct class that includes REGN10987 and COV2-2130 (Dong et al., 2021; Hansen et al., 2020). Additionally, antibodies such as S309, CR3022, and ADG-2 that cross-react with other coronaviruses comprise a more diverse group that target conserved residues in the RBD core (Pinto et al., 2020; Wec et al., 2020; Yuan et al., 2020b).
The continued transmission of SARS-CoV-2 in the human population has led to the evolution of VOCs with increased transmissibility and resistance to available medical countermeasures, including to some RBD-directed monoclonal antibodies (Alpert et al., 2021; Chen et al., 2021; Kuzmina et al., 2021). Some of the most consequential amino acid substitutions observed so far have occurred in the RBD, particularly N501Y in the B.1.1.7 (Alpha), B.1.351 (Beta), and P.1 (Gamma) lineages, and the additional combination of K417N/T and E484K in the Gamma and Beta lineages. The L452R substitution, detected in both the B.1.429 (Epsilon) and B.1.617.2 (Delta) variants also permits escape to monoclonal antibodies and a reduction in neutralization titer in comparison to USA-WA1 in vaccinees as well as individuals recovered from COVID-19 infection (McCallum et al., 2021; Mlcochova et al., 2021). Notably, the Epsilon lineage also contains substitutions (S13I and W152C) that disrupt the conformation of the NTD, resulting in the loss of numerous published NTD-directed SARS-CoV-2 neutralizing antibodies (McCallum et al., 2021). N501Y is thought to increase affinity for ACE2 (Starr et al., 2020), potentially resulting in increased infectivity, whereas E484K disrupts the antigenic landscape of the RBD that can lead to substantial decreases in neutralization titers (Hoffmann et al., 2021; Wang et al., 2021). In some cases, SARS-CoV-2 VOCs also escape neutralization by polyclonal antibodies in the serum from vaccine recipients and individuals previously infected with SARS-CoV-2 (Chen et al., 2021; McCallum et al., 2021; Mlcochova et al., 2021; Wang et al., 2021). These observations highlight the critical need for a wide range of potently neutralizing antibodies that are not sensitive to substitutions arising in VOCs.
To address this challenge, we applied linking B cell receptor to antigen specificity through sequencing (LIBRA-seq), a recently developed antibody-discovery technology (Setliff et al., 2019; Shiakolas et al., 2021), to interrogate the B cell repertoire of an individual who had recovered from COVID-19. Our efforts led to the discovery of a potently neutralizing antibody, designated 54042-4, which uses an uncommon genetic signature and distinct structural mode of SARS-CoV-2 RBD recognition to maintain neutralization potency to known VOCs. Antibody 54042-4 therefore may serve as a viable candidate for further prophylactic or therapeutic development for protection against a broad range of SARS-CoV-2 variants.
Results
Identification of SARS-CoV-2 neutralizing antibodies by LIBRA-seq
To identify SARS-CoV-2 S-directed antibodies, we utilized LIBRA-seq, a technology that enables high-throughput simultaneous determination of B cell receptor sequence and antigen reactivity at the single-cell level, expediting the process of lead candidate selection and characterization (Setliff et al., 2019; Shiakolas et al., 2021). The LIBRA-seq antigen-screening library included SARS-CoV-2 and SARS-CoV-2 D614G spikes stabilized in a prefusion conformation (Hsieh et al., 2020), along with antigens from other coronaviruses including SARS-CoV S, MERS-CoV S, HCoV-OC43 S, HCoV-229E S, HCoV-NL63 S, SARS-CoV-2 RBD, SARS-CoV RBD, and MERS-CoV RBD, as well as negative-control antigens ZM197 HIV-1 Env and influenza hemagglutinin (HA) NC99. Antigen-specific B cells were isolated from a donor with potently neutralizing antibodies in serum (1:258 NT50) 3 months after infection confirmed by nasal swab RT-PCR testing for SARS-CoV-2 (Figures S1A and S1B). Of the 73 immunoglobulin G (IgG+) B cells with high LIBRA-seq scores (≥1) for SARS-CoV-2 S (Figure 1 A), we chose nine lead candidates with diverse sequence characteristics, CDRH3 length, and germline V gene usage for characterization as recombinant monoclonal antibodies (Figures 1B and S1C). Binding to SARS-CoV-2 S by ELISA was confirmed for eight of these antibodies, with the only exception being antibody 54042-2, in agreement with its lower LIBRA-seq score (Figures 1B, S1C, and S1D). Five of these antibodies showed SARS-CoV-2 neutralization activity in a high-throughput neutralization screen using a live chimeric VSV displaying SARS-CoV-2 spike protein (Case et al., 2020) (Figure 1B). None of the antibodies showed neutralization against VSV SARS-CoV. Further, the five neutralizing antibodies did not show binding cross-reactivity to other coronavirus antigens in the screening panel, with the exception of SARS-CoV (Figure S1E). Full dose-response neutralization curves in the chimeric VSV assay were obtained for four of these five antibodies, with antibody 54042-4 showing the best potency, at a half-maximal inhibitory concentration (IC50) of 9 ng/mL (Figure 1C).
Figure 1.
Identification and characterization of SARS-CoV-2 antibodies isolated using LIBRA-seq
(A) Variable heavy gene usage (x axis) as a function of IgG+ B cells with a SARS-CoV-2 spike LIBRA-seq score (≥1) (y axis). The nine lead candidates are highlighted in purple.
(B) Sequence characteristics and antigen specificity of nine lead candidate antibodies from a recovered COVID-19 donor. Percent identity is calculated at the nucleotide level, and CDR length and sequences are displayed at the amino acid level. LIBRA-seq scores for each antigen are displayed as a heatmap with a LIBRA-seq score of −2 displayed as light yellow, 0 as white, and 2 in purple; in this heatmap scores lower or higher than that range are shown as −2 and 2, respectively. ELISA binding data for SARS-CoV-2 S are displayed as a heatmap of the area under the curve (AUC) analysis calculated from Figure S1D.
(C) RTCA VSV-SARS-CoV-2 neutralization by lead candidate antibodies. IC50 values are calculated by non-linear regression analysis by GraphPad Prism software. Neutralization assays were performed in technical triplicate; data are represented as mean ± SD.
Antibody 54042-4 targets the SARS-CoV-2 receptor-binding domain
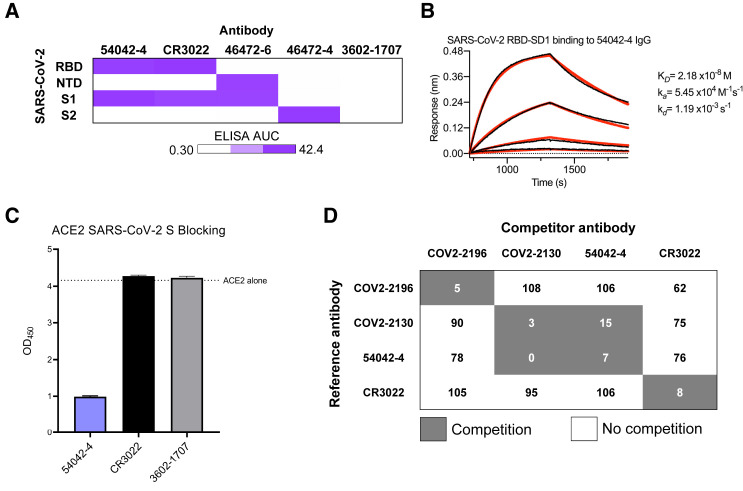
Because of the potent (≤10 ng/mL) virus neutralization observed for 54042-4, we selected this antibody for further characterization. ELISAs performed with purified RBD, NTD, S1, and S2 proteins revealed 54042-4 IgG bound to the SARS-CoV-2 S1 subunit as well as the RBD (Figures 2 A and S2). To determine the affinity of the antibody-antigen binding interaction, biolayer interferometry experiments were performed by measuring the association and dissociation kinetics of immobilized 54042-4 IgG binding to a soluble protein comprising the RBD and subdomain-1 (SD1) of the SARS-CoV-2 S protein, and curve-fitting resulted in a calculated KD of 21.8 nM (Figure 2B). Given the neutralization potency of 9 ng/mL (60 pM), these data suggest that the IgG avidly binds to the S protein on the virion surface. To assess whether 54042-4 neutralizes viral infection by directly competing with ACE2, a receptor-blocking assay was performed by testing competition of 54042-4 with soluble ACE2 for binding to SARS-CoV-2 S. The results demonstrated that 54042-4 inhibits interaction of ACE2 to SARS-CoV-2 S protein, unlike the control antibody CR3022, an extensively characterized SARS-CoV antibody that binds a cryptic epitope in the RBD of both SARS-CoV and SARS-CoV-2 (Yuan et al., 2020b) and the influenza HA-specific 3602-1707 antibody (Setliff et al., 2019) (Figure 2C). Next, we performed competition ELISAs to determine if 54042-4 competes for binding with three other RBD-directed antibodies with distinct epitopes. These antibodies included COV2-2196 and COV2-2130, which form the basis of AZD7442, an antibody cocktail currently under investigation in clinical trials for COVID-19 treatment and prevention (ClinicalTrials.gov Identifiers: NCT04625725, NCT04723394, NCT04518410, and NCT04501978) and CR3022. The competition experiment showed that 54042-4 competed for binding to SARS-CoV-2 S protein with COV2-2130 but not COV2-2196 or CR3022 (Figure 2D). Together, these results suggest that 54042-4 targets an epitope on SARS-CoV-2 RBD that at least partially overlaps with the binding sites for both ACE2 and other potently neutralizing RBD-directed antibodies.
Figure 2.
Antigenic characterization of antibody 54042-4
(A) ELISA binding values against SARS-CoV-2 subdomains are displayed as a heatmap of AUC values calculated from the data in Figure S2. Antibodies CR3022, 46472-6, and 46472-4 were used as positive controls for the RBD, NTD, and S2 antigens, respectively (Shiakolas et al., 2021; Yuan et al., 2020b). 3602-1707 was included as an influenza HA-specific negative control antibody (Setliff et al., 2019).
(B) A biolayer interferometry sensogram that shows 54042-4 IgG binding to recombinant SARS-CoV-2 RBD-SD1. Binding data are depicted by the black lines and the best fit line of the data to a 1:1 binding model is shown in red.
(C) SARS-CoV-2 spike/ACE2 inhibition ELISA is shown for 54042-4, SARS-CoV-2 antibody CR3022, and negative control HA-specific antibody 3602-1707. For each antibody, the ACE2 binding signal is depicted on the y axis, in comparison to ACE2-only binding to SARS-COV-2 spike is shown as a dotted line. ELISAs were performed in technical triplicate with 2 biological duplicates; data represented as mean ± SEM.
(D) Competition ELISA of 54042-4 with antibodies COV2-2196, COV2-2130, and CR3022. Values in white indicate no competition (presence of competing antibody reduced reference antibody binding by less than 30%) and values in dark gray indicate competition (presence of competing antibody reduced reference antibody binding by more than 60%). Competition ELISA was performed in technical quadruplicate with 2 biological duplicates; data shown as mean of quadruplicate values.
54042-4 binds the apex of the SARS-CoV-2 RBD in the down conformation
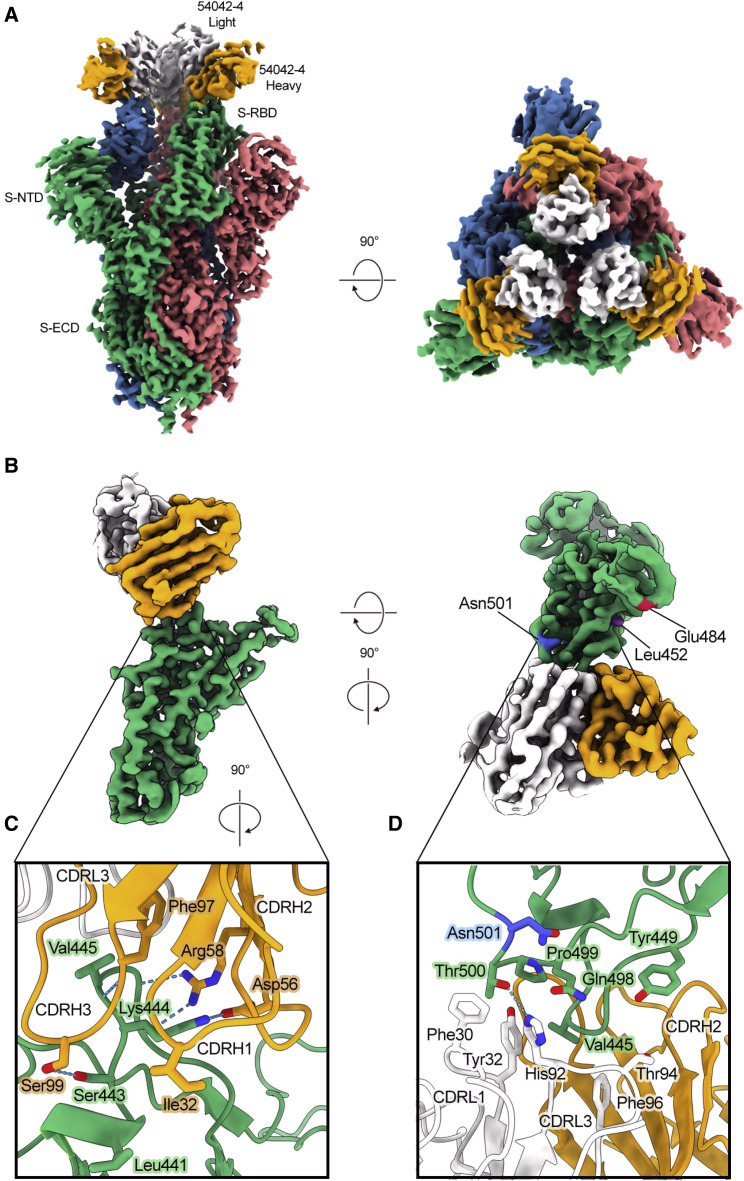
To gain a better understanding of the recognition of SARS-CoV-2 S by antibody 54042-4, we determined a 2.7 Å resolution cryo-EM structure of the 54042-4 antigen-binding fragments (Fabs) bound to the SARS-CoV-2 S extracellular domain (ECD) modified so that all three RBDs were disulfide-locked in the down conformation (Henderson et al., 2020) (Figure 3 A). Local refinement of one RBD bound to a 54042-4 Fab was performed to improve the interpretability of the map at the binding interface, resulting in a local 3D reconstruction with a resolution of 2.8 Å (Figure 3B; Table S1). The structure revealed that 54042-4 forms an extensive interface with the RBD, making contacts through the complementarity determining regions (CDRs) CDRL1, CDRL3, and all three CDRs of the heavy chain, to form a clamp around the apex of the RBM saddle (Figures 3C, 3D, S3A, and S3B). The primary interactions involve RBD residues 439–450, with a network of hydrogen bonds between the 54042-4 heavy chain and RBD residues 443–447 (Figure 3C). From CDRH3, Ser99 forms a hydrogen bond with RBD residue Ser443, and a hydrogen bond is formed between the mainchain atoms of Phe97 and Val445. From CDRH2, Asp56 forms a hydrogen bond and salt bridge with Lys444, whereas Arg58 forms hydrogen bonds with mainchain atoms from Gly446 and Gly447. The CDRH1 contributes a lone residue, Ile32, to the binding interface, forming minor contacts near Leu441. The 54042-4 light chain surrounds the opposite side of this RBD region, mediating interactions primarily through hydrophobic contacts formed by CDRL1 and CDRL3 near RBD residue Val445 (Figure 3D). Additional light chain contacts are made with residues 498–500 of the RBD, including a hydrogen bond between His92 of CDRL3 and Thr500, and hydrophobic interactions involving CDRL1 Phe30 and Tyr32. Although 54042-4 binds all three RBDs locked in the down position, the epitope region is equally accessible when the RBD is in the up position. Additionally, the epitope lies outside the RBD hinge region, makes no additional contacts with the spike trimer, and partially overlaps the ACE2 binding site. Therefore, the mechanism of neutralization likely involves inhibition of ACE2 binding rather than locking the RBDs in the down conformation.
Figure 3.
Atomic resolution of 54042-4 binding mode to SARS-CoV-2 S
(A) 3D reconstructions of side and top views of Fab 54042-4 bound to SARS-CoV-2 spike.
(B) Focused refinement maps showing the 54042-4 epitope at the apex of the RBM in the down position (left). Top-down view of the 54042-4 epitope showing heavy and light chain contacts, as well as residues outside of the binding interface that are mutated in circulating VOCs (right).
(C) The 54042-4 heavy chain binds to RBD residues 443–447 primarily through a network of hydrogen bonds involving CDRH2 and CDRH3 and hydrophobic contacts involving Ile32 of CDRH1.
(D) The 54042-4 light chain contacts RBD residues 498–500 through a hydrogen bond between Thr500 and His92 of CDRL3 and hydrophobic contacts involving Phe30 and Tyr32 of CDRL1.
Notably, the complex structure indicated that 54042-4 does not make substantial contact with a number of spike substitutions associated with current VOCs. For example, RBD residue Asn501 (present as Tyr501 in several VOCs, including Alpha, Beta, and Gamma) lies just outside of the 54042-4 epitope, whereas the Cα atoms of Glu484 (present as Lys484 or Gln484 in, e.g., Beta, Gamma, and B.1.617 [Kappa]) and Leu452 (present as Arg452 in Epsilon and Delta) are ∼18 Å and ∼14 Å away from the Cα atoms of the nearest 54042-4 residue, respectively (Figure 3B). Furthermore, the substitution Gly614, which is found in all current VOCs is outside of the RBD and is ∼75 Å from the nearest 54042-4 residue.
Antibody 54042-4 has an uncommon genetic signature and distinct structural mode of RBD recognition
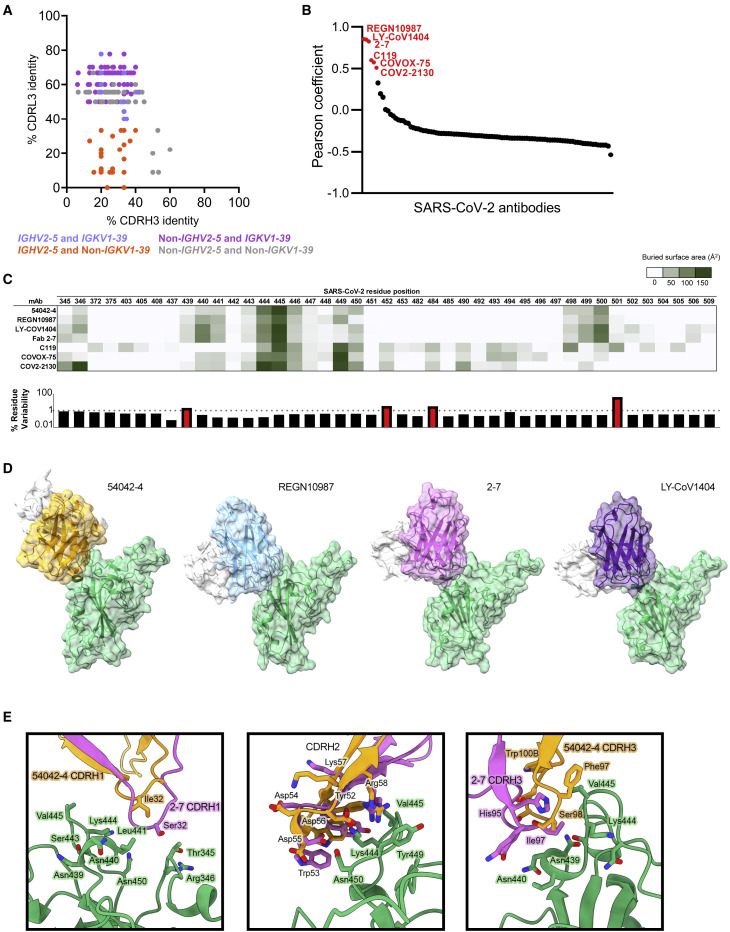
Public clonotype sequence signatures (those shared by multiple individuals recovered from COVID-19 infection) have been identified for potently neutralizing SARS-CoV-2 antibodies, including antibodies currently in clinical trials or approved for emergency use (Nielsen et al., 2020; Yuan et al., 2020a). To investigate whether antibody sequences that are closely related to 54042-4 can be identified among known SARS-CoV-2 antibodies, we searched the CoV-AbDab database that contains paired heavy-light chain sequences of coronavirus antibodies (Raybould et al., 2021). Notably, only 0.5% of antibodies in the database used the same combination of IGHV2-5 heavy and IGKV1-39 light chain germline V genes as 54042-4. Further, antibodies with high sequence identity to the 54042-4 CDRH3 and CDRL3 were not identified, whether or not the search was restricted to the IGHV2-5 heavy chain and IGKV1-39 light chain genes (Figure 4 A).
Figure 4.
Sequence and structural comparison of 54042-4 to known SARS-CoV-2 antibodies
(A) Amino acid CDRH3 identity to 54042-4 (x axis) is plotted against CDRL3 identity to 54042-4 (y axis) for paired heavy and light chain sequences obtained from the CoV-AbDab database (Raybould et al., 2021). Antibodies using the same heavy and light chain germline gene as 54042-4 (IGHV2-5 and IGKV1-39) are shown in light blue. Antibodies using the IGHV2-5 heavy chain gene and a non-IGKV1-39 light chain gene are shown in orange. Additionally, antibodies using a non-IGHV2-5 heavy chain gene and the IGKV1-39 light chain gene, with CDRH3 or CDRL3 identity to 54042-4 of at least 50%, are highlighted in purple. Finally, antibodies that do not use IGHV2-5 or IGKV1-39, but that have at least 50% identity to CDRH3 or CDRL3 of 54042-4, are shown in gray.
(B) Pearson correlation of epitopes of known SARS-CoV-2 antibodies (Table S2) in comparison to 54042-4 antibody, with the six antibodies showing a statistically significant (p < 0.05) positive correlation highlighted in red.
(C) Heatmap (top) depicting buried surface area (Å2) at the SARS-CoV-2 RBD interface for the six antibodies with highest epitope correlations with 54042-4. Bar graph (bottom) showing the frequency (%) of substitutions at each given residue position in log scale, with a dashed line at 1% and residue positions with a frequency greater than 1% highlighted in red.
(D) Distinct angles of approach of antibodies 54042-4 (heavy chain: orange, light chain: white), REGN10987 (heavy chain: blue, light chain: white) (PDB id: 6XDG), 2-7 (heavy chain: pink, light chain: white) (PDB id: 7LSS), and LY-CoV1404 (heavy chain: purple, light chain: white) (PDB:7MMO) to the SARS-CoV-2 RBD (green).
(E) Structural comparison of CDRH1, 2, and 3 of antibodies 54042-4 and 2-7. CDRH1 of 2-7 extends further than 54042-4, forming additional contacts with Thr345 and Arg346 of the RBD (left). The CDRH2 region of 2-7 approaches at a different angle, but maintains RBD contacts via Asp56 and Arg58 (center). The CDRH3 contacts of 2-7 and 54042-4 are divergent, with unique CDRH3 residues and RBD interactions (right).
Next, we compared the 54042-4 epitope to the epitopes of other known SARS-CoV-2 antibodies by computing pairwise correlations between the antibody-antigen buried surface areas for 54042-4 against a set of 99 publicly available SARS-CoV-2 antibody-antigen structures from the Protein Data Bank, as well as the structure of antibody COV2-2130 in complex with the SARS-CoV-2 RBD (Table S2). The results revealed significant positive correlations with only six other antibodies: REGN10987 (Hansen et al., 2020), 2-7 (Liu et al., 2020), C119 (Barnes et al., 2020), COVOX-75 (Dejnirattisai et al., 2021), COV2-2130 (Zost et al., 2020) (in agreement with the observed binding competition with 54042-4, Figure 2D), and LY-CoV1404 (Westendorf et al., 2021) (Figure 4B). However, of these six antibodies, COVOX-75 has been reported as not a potent neutralizer (Dejnirattisai et al., 2021). C119 makes substantial contact with residues Glu484 and Asn501, indicating potential susceptibility of this antibody to substitutions at those positions that are currently associated with relatively high substitution rates (Figure 4C) and are present in several circulating SARS-CoV-2 VOCs (Alpert et al., 2021; Tegally et al., 2021). Further, both C119 and COVOX-75, as well as COV2-2130, have substantial buried surface area interactions with a number of additional residues compared to those in the epitope of 54042-4 (Figure 4C), suggesting that these three antibodies could be susceptible to additional potential spike substitutions that would not directly affect antigen interactions with 54042-4.
We also observed that although the epitopes of antibodies 2-7, LY-CoV1404, and REGN10987 correlate well with that of 54042-4, these antibodies have distinct angles of antigen approach (Figure 4D). To quantify this observation, we aligned the RBDs from the 2-7, LY-CoV1404, and REGN10987 complex structures with the RBD from the 54042-4 structure. Using the antibody coordinates when the respective RBDs were aligned, we computed the root-mean-square deviations (RMSDs) between the Cα atoms in the FWR1-FWR3 regions of the antibody heavy and light chains. This resulted in RMSDs of 16.4 Å, 16.5Å, and 22 Å between 54042-4 versus 2-7, LY-CoV1404, and REGN10987, respectively, confirming the substantial differences in the structural mode of antigen recognition by 54042-4 compared to 2-7, LY-CoV1404, and REGN10987. Although 54042-4 and 2-7 both originate from the same IGHV2-5 germline gene and share analogous RBD contacts in the CDRH2 region, these antibodies exhibit different CDRH1 and CDRH3 interactions (Figure 4E) and use a different light chain germline gene (IGKV1-39 for 54042-4, and VL2-14 for 2-7). Interestingly, antibodies 2-7 and LY-CoV1404 use identical heavy and light chain germline genes and have a virtually indistinguishable structural mode of antigen recognition (RMSD, computed as above, of 1.7 Å). Notably, all of 2-7, LY-CoV1404, and REGN10987 have greater interactions with RBD residues 439–441 compared to 54042-4, with buried surface areas of 172, 164, 127, and 60 Å2 for 2-7, LY-CoV1404, REGN10987, and 54042-4, respectively (Figure 4C), suggesting 2-7, LY-CoV1404, and REGN10987 may be more prone to viral escape in that region. Indeed, the N439K substitution is present in several independent SARS-CoV-2 lineages and has been found to affect binding and neutralization by REGN10987 (Thomson et al., 2021).
Together, these data suggest that antibody 54042-4 utilizes an uncommon genetic signature and a distinct structural mode of antigen recognition compared to other known SARS-CoV-2 antibodies.
Antibody 54042-4 function is not affected by current SARS-CoV-2 VOC substitutions
Next, to identify substitutions capable of disrupting binding to antibody 54042-4, we performed shotgun alanine-scanning mutagenesis of the SARS-CoV-2 RBD (Davidson and Doranz, 2014). The only tested substitutions that substantially affected binding in comparison to an RBD antibody control were K444A, V445A, G446A, and P499A (Figure 5 A), which all fall within the 54042-4 epitope (Figures 3C, 3D, and S3A). To assess the functional effect of substitutions within the 54042-4 epitope, we tested neutralization against VSV-SARS-CoV-2 chimeras containing single substitutions at K444R/T/E/N, G446D, or Q498R. These mutants were generated from neutralization escape experiments using saturating concentrations of either COV2-2130 antibody (a 54042-4 competitor) (Figure 2D) or COV2-2499 (a COV2-2130 competitor) (Dong et al., 2021; Greaney et al., 2021b). These experiments revealed that the chimeric VSVs with substitutions at Lys444, Gly446, and Gln498 were resistant to neutralization by 54042-4 (Figure 5B). Together, the alanine-scanning and neutralization experiments indicated that 54042-4 may be sensitive to substitutions at spike residues Lys444, Val445, Gly446, Gln498, and Pro499. However, analysis of currently circulating SARS-CoV-2 isolates from the GISAID database as of May 6, 2021 (Elbe and Buckland-Merrett, 2017) revealed that substitutions at these five residue positions are only present at low levels (Figure 5C). Further, virtually all of the 54042-4 epitope residues (Figure S3A) are highly conserved in circulating SARS-CoV-2 lineages (Figure 5C). The only exception is residue Asn439, which has a substitution frequency of 2.1% (Figure 5C); however, this residue makes only minimal contacts with antibody 54042-4 (Figure S3A), suggesting that residue Asn439 may not be critical for 54042-4 recognition of the SARS-CoV-2 spike.
Figure 5.
Functional characterization of antibody 54042-4
(A) Binding data of 54042-4 antibody to a shotgun alanine mutagenesis screening library of the SARS-CoV-2 RBD (Wuhan-Hu-1 strain). Residues displayed are the alanine substitutions that resulted in the biggest loss of binding to 54042-4 yet still retained signal with the RBD antibody control. Data are represented as an average of at least two replicate measurements, and shown with ranges (half of the maximum minus minimum values from replicates).
(B) RTCA Neutralization of VSV SARS-CoV-2 chimera variants harboring specific substitutions. Cell sensograms are shown in boxes corresponding to mutations indicated in each row. Columns (from left to right) are each chimera treated with COV2-2381, 54042-4 and virus only control. Neutralization of 54042-4 of USA-WA1 strain and cells only are indicated on the right. COV2-2381 was chosen as a positive control due to its distinct epitope footprint from the selected substitutions. RTCA neutralization was performed in technical duplicate conditions; only one sensogram profile per condition is shown.
(C) 54042-4 epitope residues (non-zero buried surface area on SARS-CoV-2 RBD) with their associated % conservation (the percentage of deposited sequences containing the highest-frequency amino acid at that position) in the GISAID database. The only 54042-4 epitope residue with a % conservation of less than 99%, Asn439, is highlighted in red.
(D) ELISA AUC of 54042-4, CR3022, and an influenza HA-specific negative control antibody 3602-1707 calculated from the data in Figures S4A and S4B. AUC is displayed as a heatmap with a value of 0.02 corresponding to white, 50% maximum as light-purple, and maximum (42.41) AUC as purple.
(E) Authentic SARS-CoV-2 % neutralization of USA-WA1, Alpha, Beta, Delta, and Gamma strains (y axis) is depicted as a function of antibody concentration (x axis). Also shown are the respective IC50 and IC80 values for 54042-4 neutralization against each variant. Neutralization assays were performed with 2 technical duplicates; data represented as mean ± SD.
To investigate the ability of antibody 54042-4 to recognize current SARS-CoV-2 VOCs, we performed ELISAs to test binding of 54042-4 to RBD proteins containing substitutions found in one or more VOCs. These substitutions included K417N found in many isolates in the Beta lineage, as well as E484K (Beta, Gamma), N501Y (Alpha, Beta, Gamma), L452R (Delta, Epsilon), and N439K found in lineages B.1.141 and B.1.258 (Thomson et al., 2021). Notably, antibody 54042-4 bound to these RBD variants at a similar level compared to the binding to the RBD from the Wuhan-1 isolate (Figures 5D and S4A). These results are consistent with the structural observations that 54042-4 makes only minimal contacts with residue Asn439, and none of the other RBD substitutions were at residues in the 54042-4 epitope (Figure S3A). Binding of antibody 54042-4 also was not affected in the context of SARS-CoV-2 S ECD proteins that included deletions and substitutions in the S1 domain of the Alpha and Beta VOCs (Figures 5D, S4B, and S4C). Finally, we tested the ability of 54042-4 to neutralize authentic SARS-CoV-2 USA-WA1, Alpha, and Beta, Gamma and Delta SARS-CoV-2 variants. Consistent with the ELISA data, 54042-4 neutralized each virus potently with IC50 values of 3.2, 5.5, 9.7, 3.7, and 1.5, and IC80 values of 10, 48, 49, 11.4, and 3.9 ng/mL, respectively (Figure 5E). Together, these data indicate that 54042-4 may be an effective countermeasure against currently circulating SARS-CoV-2 variants.
Discussion
SARS-CoV-2 neutralizing antibody discovery efforts have produced an extensive panel of antibodies that show a wide range of functional effects, and most antibodies discovered to date cluster into several classes based on RBD-binding orientation, ACE2 antagonism, and cross-reactivity to related SARS-like coronaviruses (Greaney et al., 2021b). Here, we report the identification of 54042-4, an antibody that exhibited potent SARS-CoV-2 neutralization against USA-WA1 as well as the currently circulating Alpha, Beta, Gamma, and Delta VOCs. Interestingly, antibody 54042-4 neutralized virus at comparable IC50s to the clinical candidates LY-CoV1404 and REGN10987, despite having ∼10-fold lower affinity for the RBD (Hansen et al., 2020; Westendorf et al., 2021). Although the epitope of antibody 54042-4 showed partial overlap with that of several other known RBD-directed antibodies, our findings revealed a distinct mode of SARS-CoV-2 spike recognition, paired with an uncommon genetic signature that distinguishes 54042-4 from other SARS-CoV-2 antibodies. Notably, important differences were observed even for the six antibodies with the highest epitope correlations to 54042-4, with all six of these antibodies exhibiting substantially greater contacts with one or more known residues associated with currently circulating VOCs, as well as with other spike residues (Figure 4C). Although it is not possible to predict what SARS-CoV-2 variants will emerge in the future, having access to antibodies with differences in epitope interactions is critical to broadening the portfolio of countermeasure candidates, should virus variants emerge that are resistant to current therapies. The discovery of antibody 54042-4 is therefore a promising addition to the limited set of antibodies with a high potential for effectively counteracting current SARS-CoV-2 VOCs.
The increased spread of several SARS-CoV-2 VOCs over the past few months has emphasized the need for continued surveillance of vaccine efficacy against the evolving virus targets. The increased transmission rates of the Alpha lineage are likely a product of enhanced ACE2 affinity for the SARS-CoV-2 RBD (Starr et al., 2020) and not a result of escape from pre-existing antibodies in convalescent or vaccinated individuals (Wang et al., 2021; Xie et al., 2021). Variants that encode the E484K substitution appear to pose a significantly higher risk of neutralization escape in vaccine recipients and individuals who have recovered from COVID-19 (Wang et al., 2021). Indeed, the rise of cases associated with the Gamma variant that harbors the E484K substitution (among others) in Manaus, Brazil is on a dangerous trajectory, despite having a 76% population seropositivity rate dating back to March 2020 (Sabino et al., 2021). In the context of vaccination, early vaccine trial data for Novavax against the Beta lineage in South Africa (also encoding the E484K substitution) demonstrated a significant decrease in efficacy (Wadman, 2021). The enhanced transmission profile and recent rise in new infection cases as a result of the Delta variant (Campbell et al., 2021) is another demonstration of the need for SARS-CoV-2 therapeutics. These observations underscore the ongoing need for genomic surveillance to monitor the emergence and spread of new SARS-CoV-2 variants and their effects on population immunity.
In addition to vaccines, antibody therapeutics can play an important role for treating SARS-CoV-2 infections. Given the unknown future trajectory of the pandemic and the potential for emergence of VOCs that may escape neutralization by vaccine-elicited immunity, the development of a wide array of candidate antibody therapeutics that are insensitive to substitutions found in major VOCs may prove critical in the fight against COVID-19. Current VOCs have already shown an ability to escape neutralization by a number of antibodies in clinical development (Chen et al., 2021; Wang et al., 2021). Although the Beta variant or any lineage harboring the Glu484 substitution has yet to propagate in the United States, the nearly complete abrogation of neutralization activity of LY-CoV555 (Bamlinivimab) and REGN10933 (Cesirivimab) against the Beta variant poses a significant risk for the currently available EUA clinical candidates (Wang et al., 2021). Further, the rise in cases with viruses containing the L452R substitution (Epsilon and Delta variants) and the corresponding reduction in neutralization potency associated with the Eli Lilly cocktail (Bamlinivimab and Etesivimab), REGN10987 (Imdevimab) as well as Regdanvimab (approved for use in Europe), further motivates the continued investigation into antibodies insensitive to currently circulating VOCs (McCallum et al., 2021; Mlcochova et al., 2021). In contrast to these clinical candidates, the binding, neutralization, and structural data suggest that antibody 54042-4 maintains functional activity independent of the current major substitutions in circulating VOCs. Combined with these observations, the properties of 54042-4 in comparison to other SARS-CoV-2 antibodies motivate further clinical development of this antibody to complement the existing pool of therapeutic countermeasures. As SARS-CoV-2 virus evolution continues due to various factors, such as vaccine hesitancy and delayed vaccine rollout to underserved parts of the world, new VOCs are likely to keep emerging, with the potential to further decrease or even abrogate protection induced by current vaccines designed against the ancestral strain. Antibody therapeutic development, especially focusing on broad protection against diverse SARS-CoV-2 variants, is therefore of continued significance.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC-Cy7 Mouse Anti-Human CD14 | BD | Cat#561709; RRID: RRID: AB_10893806 |
| FITC Anti-Human CD3 (OKT3) | Tonbo Biosciences | Cat#35-0037; RRD: RRID: AB_2621662 |
| PE-Cy5 Mouse Anti-Human IgG | BD | Cat#551497; RRD: RRID: AB_394220 |
| BV711 Mouse Anti-Human CD19 | BD | • Cat#563036; RRID: AB_2737968 |
| Alexa Fluor 488-conjugated goat anti-human secondary antibody | Jackson ImmunoResearch | CAT#109-545-003 |
| CR3022 | Yuan et al., 2020a | N/A |
| Monoclonal anti-SARS-CoV/SARS-CoV-2 46472-4 | Shiakolas et al., 2021 | GenBank: MZ126647, MZ126662 |
| Monoclonal anti-SARS-CoV/SARS-CoV-2 46472-6 | Shiakolas et al., 2021 | GenBank: MZ126649, MZ126664 |
| Monoclonal anti-SARS-CoV-2 54042-2 | This paper | MZ820097, MZ820106 |
| Monoclonal anti-SARS-CoV-2 54042-3 | This paper | MZ820098, MZ820107 |
| Monoclonal anti-SARS-CoV-2 54042-4 | This paper | MZ820099, MZ820108 |
| Monoclonal anti-SARS-CoV-2 54042-5 | This paper | MZ820100, MZ820109 |
| Monoclonal anti-SARS-CoV-2 54042-7 | This paper | MZ820101, MZ820110 |
| Monoclonal anti-SARS-CoV-2 54042-10 | This paper | MZ820102, MZ820111 |
| Monoclonal anti-SARS-CoV-2 54042-11 | This paper | MZ820103, MZ820112 |
| Monoclonal anti-SARS-CoV-2 54042-14 | This paper | MZ820104, MZ820113 |
| Monoclonal anti-SARS-CoV-2 54042-15 | This paper | MZ820105, MZ820114 |
| COV2-2196 | Zost et al., 2020 | N/A |
| COV2-2130 | Zost et al., 2020 | N/A |
| Goat Anti-Human IgG Antibody (Peroxidase) | Jackson ImmunoResearch | Cat#109-035-088 |
| Monoclonal anti-influenza 3602-1707 | Setliff et al., 2019 | N/A |
| Bacterial and virus strains | ||
| VSV SARS-CoV-2 | Case et al., 2020 | N/A |
| VSV SARS-CoV-2 G446D | Greaney et al., 2021b | N/A |
| VSV SARS-CoV-2 Q498R | Greaney et al., 2021b | N/A |
| VSV SARS-CoV-2 K444R | Dong et al., 2021 | N/A |
| VSV SARS-CoV-2 K444T | Dong et al., 2021 | N/A |
| VSV SARS-CoV-2 K444E | Dong et al., 2021 | N/A |
| VSV SARS-CoV-2 K444N | Dong et al., 2021 | N/A |
| SARS-CoV-2 (Original WA isolate), | CDC | Genebank MN985325 |
| SARS-CoV-2 (Lineage B.1.1.7), UK Variant of Concern | CDC | GISAID database EPI_ISL_751801 |
| SARS-CoV-2 (Lineage B.1.351), SA Variant of Concern | Emory University | GISAID database EPI_ISL_890360 |
| SARS-CoV-2 (Lineage P1) Brazilian Variant of Concern | CDC | Genebank MW621433 |
| SARS-CoV-2 (B.1.617-2), Indian Variant of Concern; WA backbone with spike mutation | UTMB; Liu et al., 2021 | https://doi.org/10.1038/s41586-021-03693-y |
| Biological samples | ||
| PBMC from Donor | James Crowe Jr. | N/A |
| Serum From Donor | James Crowe Jr. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| A/New Caledonia/20/99 (H1N1) | Barney Graham | GenBank: ACF41878 |
| ZM197.SOSIP.664 gp140 trimer | Ivelin Georgiev | N/A |
| SARS-CoV-2 S (Hexapro) | PMID: 32703906 | N/A |
| SARS-CoV-2 S (Hexapro) RBD down | McLellan Lab | N/A |
| SARS-CoV S (S-2P) | PMID: 28807998 | N/A |
| MERS-CoV S (S-2P) | PMID: 28807998 | N/A |
| HCoV-OC43 S (S-2P) | McLellan Lab | N/A |
| HCoV-HKU1 S (S-2P) | PMID: 28807998 | N/A |
| SARS-CoV-2 S1 | Sino Biological | Cat#40591-V08H |
| SARS-CoV-2 S2 | Sino Biological | Cat#40590-V08B |
| SARS-CoV-2 RBD | Sino Biological | Cat#40592-VNAH |
| SARS-CoV-2 NTD | Sino Biological | Cat#40591-V49H |
| HCoV-229E S | Sino Biological | Cat#40605-V08B |
| HCoV-NL63 S | Sino Biological | Cat#40604-V08B |
| ACE2 | Sigma Aldrich | SAE0064-5UG |
| Streptavidin HRP | Thermo Fisher Scientific | Cat#ENN100 |
| Streptavidin R-phycoerythrin (SA-PE) | Invitrogen | Cat#S866 |
| Carboxyfluorescein diacetate N-succinimidyl ester (CFSE) | Sigma-Aldrich | Cat#21888 |
| Protein A Resin | GenScript | Cat# L00210 |
| Polyethyleniminine Linear MW 25000 | Polysciences | Cat#23966-1 |
| Ghost Dye Red 780 | Tonbo biosciences | Cat#13-0865 |
| Critical commercial assays | ||
| Biacore X100 Sensorchip NTA | GE Healthcare | Cat# BR1004 |
| EZ link Sulfo-NHS-LC-Biotin | Thermo Fisher Scientific | Cat#21327 |
| BirA-500: Bir A biotin-protein ligase standard reaction kit | Avidity | Cat# BirA500 |
| Solulink Protein-Oligonucleotide Conjugation Kit | TriLink Biotechnologies | Cat# S-9011 |
| B cell Single Cell V(D)J solution | 10X Genomics | N/A |
| Intellicyt high throughput flow cytometer | Sartorius | iQue3 |
| Deposited data | ||
| 54042 heavy chain sequences | This paper | MZ820097-MZ8200105 |
| 54042 light chain sequences | This paper | MZ8200106-MZ8200114 |
| Raw next-generation sequencing data | This paper | PRJNA754442 |
| GISAID EpiCoV SARS-CoV-2 sequence isolates | GISAID | Full list of contributing labs and accessions: https://github.com/Georgiev-Lab/GISAID_COV19_acknowledgement_May_7_2021 |
| Experimental models: cell lines | ||
| Human: Freestyle 293F cells | Thermo Fisher Scientific | Cat#A14528 |
| Human: Expi293F cells | Thermo Fisher Scientific | Cat#A14527 |
| ExpiCHO cells | Thermo Fisher Scientific | Cat#A29127 |
| Vero E6 cells | ATCC | CRL-1586 |
| Oligonucleotides | ||
| Oligonucleotides for Protein DNA-barcoding | Setliff et al., 2019. | N/A |
| Software and algorithms | ||
| Cell Ranger | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| HighV-Quest | Alamyar et al., 2012 | http://www.imgt.org/IMGTindex/IMGTHighV-QUEST.php |
| ChangeO | Gupta et al., 2015 | https://changeo.readthedocs.io/en/stable/ |
| Geneious 11.1.5 | https://www.geneious.com | N/A |
| PyMOL | The PyMOL Molecular Graphics System, Version 2.3.5 Schrödinger, LLC | N/A |
| Flowjo v10 | TreeStar | https://www.flowjo.com/ |
| GraphPad Prism 9.0.0 | https://www.graphpad.com:443/ | N/A |
| Biacore X100 Evaluation Software | GE Healthcare | V2.0.1 |
| SerialEM | Mastronarde, 2003 | https://bio3d.colorado.edu/SerialEM/ |
| cryoSPARC v3.2.0 | Punjani et al., 2017 | https://cryosparc.com/ |
| SAbPred | Dunbar et al., 2016 | http://opig.stats.ox.ac.uk/webapps/newsabdab/sabpred/ |
| COOT | Emsley and Cowtan, 2004 | http://bernhardcl.github.io/coot/ |
| Phenix | Adams et al., 2002; Afonine et al., 2018 | https://phenix-online.org/ |
| ISOLDE | Croll, 2018 | http://preview.cgl.ucsf.edu/chimerax/download.html |
| ChimeraX | Goddard et al., 2018 | https://www.rbvi.ucsf.edu/chimerax/ |
| Other | ||
| Galanthus nivalis lectin | Vector Laboratories | Cat # AL-1243-5 |
| 1-Step Ultra TMB-ELISA Substrate Solution | ThermoFisher Scientific | Cat#34029 |
| Costar High Binding Microplates | Corning | Cat#9018 |
| Saponin | VWR | TCS0019-025G |
| Paraformaldehyde | VWR | 100496-496 |
| Donor Goat Serum | Atlanta Biologicals | S13150 |
| DPBS(++) | Cytiva | SH30264.02 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Ivelin Georgiev (Ivelin.Georgiev@Vanderbilt.edu).
Materials availability
Please direct resource and reagent requests to the Lead Contact specified above, Ivelin Georgiev.
Experimental model and subject details
Human subjects
The 45 year old, male donor had previous laboratory-confirmed COVID-19, 3 months prior to blood collection. The studies were reviewed and approved by the Institutional Review Board of Vanderbilt University Medical Center. The sample was obtained after written informed consent was obtained.
Cell lines
Expi293F mammalian cells (ThermoFisher) were maintained in FreeStyle F17 expression medium supplemented at final concentrations of 0.1% Pluronic Acid F-68 and 20% 4mM L-Glutamine. These cells were cultured at 37°C with 8% CO2 saturation and shaking. FreeStyle293F cells were grown while shaking at 37 C in 8% CO2 and 80% humidity.
Method details
Antigen purification
A variety of recombinant soluble protein antigens were used in the LIBRA-seq experiment and other experimental assays.
Plasmids encoding residues 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV-2 spike HP); 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, a glycine mutation at 614, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV-2 spike HP D614G) 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, as well as mutations L18F, D80A, L242-244L del, R246I, K417N, E484K, N501Y, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV-2 spike HP Beta); 1–1208 of the SARS-CoV-2 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 817, 892, 899, 942, 986 and 987, as well as mutations 69-70del, Y144del, N501Y, A570D, P681H, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV-2 spike HP Alpha); residues 1-1190 of the SARS-CoV spike with proline substitutions at positions 968 and 969, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (SARS-CoV S-2P); residues 1-1291 of the MERS-CoV spike with a mutated S1/S2 cleavage site, proline substitutions at positions 1060 and 1061, and a C-terminal T4-fibritin trimerization motif, an AviTag, an 8x HisTag, and a TwinStrepTag (MERS-CoV S-2P Avi); residues 1-1278 of the HCoV-OC43 spike with proline substitutions at positions 1070 and 1071, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (HCoV-OC43 S-2P); residues 1-1277 of the HCoV-HKU1 spike with a mutated S1/S2 cleavage site, proline substitutions at positions 1067 and 1068, and a C-terminal T4-fibritin trimerization motif, an 8x HisTag, and a TwinStrepTag (HCoV-HKU1 S-2P); residues 319–591 of SARS-CoV-2 S with a C-terminal monomeric human IgG Fc-tag and an 8x HisTag (SARS-CoV-2 RBD-SD1); residues 306−577 of SARS-CoV S (Tor2 strain) were cloned upstream of a C-terminal HRV3C protease cleavage site, a monomeric Fc tag and an 8x HisTag (SARS-CoV RBD-SD1);residues 367–589 of MERS-CoV S with a C-terminal monomeric human IgG Fc-tag and an 8x HisTag (MERS-CoV RBD); residues 306–577 of MERS-CoV S with a C-terminal monomeric human IgG Fc-tag and an 8x HisTag (SARS-CoV RBD-SD1) were transiently transfected into FreeStyle293F cells (Thermo Fisher) using polyethylenimine. For all antigens with the exception of SARS-CoV-2 S HP, transfections were treated with 1 μM kifunensine to ensure uniform glycosylation three hours post-transfection. Transfected supernatants were harvested after 6 days of expression. SARS-CoV-2 RBD-SD1, SARS-CoV RBD-SD1, and MERS-CoV RBD were purified using Protein A resin (Pierce). SARS-CoV-2 S HP, MERS-CoV S-2P Avi, and HCoV-OC43 S-2P were purified using StrepTrap HP columns (Cytiva Life Sciences). Affinity-purified SARS-CoV-2 RBD-SD1, SARS-CoV RBD-SD1, and MERS-CoV RBD were further purified over a Superdex200 column (GE Life Sciences). SARS-CoV-2 S HP, SARS-CoV-2 S HP Beta, SARS-CoV-2 S HP Alpha, SARS-CoV S-2P, MERS-CoV S-2P, and HCoV-OC43 S-2P were purified over a Superose6 Increase column (GE Life Sciences). HCoV-NL63 and HCoV-229E alpha coronavirus spike proteins as well as the SARS-CoV-2 S1, SARS-CoV-2 S2, and SARS-CoV-2 NTD truncated proteins were purchased from the commercial vendor, Sino Biological.
Recombinant, soluble HIV-1 gp140 SOSIP trimer from strain ZM197 (clade) containing an AviTag and recombinant NC99 HA protein consisting of the HA ectodomain with a point mutation at the sialic acid-binding site (Y98F) to abolish non-specific interactions, a T4 fibritin foldon trimerization domain, AviTag, and hexahistidine-tag, were expressed in Expi 293F cells using polyethylenimine transfection reagent and cultured. FreeStyle F17 expression medium supplemented with pluronic acid and glutamine was used. The cells were cultured at 37°C with 8% CO2 saturation and shaking. After 5-7 days, cultures were centrifuged and supernatant was filtered and run over an affinity column of agarose bound Galanthus nivalis lectin. The column was washed with PBS and antigens were eluted with 30 mL of 1M methyl-a-D-mannopyranoside. Protein elutions were buffer exchanged into PBS, concentrated, and run on a Superdex 200 Increase 10/300 GL Sizing column on the AKTA FPLC system. Fractions corresponding to correctly folded protein were collected, analyzed by SDS-PAGE and antigenicity was characterized by ELISA using known monoclonal antibodies specific to each antigen. Avitagged antigens were biotinylated using BirA biotin ligase (Avidity LLC).
Spike protein used for cryo-EM was expressed by transiently transfecting plasmid encoding the HexaPro spike variant (Hsieh et al., 2020) containing additional S383C and D985C substitutions (Henderson et al., 2020) with a C-terminal TwinStrep tag into FreeStyle 293-F cells (Thermo Fisher) using polyethyleneimine. 5 μM kifunensine was added 3h post-transfection. The cell culture was harvested four days after transfection and the spike-containing medium was separated from the cells by centrifugation. Supernatants were passed through a 0.22 μm filter and passaged over StrepTactin resin (IBA). Further purification was achieved by size-exclusion chromatography using a Superose 6 10/300 column (GE Healthcare) in buffer containing 2 mM Tris pH 8.0, 200 mM NaCl and 0.02% NaN3.
DNA-barcoding of antigens
We used oligos that possess 15 bp antigen barcode, a sequence capable of annealing to the template switch oligo that is part of the 10X bead-delivered oligos, and contain truncated TruSeq small RNA read 1 sequences in the following structure: 5′-CCTTGGCACCCGAGAATTCCANNNNNNNNNNNNNCCCATATAAGA∗A∗A-3′, where Ns represent the antigen barcode. We used the following antigen barcodes: We used the following antigen barcodes: GCAGCGTATAAGTCA (SARS-CoV-2 S), AACCCACCGTTGTTA (SARS-CoV-2 S D614G), GCTCCTTTACACGTA (SARS-CoV S), GGTAGCCCTAGAGTA (MERS-CoV S), AGACTAATAGCTGAC (HCoV-OC43 S), GACAAGTGATCTGCA (HCoV-NL63 S), GTGTGTTGTCCTATG (HCoV-229E S), TACGCCTATAACTTG (ZM197 HIV EnV), TCATTTCCTCCGATT (HA NC99), TGGTAACGACAGTCC (SARS-CoV RBD-SD1), TTTCAACGCCCTTTC (SARS-CoV-2 RBD-SD1), GTAAGACGCCTATGC (MERS-CoV RBD), CAGTAAGTTCGGGAC(SARS-CoV-2 NTD), Oligos were ordered from IDT with a 5′ amino modification and HPLC purified.
For each antigen, a unique DNA barcode was directly conjugated to the antigen itself. In particular, 5′amino-oligonucleotides were conjugated directly to each antigen using the Solulink Protein-Oligonucleotide Conjugation Kit (TriLink cat no. S-9011) according to manufacturer’s instructions. Briefly, the oligo and protein were desalted, and then the amino-oligo was modified with the 4FB crosslinker, and the biotinylated antigen protein was modified with S-HyNic. Then, the 4FB-oligo and the HyNic-antigen were mixed together. This causes a stable bond to form between the protein and the oligonucleotide. The concentration of the antigen-oligo conjugates was determined by a BCA assay, and the HyNic molar substitution ratio of the antigen-oligo conjugates was analyzed using the NanoDrop according to the Solulink protocol guidelines. AKTA FPLC was used to remove excess oligonucleotide from the protein-oligo conjugates, which were also verified using SDS-PAGE with a silver stain. Antigen-oligo conjugates were also used in flow cytometry titration experiments.
Antigen-specific B cell sorting
Cells were stained and mixed with DNA-barcoded antigens and other antibodies, and then sorted using fluorescence activated cell sorting (FACS). First, cells were counted and viability was assessed using Trypan Blue. Then, cells were washed three times with DPBS supplemented with 0.1% Bovine serum albumin (BSA). Cells were resuspended in DPBS-BSA and stained with cell markers including viability dye (Ghost Red 780), CD14-APC-Cy7, CD3-FITC, CD19-BV711, and IgG-PE-Cy5. Additionally, antigen-oligo conjugates were added to the stain. After staining in the dark for 30 minutes at room temperature, cells were washed three times with DPBS-BSA at 300 g for five minutes. Cells were then incubated for 15 minutes at room temperature with Streptavidin-PE to label cells with bound antigen. Cells were washed three times with DPBS-BSA, resuspended in DPBS, and sorted by FACS. Antigen positive cells were bulk sorted and delivered to the Vanderbilt Technologies for Advanced Genomics (VANTAGE) sequencing core at an appropriate target concentration for 10X Genomics library preparation and subsequent sequencing. FACS data were analyzed using FlowJo.
Sample preparation, library preparation, and sequencing
Single-cell suspensions were loaded onto the Chromium Controller microfluidics device (10X Genomics) and processed using the B cell Single Cell V(D)J solution according to manufacturer’s suggestions for a target capture of 10,000 B cells per 1/8 10X cassette, with minor modifications in order to intercept, amplify and purify the antigen barcode libraries as previously described (Setliff et al., 2019).
Sequence processing and bioinformatics analysis
We utilized and modified our previously described pipeline to use paired-end FASTQ files of oligo libraries as input, process and annotate reads for cell barcode, unique molecular identifier (UMI), and antigen barcode, and generate a cell barcode - antigen barcode UMI count matrix (Setliff et al., 2019). BCR contigs were processed using Cell Ranger (10X Genomics) using GRCh38 as reference. Antigen barcode libraries were also processed using Cell Ranger (10X Genomics). The overlapping cell barcodes between the two libraries were used as the basis of the subsequent analysis. We removed cell barcodes that had only non-functional heavy chain sequences as well as cells with multiple functional heavy chain sequences and/or multiple functional light chain sequences, reasoning that these may be multiplets. Additionally, we aligned the BCR contigs (filtered_contigs.fasta file output by Cell Ranger, 10X Genomics) to IMGT reference genes using HighV-Quest (Alamyar et al., 2012). The output of HighV-Quest was parsed using ChangeO (Gupta et al., 2015) and merged with an antigen barcode UMI count matrix. Finally, we determined the LIBRA-seq score for each antigen in the library for every cell as previously described (Setliff et al., 2019).
Antibody expression and purification
For each antibody, variable genes were inserted into custom plasmids encoding the constant region for the IgG1 heavy chain as well as respective lambda and kappa light chains (pTwist CMV BetaGlobin WPRE Neo vector, Twist Bioscience). Antibody 54042-2 was natively an IGHG2, but was cloned into an IGHG1 Fc backbone vector for monoclonal antibody characterization. Antibodies were expressed in Expi293F mammalian cells (Thermo Fisher Scientific) by co-transfecting heavy chain and light chain expressing plasmids using polyethylenimine transfection reagent and cultured for 5-7 days. Cells were maintained in FreeStyle F17 expression medium supplemented at final concentrations of 0.1% Pluronic Acid F-68 and 20% 4mM L-Glutamine. These cells were cultured at 37°C with 8% CO2 saturation and shaking. After transfection and 5-7 days of culture, cell cultures were centrifuged and supernatant was 0.45 μm filtered with Nalgene Rapid Flow Disposable Filter Units with PES membrane. Filtered supernatant was run over a column containing Protein A agarose resin equilibrated with PBS. The column was washed with PBS, and then antibodies were eluted with 100 mM Glycine HCl at 2.7 pH directly into a 1:10 volume of 1M Tris-HCl pH 8.0. Eluted antibodies were buffer exchanged into PBS 3 times using Amicon Ultra centrifugal filter units and concentrated. Antibodies were analyzed by SDS-PAGE.
High-throughput antibody expression
For high-throughput production of recombinant antibodies, approaches were used that are designated as microscale. For antibody expression, microscale transfection were performed (∼1 ml per antibody) of CHO cell cultures using the GIBCO ExpiCHO Expression System and a protocol for deep 96-well blocks (Thermo Fisher Scientific). In brief, synthesized antibody-encoding DNA (∼2 μg per transfection) was added to OptiPro serum free medium (OptiPro SFM), incubated with ExpiFectamine CHO Reagent and added to 800 μl of ExpiCHO cell cultures into 96-deep-well blocks using a ViaFlo 384 liquid handler (Integra Biosciences). The plates were incubated on an orbital shaker at 1,000 rpm with an orbital diameter of 3 mm at 37 °C in 8% CO2. The next day after transfection, ExpiFectamine CHO Enhancer and ExpiCHO Feed reagents (Thermo Fisher Scientific) were added to the cells, followed by 4 d incubation for a total of 5 d at 37 °C in 8% CO2. Culture supernatants were collected after centrifuging the blocks at 450g for 5 min and were stored at 4°C until use. For high-throughput microscale antibody purification, fritted deep-well plates were used containing 25 μl of settled protein G resin (GE Healthcare Life Sciences) per well. Clarified culture supernatants were incubated with protein G resin for antibody capturing, washed with PBS using a 96-well plate manifold base (QIAGEN) connected to the vacuum and eluted into 96-well PCR plates using 86 μl of 0.1 M glycine-HCL buffer pH 2.7. After neutralization with 14 μl of 1 M Tris-HCl pH 8.0, purified antibodies were buffer-exchanged into PBS using Zeba Spin Desalting Plates (Thermo Fisher Scientific) and stored at 4°C until use.
ELISA
To assess antibody binding, soluble protein was plated at 2 μg/ml overnight at 4°C. The next day, plates were washed three times with PBS supplemented with 0.05% Tween-20 (PBS-T) and coated with 5% milk powder in PBS-T. Plates were incubated for one hour at room temperature and then washed three times with PBS-T. Primary antibodies were diluted in 1% milk in PBS-T, starting at 10 μg/ml with a serial 1:5 dilution and then added to the plate. The plates were incubated at room temperature for one hour and then washed three times in PBS-T. The secondary antibody, goat anti-human IgG conjugated to peroxidase, was added at 1:10,000 dilution in 1% milk in PBS-T to the plates, which were incubated for one hour at room temperature. Plates were washed three times with PBS-T and then developed by adding 3,3′,5,5′-tetramethylbenzidine (TMB) substrate to each well. The plates were incubated at room temperature for ten minutes, and then 1N sulfuric acid was added to stop the reaction. Plates were read at 450 nm. The area under the curve (AUC) was calculated using GraphPad Prism 9.0.1.
Competition ELISA
Competition ELISA was performed as done previously (Zost et al., 2020). Briefly, antibodies were biotinylated using NHS-PEG4-biotin (Thermo Fisher Scientific, cat# A39259) according to manufacturer protocol. Following biotinylation, specific binding of biotinylated antibodies was confirmed using ELISA. Wells of 384-well microtiter plates were coated with 1 μg/mL SARS-CoV-2 S HP protein at 4°C overnight. Plates were washed with PBS-T and blocked for 1 h with blocking buffer (1% BSA in PBS-T). Plates were then washed with PBS-T and unlabeled antibodies were added at a concentration of 10 μg/mL in a total volume of 25 μL blocking buffer and incubated 1 h. Without washing, biotinylated antibodies diluted in blocking buffer were added directly to each well in a volume of 5 μL per well (such that the final concentrations of each biotinylated antibody were equal to the respective EC90 of each antibody), and then incubated for 30 min at ambient temperature. Plates were then washed with PBS-T and incubated for 1 h with HRP-conjugated avidin (Sigma, 25 μL of a 1:3,500 dilution in blocking buffer). Plates were washed with PBS-T and 25 μL TMB substrate was added to each well. After sufficient development, the reactions were quenched by addition 25 μL 1M HCl and absorbance at 450 nm was quantified using a plate reader. After subtracting the background signal, the signal obtained for binding of the biotin-labeled reference antibody in the presence of the unlabeled tested antibody was expressed as a percentage of the binding of the reference antibody in the presence of 10 μg/mL of the anti-dengue antibody DENV 2D22, which served as a no-competition control. Tested antibodies were considered competing if their presence reduced the reference antibody binding by more than 60% and non-competing if the signal was reduced by less than 30%.
Real-time cell analysis (RTCA) HT neutralization assay screen
To screen for neutralizing activity in the panel of recombinantly expressed antibodies, we used a high-throughput and quantitative RTCA assay and xCelligence RTCA HT Analyzer (ACEA Biosciences) that assesses kinetic changes in cell physiology, including virus-induced cytopathic effect (CPE). Twenty μl of cell culture medium (DMEM supplemented with 2% FBS) was added to each well of a 384-well E-plate using a ViaFlo384 liquid handler (Integra Biosciences) to obtain background reading. Six thousand (6,000) Vero-furin cells in 20 μl of cell culture medium were seeded per well, and the plate was placed on the analyzer. Sensograms were visualized using RTCA HT software version 1.0.1 (ACEA Biosciences). For a screening neutralization assay, equal amounts of virus were mixed with micro-scale purified antibodies in a total volume of 40 μL using DMEM supplemented with 2% FBS as a diluent and incubated for 1 h at 37 °C in 5% CO2. At ∼17–20 h after seeding the cells, the virus–antibody mixtures were added to the cells in 384-well E-plates. Wells containing virus only (in the absence of antibody) and wells containing only Vero cells in medium were included as controls. Plates were measured every 8–12 h for 48–72 h to assess virus neutralization. Micro-scale antibodies were assessed in four 5-fold dilutions (starting from a 1:20 sample dilution), and their concentrations were not normalized. Neutralization was calculated as the percent of maximal cell index in control wells without virus minus cell index in control (virus-only) wells that exhibited maximal CPE at 40–48 h after applying virus–antibody mixture to the cells. An antibody was classified as fully neutralizing if it completely inhibited SARS-CoV-2-induced CPE at the highest tested concentration, while an antibody was classified as partially neutralizing if it delayed but did not fully prevent CPE at the highest tested concentration. Further, if the CPE curve lies between partial and the virus-only control, those mAbs were designated weak (Zost et al., 2020).
RTCA potency neutralization screening assay
To determine neutralizing activity of IgG and convalescent serum, we used real-time cell analysis (RTCA) assay on an xCELLigence RTCA MP Analyzer (ACEA Biosciences Inc.) that measures virus-induced cytopathic effect (CPE)(Suryadevara et al., 2021). Briefly, 50 μL of cell culture medium (DMEM supplemented with 2% FBS) was added to each well of a 96-well E-plate using a ViaFlo384 liquid handler (Integra Biosciences) to obtain background reading. A suspension of 18,000 Vero-E6 cells in 50 μL of cell culture medium was seeded in each well, and the plate was placed on the analyzer. Measurements were taken automatically every 15 min, and the sensograms were visualized using RTCA software version 2.1.0 (ACEA Biosciences Inc). VSV-SARS-CoV-2 (0.01 MOI, ∼120 PFU per well) was mixed 1:1 with a dilution of antibody in a total volume of 100 μL using DMEM supplemented with 2% FBS as a diluent and incubated for 1 h at 37°C in 5% CO2. At 16 h after seeding the cells, the virus-antibody mixtures were added in replicates to the cells in 96-well E-plates. Triplicate wells containing virus only (maximal CPE in the absence of antibody) and wells containing only Vero cells in medium (no-CPE wells) were included as controls. Plates were measured continuously (every 15 min) for 48 h to assess virus neutralization. Normalized cellular index (CI) values at the endpoint (48 h after incubation with the virus) were determined using the RTCA software version 2.1.0 (ACEA Biosciences Inc.). Results are expressed as percent neutralization in a presence of respective antibody relative to control wells with no CPE minus CI values from control wells with maximum CPE. RTCA IC50 and NT50 values were determined by nonlinear regression analysis using GraphPad Prism software. A full dose-response neutralization assay was not performed for antibody 54042-11 due to insufficient quantity at the time of experiment. The NT50 of the donor sample was comparable to previously reported data for other donors for SARS-CoV-2 antibody discovery efforts (Brouwer et al., 2020; Rogers et al., 2020).
Epitope mapping of antibodies by alanine scanning
Epitope mapping was performed essentially as described previously (Davidson and Doranz, 2014) using a SARS-CoV-2 (strain Wuhan-Hu-1) spike protein RBD shotgun mutagenesis mutation library, made using an expression construct for full-length spike protein. 184 residues of the RBD (between spike residues 335 and 526) were mutated individually to alanine, and alanine residues to serine and clones arrayed in 384-well plates, one mutant per well. Antibody binding to each mutant clone was determined, in duplicate, by high-throughput flow cytometry. Each spike protein mutant was transfected into HEK293T cells and allowed to express for 22 hr. Cells were fixed in 4% (v/v) paraformaldehyde (Electron Microscopy Sciences), and permeabilized with 0.1% (w/v) saponin (Sigma-Aldrich) in PBS plus calcium and magnesium (PBS++) before incubation with antibodies diluted in PBS++, 10% normal goat serum (Sigma), and 0.1% saponin. Antibody screening concentrations were determined using an independent immunofluorescence titration curve against cells expressing wild-type spike protein to ensure that signals were within the linear range of detection. Antibodies were detected using 3.75 μg/mL of AlexaFluor488-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) in 10% normal goat serum with 0.1% saponin. Cells were washed three times with PBS++/0.1% saponin followed by two washes in PBS, and mean cellular fluorescence was detected using a high-throughput Intellicyte iQue flow cytometer (Sartorius). Antibody reactivity against each mutant spike protein clone was calculated relative to wild-type spike protein reactivity by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type S-transfected controls. Mutations within clones were identified as critical to antibody binding if they did not support reactivity of the test antibody, but supported reactivity of other SARS-CoV-2 antibodies. This counter-screen strategy facilitates the exclusion of spike mutants that are locally misfolded or have an expression defect.
Plaque reduction neutralization test (PRNT)
The virus neutralization with live authentic SARS-CoV-2 virus was performed in the BSL-3 facility of the Galveston National Laboratory using Vero E6 cells (ATCC CRL-1586) following the standard procedure. Briefly, Vero E6 cells were cultured in 96-well plates (104 cells/well). Next day, 4-fold serial dilutions of antibodies were made using MEM-2% FBS, as to get an initial concentration of 100 μg/ml. Equal volume of diluted antibodies (60 μl) were mixed gently with authentic virus (60 μl containing 200 pfu) and incubated for 1 h at 37°C/5% CO2 atmosphere. The virus-serum mixture (100 μl) was added to cell monolayer in duplicates and incubated for 1 at h 37°C/5% CO2 atmosphere. Later, virus-serum mixture was discarded gently, and cell monolayer was overlaid with 0.6% methylcellulose and incubated for 2 days. The overlay was removed, and the plates were fixed in 4% paraformaldehyde twice following BSL-3 protocol. The plates were stained with 1% crystal violet and virus-induced plaques were counted. The percent neutralization and/or NT50 of antibody was calculated by dividing the plaques counted at each dilution with plaques of virus-only control. For antibodies, the inhibitory concentration at 50% (IC50) values were calculated in GraphPad Prism software by plotting the midway point between the upper and lower plateaus of the neutralization curve among dilutions. The Alpha variant virus incorporates the following substitutions: Del 69-70, Del 144, E484K, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H. The Beta variant incorporates the following substitutions: Del 24, Del 242-243, D80A, D215G, K417N, E484K, N501Y, D614G, H665Y, T1027I. The Delta variant incorporates the following substitutions: T19R, G142D, Del 156-157, R158G, L452R, T478K, D614G, P681R, Del 689-691, D950N; the deletion at positions 689-691 has not been observed in nature, and was identified upon one passage of the virus. The Gamma variant incorporates the following substitutions: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I.
BioLayer interferometry (BLI)
Purified 54042-4 IgG was immobilized to AHC sensortips (FortéBio) to a response level of approximately 1.4 nm in a buffer composed of 10 mM HEPES pH 7.5, 150 mM NaCl, 3 mM EDTA, 0.05% Tween 20 and 0.1% (w/v) BSA. Immobilized IgG was then dipped into wells containing four-fold dilutions of SARS-CoV-2 RBD-SD1 ranging in concentration from 100-1.5625 nM, to measure association. Dissociation was measured by dipping sensortips into wells containing only running buffer. Data were reference subtracted and kinetics were calculated in Octet Data Analysis software v10.0 using a 1:1 binding model.
ACE2 binding inhibition assay
96-well plates were coated with 2 μg/mL purified recombinant SARS-CoV-2 at 4°C overnight. The next day, plates were washed three times with PBS supplemented with 0.05% Tween-20 (PBS-T) and coated with 5% milk powder in PBS-T. Plates were incubated for one hour at room temperature and then washed three times with PBS-T. Purified antibodies were diluted in blocking buffer at 10 μg/mL in triplicate, added to the wells, and incubated at room temperature. Without washing, recombinant human ACE2 protein with a mouse Fc tag was added to wells for a final 0.4 μg/mL concentration of ACE2 and incubated for 40 minutes at room temperature. Plates were washed three times with PBS-T, and bound ACE2 was detected using HRP-conjugated anti-mouse Fc antibody and TMB substrate. The plates were incubated at room temperature for ten minutes, and then 1N sulfuric acid was added to stop the reaction. Plates were read at 450 nm. ACE2 binding without antibody served as a control. Experiment was done in biological replicate and technical triplicates.
RTCA neutralization assay with known antibody escape mutants
We used a real-time cell analysis assay (RTCA) and xCELLigence RTCA MP Analyzer (ACEA Biosciences Inc.) with modification of previously described assays (Greaney et al., 2021a, 2021b; Suryadevara et al., 2021). Fifty (50) μL of cell culture medium (DMEM supplemented with 2% FBS) was added to each well of a 96-well E-plate to obtain a background reading. Eighteen thousand (18,000) Vero E6 cells in 50 μL of cell culture medium were seeded per each well, and plates were placed on the analyzer. Measurements were taken automatically every 15 min and the sensograms were visualized using RTCA software version 2.1.0 (ACEA Biosciences Inc). Escape mutant VSV-SARS-CoV-2 or wild-type VSV-SARS-CoV-2 virus (5e3 plaque forming units [PFU] per well, ∼0.3 MOI) was mixed with a saturating neutralizing concentration of individual antibody (5 μg/mL) in a total volume of 100 μL and incubated for 1 h at 37°C. At 16-20 h after seeding the cells, the virus-antibody mixtures were added into 8 to 96 replicate wells of 96-well E-plates with cell monolayers. Wells containing only virus in the absence of antibody and wells containing only Vero E6 cells in medium were included on each plate as controls. Plates were measured continuously (every 15 min) for 72 h. The lack of neutralization for the individual escape mutant viruses from 54042-4 was confirmed by delayed CPE in wells containing antibody while COV2-2381 was used as positive control.
EM sample prep and data collection
To form the spike-Fab complex, a final concentration of 0.5 mg/mL spike protein and 5X molar excess of Fab were combined in buffer containing 2mM Tris-Cl pH 8.0, 200 mM NaCl, and 0.02% NaN3. The complex was incubated on ice for 30 min before 3 μL of the sample was deposited on Au-300 1.2/1.3 grids (UltrAuFoil) that had been plasma cleaned in a Solarus 950 plasma cleaner (Gatan) for 4 minutes using a 4:1 ratio of O2:H2. A force of −4 was used to blot excess liquid for 3 s using a Vitrobot Mark IV (Thermo Fisher) followed by plunge-freezing with liquid ethane. 3,762 micrographs were collected from a single grid using a Titan Krios (Thermo Fisher) equipped with a K3 detector (Gatan) with the stage set at a 30° tilt. SerialEM was used to collect movies at 29,000X nominal magnification with a calibrated pixel size of 0.81 Å/pixel. Additional details about data collection parameters can be found in Table S1.
Cryogenic electron microscopy (Cryo-EM)
Motion correction, CTF estimation, particle picking, and preliminary 2D classification were performed using cryoSPARC v3.2.0 live processing (Punjani et al., 2017). The final iteration of 2D class averaging distributed 374,669 particles into 60 classes using an uncertainty factor of 2. From that, 241,732 particles were used to perform an ab initio reconstruction with four classes followed by heterogeneous refinement of those four classes. Particles from the two highest quality classes were used for homogeneous refinement of the best volume with applied C3 symmetry. Non-uniform refinement was performed on the resulting volume using per-particle defocus and per-group CTF optimizations applied (Punjani et al., 2020; Rubinstein and Brubaker, 2015). To improve the 54042-4 Fab-RBD density, C3 symmetry expansion was performed followed by local refinement using a mask created in ChimeraX that encompassed the entire 54042-4 Fab and RBD (Pettersen et al., 2021). Local refinement was performed using a pose/shift Gaussian prior during alignment, 3° standard deviation of prior over rotation and 1 Å standard deviation of prior over shifts. Additionally, maximum alignment resolution was limited to 2.8 Å resolution to avoid over-refining. To improve map quality, the focused refinement volumes were processed using the DeepEMhancer (Sanchez-Garcia et al., 2021) tool via COSMIC2science gateway, which included the use of our refinement mask to help define noise while sharpening (Cianfrocco et al., 2017a, 2017b). An initial model was generated by docking PDBID: 6XKL (Hsieh et al., 2020) and a Fab model based on the 54042-4 sequence built using SAbPred ABodyBuilder (Dunbar et al., 2016) into map density via ChimeraX (Pettersen et al., 2021). The model was iteratively refined and completed using a combination of Phenix, Coot, and ISOLDE (Adams et al., 2002; Croll, 2018; Emsley and Cowtan, 2004). Details on structure validation and the full cryo-EM processing workflow can be found in Figures S5 and S6.
GISAID mutation frequency calculation
To evaluate the conservation of 54042-4 epitope residues, we utilized the GISAID database (Elbe and Buckland-Merrett, 2017) comprising sequences from 1,229,459 SARS-CoV-2 variants (as of May 6th, 2021). The spike glycoprotein sequences were extracted and translated, and pairwise sequence alignment with the reference sequence hCoV-19/Wuhan/WIV04/2019 was then performed. After removing incomplete sequences and sequences with alignment errors, the pairwise alignments for the remaining 1,148,887 spike protein sequences were combined to compute the conservation of each residue position using in-house perl scripts.
RMSD calculation for the differences in angle of antigen approach for different antibodies
The SARS-CoV-2 spike receptor binding domain coordinates present in each antibody-antigen complex were aligned in PyMOL (The PyMOL Molecular Graphics System, Version 2.3.5, Schrödinger, LLC.) using an all-atom alignment with 5 cycles of outlier rejection of atom pairs having an RMSD greater than 2. The alignment was performed for RBD residues 329-529 in antibody 54042-4 (PDB ID: TBD chain A), 329-529 in antibody 2-7 (PDB ID: 7LSS chain B), 333-526 in antibody REGN10987 (PDB ID: 6XDG chain A), and 334-527 in antibody LY-COV1404 (PDB ID: 7MMO chain C). This resulted in RMSD values of 0.751 Å between 54042-4 and REGN10987’s RBDs, 1.044 Å between 54042-4 and antibody 2-7’s RBDs, 0.632 Å between 54042-4 and LY-COV1404’s RBDs, 1.067 Å between REGN10987 and antibody 2-7’s RBDs, and 0.751 Å between LY-COV1404 and antibody 2-7’s RBDs with well-aligned epitope residues. Next, the residues comprising the N-termini through the end of framework region 3 were determined for the heavy and light chains of all three antibodies using IMGT Domain Gap Align (Alamyar et al., 2012). Each pair of antibodies was aligned using a pairwise sequence alignment of this region in PyMOL. Finally, the alpha carbon root mean square deviation between antibodies was calculated over this region in the heavy and light chains using residue pairs from the sequence alignment. RMSD values were calculated from 183, 183, and 180 alpha carbon pairs for the 54042-4 versus REGN1087, REGN1087 versus 2-7, and 54042-4 versus 2-7 comparisons respectively.
Quantification and statistical analysis
ELISA and neutralization error bars (standard error of the mean) were calculated using GraphPad Prism version 9.0.1.
Acknowledgments
We thank Angela Jones, Latha Raju, and Jamie Roberson of Vanderbilt Technologies for Advanced Genomics for their expertise regarding NGS and library preparation; David Flaherty and Brittany Matlock of the Vanderbilt Flow Cytometry Shared Resource for help with flow panel optimization; and members of the Georgiev laboratory for comments on the manuscript. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University (Nashville, TN). Flow cytometry experiments were performed in the VUMC Flow Cytometry Shared Resource. The VUMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). We also thank the Vanderbilt Institute for Clinical and Translational Research (VICTR), funded by the National Center for Advancing Translational Science (NCATS) Clinical Translational Science Award (CTSA) Program (5ULITR002243-03). We also thank Josh Adams for assistance with GISAID acknowledgment upload. The Vanderbilt VANTAGE Core provided technical assistance for this work. VANTAGE is supported in part by CTSA (5UL1 RR024975-03), the Vanderbilt-Ingram Cancer Center (P30 CA68485), the Vanderbilt Vision Center (P30 EY08126), and NIH/NCRR (G20 RR030956). This work was supported by NIAID/NIH (R01AI131722-S1 to I.S.G., R01 AI157155 to J.E.C., R01 AI127521 to J.S.M., and T32 AI095202 to S.J.Z.); HHSN (75N93019C00074 and DARPA HR0011-18-2-0001 to J.E.C. and 75N93019C00073 to B.J.D.); Hays Foundation COVID-19 Research Fund (to I.S.G.); Fast Grants (to I.S.G. and J.E.C.); VICTR Award (VR54494 to I.S.G.), the Dolly Parton COVID-19 Research Fund at Vanderbilt (to J.E.C.); Welch Foundation (F-0003-19620604 to J.S.M.); Mercatus Center (to J.E.C.); and George Mason University (to J.E.C.). J.E.C. is a recipient of the 2019 Future Insight Prize from Merck KGaA. The Sauer Structural Biology Laboratory is supported by The University of Texas College of Natural Sciences and the Cancer Prevention and Research Institute of Texas (CPRIT) (RR160023). The content is solely the responsibility of the authors and does not represent the official views of the U.S. government or other sponsors.
Author contributions
Methodology, K.J.K., N.V.J., J.S.M., and I.S.G.; Investigation, K.J.K., N.V.J., A.R.S., N.S., S.P., N.R., J.K.W., D.W., S.J.Z., L.M.W., S.C.W., C.M.H., C.-L.H., R.E.S., A.P., E.D., B.J.D., J.E.C., A.B., R.H.C., J.S.M., and I.S.G.; Software, A.R.S., N.R., and I.S.G.; Validation, K.J.K., N.V.J., and A.R.S.; Writing – Original Draft, K.J.K., N.V.J., J.S.M., and I.S.G.; Writing – Review & Editing, all authors; Funding Acquisition, E.D., B.J.D., J.E.C., A.B., R.H.C., J.S.M., I.S.G., A.R.S., and K.J.K.; Resources, E.D., B.J.D., J.E.C., A.B., R.H.C., J.S.M., and I.S.G.; Supervision, J.S.M. and I.S.G.
Declarations of interests
A.R.S. and I.S.G. are co-founders of AbSeek Bio. K.J.K., A.R.S., N.V.J., I.S.G., J.S.M., R.H.C., and J.E.C. are listed as inventors on patents filed describing the antibodies discovered here. R.H.C. is an inventor on patents related to other SARS-CoV-2 antibodies. J.E.C. has served as a consultant for Luna Biologics, is a member of the Scientific Advisory Board of Meissa Vaccines and is Founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received sponsored research agreements from Takeda Vaccines, IDBiologics and AstraZeneca. J.K.W., E.D., and B.J.D. are employees of Integral Molecular. B.J.D. is a shareholder of Integral Molecular. The Georgiev laboratory at Vanderbilt University Medical Center has received unrelated funding from Takeda Pharmaceuticals.
Published: September 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109784.
Supplemental information
Data and code availability
-
•
Sequences for antibodies identified and characterized in this study have been deposited to GenBank under GenBank accession numbers (MZ820097-MZ8200105) and (MZ8200106-MZ8200114). Raw sequencing data used in this study are available on the Sequence Read Archive under BioProject accession number PRJNA754442. Custom scripts used to analyze data in this manuscript are available upon request to the corresponding author. GISAID EpiCoV sequences are deposited at https://github.com/Georgiev-Lab/GISAID_COV19_acknowledgement_May_7_2021.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D. Real-Space Refinement in PHENIX for Cryo-EM and Crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamyar E., Duroux P., Lefranc M.P., Giudicelli V. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- Alpert T., Brito A.F., Lasek-Nesselquist E., Rothman J., Valesano A.L., MacKay M.J., Petrone M.E., Breban M.I., Watkins A.E., Vogels C.B.F., et al. Early introductions and transmission of SARS-CoV-2 variant B.1.1.7 in the United States. Cell. 2021;184:2595–2604.e13. doi: 10.1016/j.cell.2021.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., Pavlin B., Vandemaele K., Van Kerkhove M.D., Jombart T., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26:2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L., et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco M., Wong M., Youn C., COSMIC2 . 2017. COSMIC2 - A Science gateway for cryo-electron microscopy.https://figshare.com/articles/COSMIC2_A_science_gateway_for_cryo-electron_microscopy/5481430 [Google Scholar]
- Cianfrocco M.A., Wong-Barnum M., Youn C., Wagner R., Leschzinger A. 2017. COSMIC2: A Science Gateway for Cryo-Electron Microscopy Structure Determination; pp. 1–5. (Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact (PEARC17). Association for Computing Machinery, Article 22). [Google Scholar]
- Croll T.I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E., Doranz B.J. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143:13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M.E., Supasa P., Case J.B., Zhao Y., Walter T.S., Mentzer A.J., Liu C., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200.e22. doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Zost S.J., Greaney A.J., Starr T.N., Dingens A.S., Chen E.C., Chen R.E., Case J.B., Sutton R.E., Gilchuk P., et al. Genetic and structural basis for recognition of SARS-CoV-2 spike protein by a two-antibody cocktail. bioRxiv. 2021 doi: 10.1101/2021.01.27.428529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar J., Krawczyk K., Leem J., Marks C., Nowak J., Regep C., Georges G., Kelm S., Popovic B., Deane C.M. SAbPred: a structure-based antibody prediction server. Nucleic Acids Res. 2016;44(W1):W474-8. doi: 10.1093/nar/gkw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Petterson E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting Modern Challenges in Visualization and Analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S.M.C., Kopp M., Li D., Parks R., Hsu A.L., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in non-human primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmina A., Khalaila Y., Voloshin O., Keren-Naus A., Boehm-Cohen L., Raviv Y., Shemer-Avni Y., Rosenberg E., Taube R. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe. 2021;29:522–528.e2. doi: 10.1016/j.chom.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu Y., Xia H., Weaver S., Swanson K., Cai H., Cutler M., Cooper D., Muik A., Jansen K., et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273–275. doi: 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. A program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microscopy and Microanalysis. 2003;9(2) [Google Scholar]
- McCallum M., Bassi J., De Marco A., Chen A., Walls A.C., Di Iulio J., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlcochova P., Kemp S., Dhar M.S., Papa G., Meng B., Mishra S., Whittaker C., Mellan T., Ferreira I., Datir R., et al. SARS-CoV-2 B.1.617.2 Delta variant replication, sensitivity to neutralising antibodies and vaccine breakthrough. bioRxiv. 2021 doi: 10.1101/2021.05.08.443253. [DOI] [Google Scholar]
- Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.Y., Rustagi A., Rogers A.J., et al. Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Punjani A., Zhang H., Fleet D.J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37:734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein J.L., Brubaker M.A. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 2015;192:188–195. doi: 10.1016/j.jsb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Sabino E.C., Buss L.F., Carvalho M.P.S., Prete C.A., Jr., Crispim M.A.E., Fraiji N.A., Pereira R.H.M., Parag K.V., da Silva Peixoto P., Kraemer M.U.G., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia J.G.-B., Cuervo A., Carazo J.M., Sorzano C.O.S., Vargas J. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 2021;4:874. doi: 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setliff I., Shiakolas A.R., Pilewski K.A., Murji A.A., Mapengo R.E., Janowska K., Richardson S., Oosthuysen C., Raju N., Ronsard L., et al. High-Throughput Mapping of B Cell Receptor Sequences to Antigen Specificity. Cell. 2019;179:1636–1646. doi: 10.1016/j.cell.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiakolas A.R., Kramer K.J., Wrapp D., Richardson S.I., Schäfer A., Wall S., Wang N., Janowska K., Pilewski K.A., Venkat R., et al. Cross-reactive coronavirus antibodies with diverse epitope specificities and Fc effector functions. Cell Rep Med. 2021;2:100313. doi: 10.1016/j.xcrm.2021.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e15. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. ISARIC4C Investigators. COVID-19 Genomics UK (COG-UK) Consortium Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Di Iulio J., Wang Z., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman J.C. Novavax vaccine delivers 89% efficacy against COVID-19 in U.K.—but is less potent in South Africa. Science. 2021 doi: 10.1126/science.abg8101. [DOI] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad sarbecovirus neutralizing antibodies define a key site of vulnerability on the SARS-CoV-2 spike protein. bioRxiv. 2020 doi: 10.1101/2020.05.15.096511. [DOI] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf K., Zentelis S., Foster D., Vaillancourt P., Wiggin M., Lovett E., Hendle J., Pustilnik A., Sauder J.M., Kraft L., et al. LY-CoV1404 potently neutralizes SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.04.30.442182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27:620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Sequences for antibodies identified and characterized in this study have been deposited to GenBank under GenBank accession numbers (MZ820097-MZ8200105) and (MZ8200106-MZ8200114). Raw sequencing data used in this study are available on the Sequence Read Archive under BioProject accession number PRJNA754442. Custom scripts used to analyze data in this manuscript are available upon request to the corresponding author. GISAID EpiCoV sequences are deposited at https://github.com/Georgiev-Lab/GISAID_COV19_acknowledgement_May_7_2021.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.